Abstract
The ability to synthesize and uptake the Yersinia siderophore yersiniabactin is a hallmark of the highly pathogenic, mouse-lethal species Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica 1B. We have identified four genes, irp1, irp3, irp4, and irp5, on a 13-kb chromosomal DNA fragment of Y. enterocolitica O8, WA-314. These genes constitute the yersiniabactin biosynthetic gene cluster together with the previously defined irp2. The irp1 gene consists of 9,486 bp capable of encoding a 3,161-amino-acid high-molecular-weight protein 1 (HMWP1) polypeptide with a predicted mass of 384.6 kDa. The first 3,000 bp of irp1 show similarity to the corresponding regions of the polyketide synthase genes of Bacillus subtilis and Streptomyces antibioticus. The remaining part of irp1 is most similar to irp2, encoding HMWP2, which might be the reason for immunological cross-reactivity of the two polypeptides. Irp4 was found to have 41.7% similarity to thioesterase-like protein of the anguibactin biosynthetic genes of Vibrio anguillarum. Irp5 shows 41% similarity to EntE, the 2,3-dihydroxybenzoic acid-activating enzyme utilized in enterobactin synthesis of Escherichia coli. Irp4 and Irp5 are nearly identical to YbtT and YbtE, recently identified in Y. pestis. irp3 has no similarity to any known gene. Inactivation of either irp1 or irp2 abrogates yersiniabactin synthesis. Mutations in irp1 or fyuA (encoding yersiniabactin/pesticin receptor) result in downregulation of irp2 that can be upregulated by the addition of yersiniabactin. A FyuA-green fluorescent protein translational fusion was downregulated in an irp1 mutant. Upregulation was achieved by addition of yersiniabactin but not desferal, pesticin, or pyochelin, which indicates high specificity of the FyuA receptor and autoregulation of genes involved in synthesis and uptake of yersiniabactin.
The genus Yersinia contains at least 11 species, 3 of which are enteropathogenic for humans. Yersinia pestis is the agent of bubonic plague, while Y. pseudotuberculosis and Y. enterocolitica are pathogenic for humans. Y. enterocolitica causes a broad range of diseases ranging from acute bowel disease to extraintestinal manifestations such as reactive arthritis and uveitis. Human-pathogenic Yersinia species can be divided into highly pathogenic Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica biotype 1B (so-called American serotypes), which are mouse lethal at low doses, and low-pathogenic Y. enterocolitica biotypes 2 to 5 (so-called European serotypes), which are not mouse lethal at low doses (10).
A prerequisite for any expression of pathogenicity by Yersinia is the presence of a 70-kb pYV virulence plasmid that is found in high- and low-pathogenic strains (4, 21, 27). Differences in mouse virulence seem to be chromosomally determined. Highly pathogenic strains possess a chromosomal cluster of iron-regulated genes designated the high-pathogenicity island (HPI). This island is absent in low-pathogenic or nonpathogenic strains and was found to be unstable in Yersinia strains. Its loss leads to a marked reduction in mouse virulence (36).
Two proteins encoded by iron-repressible genes have been detected only in highly pathogenic Yersinia strains, being putatively located on the HPI: HMWP1 (high-molecular-weight protein 1; 260 kDa, encoded by irp1) and HMWP2 (190 kDa, encoded by irp2) (9, 15). Inactivation of irp2 in Y. pseudotuberculosis results in a considerable reduction of mouse virulence (8). HMWPs are suspected to be important for siderophore yersiniabactin production and therefore involved in the expression of a CAS (chrom azurol S ferric ion indicator dye)-positive phenotype in highly pathogenic Yersinia strains (31). The receptor of yersiniabactin, FyuA (ferric yersiniabactin uptake), is a receptor with dual function: it is a receptor of the siderophore and a receptor of Y. pestis bacteriocin pesticin. Thus, highly pathogenic strains are pesticin sensitive (Psts) because of such a dual nature of FyuA (44). Yersiniabactin and FyuA were shown to be produced only by mouse-lethal strains (32).
In Y. pestis, the fyuA gene, the irp2 gene, and the hms locus (encoding hemin storage) are located on a 102-kb fragment designated the pgm (pigmentation) locus. This fragment is flanked by two copies of the insertion sequence (IS) element IS100 (23, 41), which might be the reason for frequent deletions of the pgm locus. ybtA, a gene encoding a protein belonging to the AraC family of transcriptional regulators, was recently detected upstream the irp2 gene in Y. pestis. YbtA is believed to be a transcriptional activator of the yersiniabactin receptor and of the siderophore biosynthetic genes (22). Bearden et al. (3) have identified an approximately 22-kb region of the pgm locus of Y. pestis which encodes several iron-regulated proteins. Some of them (YbtT and YbtE) were shown to be involved in the biosynthesis of a putative siderophore of Y. pestis.
The HPI of Y. enterocolitica contains the fyuA and irp2 genes but does not harbor genes for the hemin storage (24). This locus is much more stable than the pgm locus of Y. pestis. No flanking IS100 elements, but at least two IS elements, IS1328 and IS1400, were identified downstream fyuA in Y. enterocolitica O8 (7, 43). Irp2 and fyuA are separated by approximately 12 kb. This fragment may contain additional irp genes involved in siderophore synthesis, including irp1 (encoding HMWP1). In this study, we have determined the nucleotide sequence of the irp1 to irp5 genes of Y. enterocolitica O8, shown their involvement in yersiniabactin biosynthesis, and demonstrated the siderophore-directed regulation of yersiniabactin synthesis and receptor genes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in the study are listed in Table 1. Strains were grown in Luria-Bertani (LB) broth or on LB agar plates (Difco Laboratories, Detroit, Mich.) at 28°C (Yersinia) or 37°C (Escherichia coli). Iron-deficient medium (NBD) was made by adding 200 μM α-α-dipyridyl (Sigma, St. Louis, Mo.) to NB medium (nutrient broth [Difco] with 5 g of NaCl per liter as described previously [44]).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype and/or phenotype | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrArelA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | 30 |
| Phi | Psts | 19 |
| S17-1 λ | pir+ tra+ | 54, 39 |
| Y. enterocolitica | ||
| WA-C | Plasmidless derivative of strain WA-134, serotype O8. Spontaneous Nalr mutant | 31 |
| WA-C irp2 | WA-C with pGPIRP2 inserted into irp2 gene | This study |
| WA-CS | Spontaneous Smr mutant derived from WA-C | This study |
| WA fyuA | fyuA mutant of WA-C, Pstr | 44 |
| WA-CS irp1::Kanr | Derivative of WA-CS, Nalr Smr Kmr | This study |
| 8081 | Serotype O8 | 42 |
| H1852 fyuA | Derivative of WA-C, fur fyuA mutant of WA-C | This study |
| Y5.27 | Clinical isolate, serotype O5.27 | 31 |
| Y-96-C | Clinical isolate, serotype O9 | 31 |
| Y-108-C | Clinical isolate, serotype O3 | 31 |
| Y. pseudotuberculosis | ||
| 346 | irp2+ serotype O3 | S. Aleksic |
| 201 | irp2 serotype O3 | S. Aleksic |
| PB1 | Serotype O1 | R. R. Brubaker |
| Y. pestis | ||
| KIM | Lcr− Pgm+ | R. R. Brubaker |
| KIM Δpgm | Lcr− ΔPgm | This study |
| KUMA | Lcr− Pgm+ | R. R. Brubaker |
| EV76 | Pesticin-producing strain | R. R. Brubaker |
| Plasmids | ||
| pLAFR2 | Tcr Mob+ | 25 |
| 17A11 | pLAFR2 carrying a 23-kb insert of WA-C chromosomal DNA, Tcr | 44 |
| pBluescript KS II | Cloning vector, Apr | Stratagene |
| pKAS 32 | Cloning vector with rpsL gene | 55 |
| pKAS-1SKan | pKAS 32 containing irp1 EcoRI-2 fragment with a kanamycin cassette introduced into SalI site | This study |
| pKAS-E1Kan | pKAS 32 containing EcoRI/SalI fragment of irp1 with a kanamycin cassette (without transcriptional terminator) inserted into the EcoRV site | This study |
| pGP704 | Cloning vector, Apr | 39 |
| pGP-CAT | Chloramphenicol cassette inserted into PstI site of pGP704, Cmr | 46 |
| pGPIRP2 | 600-bp PCR fragment of irp2 inserted into pGP-CAT vector | This study |
| pUC-4K | pUC vector containing kanamycin cassette from Tn903, Apr Kanr | 60 |
| pSB 315 | Containing kanamycin cassette without transcriptional terminator, Apr Kanr | 27 |
| pACYC 184 | Cloning vector, Cmr | 11 |
| pCJG3.3N | pACYC 184 with fyuA-gfp mut3 | This study |
| pGFP mut3 | pKS with cDNA of the mutated GFP under the lac promoter | 12 |
| SuperCos1 | Cloning vector lacking SalI restriction site, Apr Neor | Stratagene |
WA-CS is a derivative of WA-C (Y. enterocolitica serotype O8 WA-314, plasmid cured). Spontaneous streptomycin-resistant (Smr) colonies of nalidix acid-resistant (Nalr) WA-C were isolated by increasing streptomycin concentrations in LB medium (with 10, 30, 50, 70, and 100 μg/ml). The resulting strain was designated WA-CS. Strain KIM Δpgm was isolated as a spontaneous mutant unable to accumulate Congo red dye on LB medium containing 50 μg of Congo red per ml.
DNA manipulation.
Bacterial chromosomal DNA was isolated by the method of Davis et al. (14). A gene bank was prepared from Y. enterocolitica WA-314 serotype O8. The chromosomal DNA was partially digested with Sau3A and ligated into the BamHI site of vector pLAFR2 (25).
Southern blot hybridizations (56) were performed with digoxigenin (DIG)-labeled PCR probes, using the following primers: P242 (′5-AAGGATTCGCTGTTACCGGAC-3′) and P505 (′5-ATTCGTCGGGCAGCGTTTCTTCT-3′) for the start of irp2, P4801 (′5-ATTGCCGATCTGGACCTC-3′) and P5206 (′5-ATCTGGATTGGCGACTGTAG-3′) for the end of irp2, i8513 (′5-TGAATCGCGGGTGTCTTATGC-3′) and i8730 (′5-TCCCTCAATAAAGCCCACGCT-3′) for irp1 P161 (′5-CAACATCGTCACCCAGCAG-3′) and P191 (′5-CGCAGTAGGCACGATGTTGTA-3′) for fyuA, and R299 (′5-TTTACAATTACACACCCTCAA-3′) and P732 (′5-CTGGGAGATGGGAAAAACTAC-3′) for IS1328, plus DIG-11-dUTP according to the Boehringer Mannheim Biochemica protocol.
DNA sequencing and sequence comparison.
The subcloned fragments EcoRI-2 and -3 from cosmid 17A11 (Fig. 1) were treated with exonuclease III (Nested Deletion kit; Pharmacia Biotech). Vector primers for templates generated by exonuclease digestion were used.
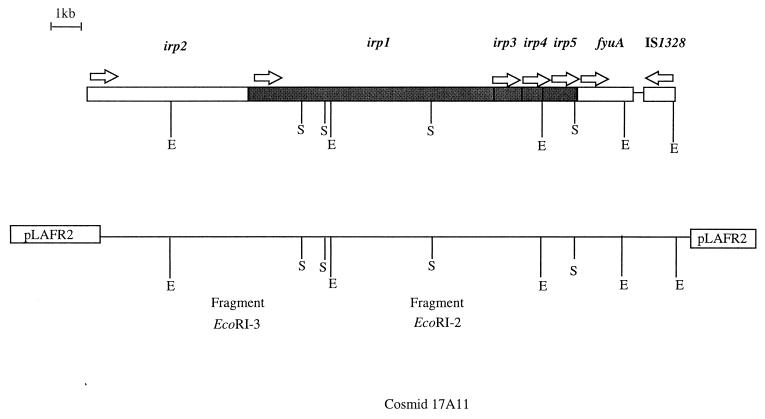
FIG. 1.
Genetic organization of the Y. enterocolitica O8 WA-314 irp2-fyuA gene cluster. The genes are depicted as boxes. Arrows above indicate the direction of transcription. E, EcoRI; S, SalI.
Sequence-specific oligonucleotides were synthesized for nonoverlapping regions and for the region downstream fragment EcoRI-2 and upstream fyuA (primer walking). DNA sequencing was performed by the chain-terminating method with model 373A and 377 DNA sequencers (ABI Prism; Perkin-Elmer). The sequences were analyzed and aligned with the HIBIO Mac DNASIS program (Hitachi Software Engineering Co.) and with the Genetics Computer Group sequence analysis software package (University of Wisconsin, Madison).
PCR conditions.
PCR amplifications were performed in an automated thermal cycler (TRIO Thermoblock; Biometra or GeneAmp PCR System 2400; Perkin-Elmer) as described by Saiki et al. (48) with TaqI polymerase and different pairs of oligonucleotides (Roth; Karlsruhe, Mannheim, Germany). The initial denaturation step (94°C, 7 min) was followed by 35 cycles of denaturing, annealing, and extension with one final extension step. Annealing and extension temperatures were set according to the primers used. PCR amplification products were separated in 1.6% agarose gels followed by purification with a QIAquick PCR purification kit or gel purified by using QIAquick gel extraction kit 250 (Qiagen GmbH, Hilden, Germany).
Comparison of the irp1 sequences of different strains was performed by using primers i965 (5′-CATCGACGACAGGCAGGTAGG-3′, bp 965 to 986) and i1233 (5′-CGGTATGGTAAAGGACTCTC-3′, bp 1233 to 1253) for the beginning and primers i8513 (5′-TGAATCGCGGGTGTCTTATGC-3′, bp 8513 to 8534) and i8730 (5′-TCCCTCAATAAAGCCCACGCT-3′, bp 8730 to 8751) for the end of irp1.
Construction of irp1 and ipr2 mutants.
The EcoRI-2 fragment from cosmid 17A11 (Fig. 1) was ligated into the EcoRI site of vector SuperCos1. Fragment EcoRI-2 harbors a SalI site in the open reading frame (ORF) of irp1. A kanamycin cassette containing a SalI fragment from plasmid pUC-4K was inserted into it. Fragment EcoRI-2 of irp1 with the kanamycin cassette was ligated into the pKAS 32 suicide vector (designated pKAS-1SKan). pKAS 32 contains the rpsL gene, which encodes the S12 protein of the ribosomes (55). Insertion of the suicide vector into the chromosome results in a Sms phenotype of a formerly Smr strain. Kanr (kanamycin resistance) Smr arose after an allelic exchange (double crossover) had taken place and the vector was lost. The construct was transformed into E. coli S17-1 λ pir+ tra+ (39, 54) followed by mobilization into WA-CS. Mutants were selected on agar plates containing kanamycin (40 μg/ml), streptomycin (100 μg/ml), and nalidixic acid (100 μg/ml), and the presence of the kanamycin cassette in irp1 was confirmed by Southern hybridization. To exclude a polar effect on the CAS phenotype, we created a second irp1 mutant by using a kanamycin cassette without transcriptional terminator. The EcoRI/SalI fragment of irp1 harboring an EcoRV cutting site (182 bases downstream of the EcoRI site) was inserted into the pKS vector, and the kanamycin cassette from pSB 315 cut by HindII was ligated into the EcoRV restriction site. The construct was excised with KpnI/SacI and inserted into the pKAS 32 suicide vector (resulting in pKAS-E1Kan) followed by mobilization and selection as described above.
Mutagenesis of irp2 was performed as described previously (46). Briefly, an internal PCR product of the irp2 gene from Y. enterocolitica O8 strain WA-C (primers UP irp2-sac1 [5′-CTCGAGCTCAAGGATTCGCTGTTACCGGAC-3′] and LP irp2-sac1 [5′-CTCGAGCTCTCGTCGGGCAGCGTTTCTTCT-3′]) was ligated into the SacI site of the suicide vector pGP-CAT and transformed into E. coli S17-1 λ pir+ tra+, generating pGPIRP2. The suicide hybrid plasmid pGPIRP2 was integrated into the irp2 gene of WA-C following conjugation and homologous recombination, giving rise to the Y. enterocolitica mutant strain WA-C irp2. The correct insertion of pGPIRP2 into the chromosomal DNA was confirmed by Southern hybridization.
FyuA-GFP reporter gene studies.
We translationally fused 267 amino acids (aa) of FyuA (including the upstream regulatory sequences and the putative YbtA binding site) and the product of the reporter gene gfp (encoding green fluorescent protein [GFP]) mut3 by using standard PCR cloning procedures and primers with designed restriction sites (HindIII-BamHI [FyuA] and BamHI-SalI [GFP mut3]). The resultant plasmid, pCJG3.3N, was transferred into WA-C and WA-CS irp1::Kanr by electroporation.
Flow cytometric measurements were performed with a Coulter Epics Flow cytometer. An argon 488-nm laser was used. Bacteria were detected by side scatter as described by Russo-Marie et al. (47). The scale was logarithmic, and fluorescence data and scatter data were collected for 50,000 bacteria.
Growth experiments: feeding assay with yersiniabactin containing culture supernatant, desferrioxamine, purified yersiniabactin, and pyochelin.
Y. enterocolitica mutant H1852 fur fyuA (siderophore hyperproducer) was cultivated aerobically in iron-deficient NBD medium for 12 h at 28°C (29). After centrifugation, the supernatant containing siderophore was sterilized by filtration and used for feeding experiments. Desferrioxamine (Desferal) was obtained from Ciby Geigy. Purified yersiniabactin and pyochelin preparations used for final confirming experiments were kindly provided by R. Reissbrodt (Wernigerode, Germany) and H. Budzikiewicz (Cologne, Germany).
Pesticin assay.
Pesticin-producing strain Y. pestis EV76 was grown overnight at 26°C, and pesticin production was induced by mitomycin C (0.3 μg ml−1) for an additional 16 h. Cells were collected by centrifugation, and the supernatant was used as a crude pesticin preparation after sterilization with 0.1% chloroform (35). Sensitivity to pesticin was monitored by serial dilution of the supernatant (1:2) on mid-log-phase bacterial cultures (106 microorganisms) in 0.6% LB agar used as an overlay (double-layer technique) with 1.2% LB agar as a support. Plates were incubated at 37°C for 18 h.
Screening for iron-chelating compounds.
Strains to be tested were plated on CAS agar (52) and incubated for 2 days at 26°C. A clearly visible red-orange halo around bacterial colonies was indicative of siderophore production (i.e., colonies were CAS positive).
SDS-PAGE and Western blotting.
The bacteria were cultured under iron-limiting conditions in NBD broth, centrifuged, washed, and solubilized by boiling in Laemmli buffer (total cell lysate) (38). Equal amounts of all strains (50 μg of protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 7.5% polyacrylamide gel at a constant current of 40 mA. The gel was stained with Roti-Blue (Roth) or electroblotted to nitrocellulose membranes (BA85; Schleicher & Schüll, Inc., Dasserl, Germany) as described previously (33). HMWP-specific antibodies were kindly provided by E. Carniel (Institut Pasteur, Paris, France). These antibodies had been obtained by using a purified HMWP fraction to immunize BALB/c mice. They specifically recognized the two HMWPs. Antibodies directed against one HMWP also recognize the other one (6).
Nucleotide sequence accession number.
The sequence determined was deposited at the EMBL/GenBank database under accession no. Y12527.
RESULTS
Cloning of irp1 and nucleotide sequence determination.
A Y. enterocolitica WA-C gene bank was screened for the presence of cosmids harboring the irp2-fyuA fragment by Southern hybridization. One of the cosmids, 17A11 with an insert of approximately 23 kb, hybridized with the irp2 terminal probe, fyuA, and the IS1328 element, indicating that this cosmid carries the DNA fragment covering the region between irp2 and fyuA. 17A11 was digested with EcoRI, resulting in six fragments of 23 (pLAFR2 vector portion), 8.2, 6.2, 3.3, 2.6, and 2.1 kb. The 3.3-kb EcoRI fragment hybridized with the fyuA probe, the 2.6-kb EcoRI fragment hybridized with the irp2 probe, and the 2.1-kb EcoRI fragment hybridized with the IS1328 probe. Double digestion with EcoRI and SalI revealed a physical restriction map of the 23-kb insert shown in Fig. 1. The 6.2-kb EcoRI fragment (designated fragment EcoRI-3) hybridized with the irp2 probe (bp 4581 to 5007 of irp2) downstream the EcoRI site. The 8.2-kb EcoRI fragment (designated fragment EcoRI-2) did not hybridize with any of the probes. It has been postulated that irp1 constitutes one operon with irp2 (8). We assumed that fragments EcoRI-2 and -3 comprise most likely the irp1 gene encoding HMWP1 with the size of 240 kDa. Therefore, both fragments were subcloned into the pKS vector and sequenced.
Fragment EcoRI-3 contains the terminal portion of irp2 downstream of the EcoRI site and the start of a new ORF 87 bp downstream the TAG stop codon of irp2 (Fig. 1). The residual portion of the new defined ORF is located on fragment EcoRI-2. Taken together, the data suggest that this new ORF, which is likely to be irp1, consists of 9,486 nucleotides, encoding a 3,161-aa polypeptide of 384.6 kDa. The ORF has the same transcriptional direction as irp2. No ORF of significant length was found on the complementary strand. The G+C content of the sequence is approximately 60 mol%, higher than the value of 47 to 50 for Yersinia (5) but similar to that for irp2.
A palindrome of 29 bases (boldface) capable of forming a secondary stem-loop structure is located between irp2 and irp1 (GGAACGCCATCGCGAACGCATGGCGTTCC). The palindrome starts 23 nucleotides after the stop codon of irp2 and lacks a poly(T) tail. Thus, it is unlikely that it is a transcriptional terminator. The GGA sequence located 6 bp upstream the first ATG codon of irp1 may serve as a weak ribosome-binding site. No promoter-operator structure could be identified upstream of irp1.
Another ORF, designated irp3, starts directly downstream irp1. The GGAG sequence 10 bp upstream the start codon can be considered a ribosome-binding site. irp3 consists of 1,098 nucleotides, encoding a 365-aa polypeptide of 40.7 kDa. irp3 has the same orientation as irp1 and irp2. The G+C content is around 60 mol%, which is in the same range as values for irp1 and irp2.
Two more ORFs, of 804 bp (designated irp4) and 1,578 bp (designated irp5), could be identified between irp3 and fyuA. These genes have the same transcriptional direction as irp2 and similar values for G+C content.
Taken together, data show that the irp gene cluster of Y. enterocolitica comprises five genes located upstream fyuA in the following order: irp2-irp1-irp3-irp4-irp5 (Fig. 1). irp1 to irp3, and irp3 and irp4, are contiguous; irp4 and irp5 are divided by three bases; irp2 and irp1 are separated by 87 bases. The gene order was confirmed by comparative Southern hybridization of cosmid 17A11 and WA-C chromosomal DNA digested with EcoRI and using PCR products corresponding to irp1 (bases 8513 to 8730) and irp2 (bases 4581 to 5007) as probes. Either probe hybridized to bands of the same size in digests of cosmid 17A11 and WA-C DNA (data not shown).
DNA and protein sequence homology.
To identify similarities of irp1 to irp5 and their deduced polypeptides to known sequences, a search in the EMBL gene data library was performed with the FastA program. The irp1 DNA sequence has highest identity to a cosmid from Mycobacterium tuberculosis (57.5% identity in a 1,049-bp overlap [bases 490 to 1530]; unpublished, accession no. Z83857), 53.1% identity over 971 bp (bp 187 to 1131) to the polyketide synthase gene of Bacillus subtilis W168 (53), 52.9% identity in a 2,763-bp overlap (bp 1 to 2678) to the eryA gene of Saccharopolyspora erythraea (17), 51.4% identity over 1,713 bp (bp 195 to 1866) to the polyketide synthase gene of Streptomyces antibioticus (58), 64.2% identity in 162 bp (bp 175 to 335) to the polyketide immunosuppressant gene of Streptomyces hygroscopicus (1, 40, 51), and 52.9% identity in a 1,481-bp overlap (bp 5716 to 7132) to irp2 of Y. enterocolitica (28). Interestingly, similarities to all of these related sequences are located within the first 3,000 nucleotides, whereas the following 6.3 kb show similarity only to irp2. A potential β-ketoacyl synthase active site could be identified in HMWP1 between aa 184 and 210. β-Ketoacyl-ACP (acyl carrier protein) synthase is the enzyme that catalyzes the condensation of malonyl-ACP with the growing fatty acid chain and is also found as a component in polyketide antibiotic synthases.
irp4 and irp5 were found to be 97.3 and 98.3% identical to ybtT (0.8 kb) and ybtE (1.5 kb), recently described for Y. pestis (3). Deduced proteins have 41.7% identity (Irp4) with a thioesterase-like protein located in the anguibactin biosynthetic gene cluster of Vibrio anguillarum (20) and 41% amino acid sequence identity (Irp5) with EntE, the 2,3-dihydroxybenzoic acid-activating enzyme utilized in the enterobactin biosynthetic pathway of E. coli (57). irp3 has no significant similarity to any known gene.
Amino acid sequence comparison of the polypeptides encoded by irp1 and irp2 showed three highly conserved motifs (Fig. 2). The presence of such motifs might be the reason for the cross-reactivity of HMWP1 and HMWP2 which was found even with monoclonal antibodies raised against the HMW proteins (37).
FIG. 2.
Common amino acids motifs found in HMWP1 and HMWP2.
Presence of irp1 in various Yersinia species and E. coli.
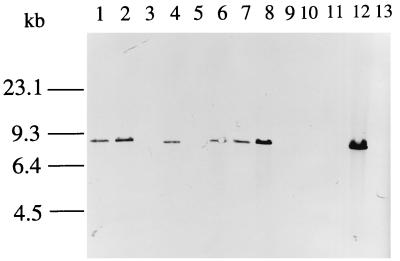
The presence of irp1 in various Yersinia and E. coli isolates was tested by Southern hybridization. The chromosomal DNAs of various strains (Table 2) were digested with EcoRI, and Southern hybridization was performed with an irp1 probe (corresponding to bp 7901 to 8139). As expected, irp2/fyuA-negative strains were also devoid of irp1. In irp2/fyuA-positive strains (Y. enterocolitica WA-CS and 8081, Y. pestis KIM and KUMA, Y. pseudotuberculosis PB1 and 346, and E. coli Phi), a band that hybridized with the irp1 probe was detected (Fig. 3).
TABLE 2.
Results of Southern hybridization with irp1, irp2, and fyuA probes
| Strain | irp1 | irp2 | fyuA |
|---|---|---|---|
| Y. enterocolitica | |||
| WA-C | + | + | + |
| 8081 | + | + | + |
| Y5.27 | − | − | − |
| Y-96-C | − | − | − |
| Y-108-C | − | − | − |
| Y. pseudotuberculosis | |||
| 346 | + | + | − |
| 201 | − | − | − |
| PB1 | + | + | + |
| Y. pestis | |||
| KIM | + | + | + |
| KUMA | + | + | + |
| KIM Δpgm | − | − | − |
| E. coli | |||
| DH5α | − | − | − |
| Phi | + | + | + |
FIG. 3.
Southern hybridization of chromosomal DNAs from Yersinia and E. coli strains with the irp1 probe. The chromosomal DNA was digested with EcoRI, and the resulting fragments were separated on a 1% agarose gel prior to Southern blotting. Hybridization was performed with a DIG-labeled PCR probe generated with primers i8513 and i8730. Lane 1, Y. pestis KUMA; lane 2, Y. pestis KIM; lane 3, Y. pestis KIM Δpgm; lane 4, Y. pseudotuberculosis 346; lane 5, Y. pseudotuberculosis 201; lane 6, Y. pseudotuberculosis PB1; lane 7, Y. enterocolitica 8081; lane 8, Y. enterocolitica WA-CS; lane 9, Y. enterocolitica Y5.27; lane 10, Y. enterocolitica Y-96-C; lane 11, Y. enterocolitica Y-108-C; lane 12, E. coli Phi; lane 13; E. coli DH5α.
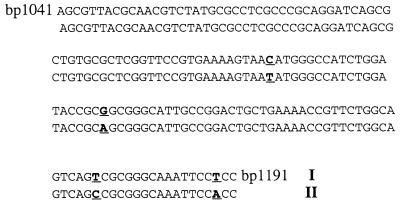
It was shown that irp2 is highly conserved between different Yersinia species (45). We analyzed the degree of variability of irp1 in the irp1-positive strains Y. enterocolitica WA-CS and 8081, Y. pestis KIM and KUMA, Y. pseudotuberculosis PB1 and 346, and E. coli Phi. PCR was performed for the start (bp 965 to 1254) and end (bp 7901 to 8139) portions of irp1; 150 bases of these amplicons were sequenced in both directions (bases 1041 to 1191 and 7948 to 8098). Comparison between these sequences and the irp1 sequence obtained for WA-C revealed 100% identity between all irp1-positive strains over bases 7948 to 8098. Four base substitutions in the amplicon (bp 1041 to 1191) were found between irp1 sequences of Y. enterocolitica (WA-C and 8081) and Y. pseudotuberculosis, Y. pestis and E. coli Phi (Fig. 4).
FIG. 4.
Comparison of the region from bp 1041 to 1191 of irp1 in Y. enterocolitica 8081, Y. enterocolitica WA-CS (I), Y. pestis KUMA, Y. pestis KIM, Y. pseudotuberculosis 346, Y. pseudotuberculosis PB1, and E. coli Phi (II). Nonmatching bases are boldfaced and underlined.
irp1- and irp2-encoded proteins are involved in yersiniabactin synthesis.
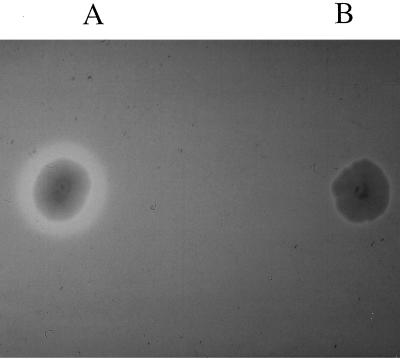
The possible relationship between the CAS phenotype and HMWP production was analyzed by mutagenesis of gene irp1. A kanamycin cassette was inserted into irp1 gene of WA-CS, and allelic exchange was performed. The resulting mutant, WA-CS irp1::Kanr, was tested on CAS agar and found to be CAS negative, indicating loss of yersiniabactin production (Fig. 5). The polar effect of the kanamycin cassette on the irp operon was ruled out by interruption of irp1 with a kanamycin cassette lacking a transcriptional terminator. This mutant was also unable to form a halo on CAS agar. The same result was achieved by inserting the total suicide vector pGPCAT carrying a fragment of irp2 (designated pGPIRP2) into the irp2 gene by homologous recombination (data not shown). These results demonstrate that mutants disrupted in either irp1 or irp2 lost the ability to synthesize the siderophore yersiniabactin.
FIG. 5.
CAS agar plate showing siderophore-producing Y. enterocolitica WA-CS (A; with halo) and mutant WA-CS irp1::Kanr (B).
Yersiniabactin-dependent expression of the irp operon.
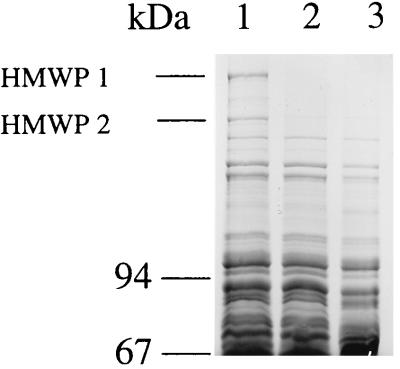
To evaluate the effect of the irp1 inactivation on the expression of HMWP1, a total-cell lysate of WA-CS and WA-CS irp1::Kanr grown under iron-poor conditions (NBD medium) was subjected to SDS-PAGE. HMWP1 could not be detected in the mutant strain, while HMWP2 was still visible as a very faint band after Roti-Blue staining (Fig. 6). The parent strain WA-CS expressed both HMWPs in comparable amounts. An additional weak protein band could be detected between HMWP1 and HMWP2. This band is thought to be a modified form of HMWP2 (5a) and reacts with the HMWP antibodies (Fig. 7). Production of HMWP1 and -2 was also decreased in an fyuA mutant impaired in its ability of ferric yersiniabactin uptake (Fig. 6). HMWP1 and HMWP2 were detected with anti-HMWP1 and anti-HMWP2 antisera in the wild-type strain but not in the mutant WA-CS irp1::Kanr (Fig. 7).
FIG. 6.
Expression of HMWP1 and -2 in WA-CS irp1::Kanr and WA fyuA mutants. SDS-PAGE (7.5% gel) of total-cell proteins from iron-starved strains WA-CS (lane 1), WA-CS irp1::Kanr (lane 2), and WA fyuA (lane 3).
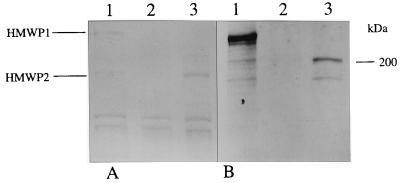
FIG. 7.
SDS-PAGE (7.5% gel) (A) and corresponding immunoblot (B) with cell lysates of WA-CS (lane 1), WA-CS irp1::Kanr (lane 2), and WA-CS irp1::Kanr with an addition of purified yersiniabactin (lane 3). The strains were grown under iron starvation in NBD medium. Western blotting was performed with HMWP polyclonal antibodies kindly provided from Elisabeth Carniel.
The pesticin sensitivity mediated by the FyuA receptor was not impaired by irp1 inactivation. The pesticin bactericidal titer was 1:512 for both strains, and no difference in the extent of sensitivity between the wild-type and mutant strains was detected.
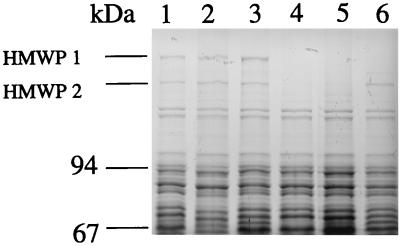
To test the possibility that inactivation of irp1 and subsequent failure in yersiniabactin production directly lead to downregulation of irp2 expression, WA-CS and WA-CS irp1::Kanr strains were grown in NBD iron-poor medium with and without addition of a culture supernatant containing yersiniabactin. A sublethal concentration of pesticin (1:1,024) was also added to bacteria grown in NBD to estimate its possible activating effect on yersiniabactin synthesis genes. Total-cell lysates were analyzed by SDS-PAGE (7.5% gel) (Fig. 8). Neither pesticin nor the supernatant had any significant effect on the expression of both HMW proteins in the wild-type strain. However, in contrast to pesticin, the yersiniabactin-containing supernatant restored expression of HMWP2 but not of HMWP1 in the mutant irp1 strain. The same results were obtained by the addition of purified yersiniabactin to the iron-deficient media (Fig. 7).
FIG. 8.
Effects of pesticin and yersiniabactin on expression of the HMWPs. SDS-PAGE (7.5% gel) of total-cell proteins of iron-starved strains WA-CS and WA-CS irp1::Kanr with addition of pesticin and culture supernatant containing yersiniabactin. Lane 1, WA-CS in NBD medium; lane 2, WA-CS in NBD medium with pesticin (1:1,024); lane 3, WA-CS in NBD medium with yersiniabactin supernatant (1:50); lane 4, WA-CS irp1::Kanr in NBD medium; lane 5, WA-CS irp1::Kanr in NBD medium with pesticin; lane 6, WA-CS irp1::Kanr in NBD medium with yersiniabactin supernatant.
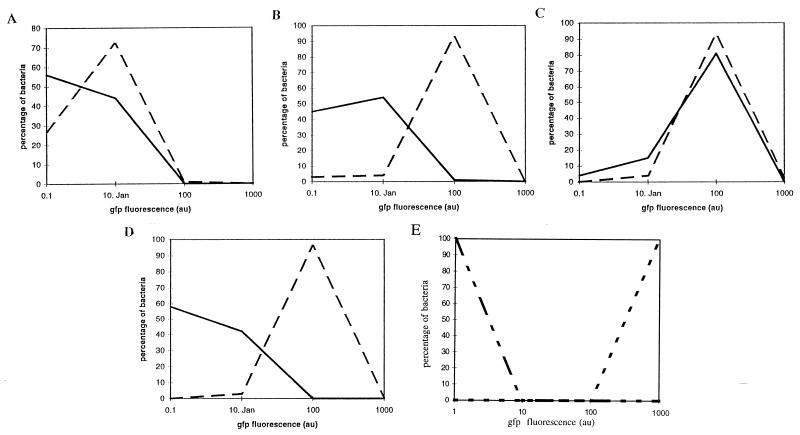
The effect of yersiniabactin on fyuA expression was examined in the irp1 mutant carrying a plasmid with fyuA translationally fused to the gfp reporter gene. The induction of FyuA-GFP leads to bright green fluorescent wild-type yersiniae in iron-deficient NBD medium. In contrast, the fluorescence of mutant WA-CS irp1::Kanr was much weaker. Fluorescent microscopy as well as FACSscan analysis revealed no increase in fluorescence when a sublethal dose of pesticin was supplied (data not shown), suggesting that pesticin does not act as an inducer of its receptor. In contrast, addition of purified yersiniabactin to the irp1 mutant results in a higher level of green fluorescence than in the wild type (Fig. 9). Addition of another siderophore (desferrioxamine B) or of a molecule structurally related to yersiniabactin (pyochelin) does not lead to an increase of the fyuA expression, indicating a specific induction of receptor expression by its own siderophore.
FIG. 9.
Fluorescence of FyuA-GFP in Y. enterocolitica WA-CS (- -) and WA-CS irp1::Kanr (—) in NB medium (A), NBD medium (B), NBD medium plus purified yersiniabactin (C), and NBD medium plus desferrioxamine B (D). (E) Positive (WA-CS[pGFP mut 3]; -- --) and negative (WA-CS; -- —— --) controls. Au, arbitrary units.
No siderophore synthesis could be detected in the irp1 mutant, but the cells were still able to grow in iron-deficient medium. The mutant strain grows in NBD medium with a final α-α-dipyridyl concentration of 400 μg/ml, indicating the presence of another iron uptake system in addition to the yersiniabactin system.
DISCUSSION
Since the rediscovery of siderophore production by highly pathogenic Yersinia species in 1987, it has been generally appreciated that this high-affinity ferric iron uptake system significantly contributes to virulence of yersiniae (31). Recently, the chemical structure of yersiniabactin has been determined (18). There was an indication that the putative genes for biosynthesis of yersiniabactin reside within the HPI on the chromosome (28). The objectives of this study were to demonstrate that the irp1 and irp2 genes, encoding HMWP1 and HMWP2, are involved in yersiniabactin biosynthesis and to characterize the irp operon. Two genes, irp2, encoding iron-repressible HMWP2, which is proposed to be involved in nonribosomal protein synthesis, and fyuA, encoding yersiniabactin/pesticin FyuA receptor, represent the yersiniabactin biosynthetic cluster. A small araC-like ybtA gene seems to be a positive regulator for irp gene expression (22) and precedes the irp operon.
In this study, we analyzed the whole irp operon and identified four additional ORFs on a 13-kb DNA fragment between the irp2 and fyuA genes. The largest one, located immediately downstream irp2, comprises 9.5 kb with the capacity to code for a 384-kDa polypeptide. The theoretical molecular mass of the polypeptide encoded by that ORF is higher than expected from the results of the SDS-PAGE (240 kDa). This can be due to the complex secondary structure of this extremely large protein as well as to its possible atypical migration in denaturing conditions. The insertional inactivation of irp1 by allelic exchange leads to loss of HMWP1 and HMWP2 in SDS-PAGE (Fig. 6).
Inactivation of irp1 or irp2 resulted in the loss of siderophore production as monitored on CAS agar (Fig. 5). Thus, irp1 and irp2 are involved in yersiniabactin biosynthesis.
HMWP1 shares a unique motif with the polyketide synthases. The β-ketoacyl-ACP synthase active site is highly conserved among the three multifunctional polypeptides of the rapamycin-producing polyketide synthases of S. hygroscopicus as well as in the polyketide synthase of S. antibioticus (1). Polyketide synthases are involved in the synthesis of a large and highly diverse group of heterocyclic compounds including antibiotics, antitumor compounds, and heterocyclic immunosuppressants. Polyketide metabolites are produced by successive condensation of simple acid units as propionate and acetate. Prolonging of the acid chain is catalyzed by the polyketide synthases (17). The similarity implies that HMWP1 could be involved in the synthesis of a siderophore or of an antibiotic.
The last 6.3 kb of irp1 have no significant similarity to any known sequence besides a 52.9% identity over 1,481 bp (bp 5716 to 7132) to irp2. HMWP2 is known to be homologous to AngR (involved in the anguibactin biosynthesis of V. anguillarum), and it was predicted to direct nonribosomal synthesis of small molecules involved in the nonribosomal synthesis of antibiotics or siderophores (28).
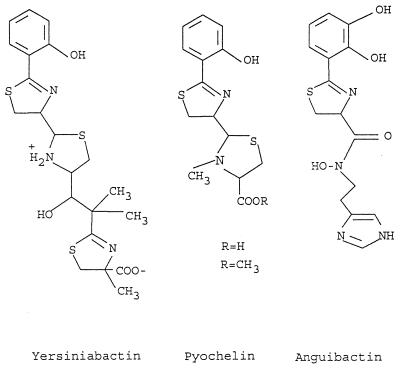
Two other defined ORFs, irp4 and irp5, have pronounced identity to a thioesterase-like protein from anguibactin biosynthetic gene cluster of V. anguillarum (20) and EntE (2,3-dihydroxybenzoic acid-activating enzyme) from E. coli (57), respectively. irp4 and irp5 are nearly identical to the ybtT and ybtE genes described as a part of a siderophore biosynthetic region in Y. pestis (3). Insertional inactivation of the ybtE gene yielded mutants unable to grow in iron-deficient medium at 37°C. A ybtE mutant could be cross-fed by a culture supernatant from a wild-type strain. It is reasonable to propose that yersiniabactin or a siderophore with a similar structure can represent this siderophore in Y. pestis. Taking into account structural similarities of yersiniabactin and anguibactin siderophores (Fig. 10), one can assume that irp2 to irp5 represent a yersiniabactin biosynthetic gene cluster with the following organization: irp2-irp1-irp3-irp4-irp5. An additional gene located between irp2 and irp1 as proposed for Y. pestis (3) could not be identified.
FIG. 10.
Chemical structures of yersiniabactin, pyochelin, and anguibactin.
Insertional inactivation of the irp1 and irp2 yersiniabactin biosynthetic genes results not only in elimination of the corresponding polypeptide bands but also in downregulation of the other proteins involved in yersiniabactin synthesis and binding; namely, irp1 inactivation was followed by considerable reduction of HMWP2 and FyuA proteins (Fig. 6 and 9). Nonsense mutation in the fyuA gene resulted in downregulation of the irp1 and irp2 genes. It was expected that inactivation of the irp2 gene, being the first gene in the polycistronic operon, would lead to a reduction of the irp1 gene product since irp1 is devoid of promoter/operator sequences. The unexpected reduction of HMWP2 and FyuA production as a result of irp1 inactivation suggests that the yersiniabactin biosynthetic operon is subjected to autoregulation by its product, the siderophore. Indeed, the addition of yersiniabactin to the siderophore-negative mutant WA-CS irp1::Kanr upregulates the production of HMWP2 (Fig. 7 and 8) and its receptor, as demonstrated by the translational fyuA-gfp fusion (Fig. 9). Consistent with these data, Y. pestis supernatant was also found to influence the expression of the ybt (yersiniabactin biosynthetic genes in Y. pestis)-encoded proteins (3).
Exogenous siderophore desferrioxamine B taken up by yersiniae through the FoxA receptor (2) did not induce fyuA. Thus, the yersiniabactin molecule specifically induces fyuA expression, while sublethal doses of neither pesticin, desferrioxamin B, nor the structurally related molecule pyochelin could serve as a signal for induction of the irp operon.
Siderophore-dependent expression of the cognate receptors was demonstrated for the iron dicitrate system in E. coli (61) and for pyoverdine, pyochelin, and enterobactin receptors in Pseudomonas aeruginosa (16, 34, 59). Moreover, phenolate siderophore pyochelin shows high structural similarity with yersiniabactin (Fig. 10). Several other features are common between yersiniabactin and pyochelin receptors. The presence of pyochelin, which exhibits a low affinity for iron in vitro, has been correlated with increased virulence and in vivo growth (13). The pyochelin receptor gene fptA is positively regulated by a pchR product, an AraC-type transcriptional regulator (34). On the basis of the similarity of ybtA to the araC-type regulators, it was proposed that the YbtA activator requires ferric yersiniabactin to interact with the palindrome sequences upstream psn (designation of the yersiniabactin receptor in Y. pestis with 99% similarity to fyuA) and irp2 for the maximum induction of these genes (22). Thus, the absence of yersiniabactin results in low expression of irp2 and fyuA genes in the irp1 mutant due to the inability of YbtA to form a complex with yersiniabactin, to bind to the palindrome sequences, and therefore to activate the yersiniabactin operon. Due to the lack of sequencing data of the pyochelin biosynthetic cluster, it is not possible to demonstrate the relationship of these two siderophore systems on the molecular level. Nevertheless, these three siderophore systems, yersiniabactin, pyochelin, and anguibactin, have a high degree of similarity in structure and function (Fig. 10).
Although it exhibited no siderophore production, the mutant WA-CS irp1::Kanr was still able to grow in iron-deficient medium. This indicates that at least one efficient iron uptake system is present in Y. enterocolitica in addition to the yersiniabactin system. The TonB-independent yfu system discovered recently in Y. enterocolitica (49) may be a candidate for such a system.
irp2 and fyuA genes present on the HPI were shown to be highly conserved (45). The same is true to the irp1 gene start and end portions, which were shown to be identical or highly similar in all three highly pathogenic species, Y. enterocolitica, Y. pseudotuberculosis O1, and Y. pestis. The irp4 and irp5 genes were also found to be nearly identical to the corresponding genes identified in Y. pestis KIM. Thus, ybtA-fyuA genes comprise a highly conserved gene cluster (HPI) present in highly pathogenic yersiniae. A high G+C content and a codon usage different from that in Yersinia housekeeping genes were found in all genes constituting the HPI. Therefore, a horizontal transfer may be responsible for the dissemination of the yersiniabactin biosynthetic operon in Yersinia. Moreover, recent studies have demonstrated that conserved HPI is widely distributed among representatives of certain pathotypes of E. coli (50). Such a wide dissemination of HPI and its impact on the virulence of Yersinia implies an important, possible multifunctional role of yersiniabactin system in vivo and the availability of an efficient mechanism for its genetic transfer.
ACKNOWLEDGMENTS
We thank Rolf Reissbrodt (Wernigerode, Germany), H. Budzikiewicz (Cologne, Germany), and Elisabeth Carniel (Paris, France) for kindly providing purified yersiniabactin, pyochelin, and the antibodies against the HMWPs, as well as Angelika Meier, Barbara Bögner, and Helmut Walter for excellent technical assistance. Furthermore, we are indebted to S. Aleksic and R. R. Brubaker for providing bacterial strains and Michael Hensel for helpful discussion.
REFERENCES
- 1.Aparicio J F, Molnar I, Schwecke T, Konig A, Haydock S F, Khaw L E, Staunton J, Leadlay P F. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 2.Baumler A, Koebnik R, Stojiljkovic I, Heesemann J, Braun V, Hantke K. Survey of newly characterized iron uptake systems of Yersinia enterocolitica. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:416–424. doi: 10.1016/s0934-8840(11)80858-3. [DOI] [PubMed] [Google Scholar]
- 3.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben Gurion R, Shafferman A. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid. 1981;5:183–187. doi: 10.1016/0147-619x(81)90019-6. [DOI] [PubMed] [Google Scholar]
- 5.Berkovier H, Mollaret H H. The family Enterobacteriaceae, genus Yersinia. In: Krieg N R, Holt J H, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 498–506. [Google Scholar]
- 5a.Carniel, E. Personal communication.
- 6.Carniel E, Antoine J C, Guiyoule A, Guiso N, Mollaret H H. Purification, location, and immunological characterization of the iron-regulated high-molecular-weight proteins of the highly pathogenic yersiniae. Infect Immun. 1989;57:540–545. doi: 10.1128/iai.57.2.540-545.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal high-pathogenicity island in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carniel E, Guiyoule A, Guilvout I, Mercereau Puijalon O. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol Microbiol. 1992;6:379–388. doi: 10.1111/j.1365-2958.1992.tb01481.x. [DOI] [PubMed] [Google Scholar]
- 9.Carniel E, Mazigh D, Mollaret H H. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter P B. Pathogenicity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang A A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 13.Cox C D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect Immun. 1982;36:17–23. doi: 10.1128/iai.36.1.17-23.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis L G, Dibner M D, Battley J F. Basic methods in molecular biology. New York, N.Y: Elsevier; 1990. [Google Scholar]
- 15.De Almeida A M, Guiyoule A, Guilvout I, Iteman I, Baranton G, Carniel E. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb Pathog. 1993;14:9–21. doi: 10.1006/mpat.1993.1002. [DOI] [PubMed] [Google Scholar]
- 16.Dean C R, Poole K. Expression of the ferric enterobactin receptor (PfeA) of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol Microbiol. 1993;8:1095–1103. doi: 10.1111/j.1365-2958.1993.tb01654.x. [DOI] [PubMed] [Google Scholar]
- 17.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 18.Drechsel H, Stephan H, Lotz R, Haag H, Zähner K, Hantke H, Jung G. Structure elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann Chem. 1995;1995:1727–1733. [Google Scholar]
- 19.Farber N M, Cantor C R. Accessibility and structure of ribosomal RNA monitored by slow tritium exchange of ribosomes. J Mol Biol. 1981;146:241–257. doi: 10.1016/0022-2836(81)90434-4. [DOI] [PubMed] [Google Scholar]
- 20.Farrell D H, Mikesell P, Actis L A, Crosa J H. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P22 cro and regulation by iron. Gene. 1990;86:45–51. doi: 10.1016/0378-1119(90)90112-5. [DOI] [PubMed] [Google Scholar]
- 21.Ferber D M, Brubaker R R. Plasmids in Yersinia pestis. Infect Immun. 1981;31:839–841. doi: 10.1128/iai.31.2.839-841.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 23.Fetherston J D, Perry R D. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol Microbiol. 1994;13:697–708. doi: 10.1111/j.1365-2958.1994.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 24.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 25.Friedmann A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 26.Galan J E, Ginnocchio C, Costeas P. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;17:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gemski P, Lazere J R, Casey T, Wohlhieter J A. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect Immun. 1980;28:1044–1047. doi: 10.1128/iai.28.3.1044-1047.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guilvout I, Mercereau Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haag H, Hantke K, Drechsel H, Stojiljkovic I, Jung G, Zähner H. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J Gen Microbiol. 1993;139:2159–2165. doi: 10.1099/00221287-139-9-2159. [DOI] [PubMed] [Google Scholar]
- 30.Hanahan D. Studies of transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 31.Heesemann J. Chromosomal-encoded siderophores are required for mouse virulence of entheropathogenic Yersinia species. FEMS Microbiol Lett. 1987;48:229–233. [Google Scholar]
- 32.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 33.Heesemann J, Gross U, Schmidt N, Laufs R. Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect Immun. 1986;54:561–567. doi: 10.1128/iai.54.2.561-567.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinrichs D E, Poole K. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol. 1996;178:2586–2592. doi: 10.1128/jb.178.9.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu P C, Yang G C, Brubaker R R. Specificity, induction, and absorption of pesticin. J Bacteriol. 1972;112:212–219. doi: 10.1128/jb.112.1.212-219.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson S, Burrows T W. The virulence enhancing effect of iron on non pigmented mutants of virulent strains of Pasteurella pestis. Br J Exp Pathol. 1956;37:577–583. [PMC free article] [PubMed] [Google Scholar]
- 37.Kooi C, Sokol P A. Characterization of monoclonal antibodies to Yersinia enterocolitica iron-regulated proteins. Can J Microbiol. 1995;41:562–571. doi: 10.1139/m95-075. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molnar I, Aparicio J F, Haydock S F, Khaw L E, Schwecke T, Konig A, Staunton J, Leadlay P F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 41.Podladchikova O N, Dikhanov G G, Rakin A V, Heesemann J. Nucleotide sequence and structural organization of Yersinia pestis insertion sequence IS100. FEMS Microbiol Lett. 1994;121:269–274. doi: 10.1111/j.1574-6968.1994.tb07111.x. [DOI] [PubMed] [Google Scholar]
- 42.Portnoy D A, Falkow S. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J Bacteriol. 1981;148:877–883. doi: 10.1128/jb.148.3.877-883.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rakin A, Heesemann J. Virulence-associated fyuA/irp2 gene cluster of Yersinia enterocolitica biotype 1B carries a novel insertion sequence IS1328. FEMS Microbiol Lett. 1995;129:287–292. doi: 10.1111/j.1574-6968.1995.tb07594.x. [DOI] [PubMed] [Google Scholar]
- 44.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Rakin A, Urbitsch P, Heesemann J. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J Bacteriol. 1995;177:2292–2298. doi: 10.1128/jb.177.9.2292-2298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roggenkamp A, Schubert S, Jacobi C A, Heesemann J. Dissection of the Yersinia enterocolitica virulence plasmid pYVO8 into an operating unit and virulence gene modules. FEMS Microbiol Lett. 1995;134:69–73. doi: 10.1111/j.1574-6968.1995.tb07916.x. [DOI] [PubMed] [Google Scholar]
- 47.Russo-Marie F, Roederer M, Sager B, Herzenberg L A, Kaiser D. β-Galactosiase activity in single differentiating bacterial cells. Proc Natl Acad Sci USA. 1993;90:8194–8198. doi: 10.1073/pnas.90.17.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 49.Saken E. Molekularbiologische Charakterisierung der Eisenassimilation in Yersinia enterocolitica. Ph.D. thesis. Wurzburg, Germany: Universität Würzburg; 1993. [Google Scholar]
- 50.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. Prevalence of the high pathogenicity island fyuA/irp of Yersinia among human pathogenic Escherichia coli. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 51.Schwecke T, Aparicio J F, Molnar I, Konig A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortes J, Lester J B, Böhm G A, Stauntonm J, Leadlay P F. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 53.Scotti C, Piatti M, Cuzzoni A, Perani P, Tognoni A, Grandi G, Galizzi A, Albertini A M. A Bacillus subtilis large ORF coding for a polypeptide highly similar to polyketide synthases. Gene. 1993;130:65–71. doi: 10.1016/0378-1119(93)90347-6. [DOI] [PubMed] [Google Scholar]
- 54.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1988;1:784–785. [Google Scholar]
- 55.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 56.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 57.Staab J F, Elkins M F, Earhart C F. Nucleotide sequence of the Escherichia coli ent E gene. FEMS Microbiol Lett. 1989;50:15–19. doi: 10.1016/0378-1097(89)90450-3. [DOI] [PubMed] [Google Scholar]
- 58.Swan D G, Rodriguez A M, Vilches C, Mendez C, Salas J A. Characterisation of a Streptomyces antibioticus gene encoding a type I polyketide synthase which has an unusual coding sequence. Mol Gen Genet. 1994;242:358–362. doi: 10.1007/BF00280426. [DOI] [PubMed] [Google Scholar]
- 59.Venturi V, Weisbeek P, Koster M. Gene regulation of siderophore mediated iron acquisition in Pseudomonas: not only the Fur repressor. Mol Microbiol. 1995;17:603–610. doi: 10.1111/j.1365-2958.1995.mmi_17040603.x. [DOI] [PubMed] [Google Scholar]
- 60.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 61.Zimmermann L, Hantke K, Braun V. Exogenous induction of the iron dicitrate transport system of Escherichia coli K-12. J Bacteriol. 1984;159:271–277. doi: 10.1128/jb.159.1.271-277.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]