Abstract
Objective: To describe the prevalence of common and clinically relevant microbial isolates before and after the migration of a 24-bed, open plan, adult intensive care unit (ICU) to a new extended design of 32 single rooms, supporting an expanded clinical oncology casemix while continuing all existing clinical services.
Design: Retrospective, observational descriptive analysis covering the period 5 May 2014 to 4 May 2018 — the 2 years before and after the ICU relocation on 5 May 2016.
Setting: A university-associated, tertiary teaching hospital and state trauma centre in Victoria, Australia.
Patients: Adult ICU patients.
Main outcome measures: Bacterial isolate frequency and incident rate ratios (IRRs) during the study period.
Results: When compared with the old ICU, the incidence rates per 1000 occupied bed-days in the new ICU were lower for bacterial isolates overall (IRR, 0.88; 95% CI, 0.83–0.93), for coagulase-negative staphylococci (IRR, 0.64; 95% CI, 0.55–0.75) and for vancomycin-resistant enterococci (IRR, 0.50; 95% CI, 0.32–0.80). The incidence rates per 1000 occupied bed-days between ICU locations were unchanged for Staphylococcus aureus (IRR, 1.1; 95% CI, 0.91–1.3), extended-spectrum beta-lactamase-producing organisms (IRR, 1.4; 95% CI, 0.78–2.6) and carbapenemase-producing Enterobacterales (IRR, 0.85; 95% CI, 0.11–6.4).
Conclusion: Within the limits of a before–after design and clinically directed sampling, relocation to a new ICU with single rooms and a growing oncological patient casemix was accompanied by no overall change in the apparent prevalence of the nosocomial pathogens S. aureus, extended-spectrum beta-lactamase-producing organisms or carbapenemase-producing Enterobacterales. These finding suggest that advanced physical infrastructure, including patient accommodation in single rooms, may play a role in overall safe delivery of critical care.
Infections acquired in intensive care units (ICUs) are frequently associated with morbidity and mortality.1, 2, 3 The epidemiology of commonly isolated pathogens in the intensive care setting shows geographical variation,4, 5 with international estimates reporting that up to 36% of organisms isolated in some intensive care settings may demonstrate multidrug resistance4 that is relevant to empirical antibiotic prescribing in those settings.3, 6 In many ICUs, methicillin-resistant Staphylococcus aureus (MRSA) and extended spectrum beta-lactamase (ESBL)-producing Enterobacterales are high-risk organisms, while vancomycin-resistant enterococci (VRE) are considered to be medium priority.7 Data from Australia associate these organisms with potentially inappropriate antimicrobial use and prolonged ICU admission.8
In most Australian states and territories, mandatory surveillance programs exist for hospital-associated infections such as central line-associated bloodstream infections (CLABSI).9 Pathogen-specific surveillance, such as testing for S. aureus bloodstream infections and monitoring for the presence of carbapenem-producing Enterobactorales (CPE), is used in some jurisdictions10, 11 but the epidemiology of other pathogens has not been characterised to the same extent. Antimicrobial burden and resistance in Australian health care settings is monitored through the National Antimicrobial Utilisation Surveillance Program and the National Alert System for Critical Antimicrobial Resistances.12, 13
The primary objective of this study was to describe the prevalence of microbial isolates of selected clinically relevant bacteria in samples taken from patients in the Royal Melbourne Hospital (RMH) ICU in the 2 years before and 2 years after an infrastructure upgrade. The upgrade replaced a 24-bed ICU with an open plan design (four single patient rooms plus 20 beds that were separated only by curtains) with a modernised 32-bed ward in which all patients are accommodated in single patient rooms. The secondary objectives were to compare the incidence of positive blood culture test results before and after ICU relocation and compare estimated community- and hospital-acquired pathogen burden.
Methods
Setting
The RMH is a university-affiliated, adult tertiary hospital in Melbourne, Australia. In addition to general and specialist surgical and medical services, it houses one of two statewide trauma services, as well as haematology, oncology, bone marrow and renal transplant services. On 5 May 2016, RMH ICU patients were moved to the new ICU. Shortly thereafter, critically ill patients at the newly opened on-campus Victorian Comprehensive Cancer Centre began receiving care at the new ICU. The RMH ICU implements a range of standardised infection control procedures that remained consistent during the 4-year study period. This includes a CLABSI reduction program, hand hygiene protocols, S. aureus bloodstream infection surveillance, cleaning with quaternary ammonium compounds, and a longstanding antimicrobial stewardship program. At the time of the study, contact precautions were implemented for patients colonised with VanA VRE, CPE or multidrug-resistant (MDR) Gram-negative pathogens. Isolation for patients with VanB VRE stopped on 25 July 2016. Isolation for other organisms, including MRSA, was managed based on individual risk assessment, in accordance with local hospital policy. Coinciding with the move to the new ICU, in-house cleaning practices were intensified. Specifically, designated ICU clinical assistants cleaned patient rooms while hospital environmental services staff cleaned communal areas such as corridors and office spaces. In accordance with College of Intensive Care Medicine of Australia and New Zealand (CICM) policies, nurse-to-patient ratios were maintained at 1:1 for ventilated patients and 1:2 for non-ventilated, high dependency patients in both old and new ICUs. There were no substantial changes in the medical staffing model throughout the study period, which also complied with CICM standards.14
Patients and data collection
The 4-year study period — 5 May 2014 to 4 May 2018 — included the 2 years before and 2 years after the RMH ICU relocation. Data summarising all positive bacterial isolates for samples collected from patients admitted for care at the ICU were retrieved from the hospital infection control database (ICNET, Baxter International). All patient samples from which these isolates were identified had been collected according to the clinical judgement of the treating clinicians, who were operating within hospital and ICU surveillance and clinical care policies at the time. This included admission screening for VRE that was introduced on 2 September 2015. Isolates obtained between 5 May 2014 and 4 May 2016 (inclusive) were attributed to the old 24-bed ICU, and isolates obtained between 5 May 2016 and 4 May 2018 (inclusive) were attributed to the new 32-bed ICU. For each 2-year period, patient admission dates, patient episode numbers, statistics on overall ICU occupied bed-days (OBD) and data on illness severity (using Acute Physiology and Chronic Health Evaluation III scores15 and the Australian and New Zealand Risk of Death prognostic system) were collected.16 The study was approved by the RMH Human Research Ethics Committee (QA2018.065).
Microbial data assessment (inclusion and exclusion criteria)
The isolates evaluated were from both clinically directed and surveillance samples. Each hospital admission for each patient during the study period was considered a discrete episode of care, so positive microbial isolates from each inpatient admission episode were counted separately. However, for individual patients, duplicate positive microbial isolates of the same microorganism across multiple ICU admissions within any one episode of hospital admission did not contribute to the count of positive cultures, as they were deemed to represent the same underlying infection during the patient’s episode of illness.
Likewise, for individual patients, duplicate positive microbial isolates from the same anatomical site in the same hospital admission episode were excluded (eg, three consecutive positive blood cultures counted as one episode of bacteraemia). However, isolation of the same organism from samples taken from two anatomically distinct sites (eg, sputum and urine) resulted in both specimens being counted. In cases where a patient had related samples (eg, sputum and bronchoscopy samples) from which the same organism was identified within 48 hours in a single hospital admission episode, only the first positive isolate was included. Where related samples were collected at the same time, the more invasive sample (eg, sample taken for tissue culture) was used.
Positive microbial isolates from samples collected < 48 hours after a patient was admitted to the hospital were classified as community-acquired, while those from samples collected ≥ 48 hours after admission to hospital were classified as hospital-acquired, in accordance with the Centers for Disease Control and Prevention definitions.17
Statistical analyses
Data from the hospital pathology database covering the 4-year study period were supplied in a single spreadsheet format. Following manual data processing as described above, this information was imported for statistical analyses into Stata 16 (StataCorp). Numeric data were summarised as mean (standard deviation), median (interquartile range) [full range] or count with accompanying proportion of a relevant total, as appropriate. Proportions representing selected results were compared between each ICU location using risk ratios with 95% confidence intervals. The cumulative counts of selected isolates of interest in each 2-year period (before and after the ICU relocation) were first expressed as incidence rates (IRs) using the relevant total ICU OBD value for each 2-year period. These IR values were then compared across the two periods as point estimates of incidence rate ratios (IRRs) with accompanying 95% confidence intervals. The uncertainty of other estimated differences was also summarised using 95% confidence intervals, supplemented by P values for selected comparisons.
Results
Over the 4-year study period, there were 9870 admissions to ICU involving 8726 patients. The patients had clinical conditions that ranged across most adult medical and surgical specialities, including general medicine, general surgery, trauma, haematology, oncology and organ transplantation (bone marrow and renal). Of these ICU patients, 1890 (21.7%) yielded at least one positive microbial isolate from any anatomical site.
While patient age, sex, illness severity and diagnostic grouping proportions were similar across the 4 years (Table 1), there was a notable increase in oncological casemix from May 2016, when the new ICU began admitting additional patients from the Victorian Comprehensive Cancer Centre. The median lengths of hospital stay before and after the relocation were 10 and 9 days, respectively. The corresponding median ICU lengths of stay were 1.9 and 1.8 days, respectively, with the lag between hospital and ICU admissions being stable at less than 0.5 days (Table 1). The subgroup of patients with microbiological isolates were rapidly transferred to ICU after hospital admission, with a median lag of 0.3 days, but these patients had substantially longer ICU stays (median, 5 days) and hospital stays (median, approaching 17 days) (Table 1).
Table 1.
Patient characteristics, overall and for the subgroup of patients with microbiological isolates*
| 5 May 2014–4 May 2016, 24-bed old ICU | 5 May 2016 – 4 May 2018, 32-bed new ICU | |
|---|---|---|
| Number of admissions | 4388 | 5482 |
| Number of occupied bed-days | 14734 | 17305 |
| Age (years), median (IQR) | ||
| Overall | 62.0 (43.9–73.5) | 61.0 (44.6–72.2) |
| Subgroup | 63.3 (47.6–73.5) | 60.9 (44.1–71.4) |
| Sex, female† | ||
| Overall | 1529 (34.9%) | 2017 (36.8%) |
| Subgroup | 281 (33.7%) | 330 (34.3%) |
| Hospital duration (days), median (IQR) | ||
| Overall | 10 (5.8–19) | 8.7 (3.8–17) |
| Subgroup | 17 (8.9–31) | 16 (8.9–29) |
| ICU duration (days), median (IQR) | ||
| Overall | 1.9 (1.0–3.8) | 1.8 (1.0–3.5) |
| Subgroup | 5.1 (2.5–9.7) | 4.7 (2.2–9.5) |
| Hospital-ICU lag (days), median (IQR) | ||
| Overall | 0.4 (0.2–1.6) | 0.3 (0.1–1.0) |
| Subgroup | 0.3 (0.2–1.2) | 0.3 (0.1–0.8) |
| Hospital-specimen lag (days), median (IQR) | ||
| Overall | na | na |
| Subgroup | 2.8 (0.9–7.2) | 2.4 (0.8–6.8) |
| ICU-specimen lag (days), median (IQR) | ||
| Overall | na | na |
| Subgroup | 1.7 (0.4–4.7) | 1.4 (0.3–4.4) |
| Medical not surgical‡ | ||
| Overall | 2727 (62.2%) | 3297 (60.2%) |
| Subgroup | 626 (75%) | 724 (75%) |
| APACHE III score, median (IQR) [range]§ | ||
| Overall | 56 (41–76) [0–190] | 54 (39–73) [3–195] |
| Subgroup | 71.5 (55–91) [13–170] | 68 (51–88) [12–169] |
| ANZROD score, median (IQR) [range]§ | ||
| Overall | 0.04 (0.01–0.17) [0–1.0] | 0.04 (0.01–0.16) [0–1.0] |
| Subgroup | 0.14 (0.05–0.34) [0.00–0.97] | 0.13 (0.04–0.32) [0.0–0.98] |
| Principal ICU diagnosis¶ | ||
| Cardiovascular | ||
| Overall | 1512 (34.5%) | 1618 (29.5%) |
| Subgroup | 168 (20.2%) | 174 (18.1%) |
| Trauma | ||
| Overall | 611 (13.9%) | 875 (16.0%) |
| Subgroup | 153 (18.4%) | 174 (25.0%) |
| Sepsis | ||
| Overall | 514 (11.7%) | 506 (9.2%) |
| Subgroup | 139 (16.7%) | 240 (15.6%) |
| Neurological | ||
| Overall | 479 (10.9%) | 568 (10.4%) |
| Subgroup | 150 (18.0%) | 137 (14.2%) |
| Respiratory | ||
| Overall | 400 (9.1%) | 562 (10.3%) |
| Subgroup | 90 (10.8%) | n (10.2%) |
| Metabolic/endocrine | ||
| Overall | 321 (7.3%) | 363 (6.6%) |
| Subgroup | 45 (5.4%) | 54 (5.6%) |
| Gastrointestinal | ||
| Overall | 256 (5.8%) | 266 (4.9%) |
| Subgroup | 49 (5.9%) | 45 (4.7%) |
| Renal/genitourinary | ||
| Overall | 123 (2.8%) | 133 (2.4%) |
| Subgroup | 19 (2.3%) | 14 (1.5%) |
| Oncological** | ||
| Overall | 90 (2.1%) | 447 (8.2%) |
| Subgroup | 11 (1.3%) | 30 (3.1%) |
| Musculoskeletal/skin | ||
| Overall | 35 (0.8%) | 46 (0.8%) |
| Subgroup | 5 (0.6%) | 6 (0.6%) |
| Other/not specified | ||
| Overall | 17 (0.4%) | 9 (0.2%) |
| Subgroup | 0 | 0 |
| Haematological** | ||
| Overall | 16 (0.4%) | 56 (1.0%) |
| Subgroup | 4 (0.5%) | 13 (1.4%) |
| Gynaecological | ||
| Overall | 14 (0.3%) | 29 (0.5%) |
| Subgroup | 0 | 1 (0.1%) |
ANZCIS CORE = Australian and New Zealand Intensive Care Society Centre for Outcome and Resource Evaluation. ANZROD = Australian and New Zealand Risk of Death. APACHE = Acute Physiology and Chronic Health Evaluation. ICU = intensive care unit. IQR = interquartile range. na = not applicable.
This table summarises data on 9870 ICU admissions for 8726 individual patients for the 2 years before and 2 years after relocation of the ICU. The data for the microbial subgroup represent the first observed ICU admission for each of the 1890 individuals who yielded at least one positive microbial isolate from any anatomical site. Up to 5% of data may be missing for some estimates.
Sex was not documented for one patient in the old ICU.
Admitting unit was not documented for one patient in the old ICU and three in the new ICU.
ICU severity of illness as measured by APACHE III scores and the ANZROD predictive tool.
Principal diagnosis for each ICU admission in accordance with the ANZCIS CORE Adult Patient Database data dictionary. Within the ANZICS CORE requirements, APACHE III diagnostic codes are allocated to each admission in the following mandatory hierarchy: (i) cardiac arrest, (ii) sepsis, (iii) trauma. If a cardiac arrest occurs before ICU admission, the diagnostic code of cardiac arrest must be selected. If the patient has sepsis and has not had a cardiac arrest and is not post-operative, a sepsis diagnostic code must be selected. If the patient has any element of trauma relating to admission, and they have not had a cardiac arrest and do not present as septic, then a trauma diagnostic code must be selected. This diagnosis hierarchy may obscure other reasons for patient admission.
Leukemia/lymphoma will appear under a haematology diagnostic code (unless cardiac arrest, sepsis or trauma is present on ICU admission). As there is no ANZICS CORE APACHE III oncological diagnostic category, this was generated for all patients with an APACHE subcode related to cancers or likely malignant neoplasms of the respiratory, gastrointestinal, neurological and renal systems. The codes used were: 1302.02, 1302.03, 1303.01, 1303.02, 1405.01, 1405.02, 1405.03, 1405.04, 1405.05, 1405.06, 1405.07, 1505.01, 1701.01, 1701.02, 1701.03, 1701.04, 1701.05, 312.01, 312.02, 312.03, 312.04, 312.05, 202.02, 202.03, 202.04, 202.05, 405.01. Excluded were two codes for likely benign neoplasms (1302.01, thoracotomy for benign tumour [eg, mediastinal chest wall mass or thymectomy]; 1505.02, transsphenoidal surgery). These oncological codes are also subordinate to the APACHE III diagnostic hierarchy of cardiac arrest, sepsis or trauma. Subgroup proportions were based on 1795 cases where an APACHE subgroup code was available in the first patient admission.
The median interval from hospital admission to positive isolate was 2.8 days before the relocation and 2.4 days after, and the corresponding lags for ICU admission were 1.7 and 1.4 days, respectively (Table 1). Per patient, the pooled 4-year median count (interquartile range) [range] of positive isolates was 1.0 (1.0–2.0) [1.0–16.0], corresponding to a mean (standard deviation) per patient of 2.05 (1.72). The median count (interquartile range) [range] of positive isolates per patient was 2.0 (1.0–3.0) [1.0–16.0] in the old ICU and 1.0 (1.0–2.0) [1.0–15.0] in the new ICU.
A total of 5862 positive isolates were identified across the 4-year study period. After applying the inclusion and exclusion criteria, 3871 unique isolates were included in the analysis, equivalent to about 121 per 1000 OBD.
Sample types
A total of 3871 samples were included in the analysis. In both the new and old ICUs, the most common sample types were sputum (1680, 43.4%), wound swabs (600, 15.5%), blood cultures (421, 10.9%), surveillance swabs (337, 8.7%) and urine (305, 7.9%). Of the 3871 total isolates, 709 (18.3%) were from samples taken from sterile sites, 3137 (81.0%) were from samples taken from non-sterile sites, and the remaining 25 (0.7%) had insufficient detail regarding sample type on the pathology request record.
New ICU versus old ICU
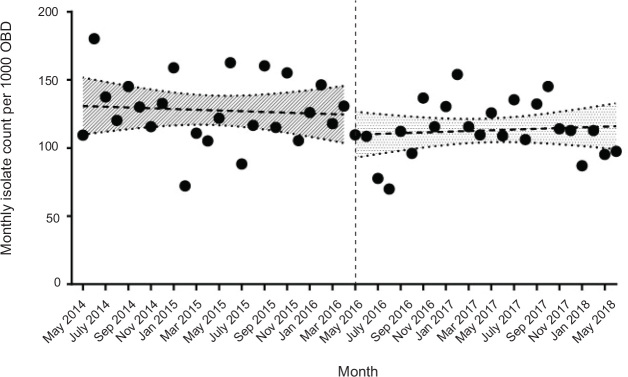
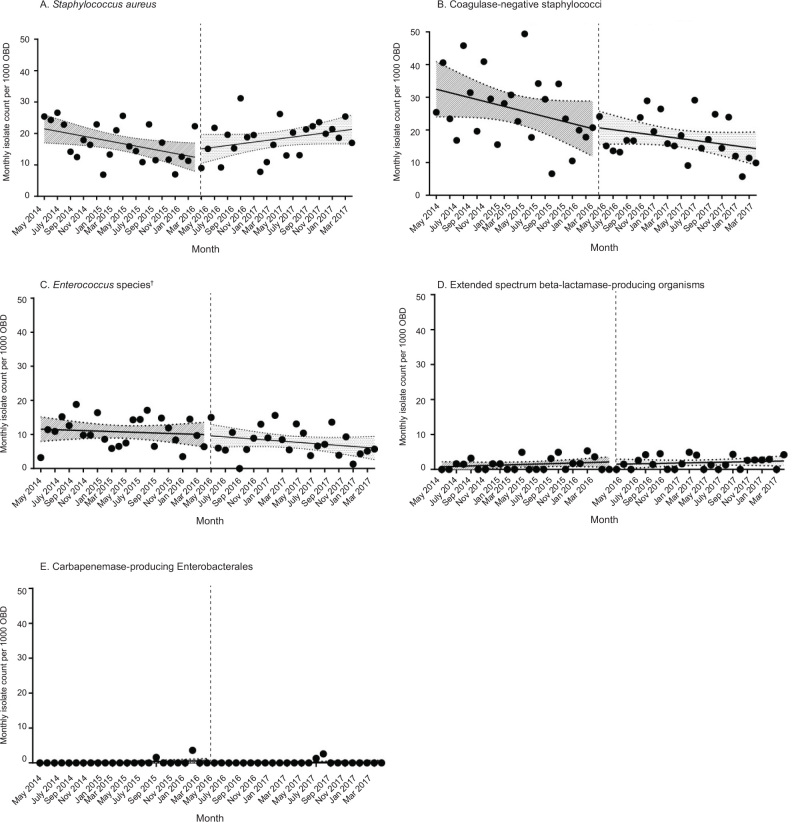
A total of 1907 positive isolates (129 per 1000 OBD) were obtained in the old ICU and 1964 (114 per 1000 OBD) were obtained in the new ICU. Overall, the incidence rate per 1000 OBD of positive isolates was estimated to be lower in the new ICU compared with the old ICU (IRR, 0.88; 95% CI, 0.82–0.93). Monthly incidence rates for the pooled data are shown in Figure 1, and monthly incidence rates for specific organisms are shown in Figure 2. In Table 2, a summary of the counts and IRRs for common isolates and selected drugresistant organisms is shown.
Figure 1.
Monthly counts of total isolates per 1000 OBD (monthly incidence rates) during the 4-year study period*
OBD = occupied bed-days. * The dashed vertical line denotes the move from the old ICU location to the newly constructed and expanded ICU on 5 May 2016. A simplifying assumption of independence of these time series data was used to derive the 95% confidence bands displayed for each linear regression in this graph.
Figure 2.
Monthly counts of isolates of several specific organisms per 1000 OBD (monthly incidence rates) during the 4-year study period*
OBD = occupied bed-days. * The dashed vertical line denotes the move from the old ICU location to the newly constructed expanded ICU on 5 May 2016. A simplifying assumption of independence of these time series data was used to derive the 95% confidence bands displayed for each linear regression in these graphs. † Excluding surveillance.
Table 2.
The ten most commonly isolated organisms, plus the uncommon drug-resistant ESBL-producing organisms and CPE, in descending order of isolate count from the old ICU
| Old ICU* |
New ICU† |
IRR (95% CI)¶ | p** | |||
|---|---|---|---|---|---|---|
| Isolate count | IR‡ | Isolate count | IR§ | |||
| Top ten organisms | ||||||
| 1. Coagulase-negative staphylococci | 399 | 27.1 | 300 | 17.3 | 0.64 (0.55–0.75) | < 0.01 |
| 2. Staphylococcus aureus | 254 | 17.2 | 320 | 18.5 | 1.07 (0.91–1.27) | 0.40 |
| Non-MRSA | 219 | 14.9 | 286 | 16.5 | 1.1 1 (0.93–1.33) | 0.24 |
| MRSA | 35 | 2.38 | 34 | 1.96 | 0.83 (0.50–1.37) | 0.43 |
| 3. Enterococcus species | 304 | 20.6 | 297 | 17.2 | 0.83 (0.71–0.98) | 0.02 |
| Non-VRE | 107 | 7.26 | 104 | 6.01 | 0.83 (0.63–1.09) | 0.10 |
| VRE (including surveillance) †† | 197 | 13.4 | 193 | 11.1 | 0.83 (0.68–1.02) | 0.07 |
| VRE (excluding surveillance) ‡‡ | 54 | 3.67 | 32 | 1.85 | 0.50 (0.32–0.80) | < 0.01 |
| 4. Escherichia coli | 155 | 10.5 | 204 | 11.8 | 1.12 (0.90–1.39) | 0.29 |
| 5. Klebsiella species | 135 | 9.16 | 117 | 6.76 | 0.74 (0.57–0.95) | 0.02 |
| 6. Enterobacter species | 104 | 7.06 | 119 | 6.88 | 0.97 (0.74–1.28) | 0.85 |
| 7. Haemophilus influenzae | 70 | 4.75 | 106 | 6.13 | 1.29 (0.94–1.77) | 0.10 |
| 8. Pseudomonas aeruginosa | 78 | 5.29 | 85 | 4.91 | 0.93 (0.67–1.28) | 0.63 |
| 9. Serratia marcescens | 39 | 2.65 | 42 | 2.43 | 0.79 (0.50–1.25) | 0.29 |
| 10. Streptococcus pneumoniae | 29 | 1.97 | 41 | 2.37 | 1.20 (0.73–2.00) | 0.45 |
| Other drug-resistant organisms | ||||||
| ESBL-producing organisms | 20 | 1.35 | 33 | 1.91 | 1.40 (0.78–2.58) | 0.23 |
| CPE | 3 | 0.20 | 3 | 0.17 | 0.85 (0.11–6.4) | 0.85 |
CPE = carbapenemase-producing Enterobacterales. ESBL = extended spectrum beta-lactamase. ICU = intensive care unit. IR = incidence rate. IRR = incidence rate ratio. MRSA = methicillin-resistant S. aureus. OBD = occupied bed days. VRE = vancomycin-resistant enterococci.
24-bed ICU used during the period 5 May 2014 to 4 May 2016.
32-bed ICU used during the period 5 May 2016 to 4 May 2018.
Incidence per 1000 OBD across 14734 OBD in the 2 years from 5 May 2014 to 4 May 2016.
Incidence per 1000 OBD across 17305 OBD in the 2 years from 5 May 2016 to 4 May 2018.
IRR of the new ICU compared with the old ICU.
Mid P values for tests of IR difference.
VRE isolates (including surveillance) in the old ICU were comprised of 35 (11.5%) VanA VRE, 142 (72.1%) VanB VRE, 13 (4.3%) VanA and VanB VRE and 7 (2.3%) unspecified VRE. VRE isolates (including surveillance) in the new ICU were comprised of 25 (13.0%) VanA VRE, 159 (81.3%) VanB VRE and 9 (4.7%) VanA and VanB VRE.
VRE isolates (excluding surveillance) in the old ICU were comprised of 11 (20.4%) VanA VRE, 41 (75.9%) VanB VRE, 1 (1.9%) VanA and VanB VRE and 1 (1.9%) unspecified VRE. VRE isolates (excluding surveillance) in the new ICU were comprised of 4 (12.5%) VanA VRE, 27 (84.4%) VanB VRE and 1 (3.1%) VanA and VanB VRE.
Blood cultures
A total of 421 unique positive blood cultures (13.1 per 1000 OBD) were obtained over the 4-year study period. There was no apparent difference between counts of positive blood cultures for the old and new ICUs: 200 (13.6 per 1000 OBD) and 221 (12.8 per 1000 OBD), respectively (IRR, 0.94; 95% CI, 0.77–1.14). The blood culture sampling rate in the new ICU (254 per 1000 OBD) appeared higher than that for the old ICU (97.3 per 1000 OBD) and the proportion of cultures drawn in ICU that became positive was substantially lower in the new ICU (221/4293 [5.1%] versus 200/1381 [14.4%]; risk ratio, 0.36; 95% CI, 0.30–0.43).
The most common blood culture isolate over the study period was coagulase-negative staphylococci (CoNS) (184/421 [43.7%]; 5.74 per 1000 OBD), followed by S. aureus (62/421 [14.7%]; 1.94 per 1000 OBD) and Enterococcus species (34/421 [8.1%]; 1.06 per 1000 OBD). Five blood cultures were positive for MRSA (1.2%; 0.16 per 1000 OBD), of which three were linked to the old ICU and two to the new ICU.
Community-acquired versus hospital-acquired organisms
Of the 3871 isolates included in this study, 1544 (39.9%) were classified as community-acquired and 2327 (60.1%) as hospital-acquired. New ICU versus old ICU incidence rates for common organisms classified as such are summarised in Table 3 and Table 4, respectively.
Table 3.
Organisms defined as community-acquired, comparing old and new ICUs, in descending order of isolate count from the old ICU
| Organism | Old ICU* |
New ICU† |
IRR (95% CI) ¶ | P** | ||
|---|---|---|---|---|---|---|
| Isolate count | IR‡ | Isolate count | IR§ | |||
| Staphylococcus aureus | 155 | 10.5 | 196 | 11.3 | 1.08 (0.87–1.34) | 0.49 |
| Non-MRSA | 133 | 9.03 | 178 | 10.3 | 1.07 (0.96–1.19) | 0.25 |
| MRSA | 22 | 1.49 | 18 | 1.04 | 0.70 (0.35–1.40) | 0.26 |
| Coagulase-negative staphylococci | 101 | 6.85 | 81 | 4.68 | 0.68 (0.50–0.92) | 0.01 |
| Enterococcus species | 74 | 5.02 | 119 | 6.88 | 1.37 (1.01–1.85) | 0.03 |
| Non-VRE | 35 | 2.38 | 28 | 1.62 | 0.68 (0.40–1.15) | 0.13 |
| VRE (including surveillance) | 39 | 2.65 | 91 | 5.26 | 1.99 (1.35–2.97) | < 0.01 |
| VRE (excluding surveillance) | 3 | 0.20 | 7 | 0.40 | 1.99 (0.45–11.9) | 0.33 |
| Pseudomonas aeruginosa | 21 | 1.43 | 20 | 1.16 | 0.81 (0.42–1.57) | 0.51 |
| Streptococcus pneumoniae | 19 | 1.29 | 35 | 2.02 | 1.57 (0.87–2.90) | 0.11 |
| Other drug-resistant organisms | ||||||
| ESBL-producing organisms | 6 | 0.41 | 12 | 0.69 | 1.70 (0.59–5.53) | 0.29 |
| CPE | 1 | 0.07 | 3 | 0.17 | 0.85 (0.11–6.36) | 0.85 |
CPE = carbapenemase-producing Enterobacterales. ESBL = extended spectrum beta-lactamase. ICU = intensive care unit. IR = incidence rate. IRR = incidence rate ratio. MRSA = methicillin-resistant S. aureus. OBD = occupied bed-days. VRE = vancomycin-resistant enterococci.
24-bed ICU used during the period 5 May 2014 to 4 May 2016.
32-bed ICU used during the period 5 May 2016 to 4 May 2018.
Incidence per 1000 OBD across 14734 OBD in the 2 years from 5 May 2014 to 4 May 2016.
Incidence per 1000 OBD across 17305 OBD in the 2 years from 5 May 2016 to 4 May 2018.
IRR of the new ICU compared with the old ICU.
Mid P values for tests of IR difference.
Table 4.
Organisms defined as hospital-acquired, comparing old and new ICUs, in descending order of isolate count from the old ICU
| Organism | Old ICU* |
New ICU† |
IRR (95% CI)¶ | P** | ||
|---|---|---|---|---|---|---|
| Isolate count | IR‡ | Isolate count | IR§ | |||
| Coagulase-negative staphylococci | 297 | 20.2 | 220 | 12.7 | 0.63 (0.53–0.75) | < 0.01 |
| Enterococcus species | 230 | 15.6 | 178 | 10.3 | 0.66 (0.54–0.80) | < 0.01 |
| Non-VRE | 72 | 4.89 | 76 | 4.39 | 0.90 (0.64–1.26) | 0.52 |
| VRE (including surveillance) | 158 | 10.7 | 102 | 5.89 | 0.34 (0.26–0.42) | < 0.01 |
| VRE (excluding surveillance) | 51 | 3.46 | 30 | 1.73 | 0.50 (0.31–0.80) | < 0.01 |
| Staphylococcus aureus | 100 | 6.79 | 124 | 7.17 | 1.06 (0.80–1.39) | 0.69 |
| Non-MRSA | 87 | 5.90 | 108 | 6.24 | 1.06 (0.79–1.41) | 0.70 |
| MRSA | 13 | 0.88 | 16 | 0.92 | 1.05 (0.47–2.37) | 0.91 |
| Pseudomonas aeruginosa | 57 | 3.87 | 65 | 3.76 | 0.97 (0.67–1.41) | 0.87 |
|
Streptococcus pneumoniae Other drug-resistant organisms |
10 | 0.68 | 7 | 0.40 | 0.60 (0.19–1.73) | 0.30 |
| ESBL-producing organisms | 14 | 0.95 | 24 | 1.39 | 1.46 (0.73–3.05) | 0.26 |
| CPE | 2 | 0.14 | 0 | 0.00 | 0.00 (0.00–4.53) | 0.21 |
CPE = carbapenemase-producing Enterobacterales. ESBL = extended spectrum beta-lactamase. ICU = intensive care unit. IR = incidence rate. IRR = incidence rate ratio. MRSA = methicillin-resistant S. aureus. OBD = occupied bed-days. VRE = vancomycin-resistant enterococci.
24-bed ICU used during the period 5 May 2014 to 4 May 2016.
32-bed ICU used during the period 5 May 2016 to 4 May 2018.
Incidence per 1000 OBD across 14734 OBD in the 2 years from 5 May 2014 to 4 May 2016.
Incidence per 1000 OBD across 17305 OBD in the 2 years from 5 May 2016 to 4 May 2018.
IRR of the new ICU compared with the old ICU.
Mid P values for tests of IR difference.
Discussion
ICU designs with single rooms have been associated with improved patient outcomes in multiple domains, including reduced rates of infections.18, 19, 20, 21 In our retrospective, descriptive, cohort study, there was no observed increase in the overall incidence rate of bacterial isolates accompanying a relocation from an older, predominantly open plan, adult ICU of 24 beds to a newly constructed ICU with single rooms for all 32 patients. Also, incidence rate estimates for MDR bacterial isolates seemed stable or decreased through this relocation and expansion process. This mirrors previous findings, which have shown decreased prevalence of MDR bacteria with the introduction of a single room policy in an ICU setting,20 despite the new ICU having a greater proportion of oncology patients (with haematological and solid tumours) and an overall increase in OBD. However, in our study, the median time from ICU admission to positive culture was short (less than 2 days), suggesting that some isolates may have been acquired before ICU admission —from another ward or the community. As well as any effect of the ICU infrastructure upgrade, changes in infection control practices in hospital settings outside of the ICU may have contributed to the observed modest reduction in overall isolate counts and static MDR isolate counts. While data on ward infection control practices outside the ICU were not collected, the very short median interval from hospital admission to ICU, not unexpected for patients requiring intensive care, suggested only a limited opportunity for ward-specific practices to affect the rate of positive isolates identified. The accompanying introduction of ICU-specific cleaning staff in the new ICU and the continuation of a well established antimicrobial stewardship program throughout the hospital campus, including in the ICU, may have also contributed, but these possibilities could not be specifically examined in our study.
The clinical implications of CoNS isolates may be challenging to interpret, however a possible reduction in the incidence rate of CoNS after the move to the new ICU was observed. While historically considered less pathogenic than other staphylococci species,22 the role of CoNS in hospital-associated infection is increasingly being recognised.23
The observation of fewer CoNS isolates over time is encouraging, particularly in the setting of an expanded haematology and oncology patient population, many of whom have long term vascular catheters that increase the risk of CoNS contamination or infection.9
The apparent incidence rate of MRSA was reassuringly low in the ICU during our 4-year study period and did not appear to change over time despite increasing community prevalence.24 Our data demonstrate no major difference between the prevalence of community- and hospital-acquired MRSA, suggesting that MRSA burden was uncommon in our ICU, which may help inform the choice of empiric antibiotic cover as part of sepsis management on campus.
Most VRE isolates counted in this study were VanB VRE, which the RMH stopped isolation precautions for in July 2016. Despite this change in protocol, the total number of VRE isolates did not appear to differ between the new ICU and old ICU, and there were fewer non-surveillance VRE isolates in the new ICU compared with the old ICU. This suggests that newer infrastructure (potentially enabling better cleaning and reduced environmental burden) and single rooms (potentially minimising patient-to-patient transmission) may not strongly influence VRE incidence rates. There is some evidence that VanB VRE may be endogenous and therefore newer facilities may not reduce its prevalence.25
Despite rising community prevalence of ESBL-producing Enterobacterales in Australia,26, 27 no apparent changes in incidence rates for these organisms were observed in our ICU during the study period. Statewide CPE surveillance guidelines were implemented in 2017 and stable CPE prevalence has been demonstrated in the community.28, 29 These organisms were uncommonly identified in samples from patients in our ICU, and found only in surveillance and non-sterile site samples. This provides reassurance that current infection prevention and screening strategies have been adequate in the face of a somewhat expanded ICU population of patients with risk factors such as immunocompromised states and prolonged hospitalisation.30
The apparent increase in blood culture sampling rate with a lower rate of positive isolates between the new and old ICUs could be explained by several factors. In November 2016, RMH introduced a hospital-wide sepsis pathway to facilitate early recognition and management, which may have encouraged a greater rate of blood culture collection. The expanded immunocompromised patient population of haematology and oncology patients with neutropenia and fever in the new ICU in the second 2 years of the study may have also contributed to this observation.
This retrospective observational study has several limitations. First, the collection of patient samples for microbial testing was not specifically protocolised or otherwise controlled. It was always based on clinical indication and done at the discretion of the treating team. Isolation of a potential pathogen does not necessarily infer infection; data from sterile site samples may give a better indication of the significance of an isolate, as would the inclusion of additional relevant clinical information. Second, surveillance programs for VRE and ESBL-producing Enterobacterales were implemented part-way through the study period in the setting of local outbreaks, so these may have influenced the numbers of these isolates recorded on the ICNET database. Third, total antimicrobial usage, which may exert selection pressure on certain bacterial pathogens in the ICU, was not quantified. Fourth, the study did not assess other factors such as hand hygiene compliance, cleaning compliance and other ward-specific cleaning protocols that may have affected the results.
Conclusion
This retrospective observational study of microbial isolates from ICU patient samples over a period of 4 years suggested stable incidence rates of several clinically important organisms without an increase in the overall incidence rate of microbial isolates. These observations coincided with the relocation of the ICU from an open plan configuration to a new, purpose-built expanded facility with individual patient rooms, facilitating an increase in ICU OBD with a modest increase in oncology patient casemix. Additional factors such as changes in hospital- and community-based infection prevention measures, surveillance protocols and overall antimicrobial use may have also contributed to the observed isolate rates. Nevertheless, as the coronavirus disease 2019 (COVID 19) pandemic increases focus on the built design of ICUs in delivering safe patient care, our study highlights the opportunity to examine how ICU infrastructure upgrades (including single rooms for patients) and dedicated ICU cleaning staff can help control the microbiological risks that accompany critical illness.
Competing interests
All authors declare that they do not have any potential conflict of interest in relation to this manuscript.
References
- 1.Doyle J.S., Buising K.L., Thursky K.A., et al. Epidemiology of infections acquired in intensive care units. Semin Respir Crit Care Med. 2011;32:115–138. doi: 10.1055/s-0031-1275525. [DOI] [PubMed] [Google Scholar]
- 2.Richards M., Thursky K., Buising K. Epidemiology, prevalence, and sites of infections in intensive care units. Semin Respir Crit Care Med. 2003;24:3–22. doi: 10.1055/s-2003-37913. [DOI] [PubMed] [Google Scholar]
- 3.Strich J.R., Palmore T.N. Preventing transmission of multidrugresistant pathogens in the intensive care unit. Infect Dis Clin North Am. 2017;31:535–550. doi: 10.1016/j.idc.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent J.L., Rello J., Marshall J., et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 5.Vincent J.L., Sakr Y., Singer M., et al. Prevalence and outcomes of infection among patients in intensive care units in 2017. JAMA. 2020;323:1478–1487. doi: 10.1001/jama.2020.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glynn C.M., Azadian B. Empiric antimicrobial therapy for severe sepsis in the intensive care unit: in early, hit hard, out early. Curr Anaesth Crit Care. 2005;16:221–230. [Google Scholar]
- 7.Rello J., Kalwaje Eshwara V., Lagunes L., et al. A global priority list of the TOp TEn resistant Microorganisms (TOTEM) study at intensive care: a prioritization exercise based on multi-criteria decision analysis. Eur J Clin Microbiol Infect Dis. 2019;38:319–323. doi: 10.1007/s10096-018-3428-y. [DOI] [PubMed] [Google Scholar]
- 8.Buising K.L., Thursky K.A., Bak N., et al. Antibiotic prescribing in response to bacterial isolates in the intensive care unit. Anaesth Intensive Care. 2005;33:571–577. doi: 10.1177/0310057X0503300504. [DOI] [PubMed] [Google Scholar]
- 9.Australian Commission on Safety and Quality in Health Care . ACSQHC; Sydney: 2019. Implementation guide: surveillance of central line-associated bloodstream infection. [Google Scholar]
- 10.Russo P.L., Cheng A.C., Richards M., et al. Healthcare-associated infections in Australia: time for national surveillance. Aust Health Rev. 2015;39:37–43. doi: 10.1071/AH14037. [DOI] [PubMed] [Google Scholar]
- 11.Velasquez Reyes D.C., Bloomer M., Morphet J. Prevention of central venous line associated bloodstream infections in adult intensive care units: a systematic review. Intensive Crit Care Nurs. 2017;43:12–22. doi: 10.1016/j.iccn.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Australian Commission on Safety and Quality in Health Care. Antimicrobial Use and Resistance in Australia Surveillance System. https://www.safetyandquality.gov.au/our-work/antimicrobial-resistance/antimicrobial-use-and-resistance-australia-surveillance-system (viewed Feb 2021).
- 13.Australian Commission on Safety and Quality in Health Care. National Alert System for Critical Antimicrobial Resistances (CARAlert). https://www.safetyandquality.gov.au/our-work/antimicrobial-resistance/antimicrobial-use-and-resistance-australia-surveillance-system/national-alert-system-critical-antimicrobial-resistances-caralert (viewed Feb 2021).
- 14.College of Intensive Care Medicine of Australia and New Zealand . CICM; Melbourne: 2016. Minimum standards for intensive care units (Cat. No. IC-1) [Google Scholar]
- 15.Knaus W.A., Wagner D.P., Draper E.A., et al. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991;100:1619–1636. doi: 10.1378/chest.100.6.1619. [DOI] [PubMed] [Google Scholar]
- 16.Pilcher D., Paul E., Bailey M., Huckson S. The Australian and New Zealand Risk of Death (ANZROD) model: getting mortality prediction right for intensive care units. Crit Care Resusc. 2014;16:3–4. [PubMed] [Google Scholar]
- 17.National Healthcare Safety Network . Centers for Disease Control and Prevention; Atlanta: 2021. Identifying healthcare-associated infections (HAI) for NHSN surveillance. [Google Scholar]
- 18.Kesecioglu J., Schneider M.M., van der Kooi A.W., Bion J. Structure and function: planning a new ICU to optimize patient care. Curr Opin Crit Care. 2012;18:688–692. doi: 10.1097/MCC.0b013e328358d4bd. [DOI] [PubMed] [Google Scholar]
- 19.Thompson D.R., Hamilton D.K., Cadenhead C.D., et al. Guidelines for intensive care unit design. Crit Care Med. 2012;40:1586–1600. doi: 10.1097/CCM.0b013e3182413bb2. [DOI] [PubMed] [Google Scholar]
- 20.Halaby T., Al Naiemi N., Beishuizen B., et al. Impact of single room design on the spread of multi-drug resistant bacteria in an intensive care unit. Antimicrob Resist Infect Control. 2017;6:117. doi: 10.1186/s13756-017-0275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olmsted R.N. Reimagining construction and renovation of health care facilities during emergence from a pandemic. Infect Dis Clin North Am. 2021;35:697–716. doi: 10.1016/j.idc.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwab F., Geffers C., Behnke M., Gastmeier P. ICU mortality following ICU-acquired primary bloodstream infections according to the type of pathogen: a prospective cohort study in 937 Germany ICUs (2O06–2O15) PLoS One. 2018;13 doi: 10.1371/journal.pone.0194210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker K., Heilmann C., Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron J.K., Hall L., Tong S.Y.C., et al. Incidence of community onset MRSA in Australia: least reported where it is most prevalent. Antimicrob Resist Infect Control. 2019;8:33. doi: 10.1186/s13756-019-0485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermott H., Skally M., O’Rourke J., et al. Vancomycin-resistant enterococci (VRE) in the intensive care unit in a nonoutbreak setting: identification of potential reservoirs and epidemiological associations between patient and environmental VRE. Infect Control Hosp Epidemiol. 2018;39:40–45. doi: 10.1017/ice.2017.248. [DOI] [PubMed] [Google Scholar]
- 26.Australian Commission on Safety and Quality in Health Care. AURA 2016 . ACSQHC; Sydney: 2016. first Australian report on antimicrobial use and resistance in human health. [Google Scholar]
- 27.Coombs G., Bell J.M., Daley D., et al. Australian Commission on Safety and Quality in Health Care; Sydney: 2018. Australian Group on Antimicrobial Resistance. Sepsis Outcome Programs 2016 report. [Google Scholar]
- 28.Australian Commission on Safety and Quality in Health Care . ACSQHC; Sydney: 2018. CARAlert summary report 1 October 2017 – 31 March 2018. [Google Scholar]
- 29.Department of Health and Human Services, Victoria . Victorian Government; Melbourne: 2017. Victorian guideline on carbapenemase-producing Enterobacteriaceae for health services, version 2. [Google Scholar]
- 30.Australian Commission on Safety and Quality in Health Care . ACSQHC; Sydney: 2017. Recommendations for the control of carbapenemase-producing Enterobacteriaceae (CPE). A guide for acute care health facilities. [Google Scholar]