Abstract
Background and Objectives:
Plasma radiation is a widely used technique for sterilization or decontamination in various industries, as well as in some healthcare settings such as dentistry. The primary aim of this study was to assess the potential of plasma radiation to create a new population of Staphylococcus aureus cells with distinct characteristics that could lead to novel healthcare challenges.
Materials and Methods:
A homemade non-thermal plasma apparatus was applied and the effects of plasma treatment on S. aureus ATCC25923 was assessed. Plasma radiation was applied under controlled conditions to ensure that some bacterial cells remained viable. The treatment was repeated 10 times, with each round followed by a recovery phase to collect any surviving bacterial cells. To assess the potential changes in the bacterial population, we examined the antibiotic susceptibility pattern, micro-structural characteristics using scanning electron microscopy (SEM), and total protein profile using the matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) technique.
Results:
The experimental results revealed slight variations in the antibiotic susceptibility patterns of certain cell wall agents (imipenem, cephalothin, and cefepime), as well as in the MALDI-TOF spectra. However, no changes were observed in the SEM images.
Conclusion:
The insufficient application of non-thermal plasma in bacterial decontamination may lead to physiological changes that could enrich or select certain subpopulations of S. aureus.
Keywords: Plasma radiation, Staphylococcus aureus, Modification, Mass spectrometry
INTRODUCTION
Most of the food-borne bacterial diseases arise from pathogens such as Salmonella, S. aureus, Escherichia coli O157:H7, Listeria monocytogenes, and Shigella spp. (1, 2). These bacteria can grow on the surface of a vast variety of foods and fresh products. However, traditional post-harvest washing and disinfection processes are only able to reduce less than 2 Log of bacterial load of foods and fresh crops (1). Furthermore, the use of heat and chemical compounds to sterilize seeds, fruits, and foods is time-consuming and may have detrimental effects on the nutritional value and quality of foods, as well as producing toxic residues harmful to the environment (3). As a result, food producers and processors are seeking alternative approaches to eradicate bacterial loads while preserving the nutrition and quality of foods (4, 5).
Generally, plasma is defined as the 4th state of matter which is composed of a partially or completely ionized gas containing some atoms in exited or natural states, free electrons, photons, radicals, active species, and a variety of ions. On the other hand, plasma has a higher energy level than gas. There are two main classes of plasma: thermal and non-thermal (6, 7). Plasma is now among the most important physicochemical techniques of sterilization or disinfection of different materials (8–10) and has been reported widely for combating pathogenic microorganisms such as S. aureus (11, 12), Streptococcus spp. (13), Salmonella spp. and Escherichia coli (14), viruses (15), yeasts, and bacterial spores (16) as well as a newly emerging technique for the treatment of cancer in some cases (17).
Non-thermal technologies such as gamma radiation, ultraviolet treatment, and ultrasound have been used for decades as safe methods of food preservation treatments with minimum thermal side effects which ensure food quality parameters (18). None thermal plasma also has gained some attention for application in food industries (18) as well as clinical purposes such as eradication of bacterial biofilms (19), and wound healing routes (20).
The emergence of multiple drug-resistant (MDR) bacteria is a major problem that has been partly attributed to the extensive and improper use of antimicrobial agents. To address this issue, new technologies must be used with caution and subjected to rigorous evaluation before being widely adopted. It is important to ensure that these technologies are effective in reducing bacterial loads while minimizing the risk of promoting antimicrobial resistance and other unintended consequences. Therefore, it is critical to carefully evaluate the safety and efficacy of new technologies before introducing them into widespread use. By doing so, we can help to mitigate the risks associated with MDR bacteria and safeguard public health. Vancomycin-resistant strains of S. aureus are now considered the most important infectious agents for humans and have resulted from natural selection and enrichment of vancomycin-resistant enterococci during prolonged feeding of livestock with antibiotic supplements. Inactivation of bacteria by plasma radiation is achieved via reactive oxygen or nitrogen species which in high doses could disrupt cell walls and cell membrane integrity (21). However physical approaches such as plasma treatment which had no selective action are not expected to induce resistance (6).
Previous studies on the inactivation of S. aureus by non-thermal or thermal plasma have shown that, in some cases, a small number of bacterial colonies can develop from materials or surfaces that have been subjected to mild plasma treatment. This raises questions about how these bacterial cells are able to survive reactive molecule bombardment, whether they possess any special characteristics, or whether the inadequate treatment was a contributing factor.
Another important question is whether there are any bacterial cells that can resist repeated plasma application. If so, are these cells different from their ancestors in ways that could lead to the development of more dangerous bacterial strains? This is particularly concerning for aggressive human pathogens that could potentially develop new pathogenic routes.
It is important to investigate these questions further to fully understand the mechanisms underlying bacterial resistance to plasma treatment and the potential risks associated with such resistance. By identifying the factors that contribute to the development of resistant bacterial strains, we can better develop strategies to mitigate the risk of bacterial infections and protect public health.
The primary objective of this study was to evaluate the effects of consecutive non-thermal plasma treatments on S. aureus and to investigate whether natural selection plays a role in shaping the characteristics of the bacterial strain. To achieve this goal, we compared the phenotypic and molecular characteristics of S. aureus cells that had undergone multiple rounds of cold plasma treatment with those of the initial, untreated cells.
By assessing the changes in the bacterial population after repeated plasma treatment, we aimed to gain a better understanding of the mechanisms underlying bacterial resistance to plasma and the potential risks associated with repeated exposure. Our findings could have important implications for the development of effective strategies for controlling bacterial infections and preventing the emergence of more dangerous bacterial strains.
MATERIALS AND METHODS
Chemicals and instruments.
All media, mineral salts, and glutaraldehyde were purchased from Merck Millipore (Darmstadt, 109 Germany). Antibiotic powders (chloramphenicol (C3175), cefixime (CDS021590) and azithromycin (PZ0007)) and disks were from Sigma and HI Media companies, respectively.
Plasma apparatus.
The plasma treatment apparatus was a capacitive coupled Radiofrequency (RF) plasma system. The system consists of a stainless-steel chamber with a diameter of 30 cm and length of 35 cm and a 10 cm water-cooled power electrode which was described in detail previously (22).
Bacterial strain and growth condition.
S. aureus ATCC25923 was kept in Nutrient broth (NB) supplemented by 15% (v/v) glycerol (as cryo-protectant) at −70°C. A nutrient agar (NA) medium was used for the preparation of fresh cultures. All incubations were performed at 37°C. NB was applied to prepare active cells in mid of logarithmic phase of growth and also in all serial dilution preparation.
Primary plasma treatments.
All experiments were carried out by a fresh liquid culture of S. aureus strain. In brief, a 5 mL NB medium was inoculated by a few fresh colonies and incubated at 37°C until reaching the mid-logarithmic phase of growth, which previously had been determined. OD600 ~0.5 was considered as turbidity that was equal to the mid-logarithmic phase of growth (according to the standard protocol of colony count in parallel with OD 600 results which were recorded for 8 hrs. with 20 minutes’ intervals).
A volume of 100 μl of fresh bacterial culture was transferred onto a sterile empty petri dish and let dry in the laminar flow hood for about 30 minutes. Then, the Petri dish was placed on the powered electrode and the treatment chamber was evacuated to the base pressure of 8 mtorr by roots and rotary vacuum pumps. 20 sccm of oxygen gas with a purity of 99.999% was injected into the chamber through a mass flow controller. A 13.56 MHz RF generator transmitted the power through a matching network to the powered electrode (cathode). The samples were immersed in 30 W and 50 W of oxygen plasma for 0.5, 1.5, 3, and 4 minutes. For each experiment, a control was prepared without plasma treatment and kept at room temperature until the next step. The schematic of the system was shown in Hosseini et al. (22). All experiments were done in triplicate.
Plasma-treated cells were washed and serially diluted using NB. Then 100 μl of each dilution was cultured on NA plates. Plates were checked after 24 h incubation at 37°C and colony forming units (CFU) per each mL of treated samples were calculated based on colony counts and dilution factors.
Consecutive plasma treatment experiments.
This part of the study was performed by rescuing colonies of the first round of plasma treatment in 30 W and oxygen plasma for 3 minutes. After each run, treated cells were washed and transferred to NA plates and 850 μL of NB simultaneously and both media were incubated at 37°C for 8 and 18 hrs., respectively. A freezer stock was made by the addition of sterile glycerol to NB medium up to 15% (v/v) and kept at −20°C, and developed colonies on NA plates were subjected to a new run of plasma treatment. For this purpose, a suspension of bacterial cells was made in NB with turbidity equal to 0.5 McFarland standard tube (about 1–2×108 cfu/mL), and 100 μl of this suspension was dried on a new empty petri dish, before the next plasma treatment. These experiments were carried out 10 times and the glycerol stock of each run was kept at mentioned condition. Treated samples were recorded as T1 to T10 samples.
Antibiotic susceptibility pattern evaluation.
Susceptibility of the first untreated sample (Ut) and 10 consecutively treated cells were evaluated by 2 methods.
a. MIC.
Minimum inhibitory concentrations against chloramphenicol, cefixime and azithromycin were determined by the standard protocol of clinical and laboratory standards (CLSI) with some modifications via broth micro-dilution method (23). In brief, a stock solution of each antibiotic (50 mg/mL) was made in sterile D.W. and were kept at −20°C in aliquots until assay. A serially diluted antibiotic samples from 512 to 0.125 μg/mL were made in 96 well plates to final volume of 100 μL using Mueller Hinton broth (MHB). Bacterial suspensions were prepared from fresh culture of the bacterium (18–20 hrs. culture on NA) in sterile normal saline, adjusted to 0.5 McFarlan turbidity and were further diluted (1:100) with Mueller Hinton broth just before adding to each well. Each experiment was done in triplicate and MIC values were recorded after 20 h incubation at 37°C, as the lowest concentration of each antibiotic which could result in inhibition of visible growth of bacteria.
b. Disk diffusion method.
A standard protocol was carried out against a collection of antibiotics (ampicillin, amoxicillin, cefepime, imipenem, cephalothin, erythromycin, nitrofurantoin, novobiocin, amikacin, trimethoprim-sulfamethoxazole, and ciprofloxacin) according to CLSI method with some modification (23). Each experiment was done in triplicate. Inhibition zones were recorded after 20 hrs. incubation at 37°C. Incubation time was extended to 72 hrs. to detect any possible resistance in form of development of colonies in inhibition zones.
Scanning electron microscopy (SEM).
A study of any structural changes in the bacterial cells after plasma treatment was performed using SEM (24). In brief selected samples of treated cells (T1, T5, and T10) and Ut samples were cultured in NB until reaching OD600 of about 0.4. 1 mL of each sample was removed and centrifuged for 5 min at 4000 g at 4°C. The precipitated cells were washed twice with phosphate-buffered saline (PBS), pH 7.4, and the pellet was again re-suspended in 1 mL PBS. Then 10 μL of bacterial suspension was coated onto a glass slide (1 cm×1 cm). Samples were fixed with 2.5% (v/v) glutaraldehyde in PBS for 16 h at 4°C and dehydrated in water-alcohol solutions in increasing Ethanol concentrations (70%, 80%, 90%, and 100%). Samples were then fixed on an SEM support and gold-covered by cathode spraying. Observation of S. aureus morphology was performed using a field emission gun scanning electron microscope (SEM, 229 Hitachi, Tokyo, Japan) in Central laboratory of Shahid Beheshti University, Tehran, Iran. The voltage of the SEM apparatus used was 20.0 kV.
Evaluation of bacterial total proteins by MALDI-TOF.
The sample preparation was carried out for some samples (Un, T1, T5, and T10) according to the previously described method with some modifications (25). In brief biomass of 1 mL, overnight cultured NB was precipitated in 6000 g for 5 minutes at 4°C. Pellets were washed with cold PBS and were dissolved in 50 μL of an organic solution containing 50% acetonitrile and 30% formic acid in Mili-Q water. The biomass samples were mixed with the mentioned solution using a vortex apparatus several times. Clarified supernatant samples were obtained via centrifugation at 8000 g for 10 minutes at 4°C and mixed with an equal amount of matrix solution for MALDI-TOF analysis. 1 μL of sample/mixture solution was transferred onto a MALDI stainless settle plate and kept to dry at room temperature.
Mass spectra were obtained using a MALDI-TOF Mass Spectrometer (AB/Sciex model 4800, in MS Spectrometry Laboratory of MPDRI, Shahid Beheshti University, Tehran, Iran) which operates in linear mode, extracting positive ions with an accelerating voltage of 25 000 V and a delay time of 350 ns. The grid voltage is set to 95%. Spectra were taken in an m/z range of 2000–15000 Da. Every spectrum was the sum of at least 1000 accumulated laser shots obtained in ten different regions and randomly selected in the same sample spot.
RESULTS
Primary plasma treatments.
Viable bacterial cells were counted after each experiment as CFU/mL in samples which had been subjected to different conditions of plasma treatments (30 W and 50 W of oxygen plasma for 0.5, 1.5, 3, and 4 minutes). As shown in Table 1, while in some cases all bacterial cells were killed and no colonies were developed after re-culturing of treated samples, in the other cases; for example, with treatment by 30 W, there were some rescued cells. So, treatment by 30 W for 3 minutes was chosen for further studies.
Table 1.
Viable S. aureus cells after plasma treatment (cfu/mL)*
| Power of oxygen plasma | Duration of treatment (minute) | |||
|---|---|---|---|---|
|
| ||||
| 0.5 min | 1.5 | 3 | 4 | |
| 30 W | 1.2×107 | 1.9×104 | 1.1×104 | 1.2×101 |
| 50 W | 1.6×104 | 1.2×102 | 0 | 0 |
* Colony-forming unit/mL.
Consecutive plasma treatment experiments.
As described before, 10 rounds of plasma treatments were performed consecutively and for each run, rescued cells of previous treatment were subjected to the same condition (30 W for 3 minutes). After culturing all treated samples (even for the 10th one) some colonies were developed and kept as freezer stock. There were some morphological changes such as decreasing size of some colonies and in some cases a light color change. However, these alterations were not consistent with treatments, and especially in the case of color changes, there were some variations between different runs of plasma bombardment.
Antibiotic susceptibility pattern evaluation.
The susceptibility pattern of 11 samples (one Ut and 10 treated) was evaluated. As indicated in Table 2, In the case of MIC determination against chloramphenicol, azithromycin, and cefixime there were some minor changes in susceptibility to assessed antimicrobial agents, but these alterations didn’t move any of the bacterial samples to the resistant or even intermediate categories of susceptibility, and all samples remained in the sensitive range of MIC.
Table 2.
Susceptibility of untreated and survived S. aureus cells after consecutive treatment by plasma (broth micro-dilution assays)
| Antibiotics | MIC* values (μg/mL) | Sensitivity† (μg/mL) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Un | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | T10 | ||
| Chloramphenicol | 2 | 2 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | ≤8 |
| Azithromycin | 0.25 | 0.5 | 0.25 | 0.25 | 0.25 | 0.125 | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | ≤2 |
| Cefixime | 0.25 | 0.5 | 0.25 | 0.25 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | ≤1 |
* Minimum inhibitory concentration.
† According to the CLSI guideline (Jorgensen & Turnidge, 2007)
Table 3 shows the inhibitory zone diameter around tested antibiotics for all samples after 20 h incubation at 37°C. As demonstrated in this Table and according to the CLSI criteria for resistant phenotype which have been specified for each antibiotic, resistance has been developed against none of the assessed antibiotics even after the 10th plasma treatment. Though, there were some fluctuations in inhibition zone diameter around some anti-cell wall agents such as imipenem. Extension of incubation time to 72 h resulted in the development of some colonies in inhibition zones around some antibiotics such as cephalothin and imipenem in all samples; treated and untreated. However, in the case of cefepime, there were some colonies in inhibition zones for T3, T4, T8, and T9 but not for the untreated sample.
Table 3.
Susceptibility pattern of untreated and survived S. aureus cells after consecutive treatment by plasma (disk diffusion assays)
| Antibiotics* | Inhibition zone diameter† (mm) | Interpretation criteria‡ (mm) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| Un | T1 | T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | T10 | S§ | I¶ | R# | |
| Ampicillin | 45 | 44 | 45 | 44 | 44 | 44 | 44 | 44 | 43 | 44 | 43 | ≥29 | - | ≤28 |
| Amoxicillin | 46 | 45 | 45 | 44 | 45 | 45 | 44 | 45 | 45 | 44 | 44 | ≥29 | - | ≤28 |
| Cefepime | 24 | 23 | 23 | 24 | 24 | 24 | 24 | 23 | 23 | 23 | 23 | ≥18 | 15–17 | ≤14 |
| Imipenem | 49 | 48 | 49 | 48 | 48 | 49 | 48 | 48 | 48 | 45 | 45 | ≥16 | 14–15 | ≤13 |
| Cephalothin | 38 | 39 | 38 | 38 | 39 | 38 | 39 | 39 | 37 | 38 | 38 | ≥18 | 15–17 | ≤14 |
| Erythromycin | 26 | 26 | 26 | 25 | 26 | 23 | 25 | 26 | 25 | 24 | 25 | ≥23 | 14–22 | ≤13 |
| Amikacin | 25 | 25 | 23 | 25 | 23 | 23 | 23 | 23 | 24 | 24 | 24 | ≥17 | 15–16 | ≤14 |
| Novobiocin†† | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | - | - | ≤16 |
| SXT‡‡ | 31 | 32 | 30 | 31 | 30 | 32 | 31 | 30 | 31 | 30 | 30 | ≥16 | 11–15 | ≤10 |
| Ciprofloxacin | 33 | 33 | 33 | 31 | 33 | 33 | 33 | 33 | 31 | 32 | 32 | ≥21 | 16–20 | ≤15 |
| Nitrofurantoin | 25 | 25 | 24 | 26 | 25 | 25 | 25 | 26 | 25 | 24 | 25 | ≥17 | 15–16 | ≤14 |
* Disk contents were as follow: 10 μg ampicillin, 10 μg Amoxicillin, 30 μg Cefepime, 10 μg imipenem, 30 μg Cephalothin, 15 μg erythromycin, 30 μg amikacin, 5 μg Novobiocin, 25 μg SXT, 5 μg ciprofloxacin and 300 μg Nitrofurantoin.
† All inhibition zones are for 6 mm diameter disks except that of Imipenem (10 mm).
‡ Adopted from CLSI guideline (Jorgensen & Turnidge, 2007).
§ Sensitive,
¶ Intermediate,
# Resistant
†† Novobiocin resistance is intrinsic to some species such as S. saprophyticus but is uncommon in the other clinically important species such as S. aureus.
‡‡ Trimethoprim-sulfamethoxazole
Scanning electron microscopy.
Analysis of bacterial cell morphology via SEM showed no apparent differences (regarding shape and dimensions) between untreated and 3 different treated bacterial cells (T1, T5, and T10). As shown in Fig. 1 there were no considerable differences between the analyzed samples.
Fig. 1.
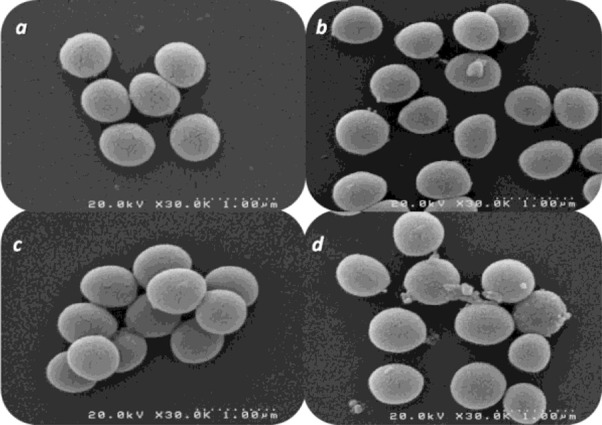
SEM analysis of untreated and plasma-treated samples. 1a: untreated S. aureus, 1b, 1c, and 1d: S. aureus cells after 1, 5, and 10 consecutive treatments by 30 W cold plasma, respectively. As apparent, there are no considerable differences regarding the shape and dimensions of treated vice none treated bacterial cells.
Evaluation of bacterial total proteins by MALDI-TOF.
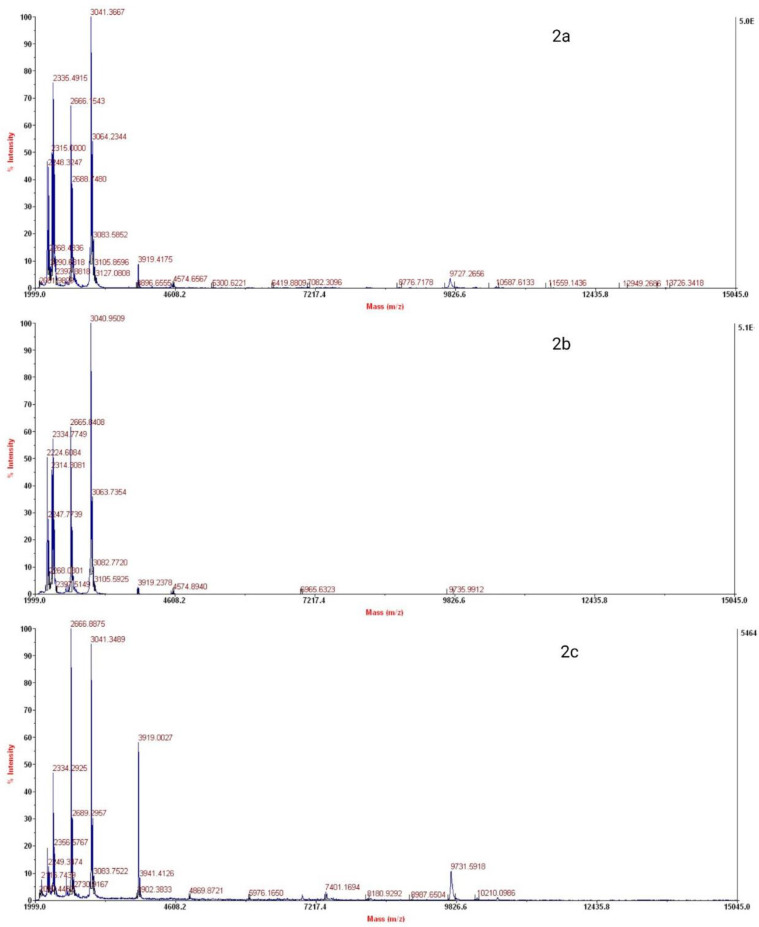
MALDI-TOF spectra of untreated, T1, T5, and T10 samples have been demonstrated in Fig. 2. While most of the major peaks are unaffected, there is some variation in the intensity of the others (for example those variations in mass (m/z) 3919.0027, 4574.6567 and 9826.5918).
Fig. 2.
MALDI-TOF spectra of untreated and treated samples of S. aureus biomass. 2a: Untreated S. aureus proteins, 2b: S. aureus proteins after 5th plasma treatment, and 2c: S. aureus proteins after 10th plasma treatment.
DISCUSSION
There are huge reports on the evaluation of antimicrobial properties of different types of plasma and this technique is now considered an applicable method for a variety of materials as well as surfaces. The homemade plasma apparatus used in this study could show antibacterial effects on S. aureus.
In the present study, we investigated the possible adaptation as well as physiological changes in plasma-treated bacterium using a non-thermal plasma apparatus and a Gram-positive bacterial model which is more resistant to plasma than Gram-negatives.
In macrostructure evaluation using SEM no minor changes in cellular morphology were seen also according to the results of the antibiogram and MIC evaluation, there were slight but not considerable changes in susceptibility patterns of treated vice untreated S. aureus. However, in the case of anti-cell wall agents such as imipenem, cephalothin, and cefepime it should be mentioned that further studies are needed. The bacterial cell wall is the first barrier against plasma bombardment and any selection in bacterial cells which could resist this danger may result in the richness of these types of bacterial sub-populations. S. aureus cell wall is a rigid structure, mainly composed of peptidoglycan and it is suggested that S. aureus cell damage caused by plasma is also the result of alteration or destruction of genetic materials as well as proteins (21).
However, it has been previously suggested that two specific mechanisms of bacterial inactivation by plasma are reaction with the cell envelope and destruction of intracellular components for Gram-negative and Gram-positive bacteria, respectively. E. coli bacterium is inactivated principally by cell leakage and low-level DNA damage, and S. aureus cells are essentially inactivated by intracellular damage via high levels of intracellular reactive oxygen species and slight envelope damage (16).
There are a few reported studies on the possible effects of plasma treatments on the physiology of bacterial cells. Winter et al. have reported some modifications in stress responses of Bacillus subtilis after treatment by a non-thermal plasma (26). Using proteomics and transcriptomic analyses they could demonstrate some changes in B. subtilis gene expression. They also could demonstrate up or down-regulation of some stress protein markers in this well-known stress-responsive bacterium. However, in the other reported study, Matthes et al. showed that repeated treatment of biofilm-embedded S. aureus did not result in habituation or resistance to a variety of plasma treatments (27). In another study which has been conducted to investigate the effects of preliminary stress on the susceptibility of S. aureus to cold plasma treatment, it was revealed that while osmosis, oxidation, heat, or cold stress could make this bacterium more susceptible, preliminary exposure to acid stress (for 24 h) surprisingly reduce susceptibility to cold plasma treatments (28).
Accordingly, in the present study total protein profile of untreated and some plasma-treated S. aureus (untreated, and some samples of cultured cells after treatments: T1 and T10) were assessed via the MALDI-TOF apparatus. Regarding the results of this preliminary study, some minor changes have been shown in spectra obtained from untreated cells and developed cells after treatments. These changes should be further investigated using this method and other techniques of mass analysis to reveal the exact identity of affected protein or peptide molecules.
CONCLUSION
It could be concluded that inadequate non-thermal plasma application in bacterial decontamination could result in enrichment or natural selection of some S. aureus subpopulations whose characteristics and abilities are slightly different from untreated bacterial cells. In consecutive applications, the situation could become worsen because of the emergence and distribution of plasma-resistant strains which may become new human pathogens with more dangerous abilities.
ACKNOWLEDGEMENTS
The authors would like to appreciate the kind collaboration of Mrs. Farzaneh Zandi and Dr. Ilnaz Soleimani-Mashhadi for some helps in bacteriologic and analytical chemistry experiments. The authors also would like to express their special thanks to the deputy of research of Shahid Beheshti University for providing some support for the work.
REFERENCES
- 1.Niemira BA. Cold plasma decontamination of foods. Annu Rev Food Sci Technol 2012; 3: 125–142. [DOI] [PubMed] [Google Scholar]
- 2.Sivapalasingam S, Friedman CR, Cohen L, Tauxe RV. Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J Food Prot 2004; 67: 2342–2353. [DOI] [PubMed] [Google Scholar]
- 3.Thirumdas R, Sarangapani C, Annapure US. Cold Plasma: A novel non-thermal technology forf ood processing. Food Biophys 2015; 10: 1–11. [Google Scholar]
- 4.Bourke P, Ziuzina D, Boehm D, Cullen PJ, Keener K. The potential of cold plasma for safe and sustainable food production. Trends Biotechnol 2018; 36: 615–626. [DOI] [PubMed] [Google Scholar]
- 5.Mir SA, Shah MA, Mir MM. Understanding the role of plasma technology in food industry. Food Bioprocess Technol 2016; 9: 734–750. [Google Scholar]
- 6.Misra NN, Tiwari BK, Raghavarao KSMS, Cullen PJ. Nonthermal plasma inactivation of foodborne pathogens. Food Eng Rev 2011; 3: 159–170. [Google Scholar]
- 7.Gomez E, Rani DA, Cheeseman CR, Deegan D, Wise M, Boccaccini AR. Thermal plasma technology for the treatment of wastes: A critical review. J Hazard Mater 2009; 161: 614–626. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, Cao Y, Yang P, Xiong Q, Xiong Z, Xian Y, et al. An RC plasma device for sterilization of root canal of teeth. IEEE Trans Plasma Sci 2009; 37: 668–673. [Google Scholar]
- 9.Zhang X, Huang J, Liu X, Peng L, Guo L, Lv G, et al. Treatment of Streptococcus mutans bacteria by a plasma needle. J Appl Phys 2009; 105: 063302. [Google Scholar]
- 10.Pasquali F, Stratakos AC, Koidis A, Berardinelli A, Cevoli C, Ragni L, et al. Atmospheric cold plasma process for vegetable leaf decontamination: A feasibility study on radicchio (red chicory, Cichorium intybus L.). Food Control 2016; 60: 552–559. [Google Scholar]
- 11.Burts ML, Alexeff I, Meek ET, McCullers JA. Use of atmospheric non-thermal plasma as a disinfectant for objects contaminated with methicillin-resistant Staphylococcus aureus. Am J Infect Control 2009; 37: 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ekem N, Akan T, Akgun Y, Kiremitci A, Pat S, Musa G. Sterilization of Staphylococcus aureus by atmospheric pressure pulsed plasma. Surf Coat Technol 2006; 201: 993–997. [Google Scholar]
- 13.Park SR, Lee HW, Hong JW, Lee HJ, Kim JY, Choi BB, et al. Enhancement of the killing effect of low-temperature plasma on Streptococcus mutans by combined treatment with gold nanoparticles. J Nanobiotechnology 2014; 12: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Critzer FJ, Kelly-Wintenberg K, South SL, Golden DA. Atmospheric plasma inactivation of foodborne pathogens on fresh produce surfaces. J Food Prot 2007; 70: 2290–2296. [DOI] [PubMed] [Google Scholar]
- 15.Terrier O, Essere B, Yver M, Barthélémy M, Bouscambert-Duchamp M, Kurtz P, et al. Cold oxygen plasma technology efficiency against different airborne respiratory viruses. J Clin Virol 2009; 45: 119–124. [DOI] [PubMed] [Google Scholar]
- 16.Lee K, Paek K-H, Ju W-T, Lee Y. Sterilization of bacteria, yeast, and bacterial endospores by atmospheric-pressure cold plasma using Helium and Oxygen. J Microbiol 2006; 44: 269–275. [PubMed] [Google Scholar]
- 17.Barekzi N, Laroussi M. Dose-dependent killing of leukemia cells by low-temperature plasma. J Phys D Appl Phys 2012; 45: 422002. [Google Scholar]
- 18.Ma R, Wang G, Tian Y, Wang K, Zhang J, Fang J. Non-thermal plasma-activated water inactivation of foodborne pathogen on fresh produce. J Hazard Mater 2015; 300: 643–651. [DOI] [PubMed] [Google Scholar]
- 19.Kovalóvá Z, Zahoran M, Zahoranová A, Machala Z. Streptococci biofilm decontamination on teeth by low-temperature air plasma of dc corona discharges. J Phys D Appl Phys 2014; 47: 224014. [Google Scholar]
- 20.Mohd Nasir N, Lee BK, Yap SS, Thong KL, Yap SL. Cold plasma inactivation of chronic wound bacteria. Arch Biochem Biophys 2016; 605: 76–85. [DOI] [PubMed] [Google Scholar]
- 21.Han L, Patil S, Boehm D, Milosavljević V, Cullen PJ, Bourke P. Mechanisms of inactivation by high-voltage atmospheric cold plasma differ for Escherichia coli and Staphylococcus aureus. Appl Environ Microbiol 2015; 82: 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosseini SI, Mohsenimehr S, Hadian J, Ghorbanpour M, Shokri B. Physico-chemical induced modification of seed germination and early development in artichoke (Cynara scolymus L.) using low energy plasma technology. Phys Plasmas 2018; 25: 013525. [Google Scholar]
- 23.Sobhani M, Abbas-Mohammadi M, Ebrahimi SN, Aliahmadi A. Tracking leading anti-Candida compounds in plant samples; Plumbago europaea. Iran J Microbiol 2018; 10: 187–193. [PMC free article] [PubMed] [Google Scholar]
- 24.Moghimi R, Aliahmadi A, McClements DJ, Rafati H. Investigations of the effectiveness of nanoemulsions from sage oil as antibacterial agents on some food borne pathogens. LWT-Food Sci Technol 2016; 71: 69–76. [Google Scholar]
- 25.Böhme K, Fernández-No IC, Barros-Velázquez J, Gallardo JM, Cañas B, Calo-Mata P. SpectraBank: An open access tool for rapid microbial identification by MALDI-TOF MS fingerprinting. Electrophoresis 2012; 33: 2138–2142. [DOI] [PubMed] [Google Scholar]
- 26.Winter T, Winter J, Polak M, Kusch K, Mäder U, Sietmann R, et al. Characterization of the global impact of low temperature gas plasma on vegetative microorganisms. Proteomics 2011; 11: 3518–3530. [DOI] [PubMed] [Google Scholar]
- 27.Matthes R, Assadian O, Kramer A. Repeated applications of cold atmospheric pressure plasma does not induce resistance in Staphylococcus aureus embedded in biofilms. GMS Hyg Infect Control 2014; 9: Doc17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao X, Li J, Suo Y, Ahn J, Liu D, Chen S, et al. Effect of preliminary stresses on the resistance of Escherichia coli and Staphylococcus aureus toward non-thermal plasma (NTP) challenge. Food Res Int 2018; 105: 178–183. [DOI] [PubMed] [Google Scholar]