Abstract
The nitrogen assimilation control protein (NAC) binds to a site within the promoter region of the histidine utilization operon (hutUH) of Klebsiella aerogenes, and NAC bound at this site activates transcription of hutUH. This NAC-binding site was characterized by a combination of random and directed DNA mutagenesis. Mutations that abolished or diminished in vivo transcriptional activation by NAC were found to lie within a 15-bp region contained within the 26-bp region protected by NAC from DNase I digestion. This 15-bp core has the palindromic ends ATA and TAT, and it matches the consensus for LysR family transcriptional regulators. Protein-binding experiments showed that transcriptional activation in vivo decreased with decreasing binding in vitro. In contrast to the NAC-binding site from hutUH, the NAC-binding site from the gdhA promoter failed to activate transcription from a semisynthetic promoter, and this failure was not due to weak binding or greatly distorted protein-DNA structure. Mutations in the promoter-proximal half-site of the NAC-binding site from gdhA allowed this site to activate transcription. Similar studies using the NAC-binding site from hut showed that two mutations in the promoter proximal half-site increased binding but abolished transcriptional activation. Interestingly, for symmetric mutations in the promoter-distal half-site, loss of transcriptional activation was always correlated with a decrease in binding. We conclude from these observations that if the binding in vitro reflects the binding in vivo, then binding of NAC to DNA is not sufficient for transcriptional activation and that the NAC-binding site can be functionally divided in two half-sites, with related but different functions.
The enteric bacterium Klebsiella aerogenes can use a variety of organic and inorganic sources of nitrogen, with ammonium being the preferred source. If the ammonium concentration in the growth medium is low, several operons are derepressed (reviewed in reference 20). The protein products of these nitrogen-regulated operons allow the catabolism of alternative nitrogen sources, such as amino acids and urea. The cellular sensitivity to a variable nitrogen concentration is provided by the Ntr system, a regulatory network built around a two-component system and using an RNA polymerase carrying the unusual sigma factor ς54 (reviewed in reference 34). All nitrogen-regulated operons in K. aerogenes require a functional Ntr system. Some operons need not only the Ntr system but also an additional transcription factor, the nitrogen assimilation control protein (NAC) (2–4). The nac gene is under positive regulation by the Ntr system and couples the ς54-dependent regulation of the Ntr system to the expression of ς70-dependent operons (11).
The NAC polypeptide is a member of the LysR family of trans-acting transcriptional regulators (31, 32), and it has been shown to be both necessary and sufficient for the activation of several operons that produce ammonia or glutamate through the catabolism of organic molecules (19, 33). Among these operons are histidine utilization (hut), proline utilization (put), urea degradation (ure), and possibly d-amino acid dehydrogenase (dad) (16). NAC also represses both its own expression (12) and the expression of operons that consume ammonia: glutamate dehydrogenase (gdhA) and glutamate syntethase (gltBD) (33). The best-characterized example of transcriptional activation by NAC is the hutU promoter (hutUp). Activation of this promoter by NAC has been demonstrated in vitro with purified components and requires a single binding site located upstream from the promoter (14). This binding site has been defined by the gel mobility shift assay and by NAC-mediated protection of the site from DNase I digestion. These studies show that NAC binds to a 26-bp fragment at hutUp. This 26-bp fragment from hutUp is sufficient to confer regulation by NAC to a variety of semisynthetic promoters (28). DNase I protection experiments showed that NAC binds in the promoter regions of other Ntr-activated promoters. Sequence alignment of the NAC-protected regions from the hut, put, and ure promoters revealed a 15-bp consensus sequence for NAC binding: 5′-ATAP-N3-WNTYGTAT-3′ (P, purine; Y, pyrimidine; W, A or T) (14). The NAC consensus fits the general LysR family consensus 5′-A-N11-T-3′ (31). In all promoters activated by NAC in K. aerogenes, this 15-bp sequence is positioned with its center at −64 relative to the transcriptional start site.
In this work, we have isolated and characterized mutations in the DNase I-protected region of hutUp that identify key nucleotides for protein-DNA interaction and transcriptional activation.
MATERIALS AND METHODS
Strains and plasmids.
K. aerogenes KC2668 (hutC515 Δ[bla]-2) was used in all experiments as a host for wild-type and mutant plasmids. Escherichia coli DH5α (Gibco BRL) was used as an intermediary host to select ligation products and for long-term storage of plasmids. The fusion vector pRJ800 (ColE1 replicon, Ampr, multiple cloning site from pUC18, promoterless lacZ reporter), a derivative of pRZ5202 (29), was used to clone all the PCR products. The cloning into the pRJ800 polylinker created transcriptional fusions of the PCR products with the plasmid-borne reporter lacZ gene.
Random mutagenesis of the NAC-binding site of hutUp.
The NAC site from the hutU promoter was mutagenized with degenerate oligonucleotides as PCR primers by the method of Chiang et al. (9). Briefly, the region to be mutagenized was synthesized under conditions in which the three non-wild-type deoxynucleoside triphosphates were present at a low frequency (9% non-wild-type nucleotides) in order to introduce degeneracy into the oligonucleotides synthesized. The sequence of the oligonucleotide was GATTACGAATTCGGACGcaatataacaaaattgtatcattctGTTAAAATCCTG, where a lowercase letter signifies a position subjected to mutagenesis. The degenerate oligonucleotides were synthesized at the University of Michigan Biomedical Research Core Facilities. These degenerate oligonucleotides were used as one primer to amplify the hutU promoter region with pRO80 (26) as the DNA template along with an opposing primer homologous to vector DNA sequence. PCR conditions are described below. The resulting PCR product was separated from other material by agarose gel electrophoresis, digested with EcoRI and HindIII, and ligated to pRJ800 digested with the same enzymes. The resulting plasmids fused the mutagenized hutUp from position −79 to +59 to the lacZ reporter gene of pRJ800. E. coli DH5α was transformed with these plasmids, and the DNA sequence of the cloned material was determined as described below. Mutant plasmids of interest were moved into K. aerogenes KC2668 by transformation for in vivo analysis.
Oligonucleotide-directed mutagenesis.
Single or multiple changes were introduced in the NAC-binding sites contained in plasmids pCB648(hutUp-lacZp chimera), pCB803 (gdhAp-lacZp chimera), and pCB886 (hutUp) by the amplification of template DNA with mutant primers. The PCR-mix contained plasmid template (0.5 mg), 0.2 mM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, and 0.5 U of Taq DNA polymerase (Gibco BRL). Each PCR yielded a major product that was purified by electroelution from agarose gels or by using Qiaquick Spin Columns (Qiagen), digested with EcoRI and XbaI, and ligated into pRJ800. The nucleotide sequences of the cloned PCR products were determined by the chain termination method by using a kit purchased from United States Biomedical. Sequenced clones were used to obtain secondary transformants of KC2668, and these secondary transformants were used for all the in vivo experiments.
DNA manipulation.
Plasmid DNA purification by alkaline lysis or Qiaprep Spin Columns (Qiagen), DNA digestion with restriction endonucleases, DNA ligation, DNA electrophoresis in agarose and polyacrylamide gels, and cell transformation were performed by standard protocols (21).
Growth conditions.
Cell cultures were grown in a roller drum at 30°C to approximately 50 Klett units in W phosphate medium (1) brought to pH 7.4 with KOH, supplemented with glucose (0.4%; Sigma) as a carbon source. Freshly made glutamine (0.2%; Calbiochem) was used as a growth-limiting nitrogen source. Ammonium sulfate (0.2%; Sigma) was added where indicated as a nitrogen-excess condition. Ampicillin (Sigma) was added at a concentration of 100 μg/ml when required.
Enzyme assays.
The β-galactosidase assays were performed as described by Miller (22) except that cells were made permeable with 0.2% hexadecyl trimethylammonium bromide and 0.02% sodium deoxycholate. Activities were reported as Miller units or specific activity (units/milligram of protein) by the method of Lowry et al. (18) to measure protein concentration.
Gel mobility shift assay.
The plasmids containing wild-type and mutant NAC sites were incubated with XbaI, labeled with Klenow fragment and [32P]dATP, and digested with EcoRI. The specific activity of labeled plasmids was 5 × 103 ± 6 × 102 cpm/fmol (± standard deviation). Labeled fragments were incubated with purified NAC or NAC dilution buffer 6 (14) and a 10-fold excess of calf thymus DNA in a total volume of 5 μl. The binding mixtures were incubated for 20 min at room temperature, and after this time 1 μl of DNA loading buffer was added. Each reaction was loaded into a Tris-EDTA 4% polyacrylamide gel, and the fragments were separated at 13.3 V/cm by using a Hoeffer electrophoresis chamber. The gel was transferred to 3MM filter paper and dried. Autoradiograms were obtained by exposing radiographic films at −70°C with an intensifying screen. Films were developed in an X-Omat developer. The radioactivity associated with a particular band was measured by excising it from the transferred gels and measuring the radioactivity in a liquid scintillation counter.
DNase I protection assay.
The general procedure we followed was described previously (5). The target DNA fragments were prepared by Qiaprep Spin Columns, digested with XbaI, labeled with Klenow fragment and [32P]dATP (ICN Biomedicals), and digested with HindIII. Labeled fragments were divided into aliquots containing 3 × 105 cpm for each binding reaction. Binding reactions were composed of 10 μl of labeled DNA (14 fmol/μl), 10 μl of calf thymus DNA (0.05 μg/μl), 10 μl of either buffer 6 or NAC protein dilution, and 20 μl of H2O and were incubated at room temperature for 20 min. Each incubated mix was exposed to DNase I at 37°C for 20 s. The digested products were resolved in an 8% polyacrylamide gel containing 7 M urea.
RESULTS
Randomized oligonucleotide mutagenesis of the hutU promoter defines key residues for activation by NAC.
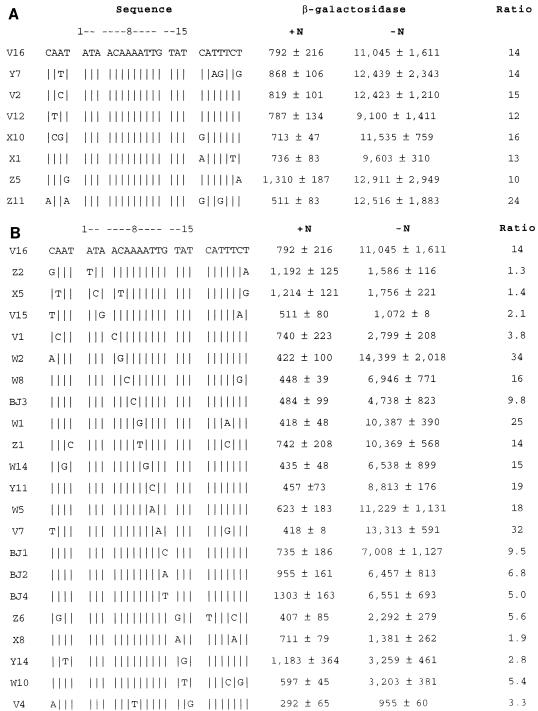
Purified NAC protects a 26-bp region of the hutU promoter from DNase I digestion (14). This NAC-protected hutUp sequence was mutagenized by PCR amplification as described in Materials and Methods, and the DNA sequence of 100 mutagenized plasmids was determined. A set of 29 clones containing mutations within the 26-bp NAC-binding site was selected for further analysis. A few changes that were absent from the random mutant set were produced by oligonucleotide-directed mutagenesis (see Materials and Methods). K. aerogenes KC2668 was transformed with each of the mutant clones, and liquid cultures of the transformants were grown under nitrogen excess (+N) or nitrogen starvation (−N) conditions as described in Materials and Methods. The β-galactosidase activity reflecting the activity of each promoter was measured in both conditions. Figure 1A shows the effect of nucleotide changes outside the consensus sequence. The sequence of clone V16 corresponds to the wild-type NAC-binding site and served as positive control. Mutations that lie to the left or right of the 15-bp sequence had little or no effect on the activated (−N) levels of transcription from hutUp. In contrast, Fig. 1B shows that several of the mutations within the 15-bp consensus did affect the activated levels of transcription from hutUp. Mutants with changes in the trimers ATA (positions 1, 2, and 3) and TAT (positions 13, 14, and 15) showed a 3- to 10-fold decrease in transcriptional activation relative to the wild type (clone V16). The A4C change in position 4 also caused a decrease in transcriptional activation (mutant V1). Mutations at positions 5 and 11 caused increased levels of transcriptional activation (mutants W2 and V7). Mutants with changes at the center of symmetry (A8G and A8T) did not show significant changes in activation (mutants W11 and Z1). Finally, changes at positions 6, 7, 9, and 12 reduced the degree of transcriptional activation about twofold.
FIG. 1.
Nucleotide changes introduced in the NAC-binding site from the hutU promoter and their effects on transcriptional activation. The clone designation is on the left. The wild-type sequence is shown at the top, and at the right are the β-galactosidase activities expressed in Miller units for each mutant grown under nitrogen excess (+N) or nitrogen limitation (−N) conditions. Values are the averages and standard deviations of at least three independent experiments. Ratio, −N/+N. Vertical lines indicate unchanged nucleotides. (A) Changes outside the 15-bp core. (B) Changes inside the 15-bp core.
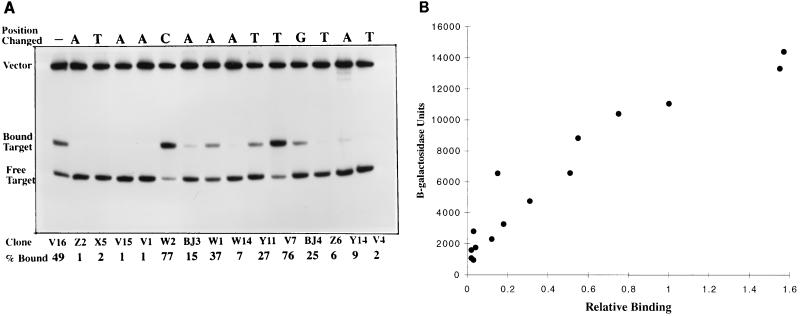
Several mutants from this set were selected for use in gel mobility shift assays. A qualitative estimate of the relative affinity of each mutant binding site was made by measuring the radioactivity associated with the shifted band as a percentage of the total target radioactivity (Fig. 2A). The amount of NAC used in the experiment was adjusted so that about 50% of the wild-type site was shifted under these conditions. Thus mutant sites that show less than 50% shifted are assumed to have weaker affinity than wild type, and those that show more than 50% shifted are assumed to have stronger affinity than wild type. In all but one of the cases analyzed (mutant W14), the percentage bound in the gel mobility shift assay was related to the level of β-galactosidase expression: the greater the percentage shifted, the higher the β-galactosidase expression under activating conditions (Fig. 2B). Although these data are qualitative estimates at best, this relationship seemed to suggest that transcriptional activation by NAC simply requires a binding site adjacent to RNA polymerase.
FIG. 2.
(A) Binding of NAC-binding site mutants from hutUp analyzed by gel mobility shift assay. The mutant NAC-binding sites were labeled with 32P and incubated with NAC protein (0.28 μM). Bound and unbound fragments were separated by polyacrylamide gel electrophoresis. The radioactivity in each band was measured as described in Materials and Methods. (B) Correlation between protein binding and transcriptional activation for hutUp NAC-binding site mutants. The graph plots the activated levels of β-galactosidase (grown under nitrogen limitation) from Fig. 1B, and the binding percentages (normalized to clone V16 which was 49%) for the analyzed NAC binding-site mutants from panel A.
Thus, we conclude that the minimal NAC-binding site is a 15-bp fragment with the palindromic ends ATA and TAT. It is important to remember that none of the changes in the ATA and TAT trimers occurred in the absence of other changes. Nevertheless, changes outside this region had small effects on transcriptional activation, whereas changes within this region could have considerable effects on transcriptional activation. Particularly interesting is the A at position 4: a change to C (mutant V1) decreased binding and activation to levels as low as mutations in the palindromic ends. Interestingly, changes at the symmetric position G12 did not have so strong an effect, suggesting that the two sides of the binding site might be contacted differently by NAC. Note that the amount of β-galactosidase expression under activation conditions is nearly the same for all three mutations at position G12. The differences in activation ratios resulted from an unexplained effect on the basal level of expression, which is particularly noticeable for the mutation G12T.
Binding to DNA by NAC is not sufficient for transcriptional activation.
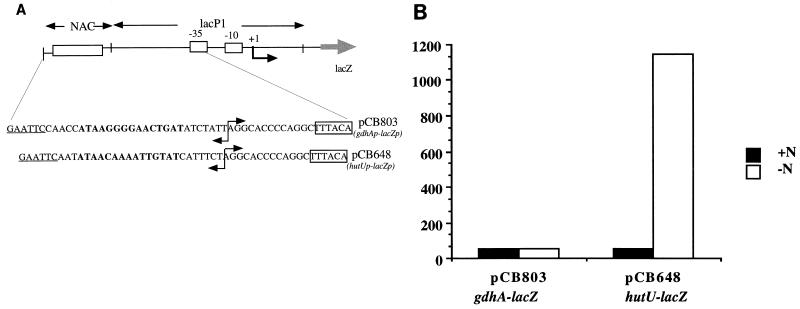
The low binding affinity of the mutant Z6 (T13G) was a surprising result, since some NAC-binding sites have the general form ATA-N9-GAT (14a, 23). Interestingly, the NAC-binding sites with this sequence pattern are involved in transcriptional repression and do not match the consensus sequence deduced from NAC-activated promoters. In order to test whether NAC could bind to a site with this sequence and activate transcription from a site with this sequence, we constructed a chimeric promoter using a NAC-binding site from the gdhA promoter (gdhAp) placed at position −64 from the transcriptional start site of the lacP1 promoter from E. coli (pCB803) (30). The construction of pCB803 is sketched in Fig. 3A. As a control, we used a chimeric promoter with the NAC-binding site from hutUp placed at position −64 from lacP1 (pCB648) (28). Both constructs were used to transform KC2668 to ampicillin resistance, and cultures of the Ampr transformants were grown under conditions of nitrogen excess and nitrogen limitation. Figure 3B shows that while the hutUp-lacp chimera was activated more than 20-fold, the gdhAp-lacp chimera was not activated by nitrogen starvation at all.
FIG. 3.
Construction and transcriptional activation from two chimeric promoters. (A) Two different NAC-binding sites were placed upstream of the lacZ promoter (centered at −64). The 15-bp core of each NAC-binding site is indicated in bold type. The EcoRI restriction sites introduced by the mutagenic primers are underlined. (B) Transcriptional activation from the chimeric hutUp-lacZp and gdhAp-lacZp promoters. The bars indicate the specific activities of β-galactosidase for each construct under either nitrogen excess (+N) or nitrogen limitation (−N) conditions. Values are the averages of at least three independent experiments.
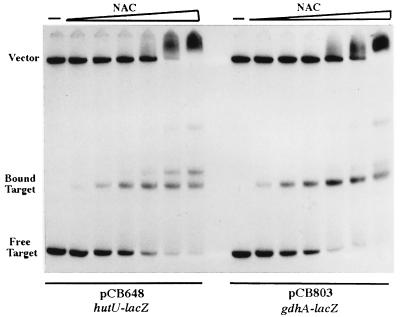
The lack of activation of the gdhAp-lacp chimera might have resulted from poor binding of NAC to this particular construct. To test this possibility, we obtained a qualitative estimate of the binding of NAC to the gdhAp-lacp and hutUp-lacp chimeras by gel mobility shift assay. Figure 4 shows that the mobility shift profiles of the two constructs were very similar, indicating that there are no major differences in the in vitro binding between these two NAC-binding sites. In a control experiment, a DNA fragment containing the wild-type lac promoter showed no mobility shift when incubated with the same NAC concentrations (data not shown).
FIG. 4.
NAC binding to the hutUp-lacZp and gdhAp-lacZp chimeric promoters. Restriction fragments from plasmids containing the NAC-binding sites were purified, labeled with 32P, and incubated with NAC protein. Bound and unbound fragments were separated by polyacrylamide gel electrophoresis. The concentrations of purified NAC used were 0, 0.02, 0.04, 0.08, 0.17, 0.34, and 0.67 μM.
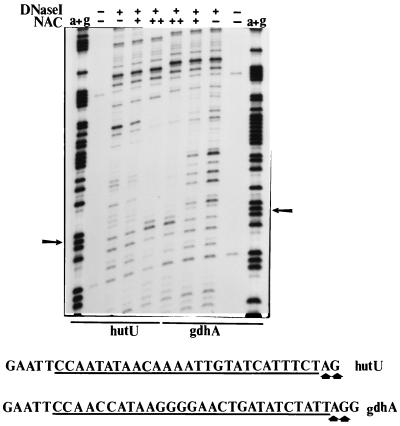
It has been previously shown that transcriptional activation by NAC is strongly dependent on correct distance between the NAC-binding site and the promoter core (28). If NAC were binding to the gdhAp site in a displaced manner, then transcriptional activation could be hindered. We tested this possibility by performing DNase I protection assays, comparing the NAC-binding sites from gdhAp and hutUp. The results are shown in Fig. 5. The NAC footprints on the two constructs were similar. The regions protected by NAC in both constructs were of equal length and equal limits. Neither footprint showed evidence of extensive DNA torsional stress, with the exception of a pair of hypersensitive sites right at the promoter-proximal end of the protected region. Therefore, it is unlikely that NAC is binding “out of register” at the gdhAp site.
FIG. 5.
DNase I footprints of NAC on the hutUp-lacZp and gdhAp-lacZp chimeric promoters. The plasmids containing the NAC-binding sites were purified, labeled, incubated with NAC protein, incubated with DNase I, and resolved in a polyacrylamide gel as explained in Materials and Methods. The final NAC concentrations used were 0, 0.134, and 0.670 μM. The first and last lanes show the products of Maxam-Gilbert sequencing reactions for the NAC targets on the hutUp-lacZp and gdhAp-lacZp promoter, respectively. Hypersensitive sites are indicated by arrows. The region protected by NAC in each construct is underlined.
We conclude from this set of experiments that the NAC-binding site from gdhAp did not allow transcriptional activation even though it seemed to have at least as high an affinity for NAC as the site from hutUp. Furthermore, the DNase I footprints failed to indicate any major structural differences between the protein-DNA complexes formed by NAC bound to the gdhAp site and NAC bound to the hutUp site.
Directed mutagenesis of the NAC-binding site from the gdhA promoter reveals residues important for transcriptional activation.
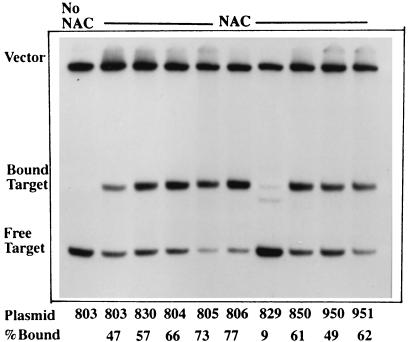
To test the hypothesis that the sequence of a NAC-binding site can modulate the ability of the NAC protein to activate transcription, we constructed a series of mutants of the gdhAp NAC-binding site. Our rationale was that if a particular set of nucleotides in the gdhAp binding site blocks activation, the replacement of these nucleotides with their positional equivalents from the hutUp NAC-binding site should release the putative barrier. With pCB803 as template, single or multiple mutations were introduced into the gdhAp NAC-binding site by PCR amplification with mutagenic primers. The PCR products were purified and cloned into pRJ800, and their nucleotide sequences were determined. The confirmed mutants were used to transform KC2668 to ampicillin resistance, and cultures of the transformants were grown under nitrogen excess and nitrogen limitation conditions. The degree of transcriptional activation in each mutant was estimated in vivo by the −N/+N ratio of β-galactosidase activities. The results are shown in Table 1. The ability of each site to bind NAC was estimated by gel mobility shift assay, and the results are shown in Fig. 6. The clone pCB830 shows that when the 15-bp core of the NAC-binding site from gdhAp was altered to match the NAC-binding site from hutUp, the resulting construct had a transcriptional activation similar to that provided by the hutUp NAC-binding site. This similarity shows that the sequence of the binding site itself regulates the ability of NAC to activate transcription, and that this regulation requires, but is not limited to, protein-DNA binding. This result also confirmed that the essential determinants for activation are contained within the 15-nucleotide core.
TABLE 1.
Transcriptional activation and binding in gdhA-lacZ chimeric promoters by NAC
| Clone | Nucleotide at positiona:
|
β-Gal sp act (U/mg of protein)b
|
Activity ratioc | Relative bindingd | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | +N | −N | |||
| 803 | A | T | A | A | G | G | G | G | A | A | C | T | G | A | T | 53 ± 38 | 54 ± 29 | 1.0 | 1.0 |
| 830 | A | T | A | A | c | a | a | a | A | t | t | g | t | A | T | 93 ± 35 | 2,048 ± 782 | 22 | 1.2 |
| 804 | A | T | A | A | G | G | G | G | A | A | C | T | t | A | T | 46 ± 21 | 46 ± 17 | 1.0 | 1.3 |
| 805 | A | T | A | A | c | a | a | a | A | A | C | T | G | A | T | 80 ± 22 | 100 ± 20 | 1.3 | 1.4 |
| 806 | A | T | A | A | c | a | a | a | A | A | C | T | t | A | T | 54 ± 26 | 106 ± 56 | 2.0 | 1.6 |
| 829 | A | T | A | A | G | G | G | G | A | t | t | g | G | A | T | 49 ± 10 | 124 ± 62 | 2.5 | 0.2 |
| 850 | A | T | A | A | G | G | G | G | A | A | C | g | t | A | T | 44 ± 3 | 303 ± 18 | 6.9 | 1.3 |
| 950 | A | T | A | A | c | a | G | G | A | A | C | g | t | A | T | 82 ± 30 | 355 ± 53 | 4.3 | 1.0 |
| 951 | A | T | A | A | G | G | G | G | A | t | C | g | t | A | T | 55 ± 5 | 807 ± 287 | 15 | 1.3 |
Altered nucleotides in the 15-bp core of the NAC-binding site are shown as lowercase letters.
Values are means ± standard deviations from three independent experiments. β-Gal, β-galactosidase.
Ratio of the activated (−N) to the basal (+N) value.
Percentage of shifted counts of each mutant normalized to the percentage of shifted counts in the wild type (pCB803).
FIG. 6.
Transcriptional activation and protein binding in gdhA-lacZ chimeric promoters by NAC. Protein binding to the gdhAp-lacZp chimeric promoters is shown. Gel mobility shift assays were performed as described for Fig. 4 with purified NAC at a concentration of 0.28 μM.
Several other changes showed little or no increase in transcriptional activation compared with pCB803, as in clones pCB804, -805, and -806. These three mutant sites bound NAC at least as well as the site in pCB803, but NAC showed virtually no activation of transcription from these sites. Thus even mutations that increase the binding of NAC to a site do not necessarily increase the ability of NAC bound there to activate transcription.
Clone pCB829 showed poor binding, consistent with the result observed for clone Z6 (T13G) in Fig. 2, and very little activation of transcription by NAC. Clone pCB850 showed that the replacement of the nucleotides TG at positions 12 and 13 with the nucleotides GT was sufficient to restore some transcriptional activation capability. Although an additional change of the GG at positions 5 and 6 with CA did not further increase transcriptional activation (clone pCB950), a change from A to T at position 10 (clone pCB951) resulted in the recovery of even more activation. Clones pCB850, -950, and -951 had affinities for NAC similar to the affinity of the gdhAp site, and thus the increased transcriptional activation observed in these clones compared to pCB803 could not be explained in terms of increased relative binding.
Mutations at the promoter-proximal half-site from hutUp uncouple NAC-mediated transcriptional activation and NAC binding.
Mutations that rendered the NAC-binding site from gdhAp functional without increasing DNA-binding affinity were clustered on the promoter-proximal side of the binding site. This asymmetry suggests that the binding site contains two types of determinants. The first type affects binding and was clustered at the two ends of the binding site (ATA and TAT). The second type might modulate transcriptional activation in addition to binding. This second type was located near the promoter-proximal end of the binding site, at positions 10, 11, and 12.
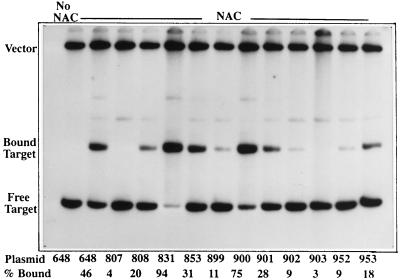
To test the possibility that positions 10, 11, and 12 might modulate transcriptional activation by a mechanism independent of binding, we constructed a series of mutant binding sites by introducing mutations in the NAC-binding site from hutUp. The mutations were introduced by oligonucleotide-directed mutagenic PCR with pCB648 as template. The resulting mutants were cloned into pRJ800, and the plasmids carrying each mutant chimeric promoter were used to transform KC2668 to Ampr. Transcriptional activation was measured by the −N/+N ratio of β-galactosidase activity of each culture. The ability of each clone to bind NAC was determined by gel mobility shift assay. The results are shown in Table 2 and Fig. 7. The site in clone pCB831 has the nucleotides at positions 10, 11, and 12 replaced by the positional equivalents from the gdhA NAC-binding site. Interestingly, it showed more binding than the site in pCB648 but reduced transcriptional activation more than fivefold. The site in clone pCB853 is a simple inversion of the 15-bp consensus region from the hutUp NAC site. This mutation reduced the ability of NAC to activate transcription from 22-fold to 2-fold, effectively abolishing activation, even though NAC bound almost as well to this site as to the site from pCB648. This result suggested that the NAC-DNA complex is functionally asymmetric and that the 5′ to 3′ polarity of the site is important for binding.
TABLE 2.
Transcriptional activation and binding in hutU-lacZ chimeric promoters by NAC
| Clone | Nucleotide at positiona:
|
β-Gal sp act (U/mg of protein)b
|
Activity ratioc | Relative bindingd | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | +N | −N | |||
| 648 | A | T | A | A | C | A | A | A | A | T | T | G | T | A | T | 58 ± 24 | 1,297 ± 137 | 22 | 1.0 |
| 807 | A | T | A | A | C | A | A | A | A | T | T | G | g | A | T | 37 ± 18 | 58 ± 12 | 1.6 | 0.1 |
| 808 | A | T | A | A | g | g | g | g | A | T | T | G | T | A | T | 52 ± 16 | 305 ± 81 | 5.9 | 0.4 |
| 831 | A | T | A | A | C | A | A | A | A | a | c | t | T | A | T | 59 ± 36 | 208 ± 34 | 3.5 | 2.0 |
| 853 | A | T | A | c | a | A | t | t | t | T | g | t | T | A | T | 95 ± 15 | 172 ± 81 | 1.8 | 0.7 |
| 899 | A | T | A | c | a | A | A | A | A | T | T | G | T | A | T | 44 ± 9 | 328 ± 186 | 7.5 | 0.2 |
| 902 | A | T | A | c | C | A | A | A | A | T | T | G | T | A | T | 67 ± 19 | 287 ± 122 | 4.3 | 0.2 |
| 901 | A | T | A | A | C | A | A | A | A | T | T | t | T | A | T | 57 ± 15 | 143 ± 26 | 2.5 | 0.6 |
| 900 | A | T | A | A | C | A | A | A | A | T | g | t | T | A | T | 65 ± 35 | 536 ± 320 | 8.2 | 1.6 |
| 903 | t | a | t | g | t | t | A | A | A | a | c | a | a | t | a | 42 ± 2 | 51 ± 5 | 1.2 | 0.1 |
| 952 | A | T | A | A | C | A | A | c | A | T | T | G | T | A | T | 57 ± 27 | 437 ± 112 | 7.7 | 0.2 |
| 953 | A | T | A | A | C | A | A | A | A | c | T | G | T | A | T | 53 ± 25 | 856 ± 276 | 16 | 0.4 |
Altered nucleotides in the 15-bp core of the NAC-binding site are shown as lowercase letters.
Values are means ± standard deviations from three independent experiments. β-Gal, β-galactosidase.
Ratio of the activated (−N) to the basal (+N) value.
Percentage of shifted counts of each mutant normalized to the percentage of shifted counts in the wild type (pCB648).
FIG. 7.
Transcriptional activation and protein binding in hutU-lacZ chimeric promoters. Protein binding to the hutUp-lacZp chimeric promoters is shown. Gel mobility shift assays were performed as described for Fig. 4 with purified NAC at a concentration of 0.34 μM.
The sites in clones pCB899 and pCB900 increase the overall symmetry of the NAC binding site by two nucleotides. Clone pCB899 (with changes in the promoter-distal half of the site) lost part of the binding affinity and also lost part of the activation. In contrast, clone pCB900 (with changes in the promoter-proximal half of the site) gained binding affinity but lost part of the activation. The constructs pCB901 and pCB902 extend the overall symmetry of the NAC-binding site by only one nucleotide. As seen with constructs pCB899 and -900, a change on the promoter-distal side that increased the overall symmetry caused a decrease in transcriptional activation with a concurrent loss of binding (pCB902). In contrast, a change on the promoter-proximal side that increased the overall symmetry caused a noticeable decrease in transcriptional activation without a large decrease in binding (pCB901). Finally, three mutant sites showed a decrease in binding that correlated with a decrease in activation. These included A8C (pCB952), which altered the central nucleotide which is conserved in many NAC sites, a mutant site that scrambled the nucleotides but kept the same base composition (pCB903, which reversed the sequence without reversing the 5′ to 3′ orientation), and T10C (pCB953).
DISCUSSION
The NAC-binding site from the hutU promoter is a 26-bp fragment protected by NAC from DNase I digestion and contains a 15-bp core that is necessary for transcriptional activation by NAC. The key nucleotides that modulate protein binding all lie within this 15-bp core, between and including the highly conserved palindromic outer ends ATA and TAT. These results are consistent with the observation that in vivo binding of NAC to the put promoter is impaired by mutations at the ATA and TAT trimers (8) and also with the consensus sequence deduced from three known sites from which NAC can activate transcription (14). The 15-bp core is also sufficient for transcriptional activation by NAC, as shown by clone pCB830, which has the 15-bp core sequence from the hutUp but the flanking sequences of gdhAp. The sequences outside the 15-bp core play at most a minor role in modulating transcriptional activation, since mutations in the flanking regions did not result in significant changes in the ability of NAC to activate β-galactosidase expression. We have established here that the binding of NAC to this site is essential for transcriptional activation. Therefore, differential binding affinity of a site for NAC probably results in differential sensitivity to in vivo regulation by NAC. However, an important limitation of these binding studies is that the binding in vitro might not fully reflect the binding in vivo. It is possible that the presence of bound RNA polymerase could modify the binding of NAC for a given binding site (14).
Several observations argue that the binding of NAC is not sufficient for transcriptional activation. First, we have demonstrated that the NAC-binding site from gdhAp does not allow transcriptional activation by NAC, despite the similarity between the gdhAp and the hutUp sites in binding affinity and footprint. The inability of the gdhA site to activate transcription upon NAC binding shows that DNA binding is not sufficient for transcriptional activation. Second, changing nucleotides at positions 10, 12, and 13 in the NAC-binding site from gdhA to their positional equivalents from hutU (pCB951 in Table 1) renders the gdhAp site functional without increasing binding affinity. Third, mutations altering the promoter-proximal side of the binding site from hutUp (pCB831 and pCB900) decreased transcriptional activation but increased binding. Similarly, the mutant pCB901, with a single substitution (G12T), showed a 10-fold decrease in transcriptional activation but a small loss of binding (less than twofold). Our results are consistent with the observations by Byerly et al. (7), who found mutations in the binding site for MetR that uncouple DNA binding and transcriptional activation of the metH gene. Gao and Gussin found similar mutations in the binding site for TrpI at the trpAB operon (13). Interestingly, two of the mutations at the MetR binding site were located in symmetric positions, adjacent to the trinucleotide ends that determine protein binding (7). It is important to note that no single substitution at the hutU NAC-binding site clearly uncoupled NAC binding and transcriptional activation (Fig. 1B and 2).
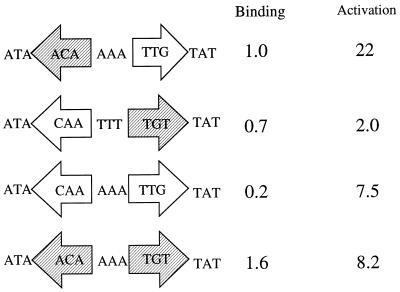
The most important conclusion from these data is that the sequence of the promoter-proximal half of the NAC-binding site from hutUp is important for transcriptional activation. The functional importance of the promoter-proximal half-site was already suggested by the activation consensus ATAP-N3-WNTYGTAT, deduced before this study. In this consensus sequence, the promoter-distal half-site has three conserved bases, while the promoter-proximal half-site has five conserved bases. Figure 8 summarizes the key observations relevant to this point. An inverted NAC site does not allow transcriptional activation and has only a slight decrease in binding. A NAC-binding site with two promoter-proximal half-sites is a very poor binder, while a site with two promoter-distal half-sites binds better than a wild-type site but shows decreased transcriptional activation. These results show that the two halves of the binding site have different sequences for different roles: the promoter-proximal half includes elements necessary for transcriptional activation as well as protein binding, while the promoter-distal half seems dedicated to protein binding.
FIG. 8.
Functional nonequivalence of the half-sites in the NAC-binding site from hutUp. Each mutant site is drawn as composed by two half-sites. In the wild-type site (pCB648), the promoter-distal half-site is shaded and the promoter-proximal half-site is not. Each mutant site is a different combination of these two half-sites. The binding and activation values were taken from Table 2.
Clones pCB831, pCB901, and pCB900 can be thought of as cis-acting positive control mutants because they contain mutations that leave the ability to form a NAC-DNA complex intact but reduce the ability to activate transcription efficiently. We propose that the most likely cause and the simplest explanation for the inability to activate transcription from the mutant NAC-binding sites is the blocking or absence of some unknown but probable contact between NAC and RNA polymerase. Several independent lines of indirect evidence suggest that transcriptional activation by NAC involves protein-protein interaction with RNA polymerase. First, the binding of RNA polymerase and NAC to the hutUp seem to be synergistic (14). Second, location of the NAC-binding site has shown a strong face-of-the-helix dependence for transcriptional activation (28). Third, the rpoA mutant E261K shows decreased in vivo activation of a hutU-lac chimeric promoter in E. coli (28a). Fourth, transcriptional activation by several other LysR family proteins (CatR, TrpI, and OxyR) has been shown to be dependent on an intact α subunit of RNA polymerase (10, 15, and 35 and 36, respectively). If NAC also requires direct physical contact with RNA polymerase for transcriptional activation, then a slight variation in NAC conformation might hinder the proper protein-protein interaction. Conformational changes induced in the protein in response to DNA binding have been demonstrated for the restriction enzyme BamHI (24) and the eukaryotic transcription factor Ets-1 (27).
Extensive genetic and biochemical work involving the catabolite activator protein (17) has shown the importance of exposed protein surfaces making contact in transcriptional complexes (25; also reviewed in reference 6). The structure of the NAC-binding site suggests that although NAC is most probably a symmetric dimer, the NAC-DNA complex might be asymmetric, and therefore the exposed surfaces of the NAC dimer might be somewhat different at either side of the bound molecule. This simple proposition explains the observations that an inverted NAC site failed to activate transcription and that mutations on the promoter-proximal side that increased the overall symmetry of the NAC-binding site actually decreased transcriptional activation by NAC.
Little is known about how NAC represses transcription from ς70-dependent promoters. However, as both activation and repression require NAC binding to DNA (14a), it is difficult to ignore the similarities and differences between the NAC sites involved in activation and the NAC sites involved in repression. NAC-binding sites involved in repression have the same promoter-distal half-sites as activation sites (ATAA), while having different promoter-proximal half-sites (NNtGAT for repression versus TYGTAT for activation (14a, 23). If the promoter-proximal half-site modulates the conformation or orientation of the NAC-DNA complex, then different sequences might determine different functions for the NAC-DNA complex.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM47156 from the National Institutes of Health to R.A.B. P.J.P. was supported by a Predoctoral Fellowship from The Horace Rackham School of Graduate Studies, University of Michigan.
We thank W. B. Muse and T. J. Goss for sharing their unpublished results.
REFERENCES
- 1.Bender R A, Janssen K A, Resnick A D, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129:1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender R A, Snyder P M, Bueno R, Quinto M, Magasanick B. Nitrogen regulation system of Klebsiella aerogenes: the nac gene. J Bacteriol. 1983;156:444–446. doi: 10.1128/jb.156.1.444-446.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender R A. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol Microbiol. 1991;5:2575–2580. doi: 10.1111/j.1365-2958.1991.tb01965.x. [DOI] [PubMed] [Google Scholar]
- 4.Best E A, Bender R A. Cloning of the Klebsiella aerogenes nac gene, which encodes a factor required for nitrogen regulation of the hut operons in Salmonella typhimurium. J Bacteriol. 1990;172:7043–7048. doi: 10.1128/jb.172.12.7043-7048.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenowitz M, Senear D. DNase I footprint analysis of protein-DNA binding. In: Ausubel F R, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1992. pp. 12.4.1–12.4.16. [DOI] [PubMed] [Google Scholar]
- 6.Busby S, Ebright R H. Promoter structure, promoter recognition, and transcription activation in Prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 7.Byerly K A, Urbanowsky M L, Stauffer G V. The MetR binding site in the Salmonella typhimurium metH gene: DNA sequence constraints on activation. J Bacteriol. 1991;173:3547–3553. doi: 10.1128/jb.173.11.3547-3553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L-M, Goss T J, Bender R A, Swift S, Maloy S. Genetic analysis, using P22 challenge phage, of the nitrogen activator protein DNA-binding site in the Klebsiella aerogenes put operon. J Bacteriol. 1998;180:571–577. doi: 10.1128/jb.180.3.571-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang L W, Kovary I, Howe M. Mutagenic oligonucleotide-directed PCR amplification (Mod-PCR): an efficient method for generating random base substitution mutations in a DNA sequence element. PCR Methods Applications. 1993;2:210–217. doi: 10.1101/gr.2.3.210. [DOI] [PubMed] [Google Scholar]
- 10.Chugani S A, Parsek M R, Hershberger C D, Murakami K, Ishihama A, Chakrabarty A M. Activation of the catBCA promoter: probing the interaction of CatR and RNA polymerase through in vitro transcription. J Bacteriol. 1997;179:2221–2227. doi: 10.1128/jb.179.7.2221-2227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng J, Goss T J, Bender R A, Ninfa A J. Activation of transcriptional initiation from the nac promoter of Klebsiella aerogenes. J Bacteriol. 1995;177:5523–5534. doi: 10.1128/jb.177.19.5523-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Goss T J, Bender R A, Ninfa A J. Repression of the Klebsiella aerogenes nac promoter. J Bacteriol. 1995;177:5535–5538. doi: 10.1128/jb.177.19.5535-5538.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Gussin G N. Mutations in TrpI binding site II that differentially affect activation of the trpBA promoter of Pseudomonas aeruginosa. EMBO J. 1991;10:4137–4144. doi: 10.1002/j.1460-2075.1991.tb04991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goss T J, Bender R A. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J Bacteriol. 1995;177:3546–3555. doi: 10.1128/jb.177.12.3546-3555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Goss, T. J., and R. A. Bender. Unpublished results.
- 15.Ishihama A. Role of the RNA polymerase α subunit in transcription activation. Mol Microbiol. 1992;6:3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 16.Janes B K, Bender R A. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J Bacteriol. 1998;180:563–570. doi: 10.1128/jb.180.3.563-570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Macaluso A, Best E A, Bender R A. Role of the nac gene product on the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J Bacteriol. 1990;172:7249–7255. doi: 10.1128/jb.172.12.7249-7255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 1344–1356. [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Muse, W. B., and R. A. Bender. The NAC gene of Escherichia coli and its role in nitrogen regulation. Ph.D. thesis. University of Michigan, Ann Arbor.
- 24.Newman M, Strzelecka T, Dorner L F, Schildkraut I, Aggarwal A K. Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 25.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Transcription activation at Class II promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osuna R, Janes B K, Bender R A. Roles of catabolite activator protein sites centered at −81.5 and −41.5 in the activation of the Klebsiella aerogenes histidine utilization operon hutUH. J Bacteriol. 1994;176:5513–5524. doi: 10.1128/jb.176.17.5513-5524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen J M, Skalicky J J, Donaldson L W, McIntosh L P, Alber T, Graves B J. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an α helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 28.Pomposiello P J, Bender R A. Activation of the Escherichia coli lacZ promoter by the Klebsiella aerogenes nitrogen assimilation control protein (NAC), a LysR family transcription factor. J Bacteriol. 1995;177:4820–4824. doi: 10.1128/jb.177.16.4820-4824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Pomposiello, P. J., and R. A. Bender. Unpublished results.
- 29.Reznikoff W, McClure W R. E. coli promoters. In: Reznikoff W S, Gold I, editors. Maximizing gene expression. Boston, Mass: Butterworths Publishers; 1986. pp. 1–33. [Google Scholar]
- 30.Reznikoff W S. Catabolite gene activator protein activation of lac transcription. J Bacteriol. 1992;174:655–658. doi: 10.1128/jb.174.3.655-658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 32.Schwacha A, Bender R A. The nac gene of Klebsiella aerogenes. J Bacteriol. 1993;175:2107–2115. doi: 10.1128/jb.175.7.2107-2115.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwacha A, Bender R A. The product of the nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J Bacteriol. 1993;175:2116–2124. doi: 10.1128/jb.175.7.2116-2124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao K, Zou C, Fujita N, Ishihama A. Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase α subunit. J Bacteriol. 1995;177:6740–6744. doi: 10.1128/jb.177.23.6740-6744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao K, Fujita N, Ishihama A. Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol Microbiol. 1993;7:859–864. doi: 10.1111/j.1365-2958.1993.tb01176.x. [DOI] [PubMed] [Google Scholar]