Abstract
Background
Charcot-Marie-Tooth type 1A (CMT1A), the most frequent type of Charcot-Marie-Tooth disease, is mainly caused by a 1.4-Mb duplication containing the PMP22 gene. There is no effective treatment other than general supportive care and symptomatic treatment. Preimplantation genetic testing for monogenic defects (PGT-M) is an alternative approach for obtaining healthy babies.
Methods
A new technology and analysis method based on next-generation sequencing (NGS) was developed to detect duplication mutations directly. Simultaneously, aneuploidy and linkage analyses were performed to achieve a comprehensive and accurate embryo diagnosis.
Results
Eight couples were recruited in this study; PMP22 duplication was validated in seven couples, and PMP22 splicing mutation was found in one. Forty-five embryos from 12 PGT cycles were successfully detected using this novel method. The direct detection results for all embryos were consistent with the linkage analyses, suggesting a 100 % accuracy rate, and the aneuploidy rate of the biopsied blastocysts was 33.3 %. Eventually, 18 of the 45 diagnosed embryos were deemed suitable for transfer. Four healthy babies from three families were delivered and their genetic status confirmed by amniocentesis. Additionally, there were no adverse effects of anesthesia or increased pregnancy complications during PGT-M in female patients with CMT1A.
Conclusions
This study provided a simple, reliable, and efficient method that can directly detect PMP22 mutations based on NGS data and does not require positive family members. A clinical workflow for CMT1A interruption in the offspring before embryo implantation is also summarized.
Keywords: PMP22, Charcot-Marie-Tooth type 1A, Preimplantation genetic testing, Clinical application
1. Introduction
Charcot-Marie-Tooth neuropathy (CMT) is the most frequently inherited peripheral nervous system disease with high phenotypic and genetic heterogeneity. It is characterized by progressive weakness, atrophy of the distal symmetrical muscle, and sensory loss [1]. Recent epidemiological studies have reported that the overall prevalence of CMT is 1/2500 to 1/10,000 worldwide [[2], [3], [4], [5]]. CMT type 1A (CMT1A; OMIM:118220) is the most common type and accounts for almost 50 % of all diagnosed CMT cases [6]. It features reduced conduction velocity, dys- or demyelination, and onion bulb formation in both motor and sensory nerves. The typical clinical manifestations of CMT1A are usually present in the first or second decade of life and include foot deformities (such as pes cavus and hammertoes), walking and running difficulties, and other classical symptoms [7,8]. The clinical diagnosis of CMT1A relies mainly on physical examination, electrophysiological evaluation, and family history [7].
With the evolution of molecular technology and investigation of pathogenesis, the causative genes have been identified. Approximately 98 % of CMT1A cases are caused by a 1.4-Mb duplication in the chromosome 17p11.2–12 region, which includes the peripheral myelin protein 22 (PMP22) gene. PMP22 encodes a 22-kDa intrinsic tetraspan transmembrane glycoprotein essential for the growth, differentiation, myelin assembly, and thickness maintenance of Schwann cells (SCs) [9,10]. PMP22 duplication leads to intracellular aggregation of the PMP22 protein in the endoplasmic reticulum, eventually resulting in apoptosis and demyelination of SCs [[11], [12], [13]]. Patients with CMT1A generally have a family history of polyneuropathy with chronic motor difficulty and sensory abnormalities and exhibit an autosomal dominant inheritance mode. However, a small proportion (approximately 10 %) of CMT1A patients are de novo or asymptomatic family members [14]. Both clinical and genetic diagnoses are crucial for the diagnosis of CMT1A. Most patients with CMT1A are initially diagnosed following typical presenting symptoms (including weakness of the feet and ankles). They are clinically diagnosed with CMT disease after general physical examination and electrophysiological evaluation. Genetic diagnosis helps identify subtypes, guide treatment, and facilitate genetic counseling and offspring interruption.
Traditional detection methods for PMP22 duplications include fluorescence in situ hybridization (FISH), restriction fragment length polymorphism-polymerase chain reaction (RFLP-PCR), competitive multiplex PCR, and multiplex ligation-dependent probe amplification (MLPA) [15,16]. Among the techniques, MLPA is a routinely used, sensitive, specific, and accurate method [17,18]. To date, there are no effective treatments for CMT1A. In the previous decade, gene and drug-based therapy have been the main treatment strategies developed to maintain or delay disease progression [[19], [20], [21]]. Although CMT1A is usually slowly progressive, patients experience sensory loss, hand, foot, and leg atrophy, and eventually rely on wheelchairs, decreasing the quality of life and burdening the family. For these families, preimplantation genetic testing for monogenic disease (PGT-M) offers an alternative approach to prevent the transmission of CMT1A and ensure the outcome of having a healthy baby [22]. For PGT-M, unaffected embryos are transferred to the uterus after intracytoplasmic sperm injection (ICSI), embryo biopsy, and targeted detection procedures [23]. Compared with prenatal interruption methods, PGT-M can prevent pregnant women from experiencing physical and psychological distress when fetuses are diagnosed with CMT1A. In particular, PGT-M shows distinct superiority in protecting the health of women with CMT1A.
A few studies have reported no significant increase in pregnancy complications in pregnant patients with CMT1A compared to the general population [[24], [25], [26]]. PGT-M has been used in several couples to interrupt the transmission of CMT1A using fluorescent PCR amplification of short tandem repeat (STR) polymorphic markers or karyomapping combined with genome-wide linkage analyses [22,23,27]. In summary, all current strategies rely on linkage analysis to identify the status of PMP22 duplication in the embryos rather than directly detecting the copy numbers of the PMP22 region in embryos. PMP22 duplication detection methods in adults, such as FISH and MPLA, are unusable for embryo detection because of the low amount of genetic material that cannot meet the detection requirements. Linkage analysis requires positive family members to help construct mutant haplotypes, which is not applicable for de novo patients with CMT1A. Furthermore, chromosomal exchanges in or adjacent to the mutation could make the diagnosis ambiguous, resulting in the formation of unaffected embryos. PMP22 duplication linkage analysis is more complicated and might be affected by interference with triple copy numbers, low resolution, and a high risk of misdiagnosis.
Many problems arise from relying only on linkage analysis for PMP22 duplication diagnosis in embryos. Therefore, we developed a novel method that could directly detect the copy numbers of the targeted 1.4-Mb region to solve the difficulties mentioned above. In this study, a new technology and analysis method was developed to directly detect copy numbers of the targeted 1.4-Mb region, combined with linkage analysis to confirm the accuracy and stability of the diagnosis in embryos. Compared to current methods, this NGS-based method is simple, reliable, cost-effective and does not require additional experimental steps (such as separate amplification of STR). We applied this technology and analyzed for eight families. To date, four healthy babies from three families have been born.
2. Materials and methods
2.1. Patients
Eight families diagnosed with CMT1A were recruited at the Center of Reproductive Medicine of Peking University Third Hospital between November 2017 and June 2022. The clinical diagnosis of CMT is based on family history and physical examination for peripheral neuropathy [28]. The pedigrees of the eight families are shown in Supplementary Fig. 1. Genetic testing showed that seven patients among the 7 recruited couples carried a PMP22 duplication mutation, and one patient in case 4 had a splicing mutation (c.319+1G > A) in the PMP22 gene. The basic information of the eight cases is summarized in Table 1. This study was approved by the Medical Science Research Ethics Committee of the Peking University Third Hospital (2018S2-002).
Table 1.
Summary of basic information and IVF-PGT cycles.
| Case ID | Female age | Male age | Patient | Clinical manifestations | PMP22 mutation | Cycles | Oocyte retrieved | Fertilized oocytes | Embryos for PGT-M |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 30 | 33 | Female | Pes cavus Lower leg atrophy |
Duplication on Exon 1_5 | 2 | 23 | 15 | 8 |
| 2 | 29 | 31 | Female | Pes cavus Feet weakness Tripping easily when walking |
Duplication on Exon 1_5 | 1 | 25 | 15 | 6 |
| 3 | 26 | 26 | Male | Pubertal onset Lower leg weakness and atrophy |
Duplication on Exon 1_5 | 1 | 13 | 9 | 6 |
| 4 | 35 | 36 | Male | Leg and hands weakness and atrophy Tripping easily when walking |
PMP22,c.319+1G > A | 1 | 27 | 19 | 6 |
| 5 | 38 | 40 | Female | Mild feet weakness | Duplication on Exon 1_5 | 1 | 8 | 3 | 0 |
| 6 | 38 | 37 | Female | Pes cavus Poor balance and finger function |
Duplication on Exon 1_5 | 3 | 24 | 18 | 7 |
| 7 | 30 | 32 | Female | Distal muscle weakness Atrophy of leg, hip and forearm |
Duplication on Exon 1_5 | 2 | 11 | 8 | 3 |
| 8 | 32 | 33 | Female | Clubfoot Lower leg weakness |
Duplication on Exon 1_5 | 1 | 38 | 20 | 9 |
2.2. Validation of the mutation and genetic counseling
Peripheral blood was collected from the patients and related family members to validate the PMP22 mutation. Genomic DNA was extracted from peripheral blood leukocytes or fetal tissues using a DNeasy Blood & Tissue Kit (Qiagen, Germany). An MLPA assay was performed to detect cases with PMP22 duplications using the commercial Probemix P405 CMT1 MLPA® kit (MRC-Holland, The Netherlands) according to the manufacturer's protocol. These probes covered the CMT/HNPP 17p12 region. Specifically, the husband of case 4 carried a de novo splicing mutation in PMP22; therefore, a single sperm was used to detect the mutation for the subsequent construction of the mutant haplotypes. Whole-genome amplification (WGA) of a single sperm sample was performed using a MALBAC® Single Cell DNA Quick-Amp Kit (Yikon Genomics Inc., China), and conventional PCR and Sanger sequencing were used to validate the c.319+1G > A mutation in PMP22.
After verifying the PMP22 mutation in family members, genetic counseling is required for all couples, including general and reproductive genetic counseling, before the ovulation cycle. General genetic counseling helps to confirm the pathogenicity of variants and familial information, and reproductive counseling aims to inform the procedures of PGT-M treatment and potential risks [29]. It is crucial for specialists, obstetricians, and reproductive physicians to evaluate reproductive and pregnancy risks in female patients with CMT1A [28].
2.3. Oocyte retrieval, in vitro fertilization (IVF), and embryo biopsy
After counseling and being fully informed of all risks, eight patients underwent routine controlled ovarian hyperstimulation, mature follicle monitoring, oocyte retrieval, and ICSI fertilization. On day 5 or 6 after fertilization, a trophectoderm (TE) biopsy was conducted on the fully expanded and hatched blastocysts to acquire 5–8 trophoblast cells per embryo. The biopsied TE cell mass was washed with DPBS (containing 0.1 % human serum albumin) and collected in 0.2 mL PCR tubes with 5 μL lysis buffer. The blastocysts were immediately frozen by vitrification after biopsy. The number of IVF cycles retrieved, fertilized oocytes, and embryos for PGT are shown in Table 1.
2.4. WGA and library preparation, and sequencing
The biopsied TE cells' WGA was performed using a commercial MALBAC Single-Cell Amplification Kit (Yikon Genomics Inc., China). WGA products were used to prepare a DNA library using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs Inc., Ipswich, MA, USA), and the libraries were sequenced on an Illumina NovaSeq platform with 2 × sequencing coverage.
2.5. Copy number variation(CNV) analysis
After basic quality control, sequencing data were aligned to the human reference genome (GRCh38/hg38). The embryos' CNV was analyzed using a previously published method [30]. A 1-Mb window was used to divide and calculate the average sequencing depth to normalize the samples. Deletions and duplications of embryo chromosomes were detected at a resolution of 10 Mb. Chromosomal mosaicism was detected when the mosaic rate was higher than 30 %.
2.6. Direct mutation detection
To directly detect the mutation, CNV of the 1.4 Mb duplication region on chromosome 17 p11.2–12 and the surrounding upstream/downstream region of approximately 2–3 Mb were analyzed. First, a reference was generated using data from 30 embryos without abnormalities in this region. The mapped reads were binned using a 100-Kb window and normalized to form the reference. The threshold values were defined as 1.4 and 2.6, which included more than 90 % of the samples in the interval. The embryos of the seven PMP22 duplication couples were compared with the reference to obtain corrected copy numbers. In the targeted 1.4-Mb region, it was identified as a duplication when there were more than 10 points (one point representing 100 kb, for a total of 14 points) exceeding 2.6. For PMP22, the c.319+1G > A variant in case 4, targeting primers were designed for specific amplification, and the products were subjected to Sanger sequencing.
2.7. Linkage analysis based on single nucleotide polymorphism (SNP) markers
The NGS results were also used to filter SNP markers surrounding the mutation sites. In this study, the filter standard for the SNPs contained more than three covered reads. The mutant and wild-type alleles were identified according to the sequenced genomes of the patients, affected villi, or single sperms using the scHaplotyper software [31]. Linkage analysis was performed by comparing the SNP information of the tested embryos with that of the mutant and wild-type alleles. SNP markers within 3 Mb of the PMP22 duplication region were calculated, and 1 Mb of the surrounding SNPs was focused on in the haplotype analysis. The identified embryo haplotypes were corrected using a Hidden Markov Model [32] to avoid allele dropout (ADO) interference.
2.8. Embryo transfer and amniotic fluid verification
Euploid embryos unaffected by PMP22 mutation were suitable for transfer. When the couples entered the embryo thawing and transfer cycles, we followed up on their pregnancy states. Amniocentesis and amniotic fluid genetic testing are recommended for couples with continuous intrauterine pregnancy to confirm the previous results. MLPA and Sanger sequencing following PCR were performed for cases 2, 6, and 4.
3. Results
3.1. Preliminary experiments for validation of PMP22 mutation
Mutations were verified in all recruited patients, who were fully informed before beginning the IVF cycle. Among the eight families diagnosed with CMT1A (Table 1), all but case 4 were confirmed to have duplications in exon 1_5 of the PMP22 gene through MLPA, and the MLPA results of case 6 are shown as an example in Fig. 1A. In case 6, the female partner carried a PMP22 duplication. In contrast, her parents had no abnormality in PMP22 copy numbers, indicating that the duplication mutation carried by the female partner might be de novo. The mutation in case 4 was validated using targeted amplification and Sanger sequencing. The results showed that only the husband carried the splicing mutation c.319+1G > A of PMP22; however, this mutation was not detected in his parents (Fig. 1B). Single sperms were used in the PCR-based evaluation to determine whether there were sperms carrying mutations, providing SNP markers to construct mutant and wild-type alleles for subsequent linkage analysis. All eight cases had clear clinical and genetic diagnoses.
Fig. 1.
Preliminary experiment for validation of the PMP22 mutation and preparation for subsequent PGT-M. A.PMP22 duplication verification of case 6 using the MLPA method. B.PMP22 mutation detection in case 4 using Sanger sequencing following PCR. Red dotted lines and red arrows indicate the mutations for case 6 and case 4, respectively.
3.2. Embryo aneuploidy detection
As shown in Table 1, 12 IVF-PGT cycles were performed with 169 retrieved oocytes, 107 fertilized oocytes, and 45 blastocysts. All 45 embryos were biopsied at the blastocyst stage for subsequent diagnosis of PGT-M. CNV analysis was performed to detect aneuploidy in the embryos, and the results for all 45 embryos are shown in Table 2. The aneuploidy rate of the biopsied blastocysts was 33.3 % (15/45). Here, we considered cases 7 and 8 as examples to illustrate the CNV analyses. As shown in Fig. 2A, chromosomal aneuploidy analyses were performed for all three embryos in case 7. E1 and E3 showed no abnormalities on the chromosome, while E2 had a 49-Mb deletion in the p-arm of the chromosome, as indicated by the red box. CNV analyses of the nine embryos in case 8 are shown in Fig. 2B. Chromosome 8 of E3 was monosomal, and E7 had a 67-Mb deletion in the q-arm of chromosome 7. In addition, monosomy mosaicism on chromosome 14 (mosaicism rate of approximately 30 %) was detected on E2. Embryos with abnormal CNV results were considered unsuitable for embryo transfer.
Table 2.
PGT-M results of all the embryos.
| Case ID | PMP22 Mutation | Cycle | Embryo ID | CNV | Direct detection | Linkage analysis | Conclusion | Clinical outcomes |
|---|---|---|---|---|---|---|---|---|
| 1 | Duplication on Exon 1_5 | 1 | E1 | 46,XN | Affected | Affected | Not suitable for ET | |
| 1 | E2 | 46,XN,-9p(pter→p13.1,∼41 M, × 1,mos,∼40 %) | Affected | Affected | Not suitable for ET | |||
| 1 | E3 | 46,XN | Normal | Normal | Suitable for ET | Implantation failure | ||
| 2 | E4 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 2 | E5 | 46,XN,-13q(q31.1→qter,∼31 M, × 1,mos) | Normal | Normal | Not suitable for ET | |||
| 2 | E6 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 2 | E7 | 46,XN,-12p(pter→p12.3,∼18 M, × 1) | Normal | Normal | Not suitable for ET | |||
| 2 | E8 | Multiple chromosomes abnormal | Affected | Affected | Not suitable for ET | |||
| 2 | Duplication on Exon 1_5 | 1 | E1 | Multiple chromosomes abnormal | Affected | Affected | Not suitable for ET | |
| 1 | E2 | 46,XN | Normal | Normal | Suitable for ET | Two live births | ||
| 1 | E3 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E4 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 1 | E5 | 45,XN,-16( × 1) | Normal | Normal | Not suitable for ET | |||
| 1 | E6 | 46,XN,-1q(q43→qter,∼10 M, × 1,mos,∼60 %) | Normal | Normal | Not suitable for ET | |||
| 3 | Duplication on Exon 1_5 | 1 | E1 | 46,XN | Affected | Affected | Not suitable for ET | |
| 1 | E2 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 1 | E3 | 46,XN,-14q(q21.3→qter,∼59 M, × 1) | Normal | Normal | Not suitable for ET | |||
| 1 | E4 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E5 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E6 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 4 |
PMP22 c.319+1G > A |
1 | E1 | 46,XN | Normal | Normal | Suitable for ET | Live birth |
| 1 | E2 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E3 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 1 | E4 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E5 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 |
E6 |
46,XN |
Normal |
Normal |
Suitable for ET |
Frozen |
||
| 6 | Duplication on Exon 1_5 | 1 | E1 | 46,XN | Affected | Affected | Not suitable for ET | |
| 1 | E2 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 2 | E3 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 2 | E4 | 47,XN,+15( × 3) | Affected | Affected | Not suitable for ET | |||
| 3 | E5 | 46,XN | Normal | Normal | Suitable for ET | Live birth | ||
| 3 | E6 | Multiple chromosomes abnormal | Affected | Affected | Not suitable for ET | |||
| 3 | E7 | 48,XN,+16( × 3),+21( × 3) | Normal | Normal | Not suitable for ET | |||
| 7 | Duplication on Exon 1_5 | 1 | E1 | 46,XN | Normal | Normal | Suitable for ET | Implantation failure |
| 1 | E2 | 46,XN,-7(pter→p12.3,∼49 M, × 1) | Affected | Affected | Not suitable for ET | |||
| 2 | E3 | 46,XN | Normal | Normal | Suitable for ET | In pregnancy | ||
| 8 | Duplication on Exon 1_5 | 1 | E1 | 46,XN | Affected | Affected | Not suitable for ET | |
| 1 | E2 | 46,XN,-14( × 1,mos,∼30 %) | Affected | Affected | Not suitable for ET | |||
| 1 | E3 | 45,XN,-8( × 1) | Affected | Affected | Not suitable for ET | |||
| 1 | E4 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E5 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E6 | 46,XN | Normal | Normal | Suitable for ET | Frozen | ||
| 1 | E7 | 46,XN,-7q(q21.11→qter,∼67 M × 1) | Normal | Normal | Not suitable for ET | |||
| 1 | E8 | 46,XN | Affected | Affected | Not suitable for ET | |||
| 1 | E9 | 46,XN | Affected | Affected | Not suitable for ET |
Fig. 2.
Copy number variation analysis. A. Copy number variation analysis for 3 embryos of case 7. B. Copy number variation analysis for 9 embryos of case 8. Red boxes indicate the abnormality of chromosomes or chromosome regions.
3.3. Direct PMP22 duplication and splicing mutation detection
Novel direct mutation detection for PMP22 duplication was carried out without an additional step, and the copy numbers of the 1.4-Mb region were analyzed based on NGS data. For all 45 blastocysts, the success and accuracy rates were 100 % (Table 2). The copy numbers of this 1.4-Mb region and its upstream and downstream regions were normalized by a 100-Kb window using normal samples, and there were a total of 14 dots in the targeted duplication region, with each dot representing 100 kb. Taking the six embryos of case 2 as an example, E1 and E4 had a continuous majority of dots exceeding the threshold value of 2.6 (refer to Methods), as indicated by the red dotted boxes, which suggest three copy numbers in this region (Fig. 3A). The same analysis was performed for case 3, with six embryos. E1 and E2 had PMP22 duplications, with more than 10 pots above the threshold of 2.6, as indicated by the red dotted boxes (Fig. 3B). Other embryos of these two cases show no abnormality in this region, with almost all dots in the interval (between 1.4 and 2.6) shown in Fig. 3A and B. In case 4, the splicing mutation (c.319+1G > A) of PMP22 gene was detected in 6 embryos by Sanger sequencing following targeted PCR. Among the six analyzed embryos, E3 carried a mutation inherited from the father, and the remaining embryos did not carry a splicing mutation (Fig. 3C).
Fig. 3.
Direct detection of PMP22 mutation in embryos. A. Direct PMP22 duplication detection and analysis for 6 embryos of case 2. Horizontal ordinates show the location of chromosome 17, and vertical coordinates show the copy numbers. A dot represents 100 kilobases, and dots in blue indicate their value between threshold values (1.4 and 2.6), while dots in red and green indicate their value more than 2.6 and lower than 1.4, respectively. Red dotted lines exhibit abnormal copy numbers. B. Direct PMP22 duplication detection and analysis for 6 embryos of case 3. Horizontal ordinates show the location of chromosome 17, and vertical coordinates show the copy numbers. A dot represents 100 kilobases, with dots in blue indicating a value between the threshold values (1.4 and 2.6) and the dots in red and green indicating a value above 2.6 and below 1.4, respectively. Red dotted lines exhibit abnormal copy numbers. C. Direct PMP22 mutation detection for 6 embryos of case 4 through Sanger sequencing following PCR. Red arrows indicate the mutations.
3.4. Linkage analysis based on SNP markers
SNP-based linkage analysis was also performed to improve the diagnostic accuracy of the embryos. A schematic of the linkage analysis is shown in Fig. 4A, which affected family members or sperm carrying the mutant allele, and was used to provide SNP markers for haplotype analysis. The principle of SNP-based linkage analysis is illustrated through several examples. As shown in Fig. 4A, C/T is a SNP marker located upstream of PMP22, and the affected family member had C/C at the same SNP site; therefore, it could be deduced that base C was associated with the mutant allele in the affected patients. Similarly, in a single sperm, base C was associated with mutations. Thus, there are two possible embryo genotypes: embryos with the C/T genotype inherit the mutant allele, whereas embryos with the T/T genotype inherit the wild-type allele. Following this principle, in case 4, sperm 7 carrying the mutation and sperm 3 without the mutation detected in the preliminary experiment were used to construct the mutant haplotype (represented in red) and the wild-type haplotype (represented in green), as shown in Fig. 4B. Among the six embryos of case 4, E3 consisted of sperm 7 (red), indicating that E3 carried the PMP22 mutation. In contrast, the other embryos of case 4 consisted of sperm 3 (green), indicating that they were free of the mutation (Fig. 4B). For case 7, the mutant haplotype represented in yellow by the female partner was constructed using the affected mother and the unaffected father (Supplementary Figure 1 and Fig. 4C). Among the three embryos in case 7, E2 inherited the mutant alleles (yellow) from the female partner, whereas E1 and E3 inherited the wild-type alleles (blue). The linkage analysis results for all 45 embryos are summarized in Table 2.
Fig. 4.
Linkage analysis results based on SNPs from NGS data. A. Schematic haplotype analyses using family members or single sperm. Letter “W” represents wild-type, while red cross represents the PMP22 mutation. Orange and gray show the mutant and wild-type alleles, respectively. B. SNP linkage analysis results for case 4 according to the haplotypes constructed by single sperm. Sperm 7 and sperm 3 are red and green, representing mutant and wild-type alleles, respectively. C. SNP linkage analysis results for case 7 according to the haplotypes constructed by the female partner and her family members. The mutant alleles of the female patient are shown in yellow, while the other alleles are in blue. The blue line indicates the PMP22 location, and the colors of embryos show 3 Mb upstream and downstream around the mutation.
3.5. Embryo transfer, clinical outcomes, and follow-up
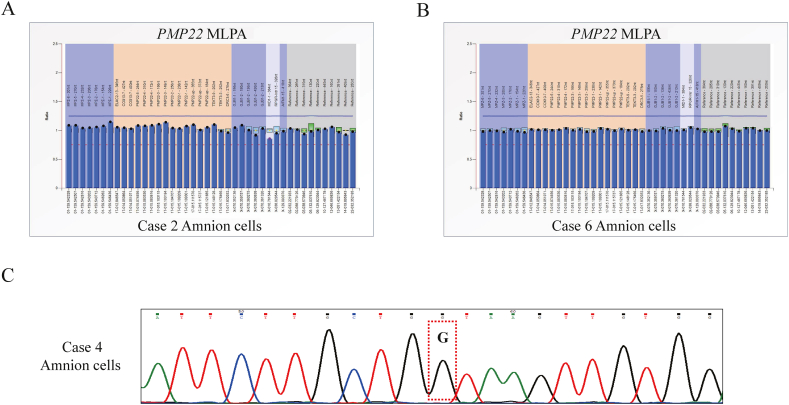
When the direct detection results of the PMP22 region and the indirect linkage analysis results with CNV results were combined, mutation-free and euploid embryos were considered for transfer. In the 12 PGT-M cycles, 18 of the 45 diagnosed embryos from eight couples were suitable for transfer (Table 2). In total, six embryos from five cases (cases 1, 2, 4, 6, and 7) underwent frozen embryo transfer (FET), and four women (cases 2, 4, 6, and 7) eventually became clinically pregnant (Table 2). Three pregnant women (cases 2, 4, and 6) underwent amniocentesis at 20 weeks of gestation, and the prenatal diagnostic results corresponded with the previous PGT-M results. Four healthy full-term babies were born. Fortunately, in case 2, identical twins were obtained through single embryo transfer. At this time, the female partner in case 7 was pregnant and waiting for prenatal validation. The prenatal diagnosis results for the three families are shown in Fig. 5. The MLPA results for cases 2 and 6 showed that both fetuses had normal copy numbers in the PMP22 region (Fig. 5A–B). For case 4, Sanger sequencing followed by specific PCR was used to detect the splicing mutation (c.319+1G > A) of PMP22 in amniotic fluid cells, and the results showed that the fetus was free of this mutation (Fig. 5C). For pregnant women with CMT1A (cases 2 and 4), during clinical follow-up, there were no adverse effects of anesthesia or increased complications observed during pregnancy and delivery. The follow-up information of the three families with live births is summarized in Table 3. All four newborns had normal Apgar scores, and three children had normal growth and development, except for one lost to follow-up. In addition, the CMT1A status of pregnant patients remained stable throughout the PGT-M, pregnancy, and delivery processes (Table 3).
Fig. 5.
Prenatal diagnosis for vitrification of the PMP22 mutation using cultured amniotic fluid cells. A.PMP22 duplication detection of the amniotic fluid cells of case 2 using the MLPA method. B.PMP22 duplication detection of amnion fluid cells of case 6 using MLPA method. C. Amnion cell PMP22 mutation of case 4 through Sanger sequencing following specific PCR. Red dotted lines show the mutation loci.
Table 3.
Follow-up for families with live birth.
| Case ID | Singleton/multiple pregnancy | Chorionicity status | Delivery mode | Birth weight | Birth length | Sex | Apgar score | Child growth and development* | Progress of CMT1A disease in female patients after pregnant |
|---|---|---|---|---|---|---|---|---|---|
| 2 | Multiple | Dichorionic | Vaginal delivery | 1850g | 43 cm | Male | Normal | Normal | – |
| 1900g | 44 cm | Male | Normal | Normal | |||||
| 3 | Singleton | Monochorionic | Cesarean section | 2000g | 40 cm | Male | Normal | / | No progress |
| 6 | Singleton | Monochorionic | Vaginal delivery | 2800g | 47 cm | Male | Normal | Normal | No progress |
*include moving, speaking, and behaving evaluations.
- Not applicable.
/lost to follow-up.
4. Discussion
CMT1A is the most frequent type of CMT, and a 1.4-Mb duplication region, including PMP22, was identified as the causative gene. There is now, except for general supportive and symptomatic treatment for CMT1A. PGT-M offers an alternative approach for obtaining unaffected newborns. However, current PGT-M strategies for CMT1A rely only on linkage analysis, which requires affected family members to construct the mutant and wild-type alleles. Furthermore, chromosome exchange in or adjacent to the mutation and triple copy numbers of the 1.4-Mb region might interfere with the linkage analysis. A novel direct detection method based on NGS was developed that is applicable for all kinds of PMP22 mutation types in CMT1A to address these difficulties. This method is simple, stable, accurate, and requires only low-depth coverage NGS sequencing after WGA of biopsied TE cells without additional experimental steps. Except for the direct detection of the PMP22 duplication region, the accuracy and stability of diagnosis in embryos can be confirmed by linkage analysis. In particular, for CMT1A cases in which positive family members cannot be found to help construct mutant haplotypes, our method can directly detect the copy numbers of PMP22 duplications in embryos. For instance, the female partner in case 6 carried a de novo PMP22 duplication that was not identified in her other family members, and through our method, the copy numbers of PMP22 were directly detected. An unaffected euploid embryo was transferred, and a healthy baby was born after amniocentesis at 20 weeks of gestation. Thus, compared to current detection methods, this NGS-based method is widely applicable, convenient, and accurate and does not require positive family members.
In addition to providing a new method for direct detection based on NGS, our study also summarizes the clinical practice for CMT1A interruption before embryo implantation. The pipeline is illustrated in Fig. 6. First, only patients with clear clinical and genetic diagnoses, who were fully informed, met the PGT-M indications. Before undergoing an IVF-PGT cycle, preliminary experiments and genetic counseling are indispensable. In female patients, multidisciplinary consultation, including specialists in this disease, obstetricians, and reproductive physicians, helps evaluate reproductive and pregnancy risks. Previous studies showed no significant increase in the miscarriage rate, postpartum bleeding rate, anesthetic complications, newborn morbidity, or mortality in pregnant patients with CMT1A compared to the general population [24–26]. Mild female patients with CMT1A show a stable status during pregnancy and delivery; nevertheless, severe female patients with CMT1A may experience adverse outcomes and high pregnancy risks, which require careful evaluation. After routine ovarian stimulation, oocyte retrieval, and ICSI, TE cell samples were acquired by blastocyst biopsy. This novel technology simultaneously performed aneuploid analysis, direct mutation detection, and haplotype analysis. Based on the above results, euploid embryos unaffected by PMP22 mutation were transferred through FET cycles. Amniocentesis and amniotic fluid genetic testing are recommended for couples with continuous intrauterine pregnancies to reduce the risk of embryo misdiagnosis. Follow-up during pregnancy and delivery is recommended to monitor the progressive status of CMT1A disease and pregnancy-related complications. In addition, long-term follow-up is required for offspring born using PGT-M. Amniocentesis and amniotic fluid genetic testing during pregnancy are the gold standards for validating this method, and further long-term follow-up of the growth and development of children is important. Compared to prenatal interruption methods, CMT1A interruption through PGT-M prevents pregnant women from suffering from physical and psychological distress when fetuses are diagnosed with CMT1A. Eight families received PGT-M using this novel method, and four healthy babies from three families were delivered.
Fig. 6.
Clinical application pipeline of PGT-M for CMT1A using a novel direct detection method.
This method provides a novel direct detection method in embryos to assist in the genetic interruption of patients with CMT1A. However, the current study has the limitation that only eight families were recruited, and the reliability and adaptation were initially proved. In the future, we expect this method and analysis to be applied to more families with PMP22 duplication mutations and can be used for monogenic disorders with similar mutation forms. For instance, this method is also applicable for PMP22 deletion, which causes hereditary neuropathy with liability to pressure palsies (HNPP), as well as other diseases caused by large duplications or deletions, such as Duchenne muscular dystrophy (DMD), which is characterized by single or multiple exon deletions or duplications of the DMD gene in approximately 80 % of cases [33], and duplication or triplication of the methyl CpG binding protein 2 (MECP2) gene, which results in MECP2 duplication syndrome (MDS) [34].
5. Conclusions
In summary, we provided a novel direct detection method for embryos based on NGS data for patients with CMT1A, which is accurate, adaptable, and cost-competitive. It has been successfully applied in eight families, and among the three families that achieved live births, further clinical practice is required in the future. This method is applicable to PMP22 deletions and other diseases caused by large duplications or deletions. In addition, the clinical PGT-M application pipeline for CMT1A is applicable to the diseases mentioned above, providing an alternative approach to interruptions in offspring.
Data availability statement
Data will be made available on request.
Ethics approval and consent to participate
This study was approved by the Medical Science Research Ethics Committee of Peking University Third Hospital (2018S2-002). Written informed consent was obtained from the participates.
Consent for publication
All the listed authors have read and approved for this manuscript, and agreed for publication.
Funding
This project is funded by National Key Research and Development Program (2017YFA0103801) and National Natural Science Foundation of China (82125013, 82288102).
CRediT authorship contribution statement
Yuqian Wang: Validation, Writing – original draft, Writing – review & editing, Investigation. Yujun Liu: Validation, Writing – original draft, Writing – review & editing, Investigation. Ying Kuo: Data curation, Methodology, Validation. Shuo Guan: Data curation, Methodology, Validation. Nan Wang: Data curation, Methodology, Validation. Ying Lian: Data curation, Resources. Jin Huang: Data curation, Resources. Xu Zhi: Data curation, Methodology, Resources. Ping Liu: Conceptualization, Supervision, Writing – review & editing. Rong Li: Conceptualization, Supervision. Liying Yan: Funding acquisition, Project administration, Supervision, Writing – review & editing. Xiaohui Zhu: Project administration, Supervision, Writing – review & editing. Jie Qiao: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank all the staffs for supporting all the procedures of IVF-PGT in the Center for Reproductive Medicine of Peking University Third Hospital.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22196.
Contributor Information
Liying Yan, Email: liying-pku@bjmu.edu.cn.
Xiaohui Zhu, Email: Zhu_Xh2023@126.com.
Jie Qiao, Email: jie.qiao@263.net.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Jung N.Y., Kwon H.M., Nam D.E., Tamanna N., Lee A.J., Kim S.B., et al. Peripheral myelin protein 22 gene mutations in charcot-marie-tooth disease type 1E patients. Genes. 2022;13(7) doi: 10.3390/genes13071219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolaou P., Zamba-Papanicolaou E., Koutsou P., Kleopa K.A., Georghiou A., Hadjigeorgiou G., et al. Charcot-Marie-Tooth disease in Cyprus: epidemiological, clinical and genetic characteristics. Neuroepidemiology. 2010;35(3):171–177. doi: 10.1159/000314351. [DOI] [PubMed] [Google Scholar]
- 3.Foley C., Schofield I., Eglon G., Bailey G., Chinnery P.F., Horvath R. Charcot-marie-tooth disease in northern England. J. Neurol. Neurosurg. Psychiatry. 2012;83(5):572–573. doi: 10.1136/jnnp-2011-300285. [DOI] [PubMed] [Google Scholar]
- 4.Braathen G.J., Sand J.C., Lobato A., Hoyer H., Russell M.B. Genetic epidemiology of Charcot-Marie-Tooth in the general population. Eur. J. Neurol. 2011;18(1):39–48. doi: 10.1111/j.1468-1331.2010.03037.x. [DOI] [PubMed] [Google Scholar]
- 5.Barreto L.C., Oliveira F.S., Nunes P.S., de Franca Costa I.M., Garcez C.A., Goes G.M., et al. Epidemiologic study of charcot-marie-tooth disease: a systematic review. Neuroepidemiology. 2016;46(3):157–165. doi: 10.1159/000443706. [DOI] [PubMed] [Google Scholar]
- 6.Rossor A.M., Polke J.M., Houlden H., Reilly M.M. Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat. Rev. Neurol. 2013;9(10):562–571. doi: 10.1038/nrneurol.2013.179. [DOI] [PubMed] [Google Scholar]
- 7.van Paassen B.W., van der Kooi A.J., van Spaendonck-Zwarts K.Y., Verhamme C., Baas F., de Visser M. PMP22 related neuropathies: Charcot-Marie-Tooth disease type 1A and Hereditary Neuropathy with liability to Pressure Palsies. Orphanet J. Rare Dis. 2014;9:38. doi: 10.1186/1750-1172-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas P.K., Marques W., Jr., Davis M.B., Sweeney M.G., King R.H., Bradley J.L., et al. The phenotypic manifestations of chromosome 17p11.2 duplication. Brain. 1997;120(Pt 3):465–478. doi: 10.1093/brain/120.3.465. [DOI] [PubMed] [Google Scholar]
- 9.Suh J.G., Ichihara N., Saigoh K., Nakabayashi O., Yamanishi T., Tanaka K., et al. An in-frame deletion in peripheral myelin protein-22 gene causes hypomyelination and cell death of the Schwann cells in the new Trembler mutant mice. Neuroscience. 1997;79(3):735–744. doi: 10.1016/s0306-4522(96)00692-6. [DOI] [PubMed] [Google Scholar]
- 10.Snipes G.J., Suter U., Welcher A.A., Shooter E.M. Characterization of a novel peripheral nervous system myelin protein (PMP-22/SR13) J. Cell Biol. 1992;117(1):225–238. doi: 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khajavi M., Shiga K., Wiszniewski W., He F., Shaw C.A., Yan J., et al. Oral curcumin mitigates the clinical and neuropathologic phenotype of the Trembler-J mouse: a potential therapy for inherited neuropathy. Am. J. Hum. Genet. 2007;81(3):438–453. doi: 10.1086/519926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Notterpek L., Ryan M.C., Tobler A.R., Shooter E.M. PMP22 accumulation in aggresomes: implications for CMT1A pathology. Neurobiol. Dis. 1999;6(5):450–460. doi: 10.1006/nbdi.1999.0274. [DOI] [PubMed] [Google Scholar]
- 13.Ryan M.C., Shooter E.M., Notterpek L. Aggresome formation in neuropathy models based on peripheral myelin protein 22 mutations. Neurobiol. Dis. 2002;10(2):109–118. doi: 10.1006/nbdi.2002.0500. [DOI] [PubMed] [Google Scholar]
- 14.Hoogendijk J.E., Hensels G.W., Gabreels-Festen A.A., Gabreels F.J., Janssen E.A., de Jonghe P., et al. De-novo mutation in hereditary motor and sensory neuropathy type I. Lancet. 1992;339(8801):1081–1082. doi: 10.1016/0140-6736(92)90668-s. [DOI] [PubMed] [Google Scholar]
- 15.Cortes H., Hernandez-Hernandez O., Bautista-Tirado T., Escobar-Cedillo R.E., Magana J.J., Leyva-Garcia N. [Detection of the PMP22 gene duplication in peripheral neuropathy patients: a study in Mexican population] Rev. Neurol. 2014;59(3):111–117. [PubMed] [Google Scholar]
- 16.Thiel C.T., Kraus C., Rauch A., Ekici A.B., Rautenstrauss B., Reis A. A new quantitative PCR multiplex assay for rapid analysis of chromosome 17p11.2-12 duplications and deletions leading to HMSN/HNPP. Eur. J. Hum. Genet. 2003;11(2):170–178. doi: 10.1038/sj.ejhg.5200920. [DOI] [PubMed] [Google Scholar]
- 17.Hung C.C., Lee C.N., Lin C.Y., Cheng W.F., Chen C.A., Hsieh S.T., et al. Identification of deletion and duplication genotypes of the PMP22 gene using PCR-RFLP, competitive multiplex PCR, and multiplex ligation-dependent probe amplification: a comparison. Electrophoresis. 2008;29(3):618–625. doi: 10.1002/elps.200700340. [DOI] [PubMed] [Google Scholar]
- 18.Slater H., Bruno D., Ren H., La P., Burgess T., Hills L., et al. Improved testing for CMT1A and HNPP using multiplex ligation-dependent probe amplification (MLPA) with rapid DNA preparations: comparison with the interphase FISH method. Hum. Mutat. 2004;24(2):164–171. doi: 10.1002/humu.20072. [DOI] [PubMed] [Google Scholar]
- 19.Gautier B., Hajjar H., Soares S., Berthelot J., Deck M., Abbou S., et al. AAV2/9-mediated silencing of PMP22 prevents the development of pathological features in a rat model of Charcot-Marie-Tooth disease 1 A. Nat. Commun. 2021;12(1):2356. doi: 10.1038/s41467-021-22593-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J.S., Lee J.Y., Song D.W., Bae H.S., Doo H.M., Yu H.S., et al. Targeted PMP22 TATA-box editing by CRISPR/Cas9 reduces demyelinating neuropathy of Charcot-Marie-Tooth disease type 1A in mice. Nucleic Acids Res. 2020;48(1):130–140. doi: 10.1093/nar/gkz1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stavrou M., Sargiannidou I., Georgiou E., Kagiava A., Kleopa K.A. Emerging therapies for charcot-marie-tooth inherited neuropathies. Int. J. Mol. Sci. 2021;22(11) doi: 10.3390/ijms22116048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Vos A., Sermon K., Van de Velde H., Joris H., Vandervorst M., Lissens W., et al. Pregnancy after preimplantation genetic diagnosis for Charcot-Marie-Tooth disease type 1A. Mol. Hum. Reprod. 1998;4(10):978–984. doi: 10.1093/molehr/4.10.978. [DOI] [PubMed] [Google Scholar]
- 23.De Vos A., Sermon K., De Rijcke M., Goossens V., Henderix P., Van Ranst N., et al. Preimplantation genetic diagnosis for Charcot-Marie-Tooth disease type 1A. Mol Hum Reprod. 2003;9(7):429–435. doi: 10.1093/molehr/gag054. [DOI] [PubMed] [Google Scholar]
- 24.Pisciotta C., Calabrese D., Santoro L., Tramacere I., Manganelli F., Fabrizi G.M., et al. Pregnancy in Charcot-Marie-Tooth disease: data from the Italian CMT national registry. Neurology. 2020;95(24):e3180–e3189. doi: 10.1212/WNL.0000000000010860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudnik-Schoneborn S., Thiele S., Walter M.C., Reinecke L., Sereda M., Schoneborn R., et al. Pregnancy outcome in Charcot-Marie-Tooth disease: results of the CMT-NET cohort study in Germany. Eur. J. Neurol. 2020;27(8):1390–1396. doi: 10.1111/ene.14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skorupinska M, Ramdharry G, Byrne B, Laurá M, Reilly MM.Pregnancy and delivery in patients with Charcot–Marie–Tooth disease and related disorders. Obstet. Med..0(0):1753495X221107328.. [DOI] [PMC free article] [PubMed]
- 27.Kim Min P.S., Ye Hong, Park Eun, Lee Yu, Choi Byung-Ok, Lee Kyung-Ah, Yu Eun, Kang Inn. Clinical application of genome-wide single nucleotide polymorphism genotyping and karyomapping for preimplantation genetic testing of Charcot–Marie–Tooth disease. J. Gene Med. 2022;19:7–13. [Google Scholar]
- 28.Bird T.D. In: GeneReviews((R)). Seattle (WA) Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., et al., editors. 1993. Charcot-marie-tooth hereditary neuropathy overview. [Google Scholar]
- 29.Group E.P.-M.W., Carvalho F., Moutou C., Dimitriadou E., Dreesen J., Gimenez C., et al. ESHRE PGT Consortium good practice recommendations for the detection of monogenic disorders. Hum Reprod Open. 2020;2020(3):hoaa018. doi: 10.1093/hropen/hoaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou Y., Fan W., Yan L., Li R., Lian Y., Huang J., et al. Genome analyses of single human oocytes. Cell. 2013;155(7):1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 31.Yan Z., Zhu X., Wang Y., Nie Y., Guan S., Kuo Y., et al. scHaplotyper: haplotype construction and visualization for genetic diagnosis using single cell DNA sequencing data. BMC Bioinf. 2020;21(1):41. doi: 10.1186/s12859-020-3381-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy S.R. What is a hidden Markov model? Nat. Biotechnol. 2004;22(10):1315–1316. doi: 10.1038/nbt1004-1315. [DOI] [PubMed] [Google Scholar]
- 33.Duan D., Goemans N., Takeda S., Mercuri E., Aartsma-Rus A. Duchenne muscular dystrophy. Nat. Rev. Dis. Prim. 2021;7(1):13. doi: 10.1038/s41572-021-00248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ta D., Downs J., Baynam G., Wilson A., Richmond P., Leonard H. A brief history of MECP2 duplication syndrome: 20-years of clinical understanding. Orphanet J. Rare Dis. 2022;17(1):131. doi: 10.1186/s13023-022-02278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.