Abstract
The specific mechanism of clear cell renal cell carcinoma (ccRCC) progression, a pathological type that accounts for the highest proportion of RCC, remains unclear. In this study, bioinformatics analysis of scRNA-seq dataset in ccRCC revealed that MIOX was a gene specifically down-regulated in tumor epithelial cells of ccRCC. Analysis of the TCGA database further validated the association between decreased MIOX mRNA levels and ccRCC malignant phenotype and poor prognosis. Immunohistochemistry indicated the down-regulation of MIOX in ccRCC tissues compared to paired adjacent renal tissues, with further down-regulation of MIOX in the primary tumors of patients with primary metastasis compared to those without metastasis. Also, patients with low expression of MIOX showed shorter metastasis-free survival (MFS) compared to those with high MIOX expression. In vitro results showed that overexpression of MIOX in ccRCC cells inhibited the proliferation, migration and invasion and promoted apoptosis. Mechanistically, up-regulation of MIOX inhibited autophagy to elevate the levels of ROS, and thus suppressed STAT3/c-Myc-mediated epithelial-mesenchymal transition in ccRCC cells. In vivo data further confirmed that increased MIOX expression suppressed the growth and proliferation of RCC cells and reduced the ability of RCC cells to form metastases in the lung. This study demonstrates that MIOX is an important regulatory molecule of ccRCC, which is conducive to understanding the potential molecular mechanism of ccRCC progression.
Keywords: Clear cell renal cell carcinoma (ccRCC), MIOX, Autophagy, Epithelial mesenchymal transition (EMT), Metastasis
1. Introduction
Renal cell carcinoma (RCC) arises from the renal tubular epithelium and is among the top 10 male and female malignancies [1]. RCC encompasses multiple pathological types, the highest percentage of which is clear cell renal cell carcinoma (ccRCC) [2]. More than 50 % of RCC patients are asymptomatic, and more than 10 % of patients present with metastases at first diagnosis [3,4]. The 5-year survival rate for metastatic RCC is less than 10 %, suggesting extremely poor prognosis [5]. Tyrosine kinase inhibitors and immune checkpoint inhibitors have shown to be effective against advanced and metastatic RCC (mRCC); yet, some patients present with low objective response rate (ORR), poor tolerance, and drug resistance, which limits further efficacy [6]. Hence, studying the underlying molecular mechanism in RCC progression and providing promising treatment strategies is necessary.
Numerous different kinds of cellular components exist within tumors, which together form a complex tumor microenvironment (TME) that mediates tumor growth, invasion, metastasis, and therapeutic response. Deep insights into TME can help identify the functional regulation of specific cell types in tumor progression [7]. Single-cell RNA sequencing (scRNA-seq) has become an advanced tool to reveal complexity and heterogeneity in TME based on the genome of single cells, which helps study the transcript changes of specific cell types in tumor tissues [8]. In this study, by bioinformatics analysis of the ccRCC scRNA-seq dataset [9], myo-inositol oxygenase (MIOX) was identified as a new regulatory gene for ccRCC. MIOX is a type of inositol metabolizing enzyme specifically expressed in the kidney [10]. A few studies have shown that MIOX regulates tumor ferroptosis in several cancer types [[11], [12], [13]]; however, relevant studies on ccRCC are lacking.
Macroautophagy, also known as autophagy, is an evolutionarily conserved and ancient cellular catabolic program that mediates quality control of proteins and organelle and cell survival under starvation or stress [14]. Autophagy has a dual role in the tumor, which can promote or inhibit tumor progression [[15], [16], [17], [18], [19]]. The anti-tumor effect is achieved mainly through inhibiting autophagy, with autophagy inhibitors chloroquine (CQ) and HCQ showing promising efficacy in clinical trials [[20], [21], [22], [23], [24], [25]].
Epithelial-mesenchymal transition (EMT) is a reversible program with the loss of the epithelial appearance of the cells, the dissolution of intercellular junctions and the loss of apical-basal cell polarity, leading to a mesenchymal phenotype [26]. Activation of EMT mediates metastasis in almost all types of cancer, including ccRCC [27,28]. In addition to classical EMT-inducing transcription factors such as ZEB, SNAIL and TWIST families [29], several cancer-promoting signals promote EMT, including STAT3 and c-Myc [30,31]. Also, studies have found that autophagy can activate or inhibit EMT to regulate tumors; yet, the regulatory effect of autophagy on EMT in ccRCC is still not fully understood [32].
In the present study, we corroborated the significant down-regulation of MIOX in ccRCC tissues relative to adjacent renal tissues, with a negative correlation between MIOX expression levels in ccRCC tissues and the malignant phenotype as well as poor prognosis of ccRCC. Overexpression of MIOX impaired the migration and invasion of ccRCC cells, and inhibited metastasis in vivo. Mechanistically, up-regulation of MIOX inhibited autophagy and elevated reactive oxygen species (ROS) levels, resulting in the suppression of the STAT3/c-Myc-mediated EMT. This study confirms that MIOX is an important regulatory molecule of ccRCC, which helps us further understand the underlying mechanism in ccRCC progression.
2. Materials and methods
2.1. Quality control and cell type recognition of scRNA-seq data
First, we obtained the scRNA-seq counting matrix from Young et al. [9]. In this study, 21 samples were involved, consisting of 12 tumor samples and 9 adjacent normal samples from 3 patients. Seurat (version 3.0.1) was used to conduct the quality control process [33]. Unique molecular identifiers (UMIs) were identified, and low-quality single cells with UMIs less than 200 or UMI counts from mitochondria greater than 10 % were removed. IntegrateData function in Seurat was used to eliminate batch effects in patients. We used the top 2000 variable genes and the top 30 principal components in this process. The regression of the effect of UMI counts and the percentage of UMI counts from mitochondrial sources was performed by using the ScaleData function. Next, Seurat's FindClusters function (resolution = 1.1) was used to identify the main cell cluster, after which the results were visualized [34]. The cell types were annotated by both R package SingleR and marker genes of each cluster according to the CellMarker database [35]. The markers corresponding to the major cell types used in the analysis are shown in Table S1. Epithelial cells from ccRCC and normal controls were extracted and differentially expressed genes (DEGs) were analyzed by the function “FindMarkers” of R package Seurat. The threshold for identifying DEGs was adj. p val<0.01 and |log2 FC|>1.0.
2.2. Analysis of RNA-seq data
A total of 533 tumor samples and 72 normal samples were included in the Cancer Genome Atlas (TCGA) ccRCC database. The expression matrix and patient clinical information were obtained from http://xena.ucsc.edu/. The RNA-seq normalized counts of TCGA were log-transformed, and according to the expression of target genes, patients were separated into high and low-expression groups for clinical correlation analysis and survival analysis.
RNA-seq data from 786-O-Vector (control group) and 786-O-MIOX (overexpression group) were provided by Biomarker Technologies. Hallmark gene sets as characteristic gene sets in Gene Set Enrichment Analysis (GSEA) were used to analyze RNA-seq data.
2.3. Patients and clinical samples
Fresh and frozen tumor tissues and paired adjacent renal tissues in ccRCC for quantitative real-time PCR detection (n = 26) were collected from patients with surgically resected specimens from 2014 to 2018. In addition, paraffin-embedded tissues from ccRCC patients (n = 153) were used to construct tissue microarrays (TMA). TMA was used for immunohistochemistry staining. All patients whose specimens were used for TMA underwent surgery in 2015, with at least 5 years of postoperative follow-up. The tumor tissues were identified as clear cell subtype at Nanjing Drum Tower Hospital based on WHO classification criteria [2]. We collected detailed clinical and pathological information of patients for further analysis and obtained informed consent from all included patients. Ethics approval was obtained from the Nanjing University Medical School affiliated Nanjing Drum Tower Hospital.
2.4. Cell lines and cell culture
Human RCC cell lines 786-O, 769-P, Caki-1, ACHN and A498 as well as human proximal tubule epithelial cell line HK-2 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai). Murine RCC cell line Renca was acquired from the American Type Culture Collection (Rockville, MD, USA). RPMI-1640 medium containing 10 % fetal bovine serum (FBS) (Gemini) was used for the culture of 786-O, 769-P and Renca. McCoy's 5A medium containing 10 % FBS was used for Caki-1 cells. DMEM medium containing 10 % FBS was used for ACHN, A498 and HK-2. All complete mediums were supplemented with 100 μg/mL streptomycin and 100 IU/mL penicillin. The cells were cultured in a humidified atmosphere containing 5%CO2/95 % air at 37 °C.
2.5. Quantitative real-time PCR (qRT-PCR)
Extraction of total RNA was performed on tissue or cell samples using TRIzol reagent (Invitrogen Biotech). Synthesis of cDNA was carried out with 5 × PrimeScript RT Master Mix (TaKaRa Biotech). The implementation of qRT-PCR relied on SYBR Premix Ex Taq (TaKaRa Biotech) binding to cDNA and generating fluorescence, and the qRT-PCR program was operated on the StepOne Real-Time PCR System. The mRNA expression of each gene relative to ACTB was measured by the 2-△△CT method. The primer sequences are shown in Table S2.
2.6. Immunohistochemistry (IHC)
IHC was performed for TMA sections and lung tissues from the murine metastatic model. After roasting at 65–80 °C for 2 h, the tissues were dewaxed, and hydrated. Next, endogenous peroxidase was removed, and antigen repair and blocking were performed. Samples were then incubated with specific primary antibodies at 4 °C overnight and secondary antibodies at room temperature for 1h. The primary antibodies used for IHC are shown in Table S3. The staining criteria were as follows: the score of 0, 1, 2 and 3 represented negative, low, moderate and high staining intensity, respectively. The score of 0, 1, 2, 3 and 4 represented 0 %, 1%–25 %, 26%–50 %, 51%–75 % and >75 % in the staining range, respectively. Staining intensity multiplied by staining range is the final IHC score (intensity score × range score). The low and high expression groups were characterized as having a final IHC score ≤4 and > 4, respectively.
2.7. Western blot
Cells and tissue samples were collected and lysed in RIPA buffer or WB/IP lysis buffer (Beyotime Biotech) supplemented with proteinase inhibitors (Biomake Biotech) and phosphatase inhibitors (Sigma), and tissue samples were homogenized in a homogenizer. SDS-PAGE was used for electrophoresis and separation of proteins, and PVDF membranes (Bio-Rad, Hercules, CA) activated by methanol were used for protein transfer. Blocking of PVDF membranes was then incubated with 5 % skim milk for 1 h and then with pre-configured primary antibodies overnight at 4 °C and then with secondary antibodies at room temperature for 1 h; the primary antibodies used for the Western blot are shown in Table S3. Proteins were finally quantified with an electrochemiluminescence (ECL) system (Tanon Co., Ltd). Quantification of gray values of protein bands was performed using ImageJ software.
2.8. siRNA and plasmid transfection
Cells were transiently transfected with siRNA and plasmids purchased from Genechem (Shanghai) using lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Briefly, siRNA or plasmid was first diluted with 100 μL of serum-free medium, and then lipofectamine 2000 was diluted with 100 μL of serum-free medium. The two liquids were mixed and left at room temperature for 15 min. After washing the cells with PBS, 1 mL of serum-free medium and the mixed liquid was added to each well. Cells were incubated in an incubator with 5 % CO2 at 37 °C for 6–8 h and then changed to the complete medium for further incubation. Cellular RNA was extracted after 24 h, while cellular protein was extracted after 48 h to test the transfection efficiency.
2.9. MTT assay
After digestion and counting, cells were plated into a 96-well plates (786-O 1000 per well, 769-P 2000 per well) and cultured for 0, 24, 48 and 72 h in RPMI 1640 containing 10 % FBS. At each time point, 10 μl of sterile MTT dye (5 mg/mL) was added to each well and incubated for another 4 h at 37 °C. After removal of the medium, 150 μl of DMSO was added to each well and properly mixed for another 10 min. The absorbance at 490 nm was determined using a microplate reader (Infinite M200 Pro, TECAN®).
2.10. Transwell assay to evaluate migration and invasion
Evaluation of migration and invasion ability in cells transfected or treated with 1 μM Colivelin (MedChemExpress) and 20 μM 10058-F4 (MedChemExpress). For migration evaluation, cells were digested, counted, uniformly distributed into serum-free medium, and then seeded into the upper transwell chamber (8 μm, Corning) (786-O: 2.5 × 104 cells, 769-P: 5 × 104 cells). The lower chamber contained a serum-containing medium. After 12 h of cell migration, the Transwell chambers were taken out. For invasion evaluation, cells were seeded in the upper chamber with matrigel (BD Biosciences) on the bottom (786-O: 2.5 × 104 cells, 769-P: 5 × 104 cells). After 24 h of cell invasion, Transwell chambers were taken out. Finally, the migrating and invading cells were washed and fixed. Staining was performed using the Crystal Violet Staining Solution (Beyotime Biotech).
2.11. Wound-healing assay
After digestion and counting, the cells were seeded into 6-well plates. After the cell reached 90 % confluence, a line was drawn using a marker on the bottom of the dish, and then a sterile 200-μl pipet tip was used to scratch three separate wounds through the cells. The cells were gently rinsed twice with PBS to remove floating cells and incubated in a serum-free medium. Images of the scratches were taken by using an inverted microscope at × 10 magnification at 0, 16, and 24 h of incubation.
2.12. Flow cytometry
Detection of apoptotic cells and intracellular ROS levels was conducted with the Apoptosis Detection Kit (Yeasen) and the ROS Assay Kit (Beyotime Biotech), respectively, by flow cytometry (NovoCyte Flow Cytometer, ACEA Biosciences). The testing process was carried out in accordance with the manufacturers' specifications. The data were analyzed with FlowJo (version 7.6.5).
2.13. Immunofluorescence
Briefly, cells were washed with PBS, fixed using 4 % paraformaldehyde, permeabilized with 0.3 % Triton X-100 and blocked using BSA. Then, pre-configured primary antibodies were added and incubated at 4 °C overnight. The next day cells were incubated with secondary antibody at room temperature for 1 h. DAPI was incubated for 5 min and then cells were washed with PBS. Photographs were taken with the THUNDER Imaging Systems (LEICA). The primary antibodies used for immunofluorescence are shown in Table S3.
2.14. Lentivirus transfection
Lentiviruses containing short hairpin RNAs (shRNAs) of MIOX, overexpressed plasmid with flag labels of human and mouse MIOX and GFP-RFP-LC3 plasmid were purchased from Genechem (Shanghai). Lentivirus transfection of cells was performed according to the protocol provided by the manufacturer. Lentivirus-transfected cells were selected by puromycin.
2.15. LC3 fluorescence assay
Cells transfected with lentivirus containing GFP-RFP-LC3 plasmid were seeded in 24-well plates. The THUNDER Imaging Systems (LEICA) was used to detect the fluorescence after 2 h of starvation.
2.16. Transmission electron microscopy (TEM)
Samples were first fixed in the electron microscope fixation solution (Servicebio), and then dehydrated, permeated, embedded and photographed under the electron microscope [36].
2.17. Autophagy flux detection
Autophagy flux was measured by detecting the expression of LC3 and SQSTM1 proteins. Cell samples were treated accordingly, and cellular proteins were then extracted. Protein expression of LC3 and SQSTM1 was assessed using Western blot assay for characterization of autophagy flux. Chloroquine (CQ) (Sigma) or rapamycin (MedChemExpress) was used to block or activate autophagy flux.
2.18. Animal experiments
A 6–8 weeks old, male BALB/c mice were purchased from SPF Biotechnology Co., Ltd. (Beijing). All the animals were housed in an environment with a temperature of 22 ± 1 °C, a relative humidity of 50 ± 1 %, and a light/dark cycle of 12/12 h. All animal studies (including the mice euthanasia procedure) were done in compliance with the regulations and guidelines of Nanjing Drum Tower Hospital institutional animal care and conducted according to the AAALAC and the IACUC guidelines.
Renca-luci (luciferase) cells stably transfected with overexpressed MIOX or vector lentivirus were digested, washed, and resuspended in PBS, followed by transplanted into the middle of the back of the mice by subcutaneous injection (1 × 105 cells per mouse). After 3 weeks, subcutaneous tumors were completely dissected and weighed, and based on the formula volume = (long diameter × short diameter^2) × 0.52 to estimate the tumor volume.
To construct pulmonary metastatic models, cells stably transfected with overexpressed MIOX or vector lentivirus were resuspended with PBS and injected to the mouse tail vein (1 × 105 cells per mouse). Two weeks after injection, in vivo imaging of mice was administrated with the IVIS spectrum imaging system (PerkinElmer) based on the luciferase label. Data analyses were conducted with LivingImage software. Three weeks after injection, murine lung tissues were harvested. Lung tissues were used for protein extraction and to make tissue sections.
2.19. Statistical analysis
GraphPad Prism 7 software and IBM SPSS Statistics 21 were used for statistical analysis. Analyses of data from normal distribution presented as mean ± SD were conducted using Student's t-test. Analyses of categorical data were performed with the Chi-square test or Yates's correction for continuity. The Pearson coefficient was performed for correlation analysis. Kaplan-Meier and Log-rank tests were carried outfor survival analysis. A p value < 0.05 indicated a statistically significant difference.
3. Results
3.1. Identification of MIOX as a potential regulatory gene in ccRCC
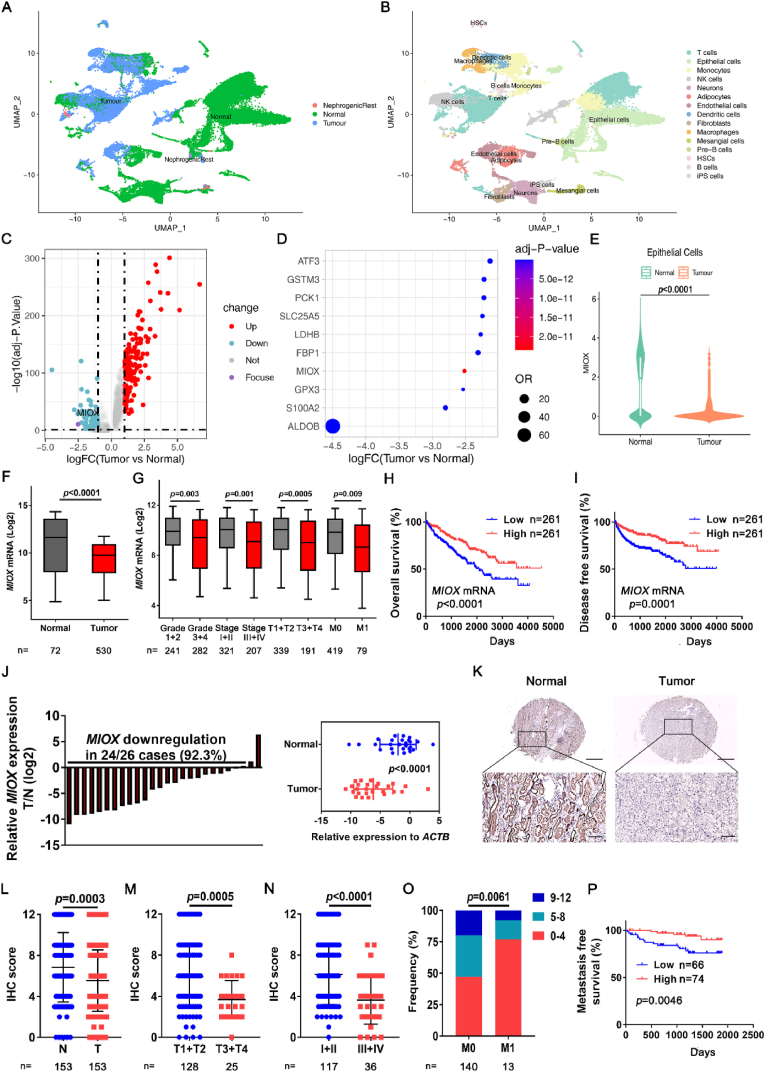
To explore the underlying regulatory gene of ccRCC, we performed further bioinformatics analysis of the ccRCC scRNA-seq dataset [9]. After quality control, 7786 single cells from normal tissues and 16,764 single cells from tumor tissues were identified, with few nephrogenic-rest cells (Fig. 1A). The cell type of each cell cluster was identified in accordance with the markers of major cell types. Eventually the cells were divided into 15 cell types (Fig. 1B).
Fig. 1.
Identification of MIOX as a suppressor gene of ccRCC. (A-B) Single cell of 12 tumor samples and 9 adjacent normal samples of ccRCC in scRNA-seq database was classified according to normal and tumor tissues (A) and cell types (B). (C) Volcano plot of DEGs between tumor epithelial cells and normal epithelial cells. Up-regulated (log2 FC > 1.0) genes are marked in red and down-regulated genes (log2 FC < −1.0) are marked in blue. (D) The top 10 down-regulated genes in tumor epithelial cells compared to normal epithelial cells. (E) The expression of MIOX in normal epithelial cells and tumor epithelial cells in scRNA-seq database. (F) The expression of MIOX in adjacent normal tissues (Normal) and tumor tissues (Tumor) in TCGA ccRCC database. (G) The expression of MIOX in subgroups of patients with different Furhman grades (Grade 1–4), clinical stage (Stage I-IV), tumor stage (T1-T4), and metastatic status (M0 or M1) in the TCGA database. (H–I) Association of MIOX expression with OS (H) and DFS (I) of ccRCC patients in the TCGA database. (J) Detection of mRNA expression levels of MIOX in 26 pairs of ccRCC tissues (T) and adjacent normal tissues (N) by qRT-PCR (left panel), and the statistical analysis of relative expression levels of the reference gene ACTB showed in the right panel. (K-L) Representative images (K) and IHC score (L) of MIOX IHC staining of paired adjacent and ccRCC tissues in TMA. Scale bar: 500 μm for images above and 100 μm for images below. (M-N) MIOX staining scores in T1-T4 tumor (M) and clinical stage I-IV (N). (O) Frequency of different MIOX staining score groups in patients of stage M0 and M1. (P) Association of MIOX expression with MFS of ccRCC patients undergoing surgery at our center. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Then, we focused on changes in gene expression profiles in tumor cells. Epithelial cells were extracted, and DEGs were screened based on the sample origin (Fig. 1C). Up-regulation of 152 genes and down-regulation of 114 genes were observed in tumor epithelial cells compared with normal epithelial cells (Supplemental file 1). We then screened out the top 10 down-regulated genes (Fig. 1D); detailed descriptions are listed in Table S4. In the top 5 down-regulated genes, the regulatory functions of ALDOB [37,38], GPX3 [39] and FBP1 [[40], [41], [42]] in RCC have been reported.
Consequently, we performed further investigation in TCGA ccRCC database. For S100A2, survival analysis revealed no significant differences in overall survival (OS) and disease-free survival (DFS) between the low and high expression groups (Logrank p values were 0.11 and 0.15, respectively) (Figs. S1A–B), which suggests that S100A2 expression in ccRCC is not be related to the prognosis of ccRCC patients. This result is also consistent with the findings of Sugiyama et al. [43].
To the best of our knowledge, there are no studies involving MIOX in ccRCC. For MIOX, analysis of scRNA-seq data indicated that MIOX was significantly down-regulated in tumor epithelial cells (Fig. 1E). In addition, analysis of the TCGA database showed that in comparison with adjacent normal tissues, MIOX was significantly downregulated in cancerous tissues. (Fig. 1F). The relevance of MIOX expression to clinical characteristics of ccRCC patients was further investigated. Decreased MIOX expression was associated with higher tumor pathologic grade (Furhman Grade 3 + 4), clinical (stage III + IV), and tumor (T3+T4) staging, and MIOX expression was significantly lower in the primary tumors of initial diagnosed metastatic patients (M1) than in non-metastatic patients (M0) (Fig. 1G). Further prognosis analysis demonstrated that both OS and DFS were significantly higher in patients with high MIOX expression than in those with low expression (OS: p < 0.0001, HR = 0.5403, 95 % CI: 0.4006–0.7288; DFS: p = 0.0001, HR = 0.4769, 95 % CI: 0.3296–0.6901) (Fig. 1H–I). Thus, this data suggests that MIOX is closely related to ccRCC progression and may be a gene with cancer suppressor function in ccRCC.
3.2. Validation of expression and clinical correlation of MIOX in ccRCC samples and cell lines
We conducted further investigation in clinical samples of ccRCC. In total, 26 pairs of cancerous and adjacent normal kidney tissues in ccRCC were gathered, and qRT-PCR was conducted to detect their MIOX mRNA expression levels. In 24/26 (92.3 %) paired tissues, MIOX was significantly reduced in ccRCC tissues relative to adjacent renal tissues. (Fig. 1J).
Next, paraffin-embedded tissues from ccRCC patients (n = 153) who underwent surgery in 2015 were collected and TMA was constructed. Consistently, the protein expression of MIOX was significantly down-regulated in ccRCC tissues relative to adjacent renal tissues measured by IHC (Fig. 1K-L). In addition, decreased MIOX expression was related to higher tumor stage (T3+T4) and TNM stage (III + IV) (Fig. 1M − N). Patients with primary metastasis (M1) also had lower MIOX expression than patients without primary metastasis (M0). (Fig. 1O). Also, patients who underwent surgery were followed up for at least 5 years after surgery. The metastasis-free survival (MFS) curve was drawn with or without postoperative metastasis as the endpoint. As shown in Fig. 1P, a significantly shorter MFS was found in patients with low MIOX expression than in those with high MIOX expression. The correlation analysis between clinical characteristics and MIOX expression of 153 patients in this study is shown in Table 1.
Table 1.
The correlation between MIOX protein expression and pathological and clinical features of 153 patients with ccRCC.
| Characteristics | MIOX expression |
||
|---|---|---|---|
| Low | High | p value | |
| Gender, n (%) | 0.906 | ||
| Male | 52 (68.4) | 52 (67.5) | |
| Female | 24 (31.6) | 25 (32.5) | |
| Age (y), median (range) | 58 (26–82) | 60 (29–82) | 0.472 |
| Grade, n (%) | 0.001 | ||
| 1 | 14 (18.4) | 17 (22.1) | |
| 2 | 28 (36.9) | 47 (61.0) | |
| 3 + 4 | 34 (44.7) | 13 (16.9) | |
| T stage, n (%) | 0.001 | ||
| T1 | 44 (57.9) | 65 (84.4) | |
| T2 | 13 (17.1) | 6 (7.8) | |
| T3+T4 | 19 (25.0) | 6 (7.8) | |
| N stage, n (%) | 0.988 | ||
| N0 | 73 (96.1) | 75 (97.4) | |
| N1 | 3 (3.9) | 2 (2.6) | |
| Distant metastasis, n (%) | 0.040 | ||
| M0 | 66 (86.8) | 74 (96.1) | |
| M1 | 10 (13.2) | 3 (3.9) | |
In addition, further detection indicated that both mRNA and protein expressions of human RCC cell lines 786-O, 769-P, ACHN, A498 and Caki-1 were significantly decreased compared to human renal tubular epithelial cell line HK-2 by qRT-PCR and western blot (Figs. S2A–B). These results suggest that MIOX is down-regulated in ccRCC tissues and cell lines, and the down-regulation of MIOX in ccRCC is related to the malignant phenotype of tumors.
3.3. Overexpression of MIOX inhibits malignant phenotype of ccRCC in vitro
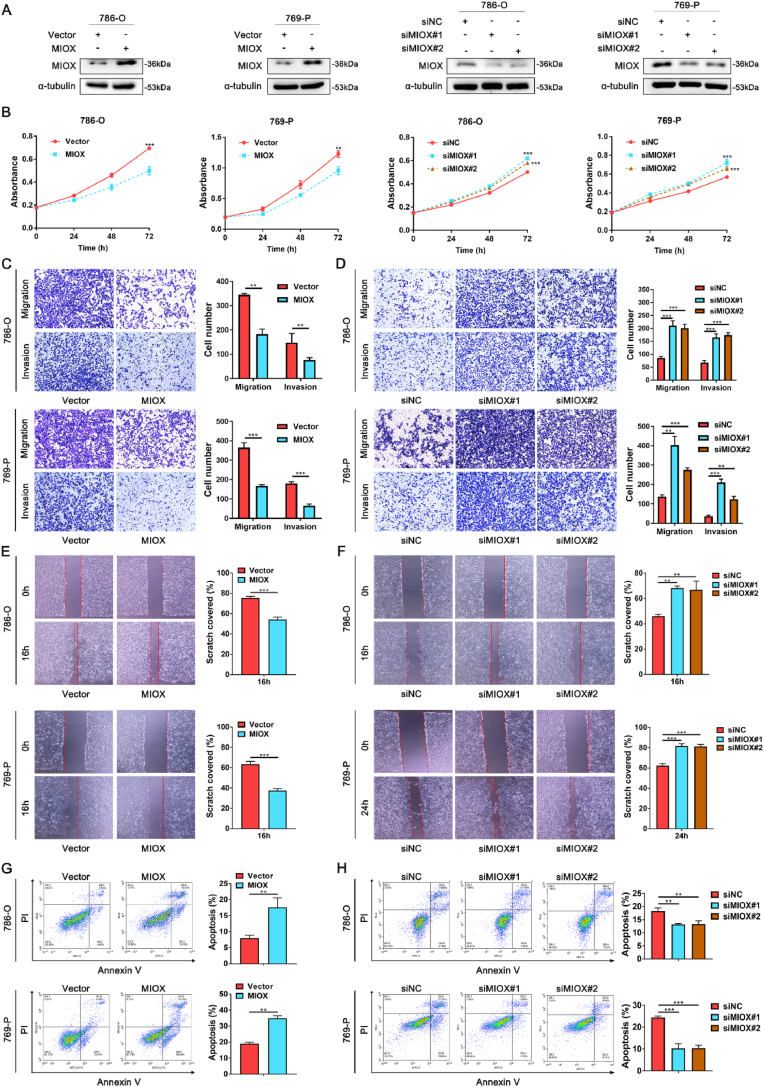
For investigation of the regulatory role of MIOX in ccRCC, we first overexpressed or knockdown MIOX in 786-O and 769-P cells with overexpressed plasmid or siRNA (Figs. S3A–D and Fig. 2A). MTT assay indicated that the proliferation was significantly inhibited by MIOX overexpression, while this process was reversed after MIOX knockdown (Fig. 2B). Then, transwell and wound-healing assays were performed for the assessment of migration and invasion of ccRCC cells. Overexpression of MIOX in 786-O and 769-P cells significantly suppressed the migration and invasion (Fig. 2C and E), while knockdown of MIOX increased migration and invasion (Fig. 2D and F). In addition, it was found that MIOX overexpression significantly facilitated apoptosis of ccRCC cells (Fig. 2G), and a remarkable decrease in the proportion of apoptotic cells was detected after MIOX knockdown, demonstrating the promotion of MIOX on the apoptosis in ccRCC cells (Fig. 2H). Apoptosis may disturb the proliferation phenotype of cells. To exclude the effect of apoptosis on proliferation, we performed Ki67 staining. We found that the positivity of Ki67 staining was reduced after overexpression of MIOX and increased after the knockdown of MIOX, confirming the effect of MIOX on proliferation (Fig. S4). Our findings prove that MIOX exerts the function of a potential cancer suppressor gene in ccRCC progression.
Fig. 2.
Overexpression of MIOX suppresses the aggressiveness of ccRCC cells in vitro. (A) The overexpression and knockdown efficiency of MIOX in 786-O and 769-P cells verified by western blot. (B) Effects of overexpression or knockdown of MIOX on proliferation of ccRCC cells (n = 3). (C-D) Detection of migration and invasion of ccRCC cells after overexpression (C) or knockdown (D) of MIOX by Transwell assay (n = 3). (E-F) Detection of migration of ccRCC cells after overexpression (E) or knockdown (F) of MIOX by wound-healing assay (n = 3). (G-H) Proportion of apoptotic cells in 786-O and 769-P cells with overexpression (G) or knockdown (H) of MIOX (n = 3). **, p < 0.01, ***, p < 0.001.
3.4. MIOX inhibits autophagy and EMT in ccRCC cells
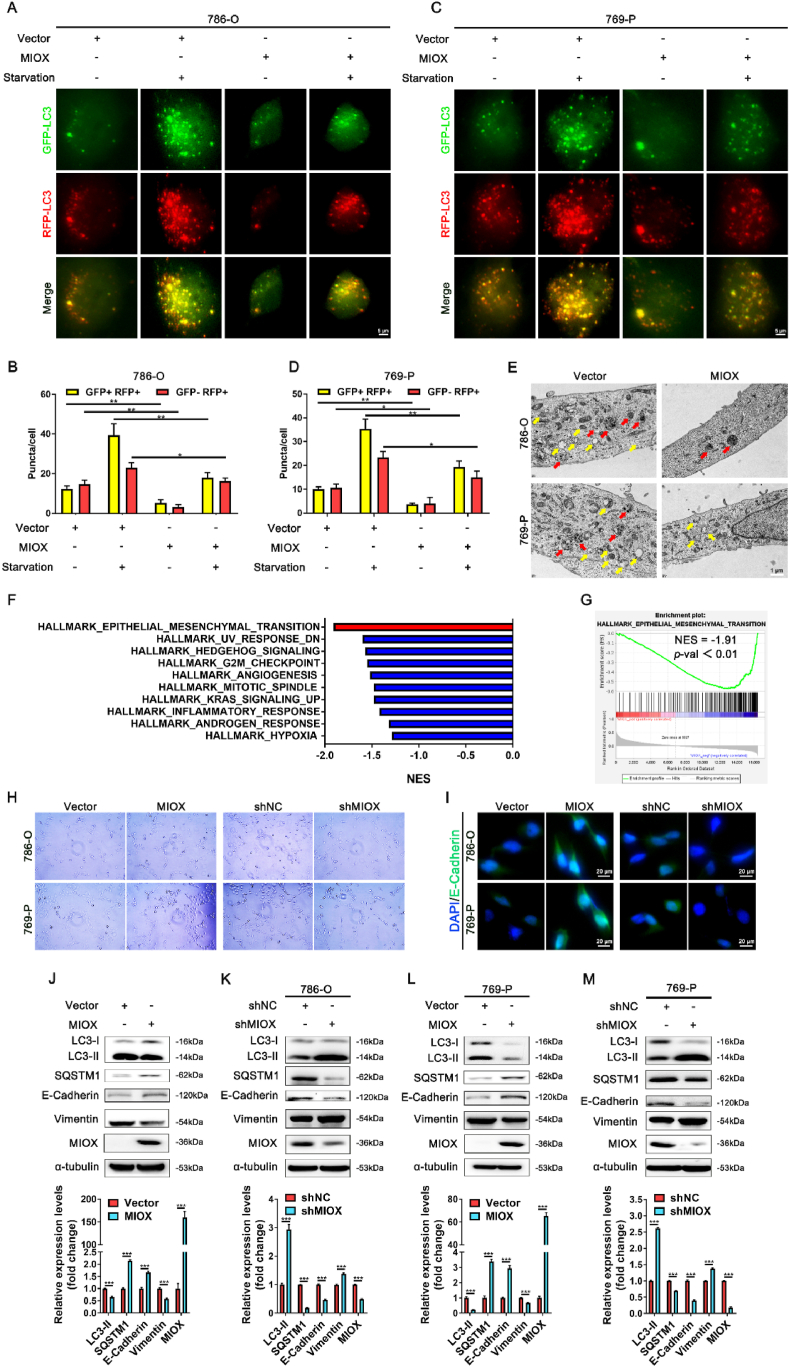
Next, we explored the regulatory mechanism of MIOX in ccRCC. It has been reported that up-regulation of MIOX leads to inhibition of autophagy in renal tubular epithelial cells [44], and autophagy inhibition could play an antitumor role in cancer progression [21]. We transfected lentivirus carrying GFP-RFP-LC3 plasmid into 786-O and 769-P cells. Under normal conditions, the number of both yellow puncta (GFP + RFP+) and red puncta (GFP- RFP+) was decreased in MIOX transiently overexpressed cells (Fig. 3A–D). With starvation (FBS free) for 2 h, the number of yellow puncta and red puncta was elevated in both Vector-transfected and MIOX-overexpressed cells, but LC3 puncta of MIOX-overexpressed cells were still less than that of Vector-transfected cells (Fig. 3A–D). In addition, TEM scanning revealed a reduction in the number of autophagosomes and autolysosomes in MIOX overexpressed 786-O and 769-P cells (Fig. 3E).
Fig. 3.
MIOX regulates autophagy and EMT in ccRCC cells. (A-D) Representative images of LC3 fluorescence of 786-O (A) and 769-P (C) cells in different treatment groups, and statistical results of LC3 puncta in 786-O (B) and 769-P (D) cells. Starvation was for 2 h. Scale bar: 5 μm. (E) TEM images to indicate the changes of autophagosome and autolysosome after MIOX overexpression in ccRCC cell lines. Yellow arrow: autophagosome, red arrow: autolysosome. Scale bar: 1 μm. (F) The top 10 gene sets significantly enriched in MIOX low expression group in the TCGA ccRCC cohort. (G) GSEA results of EMT pathway-related genes changing with MIOX expression in TCGA ccRCC cohort (normalized enrichment score (NES) = -1.91, p < 0.01). (H) Representative images of morphology of cells with the MIOX overexpression or knockdown. (I) Representative images of immunofluorescence staining for E-cadherin after MIOX overexpression or knockdown in 786-O and 769-P cells. Scale bar: 20 μm. (J-M) The protein expression and quantitative result (n = 3) of autophagy- and EMT-related markers after overexpression (J and L) or knockdown (K and M) of MIOX in ccRCC cell lines. *, p < 0.05, **, p < 0.01, ***, p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, RNA-seq data in TCGA ccRCC database were used for gene set enrichment analysis (GSEA) by MIOX expression. We found that the EMT pathway ranked first among the negatively correlated pathways of MIOX (NES = −1.91, p < 0.01) (Fig. 3F–G), demonstrating that MIOX may inhibit the EMT of ccRCC. Typically, the EMT process is characterized by altered cell morphology and changes in the expression of the epithelial marker E-cadherin [45]. Therefore, cell morphology and E-cadherin expression were analyzed to demonstrate EMT. We found that overexpression of MIOX disturbed cell polarity, restored cells to a rounded epithelial-type morphology and led to the enhanced fluorescence intensity of E-cadherin in immunofluorescence staining. In contrast knockdown of MIOX transformed cells into a spindle-shaped mesenchymal-like phenotype and attenuated the fluorescence intensity of E-cadherin, demonstrating the inhibitory effect of MIOX on EMT (Fig. 3H–I).
Then, we screened 786-O and 769-P cell lines with stable overexpression and knockdown of MIOX by lentivirus transfection (Fig. S5 A-D and Fig. 3J-M). Overexpression of MIOX resulted in up-regulation of E-cadherin and down-regulation of vimentin, indicating EMT inhibition with increased epithelial marker and decreased mesenchymal marker (Fig. 3J and L), while the knockdown of MIOX showed the opposite results (Fig. 3K and M).
Autophagy flux refers to the entire process of cargo moving through the autophagic system from phagophore formation to delivery to the lysosome for degradation [46]. We also examined the protein expression of LC3 and SQSTM1, which are commonly used to characterize autophagy flux [47]. Up-regulation of MIOX decreased LC3 II and increased SQSTM1, indicating a reduction in autophagy flux (Fig. 3J and L), and down-regulation of MIOX significantly promoted autophagy flux (Fig. 3K and M). These results confirm that MIOX inhibits autophagy and EMT of ccRCC cells.
3.5. MIOX regulates EMT by regulating autophagy
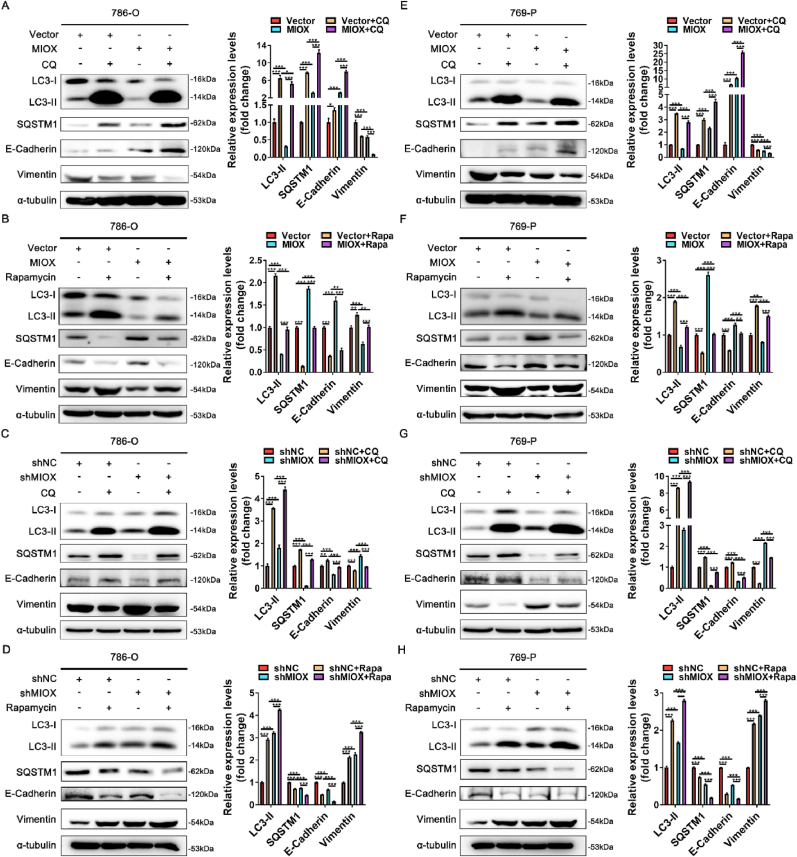
Autophagy can regulate EMT in tumor [14]. We further explored whether MIOX regulated EMT by regulating autophagy. Autophagy inhibitor chloroquine (CQ) and autophagy activator rapamycin were used to interfere with the autophagy status of ccRCC cells. In 786-O cells, CQ treatment led to the accumulation of LC3 and SQSTM1 proteins, an increase in E-cadherin, and a decrease in vimentin. Up-regulation of E-cadherin and down-regulation of vimentin caused by MIOX overexpression were accelerated by CQ treatment, suggesting a further inhibition of EMT (Fig. 4A). CQ treatment also reversed the decrease in E-cadherin and the increase in vimentin caused by MIOX knockdown (Fig. 4C). In contrast, rapamycin treatment led to autophagic flux activation, and accelerated the decrease in E-cadherin and the increase in vimentin caused by MIOX knockdown (Fig. 4D). The impact on E-cadherin and vimentin by MIOX overexpression was also reversed by rapamycin treatment (Fig. 4B). Similar results were observed in 769-P cells (Fig. 4E–H). This data confirms that the regulatory effect of MIOX on the EMT is achieved by regulating autophagy.
Fig. 4.
MIOX regulates EMT by regulating autophagy. (A-B) The protein expression and quantitative result (n = 3) of autophagy- and EMT-related markers after 50 μM CQ (A) or 400 nM Rapamycin (B) treatment for 24 h in 786-O cells with stable overexpression of MIOX. (C-D) The protein expression and quantitative result (n = 3) of autophagy- and EMT-related markers after 50 μM CQ (C) or 400 nM Rapamycin (D) treatment for 24 h in 786-O cells with stable knockdown of MIOX. (E-F) The protein expression and quantitative result (n = 3) of autophagy- and EMT-related markers after 50 μM CQ (E) or 400 nM Rapamycin (F) treatment for 24 h in 769-P cells with stable overexpression of MIOX. (G-H) The protein expression and quantitative result (n = 3) of autophagy- and EMT-related markers after 50 μM CQ (G) or 400 nM Rapamycin (H) treatment for 24 h in 769-P cells with stable knockdown of MIOX. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
3.6. The increase in ROS levels caused by MIOX-mediated autophagy inhibition led to the inhibition of STAT3/c-Myc-mediated EMT
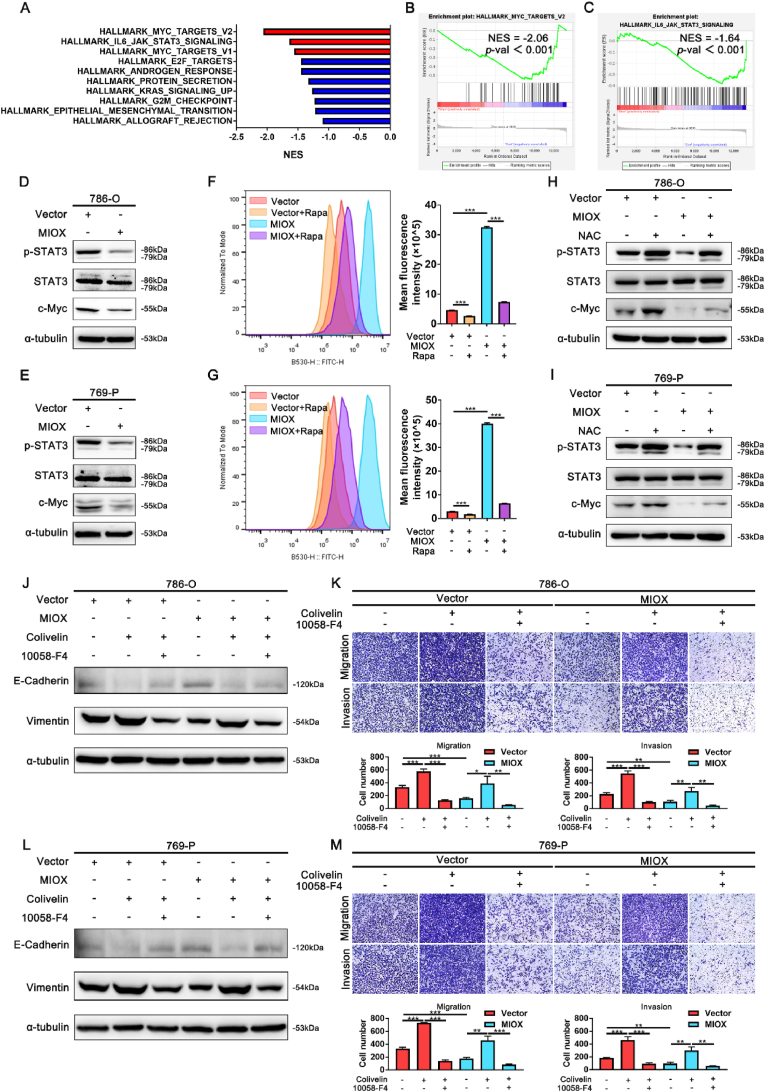
For further investigation of the mechanism in the regulation of MIOX in the progression of ccRCC, 786-O-Vector (MIOX-control) and 786-O-MIOX (MIOX-overexpression) were used to perform RNA-seq, and the expression levels of different genes in the two cells were obtained. DEGs were analyzed and GSEA was performed (Con vs MIOX). The results revealed a significant enrichment of MYC and IL6-JAK-STAT3 pathways in 786-O-Vector (Con), indicating that MIOX may be negatively correlated with MYC and IL6-JAK-STAT3 pathways (Fig. 5A–C). STAT3 and MYC (c-Myc) are two cancer-promoting signals, that can induce the EMT [30,31,48]. STAT3 could also promote the transcriptional expression of c-Myc, and the STAT3/c-Myc signaling axis exerts a key role in cancer progression [49]. Therefore, we speculated that the regulatory effect of MIOX on EMT was related to the STAT3/c-Myc signaling axis. Western blot revealed that MIOX overexpression significantly inhibited the p-STAT3 and c-Myc expression in ccRCC cell lines (Fig. 5D–E). In contrast, MIOX knockdown promoted the p-STAT3 and c-Myc expression (Fig. S6 A-B).
Fig. 5.
Elevated ROS levels resulted from inhibition of autophagy by MIOX leads to the inhibition of the STAT3/c-Myc-mediated EMT and aggressiveness in ccRCC cells. (A) The top 10 gene sets significantly enriched in MIOX-control cells from the RNA-seq data. (B–C) GSEA results of MYC (B) and IL6-JAK-STAT3 (C) pathway-related genes in MIOX-control and MIOX-overexpression cells (Con vs MIOX) form the RNA-seq data (MYC: NES = −2.06, p < 0.001; IL6-JAK-STAT3: NES = −1.64, p < 0.001). (D-E) The protein expression of p-STAT3, STAT3 and c-Myc in 786-O (D) and 769-P (E) cells with stable overexpression of MIOX. (F-G) The levels of ROS in MIOX-overexpression 786-O (F) and 769-P (G) cells detected by flow cytometry. Rapamycin: 400 nM for 24 h (n = 3). (H–I) The effect of NAC on the expression of p-STAT3, STAT3 and c-Myc in MIOX-overexpression 786-O (H) and 769-P (I) cells. NAC: 10 mM for 24 h. (J-K) The effect of Colivelin and 10058-F4 on the expression of E-cadherin and Vimentin (J) and migration and invasion (K) in MIOX-overexpression 786-O cells. Colivelin: 1 μM for 24 h. 10058-F4: 20 μM for 24 h. (L-M) The effect of Colivelin and 10058-F4 on the expression of E-cadherin and Vimentin (L) and migration and invasion (M) in MIOX-overexpression 769-P cells. Colivelin: 1 μM for 24 h. 10058-F4: 20 μM for 24 h *, p < 0.05, **, p < 0.01, ***, p < 0.001.
Previous studies indicated that autophagy inhibition could lead to ROS accumulation [50]. Moreover, ROS inhibited p-STAT3 and interfered with downstream signal transduction of STAT3 in some cancer types [51,52]. We further investigated whether MIOX upregulated ROS levels by inhibiting autophagy, thereby mediating the inhibition of the STAT3/c-Myc signaling axis. First, ROS levels were measured by flow cytometry. DCFH-DA fluorescent probe, which could reflect the overall oxidative stress state in cells [53], was used for intracellular ROS detection. We found that overexpression of MIOX significantly increased ROS levels in 786-O and 769-P cells (Fig. 5F–G). Autophagy activation by rapamycin treatment inhibited the increase of ROS levels (Fig. 5F–G). Moreover, ROS levels were decreased by the knockdown of MIOX, which was reversed by inhibition of autophagy with CQ treatment (Fig. S6 C-D). Then, NAC and H2O2 were used to explore the impact of ROS on the STAT3/c-Myc signaling axis in ccRCC cells. NAC treatment rescued the decline in p-STAT3 and c-Myc caused by MIOX overexpression (Fig. 5H–I). Consistently, H2O2 treatment reversed the increase of p-STAT3 and c-Myc in MIOX-knockdown cells (Fig. S6 E-F).
Finally, we investigated the induction of the STAT3/c-Myc signaling axis on the EMT of ccRCC cells. Colivelin is an activator of STAT3, promoting STAT3 phosphorylation [54]. 10058-F4 is an inhibitor of c-Myc [55]. Colivelin treatment reversed the increase in E-cadherin and the decrease in vimentin mediated by MIOX overexpression and rescued the migration and invasion ability in 786-O and 769-P cells (Fig. 5J-M). However, when treated with Colivelin in combination with 10058-F4, the effect of Colivelin was partially eliminated (Fig. 5J-M). We confirmed that MIOX inhibited autophagy to elevate ROS levels, thus suppressing the STAT3/c-Myc-mediated EMT in ccRCC.
3.7. Overexpression of MIOX inhibits RCC cell growth and metastasis in vivo
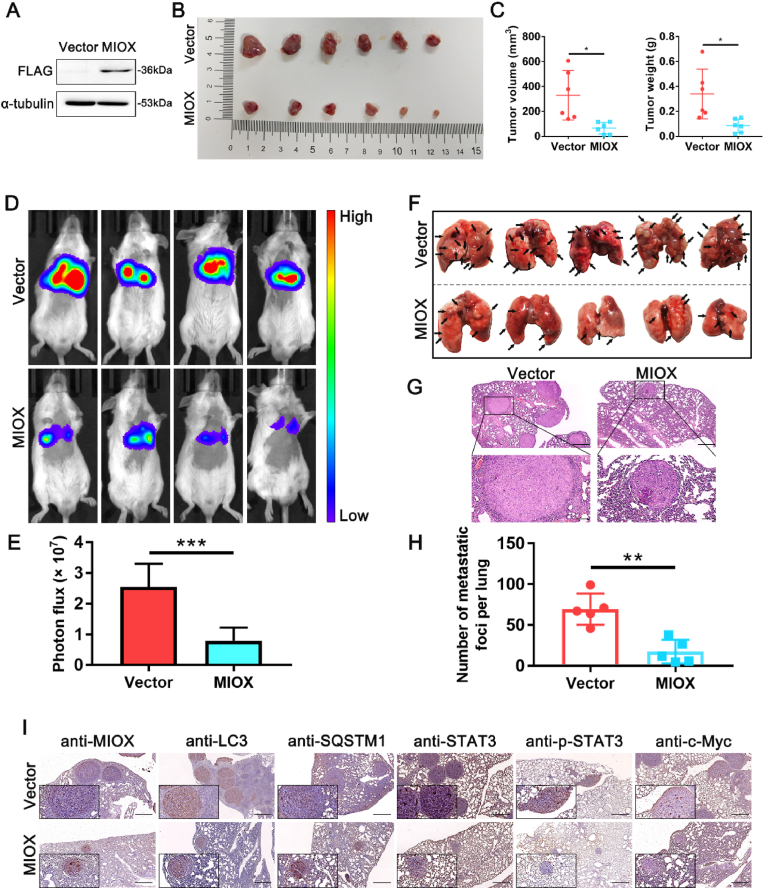
The regulatory role of MIOX on RCC was further explored in mouse models. We first constructed a murine RCC cell line Renca with stably overexpressing MIOX (Fig. 6A and Fig. S7). Subcutaneous injection of cells was administrated into the mouse middle back. Tumor-bearing mice were sacrificed at 3 weeks and subcutaneous tumors were harvested. MIOX overexpression significantly reduced tumor volume and weight compared to the Vector group (Fig. 6B–C).
Fig. 6.
Overexpression of MIOX inhibits RCC cell growth and metastasis in vivo. (A) The expression of FLAG label in Renca transfected with lentivirus carrying FLAG-labelled plasmid of MIOX overexpression by western blot. (B) Representative images of subcutaneous tumors in the MIOX control group and the overexpression group; n = 6/group. (C) The volume and weight of the subcutaneous tumors. (D) Representative living images of mice injected with Renca transfected by indicated lentivirus into tail vein. The lentivirus was Luci-labelled and therefore stably transfected Renca cell lines had in vivo luciferase activity. (E) Statistical analysis of luciferase bioluminescence intensity (n = 6). (F) Representative images of metastases in murine lung of each group; n = 5/group. The black arrow indicated the metastasis. (G) H&E staining of pulmonary tissue sections. Scale bar: 500 μm for images above and 100 μm for images below. (H) Statistical analysis of the number of pulmonary metastases of each group. (I) The protein expression of MIOX, LC3, SQSTM1, STAT3, p-STAT3 and c-Myc in pulmonary metastases of mice by IHC staining. Scale bar: 500 μm *, p < 0.05, **, p < 0.01, ***, p < 0.001.
Then, cells in each group were injected intravenously into the mice to establish a pulmonary metastasis model. Live imaging of mice was performed at 2 weeks after injection. A significant decrease in the signal intensity of lung metastasis was detected in the MIOX overexpression group compared to that in the control group (Fig. 6D–E). Mice were executed to harvest lung tissues 3 weeks after injection. A significant reduction of lung metastases was observed in the MIOX overexpression group in comparison to the control group (Fig. 6F). H&E staining also revealed a decrease in the size and number of lung metastases in the MIOX overexpression group (Fig. 6G–H).
Next, we further investigated the regulation of MIOX on the STAT3/c-Myc signaling axis in vivo. IHC staining showed that MIOX overexpression resulted in the decrease of LC3, the increase of SQSTM1, and the inhibition of p-STAT3 and c-Myc in pulmonary metastases (Fig. 6I). Furthermore, STAT3/c-Myc signaling axis was also evaluated in lung metastasis lysates by Western blot. The results showed that The expression of p-STAT3 and c-Myc was reduced in the MIOX overexpression group, which was consistent with the results of IHC staining (Fig. S8 A-B).
4. Discussion
ccRCC is the most common pathological type of RCC [2]. Existing treatment options for ccRCC have made great progress, but have limited efficacy in some patients. For this reason it is necessary to explore molecular targets related to ccRCC progression and provide new strategies for ccRCC treatment. scRNA-seq could help identify changes in gene expression in specific cell types and is currently a leading tool for exploring complex tumor microenvironments [8]. Herein, we conducted further bioinformatics analysis in the ccRCC scRNA-seq database and found that MIOX was significantly down-regulated in tumor epithelial cells compared to normal epithelial cells. Verification in the TCGA database as well as clinical samples from our center revealed a significant reduction of MIOX expression in ccRCC tissues in comparison with adjacent renal tissues. In addition, the down-regulation of MIOX in cancerous tissues was responsible for higher tumor pathological grade, higher clinical and tumor stage, and primary metastasis. In TCGA ccRCC database, significantly longer OS and DFS were observed in the high MIOX expression group than in patients with low MIOX expression. Analysis of postoperative follow-up of ccRCC patients undergoing surgery in our center showed that low MIOX expression in tumor tissues significantly shortened the MFS of patients. Our findings indicate that MIOX is closely correlated with the progression and metastasis of ccRCC.
There are currently only a few studies on the role of MIOX in tumor regulation. A close link exists between MIOX and cancer ferroptosis. For example, studies revealed that MIOX is involved in ferroptosis-related gene models in papillary thyroid carcinoma, bladder cancer and papillary renal cell carcinoma [12,13,56]. Zhang et al. reported that MIOX could suppress the progression of hepatocellular carcinoma by increasing ROS production, reducing the levels of NADPH and GSH and accelerating ferroptosis induced by erastin and RSL3 [11]. Yet, to the best of our knowledge, this is the first study reporting on MIOX in ccRCC. Our results revealed that overexpression of MIOX resulted in the suppression of the proliferation, invasion and migration and the promotion of apoptosis in ccRCC cells. In contrast, the knockdown of MIOX enhanced the aggressiveness of ccRCC cells. We confirmed that MIOX inhibited the malignant phenotype of ccRCC cell lines by in vitro assays.
Autophagy is essential in supporting cellular metabolism and survival under starvation and stress and in controlling the quantity and quality of cellular proteins and organelles [57]. Autophagy exerts a dual function in tumors, promoting or inhibiting tumor progression. In RCC, the antitumor effect of targeting autophagy is mainly achieved through inhibition of autophagy. Also, phase I/II clinical trials of CQ and hydroxychloroquine (HCQ) have been initiated [58]. Zhan et al. found that up-regulation of MIOX aggravated the oxidative stress of renal tubular epithelial cells, led to mitochondrial damage and fragmentation, triggered apoptosis and inhibited autophagy [44]. In this study, overexpression of MIOX promoted apoptosis of ccRCC cells (Fig. 2G–H); therefore, we investigated whether MIOX inhibited autophagy in ccRCC cells. In vitro assays indicated that MIOX overexpression inhibited the expression of LC3, reduced the number of autophagosomes and autolysosomes and blocked autophagy flux in ccRCC cell lines, while knockdown of MIOX significantly promoted autophagy flux. This data demonstrates that MIOX also inhibited the autophagy of ccRCC cells.
Furthermore, we performed GSEA for RNA-seq data from TCGA ccRCC database and found that the EMT pathway ranked first among the negative correlation pathways of MIOX. EMT mediates metastasis in almost all types of cancer [26]. Down-regulation of MIOX induced the suppression of E-cadherin expression as well as the promotion of vimentin expression, which is the typical molecular pattern of EMT activation, while up-regulation of MIOX reversed this process. Therefore, this data suggests that MIOX can suppress the invasiveness of ccRCC cells by inhibiting EMT.
Autophagy has dual regulatory effects on EMT. Previous studies indicated that autophagy could promote or inhibit EMT in tumor [14]. We further explored whether MIOX regulated EMT through autophagy. Activation of autophagy by rapamycin treatment suppressed E-cadherin expression, facilitated vimentin expression and reversed the increase in E-cadherin and the decrease in Vimentin caused by MIOX overexpression. Moreover, blocking autophagy by CQ treatment also reversed the effects of MIOX knockdown on the EMT molecular. Singla et al. found that EMT-like phenotypes in RCC correspond to elevated autophagy flux, where the combination of autophagy inhibitors (CQ) and current therapies may exert a synergistic role in inhibiting EMT of RCC [59], which is consistent with our results.
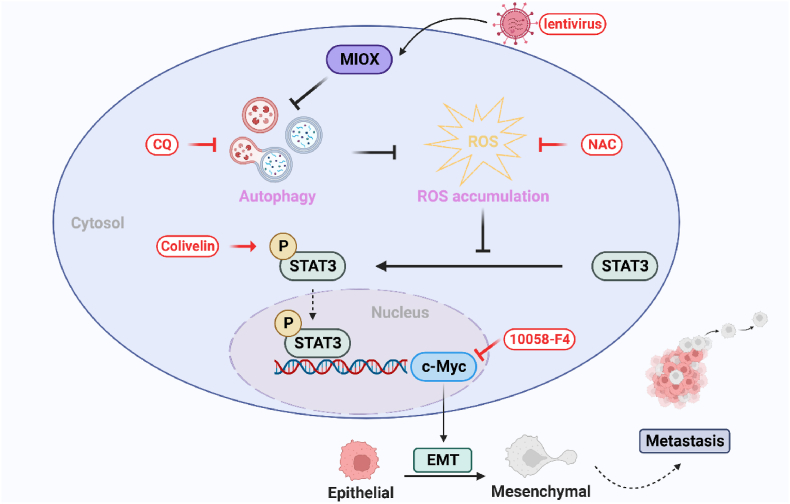
To further investigate the regulation mechanism of MIOX on autophagy and EMT, we performed RNA-seq of MIOX-control and MIOX-overexpression 786-O cells. GSEA revealed that MYC and IL6-JAK-STAT3 pathways were negatively correlated with the expression levels of MIOX. STAT3 could transcriptionally activate c-Myc [49]. Few studies have reported on the function of the STAT3/c-Myc signaling axis in ccRCC. We elucidated the regulation of MIOX on the STAT3/c-Myc signaling axis and EMT through rescue experiments: inhibition of autophagy caused by up-regulation of MIOX led to ROS accumulation and increased ROS levels, which was consistent with results reported by Zhang et al. [11]. Increased ROS inhibited the expression of p-STAT3 and c-Myc, and suppressed the STAT3/c-Myc-mediated EMT in ccRCC (Fig. 7).
Fig. 7.
Schematic illustration of the mechanism of the inhibition of MIOX on ccRCC metastasis. MIOX inhibits autophagy, and therefore leads to ROS accumulation. Elevated ROS levels suppress the expression of p-STAT3 and c-Myc and inhibit the STAT3/c-Myc-mediated EMT and metastasis. MIOX: myo-inositol oxygenase; ROS: reactive oxygen species; STAT3: signaling transducer and activator of transcription 3; EMT: epithelial-mesenchymal transition; CQ: chloroquine; NAC: N-acetyl-l-cysteine.
Finally, we verified the inhibitory effect of MIOX on ccRCC in animal models. MIOX is a metabolic enzyme specifically expressed in the kidney and is highly conserved in many species [60]. In this study, we selected BALB/c mice for the in vivo testing. MIOX overexpression suppressed the growth of Renca in vivo and reduced the volume and weight of subcutaneous tumors. Furthermore, up-regulation of MIOX reduced metastases in the lung. The STAT3/c-Myc signaling axis was also suppressed in lung metastases by IHC staining.
In conclusion, we validated that MIOX, a novel suppressor gene of ccRCC, inhibits autophagy to elevate ROS levels and suppresses the STAT3/c-Myc-mediated EMT in ccRCC. Thus, MIOX may be a potential therapeutic target for human ccRCC and may provide a novel option for the clinical treatment of ccRCC.
Author contributions
Hongqian Guo., Longxiyu Meng and Jie Gao designed the study. Longxiyu Meng and Jie Gao performed the in vitro and in vivo study. Wenjing Mo, Baojun Wang, Wenmin Cao, Jiaxin Shu and Huiqi Dai assisted with the in vitro and in vivo study. Meng Ding, Wenli Diao, Wei Chen and Qing Zhang performed the validation. Longxiyu Meng analyzed the experimental data. Longxiyu Meng wrote the manuscript. Longxiyu Meng and Hongwei Shen revised the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Disclosure of conflict of interest
The authors of this study declare no conflict of interest exist and no relationships with any companies, whose products or services may be related to the subject matter of the article.
Declaration of competing interest
The authors of this study declare no conflict of interest exist and no relationships with any companies, whose products or services may be related to the subject matter of the article.
Acknowledgements
We thank Dr. Qi Liang for her help with the experiment. Funding: This work was supported by the National Natural Science Foundation of China (81972388, 82103002).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102956.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Data availability
The data that has been used is confidential.
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., et al. Cancer statistics, 2023. CA A Cancer J. Clin. 2023;73(1):17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Warren A.Y., Harrison D. WHO/ISUP classification, grading and pathological staging of renal cell carcinoma: standards and controversies. World J. Urol. 2018;36(12):1913–1926. doi: 10.1007/s00345-018-2447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shingarev R., Jaimes E.A. Renal cell carcinoma: new insights and challenges for a clinician scientist. Am. J. Physiol. Ren. Physiol. 2017;313(2):F145–F154. doi: 10.1152/ajprenal.00480.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J., Parkin D.M., Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur. J. Cancer. 2010;46(4):765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Choueiri T.K., Motzer R.J. Systemic therapy for metastatic renal-cell carcinoma. N. Engl. J. Med. 2017;376(4):354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 6.Barata P.C., Rini B.I. Treatment of renal cell carcinoma: current status and future directions. CA A Cancer J. Clin. 2017;67(6):507–524. doi: 10.3322/caac.21411. [DOI] [PubMed] [Google Scholar]
- 7.Elhanani O., Ben-Uri R., Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell. 2023;41(3):404–420. doi: 10.1016/j.ccell.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Jovic D., Liang X., Zeng H., et al. Single-cell RNA sequencing technologies and applications: a brief overview. Clin. Transl. Med. 2022;12(3):e694. doi: 10.1002/ctm2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young M.D., Mitchell T.J., Vieira Braga F.A., et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361(6402):594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arner R.J., Prabhu K.S., Reddy C.C. Molecular cloning, expression, and characterization of myo-inositol oxygenase from mouse, rat, and human kidney. Biochem. Biophys. Res. Commun. 2004;324(4):1386–1392. doi: 10.1016/j.bbrc.2004.09.209. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Luo M., Cui X., et al. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022;29(9):1850–1863. doi: 10.1038/s41418-022-00970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L., Li C., Qin Y., et al. A novel prognostic model based on ferroptosis-related gene signature for bladder cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.686044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D., Wang J., Li C., et al. Ferroptosis-related gene model to predict overall survival of papillary thyroid carcinoma. Am. J. Otolaryngol. 2021;42(6) doi: 10.1016/j.amjoto.2021.103163. [DOI] [PubMed] [Google Scholar]
- 14.Chen H.T., Liu H., Mao M.J., et al. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer. 2019;18(1):101. doi: 10.1186/s12943-019-1030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White E., DiPaola R.S. The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 2009;15(17):5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y., Song Y., Cheng L., et al. CircCEMIP promotes anoikis-resistance by enhancing protective autophagy in prostate cancer cells. J. Exp. Clin. Cancer Res. 2022;41(1):188. doi: 10.1186/s13046-022-02381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chadet S., Allard J., Brisson L., et al. P2x4 receptor promotes mammary cancer progression by sustaining autophagy and associated mesenchymal transition. Oncogene. 2022;41(21):2920–2931. doi: 10.1038/s41388-022-02297-8. [DOI] [PubMed] [Google Scholar]
- 18.Park J.W., Kim Y., Lee S.-B., et al. Autophagy inhibits cancer stemness in triple-negative breast cancer via miR-181a-mediated regulation of ATG5 and/or ATG2B. Mol. Oncol. 2022;16(9):1857–1875. doi: 10.1002/1878-0261.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H., Zhang Y., Zhu X., et al. DEAD box protein 5 inhibits liver tumorigenesis by stimulating autophagy via interaction with p62/SQSTM1. Hepatology. 2019;69(3):1046–1063. doi: 10.1002/hep.30300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White E. The role for autophagy in cancer. J. Clin. Invest. 2015;125(1):42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sotelo J., Briceño E., López-González M.A. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 2006;144(5):337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 23.Rangwala R., Chang Y.C., Hu J., et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangwala R., Leone R., Chang Y.C., et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogl D.T., Stadtmauer E.A., Tan K.-S., et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10(8):1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 27.Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Jiang B., Chen W., Qin H., et al. TOX3 inhibits cancer cell migration and invasion via transcriptional regulation of SNAI1 and SNAI2 in clear cell renal cell carcinoma. Cancer Lett. 2019;449:76–86. doi: 10.1016/j.canlet.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Goossens S., Vandamme N., Van Vlierberghe P., et al. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim. Biophys. Acta Rev. Canc. 2017;1868(2):584–591. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Sadrkhanloo M., Entezari M., Orouei S., et al. STAT3-EMT axis in tumors: modulation of cancer metastasis, stemness and therapy response. Pharmacol. Res. 2022;182 doi: 10.1016/j.phrs.2022.106311. [DOI] [PubMed] [Google Scholar]
- 31.Zhong Y., Yang L., Xiong F., et al. Long non-coding RNA AFAP1-AS1 accelerates lung cancer cells migration and invasion by interacting with SNIP1 to upregulate c-Myc. Signal Transduct. Targeted Ther. 2021;6(1):240. doi: 10.1038/s41392-021-00562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gundamaraju R., Lu W., Paul M.K., et al. Autophagy and EMT in cancer and metastasis: who controls whom? Biochim. Biophys. Acta, Mol. Basis Dis. 2022;1868(9) doi: 10.1016/j.bbadis.2022.166431. [DOI] [PubMed] [Google Scholar]
- 33.Butler A., Hoffman P., Smibert P., et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becht E., McInnes L., Healy J., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X., Lan Y., Xu J., et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinet W., Timmermans J.-P., De Meyer G.R.Y. Methods to assess autophagy in situ--transmission electron microscopy versus immunohistochemistry. Methods Enzymol. 2014:543. doi: 10.1016/B978-0-12-801329-8.00005-2. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Wu Q., Qiu J. Accumulation of fructose 1,6-bisphosphate protects clear cell renal cell carcinoma from oxidative stress. Lab. Invest. 2019;99(6):898–908. doi: 10.1038/s41374-019-0203-3. [DOI] [PubMed] [Google Scholar]
- 38.Tan M., Pan Q., Wu Q., et al. Aldolase B attenuates clear cell renal cell carcinoma progression by inhibiting CtBP2. Front. Med. 2023 doi: 10.1007/s11684-022-0947-9. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q., Jin J., Ying J., et al. Frequent epigenetic suppression of tumor suppressor gene glutathione peroxidase 3 by promoter hypermethylation and its clinical implication in clear cell renal cell carcinoma. Int. J. Mol. Sci. 2015;16(5):10636–10649. doi: 10.3390/ijms160510636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks S.A., Khandani A.H., Fielding J.R., et al. Alternate metabolic programs define regional variation of relevant biological features in renal cell carcinoma progression. Clin. Cancer Res. 2016;22(12):2950–2959. doi: 10.1158/1078-0432.CCR-15-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X., Li S., He J., et al. TET2 suppresses VHL deficiency-driven clear cell renal cell carcinoma by inhibiting HIF signaling. Cancer Res. 2022;82(11):2097–2109. doi: 10.1158/0008-5472.CAN-21-3013. [DOI] [PubMed] [Google Scholar]
- 42.Ju D., Liang Y., Hou G., et al. FBP1/miR-24-1/enhancer axis activation blocks renal cell carcinoma progression via Warburg effect. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.928373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiyama T., Ozono S., Miyake H. Expression profile of S100A2 and its clinicopathological significance in renal cell carcinoma. Anticancer Res. 2020;40(11):6337–6343. doi: 10.21873/anticanres.14654. [DOI] [PubMed] [Google Scholar]
- 44.Zhan M., Usman I.M., Sun L., et al. Disruption of renal tubular mitochondrial quality control by Myo-inositol oxygenase in diabetic kidney disease. J. Am. Soc. Nephrol. 2015;26(6):1304–1321. doi: 10.1681/ASN.2014050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin Y., Sun G., Li E., et al. ER stress and impaired autophagy flux in neuronal degeneration and brain injury. Ageing Res. Rev. 2017;34 doi: 10.1016/j.arr.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang P., Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015;75:13–18. doi: 10.1016/j.ymeth.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 48.Chen T., Li K., Liu Z., et al. WDR5 facilitates EMT and metastasis of CCA by increasing HIF-1α accumulation in Myc-dependent and independent pathways. Mol. Ther. 2021;29(6):2134–2150. doi: 10.1016/j.ymthe.2021.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amaya M.L., Inguva A., Pei S., et al. The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation. Blood. 2022;139(4):584–596. doi: 10.1182/blood.2021013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ornatowski W., Lu Q., Yegambaram M., et al. Complex interplay between autophagy and oxidative stress in the development of pulmonary disease. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu C., Wu Z., Huang Z., et al. Nox2 impairs VEGF-A-induced angiogenesis in placenta via mitochondrial ROS-STAT3 pathway. Redox Biol. 2021;45 doi: 10.1016/j.redox.2021.102051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Z., Wang Y., Gong Y., et al. Celastrol elicits antitumor effects by inhibiting the STAT3 pathway through ROS accumulation in non-small cell lung cancer. J. Transl. Med. 2022;20(1):525. doi: 10.1186/s12967-022-03741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eruslanov E., Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 2010;594:57–72. doi: 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- 54.Zhao H., Feng Y., Wei C., et al. Colivelin rescues ischemic neuron and axons involving JAK/STAT3 signaling pathway. Neuroscience. 2019;416:198–206. doi: 10.1016/j.neuroscience.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 55.Luo Y., Yang S., Wu X., et al. Intestinal MYC modulates obesity-related metabolic dysfunction. Nat. Metab. 2021;3(7):923–939. doi: 10.1038/s42255-021-00421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Da Q., Ren M., Huang L., et al. Identification and validation of a ferroptosis-related signature for predicting prognosis and immune microenvironment in papillary renal cell carcinoma. Int. J. Gen. Med. 2022;15:2963–2977. doi: 10.2147/IJGM.S354882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X., He S., Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19(1):12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Q., Bai P. Role of autophagy in renal cancer. J. Cancer. 2019;10(11):2501–2509. doi: 10.7150/jca.29285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singla M., Bhattacharyya S. Autophagy as a potential therapeutic target during epithelial to mesenchymal transition in renal cell carcinoma: an in vitro study. Biomed. Pharmacother. 2017;94:332–340. doi: 10.1016/j.biopha.2017.07.070. [DOI] [PubMed] [Google Scholar]
- 60.Yang Q., Dixit B., Wada J., et al. Identification of a renal-specific oxido-reductase in newborn diabetic mice. Proc. Natl. Acad. Sci. U. S. A. 2000;97(18):9896–9901. doi: 10.1073/pnas.160266197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.