Abstract
The FhuA protein of Escherichia coli K-12 transports ferrichrome, the antibiotic albomycin, colicin M, and microcin 25 across the outer membrane and serves as a receptor for the phages T1, T5, φ80, and UC-1. FhuA is activated by the electrochemical potential of the cytoplasmic membrane, which probably opens a channel in FhuA. It is thought that the proteins TonB, ExbB, and ExbD function as a coupling device between the cytoplasmic membrane and the outer membrane. Excision of 34 residues from FhuA, tentatively designated the gating loop, converts FhuA into a permanently open channel. FhuA contains two disulfide bridges, one in the gating loop and one close to the C-terminal end. Reduction of the disulfide bridges results in a low in vivo reaction of the cysteines in the gating loop and no reaction of the C-terminal cysteines with biotin-maleimide, as determined by streptavidin-β-galactosidase bound to biotin. In this study we show that a cysteine residue introduced into the gating loop by replacement of Asp-336 displayed a rather high reactivity and was used to monitor structural changes in FhuA upon binding of ferrichrome. Flow cytometric analysis revealed fluorescence quenching by ferrichrome and albomycin of fluorescein-maleimide bound to FhuA. Ferrichrome did not inhibit Cys-336 labeling. In contrast, labeling of Cys-347, obtained by replacing Val-347 in the gating loop, was inhibited by ferrichrome, but ferrichrome quenching was negligible. It is concluded that binding of ferrichrome causes a conformational change of the gating loop and that Cys-347 is part of or close to the ferrichrome binding site. Fluorescence quenching was independent of the TonB activity. The newly introduced cysteines and the replacement of the existing cysteines by serine did not alter sensitivity of cells to the FhuA ligands tested (T5, φ80, T1, colicin M, and albomycin) and fully supported growth on ferrichrome as the sole iron source. Since cells of E. coli K-12 display no reactivity to thiol reagents, newly introduced cysteines can be used to determine surface-exposed regions of outer membrane proteins and to monitor conformational changes during their function.
The FhuA protein in the outer membrane of Escherichia coli serves for the uptake of ferrichrome, the antibiotic albomycin, colicin M, and microcin 25 and for infection by the phages T5, T1, φ80, and UC-1 (8, 26, 36). The activity of FhuA requires the electrochemical potential of the cytoplasmic membrane. This potential is mediated to the outer membrane by the Ton system (7, 15, 20, 32), which consists of the TonB, ExbB, and ExbD proteins. Only infection by phage T5 occurs independently of energy and the Ton system.
Isolated FhuA does not increase the conductance of artificial lipid bilayer membranes. Excision of residues 322 to 355 of the mature protein opens a channel with a conductance three times as large as the conductance of porins in lipid bilayers (16). The outer membrane of cells synthesizing FhuA Δ322–355 is permeable to ferrichrome, sodium dodecyl sulfate (SDS), and bacitracin, which diffuse through the permanently open FhuA channel independently of energy and the Ton system. This finding and the interaction of FhuA with TonB (11, 38) led to the hypothesis that the FhuA channels are opened by TonB in the energized conformation the same way a regulatory protein allosterically controls enzyme activity (7). The open state of FhuA must be short-lived, because it does not affect the permeability of the outer membrane for solutes that are not recognized by FhuA. Our current concept assumes that energy is consumed each time FhuA opens by the movement of segment 322–355.
The segment to be excised to open FhuA was deduced from a FhuA transmembrane model that was established by inserting peptides of 4 to 16 residues at 34 sites along the FhuA polypeptide (21). Most of these FhuA insertion derivatives retain activity for some or all of the ligands and are considered to be properly inserted in the outer membrane. They were used to determine proteolytic cleavage within or close to the inserted peptides, since wild-type FhuA is largely resistant to proteases. Accessibility of the cleavage sites at the cell surface or in the periplasm defined loops located in either of these compartments. The largest loop at the cell surface comprises residues 316 to 356. In one particular spontaneous fhuA mutant, FhuA is inactive with most of the ligands and lacks Asp-348 in this loop (18). This loop and the predicted loop nearby (residues 404 to 432) react in cells with monoclonal antibodies raised against isolated FhuA (28), and anti-C3 antibodies react with a C3 viral reporter epitope inserted after residues 321, 405, and 417 (27). These data are consistent with the FhuA transmembrane model, in particular with the accessibility of segment 322–355 from the cell surface. Since this segment apparently controls the permeability of FhuA, it has been designated the gating loop (16).
The gating loop also serves as the principal binding site for phages T5, T1, and φ80 and for colicin M. Cells synthesizing FhuA Δ322–355 do not respond to the FhuA ligands. Excision of segment 322–336 or 335–355 revealed that the latter determines mainly the gating and ligand-binding properties of FhuA, in particular for ferrichrome (17). Competitive peptide mapping with acetylated hexapeptide amides comprising the entire gating loop revealed three subdomains that interact with the phages (19). All these data indicate that the gating loop is an important functional region of FhuA.
FhuA contains four cysteine residues (9) which form disulfide bridges within the gating loop (C318 and C329) and close to the C-terminal end (C692 and C698) (6). The cysteines of the gating loop reacted in vivo with biotin-maleimide [N-biotinoyl-N′-(6-maleimidohexanoyl)-hydrazide] (B-M) after reduction, while the C-terminal cysteines reacted only after replacement of one of the cysteines and denaturation of FhuA in vitro (6). Virtually no protein other than overexpressed FhuA was labeled in intact cells.
In contrast to our previous study, in which FhuA was overproduced by high transcription of the fhuA gene cloned downstream of the phage T7 gene 10 promoter by the T7 RNA polymerase (6), in this study we transcribed the fhuA gene under the control of its own promoter by the E. coli RNA polymerase. Overproduction of FhuA corresponded to the medium copy number of the fhuA plasmids (about 15 copies per cell). For comparison of the results, the same FhuA expression conditions were used for all experiments. We introduced two cysteines into the gating loop by amino acid replacement. One cysteine was more reactive than the natural cysteines and was used to study conformational changes in FhuA upon binding of ferrichrome in the presence and absence of TonB. The other cysteine was suitable for determining a binding site of ferrichrome.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli K-12 strains and plasmids used in this study are listed in Table 1. FhuA(C318S), FhuA(C329S), FhuA(D336C), and FhuA(V347C) were constructed by PCR with plasmid pCB24, which contains a 1.65-kb HindIII-SalI fhuA fragment and an additional BamHI restriction site at nucleotide 1596 (numerals as in Coulton et al. [9]) in plasmid pBCKS+. The primers used were cys3, 5′-CGCCGGATCCGAGCTGACGCCG-3′ for C318S; cys4, 5′-GCTCGGATCCGGCGAATGCTTACAGCAAACAGAGTGCGGC-3′ for C329S; cys5, 5′-TACGTGCCAGATAATGGCCTTTACACGCTG-3′ for D336C; and cys6, 5′-TAAAGGCCATTATCTGGCACGTAAATACGTCTGTGATGATG-3′ for V347C (bases of the wild-type sequence were replaced by the underlined bases). cys3 and cys4 were cloned with BamHI and MluI and SalI, respectively, into pfhuA8; and cys5 and cys6 were cloned with SalI and BamHI and Van91/1, respectively, into pfhuA8 (pHK763 with a BamHI site at position 1596). All constructions were examined by DNA sequencing by the dideoxy chain termination method (37) and with the AutoRead sequencing kit and the A.L.F. Sequencer from Pharmacia Biotech (Freiburg, Germany).
TABLE 1.
E. coli K-12 strains and plasmids used in this study
| Strains and plasmids | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| UL3 | AB2847 fhuA recA Tn10 | 16 |
| BR158 | AB2847 tonB | 13 |
| HK99 | BR158 fhuA | 16 |
| Plasmids | ||
| pT7-6 | Ampr | 39 |
| pHK763 | pT7-6 FhuA wild type | 16 |
| pfhuA4 | pT7-6 FhuA(C318S) | 6 |
| pfhuA5 | pT7-6 FhuA(C329S) | 6 |
| pfhuA6 | pT7-6 FhuA(D336C) | This study |
| pfhuA7 | pT7-6 FhuA(V347C) | This study |
| pfhuA8 | pHK763 with an additional BamHI site | This study |
| pfhuA9 | pT7-6 FhuA(C318S C329S C692S C698S) | 6 |
Labeling of cells with B-M.
The procedure used to determine the specificity of labeling, shown in Fig. 2, was similar to the method described previously (3, 4). E. coli UL3 fhuA transformed with the plasmids pfhuA4 [pT7-6 FhuA(C318S)], pfhuA5 [pT7-6 FhuA(C329S)], pfhuA6 [pT7-6 FhuA(D336C)], pfhuA7 [pT7-6 FhuA(V347C)], pfhuA8 (pT7-6, FhuA-WT), pfhuA9 [pT7-6 FhuA(C318S C329S C692S C698S)], or pT7-6 (vector) was grown at 37°C in tryptone-yeast extract medium (6) containing 30 μg of ampicillin per ml. Cells (109) were harvested in the logarithmic growth phase (optical density at 578 nm, 0.5) and washed three times with 1 ml of ice-cold phosphate-buffered saline (PBS) (20 mM sodium phosphate, 0.9% NaCl, 1 mM MgCl2 [pH 7.4]). Cells were incubated at 30°C in 1 ml of 0.5 mM B-M (Sigma, St. Louis, Mo.) dissolved in PBS containing 1% dimethylsulfoxide. The reaction was stopped after 30 min by adding a 40-fold excess of dithiothreitol (DTT) (110 μl of 200 mM solution; final concentration, 20 mM). After being washed three times in PBS, the cells were suspended in 24 μl of lysis buffer (4% SDS, 20 mM Tris-HCl, 0.2 mM EDTA [pH 8.0]) and heated for 5 min at 100°C. Then, the same volume of sample buffer (4% SDS, 10% β-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue in 0.125 M Tris-HCl [pH 6.8]) was added and heated once more for 3 min in a boiling-water bath. The proteins were separated on an 11% polyacrylamide gel for 3.5 h at a constant current of 30 mA. The thioether bond between maleimide and cysteine is not affected by β-mercaptoethanol. The proteins were blotted overnight (at 24 V in Trans-Blot cell apparatus model 22E/0940; Bio-Rad) onto a nitrocellulose membrane (no. BA85, 0.45-μm pore size; Schleicher & Schüll) and stained with 0.2% Ponceau S-Red (Serva, Heidelberg, Germany) in 3% trichloroacetic acid. The surplus stain was removed by rinsing with water. The stained gel was photographed and then destained by incubation for 20 min in 20 mM Tris-HCl–0.5 M NaCl–0.005% Tween 80 (pH 7.5). To saturate nonspecific binding sites on proteins, the nitrocellulose membranes were treated for 1 h with 2% bovine serum albumin in PBS prior to incubation for 45 min with streptavidin-β-galactosidase (S-G) (6.5 μg per ml; 1.3 U of streptavidin per mg of protein, 535 U of β-galactosidase per mg of protein [Sigma] in 2% bovine serum albumin). The nitrocellulose membranes were washed four times with PBS and then incubated for about 2 min with a solution of X-Gal (1.2 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 3 mM (each) potassium hexacyanoferrate(II) and -(III) in PBS (pH 7.4). X-Gal was removed by washing with water.
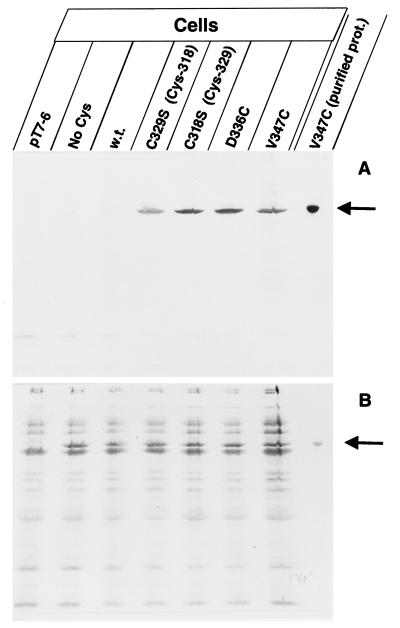
FIG. 2.
Specific labeling of the FhuA single-cysteine mutants with B-M in live cells (A). Cells of E. coli UL3 fhuA were transformed with one of the following plasmids: pT7-6 (vector without fhuA), pfhuA9 [FhuA(C318S C329S C692S C698S); No Cys], pfhuA8 [FhuA(wild type); w.t.], pfhuA5 [FhuA(C329S) Cys-318], pfhuA4 [FhuA(C318S) Cys-329], pfhuA6 [FhuA(D336C)], and pfhuA7 [FhuA(V347C)], as indicated. Cells were incubated for 30 min with 0.5 mM B-M at 30°C, and the proteins of whole cells were separated by SDS-PAGE. In addition, isolated FhuA(V347C) protein was heated for 3 min in 4% SDS and subsequently labeled with B-M (purified prot.). The proteins were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and stained with Ponceau-S-Red (B) and, after destaining and incubation with streptavidin-β-galactosidase, stained with X-Gal (A). The FhuA band is indicated by an arrow.
Determination of the degree of labeling of FhuA(D336C).
E. coli UL3 was transformed with plasmid pfhuA6 or pfhuA8. Cells were labeled with 0.5 mM B-M for 30 min at 30°C. As a control, E. coli UL3 (pfhuA8) was labeled with biotin-X-NHS (biotin amidocaproate N-hydroxysuccinimide ester; Sigma). The reagents were removed by washing the cells, and outer membranes were prepared. Streptavidin (20 μg) in 10 μl of 50 mM Tris-HCl (pH 6.8) was added to outer membranes corresponding to 4 μg of protein in 8 μl of buffer (control, buffer without streptavidin). After 30 min of incubation at 20°C, 18 μl of sample buffer was added and the proteins were dissolved during incubation for 5 min at 50°C. After SDS-polyacrylamide gel electrophoresis (PAGE) on a 9% polyacrylamide gel, the proteins were stained with Serva R and the amount of FhuA was determined by densitometry (2-D densitometer; Cybertech CS1, Berlin, Germany [software Wincam 2.2]).
FhuA labeling with F-M.
E. coli UL3 fhuA transformed with plasmids carrying the various fhuA mutant genes was grown in 20 ml of TY medium supplemented with ampicillin (30 μg per ml) to a cell density of 6 × 108 cells ml−1. Cells were washed three times with ice-cold, sterile-filtered PBS, suspended in 1 ml of 0.5 mM fluorescein-5-maleimide (F-M) (Molecular Probes Europe, Leiden, The Netherlands) dissolved in PBS–1% dimethylformamide, and incubated in darkness at 30°C for 0.5, 1.5, 5, and 15 min, after which the reaction was stopped by the addition of 20 mM DTT. The surplus F-M was removed by washing the cells five times with 1 ml of PBS. Cells were suspended in 1 ml of PBS and diluted 25-fold in PBS, and the fluorescence was determined in a flow cytometer (FACSort; Becton Dickinson, San Jose, Calif.) at an excitation wavelength of 488 nm and an emission wavelength of 530 nm. Each figure represents the mean value of 20,000 cells measured. The flow cytometer was adjusted such that only single cells, not cell aggregates or lysed cells, were counted.
Fluorescence quenching by ferrichrome.
E. coli UL3 transformed with plasmids pfhuA4 to pfhuA8 was labeled for 5 min at 30°C with F-M as described above. The labeled cells were then incubated for 10 min at 37°C in darkness, and the fluorescence was determined as described previously. Subsequently, ferrichrome (final concentration, 10 nM) or ferrioxamine B (1 μM) was added, and the fluorescence was determined. Fluorescence quenching of F–M-labeled cells of E. coli UL3 fhuA and E. coli HK99 fhuA tonB, each transformed with pfhuA6 [FhuA(D336C)], was determined with ferrichrome (1, 2.5, 5, 7.5, 10, 20, 50, and 100 nM) and albomycin (5, 10, 25, 50, and 100 nM).
Inhibition of labeling by ferrichrome.
To determine inhibition of labeling by ferrichrome, cells were incubated for 5 min with 100 μM ferrichrome at 37°C, prior to labeling for 5 min with 0.5 mM F-M in darkness.
Determination of the relative amounts of FhuA in TonB+ and TonB− cells.
The amount of FhuA was determined similarly, as described previously in the procedure of Moeck et al. (27). Cells of E. coli UL3 [FhuA(D336C)] and HK99 [FhuA(D336C)] were labeled with polyclonal FhuA antibodies raised in rabbits (13) and then incubated with affinity-purified fluorescein-labeled anti-rabbit immunoglobulin G (H and L chains) (Vector Laboratories Inc., Burlingame, Calif.). Fluorescence was determined by flow cytometry.
Phenotype assays.
Sensitivity of the E. coli fhuA mutants to the phages T1, T5, and φ80 and to colicin M and albomycin was tested on TY agar plates supplemented with ampicillin (30 μg per ml). The plates were seeded with 0.1 ml of an overnight culture grown in TY medium and suspended in 3 ml of TY low-melting agar (10 g per liter). Samples (4 μl) of 10-fold dilutions of the FhuA ligands were spotted onto the seeded plates. The plates were incubated overnight at 37°C, and the last dilution that resulted in a clear zone of growth inhibition was determined. Stimulation by ferrichrome was examined on agar plates containing the following (in grams liter−1): nutrient broth (8), NaCl (5), and ampicillin (0.03), pH 7, supplemented with 0.2 mM 2,2′-dipyridyl (12). Filter paper disks loaded with 10 μl of 0.01, 0.1, or 1.0 mM ferrichrome were placed onto the plates. After incubation overnight at 37°C, the sizes and densities of the growth zones were determined.
RESULTS
Specific labeling of FhuA.
We developed two methods to monitor the reactivity of cysteines at the cell surface of E. coli. Maleimide reacts specifically with sulfhydryl groups at the pH used (3, 4). With F-M, labeling could be determined by the very sensitive flow cytometry. F-M binding did not kill the cells, as determined by colony counting, and microscopic inspection did not reveal any changes in cell morphology. The second method involved the use of maleimide linked through a hexanoyl spacer to a biotinyl group (B-M). B-M labeling was determined with S-G, which strongly binds to biotin. No appreciable amounts of B-M diffused into the periplasm, since the periplasmic cysteines of the abundant OmpA protein were not labeled in cells after reduction of the disulfide bridge (1), but OmpA was labeled in isolated outer membranes (6).
All the FhuA derivatives used in this study conferred to the transformants wild-type sensitivity to the phages T1, T5, and φ80 and to colicin M and albomycin, and the growth zone of the transformants on nutrient agar dipyridyl plates around paper disks loaded with ferrichrome was as large as that of transformants carrying wild-type fhuA. Wild-type FhuA recovered from gels after SDS-PAGE contained the N-terminal amino acid sequence of mature FhuA (6). Activity of FhuA reflects proper insertion of FhuA in the outer membrane, an essential condition for interpreting the results of the labeling experiments.
The locations of the natural Cys-318 and Cys-329 in the gating loop and those of Cys-336 and Cys-347, which were newly inserted by replacing Asp-336 and Val-347, are illustrated in Fig. 1. Asp-336 is located in a region which serves as a binding site for the phages and colicin M, and Val-347 is close to Asp-348, whose deletion inactivates FhuA (18). E. coli Ul3 fhuA was transformed with plasmids which encode wild-type fhuA (Fig. 2, w.t.) or with fhuA mutants with single cysteine residues in the gating loop (as in Fig. 2), so that the FhuA derivatives could be labeled without exposing the cells to reducing conditions. Labeling with B-M and subsequently with S-G resulted in a single labeled band in each mutant (Fig. 2A). Transformants containing only the vector (Fig. 2B, pT7-6, no stained FhuA band), transformants with a plasmid encoding FhuA in which all four cysteines were replaced by serine (Fig. 2, No Cys), and transformants expressing wild-type FhuA (Fig. 2, w.t.) were not labeled with B-M–S-G. The electrophoretic mobility of the labeled band corresponded with FhuA(V347C) isolated from gels after SDS-PAGE (16) (Fig. 2B, V347C purified prot.). It also corresponded with the single band obtained by Western blotting with polyclonal anti-FhuA antibodies (data not shown). Labeling of purified FhuA(V347C) was enhanced fivefold after denaturation by heating in SDS (Fig. 2A). These experiments show that overexpressed FhuA containing a single cysteine at four different positions in the gating loop can be specifically labeled with B-M in viable cells. These conditions were used for the following experiments.
FIG. 1.
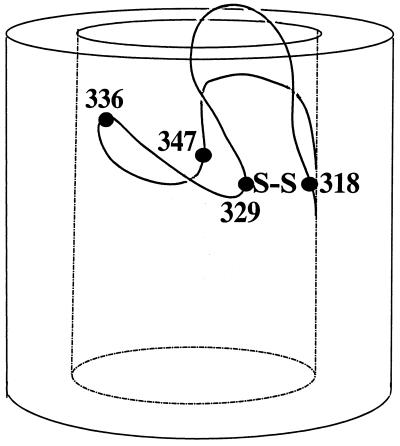
Amino acid sequence of the gating loop of FhuA in the outer membrane of E. coli, localized at the cell surface by determination of the proteolysis within genetically inserted peptides in cells and spheroplasts (21). The model illustrates the sites of the natural cysteines present at positions 318 and 329 in FhuA (○) and those of the newly inserted cysteines at sites 336 and 347 (□). The model does not imply that the surface loops (top) and periplasmic turns (bottom) extend from the outer membrane as drawn; rather, they are predicted to be highly folded (Fig. 8).
Labeling kinetics reveal different accessibilities of the gating loop cysteines.
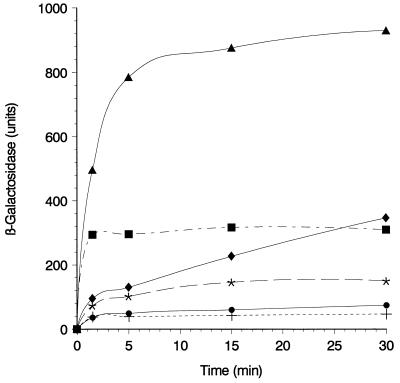
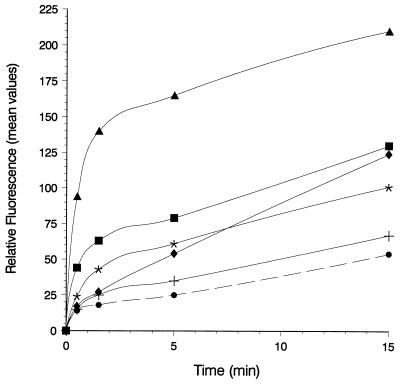
E. coli UL3 fhuA was transformed with plasmids carrying fhuA genes encoding a single cysteine residue in the gating loop. Cells were labeled with B-M for 1.5 to 30 min, after which they were incubated with S-G. Cys-318 in FhuA (C329S) displayed a low reactivity, while Cys-329 in FhuA (C318S) was more reactive (Fig. 3). Wild-type FhuA showed nearly no label, as did cells transformed with the vector. FhuA(C318S C329S, C692S, C698S) was not tested because of its low-level labeling, similar to that of wild-type FhuA (Fig. 2A and 4A). Cells synthesizing FhuA(Cys-336) were strongly labeled with B-M–S-G. Cells forming FhuA(Cys-347) were labeled faster than cells synthesizing the other FhuA gating loop derivatives, but the degree of labeling remained well below the level of Cys-336 labeling (Fig. 3).
FIG. 3.
Time course of specific labeling of gating loop cysteines with B-M in intact cells. Exponentially growing cells of E. coli UL3 fhuA transformed with plasmids pT7-6 (+), pfhuA8 [FhuA(wild type)] (•), pfhuA5 [FhuA(C329S) Cys-318] (∗), pfhuA4 [FhuA(C318S) Cys-329] (⧫), pfhuA7 [FhuA(V347C)] (▪), and pfhuA6 [FhuA(D336C)] (▴) were harvested, washed with PBS, and incubated with B-M (0.5 mM) for 1.5, 5, 15, and 30 min at 30°C. Labeling was interrupted by the addition of 0.02 M dithiothreitol. Cells were washed, treated with 2% bovine serum albumin, incubated with S-G, and washed again, and β-galactosidase activity was measured with o-nitrophenyl-β-d-galactoside (10) and related to the cell density (5% standard deviation).
FIG. 4.
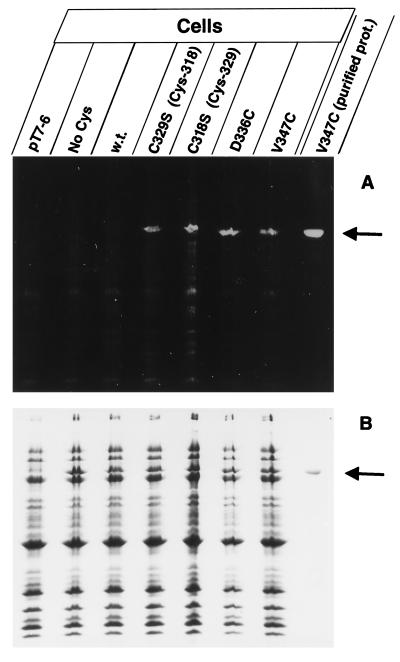
Labeling of the gating loop cysteines with F-M (A). Cells of E. coli UL3 fhuA were transformed with one of the following plasmids: pT7-6 (vector without fhuA), pfhuA9 [FhuA(C318S C329S C692S C698S); No Cys], pfhuA8 [FhuA(wild type); w.t.], pfhuA5 [FhuA(C329S) Cys-318], pfhuA4 [FhuA(C318S) Cys-329], pfhuA6 [FhuA(D336C)], and pfhuA7 [FhuA(V347C)], as indicated. Cells were incubated for 15 min with 0.5 mM F-M at 30°C in darkness, and the proteins of whole cells were separated by SDS-PAGE. In addition, purified protein of FhuA(V347C) was heated for 3 min in 4% SDS and subsequently labeled with F-M (purified prot.). Proteins were separated by SDS-PAGE, illuminated at 302 nm, and photographed with a Cybertech video camera (A). The proteins were then stained with Serva R to show that equal amounts were applied to the lanes (B).
The degree of FhuA(D336C) labeling was estimated by treating cells of E. coli UL3 transformed with plasmid pfhuA6 [FhuA(D336C)] or pfhuA8 [FhuA (wild type)] with B-M and then with streptavidin (33). Upon binding of streptavidin (60 kDa), the molecular mass of FhuA increased from 80 to 140 kDa. The outer membrane proteins were separated by SDS- PAGE and stained, and their amounts were determined by densitometry. About 60% of the total FhuA(D336C) remained at the electrophoretic position of unmodified FhuA (data not shown), meaning that about 40% ± 10% of the total FhuA(D336C) was labeled.
After treatment of FhuA(D336C) with biotin-X-NHS (which reacts with the much more abundant lysine residues) and streptavidin, the entire FhuA band disappeared (data not shown). Since biotin-X-NHS does not penetrate the cytoplasm, the complete labeling of FhuA(D336C) demonstrates that no FhuA(D336C) inclusion bodies remained in the cytoplasm.
FhuA can be specifically labeled by F-M.
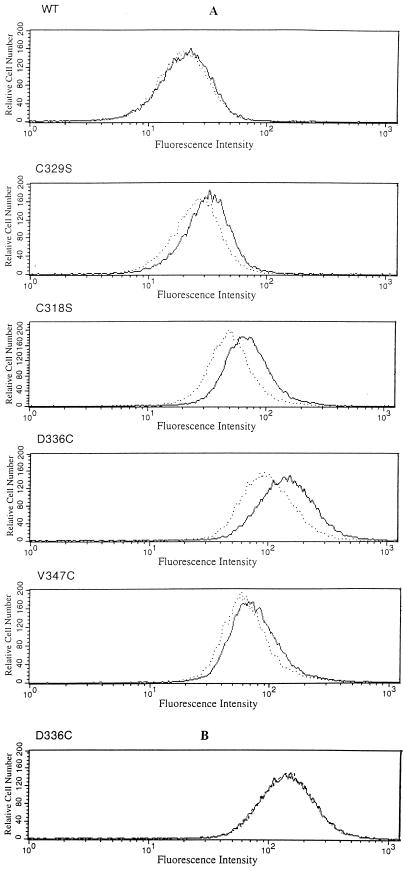
The results obtained with labeling by B-M–S-G were supported by results obtained by labeling FhuA in intact cells with F-M. Cells containing the vector (Fig. 4, pT7-6), FhuA with no cysteine (Fig. 4, No Cys), and wild-type FhuA (Fig. 4, w.t.) were not labeled; FhuA(C-318 C329S) was weakly labeled; FhuA(C-329 C318S) and FhuA(V347C) were more strongly labeled; and FhuA (D336C) was most strongly labeled. Labeling of purified FhuA (V347C) was enhanced fivefold after solubilization in SDS and heating. Care was taken that the same amounts of cells were loaded onto each lane of the gel, as shown in Fig. 4B.
Estimation of the labeling level by flow cytometry revealed a sevenfold-higher level of labeling of FhuA(D336C) than of wild-type FhuA (Fig. 5). The fluorescence increased during the entire incubation period, possibly indicating some nonspecific labeling, since F-M, with a molecular mass of 427 Da, may slowly diffuse through the outer membrane and bind to various cellular components, resulting in an unspecific increase in fluorescence. FhuA was the only single component which was labeled to an extent that could be detected on gels (Fig. 4A).
FIG. 5.
Flow cytometry of E. coli UL3 fhuA transformed with plasmids pT7-6 (vector without fhuA) (+), pfhuA8 [FhuA (wild type)] (•), pfhuA5 [FhuA(C329S) Cys-318] (∗), pfhuA4 [FhuA(C318S) Cys-329] (⧫), pfhuA7 [FhuA(V347C)] (▪), and pfhuA6 [FhuA(D336C)] (▴) after labeling with 0.5 mM F-M for 0.5, 1.5, 5, or 15 min at 30°C. The reaction was stopped by adding 20 mM DTT. The fluorescence values showed a standard deviation of 10%.
Ferrichrome binding causes a decrease in fluorescence intensity.
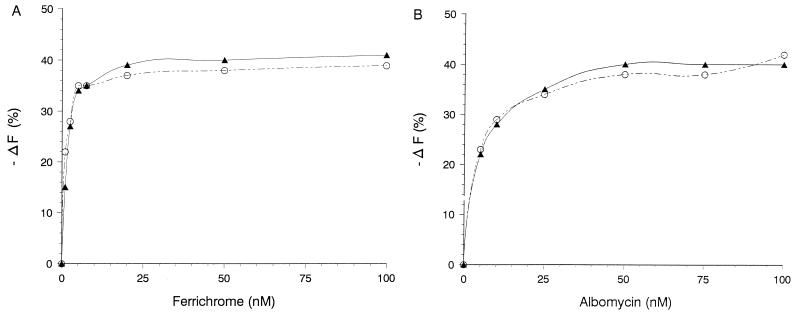
E. coli UL3 synthesizing FhuA(C329S), FhuA(C318S), FhuA(D336C), or FhuA(V347C) was labeled for 5 min with F-M. Fluorescence of the cells was measured in the flow cytometer and compared with the fluorescence upon addition of 10 nM ferrichrome after labeling. Ferrichrome quenched the fluorescence intensity most strongly in samples of F–M-labeled FhuA(Cys-336) and FhuA(Cys-329) (Fig. 6A, stippled line, with ferrichrome; solid line, without ferrichrome; and Table 2). The degree of fluorescence quenching was not determined by the degree of labeling, since V347C displayed the next-strongest labeling but nearly no quenching. Fluorescence quenching was not caused by a shift of the spectrum to lower or higher wavelengths, since there was no alteration of the excitation and emission spectra of bound F-M, which were measured with a spectrofluorimeter (data not shown). Ferrioxamine B, which does not bind to FhuA, caused no fluorescence quenching (Fig. 6B). The values calculated from the experiments shown in Fig. 6 are listed in Table 2. Ferrichrome did not quench fluorescence of free F-M in solution, which was tested at the concentrations used and at a 1,000-fold excess of ferrichrome over the F-M concentration (data not shown). These control experiments also showed that quenching was caused by ferrichrome bound to FhuA. There were no significant amounts of free ferrichrome in the samples, since ferrichrome binds very tightly to FhuA (KD, 0.05 to 0.1 μM) (25), and unbound ferrichrome was removed by the sheath fluid during flow cytometry.
FIG. 6.
Fluorescence quenching of F–M-labeled cells by ferrichrome. E. coli UL3 fhuA transformed with plasmid pfhuA8 [FhuA(wild type)], pfhuA5 [FhuA (C329S) Cys-318], pfhuA4 [FhuA(C318S) Cys-329], pfhuA6 [FhuA (D336C)], or pfhuA7 [FhuA(V347C)] was labeled with F-M. The fluorescence was determined by flow cytometry prior to (−) and after (…) the addition of 10 nM ferrichrome (A) or 1 μM ferrioxamine B {pfhuA6 [FhuA(D336C)]} (B).
TABLE 2.
Specific fluorescence quenching by ferrichrome
| FhuA | Relative fluorescence (mean values)a
|
||||
|---|---|---|---|---|---|
| Not treated with Fc | Treated with Fc | −ΔF (%) | FoB | −ΔF (%) | |
| Wild type | 48 | 45 | 6 | 50 | +2 |
| C329S (Cys-318) | 76 | 64 | 16 | 71 | 7 |
| C318S (Cys-329) | 97 | 70 | 27 | 100 | +3 |
| D336C | 405 | 262 | 35 | 385 | 5 |
| V347C | 154 | 141 | 8 | 145 | 6 |
−ΔF (%) indicates the decrease or increase (+) in the percentage of fluorescence for cells not treated or treated with 10 nM ferrichrome (Fc) or treated with 1 μM ferrioxamine B (FoB).
Determination of the concentration dependence of fluorescence quenching from 1 to 100 nM ferrichrome revealed maximum quenching between 10 and 20 nM ferrichrome (Fig. 7A, closed symbols). The same concentration dependence of ferrichrome quenching was obtained when the FhuA derivatives were synthesized in the E. coli HK99 tonB mutant (Fig. 7A, open symbols), indicating that the TonB-independent binding of ferrichrome to FhuA, not the TonB-dependent release of ferrichrome from FhuA and its transport through the FhuA channel, resulted in quenching. Incubation of the cells for 20 min at 37°C in a solution of 0.4% glucose, 20 mM HEPES buffer, 0.1 M NaCl, and 1 mM KCl (pH 7.2) to energize cells possibly deenergized during the washing procedure prior to the fluorescence measurements did not change the degree of quenching. Albomycin also caused fluorescence quenching, but the concentration of albomycin required to obtain the same degree of quenching as that with ferrichrome was about two to three times higher than the concentration needed for ferrichrome (Fig. 7B).
FIG. 7.
Fluorescence quenching of F–M-labeled TonB+ versus TonB− cells by ferrichrome (A) and albomycin (B). Cells of E. coli UL3 fhuA (▴) and E. coli HK99 fhuA tonB (○), each transformed with pfhuA6 [FhuA(D336C)], were labeled for 5 min with 0.5 mM F-M and then incubated with ferrichrome (A) and albomycin (B) at the concentrations indicated. Fluorescence was determined by flow cytometry.
Quenching could indicate either a conformational change of FhuA induced by ferrichrome binding or a reduction in the fluorescence intensity caused by the extreme closeness of ferrichrome to F-M at the various amino acid side chains. In the latter case, ferrichrome should inhibit labeling by F-M. This hypothesis was tested by incubating E. coli HK99 FhuA(C329S), FhuA(C318S), FhuA(D336C), or FhuA(V347C) for 5 min with 100 μM ferrichrome prior to labeling with F-M. F-M labeling of FhuA(D336C) in the presence of ferrichrome was as high as F-M labeling in the absence of ferrichrome (Table 3), suggesting that ferrichrome-induced quenching of F-M bound to FhuA(D336C) is caused by a conformational change that occurs in FhuA upon binding of ferrichrome.
TABLE 3.
Inhibition of cell labeling by ferrichrome
| FhuA | Relative fluorescence (mean values)a
|
||
|---|---|---|---|
| Not preincubated with Fc | Preincubated with Fc | −δF (%) | |
| C329S (Cys-318) | 45 | 38 | 16 |
| C318S (Cys-329) | 61 | 51 | 16 |
| D336C | 196 | 186 | 5 |
| V347C | 90 | 52 | 42 |
−δF (%) indicates the decrease in F-M labeling for cells not preincubated with 100 μM ferrichrome (Fc) (standard deviation, 10%).
Inhibition of Cys-347 labeling suggests a ferrichrome binding site.
In contrast to F-M labeling of cells synthesizing FhuA(D336C), F-M labeling of FhuA(V347C) was inhibited by ferrichrome. Incubation of cells with ferrichrome for 5 min prior to the addition of F-M reduced F-M labeling by 42% ± 10% (mean ± standard deviation) (Table 3). Labeling of cells that synthesized FhuA(C329S Cys-318 is free for labeling) or FhuA(C318S Cys-329 is free) was somewhat reduced (16%) by ferrichrome. It is concluded that Cys-347 is part of or close to the binding site of ferrichrome.
Cysteine labeling of E. coli UL3 tonB+ cannot be distinguished from cysteine labeling of E. coli HK99 tonB.
To examine whether different FhuA conformations in TonB+ and TonB− cells can be determined by cysteine labeling, transformants of E. coli UL3 and its tonB derivative HK99, both of which synthesized FhuA(D336C), were labeled with F-M. Flow cytometric analysis was done with 20,000 cells of each sample. The rate of labeling and the final yield was 10% higher in the TonB− cells than in the TonB+ cells. This result could be accounted for by the 10%-higher FhuA content in the TonB− cells, as determined by immunofluorescence with anti-FhuA antibodies, presumably caused by derepression of fhuA transcription due to iron limitation of the TonB− cells (data not shown).
DISCUSSION
The two methods described in this paper can be applied to any surface-exposed protein of E. coli K-12 and probably to other gram-negative bacteria. Naturally occurring cysteines as well as cysteines inserted by site-directed mutagenesis can be used for specific labeling. In all experiments, FhuA was moderately overproduced to the same extent to obtain high FhuA labeling relative to background labeling. Cells tolerate experimentally increased amounts of FhuA, as strongly increased amounts are formed under iron-limiting growth conditions. Overproduced FhuA derivatives containing foreign peptides inserted in loops at the cell surface are completely degraded by proteases added to cells (21), indicating that no appreciable amounts of FhuA remain in the cytoplasm as inclusion bodies. Overproduced FhuA did not disturb the outer membrane integrity, allowing entry of the reagents into the cells, since sensitivity of cells to SDS and bacitracin was unchanged and cells lacking LamB did not grow on maltodextrins (16, 17).
Both reagents, B-M and F-M, labeled the natural cysteines in the gating loop, but only after reduction (6) or when disulfide formation was prevented by removing one cysteine residue, as was done in this study. This result indicates that Cys-318 and Cys-329 are so close that they form a disulfide bridge (Fig. 8). In addition, both cysteine residues were not readily accessible, since they reacted slowly and incompletely with the cysteine reagents (final yield, 15%). The disulfide bridge seems to be largely buried in the gating loop.
FIG. 8.
Illustration of the natural disulfide bridge between residues 318 and 329 and of the cysteine residues introduced at sites 336 and 347 of the FhuA gating loop. The model takes into account the relatively high reactivity of Cys-336 by localizing it close to the cell surface, the lower reactivity of Cys-347, and the low reactivity of Cys-318 and Cys-329. The ferrichrome binding site is close to Cys-347. This model is not intended to predict the conformation of the gating loop.
The strong labeling of Cys-336 with B-M, which in the first 5 min of incubation was about sevenfold higher than that of the native gating loop cysteines, probably indicates a greater accessibility rather than a higher chemical reactivity of the sulfhydryl group, which depends on the local charge distribution around the cysteine residue. Under the conditions used, about 40% of the total FhuA synthesized was labeled, as determined by the decrease of the FhuA band on an SDS-polyacrylamide gel after B-M–streptavidin labeling. Also, Cys-336 does not seem to be freely accessible, or not all of the FhuA molecules in the outer membrane may be reactive. A similar observation was made with Cys-347, which was rapidly but incompletely labeled. Cys-347 was only strongly labeled after heat denaturation in SDS. An incomplete and a changing reactivity was demonstrated with the outer membrane BtuB protein. At the nonpermissive temperature, btuB amber mutants containing a temperature-sensitive suppressor became insensitive first to colicin E3 and then to phage BF23 but transported vitamin B12 much longer. The colicin E3-insensitive cells still bound colicin E3, which protected cells against phage BF23 (2).
Replacement of Asp-336 and Val-347 by cysteine had no effect on the function of FhuA. From this result, it cannot be concluded that both sites or the entire region is unimportant for FhuA activity. Deletion of Asp-348 rendered cells resistant to the phages T1 and φ80 and to albomycin, reduced sensitivity to colicin M 1,000-fold and sensitivity to phage T5 10-fold, and impaired growth on ferrichrome (18). Replacement of Asp-348 and Asp-349 by Tyr and Glu-350 by Val reduced sensitivity of cells only to colicin M and albomycin 10- to 100-fold and reduced growth on ferrichrome (18). Excision of residues 335 to 355 led to inactivation of FhuA with all the ligands, while excision of residues 322 to 336 allowed the retention to some extent of FhuA activity for the uptake of ferrichrome and colicin M and the retention of a very weak activity for infection by phages T5 and φ80 (17). Synthetic acetylated hexapeptide amides identical in sequence to this region inhibited infection by phages T5, T1, and φ80 and killing by colicin M (19). However, insertion of foreign tetrapeptides after residue 338 did not impair FhuA activity with ferrichrome, albomycin, colicin M, and the phages T5, T1, and φ80 (21). It seems that FhuA activity is affected more by deletions than by insertions. Deletions may influence FhuA conformation much more than insertions, since the missing amino acid is difficult to compensate for but the inserted peptides may form loops which leave the conformation largely intact. In FepA, single amino acid substitutions in a surface-exposed region did not diminish adsorption and transport of ferric enterobactin, but double substitutions identified two arginine residues that participate in binding of ferric enterobactin and colicins B and D (30). These data demonstrate the difficulty of assigning binding and transport functions to certain amino acids in the gating loop. Rather, these data suggest that a larger number of residues, not all necessarily located in the gating loop, are directly involved in binding and translocation of the various FhuA ligands and/or are important for the active conformations of FhuA.
At the very low concentration of 10 nM ferrichrome, the fluorescence of F–M-labeled FhuA(Cys-336) and FhuA(Cys-329) decreased by 35 and 27%, respectively. It is unlikely that ferrichrome, a rather small molecule (740 Da), came so close to the fluorescence-labeled Cys-329, Cys-336, and Cys-318 residues that a direct interaction with the label caused the decrease in fluorescence. The Cys residues are presumably too far apart and differently oriented for energy to transfer between fluorescein and ferrichrome at its specific binding site. This conclusion is supported by the failure of ferrichrome to inhibit labeling of Cys-336 by F-M. Therefore, fluorescence quenching most likely indicates a conformational change of the gating loop upon ferrichrome binding that affects mostly Cys-329 and Cys-336. The degree of quenching depends on the environment into which fluorescein is moved. A conformational change of FhuA induced by ferrichrome has also been deduced from in vitro studies with isolated FhuA in which ferrichrome inhibits cleavage of FhuA by trypsin (13). These studies were recently extended by showing protection by ferrichrome of Lys-67, but not of Lys-5, against trypsin cleavage (29). The latter study also demonstrated a ferrichrome-induced conformational change of FhuA in that anti-FhuA monoclonal antibodies that react with the periplasmic FhuA sequence 21–59 bind less to FhuA in the presence of ferrichrome than in its absence. Ferrichrome does not alter binding of FhuA antibodies that react with cell surface-exposed regions of FhuA, including the gating loop. Apparently, the antibodies do not recognize the conformational changes caused by ferrichrome in the gating loop. Circular dichroism and Fourier transform infrared spectroscopy with isolated FhuA have revealed, if any, only slight changes in FhuA secondary structure upon binding of ferrichrome, suggesting a low overall or a local conformational change of FhuA. The data presented in this study indicate a conformational change in the gating loop, which, besides the TonB box (11, 38), is an important site for FhuA activity.
Incubation of cells with ferrichrome prior to labeling with F-M decreased labeling only of Cys-347 to a significant extent, indicating that this site is part of or close to the ferrichrome binding site. The fluorescence of F–M-labeled Cys-347 changed only slightly upon addition of ferrichrome after labeling, in contrast to Cys-318, Cys-329, and Cys-336, which showed a stronger fluorescence quenching. The different degree of B-M and F-M labeling, fluorescence quenching, and labeling inhibition of the four cysteines indicates that the gating loop cannot be considered as a single structural entity but, rather, consists of subregions.
The same degree of F-M labeling of FhuA(C-336) was observed in TonB+ and TonB− cells, a result which was not unexpected if one assumes that at a given time, only a few FhuA channels are opened through the Ton system at the expense of the electrochemical potential of the cytoplasmic membrane (16). If sufficient iron is provided in the medium, the FhuA copy number is approximately 103. About 105 ferrichrome molecules are taken up per cell per generation, meaning that 100 molecules flow through a single FhuA channel in 30 min, provided all FhuA channels are active. From Vmax transport values and 100,000 FepA molecules per cell formed under iron-limiting growth conditions, it was calculated that 30 ferric enterobactin molecules are transported by a FepA monomer in 30 min (14). These transport rates are rather low, suggesting that at a given time, only a few channels are active and that these channels transport ferrichrome and ferric enterobactin much faster than the calculation suggests, based on the assumption that all receptors are active. Additional considerations favor the presence of mostly closed FhuA channels. The number of FhuA molecules under iron limitation is much higher than the number of TonB molecules. In addition, TonB energizes transport not only through a single outer-membrane receptor protein but also through several receptor proteins. Competition among receptors for TonB has been concluded from the inhibition of vitamin B12 transport by ferrichrome transport and vice versa (15). Mutual inhibition of vitamin B12 and ferrichrome transport was overcome by increased synthesis of TonB (15). Furthermore, the diffusion rate of KCl through an FhuA mutant protein lacking the gating loop is at least threefold higher than the diffusion rate through the OmpF pore (16). A permanently open FhuA channel would allow harmful substances to permeate the outer membrane.
Transport of ferrichrome through FhuA is envisaged as consisting of at least two steps: binding of ferrichrome to FhuA, which changes the conformation of FhuA, and then (energized by electrochemical potential of the cytoplasmic membrane) the opening of the FhuA channel and the release of ferrichrome into the periplasm. The ferrichrome-triggered conformational change of FhuA may facilitate interaction of FhuA with TonB (15, 29). Lack of vitamin B12 transport inhibition by free FhuA not occupied with ferrichrome suggests that unliganded FhuA binds less TonB or no TonB (15).
Recently, it was shown that isolated FhuA incorporated into lipid bilayer membranes opens a channel upon binding of phage T5 (5). The conductance of the channels formed resembles the conductance of FhuA Δ322–355 (5). In addition, T5 DNA was transferred inside the lipid vesicles (31). Ferrichrome trapped in the vesicles was released upon binding of T5 to FhuA (22). These very important in vitro data strongly support the view that FhuA forms a closed channel that is opened upon binding of phage T5 through which ferrichrome and, presumably, also the phage DNA are translocated across the outer membrane (see reference 5 for a detailed discussion of the possible molecular events underlying channel opening by T5). Binding of T5 opens the FhuA channel, but binding of ferrichrome opens the FhuA channel only through the action of the Ton system and the electrochemical potential of the cytoplasmic membrane. These events seem to be entirely different, but spontaneous host range mutants of the phages T1 and φ80 no longer require both an energized cytoplasmic membrane and the Ton system to infect cells (8). T5 and the T1 and φ80 host range phages can provide the conformational energy to open FhuA because they undergo large conformational changes upon binding to FhuA, triggering the release of DNA from the phage heads. These are individual events, and the phages die afterward. Ferrichrome cannot provide the conformational energy because it is transported unchanged into the periplasm. For this reason, FhuA must be opened by other means. The same reasoning applies to albomycin and may be true for colicin M and microcin 25. Wild-type T1 and φ80 exhibit the favorable properties of being physically more stable than the host range mutants and infecting only energized, actively growing cells which guarantee phage multiplication.
Studies on ferric enterobactin transport across the outer membrane (34) resulted in a model which is similar to the model for ferrichrome transport. Mutationally introduced cysteines at residues 280 and 310 of FepA reacted with spin labels added to the isolated protein. These spin labels were used to monitor a TonB-independent conformational change upon binding of ferric enterobactin (24). Sites 280 and 310 are located in the region whose excision (residues 202 to 340) results in a FepA derivative that forms an open channel in cells (35) and liposomes (23). Continuous electron spin resonance spectroscopy of a nitroxide spin label at Cys-280 of FepA in intact cells revealed strong motion during transport of ferric enterobactin. Since these spectral changes were not observed in a tonB mutant, under glucose deprivation or at low temperature, it was concluded that the motion of the spin label reflected energy and TonB-dependent conformation changes of FepA during ferric enterobactin transport (14). In contrast to ferric enterobactin, uptake of colicin B was associated with an immobilization of the FepA spin label, suggesting a steric restriction of the spin label motion as the colicin passes through the FepA pore (14). It was concluded that FepA fluctuates between at least two conformations during ferric enterobactin transport and that colicin uptake causes a different type of conformational dynamic (14).
The FhuA derivatives constructed in this study and the knowledge of their reactivity may be helpful for determining the three-dimensional structure of FhuA by X-ray analysis. This analysis requires heavy metal isomorphous replacements, for which cysteines are particularly suitable. The methods introduced in this study can also be used to determine the transmembrane topology of outer membrane proteins and to examine existing models.
ACKNOWLEDGMENTS
We thank H. Killmann for strains, plasmids, and experimental advice; T. F. Meyer for the use of the flow cytometer; G. Döring for the use of the protein densitometer; and K. A. Brune for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 323, Graduiertenkolleg Mikrobiologie fellowship to C.B.) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Bardwell J C A, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 2.Bassford P J, Jr, Kadner R J, Schnaitman C A. Functional stability of the bfe and tonB gene products in Escherichia coli. J Bacteriol. 1977;130:750–758. doi: 10.1128/jb.130.2.750-758.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer E A, Safars M, Wilchek M. Selective labeling of sulfhydryls and disulfides on blot transfers using avidin-biotin technology: studies on purified proteins and erythrocyte membranes. Anal Biochem. 1987;161:262–271. doi: 10.1016/0003-2697(87)90450-7. [DOI] [PubMed] [Google Scholar]
- 4.Bayer E A, Zalis M G, Wilchek M. 3-(N-Maleimido-propionyl) biocytin: a versatile thiol-specific biotinylating reagent. Anal Biochem. 1985;149:529–536. doi: 10.1016/0003-2697(85)90609-8. [DOI] [PubMed] [Google Scholar]
- 5.Bonhivers M, Ghazi A, Boulanger P, Lettelier L. FhuA, a transporter of the Escherichia coli outer membrane, is converted into a channel upon binding of bacteriophage T5. EMBO J. 1996;15:1850–1856. [PMC free article] [PubMed] [Google Scholar]
- 6.Bös C, Braun V. Specific in vivo thiol-labeling of the FhuA outer membrane ferrichrome transport protein of Escherichia coli K-12: evidence for a disulfide bridge in the predicted gating loop. FEMS Microbiol Lett. 1997;153:311–319. doi: 10.1111/j.1574-6968.1997.tb12590.x. [DOI] [PubMed] [Google Scholar]
- 7.Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Lett. 1996;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 8.Braun V, Schaller K, Wolff H. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli. Biochim Biophys Acta. 1973;323:87–97. doi: 10.1016/0005-2736(73)90433-1. [DOI] [PubMed] [Google Scholar]
- 9.Coulton J W, Mason P, Cameron D R, Carmel G, Jean R, Rode H N. Protein fusions of β-galactosidase to the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1986;165:181–192. doi: 10.1128/jb.165.1.181-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giacomini A, Corich V, Ollero F J, Suqartini A, Nuti M P. Experimental conditions may affect reproducibility of the β-galactosidase assay. FEMS Microbiol Lett. 1992;100:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- 11.Günter K, Braun V. In vivo evidence for FhuA outer membrane receptor interaction with the TonB inner membrane protein of Escherichia coli. FEBS Lett. 1990;274:85–88. doi: 10.1016/0014-5793(90)81335-l. [DOI] [PubMed] [Google Scholar]
- 12.Hantke K. Regulation of the ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann H, Fischer E, Schwarz H, Braun V. Overproduction of the proFhuA outer membrane receptor protein of Escherichia coli K-12: isolation, properties, and immunocytochemical localization at the inner side of the cytoplasmic membrane. Arch Microbiol. 1986;145:334–341. doi: 10.1007/BF00470867. [DOI] [PubMed] [Google Scholar]
- 14.Jiang X, Payne M A, Cao Z, Foster S B, Feix J B, Newton S M C, Klebba P E. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- 15.Kadner R J, Heller K J. Mutual inhibition of cobalamin and siderophore uptake systems suggests their competition for TonB function. J Bacteriol. 1995;177:4829–4835. doi: 10.1128/jb.177.17.4829-4835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Killmann H, Benz R, Braun V. Conversion of the FhuA transport protein into a diffusion channel through the outer membrane of Escherichia coli. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Killmann H, Benz R, Braun V. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J Bacteriol. 1996;178:6913–6920. doi: 10.1128/jb.178.23.6913-6920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Killmann H, Braun V. An aspartate deletion mutation defines a binding site of the multifunctional FhuA outer membrane receptor of Escherichia coli. J Bacteriol. 1992;174:3479–3486. doi: 10.1128/jb.174.11.3479-3486.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Killmann H, Videnov G, Jung G, Schwarz H, Braun V. Identification of receptor binding sites by competitive peptide mapping: phages T1, T5, and φ80 and colicin M bind to the gating loop. J Bacteriol. 1995;177:694–698. doi: 10.1128/jb.177.3.694-698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klebba P E, Rutz J M, Liu J, Murphy C L. Mechanisms of TonB-catalyzed iron uptake through the enteric bacterial cell envelope. J Bioenerg Biomembr. 1993;25:603–611. doi: 10.1007/BF00770247. [DOI] [PubMed] [Google Scholar]
- 21.Koebnik R, Braun V. Insertion derivatives containing segments of up to 16 amino acids identify surface- and periplasm-exposed regions of the FhuA outer membrane receptor of Escherichia coli K-12. J Bacteriol. 1993;175:826–839. doi: 10.1128/jb.175.3.826-839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lettelier L, Locher K P, Plancon L, Rosenbusch J. Modeling ligand-gated receptor activity. FhuA-mediated ferrichrome efflux from vesicles triggered by phage T5. J Biol Chem. 1997;272:1448–1451. doi: 10.1074/jbc.272.3.1448. , 8836. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Rutz J M, Feix J B, Klebba P E. Permeability properties of a large gated channel within the ferric enterobactin receptor, FepA. Proc Natl Acad Sci USA. 1993;90:10653–10657. doi: 10.1073/pnas.90.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Rutz J M, Klebba P E, Feix J B. A site-directed spin-labeling study of ligand-induced conformational changes in the ferric enterobactin receptor, FepA. Biochemistry. 1995;33:13274–13283. doi: 10.1021/bi00249a014. [DOI] [PubMed] [Google Scholar]
- 25.Locher K, Rosenbusch J P. Oligomeric states and siderophore binding of the ligand-gated FhuA protein that forms channels across Escherichia coli outer membranes. Eur J Biochem. 1997;247:770–775. doi: 10.1111/j.1432-1033.1997.t01-1-00770.x. [DOI] [PubMed] [Google Scholar]
- 26.Lundrigan M D, Lancaster J H, Earhart C F. UC-1, a new bacteriophage that uses the TonA polypeptide as its receptor. J Virol. 1983;45:700–707. doi: 10.1128/jvi.45.2.700-707.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moeck G S, Bazzaz S F, Gras M F, Ravi T S, Ratcliffe M J H, Coulton J W. Genetic insertion and exposure of a reporter epitope in the ferrichrome-iron receptor of Escherichia coli. J Bacteriol. 1994;176:4250–4259. doi: 10.1128/jb.176.14.4250-4259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moeck G S, Ratcliffe M J H, Coulton J W. Topological analysis of the Escherichia coli ferrichrome-iron receptor by using monoclonal antibodies. J Bacteriol. 1995;177:6118–6125. doi: 10.1128/jb.177.21.6118-6125.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moeck G S, Tawa P, Xiang H, Ismail A A, Turnbull J L, Coulton J W. Ligand-induced conformational change in the ferrichrome-iron receptor of Escherichia coli K-12. Mol Microbiol. 1996;22:459–471. doi: 10.1046/j.1365-2958.1996.00112.x. [DOI] [PubMed] [Google Scholar]
- 30.Newton S M C, Allen J S, Cao Z, Jiang X, Sprencel C, Igo J D, Foster S B, Payne M A, Klebba P E. Double mutagenesis of a positive charge cluster in the ligand-binding site of the ferric enterobactin receptor, FepA. Proc Natl Acad Sci USA. 1997;94:4560–4565. doi: 10.1073/pnas.94.9.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plancon L, Chami M, Lettelier L. Reconstitution of FhuA, an Escherichia coli outer membrane protein, into liposomes. Binding of phage T5 to FhuA triggers transfer of DNA into the proteoliposomes. J Biol Chem. 1997;272:16868–16872. doi: 10.1074/jbc.272.27.16868. [DOI] [PubMed] [Google Scholar]
- 32.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr. 1993;25:5591–5601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 33.Qiu X Q, Jakes K S, Finkelstein A, Slatin S L. Site specific biotinylation of colicin A. J Biol Chem. 1994;269:7483–7488. [PubMed] [Google Scholar]
- 34.Rutz J M, Abdallah T, Singh S, Kalve V I, Klebba P E. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J Bacteriol. 1991;173:5964–5974. doi: 10.1128/jb.173.19.5964-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rutz J M, Liu J, Lyons J A, Goranson J, Armstrong S K, McIntosh M A, Feix J B, Klebba P E. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science. 1992;258:471–475. doi: 10.1126/science.1411544. [DOI] [PubMed] [Google Scholar]
- 36.Salomon R A, Farias R N. The peptide antibiotic microcin 25 is imported through the TonB pathway and the SbmA protein. J Bacteriol. 1995;177:3323–3325. doi: 10.1128/jb.177.11.3323-3325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanger F, Coulson A R, Barrel B G, Smith A J H, Roe B A. Cloning in a single-stranded bacteriophage as an aid of rapid DNA sequencing. J Mol Biol. 1980;143:161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- 38.Schöffler H, Braun V. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol Gen Genet. 1998;217:378–383. doi: 10.1007/BF02464907. [DOI] [PubMed] [Google Scholar]
- 39.Tabor S, Richardson C C. A bacteriophage T7 polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;182:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]