Abstract
The nac gene product is a LysR regulatory protein required for nitrogen regulation of several operons from Klebsiella aerogenes and Escherichia coli. We used P22 challenge phage carrying the put control region from K. aerogenes to identify the nucleotide residues important for nitrogen assimilation control protein (NAC) binding in vivo. Mutations in an asymmetric 30-bp region prevented DNA binding by NAC. Gel retardation experiments confirmed that NAC specifically binds to this sequence in vitro, but NAC does not bind to the corresponding region from the put operon of Salmonella typhimurium, which is not regulated by NAC.
P22 challenge phages provide robust genetic tools for identifying nucleotides within a DNA site that play a critical role in specific DNA-protein interactions (reviewed in references 15 and 16). This approach is based upon the regulation of the lysis-lysogeny decision of phage P22 mnt::Kn9 arc-1605(Am). Under appropriate conditions, the decision between lysis and lysogeny is regulated by the P22 ant (antirepressor) gene product: expression of ant results in lytic growth of the phage and death of the cell, while repression of ant results in the survival of kanamycin-resistant (Kanr) lysogens. To construct a challenge phage, the operator that controls ant expression is replaced by a DNA fragment that contains a site recognized by a specific DNA-binding protein. This places ant expression under the control of the cognate DNA-binding protein. If the site is occupied by the DNA-binding protein, ant expression is repressed and the phage carrying the site can be recovered from the resulting Kanr lysogens. If a mutation in the site prevents the DNA-protein interaction, ant is expressed constitutively and the phage carrying the mutant site can be recovered from the resulting clear plaques. Thus, P22 challenge phage provide strong selections for identifying specific DNA sequences recognized by a DNA-binding protein and for mutations in the sequence that disrupt the DNA-protein interaction. In this study, we used challenge phage to characterize the DNA-binding site of the nitrogen assimilation control (NAC) protein from Klebsiella aerogenes.
NAC is a DNA-binding protein of the LysR family (reviewed in reference 1). NAC functions to couple the nitrogen-sensing mechanism of the nitrogen regulatory system (Ntr), which depends on RNA polymerase charged with ς54, to the expression of a variety of operons which are transcribed by RNA polymerase charged with ς70. When cells are starved for ammonium, the Ntr system activates nac expression and NAC accumulates and activates the expression of operons required for the catabolism of alternative nitrogen sources like proline (put), histidine (hut), and urea (ure). When ammonium is abundant, the Ntr system does not activate nac expression, so the NAC-dependent operons are expressed at low levels. Although the nac gene is present and functional in both K. aerogenes and Escherichia coli, nac is absent from Salmonella typhimurium (3). As a result, even if the NAC-binding site remains intact, there is no NAC-dependent nitrogen regulation in S. typhimurium unless NAC is supplied from another source.
The put genes of K. aerogenes, S. typhimurium, and many other bacteria allow growth on proline as a sole carbon and nitrogen source (4, 13, 26). Two genes are required for proline utilization: putP encodes a proline transport protein, and putA encodes a multifunctional enzyme that degrades proline to glutamate. In both K. aerogenes and S. typhimurium, expression of the put operon is derepressed by growth in the presence of exogenous proline and is subject to catabolite repression in the presence of glucose. However, nitrogen regulation of put differs in these closely related bacteria (4, 21). In K. aerogenes the put operon (putK) is activated in response to nitrogen starvation in a NAC-dependent manner. In contrast, in S. typhimurium the put operon (putS) is not activated in response to nitrogen starvation, even if a functional copy of the nac gene is provided in trans (3).
The differences in NAC regulation of the put operon could be due to differences in the DNA-binding sites for NAC in the put control regions of K. aerogenes and S. typhimurium. Therefore, we sought to answer the following questions: what is the DNA-binding site for NAC in putK and does putS lack this site? When these experiments were begun, the nature of the NAC-binding site was unknown, but we assumed that the NAC-binding site would be located within the putK regulatory region between the divergent putA and putP genes. Based upon this assumption, we used challenge phage to identify a small fragment from putK that contained a NAC-binding site. We then mutagenized this fragment to identify nucleotides that are essential for NAC binding in vivo. As expected, many of the nucleotides essential for NAC binding in putK are absent from the sequence of putS, explaining the failure of a functional nac gene to confer nitrogen regulation on putS.
MATERIALS AND METHODS
Bacterial strains and growth media.
The strains used in this study are listed in Table 1. Nutrient broth (NB) (Difco) with 0.5% NaCl was used for rich medium. When needed, antibiotics were added in the following concentrations: sodium ampicillin, 50 μg/ml; kanamycin sulfate (KAN), 40 μg/ml; spectinomycin (SPC), 100 μg/ml; and tetracycline HCl (TET), 10 μg/ml. Dioxane-free isopropyl-β-d-thiogalactopyranoside (IPTG) was added at the concentrations indicated. Phage lysates were prepared in phage broth, NB supplemented with 1× E salts and 0.2% glucose (6).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotypea | Reference(s) or source |
|---|---|---|
| Strain | ||
| S. typhimurium | ||
| MS1582 | leuA414(Am) Fels−supE40 ataA::(P22 sieA44 16-amH1455 tpfr49) | 9 |
| MST1762 | MS1868/pMS421 | P. Youderian |
| MS1868 | leuA414(Am) Fels−hsdSB | 9, 27 |
| MS1883 | leuA414(Am) Fels−hsdSB supE40 | 9 |
| TH564 | leuA414(Am) Fels−supE40 ataA::(P22 sieA44 Ap7 [Tn1] tpfr184 [mnt-al] 9−att+) | 12 |
| MST2778 | MS1868/pPC36 | This study |
| MST2779 | TH564/pPC36 | This study |
| MST2780 | TH564/pGW1700/pPC36 | This study |
| E. coli DH1 | thi-1 supE44 hsdR17(r− m+) endA1 recA1 relA gyr-96 | 11 |
| Plasmid | ||
| pEC205.1 | Ptac-nac+ (pBR322) | 24 |
| pGW1700 | Tetr AmprmucAB (pBR322) | 20 |
| pMS421 | Strr SpcrlacIq (pSC101) | 9 |
| pKC7 | KanrputK+ (K. aerogenes) | 4 |
| pKC9 | Ampr (subcloned pKC7 HindIII[283]-HindIII[621] into pTZ18U HindIII) | 4 |
| pPC6 | AmprputS+ (S. typhimurium) | 10 |
| pPC36 | Spcr Strrnac+ (pMS421) | This study |
| pPC37 | Tetr Ampr (subcloned pKC9 DraI[381]-SmaI[436] into pPY190 SmaI) | This study |
| pPC38 | Tetr Ampr (subcloned pKC9 SmaI[436]-DraI[381] into pPY190 SmaI) | This study |
| pPC39 | Tetr Ampr (subcloned pKC9 SmaI[436]-HincII[406] into pPY190 SmaI) | This study |
| pPC40 | Tetr Ampr (subcloned pKC9 HincII[406]-SmaI[436] into pPY190 SmaI) | This study |
| pPC41 | Ampr (subcloned pKC7 between putK oligonucleotide 1 and putK oligonucleotide 6 into pTZ18U SmaI) | This study |
| pPC42 | Ampr (subcloned pKC9 HindIII[283]-SmaI[436] into pTZ18U SmaI) | This study |
| pPC43 | Ampr (subcloned pKC9 SmaI[436]-HindIII[621] into pTZ18U SmaI) | This study |
| pPC44 | Ampr (subcloned pKC9 HindIII[283]-HincII[406] into pTZ18U SmaI) | This study |
| pPC45 | Ampr (subcloned pKC9 HincII[406]-HindIII[283] into pTZ18U SmaI) | This study |
| pPC46 | Ampr (subcloned pKC9 HindIII[283]-DraI[381] into pTZ18U SmaI) | This study |
| pPC47 | Ampr (subcloned pKC9 DraI[381]-HindIII[283] into pTZ18U SmaI) | This study |
| pPY190 | Ampr Tetr P22 mnt Pant SmaI-XmaI ant′ (pBR322) | 2 |
| pTZ18U | Ampr | 18 |
Genetic nomenclature for bacterial genes is as described in Sanderson and Roth (22). Numbers in brackets indicate the map distances in base pairs of the restriction sites from the transcriptional start site of the putP gene.
Plasmids.
The plasmids used in this study are listed in Table 1. The plasmid pPC41, which carries the entire putK regulatory region, was constructed by PCR amplification of the putK regulatory region from pKC7 and cloning of the amplified DNA (approximately 470 bp) into the HincII site on pTZ18U. PCR amplification was done as described by Ostrovsky de Spicer et al. (19). Plasmids carrying subfragments of the putK regulatory region were constructed by purifying restriction fragments from pPC41 by electroelution from 10% polyacrylamide gels (14) and then subcloning the DNA fragments into the multiple cloning site on plasmid pTZ18U.
Small fragments from the put regulatory region were subsequently subcloned into the SmaI site on pPY190 for construction of challenge phage. pPY190 is a derivative of pBR322 that carries a Pant-arc-ant′ fragment from P22 with a unique SmaI site substituted for the mnt operator between Pant and arc (2). The DNA sequences of potential clones were determined to confirm the identity of each of the cloned fragments and the orientation of the inserts.
The K. aerogenes nac gene was subcloned into pMS421 to construct a nac expression vector for the challenge phage experiments. A 2.8-kb HindIII fragment from plasmid pEC205.1 (24) containing a Tn5 tac-nac fusion (5) was eluted from a 0.8% agarose gel and subcloned into the HindIII site of pMS421.
Construction of challenge phage.
Challenge phage with potential nac-binding sites were isolated by in vivo recombination between the pPY190 clones carrying putK DNA fragments and P22 mnt::Kn9 arc-1605(Am) phage (15, 16). S. typhimurium strains carrying the pPY190 derivatives were grown overnight in NB containing TET (NB + TET), and then 0.1 ml of each overnight culture was infected with 0.1 ml of the phage (1010 PFU/ml), 5 ml of phage broth was added, and the cultures were incubated at 37°C. After 3 h the cultures were centrifuged, and the supernatant was collected and treated with chloroform. The resulting lysates were plated on strain MS1582. MS1582 contains an mnt+ prophage that prevents the parental P22 mnt::Kn9 Omnt+ arc-1605(Am) phage from growing lytically, so only recombinants or Omnt mutants form plaques on MS1582. The clear plaques were purified, and the phage DNA was isolated from the concentrated lysate as described by Silhavy et al. (25). The desired recombinants were distinguished from parental phage by a restriction fragment linked polymorphism which results from the substitution. Restriction fragment length poly-morphism mapping was done on PCR fragments from the Omnt region (15), and the DNA sequence of this region was determined for each of the recombinants.
Challenge phage assays.
Challenge phage assays were done as previously described (15, 16). Strain MST2778 was grown to mid-exponential phase in NB + SPC, and then 1-ml aliquots were subcultured into flasks containing 4 ml of NB + SPC with different concentrations of IPTG to induce NAC expression. After being incubated for 1 h at 37°C, the cultures were infected with challenge phage at a multiplicity of infection of 25. Phage were allowed to adsorb to the cells for 1 h at room temperature, and then serial dilutions of the infected cells were plated on NB + SPC + KAN plates containing the same concentration of IPTG used for preinduction. The plates were incubated at 37°C for 3 days, and then the number of Kanr lysogens was counted.
Isolation of nac challenge phage mutants.
Challenge phage with mutations in the NAC-binding site were obtained either (i) by error-prone repair after UV irradiation in a strain which expresses the mucAB gene products from the plasmid pGW1700 (20) or (ii) by isolating the plasmid from a mutS strain (15). MST2780 [pPC36 (Ptac nac) and pGW1700 (mucA+B+)] was grown to mid-exponential phase in NB + SPC + TET and then diluted 1:3 into NB + SPC + TET + IPTG (0.8 mM final concentration of IPTG). The putK challenge phage (109 PFU/ml) were irradiated with 12,000 μJ/cm2 of UV light. The UV-irradiated phage were then diluted 10−1-, 10−2-, and 10−3-fold, and 100 μl of each dilution was added to 200 μl of the MST2780 culture. After phage adsorption, 3 ml of top agar was added, and the mixtures were plated on NB + SPC + 0.8 mM IPTG medium and incubated at 37°C. Mutant phage that formed clear plaques were picked and repurified.
PCR amplification of the Pant region from challenge phage.
The Pant region from each challenge phage was amplified by PCR, and the resulting DNA fragments were used for both gel retardation assays (symmetric PCR products) and DNA sequence analysis (asymmetric PCR products). Approximately 500 ng of phage DNA was used for each PCR. The two primers used for symmetric PCR were the Omnt primer (5′-CGGCATTTTGCTCATTCC-3′), complementary to the sequence upstream of the operator region of the mnt gene, and the anti-Omnt primer (5′-GATCATCTCTAGCCATGC-3′), complementary to the sequence upstream of the −35 region of Pant on the opposite strand (2). For asymmetric PCR, 5 μl of amplified double-stranded DNA from the symmetric reaction was further amplified by using only one primer (17).
Gel retardation assays.
The K. aerogenes NAC protein was overproduced from plasmid pEC205.1 and purified as previously described (8, 24). Stock solutions containing purified NAC in storage buffer (50% glycerol, 125 mM NaCl, 50 mM NaH2PO4 [pH 7.0], 1.25 mM MgCl2, 0.5 mM 2-mercaptoethanol) were diluted in the same buffer supplemented with bovine serum albumin to a concentration of 1.0 mg/ml prior to use in gel retardation assays. To assay the relative affinities of NAC for various PCR-amplified DNA fragments, a 215-bp DNA fragment carrying a portion of the wild-type putK control region, generated by digestion of plasmid pPC47 with HincII and BamHI, was used as an internal standard. Gel retardation assays were performed essentially as follows (7). One-microliter aliquots of storage buffer with either bovine serum albumin or NAC were added to a 9-μl solution containing 0.1 pmol of digested pPC47 DNA plus 0.1 pmol of PCR-amplified experimental DNA. The protein-DNA solutions were incubated for 5 min at room temperature prior to the addition of 1.5 μl of gel-loading solution (40 mM Tris HCl [pH 8.4], 4 mM EDTA, 0.2% bromophenol blue, 0.2% xylene cyanol, and 25% glycerol), loaded onto a 4% polyacrylamide gel buffered with Tris-EDTA (pH 8.4), and subjected to electrophoresis at 200 volts for 2 h at 4°C. The extent of migration of the DNA fragments through the gel was determined by the UV fluorescence of ethidium bromide-stained gels.
DNA sequencing.
The DNA sequences of nac challenge phage were determined from the single-stranded DNA obtained from asymmetric PCRs. The PCR products were purified with Geneclean (Bio 101, La Jolla, Calif.), and the purified DNA was resuspended in 20 μl of H2O. The DNA sequence was then determined by dideoxy sequencing by using Sequenase (United States Biochemicals, Cleveland, Ohio).
RESULTS
NAC binds to specific fragments of the K. aerogenes put regulatory region in vivo.
To characterize NAC binding to the putK regulatory region in vivo, we used P22 challenge phage as a reporter for the protein-DNA interaction (2, 15, 16). To construct NAC challenge phage, the Omnt site which regulates ant expression was replaced by a fragment expected to contain the NAC-binding site from putK. Phage which carry the NAC-binding site place ant expression under the control of NAC. Thus, if the substituted operator site is occupied by NAC protein, transcription of the ant gene will be repressed, and the challenge phage will form Kanr lysogens. The frequency of lysogeny is determined by the relative affinity of NAC for the DNA-binding site in vivo.
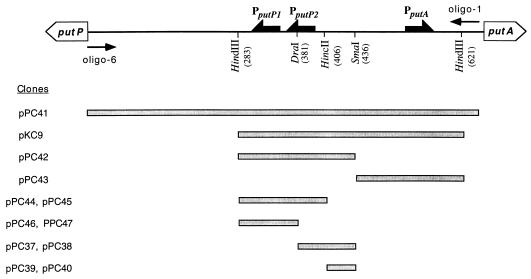
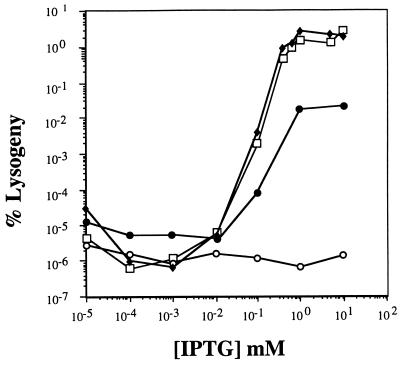
Because the precise NAC-binding site within the putK DNA fragment was not known, we blindly tested challenge phage with restriction fragments from the putK regulatory region derived from the plasmids shown in Fig. 1. Two of the restriction fragments showed positive results: pPC37 and pPC38 contain a 55-bp SmaI-DraI fragment in opposite orientations, and pPC39 and pPC40 contain a 30-bp SmaI-HincII fragment in opposite orientations. These plasmids were crossed onto P22 mnt::Kn9 arc(Am) phage to construct the challenge phages SD2-1, SD1-1, SH4-1, and SH26-10 (Fig. 2). NAC binding to these challenge phages was tested by infection of MST2778 cultures that were preinduced with varying concentrations of IPTG to express different amounts of NAC (Fig. 3). When strain MST2778 was induced by a low concentration (0.01 mM) of IPTG, and infected by phages SD2-1 and SH4-1, the frequency of lysogeny was less than 10−5%. As the IPTG concentration was increased to 0.1 mM, the frequency of lysogeny increased in proportion to the IPTG concentration. The maximal frequency of lysogeny reached 1% when the cells were induced with 0.8 mM IPTG. As a negative control, strain MST1762 (which is isogenic to MST2778 except that the plasmid pMS421 lacks the nac gene) was tested under identical conditions. The frequency of lysogeny in this NAC− strain was less than 10−7% under each of the conditions, indicating nac expression from plasmid pPC36 was required for efficient lysogeny of the challenge phage.
FIG. 1.
Physical map of the K. aerogenes put regulatory region and subcloned DNA fragments. The positions of relevant restriction sites are indicated with the distances in base pairs from the putP transcriptional start site shown in parentheses. Subcloned regions used for construction of challenge phage are shown as shaded bars directly below the corresponding region of the restriction map.
FIG. 2.
DNA sequences of the K. aerogenes put regulatory region and challenge phages. The wild-type K. aerogenes put regulatory region and DNA sequences from the wild-type K. aerogenes put regulatory region present in challenge phages SD1-1, SD2-1, SH4-1, and SH26-10 are shown. The sequence of the put DNA is shown in capital letters. The adjacent sequences in the challenge phage are shown in lowercase letters. Note that the substitution is inverted in challenge phage SD2-1 versus SD1-1 and SH4-1 versus SH26-10. Underlined nucleotides represent the −10 and −35 region of Pant.
FIG. 3.
Results of challenge phage assays with wild-type SH2-1 and SH4-1 and mutant derivatives of SH4-1 shown as follows: wild-type SH2-1, ⧫; wild-type SH4-1, □; 6G mutant, •; 8C mutant, ○. The percent lysogeny of the negative control which lacks the nac gene (MST1762) was less than 10−7 at all IPTG concentrations tested. The percent lysogeny indicates the number of Kanr lysogens formed per viable cell infected. NAC is expressed from the Ptac promoter under the control of LacIq, and thus the intracellular concentration of NAC increases as the concentration of IPTG is increased.
Phage SD1-1 carries the same insert as phage SD2-1 but in the opposite orientation. In contrast to phage SD2-1, when phage SD1-1 was used no lysogens were obtained at any of the IPTG concentrations. An explanation for this result could be that the binding site for NAC is close to one end of the inserted sequence: when the fragment is oriented as in SD2-1 the binding site is sufficiently close to the −10 region of the ant promoter that NAC binding occludes RNA polymerase binding to Pant and thus represses ant transcription; in contrast, when the fragment is oriented as in SD1-1 the binding site is too far from the ant promoter to interfer with RNA polymerase binding. Although the insert in SH26-10 is in the same orientation as SD1-1, the smaller size of the insert in SH26-10 would place the NAC-binding site close to the ant promoter. Nevertheless, because NAC efficiently represses ant transcription in phages SH4-1, SD2-1, and SH26-10, the region between the SmaI and HincII sites must contain the NAC-binding site.
NAC binds to the put regulatory region from K. aerogenes but not S. typhimurium in vitro.
Gel retardation assays with purified NAC protein confirmed that NAC binds to a 335-bp HindIII fragment from pKC7 which contains most of the putK control region (Fig. 1). Gel retardation assays with restriction digests of subclones derived from pKC7 indicated that a 30-bp HincII-SmaI fragment is necessary and sufficient for the interaction between NAC and the putK control region in vitro. In contrast, NAC did not retard a DNA fragment containing the corresponding region from the S. typhimurium put operon (data not shown).
Isolation of challenge phage mutants that prevent NAC binding in vivo.
To identify the nucleotide residues recognized by NAC, we isolated challenge phage mutants defective for NAC binding in vivo. In order to obtain a wide variety of mutations, the phage were irradiated with UV light and then grown in a strain carrying plasmid pGW1700, which enhances error-prone repair. Upon infection of cells which express high levels of NAC, the parent challenge phage formed lysogens, but challenge phage with mutations that prevent NAC binding grew lytically. The typical frequency of clear plaque mutations was approximately 10−5 per PFU of infecting phage. The clear plaques were purified and screened for challenge phage with mutations in the NAC-binding site.
Seventy-two clear plaque mutants were isolated from phage SH4-1, and the DNA sequences of the Pant region on the mutant phage were determined (Table 2). The majority of the mutants had single base pair substitutions. Two of the mutations changed the −10 region of Pant and probably increased the transcription of the ant gene by improving the promoter (27).
TABLE 2.
Challenge phage mutations that decrease NAC binding
| Mutation(s) | No. iso- lateda | DNA sequenceb | In vitro DNA bindingc |
|---|---|---|---|
| Wild type | GGGCTCATAG GGTATTTGTA TGCGTGAGTT | + | |
| 5A | 2 | GGGCACATAG GGTATTTGTA TGCGTGAGTT | ± |
| 6T | 3 | GGGCTTATAG GGTATTTGTA TGCGTGAGTT | ± |
| 6G | 2 | GGGCTGATAG GGTATTTGTA TGCGTGAGTT | ± |
| 7G | 1 | GGGCTCGTAG GGTATTTGTA TGCGTGAGTT | ND |
| 7T | 1 | GGGCTCTTAG GGTATTTGTA TGCGTGAGTT | − |
| 8C | 6 | GGGCTCACAG GGTATTTGTA TGCGTGAGTT | − |
| 8A | 10 | GGGCTCAAAG GGTATTTGTA TGCGTGAGTT | − |
| 8G | 1 | GGGCTCAGAG GGTATTTGTA TGCGTGAGTT | − |
| 9G | 8 | GGGCTCATGG GGTATTTGTA TGCGTGAGTT | − |
| 11A | 2 | GGGCTCATAG AGTATTTGTA TGCGTGAGTT | ± |
| 12T | 1 | GGGCTCATAG GTTATTTGTA TGCGTGAGTT | ± |
| 14G | 2 | GGGCTCATAG GGTGTTTGTA TGCGTGAGTT | ND |
| 15C | 2 | GGGCTCATAG GGTACTTGTA TGCGTGAGTT | ± |
| 16G | 1 | GGGCTCATAG GGTATGTGTA TGCGTGAGTT | ± |
| 19C | 1 | GGGCTCATAG GGTATTTGCA TGCGTGAGTT | ND |
| 19A | 2 | GGGCTCATAG GGTATTTGAA TGCGTGAGTT | ± |
| 20T | 4 | GGGCTCATAG GGTATTTGTT TGCGTGAGTT | − |
| 20G | 2 | GGGCTCATAG GGTATTTGTG TGCGTGAGTT | ND |
| 21G | 1 | GGGCTCATAG GGTATTTGTA GGCGTGAGTT | − |
| 21A | 2 | GGGCTCATAG GGTATTTGTA AGCGTGAGTT | ± |
| 35C | 3 | GGGCTCATAG GGTATTTGTA TGCGCGAGTT | ± |
| 12Δ | 1 | GGGCTCATAG G-TATTTGTA TGCGTGAGTT | − |
| 13Δ | 1 | GGGCTCATAG GG-ATTTGTA TGCGTGAGTT | ND |
| 19Δ | 1 | GGGCTCATAG GGTATTTG-A TGCGTGAGTT | − |
| 8C, 9Δ | 1 | GGGCTCAC-G GGTATTTGTA TGCGTGAGTT | ND |
| 9Δ, 14T | 1 | GGGCTCAT-G GGTTTTTGTA TGCGTGAGTT | − |
| 13A, 14G, 15T | 1 | GGGCTCATAG GGAGTTTTGT ATGCGTGAGTT | ND |
| 19A, 20T | 1 | GGGCTCATAG GGTATTTGAT TGCGTGAGTT | ND |
| 19G, 20T | 1 | GGGCTCATAG GGTATTTGGT TGCGTGAGTT | ND |
Number of independent mutants with the same nucleotide changes.
Nucleotide changes are shown in bold and underlined.
In vitro binding indicates Relative binding affinity of NAC based upon gel retardation assays as shown in Figure 4. +, efficient binding; ±, weak binding; −, no binding detected. ND, not determined.
Fifty-seven of the mutants had single base pair substitutions within the putK insert (Table 2). Challenge phage assays were performed for each of these mutants to determine the relative affinity of NAC for the mutant site in vivo. Compared to the parent phage SH4-1, the efficiency of lysogeny was severely decreased for each of the mutants. The frequency of lysogeny of these mutant challenge phage did not increase even when the IPTG concentration was increased to 5 mM, indicating that the mutations decrease the binding affinity so severely that high concentrations of NAC cannot overcome the mutant phenotype in vivo. These results indicate that the mutations affect nucleotides that are essential for the NAC-DNA interaction in vivo.
Another class of mutants had single base pair deletions or multiple nucleotide changes (Table 2). Most of the mutations that changed multiple nucleotides altered the same sequences in the putK fragment as the single base mutations. As expected, the in vivo binding assays for each of these mutants revealed that the NAC-DNA interaction was severely defective. Typical results are shown in Fig. 3.
Challenge phage mutants differ in their ability to interact with NAC in vitro.
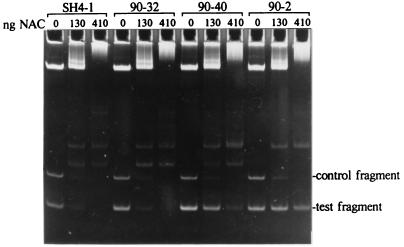
Gel retardation assays were done to determine the influence of the challenge phage mutations on the interaction of NAC with wild-type and mutant DNA sites in vitro. Typical results are shown in Fig. 4. In the presence of either limiting or excess NAC, the interaction between NAC and the 150-bp SH4-1 fragment amplified from a wild-type putK challenge phage was similar to that observed with the 215-bp HincII-BamHI control fragment carried by pPC47. Likewise, NAC binding to a derivative of SH4-1 with a Pant promoter mutation (90-32) was indistinguishable from binding to the wild-type sequence. In contrast, the interactions of NAC with fragments amplified from the mutant, NAC-insensitive challenge phages were either moderately reduced (e.g., 90-40 [6G mutation]) or severely reduced (e.g., 90-2 [8C mutation]) relative to the wild-type control. The gel retardation results are summarized in Table 2.
FIG. 4.
Gel retardation assay of the interactions between NAC and DNA fragments derived from the putAP control region. The indicated amounts of NAC were incubated with a mixture of digested pPC47 and DNA amplified from challenge phages SH4-1 (wild-type), 90-32 (Pant promoter mutation), 90-40 (6G mutation), and 90-2 (8C mutation). Digestion of pPC47 with HincII and BamHI yielded a 215-bp control fragment containing nucleotides 406 to 621 of the putAP control region and a larger fragment containing the vector pTZ18U plus nucleotides 381 to 405 of the putAP control region. The migration position of the unretarded 215-bp control and 150-bp test fragments are indicated at the right.
DISCUSSION
Expression of the put operon is regulated by nitrogen starvation in K. aerogenes but not S. typhimurium. Previous studies suggested that this lack of nitrogen regulation in S. typhimurium is due to the absence of both the trans-acting factor (NAC) and a cis-acting site in the put regulatory region (3, 4). To understand how NAC regulates expression of the put operon in K. aerogenes, we used challenge phage to genetically identify the DNA sequence from the K. aerogenes put regulatory region which is critical for NAC binding. The specific nucleotides required for NAC binding were then identified by isolating base substitution mutants that prevent NAC binding in vivo. Binding of NAC to the K. aerogenes put regulatory region was also assayed by gel retardation in vitro. Purified NAC protein specifically retarded DNA from the K. aerogenes put control region in vitro but not from the put control region from S. typhimurium.
Within the K. aerogenes put control region, NAC recognized a 55-bp SmaI-DraI fragment. The SmaI-DraI fragment was cloned onto the challenge phage in both orientations. Kanr lysogens were only obtained when the SmaI site was closest to the ant promoter (challenge phage SH2-1), indicating that NAC binds at or near this SmaI site, and interaction is strong enough to interfere with RNA polymerase binding to Pant. To narrow down the position of the NAC-binding site, the 30-bp HincII-SmaI half of this fragment was used to construct challenge phage SH4-1 and was tested for NAC binding in vivo. This 30-bp fragment alone was sufficient for efficient NAC binding.
To identify the specific nucleotides critical for NAC binding, we selected for mutations on challenge phage SH4-1 that prevented NAC binding. Fifty-seven mutants were isolated with single base substitutions in the put insert that decreased the NAC-binding affinity in vivo (Table 2). These mutations identified base substitutions in 15 different nucleotides within the DNA-binding site. Each of these mutant DNAs decreased the maximal frequency of Kanr lysogens to 1% of that of the wild-type putK challenge phage. Several of the mutants (5A, 6T, 6G, 11A, 15C, 19A, and 25C mutations) allowed weak NAC binding, producing tiny Kanr colonies at high concentrations of IPTG. NAC binding to the rest of the mutants was severely disrupted, and hence no Kanr lysogens were formed at any concentration of IPTG.
The relative binding of NAC to each of the mutant binding sites was measured by challenge phage assays in vivo (Fig. 3) and gel retardation assays in vitro (Fig. 4). Based on the results, the mutants fell into two groups (Table 2). One group of mutants severely disrupted NAC binding in vivo, and thus no Kanr lysogens were formed. Gel retardation assays confirmed that NAC did not bind to these mutants in vitro. The second group of mutants decreased NAC binding, but Kanr lysogens were obtained at a low frequency. Gel retardation assays confirmed that NAC could bind to these mutant sites but a higher concentration of NAC was required than for binding to the wild-type site. By comparing the in vivo and in vitro results, we concluded that the six nucleotides ATAN10ATN3T are critical determinants for the interaction between NAC and DNA. Base substitutions at any of these nucleotides decreased NAC binding both in vivo and in vitro. Substitutions at five additional positions only decreased NAC binding in vivo, and thus these residues may play a role in the NAC-DNA interaction. Including all of these nucleotides would extend the NAC-binding site to CATANGNTNTN3TNTN3T.
Differences between the frequencies of lysogeny of challenge phages SD2-1 and SH4-1 suggested that the flanking sequence between HincII and DraI might facilitate the interaction of NAC with the operator site in vivo. If this region plays an important role in NAC binding, we would expect that mutations in the critical nucleotides would decrease NAC binding. Therefore, we also selected for mutations that decrease NAC binding to phage SH2-1. Fifty-one challenge phage SD2-1 mutants were isolated that decreased NAC binding in vivo. All of these mutations were located within the SmaI-HincII fragment present on SH4-1. Furthermore, each of the single base substitutions obtained from challenge phage SD2-1 had also been obtained from phage SH4-1. The observation that none of these mutations were located within the DraI-HincII region suggests that these sequences do not directly interact with NAC.
The NAC-binding site in putK is between the divergent putA and putP1 promoters, which are separated by about 125 bp (4). The site is located about 20 bp upstream of the putP1 promoter, overlapping a putative cyclic AMP receptor protein-binding site, and about 90 bp upstream of the putA promoter. Previous primer extension analysis showed transcription from the putP1 promoter increases during nitrogen starvation (4). Only two of the seven critical nucleotides in the NAC-binding site are present in the put control region from S. typhimurium (CATCTGGATTTATTTTCCTCTGCGGTAGTT). This DNA sequence divergence explains why the put operon from S. typhimurium is not activated by nitrogen starvation even when NAC is provided in trans.
In the accompanying paper by Pomposiello, Janes, and Bender (20a), the NAC-binding site in the hutU promoter region from K. aerogenes was mutagenized by using synthetic oligonucleotides. The results of that study also define nucleotides required for binding and transcriptional activation by NAC. The site-directed mutants that affect NAC binding to the PhutU site are very analogous to the challenge phage mutants that affect NAC binding to the Pput site. The nucleotides identified as very important (the mutation of which results in a severe defect in NAC binding and activation) and the nucleotides that are moderately important (mutation results in a less severe defect) are similar in both the put and hut sites. An alignment of three sites known to bind NAC and to activate transcription (from PhutU, Pput, and PureD) yields similar consensus patterns of nucleotides (Table 3).
TABLE 3.
Sequence alignment of NAC-binding sites
| Nucleotide importancea | Gene(s) | Nucleotide sequence |
|---|---|---|
| Very important | putP | .ATA..........AT...T |
| hutU | .ATAA.........AT.... | |
| Moderately important | putP | CATA.G.T.T...TAT...T |
| hutU | TATAA.A.....GTAT.... | |
| Consensus | putP, hutU, ureD | .ATA......T.GTAT.... |
Nucleotides indicated as very important or moderately important reduce NAC binding severely or moderately, respectively, if mutated. The consensus sequence is for the 20-bp region from each of the NAC-binding sites of the indicated genes.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants GM34715 (S.M.) and GM47156 (R.B.) from the National Institute of General Medical Sciences.
We thank Kelly Hughes, Graham Walker, and Phil Youderian for generously providing some of the strains used in this study and Jeff Gardner for helpful comments on the manuscript.
REFERENCES
- 1.Bender R A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 2.Benson N, Sugiono P, Bass S, Mendelman L, Youderian P. General selection for specific DNA-binding activities. Genetics. 1986;114:1–14. doi: 10.1093/genetics/114.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best E, Bender R. Cloning of the Klebsiella aerogenes nac gene, which encodes a factor required for nitrogen regulation of the histidine utilization (hut) operons in Salmonella typhimurium. J Bacteriol. 1990;172:7043–7048. doi: 10.1128/jb.172.12.7043-7048.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L-M, Maloy S. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J Bacteriol. 1991;173:783–790. doi: 10.1128/jb.173.2.783-790.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow W, Berg D. Tn5tac1, a derivative of transposon Tn5 that generates conditional mutants. Proc Natl Acad Sci USA. 1988;85:6468–6472. doi: 10.1073/pnas.85.17.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis R, Botstein D, Roth J. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 7.Garner M, Rezin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: applications to components of the E. coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goss T J, Bender R A. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J Bacteriol. 1995;177:3546–3555. doi: 10.1128/jb.177.12.3546-3555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graña D, Youderian P, Susskind M. The effects of mutations in the ant promoter of Salmonella phage P22. Genetics. 1985;110:1–6. doi: 10.1093/genetics/110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn D, Myers R, Kent C, Maloy S. Regulation of proline utilization in Salmonella typhimurium: molecular characterization of the put operon and DNA sequence of the put control region. Mol Gen Genet. 1988;213:125–133. doi: 10.1007/BF00333408. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hughes K T, Youderian P, Simon M. Phase variation in Salmonella: analysis of Hin recombinase and hix recombination site interaction in vivo. Genes Dev. 1988;2:937–948. doi: 10.1101/gad.2.8.937. [DOI] [PubMed] [Google Scholar]
- 13.Maloy S. The proline utilization operon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1513–1519. [Google Scholar]
- 14.Maloy S. Experimental techniques in bacterial genetics. Boston, Mass: Jones and Bartlett; 1989. [Google Scholar]
- 15.Maloy S, Stewart V, Taylor R. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1996. [Google Scholar]
- 16.Maloy S, Youderian P. Challenge phage: a genetic tool kit for dissecting DNA-protein interactions in vivo. Methods Mol Genet. 1993;3:205–233. [Google Scholar]
- 17.McCabe P. Production of single-stranded DNA by asymmetric PCR. In: Innis M A, Gelfand D H, Snibsky J J, White T J, editors. PCR protocols: a guide to methods and Applications. New York, N.Y: Academic Press; 1990. pp. 76–83. [Google Scholar]
- 18.Mead D, Szczesna-Sorupa E, Kemper B. Single-stranded DNA ‘Blue’ T7 promoter plasmids: a versitile tandem promoter system for cloning and protein engineering. Protein Eng. 1986;1:67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- 19.Ostrovsky de Spicer P, O’Brien K, Maloy S. Regulation of proline utilization in Salmonella typhimurium: a membrane-associated dehydrogenase binds DNA in vitro. J Bacteriol. 1991;173:211–219. doi: 10.1128/jb.173.1.211-219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry K, Walker G. Identification of plasmid (pKM101)-coded proteins involved in mutagenesis and UV resistance. Nature. 1982;300:278–281. doi: 10.1038/300278a0. [DOI] [PubMed] [Google Scholar]
- 20a.Pomposiello, P. J., B. K. Janes, and R. A. Bender. Two roles for the DNA recognition site of Klebsiella aerogenes Nitrogen assimilation control protein. J. Bacteriol. 180:578–585. [DOI] [PMC free article] [PubMed]
- 21.Prival M, Magasanik B. Resistance to catabolite repression of histidase and proline oxidase during nitrogen-limited growth of Klebsiella aerogenes. J Biol Chem. 1971;246:6288–6296. [PubMed] [Google Scholar]
- 22.Sanderson K, Roth J. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988;52:485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwacha A, Bender R A. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J Bacteriol. 1993;175:2116–2124. doi: 10.1128/jb.175.7.2116-2124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwacha A, Bender R A. The nac (nitrogen assimilation control) gene from Klebsiella aerogenes. J Bacteriol. 1993;175:2107–2115. doi: 10.1128/jb.175.7.2107-2115.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silhavy T, Enquist L, Berman M. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 26.Wood J. Genetics of l-proline utilization in Escherichia coli. J Bacteriol. 1981;146:895–901. doi: 10.1128/jb.146.3.895-901.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Youderian P, Vershone A, Bouvier S, Sauer R, Susskind M. Changing the DNA-binding specificity of a repressor. Cell. 1983;35:777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]