Abstract
Glycosyl transferases which recognize identical substrates (nucleotide-sugars and lipid-linked carbohydrates) can substitute for one another in bacterial polysaccharide biosynthesis, even if the enzymes originate in different genera of bacteria. This substitution can be used to identify the substrate specificities of uncharacterized transferase genes. The spsK gene of Sphingomonas strain S88 and the pssDE genes of Rhizobium leguminosarum were identified as encoding glucuronosyl-(β1→4)-glucosyl transferases based on reciprocal genetic complementation of mutations in the spsK gene and the pssDE genes by segments of cloned DNA and by the SpsK-dependent incorporation of radioactive glucose (Glc) and glucuronic acid (GlcA) into lipid-linked disaccharides in EDTA-permeabilized cells. By contrast, glycosyl transferases which form alternative sugar linkages to the same substrate caused inhibition of polysaccharide synthesis or were deleterious or lethal in a foreign host. The negative effects also suggested specific substrate requirements: we propose that spsL codes for a glucosyl-(β1→4)-glucuronosyl transferase in Sphingomonas and that pssC codes for a glucuronosyl-(β1→4)-glucuronosyl transferase in R. leguminosarum. Finally, the complementation results indicate the order of attachment of sphingan main-chain sugars to the C55-isoprenylphosphate carrier as -Glc-GlcA-Glc-isoprenylpyrophosphate.
Many species of bacteria synthesize and secrete acidic polysaccharides if supplied with a readily convertible carbon source such as glucose (Glc) and with adequate oxygen. Xanthomonas campestris, a plant pathogen, and Rhizobium leguminosarum, a nitrogen-fixing, nodule-forming plant symbiont, secrete acidic exopolysaccharides (EPS) which promote plant-microbe interactions. Not only does X. campestris secrete xanthan gum in planta, but the polysaccharide is also produced commercially from glucose by large-scale industrial fermentation. Xanthan gum is valuable for controlling the viscosity of aqueous solutions in diverse food and industrial applications. Several members of the bacterial genus Sphingomonas (26) also secrete capsular acidic polysaccharides with commercially important rheological properties: gellan, welan, rhamsan, S-88, S-198, S-7, and NW11 (22). To recognize that they share a common core structure, we refer collectively to these polysaccharides as sphingans, after the genus. The carbohydrate structures of the repeat units of xanthan gum, the rhizobial EPS, and the sphingans S-88, S-198, and NW11 are diagramed in Fig. 1.
FIG. 1.
Biosynthetic genes and structures of repeating units for xanthan gum (14), rhizobial EPS (31), and sphingans S-88 (15), S-198 (4), and NW11 (23). Acetyl, pyruvyl, and hydroxybutanoyl groups are not shown. The reducing end of each repeat is to the right, and the IPP carrier is attached through a phosphodiester linkage at the reducing end. The order of sugars attached to IPP for the sphingans and the corresponding biosynthetic genes were inferred from the genetic and biochemical experiments reported here for strain S88. Gal, galactose; Man, mannose; Rha, rhamnose.
The assembly of the repeat unit for each of these five polymers begins by transfer of glucose-1-phosphate from UDP-glucose to a C55-isoprenylphosphate (IP) carrier (2, 13, 27). The genes that code for these glucosyl-IP transferases are spsB for Sphingomonas (27), gumD for X. campestris (34), and pssA for R. leguminosarum (35). Based on deduced amino acid sequences, the corresponding gene products are members of a large family of related glycosyl-IP transferases which are very likely to be membrane proteins. Surprisingly, although the N-terminal membrane-interacting domains of these three gene products and the cellular membranes in which they reside appear to be substantially different in composition for each bacterium, the proteins can substitute for one another (1, 27). This substitution or genetic complementation is observed when a foreign gene, which has been cloned into a plasmid and then transferred by conjugal mating into a polysaccharide-deficient mutant bacterium, restores polysaccharide synthesis to the recipient.
Previously, we used genetic complementation and DNA sequencing to characterize a large cluster of 17 genes, including spsB, which were isolated from Sphingomonas strain S88 and are required for synthesis of capsular polysaccharide (37). Within this cluster we identified three additional gene products (SpsQ, -K, and -L) as putative glycosyl transferases by comparing their deduced protein sequences to those for glycosyl transferase genes from other polysaccharide-secreting bacteria. However, the substrate specificities of SpsQ, -K, and -L could not be determined from the deduced protein sequences, since the sequence similarities to the other transferases were limited. In addition, since the repeat unit of sphingan S-88 has two noncontiguous Glc residues in the main chain, it was not possible to know which Glc was initially added to the carrier IP and, by inference, the order of addition of the remaining sugars. By contrast, the orders of assembly for xanthan gum and the rhizobial EPS are already known from structural analysis of lipid-linked carbohydrate intermediates (2, 13).
The repeat units for sphingans S-88, S-198, and NW11 share three sugar linkages, and S-88 and S-198 also have a fourth common linkage. Thus, it is reasonable to expect that these closely related strains would have complementary transferases for the common assembly steps. Similarly, by examining the structures of the repeating units of xanthan gum, the rhizobial EPS, and sphingan S-88 (Fig. 1), and assuming the order of assembly for sphingan S-88 as shown, the second glycosyl transferase reactions for R. leguminosarum and Sphingomonas would be identical and would be different from the second transferase reaction for X. campestris. Recently, three putative glycosyl transferase genes from R. leguminosarum (pssC, -D, and -E) were isolated and sequenced (35). The deduced amino acid sequence of PssD was found to be similar to that of the amino-terminal half of SpsK, while PssE was similar to the carboxyl half of SpsK. The pssDE protein-coding sequences are separated in the genome by only a single base pair (35). In the present work, we analyzed genetic complementation for specific cloned DNA segments isolated from these three genera of bacteria to identify the functions of the individual glycosyl transferase genes of Sphingomonas and R. leguminosarum and to infer the order of assembly of sphingan S-88. Not only did we observe the expected gene substitutions and restoration of EPS synthesis, but we also discovered that competing foreign glycosyl transferases that form alternative carbohydrate linkages can inhibit either EPS synthesis in the recipient bacterium or bacterial growth.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The genotypes of bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Source (reference) |
|---|---|---|
| X. campestris | ||
| X59 (ATCC 55298) | B1459S-4L Rifr Gum+ | These authors (33) |
| m31 | X59 GumD− | These authors (33) |
| Sphingomonas | ||
| S88 | Strr Sps+ | ATCC 31554 (17) |
| m265 and m260 | S88 SpsB− | These authors (27) |
| m302 | S88 SpsB−K− | These authors (37) |
| R. leguminosarum bv. trifolii | ||
| RBL5515 | LPR5 cured of Sym plasmid, Strr Rifr | C. A. Wijffelman (28) |
| RBL5523 | RBL5515 pRL1JI spc-3::Tn1831 Pss+ | C. A. Wijffelman (25) |
| RBL5807 | RBL5523 Strr RifrpssA4::Tn5 (exo4) | C. A. Wijffelman (35) |
| RBL5833 | RBL5523 Strr RifrpssD111::Tn5 (exo111) | C. A. Wijffelman (35) |
| E. coli K-12 | ||
| DH5α | F− φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 relA1 supE44 hsdR17(rK− mK+)Δ(lacZYA-argF)U169 | Bethesda Research Laboratories |
| HMS174 | F−hsdR19 (rK− mK+) recA1 rpoB331 (Rifr)IN(rrnD-rrnE)1 | W. Studier |
| Plasmids | ||
| pRK2013 | Ori(ColE1) Kanr OriT(Mob+) Tra+ | D. Helinski (9) |
| pRK311 | OriV(RK2) Tetr OriT(Mob+) Tra− λcos lacZ(α) | D. Helinski (6) |
| pSEB24 | OriV(RSF1010) Camr Ampr OriT(Mob+) Tra− | These authors (37) |
| pMP92 | OriV(RK2) Tetr OriT(Mob+) Tra− | C. A. Wijffelman (32) |
| pMP2642 | pssA (RBL5515 DNA in pMP92) | These authors (35) |
| pMP3030 | pssCDE (RBL5515 DNA in pMP92) | These authors (35) |
| pXCc8 | gumBCDEFGHIJKLMN (X59 DNA in pRK311) | These authors (33) |
| pXC1483 | gumDEF (subclone of pXCc8) | These authors (33) |
| pS88c1Δ3 | spsDCEB rhsACBD (S88 DNA in pRK311) | These authors (37) |
| pS88c2 | spsGSRQIKLJFDCEB (S88 DNA in pRK311) | These authors (37) |
| pS88c3 | spsQIKLJFDCEB rhsACBD (S88 DNA in pRK311) | These authors (37) |
| pZ206 | pS88c2-spsQ::mini-Tn10kan | These authors (37) |
| pY976 | pS88c3-spsI::mini-Tn10kan | These authors (37) |
| pY882 | pS88c3-spsK::mini-Tn10kan | These authors (37) |
| pY872 | pS88c3-spsL::mini-Tn10kan | These authors (37) |
| pB215 | spsB (subclone of pS88c3 in pSEB24) | These authors (37) |
| pB608 | spsK (S88 DNA in pMP92) | This work |
| pB554 | spsKB (S88 DNA in pMP92) | This work |
| pB610 | gumKLM SpsB (X59 and S88 DNA in pMP92) | This work |
| pB611 | gumKLM (X59 DNA in pMP92) | This work |
| pB597 | pssCDE spsB (S88 DNA and pMP3030 subclone in pMP92 | This work |
| pB599 | pssDE spsB (S88 DNA and pMP3030 subclone in pMP92) | This work |
| pB609 | pssCD spsB (S88 DNA and pMP3030 subclone in pMP92) | This work |
Culture media.
Luria-Bertani medium, YM, and M9 salts are standard media (27), and YMB was described previously (12). Glc (1 to 2% [wt/vol] final concentration) was added to media in agar plates or shake flasks to promote EPS synthesis for X. campestris and Sphingomonas. The concentrations of antibiotics (Sigma Chemical) used were as follows: rifampin, 20 to 50 μg/ml; streptomycin, 50 μg/ml; and tetracycline, 2 to 15 μg/ml.
Manipulation of cloned glycosyl transferase genes.
The glycosyl transferase genes from Sphingomonas (spsQ, -K, -L, and -B) were cloned from strain S88 by complementation of Sps− mutants m260, m265, m302, and m54 (37). Smaller segments were prepared from the original cosmid clones pS88c1Δ3, pS88c2, and pS88c3 by standard methods of DNA isolation, digestion with restriction enzymes, and ligation to multiple-cloning sites within the vector DNA (21). Plasmid pB608 was constructed by ligating the spsK-containing BglII segment (nucleotides 7038 through 9165 from the Sphingomonas DNA sequence [GenBank accession no. U51197]) obtained from pS88c2 to the BglII site in pMP92. Plasmid pB554 was assembled by ligating an EcoRI spsB-containing segment (Sphingomonas nucleotides 20249 through 24648) obtained from pS88c3 to pB608. Plasmid pB610 contained a gumKLM segment (nucleotides 11643 through 15832 of the X. campestris DNA sequence [GenBank accession no. U22511]) which was obtained from plasmid pXCc8 by restriction with ApaI and was inserted by blunt-end ligation between the BglII sites of plasmid pB554 with coincident removal of the resident spsK BglII segment. The gumKLM genes code, respectively, for glucuronosyl-(α1→2)-mannosyl transferase, mannosyl pyruvylase, and glucosyl-(β1→4)-glucosyl transferase (34). Plasmid pB611 was constructed by deleting the spsB-containing EcoRI segment from pB610. Plasmid pB597 (pssCDE spsB) was obtained by inserting the spsB-containing EcoRI segment (Sphingomonas nucleotides 20249 through 24648) into the EcoRI site of pMP3030. Adjacent to spsB in this segment is an intact rhsA gene which codes for UDP-glucose pyrophosphorylase. Plasmid pB599 (pssDE spsB) was obtained by deleting nearly the entire pssC gene between the SphI and PstI sites (35) from pB597 (pssCDE spsB). Similarly, plasmid pB609 (pssCD spsB) was obtained by deleting an essential carboxyl-terminal end of the pssE gene between the BstEII and MluI sites (35) from pB597.
Transformation, conjugation, complementation, and transposon mutagenesis.
Transfer of plasmid DNA from Escherichia coli to either Sphingomonas, X. campestris, or R. leguminosarum, or from R. leguminosarum to E. coli, was by triparental conjugal mating (7). Purified plasmid DNA was transferred from Sphingomonas or X. campestris to E. coli by transformation (11). Donor cells containing mobilizable broad-host-range tetracycline-resistant (Tetr) recombinant plasmids were mixed with helper cells containing the mobilizing pRK2013 plasmid and Sps− recipient Sphingomonas cells in a ratio of 5:2:10 and then were spotted onto YM plates lacking Glc and incubated for 6 to 16 h at 30°C. Exconjugants of Sphingomonas were isolated by spreading a loopful of the mating mixture onto YM plates containing 1% Glc, 50 μg of streptomycin per ml, and 12 μg of tetracycline per ml. Sphingomonas strains are naturally resistant to streptomycin (Strr), while the donor and helper E. coli strains are sensitive. Similarly, exconjugants of X. campestris were isolated on YM plates containing 1% glucose, 50 μg of rifampin per ml, and 12 μg of tetracycline per ml, and exconjugants of R. leguminosarum were isolated on YMB plates containing 20 μg of rifampin per ml and 2 μg of tetracycline per ml. Mutagenesis by transposition in the nonsuppressing host HMS174 was with Tn10 derivative 103 (mini-Tn10 kan/Ptac-ATS transposase) carried by λNK1316 (19).
Chemical analysis of EPS.
EPS were precipitated from culture medium with 2 to 3 volumes of isopropyl alcohol or ethanol. The precipitates were dried at 80°C and weighed. Hydrolysis mixtures containing 0.5 to 5 mg of polysaccharide and 130 to 260 μl of 2 M trifluoroacetic acid in high-performance liquid chromatography (HPLC) water were incubated for 4 to 16 h at 95°C and then dried under vacuum, resuspended in 100 μl of HPLC water, dried again, and finally resuspended in 100 μl of HPLC water. Samples and sugar standards were separated on a CarboPac PA-1 anion-exchange column, and compositions were quantitated with a Dionex DX500 HPLC system as described previously (5).
Labeling and chromatographic separation of lipid-linked saccharides.
EDTA-treated E. coli cells carrying either plasmid B608 (SpsK+) or the vector (pMP92) alone were prepared as follows. The cultures were shaken at 37°C in Luria-Bertani medium to an absorbance (at 600 nm) of about 1.8 and then were chilled on ice and washed twice by centrifugation and resuspension with 1 volume of cold 0.9% NaCl. The cell pellet was resuspended in 10 mM EDTA-Tris (10) at 1/20 of the original culture volume and frozen at −80°C. A sample was thawed at 16°C, centrifuged in the cold, resuspended in 50 mM Tris (pH 8.2)–5 mM EDTA, and then frozen and thawed two more times. The protein concentrations for the EDTA-treated cells were determined with the Bio-Rad protein assay. A standard radioactive labeling reaction mixture contained 0.3 ml of EDTA-treated cells, 0.3 ml of 50 mM Tris-HCl (pH 8.2)–20 mM MgCl2, and 15 μl (943 pmol) of [14C]UDP-Glc or [14C]UDP-glucuronic acid ([14C]UDP-GlcA) (300 nCi; 318 mCi/mmol), and the reaction mixture was incubated for 60 min at 16°C. Unlabeled UDP-Glc (300 pmol) was added with the labeled UDP-GlcA. After labeling, the permeabilized cells were centrifuged at 14,000 rpm for 2 min in a Hermle 2360K refrigerated microcentrifuge and then resuspended in 1 ml of cold 10 mM EDTA-Tris, and this washing step was repeated two more times. Lipid-linked saccharides were extracted from the cell pellets by adding 0.1 ml of chloroform-methanol-H2O (1:2:0.1), pooling three extractions, and drying (2). Saccharides were released from the lipids by cleaving the sugar-phosphate bonds with mild acid (50 μl of 0.01 M HCl at 100°C for 10 min), reneutralized, and then separated by gel filtration through a Bio-Gel P column (100 by 1 cm) with 0.1 M pyridinium acetate buffer (pH 5.0) at a rate of about 0.05 ml/min and 0.5 ml/fraction (2). Markers included blue dextran (exclusion), CoCl2 (inclusion), and maltotetraose, maltotriose, maltose, Glc, and glucuronic acid (GlcA). Unlabeled sugars were detected by the phenol-sulfuric acid method (8).
RESULTS AND DISCUSSION
Related Sphingomonas strains have gene products analogous to the SpsB, -K, and -L gene products of strain S88.
DNA sequencing of a cluster of genes required for sphingan synthesis by strain S88 and comparison of the deduced protein sequences to those of gene products involved in polysaccharide synthesis in other bacteria revealed four genes (spsQ, -K, -L, and -B) whose products were likely glycosyl transferases (37). Our strategy for determining the substrate recognition properties of these gene products in vivo was to transfer genes between related strains of Sphingomonas which synthesize similar but distinct polysaccharides and to observe whether sphingan synthesis was restored to polysaccharide-negative recipients. Random segments of the NW11 chromosome were inserted into a broad-host-range cosmid vector and screened for clones which restored sphingan synthesis to an SpsB− mutant of strain S88. One such clone (pNWc1) also restored sphingan synthesis to spsK and -L mutants of strain S88. However, mutations in spsQ in strain S88 were not complemented by pNWc1, despite the presence of sufficient coding potential on the cloned segment to the right and left of the spsKLB region. By contrast, the spsQ, -K, -L, and -B mutations in S88 were complemented by pS198c5, which carries a chromosomal segment cloned from strain S198. In addition DNA hybridization tests (data not shown) indicated that the clone from strain S198 had segments that hybridized to gene-specific probes for the spsB, -K, and -L genes of strain S88. These initial complementation and hybridization results suggested that the SpsB, -K, and -L gene products in strain S88 and the analogous enzymes in NW11 and S198 were conserved and probably catalyzed biosynthetic steps which were common to these strains. Since we knew that spsB coded for a glucosyl-IP transferase (27), the spsK and -L genes were likely to code for the other two common glycosyl transferases, the glucuronosyl-(β1→4)-glucosyl and glucosyl-(β1→4)-glucuronosyl transferases. However, which one codes for each transferase is not revealed by these complementation tests alone. By elimination, we propose that the SpsQ function, which also has a transferase-like protein sequence (37), is probably involved in adding l-rhamnose or l-mannose to the common trisaccharide core in strains S88 and S198 and is replaced by a different gene product in NW11.
Reciprocal complementation between the Sphingomonas spsK and R. leguminosarum pssDE genes.
A comparison of deduced amino acid sequences for the SpsK and PssDE products shows significant partial homology, suggesting a common function (35). Since a stable lipid-linked trisaccharide with the structure GlcA-GlcA-Glc-isoprenylpyrophosphate was isolated from R. leguminosarum, the first transferase attaches glucose-1-phosphate to the carrier IP and the second transferase attaches GlcA to Glc-PPI (2). These two reactions may be identical to assembly steps in Sphingomonas, depending on which Glc of the sphingans is added first to the carrier. However, as shown in Fig. 1, assembly of the sphingans and the rhizobial EPS must diverge with the third transferase reaction, where different substrates are recognized. Therefore, we tested whether PssDE and SpsK could complement one another.
The relative frequencies for conjugal transfer of specific plasmids carrying the spsK and pssDE genes into three recipients (Sphingomonas strain S88, X. campestris, and R. leguminosarum) and phenotypes of the exconjugants were determined (Table 2). When Sphingomonas strain S88 was the recipient, sphingan was produced by mutant m265 (SpsB−) only when a functional glucosyl-IP transferase was present (plasmids pB215, pB554, and pB599). Similarly, the pssDE genes (pB599) substituted for the SpsK− defect in mutant m302, allowing synthesis of polysaccharide which was not distinguishable by sugar composition from that made by the native spsK gene (pB554). Chromatograms of both the sphingan controls and these recombinant-derived products showed characteristic peaks of Glc, mannose, GlcA, and rhamnose (data not shown), as expected for sphingan S-88. In the reciprocal transfer, the spsK gene (pB608) at least partially restored synthesis of rhizobial EPS in a PssDE− mutant (RBL5833), and an intermediate colonial phenotype was observed. The amount of EPS produced was less than the amount secreted by wild-type R. leguminosarum (RBL5523). Complementation was eliminated by a mini-Tn10 insertion in spsK (pY882). The sugar compositions of the EPS from strain RBL5833 carrying either the normal pssCDE or foreign spsK genes appeared to be identical (data not shown) and matched the rhizobial EPS pattern. The most likely explanation is that spsK and pssDE code for functionally equivalent transferases that add GlcA to Glc-PPI. The results of the following labeling experiments are consistent with this hypothesis.
TABLE 2.
Expression of glycosyl transferase genes in Sphingomonas, X. campestris, and R. leguminosarum
| Recipient strain (phenotype) | Donor plasmid (genotype)a | Exconjugants
|
|
|---|---|---|---|
| Frequency | Morphologyb | ||
| Sphingomonas S88 | |||
| m265 (SpsB−) | pB215 (spsB) | Normal | + |
| pB554 (spsKB) | Normal | + | |
| pB599 (pssDE spsB) | Normal | + | |
| pB610 (gumM spsB) | Normal | − | |
| pB611 (gumM) | Normal | − | |
| pB597 (pssCDE spsB) | Normal | Tiny | |
| pB609 (pssC spsB) | Normal | Tiny | |
| pMP3030 (pssCDE) | Normal | − | |
| m302 (SpsK−B−) | pB554 (spsKB) | Normal | + (S-88) |
| pS88c3 (spsQIKLB) | Normal | + | |
| pY976 (spsQI::TnKLB) | Normal | + | |
| pY872 (spsQIKL::TnB) | Normal | + | |
| pY882 (spsQIK::TnLB) | Normal | − | |
| pB599 (pssDE spsB) | Normal | + (S-88) | |
| pB610 (gumM spsB) | Normal | − | |
| pB611 (gumM) | Normal | − | |
| pB597 (pssCDE spsB) | Normal | Tiny | |
| pB609 (pssC spsB) | Normal | Small | |
| pMP3030 (pssCDE) | Normal | − | |
| X. campestris X59 m31 (GumD−) | pSY1483 (gumDEF) | Normal | + |
| pS88c1Δ3 (spsB) | Normal | + | |
| pS88c2 (spsQIKLB) | Zero | ND | |
| pS88c3 (spsQIKLB) | Zero | ND | |
| pZ206 (spsQ::TnIKLB) | Zero | ND | |
| pY976 (spsQI::TnKLB) | Zero | ND | |
| pY872 (spsQIKL::TnB) | Zero | ND | |
| pY882 (spsQIK::TnLB) | Normal | + (XG) | |
| pB610 (gumM spsB) | Normal | + | |
| pB608 (spsK) | Normal | − | |
| pB554 (spsKB) | Zero | ND | |
| pB599 (pssDE spsB) | Zero | ND | |
| pB609 (pssC spsB) | Normal | + (XG) | |
| R. leguminosarum | |||
| RBL5833 (PssDE−) | pMP3030 (pssCDE) | Normal | + (rhEPS) |
| pMP2642 (pssA) | Normal | − | |
| pB608 (spsK) | Normal | Intermediate (rhEPS) | |
| pS88c3 (spsQIKLB) | Low | Intermediate/− (1/9) | |
| pY872 (spsQIKL::TnB) | Normal | Intermediate (rhEPS) | |
| pY882 (spsQIK::TnLB) | Normal | − | |
| pB610 (gumM spsB) | Zero | ND | |
| pB611 (gumM) | Zero | ND | |
| RBL5523 (Pss+) | pMP3030 (pssCDE) | Normal | + |
| pMP2642 (pssA) | Normal | + | |
| pS88c3 (spsQIKLB) | Low | + | |
| pY872 (spsQIKL::TnB) | Normal | + | |
| pY882 (spsQIK::TnLB) | Normal | +, Small | |
| RBL5807 (PssA−) | pMP3030 (pssCDE) | Normal | − |
| pMP2642 (pssA) | Normal | + | |
| pB608 (spsK) | Normal | − | |
| pB554 (spsKB) | Normal | + | |
| pS88c3 (spsQIKLB) | Low | +/− (1/9) | |
| pY872 (spsQIKL::TnB) | Normal | + | |
| pY882 (spsQIK::TnLB) | Normal | +/− (9/1) | |
| pB610 (gumM spsB) | Zero | ND | |
| pB611 (gumM) | Normal | − | |
Only the relevant genes are listed; see Table 1 for a complete list of genes on plasmids.
EPS-producing colonies of Sphingomonas, X. campestris, and R. leguminosarum are denoted as +, and nonproducers are denoted as −; intermediate for R. leguminosarum indicates a colonial appearance between smooth (+) and rough (−). XG, rhEPS, and S-88 indicate the sugar compositions typical of xanthan gum, rhizobial EPS, and sphingan S-88, respectively. Tiny and small refer to reduced colony sizes. The relative frequency of colony types is given in parentheses. Zero means that no exconjugants were observed. ND, not determinable by visual inspection. The position of each mini-Tn10kan insertion was determined by digestion with restriction endonucleases and comparison to the DNA sequence of segment S88c3 (37).
Glycosyl transferase-specific labeling of lipid-linked saccharides.
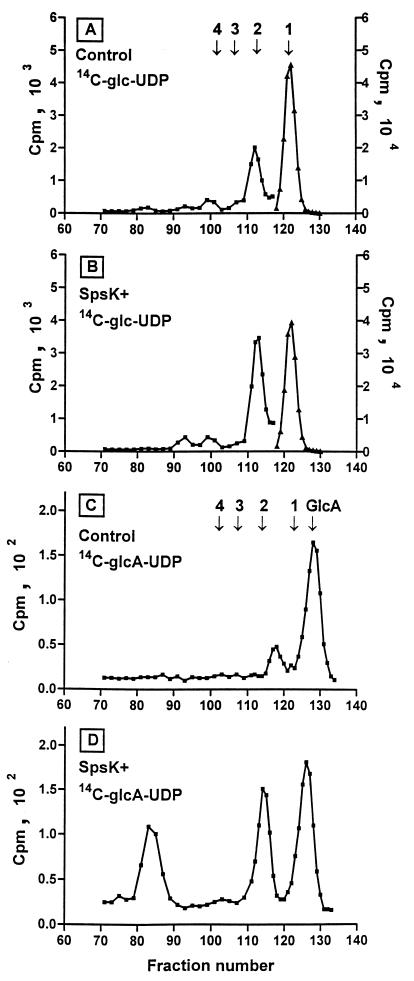
Incorporation of radioactive Glc and GlcA into lipid-linked disaccharides was used to identify the substrates recognized by the SpsK glycosyl transferase. EDTA-permeabilized cells of E. coli DH5α carrying either pB608 with the spsK gene or the vector (pMP92) alone were labeled with radioactive [14C]UDP-Glc or [14C]UDP-GlcA. The spsK fragment cloned in pB608 included the normal Sphingomonas promoter and ribosome-binding region upstream from the spsK gene and small incomplete segments of the flanking spsI and spsL genes. Therefore, the activity of the SpsK protein in E. coli depended on gene expression in the foreign host and on the production of the other putative substrate, Glc-PPI. Labeled saccharides were released from lipid-linked intermediates in the cell membranes and separated by gel filtration into mono-, di-, and oligosaccharides.
The chromatographic elution profiles are shown in Fig. 2. The permeabilized bacteria used for Fig. 2A and B were labeled with [14C]UDP-Glc. It was not surprising to find counts in the monosaccharide peak, since E. coli has at least two transferases that can attach Glc to lipids: the first transferase in colanic acid biosynthesis (16) and a potent membrane-associated activity involved in the synthesis of membrane-derived oligosaccharides (18). The activity of the SpsK gene product caused a reproducible increase in counts in the disaccharide peak in Fig. 2B. The increase was 90%, and in a replicate labeling the increase was 80%. This is consistent with Glc-PPI, produced endogenously by E. coli, as a substrate for the SpsK transferase. Figure 2C and D show that incorporation of radioactive GlcA into lipid-linked saccharides from [14C]UDP-GlcA was much lower than from UDP-Glc, but as before we observed SpsK-specific labeling of disaccharides, consistent with UDP-GlcA being a substrate for SpsK. The profile in Fig. 2D also shows a species of higher mobility that is also SpsK specific, but the size and nature of the saccharide are not known. Taken together, the in vivo genetic complementation tests and the in vitro sugar incorporation studies indicate that the repeating subunits for the sphingans are assembled from nucleotide sugars to give the intermediate structure Glc-GlcA-Glc-PPI. The first transferase (SpsB) adds glucose-1-phosphate to the IP carrier, and then SpsK adds GlcA to Glc-PPI. For the rhizobial EPS intermediate structure, GlcA-GlcA-Glc-PPI (2), PssA adds glucose-1-phosphate to IP and PssDE adds GlcA to Glc-PPI.
FIG. 2.
Gel filtration of saccharides released from lipid-linked intermediates. (A) Control plasmid vector pMP92 labeled with [14C]Glc-UDP; (B) SpsK+ plasmid pB608 labeled with [14C]Glc-UDP; (C) pMP92 labeled with [14C]GlcA-UDP; (D) pB608 labeled with [14C]GlcA-UDP. The scale on the right for panels A and B applies to fractions 117 to 130. The arrows show the positions of marker saccharides: GlcA, Glc (arrow 1), maltose (arrow 2), maltotriose (arrow 3), and maltotetraose (arrow 4).
Genetic transfers resulting in inhibition of polysaccharide synthesis or cellular growth.
By contrast with the straightforward positive complementation between two genes which are functionally equivalent, we also observed “negative” phenomena associated with the transfer of certain glycosyl transferase genes, i.e., inhibition of polysaccharide synthesis or cell growth. In earlier work we caused mutated Sphingomonas bacteria, which carried an extensive deletion of the sps genes, to secrete copious amounts of xanthan gum by transferring to the recipient a cluster of 12 biosynthetic genes (gumBCDEFGHIJKLM) from X. campestris (36). However, as we show in Table 2, we failed to achieve a reciprocal genetic transfer, and specifically, plasmids carrying Sphingomonas genes could not be recovered intact from X. campestris if the incoming DNA included a functional spsK gene. X. campestris mutant m31 was complemented by either a gumD gene (pSY1483) or an spsB gene (pS88c1Δ3, pB610, pB609, and pY882), and the exconjugants produced xanthan gum of normal composition. Notably, the spsK gene in plasmid pY882 included a gene-inactivating mini-Tn10 insertion. By contrast, plasmids carrying both an spsB gene and an spsK gene (pS88c2, pS88c3, pZ206, pY976, pY872, and pB554) were not recovered. The detrimental effect of spsK in X. campestris required the presence of a functional glucosyl-IP transferase such as SpsB, since the spsK gene alone (pB608) was not detrimental and remained intact as determined by restriction endonuclease analysis. Likewise, the pssDE genes could not be transferred and maintained in X. campestris m31 when a functional glucosyl-IP transferase was included (pB599). For the pB599 mating we did observe a single mucoid Gum+ colony after conjugal mating, but upon plasmid isolation and restriction analysis we found that the plasmid had the pssDE genes deleted and failed to complement the Sps− Sphingomonas strain m302, unlike intact plasmid pB599. These results indicate that glycosyl transferases like SpsK and PssDE, which create an unnatural lipid-linked saccharide in X. campestris, are toxic to these cells.
Transfer into Sphingomonas strain m265 (SpsB−) of the spsB and gumM genes (pB610) did not prevent cell growth but eliminated complementation by spsB, resulting in no sphingan synthesis. Similar inhibition of sphingan synthesis was observed when the gumM gene on pB610 was transferred into the SpsK−B− Sphingomonas strain m302. The gumM gene caused a large reduction in the recovery of exconjugants in R. leguminosarum, as long as the recipient carried a glucosyl-IP transferase, such as PssA or SpsB (RBL5833pB611 or RBL5807pB610, respectively). In the absence of Glc-PPI, the GumM product was not toxic [RBL5807(pB611)]. When plasmids pB610 (SpsB GumKLM) and pB611 (GumKLM) were transferred separately to an R. leguminosarum mutant which synthesizes small amounts of rhizobial EPS (data not shown), only plasmid pB610 was toxic. This suggests that the degree of toxicity may depend on the number of incorrectly linked lipid carriers that accumulate.
Plasmids carrying the pssCDE (pMP3030) and spsK (pB608) genes were transferred normally to R. leguminosarum and were stable. By contrast, plasmid pS88c3, with its additional 20 genes from Sphingomonas, was poorly transferred into the rhizobial recipient. The plasmids that were subsequently isolated from the rare pS88c3 exconjugants had severe deletions and were missing restriction sites and spsL and other genes. The plasmids with the deletions were readily transferred back into the same rhizobial recipients. This suggests that the spsL gene is detrimental to R. leguminosarum. However, normal recovery of exconjugants was obtained if either the spsL gene (pY872) or the complementing spsK gene (pY882) on plasmid pS88c3 included a gene-inactivating transposon. The pY882 plasmid caused the Pss+ RBL5523 recipient to grow as relatively small colonies, as if the plasmid spsL gene was expressed at least partially. Similarly, introduction of the pssC and spsB genes (pB597 and pB609) into Sphingomonas strain m265 or m302 caused the exconjugant colonies to be reduced in size, as if the foreign pssC gene was toxic to Sphingomonas. The detrimental effect was not observed in the absence of a functional glucosyl-IP transferase (pMP3030). The most straightforward interpretation is that both spsL and pssC code for third transferases but that their products attach different sugars to GlcA-Glc-PPI.
Deleterious effects of incomplete or unnatural lipid-linked saccharides in bacteria.
Although secreted polysaccharides are not usually essential for cell viability, accumulation of lipid-linked intermediates in the cytoplasmic membrane appears to be harmful. For example, in Salmonella enterica, absence of the O antigen gives colonies a rough appearance compared to the normal smooth phenotype, and most of the rough mutants that are spontaneous or isolated after chemical mutagenesis are defective either in wbaP (formerly rfbP; coding for a galactosyl-IP transferase as the first step in assembly of the repeat) or in synthesis of the O-antigen precursor dTDP-rhamnose (20). Interestingly, a mutation (rfbH819) that blocks the synthesis of CDP-abequose gives rise to many lipopolysaccharide-positive revertants and secondary wbaP mutations (38), and in a similar way, phosphomannoisomerase mutants which do not synthesize GDP-mannose also acquire secondary rfbP mutations (20). As a second example, in Rhizobium meliloti, mutations in exoP, -T, -Q, -L, and -M are lethal in cells that are derepressed for succinoglycan synthesis (29). Based on protein sequence similarities, ExoP, -T, and -Q appear to be involved in secretion, while ExoL and -M code for distinct glucosyl-(β1→4)-glucosyl transferases that attach the third and fourth sugars of the succinoglycan repeat (30). However, in contrast to the above-described examples, in X. campestris mutations in the glycosyl transferase genes (gumD, -M, -H, -K, and -I) are not detrimental, while mutations in genes for secretion functions (gumB, -C, and -J) and in the putative polymerase gene (gumE) are lethal (3, 34). For Sphingomonas, we also find that most recovered Sps− mutations are in the spsB gene, which codes for the glucosyl-IP transferase, and the only mutants we have so far isolated that have mutations in the spsK, spsL, and rhs operon have second mutations in spsB, as if the failure to start assembly of the repeat unit restored full viability to these mutants.
Bacteria might have mechanisms to release the IP carrier from incomplete repeat subunits so that it can be used for the synthesis of essential peptidoglycan. However, we find that synthesis of nonnative repeat structures may be especially detrimental: the spsK and pssDE genes were toxic to X. campestris, the gumM and spsL genes were detrimental to R. leguminosarum, the pssC gene was toxic to Sphingomonas, and the gumM gene inhibited sphingan synthesis in Sphingomonas and rhizobial EPS synthesis in R. leguminosarum. We suggest that the formation of a nonnative oligosaccharide linked to the lipid carrier is toxic because the carrier cannot be released for other essential cellular functions.
Overview.
As shown in this work, the substrate requirements of glycosyl transferases involved in EPS biosynthesis can be determined in vivo by cloning and expression of foreign transferase genes in a recipient host, as long as the essential nucleotide-sugar precursors are present. The applicability of this in vivo approach will broaden as the substrate specificities of additional glycosyl transferase genes are determined and the genes and corresponding polysaccharide-negative mutant bacteria become available. An alternative approach is isolation and characterization of radiolabeled lipid-linked intermediates that accumulate in the membrane fractions of permeabilized bacteria. This approach depends on labeled nucleotide sugars, some of which are not readily available, and yields definitive results when only one polysaccharide structure is assembled (16). More importantly, the requirement for an organized membrane apparatus may cause a loss of specificity, and the extracts may accumulate only a few of the incomplete subunit forms (24).
ACKNOWLEDGMENT
W. A. T. van Workum was supported by the Foundation for Life Sciences (SLW), which is subsidized by the Netherlands Organization of Scientific Research (NWO).
REFERENCES
- 1.Borthakur D, Barber C E, Lamb J W, Daniels M J, Downie J A, Johnston A W B. A mutation that blocks exopolysaccharide synthesis prevents nodulation of peas by Rhizobium leguminosarum but not of beans by R. phaseoli and is corrected by cloned DNA from Rhizobium or the phytopathogen Xanthomonas. Mol Gen Genet. 1986;203:320–323. [Google Scholar]
- 2.Bossio J C, Semino C E, Iñón de Iannino N, Dankert M A. The in vitro biosynthesis of the exopolysaccharide produced by Rhizobium leguminosarum bv. trifolii, strain NA30. Cell Mol Biol. 1996;42:737–758. [PubMed] [Google Scholar]
- 3.Capage, M. A., D. H. Doherty, M. R. Betlach, and R. W. Vanderslice. October 1987. Recombinant-DNA mediated production of xanthan gum. International patent application W087/05938.
- 4.Chowdhury T A, Lindberg B, Lindquist U, Baird J. Structural studies of an extracellular polysaccharide (S-198) elaborated by Alcaligenes ATCC 31853. Carbohydr Res. 1987;161:127–132. doi: 10.1016/0008-6215(89)84154-0. [DOI] [PubMed] [Google Scholar]
- 5.Clarke A J, Sarabia V, Keenleyside W, MacLachlan P R, Whitfield C. Compositional analysis of bacterial extracellular polysaccharides by high performance anion-exchange chromatography. Anal Biochem. 1991;199:68–74. doi: 10.1016/0003-2697(91)90270-4. [DOI] [PubMed] [Google Scholar]
- 6.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X-W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 7.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 9.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García R C, Recondo E, Dankert M. Polysaccharide biosynthesis in Acetobacter xylinum, enzymatic synthesis of lipid diphosphate and monophosphate sugars. Eur J Biochem. 1974;43:93–105. doi: 10.1111/j.1432-1033.1974.tb03389.x. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hooykaas P J J, Klapwijk P M, Nuti M P, Schilperoort R A, Rörsch A. Transfer of the Agrobacterium tumefaciens Ti plasmid to avirulent agrobacteria and to rhizobia ex planta. J Gen Microbiol. 1977;98:477–484. [Google Scholar]
- 13.Ielpi L, Couso R O, Dankert M A. Sequential assembly and polymerization of the polyprenol-linked pentasaccharide repeating unit of the xanthan polysaccharide in Xanthomonas campestris. J Bacteriol. 1993;175:2490–2500. doi: 10.1128/jb.175.9.2490-2500.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansson P-E, Kenne L, Lindberg B. Structure of the extracellular polysaccharide from Xanthomonas campestris. Carbohydr Res. 1975;45:275–282. doi: 10.1016/s0008-6215(00)85885-1. [DOI] [PubMed] [Google Scholar]
- 15.Jansson P-E, Kumar N S, Lindberg B. Structural studies of a polysaccharide (S-88) elaborated by Pseudomonas ATCC 31554. Carbohydr Res. 1986;156:165–172. doi: 10.1016/s0008-6215(00)90108-3. [DOI] [PubMed] [Google Scholar]
- 16.Johnson J G, Wilson D B. Role of sugar-lipid intermediate in colanic acid synthesis by Escherichia coli. J Bacteriol. 1977;129:225–236. doi: 10.1128/jb.129.1.225-236.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, K. S., and G. T. Veeder. August 1985. Heteropolysaccharide S-88. U.S. patent 4,535,153.
- 18.Kennedy E P. Membrane-derived oligosaccharides (periplasmic beta-d-glucans) of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1064–1071. [Google Scholar]
- 19.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 20.Mäkelä P H, Stocker B A D. Genetics of polysaccharide biosynthesis. Annu Rev Genet. 1969;3:291–322. [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 22.Moorhouse R. Structure/property relationships of a family of microbial polysaccharides. In: Yalpani M, editor. Industrial polysaccharides: genetic engineering, structure/property relations and applications. Amsterdam, The Netherlands: Elsevier Science Publishers B.V.; 1987. pp. 187–206. [Google Scholar]
- 23.O’Neill M A, Darvill A G, Albersheim P, Chou K J. Structural analysis of an acidic polysaccharide secreted by Xanthobacter sp. (ATCC 53272) Carbohydr Res. 1990;206:289–296. doi: 10.1016/0008-6215(90)80068-e. [DOI] [PubMed] [Google Scholar]
- 24.Osborn M J, Weiner I M. Biosynthesis of a bacterial lipopolysaccharide. VI. Mechanism of incorporation of abequose. J Biol Chem. 1968;243:2631–2639. [PubMed] [Google Scholar]
- 25.Pees E, Wijffelman C A, Mulders I H M, van Brussel A A N, Lugtenberg B J J. Transposition of Tn1831 to Sym plasmids of Rhizobium leguminosarum and Rhizobium trifolii. FEMS Microbiol Lett. 1986;33:165–171. [Google Scholar]
- 26.Pollock T J. Gellan-related polysaccharides and the genus Sphingomonas. J Gen Microbiol. 1993;139:1939–1945. [Google Scholar]
- 27.Pollock T J, Thorne L, Yamazaki M, Mikolajczak M, Armentrout R W. Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides. J Bacteriol. 1994;176:6229–6237. doi: 10.1128/jb.176.20.6229-6237.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priem W J E, Wijffelman C A. Selection of strains cured of the Rhizobium leguminosarum Sym plasmid pRL1JI by using small bacteriocin. FEMS Microbiol Lett. 1984;25:247–251. [Google Scholar]
- 29.Reuber T L, Long S, Walker G C. Regulation of Rhizobium meliloti exo genes in free-living cells and in planta examined by using TnphoA fusions. J Bacteriol. 1991;173:426–434. doi: 10.1128/jb.173.2.426-434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuber T L, Walker G C. Biosynthesis of succinoglycan, symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 31.Robertson B K, Åman P, Darvill A G, McNeil M, Albersheim P. Host symbiont interactions. V. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981;67:389–400. doi: 10.1104/pp.67.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 33.Thorne L, Tansey L, Pollock T J. Clustering of mutations blocking synthesis of xanthan gum by Xanthomonas campestris. J Bacteriol. 1987;169:3593–3600. doi: 10.1128/jb.169.8.3593-3600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanderslice R W, Doherty D H, Capage M A, Betlach M R, Hassler R A, Henderson N M, Ryan-Graniero J, Tecklenburg M. Genetic engineering of polysaccharide structure in Xanthomonas campestris. In: Crescenzi V, Dea I C M, Paoletti S, Stivala S S, Sutherland I W, editors. Biomedical and biotechnological advances in industrial polysaccharides. New York, N.Y: Gordon and Breach Science Publishers; 1989. pp. 145–156. [Google Scholar]
- 35.van Workum W A T, Canter Cremers H C J, Wijfjes A H M, van der Kolk C, Kijne J W. Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant-Microbe Interact. 1997;10:290–301. doi: 10.1094/MPMI.1997.10.2.290. [DOI] [PubMed] [Google Scholar]
- 36.Yamazaki M, Thorne L, Armentrout R W, Pollock T J. Production of xanthan gum by Sphingomonas bacteria carrying genes from Xanthomonas campestris. J Ind Microbiol Biotechnol. 1997;19:92–97. doi: 10.1038/sj.jim.2900449. [DOI] [PubMed] [Google Scholar]
- 37.Yamazaki M, Thorne L, Mikolajczak M, Armentrout R W, Pollock T J. Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J Bacteriol. 1996;178:2676–2687. doi: 10.1128/jb.178.9.2676-2687.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuasa R, Levinthal M, Nikaido H. Biosynthesis of cell wall lipopolysaccharide in mutants of Salmonella. J Bacteriol. 1969;100:433–444. doi: 10.1128/jb.100.1.433-444.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]