Abstract
Objective
Tumor cells hijack inflammatory mechanisms to promote their own growth. IL-6 is one of the major cytokines, and is frequently upregulated in tumors. The pentose phosphate pathway (PPP) generates the indispensable building blocks to produce various nucleotides. Here we aimed to determine whether and how PPP is timely tuned in response to IL-6 to support tumor growth.
Methods
Protein expression was examined by immunoblot. Protein interaction was examined by immunoprecipitation. Tumor cell proliferation in in vitro culture was examined by BrdU assay and colony formation assay. Tumor cell proliferation in mouse xenograft model was examined by Ki-67 staining.
Results
Here we show that the metabolic flux of PPP and enzymatic activity of glucose-6-phosphate dehydrogenase (G6PD) is rapidly induced under IL-6 treatment, without obvious changes in G6PD expression level. Mechanistically, Janus kinase 2 (JAK2) phosphorylates G6PD Y437 under IL-6 treatment, which accentuates G6PD enzymatic activity by promoting G6PD binding with its substrate G6P. Further, JAK2-dependent G6PD Y437 phosphorylation is required for IL-6-induced nucleotide biosynthesis and tumor cell proliferation, and is associated with the progression of oral squamous cell carcinoma.
Conclusions
Our findings report a new mechanism implicated in the crosstalk between tumor cells and inflammatory microenvironment, by which JAK2-dependent activation of G6PD governs nucleotide synthesis to support tumor cell proliferation, thereby highlighting its value as a potential anti-tumor target.
Keywords: G6PD, JAK2, Nucleotide metabolism, Pentose phosphate pathway, Tumorigenesis
Highlights
-
•
IL-6 rapidly induces metabolic flux of PPP and enzymatic activity of G6PD.
-
•
JAK2 phosphorylates G6PD Y437 in response to IL-6 treatment.
-
•
G6PD pY437 increases G6PD activity by facilitating G6PD binding to G6P.
-
•
G6PD pY437 is required for IL-6-induced nucleotide biosynthesis and tumor growth.
1. Introduction
Carcinogenesis is promoted by the interaction of diverse cell-intrinsic and cell-extrinsic events, including accumulation of DNA mutation, excessive oxidative stress, and aberrant expression of factors in the tissue environment [1]. It has been widely established that the surrounding tissue microenvironment has a vital impact on tumor development, driving tumor cell proliferation and survival with inhibitory effect on antitumor immune response [2]. Many cancers develop from sites of chronic inflammation, as the prolonged activation of the immune system and release of cytokines can boost the growth of cancer cells. In addition, tumor cells themselves can establish an inflammatory environment through the release of cytokines and other signaling molecules [3,4]. Therefore, comprehending the crosstalk between tumor cells and the inflammatory environment is important for the discovery of new inflammatory biomarkers to monitor tumor progression or new anti-inflammatory drugs to prevent and treat cancer.
As a major inflammatory factor, interleukin-6 (IL-6) is frequently accumulated in the tissue microenvironment of various haematopoietic malignancies or solid tumors [5]. The transduction of IL-6 signal requires the activation of transcription factor (signal transducer and activator of transcription 3) STAT3 in a Janus kinases (JAKs)-dependent manner. Specifically, through binding with IL-6 receptor (IL-6R) on cell surface, IL-6 complexes with IL-6R and glycoprotein 130 (gp130) to form a heterohexamer, thereby initiating the downstream signaling cascade by activating JAKs. JAKs, in turn, cause gp130 phosphorylation at a couple of tyrosine residues, among which four located within the C-terminal region are employed as the STAT3 docking sites. After STAT3 associates with gp130, JAKs phosphorylate STAT3 at tyrosine (Y)705 to trigger STAT3 to enter the nucleus, where STAT3 acts as a transcriptional factor to drive the transcription of its target genes and eventually modulate cell phenotype [6].
The pentose phosphate pathway (PPP) is essential for tumor growth as it generates ribose 5-phosphate (R5P), which is the indispensable building block to produce nucleotide for the rapid cell proliferation of tumor cells [7]. The rate-limiting step of PPP is governed by glucose-6-phosphate dehydrogenase (G6PD), which branches the carbon flow from glycolysis into PPP for the biosynthesis of nucleotides (Figure 1A) [8]. As a pleiotropic cytokine, IL-6 regulates not only inflammatory response but also cell metabolism, including glucose uptake, glycolysis, fatty acid oxidation and oxidative phosphorylation [9,10]. It is reported that loss of IL-6 resulted in ∼40% reduction in PPP intermediate R5P and ∼50% reduction in purines in tuberous sclerosis complex-deficient cells [11], suggesting a vital impact of IL-6 expression on the metabolic flux of PPP. However, the molecular mechanism underlying IL-6-mediated regulation of PPP remains largely elusive. In the present study, our data demonstrates that JAK2 phosphorylates G6PD at Y437 under IL-6 treatment, which activates G6PD by enhancing G6PD binding to its substrate glucose-6-phosphate (G6P). Activation of G6PD boosts PPP and DNA/RNA biosynthesis, which is required for IL-6-induced OSCC cell proliferation.
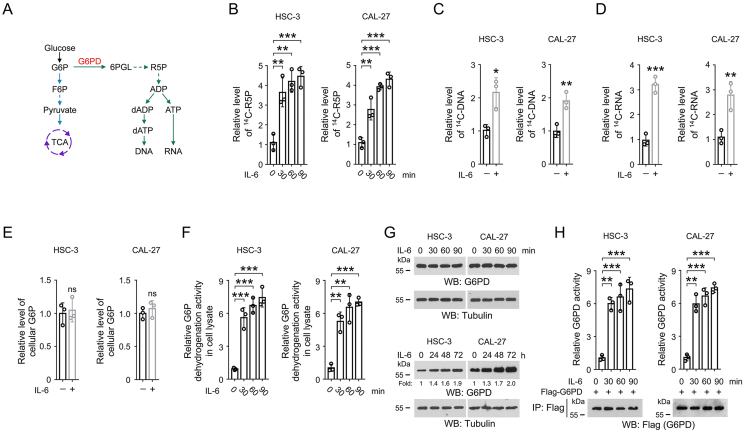
Figure 1.
IL-6 treatment increases G6PD activity in OSCC cells.
(A) A schematic of PPP and nucleotide biosynthesis pathway.
(B) HSC-3 and CAL-27 cells were challenged by 50 ng/mL IL-6 for the indicated time. Cells were washed and then incubated with [14C]-glucose (1 μCi, 0.01 mmol/L) for 1 h. The amount of cellular [14C]-R5P was determined. The data are presented as the mean ± SD from 3 independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.
(C-D) HSC-3 and CAL-27 cells were treated with 50 ng/mL IL-6 for 1 h. Cells were washed and then incubated with [14C]-glucose (1 μCi, 0.01 mmol/L) for 1 h. The amount of [14C]-DNA (C) or [14C]-RNA (D) was quantified. The data are presented as the mean ± SD from 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
(E) HSC-3 and CAL-27 cells were treated with 50 ng/mL IL-6 for 1 h. The cellular level of G6P was examined. The data are presented as the mean ± SD from 3 independent experiments. ns, not significant.
(F-G) HSC-3 and CAL-27 cells were treated with 50 ng/mL IL-6 for the indicated time. The G6P dehydrogenation activity in the cell lysates was examined (F). The expression of G6PD was determined by immunoblot (G). The data are obtained from 3 independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.(H) HSC-3 and CAL-27 cells were transfected with Flag-G6PD. 48 h after transfection, cells were incubated with 50 ng/mL IL-6 for the indicated time. Immunoprecipitation was then performed with anti-Flag M2 antibody. The G6PD activity in the precipitates was examined. The data are presented as the mean ± SD from 3 independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.
2. Materials and methods
2.1. Materials
Antibodies recognizing JAK2 (#3230), STAT3 pY705 (#9145), JAK2 pY1007 (#4406), JAK1 (#77692), JAK3 (#8827) STAT3 (#9139), and Tyk2 (#14193) were purchased from Cell Signaling Technology. Human recombinant IL-6 protein was purchased from Proteintech (HZ-1019). Antibodies recognizing Tubulin (ab7291), Flag (ab205606), His (ab18184) and G6PD (ab231828) were purchased from Abcam. [γ-32P]-ATP was purchased from PerkinElmer (BLU002Z001MC). PD98059 (HY-12028) was purchased from MedChemExpress. AM-2282 (S1421), Pyridone 6 (S6789), BMS-911543 (S7144) and SP600125 (S1460) were purchased from Selleck Chemicals. G6PD pY437 antibody was customized from Boer Biotechnology (Chengdu, China).
2.2. Cell culture and IL-6 treatment
HSC-3 cell line was obtained from Japanese Collection of Research Biosources (JCRB) Cell Bank. Human tongue squamous cell carcinoma cell line CAL-27 was obtained from American Type Culture Collection (ATCC). STR-typing-based cell line authentication was performed shortly before both cell lines were used in the experiments for this study. These cells were cultured with Dulbecco's Modified Eagle Medium supplemented with 10% fetal bovine serum. Cells were treated with active recombinate IL-6 protein at 50 ng/mL.
2.3. Immunoprecipitation
Immunoprecipitation was performed following previous report [12]. Cells were lysed in the dish or well by using a lysis buffer (0.1% SDS, 0.5 mM EDTA, 1% Triton X-100, 100 μM sodium pyrophosphate, 150 mM NaCl, 100 μM PMSF, 100 μM leupeptin, 1 μM aprotinin, 1 mM dithiothreitol, 100 μM sodium orthovanadate, 50 mM Tris-HCl [pH 7.5], and 1 mM sodium fluoride), followed by centrifugation at 12,500 g for 10 min. The precipitates were discarded, and the supernatants were mixed with specified antibodies and rotated overnight at 4°C. Agarose beads were added in the mixture, followed by rotation for another 4 h. The precipitates were washed with PBS and analyzed by immunoblot.
2.4. Immunoblot
Cells were lysed with RIPA buffer (Beyotime Biotechnology, P0013B) containing protease inhibitors (Beyotime Biotechnology) on ice for 30 min. Lysates were then centrifuged at 13,000 rpm at 4°C for 10 min, and the supernatant was collected and transferred to a clean centrifuge tube. The protein concentration was determined by using BCA assay. The protein samples were mixed with 5× loading buffer and boiled at 100°C for 10 min. The protein samples were then loaded on a SDS polyacrylamide gel electrophoresis (SDS-PAGE) gel, followed by electrophoresis for separation of the proteins. The proteins were then transferred to PVDF membranes and blocked with 5% skim milk for 30 min at room temperature. The membranes were then incubated with indicated primary antibodies (diluted with 5% BSA) at 4°C overnight, washed with TBST and followed by incubation with secondary antibodies (diluted with 5% BSA) for 1 h at 37°C. The blots were visualized by chemiluminescence reagents.
The blocking peptides were synthesized by using the following sequences: G6PD pY437, KNVKLPDApY437ERLILFCGSQ; G6PD, KNVKLPDAY437ERLILFCGSQ. The blocking peptides were added in the working solution of primary antibody at the concentration of 25 μg/mL. The primary antibody solution was then rotated at 4°C for 1 h before used for incubation with the membrane.
2.5. Constructs and shRNAs
shRNAs were synthesized using the following sequences: G6PD shRNA, TTC TTA TAG CAG AGA GGC T (targeting non-coding region); G6PD shRNA-CDS, TAA CGC AGG CGA TGT TGT C (targeting the coding sequence 682-GAC AAC ATC GCC TGC GTT A-700); JAK2 shRNA, CCC TGA CCC TAA ATA ATA CAT; JAK1 shRNA, TAT CCT CCA AGT AGC TCA G; JAK3 shRNA, GTC TCT CCT CGA AGA TCG T; Tyk2 shRNA, TAC TCC TTC AGG CAC TCT G; STAT3 shRNA, TAC CTA AGG CCA TGA ACT T (targeting non-coding region); control shRNA, GCT TCT AAC ACC GGA GGT CTT.
Human G6PD, JAK2, STAT3, IL-6 gene was cloned into pcDNA3.1-Flag, pcDNA3.1-His or pcDNA3.1 vector. QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce the indicated mutations. The rG6PD expression construct that was resistant to G6PD shRNA-CDS was prepared by introducing four nonsense mutations, including C687T, C690A, C693G and C696T, in the G6PD shRNA-CDS -targeted region.
2.6. Preparation of purified proteins
The DNA of WT or mutant G6PD was inserted into pCold I vector, which was then transferred into BL21 (DE3) bacteria as previously described [13]. Expression of G6PD proteins was stimulated by the addition of IPTG. BL21 bacterial cells were then cultured in Lysogeny Broth medium for 16 h at 16°C, followed by lysis with sonication. The DNA of WT and mutant JAK2 was inserted into pcDNA3.1-Flag vector, which was transfected into 293T cells. 72 h after transfection, cells were collected and lysed.
For the purification of His-tagged proteins, the bacterial lysates were centrifuged and the supernatants were loaded onto a Ni-NTA column (GE Healthcare Life Sciences). The column was then washed with 20 mM imidazole and the protein was eluted with 250 mM imidazole. For the purification of Flag-tagged proteins, the cell lysates were centrifuged and the supernatants were loaded onto a column containing Anti-Flag M2 agarose affinity gel. The column was washed with PBS and the protein was eluted with 100 μg/mL Flag peptide. For removal of the contaminants, the eluted protein solution was subjected to a HiPrep 16/60 Sephacryl S-200 HR gel filtration column (GE Healthcare Life Sciences), and the fractions containing the wanted protein were collected.
2.7. Metabolic influx from glycolysis into DNA/RNA synthesis
Measurement of the metabolic influx from glycolysis into DNA and RNA synthesis was performed according to previous report [14]. Briefly, cells in a well of a six-well plate were incubated in 2 mL of fresh medium containing 1 μCi of [14C]-glucose for 1 h. After wash with PBS, RNA or DNA was extracted, and the radiation signal in the RNA or DNA samples were assayed using a scintillation counter. The results were normalized to the cell numbers.
2.8. Measurement of G6PD activity
G6PD activity in cell lysates and immunoprecipitates was examined by using Glucose-6-Phosphate Dehydrogenase Activity Colorimetric Assay Kit obtained from BioVision (#K757-100). The values of G6PD activity were normalized to the cell numbers.
2.9. In vitro kinase assay for JAK2
In vitro kinase assay for JAK2 was performed as described previously [15]. Purified JAK2 protein (10 ng) was mixed with purified G6PD protein (200 ng) in 25 μL kinase reaction buffer (50 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 1 mM Na3VO4, 50 μM DTT, 5% glycerol, 100 μM ATP) at 30°C for 30 min G6PD protein was pulled down and subjected to SDS-PAGE separation, followed by immunoblotting or autoradiographic analyses. 10 μCi [γ-32P]-ATP was included in the reaction system if autoradiography was used to detect the phosphorylation.
For in vitro kinase assay using ATPγS, the experiment was performed following the Kinase Reaction and Alkylation Protocol (Abcam) according to the previous report [16]. In brief, purified JAK2 protein and G6PD protein were incubated with the reaction buffer (20 mM Hepes [pH 7.5], 100 mM NaCl, 1 mM ATPγS (ab138911, Abcam), 10 mM MgCl2) for 30 min at 30°C. The mixture was then incubated with 2.5 mM p-nitrobenzyl mesylate (ab138910, Abcam) at room temperature for 1 h. G6PD protein was then pulled down and subjected to SDS-PAGE separation, followed by immunoblot analysis with the thiophosphate ester specific antibody (ab92570, Abcam).
2.10. Analyses of protein structure
The PyMOL software was used to analyzed the protein structure data. The structure data of human G6PD protein was downloaded from PDB database. Most parts of the G6PD protein were shown in cartoon format, while Y437 site as well as NADP+ and G6P molecules were shown in sphere format.
2.11. Measurement of G6PD binding affinity with substrates
100 ng G6PD protein purified from mammalian or bacterial cells was immobilized on beads and incubated with 100 μl binding buffer (50 mM Tris-HCl [pH 7.5], 1 mM MgCl2, and 2.5 mM DTT) at 25°C for 10 min. To measure the binding between G6PD and NADP+, 0.3 mM NADP+ and 10 μCi [14C]-NADP+ was included in the binding buffer. To measure the binding between G6PD and G6P, 0.5 mM G6P and 10 μCi [14C]-G6P (MC149-250UCI, VWR) was included in the binding buffer. The protein-beads complexes were then washed with binding buffer for five times, and the radioactivity signal was then detected by liquid scintillation counting. Data was normalized to the amount of G6PD protein.
[14C]-NADP+ was enzymatically synthesized using recombinant human NAD kinase obtained from BioVision (#7560) and [14C]-NAD+ obtained from PerkinElmer (NEC831010UC), following previous report [17]. [14C]-NAD+ (50 μCi) was incubated with a reaction buffer (100 μl of 0.5 M Tris/HCl, pH 7.5; 20 μl of 0.5 M MgCl2; 240 μl of 10 mM ATP; 40 μl of 7.1 mM NAD+; 60 μg of recombinant human NAD kinase). The reaction was performed at 37°C for 120 min, and terminated by adding 1.0 mL ethanol to the mixture. The mixture was dried at 25°C, redissolved in 1.0 mL water, centrifuged at 1000 g for 15 min to remove the insoluble substance. The supernatant fluid was dried at 25°C, and redissolved in water.
2.12. Measurement of the level of cellular 14C-R5P
Measurement of the level of cellular 14C-R5P was performed by conversion of R5P to phosphoribosyl pyrophosphate and subsequent conversion of phosphoribosyl pyrophosphate to AMP, according to previous report [18]. Briefly, 20 μl cell lysate was added into 100 μl reaction buffer (5 μM Tris-HCl, pH 7.4; 50 nM disodium EDTA; 250 nM reduced GSH; 10 nM adenine; 1 μg bacterially purified phosphoribosyl pyrophosphate synthetase 1 protein; 1 μg bacterially purified adenine phosphoribosyltransferase protein; 100 nM ATP; 750 nM MgCl2; 4.2 μM Na2HPO4, pH 7.4), and incubated at 37°C for 60 min. The mixture was then loaded onto a cellulose thin layer chromatography sheet, and developed for 3 h in a solvent system containing butyl alcohol: acetone: glacial acetic acid: 5% ammonium hydroxide: water (7: 5: 3: 3: 2). The spot for AMP was collected and counted with liquid scintillation counter. The level of 14C-R5P was calculated as the difference of the amount of radioactive AMP generated in the presence and absence of phosphoribosyl pyrophosphate synthetase 1 protein. The results were normalized to the cell numbers.
2.13. Measurement of cellular 14C-ATP and 14C-ADP
Measurement of cellular 14C-ATP was performed according to the previous report [19]. Briefly, cells were treated with 5% PCA to denature proteins and extract the nucleotide components. The sample was centrifuged at 13,000 rpm for 10 min at 4°C, and the supernatant was adjusted to pH 4. The supernatant was then loaded onto chromatographic plates, and separated with the system containing isobutyl alcohol: amyl alcohol: ethoxyethanol: ammonia: water (3: 2: 6: 3: 5). The spot for ATP or ADP, respectively, was collected and counted with a liquid scintillation counter. The results were normalized to the cell numbers.
2.14. Colony formation assay
We stably expressed G6PD shRNA in HSC-3 or CAL-27 cells, and then stably expressed WT Flag-G6PD or Flag-G6PD Y437F. 100 HSC-3 or CAL-27 cells were seeded in a well of 6-well plates, and then continuously cultured for 14 days in the presence or absence of 50 ng/mL IL-6. The medium was changed with fresh medium with or without IL-6 every three days. After the 14-day culture, the clones were fixed with methanol for 3 min, and then stained with 0.05% (w/v) crystal violet for 20 min. The clones were washed with PBS for 5 times, and the number of clones were counted under microscope. Only those clones harboring more than 50 cells were selected and counted.
2.15. BrdU incorporation assay
BrdU incorporation assay was performed following previous report [20]. 2 × 104 cells were seeded in a well of 96-well plate. On the next day, cells were incubated with 50 ng/mL IL-6 for 6 h. 20 μl of the diluted 1× BrdU working solution was added in a well, and the cells were incubated for another 6 h. BrdU incorporation assay was performed by using BrdU Cell Proliferation ELISA Kit (colorimetric) purchased from Abcam (ab126556), according to the manufacture's introduction.
2.16. Mice tumor xenograft model
5 × 105 HSC-3 cells with indicated treatment were subcutaneously introduced into 6-week-old BALB/c nude mice. 27 days after implantation, mice were sacrificed and the tumor size was measured and calculated using the formula: 1/2 × length × width2. The use of animals was approved by the institutional review board of West China Hospital of Stomatology, Sichuan university. The animals were treated in accordance with relevant institutional and national guidelines and regulations.
2.17. Immunohistochemical staining of clinical samples
Immunohistochemical staining was performed following previous report [21]. Vectastain ABC kit (Vector Laboratories, CA) was used to conduct the immunohistochemical staining. Immunohistochemical staining were quantified by 2 pathologists, Dan He and Xiaofei Li, who were blinded to experimental procedures, following a previously reported scoring method [22]. Proportion scores were defined as: 0, no cells with positive staining; 1, 1–10%; 2, 11–30%; 3, 31–70%; and 4, 71–100%. Intensity scores were defined as: 0, negative; 1, weak; 2, moderate; 3, strong; 4, very strong. We then multiply the proportion and intensity scores to obtain a final score. For Figure 5H, if more than two samples have the same scores for both IL-6 and G6PD pY437, the dots representing these samples overlap each other, and are shown as only one dot in the graph. A total of 50 OSCC tissues were obtained with informed consent. OSCC clinical samples were classified as stage I (18 cases), II (11 cases), III (13 cases), or IV (8 cases) according to the oral cancer TNM standard from the Union for International Cancer Control [23], and classified as well (22 cases), moderately (16 cases) or poorly (12 cases) according to the WHO grading system. The use of human OSCC specimens was approved by the institutional review board of West China Hospital of Stomatology, Sichuan university.
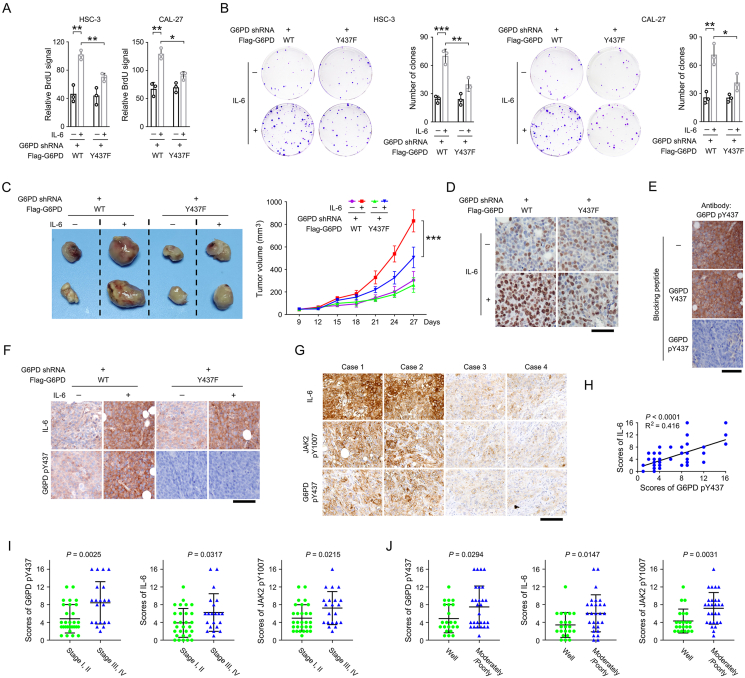
Figure 5.
JAK2-dependent G6PD Y437 phosphorylation is required for IL-6 induced OSCC cell proliferation and tumor growth.
(A-B) We stably expressed G6PD shRNA in HSC-3 and CAL-27 cells, and then stably expressed WT Flag-G6PD or Flag-G6PD Y437F. The shRNA targeted the non-coding region of G6PD mRNA, and so that expression of WT Flag-G6PD and Flag-G6PD Y437F is resistant to G6PD shRNA. Cell proliferation rate was measured by BrdU incorporation assay (A) and colony formation assay (B) in the presence or absence of IL-6. The data are presented as the mean ± SD from 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
(C) HSC-3 cells were stably expressing G6PD shRNA, WT Flag-G6PD, Flag-G6PD Y437F or/and IL-6. Cells were subcutaneously injected in mice (n = 7). The images of two representative xenografts from each group were shown (left panel). The volumes of mice tumor xenografts were measured at indicated time points (right panel). ∗∗∗P < 0.001.
(D) Mice tumor xenografts (n = 7) were analyzed by immunohistochemical staining with an anti-Ki67 antibody. Scale bar, 80 μm.
(E) The specificity of the anti-G6PD pY437 antibody was examined by immunohistochemical staining using the IL-6 and WT Flag-G6PD-expressed mice tumor xenograft samples in the presence or absence of the indicated blocking peptides. The data are obtained from 3 independent experiments. Scale bar, 80 μm.
(F) Mice tumor xenografts (n = 7) were analyzed by immunohistochemical staining with the indicated antibodies. Scale bar, 100 μm.
(G-H) 50 human OSCC samples were analyzed by immunohistochemical staining with the indicated antibodies (G). The scores of immunohistochemical staining were analyzed by linear regression (H, 50 cases). The dots representing the samples that have the same scores for both IL-6 (Y-axis) and G6PD pY437 (X-axis) overlap each other, and are shown as only one dot in the graph. Scale bar, 100 μm.
(I) The scores of immunohistochemical staining were compared between stage I/II group (29 cases in total) and stage III/IV group (21 cases in total).
(J) The scores of immunohistochemical staining were compared between well differentiated group (22 cases) and moderately/poorly differentiated group (28 cases in total).
2.18. Quantification and statistical analysis
Sample size was determined to be adequate based on the magnitude and consistency of measurable differences between groups in all experiments in this study. Statistical analyses were performed using two-sided Student's t test for comparison between two groups. All data represent the mean ± SD of at least three independent experiments/samples unless otherwise specified. Differences in means were considered statistically significant if P < 0.05. The Bonferroni correction was used for the multiple hypothesis correction (requiring P < 0.05/N, N indicates the number of comparisons), to avoid reporting false positive results.
3. Results
3.1. IL-6 treatment rapidly enhances nucleotide synthesis and G6PD activity
We set out to determine the impact of IL-6 treatment on nucleotide synthesis in OSCC cells. As both HSC-3 and CAL-27 cell lines were reported to be IL-6-responsive [[24], [25], [26], [27]], these two cell lines were selected as cell models. Cells were incubated with 14C-labeled glucose, and so that the metabolic flux from glycolysis to nucleotide synthesis could be monitored by measuring the level of 14C-labeled nucleotide metabolites. An apparently higher level of glucose-derived R5P was found when these cells were treated with IL-6 (Figure 1B). Notably, accumulation of cellular R5P could be induced by IL-6 treatment for only 30 min. R5P is used to produce the indispensable ribose for the synthesis of nucleotide, which could be further used to generate DNA and RNA. Consistently, more intense radioactive signals were detected in the genomic DNA and total RNA purified from HSC-3 and CAL-27 cells after IL-6 treatment (Figure 1C-D). In contrast, no obvious changes were found in the cellular levels of G6P (Figure 1E). These results lead to a hypothesis that PPP, which use glycolytic intermediate G6P to produce R5P, was likely activated in OSCC cells in response to IL-6.
As G6PD catalyzes the rate-limiting step in PPP to convert G6P into 6-phosphogluconolactone (6PGL) [28], we next performed time-course experiments to measure the cellular G6P dehydrogenation activity in response to IL-6 treatment. Indeed, the G6P dehydrogenation activity in the lysates of both HSC-3 and CAL-27 cells was enhanced more than 5-fold after 30-, 60- or 90-min treatment with IL-6 (Figure 1F). Nevertheless, G6PD protein expression was barely changed unless IL-6 treatment was extended to longer than 24 h (Figure 1G), suggesting that IL-6-induced cellular G6P dehydrogenation activity was not simply due to the increase in G6PD protein expression. Further, we precipitated equal amount of Flag-tagged G6PD protein from untreated or IL-6-stimualted cells, and found that exposure to IL-6 potently increased the enzymatic activity of G6PD protein (Figure 1H). These data suggest that IL-6 treatment rapidly enhances nucleotide synthesis and G6PD enzymatic activity in a G6PD expression-independent manner.
3.2. JAK2 phosphorylates G6PD at Y437 under IL-6 treatment
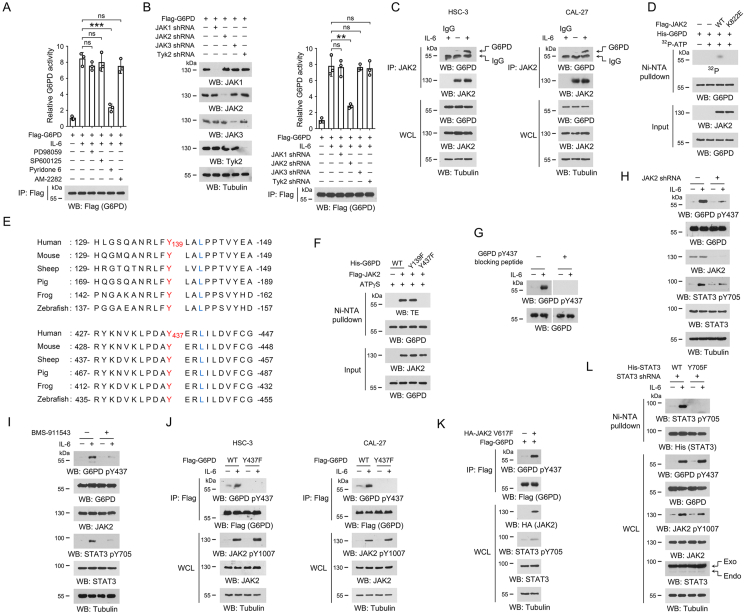
To identify the key factors that were responsible for IL-6-induced G6PD activation, a couple of inflammation-responsive pathways were blocked by small molecule inhibitors. Notably, treatment with Pyridone 6, a pan-Janus-activated kinase (JAK) inhibitor, substantially abolished IL-6-induced G6PD activity, while inhibition of MAPK by PD98059, JNK by SP600125, and PKC by AM-2282 only showed minor effects (Figure 2A). Since four JAK family members were reported as far, including JAK1, JAK2, JAK3 and Tyk2, we used specific shRNA to knock down each of them, respectively, and found that only loss of JAK2 largely alleviated IL-6-induced G6PD activity (Figure 2B). Co-immunoprecipitation analyses revealed a relatively weak association between endogenous G6PD and JAK2 in untreated OSCC cells, which could be largely enhanced after IL-6 treatment (Figure 2C). JAK2 is a non-receptor tyrosine kinase that mediates cytokine signals by phosphorylation of a wide spectrum of proteins [29,30], and this prompted us to examine if G6PD was a new phosphorylation substrate for JAK2. We thus performed an in vitro kinase experiment by incubating purified Flag-JAK2 and His-G6PD proteins with [γ-32P]-ATP. The His-G6PD protein was then isolated by nickel beads pulldown, and whether the radioactive γ-phosphate group was covalently linked to His-G6PD protein was examined by SDS-PAGE separation and autoradiography. As result, addition of [γ-32P]-ATP in the mixture of Flag-JAK2 and His-G6PD proteins resulted in a significant autoradiographic band corresponding to His-G6PD. In contrast, such autoradiograph band was not detected when His-G6PD was alone or mixed with a kinase-dead JAK2 K822E mutant protein [31] (Figure 2D). These evidences strongly suggest that JAK2 directly phosphorylates G6PD.
Figure 2.
JAK2 phosphorylates G6PD Y437 under IL-6 treatment.
(A-D, F-L) Immunoblotting analyses were performed using the indicated antibodies.
(A) HSC-3 cells were pre-treated with 20 μM PD98059, 20 μM SP600125, 2 μM Pyridone 6, or 1 μM AM-2282 for 2 h, respectively, and then treated with 50 ng/mL IL-6 for 1 h. Immunoprecipitations were performed with anti-Flag M2 antibody, and G6PD activity in the precipitates was examined. The data are presented as the mean ± SD from 3 independent experiments. ∗∗∗P < 0.001; ns, not significant.
(B) HSC-3 cells were transfected with the indicated shRNA and/or Flag-G6PD vector. 48 h after transfection, cells were treated with 50 ng/mL IL-6 for 1 h, and the cell lysates were used for immunoprecipitations with anti-Flag M2 antibody. The G6PD activity in the precipitates was examined. The data are presented as the mean ± SD from 3 independent experiments. ∗∗P < 0.01; ns, not significant.
(C) HSC-3 and CAL-27 cells were treated with or without 50 ng/mL IL-6 for 1 h, and co-immunoprecipitations were performed using the indicated antibodies. The data are obtained from 3 independent experiments. WCL, whole cell lysate.
(D) Purified His-G6PD protein was mixed with purified Flag-JAK2 or Flag-JAK2 K822E protein in the presence or absence of [γ-32P]-ATP for an in vitro kinase assay. Ni-NTA pulldown was performed, and the radioactivity in the precipitates was measured by autoradiography. The data are obtained from 3 independent experiments.
(E) Alignment analyses of G6PD Y139 or Y437 were performed using the G6PD protein sequences from the indicated species. Y139 and Y437 were shown in red, and the residues at +3 site that are required for JAK2 substrate consensus were labeled in blue.
(F) Purified WT His-G6PD protein or the indicated mutant proteins were incubated with purified Flag-JAK2 protein in the presence of ATPγS for an in vitro kinase assay. Ni-NTA pulldown was performed, and phosphorylation was detected using the thiophosphate ester (TE)-specific antibody. The data are obtained from 3 independent experiments.
(G) HSC-3 cells were treated with 50 ng/mL IL-6 for 1 h. G6PD Y437 phosphorylation was examined by immunoblot in the presence or absence of G6PD pY437 blocking peptide. The data are obtained from 3 independent experiments.
(H) HSC-3 cells with expression of JAK2 shRNA were treated with 50 ng/mL IL-6 for 1 h. The data are obtained from 3 independent experiments.
(I) HSC-3 cells were pre-treated with 2 μM BMS-911543 for 2 h, and then treated with 50 ng/mL IL-6 for 1 h. The data are obtained from 3 independent experiments.
(J) HSC-3 and CAL-27 cells with expression of WT Flag-G6PD or Flag-G6PD Y437F were incubated with 50 ng/mL IL-6 for 1 h, and immunoprecipitation was then performed with anti-Flag M2 antibody. The data are obtained from 3 independent experiments.
(K) HSC-3 cells with expression of Flag-G6PD and/or HA-JAK2 V617F, and immunoprecipitation was then performed with anti-Flag M2 antibody. The data are obtained from 3 independent experiments.
(L) We stably expressed STAT3 shRNA in HSC-3 cells, and then stably expressed WT His-STAT3 or His-STAT3 Y705F. Cells were incubated with 50 ng/mL IL-6 for 1 h, and Ni-NTA pulldown was performed. STAT3 shRNA targets the non-coding region. The data are obtained from 3 independent experiments.
JAK2 prefers to phosphorylate a tyrosine within a YxxL motif in the substrate protein [32]. We analyzed the G6PD protein sequence and revealed Y139 and Y437 with their flanking sequences, which were in accordance with JAK2 substrate consensus and highly conserved among multiple species (Figure 2E). Replacement of these two sites with non-phosphorylatable phenylalanine (F), respectively, showed that only the Y437F mutation abolished JAK2-mediated G6PD phosphorylation (Figure 2F), indicating that Y437 was the phosphorylation site.
We then generated an antibody that recognizes G6PD protein with phosphorylated Y437 (pY437), and found that G6PD Y437 phosphorylation was dramatically accumulated under IL-6 treatment (Figure 2G). The immunoblotting band corresponding to the phosphorylated G6PD was vanished when a G6PD pY437 blocking peptide was used, suggesting a good specificity of this antibody (Figure 2G). Consistently, IL6-induced G6PD Y437 phosphorylation was counteracted by JAK2 shRNA (Figure 2H), JAK2-selective inhibitor BMS-911543 (Figure 2I) or G6PD Y437F mutation (Figure 2J). Strikingly, expression of a constitutively active JAK2 V617F mutant [33] induced G6PD Y437 phosphorylation in HSC-3 cells even without IL-6 treatment (Figure 2K). STAT3 is the major downstream effector of IL-6, and could be activated by JAK2-mediated STAT3 Y705 phosphorylation [34]. Accordingly, modulation of JAK2 also changed the status of STAT3 Y705 phosphorylation (Figure 2H–I, 2K). However, mutation of STAT3 Y705F showed minor effects on G6PD Y437 phosphorylation in IL-6-stimulated HSC-3 cells, suggesting that STAT3 was not likely involved in IL-6-induced G6PD Y437 phosphorylation (Figure 2L). These results suggest that JAK2 directly phosphorylates G6PD at Y437 under IL-6 treatment.
3.3. JAK2-dependent Y437 phosphorylation activates G6PD by facilitating its binding with G6P
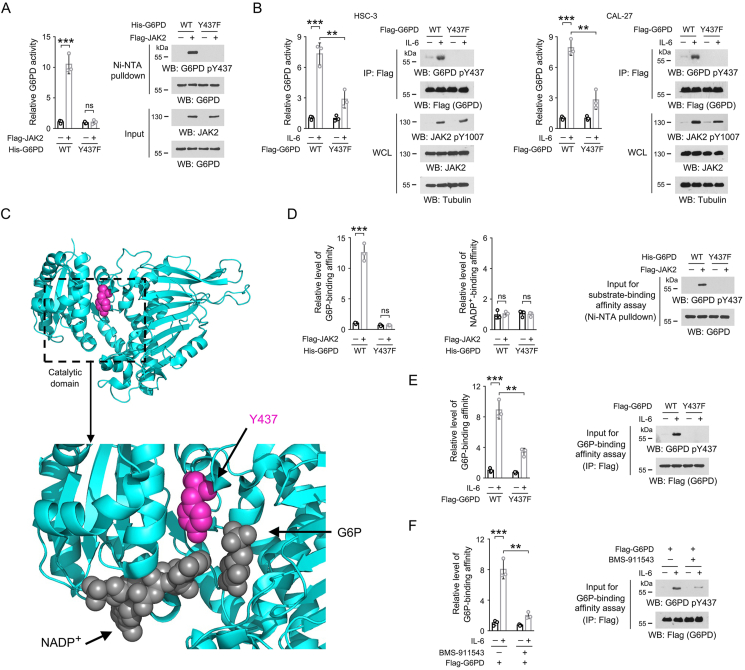
We next tested the impact of Y437 phosphorylation on G6PD enzymatic activity. We introduced Y437 phosphorylation on bacterially purified G6PD protein through JAK2-dependent in vitro kinase assay, and found that Y437 phosphorylation elevated G6PD activity by more than 10-fold (Figure 3A). In contrast, the activity of G6PD Y437F mutant protein was barely changed after the in vitro kinase assay, and remained comparable to its non-phosphorylated WT counterpart (Figure 3A). In line with this, IL-6 treatment, which induced apparent G6PD Y437 phosphorylation, markedly consolidated G6PD activity in both HSC-3 and CAL-27 cells, while this effect was abrogated by Y437F mutation (Figure 3B), suggesting that Y437 phosphorylation boosted G6PD activity.
Figure 3.
JAK2-dependent Y437 phosphorylation activates G6PD by facilitating G6PD binding with G6P.
(A, B, D-F) Immunoblotting analyses were performed using the indicated antibodies.
(A) In vitro kinase assay was performed by mixing bacterially purified WT His-G6PD or Y437F mutant protein with purified Flag-JAK2 protein. Ni-NTA pulldown was performed, and the G6PD enzymatic activity in the precipitates was examined. The data are presented as the mean ± SD from 3 independent experiments. ∗∗∗P < 0.001; ns, not significant.
(B) HSC-3 and CAL-27 cells expressing WT Flag-G6PD or Flag-G6PD Y437F were treated with 50 ng/mL IL-6 for 1 h. Flag-G6PD proteins were precipitated from cell lysates, and the G6PD enzymatic activity in the precipitates was examined. The data are presented as the mean ± SD from 3 independent experiments. WCL, whole cell lysate; ∗∗P < 0.01, ∗∗∗P < 0.001.
(C) The catalytic domain of human G6PD protein (PDB code: 7UAL) was boxed and enlarged, and the binding sites for NADP+ and G6P are shown. The side chain of Y437 was shown in lightmagenta, and NADP+ and G6P were shown in dark grey.
(D) Bacterially purified WT His-G6PD or Y437F mutant protein was incubated with purified Flag-JAK2 for an in vitro kinase assay. Ni-NTA pulldown was performed, and the binding between G6PD protein and NADP+ or G6P was measured. The data are presented as the mean ± SD from 3 independent experiments. ∗∗∗P < 0.001; ns, not significant.
(E) HSC-3 cells expressing WT Flag-G6PD or Flag-G6PD Y437F were treated with 50 ng/mL IL-6 for 1 h. Flag-G6PD proteins were precipitated from cell lysates, and the binding between G6PD protein and G6P was examined. The data are presented as the mean ± SD from 3 independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.
(F) HSC-3 cells with expression of WT Flag-G6PD were pre-treated with 2 μM BMS-911543 for 2 h, and then treated with 50 ng/mL IL-6 for 1 h. Flag-G6PD proteins were precipitated from cell lysates, and the binding between G6PD protein and G6P was assessed. The data are presented as the mean ± SD from 3 independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.
Analysis of the structure of human G6PD protein achieved in PDB database revealed that the Y437 site was located within the catalytic domain of G6PD (Figure 3C), which accommodates one G6P molecule and one NADP+ molecule to generate 6-phosphogluconolactone and NADPH [35]. Notably, previously reported cocrystallization of G6PD and G6P and NADP+ (PDB Code: 7UAL) showed that the Y437 site was close to both the NADP+ and G6P binding site (Figure 3C), which promoted us to test whether Y437 phosphorylation affect G6PD association with its substrates. By using [14C]-labeled G6P or NADP+, we found that JAK2-mediated G6PD Y437 phosphorylation palpably promoted bacterially purified G6PD protein binding with G6P, but not NADP+, and the similar effect was not observed for G6PD Y437F mutant protein (Figure 3D). In line with this, increased G6P-binding affinity was found in the Flag-G6PD protein derived from HSC-3 cells after IL-6 treatment, which could be largely abolished by G6PD Y437F mutation (Figure 3E) or BMS-911543 treatment (Figure 3F). These results suggest that JAK2-dependent Y437 phosphorylation activates G6PD by facilitating its binding with G6P.
3.4. JAK2-dependent G6PD Y437 phosphorylation is required for IL-6-enhanced nucleotide synthesis
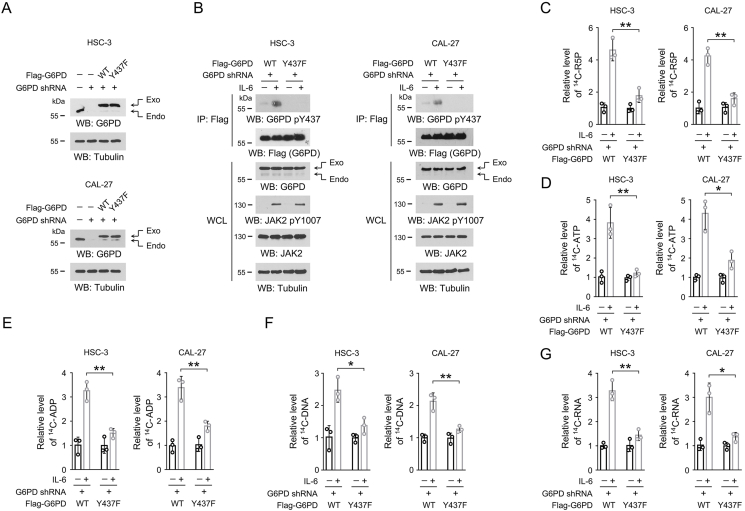
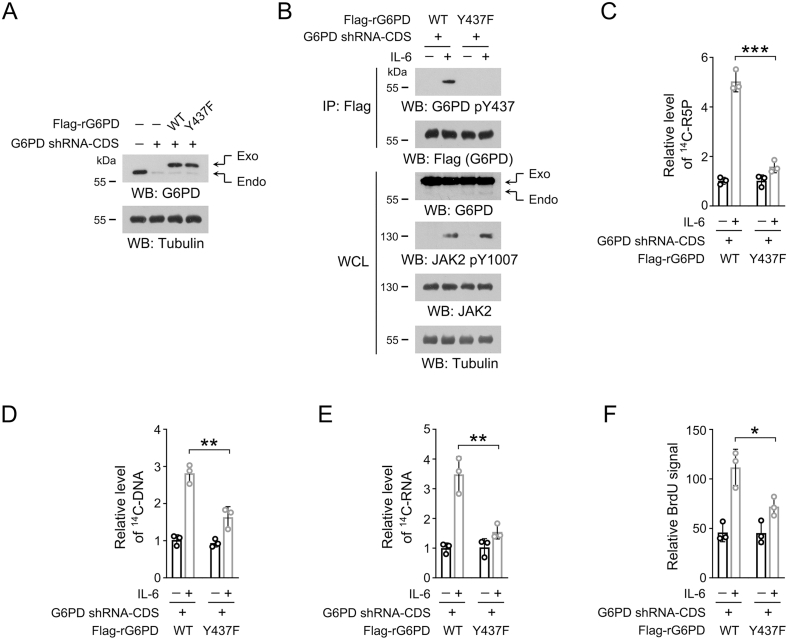
To modulate JAK2-dependent G6PD Y437 phosphorylation in OSCC cells, we used specific shRNA to silence the expression of endogenous G6PD, and exogenously expressed WT Flag-G6PD or Flag-G6PD Y437F mutant (Figure 4A). This shRNA targeted the non-coding region of G6PD mRNA, and so that it did not affect the expression of exogenous G6PD. As expected, IL-6 treatment caused apparent G6PD Y437 phosphorylation in HSC-3 and CAL-27 cells with WT G6PD, but not those cells with G6PD Y437F mutation (Figure 4B). To test the impact of G6PD Y437 phosphorylation on PPP and nucleotide synthesis, HSC-3 and CAL-27 cells were incubated with 14C-glucose, and much more 14C-labeled R5P was detected in the WT Flag-G6PD-expressed cells than the mutant cells after IL-6 treatment (Figure 4C). Adenosine triphosphate (ATP) is one of the most abundant nucleoside triphosphate in mammalian cells [36]. G6PD Y437F mutation substantially abrogated IL-6-induced synthesis of 14C-labeled ATP and its precursor, ADP (Figure 4D-E). Consistently, a markedly repressed radiation signal was detected in the genomic DNA and total RNA isolated from the mutant cells compared to the WT counterparts (Figure 4F-G). To avoid the off-target effects, we treated HSC-3 cells with another G6PD shRNA that targeted the coding sequence (labeled as G6PD shRNA-CDS in Fig. S1), and accordingly, expressed exogenous Flag-G6PD protein containing four nonsense mutations in the shRNA-targeted region (labeled as Flag-rG6PD in Fig. S1). Therefore, the expression of either WT Flag-rG6PD or Flag-rG6PD Y437F was resistant to G6PD shRNA-CDS (Fig. S1A). As expected, non-phosphorylatable G6PD Y437F mutation markedly abolished IL-6-induced R5P and DNA/RNA synthesis (Figs. S1B–S1E). These results indicate that JAK2-dependent G6PD Y437 phosphorylation is required for IL-6-enhanced PPP and nucleotide synthesis.
Figure 4.
JAK2-dependent G6PD Y437 phosphorylation is required for IL-6-induced nucleotide biosynthesis.
(A-G) We stably expressed G6PD shRNA in HSC-3 or CAL-27 cells, and then stably expressed WT Flag-G6PD or Flag-G6PD Y437F. The shRNA targeted the non-coding region of G6PD mRNA, and so that expression of WT Flag-G6PD and Flag-G6PD Y437F is resistant to G6PD shRNA.
(A) Immunoblots were performed using the indicated antibodies. The data are obtained from 3 independent experiments.
(B) Cells were treated with 50 ng/mL IL-6 for 1 h, and immunoprecipitation was performed with an anti-Flag M2 antibody. Immunoblots were performed using the indicated antibodies. The data are obtained from 3 independent experiments.
(C-G) Cells were treated with 50 ng/mL IL-6 for 1 h, and then incubated with [14C]-glucose (1 μCi, 0.01 mmol/L) for 1 h. The amount of [14C]-R5P (C), [14C]-ATP (D), [14C]-ADP (E), [14C]-DNA (F) or [14C]-RNA (G) was measured. The data are presented as the mean ± SD from 3 independent experiments. ∗P < 0.05, ∗∗P < 0.01.
3.5. JAK2-dependent G6PD Y437 phosphorylation is required for IL-6-induced OSCC cell proliferation and tumor growth
We then examined the impact of JAK2-dependent G6PD Y437 phosphorylation on OSCC cell proliferation. By BrdU incorporation assay, expression of WT or mutant G6PD resulted in comparable BrdU signal in the absence of IL-6. Notably, IL-6 treatment caused about 2-fold augment in BrdU signal in both HSC-3 and CAL-27 cells with the expression of WT G6PD, indicating an enhanced cell proliferation (Figure 5A and S1F). In contrast, those cells with expression of non-phosphoryable G6PD Y437F mutant only showed a slightly increased BrdU labeling after IL-6 treatment (Figure 5A and S1F). Similar results were observed by colony formation assay (Figure 5B). Further, we established a mouse xenograft model by subcutaneous injection of HSC-3 cells with overexpression of IL-6 and reconstituted expression of WT G6PD or G6PD Y437F. Reinforced IL-6 expression palpably accelerated the growth rate of WT G6PD-expressed tumors companied with a more intense immunohistochemical staining of Ki-67 and G6PD Y437 phosphorylation (Figure 5C-F). Notably, G6PD Y437F mutation, which annihilated G6PD Y437 phosphorylation (Figure 5F), largely abolished IL-6-induced tumor growth and Ki-67 expression (Figure 5C-D).
We next collected OSCC clinical samples derived from surgery, and examined the activation status of IL-6/JAK2 axis and the phosphorylation of G6PD Y437. Immunohistochemical staining revealed that the tumors with strong IL-6 expression and JAK2 Y1007 phosphorylation often showed a high level of G6PD Y437 phosphorylation (Figure 5G). The linear regression analyses demonstrated that the correlation between G6PD Y437 phosphorylation and IL-6 expression was statistically significant (Figure 5H). Further, OSCC clinical samples diagnosed as advanced TNM stages (stage III and IV) or moderately/poorly differentiated were found to be with higher level of G6PD Y437 phosphorylation, IL-6 expression and JAK2 Y1007 phosphorylation, compared to the samples diagnosed as early stages (stage I and II) or well differentiated, respectively (Figure 5I,J). These results suggest that JAK2-dependent G6PD Y437 phosphorylation is required for IL-6-induced OSCC cell proliferation and tumor growth.
4. Discussion
Head and neck cancer is the sixth common malignance nowadays, of which oral squamous cell carcinoma (OSCC) is the major type, with about 380,000 new cases worldwide annually [37]. Surgery, chemotherapy, and/or radiotherapy are currently the routing treatment for OSCC. Unfortunately, despite the progressions in diagnostic and therapeutic options during the past decades, the mortality of this disease is still high with a 5-year overall survival rate of 64.4% and 180,000 deaths per year [38]. In addition, up to 30% of OSCC patients experience tumor recurrence, leading to an even lower 5-year survival rate for this population [39]. Moreover, surgical treatment for OSCC often results in irreversible changes in the facial appearance and oral function, leaving physical or mental effects on survivors. Hence, a deeper deciphering of the key factors functioning in tumor development is critical to optimize the current treatment of OSCC patients.
Tumor cells hijack inflammatory mechanisms to promote their own growth and survival [40]. Tumor cells, adjacent stromal and inflammatory cells, interact with each other frequently to generate an inflammatory tumor microenvironment [3,41]. In such tumor microenvironment, rapid proliferation of tumor cells also induces homeostatic responses through increasing the number of macrophages, which is evidenced by the existence of signaling circuits composed of various cytokines mutually generated by one cell type and utilized by another [3]. IL-6, as one of the major cytokines in the tumor microenvironment, is found with high tissue concentrations and is reported to be upregulated in almost all types of tumors [42]. Therefore, IL-6-elicited signal transduction in tumor cells would be an ideal cue to investigate the crosstalk between tumor cells and inflammatory microenvironment.
Aberrant activation of IL-6 and its pro-proliferative role in OSCC have been documented. IL-6 was upregulated in OSCC tissues compared to non-cancerous adjacent tissues, which indicates IL-6 as a potential biomarker for predicting tumor recurrence and survival time for OSCC patients [43]. IL-6-mediated inflammasome activation enhanced OSCC cell proliferation via potentiating JAK2/STAT3/Sox4/NLRP3 signaling pathway [44]. Further, IL-6 desensitized OSCC cells to radiotherapy by counteracting with oxidative stress via activating Nrf2 antioxidant pathway [45]. Similarly, lipopolysaccharide stimulation-mediated IL-6 production also reduced cisplatin-induced cell death in OSCC cells [46]. Therefore, given the well-established pro-malignant properties of IL-6 during OSCC development, OSCC cells were chosen as the experimental model in this study.
The PPP, also termed as the hexose monophosphate shunt or phosphogluconate pathway, ramified from the glycolytic route at the first step and utilizes G6P as a primary source [47]. In particular, the PPP is frequently hyperactivated in tumor cells as it provides pentose phosphates to furnish their high-rate biosynthesis of nucleotides [47]. It is documented that high expression level of G6PD, the rate-limiting enzyme for PPP, was associated with high lymphatic metastasis rate and poor prognosis for OSCC patients [48,49] In addition, the level of R5P, the final product of PPP, was found to be markedly higher in the salivary samples from OSCC patients than that from non-cancerous controls [50]. In the present study, we determined the impact of IL-6 treatment on PPP in OSCC cells by using 14C-labeled glucose as a tracer. Exposure to IL-6 for only 30 min induced apparently more biosynthesis of R5P, the final product of the PPP, and increased the enzymatic activity of G6PD, the rate-limiting enzyme of the PPP. Pyridone 6-mediated inhibition of JAKs, which function as the key downstream signal transducers for IL-6, substantially abolished IL-6-induced G6PD activation. Hence, our data strongly suggest that IL-6 treatment rapidly consolidates G6PD and promotes the metabolic flux of PPP. In addition, a single concentration of 50 ng/mL IL-6 was used in all the IL-6 treatments in this study. This IL-6 concentration was widely used in previous reports to study the regulatory role of IL-6 on cancer cells [[51], [52], [53], [54]]. Considering the complexity of IL-6 signal transduction, further work is still needed to characterize G6PD Y437 phosphorylation, nucleotide metabolism as well as OSCC cell proliferation under the treatment of a wide concentration range of IL-6.
Transduction of IL-6 signal involves two cell surface proteins, IL-6R and gp130, which form a hexametric complex with IL-6 to promiscuously recruit JAK kinases, followed by activation of transcription factor STAT3 [6]. In this study, we report that G6PD is a new phosphorylation substrate of JAK2. By using purified proteins, we established an in vitro kinase assay, and showed that JAK2 directly phosphorylated G6PD at Y437 site. This phosphorylation was induced by IL-6 treatment, and could be inhibited by G6PD Y437F mutation, shRNA-mediated JAK2 knockdown or treatment with JAK2-selective antagonist BMS-911543. Interestingly, expression of a constructively active JAK2 V617F mutant was sufficient to enhance G6PD Y437 phosphorylation even without IL-6 treatment. Considering the similarity in the biological function of JAK family members, further work is needed to decipher the substrate recognition mode that enable JAK2, but not other JAKs, to phosphorylate G6PD.
It is well accepted that STAT3 serves as an important downstream effector in the transduction of IL-6 signaling. JAKs-mediated tyrosine phosphorylation promotes STAT3 dimerization and translocation into nucleus, resulting in vigorous transcription of its target genes [55]. It is reported that IL-6 stimulation induced G6PD expression via STAT3-mediated gene transcription, which played a role in prostate cancer bone metastasis [56]. Unlike the aforementioned reports, our data show that IL-6-induced G6PD Y437 phosphorylation is STAT3-independent. We replaced endogenous STAT3 in HSC-3 cells with STAT3 Y705F mutant, which was previously shown to abate STAT3 dimerization and translocation into nucleus, and found no obvious impact on IL-6-induced G6PD Y437 phosphorylation. Further, initiation of gene transcription usually takes more than 8 h to accumulate significant changes in protein level. Though we did observe an increased G6PD expression after IL-6 treatment for more than 24 h, Y437 phosphorylation and increased enzymatic activity of G6PD could be detected within only 90 min without obvious changes in G6PD protein expression. Therefore, our data report a rapid response mechanism through which JAK2-dependent G6PD Y437 phosphorylation bypass STAT3-mediated regulation and regulate PPP under IL-6 stimulation.
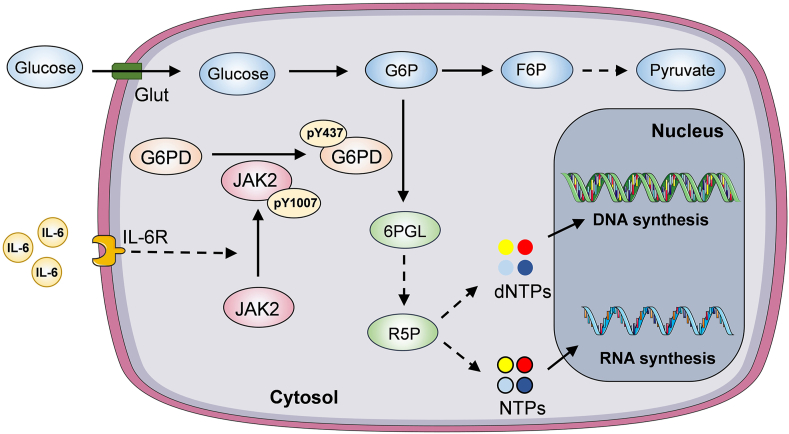
In this study, our findings demonstrate a protein posttranslational modification-based molecular event implicated in the crosstalk between tumor cells and inflammatory microenvironment. IL-6 regulates PPP to boost nucleotide biosynthesis through JAK2-dependent G6PD Y437 phosphorylation (Figure 6). Disruption of such molecular events limits tumor cell proliferation and may benefit OSCC treatment.
Figure 6.
A schematic of IL-6/JAK2 axis-dependent G6PD Y437 phosphorylation for OSCC proliferation.
IL-6 treatment rapidly activates JAK2, which in turn phosphorylates G6PD at Y437, leading to enhanced G6PD enzymatic activity. G6PD activation boosts PPP and R5P production, and so that facilitates DNA and RNA biosynthesis to support OSCC cell proliferation.
Authors' contributions
Conceptualization: R.L., L.J., Z.Z., and D.H.; methodology: X.Q., H.Y., and D.L.; investigation: X.Q., X.L., and D.L.; writing (original draft): R.L.; writing (review and editing): R.L.; funding acquisition: L.J., R.L., and D.H. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Sichuan Science and Technology Program 2023NSFSC1924 (R.L.), 2021YJ0204 (D.H.), National Natural Science Foundation of China grants 82171809 (L.J.), Scientific research project of Sichuan Provincial Health Commission 20PJ226 (D.H.), The Scientific Research Project of China Baoyuan CBYI202103 (D.H.), Science Foundation of West China Hospital of Stomatology RCDWJS2023-2 (R.L.), and Health Commission of Sichuan Province 21PJ159 (X.L.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101836.
Contributor Information
Lu Jiang, Email: jianglu@scu.edu.cn.
Rui Liu, Email: liurui_scu@hotmail.com.
Zhe Zhao, Email: 18803743168@163.com.
Dan He, Email: hd_baby81@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Figure S1.
JAK2-dependentG6PD Y437 phosphorylation is required forIL-6induced nucleotide synthesis and OSCC cell proliferation.
(A-F) We stably expressed G6PD shRNA-CDS in HSC-3 cells, and then stably expressed WT Flag-rG6PD or Flag-rG6PD Y437F. G6PD shRNA-CDS targeted the CDS sequence of G6PD mRNA. WT Flag-rG6PD and Flag-rG6PD Y437F contain four non-sense mutations in the G6PD shRNA-CDS-targeted region, and so that their expression is resistant to G6PD shRNA-CDS.
(A) Immunoblots were performed using the indicated antibodies. The data are obtained from 3 independent experiments.
(B) Cells were treated with 50 ng/mL IL-6 for 1 h, and immunoprecipitation was performed with an anti-Flag M2 antibody. Immunoblots were performed using the indicated antibodies. The data are obtained from 3 independent experiments.
(C-E) Cells were treated with 50 ng/mL IL-6 for 1 h, and then incubated with [14C]-glucose (1 μCi, 0.01 mmol/L) for 1 h. The amount of [14C]-R5P (C), [14C]-DNA (D) or [14C]-RNA (E) was measured. The data are presented as the mean ± SD from 3 independent experiments. ∗∗P < 0.01, ∗∗∗P < 0.001.
(F) Cell proliferation rate was measured by BrdU incorporation assay in the presence or absence of IL-6. The data are presented as the mean ± SD from 3 independent experiments. ∗P < 0.05.
Data availability
Data will be made available on request.
References
- 1.Elinav E., Nowarski R., Thaiss C.A., Hu B., Jin C., Flavell R.A. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 2.Bader J.E., Voss K., Rathmell J.C. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. 2020;78(6):1019–1033. doi: 10.1016/j.molcel.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greten F.R., Grivennikov S.I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51(1):27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crusz S.M., Balkwill F.R. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12(10):584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 5.Briukhovetska D., Dörr J., Endres S., Libby P., Dinarello C.A., Kobold S. Interleukins in cancer: from biology to therapy. Nat Rev Cancer. 2021;21(8):481–499. doi: 10.1038/s41568-021-00363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson D.E., O'Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du D., Liu C., Qin M., Zhang X., Xi T., Yuan S., et al. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm Sin B. 2022;12(2):558–580. doi: 10.1016/j.apsb.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang P., Du W., Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell. 2014;5(8):592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehrskov L.L., Christensen R.H. The role of interleukin-6 in glucose homeostasis and lipid metabolism. Semin Immunopathol. 2019;41(4):491–499. doi: 10.1007/s00281-019-00747-2. [DOI] [PubMed] [Google Scholar]
- 10.Li Y.S., Ren H.C., Cao J.H. Roles of Interleukin-6-mediated immunometabolic reprogramming in COVID-19 and other viral infection-associated diseases. Int Immunopharm. 2022;110 doi: 10.1016/j.intimp.2022.109005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Filippakis H., Hougard T., Du H., Ye C., Liu H.J., et al. Interleukin-6 mediates PSAT1 expression and serine metabolism in TSC2-deficient cells. Proc Natl Acad Sci U S A. 2021;118(39) doi: 10.1073/pnas.2101268118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao Y., Ren T., Huang X., Li M., Lee J.H., Chen Q., et al. Rapid phosphorylation of glucose-6-phosphate dehydrogenase by casein kinase 2 sustains redox homeostasis under ionizing radiation. Redox Biol. 2023;65 doi: 10.1016/j.redox.2023.102810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Zhang T., Ren T., Liao X., Hao Y., Lim J.S., et al. Oxygen-sensitive methylation of ULK1 is required for hypoxia-induced autophagy. Nat Commun. 2022;13(1):1172. doi: 10.1038/s41467-022-28831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Qian X., Peng L.X., Jiang Y., Hawke D.H., Zheng Y., et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol. 2016;18(5):561–571. doi: 10.1038/ncb3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J., Shao J., Zeng Z., He Y., Tang C., Park S.H., et al. Mechanosensitive turnover of phosphoribosyl pyrophosphate synthetases regulates nucleotide metabolism. Cell Death Differ. 2022;29(1):206–217. doi: 10.1038/s41418-021-00851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An L., Nie P., Chen M., Tang Y., Zhang H., Guan J., et al. MST4 kinase suppresses gastric tumorigenesis by limiting YAP activation via a non-canonical pathway. J Exp Med. 2020;217(6) doi: 10.1084/jem.20191817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaetani G.F., Ferraris A.M., Sanna P., Kirkman H.N. A novel NADPH:(bound) NADP+ reductase and NADH:(bound) NADP+ transhydrogenase function in bovine liver catalase. Biochem J. 2005;385(Pt 3):763–768. doi: 10.1042/BJ20041495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker M.A. Patterns of phosphoribosylpyrophosphate and ribose-5-phosphate concentration and generation in fibroblasts from patients with gout and purine overproduction. J Clin Invest. 1976;57(2):308–318. doi: 10.1172/JCI108282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norman G.A., Follett M.J., Hector D.A. Quantitative thin-layer chromatography of ATP and the products of its degradation in meat tissue. J Chromatogr. 1974;90(1):105–111. doi: 10.1016/s0021-9673(01)94779-x. [DOI] [PubMed] [Google Scholar]
- 20.Liao X., Huang X., Li X., Qiu X., Li M., Liu R., et al. AMPK phosphorylates NAMPT to regulate NAD(+) homeostasis under ionizing radiation. Open Biol. 2022;12(10) doi: 10.1098/rsob.220213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X., Fang J., Lai W., Hu Y., Li L., Zhong Y., et al. IL-6/STAT3 Axis activates Glut5 to regulate fructose metabolism and tumorigenesis. Int J Biol Sci. 2022;18(9):3668–3675. doi: 10.7150/ijbs.68990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R., Li J., Shao J., Lee J.H., Qiu X., Xiao Y., et al. Innate immune response orchestrates phosphoribosyl pyrophosphate synthetases to support DNA repair. Cell Metabol. 2021;33(10):2076. doi: 10.1016/j.cmet.2021.07.009. 89.e9. [DOI] [PubMed] [Google Scholar]
- 23.Cuccurullo V., Mansi L. AJCC cancer staging handbook: from the AJCC cancer staging manual (7th edition) Eur J Nucl Med Mol Imag. 2010;38(2):408. 08. [Google Scholar]
- 24.Saito K., Mitsui A., Sumardika I.W., Yokoyama Y., Sakaguchi M., Kondo E. PLOD2-driven IL-6/STAT3 signaling promotes the invasion and metastasis of oral squamous cell carcinoma via activation of integrin β1. Int J Oncol. 2021;58(6) doi: 10.3892/ijo.2021.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naher L., Kiyoshima T., Kobayashi I., Wada H., Nagata K., Fujiwara H., et al. STAT3 signal transduction through interleukin-22 in oral squamous cell carcinoma. Int J Oncol. 2012;41(5):1577–1586. doi: 10.3892/ijo.2012.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav A., Kumar B., Datta J., Teknos T.N., Kumar P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol Cancer Res. 2011;9(12):1658–1667. doi: 10.1158/1541-7786.MCR-11-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin H.Y., Hou S.C., Chen S.C., Kao M.C., Yu C.C., Funayama S., et al. (-)-Epigallocatechin gallate induces Fas/CD95-mediated apoptosis through inhibiting constitutive and IL-6-induced JAK/STAT3 signaling in head and neck squamous cell carcinoma cells. J Agric Food Chem. 2012;60(10):2480–2489. doi: 10.1021/jf204362n. [DOI] [PubMed] [Google Scholar]
- 28.Cappellini M.D., Fiorelli G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008;371(9606):64–74. doi: 10.1016/S0140-6736(08)60073-2. [DOI] [PubMed] [Google Scholar]
- 29.Hu F., Song D., Yan Y., Huang C., Shen C., Lan J., et al. IL-6 regulates autophagy and chemotherapy resistance by promoting BECN1 phosphorylation. Nat Commun. 2021;12(1):3651. doi: 10.1038/s41467-021-23923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roskoski R., Jr. Janus kinase (JAK) inhibitors in the treatment of neoplastic and inflammatory disorders. Pharmacol Res. 2022;183 doi: 10.1016/j.phrs.2022.106362. [DOI] [PubMed] [Google Scholar]
- 31.O'Leary E.E., Mazurkiewicz-Muñoz A.M., Argetsinger L.S., Maures T.J., Huynh H.T., Carter-Su C. Identification of steroid-sensitive gene-1/Ccdc80 as a JAK2-binding protein. Mol Endocrinol. 2013;27(4):619–634. doi: 10.1210/me.2011-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brien K.B., Argetsinger L.S., Diakonova M., Carter-Su C. YXXL motifs in SH2-Bbeta are phosphorylated by JAK2, JAK1, and platelet-derived growth factor receptor and are required for membrane ruffling. J Biol Chem. 2003;278(14):11970–11978. doi: 10.1074/jbc.M210765200. [DOI] [PubMed] [Google Scholar]
- 33.Wong R.A., Luo X., Lu M., An Z., Haas-Kogan D.A., Phillips J.J., et al. Cooperative blockade of PKCα and JAK2 drives apoptosis in glioblastoma. Cancer Res. 2020;80(4):709–718. doi: 10.1158/0008-5472.CAN-18-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuringa J.J., Wierenga A.T., Kruijer W., Vellenga E. Constitutive Stat3, Tyr705, and Ser727 phosphorylation in acute myeloid leukemia cells caused by the autocrine secretion of interleukin-6. Blood. 2000;95(12):3765–3770. [PubMed] [Google Scholar]
- 35.Wei X., Kixmoeller K., Baltrusaitis E., Yang X., Marmorstein R. Allosteric role of a structural NADP(+) molecule in glucose-6-phosphate dehydrogenase activity. Proc Natl Acad Sci U S A. 2022;119(29) doi: 10.1073/pnas.2119695119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traut T.W. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140(1):1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 37.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 38.Zanoni D.K., Montero P.H., Migliacci J.C., Shah J.P., Wong R.J., Ganly I., et al. Survival outcomes after treatment of cancer of the oral cavity (1985-2015) Oral Oncol. 2019;90:115–121. doi: 10.1016/j.oraloncology.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam S., Araslanova R., Low T.H., Warner A., Yoo J., Fung K., et al. Estimating survival after salvage surgery for recurrent oral cavity cancer. JAMA Otolaryngol Head Neck Surg. 2017;143(7):685–690. doi: 10.1001/jamaoto.2017.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniguchi K., Karin M.NF-κB. inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani A., Allavena P., Sica A., Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 42.Bent E.H., Millán-Barea L.R., Zhuang I., Goulet D.R., Fröse J., Hemann M.T. Microenvironmental IL-6 inhibits anti-cancer immune responses generated by cytotoxic chemotherapy. Nat Commun. 2021;12(1):6218. doi: 10.1038/s41467-021-26407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duffy S.A., Taylor J.M., Terrell J.E., Islam M., Li Y., Fowler K.E., et al. Interleukin-6 predicts recurrence and survival among head and neck cancer patients. Cancer. 2008;113(4):750–757. doi: 10.1002/cncr.23615. [DOI] [PubMed] [Google Scholar]
- 44.Xiao L., Li X., Cao P., Fei W., Zhou H., Tang N., et al. Interleukin-6 mediated inflammasome activation promotes oral squamous cell carcinoma progression via JAK2/STAT3/Sox4/NLRP3 signaling pathway. J Exp Clin Cancer Res. 2022;41(1):166. doi: 10.1186/s13046-022-02376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuoka Y., Nakayama H., Yoshida R., Hirosue A., Nagata M., Tanaka T., et al. IL-6 controls resistance to radiation by suppressing oxidative stress via the Nrf2-antioxidant pathway in oral squamous cell carcinoma. Br J Cancer. 2016;115(10):1234–1244. doi: 10.1038/bjc.2016.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Z., Luo Q., Ye D., Chen W., Chen F. Role of toll-like receptor 4 on the immune escape of human oral squamous cell carcinoma and resistance of cisplatin-induced apoptosis. Mol Cancer. 2012;11:33. doi: 10.1186/1476-4598-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39(8):347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y., Li Q., Niu L., Xu L., Guo Y., Wang L., et al. Suppression of G6PD induces the expression and bisecting GlcNAc-branched N-glycosylation of E-Cadherin to block epithelial-mesenchymal transition and lymphatic metastasis. Br J Cancer. 2020;123(8):1315–1325. doi: 10.1038/s41416-020-1007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y.C., Hsiao J.R., Jiang S.S., Chang J.Y., Chu P.Y., Liu K.J., et al. c-MYC-directed NRF2 drives malignant progression of head and neck cancer via glucose-6-phosphate dehydrogenase and transketolase activation. Theranostics. 2021;11(11):5232–5247. doi: 10.7150/thno.53417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Sá Alves M., de Sá Rodrigues N., Bandeira C.M., Chagas J.F.S., Pascoal M.B.N., Nepomuceno G., et al. Identification of possible salivary metabolic biomarkers and altered metabolic pathways in south American patients diagnosed with oral squamous cell carcinoma. Metabolites. 2021;11(10) doi: 10.3390/metabo11100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Niu X.L., Qu Y., Wu J., Zhu Y.Q., Sun W.J., et al. Autocrine production of interleukin-6 confers cisplatin and paclitaxel resistance in ovarian cancer cells. Cancer Lett. 2010;295(1):110–123. doi: 10.1016/j.canlet.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 52.Hsu C.P., Chen Y.L., Huang C.C., Chou C.C., Liu C.L., Hung C.H., et al. Anti-interleukin-6 receptor antibody inhibits the progression in human colon carcinoma cells. Eur J Clin Invest. 2011;41(3):277–284. doi: 10.1111/j.1365-2362.2010.02405.x. [DOI] [PubMed] [Google Scholar]
- 53.McLean K., Tan L., Bolland D.E., Coffman L.G., Peterson L.F., Talpaz M., et al. Leukemia inhibitory factor functions in parallel with interleukin-6 to promote ovarian cancer growth. Oncogene. 2019;38(9):1576–1584. doi: 10.1038/s41388-018-0523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delk N.A., Farach-Carson M.C. Interleukin-6: a bone marrow stromal cell paracrine signal that induces neuroendocrine differentiation and modulates autophagy in bone metastatic PCa cells. Autophagy. 2012;8(4):650–663. doi: 10.4161/auto.19226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carpenter R.L., Lo H.W. STAT3 target genes relevant to human cancers. Cancers. 2014;6(2):897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitburn J., Rao S.R., Morris E.V., Tabata S., Hirayama A., Soga T., et al. Metabolic profiling of prostate cancer in skeletal microenvironments identifies G6PD as a key mediator of growth and survival. Sci Adv. 2022;8(8) doi: 10.1126/sciadv.abf9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.