Abstract
An 8.1-kb fragment of the temperate Myxococcus xanthus phage Mx8 genome, when cloned into a plasmid vector, permits site-specific integration of the plasmid and confers superinfection immunity. Sequence analysis of a 9.5-kb region of Mx8 DNA containing this fragment reveals 19 densely packed open reading frames, four of which have predicted products with known or suspected activities. The Mx8 imm gene, required for superinfection immunity, has a sequence similar to that of Arabidopsis thaliana G-box-binding factor 1. Mx8 makes a DNA adenine methylase, Mox, and integrase, Int, related to other methylases and integrases. The int gene has two alternate translation initiation codons within the extensively overlapping uoi (upstream of int) gene. Comparison of the predicted product of the uoi gene with Salmonella phage P22 and Streptomyces plasmid Xis proteins shows that temperate phage excisionases may use variations of a helix-turn-helix motif to recognize specific DNA sequences.
Myxococcus xanthus is the best-characterized representative of the myxobacteria, gram-negative bacteria that live in the soil. The myxobacteria are unique among the eubacteria, because they undergo a complex, multicellular developmental cycle when starved for nutrients. When nutrient levels are low, hundreds of thousands of bacteria glide to aggregate into macroscopic fruiting bodies with a complex cellular organization. Within these fruiting bodies, a fraction of cells differentiate into spores that are resistant to UV light, desiccation, and heat and are capable of germination into totipotent vegetative cells once nutrients become available.
Several bacteriophages are known to infect M. xanthus. These include the lytic phages Mx1 (8), Mx4 (10), and Mx9 (28). Mx1 and Mx4 resemble coliphages T4 and P1, respectively, in morphology and have large genome sizes (7, 10). Although one mutant strain of Mx4, Mx4 ts-27 htf-1 hrm-1, is routinely used for the generalized transduction of M. xanthus (10, 15), its use is limited by its small burst sizes and low efficiencies of plating (EOPs) when grown on motile, fruiting strains of M. xanthus. Mx9 resembles Salmonella typhimurium phage P22 in morphology and has a smaller genome (28). The lytic myxophages exhibit an unusual adaptation to their host. When phage-infected cells of M. xanthus are starved, lytic phage growth arrests upon development. The infecting phage genome is trapped in mature spores in a dormant state and can escape this state to lyse vegetative cells formed by the germination of spores (8, 9).
Three serologically related myxophages, Mx8, Mx81, and Mx82, are temperate and, like Mx9, resemble P22 in morphology (28). All three phages package terminally repetitious, circularly permuted, double-stranded genomes. Among these, the best understood is Mx8, which encapsidates a genome 49 kb in length with a terminal repetition of about 4 kb (reference 43 and this report). Among the myxophages, Mx8 shows the most promise as a genetic tool to investigate the complex multicellular development of its host. Because Mx8 can lysogenize M. xanthus, and the resulting lysogen is stable during vegetative growth, Mx8 could be used as a cloning vector to construct specialized transducing phages, which would facilitate the study of vegetative M. xanthus functions. Moreover, like the lytic phages of M. xanthus, the temperate phages have adapted to respond to the unusual developmental cycle of their host. The Mx8 prophage is stable upon the passage of an M. xanthus lysogen through development (31). This is true despite the fact that M. xanthus, like Bacillus subtilis, uses a variety of different repressors, activators, and alternative sigma factors of RNA polymerase to trigger the sequential expression of different subsets of genes during successive stages of development. The question of why and how the prophage remains refractory to these dramatic changes in host gene expression during the developmental process merits investigation. Because the Mx8 prophage is stable during the developmental process, specialized transducing derivatives of Mx8 could be used to probe M. xanthus functions required for development.
Although little is known about the establishment of lysogeny by Mx8, previous work has shown that a 12-kb fragment of Mx8 DNA (EcoRI-B) includes both a trans-acting integrase gene (int) and a cis-acting attP sequence, required for prophage integration into the host chromosome (27, 43, 46). When a subfragment of EcoRI-B fragment is cloned into a plasmid, the hybrid plasmid can recombine efficiently with the bacterial attB locus and form a stable cointegrate with the M. xanthus genome (25).
Even less is known about how the Mx8 prophage maintains lysogeny. We presume that, like other temperate phages, Mx8 makes one or more repressors that inhibit lytic development and confer superinfection immunity. In this report, we show that a 9.5-kb region of the Mx8 genome includes the genes necessary for prophage integration and superinfection immunity. The sequence of this region shows that the Mx8 genes involved in immunity and integration are densely packed and transcribed in a single direction.
MATERIALS AND METHODS
Bacterial strains.
M. xanthus strains used in this work are derived from strain DZ1, a nonmotile, multiple mutant of strain DZF1 (10). CT liquid medium (14) was used for the routine growth of DZ1; electroporants of DZ1 with integrated plasmids were grown on CT medium with kanamycin sulfate (40 μg/ml). Escherichia coli strains are derivatives of JM107 (47) and were grown in LB medium supplemented with ampicillin (100 μg/ml) and/or kanamycin sulfate (40 μg/ml) (Sigma Chemical Co.)
Bacteriophage strains and methods.
Phage strains used in this work include the wild-type strains of Mx8, Mx81, and Mx82, which were reisolated from M. xanthus DK883, DK879, and DK893, respectively (28); these strains were the kind gifts of Dale Kaiser. Supernatants of exponential cultures of each strain grown in CT medium at 32°C were plated on a lawn of DZ1, by the soft (0.75%) agar overlay method on CT (1.5%) agar plates (28). The virulent mutant phage, Mx8 vir1, was isolated by selecting for a mutant in a stock of wild-type Mx8 that can form plaques on a lawn of the immune defective lysogen, DZ1(pAY50). Mx8 vir1 plates with nearly equal efficiencies on hosts DZ1, DZ1(pAY50), DZ1(Mx8), and all other immune hosts described in this report. Mx8 del1, which forms plaques and stable lysogens, has a 1,533-bp deletion of nonessential DNA in the immunity region. Preparation of high-titer stocks of wild-type and mutant phages and isolation of phage DNA have been described elsewhere (27).
Plasmid constructions.
When plasmids capable of autonomous replication in E. coli and carrying a kanamycin resistance (Kmr) determinant are introduced into M. xanthus, they confer a Kmr phenotype if and only if they can integrate into the M. xanthus genome. If a plasmid carries a region of homology with the M. xanthus genome, then general recombination can promote plasmid integration. Alternatively, if a plasmid carries the phage Mx8 integrase gene, int, and attachment site, attP, the plasmid can integrate at a preferred site on the M. xanthus genome, attB.
Plasmid pAY50 (27) carries an 8,069-bp Sau3AI-PvuII insert of Mx8 DNA with the phage-encoded site-specific recombination functions, uoi, int, and attP, as well as the primary repressor gene, imm, required for superinfection immunity. Plasmid pPLH343 has an overlapping 5,789-bp XhoI fragment of Mx8 DNA with uoi, int, and attP ligated to a DraI site of pBGS18 (17, 42). Plasmids pAY30, pAY34, pAY36, pAY39, pAY42, pAY45, and pAY48 were made by ligating each of seven EcoRI fragments of Mx8 (A, C, D, E, F, G, and H, respectively) to the EcoRI site of pPLH343. Plasmid pAY20 is the EcoRI B fragment of Mx8 ligated to the EcoRI site of pBGS18. Plasmid pAY31 carries the EcoRI A fragment ligated to the EcoRI site of pPLH343, in the opposite orientation as for pAY30. Plasmids pAY55 and pAY56 were made from plasmids pAY30 and pAY31, respectively, by cleavage with NheI and XbaI and ligation of the fragment with the plasmid origin of replication. Plasmids pAY60 and pAY62 carry the 3,484-bp MfeI-PvuII fragment of Mx8 ligated to the SmaI site of pBGS18, in opposite orientations.
Plasmid pAY54 is a deletion derivative of pAY50 made by cleavage of pAY50 with EcoRI and MfeI and ligation of the largest fragment. To assign the imm gene to a single open reading frame (ORF), three additional deletion derivatives of pAY50 were made. Plasmids pAY258 and pAY259 were constructed by ligating the partial cleavage products of pAY50 with EcoO109I. Plasmid pAY456 is the ligation product of the larger fragment of pAY50 generated by BlpI digestion.
A smaller, integration-proficient derivative of plasmid pAY62, pAY721, was made by amplifying template pAY62 DNA with primer 5′AGCGGATAACAATTTCACACAGGA and primer 5′CCCAAGCTTCCTAGGTAGCGGAAGGGCTCTC, complementary to the 3′ end of int. The 2.2-kb PCR product was cleaved with EcoRI and HindIII and then ligated to the EcoRI and HindIII sites of pBGS18. To determine which codons initiate int translation, we introduced two mutations within the int coding sequence of plasmid pAY721. Plasmid pAY979 is a derivative of pAY721 with the intVA1 mutation, a substitution of CG for TA at bp 5086. Primer pairs 5′ACGGGATAACAATTTCACACAGGA and 5′CGCGACCGCGATGCCCAGCCGTCAGGAGT and 5′CTGGGCATCGCGGTCGCGTCAAGAAGTCG and 5′ACGCGCCCCTCCATCCACTTG were used to amplify template pAY721 DNA, and products were annealed and amplified with primers 5′ACGGGATAACAATTTCACACAGGA and 5′ACGCGCCCCTCCATCCACTTG. The second-step product was cleaved with EcoRI and StuI and ligated to pLITMUS29 to make pAY978. Plasmid pAY978 was cleaved with BsiEI to confirm the presence of the intVA1 mutation. Plasmids pAY978 and pAY721 were cleaved with EcoRI and StuI, and the 835-bp fragment of pAY978 was ligated to the larger fragment of pAY721 to make plasmid pAY979. A derivative of plasmid pAY721 with the intVA42 mutation, a substitution of CG for TA at bp 5209, was made by using primers 5′GCAGCGGCC ATCCTGGCGCGGAAGGCAGGGCGGCGCCGCGGGTAACGTCTATCG CAAGAA and 5′CCCAAGCTTCCTAGGTAGCGGAAGGGCTCTC to amplify template Mx8 DNA. The product was cleaved with BglI to generate a 1,363-bp fragment, which was ligated to a mixture of three of the four BglI fragments of pAY721 to make pAY754. Cleavage with SacII was used to confirm the presence of VA42 mutation. Plasmid pAY990, with both TA-to-CG substitutions, was made by ligating the largest BglI fragment of pAY979 to three smaller fragments of pAY754. Plasmid pAY990 DNA was amplified with primers 5′ACGCGCCCCTCCATCCACTTG and 5′AGCGGATAACAATTTCACACAGGA to yield a 865-bp product; cleavage of this product with SacII and BsiEI confirms that plasmid pAY990 carries both substitutions.
Methods for plasmid constructions and DNA manipulations were adapted from those of Sambrook et al. (36). For electroporation of E. coli and M. xanthus, we used the methods of Taketo (45) and Kashefi and Hartzell (21), respectively. To measure the efficiency of electroporation of plasmids into M. xanthus DZ1, 100 to 300 ng of plasmid DNA (1 μl) was added to 5 × 108 cells (40 μl), electroporated cells were grown for 16 h, and CFU were scored on CT plates with kanamycin sulfate after 7 days.
DNA sequence analysis.
Plasmid templates used for sequence analysis by the dideoxy method (38) included pPLH343, pAY39, and smaller derivatives of pAY50 and pAY20. Sequencing runs were performed by Commonwealth Biotechnologies, Inc., Richland, Va., and resolved on an ABI Prism model 377 automated sequencing apparatus. The identity of each base in the assembled sequence was determined at least twice for each strand. Tojo et al. (46) also have sequenced the SmaI subfragment including the region upstream of the uoi gene. Their sequence of this region (GenBank no. D86464) differs from ours; it is missing 89 bp (4394 to 4482). We have determined that this deletion is not present on the wild-type Mx8 genome. Both Mx8 DNA and our subclones of this region have an MluI site absent from D86464, and primers flanking the site of the putative deletion amplify these 89 bp from both our plasmid and phage templates (data not shown).
Nucleotide sequence accession number.
The sequence of the 9.5-kb fragment of Mx8 DNA has been assigned GenBank accession no. U64984.
RESULTS
Phages Mx8 and Mx82 are independent isolates of the same temperate phage.
Three related temperate phages, Mx8, Mx81, and Mx82, are known to infect M. xanthus. To begin to characterize the genes of each phage involved in superinfection immunity, we constructed lysogens of host strain DZ1 carrying each phage as prophage and measured the EOP of each phage on each of these lysogens. As shown in Table 1, both Mx8 and Mx82 form plaques with the same high efficiency on hosts DZ1 and DZ1(Mx81) but with the same low efficiency on hosts DZ1(Mx8) and DZ1(Mx82). In contrast, phage Mx81 plates with high efficiency on hosts DZ1, DZ1(Mx8), and DZ1(Mx82) but with no measurable efficiency on DZ1(Mx81). These results show that phages Mx8 and Mx82 are homoimmune, whereas Mx81 is heteroimmune.
TABLE 1.
Myxophages Mx8 and Mx82 are homoimmunea
| Phage | EOP on indicated host
|
||
|---|---|---|---|
| DZ1(Mx8) | DZ1(Mx81) | DZ1(Mx82) | |
| Mx8 | 10−5 | 1 | 10−5 |
| Mx81 | 1 | <10−9 | 1 |
| Mx82 | 10−5 | 1 | 10−5 |
Permissive host DZ1 and lysogens of prophages Mx8, Mx81, and Mx82 were grown to an exponential density of 4 × 108 cells/ml in CT medium at 32°C, and 0.2 ml of cells was mixed with serial dilutions of high-titer stocks of each phage and plated on CT plates by the soft agar overlay method. Plates were scored for plaques after incubation at 32°C for 48 h. EOPs were calculated as the titers of each phage on each host divided by the titer on nonlysogenic host DZ1 (EOP of 1) and are the averages of at least three independent determinations.
Lysogens carrying either Mx8 or Mx82 as prophage form lawns that typically have 50 to 5,000 plaques, formed by virulent phages released spontaneously from these lysogens. This result shows that Mx8 acquires single-step mutations that confer virulence. After isolation and purification on host DZ1, the phages which form these spontaneous autoplaques form plaques with the same high EOP on both DZ1 and DZ1(Mx8). In contrast, even high-titer stocks of phage Mx81 do not include virulent mutants that form plaques on host DZ1(Mx81). Either mutation to virulence requires multiple mutational steps for Mx81, or the Mx81 prophage makes a superinfection exclusion function.
DNA extracted from Mx8 and Mx82 phage particles yields identical size distributions of products when cleaved with endonucleases EcoRI, Acc65I, AvaII, MluI, RsaI, SacI, SmaI, SphI, and StuI, suggesting that Mx8 and Mx82 are different isolates of the same phage. This result is somewhat surprising, because the natural lysogenic sources of these two phages, M. xanthus DK883 and DK893, were isolated from geographically distant places with different climates (Phoenix, Ariz., and Ames, Iowa, respectively [28]) and have different developmental phenotypes (data not shown). Cleavage of Mx81 DNA yields different sets of products, consistent with the result that Mx81 is heteroimmune.
The Mx8 immunity region maps to an 8.1-kb fragment of the Mx8 genome.
Previously, Stellwag et al. (43) showed that the 12-kb EcoRI B fragment of Mx8 DNA carries int and attP (Fig. 1). Several smaller segments of EcoRI-B also are sufficient to allow site-specific recombination. These include a 5,789-bp XhoI fragment internal to EcoRI-B (17, 25) and the 8.1-kb Sau3AI-PvuII insert of Mx8 DNA in plasmid pAY50 (27). These subfragments include an ORF, designated int, predicted to encode a product resembling other phage-encoded integrases (reference 46; see also Fig. 4).
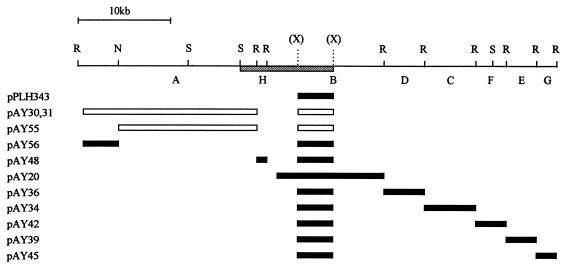
FIG. 1.
The primary determinant of Mx8 superinfection immunity, imm, lies at the right end of EcoRI-A. The physical map of phage Mx8 DNA shows restriction sites for EcoRI (R), NheI (N), and Sau3AI (S); not all XhoI (X) sites are shown. EcoRI fragments are labeled A through H, in order of decreasing size. The regions of Mx8 DNA subcloned into plasmids are shown as rectangles below the physical map. Vector pPLH343 carries the 5.8-kb XhoI subfragment of Mx8 DNA from EcoRI fragment B with the int-attP region. Mx8 forms clear plaques on DZ1(pAY45); on all other hosts, Mx8 forms turbid plaques. On hosts DZ1(pAY30), DZ1(pAY31), DZ1(pAY55), DZ1(pAY50), and DZ1(pAY54), virulent mutants in the wild-type stock of Mx8 form clear plaques. Derivatives of the permissive M. xanthus strain DZ1 were grown to exponenial density, and the EOP of Mx8 was measured on each derivative relative to that on nonlysogenic DZ1. Combinations of inserts in plasmids that confer superinfection immunity are shown as open rectangles (EOP < 10−4); inserts that do not are shown as filled rectangles (EOP = 0.3 ± 0.1). The sequenced 9.5-kb immunity region is shown as a shaded bar.
FIG. 4.
Mx8 integrase has four domains of similarity with other integrases. Regions of the predicted product of the Mx8 int gene that are similar to other phage and plasmid integrases are shown. Identical amino acids are indicated in boldface and underlined; similar residues are underlined. Numbers refer to the positions of residues within each Int protein from phages φ42 (11), D29/FRAT1 (44, 16), φ11 (48), and λ (19) and plasmid pSE211 (6). These include an N-terminal basic motif (A), a central domain (B), and two C-terminal domains (C and D) that are conserved among the family of lambdoid integrases and aligned as described elsewhere (2). A tyrosine residue within domain D (arrow) is strictly conserved among the lambdoid integrases; it is the active-site residue that forms an ester linkage with substrate DNAs (33).
To map the Mx8-encoded function(s) necessary for superinfection immunity, we constructed a series of plasmid subclones of phage Mx8 DNA. Each of these plasmids is a derivative of Kmr plasmid vector pBGS18 (42) and carries a functional Mx8 int-attP region. One plasmid, pAY20, is a subclone of the 12-kb Mx8 EcoRI B fragment. Each of the other plasmids has one of the other EcoRI fragments of the Mx8 genome inserted into plasmid pPLH343, which carries the 5.8-kb XhoI fragment internal to EcoRI-B. Each plasmid was electroporated into M. xanthus DZ1, and Kmr recombinants were selected. These recombinants carry integrated, defective Mx8 prophages with each of the eight EcoRI fragments of Mx8 DNA, representing the entire phage genome. The EOP of wild-type Mx8 on each of these hosts relative to that on the nonlysogenic host DZ1 was measured.
Mx8 plates with an EOP of 0.3 on host DZ1(pAY20), with the Mx8 EcoRI B fragment, indicating that this fragment of Mx8 does not confer immunity (Fig. 1). Phages isolated from single plaques formed by wild-type Mx8 on this host form plaques with the same relative EOP when replated on DZ1 and DZ1(pAY20). Similar results are observed with hosts carrying both the XhoI subfragment of EcoRI-B as well as each of six of the seven other EcoRI fragments of Mx8. In contrast, a host with the integrated EcoRI A fragment, DZ1(pAY30), is immune to infection and plates wild-type phage with an EOP of about 10−5. Plaques arising on this host are made by virulent mutant phages; these mutants plate with similar, high EOPs (0.3 ± 0.1) on both DZ1 and DZ1(pAY30), as does Mx8 vir1, a spontaneous virulent mutant isolated on immune host DZ1(pAY50). DZ1(pAY31), with the EcoRI A fragment cloned into pPLH343 in the opposite orientation as in pAY30, also is immune to superinfection. These results show that the primary determinant(s) of Mx8 superinfection immunity maps within its largest EcoRI fragment and/or the 5.8-kb XhoI subfragment of EcoRI-B present in pPLH343.
We iterated this mapping strategy. Two deletion derivatives of plasmids pAY30 and pAY31, pAY55 and pAY56, were constructed. Each of these carries one of the two EcoRI-NheI subfragments of EcoRI-A. Because DZ1(pAY55) retains immunity to superinfection, the primary determinant(s) of immunity must reside in the rightmost three-quarters of EcoRI-A and/or in the Mx8 DNA present in pPLH343. A derivative of host DZ1 with a much smaller integrated plasmid, pAY50, also is immune (Fig. 2). Plasmid pAY50 has the 8.1-kb PvuII-Sau3A fragment of Mx8 inserted in the ScaI-BamHI backbone of Kmr plasmid pACYC177 (12). pAY50 carries the leftmost 1.9 kb of EcoRI-A, suggesting that at least one gene necessary for immunity maps within this 1.9-kb region. Consistent with this hypothesis, we find that plasmid pAY54, a deletion derivative of pAY50 that retains this 1.9-kb region, also confers immunity when integrated into the host DZ1 genome. Conversely, plasmids pPLH343, pAY20, pAY60, and pAY62, which are missing this 1.9-kb region, do not confer superinfection immunity.
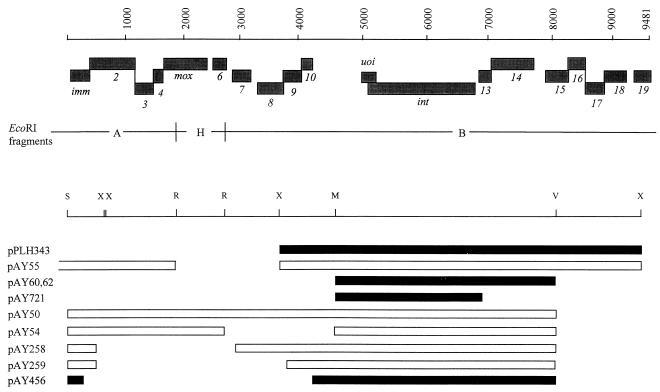
FIG. 2.
Genetic organization of the Mx8 immunity region. The 19 potential genes in the sequenced immunity region are transcribed from the conventional top strand, from left to right. The four subsets of genes, imm-2-3-4, 8-9-10, 13-14, and 15-16-17-18, are arranged so that 2 bp of the stop and start codons of adjacent genes within each cluster are shared. Below the gene map are shown the endpoints of plasmid inserts used to define the determinants of superinfection immunity and site-specific recombination. Restriction sites: Sau3AI (S; bp 1), EcoRI (R; bp 1897 and 2761), XhoI (X; bp 628, 658, 3687, and 9476), MfeI (M; bp 4585), and PvuII (V; bp 8069). The pairs of EcoO109 sites used to generate the deletions in pAY258 and pAY259 are at bp 504 and 2922 and at bp 504 and 3805, respectively. The deletion between BlpI sites in pAY456 extends from bp 278 to 4238. Bars are as described in the legend for Fig. 1.
Three additional deletion derivatives of pAY50 were constructed to assign the imm function to the first of the four ORFs within the 1.9-kb region. Derivatives of host DZ1 with integrated plasmids pAY258 and pAY259, which retain only the first of these four ORFs, are immune to superinfection. In contrast, strain DZ1(pAY456), with a deletion extending into the 3′ end of imm, is sensitive.
The Mx8 immunity region is densely packed with potential genes.
The sequence of the 9.5-kb region of Mx8 DNA represented by the inserts in plasmids pAY50 and pPLH343 was determined. Like the genome of its host, M. xanthus, this portion of the Mx8 genome is unusually rich in GC base pairs (6,375/9,481 bp = 67.2% G+C). Organisms that have GC-rich genomes accommodate this extreme of base composition in two different ways. First, codons within protein coding sequences include the base G or C in their third, most degenerate positions with high frequencies. For example, M. xanthus and Streptomyces spp. genes typically have G or C in >90% of third codon positions. Second, codons within coding sequences are also enriched for G or C in their first two positions (3). As a result, the primary sequences of both M. xanthus and Mx8 proteins include an overabundance of the four amino acids (Gly, Ala, Pro, and Arg) with codons having G and C bases in their first two positions (GGS, GCS, CCS, and CGS, respectively, where S = G or C).
As shown in Table 2 and Fig. 2, the sequence of the Mx8 immunity region includes many potential ORFs with G or C in the third positions of >75% of codons. Among these ORFs, 19 correspond to potential transcripts made from the top strand as template. The predicted products of three of the ORFs from the top strand share striking similarities with other known proteins. These ORFs, designated mox, uoi, and int, encode a nonessential DNA adenine methylase of unknown physiological function (27), a potential Mx8 excisionase, and Mx8 integrase, respectively (46).
TABLE 2.
Potential ORFs in the Mx8 imm/int regiona
| Potential gene | Designation | Frame | Interval | %G+C | RBS | Apparent Mr |
|---|---|---|---|---|---|---|
| 1 | imm(110) | 2α | 35..367 | 83.5 | TAAGGAGGG (5) | 12,426 |
| 2 | ORF 265 | 1α | 364..1158 | 88.3 | GAAGGGCGG (6) | 28,439 |
| 3 | ORF 105 | 3α | 1155..1472 | 86.6 | CGAGAGCGT (5) | 11,377 |
| 4 | ORF 59 | 2α | 1469..1648 | 84.5 | GACGAAGGG (6) | 6,532 |
| 5 | mox(258) | 1 | 1651..2427 | 86.8 | AGAGGAGGC (6) | 28,733 |
| 6 | ORF 75 | 1 | 2533..2760 | 75.7 | AGTGGGGGT (4) | 8,175 |
| 7 | orf 107 | 2 | 2870..3193 | 84.9 | CCGGGAGGA (4) | 13,751 |
| 8 | ORF 150 | 3β | 3294..3746 | 80.5 | CATGGAGCC (4) | 17,203 |
| 9 | ORF 100 | 2β | 3743..4045 | 82.8 | ACGCGAGGC (5) | 10,596 |
| 10 | ORF 80 | 1β | 4042..4284 | 82.3 | AGAGTTGGT (5) | 8,726 |
| 11 | uoi (74) | 2 | 4991..5215 | 75.3 | GAAGACGGA (4) | 7,845 |
| 12 | int(574) | 3 | 5085..6809 | 80.5 | GACGGCTGG (5) | 64,471 |
| 13 | ORF 68 | 2γ | 6857..7063 | 82.1 | AGAGGAGTC (4) | 7,219 |
| 14 | ORF222 | 1γ | 7060..7728 | 91.9 | GCGCGGGGT (5) | 25,257 |
| 15 | ORF130 | 2δ | 7898..8287 | 73.4 | CAAGGGGCA (7) | 13,913 |
| 16 | ORF93 | 1δ | 8284..8562 | 89.0 | TCCGGAGGG (4) | 10,504 |
| 17 | ORF104 | 3δ | 8559..8873 | 89.3 | GAAGGCGGT (6) | 11,785 |
| 18 | ORF106 | 2δ | 8870..9190 | 94.3 | GCAGGAGGG (6) | 11,834 |
| 19 | ORF(>68) | 2 | 9278..>9481 | 85.1 | TGAGGGAGT (7) | >7,869 |
Gene 15 is included because it begins an overlapping array of genes, even though it has <75% G or C in the third position. Four of these genes, imm, mox, uoi, and int, with functions suggested from our genetic analysis, are given three-letter names, followed by the number of amino acid residues in their predicted products. Other potential ORFs are assigned numbers corresponding to the numbers of amino acid residues in their products, including the N-terminal methionine residues. We assume that ORFs which start with GTG will have N-terminal methionines. (Although many mycophage L5 ORFs begin with TTG start codons [18], we did not find these potential starts for Mx8 genes.) The coordinates of the top strand, translation frames, and gene intervals are as in GenBank accession no. U64984. Four sets of genes are organized as overlapping arrays, designated α, β, γ, and δ. The percent G+C occurrence is for the third codon position, not including codons for N-terminal methionine residues. Ribosome-binding sites (RBS) were assigned as the best-fit similarities (underlined) to the sequence TAAGGAGGT, complementary to the 3′ end of M. xanthus 16S rRNA (41). Numbers in parentheses after each RBS indicate the spacing between the homology shown and the start codon for each ORF.
ATG or GTG start codons for all but four of the potential ORFs in the top strand are preceded 4 to 6 bp by ribosome-binding sites that share at least four bases of homology with the 3′ end of the 16S rRNA of M. xanthus (41). Genes imm, mox, and 18 have exceptional ribosome-binding sites, with eight, six, and six bases of uninterrupted homology, respectively. Most identified M. xanthus genes have less striking ribosome-binding sites.
Four subsets of ORFs in the top strand are arranged in the same way as the nin genes of phage λ (23, 37). Within each subset, the coding sequences of adjacent ORFs overlap; the first 2 bp of the TGA stop codon of one ORF are the last 2 bp of the start codon (ATG or GTG) of its successor. These overlapping arrays include genes 1(imm)-2-3-4 (upstream of mox), 8-9-10 (between mox and int), 13-14 (downstream of int), and 15-16-17-18 (Fig. 2). Each gene at the start of the three arrays has a notable ribosome-binding site with five bases of homology to the 3′ end of the 16S rRNA of M. xanthus.
The predicted product of the Mx8 imm gene is highly basic.
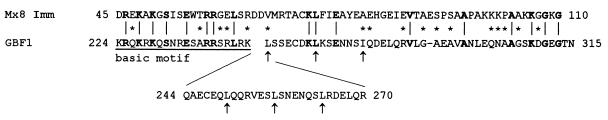
The majority of predicted products of the genes in the Mx8 immunity region have only weak similarities with the sequences of proteins of known function; among these is the product of the imm gene. Because the primary immunity function for most temperate phages is a repressor gene that encodes a specific DNA-binding protein, we used the BLAST program (1) to search for related DNA-binding proteins. As shown in Fig. 3, Imm is most similar to Arabidopsis thaliana G-box-binding factor 1 (GBF1) (40). The most striking similarities are in two regions, a central domain, corresponding to the DNA-binding basic motif of GBF1, and the C terminus. For GBF1, these regions are separated by an extensive leucine zipper. The C terminus of Imm is also similar to the C terminus of several eukaryotic histones H1 (reference 22 and data not shown).
FIG. 3.
Imm is most similar to A. thaliana GBF1. Alignment of the C termini of the predicted Imm and GBF1 sequences resulting from a BLAST search using the PAM250 weight matrix for similarity is shown. Identical residues are in boldface; similar residues are indicated by asterisks. The basic motif, essential for the DNA-binding activity of GBF1, is underlined. Periodically repeated leucine and isoleucine residues in the zipper motif of GBF1 are indicated by arrows. The two proteins also show limited similarity in their proline-rich N termini (not shown).
Immediately distal to the imm array is the mox gene. None of the five genes (6 to 10) between mox and uoi-int is essential for either the lytic or lysogenic development of Mx8. A deletion mutant missing these genes, Mx8 del1, forms plaques and stable lysogens with host DZ1 (27).
The Mx8 int gene has two alternate translation start codons.
The longest ORF within the 8.1-kb Mx8 fragment, int, encodes Mx8 integrase. The int gene has several potential translation initiation codons. The first of these, GTG-5085, initiates a predicted product with an N-terminal sequence similar to that of temperate Staphylococcus aureus phage φ42 integrase (Fig. 4 and reference 11). Both the first and second (GTG-5208) potential starts lie within the uoi coding sequence (bp 4991 to 5215). To determine which of these two GTG start codons is used, we made site-directed changes of each GTG codon to GCG and tested whether these mutations abolish integrase function.
Results in Table 3 show that plasmid pAY721, which carries a 2.2-kb fragment of Mx8 DNA with the wild-type uoi and int genes, gives rise to Kmr recombinants after electroporation of sensitive host DZ1 with a high efficiency. Otherwise isogenic plasmids pAY979 and pAY754, with changes of GTG-5085 and GTG-5208 codons to GCG, respectively, also give rise to Kmr electroporants. In contrast, plasmid pAY990, with changes of both GTG codons to GCG, does not give rise to Kmr electroporants. These results show that at least one of these first two GTG codons is necessary, and either one of these first two GTG codons is sufficient, to initiate the translation of a functional int gene product.
TABLE 3.
The Mx8 int gene has two alternate start codonsa
| Plasmid | Genotype | Avg EOE (103 μg−1) ± SD | Phenotype |
|---|---|---|---|
| pAY721 | Wild type | 2.1 ± 1.2 | Int+ |
| pAY979 | intVA1 | 1.0 ± 0.8 | Int+ |
| pAY754 | intVA42 | 0.5 ± 0.4 | Int+ |
| pAY990 | intVA1VA42 | <0.001 | Int− |
Plasmid pAY721 with the wild-type int gene or its otherwise isogenic derivatives with each or both changes of potential GTG-5085 and GTG-5208 start codons to GCG, VA1, and VA42, respectively, were electroporated into host DZ1. The efficiency of electroporation (EOE) is the frequency of Kmr electroporants per microgram of plasmid DNA; values are the averages of four independent experiments. The double-mutant plasmid fails to integrate.
As shown in Fig. 4, Mx8 Int protein can be divided into four domains, each with similarity to other proteins. The first (domain A) is its highly basic N terminus. Between this N terminus and two conserved domains C and D that Mx8 Int shares with other integrases (2) is domain B, which includes stretches similar to Staphylococcus phage ϕ42 (11), Mycobacterium phage FRAT1/D29 (16, 44), and Streptomyces plasmid pSE211 (6) integrases.
The Mx8 uoi gene encodes a helix-turn-helix (HTH) protein.
Overlapping the beginning of int is the second-longest potential ORF in the Mx8 immunity region, uoi. This ORF has 12 potential start codons, only 2 of which are preceded by recognizable ribosome-binding sites. One, a GTG codon at bp 4490, would initiate a product of 241 amino acids, encoded by a gene with relatively poor codon usage (66% G+C in the third position), whereas the other, a GTG codon at bp 4991, would initiate a product of 74 amino acids with more typical codon usage (75.3% G+C in the third position). The N terminus of the latter product, Uoi, shares significant sequence similarity with P22 Xis protein (26) and Methylobacterium DcmR repressor (24). Uoi has two additional domains, (i) a central domain that also shares similarity with DcmR repressor and (ii) a basic C terminus (Fig. 5).
FIG. 5.
Excisionases are likely HTH proteins. Alignment of the potential Mx8 excisionase, Uoi, with other phage, plasmid, and transposon excisionases is shown. The alignment of P22 Xis with Uoi was found by using the BLAST program (1); additional alignments were made by using iterations of the BLAST program with published Xis sequences. Residues identical to those of Uoi are in boldface; similar residues are underlined. The N terminus of Mx8 Uoi (domain A) scores extremely well as an HTH protein, as does its closest relative, Methylobacterium DcmR repressor (24), using the weight matrix of Dodd and Egan (13). The SD (standard deviation) scores for these proteins, as well as for P22 (26), pSE101 (5), pSE211 (6), pSAM2 (4), and Tn1545 (34) excisionases, are given; a score of 2.5 or better signifies that a protein has an HTH motif with near certainty (13). After the HTH motif in Mx8 Uoi is a second region of similarity with DcmR (domain B). Uoi terminates with a basic motif. We note that only one other phage excisionase, λ Xis (19), has a highly basic C terminus; this region of λ Xis is thought to be involved in specific interactions with Int (30).
Uoi begins with an HTH sequence motif that scores dramatically well by the metric of Dodd and Egan (13) for recognizing these motifs. As shown in Fig. 5, alignment of the N terminus of Uoi with P22 Xis and integrative plasmid and transposon excisionases from Streptomyces spp. allows us to identify several highly conserved residues which span the HTH motif of Uoi. These residues are conserved, not only among this family of proteins, but also among many HTH proteins. This comparison suggests that temperate phage excisionases use variations of the HTH motif to bind DNA.
DISCUSSION
A 9.5-kb region of the 49-kb Mx8 genome with a base composition of 67.2% GC is densely packed with 19 potential ORFs. These genes are transcribed in one direction from the conventional top strand of the sequence and, with the exception of uoi and int, do not overlap extensively. More than half (13 of 19) of these potential genes are arranged in overlapping arrays, such that the first 2 bp of the termination codon of one gene are the last 2 bp of the start codon of its successor within each array.
With one exception, when genes are not arranged in overlapping arrays, they are separated from one another by short intervals, which tend to be AT rich and may contain promoters. The exception is the large (707-bp) gap between the second array (8-9-10) and uoi, which includes no long ORFs on either strand. Although uoi has no fewer than 12 potential start codons, only start codons after GTG-4829 define ORFs with >75% G or C in the third codon position. Only one of these, GTG-4991, has a good ribosome-binding site.
The Mx8 uoi and int genes overlap extensively. Although Tojo et al. (46) have assigned the GTG codon at bp 5208 as the start codon for int, we find that the int gene has two potential starts, either one of which gives rise to a functional product. Furthermore, we find the similarity between the sequences of the predicted product of uoi and other phage excisionases particularly instructive. A subset of this family of proteins may use an N-terminal HTH motif to bind specific DNA sites.
We have chosen genes in the Mx8 immunity region as those with codons having >75% G+C in the third codon position. Initially, we thought that this measure would be conservative, because M. xanthus genes typically have >90% G or C in this position. Surprisingly, only 2 of 19 Mx8 genes, 14 and 18, meet this stringent requirement. Because genomes with high GC base compositions are enriched for G+C in all three codon positions (3), the Mx8 genome accommodates this slack in the third position by enrichment for G+C in the first two codon positions, resulting in enrichment for the amino acids Gly, Pro, Ala, and Arg in the primary sequences of potential Mx8 proteins.
Sequences of the predicted products of most genes in the Mx8 immunity region, with the exceptions of mox, uoi, and int, have little similarity with those of other known proteins. One notable exception is the product of gene 14, which has a conserved domain shared by gram-positive endo-β-1,4-glucanases (20, 39). Quillet et al. (35) have shown that the M. xanthus celA gene encodes an endo-β-1,4-glucanase closely related to the licheninases from Actinomyces spp., suggesting that the coding sequences for these enzymes have been transferred horizontally from actinomycetes to myxobacteria. Phages such as Mx8 are possible vehicles for this transfer. Like many phages, the host range of Mx8 extends across different species of Myxococcus, including M. virescens and M. fulvus in addition to M. xanthus (data not shown), and may extend to different genera with genomes having a range of base compositions.
Prokaryotic viruses are among the most deeply rooted organisms in evolutionary history. At least one archaeal virus, SSV1, makes an integrase closely related to phage integrases (29, 32). Whether this and other similarities argue that viruses existed prior to the archaea-eubacteria split, or that viruses mediate extensive lateral transfer between domains, remains open to debate. Independent of these evolutionary questions, we may ask whether phages represent the finite permutations of a relatively limited repertoire of genes. If so, then the study of phages of diverse prokaryotes may hold few new surprises. Are the phages simply variations on a small number of baroque genetic themes, or do they offer new, classical paradigms for our understanding of genes and their regulation?
The lesson that we have learned from studying the genomes of diverse phages is that the repertoire of their genes is far from limited. Thus, the sequence of mycophage L5 yields only about a dozen of 88 genes with functions resembling lambdoid or other phage homologues (18). The sequence of one-fifth of the myxophage Mx8 genome tells much the same story. Few Mx8 gene products are similar to other phage proteins or, for that matter, other known proteins. Clearly, if phages have only finite permutations of a limited number of genes, then only a small subset of phages have been characterized, and many surprises are yet to come from the study of phage genetics.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V, Pierson III L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 4.Boccard F, Smokvina T, Pernodet J L, Friedmann A, Guerineau M. The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J. 1989;8:973–980. doi: 10.1002/j.1460-2075.1989.tb03460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown D P, Idler K B, Backer D M, Donadio S, Katz L. Characterization of the genes and attachment sites for site-specific integration of plasmid pSE101 in Saccharopolyspora erythraea and Streptomyces lividans. Mol Gen Genet. 1994;242:185–193. doi: 10.1007/BF00391012. [DOI] [PubMed] [Google Scholar]
- 6.Brown D P, Idler K B, Katz L. Characterization of the genetic elements required for site-specific integration of plasmid pSE211 in Saccharopolyspora erythraea. J Bacteriol. 1990;172:1877–1888. doi: 10.1128/jb.172.4.1877-1888.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown N L, Morris D W, Parrish J H. DNA of Myxococcus bacteriophage Mx1: macromolecular properties and restriction fragments. Arch Microbiol. 1976;108:221–226. doi: 10.1007/BF00428955. [DOI] [PubMed] [Google Scholar]
- 8.Burchard R P, Dworkin M. A bacteriophage for Myxococcus xanthus: isolation, characterization and relation of infectivity to host morphogenesis. J Bacteriol. 1966;91:1305–1313. doi: 10.1128/jb.91.3.1305-1313.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burchard R P, Voelz H. Bacteriophage infection of Myxococcus xanthus during cellular differentiation and vegetative growth. Virology. 1972;48:555–566. doi: 10.1016/0042-6822(72)90066-9. [DOI] [PubMed] [Google Scholar]
- 10.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage Mx4, a generalized transducing phage of Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 11.Carroll D, Kehoe M A, Cavanagh D, Coleman D C. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages phi 13 and phi 42. Mol Microbiol. 1995;16:877–893. doi: 10.1111/j.1365-2958.1995.tb02315.x. [DOI] [PubMed] [Google Scholar]
- 12.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dworkin M. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol. 1962;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisselsoder J, Campos J M, Zusman D R. Physical characterization of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:179–189. doi: 10.1016/0022-2836(78)90432-1. [DOI] [PubMed] [Google Scholar]
- 16.Haeseleer F, Pollet J F, Bollen A, Jacobs P. Molecular cloning and sequencing of the attachment site and integrase gene of the temperate mycobacteriophage FRAT1. Nucleic Acids Res. 1992;20:1420. doi: 10.1093/nar/20.6.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartzell P, Kaiser D. Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus. J Bacteriol. 1991;173:7625–7635. doi: 10.1128/jb.173.23.7625-7635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfull G F, Sarkis G J. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 19.Hoess R H, Foeller C, Bidwell K, Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci USA. 1980;77:2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofemeister J, Kurtz A, Borriss R, Knowles J. The beta-glucanase gene from Bacillus amyloliquefaciens shows extensive homology with that of Bacillus subtilis. Gene. 1986;49:177–187. doi: 10.1016/0378-1119(86)90278-7. [DOI] [PubMed] [Google Scholar]
- 21.Kashefi K, Hartzell P L. Genetic suppression and phenotypic masking of Myxococcus xanthus frzF− defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 22.Knowles J A, Childs G J. Comparison of the late H1 histone genes of the sea urchins Lytechinus pictus and Strongelocentrotus purpuratus. Nucleic Acids Res. 1986;14:8121–8133. doi: 10.1093/nar/14.20.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroger M, Hobom G. A chain of interlinked genes in the ninR region of bacteriophage lambda. Gene. 1982;20:25–38. doi: 10.1016/0378-1119(82)90084-1. [DOI] [PubMed] [Google Scholar]
- 24.La Roche S D, Leisinger T. Sequence analysis and expression of the bacterial dichloromethane dehalogenase structural gene, a member of the glutathione S-transferase supergene family. J Bacteriol. 1990;172:164–171. doi: 10.1128/jb.172.1.164-171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S F, Shimkets L J. Site-specific integration and expression of a developmental promoter in Myxococcus xanthus. J Bacteriol. 1988;170:5552–5556. doi: 10.1128/jb.170.12.5552-5556.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leong J M, Nunes-Duby S E, Oser A B, Lesser C F, Youderian P, Susskind M M, Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986;189:603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- 27.Magrini V, Salmi D, Thomas D, Herbert S K, Hartzell P L, Youderian P. Temperate Myxococcus xanthus phage Mx8 encodes a DNA adenine methylase, Mox. J Bacteriol. 1997;179:4254–4263. doi: 10.1128/jb.179.13.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin S, Sodergren E, Masuda T, Kaiser D. Systematic isolation of transducing phages of Myxococcus xanthus. Virology. 1978;88:44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 29.Muskhelishvili G, Palm P, Zillig W. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol Gen Genet. 1993;237:334–342. doi: 10.1007/BF00279436. [DOI] [PubMed] [Google Scholar]
- 30.Numrych T E, Gumport R I, Gardner J F. Characterization of the bacteriophage lambda excisionase (Xis) protein: the C-terminus is required for Xis-integrase cooperativity but not for DNA binding. EMBO J. 1992;11:3797–3806. doi: 10.1002/j.1460-2075.1992.tb05465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orndorff P, Stellwag E, Starich T, Dworkin M, Zissler J. Genetic and physical characterization of lysogeny by bacteriophage Mx8 in Myxococcus xanthus. J Bacteriol. 1983;154:772–779. doi: 10.1128/jb.154.2.772-779.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter W D, Zillig W. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology. 1991;185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 33.Pargellis C A, Nunes-Duby S E, de Vargas L M, Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 34.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989;8:2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quillet L, Barray S, Labedan B, Petit F, Guespin-Michel J. The gene encoding the beta-1,4-endoglucanase (CelA) from Myxococcus xanthus: evidence for independent acquisition by horizontal transfer of binding and catalytic domains from actinomycetes. Gene. 1995;158:23–29. doi: 10.1016/0378-1119(95)00091-j. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatas T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 38.Sanger F S, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schimming S, Schwarz W H, Staudenbauer W L. Structure of the Clostridium thermocellum gene licB and the encoded beta-1,3-1,4-glucanase. A catalytic region homologous to Bacillus lichenases joined to the reiterated domain of clostridial cellulases. Eur J Biochem. 1992;204:13–19. doi: 10.1111/j.1432-1033.1992.tb16600.x. [DOI] [PubMed] [Google Scholar]
- 40.Schindler U, Menkens A E, Beckmann H, Ecker J R, Cashmore A R. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimkets L J, Woese C R. A phylogenetic analysis of the myxobacteria: basis for their classification. Proc Natl Acad Sci USA. 1992;89:9459–9463. doi: 10.1073/pnas.89.20.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 43.Stellwag E, Fink J M, Zissler J. Physical characterization of the genome of the Myxococcus xanthus bacteriophage Mx8. Mol Gen Genet. 1985;199:123–132. doi: 10.1007/BF00327521. [DOI] [PubMed] [Google Scholar]
- 44.Suissa, M., J. Wyse, T. Bar-On, and J. Kuhn. 1994. DNA sequence and analysis of the mycobacteriophage D29 origin and its integrase-like protein. GenBank no. X70352.
- 45.Taketo A. DNA transfection of Escherichia coli by electroporation. Biochim Biophys Acta. 1988;949:318–324. doi: 10.1016/0167-4781(88)90158-3. [DOI] [PubMed] [Google Scholar]
- 46.Tojo N, Sanmiya K, Sugawara H, Inouye S, Komano T. Integration of bacteriophage Mx8 into the Myxococcus xanthus chromosome causes a structural alteration at the C-terminal region of the IntP protein. J Bacteriol. 1996;178:4004–4011. doi: 10.1128/jb.178.14.4004-4011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 48.Ye Z-H, Buranen S L, Lee C Y. Sequence analysis and comparison of int and xis genes from Staphylococcal bacteriophages L54a and phi-11. J Bacteriol. 1990;172:2568–2575. doi: 10.1128/jb.172.5.2568-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]