Abstract
In budding yeast, the transcriptional repressor Opi1 regulates phospholipid biosynthesis by repressing expression of genes containing inositol-sensitive upstream activation sequences. Upon genotoxic stress, cells activate the DNA damage response to coordinate a complex network of signaling pathways aimed at preserving genomic integrity. Here, we reveal that Opi1 is important to modulate transcription in response to genotoxic stress. We find that cells lacking Opi1 exhibit hypersensitivity to genotoxins, along with a delayed G1-to-S-phase transition and decreased gamma-H2A levels. Transcriptome analysis using RNA sequencing reveals that Opi1 plays a central role in modulating essential biological processes during methyl methanesulfonate (MMS)–associated stress, including repression of phospholipid biosynthesis and transduction of mating signaling. Moreover, Opi1 induces sulfate assimilation and amino acid metabolic processes, such as arginine and histidine biosynthesis and glycine catabolism. Furthermore, we observe increased mitochondrial DNA instability in opi1Δ cells upon MMS treatment. Notably, we show that constitutive activation of the transcription factor Ino2-Ino4 is responsible for genotoxin sensitivity in Opi1-deficient cells, and the production of inositol pyrophosphates by Kcs1 counteracts Opi1 function specifically during MMS-induced stress. Overall, our findings highlight Opi1 as a critical sensor of genotoxic stress in budding yeast, orchestrating gene expression to facilitate appropriate stress responses.
Keywords: DNA damage response, genotoxic stress, methyl methanesulfonate, Opi1, inositol pyrophosphates, mitochondrial function, transcriptional regulation
Introduction
The DNA molecule is constantly exposed to endogenous and exogenous genotoxins, leaving it subjected to a multitude of lesions that, if left unchecked, can lead to genomic instability, a hallmark of cancer and other diseases (Jackson and Bartek 2009). Methyl methanesulfonate (MMS) is a widely used genotoxic agent that induces DNA damage by alkylating DNA bases, resulting in the formation of DNA adducts. The most prevalent adducts formed by MMS are N7-methylguanine (N7-MeG) and N3-methyladenine (N3-MeA) (Beranek 1990), while N1-methyladenine (N1-MeA) and N3-methylcytosine (N3-MeC) can also be generated during DNA replication (Wyatt and Pittman 2006). These DNA adducts can interfere with DNA synthesis and replication by inducing replication fork stalling and subsequent DNA strand breaks (Branzei and Foiani 2010). Additionally, MMS has been shown to cause damage to mitochondrial DNA (mtDNA), which can disrupt oxidative phosphorylation and contribute to increased generation of reactive oxygen species (ROS) (Salmon et al. 2004; Kitanovic and Wölfl 2006; Kitanovic et al. 2009). Under such circumstances, a comprehensive DNA damage response (DDR) is activated to tackle the consequences of DNA lesion accumulation, such as through DNA repair, telomere maintenance, chromatin remodeling, inhibition of DNA synthesis and cell cycle progression, and transcription reprogramming, among others (Hanawalt 2015; Lanz et al. 2019; Cussiol et al. 2020).
Although the DDR has been extensively studied at the molecular level, recent studies have proposed that the DDR is connected with metabolism of biomolecules such as carbohydrates and lipids (Simpson-Lavy et al. 2015; Ferrari et al. 2017; Yi et al. 2017). Remarkably, the existence of crosstalk between the DDR and phospholipid metabolism in eukaryotes was proposed several years ago (Zewail et al. 2003). Supporting this notion, numerous proteins involved in the phosphatidylinositol (PI) pathway were found to undergo phosphorylation in response to DNA damage induced by MMS (Zhou et al. 2016; Lanz et al. 2021). Inositol metabolites such as inositol polyphosphates (IPs) and inositol pyrophosphates (PP-IPs) have been implicated in cell cycle regulation and DNA damage repair (Banfic et al. 2013, 2016; Jadav et al. 2013), but the molecular basis for these effects is unknown. Furthermore, it was proposed that PI phosphate lipids (PIPs) are enriched in the nucleus after DNA damage, serving as important mediators of ATR signaling in mammalian cells (Wang et al. 2017). Nonetheless, little is known about how DNA damage can regulate inositol metabolism and vice versa.
In budding yeast, inositol biosynthesis is fine-tuned by the transcriptional repressor Opi1. In the absence of inositol, Opi1 is localized to the perinuclear endoplasmic reticulum (ER) membrane, establishing interactions with the integral membrane protein Scs2 and the phospholipid precursor phosphatidic acid (PA) (Craven and Petes 2001; Brickner and Walter 2004; Gaspar et al. 2017; Hofbauer et al. 2018). As a result, the heterodimeric transcriptional activator Ino2-Ino4 binds to cis-acting inositol-sensitive upstream activation sequences (UASINO) and upregulates expression of several genes related to phospholipid metabolism including the inositol-3-phosphate synthase (INO1), which promotes inositol de novo synthesis. Full repression of genes containing UASINO is also dependent on choline, leading to the designation of this sequence as inositol/choline-responsive elements (ICRE) (Schüller et al. 1992). Once intracellular inositol concentration increases, PA is redirected to the synthesis of PI leading to its exhaustion. Subsequently, Opi1 migrates to the nucleus where it binds to the heterodimeric transcriptional factor Ino2-Ino4 and promotes transcriptional repression through the interaction with the proteins Sin3 and Cyc8, creating a scaffold for the recruitment of histone deacetylase (HDAC) complexes (Wagner et al. 2001; Jäschke et al. 2011; Kliewe et al. 2017) (Fig. 1a). Importantly, deletion of Opi1 leads to constitutive expression of INO1 with overproduction of inositol even when cells are supplemented with inositol (Graves and Henry 2000). Moreover, cells lacking Opi1 show constitutive activation of a large number of genes, most of which are regulated by Ino2-Ino4 (Santiago and Ben Mamoun 2003; Hoppen et al. 2005; Jesch et al. 2005). Many of these genes are involved in phospholipid biosynthesis, although UASINO motifs are found in several genes related to other metabolic processes, which implicates Ino2-Ino4 in the control of the expression of distinct biological processes (Wimalarathna et al. 2011). Interestingly, phosphoproteomic analysis in budding yeast showed that the transcriptional repressor Opi1 is phosphorylated in a Mec1/Tel1-dependent manner after MMS exposure (Balint et al. 2015; Bastos de Oliveira et al. 2015; Lanz et al. 2021), which might suggest that Opi1 is involved in the DDR.
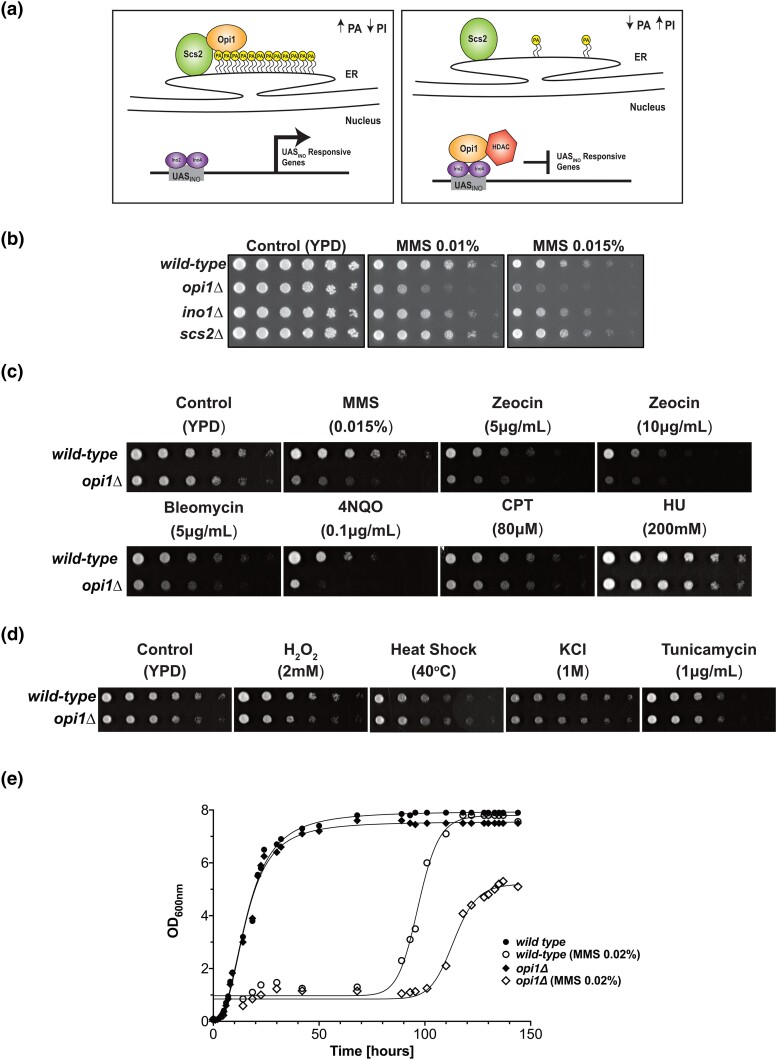
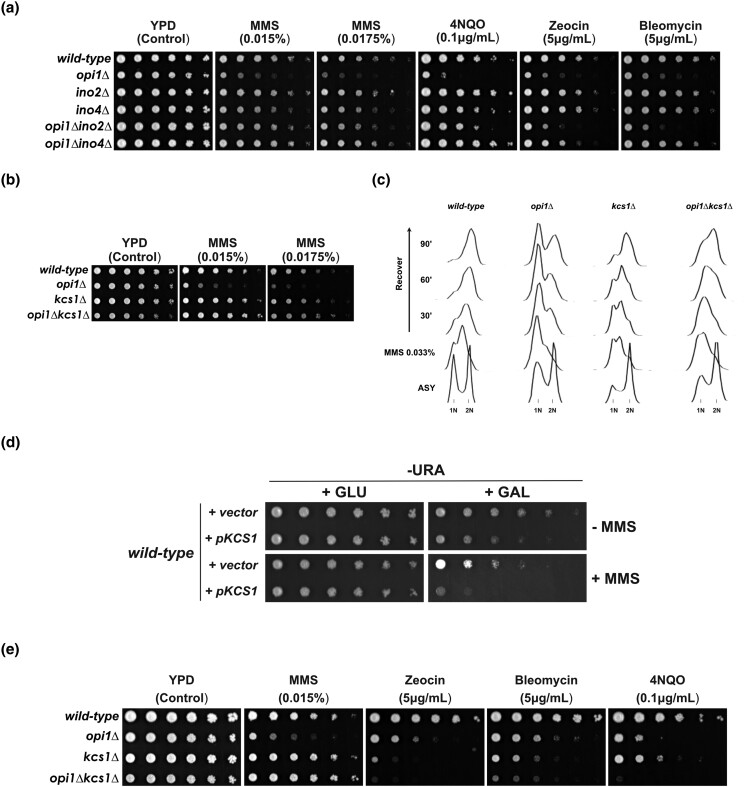
Fig. 1.
Cellular response of Opi1-deficient cells to genotoxic and proteotoxic stress. a) Working model for how Opi1 represses transcription of target genes. b) Effect of deletion of genes involved in inositol metabolism in sensitivity to genotoxins. c) Cells lacking Opi1 show sensitivity to different genotoxins. d) Cells lacking Opi1 do not show sensitivity to proteotoxic stress agents. e) Growth rate of wild-type and opi1Δ cells determined by monitoring OD600 over time. The data points (geometrical symbols) were fitted to a nonlinear regression curve using GraphPad Prism version 9.0.0 to analyze the growth kinetics. For b), c), and d), 4-fold serial dilutions were spotted on the media indicated in the figure, and plates were grown for 2–3 days at 30°C.
In this study, we employ an integrative approach combining RNA sequencing (RNA-seq) transcriptome analysis, yeast genetics, molecular biology, cell biology, and biochemistry techniques to investigate the role of the transcriptional repressor Opi1 in the response to genotoxic stress. Our findings demonstrate that under MMS-induced stress, cells lacking Opi1 exhibit enhanced sensitivity and cell cycle defects characterized by a delayed G1-to-S-phase transition, resulting in reduced histone H2A phosphorylation. Notably, RNA-seq analysis revealed that during MMS-induced genotoxic stress, Opi1 is important to modulate expression of genes involved in important biological processes. Additionally, our study reveals that treatment with MMS leads to an increase in mtDNA instability in opi1Δ cells. This observation suggests a potential link between mtDNA instability and the genotoxin sensitivity exhibited by cells lacking Opi1. Importantly, deletion of the transcriptional activator Ino2-Ino4 rescues the MMS sensitivity of opi1Δ cells showing that constitutive activation of Ino2-Ino4 is the cause of MMS sensitivity. Finally, while deletion of the PP-IP kinaseKcs1 in an opi1Δ strain rescues MMS sensitivity and cell cycle defects, overexpression of Kcs1 in wild-type cells phenocopies the MMS sensitivity of an opi1Δ strain. These findings suggest that PP-IPs may counteract the function of Opi1 during MMS-induced stress in yeast.
Collectively, these results highlight the crucial role of Opi1 as a key sensor of genotoxic stress and emphasize the significance of modulating PP-IP synthesis as an integral component of the DDR and a critical factor for proper cell function in budding yeast.
Materials and methods
Yeast strains and plasmids
Strains generated in this study are isogenic from BY4741 and S288C (where indicated). Detailed information on all yeast strains and plasmids used in this study can be found in Supplementary Tables 1 and 2, respectively. To generate knockout and epitope-tagged strains, the 1-step gene disruption method, as described by Rothstein (Rothstein 1983, 1991; Longtine et al. 1998), was employed. All yeast transformations were performed using the lithium acetate method (Gietz et al. 1992; Gietz and Woods 2006). PCR genotyping using specific primers (Supplementary Table 3) was conducted to confirm the successful generation of knockout strains, while Western blotting was performed to verify the presence of epitope tags in the respective strains.
Growth conditions
Cells were grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose), and plasmid-bearing cells were grown in synthetic complete media lacking uracil (SC-URA) (0.17% YNB without amino acids, Difco), 0.5% ammonium sulfate (MP Biomedicals), 0.07% CSM-URA dropout mix (Sunrise Science), and 2% dextrose (Sigma). For experiments performed in the absence of inositol, SC − INO medium was prepared using YNB without inositol (MP Biomedicals). For expression of INO1 and KCS1 under the control of the GAL1 promoter, cells were grown overnight in SC-URA medium with 2% dextrose and were plated in SC-URA plates in the presence of 2% dextrose (Sigma) or 2% galactose (Difco). For galactose induction in liquid cultures, the strains were initially grown in SC-URA medium supplemented with lactate (pH 5.5) as described by the Haber lab (Haber and Leung 1996). Once the cell density reached an optical density (OD600nm) of 1.0, galactose was introduced into the culture to achieve a final concentration of 2%. To evaluate growth under respiratory conditions, cells were cultured in either YPGal (1% yeast extract, 2% peptone, and 2% galactose) or YPG medium (1% yeast extract, 2% peptone, and 3% glycerol). For the detailed concentrations and durations of specific chemical agents, including genotoxins and other drugs, please refer to the respective figure legends associated with the experimental data.
Serial dilution assays
Overnight cultures were inoculated in the appropriate medium to an OD600nm of 0.1, and cells were grown until they reached the logarithmic growth phase and were subsequently normalized to an OD600nm of 1.0. Four-fold serial dilutions were spotted on yeast plates and grown for 1–3 days at 30°C in the presence or absence of specific chemical agents, including genotoxins and other drugs. The specific concentrations of these agents used in the assay are provided in the corresponding figure and/or figure legends, serving as a clear reference for the experimental setup. Following the incubation period, the plates were manually inspected for growth changes and digitalized in an Uvitec Alliance 4.7 imaging system.
Western blotting
Proteins were transferred onto PVDF membranes (Amersham Hybond P, GE Healthcare). Phosphorylated histone H2A (gamma-H2A), total histone H2A, Rad53, and epitope-tagged proteins were probed using specific antibodies: anti-gamma-H2A (ab17353, Abcam, 1:2,500 dilution in TBST and 5% nonfat dry milk), anti-histone H2A (ab188312, Abcam, 1:500 dilution in TBST and 5% nonfat dry milk), anti-HA (12CA5, Roche, 1:10,000 in TBST and 2% nonfat dry milk), and anti-Rad53 (ab104232, Abcam, 1:3,000 dilution in TBST and 5% nonfat dry milk). Anti-PGK1 was used as a housekeeping control (22C5D8, Abcam, 1:15,000 dilution in TBST and 2% nonfat dry milk). After primary antibodies, PVDF membranes were incubated in the presence of ECL HRP-linked secondary antibodies (mouse: NA931-GE or rabbit: NA934-GE, 1:10,000 dilution in TBST and 2% nonfat dry milk). Blots were developed using the Amersham ECL Prime detection reagent and imaged in an Uvitec Alliance 4.7 imaging system.
Cell cycle synchronization
Yeast cells were synchronized in the G1 phase as described before (Jablonowski et al. 2015). In brief, cells were initially grown in YPD medium at 30°C until they reached the logarithmic growth phase. Subsequently, α-factor (Zymo Research) treatment was applied at a concentration of 30 ng/mL (for bar1Δ background strains) for 2 h. To release the cells from G1 arrest, they were centrifuged and resuspended in fresh medium. In order to assess the impact of MMS-induced genotoxic stress on S-phase progression, MMS was added at a concentration of 0.033% during the release step. To induce an intra-S-phase arrest of the cell cycle, cells were exposed to MMS at a concentration of 0.033% for 2 h, which is sufficient to activate the intra-S-phase checkpoint (Shirahige et al. 1998). For recovery of cells following MMS treatment, cells were exposed to MMS at a concentration of 0.033% for 2 h, harvested, and subsequently resuspended in fresh YPD medium.
Flow cytometry
Cell cycle analysis was performed as described before (Jablonowski et al. 2015). In brief, logarithmic growing yeast cells were collected, harvested, fixed in 1 mL of 70% ethanol (Sigma-Aldrich), and incubated overnight at 4°C. Cells were then centrifuged, and residual ethanol was dried in SpeedVac. After that, samples were solubilized in sodium citrate buffer (Labsynth) (50 mM, pH 7.2) and sonicated (3 cycles of 3 s, amplitude 30%) to release cell clumps. Samples were then incubated with 100 μg of RNAse A (Invitrogen) for 1 h at 37°C, followed by incubation with 500 μg of Proteinase K (Invitrogen) for 1 h at 42°C. Then, 1 μL of SYTOX Green (Thermo Fisher Scientific) was added to the samples and incubated for 2 h at 4°C protected from the light. Data were acquired using a BD Accuri C6 Flow Cytometer. For viability assessment, wild-type and opi1Δ cells were cultured in YPD liquid medium until the logarithmic phase of growth and treated with 0.1% MMS for 4 h. After treatment, the samples were normalized to a concentration of 1 × 106 cells/mL, harvested, and resuspended in 1 mL of 50 mM sodium citrate. Next, 0.4 μM propidium iodide (Thermo Fisher Scientific) was added, and the cells were immediately analyzed using a BD Accuri C6 Flow Cytometer. To distinguish between dead and live cells, we used boiled cells (100°C for 10 min) as a control for dead cells and untreated cells to obtain a population of live cells. The data were obtained from 3 biological replicates, and multiple t-tests were used to determine statistical significance, comparing the mean viability values of the strains with the significance set at α = 0.05.
Real-time quantitative PCR analysis
Cells were grown overnight in YPD and diluted the next day to an OD600nm of 0.1 in synthetic complete medium either supplemented or lacking inositol (SC + INO or SC − INO). Cells were grown for at least 2 cell cycle divisions and immediately harvested at 4°C, washed with sterile ultrapure H2O, and kept at −80°C. Alternatively, log-phase cells were treated with MMS (Sigma-Aldrich) 0.1% for 1 h. Total RNA from 5 × 107 cells was extracted using an RNeasy Mini Kit (Qiagen). One microgram of total RNA was used for cDNA synthesis using a QuantiNova Reverse Transcription Kit (Qiagen). Real-time PCR was performed in an ABI Prism 7500 (Applied Biosystems) using a QuantiNova Probe PCR Kit (Qiagen) in the presence of TaqMan probes for INO1 (Sc04136910_s1) and ACT1 (Sc04120488_s1) (Thermo Fisher Scientific). The ACT1 gene served as an internal standard for normalization as it was previously shown that there is no change in mRNA levels after either inositol depletion (Ye et al. 2013; Gaspar et al. 2017) or MMS treatment (Gasch et al. 2001). Relative values of mRNA are described as fold change relative to a control condition indicated in the experiment. The data were obtained from 3 biological replicates, and statistical analysis was performed using 1-way analysis of variance (ANOVA) with Tukey's post hoc test to determine the significant differences between the groups.
RNA-seq sample preparation and analysis
Cells were grown overnight in SC + INO media and diluted the next day to an OD600nm of 0.1 in 250 mL of SC + INO media in biological duplicates. Cells were grown for at least 2 cell cycle divisions (∼OD600nm = 0.4), then vacuum filtered in 0.22-µm nitrocellulose membranes to remove media and immediately “flash frozen” in liquid nitrogen. One milliliter of frozen 1× RIPA buffer (Millipore) was combined into each frozen pellet and lysed by mechanical grinding with a Spex 6750 Freezer Mill at a rate of 10 Hz for 2 min. The lysate was then thawed on ice and centrifuged at 2,000 g for 2 min at 4°C to remove debris; 250 µL of cleared lysate was used for RNA extraction by combining it with 250 µL of 2× AE Buffer (100 mM sodium acetate pH 5.2, 20 mM EDTA), 50 µL of 10% SDS, and 500 µL of acid phenol:chloroform pH 5.2. The mixture was incubated at 65°C for 10 min under 1,400 rpm agitation, then transferred to ice for 5 min. The solution was then spun at 15,000 g for 15 min at 4°C. The supernatant was then transferred to new microcentrifuge tubes, and 550 µL of chloroform was added, followed by centrifugation at 15,000 g for 5 min at 4°C. The aqueous phase was then transferred to new microcentrifuge tubes, and RNA was precipitated in 0.3 M sodium acetate and 400 µL of isopropanol after centrifugation at 15,000 g for 20 min at 4°C. RNA pellets were washed with 75% ethanol and centrifuged at 15,000 g for 5 min at 4°C. Finally, RNA pellets were resuspended in nuclease-free water and submitted to Bioanalyzer High Sensitivity DNA Assay (Agilent) for quality check and quantified using Qubit RNA High Sensitivity (Invitrogen). One microgram of total RNA was used as input for preparing the libraries for RNA-seq with Ultra RNA Library Prep II (New England Biolabs), following the recommended workflow. Final cDNA libraries after PCR were quantified by Qubit dsDNA High Sensitivity (Invitrogen). Samples were then equimolarly pooled and distributed on Illumina NextSeq High 500/550, resulting in ∼7–8 million reads per sample. Reads were demultiplexed using bcl2fastq (version 2.20), and quality of reads was assessed using FastQC (version 0.11.8). Reads were then aligned to the S288C reference genome (release R64-2-1) using STAR (version 2.5.4b) (Dobin et al. 2013). Differential gene expression was calculated with DESeq2 (version 1.40.0) (Love et al. 2014) using the design “∼genotype + condition + genotype: condition.” A threshold of 0.8 log2 fold change with an adjusted P-value of 0.05 was used to analyze differentially expressed genes (DEGs) between samples. A principal component analysis (PCA) plot for the 2 biological replicates across genotypes and treatments is shown in Supplementary Fig. 1. Supplementary Table 4 (“genotype_OPI1_vs_WT.csv”) shows the DEGs between the opi1Δ and wild-type strains in an untreated condition. Supplementary Table 5 (“treatment_MMS_vs_unt.csv”) shows the DEGs in the wild-type strain treated with MMS relative to an untreated condition. Supplementary Table 6 (“interaction.csv”) shows the differential effect of MMS treatment in the opi1Δ strain relative to the wild type (interaction term). Supplementary Table 7 (“treatment_MMS_vs_unt_OPI1.csv”) shows the DEGs in the opi1Δ strain treated with MMS relative to the opi1Δ without treatment. Supplementary Table 8 (“OPI1_vs_WT_treatment.csv”) shows the DEGs between the opi1Δ and wild-type strains both treated with MMS.
To identify enriched biological processes and cellular components among the list of DEGs, we utilized the PANTHER database (http://pantherdb.org) and the Saccharomyces Genome Database (SGD) with the following parameters: PANTHER version 17.0 overrepresentation test, FISHER test with false discovery rate (FDR) correction (Thomas et al. 2022), and Gene Ontology (GO) Slim term mapper (https://www.yeastgenome.org/goSlimMapper), respectively. Additionally, interaction networks of DEGs were generated using the STRING v11.5 database (http://string-db.org) (Szklarczyk et al. 2019).
Respiratory capacity assay
Oxygen consumption rates were measured using a modified protocol based on the method described by Zulkifli et al. (2020) using a high-resolution O2k-FluoRespirometer (Oroboros, Innsbruck, Austria). For each assay, 106 cells were added to the Oroboros chamber containing a final volume of 2.0 mL of YPEG medium at 30°C under agitation. Briefly, exponentially growing cells in YPD were harvested, resuspended in respiratory medium (YPEG; 2% glycerol and 1% ethanol), and incubated for at least 3 h at 30°C. Finally, cells were transferred to O2k chambers for measurement of O2 consumption rates. After basal respiration assessment, 5 μM of the mitochondrial uncoupler FCCP (Sigma-Aldrich) was added to measure maximal respiration. At the end, 2 μM of antimycin A (Sigma-Aldrich) was added to inhibit mitochondrial respiration. Nonmitochondrial oxygen consumption rates were subtracted from all measurements. Optimal concentrations of FCCP and antimycin A were previously determined by titration. The data were obtained from 2 biological replicates, and statistical analysis was performed using 1-way ANOVA with Tukey's post hoc test to determine the significant differences between the groups.
Results
Opi1 is important for cell survival during genotoxic stress
To determine whether cells lacking Opi1 are more sensitive to DNA damage, we performed sensitivity assays in the presence of genotoxins. We found that opi1Δ cells showed sensitivity to genotoxic stress induced by the DNA-alkylating agent MMS (Fig. 1b), indicating that Opi1 is important for the response to genotoxic stress. To confirm that this sensitivity to MMS was specific to OPI1 deletion, we constructed a pRS416 plasmid expressing Opi1-HA from its endogenous promoter and show that Opi1-HA expression rescues the MMS sensitivity in opi1Δ (Supplementary Fig. 2a and b, respectively). Furthermore, deletion of genes that counteract Opi1-repressing functions, such as INO1 and SCS2, did not lead to MMS sensitivity (Fig. 1b).
Besides MMS, we performed a screening of several known DNA-damaging agents to assess for genotoxicity in opi1Δ cells (Fig. 1c). Interestingly, we found that opi1Δ cells are sensitive to different genotoxins such as the radiomimetic drugs bleomycin and zeocin and 4-nitroquinoline 1-oxide (4NQO), while they did not show sensitivity to the replication stress inducers hydroxyurea (HU) and camptothecin (CPT). Importantly, cells lacking Opi1 did not show sensitivity to agents that activate the environmental stress response (ESR) and the unfolded protein response (UPR), indicating that Opi1 shows specificity to cope with increased DNA damage inflicted by genotoxins (Fig. 1d).
To determine if the increased sensitivity to genotoxins observed in cells lacking Opi1 is due to loss of viability rather than a decreased growth rate, we used colony-forming unit assay and flow cytometry to monitor cell survival (Supplementary Fig. 2c and d, respectively). Our results show that opi1Δ cells do not exhibit a significant decrease in cell viability compared to wild-type cells, indicating that the hypersensitivity to genotoxins is likely due to delayed cell proliferation rather than cell death. Consistent with that, opi1Δ cells show growth defects in YPD liquid medium in the presence of 0.02% MMS, suggesting an increased arrest of the cell cycle in opi1Δ cells (Fig. 1e).
Cells lacking Opi1 show delayed G1-to-S-phase progression and decreased levels of gamma-H2A
Since opi1Δ mutants are sensitive to genotoxic stress (Fig. 1c) and show a prolonged arrest of growth (Fig. 1e), we sought to investigate if these cells have defects in the DDR. To investigate the potential involvement of Opi1 in the regulation of the DDR, we subjected asynchronous yeast cells to genotoxic agents, such as MMS, zeocin, and 4NQO, and monitored the activation of Rad53, a pivotal kinase in the signal transduction pathway responsible for DDR regulation (Pellicioli and Foiani 2005; Branzei and Foiani 2006; Cussiol et al. 2020). Our findings revealed no significant differences in Rad53 activation between wild-type and opi1Δ cells in asynchronous conditions, suggesting that Opi1 may not play a critical role in the regulation of the Rad53 axis within the DDR (Supplementary Fig. 3).
Given the indications from our results that genotoxic stress leads to delayed cell proliferation in opi1Δ cells (Fig. 1e), we proceeded to investigate the role of Opi1 in the regulation of the cell cycle. To accomplish this, we utilized flow cytometry to monitor cell cycle progression. Initially, we arrested cells in the G1 phase using α-factor and subsequently released them into fresh YPD medium in the presence or absence of 0.033% MMS, a concentration known to reduce S-phase progression and induce a robust activation of the intra-S-phase checkpoint (Tercero and Diffley 2001). Our findings reveal that cells lacking Opi1 exhibit a noticeable delay in S-phase progression under both unchallenged conditions and in the presence of MMS, as depicted in Fig. 2a. Furthermore, it is noteworthy that cells lacking Opi1 consistently display a discernible G1 peak, indicating a G1/S arrest in the cell cycle after release from pheromone arrest. Interestingly, cells lacking Opi1 show a delayed phosphorylation of histone H2A (gamma-H2A) and a reduced intensity of the gamma-H2A signal at later time points (Fig. 2b). Importantly, a previous study has demonstrated that gamma-H2A is enriched in regions containing newly replicated DNA, particularly behind the replication forks at early-firing replication origins following MMS treatment (Balint et al. 2015). The observed decrease in gamma-H2A levels in cells lacking Opi1 may be attributed to a decrease in the firing of replication origins. Moreover, our results reveal a delayed activation of Rad53 in Opi1-deficient cells (Fig. 2c), which aligns with the experimental observations regarding gamma-H2A (Fig. 2b). Specifically, we observed the appearance of a phosphorylated isoform of Rad53 at 30 min of MMS treatment in wild-type cells, while this band was not detected at this time point in opi1Δ cells (Fig. 2c).
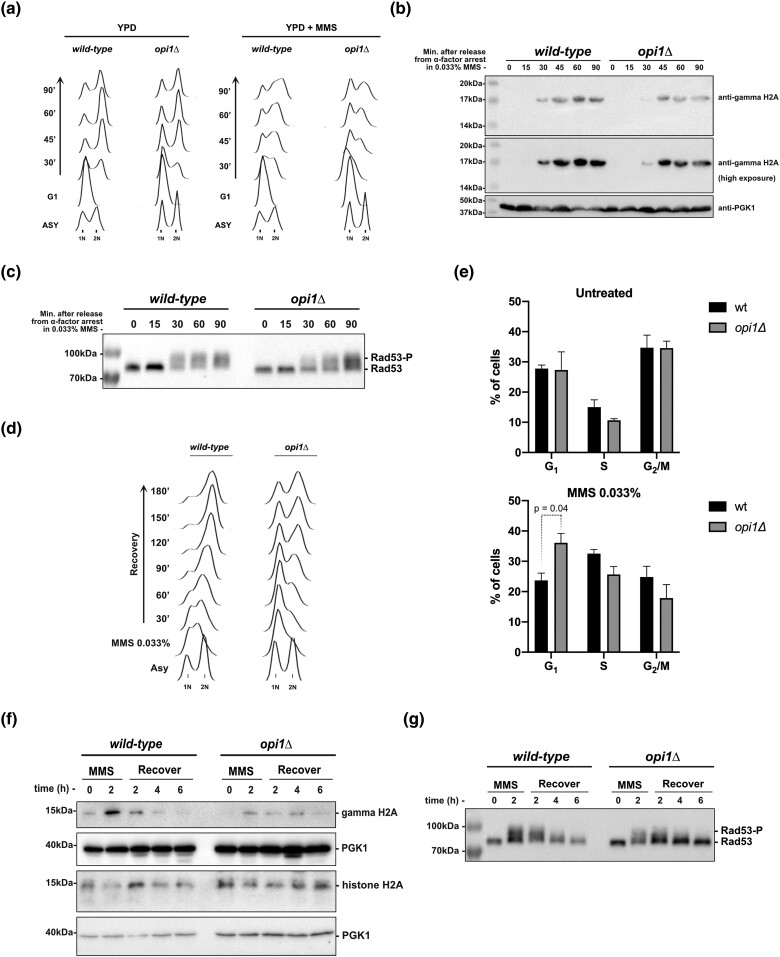
Fig. 2.
Cells lacking Opi1 show a delayed G1-to-S-phase transition and activation of the DDR upon genotoxic stress induced by MMS. a) Flow cytometry analysis depicting S-phase progression in wild-type and opi1Δ strains. b) Western blot analysis of gamma-H2A (histone H2A phosphorylation) in the indicated strains. c) Western blot analysis showing the MMS-induced Rad53 phospho-shift in the indicated strains. d) Flow cytometry analysis illustrating the resumption of S-phase progression in wild-type and opi1Δ strains after MMS treatment (0.033% for 2 h). e) Quantification of the percentage of cells in the G1, S, and G2/M phases in untreated cells and after MMS treatment (0.033% for 2 h). The results shown are representative of 3 independent experiments, and error bars indicate the standard deviation. Two-way ANOVA with Tukey's post hoc testwas used for significance analysis between samples. f) Western blot analysis of gamma-H2A and total histone H2A in the indicated strains. g) Western blot analysis showing the MMS-induced Rad53 phospho-shift in the indicated strains. Note: For a) to c), cells were arrested in G1 after 2-h treatment with α-factor and then released in fresh YPD medium ± 0.033% MMS. All strains are bar1Δ. For d) to g), cells were treated with 0.033% MMS for 2 h and then released in fresh YPD medium. Western blots were probed with anti-gamma-H2A, anti-histone H2A, anti-Rad53, and anti-PGK1 (loading control) antibodies as described in Materials and methods.
Next, we assessed the ability of cells to recover from genotoxic stress induced by MMS. Exponentially growing cells were treated with 0.033% MMS for 2 h to induce an intra-S-phase arrest (Gasch et al. 2001). Afterward, cells were released into fresh medium, and cell cycle progression and DDR deactivation were monitored. We observed that MMS-treated opi1Δ cells had a significantly increased population of G1 cells, indicating a persistent G1/S arrest of the cell cycle (Fig. 2d and e). In contrast, wild-type cells displayed an intra-S phase arrest, which is consistent with the replication stress induced by MMS (Fig. 2d). It is noteworthy that, consistent with the result from Fig. 2b, gamma-H2A levels were significantly lower in opi1Δ cells relative to wild-type cells after 2 h of MMS treatment. However, no significant difference was observed in the disappearance of gamma-H2A during the recovery period (Fig. 2f). Notably, the decrease in gamma-H2A levels observed in opi1Δ cells was not due to histone depletion since total histone H2A levels were not decreased in opi1Δ cells after genotoxic stress (Fig. 2f). Also, we found that both wild-type and opi1Δ cells exhibited comparable levels of Rad53 activation after 2 h of MMS treatment, and there was no difference in Rad53 deactivation during recovery (Fig. 2g). Our results suggest that DNA damage signaling downregulation is not altered in opi1Δ cells but indicate that MMS treatment induces cell cycle defects related to delayed G1-to-S-phase progression.
Opi1 migrates to the nucleus upon genotoxic stress induced by MMS
Knowing that transcriptional repression function of Opi1 is dependent on its cellular localization (Fig. 1a) and because its absence causes genotoxin sensitivity and cell cycle defects (Figs. 1c and 2, respectively), we investigated Opi1 localization after treatment with 0.1% MMS since this concentration was previously used to monitor global changes in gene expression (Jelinsky and Samson 1999). Using a strain expressing Opi1-GFP, we recapitulated Opi1 cellular localization during inositol metabolism (Fig. 3a panels I and II). MMS treatment of cells supplemented with 100 μM of inositol (+INO), a concentration known to sufficiently repress INO1 (Hirsch and Henry 1986), did not cause any significant alteration in Opi1-GFP intensity or localization (data not shown). Surprisingly, MMS treatment of cells kept without inositol supplementation (−INO) induced a rapid translocation of Opi1-GFP to the nucleus (Fig. 3a and b and Supplementary Fig. 4). Since Opi1 mediates gene repression when it translocates to the nucleus, this observation might suggest that the nuclear localization of Opi1 is important to mediate MMS-induced stress resistance. Importantly, we found that yeast cells treated with MMS in SC − INO remained viable (Fig. 3c), which implies that Opi1 does not fully repress INO1 in these conditions; otherwise, cells would die.
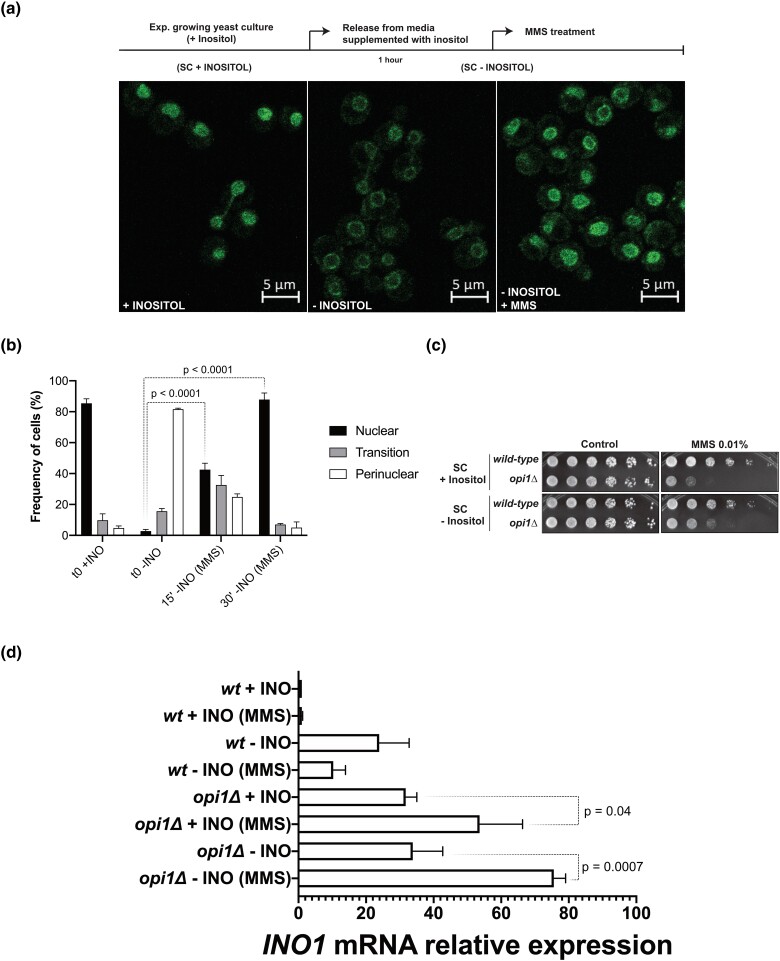
Fig. 3.
Opi1 translocates to the nucleus after MMS treatment and is required to repress INO1 expression during genotoxic stress. a) and b) Effect of inositol availability and MMS treatment on Opi1-GFP localization. Cellular localization of Opi1-GFP was scored manually, and the means and standard deviations from 2 independent experiments are shown. c) Cells treated with MMS in medium lacking inositol are still viable. d) Effect of inositol availability and MMS treatment on INO1 expression by RT-qPCR. Relative values of mRNA are described as fold change relative to a control condition (wt + INO). For c), 4-fold serial dilutions were spotted on the media indicated in the figure, and plates were grown for 2–3 days at 30°C. For d), experiments were made with 3 biological replicates, and statistical significance of the differences observed was calculated using 1-way ANOVA with Tukey's multiple comparison test.
Subsequently, to understand if Opi1 migration to the nucleus in the presence of MMS causes gene repression, we looked at INO1 expression during MMS treatment by real-time quantitative PCR (RT-qPCR) analysis (Fig. 3d). While MMS treatment does not change INO1 expression in wild-type cells supplemented with inositol (+INO), there is a substantial decrease in INO1 expression in the absence of inositol (−INO). Strikingly, INO1 expression significantly increases in an opi1Δ strain upon MMS treatment in both +INO and −INO conditions (Fig. 3d). These results show that under genotoxic stress, Opi1 migrates to the nucleus to repress INO1 expression in order to downregulate inositol synthesis. Because Opi1 represses genes containing UASINO, it is possible that in its absence, several other genes are also upregulated upon MMS treatment.
Opi1 modulates gene expression during genotoxic stress induced by MMS
In order to gain a deeper understanding of the impact of Opi1-mediated gene expression regulation in response to MMS treatment on a transcriptome-wide scale, we conducted RNA-seq experiments using exponentially growing cells (wild-type vs opi1Δ) supplemented with 100 μM inositol (SC + INO) in the presence of ±0.1% MMS for 60 min (Fig. 4a). We selected this specific experimental condition based on 2 primary reasons. First, our experiments have demonstrated the heightened sensitivity of Opi1-deficient cells to MMS when grown in medium supplemented with inositol (Fig. 3c). Second, our own observation revealed that MMS-induced upregulation of INO1 expression in opi1Δ cells is independent of inositol supplementation (Fig. 3d).
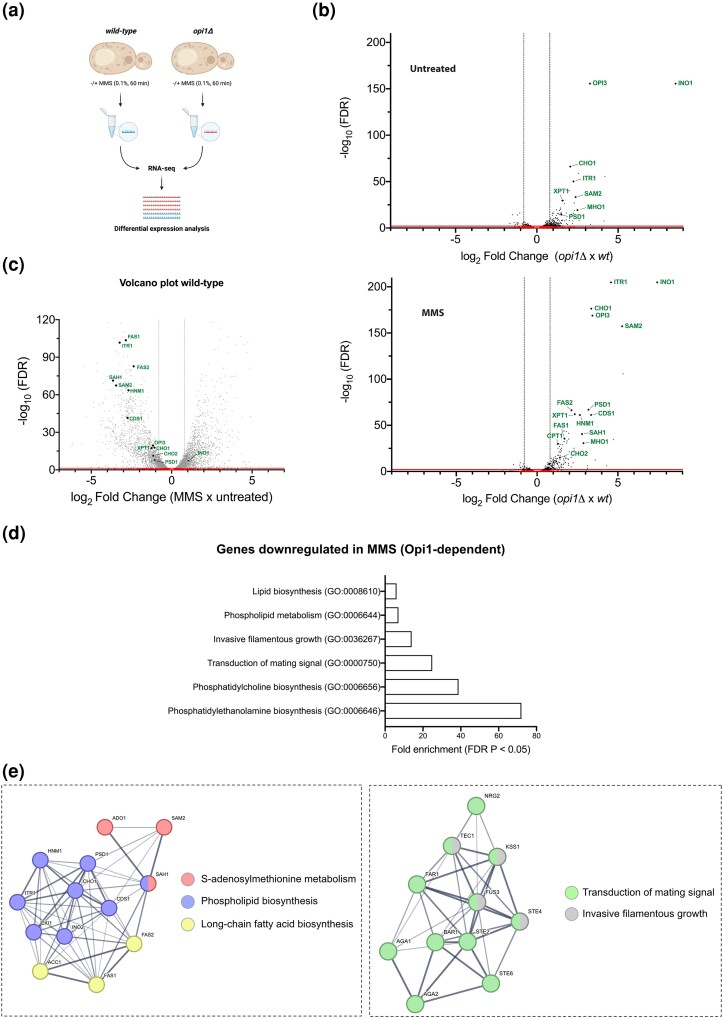
Fig. 4.
Opi1 is important to modulate gene expression during MMS-induced genotoxic stress. a) Schematic representation of RNA-seq performed in wild-type and opi1Δ cells cultured in minimal medium (SC + INO) with or without 0.1% MMS treatment for 60 min. Created with BioRender.com. b) Volcano plot illustrating the differential gene expression between opi1Δ and wild-type cells (top panel) and between opi1Δ + MMS and wild-type + MMS cells (bottom panel). c) Volcano plot displaying the differential gene expression between wild-type + MMS and wild-type cells. For b) and c), genes with significant differential expression are plotted based on a threshold of ±0.8 log2 fold change and an adjusted P-value of 0.05. The red line (x-axis) and dotted line (y-axis) represent the significant cutoff. Known genes regulated by Opi1 are labeled in both plots. d) Bar plot illustrating the top enriched biological processes identified in the gene set that are repressed during MMS treatment in an Opi1-dependent manner (Supplementary Table 6). Each bar represents a specific biological process, and the height of the bar corresponds to the fold enrichment with a P-adjusted value of 0.05. GO analysis was conducted in PANTHER using the overrepresentation test with S. cerevisiae serving as the reference list, which includes all genes in the database. The test type used was FISHER, and the FDR correction was applied. e) GO term enrichment network depicting the relationships between genes grouped in clusters that are repressed during MMS treatment in an Opi1-dependent manner (Supplementary Table 6). The enrichment analysis was performed using the STRING database, which integrates various sources of functional information with a confidence score of >0.4 (medium confidence). Nodes in the network represent genes, while edges indicate the connections between them. The color of the nodes represents a biological process, and the thickness of the edges represents the increased confidence of the interaction between nodes.
Previous studies utilizing transcriptome analysis have characterized genes whose expression is modulated by Opi1 (Santiago and Ben Mamoun 2003; Jesch et al. 2005). As anticipated, the results of our differential expression analysis revealed the upregulation of several canonical Opi1 targets in opi1Δ cells (Fig. 4b top panel and Supplementary Table 4). Interestingly, our results demonstrated that the expression of several Opi1 canonical targets was further augmented following MMS treatment in opi1Δ cells (Supplementary Table 8 and Fig. 4b, compare top and bottom panels), indicating the crucial role of Opi1 in modulating gene expression during such conditions. Moreover, to reinforce this hypothesis, we observed that genes highlighted in Fig. 4b were repressed in wild-type cells exposed to MMS (Fig. 4c and Supplementary Table 5). This finding further supports the notion that Opi1 plays a critical role in regulating gene expression in response to MMS treatment and that its absence leads to misregulation of several genes, ultimately resulting in MMS sensitivity. Curiously, contrary to the observed trend in INO1 expression determined by RT-qPCR (Fig. 3d), INO1 expression did not follow the same pattern as other Opi1 targets, as MMS treatment did not increase INO1 expression in opi1Δ cells (Fig. 4b, compare top and bottom panels). The observed discrepancy in INO1 expression between the RT-qPCR and RNA-seq analyses could stem from various factors, including technical variability inherent to the methods, biological variability between samples, and differences in the sensitivity and dynamic range.
To clarify the role of Opi1 during MMS treatment, we sought to identify genes exhibiting differential expression in an Opi1-dependent manner (Supplementary Table 6). We then selectively screened genes showing a ±0.8 log2 fold change with an adjusted P-value of ≤0.05 and conducted a statistical overrepresentation analysis using the PANTHER database (http://pantherdb.org) to investigate biological processes that were enriched within our gene list. With that, we were able to see which biological processes are misregulated upon MMS treatment in an Opi1-dependent manner. Notably, we observe a significant enrichment of phospholipid biosynthesis genes within our dataset (Fig. 4d). Moreover, we observe that Opi1 is pivotal in repressing the expression of proteins associated with MAPK signaling such as cellular conjugation, transduction of mating signaling (pheromone response), and filamentous growth (Fig. 4d), thereby indicating its significance in modulating these biological processes during genotoxic stress. In addition, we generated an interactome of the DEGs, which aligns in clusters using the STRING database (http://string-db.org) (Fig. 4e). Importantly, the upregulation of genes involved in the mating signaling pathway is associated with the inhibition of the G1-to-S-phase transition. This finding is particularly relevant since we observed that cells lacking Opi1 exhibit a delayed transition from the G1 phase to the S phase under genotoxic stress induced by MMS (Fig. 2a, d, and e), which can be correlated with the observed delay in cell growth in both solid and liquid cultures (Fig. 1c and e, respectively).
Finally, our findings challenge the canonical role of Opi1 as a gene repressor by also demonstrating its involvement in the upregulation of several genes related to amino acid biosynthesis, including arginine and histidine biosynthesis, the mitochondrial glycine cleavage system, and sulfate assimilation (Supplementary Fig. 5a and b). However, it is important to emphasize that we cannot exclude the possibility that Opi1 indirectly regulates many of these targets by influencing the expression of genes involved in transcriptional regulation.
The constitutive activation of Ino2-Ino4-responsive genes is the cause of MMS sensitivity in cells lacking Opi1
A previous study by Heyken et al. (2005) provided evidence for a physical interaction between Opi1 and a functional domain within the transcriptional factor Ino2. This interaction was shown to result in the repression of Ino2's ability to promote gene expression. Moreover, our RNA-seq analysis revealed that INO2 and INO4 expression is upregulated in opi1Δ cells following MMS treatment (Supplementary Table 8), indicating a role for Opi1 in INO2 repression during MMS-induced stress. In line with that, we investigated if INO2 and INO4 deletions could rescue the genotoxin sensitivity of cells lacking Opi1. Remarkably, opi1Δino2Δ and opi1Δino4Δ strains are rescued for the sensitivity to MMS and 4NQO, which indicates that constitutive expression of Ino2-Ino4-responsive genes is causing the hypersensitivity to these genotoxins (Fig. 5a). Interestingly, our findings demonstrate that the deletion of INO4 alleviates the sensitivity to zeocin and bleomycin, while the deletion of INO2 does not have the same effect. Moreover, we notice that ino4Δ cells display sensitivity to heat shock but not to other types of proteotoxic stress inducers (Supplementary Fig. 6). Intriguingly, ino2Δ cells do not exhibit sensitivity to heat shock, and the deletion of Opi1 rescues the heat shock sensitivity of ino4Δ cells while conferring sensitivity to ino2Δ cells (Supplementary Fig. 6). These findings provide further support for previous evidence demonstrating the distinct roles of Ino2 and Ino4 in transcriptional regulation under various stress conditions (Chumnanpuen et al. 2013). These consistent findings underscore the unique and context-dependent roles of Ino2 and Ino4 in transcriptional regulation.
Fig. 5.
Opi1 counteracts the function of the transcriptional repressor Ino2-Ino4 and the PP-IP kinase Kcs1 during MMS-induced genotoxic stress. a) Deletion of INO2 and INO4 rescues the genotoxin sensitivity of cells lacking Opi1. b) and c) Deletion of the PP-IP kinase KCS1 rescues the MMS sensitivity b) and the delayed G1-to-S-phase transition c) of cells lacking Opi1. d) Overexpression of Kcs1 phenocopies the MMS sensitivity of an opi1Δ strain. e) Different from the effect on MMS, deletion of Kcs1 causes a synergistic increase in sensitivity of opi1Δ cells in the presence of several DNA damage agents. For a), b), d), and e), 4-fold serial dilutions were spotted on the media indicated in the figure, and plates were grown for 2–3 days at 30°C.
The transcriptional activator Ino2-Ino4 upregulates the expression of several other genes counteracting Opi1, suggesting that constitutive expression of another Ino2-Ino4 target in opi1Δ cells is causing MMS sensitivity. Thus, we conducted a screening of a small set of genes whose expression is dependent on Ino2-Ino4 and that are upregulated in opi1Δ cells treated with MMS (Supplementary Table 8). We selected genes involved in PI (INO1, inositol-3-phosphate synthase; ITR1, inositol transporter) and phosphatidylcholine (PC) biosynthesis (OPI3, phospholipid methyltransferase; CKI1, choline kinase) since these pathways are upregulated in opi1Δ cells under genotoxic stress (Fig. 4d). Consequently, we deleted these genes in an opi1Δ background with the expectation that it would phenocopy the MMS resistance of opi1Δino2Δ and opi1Δino4Δ strains. Surprisingly, all double mutants showed growth defects and increased MMS sensitivity (Supplementary Fig. 7a and b). This negative genetic interaction between Opi1 and genes from PI and PC biosynthesis suggests that upregulation of 1 or more genes repressed by Opi1 in the absence of Ino1, Itr1, Opi3, and Cki1 negatively impacts the cell.
It is well established that the expression of INO1 is positively regulated by Ino2-Ino4 and massively increases in the absence of Opi1 (Figs. 3d and 4b) (Graves and Henry 2000; Santiago and Ben Mamoun 2003; Jesch et al. 2005). To test if constitutive expression of INO1 in cells lacking Opi1 is causing MMS sensitivity, we overexpressed INO1 by cloning it under the control of the strong inducible GAL1 promoter. Overexpression of INO1 from the GAL1 promoter rescues the inositol auxotrophy of an ino1Δ strain (Supplementary Fig. 7c) but does not show sensitivity to MMS (Supplementary Fig. 7d), which is an indication that increased levels of inositol do not affect the response to MMS-induced stress.
PP-IPs counteract the effects of Opi1 on gene expression during MMS-induced stress
In budding yeast, Kcs1 catalyzes the pyrophosphorylation of the C5 α-phosphate of IPs to form 5-PP-IP4 and 5-PP-IP5, respectively (Saiardi et al. 1999). Miriam Greenberg's group has proposed that inositol pyrophosphate 5-PP-IP4 may play a role in recruiting transcriptional activators, such as Ino2-Ino4, to the promoter region of INO1, thus regulating its expression (Ye et al.’s (2013). This suggests that deletion of Kcs1, involved in the synthesis of PP-IPs, could lead to a reduction in the expression of genes that are upregulated by Ino2-Ino4 in cells lacking Opi1. We consequently conducted a genetic rescue experiment by deleting Kcs1 in an opi1Δ strain to determine whether it would ameliorate the MMS sensitivity observed in the absence of Opi1. Remarkably, Kcs1 deletion significantly rescues the MMS sensitivity observed in the opi1Δ strain (Fig. 5b). Additionally, Kcs1 deletion also rescues the G1/S arrest observed in opi1Δ cells during recovery from MMS treatment (Fig. 5c). These findings might indicate that in the presence of stress induced by MMS, Opi1 functions in opposition to PP-IPs to facilitate the modulation of gene expression in an Ino2-Ino4-dependent manner. In agreement with that, cells lacking the Siw14 phosphatase that counteracts Kcs1-dependent PP-IP synthesis have a mild sensitivity to MMS (Supplementary Fig. 8a). It has been shown that siw14Δ mutants have increased levels of 5-PP-IP5 (Steidle et al. 2016).
To assess the impact of elevated PP-IP levels on MMS sensitivity, we constructed a strain in which KCS1 was placed under the control of the GAL1 promoter. Our data demonstrate that induction with galactose results in elevated levels of Kcs1 mRNA and protein (see Supplementary Fig. 8b and c). Remarkably, wild-type cells overexpressing Kcs1 phenocopy an opi1Δ strain with cell growth delay and sensitivity to MMS (Fig. 5d). Taken together, our results suggest that increased PP-IP levels due to the lack of the transcriptional repressor Opi1 lead to impaired capacity to cope with stress induced by MMS.
Intriguingly, while the deletion of Ino2-Ino4 rescues the sensitivity to other genotoxic agents (Fig. 5a), the deletion of Kcs1 in opi1Δ cells instead increases sensitivity to zeocin/bleomycin and 4NQO (Fig. 5e). Furthermore, opi1Δkcs1Δ cells show sensitivity to HU and CPT (Supplementary Fig. 8d). These findings suggest a complex interplay between Opi1 and PP-IPs in the cellular response to different sources of stress. The specific mechanisms underlying the distinct responses to genotoxic agents warrant further investigation.
Opi1 regulates mitochondrial function upon MMS-induced stress
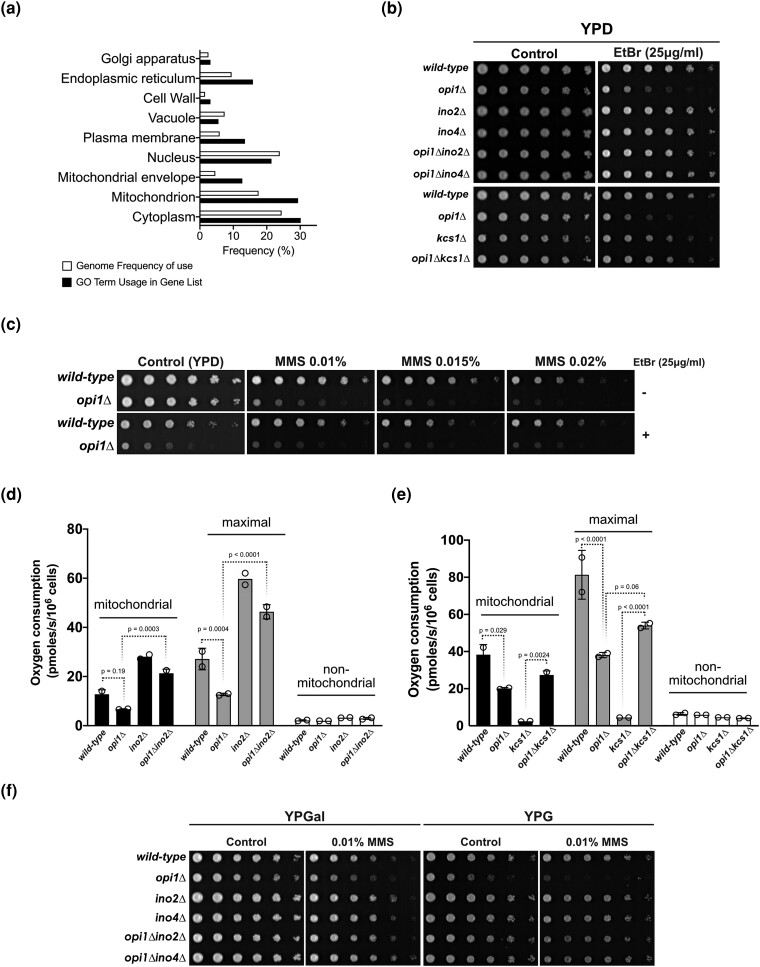
Remarkably, our RNA-seq data, combined with GO analysis, demonstrate a significant enrichment of genes with mitochondrial localization that show significant upregulation or downregulation (log2 fold change ≥0.8 or ≤0.8, respectively) in response to MMS treatment in opi1Δ cells (Fig. 6a). Of note, it has been shown that the lack of Opi1 causes a decrease in mitochondrial membrane cardiolipin content leading to respiratory defects and a petite-negative phenotype (pet-) due to the inability to survive in the presence of damaged mtDNA (Dunn et al. 2006; Luévano-Martínez et al. 2013). Interestingly, the expression of Opi1 is upregulated in cells lacking mtDNA (rho0), emphasizing the critical role of Opi1 in managing mitochondrial dysfunction (Singh et al. 2004). Our results confirm the sensitivity of Opi1-deficient cells to ethidium bromide (EtBr), a known inducer of mtDNA damage, and we demonstrate that deletion of Ino2-Ino4 and Kcs1 rescues the pet- phenotype observed in opi1Δ cells (Fig. 6b). Moreover, cells lacking Opi1 are sensitive to antimycin A, which can also be reverted by deletion of INO2 and INO4 (Supplementary Fig. 9). Antimycin A, an inhibitor of complex III of the electron transport chain, can induce oxidative damage to mtDNA by increasing ROS leakage (Doudican et al. 2005).
Fig. 6.
Opi1 is important to prevent mtDNA instability and to promote respiratory function during MMS-induced genotoxic stress. a) GO Slim term analysis using the Yeast Genome Database's GO Slim Mapper revealing enrichment of genes associated with specific cellular components that are misregulated in cells lacking Opi1 upon MMS treatment (Supplementary Table 8). All genes from S. cerevisiae were included as a reference list. b) Deletion of INO2, INO4, and KCS1 rescues the EtBr sensitivity of cells lacking Opi1. c) Combined treatment of MMS and EtBr does not synergistically enhance the sensitivity phenotype of an opi1Δ strain beyond the sensitivity conferred by each drug independently. d) Deletion of INO2 completely restores mitochondrial respiratory capacity of cells lacking Opi1. e) The opi1Δkcs1Δ double mutant exhibits restored mitochondrial respiratory capacity, which is impaired in the single opi1Δ and kcs1Δ mutants. f) Deletion of INO2 and INO4 rescues the MMS sensitivity of cells lacking Opi1 in medium containing respiratory carbon sources (galactose and glycerol). For b), c), and f), 4-fold serial dilutions of the samples were spotted onto the designated growth medium as indicated in the figure. The plates were then incubated at 30°C for 2–3 days, except for plates containing YPG medium, which were photographed after 4 days of incubation.
Together, these data strongly suggest that constitutive activation of Ino2-Ino4 is also involved in mitochondrial dysfunction. Therefore, we hypothesize that the sensitivity of opi1Δ cells to MMS is more likely due to increased mtDNA instability rather than direct damage to nuclear DNA, as MMS is also known to induce damage to mtDNA (Kitanovic et al. 2009). Notably, the combined treatment of MMS and EtBr did not result in an additive or synergistic increase in the sensitivity of opi1Δ cells, suggesting that the MMS sensitivity of these cells may be attributed to increased mtDNA damage rather than direct nuclear DNA damage (Fig. 6c).
To elucidate if the rescue of MMS sensitivity is related to a recovery of mitochondrial function, we sought to investigate if INO2 deletion would also rescue the respiratory defect and MMS sensitivity of opi1Δ cells in the presence of a respiratory carbon source. Utilizing the Oroboros system, we measured the rate of oxygen consumption in respiratory medium (YPEG; 2% glycerol and 1% ethanol) and found that deletion of Opi1 led to a reduction in mitochondrial respiration, which is consistent with a defect in oxidative phosphorylation. Interestingly, the respiratory defect in Opi1-deficient cells was fully restored upon the deletion of INO2 (Fig. 6d). Surprisingly, the opi1Δkcs1Δ strain showed a significant rescue of mitochondrial respiration (Fig. 6e). It is worth noting that cells lacking Kcs1 are typically unable to survive in a nonfermentable carbon source medium due to the critical role of PP-IPs in mediating the diauxic shift through the modulation of Gcr1, a key transcription factor involved in glycolysis regulation (Szijgyarto et al. 2011). Furthermore, we tested if the deletion of Ino2-Ino4 would rescue the MMS sensitivity of opi1Δ cells in the presence of a respiratory carbon source (Fig. 6f). In the presence of galactose, which is known to induce simultaneous respiration and fermentation (Fendt and Sauer 2010), deletion of Ino2 and Ino4 completely restored the MMS sensitivity of opi1Δ cells. Importantly, in the presence of glycerol, there was a significant rescue of MMS sensitivity, which indicates that constitutive expression of genes regulated by Ino2-Ino4 increases mtDNA instability in the presence of MMS.
Overall, our findings indicate that under MMS-induced stress, Opi1 is important to the fine-tuning of gene expression to ensure appropriate cellular responses. Future investigations into these mechanisms will deepen our understanding of how cells maintain homeostasis and cope with genotoxic stress.
Discussion
Through transcriptomic and functional analyses, our study provides evidence supporting the role of the transcriptional repressor Opi1 as a crucial regulator of gene expression during MMS-induced stress. Our findings indicate that the constitutive activation of the transcriptional activator Ino2-Ino4 is associated with increased MMS sensitivity, cell cycle defects, and mitochondrial dysfunction. Importantly, reduction of PP-IP synthesis resulting from the deletion of Kcs1 can ameliorate the MMS-induced defects observed in opi1Δ cells. In contrast, the overexpression of Kcs1 in wild-type cells mimics the phenotype of cells lacking Opi1, suggesting that PP-IPs compete with Opi1 for the expression of genes in response to MMS treatment.
Opi1 modulates different biological processes during MMS-induced stress
The regulation of multiple metabolic pathways by Opi1 poses a significant challenge in identifying the underlying cause of MMS sensitivity. In this study, we employed RNA-seq analyses to uncover Opi1's modulation of diverse biological processes during MMS treatment. Importantly, our findings underscore the pivotal role of Opi1 in the repression of phospholipid biosynthesis after MMS-induced stress. It is worth noting that despite the involvement of Opi1 in regulating genes involved in phospholipid biosynthesis, the deletion of several genes related to this process did not rescue the MMS sensitivity of opi1Δ cells. Surprisingly, Opi1 exhibited negative genetic interactions with genes involved in PI (Ino1 and Itr1) and PC synthesis (Opi3 and Cki1), as revealed by Supplementary Fig. 7a and b. One explanation is that impairment of PC synthesis leading to an unbalance of the phosphatidylethanolamine (PE)/PC ratio causes ER stress (Thibault et al. 2012). Furthermore, accumulation of the PC metabolic intermediate phosphatidylmonomethylethanolamine (PMME) has been shown to exacerbate ER stress in yeast cells (Ishiwata-Kimata et al. 2022). Given the increased expression of genes upstream of Opi3, such as CDS1, CHO1, PSD1, and CHO2 (Supplementary Table 8), it is conceivable that PMME levels are even higher in opi1Δ cells. Significantly, previous research has demonstrated that exposure to MMS leads to lipid stress specifically at the inner nuclear membrane of RPE-1 cells. This stress response is mitigated through a combination of nuclear membrane deformation and the emission of nuclear lipid droplets (Ovejero et al. 2021). Given that alterations in the nuclear membrane can impact critical nuclear processes including DNA replication, transcription, and repair (Mekhail and Moazed 2010), it is of particular interest to explore the potential involvement of Opi1 in the preservation of nuclear membrane integrity. Further investigation could help to clarify the specific mechanisms by which Opi1 and proteins involved in phospholipid biosynthesis are involved in the response to MMS-induced stress.
Furthermore, our study revealed that multiple proteins involved in the transduction of mating signaling with cellular conjugation and invasive filamentous growth are significantly downregulated during genotoxic stress in an Opi1-dependent manner (Fig. 4d). Filamentous growth in yeast is triggered by nutrient limitation and involves various pathways including MAPK, Ras/PKA, Snf1, and TOR. A comprehensive review of filamentous growth response is available in Cullen and Sprague (2012). Interestingly, it has been shown that genotoxic stress induces filamentous growth in budding yeast (Jiang and Kang 2003) and in Candida species (Shi et al. 2007; Bravo Ruiz et al. 2020). Noteworthy, it has been proposed that Saccharomyces cerevisiae Opi1 regulates filamentous growth through the upregulation of Flo11, a glycosylphosphatidylinositol (GPI) anchored cell surface glycoprotein that determines colony morphology in yeast (Reynolds 2006). While we did not observe significant changes in Flo11 expression in opi1Δ cells, our RNA-seq analysis revealed significant upregulation of several genes that inhibit filamentous growth, especially after MMS treatment, including the Fus3 MAP kinase, the Nrg2 transcriptional repressor, and Mho1, a protein of unknown function. Interestingly, while Fus3 inhibits filamentous growth by causing degradation of the transcription factor Tec1 via phosphorylation (Bao et al. 2004; Chou et al. 2004), it promotes mating signaling by phosphorylating the CDK inhibitor Far1, leading to its association with Cln-Cdc28 (Peter et al. 1993; Tyers and Futcher 1993; Peter and Herskowitz 1994). Consequently, Far1 impedes the function of the Cln-Cdc28 complex in the G1 phase, thus preventing cells from advancing from G1 to START (reviewed in Bardwell 2005; Sieber et al. 2023). Our findings are intriguing as they provide evidence for the involvement of Opi1 in promoting the suppression of mating signaling under MMS-induced stress conditions. To begin with, FAR1 and FUS3 expression are downregulated in an Opi1-dependent manner (Fig. 4e). Moreover, we observed an increase in the percentage of cells in the G1 phase following a 2-h MMS treatment (Fig. 2d and e). Furthermore, upon release from MMS treatment, opi1Δ cells exhibited a delayed G1-to-S-phase transition, indicating that mating signaling is hyperactivated (Fig. 2e).
It is important to note that all of our results were obtained using BY4741, which is known to be a filamentation-deficient yeast strain. Thus, any phenotypes related to filamentous growth, such as biofilm formation and invasive growth on agar, were not observable in our study.
PP-IPs generated by Kcs1 dampen Opi1-mediated gene expression during genotoxic stress
Our findings reveal that the MMS sensitivity and cell cycle defects observed in opi1Δ cells can be rescued by deletion of the PP-IP kinase Kcs1 (Fig. 5b and c), which provides compelling evidence that PP-IP synthesis in the absence of Opi1 is detrimental for cells under MMS-induced stress. It has been previously shown that KCS1 possesses UASINO in its promoter region and that its expression is upregulated when inositol concentration increases (Wimalarathna et al. 2011). Additionally, the IP multikinase Ipk2 (ARG82) that catalyzes the synthesis of IP4 and IP5 also contains UASINO in its promoter region, and its expression is dependent on Ino2-Ino4 (Wimalarathna et al. 2011). However, while we did not observe changes in KCS1 and ARG82 expression levels in our dataset for opi1Δ cells (Supplementary Tables 4, 6, and 8), it is plausible that PP-IP synthesis is elevated in these cells, as previous studies have demonstrated an increase in Kcs1 protein levels in the absence of Opi1 (Ye et al. 2013). It is interesting to note that while INO1 overexpression does not sensitize cells to MMS, that of KCS1 does (Supplementary Fig. 7d and Fig. 5d, respectively), suggesting that INO1 overexpression probably does not lead to an increase in PP-IP synthesis, which might indicate that there is a rate-limiting step reaction downstream of inositol-3-phosphate synthesis restraining PP-IP concentration in the cell.
How do PP-IPs affect the DDR leading to MMS sensitivity in the absence of Opi1? In budding yeast, PP-IPs are synthetized from IP5 and IP6 in a reaction catalyzed by the PP-IP kinases Kcs1 and Vip1 (Saiardi et al. 1999; Mulugu et al. 2007; Lee et al. 2008). Kcs1 catalyzes the conversion of IP5 and IP6 to different isomers of PP-IPs such as 5-PP-IP4 and 5-PP-IP5 (Saiardi et al. 1999). It is established that PP-IPs modulate protein function through either nonenzymatic pyrophosphorylation of proteins or allosteric regulation through its interaction with protein domains (Wilson et al. 2013). However, up until now, only a few targets have been characterized. Kcs1 and its mammalian orthologs IP6Ks have been implicated in several biological processes such as cell cycle progression, DNA repair, telomere length, nutrient signaling, ESR, and polyphosphate metabolism, among others (Auesukaree et al. 2005; Saiardi et al. 2005; Szijgyarto et al. 2011; Banfic et al. 2013, 2016; Wilson et al. 2013; Worley et al. 2013). In light of these considerations, our findings suggest a potential role for PP-IPs in modulating the function of Opi1-dependent genes during MMS-induced stress. One plausible explanation is the previous proposal that PP-IPs generated by Kcs1 are important to induce INO1 expression, indicating their potential involvement in recruiting Ino2-Ino4 to UASINO regions (Ye et al. 2013). However, this hypothesis requires further investigation, as there is currently no evidence suggesting that PP-IPs directly target Ino2-Ino4 for either allosteric regulation or pyrophosphorylation. It is also important to note that while Kcs1 deletion rescues MMS sensitivity of opi1Δ cells, it shows a synergistic increase in sensitivity to other genotoxins (Fig. 5e). It has been shown that Kcs1 is required for the DNA hyperrecombination phenotype in yeast cells with defects in protein kinase C1 (Pkc1) (Luo et al. 2002). Moreover, it was shown in mammalian cells that the orthologs of Kcs1 (IP6K1/2/3) are important for homologous recombination repair (Jadav et al. 2013). Hence, it is important to conduct future investigations to examine the potential cooperative role between Opi1 and Kcs1 in dealing with specific types of DNA lesions and their impact on DNA repair pathways. By looking into these aspects, we can gain a deeper understanding of the role of Opi1 and Kcs1 for cellular responses to genotoxic stress.
Genotoxin sensitivity in opi1Δ is associated with mtDNA damage
Deletion of the transcriptional repressor Opi1 can induce pleiotropic effects in the cell. First, we ruled out the possibility that the heightened sensitivity of opi1Δ cells to MMS is linked to deregulation of the ESR (Fig. 1d). Besides that, tunicamycin treatment did not sensitize opi1Δ cells (Fig. 1d), which is in line with previous evidence that the absence of Opi1 causes an expansion of the ER membrane, alleviating the ER stress induced by dithiothreitol (DTT) or tunicamycin in an UPR-independent manner (Schuck et al. 2009). Furthermore, it was previously shown that cells lacking Opi1 possess short telomeres (Askree et al. 2004). In contrast, cells lacking Kcs1 have longer telomeres (York et al. 2005). This finding could indicate that deletion of Kcs1 rescues MMS sensitivity due to restoration of telomere length. However, this is probably not the case as it was shown that telomere length does not affect cellular fitness or yeast sensitivity to DNA damage (Harari et al. 2017).
MMS can also damage mtDNA leading to direct inhibition of the respiratory chain with increased ROS leakage, oxidative inactivation of glycolytic enzymes, and cell cycle arrest (Kitanovic et al. 2009). Importantly, cells lacking Opi1 exhibit a decrease in mitochondrial cardiolipin content and are unable to survive in the absence of mtDNA (pet- phenotype) (Luévano-Martínez et al. 2013). Here, we have discovered that the sensitivity of cells lacking Opi1 to MMS may be linked to mtDNA instability. This conclusion is supported by 2 observations: (1) deletion of Ino2-Ino4 genes rescues the pet- phenotype, antimycin A hypersensitivity, and respiratory capacity of cells lacking Opi1; and (2) the combined treatment of MMS and EtBr does not increase the sensitivity of opi1Δ cells. It is worth noting that a previous study has demonstrated that cells lacking mtDNA exhibit defects in the G1-to-S-phase progression (Crider et al. 2012). Therefore, it would be important to investigate whether Opi1 is involved in a mitochondria-to-nucleus retrograde signaling pathway that promotes the transition from the G1 phase to the S phase.
Furthermore, we observed that there is a significant enrichment of mitochondrial proteins among the genes that are differentially expressed in opi1Δ cells following MMS treatment (Fig. 6a). For instance, we observed upregulation of Psd1 in opi1Δ cells upon genotoxic stress, as depicted in Fig. 4d and Supplementary Tables 6 and 7. Psd1 is an enzyme located in the mitochondrial inner membrane that converts phosphatidylserine to PE and plays a crucial role in regulating mitochondrial fusion and morphology (Chan and McQuibban 2012). In contrast, Aim17, a protein with a mitochondrial localization and unknown function, was found to be downregulated in opi1Δ cells upon genotoxic stress, as shown in Supplementary Fig. 5b and Supplementary Tables 6 and 7. Although limited information is available regarding the precise role of Aim17 in mitochondrial homeostasis, it seems to be important for mtDNA integrity (Hess et al. 2009). Considering that Psd1 and Aim17 are among the top hits of DEGs in our screening (Supplementary Table 6), it would be valuable to explore in future investigations the specific contributions of these genes to the observed genotoxin sensitivity and mitochondrial dysfunction in opi1Δ cells.
A working model for Opi1 function during genotoxic stress
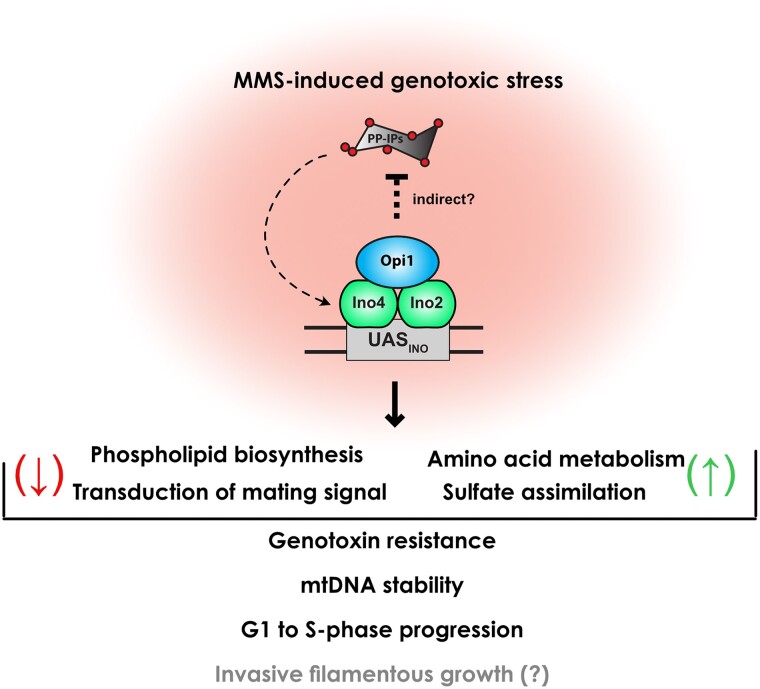
We propose a model in which Opi1 functions as a crucial sensor of genotoxic stress in budding yeast (Fig. 7). Upon genotoxic stress, Opi1 translocates to the nucleus and regulates the expression of multiple genes either directly or indirectly. Specifically, during MMS-induced genotoxic stress, Opi1 represses phospholipid biosynthesis and MAPK signaling, potentially suppressing cellular conjugation with mating while promoting invasive filamentous growth. Additionally, Opi1 fosters the expression of genes involved in amino acid metabolism and sulfate assimilation. In cells lacking Opi1, the constitutive activation of Ino2-Ino4 promotes gene transcription, which is further potentiated by PP-IPs produced by Kcs1. Consequently, the deletion of Opi1 results in pleiotropic defects affecting cell cycle progression and mitochondrial function, ultimately reducing cellular fitness. Understanding the interconnections between these processes and determining their relative contributions to MMS resistance will be crucial for future investigations.
Fig. 7.
Model summarizing the Opi1 role as a key sensor of genotoxic stress in budding yeast. Briefly, upon genotoxic stress induced by MMS, Opi1 physically interacts with the Ino2-Ino4 transcriptional activator and counteracts PP-IPs, thereby modulating the expression of several genes. Opi1 plays an important role in repressing phospholipid biosynthesis and transduction of mating signaling while promoting the expression of genes involved in amino acid metabolism and sulfate assimilation. The coordinated modulation of these genes contributes to genotoxin resistance by preventing mtDNA instability, delaying G1-to-S-phase progression, and potentially promoting invasive filamentous growth. See Discussion for more details.
In conclusion, this study provides insights into the intricate molecular mechanisms underlying Opi1's role in cellular response to genotoxic stress. The significance of these findings is magnified by the presence of Opi1 in various pathogenic fungal species, which have become a global concern due to their widespread distribution and increasing antifungal resistance. Importantly, as Opi1 is absent in mammals, our results highlight its potential as a target for the development of more effective treatments against pathogenic fungal infections.
Supplementary Material
Acknowledgments
We thank Jacilene Barbosa and Natalia Bromberg for technical support; Dr. Aparecida Sadae Tanaka and Dr. Maria Luiza Vilela Oliva for kindly allowing access to their laboratory and equipment; Dr. Francisco Bastos de Oliveira and Dr. Gustavo Monteiro Silva for comments and suggestions; and Dr. Francisco Bastos de Oliveira, Dr. Marcus Bustamante Smolka, and Dr. Luis Eduardo Soares Netto for donating plasmid and yeast strains.
Contributor Information
Giovanna Marques Panessa, Departamento de Bioquímica, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP 04023-900, Brazil.
Eduardo Tassoni-Tsuchida, Department of Biology, Stanford University, Stanford, CA 94305, USA; Department of Biochemistry, Stanford University, Stanford, CA 94305, USA.
Marina Rodrigues Pires, Departamento de Bioquímica, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP 04023-900, Brazil.
Rodrigo Rodrigues Felix, Departamento de Bioquímica, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP 04023-900, Brazil.
Rafaella Jekabson, Departamento de Bioquímica, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP 04023-900, Brazil.
Nadja Cristhina de Souza-Pinto, Departamento de Bioquímica, Instituto de Química, Universidade de São Paulo, São Paulo, SP 05508-900, Brazil.
Fernanda Marques da Cunha, Departamento de Bioquímica, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP 04023-900, Brazil.
Onn Brandman, Department of Biochemistry, Stanford University, Stanford, CA 94305, USA.
José Renato Rosa Cussiol, Departamento de Bioquímica, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP 04023-900, Brazil.
Data availability
The raw and processed RNA-seq transcriptome data were deposited to the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/biostudies/) under the accession number E-MTAB-13057. Strains and plasmids are available upon request. Supplementary Tables 1 to 3 contain the list of yeast strains, plasmids, and primers used in this study, respectively. Supplementary Tables 4 to 8 contain the RNA-seq dataset as described in Materials and methods. Supplementary Fig. 1 contains the PCA showing the variation among biological replicates in RNA-seq. Supplementary Fig. 2 contains supplemental data in support of Fig. 1. Supplementary Fig. 3 contains supplemental data in support of Fig. 2. Supplementary Fig. 4 contains supplemental data in support of Fig. 3a and b. Supplementary Fig. 5 contains supplemental data in support of Fig. 4. Supplementary Figs. 6 and 7 contain supplemental data in support of Fig. 5. Supplementary Fig. 8 contains supplemental data in support of Fig. 5d and e. Supplementary Fig. 9 contains supplemental data in support of Fig. 6b.
Supplemental material available at GENETICS online.
Funding
This work is supported by a grant from FAPESP (2018/05417-0) to JRRC, a grant from FAPESP (2021/04887-6) to GMP, a grant from FAPESP (2019/25497-1) to RJ, a grant from FAPESP (2019/09732-0) to FMdC, a grant from FAPESP (2017/04372-0) to NCdS-P, and a grant from NIH (R01GM148526) to OB.
Author contributions
JRRC conceived the project and wrote the paper. JRRC, GMP, ET-T, MRP, RRF, and RJ designed the experiments, analyzed the data, and helped revise the manuscript. FMdC designed and performed experiments for measuring oxygen consumption and helped revise the manuscript. NCdS-P and OB provided reagents and infrastructure and helped revise the manuscript.
Literature cited
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101(23):8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J Biol Chem. 2005;280(26):25127–25133. doi: 10.1074/jbc.M414579200. [DOI] [PubMed] [Google Scholar]
- Balint A, Kim T, Gallo D, Cussiol JR, Bastos De Oliveira FM, Yimit A, Ou J, Nakato R, Gurevich A, Shirahige K, et al. Assembly of Slx4 signaling complexes behind DNA replication forks. EMBO J. 2015;34(16):2182–2197. doi: 10.15252/embj.201591190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfic H, Bedalov A, York J, Visnjic D. Inositol pyrophosphates modulate S phase progression after pheromone-induced arrest in Saccharomyces cerevisiae. J Biol Chem. 2013;288(3):1717–1725. doi: 10.1074/jbc.M112.412288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfic H, Crljen V, Lukinovic-Skudar V, Dembitz V, Lalic H, Bedalov A, Visnjic D. Inositol pyrophosphates modulate cell cycle independently of alteration in telomere length. Adv Biological Regul. 2016;60:22–28. doi: 10.1016/j.jbior.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Bao MZ, Schwartz MA, Cantin GT, Yates JR, Madhani HD. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119(7):991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26(2):339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos de Oliveira FM, Kim D, Cussiol JR, Das J, Jeong MC, Doerfler L, Schmidt KH, Yu H, Smolka MB. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol Cell. 2015;57(6):1124–1132. doi: 10.1016/j.molcel.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231(1):11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The Rad53 signal transduction pathway: replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 2006;312(14):2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Bravo Ruiz G, Ross ZK, Gow NAR, Lorenz A. Pseudohyphal growth of the emerging pathogen Candida auris is triggered by genotoxic stress through the S phase checkpoint. mSphere. 2020;5(2):e00151-20. doi: 10.1128/mSphere.00151-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2(11):e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EYL, McQuibban GA. Phosphatidylserine decarboxylase 1 (Psd1) promotes mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and alternative topogenesis of mitochondrial genome maintenance protein 1 (Mgm1). J Biol Chem. 2012;287(48):40131–40139. doi: 10.1074/jbc.M112.399428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119(7):981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Chumnanpuen P, Nookaew I, Nielsen J. Integrated analysis, transcriptome-lipidome, reveals the effects of INO- level (INO2 and INO4) on lipid metabolism in yeast. BMC Syst Biol. 2013;7(S3):S7. doi:10.1186/1752-0509-7-Supplementary 3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD. The Saccharomyces cerevisiae suppressor of choline sensitivity (SCS2) gene is a multicopy suppressor of mec1 telomeric silencing defects. Genetics. 2001;158(1):145–154. doi: 10.1093/genetics/158.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider DG, García-Rodríguez LJ, Srivastava P, Peraza-Reyes L, Upadhyaya K, Boldogh IR, Pon LA. 2012. Rad53 is essential for a mitochondrial {DNA} inheritance checkpoint regulating G1 to S progression. J. Cell Biol. 198(5): 793–798. doi: 10.1083/jcb.201205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF. The regulation of filamentous growth in yeast. Genetics. 2012;190(1):23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cussiol JRR, Soares BL, de Oliveira FMB. From yeast to humans: understanding the biology of DNA damage response (DDR) kinases. Genet Mol Biol. 2020;43(1 suppl 1):e20190071. doi: 10.1590/1678-4685-gmb-2019-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(12):5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CD, Lee MS, Spencer FA, Jensen RE. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol Biol Cell. 2006;17(1):213–226. doi: 10.1091/mbc.e05-06-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt S-M, Sauer U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst Biol. 2010;4(1):12. doi: 10.1186/1752-0509-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E, Bruhn C, Peretti M, Cassani C, Carotenuto WV, Elgendy M, Shubassi G, Lucca C, Bermejo R, Varasi M, et al. PP2A controls genome integrity by integrating nutrient-sensing and metabolic pathways with the DNA damage response. Mol Cell. 2017;67(2):266–281.e4. doi: 10.1016/j.molcel.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12(10):2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar ML, Chang Y-F, Jesch SA, Aregullin M, Henry SA. Interaction between repressor Opi1p and ER membrane protein Scs2p facilitates transit of phosphatidic acid from the ER to mitochondria and is essential for INO1 gene expression in the presence of choline. J Biol Chem. 2017;292(45):18713–18728. doi: 10.1074/jbc.M117.809970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, Jean SA, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20(6):1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- Graves JA, Henry SA. Regulation of the yeast INO1 gene: the products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics. 2000;154(4):1485–1495. doi: 10.1093/genetics/154.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Leung WY. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci U S A. 1996;93(24):13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC. Historical perspective on the DNA damage response. DNA Repair (Amst). 2015;36:2–7. doi: 10.1016/j.dnarep.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y, Zadok-Laviel S, Kupiec M. Long telomeres do not affect cellular fitness in yeast. mBio. 2017;8(4):e01314-17. doi: 10.1128/mBio.01314-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5(3):e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyken W-TT, Repenning A, Kumme J, Schüller H-JJ. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol Microbiol. 2005;56(3):696–707. doi: 10.1111/j.1365-2958.2004.04499.x. [DOI] [PubMed] [Google Scholar]
- Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6(10):3320–3328. doi: 10.1128/mcb.6.10.3320-3328.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer HF, Gecht M, Fischer SC, Seybert A, Frangakis AS, Stelzer EHK, Covino R, Hummer G, Ernst R. The molecular recognition of phosphatidic acid by an amphipathic helix in Opi1. J Cell Biol. 2018;217(9):3109–3126. doi: 10.1083/jcb.201802027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppen J, Repenning A, Albrecht A, Geburtig S, Schüller H-J. Comparative analysis of promoter regions containing binding sites of the heterodimeric transcription factor Ino2/Ino4 involved in yeast phospholipid biosynthesis. Yeast. 2005;22(8):601–613. doi: 10.1002/yea.1209. [DOI] [PubMed] [Google Scholar]
- Ishiwata-Kimata Y, Le QG, Kimata Y. Induction and aggravation of the endoplasmic-reticulum stress by membrane-lipid metabolic intermediate Phosphatidyl-N-Monomethylethanolamine. Front Cell Dev Biol. 2022;9:743018. doi: 10.3389/fcell.2021.743018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski CM, Cussiol JR, Oberly S, Yimit A, Balint A, Kim T, Zhang Z, Brown GW, Smolka M. Termination of replication stress signaling via concerted action of the Slx4 scaffold and the PP4 phosphatase. Genetics. 2015;201(3):937–949. doi: 10.1534/genetics.115.181479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadav RS, Chanduri MVL, Sengupta S, Bhandari R. Inositol pyrophosphate synthesis by inositol hexakisphosphate kinase 1 is required for homologous recombination repair. J Biol Chem. 2013;288(5):3312–3321. doi: 10.1074/jbc.M112.396556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäschke Y, Schwarz J, Clausnitzer D, Müller C, Schüller H-J. Pleiotropic corepressors Sin3 and Ssn6 interact with repressor Opi1 and negatively regulate transcription of genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol Genet Genomics. 2011;285(2):91–100. doi: 10.1007/s00438-010-0589-5. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci U S A. 1999;96(4):1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch SA, Zhao X, Wells MT, Henry SA. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280(10):9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YW, Kang CM. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol Biol Cell. 2003;14(12):5116–5124. doi: 10.1091/mbc.e03-06-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanovic A, Walther T, Loret MO, Holzwarth J, Kitanovic I, Bonowski F, Van Bui N, Francois JM, Wölfl S. Metabolic response to MMS-mediated DNA damage in Saccharomyces cerevisiae is dependent on the glucose concentration in the medium. FEMS Yeast Res. 2009;9(4):535–551. doi: 10.1111/j.1567-1364.2009.00505.x. [DOI] [PubMed] [Google Scholar]
- Kitanovic A, Wölfl S. Fructose-1,6-bisphosphatase mediates cellular responses to DNA damage and aging in Saccharomyces cerevisiae. Mutat Res. 2006;594(1–2):135–147. doi: 10.1016/j.mrfmmm.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Kliewe F, Engelhardt M, Aref R, Schüller H-J. Promoter recruitment of corepressors Sin3 and Cyc8 by activator proteins of the yeast Saccharomyces cerevisiae. Curr Genet. 2017;63(4):739–750. doi: 10.1007/s00294-017-0677-8. [DOI] [PubMed] [Google Scholar]
- Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38(18):e101801. doi: 10.15252/embj.2019101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Yugandhar K, Gupta S, Sanford EJ, Faça VM, Vega S, Joiner AMN, Fromme JC, Yu H, Smolka MB. In-depth and 3-dimensional exploration of the budding yeast phosphoproteome. EMBO Rep. 2021;22(2):e51121. doi: 10.15252/embr.202051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Huang K, Quiocho FA, O'Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4(1):25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi:https://doi.org/10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luévano-Martínez LA, Appolinario P, Miyamoto S, Uribe-Carvajal S, Kowaltowski AJ. Deletion of the transcriptional regulator opi1p decreases cardiolipin content and disrupts mitochondrial metabolism in Saccharomyces cerevisiae. Fungal Genet Biol. 2013;60:150–158. doi: 10.1016/j.fgb.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase C1 mutant yeast. Biochemistry. 2002;41(8):2509–2515. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11(5):317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316(5821):106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Ovejero S, Soulet C, Moriel-Carretero M. The alkylating agent methyl methanesulfonate triggers lipid alterations at the inner nuclear membrane that are independent from its DNA-damaging ability. Int J Mol Sci. 2021;22(14):7461. doi: 10.3390/ijms22147461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Foiani M. Signal transduction: how Rad53 kinase is activated. Curr Biol. 2005;15(18):R769–R771. doi: 10.1016/j.cub.2005.08.057. [DOI] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73(4):747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265(5176):1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Reynolds TB. The Opi1p transcription factor affects expression of FLO11, Mat formation, and invasive growth in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5(8):1266–1275. doi: 10.1128/EC.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9(22):1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci U S A. 2005;102(6):1911–1914 doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon TB, Evert BA, Song B, Doetsch PW. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32(12):3712–3723. doi: 10.1093/nar/gkh696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago TC, Ben Mamoun C. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J Biol Chem. 2003;278(40):38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187(4):525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller HJ, Hahn A, Tröster F, Schütz A, Schweizer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992;11(1):107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q-M, Wang Y-M, Zheng X-D, Teck Ho Lee R, Wang Y. Critical role of DNA checkpoints in mediating genotoxic-stress–induced filamentous growth in Candida albicans. Mol Biol Cell. 2007;18(3):815–826. doi: 10.1091/mbc.e06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395(6702):618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Sieber B, Coronas-Serna JM, Martin SG. A focus on yeast mating: from pheromone signaling to cell-cell fusion. Semin Cell Dev Biol. 2023;133:83–95. doi: 10.1016/j.semcdb.2022.02.003. [DOI] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Bronstein A, Kupiec M, Johnston M. Cross-talk between carbon metabolism and the DNA damage response in S. cerevisiae. Cell Rep. 2015;12(11):1865–1875. doi: 10.1016/j.celrep.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Rasmussen AK, Rasmussen LJ. Genome-wide analysis of signal transducers and regulators of mitochondrial dysfunction in Saccharomyces cerevisiae. Ann N Y Acad Sci. 2004;1011(1):284–298. doi: 10.1007/978-3-662-41088-2_27. [DOI] [PubMed] [Google Scholar]
- Steidle EA, Chong LS, Wu M, Crooke E, Fiedler D, Resnick AC, Rolfes RJ. A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 protein selectively cleaves the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J Biol Chem. 2016;291(13):6772–6783. doi: 10.1074/jbc.M116.714907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szijgyarto Z, Garedew A, Azevedo C, Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334(6057):802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING V11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JFX. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412(6846):553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Thibault G, Shui G, Kim W, McAlister GC, Ismail N, Gygi SP, Wenk MR, Ng DTW. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol Cell. 2012;48(1):16–27. doi: 10.1016/j.molcel.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou L, Mi H. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 2022;31(1):8–22. doi: 10.1002/pro.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13(9):5659–5669. doi: 10.1128/mcb.13.9.5659-5669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C, Dietz M, Wittmann J, Albrecht A, Schüller HJ. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol Microbiol. 2001;41(1):155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- Wang Y-H, Hariharan A, Bastianello G, Toyama Y, Shivashankar GV, Foiani M, Sheetz MP. DNA Damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat Commun. 2017;8(1):2118. doi: 10.1038/s41467-017-01805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MSC, Livermore TM, Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem J. 2013;452(3):369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- Wimalarathna R, Tsai C-H, Shen C-H. Transcriptional control of genes involved in yeast phospholipid biosynthesis. J Microbiol. 2011;49(2):265–273. doi: 10.1007/s12275-011-1130-1. [DOI] [PubMed] [Google Scholar]
- Worley J, Luo X, Capaldi AP. Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 2013;3(5):1476–1482. doi: 10.1016/j.celrep.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19(12):1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Bandara WMMS, Greenberg ML. Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J Biol Chem. 2013;288(34):24898–24908. doi: 10.1074/jbc.M113.493353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Tong J, Lu P, Wang Y, Zhang J, Sun C, Yuan K, Xue R, Zou B, Li N, et al. Formation of a Snf1-Mec1-Atg1 module on mitochondria governs energy deprivation-induced autophagy by regulating mitochondrial respiration. Dev Cell. 2017;41(1):59–71.e4. doi: 10.1016/j.devcel.2017.03.007. [DOI] [PubMed] [Google Scholar]
- York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280(6):4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- Zewail A, Xie MW, Xing Y, Lin L, Zhang PF, Zou W, Saxe JP, Huang J. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc Natl Acad Sci U S A. 2003;100(6):3345–3350. doi: 10.1073/pnas.0530118100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Elia AEH, Naylor ML, Dephoure N, Ballif BA, Goel G, Xu Q, Ng A, Chou DM, Xavier RJ, et al. Profiling DNA damage-induced phosphorylation in budding yeast reveals diverse signaling networks. Proc Natl Acad Sci U S A. 2016;113(26):E3667–E3675. doi: 10.1073/pnas.1602827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulkifli M, Neff JK, Timbalia SA, Garza NM, Chen Y, Watrous JD, Murgia M, Trivedi PP, Anderson SK, Tomar D, et al. Yeast homologs of human MCUR1 regulate mitochondrial proline metabolism. Nat Commun. 2020;11(1):4866. doi: 10.1038/s41467-020-18704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed RNA-seq transcriptome data were deposited to the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/biostudies/) under the accession number E-MTAB-13057. Strains and plasmids are available upon request. Supplementary Tables 1 to 3 contain the list of yeast strains, plasmids, and primers used in this study, respectively. Supplementary Tables 4 to 8 contain the RNA-seq dataset as described in Materials and methods. Supplementary Fig. 1 contains the PCA showing the variation among biological replicates in RNA-seq. Supplementary Fig. 2 contains supplemental data in support of Fig. 1. Supplementary Fig. 3 contains supplemental data in support of Fig. 2. Supplementary Fig. 4 contains supplemental data in support of Fig. 3a and b. Supplementary Fig. 5 contains supplemental data in support of Fig. 4. Supplementary Figs. 6 and 7 contain supplemental data in support of Fig. 5. Supplementary Fig. 8 contains supplemental data in support of Fig. 5d and e. Supplementary Fig. 9 contains supplemental data in support of Fig. 6b.
Supplemental material available at GENETICS online.