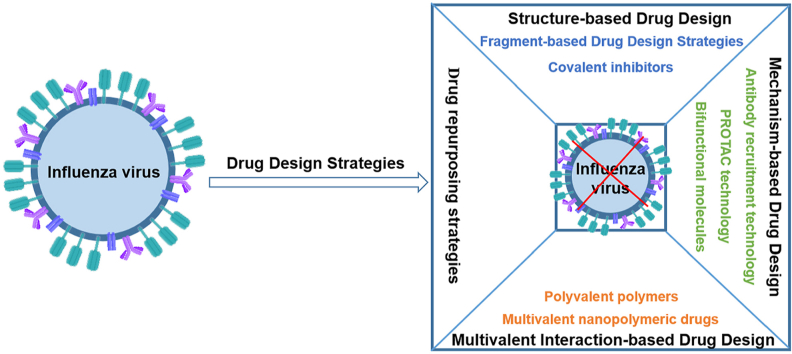
Abstract
Influenza is an acute respiratory infection caused by influenza viruses (IFV), According to the World Health Organization (WHO), seasonal IFV epidemics result in approximately 3–5 million cases of severe illness, leading to about half a million deaths worldwide, along with severe economic losses and social burdens. Unfortunately, frequent mutations in IFV lead to a certain lag in vaccine development as well as resistance to existing antiviral drugs. Therefore, it is of great importance to develop anti-IFV drugs with high efficiency against wild-type and resistant strains, needed in the fight against current and future outbreaks caused by different IFV strains. In this review, we summarize general strategies used for the discovery and development of antiviral agents targeting multiple IFV strains (including those resistant to available drugs). Structure-based drug design, mechanism-based drug design, multivalent interaction-based drug design and drug repurposing are amongst the most relevant strategies that provide a framework for the development of antiviral drugs targeting IFV.
Key words: Influenza virus, Drug design strategies, Anti-drug resistance, High efficiency, Broad spectrum
Graphical abstract
This article reviews the application of emerging drug design strategies in the discovery of anti-influenza virus drugs, and lists the representative inhibitors based on different drug design strategies.
1. Introduction
Influenza is an acute respiratory infection caused by a virus of the family Orthomyxoviridae known as the IFV (influenza virus)1. IFV are classified into four types: A, B, C, D, representing the Alpha-, Beta-, Gamma- and Deltainfluenzavirus genera, respectively. IFV A and B are the main pathogens causing respiratory diseases, while seasonal influenza is caused by influenza A viruses of the H1N1 and H3N2 subtypes2.
During the 20th century, there were three influenza pandemics: Spanish flu outbreak in 1918 (A, H1N1), Asian flu outbreak in 1957 (A, H2N2), and Hong Kong flu in 1968–69 (A, H3N2). The death toll of the last two pandemics has been estimated at 1–4 million people, while the infamous Spanish flu claimed millions of lives, from 17 to 100 million, depending on different estimates3. In addition to the global pandemics, it is estimated that 5%–15% of the global population is infected by seasonal IFV annually, with about 3–5 million cases of severe disease and 500,000 deaths, according to the World Health Organization4, 5, 6. In addition, the infectious disease is responsible for severe economic losses and social distress. In this scenario, there is an urgent need for prevention measures and suitable treatments.
Influenza A and B viruses contain eight segments of linear negative-sense, single-stranded RNA7. Two viral glycoproteins (hemagglutinin and neuraminidase) encoded in different RNA segments play key roles in infection and viral release, and are used in the classification of different serotypes8.
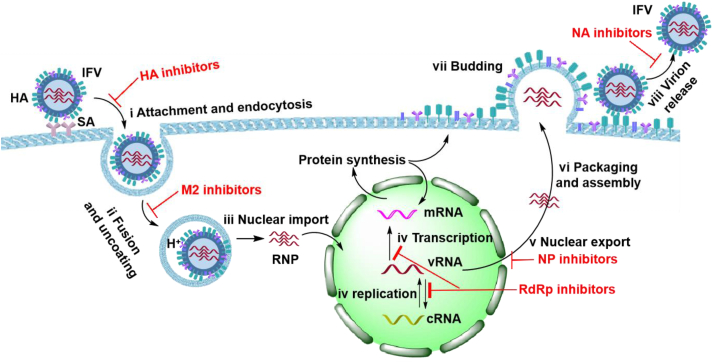
Researchers have extensively studied the replicative cycle of IFV (Fig. 1)9,10. The replicative cycle of IFV is divided into the following steps.
-
(i)
Attachment and endocytosis: Sialic acid (SA) receptors on the surface of host cells are recognized and bound by hemagglutinin (HA) on the surface of viral particles, and then virions are internalized in the host cell cytoplasm through the formation of an endosome11. Therefore, small molecule inhibitors targeting HA can prevent virus from entering host cells.
-
(ii)
Fusion and uncoating: After fusion of the cell membrane, viral particles are transported in endocytic vesicles, whose acidification facilitates the release of the viral ribonucleoprotein (RNP) into the cytoplasm12. The acidification process is mediated by the viral M2 ion channel protein. Small molecule inhibitors targeting M2 can prevent the fusion of viral envelope and host endosomal membrane.
-
(iii)
Nuclear import: The RNP in the cytoplasm is then transferred to the nucleus of the host cell through the nuclear localization sequence found in the viral nucleoprotein (NP)13.
-
(iv)
Transcription and replication: The IFV capped and polyadenylated mRNAs are synthesized by the viral RNA-dependent RNA polymerase (RdRp), using 5′ capped RNA primers and the viral RNA template (vRNA). The 5′ capped RNA primers have a length of 10–20 nucleotides and derive from cleavage of cellular mRNAs by the viral endonuclease (in a process known as cap-snatching mechanism). During the replication process, new copies of vRNA can be synthesized using the complementary RNA (cRNA) as template, which a replication intermediate produced by the viral RNA polymerase, in the presence of viral NP. The newly transcribed mRNAs are translated into proteins14, required to produce progeny viral particles. Therefore, inhibitors targeting RdRp can block transcription and replication.
-
(v)
Nuclear export: The newly synthesized vRNA, polymerase acidic protein (PA), polymerase basic 1 (PB1), polymerase basic 2 (PB2) and NP are transported back into the nucleus, where they are packaged into RNPs15. In this process, compounds binding NP could prevent its export from the nucleus into the cytoplasm.
-
(vi)
Packaging and assembly: RNPs are transported to the vicinity of the cell membrane through the nuclear pore complex and assembled into progeny virus particles16.
-
(vii)
Budding: The generated virions bud on the cell membrane, where they remain attached on the host cell surface due to the interaction between SA receptors and the viral HA17.
-
(viii)
Virion release: Cleavage of the glycosidic bonds of SA by the viral neuraminidase (NA) facilitate virus release into the extracellular medium. Therefore, inhibitors targeting NA can inhibit the release of new virus particles.
Figure 1.
IFV replication cycle and relevant targets for antiviral therapy.
In summary, proteins and biochemical events occurring during infection and viral replication and propagation can all serve as important targets for antiviral drug development18.
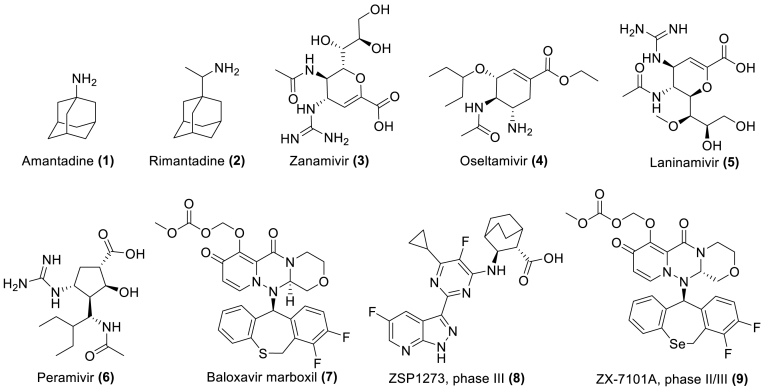
Currently, influenza is primarily prevented and treated with vaccines and antiviral drugs19. Clinically available drugs (Fig. 2) (approved by the U.S. Food and Drug Administration and the European Medicines Agency) include M2 ion channel blockers (1 and 2), NA inhibitors (3–6), and PA inhibitors (7)20,21. In addition, with the continuous efforts of researchers, several candidate drugs with inhibitory effects on IFV have also been found. At present, there are two representative drug candidates in the clinical research stage, namely ZSP1273 (phase III, 8) and ZX-7101A (phase II/III, 9)22,23. ZSP1273 is a potent antiviral inhibitor of cap binding to the PB2 subunit of influenza A polymerase22, while ZX-7101A is an analogue of baloxavir marboxil, and therefore a cap-dependent endonuclease inhibitor23.
Figure 2.
Approved anti-influenza drugs and representative drug candidates at different clinical stages of development.
Although approved drugs have represented major advances in the treatment of influenza, mutations in the IFV genome have led to different degrees of resistance to existing drugs24,25. For example: adamantanes (i.e., 1 and 2) are no longer recommended for the treatment of seasonal influenza because of their extensive resistance to the prevalent influenza A virus26. Oral oseltamivir (4) has become the first choice for clinical treatment, but oseltamivir is prone to drug resistance due to the H274Y mutation in NA of H5N1 and other strains27. Baloxavir marboxil (7) is currently the only PA inhibitor in the market. Unfortunately, the PA I38T substitution is observed in circulating viruses and confers resistance to baloxavir marboxil, while E23R has a synergistic effect and increases baloxavir EC50 values by > 10-fold28. In addition, cross-resistance has been observed in a number of drugs. Thus, G147R and H274Y substitutions in the influenza A H1N1 pdm09 strain confer resistance to NA inhibitors oseltamivir and peramivir (6)29. The A (H1N1) pdm09 virus containing H274Y and other substitutions (I222R, E119D/G) in the NA are also resistant to both oseltamivir and zanamivir (3)30. Single amino acid substitutions at positions 26, 27, 30, 31 or 34 in the viral M2 protein confer cross-resistance to amantadine and rimantadine31.
Variability is a hallmark of RNA viruses. High rates of mutation and recombination help RNA viruses to evade the immune system and develop resistance to antiviral drugs32, 33, 34. In IFV, genetic exchange through reassortment has the potential to accelerate viral evolution35. Gene reassortment occurs mainly in segmented viruses such as IFV. The genome of IFV is composed of an RNA set made of eight individual single-stranded RNAs. These RNAs have to be copied during the IFV replication cycle. Gene reassortment involves the combination of different RNA molecules when cells are simultaneously infected by different IFV strains, originating different serotypes (e.g., H1N1, H2N2, H3N2, etc.) that can become predominant in seasonal flu. Despite major efforts towards the discovery of a “universal” influenza vaccine, the manufacturing process for current vaccines against seasonal flu has to be delayed to ensure protection against closely matched circulating strains36. It is estimated that the antigenicity drift of the virus significantly reduces the effectiveness of the treatment of influenza, and the cure rate for symptomatic disease is merely 50%–60%37.
So far, there are comparatively few drugs available to treat influenza. In addition, mutation, recombination and reassortment, all contribute to reduce the antiviral efficacy of approved drugs. In this context, there is an urgent need to develop novel and efficient IFV inhibitors to block the replication of current and emerging IFV strains and serotypes38.
In this review, we summarize general strategies for the development of antiviral drugs effective against resistant strains, whose discovery and development has been based on different strategies: structure-based drug design, mechanism-based drug design, multivalent interaction-based drug design and drug repurposing.
2. Structure-based drug design (SBDD) strategies
2.1. Fragment-based drug design (FBDD) strategies
FBDD has been one of the mainstream methods for discovering lead compounds in recent years. This strategy uses nuclear magnetic resonance (NMR), surface plasmon resonance (SPR), X-ray single crystal diffraction (X-ray) or thermal migration analysis to screen out small molecular fragments that interact weakly with the target protein, and then optimize the active fragments according to the structural information, in order to obtain more active and high affinity lead compounds for further development39. Compared with high-throughput screening (HTS) methods, FBDD has higher screening efficiency. The active fragments used in FBDD are more easily modified, more active and show improved druglikeness40. Druglikeness is a qualitative concept used in drug design that relates to bioavailability. A drug-like molecule should have good solubility, high potency at the biological target, strong ligand efficiency and a relatively small molecular weight (i.e., 200–600 Da).
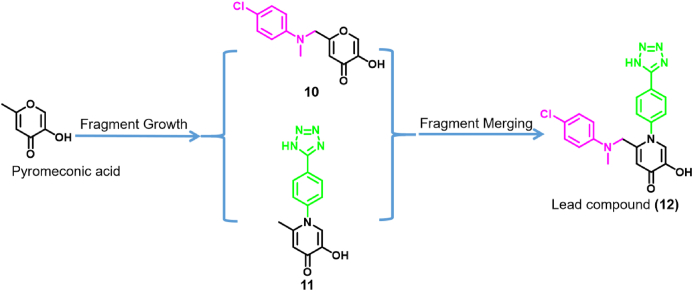
There are a few examples of compounds obtained through FBDD strategies that target key steps in IFV replication and propagation. A notable example is the PA subunit of the viral RdRp. Credille et al.41 screened a privileged metal-binding pharmacophore library to identify inhibitors of the PA subunit of IFV RdRp. After performing enzyme activity inhibition assays, it was found that pyromeconic acid (Fig. 3) was an effective inhibitor of the PA endonuclease activity (IC50 = 22.5 μmol/L). Under the guidance of previously reported modeling and structural data, a molecular library was elaborately constructed from the initial hits. With pyromeconic acid as the privileged fragment, the synthesized compounds 10 and 11 obtained by fragment growth strategy showed strong antiviral biological activity against H1N1. The IC50 values of compounds 10 and 11 were 0.94 ± 0.08 μmol/L and 36 ± 0.08 nmol/L, respectively. Then, according to established structure–activity relationship analysis, the privileged fragments of compounds 10 and 11 were combined using a fragment merging strategy to design and synthesize the lead compounds with the best biological activity. Finally, compound 12 with the best antiviral activity was obtained, and its IC50 for inhibiting PA endonuclease activity was 14 ± 3 nmol/L. In vitro, the EC50 value of compound 12 against influenza A (H1N1) virus was 2.1 μmol/L, and the cytotoxicity was low (CC50 = 280 μmol/L). Molecular docking studies showed that compound 12 could chelate with Mn2+ in the endonuclease active site.
Figure 3.
Design and synthesis of effective molecules using fragment growth and fragment merging strategies41.
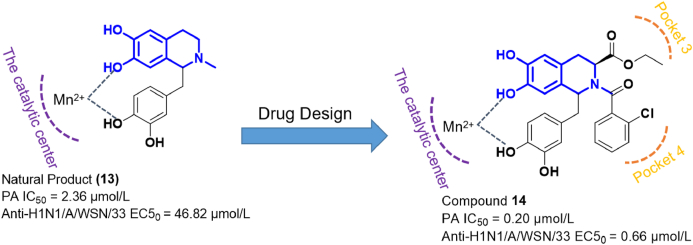
In a previous work, Arnold et al.42 found that the natural product d,l-laudanosoline (13) could inhibit H1N1 IFV in antiviral assays (EC50 = 46.82 μmol/L) (Fig. 4). Mechanistic experiments showed that compound 13 exerted anti-influenza activity by inhibiting the PA endonuclease (IC50 = 2.63 μmol/L). After analyzing the binding mode of 13 and PA, Song et al.43 decided to use compound 13 as the privileged fragment, and optimized its structure through the synthesis of a series of novel compounds. Among them, compound 14 showed the strongest inhibitory effect against the IFV H1N1/A/WSN/33 strain, with an EC50 value of 0.66 μmol/L. In addition, the IC50 value of compound 14 in PA inhibition assays was 0.2 μmol/L. Antiviral experiments carried out in mice showed that compound 14 had a strong protective effect. In relation to the binding mode of compound 14 to PA, researchers found that it maintained the original Mn2+-binding properties of 13, while acting on pockets 3 and 4 of the PA subunit.
Figure 4.
Structure-based drug design strategy to discover effective PA inhibitors.
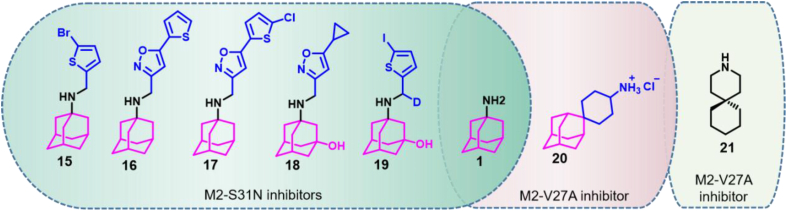
Amantadine resistance mutations V27A, L26F and S31N in the IFV M2 protein have been widely spread in human populations44. Amantadine has been used as a privileged fragment and its structure optimized to identify compounds showing efficacy against IFV strains resistant to classical M2 ion channel inhibitors. Thus, Wu et al.45 designed a series of M2 ion channel inhibitors based on the amantadine skeleton that showed significant inhibitory activity against mutants containing the S31N substitution (Fig. 5). Among them, compound 15 showed an EC50 value of 1.8 μmol/L against the A/WSN/33 (S31N) mutant strain, while retaining significant inhibitory activity against wild type strains (EC50 value of 4.6 μmol/L for the wild-type A/Udorn/72 strain)45. Further efforts in the discovery of novel inhibitory compounds led to the discovery of compound 16 that showed an EC50 value of 0.353 μmol/L for the mutant strains A/WSN/33 (S31N)46. Amantadine derivatives such as compound 17 were also good inhibitors of wild-type and oseltamivir-resistant strains. The EC50 values obtained for compound 17 and strains A/California/07/2009 (H1N1), A/Switzerland/9715293/2013 (H1N1), A/Washington/29/2009 (H1N1) H275Y, and A/Denmark/528/2009 (H1N1) H275Y were 0.1 ± 0.03 0.2 ± 0.04, 0.2 ± 0.02 and 0.1 ± 0.01 μmol/L, respectively47.
Figure 5.
Structure-based drug design strategy to discover M2 inhibitors effective against drug-resistant IFV strains.
Compound 16 showed poor stability in mouse liver microsomes (t1/2 = 1 min), and its structure was optimized for improved stability. Thus, compound 18 showed a half-life greater than 145 min both in mouse and human liver microsomes48. Further studies led to the discovery of compound 19 that showed improved pharmacokinetics. After intraperitoneal administration in mice, the time for 19 to reach the maximum plasma concentration was 0.42 h, and the peak plasma concentration was 5610 nmol/L. The half-life of 19 was 2.34 h, the area under the concentration–time curve was 11,459 ± 1175 nmol/L·h, the apparent volume of distribution was 38.0 ± 4.4 L/kg, and the apparent clearance rate was 11.2 ± 1.1 L/h/kg49.
Compound 20 was identified as the first amantadine derivative showing efficacy against resistant strains containing the amino acid substitution V27A in the M2 protein. This compound showed EC50 values of 0.3 ± 0.1 and 1.8 ± 0.2 μmol/L for wild-type IFV strain A/Udorn/72 (H3N2) and mutant A/WSN/33 (M2-N31S/V27A) (H1N1), respectively50. In addition, Wang et al.51 reported that spiro-piperidine (compound 21) could inhibit influenza A virus in the two-electrode voltage clamp (TEV) experiment, and its IC50 was 0.92 μmol/L. Nuclear magnetic resonance experiments revealed that 21 binds to the transmembrane region of the influenza A virus M2 protein. This region contains residues Val27, Ala30, Ser31 and Gly34, involved in amantadine resistance. It is possible that a combination of active amantadine derivatives and the spiro-piperidine binding to the M2 transmembrane region could inhibit the replication of IFV drug-resistant strains51.
2.2. Covalent inhibitors
The binding of traditional small molecule drugs to biological targets is usually a reversible process that affects treatment effectiveness for a specific period of time. By prolonging the time of drug–target interaction, the therapeutic effect of drugs can be significantly improved. A long-term interaction can be achieved through covalent binding52. Covalent inhibitors are compounds that form covalent bonds with specific molecular targets. A covalent inhibitor is structurally composed of a seeker and a warhead. The seeker interacts with the target protein in a non-covalent manner, while the warhead covalently binds to the amino acid residues in the target protein53. Depending on the selected warheads, covalent bonds can be classified into reversible and irreversible.
At present, researchers have discovered many different types of warheads that covalently bind to many different amino acid residues54. Highly potent covalent inhibitors show strong specificity with low IC50 values, and long binding duration. As a result, their doses can be reduced, thereby facilitating compliance due to less-frequent dosing. Other interesting advantages of covalent inhibitors are their capacity to target shallow binding sites, while reducing the potential for emergence of drug resistance55. Currently, covalent inhibitors are being used to combat cancers, neurological, cardiovascular and cerebrovascular diseases, as well as infections disorders.
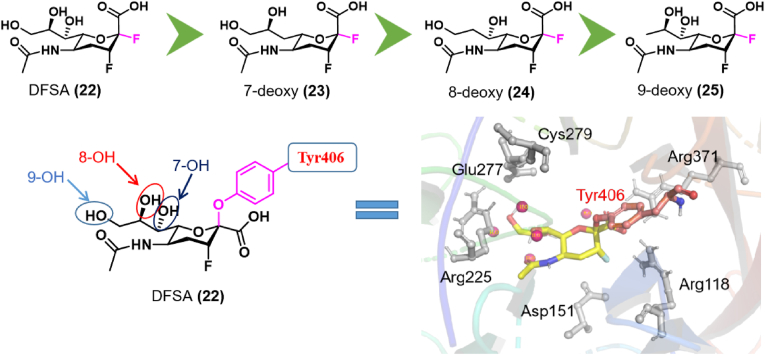
A number of covalent inhibitors targeting IFV NA have been described. As mentioned above, zanamivir (ZA) and oseltamivir approved by the FDA are used to treat influenza. These drugs interact with the IFV NA enzyme. They are transition analogues based on SA substrates. In addition, a series of difluorinated derivatives, including the 5-N-(acetylamino)-2,3,5-trideoxy-2,3-difluoro-d-erythro-β-l-manno-2-nonulopyranosonic acid (DFSA) have been characterized as IFV NA inhibitors (Fig. 6)56, 57, 58. DFSA derivatives form covalent bonds between the C2 atom of the sugar ring and the side chain of Tyr406 in the NA active site, leading to enzyme inactivation56,57. For this reason, McKimm-Breschkin et al.58 designed and synthesized a set of deoxy-DFSA derivatives (22–25) to test the effect of each –OH in DFSA in their interaction with IFV NA. The NA enzyme inhibition assay showed that the IC50 values of compound 22 against wild-type H1N1 and mutant H1N1 were 0.069 and 0.133 μmol/L, respectively. The IC50 values of compounds 23–25 against wild-type H1N1 were 0.037 2.61 and 4.50 μmol/L, respectively, and the IC50 values against mutant H1N1 were 0.104 0.28 and 4.69 μmol/L. At the cellular level, the EC50 value of compound 22 against the IFV H1N1 A/Mississippi strain was 1 μmol/L, while the EC50 values for compounds 23–25 were 0.1–1, 10 and 10–100 μmol/L, respectively. Enzymatic assays together with cell culture and X-ray crystallography studies showed that the compounds without 8-OH and 9-OH had the greatest impact on binding affinity and the largest antiviral effect.
Figure 6.
Covalent binding diagram of DFSA derivatives with NA. Compounds 22–25 are shown in decreasing order of turnover, binding affinity and antiviral potency57.
3. Mechanism-based drug design strategies
3.1. Bifunctional molecules
Drug combinations can produce additional or synergistic effects while improving their therapeutic effects59. However, combination therapies often cause drug–drug interactions, responsible for adverse reactions and side effects60. In recent years, more and more drug researchers have begun to pay attention to the development of bifunctional drugs, which generally refer to a molecule acting on two different targets at the same time. In the process of drug development, bifunctional drugs are often superior to the corresponding single functional drugs in preclinical pharmacodynamics, while delaying the emergence of drug resistance61.
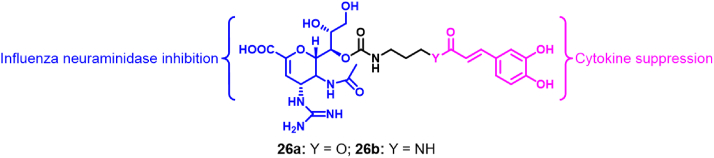
In order to overcome the problem of drug resistance, Liu et al.62 designed bifunctional molecules targeting IFV. This was achieved through the combination of antiviral drugs with anti-inflammatory agents (Fig. 7). Specifically, ZA conjugates containing caffeic acid (CA) 26a and 26b can significantly inhibit IFV NA, while reducing the production of pro-inflammatory cytokines. Enzymatic assays showed that compounds 26a and 26b exhibited H1N1 NA inhibitory activity similar to that of ZA (IC50 = 2.4–7.0 nmol/L), with IC50 values of 2.9–7.4 and 41.3–60.3 nmol/L, respectively. However, they showed slightly better anti-IFV activity than ZA with EC50 values of 1.4–10.6 and 5.1 nmol/L, respectively. In addition, the release of IL-6 and TNF-α was also inhibited by 26a and 26b. Antiviral activity assays carried out in vivo showed that the ZA conjugate is more effective than the combination of ZA and CA in protecting mice infected by H1N1 IFV, being effective at less than 1.2 μmol/kg/day.
Figure 7.
Bifunctional molecules made of ZA and CA62.
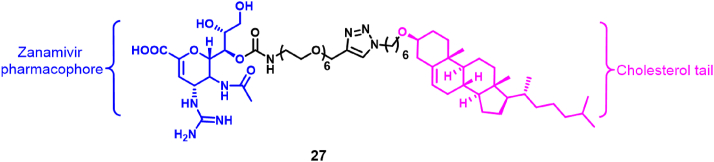
In addition, Lv et al.63 designed a novel NA inhibitor (27), which is a conjugate of ZA and cholesterol (Fig. 8). In NA inhibition experiments, the IC50 values obtained for this molecule were in the range of 22.0–28.0 nmol/L, weaker than the values obtained for ZA (IC50s in the range 0.3–1.0 nmol/L). However, the antiviral activity of the conjugate was better than for ZA (EC50 values of 22.0–36.8 nmol/L for the conjugate versus 26.6–123.4 nmol/L for ZA). More importantly, the plasma half-life of the ZA-cholesterol conjugate reached 7.6 h, which was about 25 times higher than that of ZA alone (0.3 h). Compared with ZA, the conjugate improved the antiviral effect. A single-dose administration of ZA-cholesterol protected mice from lethal attack of influenza A virus (wild-type or H1N1-H274Y), conferring resistance to oseltamivir. Mechanistic studies have shown that the cholesterol moiety of compound 27 binds to the host cell membrane and enters the host cell. After entering the host cell, the ZA moiety of compound 27 binds to the NA of the virus, thereby inhibiting the assembly of viral particles.
Figure 8.
Dual-target antiviral drug containing ZA and cholesterol63.
3.2. Drug design strategy based on protein hydrolysis targeting chimera (PROTAC) technology
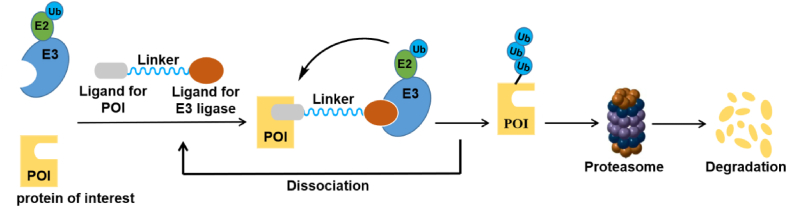
Compared with the mechanism of classical small-molecule inhibitors, the rapidly developing PROTAC technology uses the body's own protein removal system to eliminate pathogenic target proteins64,65. PROTAC is a bifunctional molecule composed of two ligands (also known as warheads), which are connected by a flexible chemical linker, that facilitates the connection of E3 ubiquitin ligase and the target protein (also known as the protein of interest (POI)). PROTAC molecules can induce poly-ubiquitination of target molecules by recruiting E3 ubiquitin ligase, and then the ubiquitin proteasome system (UPS), therefore degrading the POI (Fig. 9)66. PROTAC technology has become a powerful tool in modern drug discovery and development, with important advantages particularly favorable for overcoming drug resistance and for acting against "non-druggable” targets67,68. More importantly, the PROTAC molecules interact with the target protein without requiring high affinity binding to degrade the target protein.
Figure 9.
Scheme of the PROTAC technology.
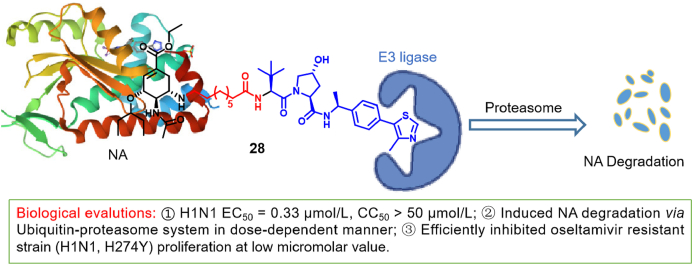
In the past five years, PROTAC technology has been widely used in antitumoral therapy, in order to overcome problems of drug resistance and difficult drug targets68, 69, 70, 71. The emergence of drug-resistant IFV strains has triggered studies towards the development of new therapeutical methods against IFV. Recently, PROTAC strategies aimed at degrading IFV-associated proteins have emerged in a successive manner. Xu et al.72 designed a series of novel PROTAC molecules with oseltamivir as a ligand. Most of the compounds degraded NA effectively and showed anti-H1N1 activity. Among them, compound 28, containing nine carbon alkyl chains, showed the strongest inhibitory effect on H1N1 (chemical structure of 28 given in Fig. 10). It showed an EC50 value of 0.33 μmol/L, better than that of oseltamivir phosphate. The activity of the PROTAC molecule was demonstrated by Western blotting, revealing the degradation of the viral NA in a dose-dependent manner. Researchers showed that its degradation occurred through the ubiquitin proteasome pathway. In addition, compound 28 showed significant inhibitory activity and degradatory effects against oseltamivir-resistant strains (H1N1-H274Y). These results indicate that 28 can degrade mutant NA without showing strong affinity. So far, this is the first report of a PROTAC successfully targeting IFV, while opening new avenues in antiviral drug discovery.
Figure 10.
IFV NA-degrading agent containing oseltamivir, based on PROTAC technology72.
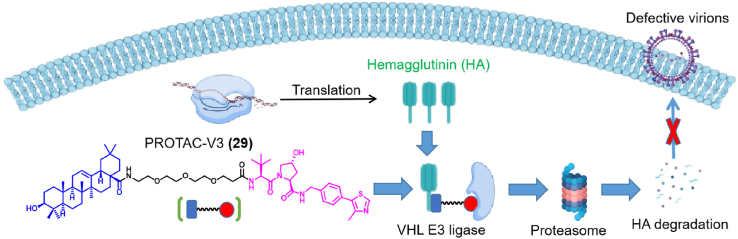
In addition, Zhou et al.73 reported a novel class of PROTAC molecules based on pentacyclic triterpenoids, which can degrade the HA on the surface of the virus (Fig. 11). Among them, PROTAC-V3 (29) showed the best degradation efficiency of HA, and its half maximum degradation concentration was 1.44 μmol/L. The degradation process was ubiquitin- and proteasome-dependent, while the drug candidate showed broad-spectrum anti-influenza A virus activity, without affecting IFV entry. More interestingly, the oral bioavailability of compound 29 was only 6.8%. Besides, compound 29 protected mice from influenza A virus-induced disease manifestations. Compound 29 contains an oleanolic acid-based moiety that binds HA linked to a ligand of the von Hippel-Lindau (VHL) E3 ligase. The preclinical success of this approach provides new directions towards the discovery of novel anti-influenza drugs.
Figure 11.
IFV HA-degrading agent containing an oleanolic acid derivative, based on PROTAC technology73.
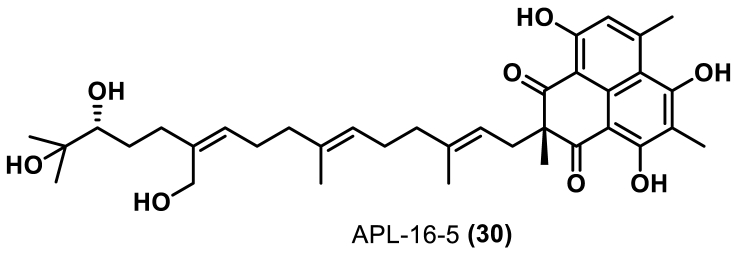
APL-16-5 (30) was isolated from the endophytic fungus Aspergillus sp. CPCC400735 (Fig. 12). Cen et al.74 found that 30 had inhibitory activity against influenza A (EC50 = 0.28 μmol/L) and influenza B (EC50 = 1.22 μmol/L) viruses, and low cytotoxicity (CC50 > 100 μmol/L, MDCK cells). It was found that 30 could bind to the E3 ligase TRIM25 and the influenza A virus polymerase PA subunit at the same time, resulting in TRIM25 ubiquitination of PA, which facilitated the degradation of PA by proteasome, through a pathway similar to that used by PROTAC molecules. Furthermore, Cen et al.74 also showed that 30 protected mice from lethal influenza A virus infection, thereby constituting an effective antiviral drug.
Figure 12.
Chemical structure of APL-16-574.
This study also suggests that microbial natural products can also degrade target proteins, while showing that their degradation mechanism is similar to that used by PROTACs, opening the possibility of identifying novel protein degradation agents.
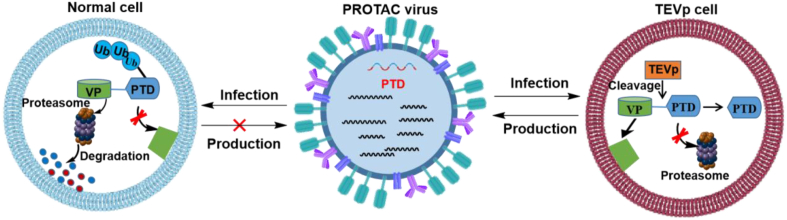
Attenuated live vaccines are limited by safety issues, poor immunogenicity, and complex manufacturing processes. Interestingly, an attenuated influenza A virus vaccine has been made by PROTAC technology75. Researchers have prepared a novel PROTAC virus by fusing the proteasome targeting domain (PTD) to the protein of the IFV (VP) (Fig. 13). The PTD contains a proteasome targeting peptide (ALAPYIP) and a tobacco etch virus (TEV) cleavage site linker. Previous studies had demonstrated that the VHL tumor suppressor protein can recognized ALAPYIP. Therefore, ALAPYIP is bound to E3 ubiquitin ligase and then degraded by proteasome76,77. In addition, PTD lacks sufficient sequence homology with human proteins, reducing the possibility of a PTD-induced autoimmune response.
Figure 13.
Scheme showing the principles for constructing a PROTAC-based IFV vaccine75.
The process used to generate the vaccine involves growing the modified virus in permissive cells (e.g., human embryonic kidney (HEK) 293T and MDCK cells) and in TEV protease-expressing stable cell lines. Virus production in conventional cells is blocked due to degradation of the viral proteins, while the expression of TEV protease spares the viral protein from proteasome degradation. In a comprehensive analysis, Si et al.76 tested the effects of incorporating the PTD sequence in each of the eight proteins of influenza A virus, and found that the best effects were obtained with M1, a bifunctional membrane/RNA-binding protein that mediates encapsidation of NP cores into the viral envelope. Experiments carried out in vivo showed that PROTAC virus vaccines can enhance cellular immunity in body fluids and mucus, thereby circumventing infection by homologous and heterologous viruses. This study also demonstrated the feasibility of the concept of PROTAC virus vaccine in cells and animal models, and provided new ideas for the development of virus vaccines.
3.3. Drug design based on antibody recruitment technology
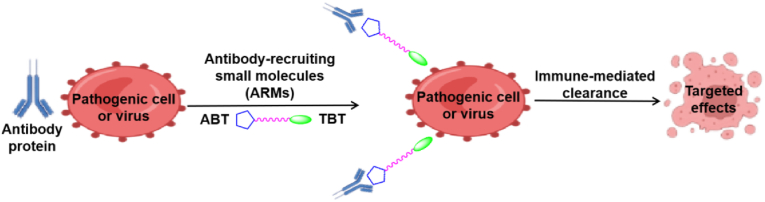
In recent years, synthetic systems (synthetic immunology) that regulate immune responses have become a hot research field. The focus of this field is the rational design and construction of synthetic molecular complexes performing sophisticated immunological functions. Thus, some small molecules could enhance antibody binding to disease-related cells or viruses, resulting in immune-mediated elimination of cells or viruses (Fig. 14). These compounds are known as antibody recruitment molecules (ARMs)78,79. ARMs are composed of three basic components: a target-binding terminal (TBT) moiety, an antibody-binding terminal (ABT) moiety and a chemical linker. Compared with traditional small molecules, ARMs have the advantage of relatively small side effects80. ARMs have been used in the development of antitumoral81,82, antibacterial83 and antiviral agents84, as well as other drugs. In antiviral therapy, ARMs have been used for recruit antibodies in plasma to viral particles and virus-infected cells, as well as for inhibiting virus binding to host cells85.
Figure 14.
Mechanism of antibody recruitment molecules in antiviral therapy79.
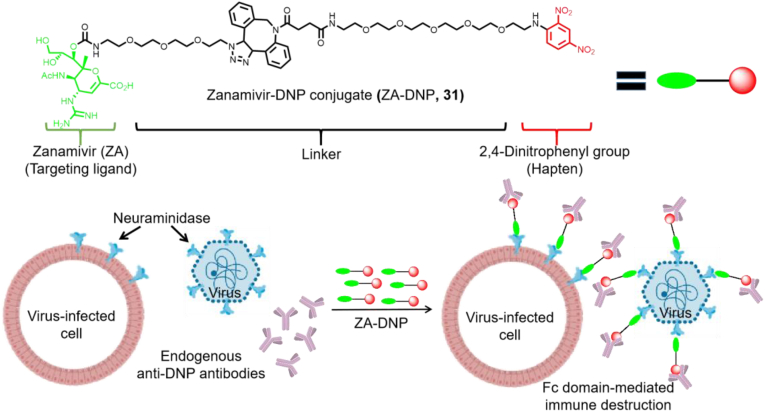
The antibody recruitment strategy has been applied in the fight against influenza. Liu et al.86 designed and synthesized a bifunctional small molecule by joining the NA inhibitor ZA with the highly immunogenic hapten, dinitrobenzene (DNP). The obtained molecule targets free viruses and virus-infected cells (Fig. 15). The inhibitory effect of ZA-DNP (31) on influenza A virus (H1N1 and H3N2 serotypes) was comparable to that of ZA alone (EC50 values of 1.7 and 7.6 nmol/L, respectively). A single dose of nasal or intraperitoneal administration of the compound to mice infected with 100 × MLD50 (median lethal doses) was found to be sufficient to eradicate advanced infections of representative influenza A and B virus strains. The treatment was still effective three days after lethal inoculation, suggesting that this method could be successful to treat infections that are difficult to cure at present.
Figure 15.
Dual mechanism immunotherapy based on the use of NA-targeted conjugates86.
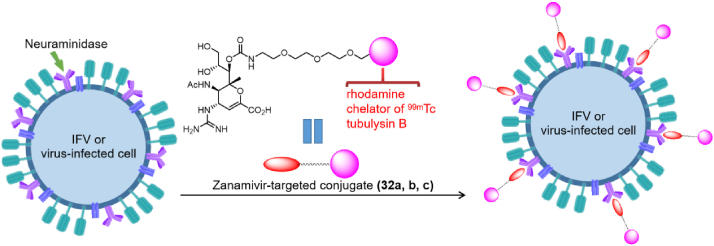
In another study, Liu et al.87 obtained a bi-functional molecule containing ZA and a rhodamine dye (32a). This conjugate allows the visualization of virus-cell binding, and the internalization and intracellular transport of the virion (Fig. 16). The researchers also synthesized a 99mTc-ZA-conjugated dimer molecule (32b) that monitors the distribution of IFVs in mice by radio imaging. Finally, they synthesized a novel IFV inhibitor (32c) by coupling the NA inhibitor ZA with the cytotoxic drug tubulysin B. The EC50 values of 32c and free ZA against NA-infected HEK293 cells were 5.1 and 9.9 nmol/L, respectively. In addition, compound 32c can kill virus-infected cells without damaging healthy cells. In conclusion, these conjugates can be used for diagnosis and treatment of IFV infection.
Figure 16.
Anti-IFV mechanism diagram of ZA-targeted conjugates86.
4. Multivalency-based drug design strategies
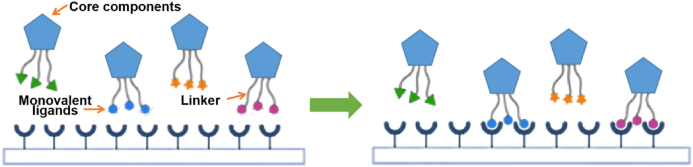
Multivalent binding is ubiquitous and occurs in many important biological processes (e.g., cell signal transduction, cell–cell interaction, pathogen recognition, etc.)88. Based on the importance of multivalent binding in biological systems, more and more drug researchers are committed to exploring the mechanism of multivalent binding, and designing and synthesizing multivalent binding drugs facilitating the interaction between multivalent ligands and multivalent receptors (Fig. 17)89. Typical synthetic polyvalent drugs are composed of dendritic core components or nanoparticles connected to monovalent ligands (agonists or inhibitors) through flexible linkers90,91. Compared with monovalent drugs, the main advantages of multivalent drugs relate to their strong binding ability with receptor molecules promoting a strong biological activity and that binding of multivalent drugs to receptor molecules is not easy to dissociate, and therefore their biological activity lasts longer92. In view of the advantages of multivalent drugs, multivalent binding theory has been used in the design of a variety of targeted drugs, including antibody–drug conjugates (ADCs), drug-loaded nanoparticles, and even chimeric antigen receptor (CAR) T cells93. Examples of synthetic or modified polymers and nanoparticles with good anti-IFV activity that can be used to inhibit the binding of viruses to cellular receptors are described in the following sections.
Figure 17.
Binding mode of multivalent ligands and receptors92.
4.1. Drug design of polyvalent polymers
Polyvalent polymers constitute a type of dendritic three-dimensional molecules. Their structure is composed of core multi-branched small molecules and monovalent ligands (agonists or inhibitors), joined through suitable linkers. Each branch of the core is connected with the monovalent ligand through the linker to form a multivalent polymer94. Since the polymer molecular structure contains multiple ligands (agonists or antagonists), the probability of binding of the polymer to the receptor molecule increases, which in turn increases its biological activity95. For now, the polyvalent polymer molecules are used in diagnostic, antibacterial, antitumor, and biomedical applications96.
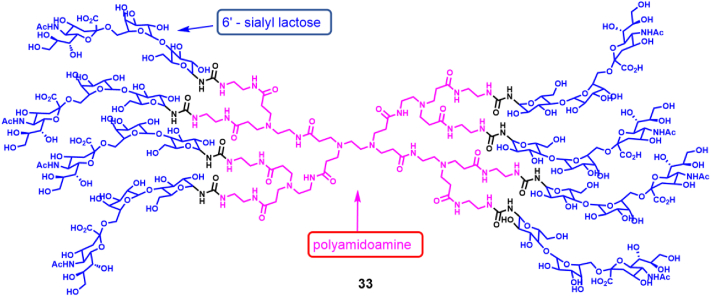
This design has been applied to block virus-host cell attachment to provide early and effective blocking of viral infection. Therefore, receptor analogues are considered to be an option for early blocking of virus-receptor binding. By coupling polyamidoamine (PAMAM) and 6′-sialyllactose (6SL) to form a dendritic polymer molecule 33 (Fig. 18), researchers obtained polymers that block the infection a variety of human and avian IFV strains97. In hemagglutination inhibition tests, they tested the HA inhibitory activity of the compounds against H3N2 subtype influenza strains (A/Panama/2007/1999 (PA/99), A/Brisbane/10/2007 (BR/10) and (A/Hong Kong/1/1968 (HK/68)). It was found that (6SL)8-PAMAM had the strongest HA inhibitory activity against influenza strain PA/99 with an IC50 value of <200 μmol/L. In addition, PAMAM polymers containing 6SL inhibited a representative series of human influenza A virus strains. Cell neutralization assays confirmed that the polymer had an inhibitory effect on human and avian influenza A virus. Based on those results, further optimization of the PAMAM polymer containing 6SL has been encouraged as an alternative to produce novel anti-IFV inhibitors.
Figure 18.
PAMAM polymer molecules containing 6SL97.
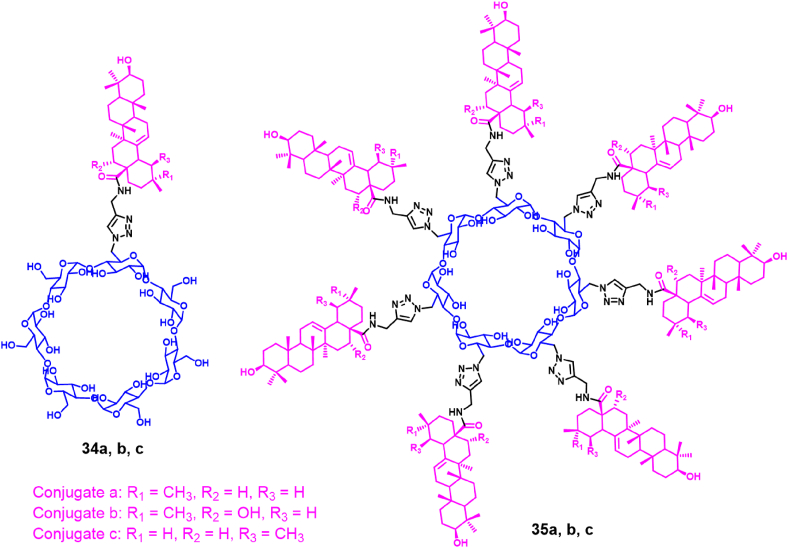
Alternative multivalent polymers designed and synthesized by researchers involved the combination of a pentacyclic triterpene and cyclodextrin (Fig. 19)98. Several polymer molecules with pentacyclic triterpenoid pharmacophores were prepared by coupling pentacyclic triterpenoids with cyclodextrins by click chemistry. Some polymers showed significant inhibitory effect on H1N1 virus, which was much better than the antiviral activity obtained in control experiments carried out with oseltamivir. The strongest anti-influenza activity was obtained with the β-cyclodextrin polymer containing seven oleanolic acids (35a–c). Its anti-influenza activity was 125 times stronger than the one obtained with the monovalent conjugate (34a–c) and oleanolic acid, revealing a significant multivalent effect. In addition, these polymers can also inhibit A/JX/312 (H3N2) and A/HN/1222 (H3N2), with EC50 values of 2.47–14.90 μmol/L and no cytotoxicity at 100 μmol/L. Most importantly, the β-cyclodextrin polymer containing seven oleanolic acids can be tightly bound to HA (dissociation constant, KD = 2.08 μmol/L), blocking HA-SA receptors binding. Thereby destroying the adsorption of the virus to host cells. This study demonstrates that this novel inhibitor based on multivalent binding can efficiently block the binding of IFV to host cells.
Figure 19.
β-Cyclodextrin polymers containing pentacyclic triterpenoids98.
4.2. Design of multivalent nanopolymeric drugs
More and more researches in the medical field are paying attention to multivalent nanopolymeric drugs. Due to the small size of nanoparticles (10–100 nm), this approach might be beneficial for entering capillaries and cells, thereby increasing the availability of the drug for binding to biological targets. In addition, nanopolymers have a high surface-to-volume ratio, which helps to increase the binding area of the polymer to the virus surface receptor99. Nanopolymers typically use nanoparticles as carriers and agonists or inhibitors as monovalent ligands, both of which are simultaneously connected by flexible linkers100. Nanopolymers have been widely used in the development of antiviral drugs due to their many advantages101.
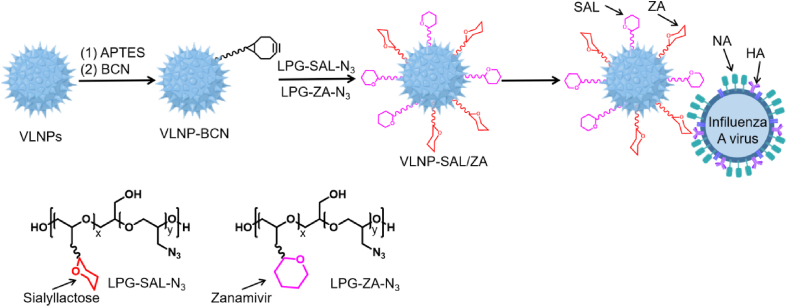
The concept of "topological matching design” has been used to develop nanoparticle inhibitors targeting influenza A virus infection. Nie et al.102 designed and synthesized virus-like nanoparticles (VLNPs) with nanospikes. First, an intermediate was obtained through the chemical reaction of VLNPs with 3-aminopropyltriethoxysilane (APTES), that rendered VLNP-BCN after addition of (1R,8S,9S)-bicyclo [6.1.0]non-4-yn-9-yl-methyl N-succinimidyl carbonate (BCN–NHS). VLNP-BCN was then reacted with the azide-containing linear polyglycerol-sialolactose (LPG-SAL-N3) and LPG-ZA (LPG-ZA-N3) by click chemistry to obtain VLNP-SAL/ZA, as shown in Fig. 20. Results showed that the IC50 value of VLNP-SAL/ZA was 5.38 ± 1.37 μg/mL in NA inhibition assays, while in plaque-reduction assays, the IC50 value was 1.33 ± 0.14 mg/mL. In summary, VLNP-SAL/ZA inhibitor not only had a dual inhibitory effect on HA binding and NA, but also showed multivalent inhibition of both functions. In addition, the nanoparticles provided a topological structure that could match the spatial structure of influenza A virions. The obtained nanoinhibitors can bind to extracellular viral particles and prevent virion attachment to host cells. More importantly, even 24 h after virus infection, reduction of virus propagation was still above 99.999%, demonstrating the efficacy of this novel antiviral strategy.
Figure 20.
Construction of heterologous multivalent nanoinhibitors and their interaction with IFV.
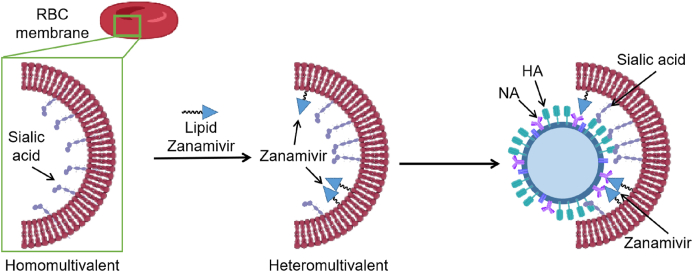
Virus binding kinetics confirmed that the heterogeneous multivalent nanoinhibitors had better binding properties than homologous multivalent nanoinhibitors. The binding moiety of heterogeneous multivalent nanoinhibitors matched the surface of the virus, forming bowl-like nanostructures similar to the viral spherical surface103. In addition, compared with homologous multivalent inhibitors, heterogeneous multivalent nanoinhibitors have synergistic multivalent effects. Most human influenza A virus strains bind to red blood cells, since their phospholipid bilayer contains SA which is the ligand of IFV HA. Based on this evidence, researchers made covalent bonds between the SA-containing membrane of red blood cells and ZA. This way, they obtained a heterogeneous multivalent nanoinhibitor (Fig. 21), whose EC50 value in antiviral assays was 32.4 ± 13.7 g/mL, while virus transmission was reduced by more than 99.99% at a dose that was not cytotoxic.
Figure 21.
Construction of heterogeneous multivalent inhibitors based on erythrocyte membrane and their interaction with IFV.
In addition to agents described above, researchers have synthesized nanogels containing flexible SA (Fig. 22)104. SA residues in those gels bind to HA on the surface of the influenza virion in a multivalent manner. Firstly, researchers prepared dendritic and linear polyglycerol sialosides (dPG (SA)15% (N3)10% and LPG (SA)15% (N3)10%) containing 10% azide and containing 15% and 40% SA residues. Then, dendritic and linear polyglycerols (dPG (cyclooctyne)10%) and (LPG (cyclooctyne)5%) containing 5% and 10% cyclooctyne were also prepared. Finally, dendritic and linear polyglycerols containing alkynes and azides were subjected to click chemistry to obtain R-NG 1, F-NG 2 and F-NG 3 nanogels. In the antiviral experiment of MDCK, only F-NG 3 showed antiviral activity against A/X31 virus, with an IC50 value of 23 μg/mL, which is equivalent to the concentration of 2.3 pmol/L concentration. F-NG 3 could inhibit the binding of virus to host cells with 98% blocking. In summary, in HA binding inhibition assays, flexible nanogels (F-NG 3) had stronger inhibitory effects on influenza A virus than hard gels. At picomolar concentrations, the flexible sialylated nanogel can effectively inhibit the infection of IFV strain A/X31 (H3N2) and block virus from entering MDCK cells.
Figure 22.
Schematic diagram of multivalent gel preparation.
In addition, the design and synthesis of multivalent binders is an effective method to interfere with virus attachment to host cells (Fig. 23). Researchers connected SA containing polyethylene glycol with different lengths and azides (SA1, SA3, SA5 adn SA8) to the Qβ phage capsid containing alkynyl groups (Qβ) by click chemistry, and finally synthesized a multivalent binder molecule (Qβ[SA1], Qβ[SA3], Qβ[SA5], and Qβ[SA8])105. Furthermore, a triethylene glycol linker (Gal3) was used as a ligand to synthesize a multivalent binder as a negative control (Qβ[Gal3]. In the antiviral activity test, Qβ[SA1] showed the best antiviral activity, and its EC50 value for inhibiting A/X31 virus was 0.04 nmol/L. As demonstrated using cryo-electron tomography, the modified capsids cover the entire IFV envelope and prevent virus binding to the host cell. The highly functionalized capsids prevent virus infection in vitro, ex vivo and in vivo.
Figure 23.
Preparation of multivalent inhibitors of Qβ phage capsid containing SA.
5. Drug repurposing strategies
In view of the many problems confronted by de novo drug discovery, finding new indications for existing drugs has become an attractive new strategy106,107. Drug repurposing is a strategy to identify new uses for drugs that have been approved or are being studied108, 109, 110, 111. Compared with the traditional discovery of new drugs, drug repurposing has the following advantages: simpler research and development process, shorter research and development cycle, low cost, low toxicity, clear mechanism of action, clear side effects and less adverse reactions112,113. Therefore, drug repurposing strategies have become the focus of new drug discovery.
In recent years, drug repurposing as a new drug development strategy has received more and more attention, and a large number of new indications drugs have been born. For example, celecoxib was initially used to treat rheumatoid arthritis and is now also used to treat colon and breast cancer114; Methotrexate, which has been used to treat cancer, is also now being used to treat rheumatoid arthritis115; Lidocaine was initially used for local anesthesia and later also for the treatment of arrhythmia116.
In addition, drug repurposing has also been widely used in anti-influenza drug discovery. Initially, nitazoxanide was used as an anti-parasitic treatment. Later, it was found that nitazoxanide can have anti-IFV activity by preventing the maturation and intracellular transport of HA117. Mifsud et al.118 studied the in vitro anti-influenza activity of nitazoxanide against various human and avian influenza A viruses (H1N1, H3N2, H5N9, H7N1) including amantadine-resistant and oseltamivir-resistant strains. Notably, nitazoxanide showed a significant inhibitory effect on a variety of IFV, with EC50s in the range of 0.9–3.2 μmol/L. Besides, they studied the anti-IFV activity of nitazoxanide combined with oseltamivir against the IFV strains A/Puerto Rico/8/1934 (H1N1) and A/WSN/1933 (H1N1), showing additive to synergistic effects depending on their concentrations. Similar results were obtained when nitazoxanide was combined with zanamivir.
Recent studies have found that naproxen, a non-steroidal anti-inflammatory drug, can inhibit the replication of influenza A and B viruses. The EC50 value of naproxen against H1N1 A/WSN/33 virus was 16 ± 5 μmol/L. Further studies have found that naproxen can block the binding of the host's nuclear export-related protein chromosome maintenance protein 1 (CRM1) to viral NP and inhibit CRM1-mediated NP export from the nucleus, thereby exerting anti-IFV activity119,120.
Aprotinin, a protease inhibitor, is clinically used to prevent and treat acute pancreatitis, bleeding caused by fibrinolysis and diffuse intravascular coagulation. It can also be used for anti-shock treatment. In addition, studies have found that aprotinin can significantly inhibit the replication of IFV. In an in vitro antiviral test, the EC50 values of aprotinin against A/PR/8/34 (H1N1), A/CA/04/09 (H1N1, 2009 pandemic), A/PH/2/82 (H3N2), A/AB/Kor/CN5/09 (H6N5), A/Ck/Kor/01310/01 (H9N2) and A/Bris/10/07 (H3N2, oseltamivir-resistant) were 14 11, 21, 87, 57 and 110 nmol/L, respectively. In mice infected with lethal doses of the virus, aprotinin improved the survival rate of mice. The anti-IFV activity of aprotinin relates to its capacity to inhibit the proteolytic cleavage and structural rearrangement of HA required for successful fusion with host endosomes during IFV infection121.
6. Conclusions and prospects
Based on the understanding of each step of the IFV replication cycle, researchers have developed IFV inhibitors acting on different targets. Some of these inhibitors have been approved and successfully used in the treatment and prevention of influenza. However, due to the emergence of resistance mutations in IFV, the effectiveness of existing drugs has diminished, and therefore, there is an urgent need to develop highly effective anti-influenza agents, active against drug-resistant strains.
This article describes recent advances using four common drug design strategies focused on fighting drug resistance. The main reason for IFV resistance and cross-resistance is that mutations in IFV often lead to loss of binding affinity between antiviral drugs and target proteins. Therefore, improving the affinity between drugs and target proteins is one of the main strategies to improve their drug resistance profiles.
Strategies based on the use of covalent agents, multivalent drugs and FBDD introduced in this paper can be used to improve the affinity between the ligand and the target protein. In addition, bifunctional molecules, PROTACs and antibody recruitment techniques can also help in solving the problem of fighting drug resistance. These techniques use antibodies, proteasomes and other factors in the endosome to kill viruses or degrade viral proteins, and do not require a high affinity of the ligand for the target protein. In recent years, new mechanisms and targets of existing drugs have also been explored through drug repurposing strategies.
At present, the development of anti-drug resistant IFV inhibitors still confronts many difficulties. Firstly, drug design strategies targeting IFV proteins also have their respective disadvantages, such as: multivalent inhibitors usually have very high molecular weight, and their anti-influenza activity is usually tested in cell-based studies, so multivalent molecules are difficult to apply clinically as oral drugs. It is well known that covalent inhibitors usually have potential cytotoxicity. Bioavailability is the bottleneck for the development of bifunctional molecules, PROTAC molecules and ARMs. The limitation of the drug repurposing strategies is that the number of drugs on the market is limited.
On the whole, drug design strategies summarized in this paper are only applied for a limited number of drug targets, including IFV NA, HA, PA and NP proteins. We expect these strategies to be more promising due to their versatility. We also envisioned that some of the newly emerging drug design strategies in other fields will gradually be used in the field of anti-influenza drug discovery, such as ribonuclease-targeted chimeras122, emerging degrader technologies engaging lysosomal pathways123, substrate envelope hypothesis124, and other developments assisted by the use of bioinformatics and artificial intelligence technologies125.
Acknowledgments
We gratefully acknowledge financial support from the National Natural Science Foundation of China (NSFC No. 82173677), the Science Foundation for Outstanding Young Scholars of Shandong Province (ZR2020JQ31, China), and the China Postdoctoral Science Foundation (2022M711938). This work was supported in part by the Ministry of Science and Innovation of Spain through grant PID2019-104176RB-I00/AEI/10.13039/501100011033 awarded to Luis Menéndez-Arias-A. An institutional grant of the Fundación Ramón Areces to the CBMSO is also acknowledged. The team at CBMSO is affiliated to the Global Virus Network.
Author contributions
The writing of the first draft was initiated by Chuanfeng Liu and Lide Hu. Guanyu Dong and Ying Zhang put forward constructive suggestions. Edeildo Ferreira da Silva-Júnior, Xinyong Liu, Luis Menéndez-Arias, Peng Zhan edited the manuscript. Luis Menéndez-Arias and Peng Zhan reviewed and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Xinyong Liu, Email: xinyongl@sdu.edu.cn.
Luis Menéndez-Arias, Email: lmenendez@cbm.csic.es.
Peng Zhan, Email: zhanpeng1982@sdu.edu.cn.
References
- 1.Flerlage T., Boyd D.F., Meliopoulos V., Thomas P.G., Schultz-Cherry S. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat Rev Microbiol. 2021;19:425–441. doi: 10.1038/s41579-021-00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gostic K.M., Bridge R., Brady S., Viboud C., Worobey M., Lloyd-Smith J.O. Childhood immune imprinting to influenza A shapes birth year-specific risk during seasonal H1N1 and H3N2 epidemics. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington W.N., Kackos C.M., Webby R.J. The evolution and future of influenza pandemic preparedness. Exp Mol Med. 2021;53:737–749. doi: 10.1038/s12276-021-00603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichberg J., Maiworm E., Oberpaul M., Czudai-Matwich V., Lüddecke T., Vilcinskas A., et al. Antiviral potential of natural resources against influenza virus infections. Viruses. 2022;14:2452. doi: 10.3390/v14112452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faust J.S., Lin Z.Q., Rio C.D. Comparison of estimated excess deaths in New York City during the COVID-19 and 1918 influenza pandemics. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.17527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillezeau C., Alpert N., Caltabiano M., Flores R., Taioli E. Informing the global COVID-19 response by estimating excess deaths in Italy during the COVID-19 and 1918 influenza pandemics. J Med Virol. 2021;93:5239–5240. doi: 10.1002/jmv.27052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krischuns T., Lukarska M., Naffakh N., Cusack S. Influenza virus RNA-dependent RNA polymerase and the host transcriptional apparatus. Annu Rev Biochem. 2021;90:321–348. doi: 10.1146/annurev-biochem-072820-100645. [DOI] [PubMed] [Google Scholar]
- 8.Gu Y., Hsu A.C., Pang Z., Pan H., Zuo X., Wang G., et al. Role of the innate cytokine storm induced by the influenza A virus. Viral Immunol. 2019;32:244–251. doi: 10.1089/vim.2019.0032. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson E.C. Influenza virus. Trends Microbiol. 2018;26:809–810. doi: 10.1016/j.tim.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Z., Fodor E., Keown J.R. A structural understanding of influenza virus genome replication. Trends Microbiol. 2023;31:308–319. doi: 10.1016/j.tim.2022.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Gaymard A., Le Briand N., Frobert E., Lina B., Escuret V. Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin Microbiol Infect. 2016;22:975–983. doi: 10.1016/j.cmi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Blijleven J.S., Boonstra S., Onck P.R., van der Giessen E., van Oijen A.M. Mechanisms of influenza viral membrane fusion. Semin Cell Dev Biol. 2016;60:78–88. doi: 10.1016/j.semcdb.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Hutchinson E.C., Fodor E. Nuclear import of the influenza A virus transcriptional machinery. Vaccine. 2012;30:7353–7358. doi: 10.1016/j.vaccine.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 14.Fodor E., Te Velthuis A.J.W. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb Perspect Med. 2020;10:a038398. doi: 10.1101/cshperspect.a038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber A., Boff L., Anhlan D., Krischuns T., Brunotte L., Schuberth C., et al. Dissecting the mechanism of signaling-triggered nuclear export of newly synthesized influenza virus ribonucleoprotein complexes. Proc Natl Acad Sci U S A. 2020;117:16557–16566. doi: 10.1073/pnas.2002828117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haralampiev I., Prisner S., Nitzan M., Schade M., Jolmes F., Schreiber M., et al. Selective flexible packaging pathways of the segmented genome of influenza A virus. Nat Commun. 2020;11:4355. doi: 10.1038/s41467-020-18108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayak D.P., Balogun R.A., Yamada H., Zhou Z.H., Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe T., Watanabe S., Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Sun Y., Liu S. Emerging antiviral therapies and drugs for the treatment of influenza. Expet Opin Emerg Drugs. 2022:1–15. doi: 10.1080/14728214.2022.2149734. [DOI] [PubMed] [Google Scholar]
- 20.Wu X., Wu X.L., Sun Q.Z., Zhang C.H., Yang S.Y., Li L., et al. Progress of small molecular inhibitors in the development of anti-influenza virus agents. Theranostics. 2017;7:826–845. doi: 10.7150/thno.17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terrier O., Slama-Schwok A. Anti-influenza drug discovery and development: targeting the virus and its host by all possible means. Adv Exp Med Biol. 2021;1322:195–218. doi: 10.1007/978-981-16-0267-2_8. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y., Li H., Wu M., Zhang H., Ding Y., Peng Y., et al. Single and multiple dose pharmacokinetics and safety of ZSP1273, an RNA polymerase PB2 protein inhibitor of the influenza A virus: a phase 1 double-blind study in healthy subjects. Expet Opin Invest Drugs. 2021;30:1159–1167. doi: 10.1080/13543784.2021.1994944. [DOI] [PubMed] [Google Scholar]
- 23.Luo D., Ye Q., Li R.T., Zhou H.Y., Guo J.J., Zhao S.Q., et al. PA-E18G substitution in influenza A virus confers resistance to ZX-7101, a cap-dependent endonuclease inhibitor. Virol Sin. 2023;S1995-820X:62–67. doi: 10.1016/j.virs.2023.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou L., Zhang Y., Ju H., Cherukupalli S., Jia R., Zhang J., et al. Contemporary medicinal chemistry strategies for the discovery and optimization of influenza inhibitors targeting vRNP constituent proteins. Acta Pharm Sin B. 2022;12:1805–1824. doi: 10.1016/j.apsb.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Z., Lou K., Wang W. New small-molecule drug design strategies for fighting resistant influenza A. Acta Pharm Sin B. 2015;5:419–430. doi: 10.1016/j.apsb.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurt A.C. The epidemiology and spread of drug resistant human influenza viruses. Curr Opin Virol. 2014;8:22–29. doi: 10.1016/j.coviro.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Yusuf M., Mohamed N., Mohamad S., Janezic D., Damodaran K.V., Wahab H.A. H274Y's Effect on oseltamivir resistance: what happens before the drug enters the binding site. J Chem Inf Model. 2016;56:82–100. doi: 10.1021/acs.jcim.5b00331. [DOI] [PubMed] [Google Scholar]
- 28.Jones J.C., Rovito S.W., Penaflor M.K., Webby R.J., Govorkova E.A. Influenza A virus polymerase acidic protein E23R substitution is a marker of reduced susceptibility to baloxavir. Antivir Res. 2022;204 doi: 10.1016/j.antiviral.2022.105369. [DOI] [PubMed] [Google Scholar]
- 29.Takashita E., Fujisaki S., Shirakura M., Nakamura K., Kishida N., Kuwahara T., et al. Influenza A(H1N1)pdm09 virus exhibiting enhanced cross-resistance to oseltamivir and peramivir due to a dual H275Y/G147R substitution, Japan, March 2016. Euro Surveill. 2016:21. doi: 10.2807/1560-7917.ES.2016.21.24.30258. [DOI] [PubMed] [Google Scholar]
- 30.Abed Y., Boivin G. A review of clinical influenza A and B infections with reduced susceptibility to both oseltamivir and zanamivir. Open Forum Infect Dis. 2017;4 doi: 10.1093/ofid/ofx105. ofx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pabbaraju K., Ho K.C., Wong S., Shokoples S., Pang X.L., Fonseca K., et al. Adamantane resistance in circulating human influenza A viruses from Alberta, Canada (1970–2007) Antivir Res. 2008;79:81–86. doi: 10.1016/j.antiviral.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Hadj Hassine I., Ben M'hadheb M., Menéndez-Arias L. Lethal mutagenesis of RNA viruses and approved drugs with antiviral mutagenic activity. Viruses. 2022;14:841. doi: 10.3390/v14040841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Y., Frutos-Beltrán E., Kang D., Pannecouque C., De Clercq E., Menéndez-Arias L., et al. Medicinal chemistry strategies for discovering antivirals effective against drug-resistant viruses. Chem Soc Rev. 2021;50:4514–4540. doi: 10.1039/d0cs01084g. [DOI] [PubMed] [Google Scholar]
- 34.Domingo E., Mas A., Yuste E., Pariente N., Sierra S., Gutiérrez-Riva M., et al. Virus population dynamics, fitness variations and the control of viral disease: an update. Prog Drug Res. 2001;57:77–115. doi: 10.1007/978-3-0348-8308-5_2. [DOI] [PubMed] [Google Scholar]
- 35.Greenbaum B.D., Li O.T., Poon L.L., Levine A.J., Rabadan R. Viral reassortment as an information exchange between viral segments. Proc Natl Acad Sci U S A. 2012;109:3341–3346. doi: 10.1073/pnas.1113300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyk J.M., Szydłowska N., Szulc W., Majewska A. Evolution of influenza viruses-drug resistance, treatment options, and prospects. Int J Mol Sci. 2022;23 doi: 10.3390/ijms232012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sautto G.A., Kirchenbaum G.A., Ross T.M. Towards a universal influenza vaccine: different approaches for one goal. Virol J. 2018;15:17. doi: 10.1186/s12985-017-0918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robert J Geraghty, Matthew T Aliota, Laurent F Bonnac. Broad-spectrum antiviral strategies and nucleoside analogues. Viruses. 2021;13:667. doi: 10.3390/v13040667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bissaro M., Sturlese M., Moro S. The rise of molecular simulations in fragment-based drug design (FBDD): an overview. Drug Discov Today. 2020;25:1693–1701. doi: 10.1016/j.drudis.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q., Kang C. Perspectives on fragment-based drug discovery: a strategy applicable to diverse targets. Curr Top Med Chem. 2021;21:1099–1112. doi: 10.2174/1568026621666210804115700. [DOI] [PubMed] [Google Scholar]
- 41.Credille C.V., Chen Y., Cohen S.M. Fragment-based identification of influenza endonuclease inhibitors. J Med Chem. 2016;59:6444–6454. doi: 10.1021/acs.jmedchem.6b00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauman J.D., Patel D., Baker S.F., Vijayan R.S., Xiang A., Parhi A.K., et al. Crystallographic fragment screening and structure-based optimization yields a new class of influenza endonuclease inhibitors. ACS Chem Biol. 2013;8:2501–2508. doi: 10.1021/cb400400j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao Y., Ye Y., Liu M., Liu Z., Wang J., Li B., et al. Identification of N- and C-3-modified laudanosoline derivatives as novel influenza PAN endonuclease inhibitors. J Med Chem. 2023;66:188–219. doi: 10.1021/acs.jmedchem.2c00857. [DOI] [PubMed] [Google Scholar]
- 44.Jalily P.H., Duncan M.C., Fedida D., Wang J., Tietjen I. Put a cork in it: plugging the M2 viral ion channel to sink influenza. Antivir Res. 2020;178 doi: 10.1016/j.antiviral.2020.104780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y., Canturk B., Jo H., Ma C., Gianti E., Klein M.L., et al. Flipping in the pore: discovery of dual inhibitors that bind in different orientations to the wild-type versus the amantadine-resistant S31N mutant of the influenza A virus M2 proton channel. J Am Chem Soc. 2014;136:17987–17995. doi: 10.1021/ja508461m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F., Ma C., DeGrado W.F., Wang J. Discovery of highly potent inhibitors targeting the predominant drug-resistant S31N mutant of the influenza A virus M2 proton channel. J Med Chem. 2016;59:1207–1216. doi: 10.1021/acs.jmedchem.5b01910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F., Hu Y., Wang Y., Ma C., Wang J. Expeditious lead optimization of isoxazole-containing influenza A virus M2-S31N inhibitors using the suzuki-miyaura cross-coupling reaction. J Med Chem. 2017;60:1580–1590. doi: 10.1021/acs.jmedchem.6b01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Hu Y., Xu S., Zhang Y., Musharrafieh R., Hau R.K., et al. In vitro pharmacokinetic optimizations of AM2-S31N channel blockers led to the discovery of slow-binding inhibitors with potent antiviral activity against drug-resistant influenza A viruses. J Med Chem. 2018;61:1074–1085. doi: 10.1021/acs.jmedchem.7b01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cáceres C.J., Hu Y., Cárdenas-García S., Wu X., Tan H., Carnaccini S., et al. Rational design of a deuterium-containing M2-S31N channel blocker UAWJ280 with in vivo antiviral efficacy against both oseltamivir sensitive and -resistant influenza A viruses. Emerg Microb Infect. 2021;10:1832–1848. doi: 10.1080/22221751.2021.1972769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y., Musharrafieh R., Ma C., Zhang J., Smee D.F., DeGrado W.F., et al. An M2-V27A channel blocker demonstrates potent in vitro and in vivo antiviral activities against amantadine-sensitive and -resistant influenza A viruses. Antivir Res. 2017;140:45–54. doi: 10.1016/j.antiviral.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Cady S.D., Balannik V., Pinto L.H., DeGrado W.F., Hong M. Discovery of spiro-piperidine inhibitors and their modulation of the dynamics of the M2 proton channel from influenza A virus. J Am Chem Soc. 2009;131:8066–8076. doi: 10.1021/ja900063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Cesco S., Kurian J., Dufresne C., Mittermaier A.K., Moitessier N. Covalent inhibitors design and discovery. Eur J Med Chem. 2017;138:96–114. doi: 10.1016/j.ejmech.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 53.Lonsdale R., Ward R.A. Structure-based design of targeted covalent inhibitors. Chem Soc Rev. 2018;47:3816–3830. doi: 10.1039/c7cs00220c. [DOI] [PubMed] [Google Scholar]
- 54.Gehringer M., Laufer S.A. Emerging and re-Emerging warheads for targeted covalent inhibitors: applications in medicinal chemistry and chemical biology. J Med Chem. 2019;62:5673–5724. doi: 10.1021/acs.jmedchem.8b01153. [DOI] [PubMed] [Google Scholar]
- 55.Sutanto F., Konstantinidou M., Dömling A. Covalent inhibitors: a rational approach to drug discovery. RSC Med Chem. 2020;11:876–884. doi: 10.1039/d0md00154f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vavricka C.J., Liu Y., Kiyota H., Sriwilaijaroen N., Qi J., Tanaka K., et al. Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors. Nat Commun. 2013;4:1491. doi: 10.1038/ncomms2487. [DOI] [PubMed] [Google Scholar]
- 57.Kim J.H., Resende R., Wennekes T., Chen H.M., Bance N., Buchini S., et al. Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity. Science. 2013;340:71–75. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- 58.McKimm-Breschkin J.L., Barrett S., Pilling P.A., Hader S., Watts A.G., Streltsov V.A. Structural and functional analysis of anti-influenza activity of 4-, 7-, 8- and 9-deoxygenated 2,3-difluoro-N-acetylneuraminic acid derivatives. J Med Chem. 2018;61:1921–1933. doi: 10.1021/acs.jmedchem.7b01467. [DOI] [PubMed] [Google Scholar]
- 59.Makhoba X.H., Viegas C., Jr., Mosa R.A., Viegas F.P.D., Pooe O.J. Potential impact of the multi-target drug approach in the treatment of some complex diseases. Drug Des Dev Ther. 2020;14:3235–3249. doi: 10.2147/DDDT.S257494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohamed M.R., Mohile S.G., Juba K.M., Awad H., Wells M., Loh K.P., et al. Association of polypharmacy and potential drug–drug interactions with adverse treatment outcomes in older adults with advanced cancer. Cancer. 2023;129:1096–1104. doi: 10.1002/cncr.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu R.G., Sun Y., Sheng W.B., Liao D.F. Designing multi-targeted agents: an emerging anticancer drug discovery paradigm. Eur J Med Chem. 2017;136:195–211. doi: 10.1016/j.ejmech.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 62.Liu K.C., Fang J.M., Jan J.T., Cheng T.J., Wang S.Y., Yang S.T., et al. Enhanced anti-influenza agents conjugated with anti-inflammatory activity. J Med Chem. 2012;55:8493–8501. doi: 10.1021/jm3009844. [DOI] [PubMed] [Google Scholar]
- 63.Lv X., Wang P., Li C., Cheng S., Bi Y., Li X. Zanamivir-cholesterol conjugate: a long-acting neuraminidase inhibitor with potent efficacy against drug-resistant influenza viruses. J Med Chem. 2021;64:17403–17412. doi: 10.1021/acs.jmedchem.1c01531. [DOI] [PubMed] [Google Scholar]
- 64.Békés M., Langley D.R., Crews C.M. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21:181–200. doi: 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li K., Crews C.M. PROTACs: past, present and future. Chem Soc Rev. 2022;51:5214–5236. doi: 10.1039/d2cs00193d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X., Song Y.C. Proteolysis-targeting chimera (PROTAC) for targeted protein degradation and cancer therapy. J Hematol Oncol. 2020;13:50. doi: 10.1186/s13045-020-00885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding Y., Fei Y.Y., Lu B.X. Emerging new concepts of degrader technologies. Trends Pharmacol Sci. 2020;41:464–474. doi: 10.1016/j.tips.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martín-Acosta P., Xiao X.S. PROTACs to address the challenges facing small molecule inhibitors. Eur J Med Chem. 2021;210 doi: 10.1016/j.ejmech.2020.112993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dale B., Cheng M., Park K.S., Kaniskan H.Ü., Xiong Y., Jin J. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer. 2021;21:638–654. doi: 10.1038/s41568-021-00365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barghout S.H. Targeted protein degradation: an emerging therapeutic strategy in cancer. Anti Cancer Agents Med Chem. 2021;21:214–230. doi: 10.2174/1871520620666200410082652. [DOI] [PubMed] [Google Scholar]
- 71.Qi S.M., Dong J., Xu Z.Y., Cheng X.D., Zhang W.D., Qin J.J. PROTAC: an effective targeted protein degradation strategy for cancer therapy. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.692574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Z.C., Liu X.J., Ma X.Y., Zou W.T., Chen Q., Chen F.F., et al. Discovery of oseltamivir-based novel PROTACs as degraders targeting neuraminidase to combat H1N1 influenza virus. Cell Insight. 2022;1 doi: 10.1016/j.cellin.2022.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H., Wang S., Ma W., Cheng B., Yi Y., Ma X., et al. Discovery of pentacyclic triterpenoid PROTACs as a class of effective hemagglutinin protein degraders. J Med Chem. 2022;65:7154–7169. doi: 10.1021/acs.jmedchem.1c02013. [DOI] [PubMed] [Google Scholar]
- 74.Zhao J., Wang J., Pang X., Liu Z., Li Q., Yi D., et al. An anti-influenza A virus microbial metabolite acts by degrading viral endonuclease PA. Nat Commun. 2022;13:2079. doi: 10.1038/s41467-022-29690-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Si L., Shen Q., Li J., Chen L., Shen J., Xiao X., et al. Generation of a live attenuated influenza A vaccine by proteolysis targeting. Nat Biotechnol. 2022;40:1370–1377. doi: 10.1038/s41587-022-01381-4. [DOI] [PubMed] [Google Scholar]
- 76.Hon W.C., Wilson M.I., Harlos K., Claridge T.D., Schofield C.J., Pugh C.W., et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 77.Gu S., Cui D., Chen X., Xiong X., Zhao Y. PROTACs: an emerging targeting technique for protein degradation in drug discovery. Bioessays. 2018;40 doi: 10.1002/bies.201700247. [DOI] [PubMed] [Google Scholar]
- 78.Jakobsche C.E., Parker C.G., Tao R.N., Kolesnikova M.D., Douglass E.F., Jr., Spiegel D.A. Exploring binding and effector functions of natural human antibodies using synthetic immunomodulators. ACS Chem Biol. 2013;8:2404–2411. doi: 10.1021/cb4004942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spiegel D.A. Grand challenge commentary: synthetic immunology to engineer human immunity. Nat Chem Biol. 2010;6:871–872. doi: 10.1038/nchembio.477. [DOI] [PubMed] [Google Scholar]
- 80.Sasaki K., Muguruma K., Osawa R., Fukuda A., Taniguchi A., Kishimura A., et al. Synthesis and biological evaluation of a monocyclic Fc-binding antibody-recruiting molecule for cancer immunotherapy. RSC Med Chem. 2021;12:406–409. doi: 10.1039/d0md00337a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schrand B., Clark E., Levay A., Capote A.R., Martinez O., Brenneman R., et al. Author correction: hapten-mediated recruitment of polyclonal antibodies to tumors engenders antitumor immunity. Nat Commun. 2021;12:6939. doi: 10.1038/s41467-021-25400-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rullo A.F., Fitzgerald K.J., Muthusamy V., Liu M., Yuan C., Huang M., et al. Re-engineering the immune response to metastatic cancer: antibody-recruiting small molecules targeting the urokinase receptor. Angew Chem Int Ed Engl. 2016;55:3642–3646. doi: 10.1002/anie.201510866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feigman M.S., Kim S., Pidgeon S.E., Yu Y., Ongwae G.M., Patel D.S., et al. Synthetic immunotherapeutics against gram-negative pathogens. Cell Chem Biol. 2018;25:1185–1194.e5. doi: 10.1016/j.chembiol.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferrari G., Haynes B.F., Koenig S., Nordstrom J.L., Margolis D.M., Tomaras G.D. Envelope-specific antibodies and antibody-derived molecules for treating and curing HIV infection. Nat Rev Drug Discov. 2016;15:823–834. doi: 10.1038/nrd.2016.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parker C.G., Dahlgren M.K., Tao R.N., Li D.T., Douglass E.F., Jr., Shoda T., et al. Illuminating HIV gp120-ligand recognition through computationally-driven optimization of antibody-recruiting molecules. Chem Sci. 2014;5:2311–2317. doi: 10.1039/C4SC00484A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu X., Zhang B., Wang Y., Haymour H.S., Zhang F., Xu L.C., et al. A universal dual mechanism immunotherapy for the treatment of influenza virus infections. Nat Commun. 2020;11:5597. doi: 10.1038/s41467-020-19386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu X., Luo W., Zhang B., Lee Y.G., Shahriar I., Srinivasarao M., et al. Design of neuraminidase-targeted imaging and therapeutic agents for the diagnosis and treatment of influenza virus infections. Bioconjugate Chem. 2021;32:1548–1553. doi: 10.1021/acs.bioconjchem.1c00255. [DOI] [PubMed] [Google Scholar]
- 88.Qian E.A., Han Y., Messina M.S., Maynard H.D., Král P., Spokoyny A.M. Multivalent cluster nanomolecules for inhibiting protein-protein interactions. Bioconjugate Chem. 2019;30:2594–2603. doi: 10.1021/acs.bioconjchem.9b00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen B., Gianolio D.A., Stefano J.E., Manning C.M., Gregory R.C., Busch M.M., et al. Design, synthesis, and in vitro evaluation of multivalent drug linkers for high-drug-load antibody-drug conjugates. ChemMedChem. 2018;13:790–794. doi: 10.1002/cmdc.201700722. [DOI] [PubMed] [Google Scholar]
- 90.Wittmann V., Pieters R.J. Bridging lectin binding sites by multivalent carbohydrates. Chem Soc Rev. 2013;42:4492–4503. doi: 10.1039/c3cs60089k. [DOI] [PubMed] [Google Scholar]
- 91.Kitov P.I., Sadowska J.M., Mulvey G., Armstrong G.D., Ling H., Pannu N.S., et al. Shiga-like toxins are neutralized by tailored multivalent carbohydrate ligands. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]
- 92.Liese S., Netz R.R. Quantitative prediction of multivalent ligand-receptor binding affinities for influenza, cholera, and anthrax inhibition. ACS Nano. 2018;12:4140–4147. doi: 10.1021/acsnano.7b08479. [DOI] [PubMed] [Google Scholar]
- 93.Csizmar C.M., Petersburg J.R., Perry T.J., Rozumalski L., Hackel B.J., Wagner C.R. Multivalent ligand binding to cell membrane antigens: defining the interplay of affinity, valency, and expression density. J Am Chem Soc. 2019;141:251–261. doi: 10.1021/jacs.8b09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martínez-Gualda B., Sun L., Rivero-Buceta E., Flores A., Quesada E., Balzarini J., et al. Structure-activity relationship studies on a Trp dendrimer with dual activities against HIV and enterovirus A71. Modifications on the amino acid. Antivir Res. 2017;139:32–40. doi: 10.1016/j.antiviral.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 95.Overeem N.J., van der Vries E., Huskens J. A dynamic, supramolecular view on the multivalent interaction between influenza virus and host cell. Small. 2021;17 doi: 10.1002/smll.202007214. [DOI] [PubMed] [Google Scholar]
- 96.Lin M., Zhang J., Wan H., Yan C., Xia F. Rationally designed multivalent aptamers targeting cell surface for biomedical applications. ACS Appl Mater Interfaces. 2021;13:9369–9389. doi: 10.1021/acsami.0c15644. [DOI] [PubMed] [Google Scholar]
- 97.Günther S.C., Maier J.D., Vetter J., Podvalnyy N., Khanzhin N., Hennet T., et al. Antiviral potential of 3′-sialyllactose- and 6′-sialyllactose-conjugated dendritic polymers against human and avian influenza viruses. Sci Rep. 2020;10:768. doi: 10.1038/s41598-020-57608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiao S., Si L., Tian Z., Jiao P., Fan Z., Meng K., et al. Pentacyclic triterpenes grafted on CD cores to interfere with influenza virus entry: a dramatic multivalent effect. Biomaterials. 2016;78:74–85. doi: 10.1016/j.biomaterials.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 99.Vonnemann J., Liese S., Kuehne C., Ludwig K., Dernedde J., Böttcher C., et al. Size dependence of steric shielding and multivalency effects for globular binding inhibitors. J Am Chem Soc. 2015;137:2572–2579. doi: 10.1021/ja5114084. [DOI] [PubMed] [Google Scholar]
- 100.Kanfar N., Bartolami E., Zelli R., Marra A., Winum J.Y., Ulrich S, et al. Emerging trends in enzyme inhibition by multivalent nanoconstructs. Org Biomol Chem. 2015;13:9894–9906. doi: 10.1039/c5ob01405k. [DOI] [PubMed] [Google Scholar]
- 101.Zelikin A.N., Stellacci F. Broad-spectrum antiviral agents based on multivalent inhibitors of viral infectivity. Adv Healthcare Mater. 2021;10 doi: 10.1002/adhm.202001433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nie C., Parshad B., Bhatia S., Cheng C., Stadtmüller M., Oehrl A., et al. Topology-matching design of an influenza-neutralizing spiky nanoparticle-based inhibitor with a dual mode of action. Angew Chem Weinheim Bergstr Ger. 2020;132:15662–15666. doi: 10.1002/ange.202004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nie C., Stadtmüller M., Parshad B., Wallert M., Ahmadi V., Kerkhoff Y., et al. Heteromultivalent topology-matched nanostructures as potent and broad-spectrum influenza A virus inhibitors. Sci Adv. 2021;7 doi: 10.1126/sciadv.abd3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bhatia S., Hilsch M., Cuellar-Camacho J.L., Ludwig K., Nie C., Parshad B., et al. Adaptive flexible sialylated nanogels as highly potent influenza a virus inhibitors. Angew Chem Int Ed Engl. 2020;59:12417–12422. doi: 10.1002/anie.202006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lauster D., Klenk S., Ludwig K., Nojoumi S., Behren S., Adam L., et al. Phage capsid nanoparticles with defined ligand arrangement block influenza virus entry. Nat Nanotechnol. 2020;15:373–379. doi: 10.1038/s41565-020-0660-2. [DOI] [PubMed] [Google Scholar]
- 106.Dominguez L.W., Willis J.S. Research and development costs of new drugs. JAMA. 2020;324:516. doi: 10.1001/jama.2020.8645. [DOI] [PubMed] [Google Scholar]
- 107.Dickson M., Gagnon J.P. Key factors in the rising cost of new drug discovery and development. Nat Rev Drug Discov. 2004;3:417–429. doi: 10.1038/nrd1382. [DOI] [PubMed] [Google Scholar]
- 108.He H., Duo H., Hao Y., Zhang X., Zhou X., Zeng Y., et al. Computational drug repurposing by exploiting large-scale gene expression data: strategy, methods and applications. Comput Biol Med. 2023;155 doi: 10.1016/j.compbiomed.2023.106671. [DOI] [PubMed] [Google Scholar]
- 109.Dos Santos Nascimento I.J., de Aquino T.M., da Silva-Júnior E.F. Drug repurposing: a strategy for discovering inhibitors against emerging viral infections. Curr Med Chem. 2021;28:2887–2942. doi: 10.2174/0929867327666200812215852. [DOI] [PubMed] [Google Scholar]
- 110.Jain P., Jain S.K., Jain M. Harnessing drug repurposing for exploration of new diseases: an insight to strategies and case studies. Curr Mol Med. 2021;21:111–132. doi: 10.2174/1566524020666200619125404. [DOI] [PubMed] [Google Scholar]
- 111.Roessler H.I., Knoers N.V.A.M., van Haelst M.M., van Haaften G. Drug repurposing for rare diseases. Trends Pharmacol Sci. 2021;42:255–267. doi: 10.1016/j.tips.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 112.Schcolnik-Cabrera A., Juárez-López D., Duenas-Gonzalez A. Perspectives on drug repurposing. Curr Med Chem. 2021;28:2085–2099. doi: 10.2174/0929867327666200831141337. [DOI] [PubMed] [Google Scholar]
- 113.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 114.Rosas C., Sinning M., Ferreira A., Fuenzalida M., Lemus D. Celecoxib decreases growth and angiogenesis and promotes apoptosis in a tumor cell line resistant to chemotherapy. Biol Res. 2014;47:27. doi: 10.1186/0717-6287-47-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tabata R., Tabata C., Uesugi H., Takei Y. Highly aggressive plasmablastic neoplasms in patients with rheumatoid arthritis treated with methotrexate. Int Immunopharm. 2019;68:213–217. doi: 10.1016/j.intimp.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 116.Wu K.C., Liao K.S., Yeh L.R., Wang Y.K. Drug repurposing: the mechanisms and signaling pathways of anti-cancer effects of anesthetics. Biomedicines. 2022;10:1589. doi: 10.3390/biomedicines10071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rossignol J.F., La Frazia S., Chiappa L., Ciucci A., Santoro M.G. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J Biol Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mifsud E.J., Tilmanis D., Oh D.Y., Ming-Kay Tai C., Rossignol J.F., Hurt A.C. Prophylaxis of ferrets with nitazoxanide and oseltamivir combinations is more effective at reducing the impact of influenza a virus infection compared to oseltamivir monotherapy. Antivir Res. 2020;176 doi: 10.1016/j.antiviral.2020.104751. [DOI] [PubMed] [Google Scholar]
- 119.Lejal N., Tarus B., Bouguyon E., Chenavas S., Bertho N., Delmas B., et al. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob Agents Chemother. 2013;57:2231–2242. doi: 10.1128/AAC.02335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zheng W., Fan W., Zhang S., Jiao P., Shang Y., Cui L., et al. Naproxen exhibits broad anti-influenza virus activity in mice by impeding viral nucleoprotein nuclear export. Cell Rep. 2019;27:1875–1885.e5. doi: 10.1016/j.celrep.2019.04.053. [DOI] [PubMed] [Google Scholar]
- 121.Song E.J., Españo E., Shim S.M., Nam J.H., Kim J., Lee K., et al. Inhibitory effects of aprotinin on influenza A and B viruses in vitro and in vivo. Sci Rep. 2021;11:9427. doi: 10.1038/s41598-021-88886-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Haj-Yahia S., Nandi A., Benhamou R.I. Targeted degradation of structured RNAs via ribonuclease-targeting chimeras (RiboTacs) Expet Opin Drug Discov. 2023;18 doi: 10.1080/17460441.2023.2224960. 9290–42. [DOI] [PubMed] [Google Scholar]
- 123.Ding Y., Xing D., Fei Y., Lu B. Emerging degrader technologies engaging lysosomal pathways. Chem Soc Rev. 2022;51:8832–8876. doi: 10.1039/d2cs00624c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chellappan S., Kairys V., Fernandes M.X., Schiffer C., Gilson M.K. Evaluation of the substrate envelope hypothesis for inhibitors of HIV-1 protease. Proteins. 2007;68:561–567. doi: 10.1002/prot.21431. [DOI] [PubMed] [Google Scholar]
- 125.Li S., Wu S., Wang L., Li F., Jiang H., Bai F. Recent advances in predicting protein-protein interactions with the aid of artificial intelligence algorithms. Curr Opin Struct Biol. 2022;73 doi: 10.1016/j.sbi.2022.102344. [DOI] [PubMed] [Google Scholar]