Abstract
Metazoans guard their germlines against transposons and other foreign transcripts with PIWI-interacting RNAs (piRNAs). Due to the robust heritability of the silencing initiated by piRNAs in Caenorhabditis elegans (C. elegans), previous screens using C. elegans were strongly biased to uncover members of this pathway in the maintenance process but not in the initiation process. To identify novel piRNA pathway members, we have utilized a sensitized reporter strain which detects defects in initiation, amplification, or regulation of piRNA silencing. Using our reporter, we have identified Integrator complex subunits, nuclear pore components, protein import components, and pre-mRNA splicing factors as essential for piRNA-mediated gene silencing. We found the small nuclear processing cellular machine termed the Integrator complex is required for both type I and type II piRNA production. Notably, we identified a role for nuclear pore and nucleolar components NPP-1/Nup54, NPP-6/Nup160, NPP-7/Nup153, and FIB-1 in promoting the perinuclear localization of anti-silencing CSR-1 Argonaute, as well as a role for Importin factor IMA-3 in nuclear localization of silencing Argonaute HRDE-1. Together, we have shown that piRNA silencing in C. elegans is dependent on evolutionarily ancient RNA processing machinery that has been co-opted to function in the piRNA-mediated genome surveillance pathway.
Keywords: piRNA silencing, reporter screen, nuclear pore, nucleolus, integrator, nuclear import
Brown et al. use a sensitized piRNA reporter to identify factors involved in piRNA silencing in C. elegans. The authors reveal diverse factors critical for the localization and function of piRNA pathway factors, as well as novel connections between nucleoli, the nuclear pore, and germ granules in piRNA silencing. The results also support an emerging model that the proper balance between gene silencing and gene licensing pathways is critical for optimal piRNA-guided genome surveillance.
Introduction
To defend against invading nucleic acids, organisms must identify and silence foreign RNAs while preserving the expression of endogenous RNAs. When organisms fail to combat virus and transposon derived invasive nucleic acids, these elements can destabilize the genome and lead to widespread mutagenesis and infertility (Brennecke et al. 2007). One system animals have evolved to combat foreign nucleic acids is comprised of PIWI-clade Argonaute and its associated noncoding PIWI-interacting RNAs (piRNAs) (Saito et al. 2006; Batista et al. 2008; Das et al. 2008). In Caenorhabditis elegans, piRNAs are able to initiate a robust silencing signal against mRNAs deemed as non-self (GFP, for example) which can be inherited for as many as 20 generations after the loss of the initiating PIWI protein complex (Ashe et al. 2012). Because piRNA-initiated gene silencing is heritable independent of the initiation factors, piRNA reporters that rely on activation of a piRNA-silenced GFP transgene are biased to detect components that are required for maintaining and amplifying the already initiated silencing signal. For this reason, some of the requirements for the piRNA pathway in C. elegans likely remain unknown.
Here, we present a candidate RNA interference (RNAi) screen in C. elegans using a sensitized reporter that relies on the previously characterized phenomenon that particular GFP transgenes have distinct propensities to be silenced when challenged with a perfectly complementary piRNA (Seth et al. 2018). We used this piRNA reporter to test for the involvement of small nuclear (snRNA) processing factors, mRNA processing factors, germ granules components, and protein transport factors in piRNA-mediated silencing. We further characterized components from each of these categories to better understand how they mediate silencing in C. elegans. Our data suggest that the more recently evolved piRNA pathway utilizes highly evolutionarily conserved snRNA processing, pre-mRNA splicing, and protein import factors to carry out small RNA silencing, including the biogenesis of piRNAs and the regulation of Argonaute localization. Taken together, our results demonstrate that our piRNA reporter can identify various factors involved in distinct steps of the piRNA pathway. Our results also showed that proper piRNA-mediated transcriptome-wide surveillance relies on various regulatory factors that are not directly required for gene silencing.
Materials and methods
C. elegans strains
Animals were grown on standard nematode growth media (NGM) plates seeded with the Escherichia coli OP50 strain at 20°C. COP262 (knuSi221 [fib-1p::fib-1(genomic)::eGFP::fib-1 3′ UTR + unc-119(+)] II.), SS747 (bnIs1[pie-1::GFP::pgl-1 + unc-119(+)]), YY538 (hrde-1 (tm1200) III.), and YY1492 (mut-16(cmp3[mut-16::gfp::flag + loxP] I; znfx-1(gg634[HA::tagRFP::znfx-1]) II; pgl-1(gg640[pgl-1::3xflag::mCardinal]) IV.) were obtained from the Caenorhabditis Genetics Center. YY584 (ggSi1[hrde-1p::3xflag::gfp::hrde-1] II.) was a gift from Scott Kennedy. HCL199 (hrde-1 (tm1200) III; wago-10 (tm1186) V.) was outcrossed from WM191. HCL105 (gfp::csr-1 IV.) and HCL125 (gfp::prg-1 flag::mCherry::glh-1 I.) are described in (Chen et al. 2020). Creation of HCL202 (mCherry::npp-7(uoc21) I.) and HCL135 (prg-1(uoc8) oma-1::gfp I; cdk-1::gfp II; 21ur-anti-gfp(type I) IV.) are described below. Crosses used to generate the type I and type II piRNA reporter strains are depicted in Supplementary Fig. 1a.
RNAi
RNAi was performed by feeding animals with E. coli HT115 (DE3) strains expressing the appropriate double-stranded RNA. RNAi bacterial strains were obtained from the Ahringer C. elegans RNAi Collection (Source BioScience). Bacterial cultures were grown in Luria broth supplemented with 100 μg/mL ampicillin and 50 μg/mL tetracycline overnight at 37°C. Cultures were seeded on NGM plates containing 100 μg/mL ampicillin, 50 μg/mL tetracycline, and 1 mM IPTG and incubated at room temperature until dry. L4 hermaphrodites were picked onto the plates for feeding at 20°C and removed after 24 h. Adult progeny were screened. In instances where L4 treatment resulted in early next generation arrest, L1s were plated and screened as adults. Experiments where same generation screening was necessary are indicated in the relevant figure legend or data table. HT115 (DE3) expressing empty RNAi vector L4440 was used as the control. To assign significance to treatments in the piRNA reporter activation screen, a threshold of 20% activation was used as control treatment sometime resulted in spurious activation, but never resulted in over 20% naïve activation at 20°C for type I or type II piRNA reporters.
CRISPR
Cas9/sgRNA constructs. We used the online tool sgRNA Scorer 2.0 (https://crispr.med.harvard.edu/) to design sgRNAs. The sgRNAs were cloned into pDD162 (Dickinson et al. 2013) by overlapping PCR using pDD162 as the PCR template and the appropriate primers. Overlapping PCR products were inserted into pDD162 linearized with SpeI/BsrBI digestion by the seamless ligation cloning extract (SLiCE) method (Zhang and Glotzer 2015).
Donor constructs
To generate the flag::mCherry::NPP-7 donor construct, 500 bp upstream and 500 bp downstream of the npp-7 TSS, and the flag::mCherry coding sequences were amplified by PCR using N2 genomic DNA or plasmids containing flag::mCherry as templates. PCR fragments were inserted into pUC19 linearized with HindIII/KpnI digestion by SLiCE. Silent mutations were introduced in guide RNA targeting sites by site-directed mutagenesis in donor constructs using Phusion High-Fidelity DNA Polymerases (Thermo Fisher Scientific).
Fluorescence imaging
GFP- and RFP/mCherry-tagged fluorescent proteins were visualized in living nematodes by mounting young adult animals on 2% agarose pads with M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, and 86 mM NaCl) with 10 mM levamisole. Fluorescent images used for localization studies were captured using a Zeiss LSM800 confocal microscope with a Plan-Apochromat 40×/1.4 Oil objective. Fluorescent images used for screening for GFP expression were captured using a Zeiss Axio Imager M2 compound microscope with a Plan-Apochromat 40×/1.4 Oil objective. NPP-7 nucleolar localization in Supplementary Fig. 3f was inferred by observing NPP-7 expression to be within nuclei distinct from nuclear membranes in 3D space.
Fluorescent image quantification
Confocal images were processed and mean gray intensity values were quantified in ImageJ. Using a fixed circle area, three non-overlapping mean gray intensity measurements were made per gonad. Images of 7–8 gonads were quantified for each RNAi condition.
Quantitative real-time PCR
For mRNA expression
1 μg of total RNA was reverse transcribed with SuperScript IV Reverse Transcriptase (Invitrogen) in 1 × reaction buffer, 2 U SUPERase-In RNase Inhibitor (Invitrogen), 0.5 mM dNTPs, and 2.5 μM random hexamers. Each real-time PCR reaction consisted of 3 μL of cDNA, 1 μM forward gene-specific primer and 1 μM reverse gene-specific primer. The amplification was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) on the Bio-Rad CFX96 Touch Real-Time PCR Detection System. The experiments were repeated for a total of three technical replicates. tba-1 expression was used as a housekeeping gene for experiments shown in S1B and prg-1 for experiments shown in S3D.
For piRNA expression
For stem-loop real-time PCR to measure piRNA levels, 50 pM stem-loop reverse primer 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAG-n8-3′ (n8 = reverse complement sequences of last eight nucleotide acids in piRNA or miRNA). Each real-time PCR reaction consisted 4 μL of cDNA, 1 μM forward primer 5′-ACACTC-CAGCTGGG-n16-3′ (n16 = first 16 nucleotide acids in piRNA or miRNA), and 1 μM universal reverse primer 5′-CTCAAGTGTCGTGGAGTCGGCAA-3′. The amplification was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) on the Bio-Rad CFX96 Touch Real-Time PCR Detection System. The experiments were repeated for a total of four technical replicates. mir-35 expression was used as a housekeeping gene.
Western blotting
Lysates were prepared from ∼100 worms in M9. 4X NuPAGE LDS Sample Buffer and a final concentration of 50 mM of DTT were added. Samples were boiled (10 min), and proteins were separated by standard SDS-PAGE (NuPAGE precasted Bis-Tris Gel in MOPS running buffer), and transferred to nitrocellulose membrane using the semi-dry transfer system (Bio-Rad). The membrane was blocked in 5% non-fat milk in TBST for 1 h at room temperature and incubated with primary antibodies (1:1,000 anti PRG-1 (gift from C. Mello) and 1:1,000 anti GFP (Santa Cruz sc-9996) diluted in 5% milk) for 1 h at room temperature. Antibodies were washed off with TBST for 5 min/5 times, incubate with secondary antibodies (1:15,000 anti mouse and 1:100,000 anti rabbit diluted in 5% milk) for 1 h at room temperature, wash off antibodies with TBST for 5 min/5 times. Bands were visualized by ECL Select Western Blotting Detection Reagent (Amersham) and imaged by Amersham Imagequant 800. Experiments were performed twice and results were quantified using ImageJ to measure band intensities. PRG-1 was used as an internal control.
Small RNA sequencing
Library preparation
Total RNA was extracted from whole animals of ∼100,000 synchronized young adults. Small (<200 nt) RNAs were enriched with mirVana miRNA Isolation Kit (Ambion). In brief, 80 μL (200–300 μg) of total RNA, 400 μL of mirVana lysis/binding buffer and 48 μL of mirVana homogenate buffer were mixed well and incubated at room temperature for 5 min. Then, 176 μL of 100% ethanol was added and samples were spun at 2500×g for 4 min at room temperature to pellet large (>200 nt) RNAs. The supernatant was transferred to a new tube and small (<200 nt) RNAs were precipitated with pre-cooled isopropanol at -70°C. Small RNAs were pelleted at 20,000×g at 4°C for 30 min, washed once with 70% pre-cooled ethanol, and dissolved with nuclease-free water. Ten micrograms of small RNAs were fractionated on a 15% PAGE/7M urea gel, and RNA from 17 nt to 40 nt was excised from the gel. RNA was extracted by soaking the gel in 2 gel volumes of NaCl TE buffer (0.3 M NaCl, 10 mM Tris-HCl, 1 mM EDTA pH 7.5) overnight. The supernatant was collected through a gel filtration column. RNA was precipitated with isopropanol, washed once with 70% ethanol, and resuspended with 15 μL nuclease-free water. RNA samples were treated with RppH to convert 22G-RNA 5′ triphosphates to monophosphates in 1 × reaction buffer, 10 U RppH (New England Biolabs), and 20 U SUPERase-In RNase Inhibitor (Invitrogen) for 3 h at 37°C, followed by 5 min at 65°C to inactivate RppH. RNA was then concentrated with the RNA Clean and Concentrator-5 Kit (Zymo Research). Small RNA libraries were prepared according to the manufacturer's protocol of the NEBNext Multiplex Small RNA Sample Prep Set for Illumina-Library Preparation (New England Biolabs). NEBNext Multiplex Oligos for Illumina Index Primers were used for library preparation (New England Biolabs). Libraries were sequenced using an Illumina HiSeq4000 to obtain single-end 50 nt sequences at the University of Chicago Genomic Facility.
Analysis
Fastq reads were trimmed using custom perl scripts. Trimmed reads were aligned to the C. elegans genome build WS230 using bowtie ver 1.2.1.1 (Langmead et al. 2009) with options -v 0 --best --strata. After alignment, reads that were between 17 and 40 nucleotides in length were overlapped with genomic features (rRNAs, tRNAs, snoRNAs, miRNAs, piRNAs, protein-coding genes, pseudogenes, transposons) using bedtools intersect (Quinlan and Hall 2010). Sense and antisense reads mapping to individual miRNAs, piRNAs, protein-coding genes, pseudogenes, RNA/DNA transposons, simple repeats, and satellites were totaled and normalized to reads per million (RPM) by multiplying be 1e6 and dividing read counts by total mapped reads, minus reads mapping to structural RNAs (rRNAs, tRNAs, snoRNAs) because these sense reads likely represent degraded products. Reads mapping to multiple loci were penalized by dividing the read count by the number of loci they perfectly aligned to. Reads mapping to miRNAs and piRNAs were only considered if they matched to the sense annotation without any overlap. Annotations for miRNAs were according to miRBase 16, and piRNA annotations were as previously described using PRG-1 IP data (Gu et al. 2012). In other words, piRNA and miRNA reads that contained overhangs were not considered as mature piRNAs or miRNAs, respectively. piRNA precursors were defined as sequences containing a full mature piRNA sequence plus a 2 nucleotide 5′ overhang corresponding to the genomic sequence 2 nucleotides upstream of that piRNA's mature 5′ end. 22G-RNAs were defined as 21 to 23 nucleotide long reads with a 5′G that aligned antisense to protein-coding genes, pseudogenes, or transposons. 22G-RNAs against Worm-specific ArGOnaute (WAGO) targets were considered if they met the above criteria and also aligned antisense to the coding region of WAGO-targeted genes, defined as genes whose mapped 22G-RNAs exhibit over two-fold enrichment from either WAGO-1 IP than that from input 22G-RNAs (Gu et al. 2009) or WAGO-9 IP than that from input 22G-RNAs (Shirayama et al. 2012). 22G-RNAs against CSR-1 targets were considered if they met the above criteria and algo aligned antisense to the coding region of CSR-1 targeted genes, defined as genes whose mapped 22G-RNAs exhibit over two-fold enrichment from CSR-1 IP than that from input 22G-RNAs (Claycomb et al. 2009). To control for the effects of adult gonad size variability, RNA extraction, and sequencing depth, read counts were normalized by total read counts against germline expressed miRNAs as defined previously (Minogue et al. 2018). RPM mapped germline expressed miRNAs (RPM) values were computed and used in all downstream analyses using custom R scripts using R version 4.0.3 (R Core Team 2020), which rely on packages ggplot2 (Wickham 2016), reshape2 (Wickham 2007), ggpubr (Kassambara 2020), and dplyr (Wickham et al. 2021). piRNA loci were defined as most snpc-4 dependent using mature type I normalized reads in snpc-4 RNAi and empty vector control libraries. A Bayesian approach was used to select loci with at least a two-fold reduction in expression compared to control and a significant difference in expression (adjusted P < 0.05) (Maniar and Fire 2011)
Cap-dependent RNA sequencing
Library preparation
Total RNA isolation was performed as described above for small RNA sequencing. Capped RNA molecules were enriched as described previously (Gu et al. 2012). Terminator exonuclease (Epicentre) was used to selectively degrade monophosphorylated RNAs. Quick CIP (New England Biolabs) was used to dephosphorylate non-capped triphosphorylated RNAs. RppH was used with Thermopol Buffer (New England Biolabs) to decap RNAs for ligation. Libraries were prepared according to the manufacturer's protocol of the NEBNext Multiplex Small RNA Sample Prep Set for Illumina-Library Preparation (New England Biolabs). NEBNext Multiplex Oligos for Illumina Index Primers were used for library preparation (New England Biolabs). Libraries were sequenced using an Illumina NovaSeq6000 to obtain single-end 100 nt sequences at the University of Chicago Genomic Facility.
Analysis
Adaptor trimming and alignment were performed as described above for small RNA sequencing. Reads that were between 17 and 100 nucleotides were retained. Precursor length histograms were constructed based on lengths of unique piRNA precursor sequences (molecules containing a full mature piRNA sequence plus the 2 nucleotides upstream of the mature piRNA 5′ end, as described above) from each library. Success of cap-dependent isolation was determined by comparing the enrichment for piRNA precursors vs mature piRNAs in the cap-dependent libraries compared to the previously constructed traditional small RNA libraries.
Results
piRNA reporter detects loss of piRNA-dependent silencing factors
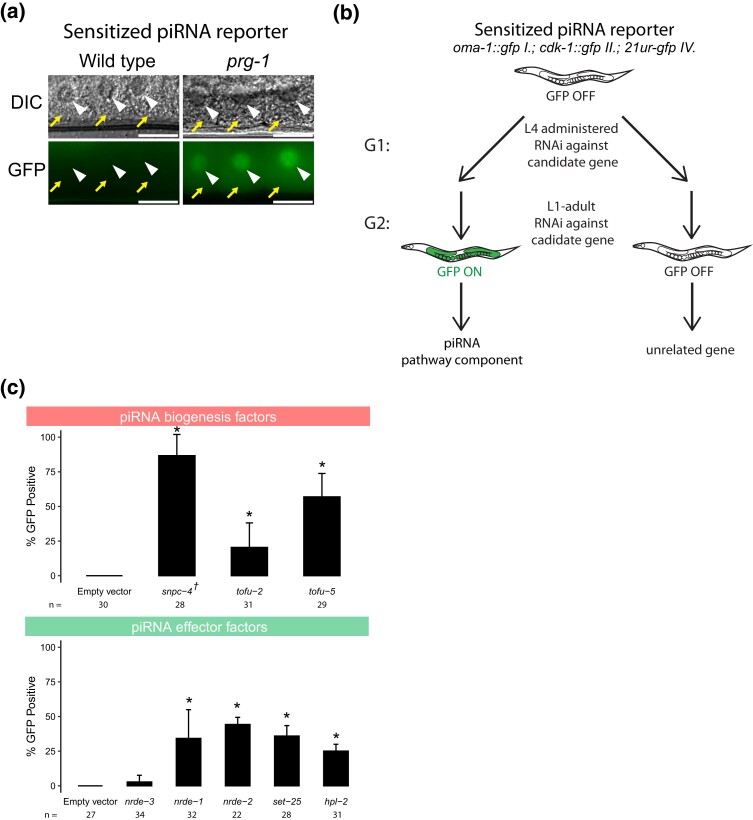
As described above, because piRNA targeting initiates a robust and stable silencing signal that persists even in the absence of the PIWI-clade Argonaute protein PRG-1, previous reporter-based strategies have been biased to detect downstream components of the piRNA pathway (Ashe et al. 2012; Bagijn et al. 2012). To remove this bias and increase the sensitivity of our screen, we aimed to establish a GFP reporter that is silenced in a piRNA dependent manner. We relied on previous observations that different GFP transgenes show distinct capacities to become silenced by the piRNA pathway (Seth et al. 2018). Our reporter strain contains two separate GFP transgenes with contrasting silencing propensities. One transgene, cdk-1::gfp, becomes stably silenced when challenged by an artificial piRNA (integrated into endogenously encoded piRNA locus 21ur-5499) that perfectly complements a 21nt site within the gfp coding sequence (Zhang et al. 2018). cdk-1::gfp remains very stably silenced for many generations, even after loss of the GFP-targeting piRNA (Shirayama et al. 2012). A second transgene, oma-1::gfp, is reported to carry anti-silencing capabilities. Not only does oma-1::gfp fail to be silenced by a perfectly complementary piRNA (Seth et al. 2018), it has also been shown previously that the piRNA resistance properties of oma-1::gfp can allow other GFP transgenes to escape piRNA silencing, a process referred to as RNA activation (Seth et al. 2018). When we crossed these two lines to one another to obtain a strain that contains both GFP transgenes as well as the perfectly complementary piRNA, both transgenes are silenced (Supplementary Fig. 1a). We hypothesized that because this strain contains one transgene that is vulnerable to piRNA silencing and another that resists piRNA silencing, then perhaps the silenced state of both GFP transgenes are not robust due to the constant competition of silencing and anti-silencing signals against GFP and can be easily perturbed by loss of the GFP-targeting piRNA. To test this hypothesis, we introduced a frameshift mutation to the sole PIWI-clade Argonaute prg-1 using the CRISPR/Cas9 system. We found that both transgenes became expressed at the first generation of prg-1 homozygosity (F2 generation) after injection (Fig. 1a and Supplementary Fig. 1b). Therefore, this reporter can successfully detect loss of prg-1, suggesting that a screen using this sensitized reporter would be sensitive to loss of upstream factors in the piRNA pathway.
Fig. 1.
piRNA reporter detects loss of piRNA biogenesis and effector factors. a) Fluorescent micrographs show the expression of GFP transgenes in oocyte cytoplasm and nuclei in the presence and absence of prg-1. The arrowheads indicate germ cell nuclei and yellow arrows indicate cytoplasm of oocytes. Scale bar, 10 μm. b) Schematic shows the screening strategy using the sensitized piRNA reporter. c) GFP expression in the piRNA reporter in indicated RNAi knockdowns. † indicates L1 to adult same generation RNAi treatment. Asterisks indicate significant reporter activation, defined as having at least 20% of worms screened showing visible GFP signals. Total worms screened shown below each bar. Error bars represent standard deviation of activation between replicate RNAi plates.
Although many factors in the C. elegans piRNA pathway have been characterized, there are likely unknown components of this pathway that have not yet been discovered due to the use of less sensitive reporters in genetic screens. We, therefore, sought to use our sensitized reporter to identify components involved in the pathway both at the piRNA biogenesis level and at the effector level (Supplementary Fig. 1c). We used a candidate approach by selecting specific factors to knockdown using RNAi (in L4 animals) and monitored the piRNA reporter expression in the next generation (Fig. 1b). We see both of GFP transgenes are co-silenced in a wild type background and are co-expressed in prg-1 mutants, and also in many other RNAi-treated animals reported here (Fig. 1a). Nonetheless, because OMA-1::GFP signal is only visible in oocytes, in experiments involving RNAi treatments where oocytes are not found, we assigned GFP activation state based on CDK-1::GFP expression alone. To ensure that this approach can successfully identify the loss of factors at both of these levels, we performed RNAi against known piRNA biogenesis and effector components. We saw that depletion of piRNA biogenesis factors snpc-4, tofu-2, and tofu-5 (Goh et al. 2014; Kasper et al. 2014) resulted in significant GFP expression in the reporter, as did effector factors nrde-1, nrde-2, set-25, and hpl-2 (Buckley et al. 2012; Spracklin et al. 2017; Fields and Kennedy 2019) (Fig. 1c). As expected, we failed to observe significant GFP expression when a somatic RNAi factor, nrde-3, was depleted with RNAi (Guang et al. 2008). These results suggested that our sensitized reporter can be used to detect loss of piRNA components using an RNAi based approach. The full list of candidates tested using our reporter can be found in Supplementary Table 1.
RNAi screen identifies P granule factors, mRNA processing factors, and protein transport factors as piRNA silencing components
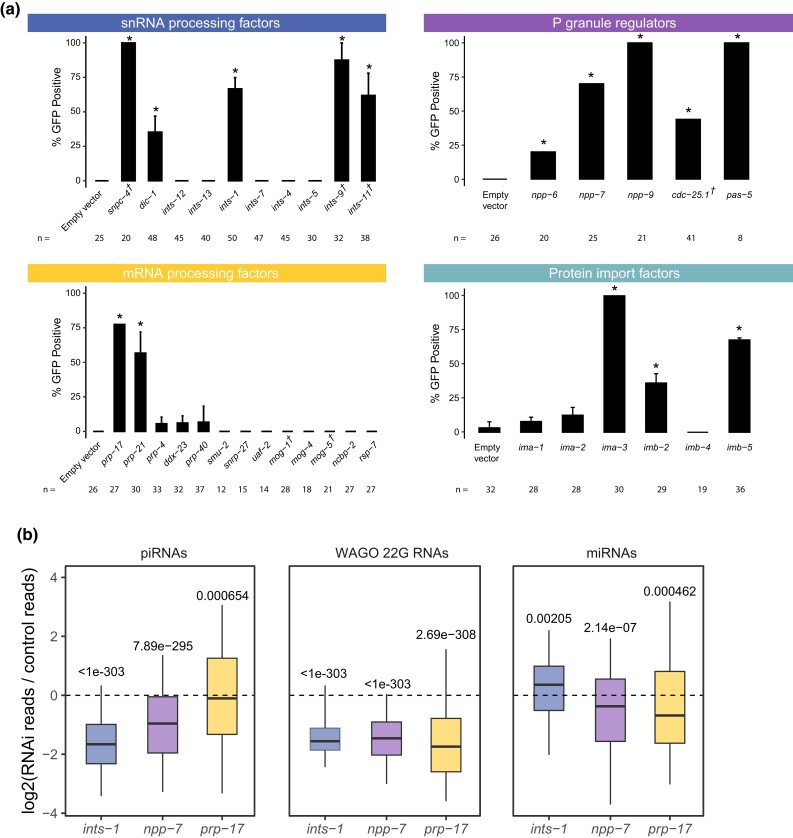
piRNA transcription in C. elegans is accomplished by RNA pol II. Since piRNA precursor transcription requires the SNPC-4 complex, which is also involved in initiation of snRNA transcription (Kasper et al. 2014), we hypothesized that the Integrator complex, a multimeric snRNA processing complex required for snRNA 3′ end processing, may be involved in the termination of piRNA transcription. Our model is consistent with the recent report that termination of piRNA precursor transcription involves the Integrator complex (Beltran et al. 2021; Berkyurek et al. 2021). We performed RNAi against nine different components of the Integrator complex in the sensitized reporter and found four of the components, dic-1, ints-1, ints-9, and ints-11, triggered GFP expression in the reporter following knockdown (Fig. 2a). We failed to observe GFP expression following knockdown of ints-4, ints-5, ints-7, ints-12, or ints-13 despite the gross morphological phenotypes resulting from most of these treatments (Supplementary Table 1). As individual knockdown of ints-1, ints-2, ints-4, ints-5, dic-1, ints-7, ints-8, ints-9, or ints-11 has been shown to disrupt snRNA 3′ end processing in C. elegans (Gómez-Orte et al. 2019), our finding suggests that a subset of the Integrator complex could be functioning at piRNA loci in a fashion distinct from its function at snRNA loci and coding genes. It has been shown recently that the mammalian Integrator complex form structurally distinct subcomplexes (Pfleiderer and Galej 2021). Although a subcomplex composed of ints-1, dic-1, ints-9, and ints-11 was not reported, our results suggest that these four components of the Integrator complex promote piRNA biogenesis.
Fig. 2.
Loss of mRNA processing factors, snRNA processing factors, protein import factors, and P granule regulators trigger piRNA reporter activation. a) GFP expression in the piRNA reporter in indicated RNAi knockdowns. † indicates L1 to adult same generation RNAi treatment. Asterisks indicate significant reporter activation, defined as having at least 20% of worms screened showing visible GFP signals. Total worms screened shown below each bar. Error bars represent standard deviation of activation between replicate RNAi plates. b) Small RNA fold changes in the indicated RNAi knockdowns relative to empty vector control knockdown. Statistical analysis was performed using a two-tailed Student's t-test against a null hypothesis that the fold change between treatment and control is 0. For all boxplots, lines display median values, boxes display first and third quartiles, and whiskers display 5th and 95th percentiles.
piRNA pathway components are enriched in phase-separated perinuclear organelles called P granules. Previous studies have shown that when P granules are lost, RNAi is compromised (Spike et al. 2008). We recently reported that loss of both glh-1 and glh-4 or of meg-3 and meg-4 led to GFP expression in the piRNA reporter (Chen et al. 2022). It remains unclear, however, whether specific P granule regulators are important for gene silencing or whether the structures themselves are indispensable. We addressed this question by treated the sensitized reporter with RNAi against genes which have previously been shown to be enriched in or important for the formation of P granules (Updike and Strome 2009). We found that knockdown of nuclear pore components npp-6, npp-7, and npp-9, phosphatase cdc-25.1, and proteasome component pas-5 all led to GFP expression in the reporter (Fig. 2a). The specific role of npp-6/7 in promoting piRNA silencing is further explored below.
Pre-mRNA splicing has recently been shown to act as a signal for endogenous RNAi pathways in C. elegans (Akay et al. 2017; Tyc et al. 2017; Newman et al. 2018). In addition, knockdown of some splicing factors leads to defects in P granule formation (Updike and Strome 2009), and splicing factor EMB-4 is essential for piRNA-mediated silencing for reporters containing multiple introns (Akay et al. 2017; Tyc et al. 2017). We, therefore, wondered whether loss of particular splicing factors would trigger GFP expression in our sensitized reporter. We found that of the 13 splicing factors we tested, only loss of the second step factors prp-17 and prp-21 triggered GFP expression (Fig. 2a), despite the gross morphological phenotypes resulting from treatment with most other RNAi constructs (Supplementary Table 1). This finding suggests some specific pre-mRNA splicing factors contribute to piRNA silencing.
piRNA targeting triggers a silencing signal that relies on siRNAs to function with secondary Argonaute proteins called WAGOs both in the cytoplasm and in the nucleus (Billi et al. 2014). The RNA-dependent RNA polymerases that manufacture these siRNAs, termed 22G-RNAs due to their possession of a 5′ guanosine and 22nt length, all exist and function in the cytoplasm (Gu et al. 2009; Phillips et al. 2012). The nuclear Argonaute HRDE-1 (WAGO-9) is critical for the inheritance of gene silencing over generations (Buckley et al. 2012). However, it remains unknown how HRDE-1 and piRNA-dependent 22G-RNAs translocate into the nucleus in germ cells. We hypothesized that HRDE-1 binds 22G-RNAs in the cytoplasm and subsequently translocates into the nucleus as has been shown with the nuclear PIWI-clade Argonaute in Drosophila and the somatic nuclear Argonaute in C. elegans NRDE-3 (Guang et al. 2008; Yashiro et al. 2018). To identify the protein import machinery required for nuclear localization of HRDE-1, we depleted several Importin factors in our sensitized reporter and observed GFP expression following ima-3, imb-2, and imb-5 knockdown, but not following ima-1 or ima-2 knockdown (Fig. 2a). Since ima-1 RNAi knockdown led to sterility but not piRNA reporter activation, this suggests that specific protein import machinery is essential for piRNA-mediated gene silencing (see more details on the role of ima-3 in HRDE-1 nuclear localization below).
Because our sensitized reporter can detect piRNA pathway defects at any level of the pathway, such as piRNA biogenesis or production of WAGO-associated 22G-RNAs (Fig. 1c), we next sought to characterize the defects contributed by loss of the factors reported above. To accomplish this goal, we performed RNAi against a subset of these factors and sequenced the small RNAs from a population of depleted animals. While loss of npp-7, prp-17, or ints-1 all led to a reduction in 22G accumulation on WAGO-targeted genes (Fig. 2b), RNAi of Integrator complex component ints-1 caused an extreme reduction in piRNAs as well. Reduction of both piRNA and 22G-RNA levels is consistent with the Integrator complex's involvement in piRNA biogenesis, as compromised piRNA production can also lead to reduced 22G-RNA accumulation as is the case in prg-1 mutants (Reed et al. 2020). In contrast, the reduction in 22G-RNA but not piRNA levels suggests that prp-17 likely functions as an effector in the piRNA pathway and does not function in piRNA biogenesis. We then characterized the defects in each of these cases further.
Integrator complex resolves piRNA precursor 3′ ends and promotes the production of both type I and type II piRNAs
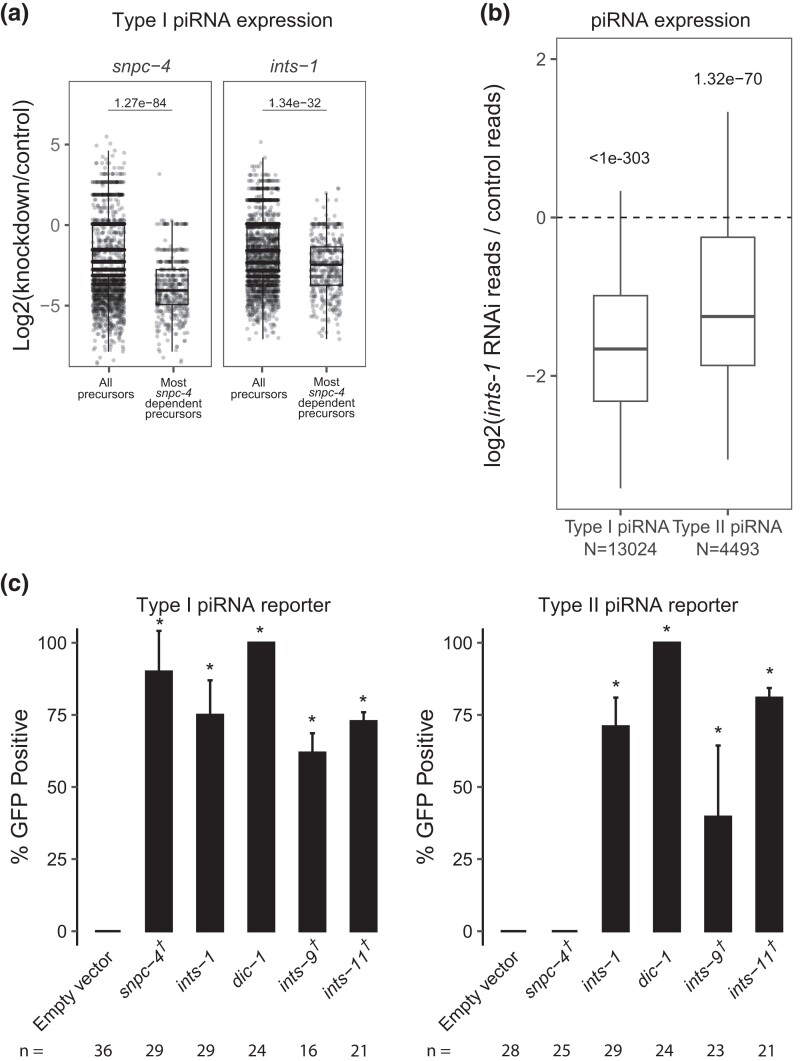
As described above, we found that loss of a subset of Integrator complex components caused GFP expression in our sensitized piRNA reporter, consistent with a recent report (Beltran et al. 2021). In addition, animals depleted for Integrator component ints-1 exhibited a global reduction in mature piRNA production (Fig. 2b and Supplementary Fig. 2a). Furthermore, piRNA precursor levels are strongly decreased following ints-1 knockdown (Supplementary Fig. 2b). In mammals and Drosophila, the SNAPc complex is required for Integrator complex action to bind the promoter of snRNA genes (Baillat and Wagner 2015). The SNAPc homolog SNPC-4 in C. elegans is also required for piRNA biogenesis and interacts with piRNA promoters (Kasper et al. 2014). Upon snpc-4 RNAi (a partial depletion of SNPC-4), some piRNAs exhibit more depletion than others (Supplementary Fig. 2a). While the biological reasons for this difference in sensitivity are unknown to us, we reasoned that if SNPC-4 recruits the Integrator complex to piRNA loci, then piRNAs that rely most on SNPC-4 for their accumulation should also rely most on the Integrator. We found that this was the case for INTS-1 (Fig. 3a). Together, these observations suggest that the Integrator plays a role in regulating the production or processing of piRNA precursors.
Fig. 3.
The integrator complex promotes biogenesis of both type I and type II piRNAs. a) piRNA precursor fold changes in indicated knockdowns relative to empty vector control knockdown. All piRNA precursors are compared to precursors originating from loci which most depend on snpc-4 for the accumulation of their mature sequences. Statistical analysis was performed using a two-tailed Mann–Whitney Wilcoxon test. For all boxplots, lines display median values, boxes display first and third quartiles, and whiskers display 5th and 95th percentiles. b) piRNA fold changes in ints-1 knockdown relative to empty vector control knockdown. Type I and type II piRNAs are compared separately. Statistical analysis was performed using a two-tailed Student's t-test against a null hypothesis that the fold change between treatment and control is 0. For all boxplots, lines display median values, boxes display first and third quartiles, and whiskers display 5th and 95th percentiles. c) GFP expression in the type I and type II piRNA reporters in indicated RNAi knockdowns. † indicates L1 to adult same generation RNAi treatment. Asterisks indicate significant reporter activation, defined as having at least 20% of worms screened showing activated GFP. Total worms screened shown below each bar. Error bars represent standard deviation of activation between replicate RNAi plates.
At snRNA loci in C. elegans and Drosophila, loss of the Integrator complex results in transcriptional readthrough and consequently elongated snRNA transcripts (Ezzeddine et al. 2011; Gómez-Orte et al. 2019). Therefore, we predicted that Integrator depletion would result in readthrough of piRNA precursors at piRNA loci. Since piRNA precursor molecules are rare compared to mature piRNAs and other abundant small RNA species, we performed CapSeq to enrich for capped piRNA precursor molecules (Gu et al. 2012). In empty vector-treated control animals, we found the size of piRNA precursor lengths peak at 25 with minor peaks at 41 and 61 nucleotides. Following ints-1 depletion, animals showed less defined peaks and elongated piRNA precursors (Supplementary Fig. 2c), consistent with a role for the Integrator complex in transcriptional termination of piRNA precursors. These findings support a model where at type I piRNA loci, SNPC-4 recruits a subset of the Integrator complex to elongating RNA pol II, leading to the endonucleolytic cleavage and subsequent termination of nascent piRNA precursors (Supplementary Fig. 2d).
SNPC-4 interacts with an upstream sequence called the Ruby motif, which is only found at type I piRNA loci (Batista et al. 2008). Surprisingly, we found that both type I and type II piRNA accumulation were globally reduced following ints-1 knockdown, although type II accumulation was less affected than type I following depletion of either component (Fig. 3b). Our finding contradicts the recent report that INTS-11 does not function at type II piRNA loci (Beltran et al. 2021), so we sought to clarify this point. To investigate whether the Integrator is required for gene silencing by type II piRNAs, we built a type II piRNA reporter using the same GFP transgenes as our sensitized type I piRNA reporter, but in this case, the GFP-targeting piRNA is made from a type II piRNA locus (Gu et al. 2012). We found that snpc-4 depletion leads to only type I reporter activation but not type II reporter activation, consistent with the role of snpc-4 in the biogenesis of type 1 but not type 2 piRNAs. However, RNAi depletion of ints-1, dic-1, ints-9, or ints-11 resulted in GFP expression in both type I and type II reporters (Fig. 3c). This suggests that at type I piRNA loci, the Integrator functions with SNPC-4 to promote piRNA accumulation while at type II piRNA loci, the Integrator can function in a SNPC-4 independent manner to promote piRNA biogenesis.
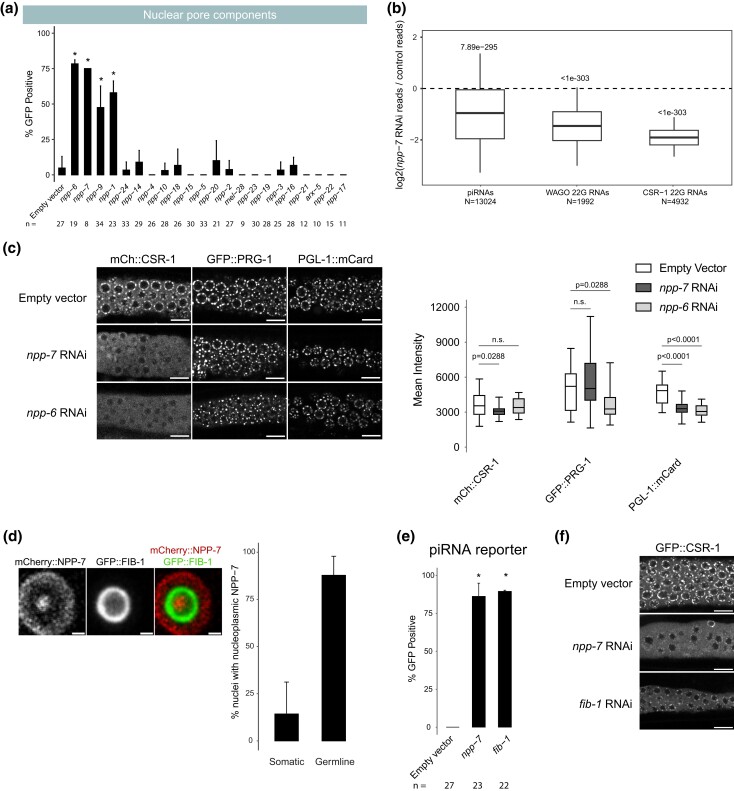
Nuclear pore components NPP-1/6/7 maintain CSR-1 perinuclear accumulation and promote piRNA silencing
Because we found that knockdown of three nuclear pore components in our initial screen activated piRNA reporter GFP expression, we expanded our screen to include additional nuclear pore components. Out of over 20 nuclear pore components tested, only four nuclear pore components (npp-1, npp-6, npp-7, and npp-9) caused over 40% GFP expression in our reporter when depleted (Fig. 4a), despite most treatments resulting in morphologically effected worms (Supplementary Table 1). This suggests that only a subset of the factors that make up the nuclear pore are essential for piRNA silencing. As describe above, we found that loss of npp-7 resulted in a reduction in piRNAs and in 22G-RNAs that target WAGO-associated genes (Fig. 2b). To better understand the role NPP-7 might play in piRNA biogenesis, we took advantage of the GFP-targeting piRNA present in our sensitized reporter. We used RT-qPCR to measure the relative expression of the GFP-targeting piRNA during control knockdown and during npp-7 knockdown. Surprisingly, we found that the GFP-targeting piRNA does not show decreased expression compared to control (Supplementary Fig. 3a). This suggests that while piRNA biogenesis may be affected, the reason we observed reporter activation may not be due to a function for NPP-7 in piRNA biogenesis. Furthermore, when we performed RNAi against npp-1, npp-6, or npp-7 using the type II piRNA reporter established above, each treatment led to type II reporter activation (Supplementary Fig. 3b), consistent with nuclear pore components functioning downstream of piRNA biogenesis. Notably, 22G-RNAs that map to CSR-1 targeted genes were significantly more reduced than 22G-RNAs that map to WAGO-targeted genes following npp-7 knockdown (Fig. 4b), indicating that NPP-7 may play a critical role in regulating CSR-1 function.
Fig. 4.
Nuclear pore and nucleolar components NPP-6/7 and FIB-1 regulate CSR-1 localization and promote piRNA dependent silencing. a) GFP expression in the piRNA reporter in indicated RNAi knockdowns. Asterisks indicate significant reporter activation, defined as having at least 20% of worms screened showing activated GFP. Total worms screened shown below each bar. Error bars represent standard deviation of activation between replicate RNAi plates. b) Small RNA fold changes in npp-7 RNAi knockdowns relative to empty vector control knockdown. Statistical analysis was performed using a two-tailed Student's t-test against a null hypothesis that the fold change between treatment and control is 0. For all boxplots, lines display median values, boxes display first and third quartiles, and whiskers display 5th and 95th percentiles. c) (Left) Fluorescent micrographs of live adult worm gonads show the localization of mCherry::CSR-1, GFP::PRG-1, and PGL-1::mCardinal in indicated RNAi knockdowns. Scale bar, 10 μm. (Right) Quantification of fluorescent signal from 7 to 8 gonads per condition. Statistical analysis was performed using a two-tailed Mann–Whitney Wilcoxon test. For all boxplots, lines display median values, boxes display first and third quartiles, and whiskers display minimum and maximum values. d) Fluorescent micrograph of individual live adult worm germline nucleus shows the localization of mCherry::NPP-7 and GFP::FIB-1. Panels display a single slice through the nucleus center. Percentage of soma and germline nuclei with nucleoli that contain NPP-7 are quantified to the right. Ten worms were quantified and calculations were pooled. >30 germline and somatic nuclei were scored per worm. Scale bar, 1 μm. e) GFP expression in piRNA reporter in indicated RNAi knockdowns. Asterisks indicate significant reporter activation, defined as having at least 20% of worms screened showing visible GFP signals. Total worms screened shown below each bar. Error bars represent standard deviation of activation between replicate RNAi plates. f) Fluorescent micrographs of live adult worm gonads show the localization of GFP::CSR-1 in indicated RNAi knockdowns. Scale bar, 10 μm.
CSR-1 is an Argonaute protein that prevents PRG-1 from targeting endogenous germline genes (Wedeles et al. 2013; Shen et al. 2018; Wu et al. 2023). Consistent with a more pronounced loss of CSR-1 bound 22G-RNAs following npp-7 knockdown, we also observed mis-localization of CSR-1 protein in npp-7 and npp-6 RNAi-treated animals, and we failed to observe mis-localization of two other P granule proteins, PRG-1 and PGL-1, under the same conditions (Fig. 4c, left). We found that the total fluorescent signal from tagged CSR-1 decreased significantly following npp-7 treatment in addition to the observed localization defect. However, tagged PRG-1 and PGL-1 fluorescent intensities were also significantly decreased following npp-6 and npp-7 or npp-6 knockdown, respectively, despite showing no localization defect (Fig. 4c, right). To more robustly characterize the change in CSR-1 protein expression following depletion of npp-6 or npp-7, we measured the expression of csr-1 mRNA and CSR-1 protein using RT-qPCR and Western blot, respectively (Supplementary Fig. 3c). We found that loss of npp-6 or of npp-7 does not lead to a change in csr-1 mRNA expression, but each treatment does lead to a significant reduction in CSR-1 protein expression relative to PRG-1 protein. This suggests that the localization defect apparent in our microscopy data may also be accompanied by a protein stability defect for CSR-1 following npp-6 or npp-7 knockdown. We hypothesize that the stability defect is related to the observed localization defect, but this remains to be tested. Together, these results suggest that the localization and stability of CSR-1 are compromised in npp-6 and npp-7 knockdown animals. If the GFP expression that we observed in our piRNA reporter following npp-7 knockdown was caused by a CSR-1 defect, then we reasoned that knockdown of csr-1 itself by RNAi should also result in GFP expression. Indeed, we found that knockdown of csr-1 or H3K36 methyl transferase mes-4 both caused GFP expression in our reporter (Supplementary Fig. 3d). This suggests that disruption of CSR-1 or of chromatin modifiers associated with gene expression can also impact piRNA-mediated gene silencing. Therefore, our observations suggest that npp-6 and npp-7 may contribute to piRNA silencing by regulating the CSR-1 pathway.
We wondered how predictive dispersal of CSR-1 was on piRNA reporter activation for the remaining nuclear pore components. Therefore, we monitored the expression of CSR-1 and PRG-1 in live adult animals following knockdown of all previously screened nuclear pore components (Supplementary Fig. 3e). We found that knockdown of npp-1 caused CSR-1 but not PRG-1 to become mis-localized as with npp-7 and npp-6. These observations suggest that NPP-1, NPP-6, and NPP-7 specifically impact the localization of CSR-1 but not that of other P granule factors at the nuclear periphery, and these three factors also promote piRNA-dependent gene silencing.
On the other hand, npp-9 knockdown led to piRNA reporter activation but did not cause dispersal of CSR-1 or of PRG-1 (Supplementary Fig. 3e). This result was surprising, as it has been previously shown that RNAi knockdown of npp-9 does cause CSR-1 dispersal in C. elegans embryos (Updike and Strome 2009). Conversely, knockdown of npp-24 and npp-2 cause CSR-1 dispersal but do not lead to piRNA reporter activation. Therefore, there is an imperfect correlation between nuclear pore component loss leading to piRNA reporter activation and CSR-1 dispersal. Nonetheless, our study reveals that select nuclear pore components, including NPP-1/6/7, promote CSR-1 perinuclear localization and are required for proper piRNA silencing.
Nucleolar component FIB-1 contribute to piRNA silencing and regulate the localization of NPP-7 and CSR-1
The Drosophila homolog of NPP-7, Nup153, has been shown to function both at the nuclear membrane as a component of the nuclear pore and as a soluble component within the nucleoplasm to promote both gene expression and silencing (Vaquerizas et al. 2010; Jacinto et al. 2015). We, therefore, wondered whether NPP-7 in C. elegans may also be expressed intranuclearly to help CSR-1 define transcriptionally active regions of the genome (Cecere et al. 2014). We used the CRISPR/Cas9 system to tag endogenous NPP-7 with mCherry and observed its expression in adult animals, and we found that in addition to its strong expression at the nuclear periphery, NPP-7 is also expressed as a single distinct focus in the nucleoplasm (Fig. 4d). Surprisingly, we found that the single focus of intranuclear NPP-7 in germ cells resides in the nucleolus, and the constitutive nucleolar component FIB-1/fibrillarin surrounds the intranuclear NPP-7 focus. We found that nucleolar NPP-7 accumulates in nearly every germline nucleus but only rarely in somatic nuclei (Fig. 4d). Additionally, nucleolar but not membrane-associated accumulation of NPP-7 relies on FIB-1 (Supplementary Fig. 3f). We wondered whether reporter GFP expression following npp-7 RNAi occurred due to loss of membrane-associated or nucleolar NPP-7. If nucleolar NPP-7 is required for piRNA silencing, then we expected that loss of FIB-1, which disrupts nucleolar NPP-7 accumulation, would cause piRNA reporter activation. Indeed, fib-1 RNAi led to GFP expression in the piRNA reporter, consistent with involvement of the nucleolus in the piRNA pathway (Fig. 4e). This result does not exclude the possibility that both nucleolar as well as membrane bound NPP-7 are mutually required for piRNA silencing. In vertebrae, the NPP-2 homologue Nup85 is required for nuclear pore assembly following cell division (Harel et al. 2003). We reasoned that depletion of NPP-2 might disrupt the membrane bound fraction of NPP-7 while leaving the nucleolar fraction unaffected, giving us an opportunity to observe the consequences of membrane bound but not nucleolar loss of NPP-7. Indeed, we found that npp-2 knockdown affected the enrichment of NPP-7 in the nuclear membrane, resulting in a greater proportion of NPP-7 signal in the nucleolus (Supplementary Fig. 3f). As shown above, npp-2 knockdown also leads to CSR-1 dispersal from perinuclear granules (Supplementary Fig. 3e). The presence of nucleolar NPP-7 may explain why npp-2 knockdown does not compromise piRNA reporter silencing. Furthermore, npp-1 and npp-6 knockdown selectively disrupted NPP-7 nucleolar accumulation and also led to piRNA reporter GFP expression and CSR-1 dispersal. However, the correlation of nucleolar NPP-7 localization and piRNA silencing was not perfect as npp-5 knockdown disrupted nucleolar NPP-7 accumulation but did not lead to reporter GFP expression or CSR-1 dispersal.
We next sought to reconcile the two observations concerning NPP-7 presented here: (1) the involvement of NPP-7 in CSR-1 localization and function and (2) the role of nucleolar NPP-7 in piRNA pathway function. We hypothesized that these two observations may be directly connected if nucleolar NPP-7 is required for CSR-1 localization. To test this, we performed fib-1 RNAi in a GFP::CSR-1 tagged strain. Remarkably, fib-1 knockdown disrupts perinuclear CSR-1 accumulation (Fig. 4f), but not PRG-1 perinuclear accumulation similarly to npp-7 knockdown (Supplementary Fig. 3e). Together, our study uncovers roles for select nuclear pore and nucleolus components in promoting CSR-1 localization and piRNA silencing.
Importin component IMA-3 promotes HRDE-1 silencing and localization
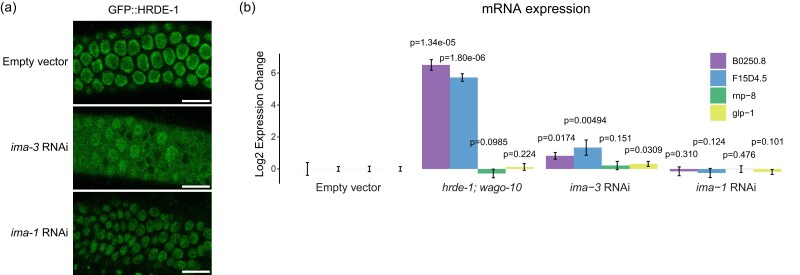
We found that depletion of three Importin family components, ima-3, imb-2, and imb-5, caused GFP expression in the sensitized reporter (Fig. 2a). Activation of the type II piRNA reporter was also found in ima-3 and imb-5 knockdown, supporting a role for the Importin family downstream of piRNA biogenesis (Supplementary Fig. 4a). Since knockdown of HRDE-1 cofactors NRDE-1 and NRDE-2 led to activation of our piRNA reporter, we hypothesized that HRDE-1 nuclear import might be compromised following depletion of these factors, which would in turn impact piRNA-mediated silencing. To test this, we treated a GFP::HRDE-1 tagged animal with ima-3 and observed HRDE-1 localization in dissected adult gonads using confocal microscopy. Indeed, in the ima-3 depleted germline, HRDE-1 failed to localize to nuclei. On the contrary, in the ima-1 depleted germline, which continues to silence the piRNA reporter, HRDE-1 localization was unaffected (Fig. 5a). These results are consistent with a model in which IMA-3 promotes piRNA-mediated gene silencing by translocating 22G-RNA-bound HRDE-1 from the cytoplasm to the nucleus where it can interact with nascent RNAs to transcriptionally silence piRNA targets. Interestingly, the nuclear Argonaute protein downstream of the piRNA pathway in Drosophila, Piwi, also relies on IMA-3 for translocation into the nucleus following piRNA binding (Yashiro et al. 2018).
Fig. 5.
IMA-3 promotes HRDE-1 nuclear import. a) Fluorescent micrographs of live adult worm gonads show the localization of GFP::HRDE-1 in indicated RNAi knockdowns. Scale bar, 10 μm. b) RT-qPCR results show expression change in indicated genotypes (hrde-1; wago-10) or RNAi knockdowns (Empty vector, ima-3, and ima-1). Statistical significance from two-tailed Student's t-test compared to control is displayed above treatments. Error bars represent standard deviation of expression between technical replicates.
To further support this model, we asked whether endogenous genes that are silenced by HRDE-1 also rely on IMA-3 to maintain that silenced state. Using RT-qPCR, we found that B0250.8 and F15D4.5 are significantly upregulated both in hrde-1 mutants and following ima-3 RNAi treatment (Fig. 5b). Therefore, we conclude that HRDE-1 translocates from the cytoplasm to the nucleus using the Importin α member IMA-3. Importin α members are known to function as adaptor molecules between protein cargo that contains a nuclear localization sequence (NLS) and Importin β members (Mattaj and Englmeier 1998). In C. elegans, the somatic nuclear Argonaute protein NRDE-3 contains a NLS that, when mutated, results in failure of NRDE-3 to translocate into nuclei (Guang et al. 2008). Surprisingly, despite sharing significant sequence identity with HRDE-1, the NLS within NRDE-3 is not fully conserved in HRDE-1 (Supplementary Fig. 4b). Therefore, how IMA-3 recognizes HRDE-1 for nuclear translocation requires further study.
Discussion
In this study, we have utilized a sensitized reporter strain which detects defects in both piRNA biogenesis and effector factors to identify and characterize factors involved in piRNA silencing. Using our reporter, we have identified pre-mRNA splicing factors, Integrator complex subunits, protein import components, and nuclear pore components as essential for piRNA-mediated gene silencing. While we have currently used our reporter in a candidate screen, it is likely possible to use this strain in a forward screen, which would allow for even less biased detection of piRNA pathway components. Whether the expression of GFP can be distinguished from the significant auto-fluorescence contributed by intestinal granules using a worm sorter remains to be seen and could represent a significant barrier to a high throughput approach.
We found that a subset of the Integrator complex contributes to both type I and type II piRNA biogenesis. Because both snRNA loci and type I piRNA loci require the SNAPc complex for biogenesis (Kasper et al. 2014; Baillat and Wagner 2015), we expected that the Integrator complex would be involved in piRNA biogenesis as described by recent reports (Beltran et al. 2021; Berkyurek et al. 2021). Our reporter assay and sequencing data support this model, but unexpectedly and inconsistent with the previously published findings, we also noticed that type II piRNA biogenesis and silencing were compromised following Integrator complex knockdown. Because SNPC-4 is recruited to Ruby motif-containing type I piRNA loci and not type II piRNA loci (Batista et al. 2008; Weng et al. 2019), our data suggest that the Integrator also functions at type II piRNA loci in a SNPC-4 independent manner. It has been shown that the Integrator can also function at protein coding genes to terminate transcription, promote RNAP II transcription proximal pausing, and promote enhancer RNA accumulation (Gardini et al. 2014; Stadelmayer et al. 2014; Lai et al. 2015; Skaar et al. 2015; Tatomer et al. 2019). While the canonical function of the Integrator at snRNA loci requires recognition of a conserved 3′ box sequence (Baillat et al. 2005), protein-coding genes which rely on the Integrator complex to cleave nascent RNAs do not contain a recognizable 3′ box sequence (Tatomer et al. 2019). Intriguingly, C. elegans snRNA loci are devoid of a 3′ box sequence as well (Thomas et al. 1990). Our data suggest that type II piRNA biogenesis represents yet another non-canonical role for the Integrator complex. Further investigation into the commonalities between snRNA loci, type I piRNA loci, type II piRNA loci, and protein coding genes may help to explain how the Integrator complex recognizes and cleaves nascent RNA molecules in C. elegans, and also may shed light on the Integrator's non-canonical roles in other organisms.
We found that a nuclear pore component, NPP-7, localizes to both the nuclear envelope and the nucleolus, and that depletion of both NPP-7 and of nucleolar component FIB-1 both activate our piRNA reporter. This suggests a potential role for the nucleolus in the piRNA pathway. In Drosophila ovarian somatic cells, nuclear Argonaute Piwi has been shown to localize to nucleoli under heat shock, correlating with the expression of retrotransposons which have integrated into rDNA copies (Mikhaleva and Leinsoo 2018). Additionally, it has been shown in C. elegans that nuclear RNAi machinery targets ribosomal RNAs (rRNAs) which are degraded by exosomes in the nucleolus (Liao et al. 2021). Both of these observations together with our own data suggest that the nucleolus may be involved in specialized small RNA-mediated silencing events. Additionally and possibly connected to nucleolar NPP-7, we found that NPP-7 promotes the localization of germ granule component CSR-1, but not germ granule components PRG-1 or PGL-1. We recently showed that CSR-1 uniquely retains its perinuclear localization when VASA helicases are mutated while PRG-1 and PGL-1 each show significant dispersal, suggesting that CSR-1 is recruited to the perinucleus in a VASA-independent manner (Chen et al. 2022). Our results suggest that the nuclear pore itself may represent a part of this VASA-independent pathway, and further that CSR-1 recruitment may require involvement of the nucleolus as fib-1 knockdown similarly led to specifically CSR-1 dispersal. It will be interesting to further investigate the mechanism by which select nuclear pore and nucleolar components promote the recruitment of CSR-1 to P granules in the future.
We observed that knockdown of licensing Argonaute csr-1 and of the H3K36 methyl transferase mes-4 both caused GFP expression in our piRNA reporter (Supplementary Fig. 2b). It has been shown that H3K36me3 occupancy and CSR-1 22G-RNA targeting are highly correlated (Cecere et al. 2014). However, it has also been shown that depletion CSR-1 does not reduce MES-4 localization to chromatin (Kreher et al. 2018). Therefore, we hypothesize that our piRNA reporter is activated when pathways that promote gene expression as well as those that control gene repression are disrupted. If correct, then this model would suggest that a balance between gene silencing and expression exists within the germline and disruption of pathways that control either gene silencing or gene expression can impinge on the alternative. Consistent with this notion, it has been shown that “resetting” germline RNAi pathways by re-introducing RNAi machinery to RNAi deficient C. elegans in the absence of piRNAs led to potent sterility due to the silencing of essential genes (de Albuquerque et al. 2015; Phillips et al. 2015). This finding suggests that silencing machinery is essential for keeping pathways that control silencing and expression distinct from one another and balanced in the germline. Our finding, conversely, suggests that factors which promote gene expression are also essential for maintaining this balance.
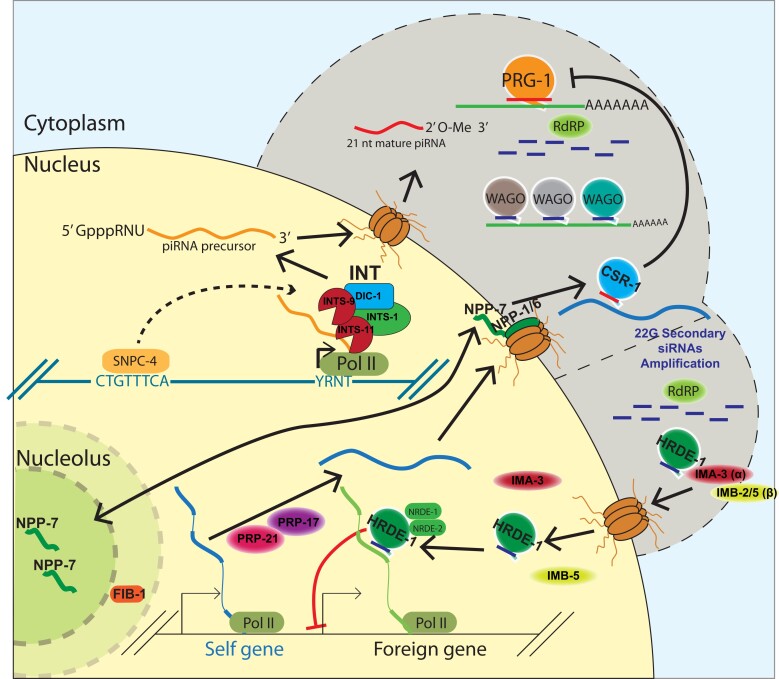
Altogether, we used a piRNA reporter assay to identify and characterize factors that act at multiple levels of gene silencing, revealing a role for snRNA processing machinery, pre-mRNA splicing factors, the nuclear pore, the nucleolus, and protein import machinery in initiating and maintaining small RNA-mediated gene regulation (Fig. 6).
Fig. 6.
Multifaceted involvement of conserved and essential machinery work in concert with small RNA-mediated gene silencing in germ cells. A model depicting a role for factors identified in our screen. The Integrator complex promotes piRNA biogenesis, a subset of nuclear pore factors promotes CSR-1 localization and interface with the nucleolus, second step splicing factors ensure proper endogenous splicing occurs, and protein import machinery allow for the nuclear Argonaute HRDE-1 to translocate into the nucleus to transcriptionally silence target genes.
Supplementary Material
Acknowledgements
We thank members of the Lee Lab for critical comments on the manuscript. Some strains used in this study were provided by the Caenorhabditis Genetics Center (CGC), which is funded by Office of Research Infrastructure Programs, National Institutes of Health (P40 OD010440).
Contributor Information
Jordan S Brown, Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL 60637, USA.
Donglei Zhang, Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL 60637, USA.
Olivia Gaylord, Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL 60637, USA.
Wenjun Chen, Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL 60637, USA; Department of Laboratory Medicine, Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong Province 510000, China.
Heng-Chi Lee, Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL 60637, USA.
Data availability
All sequencing data generated in this study (small RNA-seq and Cap-seq) area available at the NCBI SRA database with accession number PRJNA928769. All scripts described in the methods are available at https://github.com/Uchicago-BSD-hlee-lab/Brown_2023. All strains generated in this study are available upon request.
Supplemental material available at GENETICS online.
Funding
This work is supported in part by NIH predoctoral training grant T32 GM07197 to J.B.; the NIH grant R01-GM132457 to H.-C.L.
Literature cited
- Akay A, Di Domenico T, Suen KM, Nabih A, Parada GE, Larance M, Medhi R, et al. The helicase aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev Cell. 2017;42(3):241–255.e6. doi: 10.1016/j.devcel.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E-M, Mitchell J, Bagijn MP, Cording AC, Doebley A-L, et al. PiRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150(1):88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick E-M, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans PiRNAs. Science. 2012;337(6094):574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi M-A, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Baillat D, Wagner EJ. Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci. 2015;40(5):257–264. doi: 10.1016/j.tibs.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Graham Ruby J, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, et al. PRG-1 and 21U-RNAs interact to form the PiRNA complex required for fertility in C. elegans. Mol Cell. 2008;31(1):67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran T, Pahita E, Ghosh S, Lenhard B, Sarkies P. Integrator is recruited to promoter-proximally paused RNA Pol II to generate Caenorhabditis elegans PiRNA precursors. EMBO J. 2021;40(5):1–17. doi: 10.15252/embj.2020105564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkyurek AC, Furlan G, Lampersberger L, Beltran T, Weick E-M, Nischwitz E, Cunha Navarro I, et al. The RNA polymerase II subunit RPB-9 recruits the integrator complex to terminate Caenorhabditis elegans PiRNA transcription. EMBO J. 2021;40(5):e105565. doi: 10.15252/embj.2020105565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SEJ, Kim JK. Endogenous RNAi pathways in C. elegans. WormBook. 2014:1–49. doi: 10.1895/wormbook.1.170.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489(7416):447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Hoersch S, O’keeffe S, Sachidanandam R, Grishok A. Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat Struct Mol Biol. 2014;21(4):358–365. doi: 10.1038/nsmb.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Brown JS, He T, Wu WS, Tu S, Weng Z, Zhang D, Lee HC. GLH/VASA helicases promote germ granule formation to ensure the fidelity of PiRNA-mediated transcriptome surveillance. Nat Commun. 2022;13(1):5306. doi: 10.1038/s41467-022-32880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Hu Y, Lang CF, Brown JS, Schwabach S, Song X, Zhang Y, et al. The dynamics of P granule liquid droplets are regulated by the Caenorhabditis elegans germline RNA helicase GLH-1 via its ATP hydrolysis cycle. Genetics. 2020;215(2):421–434. doi: 10.1534/genetics.120.303052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139(1):123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BFM, Placentino M, Ketting RF. Maternal PiRNAs are essential for germline development following de novo establishment of Endo-SiRNAs in Caenorhabditis elegans. Dev Cell. 2015;34(4):448–456. doi: 10.1016/j.devcel.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, et al. Piwi and PiRNAs act upstream of an endogenous SiRNA pathway to suppress tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31(1):79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10(10):1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ. A subset of Drosophila integrator proteins is essential for efficient U7 SnRNA and spliceosomal SnRNA 3′-End formation. Mol Cell Biol. 2011;31(2):328–341. doi: 10.1128/mcb.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BD, Kennedy S. Chromatin compaction by small RNAs and the nuclear RNAi machinery in C. elegans. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-45052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, Lazar MA, Shilatifard A, Shiekhattar R. Integrator regulates transcriptional initiation and pause release following activation. Mol Cell. 2014;56(1):128–139. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh W-SS, Seah JWE, Harrison EJ, Chen C, Hammell CM, Hannon GJ. A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans PiRNA biogenesis. Genes Dev. 2014;28(7):797–807. doi: 10.1101/gad.235622.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Orte E, Sáenz-Narciso B, Zheleva A, Ezcurra B, de Toro M, López R, Gastaca I, et al. Disruption of the Caenorhabditis elegans integrator complex triggers a non-conventional transcriptional mechanism beyond SnRNA genes. PLoS Genet. 2019;15(2):e1007981. doi: 10.1371/journal.pgen.1007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee H-C, Chaves D, Youngman EM, Pazour GJ, Conte D, Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans PiRNA precursors. Cell. 2012;151(7):1488–1500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36(2):231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An argonaute transports SiRNAs from the cytoplasm to the nucleus. Science. 2008;321(5888):537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11(4):853–864. doi: 10.1016/S1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29(12):1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper DM, Wang G, Gardner KE, Johnstone TG, Reinke V. The C. Elegans SNAPc component SNPC-4 coats PiRNA domains and is globally required for PiRNA abundance. Dev Cell. 2014;31(2):145–158. doi: 10.1016/j.devcel.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. 2020. “Ggpubr: ‘ggplot2’ Based Publication Ready Plots.”.
- Kreher J, Takasaki T, Cockrum C, Sidoli S, Garcia BA, Jensen ON, Strome S. Distinct roles of two histone methyltransferases in transmitting H3K36me3-based epigenetic memory across generations in Caenorhabditis elegans. Genetics. 2018;210(3):969–982. doi: 10.1534/genetics.118.301353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Gardini A, Zhang A, Shiekhattar R. Integrator mediates the biogenesis of enhancer RNAs. Nature. 2015;525(7569):399–403. doi: 10.1038/nature14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Chen X, Xu T, Jin Q, Xu Z, Xu D, Zhou X, Zhu C, Guang S, Feng X. Antisense ribosomal SiRNAs inhibit RNA polymerase I-directed transcription in C. elegans. Nucleic Acids Res. 2021;49(16):9194–9210. doi: 10.1093/nar/gkab662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ. EGO-1, a C. Elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol. 2011;21(6):449–459. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Mikhaleva EA, Leinsoo TA. 2018. “The nucleolar transcriptome regulates Piwi shuttling between the nucleolus and the nucleoplasm.”.
- Minogue AL, Tackett MR, Atabakhsh E, Tejada G, Arur S. Functional genomic analysis identifies MiRNA repertoire regulating C. elegans oocyte development. Nat Commun. 2018;9(1):1–11. doi: 10.1038/s41467-018-07791-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Ji F, Fischer SEJ, Anselmo A, Sadreyev RI, Ruvkun G. The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes. Genes Dev. 2018;32(9–10):670–681. doi: 10.1101/gad.311514.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleiderer MM, Galej WP. Structure of the catalytic core of the integrator complex. Mol Cell. 2021;81(6):1246–1259.e8. doi: 10.1016/j.molcel.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Brown KC, Montgomery BE, Ruvkun G, Montgomery TA. PiRNAs and PiRNA-dependent SiRNAs protect conserved and essential C. elegans genes from misrouting into the RNAi pathway. Dev Cell. 2015;34(4):457–465. doi: 10.1016/j.devcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery TA, Breen PC, Ruvkun G. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012;26(13):1433–1444. doi: 10.1101/gad.193904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A Language and Environment for Statistical Computing. Vienna: R Core Team. [Google Scholar]
- Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, Vijayasarathy T, Parker DM, Nishimura EO, Updike DL, Montgomery TA. Widespread roles for PiRNAs and WAGO-class SiRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 2020;48(4):1811–1827. doi: 10.1093/nar/gkz1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20(16):2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Tang W, Shen E-Z, Tu S, Lee H-C, Weng Z, Mello CC. The coding regions of germline mRNAs confer sensitivity to argonaute regulation in C. elegans. Cell Rep. 2018;22(9):2254–2264. doi: 10.1016/j.celrep.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E-Z, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding Y-H, Dai S-Y, Weng Z, Mello CC. Identification of PiRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell. 2018;172(5):937–951.e18. doi: 10.1016/j.cell.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H-C, Gu W, Ishidate T, Conte D, Mello CC. PiRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150(1):65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, Washburn MP, Hughes SH, Pagano M. The integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015;25(3):288–305. doi: 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike CA, Bader J, Reinke V, Strome S. DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development. 2008;135(5):983–993. doi: 10.1242/dev.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spracklin G, Fields B, Wan G, Becker D, Wallig A, Shukla A, Kennedy S. The RNAi inheritance machinery of Caenorhabditis elegans. Genetics. 2017;206(3):1403–1416. doi: 10.1534/genetics.116.198812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatomer DC, Elrod ND, Liang D, Xiao M-S, Jiang JZ, Jonathan M, Huang K-L, Wagner EJ, Cherry S, Wilusz JE. The integrator complex cleaves nascent mRNAs to attenuate transcription. Genes Dev. 2019;33(21–22):1525–1538. doi: 10.1101/gad.330167.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Lea K, Zucker-Aprison E, Blumenthal T. The spliceosomal SnRNAs of Caenorhabditis elegans. Nucleic Acids Res. 1990;18(9):2633–2642. doi: 10.1093/nar/18.9.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc KM, Nabih A, Wu MZ, Wedeles CJ, Sobotka JA, Claycomb JM. The conserved intron binding protein EMB-4 plays differential roles in germline small RNA pathways of C. elegans. Dev Cell. 2017;42(3):256–270.e6. doi: 10.1016/j.devcel.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Updike DL, Strome S. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics. 2009;183(4):1397–1419. doi: 10.1534/genetics.109.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6(2):e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans argonaute CSR-1. Dev Cell. 2013;27(6):664–671. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- Weng C, Kosalka A, Berkyurek AC, Stempor P, Feng X, Mao H, Zeng C, et al. The USTC Complex co-opts an ancient machinery to drive PiRNA transcription in C. elegans. Genes Dev. 2019;33(1-2):90–102. doi: 10.1101/gad.319293.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Reshaping data with the {reshape} package. J Stat Softw. 2007;21(12):1–20. doi: 10.18637/jss.v021.i12 [DOI] [Google Scholar]
- Wickham H. Ggplot2: Elegant Graphics for Data Analysis Title. New York: Springer; 2016. [Google Scholar]
- Wickham H, François R, Henry L, Müller K. “Dplyr: A Grammar of Data Manipulation”, 2021.
- Wu W-S, Brown JS, Shiue S-C, Chung C-J, Lee D-E, Zhang D, Lee H-C. Transcriptome-wide analyses of PiRNA binding sites suggest distinct mechanisms regulate PiRNA binding and silencing in C. elegans. RNA. 2023;29(5):557–569. doi: 10.1261/rna.079441.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro R, Murota Y, Nishida KM, Negishi L, Siomi H, Siomi MC, Yashiro R, et al. Piwi nuclear localization and its regulatory mechanism in Drosophila ovarian somatic cells. Cell Rep. 2018;23(12):3647–3657. doi: 10.1016/j.celrep.2018.05.051. [DOI] [PubMed] [Google Scholar]
- Zhang D, Glotzer M. The RhoGAP activity of CYK-4/MgcRacGAP functions non-canonically by promoting RhoA activation during cytokinesis. ELife. 2015;4(AUGUST2015):1–25. doi: 10.7554/eLife.08898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu W-S, Huang W-C, Weng Z, Lee H-C. The PiRNA targeting rules and the resistance to PiRNA silencing in endogenous genes. Science. 2018;359(6375):587–592. doi: 10.1126/science.aao2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated in this study (small RNA-seq and Cap-seq) area available at the NCBI SRA database with accession number PRJNA928769. All scripts described in the methods are available at https://github.com/Uchicago-BSD-hlee-lab/Brown_2023. All strains generated in this study are available upon request.
Supplemental material available at GENETICS online.