Abstract
The effect of DNA supercoiling on gene expression is dependent not only on specific genes but also on the sequence context of the genes. This position-dependent supercoiling effect on gene activation is best illustrated in the study of the suppression of the leu-500 mutation of the leuABCD operon in a Salmonella typhimurium topA mutant. In this communication, we report a novel promoter relay mechanism whereby several genes are sequentially expressed in a position-dependent manner: the ilvIH promoter (pilvIH) activates a cryptic leuO promoter (pleuO) located between the two divergently arrayed ilvIH and leu-500 promoters. Both the cis-acting pleuO activity and the trans-acting LeuO protein are necessary for subsequent activation of the leu-500 promoter (pleu-500). Furthermore, pleuO can be functionally replaced with the inducible tac promoter (ptac) for leu-500 activation, suggesting that transcription-driven DNA supercoiling underlies the relay mechanism. This is the first example of several related genes communicating via a promoter relay mechanism for their coordinated expression.
Numerous genetic and biochemical studies have documented that negative supercoiling affects expression of many prokaryotic and eukaryotic genes 14, 30, 37; for reviews, see references 10, 27, 34, and 39). However, the effect of negative supercoiling on gene expression is dependent not only on specific genes but also on the locations and therefore the sequence context of the genes (9, 16, 21). This position-dependent supercoiling effect on gene expression indicates that local DNA supercoiling is important in gene expression regulation.
The suppression of the leu-500 mutation of the leuABCD operon in a Salmonella typhimurium topA mutant is one of the best examples of such a position-dependent supercoiling effect. The leu-500 promoter (pleu-500) is activated in topA mutants (8, 20, 22, 37). However, leu-500 activation correlates with ΔtopA but not with reduced levels of negative supercoiling (28, 29). Strikingly, activation of pleu-500 is lost when the promoter is removed from its original chromosomal location (29). These results suggest that local supercoiling may be responsible for leu-500 activation. On the basis of the twin supercoiled-domain model of transcription (19), Lilley and Higgins postulated that transcription-induced DNA supercoiling may be the source of the local DNA supercoiling for leu-500 activation (18). Since then, a number of studies have experimentally demonstrated the involvement of one or more adjacent transcription activities in activating pleu-500 on a plasmid (3–5, 36). While several parameters, including a topological domain flanked by two divergent transcription units, an adjacent transcription unit encoding a membrane anchorage peptide, the promoter strength, and the size of the adjacent transcription unit, may affect the supercoiling effect, studies have indicated that the transcription-driven supercoiling effect on leu-500 activation is usually limited to a short distance (<250 bp) (4, 36). However, a long-distance effect was demonstrated in a recent study (35). The expression of a transcription unit located 3,000 bp downstream of pleu-500 resulted in leu-500 activation.
To address whether an adjacent transcription activity is also involved in leu-500 activation in the context of the chromosome, we have demonstrated the long-range interaction between the ilvIH and leu-500 promoters. We have found that the plasmid-borne leu-500 promoter cannot be activated in a topA mutant unless a 1.9-kb upstream sequence of pleu-500 is present (41). This 1.9-kb sequence contains pilvIH, whose activity is crucial for leu-500 activation. Replacement of pilvIH with the inducible lac promoter (plac) leads to inducible activation of leu-500, indicating that the promoter activity, but not the specific promoter, is important for leu-500 activation (41). While promoter activity is important for leu-500 activation, the associated transcript from pilvIH is not. We have explained this long-range promoter-promoter interaction in terms of the transcription-driven supercoiling effect (41). However, the large distance between the two interacting promoters together with the requirement for the 1.9-kb sequence suggests that a more complicated supercoiling transmission mechanism is involved. In this study, we have found a novel relay mechanism which is crucial for transmitting the supercoiling effect from pilvIH to pleu-500 via an intermediary promoter, pleuO, located between the two interacting promoters in S. typhimurium topA mutant CH582. Activation of the cryptic pleuO is strictly dependent on pilvIH activity. Expression of the leuO gene is required for subsequent leu-500 activation. pleuO can be functionally replaced with the inducible ptac, provided that the LeuO protein is supplied in trans. This long-range promoter-promoter interaction via another promoter characterizes a promoter relay mechanism for gene regulation.
MATERIALS AND METHODS
Plasmid constructs.
pWU804 and pWU805 have been previously described (41). pWU807 is identical to pWU804 except that 2-bp mutations were introduced in the −10 region of pilvIH by a double-stranded site-directed mutagenesis procedure (7). The mutagenic primer is 5′-CGGTTTGACGGACAGCTGAACCGCTCGC-3′ (mutations located at positions -8 and -11 of pilvIH are underlined). pWU804M is the same as pWU804 except that pleuO was mutated by the site-directed mutagenesis procedure. The mutagenic primer (5′-GTTTAAATTACGCAAGCTCTAGAACCATAACTATG-3′) introduced T, C, and G to replace A, A, and T at positions -7, -8, and -11 of pleuO, respectively, and generated a unique XbaI site on the plasmid. pWU804T and pWU804TR were constructed by insertion of ptac into the unique XbaI site of pWU804M. ptac was generated by annealing two complementary oligonucleotides: 5′-CTAGCTGTTGACAATTAATCATCGGCTCGTATAATGTGTGGAATTGTGAGCGGATAACAATTTCACACA-3′ and 5′-CTAGTGTGTGAAATTGTTATCCGCTCACAATTCCACACATTATACGAGCCGATGATTAATTGTCAACAG-3′ (where underlined sites show the −35 and −10 regions and the lac operator, respectively, from left to right; modified from reference 6). The resulting plasmid with ptac oriented toward the leuO gene was pWU804T, with the opposite orientation being pWU804TR. pWU802T was obtained by deletion of the NsiI-BamHI fragment including the leuO coding region and the downstream pilvIH from pWU804T. pWU804H was constructed by introducing 2-bp mutations in pWU804 by the site-directed mutagenesis procedure. The mutagenic primer (5′-GTGTGGGCGGCCGGCGTAATATTCTG-3′) replaced GC with CG, which resulted in the replacement of the arginine residue with a proline at position 3 of the LeuO protein helix-turn-helix motif (12). Theoretically, this point mutation severely distorts the first helical structure of the DNA-binding motif since proline cannot form a hydrogen bond from its main-chain nitrogen. The coding region of lacZ, containing its own ShineDalgarno sequence and ATG translation start codon obtained from pSVβ (CLONTECH Laboratories, Inc.), was transcriptionally fused with ptac in pWU802T to generate pWU802TLZ. Cells harboring the fusion plasmid showed dark blue colonies on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction, confirming that ptac activity expresses the downstream lacZ coding sequence. pWU904 and pWU907 are identical to pWU804 and pWU807, respectively, except that pleu-500 was replaced by the wild-type leu promoter (pleu) in pWU904 and pWU907.
The expression vector pEV101 is a derivative of pSO1000 (24) carrying a pACYC origin of replication (ori) so that it can coexist with a ColE1 ori-based testing plasmid in a bacterium. pSO1000 also contains the Iq promoter-controlled lacI gene for expression of the lac repressor. The 1,550-bp BclI-BamHI fragment containing the ptac-controlled leuO coding region was isolated from pWU804T and subcloned into the compatible BglII site of pSO1000 to yield pEV101. All plasmid constructs were verified by DNA sequencing.
Bacterial strains.
CH582, a ΔtopA2726 leu-500 ara9 derivative of S. typhimurium LT2, was used in all experiments. Cells were grown aerobically at 32°C in synthetic SSA medium without supplemented leucine (2). Plasmids were transformed into bacteria by electroporation. Since S. typhimurium does not contain the regulatory system of the lac operon, pSO1000 or pEV101 is used to provide the lac repressor exogenously. IPTG was added to a final concentration of 1 mM 1 h prior to cell harvest, as required in some experiments.
RNA isolation.
RNA was isolated from freshly inoculated cultures in log-phase growth (optical density at 650 nm of 0.8) in SSA medium lacking leucine. RNA concentrations were measured by absorbance at 260 nm. A 100-μg amount of total RNA was used in each primer extension reaction.
Primer extension.
Primer extensions were carried out as previously described (36). Several primers were designed to search for possible transcription initiations in the 1.9-kb intervening sequence. These primers consist of sequences of chromosome context and therefore detect transcription activities arising from both the plasmid and the chromosome. However, due to the high copy number of the plasmid, transcription activity from primarily the plasmid-borne 1.9-kb region was detected. This is evidenced by the fact that leuO transcription activity was not detectable in RNAs isolated from pWU805-harboring CH582 (Fig. 1B, lanes 1 and 4). Among these primers, two primers, O1 and O2, were located close enough to allow detection of leuO transcription initiation. O1 (31-mer, 5′-GCAGAAATAATTCCTGAAAATATGATTTACC-3′) was used only in the experiment whose results are shown in Fig. 1. O2 (28-mer, 5′-CGGAAAACATAAAGACGCTGACAGAGAC-3′) was closer to the leuO transcription start site and was used in all experiments. Transcription activities of the ilvIH, leu-500, and bla promoters were detected by use of a pBR322 EcoRI clockwise primer, a pBR322 HindIII clockwise primer, and a synthetic DNA oligomer, respectively (41). These three primers hybridize with the vector sequence and detect only the transcription activities arising from the plasmid-borne promoters. The bla promoter activity was included in all primer extension experiments as an internal control. The bla transcript control of the primer extension shown in Fig. 6B was done in a separate reaction using the same RNA sample. When wild-type leu promoter activity was tested, the high activity of the leu promoter often caused a background in the original position of the bla promoter in primer extension. To avoid this problem, another primer (5′-CTGATCTTCAGCATCTTTTACTTTCACC-3′) was chosen to detect the bla promoter activity in Fig. 6A. All primers were end labeled with [γ-32P]ATP by using T4 polynucleotide kinase before they were added to the reaction mixture.
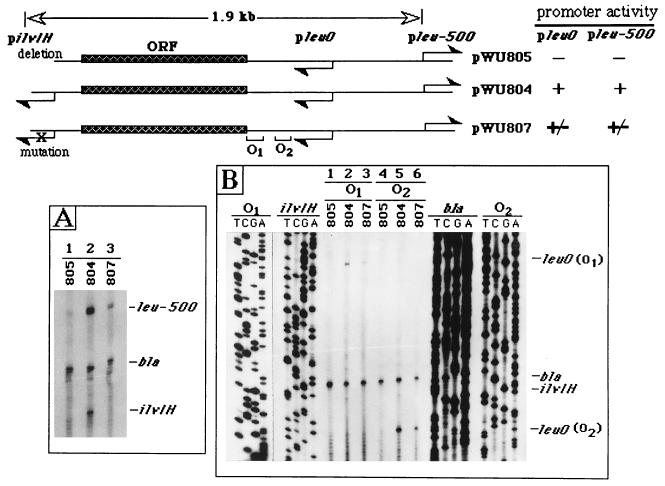
FIG. 1.
pilvIH activity-dependent transcription initiation in the 1.9-kb intervening sequence. Only the relevant regions in pWU804, pWU805, and pWU807 are illustrated. The vector, pWU800, was previously described (41) and is not shown here. The orientations of promoters are indicated (arrows). pilvIH was deleted in pWU805 and was mutated (X) in pWU807; otherwise, these three plasmids are identical. O1 and O2, the two primers for detection of pleuO activity. The primer extension results are summarized at the top right (−, no activity; +, activity; +/−, weak activity). (A) RNAs isolated from CH582 harboring pWU804, pWU805, or pWU807 were used in the primer extension analysis. A mixture of the three primers was used to detect transcripts from pleu-500, pbla, and pilvIH simultaneously. The initiation sites of leu-500, bla, and ilvIH transcripts are indicated on the right. (B) Activities of pleuO, pilvIH, and pbla were analyzed with the same RNAs as for panel A. leuO promoter activity was detected with either the O1 or the O2 primer [leuO(O1) (lanes 1 to 3) or leuO(O2) (lanes 4 to 6)]. The sequence ladders prepared with primers O1 and O2 were included to indicate the start site of the leuO transcript.
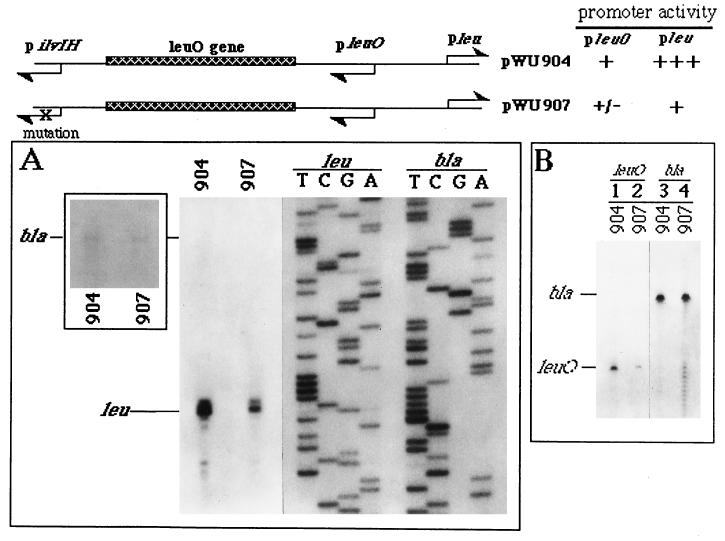
FIG. 6.
The wild-type promoter of the leuABCD operon is regulated by the promoter relay mechanism. pWU904 and pWU907 contain pleu instead of pleu-500; otherwise they are identical to pWU804 and pWU807, respectively. (A) Transcripts initiated from pleu and pbla were detected by primer extension. A longer-exposure autoradiograph is included on the left to show bla transcripts as the internal controls, which are too light to be visualized in the original exposure. (B) Transcripts initiated from pleuO of pWU904 and pWU907 (lanes 1 and 2, respectively) and pbla of pWU904 and pWU907 (lanes 3 and 4, respectively) were detected in separate primer extension reactions. The primer extension results are summarized at the top right (+, activity; +++, strong activity; +/−, weak activity).
RESULTS AND DISCUSSION
Activation of the cryptic pleuO by pilvIH activity.
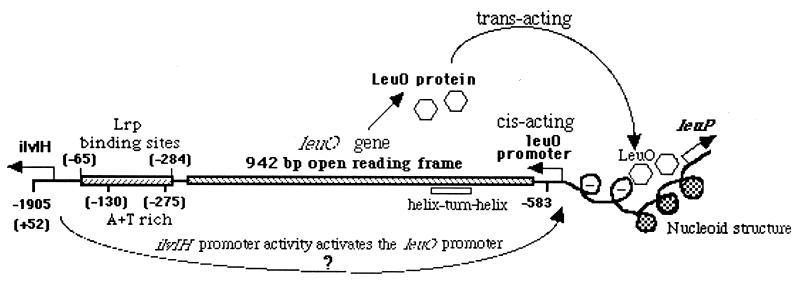
We have shown previously that the 1.9-kb upstream sequence of pleu-500 is crucial for leu-500 activation (41). So far, we have identified pilvIH, located at the pleu-500-distal end of the 1.9-kb sequence, to be necessary for leu-500 activation. The rest of the sequence is also essential for the long-range promoter-promoter interaction (41). Within this region, a 942-bp open reading frame (ORF), the putative leuO gene, has been identified (11). However, the transcript associated with this ORF has not been detected (11, 12). To search for a possible transcript from this ORF, primer extensions were performed with a series of oligomers along the 1.9-kb sequence. Only one transcription initiation site, which was located upstream of the ORF and presumably was associated with a promoter, the putative pleuO, was detected (Fig. 1B). The two primers used to detect the transcript by primer extension were located downstream from the putative pleuO, as shown in Fig. 1. The two transcripts detected by the two primers [Fig. 1B, leuO(O1) and leuO(O2)] originated from the same initiation site specified by the putative pleuO (sequence ladders of O1 and O2 in Fig. 1B). Strikingly, leuO transcription activity was strictly dependent on the functional pilvIH, since deletion of pilvIH abolished pleuO activity (Fig. 1B, compare lanes 1 and 2 or lanes 4 and 5). The 2-bp mutations in the −10 region of pilvIH which almost completely eliminated the promoter activity also significantly decreased pleuO activity (Fig. 1B, compare lanes 2 and 3 or lanes 5 and 6). As expected, activation of pleu-500 was also dependent on the functional pilvIH (Fig. 1A, compare lanes 1 to 3). The pilvIH activity-dependent activation of both pleuO and pleu-500 suggested a possible involvement of the leuO gene in the long-range interaction between pilvIH and pleu-500.
Role of expression of the leuO gene in mediating the long-range promoter-promoter interaction.
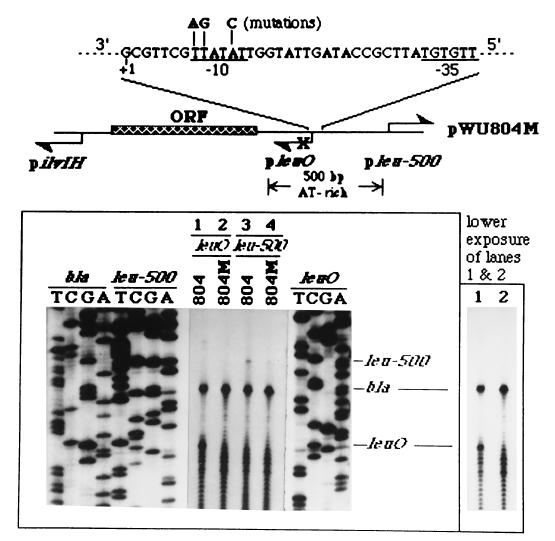
On the basis of computer analysis of the entire 1.9-kb sequence, we identified one potential Pribnow box (−10) sequence (deviating by 1 bp from the consensus −10 sequence) and −35 sequence (deviating by 3 bp from the consensus −35 sequence) located at the expected positions upstream of the leuO transcription initiation (+1) site. The distance between the −10 and −35 sequences is 18 bp, which is characteristic of the 16- to 18-bp spacer of a typical prokaryotic promoter recognized by RNA polymerase containing the ς70 subunit (15, 33). In order to test whether this promoter sequence was responsible for ilvIH-dependent leuO transcription initiation, we introduced a 3-bp mutation in the −10 region, using site-directed mutagenesis. This mutation indeed abolished leuO transcription initiation (Fig. 2, compare lanes 1 and 2). Hence, this promoter sequence is responsible for ilvIH activity-dependent leuO transcription initiation. Using a lacZ reporter fused with the downstream ORF, we have also demonstrated that this promoter sequence is responsible for the expression of the downstream ORF, the putative leuO gene (data not shown). Northern analysis confirmed that the leuO gene was active in an S. typhimurium ΔtopA strain but was normally silent in a topA+ strain (8a). However, the silent leuO gene was activated when ilvIH transcription activity was turned on due to the low growth rate of cells in nutrient-limited conditions, such as in minimal synthetic medium, or in stationary phase in rich medium (8a). It seems that leuO is a functional gene. Interestingly, the 3-bp mutation in pleuO also abolished leu-500 activation (Fig. 2, compare lanes 3 and 4), suggesting that leuO gene expression is crucial in mediating the long-range interaction between pilvIH and pleu-500.
FIG. 2.
Expression of the leuO gene in mediating the long-range promoter-promoter interaction. The nucleotide sequence of the leuO promoter, which is located in the 500-bp evolutionarily conserved AT-rich sequence between pleu-500 and the ORF (the coding region of leuO), is illustrated. The −35 and −10 sequences of the promoter (underlined), the transcription start site (+1), and a 3-bp mutation which inactivates pleuO in pWU804M are indicated. The pleuO activities from RNAs obtained from pWU804- or pWU804M-harboring CH582 were detected by using O2 primer (lanes 1 and 2, respectively). The leuO band migrates near the bottom of the primer extension electrophoresis gel, where several nonspecific bands are often located. For better resolution of the transcription initiation of leuO, a lower exposure of lanes 1 and 2 is included on the right. The pleu-500 activity in pWU804- or pWU804M-harboring CH582 is shown in lanes 3 and 4, respectively.
Effects of leuO gene expression on leu-500 activation.
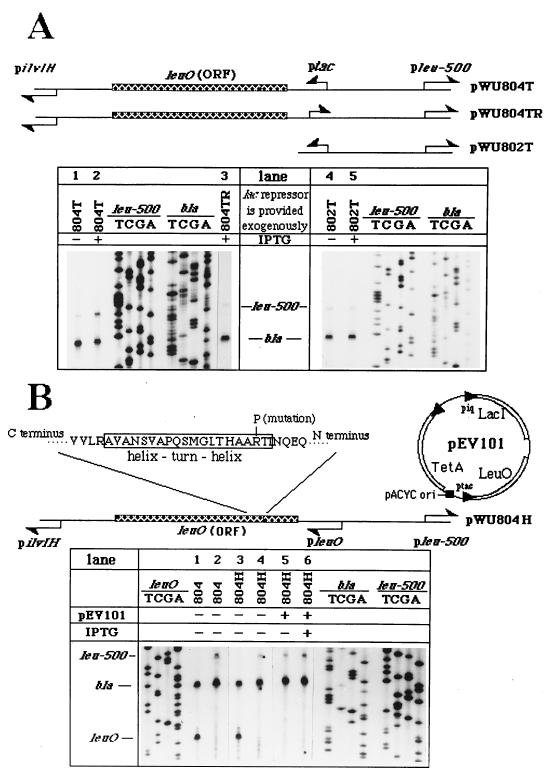
The finding that pilvIH activity-dependent leuO transcription, which occurs divergently 410 bp away from pleu-500, was required for mediation of the long-range interaction led us to consider two possible effects of leuO gene expression on leu-500 activation. One was that the translation product of leuO plays a trans-acting role in activating pleu-500, and the second was that the transcriptional process itself is important in activating the upstream pleu-500, perhaps via a mechanism of transcription-driven supercoiling as in the short-range promoter-promoter interaction (36). To examine these two possibilities, we replaced pleuO of pWU804 with an inducible ptac, which resulted in pWU804T. With ptac in place, leu-500 activation was shown to depend on IPTG induction (Fig. 3A, compare lanes 1 and 2). The following experimental results suggested that the leuO gene product (LeuO) is required to provide a trans-acting function for leu-500 activation. (i) Deletion of the leuO coding sequence including the rest of the downstream sequence from pWU804T eliminated IPTG-induced leu-500 activation, suggesting that the promoter activity alone was insufficient to activate pleu-500 at this 410-bp distance (Fig. 3A, compare lanes 4 and 5). (ii) Inversion of ptac also eliminated leu-500 activation, consistent with the importance of transcription of leuO (Fig. 3A, lane 3). (iii) A point mutation (R to P at position 41 of the deduced LeuO peptide) in the helix-turn-helix motif of LeuO (12), which presumably impaired LeuO binding to DNA, also significantly reduced leu-500 activation (Fig. 3B, compare lanes 2 and 4), while pleuO activity remained unchanged (Fig. 3B, compare lanes 1 and 3). This reduced leu-500 activation in pWU804H was restored when the wild-type LeuO protein was provided in trans upon IPTG induction (Fig. 3B, compare lanes 5 and 6). The wild-type leuO coding region was expressed from another, coexisting plasmid, pEV101, under the control of the IPTG-inducible ptac promoter. This result clearly demonstrated that LeuO plays a trans-acting role in leu-500 activation.
FIG. 3.
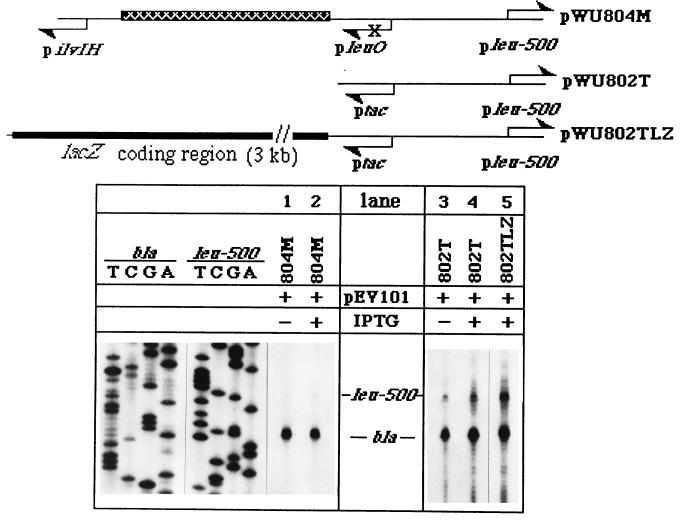
The LeuO protein plays a trans-acting role in leu-500 activation. (A) The relevant regions in the linear maps of pWU804T, pWU804TR, and pWU802T in which pleuO was replaced by ptac are shown. The orientations of the promoters are indicated (arrows). pSO1000 was used in this experiment to provide lac repressor (24). Primer extension results for transcripts initiated from pleu-500 with RNAs isolated from CH582 are shown (lanes 1 to 5). The testing conditions are indicated above the lanes. (B) Part of the peptide sequence of the 942-bp leuO gene coding region product is illustrated in the linear map of pWU804H. The helix-turn-helix motif located at positions 39 to 58 of the LeuO peptide (12) (boxed) and the R41P mutation within this motif are indicated. The map of expression vector pEV101, which is used to supply wild-type LeuO in trans upon IPTG induction, is also shown. Primer extension results obtained with pWU804 and pWU804H in the absence of pEV101 are shown for transcripts initiated from pleuO (lanes 1 and 3, respectively) and transcripts from pleu-500 (lanes 2 and 4, respectively). In the presence of pEV101, pleu-500 activity was detected by using RNAs isolated from pWU804H-harboring CH582 without or with IPTG treatment (lanes 5 and 6, respectively).
The importance of LeuO in leu-500 activation appears to support the first possibility. If so, the LeuO protein provided in trans should also restore leu-500 activation in pWU804M, in which pleuO was inactivated due to the mutation in the −10 region of the promoter. Surprisingly, the LeuO protein supplied in trans failed to restore leu-500 activation in pWU804M upon IPTG induction (Fig. 4, compare lanes 1 and 2). The fact that trans-acting LeuO was able to restore leu-500 activation in pWU804H (Fig. 3B, lanes 5 and 6) but not in pWU804M indicated that something important for leu-500 activation was missing in pWU804M. In the presence of wild-type LeuO provided in trans, the major difference between these two conditions was that the pleuO activity was diminished in pWU804M, while the promoter remained active in pWU804H. This result suggested that in addition to the gene product, LeuO, the functional pleuO was also important for leu-500 activation. Furthermore, when pleuO was replaced with the IPTG-inducible ptac, the LeuO protein provided in trans significantly activated pleu-500 upon IPTG induction (Fig. 4, compare lanes 3 and 4), suggesting that the transcriptional process itself, but not the specific promoter, is important for leu-500 activation. Thus, both LeuO in trans and an active divergent promoter in cis are required for leu-500 activation. Note that pilvIH was deleted in the pWU802T construct (Fig. 4). Thus, in the absence of pilvIH, the leu-500 promoter was activated by another promoter located 410 bp upstream, provided that LeuO was present.
FIG. 4.
The cis-acting transcription process is also required for leu-500 activation. The plasmids used in this experiment are illustrated. pEV101 (Fig. 3B) was used to provided LeuO protein in trans upon IPTG induction. Transcription activity of pleu-500 was detected by primer extension (lanes 1 to 5). pbla activity was detected simultaneously in all reactions as an internal control.
Promoter relay model for sequential gene activation.
Our results can best be explained in terms of a promoter relay model in which transcriptional activity from one promoter (i.e., pilvIH) can activate a distant promoter (i.e., pleu-500) via an intermediary promoter (i.e., pleuO) (Fig. 5). The interactions among the promoters are difficult to understand. The fact that the transcriptional process itself but not the specific promoter (e.g., both pilvIH and pleuO can be replaced by an inducible promoter) is important for leu-500 activation suggests that transcription-driven local supercoiling could underlie the mechanism for promoter-promoter interactions. Previous studies have demonstrated that promoter-promoter interaction via localized DNA supercoiling generated by RNA transcriptional processes is normally short-range (<250 bp) (4, 36). The distance between pleu-500 and pilvIH is about 1.9 kb. The relay mechanism for such a long-range interaction is therefore reasonable.
FIG. 5.
Promoter relay mechanism for sequential gene activation at a distance. See the text for details.
Chromosome supercoiling dynamics may play a role in this type of gene expression regulation. Under stringent growth conditions or other environmental stresses, ilvIH transcription-driven local supercoiling could serve as the signal which triggers activation of the normally silent leuO gene. The leuO gene in turn activates pleu-500 via two signals, the LeuO protein and transcription-driven supercoiling from pleuO.
The function of the LeuO protein is unknown. However, it contains a helix-turn-helix motif at the N terminus, suggesting a potential role for DNA binding (12). LeuO has previously been identified as a multicopy suppressor of the hns mutant (32). The product of the hns gene is the H-NS protein. As with other histone-like proteins, such as HU, integration host factor (IHF), etc., H-NS is one of the chromosome architecture proteins in bacteria (26). These proteins have been shown to organize DNA into nucleoid structures and restrain negative DNA supercoils (1, 13, 25, 42). The LeuO protein may affect nucleoid protein-DNA interactions and therefore alter the chromosome supercoiling dynamic in a local region. From this, it seems possible that LeuO may exert its function through remodeling of the chromosome structure (Fig. 5).
The effect of cis-acting pleuO on leu-500 activation could be mediated by transcription-driven supercoiling. We have shown that the transcriptional process but not the specific promoter is important for leu-500 activation. This transcriptional process can generate local DNA supercoiling variation according to the twin-domain model of transcription (19). If transcription-driven local supercoiling contributes to leu-500 activation, we can anticipate that the longer the transcription unit, the more supercoiling is generated from the transcription process, leading to increasingly stronger leu-500 activation. As expected, increasing the size of transcription-translation coupling from ptac due to the fusion of a 3-kb coding region of the lacZ gene with ptac further enhanced leu-500 activation in pWU802TLZ (Fig. 4 map and compare lanes 4 and 5), suggesting that the role of cis-acting pleuO activity is to provide transcription-driven supercoiling for leu-500 activation. In the presence of LeuO protein, the negative supercoiling originating from pleuO could activate pleu-500 directly and/or influence the chromosomal structural remodeling activity of LeuO. Further studies are necessary to reveal the molecular details of the combined actions of transcription-driven supercoiling and LeuO protein binding.
Using the supercoiling-sensitive leu-500 promoter, we have identified sequential long-range interactions among multiple promoters. However, it is unclear whether the wild-type promoter of the leuABCD operon (pleu) is under the same kind of regulation. In order to answer this question, the effect of pilvIH on pleu was studied. As shown in Fig. 6A, pleu activity was reduced approximately threefold in pWU907, in which pilvIH was mutated. In addition, pleuO activity was also significantly reduced (Fig. 6B, compare lanes 1 and 2). The fact that the leuABCD operon is regulated by the distant pilvIH promoter located 1.9 kb upstream suggests that in addition to being regulated by attenuation (31), the leuABCD operon is also controlled at the transcriptional level via the promoter relay mechanism.
The sequential gene activation may be part of a nutritional environmental stress response cascade. It has been shown that pilvIH activity is under the control of Lrp (leucine-responsive regulatory protein), which is a global transcription regulator whose cellular level is up-regulated by cellular guanosine 3′,5′-bispyrophosphate (ppGpp) in response to nutrient limitation (17, 23, 38, 40). Therefore, it is reasonable that under nutrient-limited growth conditions, the ilvIH operon is turned on and its transcription-driven supercoiling serves as a signal to turn on the leuO gene, which subsequently enhances expression of the leuABCD operon.
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health grant GM53617 and by a university research award from Wayne State University.
We thank Jason Schnepf for technical assistance and critical reading of the manuscript. We also thank Bonnie Sloane, Ronald Hines, Roy McCauley, and George Dambach for their support and encouragement during the course of this work.
REFERENCES
- 1.Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. Cross-talk between topoisomerase I and HU in Escherichia coli. J Mol Biol. 1996;256:292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- 2.Calvo J M, Freundlich M, Umbarger H E. Regulation of branched-chain amino acid biosynthesis in Salmonella typhimurium: isolation of regulatory mutants. J Bacteriol. 1969;97:1272–1282. doi: 10.1128/jb.97.3.1272-1282.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Bowater R, Dorman C J, Lilley D M J. Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc Natl Acad Sci USA. 1992;89:8784–8788. doi: 10.1073/pnas.89.18.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen D, Bowater R P, Lilley D M J. Activation of the leu-500 promoter: a topological domain generated by divergent transcription in a plasmid. Biochemistry. 1993;32:13162–13170. doi: 10.1021/bi00211a027. [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Bowater R P, Lilley D M J. Topological promoter coupling in Escherichia coli: ΔtopA-dependent activation of the leu-500 promoter on a plasmid. J Bacteriol. 1994;176:3757–3764. doi: 10.1128/jb.176.12.3757-3764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Boer H A, Comstock L J, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 8.Dubanau E, Margolin P. Suppression of promoter mutations by the pleiotropic supX mutations. Mol Gen Genet. 1972;117:91–112. doi: 10.1007/BF00267607. [DOI] [PubMed] [Google Scholar]
- 8a.Fang, M., and H.-Y. Wu. Unpublished data.
- 9.Franco R J, Drlica K. Gyrase inhibitors can increase gyrA expression and DNA supercoiling. J Bacteriol. 1989;171:6573–6579. doi: 10.1128/jb.171.12.6573-6579.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- 11.Haughn G W, Wessler S R, Gemmill R M, Calvo J M. High A+T content conserved in DNA sequences upstream of leuABCD in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1986;166:1113–1117. doi: 10.1128/jb.166.3.1113-1117.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff S, Haughn G W, Calvo J M, Wallace J C. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillyared D, Edlund M, Hughes K, Marsh M, Higgins N. Subunit phenotypes of Salmonella typhimurium HU mutants. J Bacteriol. 1990;172:5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirose S, Suzuki Y. In vitro transcription of eukaryotic genes is affected differently by the degree of DNA supercoiling. Proc Natl Acad Sci USA. 1988;85:718–722. doi: 10.1073/pnas.85.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirkegaard K, Buc H, Spassky A, Wang J C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-stranded-specific cytosine methylation in RNA polymerase-promoter complex. Proc Natl Acad Sci USA. 1983;80:2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamond A I. Supercoiling response of a bacterial tRNA gene. EMBO J. 1985;4:501–507. doi: 10.1002/j.1460-2075.1985.tb03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landgraf J R, Wu J, Calvo J M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilley D M J, Higgins C F. Local DNA topology and gene expression: the case of the leu-500 promoter. Mol Microbiol. 1991;5:779–783. doi: 10.1111/j.1365-2958.1991.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu L F, Wang J C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margolin P, Zumstein L, Sternglanz R, Wang J C. The Escherichia coli supX locus is topA, the structural gene for DNA topoisomerase I. Proc Natl Acad Sci USA. 1985;82:5437–5441. doi: 10.1073/pnas.82.16.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menzel R, Gellert M. Fusions of the Escherichia coli gyrA and gyrB control region to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol. 1987;169:1272–1278. doi: 10.1128/jb.169.3.1272-1278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukai F H, Margolin P. Analysis of unlinked suppressors of an Oo mutation in Salmonella. Proc Natl Acad Sci USA. 1963;50:140–148. doi: 10.1073/pnas.50.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman E B, Lin R T, D’Ari R. The leucine/Lrp regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1513–1525. [Google Scholar]
- 24.Oehler S, Eismann E R, Kramer H, Muller-Hill B. The three operators of the lac operon co-operate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettijohn D. Histone-like proteins and bacterial chromosome structure. J Biol Chem. 1988;263:12793–12796. [PubMed] [Google Scholar]
- 26.Pettijohn D. The nucleoid. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 158–166. [Google Scholar]
- 27.Pruss G J, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 28.Richardson S M H, Higgins C F, Lilley D M J. The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J. 1984;3:1745–1752. doi: 10.1002/j.1460-2075.1984.tb02041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson S M H, Higgins C F, Lilley D M J. DNA supercoiling and the leu-500 promoter mutation of Salmonella typhimurium. EMBO J. 1988;7:1863–1869. doi: 10.1002/j.1460-2075.1988.tb03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnetz K, Wang J C. Silencing of the Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 1996;24:2422–2428. doi: 10.1093/nar/24.12.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Searles L L, Wessler S R, Calvo J M. Transcription attenuation is the major mechanism by which the leu operon of Salmonella typhimurium is controlled. J Mol Biol. 1983;163:377–394. doi: 10.1016/0022-2836(83)90064-5. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Bennett G N. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J Bacteriol. 1995;177:810–814. doi: 10.1128/jb.177.3.810-814.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siebenlist U, Simpson R B, Gilbert W. Escherichia coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 34.Smith G R. DNA supercoiling: another level for regulating gene expression. Cell. 1981;24:599–600. doi: 10.1016/0092-8674(81)90085-4. [DOI] [PubMed] [Google Scholar]
- 35.Spirito F, Bossi L. Long-distance effect of downstream transcription on activity of the supercoiling-sensitive leu-500 promoter in a topA mutant of Salmonella typhimurium. J Bacteriol. 1996;178:7129–7137. doi: 10.1128/jb.178.24.7129-7137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan J, Shu L, Wu H-Y. Activation of the leu-500 promoter by adjacent transcription. J Bacteriol. 1994;176:1077–1086. doi: 10.1128/jb.176.4.1077-1086.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trucksis M, Golub E I, Zabel D J, Depew R E. Escherichia coli and Salmonella typhimurium supX genes specify deoxyribonucleic acid topoisomerase I. J Bacteriol. 1981;147:679–681. doi: 10.1128/jb.147.2.679-681.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1987. pp. 352–367. [Google Scholar]
- 39.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Sacco M, Ricca E, Lago C T, DeFelice M, Calvo J M. Organization of Lrp-binding sites upstream of ilvIH in Salmonella typhimurium. Mol Microbiol. 1993;7:883–891. doi: 10.1111/j.1365-2958.1993.tb01179.x. [DOI] [PubMed] [Google Scholar]
- 41.Wu H-Y, Tan J, Fang M. Long-range interaction between two promoters: activation of the leu-500 promoter by a distant upstream promoter. Cell. 1995;82:445–451. doi: 10.1016/0092-8674(95)90433-6. [DOI] [PubMed] [Google Scholar]
- 42.Yasuzawa K, Hayashi N, Goshima N, Kohno K, Imamoto F, Kano Y. Histone-like proteins are required for cell growth and constraint of supercoils in DNA. Gene. 1992;122:9–15. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]