Abstract
Bromodomain and extraterminal domain protein inhibitors have shown therapeutic promise in hepatocellular carcinoma. However, resistance to bromodomain and extraterminal domain protein inhibitors has emerged in preclinical trials, presenting an immense clinical challenge, and the mechanisms are unclear. In this study, we found that overexpression of SIRT1 induced by JQ-1, a bromodomain and extraterminal domain protein inhibitor, may confer resistance to JQ-1 in hepatocellular carcinoma. SIRT1 protein expression was higher in hepatocellular carcinoma tissues than in normal tissues, and this phenotype was correlated with a poor prognosis. Cotreatment with JQ-1 and the SIRT1 inhibitor EX527 synergistically suppressed proliferation and blocked cell cycle progression in hepatocellular carcinoma cells. Combined administration of JQ-1 and EX527 successfully reduced the tumor burden in vivo. In addition, JQ-1 mediated AMPK/p-AMPK axis activation to upregulate SIRT1 protein expression and enhanced autophagy to inhibit cell apoptosis. Activation of AMPK could alleviate the antitumor effect of the combination of JQ-1 and EX527 on hepatocellular carcinoma cells. Furthermore, inhibition of SIRT1 further enhanced the antitumor effect of JQ-1 by blocking protective autophagy in hepatocellular carcinoma. Our study proposes a novel and efficacious therapeutic strategy of a BET inhibitor combined with a SIRT1 inhibitor for hepatocellular carcinoma.
Keywords: Hepatocellular carcinoma, JQ-1, SIRT1, AMPK, Autophagy
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most aggressive solid malignancies, with a rapidly increasing incidence worldwide [1]. Despite tremendous progress in surgical treatments, liver transplantation and other interventions, the prognosis of HCC patients is still unsatisfactory, and the 5-year overall survival rate is less than 20 % [2]. Members of the bromodomain and extracellular domain (BET) family, which contains 4 proteins (BRD2, BRD3, BRD4, and BRDT), act as epigenetic readers to recognize acetylated lysine residues on histone tails and other nucleoproteins [3]. Once BRD4 is activated, positive transcription elongation factor b is recruited, and RNA polymerase II is phosphorylated to promote transcriptional elongation and the expression of associated oncogenes, such as c-Myc, BCL2, and MYCN, leading to abnormal cell proliferation, tumor progression or metastasis [4]. To date, several small-molecule BET inhibitors, such as CPI-203 and JQ-1, have been designed to inhibit tumor growth and angiogenesis. Promising therapeutic results were observed in hematological tumors treated with BET inhibitors, but the efficacy of BET inhibitors in solid tumors remains unsatisfactory [5,6]. According to our previous report, the effect of a single administration of JQ-1 was limited to suppressing HCC progression in a c-Myc/N-Ras plasmid-induced mouse liver cancer model [7]. Disappointingly, several preclinical trials on JQ-1 monotherapy have also exhibited unsatisfactory outcomes [8]. Because of the more complex immune microenvironment in solid tumors, inherent and acquired resistance inevitably occurs during treatment with JQ-1, possibly explaining their poor therapeutic effectiveness, and further exploration of the underlying mechanism is urgently needed to overcome resistance [6,9].
Currently, accumulating evidence suggests that combination therapy may offer several advantages to delay the emergence of drug resistance, reduce selected adverse toxic effects and be more effective than single-agent treatments [10,11]. Our previous research showed that JQ-1 decreased the expression levels of YAP/TAZ and that the combination of JQ-1 and tadalafil more effectively reduced the HCC tumor burden [7]. JQ-1 was also proposed to have a great ability to enhance sensitivity to BEZ235, a dual PI3K/mTOR inhibitor, in intrahepatic cholangiocarcinoma [12]. There are also some reports about combination treatment with JQ-1 and histone deacetylase (HDAC) inhibitors, anti-PD-1 antibodies or PARP inhibitors, which exhibited synergistic effects in multiple cancer models and showed anticancer effects superior to those of BET inhibitor monotherapy [[13], [14], [15], [16]]. Considering these encouraging results, we continued our research on JQ-1 treatment and explored more novel combination strategies for treating HCC.
Sirtuin-1 (SIRT1), a class III HDAC, is an enzyme that deacetylates histones and other intracellular substrates. Unlike the classical HDAC family members, SIRT1 does not rely on the activating effect of Zn2+ but needs NAD + as a cofactor to exert essential biological functions in cancer, inflammation, aging, and autoimmune diseases [17,18]. Contradictory opinions exist about the role of SIRT1 in tumorigenesis. Generally, SIRT1 is recognized as a tumor suppressor gene in oral squamous cell carcinoma and gastric cancer but is considered an oncogene in prostate, colon, and pancreatic cancer [[19], [20], [21]]. Moreover, high expression of SIRT1 was widely observed in HCC tissues from patients with poor prognosis, indicating the critical role of SIRT1 in hepatocarcinogenesis [22]. Surprisingly, we found that JQ-1 treatment enhanced the expression of SIRT1, suggesting that SIRT1 may confer resistance to JQ-1 in HCC. In this study, we further investigated the efficacy and mechanism of combination treatment with JQ-1 and the SIRT1 inhibitor EX527 and proposed a novel therapeutic strategy for HCC.
2. Materials and methods
2.1. Cells and reagents
The human HCC cell lines Hep3B, HCCLM3, and Huh-7 and the mouse HCC cell line Hep 1–6 were obtained from the Stem Cell Bank of the Chinese Academy of Sciences. The drugs used in our research were obtained from Selleck Chemicals (Houston, USA): JQ-1 (S7110), EX 527 (S1541), AICAR (S1802), Compound C (S7840) and 3-MA (S2767). The working concentrations of JQ-1, EX527, AICAR, Compound C and 3-MA in the in vitro assays were 2 μM, 5 μM, 2 mM, 2 μM and 5 mM, respectively.
3. Animals
Male wild-type C57BL/6 (6cc8 weeks) mice and male nude mice (6 weeks) were purchased from Shanghai Slack Laboratory Animal Co., Ltd. (Shanghai, China). JQ-1 (50 mg/kg/day, intraperitoneally (i.p.)) and EX527 (30 mg/kg/day, subcutaneously (s.c.)) were administered to the mice. All animal studies were approved by the Institutional Animal Care and Use Committee of the Second Affiliated Hospital, School of Medicine, Zhejiang University.
3.1. Murine tumor models
The transgenic HCC mouse model established by transfection with the c-Myc/N-Ras/SB plasmid was described previously [8]. The orthotopic liver cancer model was established by orthotopic injection of mouse Hep 1–6 cells (1 × 105) into the livers of mice. The subcutaneous xenograft tumor models were established by subcutaneously transplanting lentivirally transduced Hep3B (5 × 106) or HCCLM3 (5 × 106) cells.
3.2. Cell proliferation, EdU incorporation assay and TUNEL assay
The cell proliferation and EdU incorporation assays and TUNEL assays were conducted using the Cell Counting Kit-8 (CCK-8) (CK04, Dojindo, Japan), BeyoClick™ EdU Cell Proliferation Kit (C0071S, Beyotime, China) and TUNEL Apoptosis Assay Kit (C1090, Beyotime, China) according to the instructions. HCC cells were treated with drugs for 24 h. A microplate reader (ELX-808, BioTek, USA) was used to measure the absorbance at 450 nm, and a fluorescence microscope (Leica DMi8, Leica, Germany) was applied to visualize and image the cells.
3.3. Cell cycle and apoptosis analysis
HCC cells seeded in 6-well plates were treated with drugs for 24 h. The supernatant medium containing suspended cells and adherent cells was collected. After washing, the cells were incubated with reagents from a Cell Cycle and Apoptosis Analysis Kit (C1052, Beyotime, China). Samples were analyzed by flow cytometry (Cytoflex LX, Beckman, USA).
3.3.1. Colony formation, cell migration and cell invasion assays
For the colony formation assay, HCC cells were plated in a 6-well plate at 2 × 103 cells/well and incubated for 14 days. Different treatments were added when the cells became adherent. Then, the HCC cells were fixed and stained with crystal violet (C0121, Beyotime, China) for imaging. For the cell migration and invasion assays, HCC cells were treated for 24 h and 48 h, respectively. Next, cell migration and invasion assays were performed using conventional techniques.
3.4. Western blot (WB) analysis
Protein was extracted from samples with RIPA buffer (R0010, Solarbio, China) supplemented with a protease inhibitor (P0100, Solarbio, China) and phosphatase inhibitors (P1082, Beyotime, China). All procedures were performed according to a standard protocol. Primary antibodies specific for the following proteins were used: SIRT1 (#9475, Cell Signaling Technology, USA), AMPK (#5831, Cell Signaling Technology, USA), phospho-AMPK (#2535, Cell Signaling Technology, USA), cleaved caspase-3 (#9661, Cell Signaling Technology, USA), ATG5 (#12994, Cell Signaling Technology, USA), Beclin-1 (#3495, Cell Signaling Technology, USA), LC3A/B (#12741, Cell Signaling Technology, USA), p-ULK1 (#14202, Cell Signaling Technology, USA), and β-actin (#4970, Cell Signaling Technology, USA).
3.5. Lentiviral transduction
The lentivirus-Flag-SIRT1-expression vector and lentivirus-sh-SIRT1-expression vector were synthesized by ViGene BioSciences (China). The pLent-EF1a–FH–CMV-GFP-P2A-Puro lentiviral vector and pLent-U6-shRNA-CMV-GFP-P2A-Puro lentiviral vector were used as controls. The lentivirus titer was higher than 1 × 108 TU/mL, and transduction was performed according to the manufacturer's protocol.
3.6. Hematoxylin and eosin (HE) staining, immunohistochemical (IHC) analysis and immunofluorescence analysis
For IHC analysis, the samples were incubated first with an anti-Ki67 antibody (ab15580, Abcam, USA) overnight at 4 °C and then with a secondary antibody at 37 °C for 1 h. Then, the sections were stained with hematoxylin. For immunofluorescence analysis, anti-SIRT1 and anti-LC3A/B antibodies were used. Then, the slides were examined by a fluorescence microscope (Leica DMi8, Wetzlar, Germany).
3.7. Quantitative real-time polymerase chain reaction assay
Quantitative real-time polymerase chain reaction (PCR) was conducted according to the manufacturers' instructions. The mRNA expression level of SIRT1 was normalized to that of β-actin. The primers used for PCR were as follows: SIRT1: forward 5′-TGCCGGAAACAATACCTCCA-3′, reverse 5′-AGACACCCCAGCTCCAGTTA-3'; actin: forward 5′-CATGTACGTTGCTATCCAGGC-3′, reverse 5′-CTCCTTAATGTCACGCACGAT-3'.
3.8. RNA-sequencing
Hep3B cells were harvested for total RNA extraction after treatment with 2 μM JQ-1 for 24 h. Vehicle (DMSO) was added to the control group as a negative control. The raw sequencing data were processed on the Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
3.9. Bioinformatic analysis
The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) database, and Human Protein Atlas (https://www.proteinatlas.org/) were used to validate the results of our experiments. GEPIA (http://gepia.cancer-pku.cn/) was applied to analyze the TCGA and CPTAC sample data.
3.10. Statistical analysis
All quantitative data are expressed as the means ± SDs. Unpaired, two-tailed Student's t-test was applied to evaluate the statistical significance between groups. A p value of less than 0.05 was considered to indicate statistical significance (*), a p value of less than 0.01 also indicated statistical significance (**), and a p value of less than 0.001 indicated high statistical significance (***).
4. Results
4.1. JQ-1 suppressed HCC cell proliferation in vitro but performed poorly against liver tumors in a transgenic HCC mouse model
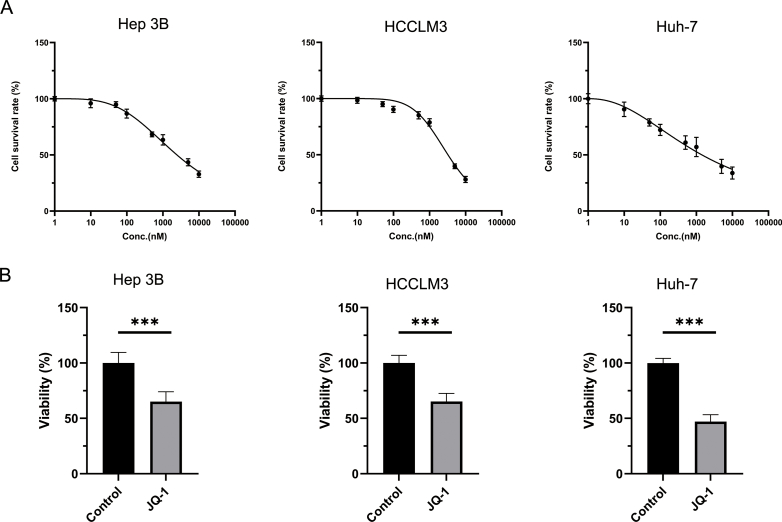
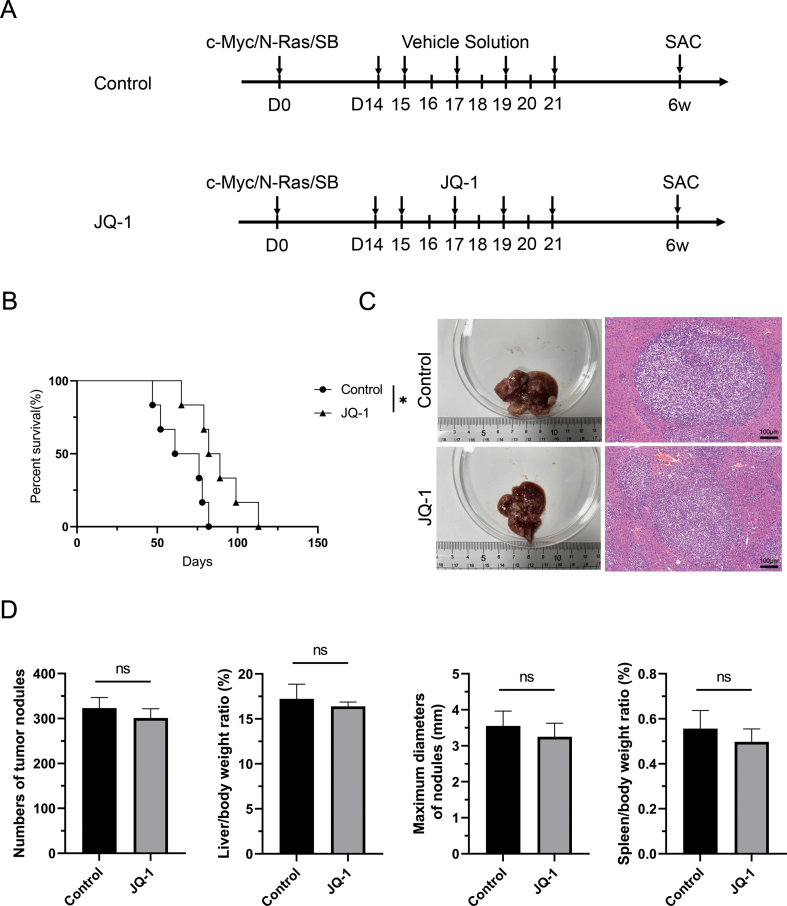
To explore the effect of JQ-1 on the progression of HCC, we performed CCK-8 and EdU incorporation assays to assess Hep3B, HCCLM3 and Huh-7 cell proliferation. First, the IC50 of JQ-1 in these three cell lines was determined independently. The IC50 values of JQ-1 were 2.449 μM, 3.235 μM and 1.422 μM in Hep3B, HCCLM3 and Huh-7 cells, respectively (Fig. S1A). Thus, the appropriate concentration of 2 μM in vitro was utilized for subsequent experiments. As shown in fig. S1B and fig. 1A, JQ-1 significantly inhibited HCC cell proliferation, accompanied by lower cell viability and a lower percentage of EdU-positive cells. In the cell migration and invasion assays, JQ-1 also exerted a similar inhibitory effect on invasion and metastasis abilities compared to those in the control group (Fig. 1B). The concentration of JQ-1 affected cell survival, which may have interfered with the accuracy of the cell migration and cell invasion assays. We repeated the CCK-8 assay. As shown in Figs. S2A and 50 nM was selected since this dose had a small impact on the cell survival rate. Similar to previous results, JQ-1 also showed a significant effect on the ability of HCC cells to migrate and invade (Fig. S2B). However, the therapeutic efficacy of JQ-1 in the transgenic HCC mouse model seemed poor. The transgenic HCC mouse model was established by hydrodynamic transfection to mimic diffuse hepatocellular carcinoma in humans. Diffuse hepatocellular carcinoma is more malignant, more invasive and has a worse prognosis than other types of liver cancer [23]. Unfortunately, JQ-1 significantly increased the survival time (Fig. S3B), with no significant effect on H&E staining and gross morphological indicators, such as the number of tumor nodules, the liver/body weight (LW/BW) ratio, the maximum diameters of nodules and the spleen/body weight (SW/BW) ratio (Figs. S3C and D). Therefore, JQ-1 exhibited robust antiproliferative and antimigratory abilities in vitro, but unsatisfactory effects on HCC progression were observed in mice, indicating that more drug resistance mechanisms need to be investigated.
Fig. 1.
JQ-1 significantly reduced HCC cell viability in vitro but performed poorly against liver tumors in the mouse HCC model. (A) EdU incorporation assay of Hep3B, HCCLM3, and Huh-7 cell lines treated with JQ-1 (2 μM) or vector (DMSO) control for 24 h. Scale bar, 100 μm. (B) The transwell migration assay showed significant inhibition of the invasion and metastasis abilities after treatment with JQ-1 (2 μM) for 24 h in these three HCC cell lines compared to the control group. Scale bar, 100 μm.
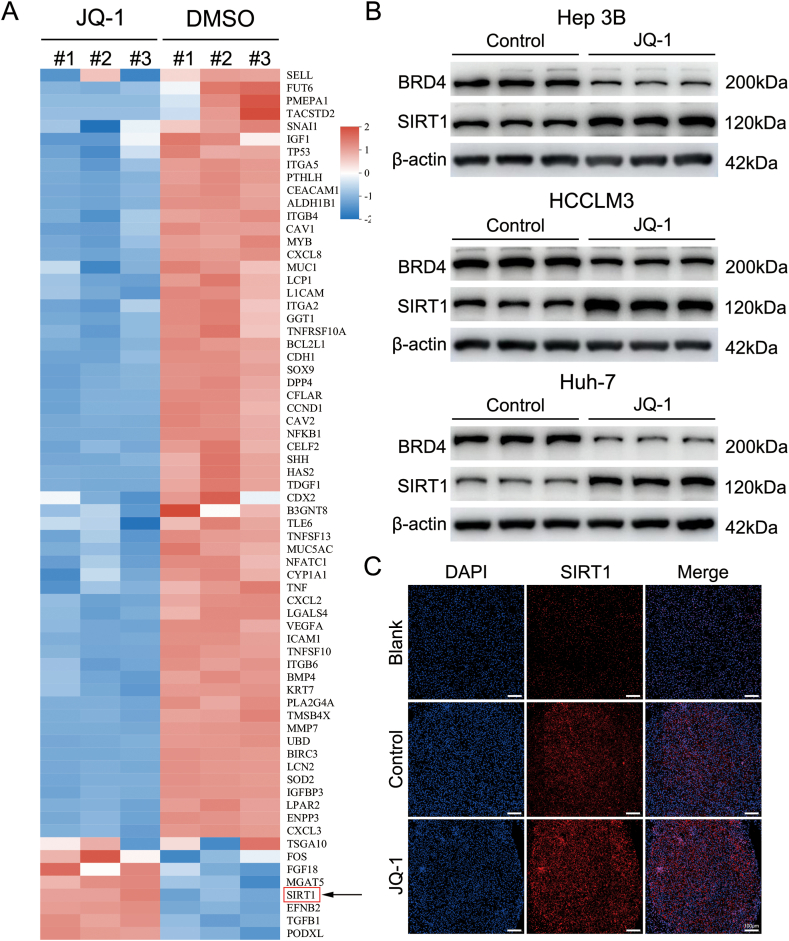
4.2. JQ-1 increases SIRT1 expression in vitro and in vivo
Many patients who are administered BET inhibitors experience drug resistance or decreased sensitivity, as noted in previous clinical reports [6,7]. To further explore the potential mechanisms of JQ-1, RNA sequencing was applied using Hep3B cells. After disease ontology (DO) enrichment analysis, the key genes in liver cancer were analyzed. As indicated in the heatmap, many oncogenes were downregulated, while the SIRT1 genes were aberrantly upregulated (Fig. 2A). WB analysis was used to measure the protein expression levels of the SIRT1 gene in Hep3B cells. The WB results revealed that compared to that in the control group, the expression of the SIRT1 protein was increased in Hep3B cells treated with JQ-1. HCCLM3 and Huh-7 cells were also investigated by WB analysis again to demonstrate that SIRT1 protein expression was elevated during JQ-1 treatment (Fig. 2B). Furthermore, the protein expression of SIRT1 in transgenic HCC model mice treated with JQ-1 was examined by immunofluorescence staining (Fig. 2C). A similar result was obtained in the immunofluorescence assay, suggesting that aberrant expression of SIRT1 may be partially responsible for resistance to JQ-1.
Fig. 2.
JQ-1 increases SIRT1 expression in vitro and in vivo (A) RNA-sequencing analysis of Hep3B cells with and without JQ-1 treatment. (B) Significant increase in SIRT1 expression in JQ-1-treated Hep3B, HCCLM3, and Huh-7 cells compared to control cells, as measured by western blotting. (C) Representative immunofluorescence staining of SIRT1 (red) in HCC tissue sections from transgenic mice. Magnification, 100 × ; scale bar, 100 μm.
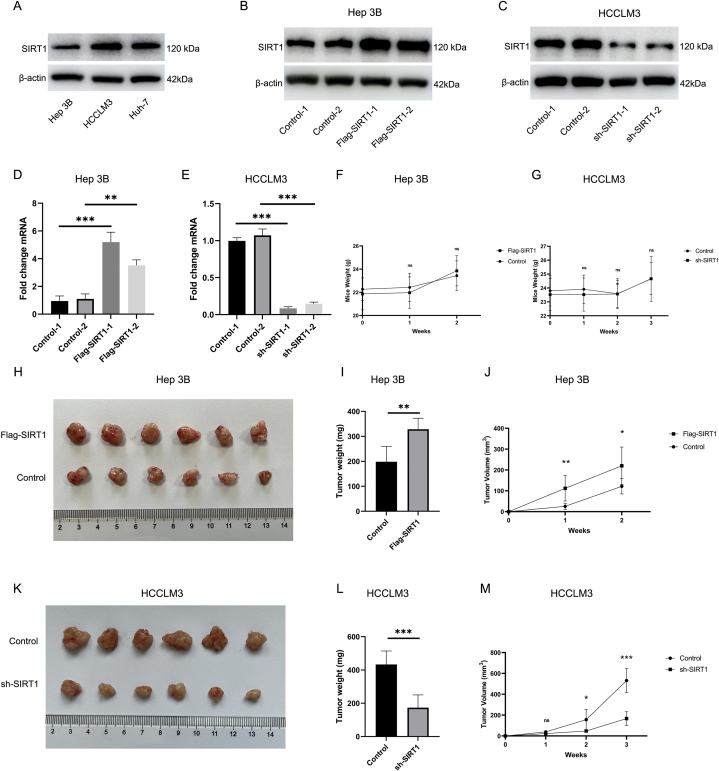
4.3. Upregulation of SIRT1 contributes to the acceleration of hepatocellular carcinoma progression
Several studies have reported the augmentation of SIRT1 expression in hepatocellular carcinoma patients with poor clinical outcomes and may be one of the underlying mechanisms causing resistance to sorafenib and other drugs, suggesting that aberrant expression of SIRT1 may be partially responsible for resistance to JQ-1 [[24], [25], [26]]. To validate the role of SIRT1 in the progression of hepatocellular carcinoma, bioinformatic analysis was first performed to explore our hypothesis. As shown in Fig. S4, HCC samples from CPTAC were analyzed, including subgroup analyses based on the distributions of age and sex, to identify differences between the normal controls and primary tumor patients. In the analyses incorporating age and sex data, the protein expression of SIRT1 in HCC patients was statistically higher than that in the normal group (Figs. S4A–C). Analyses of TCGA data produced similar results (Fig. S4D). Then, we downloaded the GSE45114 dataset from the GEO database and showed that SIRT1 expression was markedly augmented in HCC tissue compared with normal tissue and that increased SIRT1 expression was correlated with poor prognosis. The gene expression differences, survival curves, and surv_cutpoint function output are provided in Figs. S4E and F. Next, SIRT1 protein expression was evaluated in the Hep3B, HCCLM3, and Huh-7 cell lines, and relatively high and low expression levels of SIRT1 were found in HCCLM3 and Hep3B cells, respectively (Fig. 3A). Thus, these two cell lines were used in follow-up studies to explore the biological role of SIRT1 in HCC. Based on the results of bioinformatic analysis, lentiviral overexpression and knockdown of SIRT1 were conducted. The results of WB and PCR verification analyses are shown in Fig. 3B–E. After successful construction of the overexpression and knockdown vectors, transduced cells were implanted subcutaneously into nude mice to establish a xenograft tumor model. The tumor weight, tumor volume, and mouse weight were recorded. As shown in Fig. 3F-M, overexpression of SIRT1 in Hep3B cells promoted HCC tumorigenesis, while knockdown of SIRT1 in HCCLM3 cells delayed HCC progression. To observe the response of HCC cells to JQ-1 treatment after overexpression or knockdown of SIRT1, CCK-8 and TUNEL assays were performed. As reported in the CCK-8 and TUNEL assays, JQ-1 inhibited the proliferation of HCC cells and induced their apoptosis, and the tumor cell-killing ability was significantly enhanced after the knockdown of SIRT1 but reversed by the overexpression of SIRT1 (Fig. S5).
Fig. 3.
Upregulation of SIRT1 contributes to the acceleration of hepatocellular carcinoma progression (A) The relative expression levels of SIRT1 were examined in Hep3B cells, HCCLM3 cells, and Huh-7 cells by western blotting for screening. (B) and (C) Representative immunoblots showing the protein level of SIRT1 in SIRT1-overexpressing Hep3B cells and SIRT1-silenced HCCLM3 cells. (D) and (E) Gene expression of SIRT1 was normalized to that of β-actin, and the data are presented as the fold changes in the mRNA levels. (F) and (G) The mouse weights in the SIRT1-overexpressing, SIRT1-silenced and control groups were not substantially different. Due to differences in the inherent biological properties of these 2 xenograft models, the observation time was changed accordingly (2 weeks and 3 weeks). (H) Gross images of the tumors from nude mice subcutaneously transplanted with SIRT1-overexpressing Hep3B cells (5 × 106) or vector control cells. (I) and (J) Tumor weights and tumor volumes in the Flag-SIRT1 group and control group (n = 6 mice per group). (K) Gross images of tumors from nude mice subcutaneously transplanted with SIRT1-silenced HCCLM3 cells (5 × 106) or vector control cells. (L) and (M) Tumor weights and tumor volumes in the sh-SIRT1 group and control group (n = 6 mice per group).
4.4. Inhibition of SIRT1 enhances the antitumor effect of JQ-1 in vitro
Given the important role of SIRT1 in hepatocarcinogenesis, we hypothesized that the resistance to JQ-1 may be caused partially by the increase in SIRT1 expression. EX527, a selective SIRT1 inhibitor, was used to decrease the expression of endogenous SIRT1. The suitable concentration of EX527 in combination with JQ-1 was determined by a CCK-8 assay. As shown in Fig. 4A, compared to JQ-1 monotherapy, combination treatment with JQ-1 and 1 μM EX527 exhibited significant synergistic effects on HCCLM3 cells. However, no significant synergistic effect was observed in the other two cell lines at a concentration of 1 μM. As the concentration of EX527 increased, the drug combination finally exerted a greater therapeutic effect than monotherapy in all three HCC cell lines. The combination index is shown in Table 1 based on the formula described by Chou (CI = D A/ICX A + D B/ICX B) [27]. When the drug concentration of JQ-1 was 2 μM and that of EX527 was 5 μM, the combination index in the three cell lines was lowest among the different doses assessed. To further validate the synergistic effect of JQ-1 and EX527, the combination index formula (CI = D A/ICX A + DB/ICX B + DA*D B/ICX A *ICX B) was also employed, and the results are shown in Table S1: the results indicated the significant synergistic effect between JQ-1 and EX527 [27]. Therefore, 5 μM EX527 was used in subsequent experiments. The colony formation assay results confirmed that the combination of EX527 and JQ-1 attenuated the proliferative and metastatic capacities of HCC cells (Fig. 4B). The cell migration and invasion assays also suggested that, compared to the control group, the combination group of HCC cells exhibited significantly reduced migration and invasion abilities (Fig. S6). Furthermore, similar results were observed in the EdU incorporation assays: JQ-1 inhibited the proliferation of HCC cells after SIRT1 inhibition (Fig. S7). In addition, BET inhibitors are widely thought to be modulators of cell cycle regulators. Thus, we evaluated the effect of the combination strategy compared to that of JQ-1 or EX527 alone on cell cycle progression. Flow cytometric analysis revealed that the proportion of HCC cells in G0/G1 phase was increased and that the proportion in G2/M phase was reduced, accompanied by a concomitant decrease in the proportion of cells in S phase (Fig. 4C). Collectively, these results demonstrate that the combination of JQ-1 and EX527 significantly promotes synergistic antitumor benefits in HCC cells.
Fig. 4.
Cotreatment with JQ-1 and EX527 synergistically suppresses proliferation and blocks the cell cycle progression of HCC cells (A) The appropriate concentration of EX527, a selective SIRT1 inhibitor, in combination with JQ-1 was determined by a CCK-8 assay. A concentration of 5 μM EX527 was used in the subsequent experiments. (B) A colony formation assay was performed in the three HCC cell lines treated with JQ-1 (2 μM), EX527 (5 μM), their combination or the vehicle solution for 14 days. (C) Cell cycle profiles of HCC cells treated with JQ-1 and EX527 alone or in combination. The bar plot shows that the proportion of HCC cells in G0/G1 phase was increased and the proportion in G2/M phase was reduced, accompanied by a concomitant decrease in cells in S phase.
Table 1.
The combined index of the drug combination with different concentrations in three HCC cell lines.
| Combination strategy | The combination index CI = D A/ICX A + D B/ICX B |
||
|---|---|---|---|
| Hep 3B | HCCLM3 | Huh-7 | |
| JQ-1 2 μM + EX527 1 μM | 0.95 | 0.79 | 1.13 |
| JQ-1 2 μM + EX527 2 μM | 0.78 | 0.71 | 0.49 |
| JQ-1 2 μM + EX527 5 μM | 0.60 | 0.49 | 0.37 |
| JQ-1 2 μM + EX527 10 μM | 0.75 | 0.57 | 0.52 |
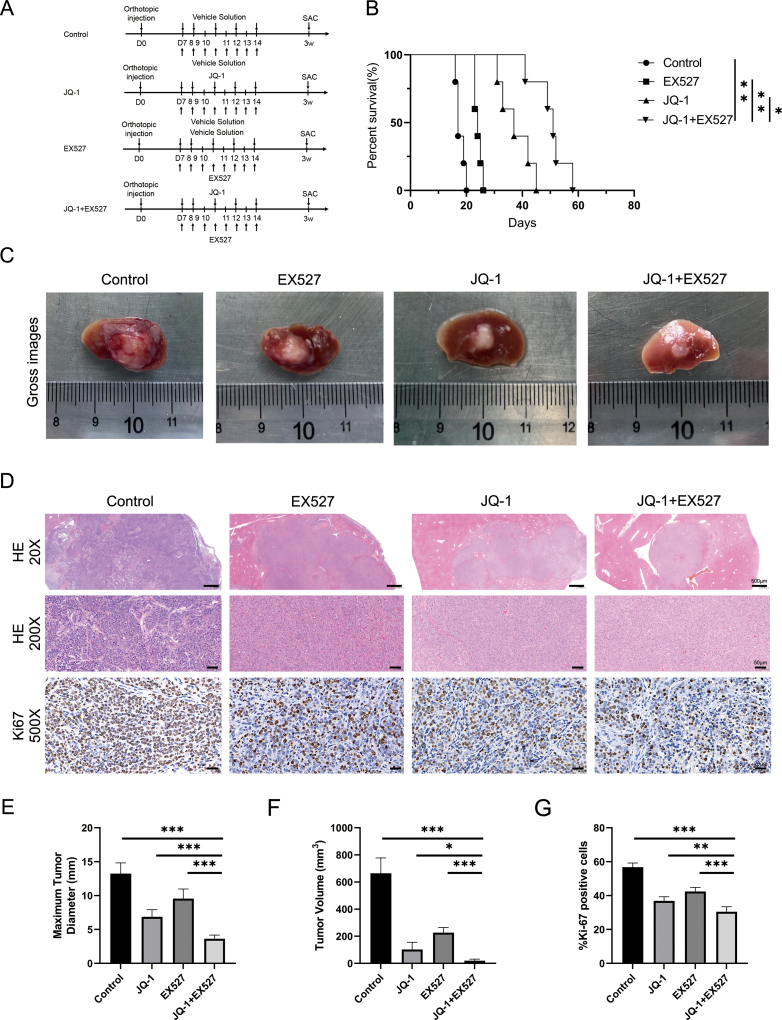
4.5. The combination of JQ-1 and EX527 efficiently suppressed HCC progression in vivo
Encouraged by the successful effects observed in cell experiments, the effect of combination treatment with JQ-1 and EX527 at the animal level became the main focus of our biological investigations. The experimental process of drug administration is presented in Fig. 5A. The results of the animal model comparisons were consistent with the results of the cell experiments, and both suggest that the combination of JQ-1 and EX527 significantly reduced the HCC tumor burden, as evaluated by the number of tumor nodules, the LW/BW ratio, maximum diameter of the nodules and the SW/BW ratios, and prolonged survival (Fig. 5B–E). Moreover, the combination therapy significantly decreased the ratio of Ki-67-positive cells compared to those in the control and single-drug groups, as determined by Ki-67 immunohistochemical staining (Fig. 5F and G). As reported by Kyungjoo Cho et al. [28], murine models of HCC, xenograft models, and genetically engineered mouse models might have different biological properties, which may explain our previous finding that JQ-1 alone did not significantly suppress tumor progression in a mouse model of HCC induced by c-Myc/N-Ras/SB plasmid treatment(Fig. 5H). To further validate the effects of JQ-1 and the combination strategy in other mouse HCC models, we established a Hep 1–6 cell-derived orthotopic mouse model (Fig. S8). Interestingly, we found that JQ-1 alone significantly suppressed the growth of Hep 1–6 cell-derived tumors. Moreover, the combination of JQ-1 and EX527 boosted the inhibitory effect in the orthotopic liver cancer model. To further investigate the relationship between SIRT1 and JQ-1, representative immunofluorescence staining of SIRT1 in liver sections from the transgenic liver cancer model and orthotopic liver cancer model after treatment with these drugs was performed (Fig. S9). In conclusion, these data indicated that EX527, a selective SIRT1 inhibitor, can enhance the antitumor effect of JQ-1 in HCC and significantly suppress xenograft tumor progression.
Fig. 5.
Combination treatment with JQ-1 and EX527 showed significant therapeutic benefits in inhibiting HCC progression (A) Schematic showing the drug administration design. (B) Survival curve of mice in the transgenic model of HCC after treatment with JQ-1 (50 mg/kg/day, i.p.), EX527 (30 mg/kg/day, s.c.), their combination or the vehicle solution. (C) Gross appearance and (D) H&E staining of tumor tissues from the four groups of mice. Magnification, 100 × ; scale bar, 100 μm. (E) The gross morphological indicators of the number of tumor nodules, the LW/BW ratio, the maximum diameter of the nodules, and the SW/BW ratio showed significant differences upon combination treatment compared with the other treatments (n = 6 mice per group). (F) Ki-67 staining of tumor tissues from the four groups of mice and (G) histogram of the Ki-67-positive cell data. Magnification, 200 × ; scale bar, 50 μm. (H) The mouse weight did not differ significantly among the four groups.
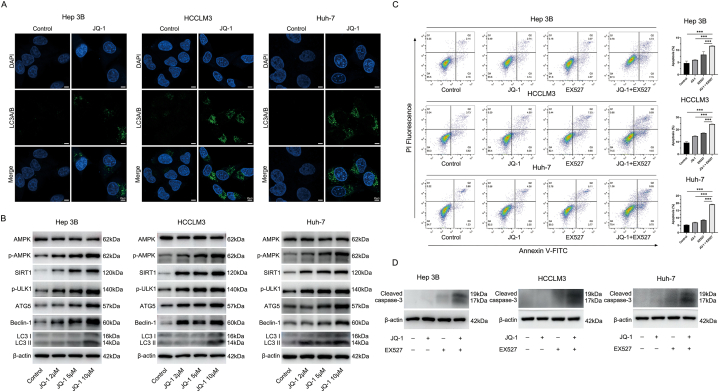
4.6. Targeting SIRT1 confers resistance to JQ-1 at least partially via the AMPK pathway in HCC cell lines
We further explored the molecular mechanism underlying the increase in SIRT1 expression caused by JQ-1. As reported in our previous research, we found that JQ-1 could protect the survival of cardiac grafts by activating the AMPK pathway [29]. Due to the strong relative relationship between SIRT1 and AMPK, we hypothesized that the increased expression of SIRT1 might be caused by the activation of the AMPK pathway after treatment with JQ-1 [[30], [31], [32]]. Not unexpectedly, the SIRT1 inhibitor Compound C and SIRT1 activator AICAR altered the AMPK pathway, with effects similar to those of JQ-1 (Fig. 6A). Moreover, we used lentivirus or Compound C to knock down SIRT1 expression in Hep3B, HCCLM3, and Huh-7 cell lines. As shown in Fig. 6B and C, WB results indicated that the activating effect of JQ-1 on the AMPK/SIRT1 axis was abolished in SIRT1-knockdown and Compound C-treated HCC cell lines. Next, the combined use of JQ-1, EX527, AICAR, and Compound C was evaluated by CCK-8 assay. The concentrations of these drugs are described in the Materials. As shown in Fig. 6D, we only found that the inhibitory effect of JQ-1+EX527+Compound C was better than that of JQ-1+EX527 in the HCCLM3 cell line, but the antitumor activities were enhanced to a very limited extent in the Hep3B and Huh-7 cell lines. Compared with the JQ-1+EX527 group, the tumor cell-killing ability in the JQ-1+EX527 +AICAR group was significantly reversed by AICAR in HCC cell lines. To further explore the reasons for the limited effects of JQ-1+EX527+Compound C, we examined the effect of JQ-1 combined with Compound C on the function of HCC cells. The combined use of JQ-1 and Compound C also showed better tumor cell-killing ability than the single agent alone, which enhances the antiproliferative ability of HCC cells (Fig. 6E). In addition, when the concentration of JQ-1 was increased to 5 μM, the drug combination of JQ-1+EX527+Compound C had a significantly stronger inhibitory effect on all three HCC cell lines than the combination of JQ-1+EX527 (Fig. 6F). Taken together, these results confirmed that the increase in SIRT1 expression confers resistance to JQ-1 at least partially via the AMPK pathway.
Fig. 6.
Targeting SIRT1 confers resistance to JQ-1 at least partially via the AMPK pathway in HCC cell lines (A) The protein levels of SIRT1, AMPK, and p-AMPK were measured in the JQ-1 (2 μM)-, AICAR (2 mM)-, and Compound C (10 μM)-treated groups of HCC cells after treatment for 24 h. (B) The protein levels of SIRT1, AMPK, and p-AMPK were measured in SIRT1-silenced and (C) Compound C (10 μM)-treated HCC cells after treatment with JQ-1 for 24 h. (D) Cell proliferation rate of HCC cell lines treated with JQ-1 (2 μM), EX527 (5 μM), AICAR (2 mM), Compound C (10 μM), their combination or the vehicle solution for 24 h. (E) Cell proliferation rate of HCC cell lines treated with JQ-1 (2 μM), compound C (10 μM), their combination or the vehicle solution for 24 h. (F) Cell proliferation rate of HCC cell lines treated with JQ-1 (5 μM), EX527 (5 μM), Compound C (10 μM), their combination or the vehicle solution for 24 h.
4.7. The AMPK/SIRT1 pathway could mediate protective autophagy activation to suppress the antitumor effect of JQ-1
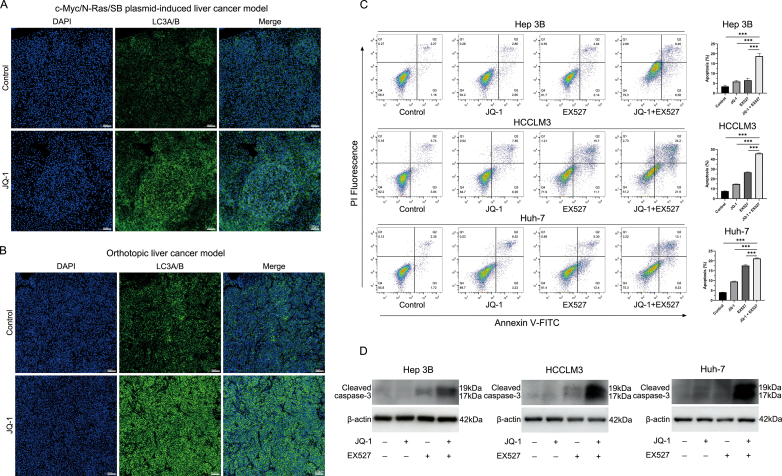
After verifying that JQ-1 could cause an increase in SIRT1 expression by activating the AMPK pathway, we continued to explore the synergistic mechanism of JQ-1 combined with EX527. After reviewing the literature, we found that the BET protein and AMPK/SIRT1 pathway were closely related to autophagy [33,34]. Our previous research also indicated that enhanced autophagy induced by JQ-1 had a protective effect in terms of cell survival [29]. Therefore, we speculated that the protective autophagy induced by JQ-1 might limit its antitumor effect and that the targeted inhibition of SIRT1 downregulates autophagy and exerts a synergistic effect. As shown in Fig. 7A and Figs. S10A and B, the immunofluorescence results showed that the use of JQ-1 could significantly enhance the production of the autophagy marker LC3A/B in vitro and in vivo. Moreover, JQ-1 treatment increased the expression level of autophagy marker proteins in a dose-dependent manner (Fig. 7B). Subsequently, we explored the effect of JQ-1 combined with EX527 on cell apoptosis. Compared with the control group, although JQ-1 could promote cell apoptosis, its ability to promote cell apoptosis was not as good as EX527. Moreover, compared with the control or either drug alone, the combined drug had a more significant cell-killing effect, resulting in a higher rate of cell apoptosis (Fig. 7C). The WB results showed a similar trend as the cell apoptosis assay (Fig. 7D). In addition, since the AMPK/SIRT1 pathway is closely related to the cellular energy supply, in the case of cell starvation or energy depletion, the AMPK/SIRT1 pathway is significantly activated to protect cells from survival under extreme conditions. We repeated the flow cytometry apoptosis assays under serum-free culture conditions. As shown in Figs. S10C and D, compared with culture with serum (10 % FBS + DMEM), serum-free culture (0 % FBS + DMEM) had a slight effect on the apoptosis of normal HCC cells and HCC cells treated with JQ-1, while SIRT1 inhibition could significantly promote apoptosis in the case of serum-free culture conditions.
Fig. 7.
The AMPK/SIRT1 pathway could mediate protective autophagy activation against the antitumor effect of JQ-1 (A) Representative immunofluorescence staining of LC3A/B (green) in HCC cells treated with JQ-1 for 24 h. Magnification, 400 × ; scale bar, 25 μm. (B) JQ-1 treatment increased the expression level of SIRT1 and autophagy in a dose-dependent manner. (C) The rate of apoptosis and (D) protein levels of cleaved caspase-3 in Hep3B, HCCLM3, and Huh-7 cell lines treated with JQ-1 (2 μM), EX527 (5 μM), their combination or the vehicle solution for 24 h.
4.8. Inhibition of SIRT1 could overcome resistance to JQ-1 primarily through blocking protective autophagy
We further verified that SIRT1 inhibition synergized with JQ-1 by inhibiting autophagy. The classic autophagy inhibitor 3-MA was applied to block autophagy. A similar synergistic effect was observed with the combination of JQ-1 and 3-MA (Fig. 8A and B). Then, we used lentivirus and EX527 to downregulate the expression of SIRT1 in HCC cells. As shown in Fig. 8C and D, WB results suggested that the knockdown of SIRT1 significantly inhibited the activation of the JQ-1-induced autophagy pathway. In general, our research indicated that JQ-1 could enhance autophagy by activating the AMPK/SIRT1 pathway and preserving HCC survival. Combined inhibition of BET protein and SIRT1 protein expression could significantly inhibit protective autophagy and promote cell apoptosis to exert a stronger antitumor effect in HCC.
Fig. 8.
Inhibition of SIRT1 could overcome drug resistance to JQ-1 by blocking protective autophagy (A) The rate of apoptosis of Hep3B, HCCLM3, and Huh-7 cell lines treated with JQ-1 (2 μM), 3-MA (5 mM), their combination or the vehicle solution for 24 h. (B) Cell proliferation rate of Hep3B, HCCLM3, and Huh-7 cell lines treated with JQ-1 (2 μM), 3-MA (5 mM), AICAR (2 mM), their combination or the vehicle solution for 24 h. The protein levels of autophagy were measured in (C) SIRT1-silenced and (D) EX527 (5 μM)-treated HCC cells after treatment with JQ-1 for 24 h.
5. Discussion
Currently, hepatocellular carcinoma is among the most common malignant solid tumors and major causes of cancer-related deaths worldwide. However, even with new first-line agents and second-line agents, HCC remains a concerning human health burden [35]. JQ-1, a BET inhibitor, is a potential candidate drug to treat patients with early-stage hematological malignancies or solid tumors [36]. As an epigenetic inhibitor that targets the bromodomain, JQ-1 has been reported to correlate with interferon signaling [37], PD-1/PD-L1 signaling [38], GSDMD signaling pathways [39], etc. However, the emergence of acquired drug resistance during JQ-1 treatment has limited the further use of JQ-1.
After RNA sequencing was performed, several key genes that may be responsible for acquired drug resistance were identified, and we selected the SIRT1 gene for further analysis via a second verification by western blotting. Although many differentially expressed genes were found by RNA sequencing, we focused on oncogenes in the field of cancer through DO enrichment analysis. Similar to the traditional view, after JQ-1 treatment, the expression of many oncogenes was downregulated, but we also found abnormally high expression of several key genes that may lead to acquired resistance to JQ-1. PubMed and Web of Science were used to explore the biological roles of TSGA10, FOS, FGF18, MGAT5, EFNB2, and PODXL in cancer research [[40], [41], [42]]. However, only a small number of cancer studies have focused on these specific genes, which limits our further research. For TGF-β, WB was performed to confirm whether JQ-1 treatment affects the expression of TGF-β. However, the WB results showed that the effect of JQ-1 on TGF-β was not obvious (Fig. S11).
Notably, SIRT1, as an important epigenetic regulatory gene, plays an important role in the occurrence and development of cancer, but there is still some controversy about whether it promotes the progression of HCC [43]. Previous studies have shown that SIRT1 plays dual roles in different types of diseases [17,18]. For example, SIRT1 is highly expressed in breast cancer, colon cancer, liver cancer, etc., while it is expressed at low levels in luminal squamous cell carcinoma and gastric cancer [[19], [20], [21]]. This attracted our intense interest. Through a literature search, we found that SIRT1 is closely related to the development and poor prognosis of HCC, and recent research supports its role as a proto-oncogene in promoting the progression of HCC [22,44,45]. Therefore, it seems reasonable to hypothesize that high expression of SIRT1 may be a potential mechanism of JQ-1 resistance [22,24].
Overexpression of SIRT1 was also indicated to be an underlying mechanism of resistance to radiotherapy [46] and sorafenib [26] in some studies. However, the role of SIRT1 in JQ-1 treatment remains unclear. Notably, our findings confirmed that SIRT1 expression was increased via the AMPK pathway during JQ-1 treatment and that combined inhibition of BET and SIRT1 could overcome resistance to JQ-1 by blocking protective autophagy, thus exerting a stronger antitumor effect in HCC. However, some limitations existed. As we reported, the BET inhibitor JQ-1 significantly prolonged survival but had few effects on other tumor indices in the c-Myc/N-Ras/SB-driven HCC mouse model. In our study, we confirmed that JQ-1 exerted anticancer effects in vitro, but JQ-1 monotherapy did not manifest significant therapeutic effects in vivo. Compared with the relatively simple in vitro conditions, the more complex environment in vivo may allow cancer cells to have stronger drug resistance and drug efflux abilities that cells adapted to the simpler cellular context in vitro cannot [47]. The different mouse tumor models possess unique biological properties that result in different responses to JQ-1 treatment, which may explain the different effects of JQ-1 application in these two murine tumor models.
Overall, our findings provide a new mechanism by which SIRT1 plays a key role in enhancing JQ-1 resistance and propose a novel combination therapeutic strategy by targeting the AMPK/SIRT1 axis to improve its antitumor activity. However, the combination therapeutic strategy of a BET inhibitor and SIRT1 inhibition is preferentially recommended and must be evaluated in clinical trials of HCC patients.
Funding information
This project was supported by the National Natural Science Foundation of China (No. 81870306).
Ethics statement
All animal studies were approved by the Institutional Animal Care and Use Committee of the Second Affiliated Hospital, School of Medicine, Zhejiang University 2020–109. During the preparation of this work the authors have not used AI and AI-assisted technologies.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Yuancong Jiang: Writing – original draft. Xiaolong Miao: Writing – original draft. Zelai Wu: Writing – review & editing, Writing – original draft. Weixun Xie: Writing – original draft. Li Wang: Writing – original draft. Han Liu: Writing – original draft. Weihua Gong: Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22093.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Fig. S1 (A) The IC50 values of JQ-1 were 2.449 μM, 3.235 μM and 1.422 μM in Hep3B, HCCLM3 and Huh-7 cells, respectively. (B) Cell proliferation rate of HCC cell lines treated with JQ-1 (2 μM) or vector (DMSO) control for 24 h.
Fig. S2 (A)
The concentrations of JQ-1 at 10 nM, 25 nM, 50 nM and 100 nM in Hep3B, HCCLM3 and Huh-7 cells were used to detect cell viability. (B) The transwell migration assay showed significant inhibition of the invasion and metastasis abilities after treatment with JQ-1 (50 nM) for 24 h in these three HCC cell lines compared to the control group. Scale bar, 100 μm.
Fig. S3 (A) Schematic design of the experimental procedure. Transgenic HCC mice were transfected with the c-Myc/N-Ras/SB plasmid and treated with JQ-1 (50 mg/kg/day, i.p.) or vehicle solution. The mice were sacrificed at 6 weeks for phenotypic analysis. (B) JQ-1 significantly increased the survival time compared with that in the control group. (C) Representative gross images and H&E staining images are provided. Magnification, 100 × ; scale bar, 100 μm. (D) The gross morphological indicators of the number of tumor nodules, the LW/BW ratio, the maximum diameter of the nodules and the SW/BW ratio were calculated based on the macroscopic images (n = 6 mice per group).
Fig. S4 (A)
Bioinformatic analysis of SIRT1 expression in normal controls and primary tumor patients using CPTAC HCC samples. (B) and (C) Subgroup analysis of CPTAC HCC samples based on age and sex distributions. (D) Bioinformatic analysis of SIRT1 expression in TCGA data. (E) and (F) Survival curves based on SIRT1 expression in HCC tissues compared with normal tissues from the GEO database (GSE45114).
Fig. S5 (A) Cell proliferation rate of Hep3B cells with SIRT1 overexpression or HCCLM3 cells with knockdown of SIRT1 after treatment with JQ-1 (2 μM) for 24 h. (B) and (C) TUNEL assay in Hep3B cells with SIRT1 overexpression or HCCLM3 cells with knockdown of SIRT1 after treatment with JQ-1 (2 μM) for 24 h. The percentage of TUNEL-positive cells was decreased after overexpression of SIRT1 in Hep3B cell line and increased after knockdown of SIRT1 in HCCLM3 cell line compared to treatment with the single use of JQ-1. Scale bar, 100 μm.
Fig. S6 Migration and invasion potential of Hep3B, HCCLM3, and Huh-7 cells upon treatment with JQ-1 (2 μM), EX527 (5 μM), the combination of these two agents or the vehicle solution. Cells were treated for 24 h in the migration assay and for 48 h in the invasion assay. Scale bar, 100 μm.
Fig. S7 EdU incorporation assay in Hep3B, HCCLM3, and Huh-7 cell lines treated with JQ-1 (2 μM), EX527 (5 μM), their combination or the vehicle solution for 24 h. The percentage of EdU-positive cells was decreased by treatment with the combination of the two drugs compared to treatment with the single agents or the vehicle control. Scale bar, 100 μm.
Fig. S8 (A) Scheme of drug administration. (B) Survival curve of the orthotopic liver cancer model after treatment with JQ-1 (50 mg/kg/day, i.p.), EX527 (30 mg/kg/day, s.c.), their combination or the vehicle solution. (C) Gross appearance, (D) H&E staining, and Ki-67 staining of tumor tissues from the four groups of mice. Magnification, 20 × ; scale bar, 500 μm and magnification, 200 × ; scale bar, 50 μm in H&E staining and magnification, 500 × ; scale bar, 20 μm in Ki-67 staining. Comparison of the (E) maximum tumor diameter and (F) tumor volume among the four groups. (G) The histogram of the Ki-67-positive cell data showed significant differences between the combination treatment group and the other groups (n = 5 mice per group).
Fig. S9 Representative immunofluorescence staining of SIRT1 (red) in liver sections in the (A) c-Myc/N-Ras/SB plasmid-induced liver cancer model and (B) orthotopic liver cancer model after treatment with JQ-1 (50 mg/kg/day, i.p.), EX527 (30 mg/kg/day, s.c.), their combination, and vehicle solution. Magnification, 100 × ; scale bar, 100 μm.
Fig. S10 Representative immunofluorescence staining of (A) the c-Myc/N-Ras/SB plasmid-induced liver cancer model and (B) the orthotopic liver cancer model after treatment with JQ-1 (50 mg/kg/day, i.p.). Magnification, 100 × ; scale bar, 100 μm. (C) The rate of apoptosis and (D) protein levels of cleaved caspase-3 in Hep3B, HCCLM3, and Huh-7 cells treated with JQ-1 (2 μM), EX527 (5 μM), their combination or the vehicle solution in a serum-free (0 % FBS + DMEM) environment for 24 h.

Fig. S11 The expression of TGF-β was measured in JQ-1-treated Hep3B cells and DMSO-treated Hep3B cells by western blotting.
References
- 1.Llovet J.M., Castet F., Heikenwalder M., Maini M.K., Mazzaferro V., Pinato D.J., et al. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2022;19:151–172. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 2.Kim D.H., Choi S.H., Shim J.H., Kim S.Y., Lee S.S., Byun J.H., et al. Meta-analysis of the accuracy of abbreviated magnetic resonance imaging for hepatocellular carcinoma surveillance: non-contrast versus hepatobiliary phase-abbreviated magnetic resonance imaging. Cancers. 2021;13 doi: 10.3390/cancers13122975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng L., Wang G., Chen Y., He G., Liu B., Liu J., et al. Dual-target inhibitors of bromodomain and extra-terminal proteins in cancer: a review from medicinal chemistry perspectives. Med. Res. Rev. 2022;42:710–743. doi: 10.1002/med.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards D.S., Maganti R., Tanksley J.P., Luo J., Park J.J.H., Balkanska-Sinclair E., et al. BRD4 prevents R-loop formation and transcription-replication conflicts by ensuring efficient transcription elongation. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y., Han J., Wang Z., Li X., Sun Y., Hu Z. Safety and efficacy of bromodomain and extra-terminal inhibitors for the treatment of hematological malignancies and solid tumors: a systematic study of clinical trials. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.621093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J., Liu Y., Lv J., Zou B., Chen Z., Li K., et al. BCL6 confers KRAS-mutant non-small-cell lung cancer resistance to BET inhibitors. J. Clin. Invest. 2021:131. doi: 10.1172/JCI133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong D., Jiang Y., Miao X., Wu Z., Liu H., Gong W. Tadalafil enhances the therapeutic efficacy of BET inhibitors in hepatocellular carcinoma through activating Hippo pathway. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2021;1867 doi: 10.1016/j.bbadis.2021.166267. [DOI] [PubMed] [Google Scholar]
- 8.Liu C., Miao X., Wang Y., Wen L., Cheng X., Kong D., et al. Bromo- and extraterminal domain protein inhibition improves immunotherapy efficacy in hepatocellular carcinoma. Cancer Sci. 2020;111:3503–3515. doi: 10.1111/cas.14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Y., Sun M., Zhan X., Wu C., Geng P., Sun X., et al. EGFR signaling confers resistance to BET inhibition in hepatocellular carcinoma through stabilizing oncogenic MYC. J. Exp. Clin. Cancer Res. : CR. 2019;38:83. doi: 10.1186/s13046-019-1082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang P., Chang Y., Zhang J., Gao F., Liu X., Wei Q., et al. The combination of in situ photodynamic promotion and ion-interference to improve the efficacy of cancer therapy. J. Colloid Interface Sci. 2023;629:522–533. doi: 10.1016/j.jcis.2022.08.125. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Miao X., Jiang Y., Wu Z., Zhu X., Liu H., et al. The synergistic antitumor effect of IL-6 neutralization with NVP-BEZ235 in hepatocellular carcinoma. Cell Death Dis. 2022;13:146. doi: 10.1038/s41419-022-04583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao X., Liu C., Jiang Y., Wang Y., Kong D., Wu Z., et al. BET protein inhibition evidently enhances sensitivity to PI3K/mTOR dual inhibition in intrahepatic cholangiocarcinoma. Cell Death Dis. 2021;12:1020. doi: 10.1038/s41419-021-04305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slaughter M.J., Shanle E.K., Khan A., Chua K.F., Hong T., Boxer L.D., et al. HDAC inhibition results in widespread alteration of the histone acetylation landscape and BRD4 targeting to gene bodies. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y., Fei Q., Xiong P., Yang J., Zhang Z., Lin X., et al. Synergistic inhibition of pancreatic cancer with anti-PD-L1 and c-Myc inhibitor JQ1. OncoImmunology. 2019;8 doi: 10.1080/2162402X.2019.1581529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehling S.C., Miller A.L., Garcia P.L., Vance R.B., Yoon K.J. The combination of BET and PARP inhibitors is synergistic in models of cholangiocarcinoma. Cancer letters. 2020;468:48–58. doi: 10.1016/j.canlet.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gusyatiner O., Bady P., Pham M.D.T., Lei Y., Park J., Daniel R.T., et al. BET inhibitors repress expression of interferon-stimulated genes and synergize with HDAC inhibitors in glioblastoma. Neuro Oncol. 2021;23:1680–1692. doi: 10.1093/neuonc/noab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves-Fernandes D.K., Jasiulionis M.G. The role of SIRT1 on DNA damage response and epigenetic alterations in cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20133153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Q., Dong L., Li Y., Liu G. SIRT1 and HIF1α signaling in metabolism and immune responses. Cancer letters. 2018;418:20–26. doi: 10.1016/j.canlet.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S., Yang Y., Huang S., Deng C., Zhou S., Yang J., et al. SIRT1 inhibits gastric cancer proliferation and metastasis via STAT3/MMP-13 signaling. J. Cell. Physiol. 2019;234:15395–15406. doi: 10.1002/jcp.28186. [DOI] [PubMed] [Google Scholar]
- 20.Kang Y.Y., Sun F.L., Zhang Y., Wang Z. SIRT1 acts as a potential tumor suppressor in oral squamous cell carcinoma. J. Chin. Med. Assoc. : JCMA. 2018;81:416–422. doi: 10.1016/j.jcma.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Leng S., Huang W., Chen Y., Yang Y., Feng D., Liu W., et al. SIRT1 coordinates with the CRL4B complex to regulate pancreatic cancer stem cells to promote tumorigenesis. Cell Death Differ. 2021;28:3329–3343. doi: 10.1038/s41418-021-00821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Liu J., Xiao W., Zeng Q., Bo H., Zhu Y., et al. SIRT1 regulates N(6) -methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatology. 2020;72:2029–2050. doi: 10.1002/hep.31222. [DOI] [PubMed] [Google Scholar]
- 23.Torbenson M.S. Morphologic subtypes of hepatocellular carcinoma. Gastroenterol. Clin. N. Am. 2017;46:365–391. doi: 10.1016/j.gtc.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Wang M.J., Chen J.J., Song S.H., Su J., Zhao L.H., Liu Q.G., et al. Inhibition of SIRT1 limits self-renewal and oncogenesis by inducing senescence of liver cancer stem cells. J. Hepatocell. Carcinoma. 2021;8:685–699. doi: 10.2147/JHC.S296234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farcas M., Gavrea A.A., Gulei D., Ionescu C., Irimie A., Catana C.S., et al. SIRT1 in the development and treatment of hepatocellular carcinoma. Front. Nutr. 2019;6:148. doi: 10.3389/fnut.2019.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garten A., Grohmann T., Kluckova K., Lavery G.G., Kiess W., Penke M. Sorafenib-induced apoptosis in hepatocellular carcinoma is reversed by SIRT1. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20164048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 28.Cho K., Ro S.W., Seo S.H., Jeon Y., Moon H., Kim D.Y., et al. Genetically engineered mouse models for liver cancer. Cancers. 2019;12 doi: 10.3390/cancers12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., Miao X., Liu C., Liu B., Wu X., Kong D., et al. BET protein inhibition prolongs cardiac transplant survival via enhanced myocardial autophagy. Transplantation. 2020;104:2317–2326. doi: 10.1097/TP.0000000000003319. [DOI] [PubMed] [Google Scholar]
- 30.Ge Y., Zhou M., Chen C., Wu X., Wang X. Role of AMPK mediated pathways in autophagy and aging. Biochimie. 2022;195:100–113. doi: 10.1016/j.biochi.2021.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Sánchez B., Muñoz-Pinto M.F., Cano M. Irisin enhances longevity by boosting SIRT1, AMPK, autophagy and telomerase. Expet Rev. Mol. Med. 2022;25:e4. doi: 10.1017/erm.2022.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ammar R.A., Mohamed A.F., Kamal M.M., Safar M.M., Abdelkader N.F. Neuroprotective effect of liraglutide in an experimental mouse model of multiple sclerosis: role of AMPK/SIRT1 signaling and NLRP3 inflammasome. Inflammopharmacology. 2022;30:919–934. doi: 10.1007/s10787-022-00956-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo G., Jian Z., Zhu Y., Zhu Y., Chen B., Ma R., et al. Sirt1 promotes autophagy and inhibits apoptosis to protect cardiomyocytes from hypoxic stress. Int. J. Mol. Med. 2019;43:2033–2043. doi: 10.3892/ijmm.2019.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C., Wang L., Fozouni P., Evjen G., Chandra V., Jiang J., et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat. Cell Biol. 2020;22:1170–1179. doi: 10.1038/s41556-020-00579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel A., Meyer T., Sapisochin G., Salem R., Saborowski A. Hepatocellular carcinoma. Lancet (London, England) 2022;400:1345–1362. doi: 10.1016/S0140-6736(22)01200-4. [DOI] [PubMed] [Google Scholar]
- 36.Stathis A., Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8:24–36. doi: 10.1158/2159-8290.CD-17-0605. [DOI] [PubMed] [Google Scholar]
- 37.Gopalakrishnan R., Matta H., Tolani B., Triche T., Jr., Chaudhary P.M. Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene. 2016;35:1797–1810. doi: 10.1038/onc.2015.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Yu B., Cao B., Zhou J., Deng Y., Wang Z., et al. A chemical conjugation of JQ-1 and a TLR7 agonist induces tumoricidal effects in a murine model of melanoma via enhanced immunomodulation. Int. J. Cancer. 2021;148:437–447. doi: 10.1002/ijc.33222. [DOI] [PubMed] [Google Scholar]
- 39.Hao K., Jiang W., Zhou M., Li H., Chen Y., Jiang F., et al. Targeting BRD4 prevents acute gouty arthritis by regulating pyroptosis. Int. J. Biol. Sci. 2020;16:3163–3173. doi: 10.7150/ijbs.46153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G., Liu J., Cai Y., Chen J., Xie W., Kong X., et al. Loss of Barx1 promotes hepatocellular carcinoma metastasis through up-regulating MGAT5 and MMP9 expression and indicates poor prognosis. Oncotarget. 2017;8:71867–71880. doi: 10.18632/oncotarget.18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H., Kong J.S., Lee S.S., Kim A. Radiation-induced overexpression of TGFβ and PODXL contributes to colorectal cancer cell radioresistance through enhanced motility. Cells. 2021;10 doi: 10.3390/cells10082087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei W., Mok S.C., Oliva E., Kim S.H., Mohapatra G., Birrer M.J. FGF18 as a prognostic and therapeutic biomarker in ovarian cancer. J. Clin. Invest. 2013;123:4435–4448. doi: 10.1172/JCI70625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X., Su Q., Xie H., Song L., Yang F., Zhang D., et al. SIRT1 deacetylates WEE1 and sensitizes cancer cells to WEE1 inhibition. Nat. Chem. Biol. 2023;19:585–595. doi: 10.1038/s41589-022-01240-y. [DOI] [PubMed] [Google Scholar]
- 44.Chen X., Huan H., Liu C., Luo Y., Shen J., Zhuo Y., et al. Deacetylation of β-catenin by SIRT1 regulates self-renewal and oncogenesis of liver cancer stem cells. Cancer letters. 2019;463:1–10. doi: 10.1016/j.canlet.2019.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Liu L., Liu C., Zhang Q., Shen J., Zhang H., Shan J., et al. SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology. 2016;64:814–827. doi: 10.1002/hep.28690. [DOI] [PubMed] [Google Scholar]
- 46.Yang M., Liu Q., Dai M., Peng R., Li X., Zuo W., et al. FOXQ1-mediated SIRT1 upregulation enhances stemness and radio-resistance of colorectal cancer cells and restores intestinal microbiota function by promoting β-catenin nuclear translocation. J. Exp. Clin. Cancer Res. : CR. 2022;41:70. doi: 10.1186/s13046-021-02239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLaughter D.M., Christodoulou D.C., Robinson J.Y., Seidman C.E., Baldwin H.S., Seidman J.G., et al. Spatial transcriptional profile of the chick and mouse endocardial cushions identify novel regulators of endocardial EMT in vitro. J. Mol. Cell. Cardiol. 2013;59:196–204. doi: 10.1016/j.yjmcc.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.