Abstract
We have identified and characterized the protease-resistant SecA fragments (X. Chen, H. Xu, and P. C. Tai, J. Biol. Chem. 271:29698–29706, 1996) through immunodetection with region-specific antibodies, chemical extraction, and sequencing analysis. The 66-, 36-, and 27-kDa proteolytic fragments in the membranes all start at Met1, whereas the 48-kDa fragment starts at Glu361. The overlapping of the sequences of the 66- and 48-kDa fragments indicates that they are derived from different SecA molecules. These two fragments were generated differently in response to ATP hydrolysis and protein translocation. Furthermore, the presence of membrane is required for the generation of the 48-kDa fragment but not for that of the 66-kDa fragment. These data suggest that there are two different integral forms of SecA in the membrane: SecAS and SecAM. The combination of these two forms of SecA has several membrane-interacting domains. Both forms of SecA are integrated in the membrane, since both the 48- and 66-kDa fragments could be derived from urea- or Na2CO3-washed membranes. Moreover, all fragments are resistant to extraction with a high concentration of salt or with heparin, but the membrane-specific 48-kDa SecA domain is more sensitive to Na2CO3 or urea extraction. This suggests that this domain may interact with other membrane proteins in an aqueous microenvironment and therefore may form a part of the protein-conducting channel.
SecA is an essential component of the protein translocation machinery in Escherichia coli (3, 8, 21, 38). It hydrolyzes ATP and uses the energy of this hydrolysis to translocate precursor proteins across the cytoplasmic membrane (5, 6, 24, 25). SecA is composed of 901 amino acids (31) and was initially identified as a soluble and peripheral membrane protein (4, 26). It has been reported that SecA cycles on and off the membrane and that a 30-kDa SecA domain undergoes cycles of membrane insertion and deinsertion during protein translocation (11, 12). Recent studies have found, however, that a significant fraction of SecA behaves like an integral membrane protein (4, 7, 22, 38). This fraction of SecA is resistant to extraction with heparin, Na2CO3, alkaline, or urea, all of which are widely used to extract peripheral membrane proteins (4, 7, 22, 38). In a SecDF-overproducing strain, SecA was found almost entirely in an integral membrane form and part of SecA was exposed to the periplasm (22). Despite these apparently unusual findings, this strain still displayed normal protein translocation, as measured by rapid processing of preproteins in vivo. Membranes washed with heparin, which removes all but the integral SecA from the membrane (38), were also active in protein translocation, although Na2CO3 or urea treatment partially inactivated this activity (7, 38). However, supplementing the urea-washed membranes with F1 protein restored the translocation activity (38). These findings indicate that the integral form of SecA is functional.
Electrophysiological measurements have suggested that protein translocation across membranes occurs through protein-conducting channels in both prokaryotes and eukaryotes (33, 34). Such channels have been shown to consist of a heterotrimeric Sec61p complex in yeast and mammalian endoplasmic reticulum membranes (17). SecY and SecE are the homologs of Sec61α and Sec61γ (16, 18), which are components of the Sec61p complex in yeast and mammalian cells. Therefore, SecY and SecE might be part of the protein-conducting channel in E. coli. However, it is not clear whether SecA is also a part of the channels, since SecA does not have homologs in endoplasmic reticulum. More recent studies have strongly suggested that SecA might be a part of the channels. SecA and SecY were the only proteins which were cross-linked to translocating proOmpA molecules (20). Furthermore, a fraction of SecA is permanently embedded in the membrane (7), and it can be accessed from the periplasmic side (22, 29, 37). Moreover, the membrane-embedded SecA gave rise to several major fragments after proteinase K digestion, and some of these fragments were obtained in the absence of protein translocation (7). These translocation-independent SecA fragments may form the constant part of the channels. Here we present the identification, through sequencing analysis and immunodetection using region-specific antibodies, of these SecA fragments and the characterization of their interactions with membranes by a variety of chemical treatments. Our findings suggest that there are two different forms of membrane-integral SecA and that this combination of SecA has at least two membrane-interacting domains (MID), which have different interactions with the membrane. Moreover, a 48-kDa fragment represents a SecA domain induced specifically by interaction with the membrane. This fragment may form a part of the protein-conducting channels.
MATERIALS AND METHODS
Materials.
The following solutions were used where indicated: TK buffer (10 mM Tris-HCl [pH 7.6], 50 mM KCl), TAKM buffer {50 mM Tris-HCl [pH 7.6], 20 mM NH4Cl, 40 mM KCl, 10 mM magnesium acetate [Mg(OAc)2], 2 mM dithiothreitol}, translocation buffer (TAKM buffer with 1 mM spermidine trihydrochloride, 8 mM putrescine dihydrochloride), energy source [1 mM ATP, 20 μM GTP, 5 mM phospho(enol)pyruvate, 30 μg of pyruvate kinase/ml], and cushion solution [10 mM Tris-HCl (pH 7.6), 10 mM Mg(OAc)2, 0.5 M NaCl, and 0.5 M sucrose]. Stop solution [2 mM phenylmethylsulfonyl fluoride (PMSF) in 10 mM Tris-HCl (pH 7.6), 500 mM KCl, and 10 mM Mg(OAc)2] was prepared immediately before use. MinA medium was prepared as described elsewhere (36). S-Sepharose FF and Sephacryl S-200 were obtained from Pharmacia Biotech. Proteinase K was obtained from Boehringer Mannheim. ATP, GTP, PMSF, spermidine trihydrochloride, putrescine dihydrochloride, trypsin (treated with Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK]), and soybean trypsin inhibitor were obtained from Sigma. [35S]Met (1,175 Ci/mmol), [3H]Gly (51 Ci/mmol), and [3H]Leu (140 Ci/mmol) were obtained from Dupont-New England Nuclear. Heparin was obtained from CalBiochem.
Preparations of membrane vesicles and protein components for in vitro translocation assays.
Membrane vesicles were prepared from SecA-depleted CK1801.4 and D10.2, an Unc− derivative of D10, by following procedures described previously (7, 36). proOmpA, SecB, and anti-SecA antibodies were prepared as described elsewhere (7, 36). Radioactive SecA was prepared from BL21 (λDE3)/pT7-secA as previously described (3, 7) with the following modifications in order to obtain radioactive SecA with a high specific activity. Cells were grown in 50 ml of MinA medium supplemented with 0.5% glucose and an amino acid mixture (50 μg/ml) lacking either Met, Gly, or Leu. Five millicuries of either [35S]Met, [3H]Gly, or [3H]Leu was used to label the proteins. Labeled SecA was purified by stepwise elution from a 1-ml column packed with SP-Sepharose FF followed by gel filtration chromatography on a Sephacryl S-200 column (1.6 by 60 cm). The final preparations contained more than 98% SecA as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein translocation assay and proteolysis.
SecA was reconstituted into SecA-depleted CK-1801.4 membranes according to procedures described previously (7). The reconstituted membranes were incubated at 37°C for 15 min in 100 μl of translocation mixture containing 2 μg of SecB and 1 μg of proOmpA, followed by incubation with 1 mg of proteinase K or trypsin/ml on ice for 15 min. After addition of 0.7 ml of stop solution containing 1 mM PMSF or soybean trypsin inhibitor (final concentration, 2 mg/ml) to stop the proteolysis, the membranes were recovered by centrifugation at 95,000 rpm for 20 min over a 0.2-ml sucrose cushion with a Beckman TL100 centrifuge. Two-thirds of the resulting supernatant was mixed with an equal volume of 16% cold trichloroacetic acid (TCA) and incubated on ice for 30 min. The precipitates were recovered by centrifugation at 14,000 rpm for 10 min in a Jovan A14 centrifuge, washed with 1 ml of cold acetone, and air dried. The radioactive SecA fragments recovered from both the membrane and supernatant fractions were then separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane sheets (ProBlott; Applied Biosystems), and visualized either by Coomassie blue staining or by Western blotting with anti-SecA rabbit serum.
Sequencing of SecA fragments.
SecA labeled with [35S]Met, [3H]Gly, or [3H]Leu was reconstituted into SecA-depleted CK-1801.4 membranes (2 × 106 to 20 × 106 cpm/mg of membrane protein) by following procedures described previously (7). After translocation reaction and proteolysis, the SecA fragments were separated by SDS-PAGE and transferred to PVDF membranes. Individual bands were excised from the Coomassie blue-stained PVDF membrane sheets with a razor blade and were subjected to Edman degradation with a Beckman LF3200 protein/peptide sequencer in the Biology Department’s Molecular Biology Core Facility. The samples from each cycle were collected and counted for their radioactivity with a Beckman LS 6500 liquid scintillation counter, with counting efficiencies of 80% for 14C and 35S and 35% for 3H. The radioactive peaks identified the positions of labeled amino acids. Since the cleavage sites for trypsin are the carboxyl ends following either Lys or Arg, the N-terminal sequence of each fragment was identified by matching the cycle number of the radioactivity peak(s) to the occurrence between the labeled amino acid and its nearest upstream Arg or Lys of the SecA sequence deduced from its DNA sequence (31). This first identification was confirmed by repeating the radiosequencing of the fragment by using SecA labeled with a different amino acid.
Normal chemical peptide sequencing was also used to directly identify the tryptic SecA fragments in some cases. D10.2 membranes were incubated with 6 M urea on ice for 60 min, recovered by centrifugation in a Beckman TL centrifuge at 70,000 rpm for 40 min, and then resuspended in 20 mM HEPES-KOH buffer (pH 8). Nonradioactive SecA was incubated with the urea-washed D10.2 membranes under translocation conditions, followed by incubation with trypsin at 1 mg/ml on ice for 15 min. The reaction mixture was then separated into supernatant and membrane fractions. The resulting fragments were extracted from the membranes by incubation with 6 M urea on ice for 30 min or were recovered from the top two-thirds of the supernatant fraction. These SecA fragments, as well as those generated without membranes (by incubation with 20 μg of trypsin/ml on ice for 15 min in TAKM buffer), were precipitated with 8% TCA, separated by SDS-PAGE, transferred to PVDF membrane sheets, and visualized by Coomassie blue staining. Individual bands were then excised from the PVDF membrane sheets and subjected to chemical peptide-sequencing analysis. The amino acids identified were in the range of 1 to 10 pmol for the urea-extracted fragments and were more than 50 pmol for SecA fragments recovered from the supernatant fraction or generated in the absence of membranes. In cases where the excised band contained multiple peptides, all possible amino acids were called and were aligned against the known sequences of SecA, trypsin, and trypsin inhibitor.
Identification of SecA fragments by using region-specific antibodies.
The region-specific antibodies were a generous gift from D. Oliver (Wesleyan University, Middletown, Conn.). Six SecA fragments (A1 [SecA1-209], A2 [SecA211-350], A3 [SecA351-509], A4 [SecA519-664], A5 [SecA665-820], and A6 [SecA822-901]) fused to the C-terminal end of maltose-binding protein were purified from E. coli strains overexpressing these chimeric proteins and were used to raise region-specific antibodies against A1 to A6, respectively (29). The antibodies were diluted 5,000- to 20,000-fold so that they gave similar band densities when the same amount of purified SecA was analyzed. Reconstituted membranes were incubated at 37°C for 15 min (7) and were digested with proteinase K or trypsin at 1 mg/ml on ice for 15 min. After addition of either stop solution or soybean trypsin inhibitor (final concentration, 2 mg/ml) to stop the proteolysis, the reaction mixture was centrifuged at 95,000 rpm for 20 min over a 0.2-ml sucrose cushion with a Beckman TL100 ultracentrifuge. SecA fragments were recovered from both membrane and supernatant fractions, separated by SDS-PAGE, and transferred to PVDF membrane sheets as described above. Individual lanes were then cut from the PVDF membrane sheets and incubated with antibodies against full-length SecA and specific SecA regions separately, followed by incubations with alkaline phosphatase-conjugated secondary antibody and chemiluminescence detection using a kit from Bio-Rad (Hercules, Calif.). The developed membrane sheets were then air dried and exposed to Kodak (Rochester, N.Y.) BioMax MR film to obtain an autoradiogram.
RESULTS
Comparison of proteolysis of membrane-associated SecA with proteinase K and trypsin.
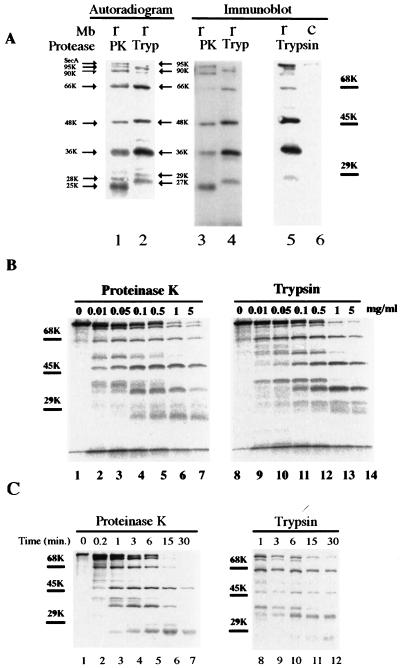
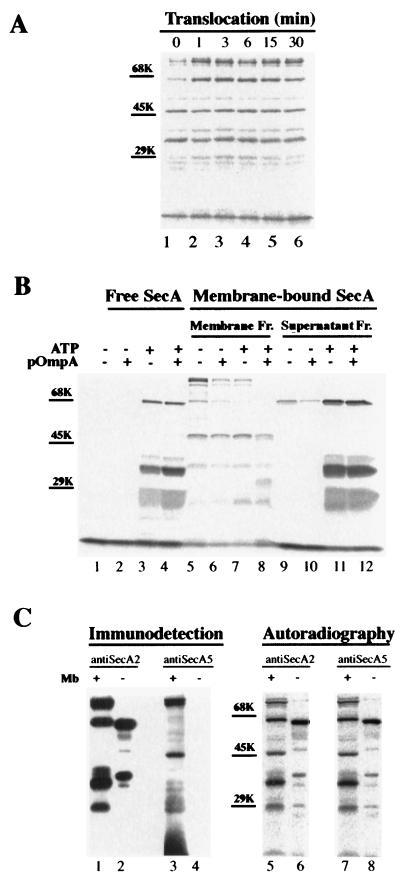
It has been shown (7) that the membrane-embedded SecA gives rise to several major fragments after incubation with proteinase K (at 1 mg/ml, on ice, for 15 min). To further characterize these fragments (also see below), we compared the proteolytic profiles of SecA with proteinase K and trypsin at various concentrations and for various periods (Fig. 1). The sizes of the corresponding fragments generated by these two proteases were slightly different, as revealed by both the autoradiograms (Fig. 1A, lanes 1 and 2) and the immunoblots (Fig. 1A, lanes 3 and 4). Although the 66-, 48-, and 36-kDa fragments remained unchanged, the fragments that were 28 and 25 kDa with proteinase K digestion moved up to 29 and 27 kDa, respectively, with trypsin digestion (Fig. 1A, lanes 1 and 2). From now on, these two fragments will be designated the 29- and 27-kDa fragments for consistency and simplicity.
FIG. 1.
Proteolysis of membrane-integrated SecA with proteinase K and trypsin. (A) 35S-labeled SecA fragments in membranes generated by proteolysis with proteinase K or trypsin. Reconstituted membranes (r) or SecA-depleted control membranes (c) from CK1801.4 were subjected to protein translocation reaction and proteolysis as described in Materials and Methods. The resulting SecA fragments in the membrane fraction were analyzed by SDS-PAGE and autoradiography (lanes 1 and 2) or by immunoblotting using anti-SecA antibodies (lanes 3 to 6). Lanes 1 and 2 and lanes 3 and 4 were from the same gel but were developed differently. The positions of the molecular-marker proteins bovine serum albumin (68 kDa), ovalbumin (45 kDa), and carbonic anhydrase (29 kDa) are shown by bars. The sizes of the protease-resistant SecA fragments are indicated by arrows. (B) Proteolysis of membrane-associated SecA with proteinase K or trypsin was carried out as for panel A, except that the different protease concentrations indicated were used. (C) Kinetics of proteolysis of membrane-integrated 35S-labeled SecA. Proteolysis was performed as for panel A except that the incubation was carried out for the times indicated.
Generally, larger fragments were obtained with lower concentrations of protease (Fig. 1B) or shorter incubation times (Fig. 1C). However, all fragments except the 27-kDa fragment were observed with concentrations of proteinase K as low as 0.01 mg/ml and as early as after 1 min of incubation. The 66-kDa fragment and an intermediate 40-kDa fragment decreased with time, with a concomitant increase in the 36- and 27-kDa fragments, suggesting that the latter two fragments are degradation products of the 66- and 40-kDa fragments. The 40-kDa fragment was completely digested after 15 min. In contrast, the 48-kDa fragment was relatively constant and stable. After prolonged proteolysis (at 1 mg/ml for 30 min), only the 48- and 27-kDa fragments remained (Fig. 1C, lane 7). The 29-kDa band was relatively weak, as judged by both autoradiography and immunoblotting (Fig. 1A). Furthermore, the majority of this fragment was normally found in the supernatant fraction (7). Proteolysis with trypsin gave a similar pattern but slower kinetics (Fig. 1B, lanes 8 to 14, and Fig. 1C, lanes 8 to 12). These results suggest that protease-resistant SecA fragments represent individual SecA domains. The SecA fragments visualized by anti-SecA antibodies corresponded well to those on the autoradiograms and were absent in the SecA-depleted control membranes (Fig. 1A, lanes 5 and 6). Thus, the bands of the radioactive SecA fragments visualized by anti-SecA antibodies could be excised directly from the PVDF membrane sheets in a radiosequencing experiment described below. This eliminated the need for alignment of radiograms to the PVDF membrane sheets.
Identification of the major protease-resistant SecA fragments by radiosequencing.
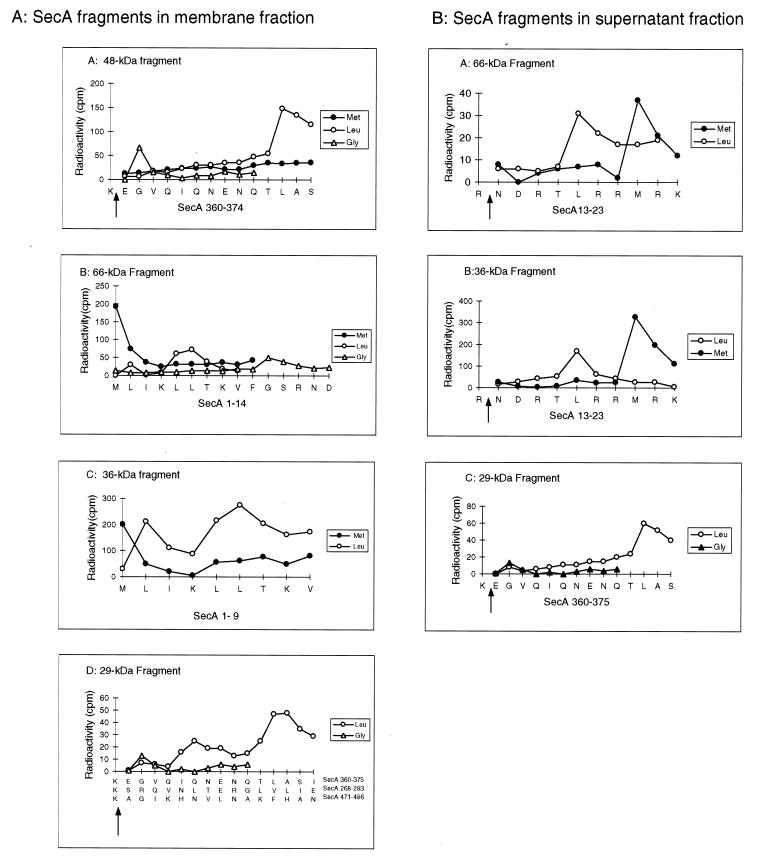
Radiosequencing was used to identify the protease-resistant SecA fragments without purification. Trypsin was used instead of proteinase K in proteolysis because of its specificity in cleaving only the peptide bonds after Arg or Lys residues. The specificity of trypsin cleavage offers the advantage of providing an additional marker or index in aligning amino acids after radiosequencing. Figure 2 shows the radiosequencing profiles of the major trypsinized SecA fragments. The 48-kDa fragment recovered from the membrane fraction gave a radioactive peak at cycle 2 with [3H]Gly-labeled SecA and one at cycle 12 with [3H]Leu-labeled SecA but showed no peaks with [35S]Met-labeled SecA in the first 14 cycles (Fig. 2A). Searching the SecA sequence for (K/R)XG(9X)L determined that this sequence only matches Lys360 to Ser374. Thus, the 48-kDa fragment was identified as an internal fragment starting after Lys360 at Glu361. The 66-, 36-, and 27-kDa fragments were similarly identified as N-terminal fragments starting from Met1 by the same method (Fig. 2A and data not shown). The 95- and 90-kDa fragments were not sequenced because of their small amounts. The 29-kDa fragment, which potentially corresponds to the translocation-dependent 30-kDa fragment identified previously (7, 11, 12), apparently contained at least three species, starting at Glu361, Ser269, and Ala472. All three fragments are different from the reported translocation-dependent 30-kDa fragment, which starts at Leu610 (27). The 29-kDa fragment starting at Glu361 was named the s29-kDa fragment because the supernatant 29-kDa band constituted about 80% of this species (Fig. 2) (7) and contained a single fragment which starts at Glu361 (Fig. 2B). On the other hand, the 66-, 36-, and 27-kDa fragments from the supernatant fraction were shown to start at Asn14 (Fig. 2B and data not shown). These fragments clearly correlate to their counterparts on the membranes, which start at Met1. Thus, the N-terminal 13 amino acids might play a role in anchoring these fragments on the membrane. However, these fragments are not released from the membranes by extensive digestion of their receptors on the membrane (14), since similar fragments could be generated in the absence of membrane (described below). The significance of the presence of these fragments in both fractions and of the absence of the 48-kDa fragment in the supernatant fraction is discussed below where the requirement for the formation of each fragment is examined.
FIG. 2.
Radiosequencing of the protease-resistant SecA fragments. After translocation and proteolysis as described in Materials and Methods, SecA fragments recovered from both membrane pellet (A) and supernatant (B) fractions were separated by SDS-PAGE, transferred to PVDF membrane sheets, and visualized by Coomassie blue staining or immunodetection. Individual bands were then excised from the PVDF membrane sheets and subjected to radiosequencing as described in Materials and Methods. The radiosequencing profiles for each fragment are aligned with the identified sequences. The cleavage sites of trypsin are indicated by arrows. The sequences identified were unique. There is no other match in the SecA sequence except for the 29-kDa fragments, for which all possible sequences are shown.
Identification of SecA fragments by using region-specific antibodies.
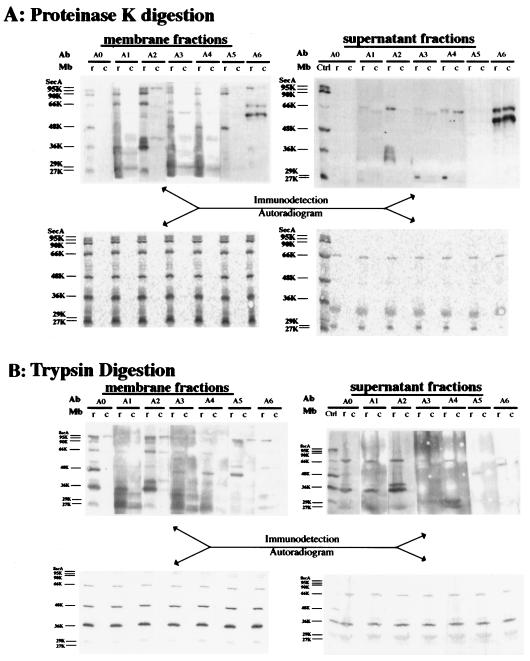
To confirm the radiosequencing identification of these tryptic fragments of SecA and their relationship to the proteinase K fragments, reconstituted membranes were treated with proteinase K (Fig. 3A) or trypsin (Fig. 3B), separated by SDS-PAGE, and developed by immunoblotting using the region-specific antibodies against six consecutive SecA regions (A1 to A6). SecA-depleted CK1801.4 membranes treated in the same way were used as a control to detect nonspecific bands. The data from both protease treatments clearly showed that the 66- and 36-kDa fragments were the N-terminal fragments (A1 and A2), while the 48-kDa fragment represents a center domain (regions A3 to A5). This supports the radiosequencing identification of these fragments (Fig. 2). Several bands, including a 66-kDa band detected by anti-A6 (Fig. 3A), are probably nonspecific, since these bands were also detected in the SecA-depleted control membranes. The 90- and 95-kDa fragments generated by proteinase K were recognized by antibodies against all regions except the A6 region. Therefore, these fragments represented forms of SecA with C-terminal deletions, which have been reported previously (37). The 95- and 90-kDa bands generated with trypsin were not as clear but were detected by antibodies against SecA2 and SecA5. Taking the sizes (A1 to A5, 820 amino acids; A2 to A6, 700 amino acids) into consideration, these fragments should also be the forms of SecA with C-terminal deletions. Immunodetection of the 29- and 27-kDa fragments was not clear due to nonspecific bands. There are probably many species in this area, but most of them were found within regions A1 to A4, although some faint bands were also observed in regions A5 to A6. SecA fragments corresponding to the 66-, 36-, 29-, and 27-kDa fragments in the membrane were also found in the supernatant fraction (7). The 66-, 36-, and 29-kDa fragments obtained with both protease treatments were identified as fragments corresponding to regions A1 to A4, A1 to A2, and A3 to A4 of SecA (see Fig. 7B), respectively. The 27-kDa band was not observed with proteinase K digestion in Fig. 3A but was identified as a fragment corresponding to regions A1 to A2 in a separate experiment (data not shown). A minor 48-kDa band in the supernatant fraction was recognized by anti-A5, but it was not detected by antibodies against A3 and A4 or by autoradiogram (Fig. 3B). Thus, this 48-kDa band is probably nonspecific or might represent a small amount of the 48-kDa SecA fragment released from the membranes. In conclusion, immunodetection with region-specific antibodies demonstrated that SecA fragments generated by both proteases represented the same SecA domains and confirmed the radiosequencing results. Moreover, the 66- and 48-kDa fragments overlapped in the central region (SecA361-610), as both of them were detected by anti-SecA3 and anti-SecA4. On the other hand, each was recognized specifically by anti-SecA1 (and anti-SecA2) and anti-SecA5, respectively. Therefore, these two fragments are derived from different SecA molecules.
FIG. 3.
Identification of SecA fragments on membrane and the supernatant fractions by region-specific antibodies. CK1801.4 membranes (c) and reconstituted membranes (r) were subjected to translocation and digestion with proteinase K (A) or trypsin (B) as described in Materials and Methods. The SecA fragments in both the membrane fractions and the supernatant fractions were separated by SDS-PAGE, transferred to PVDF membrane sheets, and developed both by autoradiography and by immunoblotting with specific antibodies against different regions of SecA as indicated. Mb, membrane; A0, whole SecA; Ctrl, SecA fragments recovered from membrane fraction. For A1 to A6, see Materials and Methods.
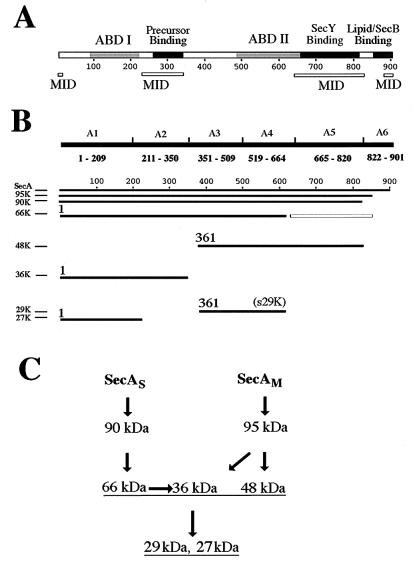
FIG. 7.
Summary of identification and characterization of SecA fragments. (A) Schematic presentation of proposed SecA domains. The two ABDs of SecA (ABD I and ABD II) (25) are shown as shaded rectangles, and the domains interacting with precursor (23, 37) or with lipid and SecB (2) are shown as solid rectangles, on the SecA sequence. The known and proposed MID are shown by open bars below the SecA sequence. (B) Schematic presentation of the identification of SecA fragments. The identified SecA fragments and the six SecA fragments fused to maltose-binding protein to produce the region-specific antibodies are aligned with the SecA sequence illustrated in panel A. Starting amino acid numbers are given for the major fragments of the integral SecA. The open box represents the potential cleavage fragment corresponding to the reported 30-kDa fragment starting at Leu610 (27) but not identified here. (C) Proposed proteolysis pathway for SecAS and SecAM. SecAS, membrane-integral SecA with a conformation similar to that of soluble SecA; SecAM, membrane-integral SecA with a membrane-induced conformation.
Characterization of the formation of the SecA fragments.
To elucidate the relationship between the formation of these SecA domains and protein translocation, we examined the formation of the SecA fragments in various translocation stages (Fig. 4A). Reconstituted membranes were incubated at 37°C under protein translocation conditions for the times indicated and then trypsinized on ice for 15 min. As observed previously with proteinase K digestion (7), all the major fragments were detected under no-translocation conditions (Fig. 4A, lane 1). After 3 min of incubation, the 95-, 66-, and 29-kDa fragments increased significantly while the other fragments remained unchanged. There were no obvious changes in the proteolysis profile of SecA after longer translocation periods (3 to 30 min). Taking into consideration that release of SecA from the membrane was completed within the first 3 min (7), these data suggest that SecA adapts its conformation in the membrane, which causes the formation of the individual domains, in the early stages of protein translocation.
FIG. 4.
Characterization of the protease-resistant SecA domains. (A) Proteolysis of membrane-associated SecA at different translocation stages. After translocation and proteolysis as described in Materials and Methods, SecA fragments recovered from the membrane fraction were separated by SDS-PAGE and visualized by autoradiography. Positions of molecular-marker proteins are shown by bars. (B) Effects of ATP, proOmpA, and membrane on the formation of the proteolytic SecA fragments. 35S-labeled SecA was incubated at 37°C for 15 min under the conditions indicated. After the incubation, the reaction mixture was incubated with 1 mg of trypsin/ml on ice for 15 min. The SecA fragments generated in the absence of membrane were TCA precipitated and were analyzed by SDS-PAGE and autoradiography (lanes 1 to 4). The proteolytic SecA fragments generated in the presence of membranes were separated into membrane and supernatant fractions (Fr.), followed by SDS-PAGE and autoradiography (lanes 5 to 12). Positions for molecular-weight marker proteins are indicated. (C) Limited proteolysis of SecA in the absence of membranes (Mb). After incubation in translocation mixture minus membranes, 35S-labeled SecA was incubated with 60 μg of trypsin/ml on ice for 15 min and precipitated by 8% TCA. The pelleted 35S-labeled SecA fragments were separated by SDS-PAGE and visualized by autoradiography (lanes 6 and 8) and by immunodetection with anti-SecA2 (lane 2) and anti-SecA5 (lane 4). SecA fragments from trypsinized (1 mg/ml; 15 min) membranes were analyzed for comparison (lanes 1, 3, 5, and 7).
ATP, precursor, phospholipids, and urea-washed membranes were shown to change SecA conformations in free solution, as revealed by their sensitivity to limited proteolysis with V8 protease at low concentration (32). Therefore, we examined the effects of membranes, as well as ATP and precursor, on the formation of these SecA fragments when treated with trypsin (Fig. 4B). With 1 mg of trypsin/ml, SecA in buffer alone was completely digested (Fig. 4B, lanes 1 and 2), but several bands at 66, 36, 29, and 27 kDa were observed in the presence of ATP (Fig. 4B, lanes 3 and 4). Similar bands were also found in both the membrane and supernatant fractions when SecA was digested with trypsin in the presence of membrane (Fig. 4B, lanes 5 to 12). This indicates that the formation of the 66-, 36-, 29-, and 27-kDa fragments is dependent on ATP. In contrast, the 48-kDa fragment was observed only in the membrane fraction (Fig. 4B, lanes 5 to 8). ATP and/or proOmpA had little effect on the amount of the 48-kDa fragment generated. These results indicate that the presence of membranes is necessary and sufficient for the formation of the 48-kDa fragment. Since the presence of membrane reportedly increased the protease resistance of SecA by 10- to 100-fold (27), it is possible that the 48-kDa fragment or its related fragments could be generated by trypsinization at low concentrations in the absence of membrane. To examine this possibility, 35S-labeled SecA was treated with 60 μg of trypsin/ml after incubation in translocation mixture without membrane. Two major bands at 65 and 40 kDa and several minor bands, including one at 50 kDa, were generated, as revealed by autoradiography (Fig. 4C, lanes 5 to 8). However, none of these fragments was detected by anti-SecA5, which specifically reacts with the 48-kDa fragment generated from membrane-associated SecA (Fig. 4C, lanes 3 and 4). Similar identifications were obtained by sequencing analysis of SecA fragments generated with 20 μg of trypsin/ml (Table 1). Therefore, the 48-kDa SecA fragment is membrane specific: formation of this SecA domain is induced only by interaction with membranes.
TABLE 1.
Sequence analysis of SecA fragmentsa
| SecA fragment (kDa) | N-terminal sequence |
|---|---|
| Generated in the presence of membrane | |
| Extracted from membrane | |
| 66 | 14NDRT |
| 48 | 361EGVQIQNENQ |
| 36 | 1MLIKLLtKVF |
| 9vFGSRNDRTL | |
| 14nDRTLRRMRK | |
| 31 | 361EGVQIQNENQ |
| 575SGRQGdaGS | |
| s29 | 361EGVQIQNENQ |
| Recovered from supernatant | |
| 66 | 9VFGSRNDRTL |
| 14NDRTLRRMRK | |
| 36 | 9VFGSRNDRTL |
| 14NDRTLRRMRK | |
| s29 | 361EGVQIQNENQ |
| Generated in the absence of membrane | |
| In buffer containing ATP and precursor | |
| 66 | 9VFGSRNDRTL |
| 14NDRTLRRMRK | |
| 36 | 9VFGSRNDRTL |
| 14NDRTLRRMRK | |
| s29 | 361EGVQIQNENQ |
| In buffer with low trypsin | |
| 66 | 9VFGSRNDRTL |
| 50 | 9VFGsRNDRTL |
| 36 | 104TGEGKTLTAT |
| 30 | 644QLLEQQDVA |
SecA fragments were generated in the presence of membranes as described in Materials and Methods and were subjected to peptide-sequencing analysis. Soluble SecA (1 mg/ml) was digested with trypsin on ice for 15 min in buffer in the presence of energy source and proOmpA (1 mg of trypsin/ml) or in the absence of energy source and proOmpA (20 μg of trypsin/ml). The resulting SecA fragments were then analyzed in the same manner as the SecA fragments generated in the presence of membranes. Identified SecA sequences are shown in single-letter code. Uppercase letters represent amino acids of the analyzed sequences which match the known SecA sequence (31), whereas lowercase letters represent unconfirmed residues.
Most of the stable SecA fragments generated in the absence of membrane at low trypsin concentrations are N-terminal fragments, as revealed by both peptide sequencing (Table 1) and immunodetection with anti-SecA2 and anti-SecA5 (Fig. 4C). These N-terminal fragments resemble those generated from reconstituted membrane, although the sizes are slightly different (Fig. 4C). This finding indicates that some SecA on the membrane forms a similar conformation to that of soluble SecA (27). The proteolytic patterns of free SecA (Fig. 4B, lanes 1 to 4) and supernatant SecA (Fig. 4B, lanes 9 to 12) are similar. Chemical peptide sequencing revealed identical sequences of the corresponding fragments (Table 1). This result indicates that the enhanced protease resistance of the 66-kDa fragment is not dependent on membranes, as reported elsewhere (27), but instead is dependent on ATP. It is worth noting that the 66-kDa (A1 to A4) fragments could give rise to the 27-kDa (A1 to A2) and 29-kDa (A3 to A4) fragments, which contain ATP-binding domain I (ABD I) and ABD II, respectively (12, 28). These observations explain why these domains are extremely resistant to proteolysis in the presence of ATP. Indeed, the 27- and 29-kDa fragments were the two major species detected in the supernatant after proteinase K digestion (7). It has been suggested that SecA in buffer possesses an N-terminal ATPase domain and a C-terminal domain (27). The coexistence of the 27- and 29-kDa fragments in the presence of ATP indicates that ATP binding to SecA induces conformational changes that form ABD I and ABD II. These two fragments were not observed when SecA was treated with a low concentration of trypsin in the absence of ATP (Table 1).
Characterization of interactions between the SecA fragments and the membrane.
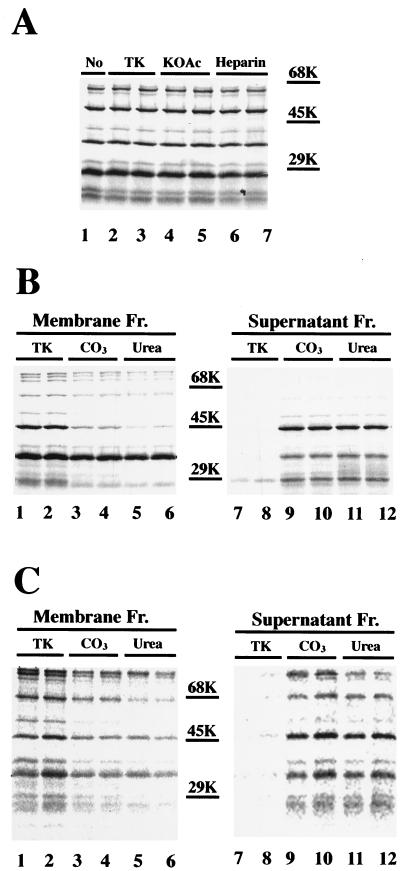
To determine the nature of the interactions between the SecA fragments and the membrane, the stability of these fragments was examined with extraction by acid (1% acetic acid), alkali (0.1 M NaOH), a high concentration of salt (1 M potassium acetate [KOAc]), heparin (10 mg/ml), 0.1 M Na2CO3, and 6 M urea. Extraction with TK buffer was used as a control. All fragments were resistant to extraction by a high concentration of salt or by heparin (Fig. 5A). Since heparin specifically removes peripheral SecA (7, 38), these results indicate that all fragments in the membrane are integral and that the membrane integration is independent of ionic interactions. The 48- and 27-kDa fragments were partially extracted by Na2CO3 and were extensively extracted by urea (Fig. 5B), suggesting a more hydrophilic environment. In contrast, the majority of the 66- and 36-kDa fragments were not extracted with Na2CO3 or urea (Fig. 5B). Therefore, these fragments might interact with lipids or proteins in the membrane through hydrophobic interactions. Extraction with acetic acid or alkali gave profiles similar to those given by extraction with a high concentration of salt or with Na2CO3, respectively (data not shown). All these fragments were also observed when the reconstituted membranes were extracted with urea or Na2CO3 prior to trypsinization (Fig. 5C), indicating that all these fragments were derived from integral SecA.
FIG. 5.
Nature of protease-resistant SecA domains in the membrane. (A) Extraction of protease-resistant SecA fragments with a high concentration of salt and with heparin. After translocation and proteolysis, the reconstituted membranes were recovered by centrifugation and either solubilized with SDS-sample buffer (no extraction) or resuspended in 100 μl of TK buffer (TK), 1 M KOAc, or 10 mg of heparin/ml in duplicates. After incubation on ice for 30 min, the extracted membranes were recovered by centrifugation at 95,000 rpm for 40 min in a Beckman TL100 centrifuge and were analyzed by SDS-PAGE and autoradiography. (B) Extraction of protease-resistant SecA fragments with Na2CO3 or urea. The procedure was the same as for panel A except that the extraction was performed with 0.1 M Na2CO3 (CO3) or 6 M urea and both the membrane fraction (Fr.) and the supernatant fraction (after TCA precipitation) were analyzed. (C) Proteolysis of membrane-associated SecA after extraction with Na2CO3 or urea. Reconstituted membranes were recovered by centrifugation after translocation, incubated with 0.1 M Na2CO3 (CO3) or 6 M urea in duplicates on ice for 30 min, recovered again by centrifugation, resuspended in translocation buffer, and incubated with 1 mg of trypsin/ml on ice for 15 min. Both the membrane fraction (Fr.) and the supernatant fraction were analyzed. The positions for the molecular-weight marker proteins are indicated.
Chemical sequencing analysis of SecA fragments.
Since some of the membrane-associated SecA fragments could be extracted by urea (Fig. 5), we sequenced these fragments after extraction, as well as those recovered from the supernatant and those generated in the absence of membranes (Table 1). In order to compare our results to the reported sequencing results of SecA fragments extracted from urea-washed membranes (27), SecA was reconstituted into urea-washed D10.2 membranes and CK1081.4 membranes. Proteolysis of these two kinds of reconstituted membranes generated similar SecA fragments, as revealed by immunodetection using region-specific antibodies (data not shown). Chemical peptide sequencing of the 48-kDa fragment extracted from the D10.2 membranes by urea revealed a sequence starting at Glu361 (Table 1), the same starting sequence as for the 48-kDa fragment from the reconstituted membranes identified by radiosequencing (Fig. 2) and immunodetection (Fig. 3). This fragment was not observed in the supernatant fraction and was not generated in the absence of membrane. The 50-kDa fragment generated in the absence of membrane, which is close to the 48-kDa fragment in size, was an N-terminal fragment starting at Val9. The 29-kDa fragments from both the supernatant fraction and the urea extracts also start at Glu361, matching the sequence of the s29-kDa fragment obtained by radiosequencing (Fig. 2A). The other two possible 29-kDa fragments identified by radiosequencing (Fig. 2A) were not detected in the urea extracts. The one starting at Ser269 might have remained on the membrane because of its hydrophobicity (see Discussion), and the other one, which starts at Ala472, might be too scarce to be detected. The 66-kDa fragment in the urea extracts is the same as the 66-kDa fragment recovered from the supernatant, since both fragments start at Asn14. These results confirmed that the 66-kDa SecA fragment starting at Met1 was not extracted by urea. Trypsin digestion of SecA in the absence of membrane generated a 66-kDa fragment starting at Val9. This fragment was not observed in the membrane fraction. In contrast, sequencing analysis of the 36-kDa band in the urea extracts revealed three sequences starting at Met1, Val9, and Asn14. The latter two fragments were also found in the supernatant fraction. These two 36-kDa fragments either contained no Leu or Met in their first 9 residues or were minor on the membrane, so that only the 36-kDa fragment starting from Met1 was detected by radiosequencing (Fig. 2A). On the other hand, all three 36-kDa fragments (Table 1) are present in similar amounts in urea extracts. These results indicate that the majority of the 36-kDa fragment starting from Met1 was not extracted by urea; indeed, most 36-kDa fragments were resistant to urea extraction (Fig. 5). This observation indicates that the urea-resistant 36-kDa fragment must be the one starting from Met1. This major 36-kDa fragment and the 66-kDa fragment starting at Met1 were not found in the supernatant fraction. Collectively, these results suggest that the N-terminal 8 or 13 amino acids may serve as one of the anchors for membrane-integral SecA. Therefore, removal of these peptides resulted in the release of the N-terminal SecA fragments from the membrane. Most importantly, the chemical sequencing data confirmed that the 48- and 66-kDa fragments are overlapping fragments and thus are derived from different SecA molecules.
Characterization of 30-kDa SecA fragments.
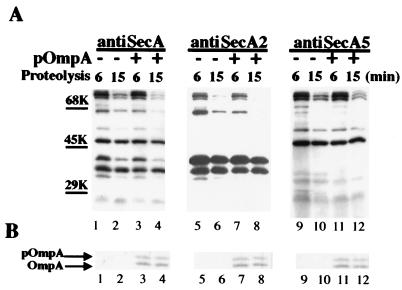
A C-terminal 30-kDa fragment was reported to undergo a translocation-dependent insertion-deinsertion cycle during protein translocation (11, 12, 27). However, we were unable to detect this 30-kDa fragment by radiosequencing. On the other hand, several weak bands around 30 kDa were observed when the SecA fragments were visualized by anti-A5 or anti-A6 (Fig. 3 and 4). Therefore, we examined the translocation dependence of these fragments by using anti-SecA5 (Fig. 6). Normal anti-SecA antibody and anti-SecA2 were used for comparison. Nonradioactive SecA and 35S-labeled proOmpA were used to allow simultaneous detection of SecA fragments and OmpA translocation. Two SecA fragments around 30 kDa were detected by anti-SecA5, but none of them increased upon translocation of proOmpA. Since the translocation-dependent 30-kDa fragment could be extracted by urea (27), trypsinized membranes were treated with urea by following procedures described previously (27), and the extracted SecA fragments were separated by SDS-PAGE and transferred to PVDF membrane sheets. Several bands were observed around 30 kDa by Coomassie staining. Protein sequence analysis of these bands identified SecA fragments starting from Met1, Val9, Asn14, Glu361, and possibly Ser575 (Table 1). Furthermore, a C-terminal 30-kDa fragment was also identified as starting at SecA644 when SecA was digested with 0.02 mg of trypsin/ml in TAKM buffer (Table 1). However, we have not been able to detect the reported translocation-dependent C-terminal 30-kDa fragment starting at Leu610 (27). These results suggest that this C-terminal 30-kDa fragment probably represents an unstable or minor species in the membrane after proteolysis at high protease concentrations, and thus it is undetectable in our system.
FIG. 6.
Characterization of 30-kDa fragments by region-specific antibodies. Nonradioactive SecA was reconstituted into SecA-depleted CK1801.4 membranes. The reconstituted membranes were incubated with or without 35S-labeled proOmpA in translocation buffer with the energy source at 37°C for 15 min and were digested with 1 mg of trypsin/ml on ice for 15 min. The resulting SecA fragments were separated by SDS-PAGE, transferred to PVDF membrane sheets, and visualized by immunodetection with region-specific antibodies (A). Arrows indicate the two “30-kDa” bands detected by anti-A5. The same PVDF membrane sheets were also visualized by autoradiography to show the translocated OmpA (B).
DISCUSSION
A significant fraction of SecA has been shown to be permanently embedded in the membranes and does not cycle on and off the membrane during protein translocation (7). In contrast to findings that only a 30-kDa domain of SecA is resistant to proteolysis (11, 12, 27), several SecA domains in the membrane were found to be resistant to proteolysis, and some of them were independent of protein translocation (7). We have now identified and characterized these protease-resistant SecA fragments through immunodetection using region-specific antibodies, chemical extraction, and sequencing analysis. The observed proteolytic patterns suggested that the SecA proteins in the membrane are cleaved at two main sites, in addition to the C-terminal cleavage (37). One of these sites is at Lys360, giving rise to the N-terminal 36-kDa fragment and the central 48-kDa fragment. The other site is located around SecA610, giving rise to the N-terminal 66-kDa fragment and perhaps a C-terminal 30-kDa fragment which has not been identified in the present study (Fig. 7B). The 66- and 48-kDa fragments partially overlap in sequences (Fig. 7B), indicating that they originate from different SecA molecules. The most obvious explanation is that there are two different membrane-integral forms of SecA, SecAS and SecAM, based on the different behaviors of the overlapping 66- and 48-kDa fragments. The 66-kDa fragment increases significantly in the presence of ATP hydrolysis and protein translocation (Fig. 4) and can be chased off the membrane by excess nonradioactive SecA (7) but is resistant to Na2CO3 extraction (Fig. 5). In contrast, the 48-kDa fragment is not affected significantly by ATP hydrolysis and protein translocation (Fig. 4) and is relatively more sensitive to Na2CO3 extraction (Fig. 5) but is resistant to chasing by excess nonradioactive SecA (7). SecAS may have the same conformation as soluble SecA in buffer, which also gives rise to an N-terminal 65-kDa fragment (27), although the protease resistance is 10- to 100-fold greater than that for free SecA. On the other hand, SecAM is induced by interaction with the membrane, since the formation of a specific 48-kDa fragment requires the presence of membranes (Fig. 4B). Therefore, the 66- and 48-kDa fragments are specifically derived from SecAS and SecAM, starting at Met1 and Glu361, respectively. The 36-, s29-, and 27-kDa fragments could come from either SecAS or SecAM or both. However, they most likely come from SecAS because, like the 66-kDa fragment, they are dependent on ATP and independent of membranes (Fig. 4 and Table 1). It has been suggested that there is an equilibrium between a soluble form and a hydrophobic form of SecA, and that the latter is stable only on the membrane (28). Here, we present evidence that the membrane-integral SecA probably possesses two different conformations as revealed by proteolysis. Our previous findings, however, showed that the proteolytic 48-kDa fragment can still be generated from membranes even after incubation with excess nonradioactive SecA under protein translocation conditions (7); therefore, at least a significant fraction of SecAM does not participate in this equilibrium. Since SecA is known to be functional as a dimer (1, 10), it may form a conformational heterodimer of SecAS-SecAM in the membrane. This is supported by the observation that the molar ratio of the 66-kDa fragment and the 48-kDa fragment was close to 1:1 (Fig. 5B) when membranes were treated with proteases after urea or Na2CO3 extraction, which removes peripheral SecA. Alternatively, there may be two populations of SecA homodimers. The different proteolysis patterns of these two forms of SecA may also reflect their differences in interacting with the membrane. Work is under way to differentiate these possibilities and determine the different interacting components.
The limited number of SecA fragments observed in proteolysis with two different proteases under various conditions suggests that SecA assumes definite conformations on the membrane, in which only limited sites are exposed to protease digestion. The observed SecA fragments thus represent individual SecA domains which are resistant to proteolysis in reconstituted membranes as well as in wild-type membranes (data not shown) (see also reference 9). The major protease-resistant SecA fragments can be divided into two groups: the “N-terminal” group, containing the 66-, 36-, and 27-kDa fragments, and the “internal” group, containing the 48- and s29-kDa fragments. These results suggest that SecA has at least two MID (Fig. 7), one of which is located within SecA240-330. Removal of this region from the 36-kDa fragment, which is resistant to urea extraction, gave rise to the 27-kDa fragment, which could be extracted by urea; therefore, this region plays an important role in anchoring SecA on the membrane. In support of this notion, SecA300-350 has been shown to be periplasmically accessible (29). The other MID is possibly located within the C-terminal 20-kDa region of the 48-kDa fragment, or SecA610-800, since removal of this region could give rise to the soluble s29-kDa fragment. The extreme N-terminal 8 or 13 amino acids may also possess a third MID, since the N-terminal fragments found in the supernatant fraction are missing these 8 or 13 amino acids (Fig. 2B). In addition, the extreme C terminus of SecA may possess yet another MID. This region is known to interact with lipids (2) and is exposed to the periplasm (29, 37).
Proteolysis of SecAS in the membrane could give rise to the N-terminal 66-kDa fragment and the C-terminal 30-kDa fragment, as is apparently the case for the limited proteolysis of free SecA in buffer (27). We have identified the translocation-enhanced 66-kDa fragment but have been unable to detect a major stable C-terminal 30-kDa fragment. One possibility is that this domain, if it exists, represents an unstable or minor species and therefore its presence may be masked by others in this range. Indeed, we found a 31-kDa fragment which might correspond to a C-terminal SecA fragment starting at Ser575 (Table 1). The observation of this 30-kDa fragment as a major species (11, 12, 27) is due to the uneven artificial labeling of SecA by 125I, as predicted (7). It has recently been confirmed that the radioactivity in 125I-labeled SecA is essentially confined to the C-terminal 30-kDa region (13) and that proteolysis of metabolically labeled 35S-SecA gave rise to an N-terminal 65-kDa fragment and several other fragments in addition to the C-terminal 30-kDa domain of SecA that were protected from proteolysis (14). This N-terminal 65-kDa fragment apparently corresponds to the N-terminal 66-kDa fragment here. Protection of this fragment appears to depend on ATP, not on membranes, because similar amounts of this fragment were observed in the presence of ATP with or without membrane, and the majority of the 66-kDa fragments generated in the presence of membranes were found in the supernatant fraction (Fig. 4). TCA precipitation could bring down the 66-kDa fragment from the supernatant fraction, thus producing a stronger 66-kDa band and an apparently greater increase with addition of proOmpA (data not shown). Therefore, the reported large increase of a 65-kDa N-terminal SecA fragment upon translocation of proOmpA was probably due to analyzing the total SecA fragments precipitated by TCA (14). This 66- or 65-kDa domain can be further cleaved at around SecA330 into the N-terminal 36-kDa domain, which contains ABD I, and the s29-kDa domain, which contains ABD II (Fig. 7). Since proOmpA binds to SecA at SecA267-340 (23), such binding may shield the cleavage site on the 66-kDa domain, thus increasing the amount of the 66-kDa fragment. Indeed, a modest increase of the 66-kDa fragment in the membrane fraction was observed (7). Therefore, protection of the 66- or 65-kDa N-terminal SecA domain is mainly due to ATP-induced conformational change, not merely due to membrane insertion. The apparent translocation dependence of this domain is probably due to shielding of the cleavage site by proOmpA, preventing further digestion of the 66-kDa fragment.
Proteolysis of SecAM gave rise to the major 48-kDa fragment. Several lines of evidence suggest that the 48-kDa domain forms part of the protein-conducting channel. First, this domain is located immediately after the precursor binding region (23) and contains ABD II (25), which is considered important for coupling the insertion-deinsertion cycle of the 30-kDa SecA domain to protein translocation (11). Second, formation of this domain is induced by interactions with membranes (Fig. 4). Third, this domain is embedded in the membrane. There are 18 lysines and arginines in the 48-kDa domain. It is unlikely that all these sites are embedded inside the SecA molecule and are thus inaccessible to proteases. In fact, this fragment disappeared when the membranes were incubated with 1 mg of proteinase K/ml in the presence of 1% Triton X-100, which disrupts the membranes (7). Fourth, this domain is permanently embedded in the membrane. It is insensitive to ATP or protein translocation (Fig. 4) and cannot be chased out from the membrane by excess nonradioactive SecA (7). Fifth, this domain is in an aqueous environment like the translocating precursor proteins (15), since it was partially extracted by urea, Na2CO3, or alkali but was resistant to extraction with a high concentration of salt. Sixth, this domain (SecA610-800) might possess an important SecY-binding determinant (35), thus coupling the SecA channel and the other Sec machinery. This major 48-kDa fragment was not observed in a previous study (14). It is not clear whether this is related to the use of the urea-treated membranes there. The authors did find a major SecA fragment of about 48 kDa in the absence of membrane. This fragment probably correlates to our 50-kDa N-terminal fragment (Fig. 4C and Table 1), since both of them were generated only with low concentrations of trypsin. In contrast, the membrane-specific 48-kDa domain was resistant to protease ranging from 0.1 to 10 mg/ml (Fig. 1 and data not shown). Such protection is not likely to be due to nonspecific association of SecA with lipids. This 48-kDa domain and SecAM play an important structural role as translocation core or channel, rather than being nonfunctional (14). The structural role for SecAM explains why it is insensitive to ATP hydrolysis and protein translocation.
The current dogma depicts SecYEG as forming the core of the protein translocase, while SecA is the peripheral subunit that goes through the cycles of membrane insertion-deinsertion and that cycles on and off the membrane during translocation (39). However, it is not certain how the SecA dimer (1, 10) functions in the model, and the perpetual recycling of SecA is extremely inefficient physiologically. Moreover, recent studies have found that SecYG-deficient (38, 41) or SecE-deficient (40) membranes still had more than 50% translocation activity of proOmpA and other precursors with processing of the signal peptides. Thus, the SecYEG complex is not obligatorily required for all precursors but may enhance the efficiency and specificity. (The requirement of SecYEG for a subset of precursor proteins, like prePhoA, also is consistent with results of genetic studies showing that secY and secE are the essential genes [19, 30].) If SecYEG is not essential for the translocation of all proteins, and thus is not the core or channel for protein translocation, then what? The results presented here suggest that SecA might be the intrinsic core or channel. Membranes treated with urea have been shown to contain functional SecA (38). The majority is probably SecAM, since the 48-kDa fragment became a major band when both the reconstituted and the native membranes were digested with trypsin after urea extraction (Fig. 5 and data not shown). Taking into account that some SecA goes through cycles of membrane insertion-deinsertion (39), there may be two forms of SecA in the channel, one that is dynamic and one that is static. The finding of two conformational forms of integral SecA, SecAS and SecAM, perhaps as a heterodimer, fulfills this requirement. It is tempting to speculate that the dynamic SecAS may go through cycles of translocation-dependent insertion-deinsertion as proposed elsewhere (14, 39) while the static SecAM is permanently embedded in the membrane, interacting with SecY and other membrane proteins to form the protein-conducting channel. Taking into account all the available data, the simplest model puts the integral SecA (both SecAS and SecAM) in the core and moves SecYEG from the core to a role as accessory proteins like SecDFYajC, important but not essential components. In this scenario, the roles of SecA and SecYEG are analogous to those of ATP hydrolysis and proton motive force in protein translocation, in which one is essential and the other contributes significantly to efficiency (5, 40). The roles of SecYEG and SecA in protein translocation are thus uncertain and controversial. Further work is needed to reveal the details and resolve differences in interpretation.
ACKNOWLEDGMENTS
We thank D. Oliver and V. Ramamurthy for strains, plasmids, and the region-specific antibodies and for communicating unpublished observations; Y. Yang for stimulating discussions; and J. Ingraham, J. Houghton, A. Boyer, and G. Buck for comments.
This work is supported by a grant from the National Institutes of Health (GM34766) and equipment grants from the Georgia Research Alliance.
REFERENCES
- 1.Akita M, Shinkai A, Matsuyama S, Mizushima S. SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer. Biochem Biophys Res Commun. 1991;174:211–216. doi: 10.1016/0006-291x(91)90507-4. [DOI] [PubMed] [Google Scholar]
- 2.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C-terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 3.Cabelli R J, Chen L, Tai P C, Oliver D B. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 4.Cabelli R J, Dolan K M, Qian L P, Oliver D B. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J Biol Chem. 1991;266:24420–24427. [PubMed] [Google Scholar]
- 5.Chen L, Tai P C. ATP is essential for protein translocation into Escherichia coli membrane vesicles. Proc Natl Acad Sci USA. 1985;82:4384–4388. doi: 10.1073/pnas.82.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L L, Tai P C. Evidence for the involvement of ATP in co-translational protein translocation. Nature. 1987;328:164–166. doi: 10.1038/328164a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Xu H, Tai P C. A significant fraction of functional SecA is permanently embedded in the membrane. SecA cycling on and off the membrane is not essential during protein translocation. J Biol Chem. 1996;271:29698–29706. doi: 10.1074/jbc.271.47.29698. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham K, Lill R, Crooke E, Rice M, Moore K, Wickner W, Oliver D. SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Blaauwen T, Fekkes P, de Wit J G, Kuiper W, Driessen A J. Domain interactions of the peripheral preprotein translocase subunit SecA. Biochemistry. 1996;35:11994–12004. doi: 10.1021/bi9605088. [DOI] [PubMed] [Google Scholar]
- 10.Driessen A J. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 11.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 12.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 13.Eichler J, Brunner J, Wickner W. The protease-protected 30 kDa domain of SecA is largely inaccessible to the membrane lipid phase. EMBO J. 1997;16:2188–2196. doi: 10.1093/emboj/16.9.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichler J, Wickner W. Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc Natl Acad Sci USA. 1997;94:5574–5581. doi: 10.1073/pnas.94.11.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilmore R, Blobel G. Translocation of secretory proteins across the microsomal membrane occurs through an environment accessible to aqueous perturbants. Cell. 1985;42:497–505. doi: 10.1016/0092-8674(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich D, Prehn S, Hartmann E, Kalies K U, Rapoport T A. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- 17.Hanein D, Matlack K E, Jungnickel B, Plath K, Kalies K U, Miller K R, Rapoport T A, Akey C W. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann E, Sommer T, Prehn S, Gorlich D, Jentsch S, Rapoport T A. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Cerretti D P, Nashimoto H, Nomura M. Characterization of an amber mutation in the structural gene for ribosomal protein L15, which impairs the expression of the protein export gene, secY, in Escherichia coli. EMBO J. 1984;3:2319–2324. doi: 10.1002/j.1460-2075.1984.tb02133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joly J C, Wickner W. The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J. 1993;12:255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki H, Matsuyama S, Sasaki S, Akita M, Mizushima S. SecA protein is directly involved in protein secretion in Escherichia coli. FEBS Lett. 1989;242:431–434. doi: 10.1016/0014-5793(89)80516-2. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y J, Rajapandi T, Oliver D. SecA protein is exposed to the periplasmic surface of the E. coli inner membrane in its active state. Cell. 1994;78:845–853. doi: 10.1016/s0092-8674(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 23.Kimura E, Akita M, Matsuyama S, Mizushima S. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli. J Biol Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- 24.Lill R, Cunningham K, Brundage L A, Ito K, Oliver D, Wickner W. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 26.Oliver D B, Beckwith J. Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli. J Bacteriol. 1982;150:686–691. doi: 10.1128/jb.150.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price A, Economou A, Duong F, Wickner W. Separable ATPase and membrane insertion domains of the SecA subunit of preprotein translocase. J Biol Chem. 1996;271:31580–31584. doi: 10.1074/jbc.271.49.31580. [DOI] [PubMed] [Google Scholar]
- 28.Rajapandi T, Oliver D. Integration of SecA protein into the Escherichia coli inner membrane is regulated by its amino-terminal ATP-binding domain. Mol Microbiol. 1996;20:43–51. doi: 10.1111/j.1365-2958.1996.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 29.Ramamurthy V, Oliver D. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J Biol Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 30.Schatz P J, Bieker K L, Ottemann K M, Silhavy T J, Beckwith J. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 1991;10:1749–1757. doi: 10.1002/j.1460-2075.1991.tb07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt M G, Rollo E E, Grodberg J, Oliver D B. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J Bacteriol. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinkai A, Mei L H, Tokuda H, Mizushima S. The conformation of SecA, as revealed by its protease sensitivity, is altered upon interaction with ATP, presecretory proteins, everted membrane vesicles, and phospholipids. J Biol Chem. 1991;266:5827–5833. [PubMed] [Google Scholar]
- 33.Simon S M, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- 34.Simon S M, Blobel G. Signal peptides open protein-conducting channels in E. coli. Cell. 1992;69:677–684. doi: 10.1016/0092-8674(92)90231-z. [DOI] [PubMed] [Google Scholar]
- 35.Snyders S, Ramamurthy V, Oliver D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J Biol Chem. 1997;272:11302–11306. doi: 10.1074/jbc.272.17.11302. [DOI] [PubMed] [Google Scholar]
- 36.Tai P C, Tian G, Xu H, Lian J P, Yu J N. In vitro protein translocation into Escherichia coli inverted membrane vesicles. Methods Cell Biol. 1991;34:167–187. [PubMed] [Google Scholar]
- 37.van der Does C, den Blaauwen T, de Wit J G, Manting E H, Groot N A, Fekkes P, Driessen A J. SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxyl terminus to the periplasm. Mol Microbiol. 1996;22:619–629. doi: 10.1046/j.1365-2958.1996.d01-1712.x. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe M, Blobel G. SecA protein is required for translocation of a model precursor protein into inverted vesicles of Escherichia coli plasma membrane. Proc Natl Acad Sci USA. 1993;90:9011–9015. doi: 10.1073/pnas.90.19.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickner W, Leonard M R. Escherichia coli preprotein translocase. J Biol Chem. 1996;271:29514–29516. doi: 10.1074/jbc.271.47.29514. [DOI] [PubMed] [Google Scholar]
- 40.Yang Y B, Yu N, Tai P C. SecE-depleted membranes of Escherichia coli are active. SecE is not obligatorily required for the in vitro translocation of certain protein precursors. J Biol Chem. 1997;272:13660–13665. doi: 10.1074/jbc.272.21.13660. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y B, Lian J, Tai P C. Differential translocation of protein precursors across SecY-deficient membranes of Escherichia coli: SecY is not obligatorily required for translocation of certain secretory proteins in vitro. J Bacteriol. 1997;179:7386–7393. doi: 10.1128/jb.179.23.7386-7393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]