Abstract
Background
Epigenetic age acceleration (EAA), a measure of accelerated biological aging, has been associated with an increased risk of several age-related chronic conditions. This is the first study to prospectively examine the relationship between EAA and both multimorbidity count and a weighted multimorbidity score among long-lived postmenopausal women.
Methods
We included 1 951 women from the Women’s Health Initiative who could have survived to age 90. EAA was estimated using the Horvath pan-tissue, Hannum, PhenoAge, and GrimAge “clocks.” Twelve chronic conditions were included in the multimorbidity count. The multimorbidity score was weighted for each morbidity’s relationship with mortality in the study population. Using mixed-effects Poisson and linear regression models that included baseline covariates associated with both EAA and multimorbidity, we estimated relative risks (RRs) and 95% confidence intervals (CIs) for the relationships between each EAA measure at the study baseline with both multimorbidity count and weighted multimorbidity score at age 90, respectively.
Results
For every one standard deviation increase in AgeAccelPheno, the rate of multimorbidity accumulation increased 6% (RR = 1.06; 95% CI = 1.01–1.12; p = .025) and the multimorbidity score by 7% (RR = 1.07; 95% CI = 1.01–1.13; p = .014) for women who survived to age 90. The results for a one standard deviation increase in AgeAccelHorvath, AgeAccelHannum, and AgeAccelGrim with multimorbidity accumulation and score were weaker compared to AgeAccelPheno, and the latter 2 did not reach statistical significance.
Conclusion
AgeAccelPheno and AgeAccelHannum may predict multimorbidity count and score at age 90 in older women and, thus, may be useful as a biomarker predictor of multimorbidity burden in the last decades of life.
Keywords: Biomarkers, Epidemiology, Multimorbidities, Successful aging
In the United States (U.S.), the number of individuals who are 90 or older is expected to quadruple from 1.9 million in 2016 to 7.6 million individuals in 2050 (1). Women outnumber men 3 to 1 among those 90 or older (2). Among Medicare beneficiaries in 2008, 82.3% of women ages 85 or older had multimorbidity, defined by the U.S. Department of Health and Human Services as the presence of 2 or more chronic conditions (3). There is substantial evidence supporting the relationship between multimorbidity and reduced functional status and quality of life, as well as an increased risk of mortality (4–10).
Recently, the National Institutes of Health (NIH) developed a framework highlighting the influence of factors that may cause, increase the risk for, or exacerbate multimorbidity, and the potential for these factors to inform prevention strategies to achieve significant public health impact (11). Relatedly, biological aging focuses on biological mechanisms that are fundamental to aging-related increases in disease and disability as one ages and may serve as targets for prevention (12). Individuals with the same chronological age may experience different rates of biological aging, and faster biological aging is associated with chronic disease onset, morbidity, and mortality (13). Healthy longevity can be characterized as having a biological age less than one’s chronological age and is closely linked with the concept of healthspan. Healthspan prioritizes physical and cognitive functioning with advancing age, and preservation of healthspan targets both primary and secondary prevention of impaired function (14).
For a biomarker to be a useful indicator or predictor of healthy longevity and healthspan, it should move beyond the prediction of all-cause mortality and be capable of predicting multimorbidity burden at an advanced age. Epigenetic age is a composite measure of DNA methylation (DNAm) levels across specific cytosine-guanine dinucleotides (CpG) sites that are associated with chronologic and phenotypic age. These DNAm signatures are associated with age-related diseases and all-cause mortality, independent of chronologic age (15–17). Epigenetic age acceleration (EAA) is then the residual variation between one’s measured epigenetic age and their epigenetic age predicted by their chronological age and is indicative of whether one is aging slower or faster than their chronological age.
To the best of our knowledge, all other evaluations of EAA with multimorbidity count were cross-sectional (18). A meta-analysis including 9 studies from 4 unique cohorts was conducted to assess the relationship of a 1-year increase in EAA and multimorbidity count at the time of blood draw. Overall, all EAA measures showed a statistically significant association with cross-sectional multimorbidity count. The previous study limited the multimorbidities that were included in the age-related conditions available in each cohort, did not consider the risk of mortality associated with each condition, was not restricted to older age groups, and did not examine associations with multimorbidity at a specific older age when all participants would have the same amount of chronological aging.
The NIH report on multimorbidity additionally recommended the use of prospective, age-based, epidemiologic studies to examine potential mechanisms that may be intervened in to target multimorbidity among older adults (11). No prospective studies have examined the relationship between EAA and multimorbidity among women who survive to older ages (90+ years). The aims of this study, therefore, were to examine the relationships between EAA and both multimorbidity count and weighted multimorbidity score at age 90. We hypothesized that women who experienced accelerated biological aging, as measured by epigenetic age, would have higher multimorbidity counts and weighted multimorbidity scores at age 90.
Method
Study Population
In 1993, the Women’s Health Initiative (WHI) was created in order to identify strategies to prevent heart disease, osteoporosis, and breast and colorectal cancers among postmenopausal women (19,20). The current study included three WHI ancillary studies, 2 designed as nested case–control studies and 1 nested cohort of WHI clinical trial participants, that previously assayed genome-wide DNAm. The Bladder Cancer and Leukocyte Methylation Ancillary Study identified methylation profiles associated with bladder cancer risk among 468 women with and 468 women without bladder cancer (Study 1) (21). The Epigenetic Mechanisms of Particulate Matter-Mediated Cardiovascular Disease Ancillary Study identified the pathophysiological mechanisms that underlie particulate matter-related cardiovascular disease using a random sample of 2 200 WHI clinical trial participants (Study 2) (22). The Integrative Genomics for Risk of Coronary Heart Disease and Related Phenotypes in the WHI Cohort Ancillary Study included 1 070 women with and 1 070 women without coronary heart disease (Study 3) (23). DNA methylation was evaluated before the diagnosis of incident bladder cancer and incident coronary heart disease (CHD).
The current study included women that were eligible to survive to age 90 between baseline and the end of the most recent observation period (September 30, 2020) and also had baseline assays of genome-wide DNAm. There were a total of 2 079 women who were eligible to survive to age 90 (443 from Study 1, 694 from Study 2, and 942 from Study 3). After excluding women who had an unknown vital status the final analytic sample included 1 951 women (94%) of which 1 022 women survived to age 90, and 929 women died before reaching age 90. The study protocols were approved by the WHI Papers and Publication Committee, and all women provided informed consent in writing or by phone.
Measures
Epigenetic age
In each ancillary study, DNAm was measured using the Illumina Infinium 450K platform (Illumina, San Diego, CA). The minfi R package was used to read in all DNAm data files, check for failed samples, and implement normalization and quality control steps. The basic quality controls excluded probes targeting cytosine-guanine (CpG) sites on the Y chromosome, probes with detection p values > .01 in > 1% of samples, probes with a bead count <3 in >10% of samples, and probes that measure non-CpG methylation. Normalization was completed using beta-mixture quantile normalization and implemented in beta-mixture quantile dilation (BMIQ) (24).
Epigenetic age was estimated using four established “clocks,” including the Horvath pan-tissue, Hannum, PhenoAge, and GrimAge. Hannum et al. (17) used 71 CpG sites in the blood to predict age and Horvath (16) used 353 CpG sites to predict age across several different tissues. PhenoAge (25) used 513 CpG sites and was trained on a “phenotypic age” measure created using 9 clinical biomarkers associated with time-to-death. GrimAge (18) used 1 030 CpG sites and was developed by predicting time-to-death using age, sex, DNAm-based surrogate biomarkers of plasma protein levels, and a DNAm-based estimator of smoking pack-years. A description of each clock estimate is summarized in Supplementary Table 1, and these clocks have also been previously compared in greater detail (26).
Multimorbidity outcomes
There were 12 chronic conditions included in our multimorbidity count and weighted multimorbidity score (Table 1). These conditions were selected due to their prevalence among older women in the United States, their strong influence on physical functioning and quality of life, as well as guidance from current literature (27,28). Certain conditions such as sensory impairment, cognitive impairment, hip fracture, frequent faller, and urinary incontinence were added due to their high prevalence in this study of older women and previous examination in WHI studies of multimorbidity. While conditions such as hypertension, hyperlipidemia, and obesity were considered, they were not included due to their role as major risk factors for many conditions included in the definition and to maintain focus on conditions that were disease endpoints.
Table 1.
Definition of 12 Chronic Conditions and Assigned Weighted Score in Multimorbidity Count and Multimorbidity Score Outcomes
| Chronic Condition | Definition | Weighted Score |
|---|---|---|
| Stroke | One or more of the following: carotid artery disease, stroke, and transient ischemic attack | 18.4 |
| Coronary disease | One or more of the following: coronary heart disease, clinical myocardial infarction, congestive heart failure, coronary artery bypass graft, or percutaneous transluminal coronary angioplasty | 16.5 |
| Cancer | Any cancer (excluding nonmelanoma skin cancer) | 15.8 |
| Chronic obstructive pulmonary disease | Self-reported physician diagnosis | 14.6 |
| Sensory impairment | Self-reported moderate to severe trouble with vision or hearing loss | 13.0 |
| Diabetes | Self-reported physician diagnosis of diabetes and treatment for diabetes (pills and insulin) | 11.7 |
| Frequent faller | Self-reported ≥2 falls within 1 year | 7.5 |
| Cognitive impairment | Self-reported physician diagnosis with dementia or Alzheimer’s | 6.4 |
| Hip fracture | Broken hip | 4.5 |
| Osteoarthritis | Self-reported physician diagnosis | 4.0 |
| Depression | Self-reported treatment for depression (pills or therapy) | 3.1 |
| Urinary incontinence | Self-reported very or extremely bothersome urinary leakage | 1.0 |
These conditions were identified as part of the WHI follow-up protocol using both self-report on annual or semiannual outcome forms, followed by physician adjudication for selected outcomes of major interest within WHI. For self-reported items, the following question was used for ascertainment, “Since the date on the front of this form, has a doctor told you that you have any of the following conditions or have you had any of the following procedures?” The following conditions were self-reported: Alzheimer’s disease, diabetes characterized by self-reported use of diabetic medications, depression characterized by self-reported treatment of medication or therapy, sensory impairment self-reported as moderate to severe vision or hearing loss, urinary incontinence self-reported as ever leaking urine and feeling extremely bothered by it and frequently falling self-reported as falling at least 2 times in the past 12 months. The primary outcomes of the WHI study were adjudicated throughout the study by a physician using medical records, including incident CHD, cerebrovascular disease, cancer, and hip fracture. Participants who reported “yes” for any of the multimorbidity conditions or were classified as such through adjudication from baseline to follow-up through reaching age 90 were classified as having the condition at age 90. This approach was taken in recognition of the chronicity of the conditions under study.
There were 2 outcomes for this study: multimorbidity count and weighted multimorbidity score. Multimorbidity count was defined as the total number of morbidities from baseline to follow-up to age 90 or the last follow-up for those who did not reach age 90. Weighted multimorbidity score was a derived score based on the association of each morbidity with survival status among women eligible to survive to age 90. The weights were created using the subset of women from WHI included in the analytic sample. Each condition was placed in an univariate model with survival status at age 90, and the weight was calculated as the beta of each condition over the beta of urinary incontinence, which had the lowest beta and served as a reference weight of 1 (Table 1). This method was developed and applied in a previous study that examined multimorbidity among hospitalized individuals (29). The final multimorbidity score was the sum of the relative weights based on all of the conditions a woman had acquired from baseline to age 90 or her last study visit before the date of death. The purpose of the weighted multimorbidity score was to capture the degree to which each disease was life-threatening, and assign value accordingly using a weighted versus absolute count.
Covariates
Covariates were measured at WHI baseline and selected due to their associations with both EAA and multimorbidity. Covariates included age at blood draw, estimated blood cell composition using the Houseman method (30) (CD8+ T Cells, CD4 T cells, Natural Killer cells, B lymphocyte cells, monocytes, and granulocytes), race/ethnicity (Black/African American), Hispanic (Latino), White not of Hispanic origin, Unknown (not one of the above), education (high school/general education development or less, some college, and college graduate or more), walking frequency (rarely or never, 1–3 times/mo, 1 time/wk, 2–3 times/wk, 4–6 times/wk, and 7+ times/wk), body mass index categories (underweight [<18.5 kg/m2], normal [18.5–24.9 kg/m2], overweight [25.0–29.9 kg/m2], and obese [≥30 kg/m2]), alcohol consumption (nondrinker, past drinker, <1 drink/mo, <1 drink/wk, 1–<7 drinks/wk, and 7+ drinks/wk), pack-years smoking (never smoker, <5, 5–20, and 20+), and physical function score (RAND-36 10-item physical function subscale (31), range 0–100, higher score reflects higher function).
Statistical analysis
Baseline characteristics were examined by PhenoAgeAccel quartiles, and differences across quartiles were tested using Pearson’s chi-squared tests for categorical variables and F-tests for continuous variables. Unadjusted and fully-adjusted Poisson and linear regression models with a random intercept for the ancillary study were used to estimate relative risks (RRs) and 95% confidence intervals (CIs) for the associations between each EAA measure (one standard deviation increase) with multimorbidity count and multimorbidity score, respectively. For the analysis that included all women who were eligible to survive to age 90, a log link offset was used to account for differential follow-up times (ie, differential time to accumulate comorbidities). Adjusted models included all covariates as described earlier and inverse probability weights to account for the case–control sampling of two ancillary studies and the oversampling of racial/ethnic minorities in the third ancillary study to create an analytic study population more representative of the WHI overall. The weights were the inverse of the selection probability in Study 1–3 for each individual in order to downweight cases. The sample was reweighted, so the sum of the weights approximated the original sample size of the analytic sample using the WHI arm for which the participant was originally sampled.
The primary analyses were conducted among women who survived to age 90. The rationale for the primary analysis restricted to women who survived to age 90 is to understand how useful these epigenetic clocks may be in predicting a woman’s health status as defined by their comorbidity burden at age 90. A sensitivity analysis was conducted in all women eligible to survive to age 90, regardless of their survival to age 90, to evaluate the robustness of the findings to selective mortality. A second sensitivity analysis repeated the primary analysis adding an adjustment for baseline multimorbidity count. This analysis was done to account for the multiple morbidity count at the time of blood draw that could influence the prediction of total morbidity count at age 90 and also evaluates the prediction of incident multimorbidity. Additionally, a sensitivity analysis was conducted using a modified Charlson Comorbidity Index. The rationale for the sensitivity analysis was to assess the relationship of EAA with a frequently utilized measure of multimorbidity as well as one that primarily focuses on severe and life-threatening conditions. All analyses were conducted using R Version 1.4.1106 (R Foundation for Statistical Computing, Vienna, Austria).
Results
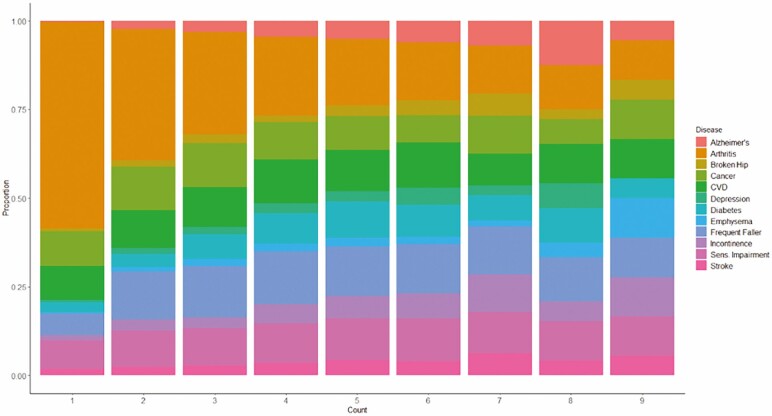
The 1 022 women who survived to age 90 were followed for 20.7 years on average (range = 10.4–25.4 years) from WHI baseline to age 90. The 929 women who did not survive to age 90 were followed for 12.8 years on average (range = 0.1–24.7 years). Women who survived to age 90 had an average of 1.1 multimorbidities at baseline (range = 0–5 multimorbidities) and 2.8 at age 90 (range = 0–8 multimorbidities). Women who did not survive to age 90 had an average of 1.3 multimorbidities at baseline (range = 0–5 multimorbidities) and 3.2 by last-follow up on study (range = 0–9 multimorbidities). The distributions of conditions by multimorbidity count among women eligible to survive to age 90 are shown in Figure 1. In brief, a large proportion of women who had 3 or fewer chronic conditions had arthritis, cancer, cardiovascular disease, sensory impairment or were frequent fallers. Additionally, among women who had greater counts of morbidities, the conditions tended to be more consistently distributed.
Figure 1.
The following is the distribution of women in each multimorbidity count group: 0 (n = 60); 1 (n = 264); 2 (n = 446); 3 (n = 512); 4 (n = 355); 5 (n = 197); 6 (n = 90); 7 (n = 16); 8 (n = 9); 9 (n = 2).
Women with higher EAA (accelerated biological aging) as measured by the Pheno clock were more likely to be Black or Hispanic than non-Hispanic White, have lower education, be obese, drink less alcohol, have a lower physical functioning score and accumulate a greater number of multimorbidities. In addition, those with higher AgeAccelPheno measures were more likely to have severe conditions such as cardiovascular disease and diabetes (Table 2). Compared to all women in the WHI the women in the analytic sample had lower levels of education, higher BMI, lower alcohol consumption, lower physical functioning scores and were also older on average (Supplementary Table 2).
Table 2.
Baseline Characteristics by PhenoAgeAccel Quartile (n = 1 951)
| Decelerated Aging | Accelerated Aging | ||||
|---|---|---|---|---|---|
| −31–−4.5 | −4.4–0.0 | 0.1–4.0 | 4.1–29.4 | p | |
| (n = 493) | (n = 516) | (n = 411) | (n = 529) | ||
| Age, mean (SD) | 71.0 (3.5) | 71.2 (3.4) | 71.0 (3.6) | 70.5 (3.3) | .026* |
| Race/ethnicity, n (%) | .005* | ||||
| Black (African American) | 77 (15.7) | 73 (14.3) | 70 (17.1) | 123 (23.5) | |
| Hispanic (Latino) | 32 (6.5) | 41 (8.0) | 34 (8.3) | 47 (9.0) | |
| White not of Hispanic origin | 365 (74.6) | 378 (73.8) | 289 (70.5) | 342 (65.3) | |
| Unknown (not one of the above) | 15 (3.1) | 20 (3.9) | 17 (4.1) | 12 (2.3) | |
| Education, n (%) | .027* | ||||
| HS/GED or less | 119 (24.3) | 133 (25.9) | 117 (28.6) | 174 (33.1) | |
| Some college | 195 (39.8) | 214 (41.7) | 170 (41.6) | 204 (38.8) | |
| College grad or more | 176 (35.9) | 166 (32.4) | 122 (29.8) | 148 (28.1) | |
| Walking frequency, n (%) | .062 | ||||
| Rarely or never | 81 (16.5) | 87 (17.0) | 75 (18.5) | 131 (25.0) | |
| 1–3 times/month | 74 (15.1) | 74 (14.5) | 55 (13.5) | 77 (14.7) | |
| 1 time/week | 53 (10.8) | 59 (11.5) | 40 (9.9) | 53 (10.1) | |
| 2–3 times/week | 133 (27.1) | 131 (25.6) | 123 (30.3) | 145 (27.6) | |
| 4–6 times/week | 104 (21.2) | 113 (22.1) | 84 (20.7) | 84 (16.0) | |
| 7+ times/week | 45 (9.2) | 47 (9.2) | 29 (7.1) | 35 (6.7) | |
| BMI category (kg/m2), n (%) | <.001* | ||||
| Underweight (<18.5) | 5 (1.0) | 4 (0.8) | 5 (1.2) | 5 (1.0) | |
| Normal (18.5-24.9) | 177 (36.1) | 156 (30.3) | 108 (26.4) | 118 (22.5) | |
| Overweight (25.0–29.9) | 182 (37.1) | 184 (35.7) | 148 (36.2) | 170 (32.4) | |
| Obese (≥30) | 126 (25.7) | 171 (33.2) | 148 (36.2) | 231 (44.1) | |
| Alcohol consumption, n (%) | .014* | ||||
| Nondrinker | 79 (16.1) | 56 (11.1) | 48 (11.8) | 80 (15.3) | |
| Past drinker | 94 (19.1) | 112 (22.1) | 87 (21.4) | 126 (24.1) | |
| <1 drink/month | 57 (11.6) | 60 (11.9) | 51 (12.6) | 86 (16.4) | |
| <1 drink/week | 96 (19.6) | 116 (22.9) | 82 (20.2) | 93 (17.8) | |
| 1–<7 drinks/week | 112 (22.8) | 114 (22.5) | 89 (21.9) | 81 (15.5) | |
| 7+ drinks/week | 53 (10.8) | 48 (9.5) | 49 (12.1) | 57 (10.9) | |
| Smoking pack-years, n (%) | .961 | ||||
| Never smoker | 265 (55.4) | 270 (54.3) | 209 (52.9) | 283 (55.6) | |
| <5 | 58 (12.1) | 66 (13.3) | 45 (11.4) | 57 (11.2) | |
| 5–<20 | 62 (13.0) | 60 (12.1) | 52 (13.2) | 60 (11.8) | |
| 20+ | 93 (19.5) | 101 (20.3) | 89 (22.5) | 109 (21.4) | |
| Age-related condition | |||||
| Alzheimer’s | 58 (11.8) | 63 (12.2) | 52 (12.7) | 65 (12.3) | .982 |
| Arthritis | 387 (78.5) | 406 (78.7) | 320 (77.9) | 426 (80.5) | .760 |
| Broken hip | 43 (8.7) | 36 (7.0) | 42 (10.2) | 25 (4.7) | .009* |
| Cancer | 155 (31.4) | 154 (29.8) | 130 (31.6) | 187 (35.3) | .272 |
| Cardiovascular disease | 149 (30.2) | 168 (32.6) | 154 (37.5) | 198 (37.4) | .037* |
| Depression | 36 (7.3) | 30 (5.8) | 36 (8.8) | 48 (9.1) | .191 |
| Diabetes | 87 (17.6) | 100 (19.4) | 103 (25.1) | 143 (27.0) | .001* |
| Emphysema | 25 (5.1) | 33 (6.4) | 25 (6.1) | 34 (6.4) | .784 |
| Frequent faller | 198 (40.2) | 218 (42.2) | 173 (42.1) | 222 (42.0) | .901 |
| Sensory impairment | 154 (31.2) | 179 (34.7) | 136 (33.1) | 168 (31.8) | .648 |
| Stroke | 42 (8.5) | 66 (12.8) | 34 (8.3) | 52 (9.8) | .070 |
| Urinary incontinence | 66 (13.4) | 63 (12.2) | 63 (15.3) | 80 (15.1) | .442 |
| Physical function score, mean (SD) | 76.7 (22.7) | 75.0 (22.3) | 72.5 (23.8) | 70.0 (25.3) | <.001* |
| Baseline multimorbidity count | 1.1 (0.9) | 1.2 (1.0) | 1.3 (1.0) | 1.4 (1.1) | <.001* |
| Follow-up multimorbidity count | 2.3 (1.5) | 2.4 (1.5) | 2.6 (1.5) | 2.6 (1.5) | .040* |
| Total multimorbidity count | 2.8 (1.5) | 2.9 (1.5) | 3.1 (1.6) | 3.1 (1.6) | .015* |
| Multimorbidity score | 26.0 (15.9) | 27.8 (17.0) | 28.8 (16.9) | 30.0 (1.6) | .015* |
| AgeAccelHorvath | −3.0 (4.8) | −0.9 (4.4) | 0.4 (4.8) | 2.9 (5.4) | <.001* |
| AgeAccelHannum | −3.2 (4.9) | −1.1 (4.4) | 0.9 (4.2) | 2.9 (4.6) | <.001* |
| AgeAccelGrim | −1.8 (3.3) | −0.8 (3.7) | 0.4 (3.5) | 1.9 (4.1) | <.001* |
Notes: AgeAccel measures are the residual between chronological age and epigenetic age as measured by each individual epigenetic clock. HS = high school; GED = general educational development; BMI = body mass index; kg = kilograms; m = meters; SD = standard deviation.
*Conditions include cardiovascular disease, cancer, cognitive impairment, depression, osteoarthritis, history of falls, chronic obstructive pulmonary disease, hypertension, diabetes, hip fracture, and cerebrovascular disease.
*p < .05.
The associations between baseline EAA and multimorbidity count and multimorbidity score at age 90 based on our adjusted models are reported in Table 3. For every one standard deviation increase in AgeAccelHorvath (5.1 years) the relative multimorbidity count at age 90 was 4% higher (RR = 1.04; 95% CI = 1.00–1.09; p = .074) and the relative weighted multimorbidity score was 3% higher (RR = 1.03, 95% CI = 0.98–1.07; p = .174). For every one standard deviation increase in AgeAccelPheno (7.0 years), the rate of multimorbidity accumulation increased 6% (RR = 1.06; 95% CI = 1.01–1.12; p = .025) and the weighted multimorbidity score 7% (RR = 1.07; 95% CI = 1.01–1.13; p = .014) for women at age 90. The results for both AgeAccelHannum and AgeAccelGrim with multimorbidity count and score were null. The results were slightly attenuated for the covariate unadjusted analysis and remained similar for the sensitivity analyses that additionally adjusted for baseline multimorbidity count (Supplementary Table 4). Additionally, the results for AgeAccelPheno were similar in direction and magnitude when examined separately among cases and controls from Study 1 and Study 3.
Table 3.
Association of Epigenetic Age Acceleration With Multimorbidity Count and Multimorbidity Score Among Women Who Survived to Age 90 (N = 1 022)
| Multimorbidity Count | Multimorbidity Score | |||
|---|---|---|---|---|
| RR (95% CI)* | p | RR (95% CI)* | p | |
| AgeAccelHorvath | 1.04 (1.00–1.09) | .074 | 1.03 (0.98–1.07) | .174 |
| AgeAccelHannum | 1.02 (0.97–1.07) | .441 | 1.00 (0.95–1.05) | .947 |
| AgeAccelPheno | 1.06 (1.01–1.12) | .025 | 1.07 (1.01–1.13) | .014* |
| AgeAccelGrim | 0.98 (0.93–1.03) | .436 | 0.96 (0.92–1.01) | .117 |
Notes: All models were adjusted for the following baseline covariates: blood cell composition (CD8T, CD4T, NK, Bcell, Mono, and Gran), age, race/ethnicity, education, walking frequency, BMI, alcohol consumption, pack-years smoking, broken hip, emphysema, arthritis, depression, urinary incontinency, and visual/auditory sensory impairment; and RAND physical functioning score. RR = relative risk; CI = confidence interval; SD = standard deviation; BMI = body mass index.
*Results are presented for one standard deviation increase in DNAmAge measure: AgeAccelHorvath (SD = 5.1 years), AgeAccelHannum (SD = 5.3 years), AgeAccelPheno (SD = 7.0 years), and AgeAccelGrim (SD = 3.9 years).
*p < .05.
The results of the sensitivity analyses in which we examined the adjusted associations between baseline EAA and multimorbidity count and multimorbidity score among all women eligible to survive to age 90 (n = 1 951 total) are reported in Table 4. For every standard deviation increase in AgeAccelPheno (7.0 years), the rate of multimorbidity accumulation increased 4% (RR = 1.04; 95% CI = 1.00–1.07; p = .040) and the weighted multimorbidity score 7% (RR = 1.07; 95% CI = 1.04–1.10; p < .001). The results for AgeAccelHannum, AgeAccelHorvath, and AgeAccelGrim with multimorbidity count and score were null. The results were similar for the covariate unadjusted analysis and slightly attenuated for the sensitivity analysis that additionally adjusted for baseline multimorbidity count (Supplementary Table 3). In the final sensitivity analysis there was a similar relationship between one standard deviation increase in EAA and a modified Charlson Comorbidity Index for each of the 4 measures: AgeAccelHorvath (RR = 1.09; 95% CI = 0.99–1.19; p = .065), AgeAccelHannum (RR = 0.93; 95% CI = 0.84–1.04; p = .209), AgeAccelPheno (RR = 1.16; 95% CI = 1.04–1.30; p = .006), and AgeAccelGrim (RR = 1.06; 95% CI = 0.95–1.19; p = .303).
Table 4.
Association of Epigenetic Age Acceleration With Multimorbidity Count and Multimorbidity Score Among All Women Eligible to Survive to Age 90 (N = 1 951)
| Multimorbidity Count | Multimorbidity Score | |||
|---|---|---|---|---|
| RR (95% CI)* | p | RR (95% CI)* | p | |
| AgeAccelHorvath | 0.99 (0.96–1.02) | .651 | 1.01 (0.98–1.04) | .448 |
| AgeAccelHannum | 0.97 (0.94–1.01) | .105 | 0.98 (0.95–1.02) | .360 |
| AgeAccelPheno | 1.04 (1.00–1.07) | .040 | 1.07 (1.04–1.10) | <.001* |
| AgeAccelGrim | 1.00 (0.97–1.04) | .917 | 0.98 (0.95–1.01) | .252 |
Notes: All models were adjusted for the following baseline covariates: blood cell composition (CD8T, CD4T, NK, Bcell, Mono, Gran), age, race/ethnicity, education, walking frequency, BMI, alcohol consumption, pack-years smoking, broken hip, emphysema, arthritis, depression, urinary incontinency, and visual/auditory sensory impairment; and RAND physical functioning score. There were 1 022 women who survived to age 90 and 929 women who died before age 90. All models included an offset for age to account for differing lengths of follow-up. RR = relative risk; CI = confidence interval; SD = standard deviation; BMI = body mass index.
*Results are presented for one standard deviation increase in DNAmAge measure: AgeAccelHorvath (SD = 5.1 years), AgeAccelHannum (SD = 5.3 years), AgeAccelPheno (SD = 7.0 years), and AgeAccelGrim (SD = 3.9 years).
*p < .05.
Discussion
To the best of our knowledge, this is the first study to examine the relationship between EAA and multimorbidity among older women at the time they reach age 90. In this racially and ethnically-diverse group of older women, this prospective study showed that increased EAA as measured by AgeAccelPheno was associated with an increased risk of acquiring additional multimorbidities and more deadly multimorbidities among women who survived to age 90. EAA as measured by AgeAccelHorvath was also associated with an increase in multimorbidity count. A similar association was observed among all women eligible to survive to age 90 for AgeAccelPheno. The results also remained similar in the covariate unadjusted models and the fully covariate adjusted models that included baseline multimorbidity count. EAA measured by AgeAccelHannum and AgeAccelGrim were not associated with either multimorbidity count or multimorbidity score over time.
To date, the few studies that have examined the relationship between EAA and multimorbidity count have been cross-sectional. Lu et al. conducted a cross-sectional meta-analysis between each of the same four EAA measures utilized in this study and multimorbidity count at the time of blood draw. Participants were from the WHI, Framingham Heart Study (FHS), Invecchiare in Chianti (InChianti), and Jackson Heart Study (JHS). This study as well as Lu et al.’s benefitted from a large sample size and racial and ethnic diversity in the sample. Additionally, the magnitude of the results were similar. Each 1-year increase in EAA was statistically significantly associated with multimorbidity count at the time of blood draw in Lu et al.’s cross-sectional analysis. Specifically, in Lu et al.’s study the regression estimates for a 1-year increase in AgeAccelPheno ranged from 0.01 to 0.03 and were significant for InChianti, JHS, FHS, and WHI Study 2. In our study, the betas for a 1-year increase from AgeAccelPheno and both multimorbidity count and multimorbidity score were also close to 0.01 for all analyses.
There were also some differences to note. Lu et al.’s study and ours both included the following conditions: stroke, coronary disease, cancer, chronic obstructive pulmonary disease, visual impairment, diabetes, and cognitive impairment. However, the current study also included: hearing impairment, frequently falling, hip fracture, osteoarthritis, and depression. Within Lu et al.’s analysis, it is unclear which covariates were included in the analysis of each study cohort and if they differed across cohorts in availability and measurement. Another major difference in the Lu et al. analysis was the inclusion of a broad age range of adult men and women ranging from ages 20 to 102. These differences in study design and populations may explain the difference in results found for AgeAccelHorvath, AgeAccelHannum, and AgeAccelGrim in relation to multimorbidity count and score at age 90 in the present study.
Epigenetic clocks are thought to be a promising measure of biological age (32). Having accelerated biological age as measured by these epigenetic clocks has been associated with increased risks of several age-related conditions such as Alzheimer’s disease, cancer, coronary heart disease, cognitive performance, frailty, osteoarthritis, and Parkinson’s disease among others (16–18,25). The CpGs that were included in the clocks during the model building phase are thought to have a relationship with the epigenetic maintenance system, especially at promoters and enhancers throughout the genome. More specifically for the PhenoAge clock, the CpG sites that were more prevalent among individuals with accelerated aging were associated with several proinflammatory signaling pathways, while those that were less prevalent among those with accelerated aging were involved in transcriptional and translational machinery and DNA damage recognition and repair (25). Although the specific mechanisms are still under examination, the change in DNAm with age is most likely linked to declines in tissue function related to both intracellular changes that lead to a loss of cellular identity and small changes in cell composition over time (26). Only 41 of the 513 CpGs in the Horvath pan-tissue clock and only 5 CpGs in the Hannum clock are shared with the PhenoAge clock. DNAm PhenoAge was unique among the clocks examined because it was developed to predict phenotypic age rather than chronological age using biomarkers and risk factors related to all-cause mortality. In addition, PhenoAge was trained using longitudinal data that may better account for changes in health status over time. These differences may explain the associations between EAA measured by PhenoAge with both multimorbidity count and score that were not seen with some of the other epigenetic clocks. In the sensitivity analysis, all four epigenetic clocks had a similar relationship to that of the primary analysis using a modified Charlson Comorbidity Index.
This study had several strengths. The study population included a large number of women who survived to age 90 and was also racially and ethnically diverse. On average, women were followed for 2 decades with low rates of loss-to-follow-up. The WHI had information on relevant baseline characteristics and potential confounders. Epigenetic age was measured using several different clocks, which is currently considered best practice due to the low overlap in CpG sites and associated genes between the clocks. Each clock may be capturing potentially different biological pathways (32). Finally, there was also longitudinal measurement of several age-related chronic conditions, some of which were also adjudicated by trained physicians. There were also some limitations to note in this study. This study population included 2 ancillary studies that utilized nested case–control sampling, and thus the women included in this study were not representative of the larger WHI cohort; however, inverse probability selection weights were used to account for differences in the selection criteria, which is currently the recommended approach (33). As this study was limited to women, it will be important to replicate the findings among both men and women with diverse racial, ethnic and geographical representation. Another limitation to note is that biological aging can be measured in several different ways (genomics, metabolomics, proteomics, microbiomics, transcriptomics, etc). Although EAA is the focus of this study, several biological processes are likely to simultaneously contribute to age-related disease onset and progression (34). There is currently no gold standard to measure biological aging, and thus this research should be interpreted within a larger systems biology framework, that acknowledges the influence and interaction of many underlying processes.
In this study, we report that increased EAA measured by DNAm PhenoAge was associated with an increased number and more life-threatening multimorbidities at age 90 among older women. These results suggest that PhenoAgeAccel is a promising biomarker of multimorbidity burden among older women that captures the biological age and functional state of several organ systems and tissues beyond one’s chronological age. These findings should be replicated among independent populations of long-lived individuals to validate the observed findings. Additional studies are required to identify specific CpG sites that may be capturing cellular changes related to certain biological pathways and to establish PhenoAgeAccel as a biomarker for the prediction of multimorbidity burden among long-lived women. As women continue to live to more advanced ages, it will be increasingly important to predict the risk of age-related diseases, especially those that are life-threatening and focus on the potential for public health interventions to counteract the effect of EAA and lower overall disease burden.
Supplementary Material
Acknowledgments
All authors have contributed importantly to the conception of the study design, data collection, analysis, interpretation, and/or critical revision of the manuscript and have approved the manuscript. In addition, we would like to acknowledge the Women’s Health Initiative (WHI) participants and staff for their participation in this important scientific endeavor. We also acknowledge the WHI investigators.
Contributor Information
Purva Jain, The Herbert Wertheim School of Public Health and Human Longevity Science, University of California San Diego, La Jolla, California, USA.
Alexandra Binder, Cancer Epidemiology Program, University of Hawaii Cancer Center, Honolulu, Hawaii, USA; Department of Epidemiology, Fielding School of Public Health, University of California, Los Angeles, California, USA.
Brian Chen, The Herbert Wertheim School of Public Health and Human Longevity Science, University of California San Diego, La Jolla, California, USA.
Humberto Parada, Jr., Division of Epidemiology and Biostatistics, School of Public Health, San Diego State University, San Diego, California, USA; San Diego Moores Cancer Center, University of California, San Diego, California, La Jolla, California, USA.
Linda C Gallo, Division of Epidemiology and Biostatistics, School of Public Health, San Diego State University, San Diego, California, USA.
John Alcaraz, San Diego Moores Cancer Center, University of California, San Diego, California, La Jolla, California, USA.
Steve Horvath, Department of Human Genetics, David Geffen School of Medicine, UCLA, Los Angeles, California, USA; Department of Biostatistics, School of Public Health, University of California, Los Angeles, California, USA.
Parveen Bhatti, Cancer Control Research, BC Cancer, Vancouver, British Columbia, Canada.
Eric A Whitsel, Department of Epidemiology, Gillings School of Public Health and Department of Medicine, School of Medicine, University of North Carolina, Chapel Hill, North Carolina, USA.
Kristina Jordahl, Department of Epidemiology, School of Public Health, University of Washington, Seattle, Washington, USA.
Andrea A Baccarelli, Department of Environmental Health Sciences, Mailman School of Public Health, Columbia University Irving Medical Center, New York, New York, USA.
Lifang Hou, Institute for Public Health and Medicine, Northwestern University, Chicago, Illinois, USA.
James D Stewart, Department of Epidemiology, Gillings School of Public Health and Department of Medicine, School of Medicine, University of North Carolina, Chapel Hill, North Carolina, USA.
Yun Li, Department of Genetics, University of North Carolina, Chapel Hill, North Carolina, USA; Department of Biostatistics, University of North Carolina, Chapel Hill, North Carolina, USA; Department of Computer Science, University of North Carolina, Chapel Hill, North Carolina, USA.
Michael J LaMonte, Department of Epidemiology and Environmental Health, School of Public Health and Health Professions, University at Buffalo―SUNY, Buffalo, New York, USA.
JoAnn E Manson, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Andrea Z LaCroix, The Herbert Wertheim School of Public Health and Human Longevity Science, University of California San Diego, La Jolla, California, USA.
Funding
This work was supported by the following sources of funding: The Women’s Health Initiative (WHI) program is funded by the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C.
Funding also came from a grant supported by the National Institute on Aging: T32 Predoctoral Training Fellowship T32 AG058529 to (P.J.).
Bladder Cancer and Leukocyte Methylation was supported by American Cancer Society award 125299-RSG-13-100-01-CCE to (P.B.). The Epigenetic Mechanisms of Particulate Matter-Mediated Cardiovascular Disease was supported by National Institute of Environmental Health Sciences grant R01-ES020836 to (L.H., A.B., and E.A.W.). The Integrative Genomics for Risk of Coronary Heart Disease and Related Phenotypes in the WHI Cohort was supported by NHLBI Broad Agency Announcement contract HHSN268201300006C to (S.H.).
Finally, this work was supported by the National Cancer Institute (K01 CA234317), the SDSU/UCSD Comprehensive Cancer Center Partnership (U54 CA132384 and U54 CA132379), and the Alzheimer’s Disease Resource Center for advancing Minority Aging Research at the University of California San Diego (P30AG059299) to (H.P.).
Conflict of Interest
None declared.
References
- 1. Bureau USC. National Demographic Analysis Tables: 2020. https://www.census.gov/data/tables/2020/demo/popest/2020-demographic-analysis-tables.html. Published 2020. Accessed 2 January 2021.
- 2. He W, Muenchrath MN.. 90+ in the United States: 2006–2008.Census.gov. 2021. https://www.census.gov/library/publications/2011/acs/acs-17.html. Accessed 3 October 2022. [Google Scholar]
- 3. Parekh AK, Goodman RA, Gordon C, Koh HK. Managing multiple chronic conditions: a strategic framework for improving health outcomes and quality of life. Public Health Rep. 2011;126(4):460–471. doi: 10.1177/003335491112600403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verbrugge LM, Lepkowski JM, Imanaka Y. Comorbidity and its impact on disability. Milbank Q. 1989;67(3–4):450–484. doi: 10.2307/3350223 [DOI] [PubMed] [Google Scholar]
- 5. Gijsen R, Hoeymans N, Schellevis FG, Ruwaard D, Satariano WA, van den Bos GA. Causes and consequences of comorbidity: a review. J Clin Epidemiol. 2001;54(7):661–674. doi: 10.1016/s0895-4356(00)00363-2 [DOI] [PubMed] [Google Scholar]
- 6. Tinetti ME, McAvay GJ, Chang SS, et al. Contribution of multiple chronic conditions to universal health outcomes. J Am Geriatr Soc. 2011;59(9):1686–1691. doi: 10.1111/j.1532-5415.2011.03573.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fortin M, Lapointe L, Hudon C, Vanasse A, Ntetu AL, Maltais D. Multimorbidity and quality of life in primary care: a systematic review. Health and Qual Life Outcomes. 2004;2:51. doi: 10.1186/1477-7525-2-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen HY, Baumgardner DJ, Rice JP. Health-related quality of life among adults with multiple chronic conditions in the United States, Behavioral Risk Factor Surveillance System, 2007. Prev Chronic Dis. 2011;8(1):A09. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3044020/pdf/PCD81A09.pdf. Published December 17, 2010. [PMC free article] [PubMed] [Google Scholar]
- 9. Wikman A, Wardle J, Steptoe A. Quality of life and affective well-being in middle-aged and older people with chronic medical illnesses: a cross-sectional population based study. PLoS One. 2011;6(4):e18952. doi: 10.1371/journal.pone.0018952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Working Group on Health Outcomes for Older Persons with Multiple Chronic Conditions. Universal health outcome measures for older persons with multiple chronic conditions. J Am Geriatr Soc. 2012;60(12):2333– 2341. doi: 10.1111/j.1532-5415.2012.04240.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Salive ME, Suls J, Farhat T, Klabunde CN. National institutes of health advancing multimorbidity research. Med Care. 2021;59(7):622–624. doi: 10.1097/mlr.0000000000001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berlau DJ, Corrada MM, Kawas C. The prevalence of disability in the oldest-old is high and continues to increase with age: findings from The 90+ Study. Int J Geriatr Psychiatry. 2009;24(11):1217–1225. doi: 10.1002/gps.2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li X, Ploner A, Wang Y, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife. 2020;9:e51507. doi: 10.7554/eLife.51507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner K-H, Cameron-Smith D, Wessner B, Franzke B. Biomarkers of aging: from function to molecular biology. Nutrients. 2016;8(6):338. doi: 10.3390/nu8060338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seals DR, Justice JN, LaRocca TJ. Physiological geroscience: targeting function to increase healthspan and achieve optimal longevity. J Physiol. 2016;594(8):2001–2024. doi: 10.1113/jphysiol.2014.282665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115–R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sibbett RA, Altschul DM, Marioni RE, Deary IJ, Starr JM, Russ TC. DNA methylation-based measures of accelerated biological ageing and the risk of dementia in the oldest-old: a study of the Lothian Birth Cohort 1921. BMC Psychiatry. 2020;20(1):91. doi: 10.1186/s12888-020-2469-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anderson GL, Manson J, Wallace R, et al. Implementation of the Women’s Health Initiative study design. Ann Epidemiol. 2003;13(suppl. 9):S5–S 17. doi: 10.1016/S1047-2797(03)00043-7 [DOI] [PubMed] [Google Scholar]
- 21. Bhatti P. AS311―DNA Methylation Measured in Prospectively Collected Blood Samples and Risk of Bladder Cancer Among Post-menopausal Women. https://sp.whi.org/researchers/data/WHIStudies/StudySites/AS311/Pages/home.aspx. Accessed 2 January 2021.
- 22. Whitsel EA. AS315―Epigenetic Mechanisms of PM-mediated CVD Risk. https://sp.whi.org/researchers/data/WHIStudies/StudySites/AS315/Pages/home.aspx. Accessed 2 January 2021.
- 23. Assimes TL, Tsao PS, Abhser D, Horvath S. BA23―Integrative Genomics and Risk of CHD and Related Phenotypes in the Women’s Health Initiative. https://sp.whi.org/researchers/data/WHIStudies/StudySites/BA23/pages/home.aspx. Accessed 2 January 2021.
- 24. Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 27. Salive ME. Multimorbidity in older adults. Epidemiol Rev. 2013;35:75–83. doi: 10.1093/epirev/mxs009 [DOI] [PubMed] [Google Scholar]
- 28. Goodman RA, Posner SF, Huang ES, Parekh AK, Koh HK. Defining and measuring chronic conditions: imperatives for research, policy, program, and practice. Prev Chronic Dis. 2013;10:E66. doi: 10.5888/pcd10.120239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Walraven C, Austin PC, Jennings A, Quan H, Forster AJ. A modification of the Elixhauser comorbidity measures into a point system for hospital death using administrative data. Med Care. 2009;47(6):626–633. doi: 10.1097/MLR.0b013e31819432e5 [DOI] [PubMed] [Google Scholar]
- 30. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13(1):86. doi: 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corporation R. 36-Item Short Form Survey (SF-36) Scoring Instructions. rand.org/health-care/surveys_tools/mos/36-item-short-form/scoring.html. Published 2021. Accessed 2 January 2021.
- 32. Levine ME. Assessment of epigenetic clocks as biomarkers of aging in basic and population research. J Gerontol A Biol Sci Med Sci. 2020;75(3):463–465. doi: 10.1093/gerona/glaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richardson DB, Rzehak P, Klenk J, Weiland SK. Analyses of case–control data for additional outcomes. Epidemiology. 2007;18(4):441–445. doi: 10.1097/EDE.0b013e318060d25c [DOI] [PubMed] [Google Scholar]
- 34. Zierer J, Menni C, Kastenmüller G, Spector TD. Integration of “omics” data in aging research: from biomarkers to systems biology. Aging Cell. 2015;14(6):933–944. doi: 10.1111/acel.12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.