Abstract
Cancer stem cells (CSCs) drive tumor growth, metastasis, and chemoresistance. While emerging evidence suggests that CSCs have a unique dependency on lipid metabolism, the functions and regulation of distinct lipid species in CSCs remain poorly understood. Here, we developed a stem cell factor SOX9-based reporter for isolating CSCs in primary tumors and metastases of spontaneous mammary tumor models. Transcriptomic analyses uncover that SOX9high CSCs up-regulate the ABCA12 lipid transporter. ABCA12 down-regulation impairs cancer stemness and chemoresistance. Lipidomic analyses reveal that ABCA12 maintains cancer stemness and chemoresistance by reducing intracellular ceramide abundance, identifying a CSC-associated function of ABCA subfamily transporter. Ceramide suppresses cancer stemness by inhibiting the YAP-SOX9 signaling pathway in CSCs. Increasing ceramide levels in tumors enhances their sensitivity to chemotherapy and prevents the enrichment of SOX9high CSCs. In addition, SOX9high and ABCA12high cancer cells contribute to chemoresistance in human patient-derived xenografts. These findings identify a CSC-suppressing lipid metabolism pathway that can be exploited to inhibit CSCs and overcome chemoresistance.
CSCs maintain stemness and enhance chemoresistance via the ABCA12-mediated expulsion of ceramide lipids that inhibit YAP-SOX9.
INTRODUCTION
Breast cancer displays high degrees of intratumor heterogeneity (ITH), containing distinct subpopulations of cancer cells with diverse genetic, epigenetic, phenotypic, or functional characteristics in each tumor (1, 2). ITH is one of the leading causes of therapeutic resistance to various treatments (3). One major source of ITH is the ability of cancer cells to acquire functionally diverse cell states through differentiation and dedifferentiation processes (4). In breast cancer, often only a small population of cancer cells are highly tumorigenic and show stem-like properties capable of both self-renewing and differentiating into less tumorigenic cells. These cancer stem cells (CSCs), defined by their tumor-initiating capacity, drive tumor growth, metastasis, and relapse (5–8). However, current therapeutic strategies do not effectively target CSCs. Therefore, a better understanding of the mechanisms controlling CSC induction and maintenance is needed, particularly in spontaneously developing primary tumors and metastases that recapitulate ITH. However, these studies have been hampered by a lack of specific CSC markers for spontaneous tumor models. Although combinations of cell-surface markers have been used to enrich CSCs, they are not unique to CSCs and identify rather heterogeneous cell populations (9, 10).
Metabolic reprogramming is one of the hallmarks of cancer (11–13). Emerging studies demonstrated that CSCs have unique metabolic phenotypes and exhibit metabolic plasticity between high levels of glycolysis and oxidative phosphorylation, depending on the tumor microenvironment (14–16). Furthermore, certain CSCs rely on high levels of fatty acid oxidation and cholesterol synthesis (17–19). However, whether any lipid pathways can inhibit cancer stemness remains poorly understood. As a prominent component of membrane lipids and a main class of bioactive lipid molecules, various sphingolipids play complex and often opposing functions in cancer. For example, ceramide, the central molecule of sphingolipid metabolism, can cause cell death and growth arrest. In contrast, its metabolic derivative, sphingosine1phosphate, has anti-apoptotic and tumor-promoting functions (20). Thus far, it is unclear whether sphingolipids play a role in regulating cancer stemness.
Master regulators of normal stem cell programs are often coopted by CSCs to promote cancer stemness. We and others have demonstrated that the transcription factor SOX9 acts as a key regulator of mammary stem and progenitor cells (21–26). In breast cancer cells, SOX9 down-regulation inhibits tumor-initiating activity and metastatic ability (21, 23, 26). Furthermore, SOX9 up-regulation promotes lineage plasticity and drives the dedifferentiation of luminal progenitor cells to generate basal-like breast cancer (BLBC) (26). These findings suggest that SOX9 is a key regulator of breast cancer stemness and may serve as a specific marker for breast CSCs. In this study, we developed spontaneous mouse tumor models expressing a Sox9–green fluorescent protein (GFP) transgenic reporter and demonstrated that cancer cells expressing high levels of SOX9 functioned as CSCs in both primary tumors and metastases in distinct mammary tumor models. Furthermore, through transcriptomic and lipidomic analyses of SOX9high CSCs, we uncovered a critical dependency of CSCs on ceramide homeostasis, providing a strategy for targeting CSCs.
RESULTS
Sox9-GFP reporter recapitulates heterogeneous SOX9 expression in spontaneous primary tumors and metastases
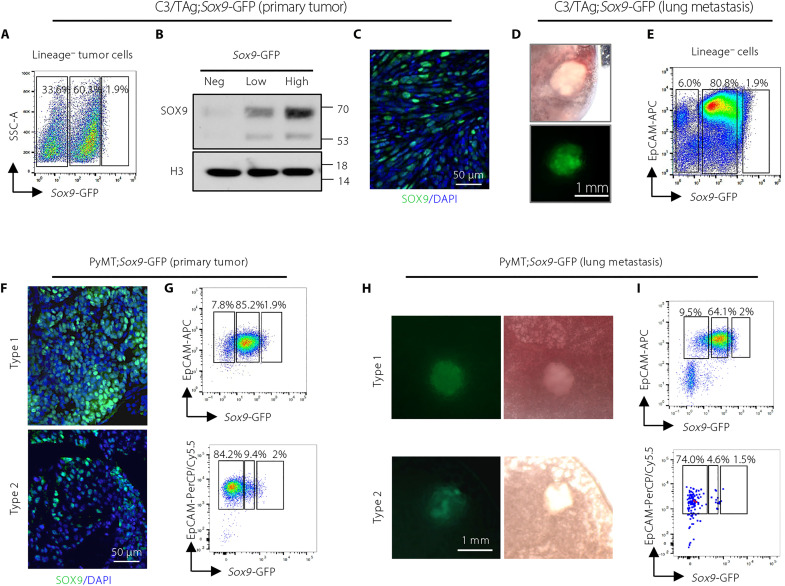
Evaluation of SOX9 expression using a single-cell RNA sequencing (RNA-seq) dataset (27) revealed substantial ITH of SOX9 levels in human breast tumors of various subtypes (fig. S1A). To investigate the function of cancer cells with different levels of SOX9, we used the C3/TAg BLBC mouse model carrying a Sox9-GFP reporter that expresses GFP under the control of the Sox9 enhancer and promoter region (26). C3/TAg mice develop a small population (~2%) of Sox9-GFPhigh cells in the premalignant mammary epithelium, and these cells serve as the precursors for tumor development (26). However, SOX9 expression patterns in invasive C3/TAg tumors and distant metastases have not been determined. We found that C3/TAg tumors contained heterogeneous Sox9-GFP–negative and Sox9-GFP–positive cancer cells, and the GFP-positive cells can be further stratified into GFP-low and GFP-high subpopulations (Fig. 1A). The Sox9-GFP expression faithfully reflected the endogenous SOX9 protein levels (Fig. 1B). The spatially heterogeneous SOX9 expression in cancer cells was further confirmed by immunofluorescence on primary tumor sections (Fig. 1C). Analysis of lung metastases in C3/TAg; Sox9-GFP mice showed that metastatic tumor cells also expressed variable levels of Sox9-GFP (Fig. 1, D and E).
Fig. 1. Sox9-GFP reporter identifies cancer cells expressing distinct levels of SOX9 in spontaneous primary tumors and metastases.
(A) Flow cytometry showing the expression of Sox9-GFP in C3/TAg;Sox9-GFP tumors (n = 22). CD31, CD45, and Ter-119 were used as lineage markers. (B) SOX9 Western blot of cells sorted from C3/TAg;Sox9-GFP tumors as in (A). One representative experiment of three repeats was shown (n = 3). (C) Representative immunofluorescence of SOX9 in C3/TAg;Sox9-GFP primary tumors (n = 3). (D) Representative images of lung metastases in C3/TAg;Sox9-GFP mice (n = 4). (E) Flow cytometry showing the expression of Sox9-GFP in lung metastases in C3/TAg;Sox9-GFP mice (n = 4). (F) Representative immunofluorescence of SOX9 in PyMT;Sox9-GFP tumors (type 1: n = 6; type 2: n = 2). (G) Flow cytometry measuring Sox9-GFP in PyMT;Sox9-GFP tumors (type 1: n = 6; type 2: n = 2). (H) Representative images of lung metastases in PyMT;Sox9-GFP mice (type 1: n = 4; type 2: n = 2). (I) Flow cytometry measuring Sox9-GFP in lung metastases from PyMT;Sox9-GFP mice (type 1: n = 4; type 2: n = 2). DAPI, 4′,6-diamidino-2-phenylindole; PerCP, peridinin-chlorophyll-protein; APC, allophycocyanin; SSC, side scatter; EpCAM, epithelial cell adhesion molecule.
We further evaluated the SOX9 expression in MMTV-PyMT (PyMT) tumors that model luminal B breast cancer. We observed two types of PyMT primary tumors and metastases based on SOX9 expression. Type 1 tumors were composed of a majority of SOX9low tumor cells, while type 2 contained a majority of SOX9neg tumor cells (Fig. 1, F to I). In both cases, SOX9 protein and Sox9-GFP were heterogeneously expressed in cancer cells of PyMT;Sox9-GFP primary tumors and lung metastases (Fig. 1, F to I).
SOX9high cancer cells were enriched in tumor-initiating activity
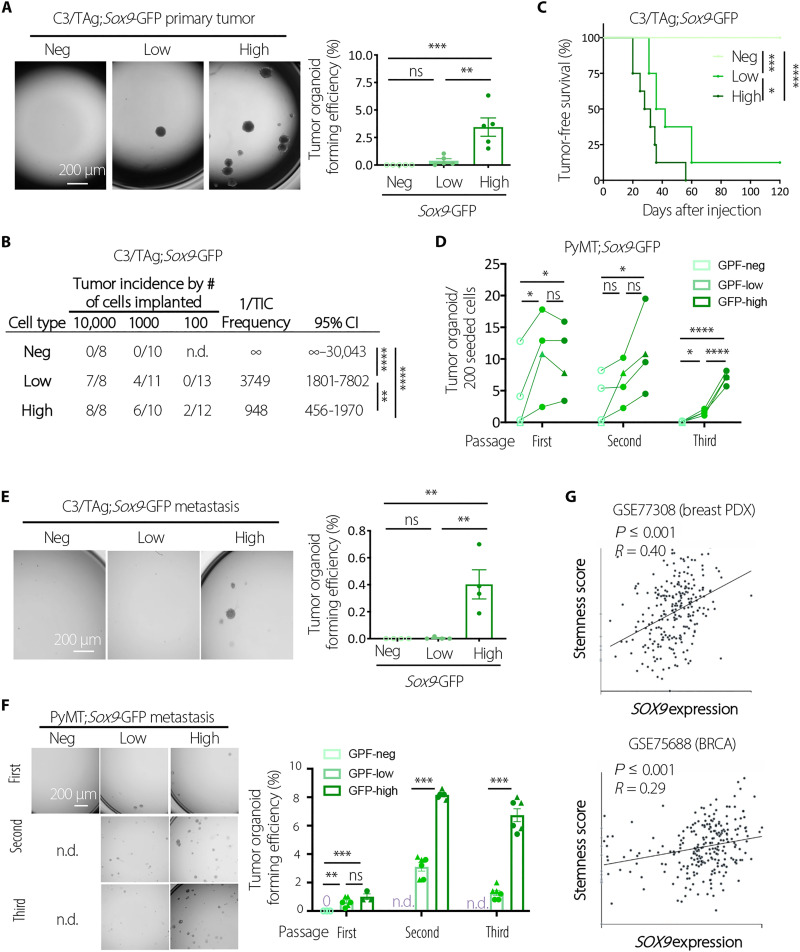
To determine the functional difference of SOX9high cells from other cancer cells, we sorted the top 2% Sox9-GFPhigh cells from C3/TAg tumors by fluorescence-activated cell sorting (FACS) using the Sox9-GFP reporter based on the similar frequency of Sox9-GFPhigh cells in hyperplastic glands (26) and compared them with other cells by tumor organoid culture and in vivo transplantation. We observed that SOX9high cells have a markedly higher ability to form tumor organoids than other cells (Fig. 2A). In organoid culture, SOX9high cells generated cells with heterogeneous levels of SOX9 same as in the original tumor (fig. S2A). Furthermore, SOX9high cells in organoids were enriched in secondary organoid-forming ability, indicating that SOX9high cells have long-term clonogenic potential and can differentiate to SOX9neg/low cells in culture (fig. S2B). Using in vivo limiting dilution transplantation assay, we showed that only SOX9-expressing C3/TAg tumor cells could initiate new tumors, and the SOX9high cells had a further ~4-fold increase in tumor-initiating capacity compared to the SOX9low cells (Fig. 2, B and C).
Fig. 2. SOX9high cancer cells are enriched in tumor-initiating activity.
(A) Representative images and quantification of organoid-forming efficiency of Sox9neg, Sox9low, and Sox9high cells sorted from C3/TAg;Sox9-GFP primary tumors (n = 5). (B) Tumor-initiating cell (TIC) frequency tumor cells sorted from C3/TAg;Sox9-GFP primary tumors. Combination of two experiments using two different spontaneous tumors was shown. CI, confidence interval. (C) Tumor onset in NOD/SCID mice transplanted with 10,000 tumor cells sorted from C3/TAg;Sox9-GFP tumors. Combination of two experiments using two different spontaneous tumors was shown (n = 8 per group). (D) Organoid forming ability during serial replating of tumor cells sorted from PyMT;Sox9-GFP primary tumors (n = 4; circles indicate type-1 tumors, and triangles indicate type-2 tumors). (E) Representative images and quantification of organoid-forming efficiency of cells sorted from lung metastases in C3/TAg;Sox9-GFP mice (n = 4). (F) Representative images and quantification of organoid-forming efficiency during serial replating of cells sorted from lung metastases in PyMT;Sox9-GFP (n.d., not determined). Combination of two experiments using two different lung metastases was shown. (G) Correlation of stemness score and SOX9 expression in breast tumors at the single-cell level as determined by CancerSEA [GSE77308 (29) and GSE75688 (27)]. All data are represented as means ± SEM. P values were determined by one-way ANOVA with Tukey’s test [(A), (E), and (F), first passage], Chi-square test in Extreme Limiting Dilution Analysis (ELDA) (B), log-rank test (C), one-way ANOVA with Tukey’s test (D), or unpaired two-tailed t test [(F), second and third passages]. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05; ns, not significant.
We further characterized SOX9high cells in PyMT tumors by organoid culture. By subjecting freshly sorted primary tumor cells to a serial organoid replating assay, we found that only SOX9high cells maintained robust organoid-forming capacity during the serial replating, while SOX9neg and SOX9low cells gradually lost the ability to generate organoids after two to three passages (Fig. 2D).
We next compared SOX9high and other cancer cells FACS sorted from lung metastases in C3/TAg and PyMT models as shown in Fig. 1 (E and I). Such as in primary tumors, SOX9high cells in C3/TAg metastases exhibited markedly higher organoid-forming capacity than other cell types (Fig. 2E). Markedly increased organoid-forming ability was also found for SOX9high cells in PyMT metastases (Fig. 2F). Together, these data suggested that SOX9high cancer cells in both primary tumors and metastases are enriched in CSCs, defined by their tumor-initiating ability. This notion is further supported by the correlation of high SOX9 expression with increased stemness scores at the single-cell level in multiple human cancer types, including breast, kidney, brain, and lung cancers, as revealed by CancerSEA analysis (Fig. 2G and fig. S2C) (27–29).
SOX9high tumor cells have increased metastasis-initiating capacity
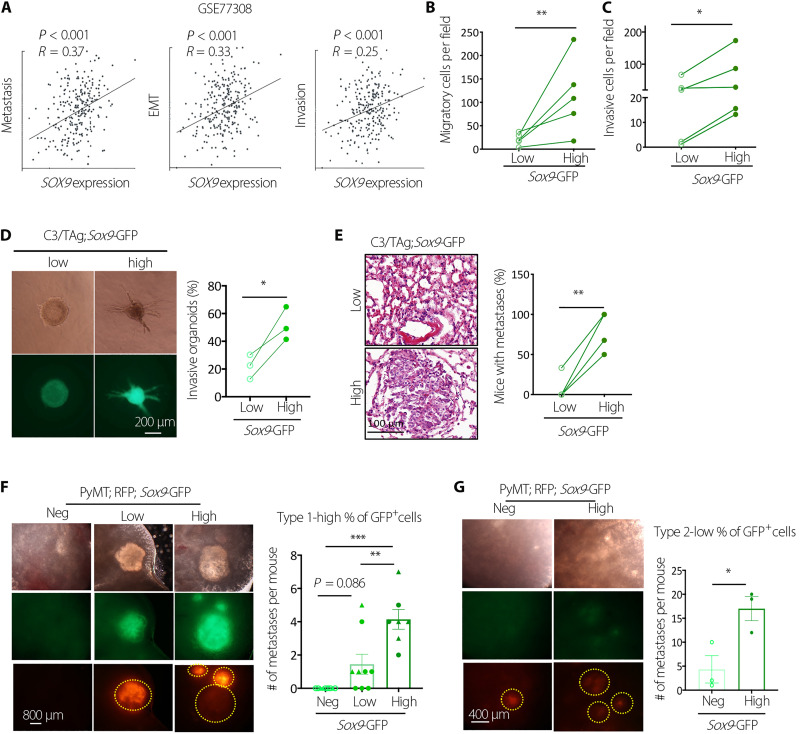
Analysis of breast patient-derived xenograft (PDX) single-cell RNA-seq data showed that SOX9 levels were positively correlated with metastasis, epithelial-mesenchymal transition, and invasion signatures, suggesting that SOX9high cancer cells have higher metastatic potential (Fig. 3A). Consistently, freshly sorted SOX9high C3/TAg primary tumor cells exhibited increased transwell migration and invasion capacity compared to SOX9low cells (Fig. 3, B and C). Organoids formed by SOX9high cells also showed more invasive morphology compared to that by SOX9low cells when cultured in three-dimensional (3D) collagen gel (Fig. 3D). Furthermore, in the experimental metastasis assay via tail vein injection, SOX9high C3/TAg cells developed significantly more lung metastases than SOX9low cells (Fig. 3E). SOX9high cells sorted from PyMT tumors similarly exhibited significantly higher metastatic ability compared to SOX9neg/low cells (Fig. 3, F and G). In these experiments, we isolated tumors from PyMT mice carrying a constitutively expressing CAG::mRFP1 (monomeric red fluorescent protein 1) transgene to facilitate the detection of metastases, although the mRFP expression was suppressed in some metastatic nodules. Together, these results suggest that SOX9high CSCs contribute to metastasis in different breast tumor models.
Fig. 3. SOX9high tumor cells have increased metastasis-initiating capacity.
(A) Correlation between SOX9 levels and functional states in breast tumors at the single-cell level as determined by CancerSEA. EMT, Epithelial-Mesenchymal Transition. (B) Transwell migration assay of C3/TAg primary tumor cells (n = 5 tumors). (C) Transwell invasion assay of C3/TAg primary tumor cells (n = 5 tumors). (D) Representative images and frequencies of organoids with the invasive morphology in collagen gel (n = 3 tumors). (E) Lung metastasis-initiating ability of Sox9low and Sox9high C3/TAg primary tumor cells by tail vein injection. Each line indicates one experiment with a distinct tumor (n = 4). (F) Lung metastasis-initiating ability of Sox9neg, Sox9low, and Sox9high PyMT tumor cells by tail vein injection. Metastatic nodules were determined after 14 weeks (triangles and circles representing two different donors). (G) Lung metastasis-initiating ability of Sox9neg, Sox9low, and Sox9high PyMT tumor cells by tail vein injection. Metastatic nodules were determined 14 weeks after injection (n = 3). All data are represented as means ± SEM. P values were determined by two-tailed t test [(B), (C), (D), (E), and (G)] or one-way ANOVA with Tukey’s test (F). ***P < 0.001, **P < 0.01, and *P < 0.05.
SOX9high tumor cells are enriched after chemotherapy
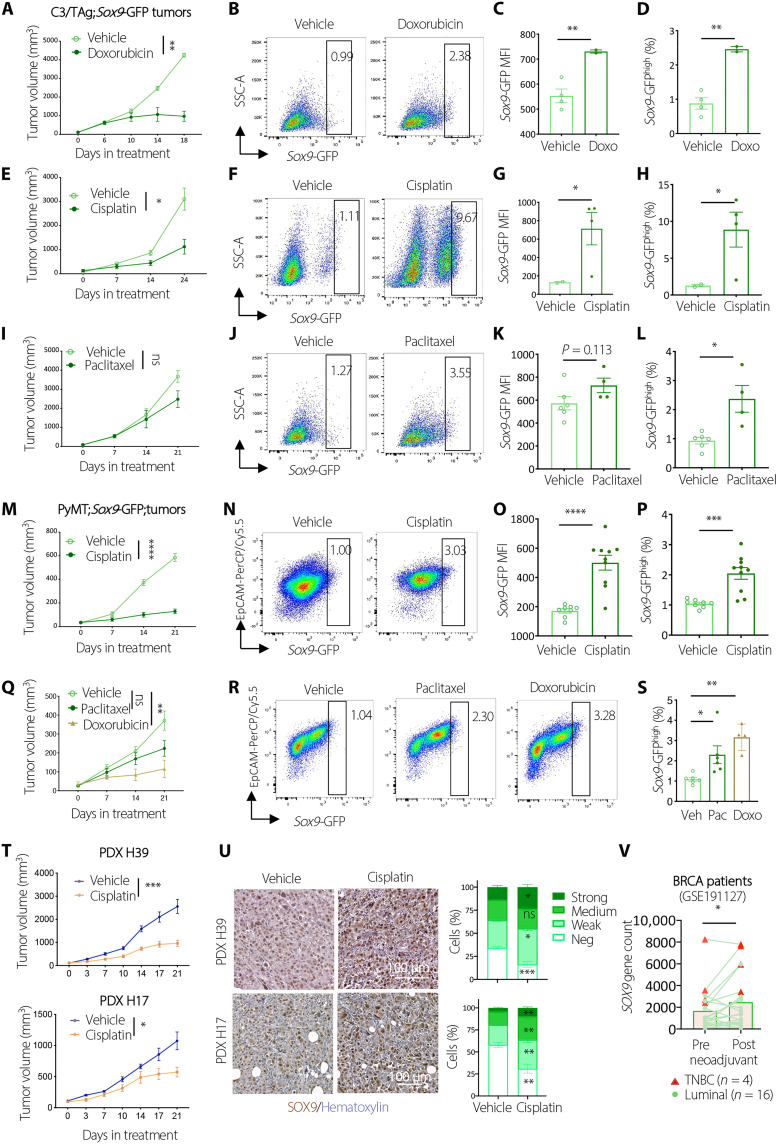
Resistance to chemotherapy is a cardinal feature of CSCs (6, 30–32). To test whether SOX9high tumor cells are chemoresistant, we treated tumor organoids derived from C3/TAg;Sox9-GFP tumors with vehicle or chemotherapeutic drugs, including doxorubicin, cisplatin, or paclitaxel. Each of the drugs induced substantial cell death after a 3-day treatment. However, the frequency of Sox9-GFPhigh cells was significantly increased among tumor cells that survived the drug treatment (fig. S3, A and B). We then tested the response of SOX9high cells in vivo by treating C3/TAg;Sox9-GFP allograft tumors with the chemotherapy drugs. While doxorubicin and cisplatin markedly inhibited tumor growth (Fig. 4, A and E), the overall Sox9-GFP expression levels and the frequency of Sox9-GFPhigh cells were significantly increased in doxorubicin or cisplatin-treated tumors (Fig. 4, B to D and F to H). The frequency of SOX9high cells was also increased significantly in paclitaxel-treated tumors, although it had a modest effect on tumor growth (Fig. 4, I to L).
Fig. 4. Chemotherapy enriches SOX9high tumor cells.
(A) Growth of tumors treated with Vehicle (n = 4) or doxorubicin (n = 2). (B to D) Flow cytometric profiles (B), Sox9-GFP mean fluorescence intensity (MFI) (C), and the percentage of SOX9high cells (D) in tumors treated in (A). (E) Growth of tumors treated with vehicle (n = 2) or cisplatin (n = 4). (F to H) Flow profiles (F), Sox9-GFP MFI (G), and the percentage of SOX9high cells (H) in tumors treated in (E). (I) Growth of tumors treated with vehicle (n = 6) or paclitaxel (n = 4). (J to L) Flow profiles (J), Sox9-GFP MFI (K), and the percentage of SOX9high cells (L) in tumors treated in (I). (M) Growth of tumors treated with vehicle (n = 8) or cisplatin (n = 10). (N to P) Flow cytometric profiles (N), Sox9-GFP MFI (O), and the percentage of SOX9high cells (P) in tumors treated in (M). (Q) Growth of tumors treated with vehicle (n = 6), doxorubicin (n = 4), or paclitaxel (n = 6). (R and S) Flow cytometric profiles (R) and the percentage of SOX9high cells in tumors treated in (Q). (T) Growth of PDXs (H39: vehicle, n = 6; cisplatin, n = 8; H17: vehicle, n = 9; cisplatin, n = 10). (U) IHC and quantification of SOX9 expression in PDXs treated in (T). (V) SOX9 expression in matched pre- and post-neoadjuvant tumors. All data are represented as means ± SEM. P values were determined by two-way ANOVA [(A), (E), (I), (M), (Q), and (T)], multiple unpaired t test (U), unpaired [(C), (D), (G), (H), (K), (L), (O), and (P)] or paired (V) t test, or one-way ANOVA (S). ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05.
To assess the chemoresistant property of SOX9high cells in additional tumor models, we treated PyMT;Sox9-GFP tumor organoids with doxorubicin, paclitaxel, cisplatin, or vehicle. Consistent with the observations in C3/TAg organoids, the drug treatment caused substantial cell death but greatly increased the frequency of Sox9-GFPhigh cells in the surviving tumor cells (fig. S3, C and D). We then treated PyMT;Sox9-GFP tumors with cisplatin in vivo. In two cohorts of allografts generated from different PyMT spontaneous tumors, tumor growth was markedly inhibited by cisplatin, whereas Sox9-GFP expression and Sox9-GFPhigh cell frequency were increased significantly (Fig. 4, M to P, and fig. S3, E to G). Likewise, doxorubicin or paclitaxel treatment resulted in a significant enrichment of Sox9-GFPhigh cells in PyMT tumors (Fig. 4, Q to S).
We further assessed the effect of chemotherapy on SOX9 expression in two human triple-negative breast cancer PDX models established previously (33). Cisplatin inhibited the tumor growth of both PDX models (Fig. 4T). However, SOX9 levels were significantly increased in both PDXs after cisplatin treatment, consistent with mouse tumor models (Fig. 4U). Furthermore, by analyzing RNA-seq data of 20 matching pre- and post-neoadjuvant chemotherapy breast cancer patient samples (34), we found that SOX9 levels significantly increased in the post-treatment samples (Fig. 4V). Together, these data showed that SOX9high cancer cells are chemoresistant and enriched by chemotherapy in both mouse tumor models and human breast cancer, further supporting the notion that SOX9high cells have properties of CSCs.
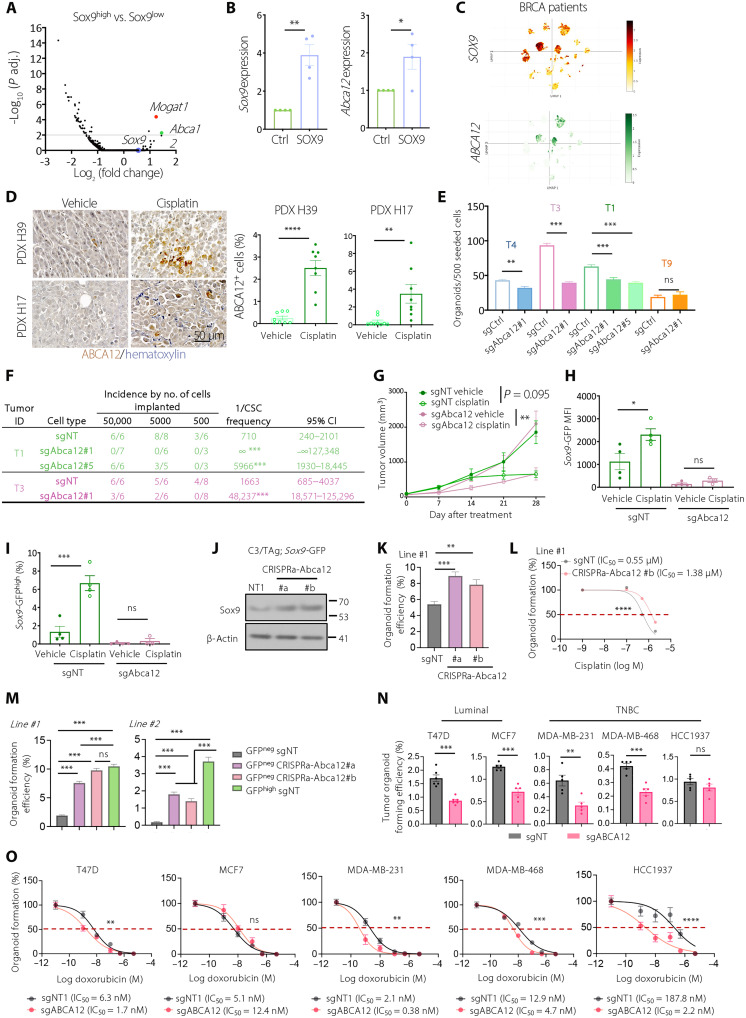
SOX9high CSCs up-regulate the ABCA12 lipid transporter
Specific identification of breast CSCs in spontaneous tumor models allowed us to investigate molecular determinants of cancer stemness in vivo. We performed RNA-seq comparing SOX9high cells to other cancer cells freshly isolated from C3/TAg;Sox9-GFP tumors. While >2000 genes were significantly differentially expressed between SOX9+ and SOX9− cells, only 9 genes were found significantly different between SOX9high and SOX9low cells (Fig. 5A and fig. S4A). Among them, only Abca12 and Mogat1 were consistently correlated with the Sox9 levels, as confirmed by reverse transcription quantitative polymerase chain reaction (RT-qPCR) (fig. S4B). Abca12 was also the most up-regulated gene in SOX9high luminal/basal hybrid cells compared to SOX9low cells in C3/TAg premalignant lesions, whereas Mogat1 was not significantly changed (fig. S4C) (26). In the normal mammary gland, Abca12 is exclusively expressed in fetal mammary stem cells, but not in adult lineage-committed epithelial cells, suggesting that increased Abca12 expression in C3/TAg tumors is associated with reactivation of embryonic multipotency (fig. S4D) (35). FACS analysis confirmed that ABCA12 protein levels are up-regulated along with the increased SOX9 expression in C3/TAg tumor organoid cells (fig. S4, E and F). Furthermore, Abca12 expression was positively correlated with the Sox9 levels in PyMT tumors (fig. S4G). SOX9 overexpression in C3/TAg tumor cells also significantly elevated Abca12 levels (Fig. 5B), although we cannot distinguish whether this is due to direct transcriptional regulation or an indirect effect of increasing CSC frequency by SOX9. Together, these data showed that ABCA12 is up-regulated in SOX9high breast CSCs.
Fig. 5. ABCA12 lipid transporter is up-regulated in SOX9high cells and required for CSC activity.
(A) Volcano plot of RNA-seq comparing SOX9low and SOX9high cells from C3/TAg;Sox9-GFP tumors (n = 4). (B) Sox9 and Abca12 mRNA levels in pLVX-ctrl and pLVX-SOX9 transduced C3/TAg organoids (n = 4). (C) UMAP showing SOX9 and ABCA12 levels in tumor cells in patients with breast cancer (BRCA) (GSE176078). (D) ABCA12 expression in PDX tumors treated with vehicle or cisplatin as in Fig. 4T (H39: n = 8 each group; H17: vehicle, n = 8; cisplatin, n = 9). (E) Organoid-forming efficiency of sgNT and sgAbca12 C3/TAg;Sox9-GFP tumor cells (n = 4). (F) Tumor-initiating cell frequency of sgNT and sgAbca12 C3/TAg;Sox9-GFP organoid cells, as determined by ELDA (n = 2). (G) Growth of sgNT and sgAbca12 C3/TA;Sox9-GFP tumor (sgAbca12/cisplatin, n = 3; other groups, n = 4). (H and I) Sox9-GFP MFI (H) and percentage of Sox9-GFPhigh cells (I) in sgNT and sgAbca12 C3/TAg;Sox9-GFP tumors treated in (G). (J) SOX9 Western blot of the indicated cells. (K) Organoid-forming efficiency of control (sgNT) or Abca12-overexpressing (CRISPRa-Abca12) C3/TAg;Sox9-GFP tumor cells. (L) Effect of cisplatin on organoid formation in control (sgNT) or Abca12-overexpressing (CRISPRa-Abca12 #b) C3/TAg;Sox9-GFP tumor cells. IC50, median inhibitory concentration. (M) Organoid-forming efficiency of the indicated C3/TAg;Sox9-GFP tumor cells. (N) Organoid-forming efficiency of sgNT or sgABCA12-transduced human cell lines. (O) Doxorubicin dose response of sgNT and sgABCA12-transduced breast cancer cells in organoid culture. All data are represented as means ± SEM. P values were determined by unpaired two-tailed t test [(B), (D), (E), and (N)], one-way ANOVA with Dunnett’s (K) or Tukey’s test [(H) to (I) and (M)], Chi-square test in ELDA (F), ordinary two-way ANOVA [(L) and (O)], or two-way RM ANOVA (G). ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05.
We investigated the expression of ABCA12 in human breast cancer. Single-cell RNA-seq analysis of 26 human breast tumors (GSE176078) (36) revealed a positive correlation between SOX9 and ABCA12 expression at the single-cell level (Fig. 5C). Analysis of The Cancer Genome Atlas data using GEPIA2 (37) showed that ABCA12 is overexpressed in human breast cancer compared to normal breast tissue (fig. S4H). In addition, ABCA12 is one of the most highly up-regulated adenosine 5′-triphosphate (ATP)–binding cassette transporter genes in breast tumors compared to normal breast tissue (fig. S4I) (38). ABCA12 level is markedly higher in breast tumors that retained residual disease after neoadjuvant chemotherapy than those that had pathologically complete responses (fig. S4J) (39). Further supporting the role of ABCA12 in chemoresistance, the frequencies of ABCA12+ cells were markedly increased in PDX tumors after cisplatin treatment (Fig. 5D).
ABCA12 promotes cancer stemness and chemoresistance of SOX9high cells
To determine the function of ABCA12 in CSCs, we deleted Abca12 in C3/TAg;Sox9-GFP tumor organoids using the lentiCRISPRv2 vector, which led to a marked decrease in both ABCA12 RNA and protein levels (fig. S5, A and B). We found that, in three of the four tumor organoid lines, Abca12 deletion significantly inhibited organoid formation (Fig. 5E). We further assessed the impact of Abca12 deletion on tumor-initiating ability by in vivo limiting dilution transplantation assay. In two independent tumor organoid lines, Abca12 deletion markedly reduced the tumor-initiating ability (8- to 30-fold) (Fig. 5F).
We next evaluated whether ABCA12 contributed to chemoresistance in SOX9high cells. Cisplatin treatment of both wild-type (WT) and Abca12-deleted C3/TAg;Sox9-GFP tumor organoids led to pronounced cell death. However, SOX9high cells were no longer enriched by cisplatin treatment in the Abca12-deleted organoids (fig. S5, C to E). Furthermore, the loss of Abca12 enhanced the inhibitory effects of cisplatin on tumor organoid formation (fig. S5F). To extend this finding in vivo, we transplanted a saturated number (3 × 105) of WT and Abca12-deleted C3/TAg tumor organoid cells to allow them to form tumors at similar take rates. Under this condition, WT and Abca12-deleted tumors grew at the same speed and responded to cisplatin similarly (Fig. 5G). It is possible that CSCs can activate ABCA12-independent compensatory mechanisms to maintain stemness and chemoresistance when ABCA12 is chronically knocked out. Injecting large numbers of tumor cells may have facilitated the generation and/or selection of these ABCA12-independent CSCs. Nevertheless, in Abca12-deleted tumors, SOX9high cells were no longer enriched by cisplatin treatment (Fig. 5, H and I, and fig. S5G).
We then tested the effect of ABCA12 overexpression using a CRISPR activation (CRISPRa) approach (40). Coexpression of the dCas9-VP64-p65-Rta (VPR) transcriptional activator and single guide RNAs (sgRNAs) targeting the Abca12 promoter region markedly increased Abca12 expression in C3/TAg tumor cells (fig. S5H). Abca12 overexpression led to a marked up-regulation of SOX9 and significantly increased organoid formation and resistance to cisplatin (Fig. 5, J to L, and fig. S5, I and J). More remarkably, Abca12 overexpression was sufficient to induce organoid-forming ability in SOX9neg non-CSCs (Fig. 5M).
To assess the ABCA12 function in human cells, we deleted ABCA12 in a panel of human breast cancer cell lines, including two luminal lines (T47D and MCF7) and three triple-negative lines (MDA-MB-231, MDA-MB-468, and HCC1937), with the lentiCRISPRv2 vector (fig. S5K). Notably, loss of ABCA12 significantly impaired the organoid formation ability in four of the five cell lines except for HCC1937 (Fig. 5N and fig. S5L). Furthermore, ABCA12 knockout significantly increased the sensitivity to doxorubicin in most cell lines except MCF7 (Fig. 5O). These results reinforced the role of ABCA12 in promoting breast cancer stemness, although various cell lines have slightly different dependencies on ABCA12, likely due to heterogeneous genetic and epigenetic makeup of these cell lines.
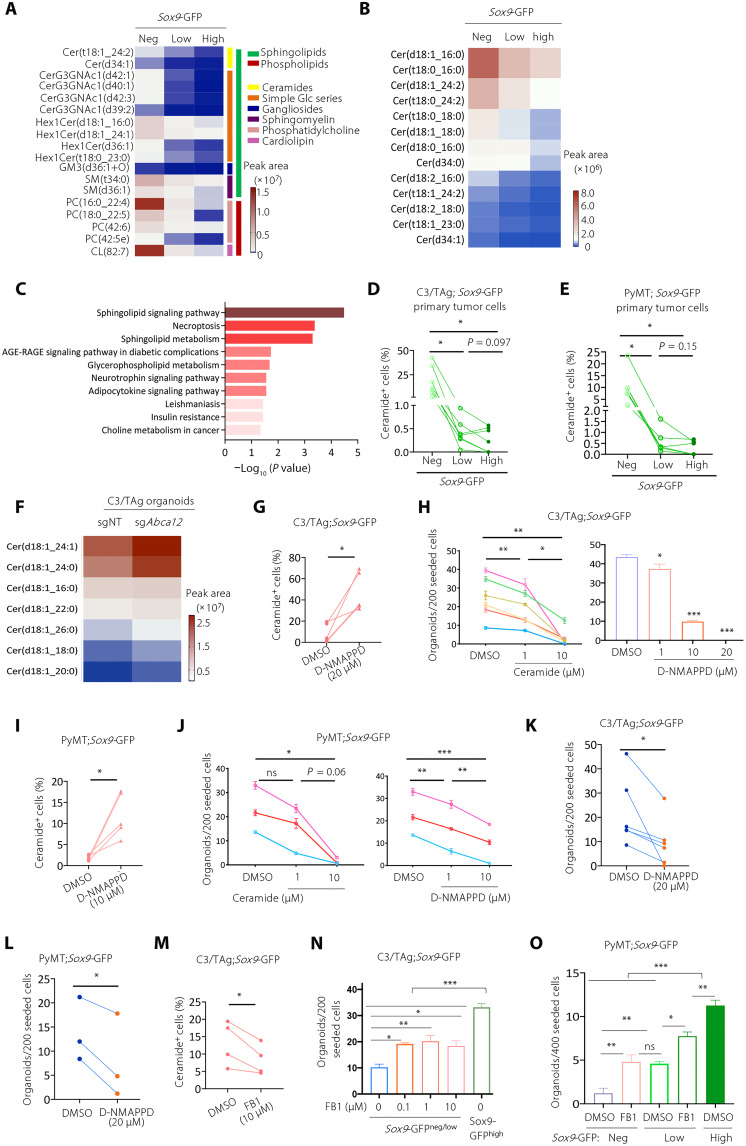
Lipidomic analyses uncovered that ABCA12 regulates SOX9 expression and cancer stemness by controlling ceramide abundance
ABCA12 is a member of the ATP-binding cassette transporter family and functions as a lipid transporter (41–45), suggesting that SOX9high CSCs may have unique lipid metabolism. Therefore, we performed untargeted liquid chromatography–mass spectrometry (LC-MS)–based lipidomics to compare SOX9neg, SOX9low, and SOX9high cells sorted from C3/TAg;Sox9-GFP tumors. Overall, 1144 lipid species from 30 lipid classes were detected, indicating high sensitivity of lipidomics using small numbers of sorted cells (fig. S6A). The abundances of 230 lipid species from 19 lipid classes were significantly altered between SOX9neg and SOX9high cells (fig. S6B). Most of these lipid species were down-regulated in SOX9high cells, except for 12 species that were up-regulated (fig. S6C). Among the 230 down-regulated lipid species in SOX9high cells, 18 of them also showed a consistent down-regulation comparing SOX9high to SOX9low and SOX9low to SOX9neg cells (fold change > 2, P < 0.05) (Fig. 6A). Most of these lipid species (12 of 18) were ceramides and their derivatives, such as Hex1Cer, CerG3GNAc1, and sphingomyelin. Furthermore, all the significantly altered ceramide species between SOX9neg and SOX9high cells (P < 0.05) showed a gradually decreasing trend when SOX9 expression increased (Fig. 6B). Lipid pathway enrichment analysis using Lipid Pathway Enrichment Analysis (LIPEA) showed significant down-regulation of sphingolipid signaling and metabolism pathways in SOX9high CSCs, concordant with the role of ceramide as a central component of sphingolipid metabolism (Fig. 6C).
Fig. 6. Lipidomic analyses reveal that ABCA12 reduces intracellular ceramide abundance to maintain cancer stemness.
(A) Lipid species significantly down-regulated in SOX9high cells (fold change > 2) in C3/TAg;Sox9-GFP tumor (n = 5). (B) Level of ceramide lipids in Sox9neg, Sox9low, and Sox9high C3/TAg tumor cells. (C) Lipid pathway enrichment analyses of the lipids identified in (A). AGE, advanced glycation end products; RAGE, receptor for AGE. (D) Flow cytometry measuring the abundance of ceramide in the SOX9neg, Sox9low, and Sox9high C3/TAg tumor cells (n = 7). (E) Abundance of ceramide in the SOX9neg, Sox9low, and Sox9high PyMT tumor cells (n = 5). (F) Abundance of ceramide species in sgNT and sgAbca12 C3/TAg organoids (n = 3). (G) Ceramide levels in C3/TAg organoids treated with dimethyl sulfoxide (DMSO) or D-NMAPPD for 5 days as measured by flow cytometry (n = 5). (H) Effect of C6-ceramide or D-NMPAAD on C3/TAg organoid formation (n = 6). (I) Ceramide levels in PyMT organoids treated with DMSO or D-NMAPPD (10 μM) for 5 days (n = 5). (J) Effect of C6-ceramide or D-NMPAAD on PyMT organoid formation (n = 3). (K and L) Organoid-forming efficiency of C3/TAg [(K), n = 5] or PyMT [(L), n = 3] tumor cells pretreated with D-NMAPPD (20 μM) for 3 days. (M) Ceramide levels in C3/TAg organoids treated with DMSO or Fumonisin B1 (FB1; 10 μM) for 5 days (n = 4). (N and O) Effect of FB1 on organoid formation by SOX9neg/low C3/TAg [(N), n = 2] or SOX9neg/low PyMT [(O), n = 3] tumor cells. All data are represented as means ± SEM. P values were determined by one-way ANOVA [(D) and (E)], or with Tukey’s test [(H), (J), (N), and (O)], or paired two-tailed t test [(G), (I), and (K) to (M)]. ***P < 0.001, **P < 0.01, and *P < 0.05.
We then performed flow cytometry to measure ceramide levels in SOX9neg, SOX9low, and SOX9high tumor cells from C3/TAg and PyMT tumors. Consistent with the lipidomics analysis, the ceramide level was markedly decreased in SOX9high cells in both tumor models (Fig. 6, D and E, and fig. S6, D and E). Similar differences were observed in C3/TAg tumor organoid cells (fig. S6F). Previous studies showed that ABCA12 is required for the secretion of ceramide by keratinocytes to form extracellular lipid layers that are essential for skin barrier function (44). This suggests that high ABCA12 expression in SOX9high CSCs maintains low intracellular ceramide levels by efflux. To test this, we compared lipid profiles of WT and Abca12-deleted C3/TAg tumor cells by nontargeted LC-MS–based lipidomics and found that intracellular ceramides levels were indeed increased after Abca12 deletion (Fig. 6F). These results suggest that SOX9high CSCs maintain low ceramide levels via ABCA12-mediated efflux.
We then explored whether ceramide levels affect CSC activity. We treated C3/TAg and PyMT tumor organoids with exogenous C6-ceramide or with the acid ceramidase inhibitor D-NMAPPD {N-[(1R,2R)-2-hydroxy-1-(hydroxymethyl)-2-(4-nitrophenyl)ethyl]-tetradecanamide; (1R,2R)-B13} to increase intracellular ceramide levels (Fig. 6, G and I, and fig. S6, G and H) (46). Both treatments markedly inhibited tumor organoid-forming ability (Fig. 6, H and J). To further evaluate the long-term effect of ceramide on tumor stemness, we pretreated C3/TAg and PyMT tumor cells with D-NMAPPD and then measured their organoid-forming efficiency without further treatment. We observed that D-NMAPPD pretreatment inhibited organoid formation in both tumor models, suggesting that D-NMAPPD led to irreversible inhibition of cancer stemness (Fig. 6, K and L). Conversely, we used fumonisin B1 (FB1), a ceramide synthase inhibitor, to reduce the intracellular accumulation of ceramides in C3/TAg cells (Fig. 6M and fig. S6G). We found that FB1 treatment significantly increased the organoid-forming efficiency of SOX9neg/low cells (Fig. 6N). Similar effects of FB1 were confirmed in PyMT tumor organoids (Fig. 6O).
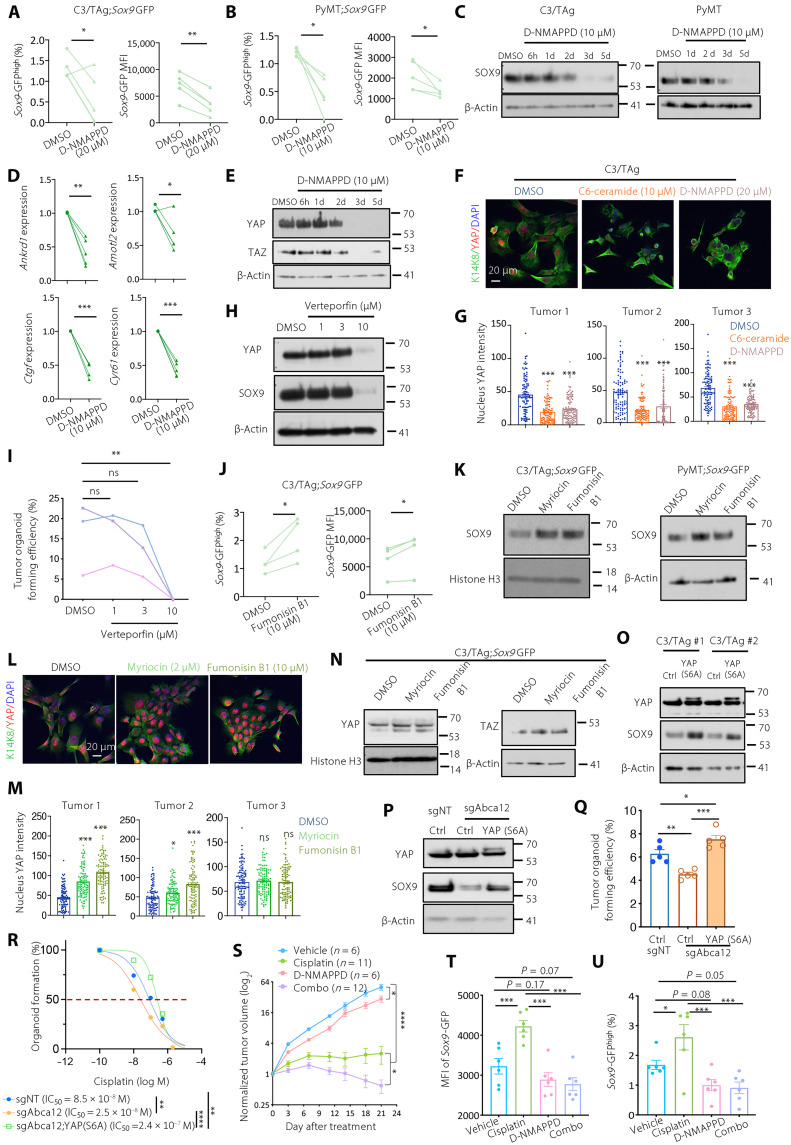
Ceramide suppresses SOX9 expression in tumor cells by inhibiting the YAP/TAZ pathway
Increasing intracellular ceramide levels by D-NMAPPD greatly reduced the expression of Sox9-GFP reporter and endogenous SOX9 protein levels in C3/TAg and PyMT organoid cells, consistent with the decreased organoid-forming ability (Fig. 7, A to C, and fig. S6, G to I). Ceramide has been shown as an inhibitor for the Yes-associated protein 1 (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) signaling in hepatic stellate cells (47). Therefore, we asked whether ceramide regulates the YAP/TAZ pathway to control breast cancer stemness. The elevation of ceramide abundance with C6-ceramide or D-NMAPPD treatment greatly inhibited YAP/TAZ signaling in C3/TAg and PyMT tumor organoids (Fig. 7, D to G, and fig. S7, A and B). Furthermore, inhibiting YAP by verteporfin caused a marked reduction of the SOX9 protein level (Fig. 7H) and organoid-forming ability of C3/TAg organoids (Fig. 7I and fig. S7C).
Fig. 7. Ceramide down-regulates SOX9 expression by inhibiting the YAP/TAZ pathway.
(A and B) Sox9-GFP expression in C3/TAg (A) or PyMT (B) organoids treated for 5 days (n = 5). (C) SOX9 levels in D-NMAPPD–treated organoids (n = 3). (D) RT-qPCR measuring YAP target genes in C3/TAg organoids (n = 4) treated for 6 hours. (E) YAP and TAZ levels in D-NMAPPD–treated C3/TAg organoids (n = 3). (F and G) Immunofluorescence (F) and quantification of nuclear YAP (G) in C3/TAg cells treated for 2 days (n = 100 cells per group). (H) SOX9 and YAP levels in C3/TAg organoids treated for 3 days. (I) Organoid-forming efficiency of C3/TAg cells (n = 3). (J) Sox9-GFP level in C3/TAg organoids treated for 5 days. (K) SOX9 levels in organoids treated with myriocin (2 μM) or FB1 (10 μM) for 5 days. (L and M) Immunofluorescence (L) and quantification (M) of nuclear YAP in C3/TAg cells as treated for 2 days (n = 100 cells per group). (G) and (M) share the same DMSO group. (N) YAP and TAZ levels in C3/TAg organoids as treated for 5 days. (O) Effect of YAP (S6A) on SOX9 levels in C3/TAg organoids. (P and Q) Effect of YAP (S6A) on SOX9 levels (P) and organoid-forming efficiency (Q) in sgAbca12 C3/TAg organoids. (R) Cisplatin dose-response curve of the indicated cells. (S) Growth of C3/TAg;Sox9-GFP tumors treated as indicated. (T and U) Sox9-GFP MFI (T) and percentage of SOX9high cells (U) in tumors as shown in (S). All data are represented as means ± SEM. P values were determined by paired t test [(A), (B), (D), and (J)], one-way ANOVA with Dunnett’s test [(G), (I), and (M)], Tukey’s test (Q), ordinary one-way ANOVA [(T) and (U)], or two-way ANOVA [(R) and (S)]. ****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05.
In contrast, reducing ceramide levels by FB1 or myriocin (a serine palmitoyltransferase inhibitor) markedly increased SOX9 and YAP/TAZ levels (Fig. 7, J to N, and figs. S6, G and J, and S7D). Expressing constitutively active YAP (S6A) (48) led to the up-regulation of SOX9 protein levels (Fig. 7O), supporting the role of YAP in SOX9 up-regulation. This is likely achieved by the binding of the YAP/TEA domain transcription factor (TEAD) complex to the SOX9 promoter, as previously reported in esophageal cancer (49, 50). YAP activation also significantly increased organoid formation and chemoresistance (fig. S7, E and F). Furthermore, YAP activation restored SOX9 expression, organoid formation, and cisplatin resistance in ABCA12 knockdown cells (Fig. 7, P to R, and fig. S7G), supporting the notion that ABCA12 enhances cancer stemness through activating the YAP pathway.
We then investigated whether increasing ceramide levels by D-NMAPPD could suppress chemotherapy-induced enrichment of SOX9high CSCs and inhibit tumor progression synergistically with cisplatin in C3/TAg allograft models. D-NMAPPD or cisplatin treatment alone led to a significant reduction in tumor growth, although neither could induce tumor regression (Fig. 7S). The combination of two drugs led to marked tumor regression (Fig. 7S). Furthermore, combinatory treatment completely blunted the increase of Sox9-GFP expression and Sox9-GFPhigh cell frequency that would be induced by cisplatin treatment alone (Fig. 7, T to U). We note that D-NMAPPD treatment showed a more pronounced effect than ABCA12 knockdown (Fig. 5G) on tumor growth and response to cisplatin. It is possible that acute D-NMAPPD treatment directly increases ceramide levels that are less likely to be overcome by compensatory mechanisms that may occur in ABCA12 knockdown cells. Together, our results showed that high ceramide levels inhibit SOX9 expression by inactivation of the YAP/TAZ pathway and that increasing ceramide levels pharmacologically inhibits CSCs and overcomes chemoresistance.
DISCUSSION
Understanding the determinants of CSCs in spontaneous tumors will help develop targeted strategies against these clinically important cell types. Using a CSC reporter based on the stemness regulator SOX9, we established an effective method for identifying CSCs in both primary tumors and metastases of different mammary tumor models. We showed that SOX9high CSCs were metastasis initiating and chemoresistant, highlighting the need to target these cells. Specific isolation of CSCs from spontaneous tumors enabled molecular characterization of these cells by transcriptomic and lipidomic profiling. We found an important role of ceramide homeostasis in controlling breast cancer stemness and chemoresistance. SOX9high CSCs expressed high levels of ABCA12, which promoted ceramide efflux to maintain low cellular ceramide. Elevating ceramide levels down-regulated SOX9 by suppressing YAP/TAZ signaling, inhibited cancer stemness, and overcame chemoresistance. These results uncovered a critical lipid metabolism mediated signaling circuitry for sustaining breast cancer stemness (fig. S7H).
Reliable identification of the CSC state in spontaneously developing tumors has been challenging (6, 51, 52). Several studies have used cell surface markers for enriching CSCs in transgenic tumor models, such as CD29hiCD24hi or CD29hiCD24med (53, 54), Thy1+CD24+ (55), and CD61+ (56). However, these markers are not specific or functionally linked to cancer stemness (9, 10). Reporters controlled by core embryonic stem cell regulators, such as OCT4/SOX2 and NANOG, have been developed to identify breast CSCs in cancer cell lines (57, 58). However, their utilities in spontaneous tumors remain unclear. Recently, the stem-SH2-containing 5′-inositol phosphatase (s-SHIP) promoter activity has been used to enrich CSCs in C3/TAg tumors, but substantial tumor-initiating activity can still be found in s-SHIP promoter inactive cells, suggesting that it only marks a subset of CSCs (59). Here, using the mammary stem cell regulator SOX9, we developed a specific CSC reporter for various spontaneous mammary tumor models. We found a strict association of cancer stemness with Sox9-GFP expression. Only Sox9-GFP+ cancer cells could generate organoids in vitro and reinitiate tumors in vivo, whereas Sox9-GFP− cells completely lacked organoid-forming and tumor-initiating activity. Among Sox9-GFP+ cells, Sox9-GFPhigh cells showed markedly higher CSC activity than Sox9-GFPlow cells. The SOX9 reporter can identify CSCs from primary tumors and metastases of different mammary tumor models. SOX9high CSCs contributed to metastasis and chemoresistance, highlighting the importance of these cells. Thus, our Sox9-GFP reporter provides a much-needed experimental tool for studying CSCs in spontaneous tumors.
Accumulating evidence shows that CSCs rely on altered lipid metabolism. However, current understanding is limited to the CSC-promoting role of fatty acid oxidation and cholesterol synthesis (17–19, 60, 61). In this study, we found a critical need for CSCs to control their intracellular ceramide level. Ceramide serves as the hub for sphingolipid metabolism and has well-established roles in inducing apoptosis, autophagy, and cell cycle arrest (62, 63). Here, we identified a function of ceramide in inhibiting stemness by suppressing stem cell regulator expression. In C3/TAg tumor cells, ceramide abundance was negatively correlated with SOX9 levels. Increasing ceramide levels markedly decreased SOX9 expression in tumor organoids, which, in turn, impaired tumor organoid formation. Conversely, lowering intracellular ceramide levels by blocking its synthesis up-regulated SOX9 expression and promoted organoid formation. We further demonstrated that ceramide inhibits the YAP/TAZ signaling that is required for SOX9 expression in breast CSCs.
We uncovered a unique mechanism by which SOX9high CSCs down-regulate intracellular ceramide levels via ABCA12 lipid transporter-mediated efflux. SOX9high CSCs overexpressed ABCA12 and had low levels of ceramide. Abca12 knockout increased intracellular ceramide levels. This is consistent with previous studies showing that ABCA12 is required for extracellular ceramide secretion by keratinocytes to form a proper skin barrier (43, 45). ABCA12 is one of the most significantly up-regulated ATP-binding cassette transporter genes in cancer compared to normal tissues (38). High ABCA12 levels also correlate with reduced response to neoadjuvant chemotherapy in breast cancer patients (39). Chemotherapy is known to induce ceramide generation, which sensitizes cells to cytotoxic agents (20, 64, 65). Therefore, high ABCA12 expression could protect cells from chemotherapy by reducing the level of chemotherapy-induced ceramide. This is also likely to endow survival advantage of SOX9high CSCs against chemotherapy. Supporting this notion, we showed that loss of ABCA12 abolished the enrichment of SOX9high CSCs by chemotherapy in vivo. Furthermore, increasing ceramide levels using acid ceramidase inhibitor D-NMAPPD sensitized tumors to chemotherapy and prevented enrichment of SOX9high CSCs. These data suggest a potential strategy for targeting CSCs and overcoming chemoresistance.
MATERIALS AND METHODS
Mice
NOD/SCID (#001303), NSG (#005557), and MMTV-PyMT (#002374) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and bred in-house. Sox9-GFP;C3/TAg mice were generated as described previously (26). Sox9-GFP transgenic mice [Tg(Sox9-EGFP)EB209Gsat/Mmucd] were obtained from Mutant Mouse Resource & Research Centers and backcrossed to the FVB/n background. CAG::mRFP1 (#005884) mice were obtained from the Jackson Laboratory and backcrossed to the FVB/n background. Mice were housed in microisolator cages and a pathogen-free condition. All animal experiments were performed according to protocols (20170607 and 00001266) approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
Tumor cell isolation
Tumor cells were isolated and cultured as described previously (66). Tumor samples were dissected, minced, and then digested with collagenase type 3 (300 U/ml; Worthington Biochemical, LS004182), hyaluronidase (100 U/ml; Worthington Biochemical, LS002592), 5 μM Y-27632 (Cayman Chemical Company, 10005583-10), and deoxyribonuclease I (DNase I) (10 μg/ml; Roche, 104159) in the Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium at 37°C for 2 hours on an orbital shaker. Afterward, the cell pellet was washed with 1× phosphate-buffered saline (PBS). Red blood cells (RBCs) were then lysed by RBC lysis buffer (eBioscience, 00-4300-54). Cells were further digested with 0.05% trypsin-EDTA for 5 min and neutral protease (1 U/ml; dispase) (Worthington Biochemical, LS02109) plus DNase I (100 μg/ml; Roche, 104159) for 5 min. The dissociated cells were then filtered through 40-μm cell strainers to obtain single cells.
Tumor organoid culture
Tumor organoids were generated and cultured as described previously (66). For measuring tumor organoid-forming efficiency, 200 to 500 sorted primary tumor cells per well or 600 to 1000 sorted lung metastasis tumor cells per well were seeded in ultralow attachment 96-well plates (Corning Life Sciences), with organoid culture medium containing Epicult (R)-B medium (STEMCELL Technologies) or Advanced DMEM/F-12 (Life Technology) supplemented with 1% l-glutamine, 5% heat-inactivated fetal bovine serum (FBS) (Sigma-Aldrich, F2442), 5% Matrigel (Corning Life Sciences, 354234), epidermal growth factor (10 ng/ml; Sigma-Aldrich, E9644), basic fibroblast growth factor (20 ng/ml; EMD Millipore, GF003), heparin (4 μg/ml; Sigma-Aldrich, H4784), and 5 μM Y-27632 (Cayman Chemical Company, 10005583-10). After 7-day culture, organoids greater than 100 μm in diameter were counted. For passaging organoids, 300,000 cells in 2 ml of organoid medium per well were seeded in six-well plates.
Cell lines and adherent cell culture
MCF7, T47D, MDA-MB-231, and MDA-MB-468 cell lines were cultured in DMEM (Corning) with 10% FBS (VWR) and 1% (v/v) antibiotic-antimycotic solution (GE Healthcare). HCC1937 were maintained in RPMI 1640 (Corning) with 10% FBS (VWR) and 1% (v/v) antibiotic-antimycotic solution (GE Healthcare). Cell lines were authenticated by short tandem repeat analysis, matching to the American Type Culture Collection database.
Orthotropic tumor cell implantation
Female NOD/SCID (NOD.Cg-Prkdcscid/J) mice at 6 to 8 weeks of age were injected in the inguinal mammary fat pad with 3 × 105 or 5 × 105 Sox9-GFP;C3/TAg tumor cells or 1 × 106 PyMT;Sox9-GFP;RFP tumor cells suspended in 50 μl of 25% Matrigel diluted in PBS. Tumor onset was monitored by weekly palpation. Tumor volume was measured by caliper and calculated as V = (4/3) × π × (L/2) × (D/2) × (D/2). For in vivo limiting dilution assay of SOX9neg, SOX9low, or SOX9high cells, 100, 1000, and 10,000 sorted tumor cells were injected into the third and fourth mammary fat pad of NOD/SCID mice. Mice were monitored twice a week for a total of 4 months for tumor onset. For in vivo limiting dilution assay of nontargeting sgRNA (sgNT) and sgAbca12 cells, 500, 5000, and 50,000 cells were injected into the third and fourth mammary fat pads of NOD/SCID mice. Mice were monitored twice a week for a total of 2 to 2.5 months for tumor onset.
PDX model
Human PDX models were a gift from A. Patsialou and J. Condeelis (33). Each tumor fragment (1 mm in diameter) was implanted in the fourth mammary pad of NSG recipient mice. The resulting tumors can be palpated 4 to 6 weeks following the transplantation. The PDX experiments were approved by the Institutional Review Board (IRB) of Albert Einstein College of Medicine (IRB# 2018-9307). Informed consent is not relevant because the PDX models were previously established, and we did not gather the tissue ourselves.
In vivo chemotherapy treatment
When tumors reached 3 to 5 mm in diameter, mice received weekly intraperitoneal injections with cisplatin (6 mg/kg), paclitaxel (15 mg/kg), doxorubicin (5 mg/kg), or vehicle [PBS or ethanol/cremophor EL/PBS (1:1:8)] for 3 weeks. Tumor volume was measured by caliper and calculated as V = (4/3) × π × (L/2) × (D/2) × (D/2) every week or twice a week.
In vitro drug treatment
A total of 300,000 cells were seeded in poly-(2-hydroxyethyl methacrylate) (poly-HEMA)–coated ultralow attachment six-well plate, or 500 cells were seeded in ultralow attachment 96-well plate. The test compounds—including doxorubicin (1 μM), cisplatin (0.5, 1, or 2 μM), paclitaxel (5 nM), D-NMAPPD (1, 10, or 20 μM), C6-ceramide (1 or 10 μM), verteporfin (1, 3, or 10 μM), myriocin (2 μM), FB1 (10 μM), or vehicle—were applied.
Experimental metastasis assay
A total of 80,000 to 200,000 sorted SOX9low or SOX9high cells from Sox9-GFP;C3/TAg tumors or 10,000 to 20,000 sorted SOX9neg, SOX9low, or SOX9high cells from Sox9-GFP;PyMT;CAG::mRFP1 tumors were injected via tail vein into 4- to 6-week-old female NOD/SCID mice. Fourteen to 16 weeks later, mice were perfused with 5 ml of PBS and lungs were harvested and imaged under the fluorescence microscope ex vivo.
Transwell cell migration and invasion assay
For invasion assay, 30,000 cells were plated in Matrigel-coated (40 μl, 2 mg/ml; Corning Life Sciences, 354234) upper chambers (pore diameter of 8 μm; Greiner Bio-One, 662638) with serum-free Epicult medium. For migration assay, 20,000 cells were plated in uncoated upper chambers with serum-free Epicult medium. In both assays, the lower chamber was added with Epicult medium plus 10% FBS. After incubation for 24 hours, cells on the upper side of the membrane were removed by wiping with a cotton swab. The membranes were stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured using a confocal microscope (×20 magnification).
3D collagen gel migration assay
3D collagen gel migration assay was done on the basis of the protocol described previously (67). Briefly, acid-solubilized rat-tail collagen I gels [collagen I (3 mg/ml), pH 7 to 7.5) were prepared. Eight-well chamber slides were coated with 200 μl of collagen I gel (3 mg/ml) per well for 1 hour at 37°C. Sorted Sox9-GFPlow and GFPhigh cells were cultured in the Epicult/Matrigel organoid culture for 1 week to form organoids and then transferred into collagen-coated chamber slides for 3 days. The images were taken, and the numbers of organoids with or without branching morphology were counted.
Western blot
Cells were lysed with the radioimmunoprecipitation assay buffer containing 25 mM tris-HCL (pH 7.5), 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, and 1.0% NP-40 on ice. Samples were resolved by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. The Western blots were performed with antibodies against SOX9 (MilliporeSigma, AB5535), YAP (Cell Signaling Technology, 14074S), pYAP (S397) (Cell Signaling Technology, 13619S), TAZ (Cell Signaling Technology, 72804S), β-actin (BD Biosciences, 612656), or histone H3 (Cell Signaling Technology, 14269S). Original scans of the Western blot results are provided in the Supplementary Materials.
Flow cytometry and FACS sorting
Multiparametric flow cytometric analyses were performed on an LSR II equipped with FACS Diva 6.1 software (BD Biosciences) and analyzed with FlowJo software v9.3 or 10.0.8 (FlowJo, LLC). Dead cells were excluded by forward scatter, side scatter, and DAPI (Sigma-Aldrich) staining. Lineage markers were used to exclude immune cells and endothelial cells in tumor cell analyses. Cell sorting was performed using MoFlow Astrios Cell Sorter (Beckman Coulter). The following antibodies were used in the current study: anti–epithelial cell adhesion molecule (EpCAM)–allophycocyanin (clone G8.8, BioLegend, 118214), anti–EpCAM–peridinin-chlorophyll-protein/Cy5.5 (clone G8.8, BioLegend, 118220), biotinylated antibodies against lineage markers (CD45 (clone 30-F11, BioLegend, 103104), CD31 (clone 390, BioLegend, 102404) and TER-119 (clone TER-119, BioLegend, 116204), anti-ABCA12 (Novus Biologicals, NB100-93466), and anti-ceramide (clone MID 15B4, Sigma-Aldrich, C8104) antibody. Biotinylated antibodies were further labeled with Strepavidin-V450 (BD Biosciences, 560797). Unless otherwise specified, all antibodies were used at a 1:100 dilution. Anti-goat or anti-mouse secondary antibodies conjugated with Alexa Fluor 633 or 549 and anti–immunoglobulin M (IgM) secondary antibody conjugated with Alexa Fluor 549 or DyLight 755 were used.
Immunofluorescence and immunohistochemistry
Formalin-fixed and paraffin-embedded tissue sections or Optimal Cutting Temperature compound (OCT)–embedded frozen sections were stained with standard protocols. The following antibodies were used in this study: anti-YAP (1:200; Cell Signaling Technology, 14074S), anti-SOX9 (1:200; MilliporeSigma, AB5535), anti-Keratin 8 (1:60; Developmental Studies Hybridoma Bank, TROMA-I), anti–Keratin 14 (1:1000; BioLegend, 905304), anti–Keratin 14 (1:800; BioLegend, 906004), and anti-ABCA12 (1:100; Sigma-Aldrich, HPA043194). The ImmPRESS horseradish peroxidase (HRP) goat anti-rabbit IgG polymer reagent (Vector Laboratories, MP-7451) and the DAB/HRP Detection IHC Kit (Vector Laboratories, SK-4100) were used for immunohistochemistry. Fluorescent slides were imaged with an AXIO Examiner D1 microscope (Zeiss) with a confocal scanner unit (Yakagawa). SlideBook software 6.0 was used for the acquisition of images. Immunohistochemistry slides were scanned with a P250 High-Capacity Slide Scanner (3DHISTECH).
Quantitative RT-PCR
Total RNA was isolated from cells using the Nucleo Spin RNA Kit (Macherey-Nagel AG), and cDNA synthesis was performed by the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems) with Rpl13a as internal controls in an Applied Biosystems 7900HT real-time PCR machine. Primer sequences are listed in table S1.
RNA-seq gene expression analyses
RNA-seq libraries were prepared using the NEBNext Ultra II Directional Library Prep Kit (New England Biolabs, E7765) and sequenced by GENEWIZ. RNA-seq reads were aligned using Hisat2 (68) to the reference mouse genome (GRCm38) using the M14 Gencode Basic CHR Annotations. Stringtie and the prepDE.py script were used to generate raw counts from aligned reads (69). Differential expression was determined using DESeq2 (70) (from Galaxy version 2.11.40.7+galaxy2). All data were accessible at Gene Expression Omnibus (GEO; GSE220178).
Lentivirus production and lentiviral CRISPR-mediated gene knockout
Lentivirus was prepared as described previously (26). The mouse Sox9 cDNA was cloned in the pLVX-puro (Takara, 632164) lentiviral vector. The sgRNAs targeting ABCA12 were cloned into the lentiCRISPRv2 vector (Addgene, #52961, a gift from F. Zhang). The sgRNA targeting mouse sequences are as follows: sgABCA12 #1-AAATGTAAAGACACGCCCTA, sgABCA12 #5-CAATTGCTTACACGCATCCT, and sgNT-GCGAGGTATTCGGCTCCGCG. The sgRNA targeting human ABCA12 is as follows: sgABCA12-ATAGAAGGCTGTCTGTTCGG. Cells were transduced with the lentiviral vectors and selected by puromycin.
CRISPR targeting efficiency measurement by TIDE assay
A TIDE assay was used to assess editing efficiency by CRISPR-Cas9 as in the previous study (71). Genomic DNA was extracted, and 100 ng of genomic DNA was used in a PCR amplification using SapphireAMP Fast PCR Master Mix (Clontech, catalog no. RR350B) and appropriate primers surrounding the sgRNA target region. The primer sequences for human ABCA12 are as follows: forward primer: -AAAGACACAGTCTCAGGAAAAGA, reverse primer-CTGTTGCTGAAGAAGATCATGGC.
Lentiviral CRISPR-mediated gene activation
Tumor organoid cells were transduced with the lentiviral vector lenti-ER1a-dCas9-VPR-Puro (Addgene, #99373, a gift from K. Brennand), and puromycin was selected for 3 days. Cells were then transduced with sgNT or CRISPRa.Abca12 cloned into pLKO5.sgRNA.EFS.tRFP and sorted for RFP-positive cells. Guide RNAs were designed using the CRISPR-ERA (http://crispr-era.stanford.edu) web tools. The sgRNA targeting sequences are as follows: sgAbca12 #a-GGGACCCTGGAAGGGCCAGT, sgAbca12 #b-GTGGCAGGCGCTGGGTACTG, and sgNT-GCGAGGTATTCGGCTCCGCG.
YAP overexpression
YAP1 (S6A)-V5 in pLX304 (Addgene, #42562, a gift from W. Hahn) and control pLX304 (Addgene, #25890, a gift from D. Root) plasmids were used. Lentivirus was prepared as described previously (26). For the selection of virally infected cells, blasticidin (5 μg/ml) was used 24 hours after infection.
Lipidomics analysis
MS-based lipidomics was performed by Biological Mass Spectrometry Core Facility at University of Colorado School of Medicine as described previously (72). Freshly sorted tumor cells (100,000) were centrifuged, and the pellet was flash-frozen in liquid nitrogen. Cells were extracted at 4°C in the presence of cold methanol at a concentration of 2 × 106 cells/ml. The samples were vortexed for 10 min, placed at −20°C for 20 min, and then centrifuged at 18,213g for 10 min at 4°C. The resulting supernatant was analyzed on a Thermo Vanquish Ultra High Performance Liquid Chromatography (UHPLC) coupled to a Thermo Q Exactive MS. The UHPLC was equipped with a Waters ACQUITY HSS T3 column and instrument methods were 15-min positive ion mode and 17-min negative ion mode (both ESI) with top 15 ddMS2 data acquisition. Features were annotated and integrated using LipidSearch (Thermo Fisher Scientific). Lipid metabolic pathway enrichment analysis was performed using the LIPEA software.
Quantification and statistical analysis
Data are represented as means ± SEM. Statistical tests were performed with GraphPad Prism V8.0 (San Diego, CA). A two-tailed Student’s t test was used for comparisons of continuous variables between two groups. One-way analysis of variance (ANOVA) with Tukey’s or Dunnett’s test was used when three or more groups were compared. Two-way ANOVA was used to find the comparison between two independent variables. Kaplan-Meier survival curves were analyzed by using GraphPad Prism V8.0, V9.4.1, and V10.0.0 (San Diego, CA), and P values were calculated using the log-rank test.
Acknowledgments
We thank the technical assistance from Flow Cytometry, Histopathology, Analytical Imaging, and Stem Cell Isolation core facilities of Albert Einstein College of Medicine, supported by Einstein Cancer Center Support Grant (P30 CA013330); the NYSTEM shared facility grant support (C029154); and P250 high-capacity slide scanner (1S10OD019961-01). We thank Y. Liu for the assistance with mouse colony management. The University of Colorado School of Medicine Mass Spectrometry Facility is a shared resource supported in part by the University of Colorado Cancer Center Support Grant P30 CA046934. W.G. is a V Scholar of the V Foundation for Cancer Research and supported by the Mary Kay Foundation.
Funding: This work was supported by DOD BCRP grants BC171714 (W.G.) and BC190403/P1 (to W.G.) and NIH grant 1P01CA257885-01A1 (to W.G.).
Author contributions: J.C. designed and performed the experiments and acquired and analyzed data. J.R.C. performed the RNA-seq analyses. R.S. and M.C.D.M. contributed to the SOX9 overexpression and knockdown experiments. F.I.C. and J.A.R. performed lipidomics. A.D. coordinated the lipidomic analysis and contributed to manuscript editing. J.C. and W.G. wrote the manuscript with inputs from all other authors. W.G. supervised the study.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA-seq data have been deposited at GEO (GSE220178). Accession numbers are listed in the key resources table (data S2). This paper does not report the original code. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Data S1 and S2
Legend for data S3
Other Supplementary Material for this manuscript includes the following:
Data S3
REFERENCES AND NOTES
- 1.L. G. Martelotto, C. K. Ng, S. Piscuoglio, B. Weigelt, J. S. Reis-Filho, Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 16, 210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.K. Polyak, Heterogeneity in breast cancer. J. Clin. Invest. 121, 3786–3788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Marusyk, M. Janiszewska, K. Polyak, Intratumor Heterogeneity: The rosetta stone of therapy resistance. Cancer Cell 37, 471–484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. Kreso, J. E. Dick, Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–291 (2014). [DOI] [PubMed] [Google Scholar]
- 5.C. E. Meacham, S. J. Morrison, Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. Batlle, H. Clevers, Cancer stem cells revisited. Nat. Med. 23, 1124–1134 (2017). [DOI] [PubMed] [Google Scholar]
- 7.P. R. Prasetyanti, J. P. Medema, Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 16, 41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. Beck, C. Blanpain, Unravelling cancer stem cell potential. Nat. Rev. Cancer 13, 727–738 (2013). [DOI] [PubMed] [Google Scholar]
- 9.J. P. Medema, Cancer stem cells: The challenges ahead. Nat. Cell Biol. 15, 338–344 (2013). [DOI] [PubMed] [Google Scholar]
- 10.J. E. Visvader, G. J. Lindeman, Cancer stem cells: Current status and evolving complexities. Cell Stem Cell 10, 717–728 (2012). [DOI] [PubMed] [Google Scholar]
- 11.D. Hanahan, R. A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 12.D. Hanahan, Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 13.N. N. Pavlova, J. J. Zhu, C. B. Thompson, The hallmarks of cancer metabolism: Still emerging. Cell Metab. 34, 355–377 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. Ciavardelli, C. Rossi, D. Barcaroli, S. Volpe, A. Consalvo, M. Zucchelli, A. De Cola, E. Scavo, R. Carollo, D. D'Agostino, F. Forli, S. D'Aguanno, M. Todaro, G. Stassi, C. Di Ilio, V. De Laurenzi, A. Urbani, Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 5, e1336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.E. Vlashi, C. Lagadec, L. Vergnes, T. Matsutani, K. Masui, M. Poulou, R. Popescu, L. Della Donna, P. Evers, C. Dekmezian, K. Reue, H. Christofk, P. S. Mischel, F. Pajonk, Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. U.S.A. 108, 16062–16067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. Diehn, R. W. Cho, N. A. Lobo, T. Kalisky, M. J. Dorie, A. N. Kulp, D. Qian, J. S. Lam, L. E. Ailles, M. Wong, B. Joshua, M. J. Kaplan, I. Wapnir, F. M. Dirbas, G. Somlo, C. Garberoglio, B. Paz, J. Shen, S. K. Lau, S. R. Quake, J. M. Brown, I. L. Weissman, M. F. Clarke, Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. M. Intlekofer, L. W. S. Finley, Metabolic signatures of cancer cells and stem cells. Nat. Metab. 1, 177–188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.T. Wang, J. F. Fahrmann, H. Lee, Y. J. Li, S. C. Tripathi, C. Yue, C. Zhang, V. Lifshitz, J. Song, Y. Yuan, G. Somlo, R. Jandial, D. Ann, S. Hanash, R. Jove, H. Yu, JAK/STAT3-regulated fatty acid β-Oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 27, 136–150.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. Gabitova-Cornell, A. Surumbayeva, S. Peri, J. Franco-Barraza, D. Restifo, N. Weitz, C. Ogier, A. R. Goldman, T. R. Hartman, R. Francescone, Y. Tan, E. Nicolas, N. Shah, E. A. Handorf, K. Q. Cai, A. M. O'Reilly, I. Sloma, R. Chiaverelli, R. A. Moffitt, V. Khazak, C. Y. Fang, E. A. Golemis, E. Cukierman, I. Astsaturov, Cholesterol pathway inhibition induces TGF-β signaling to promote basal differentiation in pancreatic cancer. Cancer Cell 38, 567–583.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. Ogretmen, Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 18, 33–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.W. Guo, Z. Keckesova, J. L. Donaher, T. Shibue, V. Tischler, F. Reinhardt, S. Itzkovitz, A. Noske, U. Zurrer-Hardi, G. Bell, W. L. Tam, S. A. Mani, A. van Oudenaarden, R. A. Weinberg, Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Wang, J. R. Christin, M. H. Oktay, W. Guo, Lineage-biased stem cells maintain estrogen-receptor-positive and -negative mouse mammary luminal lineages. Cell Rep. 18, 2825–2835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.G. Domenici, I. Aurrekoetxea-Rodriguez, B. M. Simoes, M. Rabano, S. Y. Lee, J. S. Millan, V. Comaills, E. Oliemuller, J. A. Lopez-Ruiz, I. Zabalza, B. A. Howard, R. M. Kypta, M. D. Vivanco, A Sox2-Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene 38, 3151–3169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.J. C. Larsimont, K. K. Youssef, A. Sanchez-Danes, V. Sukumaran, M. Defrance, B. Delatte, M. Liagre, P. Baatsen, J. C. Marine, S. Lippens, C. Guerin, V. Del Marmol, J. M. Vanderwinden, F. Fuks, C. Blanpain, Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 17, 60–73 (2015). [DOI] [PubMed] [Google Scholar]
- 25.A. Wuidart, M. Ousset, S. Rulands, B. D. Simons, A. Van Keymeulen, C. Blanpain, Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261–1277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. R. Christin, C. Wang, C. Y. Chung, Y. Liu, C. Dravis, W. Tang, M. H. Oktay, G. M. Wahl, W. Guo, Stem cell determinant SOX9 promotes lineage plasticity and progression in basal-like breast cancer. Cell Rep. 31, 107742 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.W. Chung, H. H. Eum, H. O. Lee, K. M. Lee, H. B. Lee, K. T. Kim, H. S. Ryu, S. Kim, J. E. Lee, Y. H. Park, Z. Kan, W. Han, W. Y. Park, Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 8, 15081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.H. Yuan, M. Yan, G. Zhang, W. Liu, C. Deng, G. Liao, L. Xu, T. Luo, H. Yan, Z. Long, A. Shi, T. Zhao, Y. Xiao, X. Li, CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 47, D900–D908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.E. B. Braune, Y. L. Tsoi, Y. P. Phoon, S. Landor, H. Silva Cascales, D. Ramskold, Q. Deng, A. Lindqvist, X. Lian, C. Sahlgren, S. B. Jin, U. Lendahl, Loss of CSL unlocks a hypoxic response and enhanced tumor growth potential in breast cancer cells. Stem Cell Reports 6, 643–651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Dean, T. Fojo, S. Bates, Tumour stem cells and drug resistance. Nat. Rev. Cancer 5, 275–284 (2005). [DOI] [PubMed] [Google Scholar]
- 31.C. J. Creighton, X. Li, M. Landis, J. M. Dixon, V. M. Neumeister, A. Sjolund, D. L. Rimm, H. Wong, A. Rodriguez, J. I. Herschkowitz, C. Fan, X. Zhang, X. He, A. Pavlick, M. C. Gutierrez, L. Renshaw, A. A. Larionov, D. Faratian, S. G. Hilsenbeck, C. M. Perou, M. T. Lewis, J. M. Rosen, J. C. Chang, Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. U.S.A. 106, 13820–13825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. Shibue, R. A. Weinberg, EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. Patsialou, Y. Wang, J. Lin, K. Whitney, S. Goswami, P. A. Kenny, J. S. Condeelis, Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 14, R139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Hoogstraat, E. H. Lips, I. Mayayo-Peralta, L. Mulder, P. Kristel, I. van der Heijden, S. Annunziato, M. van Seijen, P. M. Nederlof, G. S. Sonke, W. Zwart, J. Wesseling, L. F. A. Wessels, Comprehensive characterization of pre- and post-treatment samples of breast cancer reveal potential mechanisms of chemotherapy resistance. NPJ Breast Cancer 8, 60 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Y. Chung, Z. Ma, C. Dravis, S. Preissl, O. Poirion, G. Luna, X. Hou, R. R. Giraddi, B. Ren, G. M. Wahl, Single-cell chromatin analysis of mammary gland development reveals cell-state transcriptional regulators and lineage relationships. Cell Rep. 29, 495–510.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. Z. Wu, G. Al-Eryani, D. L. Roden, S. Junankar, K. Harvey, A. Andersson, A. Thennavan, C. Wang, J. R. Torpy, N. Bartonicek, T. Wang, L. Larsson, D. Kaczorowski, N. I. Weisenfeld, C. R. Uytingco, J. G. Chew, Z. W. Bent, C. L. Chan, V. Gnanasambandapillai, C. A. Dutertre, L. Gluch, M. N. Hui, J. Beith, A. Parker, E. Robbins, D. Segara, C. Cooper, C. Mak, B. Chan, S. Warrier, F. Ginhoux, E. Millar, J. E. Powell, S. R. Williams, X. S. Liu, S. O'Toole, E. Lim, J. Lundeberg, C. M. Perou, A. Swarbrick, A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 53, 1334–1347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Z. Tang, B. Kang, C. Li, T. Chen, Z. Zhang, GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.V. Hlavac, V. Brynychova, R. Vaclavikova, M. Ehrlichova, D. Vrana, V. Pecha, R. Kozevnikovova, M. Trnkova, J. Gatek, D. Kopperova, I. Gut, P. Soucek, The expression profile of ATP-binding cassette transporter genes in breast carcinoma. Pharmacogenomics 14, 515–529 (2013). [DOI] [PubMed] [Google Scholar]
- 39.S. Park, C. Shimizu, T. Shimoyama, M. Takeda, M. Ando, T. Kohno, N. Katsumata, Y. K. Kang, K. Nishio, Y. Fujiwara, Gene expression profiling of ATP-binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res. Treat. 99, 9–17 (2006). [DOI] [PubMed] [Google Scholar]
- 40.A. Chavez, J. Scheiman, S. Vora, B. W. Pruitt, M. Tuttle, E. P. R. Iyer, S. Lin, S. Kiani, C. D. Guzman, D. J. Wiegand, D. Ter-Ovanesyan, J. L. Braff, N. Davidsohn, B. E. Housden, N. Perrimon, R. Weiss, J. Aach, J. J. Collins, G. M. Church, Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.T. Annilo, S. Shulenin, Z. Q. Chen, I. Arnould, C. Prades, C. Lemoine, C. Maintoux-Larois, C. Devaud, M. Dean, P. Denefle, M. Rosier, Identification and characterization of a novel ABCA subfamily member, ABCA12, located in the lamellar ichthyosis region on 2q34. Cytogenet. Genome Res. 98, 169–176 (2002). [DOI] [PubMed] [Google Scholar]
- 42.F. Peelman, C. Labeur, B. Vanloo, S. Roosbeek, C. Devaud, N. Duverger, P. Denefle, M. Rosier, J. Vandekerckhove, M. Rosseneu, Characterization of the ABCA transporter subfamily: Identification of prokaryotic and eukaryotic members, phylogeny and topology. J. Mol. Biol. 325, 259–274 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Y. Zuo, D. Z. Zhuang, R. Han, G. Isaac, J. J. Tobin, M. McKee, R. Welti, J. L. Brissette, M. L. Fitzgerald, M. W. Freeman, ABCA12 maintains the epidermal lipid permeability barrier by facilitating formation of ceramide linoleic esters. J. Biol. Chem. 283, 36624–36635 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. Akiyama, The roles of ABCA12 in epidermal lipid barrier formation and keratinocyte differentiation. Biochim. Biophys. Acta 1841, 435–440 (2014). [DOI] [PubMed] [Google Scholar]
- 45.M. Akiyama, Y. Sugiyama-Nakagiri, K. Sakai, J. R. McMillan, M. Goto, K. Arita, Y. Tsuji-Abe, N. Tabata, K. Matsuoka, R. Sasaki, D. Sawamura, H. Shimizu, Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J. Clin. Invest. 115, 1777–1784 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. Selzner, A. Bielawska, M. A. Morse, H. A. Rudiger, D. Sindram, Y. A. Hannun, P. A. Clavien, Induction of apoptotic cell death and prevention of tumor growth by ceramide analogues in metastatic human colon cancer. Cancer Res. 61, 1233–1240 (2001). [PubMed] [Google Scholar]
- 47.S. Alsamman, S. A. Christenson, A. Yu, N. M. E. Ayad, M. S. Mooring, J. M. Segal, J. K. Hu, J. R. Schaub, S. S. Ho, V. Rao, M. M. Marlow, S. M. Turner, M. Sedki, L. Pantano, S. Ghoshal, D. D. S. Ferreira, H. Y. Ma, C. C. Duwaerts, R. Espanol-Suner, L. Wei, B. Newcomb, I. Mileva, D. Canals, Y. A. Hannun, R. T. Chung, A. N. Mattis, B. C. Fuchs, A. M. Tager, D. Yimlamai, V. M. Weaver, A. C. Mullen, D. Sheppard, J. Y. Chen, Targeting acid ceramidase inhibits YAP/TAZ signaling to reduce fibrosis in mice. Sci. Transl. Med. 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J. Rosenbluh, D. Nijhawan, A. G. Cox, X. Li, J. T. Neal, E. J. Schafer, T. I. Zack, X. Wang, A. Tsherniak, A. C. Schinzel, D. D. Shao, S. E. Schumacher, B. A. Weir, F. Vazquez, G. S. Cowley, D. E. Root, J. P. Mesirov, R. Beroukhim, C. J. Kuo, W. Goessling, W. C. Hahn, β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457–1473 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. Wang, Z. Zhang, X. Yu, X. Huang, Z. Liu, Y. Chai, L. Yang, Q. Wang, M. Li, J. Zhao, J. Hou, F. Li, Unbalanced YAP-SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene 38, 2042–2055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.S. Song, J. A. Ajani, S. Honjo, D. M. Maru, Q. Chen, A. W. Scott, T. R. Heallen, L. Xiao, W. L. Hofstetter, B. Weston, J. H. Lee, R. Wadhwa, K. Sudo, J. R. Stroehlein, J. F. Martin, M. C. Hung, R. L. Johnson, Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 74, 4170–4182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. R. Pattabiraman, R. A. Weinberg, Tackling the cancer stem cells—What challenges do they pose? Nat. Rev. Drug Discov. 13, 497–512 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H. Clevers, The cancer stem cell: Premises, promises and challenges. Nat. Med. 17, 313–319 (2011). [DOI] [PubMed] [Google Scholar]
- 53.M. Zhang, F. Behbod, R. L. Atkinson, M. D. Landis, F. Kittrell, D. Edwards, D. Medina, A. Tsimelzon, S. Hilsenbeck, J. E. Green, A. M. Michalowska, J. M. Rosen, Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 68, 4674–4682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.N. Shafee, C. R. Smith, S. Wei, Y. Kim, G. B. Mills, G. N. Hortobagyi, E. J. Stanbridge, E. Y. Lee, Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 68, 3243–3250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R. W. Cho, X. Wang, M. Diehn, K. Shedden, G. Y. Chen, G. Sherlock, A. Gurney, J. Lewicki, M. F. Clarke, Isolation and molecular characterization of cancer stem cells in MMTV-Wnt-1 murine breast tumors. Stem Cells 26, 364–371 (2008). [DOI] [PubMed] [Google Scholar]
- 56.F. Vaillant, M. L. Asselin-Labat, M. Shackleton, N. C. Forrest, G. J. Lindeman, J. E. Visvader, The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 68, 7711–7717 (2008). [DOI] [PubMed] [Google Scholar]
- 57.B. Tang, A. Raviv, D. Esposito, K. C. Flanders, C. Daniel, B. T. Nghiem, S. Garfield, L. Lim, P. Mannan, A. I. Robles, W. I. Smith Jr., J. Zimmerberg, R. Ravin, L. M. Wakefield, A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports 4, 155–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.P. S. Thiagarajan, M. Hitomi, J. S. Hale, A. G. Alvarado, B. Otvos, M. Sinyuk, K. Stoltz, A. Wiechert, E. Mulkearns-Hubert, A. Jarrar, Q. Zheng, D. Thomas, T. Egelhoff, J. N. Rich, H. Liu, J. D. Lathia, O. Reizes, Development of a fluorescent reporter system to delineate cancer stem cells in triple-negative breast cancer. Stem Cells 33, 2114–2125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L. Tian, M. J. Truong, C. Lagadec, E. Adriaenssens, E. Bouchaert, H. Bauderlique-Le Roy, M. Figeac, X. Le Bourhis, R. P. Bourette, s-SHIP promoter expression identifies mouse mammary cancer stem cells. Stem Cell Reports 13, 10–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. Mukherjee, H. A. Kenny, E. Lengyel, Unsaturated fatty acids maintain cancer cell stemness. Cell Stem Cell 20, 291–292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.K. M. Nieman, H. A. Kenny, C. V. Penicka, A. Ladanyi, R. Buell-Gutbrod, M. R. Zillhardt, I. L. Romero, M. S. Carey, G. B. Mills, G. S. Hotamisligil, S. D. Yamada, M. E. Peter, K. Gwin, E. Lengyel, Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 17, 1498–1503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. A. Morad, M. C. Cabot, Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 13, 51–65 (2013). [DOI] [PubMed] [Google Scholar]
- 63.V. Garcia-Gonzalez, J. F. Diaz-Villanueva, O. Galindo-Hernandez, I. Martinez-Navarro, G. Hurtado-Ureta, A. A. Perez-Arias, Ceramide metabolism balance, a multifaceted factor in critical steps of breast cancer development. Int. J. Mol. Sci. 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.C. Swanton, M. Marani, O. Pardo, P. H. Warne, G. Kelly, E. Sahai, F. Elustondo, J. Chang, J. Temple, A. A. Ahmed, J. D. Brenton, J. Downward, B. Nicke, Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell 11, 498–512 (2007). [DOI] [PubMed] [Google Scholar]
- 65.R. Kolesnick, The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J. Clin. Invest. 110, 3–8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Cui, W. Guo, Establishment and long-term culture of mouse mammary stem cell organoids and breast tumor organoids. STAR Protoc 2, 100577 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.K. V. Nguyen-Ngoc, K. J. Cheung, A. Brenot, E. R. Shamir, R. S. Gray, W. C. Hines, P. Yaswen, Z. Werb, A. J. Ewald, ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc. Natl. Acad. Sci. U.S.A. 109, E2595–E2604 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D. Kim, J. M. Paggi, C. Park, C. Bennett, S. L. Salzberg, Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.M. Pertea, G. M. Pertea, C. M. Antonescu, T. C. Chang, J. T. Mendell, S. L. Salzberg, StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.E. K. Brinkman, T. Chen, M. Amendola, B. van Steensel, Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.J. A. Reisz, C. Zheng, A. D'Alessandro, T. Nemkov, Untargeted and semi-targeted lipid analysis of biological samples using mass spectrometry-based metabolomics. Methods Mol. Biol. 1978, 121–135 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Data S1 and S2
Legend for data S3
Data S3