Abstract
Coronavirus disease 2019 (COVID‐19), a global pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has posed a catastrophic threat to human health worldwide. Human stem cell‐derived organoids serve as a promising platform for exploring SARS‐CoV‐2 infection. Several review articles have summarized the application of human organoids in COVID‐19, but the research status and development trend of this field have seldom been systematically and comprehensively studied. In this review, we use bibliometric analysis method to identify the characteristics of organoid‐based COVID‐19 research. First, an annual trend of publications and citations, the most contributing countries or regions and organizations, co‐citation analysis of references and sources and research hotspots are determined. Next, systematical summaries of organoid applications in investigating the pathology of SARS‐CoV‐2 infection, vaccine development and drug discovery, are provided. Lastly, the current challenges and future considerations of this field are discussed. The present study will provide an objective angle to identify the current trend and give novel insights for directing the future development of human organoid applications in SARS‐CoV‐2 infection.
Keyword co‐occurrence overlay visualization of the research of organoids for SARS‐CoV‐2 infection. More studies focused on pulmonary infection using organoid models at the early stages of the COVID‐19 pandemic. Recently, keywords included ‘drug repurposing’, ‘personalized medicine’, ‘repair’, ‘therapy’, ‘resistance’ and ‘therapeutics’ are more popular. Node size indicates the occurrence frequency; Node colour represents the keyword co‐occurrence based on their average publication year, purple means that keywords occurred earlier and marked with yellow stands that keywords are vigorous recently.
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection, has posed a catastrophic threat to human health worldwide. Since its widely spread, as of 30 November 2022, roughly 639.1 million global cases were confirmed and 6.6 million deaths from COVID‐19. 1 COVID‐19 cases could be asymptomatic or symptomatic, including fever or chills, cough, fatigue, shortness of breath and a loss of taste or sense of smell. Respiratory injury is the most common outcome in COVID‐19 patients, but SARS‐CoV‐2 can cause widespread damage to multi‐organs, including the eye, brain, heart, kidney, liver, lung and gastrointestinal tracts. 2 To date, multiple SARS‐CoV‐2 variants have been described. For instance, the SARS‐CoV‐2 Omicron variant (B.1.1.529) was initially reported in South Africa and Botswana in November 2021. 3 , 4 Compared with the original coronavirus, the Omicron variant exhibits enhanced infectivity and transmissibility, high immune evasion ability and reduced pathogenicity. 3 , 5 , 6 Although several COVID‐19 vaccines can prevent viral infections, new coronavirus variant may escape from immunity induced by the existing vaccines, leading to a great number of breakthrough infections. 7 , 8 , 9 In this case, more efforts are needed to comprehensively decipher the pathophysiological processes of coronavirus infection and evaluate vaccine efficacy and safety.
Most current data on SARS‐CoV‐2 infection are limited to clinical data, classic cell experiments and restricted animal models. 10 , 11 Given angiotensin‐converting enzyme 2 (ACE2) functions as the key SARS‐CoV‐2 receptor, transgenic animals expressing human ACE2 are good options. However, animal‐based studies fail to faithfully mimic human physiology and recapitulate the human response to SARS‐CoV‐2 infection because of species variants. In addition, limited data reported how genetic diversity will influence the responses to SARS‐CoV‐2. 12 , 13 Human cell lines, including Caco‐2, Calu‐3, HEK293T, Huh7 and A549, have been utilized in viral infection experiments, whereas they are unable to adequately recapitulate the in vivo infection processes because monolayer cells lack cellular microenvironment, including cell–cell and cell–matrix interactions. Moreover, cancer cell lines carry the potential for malignant proliferation and uncontrollable mutations. Therefore, building human physiologically relevant model systems is dramatically demanded in the studies of viral infections.
Advanced stem cell technology has provided promising models in vitro to recapitulate human sophisticated, multicellular and physiological organs, namely organoids. They are three‐dimensional (3D) self‐organized tissues from human adult stem cells (ASCs) and pluripotent stem cells (PSCs), including human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs). Human adult stem cell‐ or progenitor cell‐derived organoids could retain their organ identity, such as containing various cell types present in the native organ, recapitulating the cellular architecture and spatial organization of the organ and maintaining genetic stability over a long period of time. 14 In contrast to the complicated and variable process of PSC differentiation, ASCs can be generated from biopsies isolated directly from healthy or patient tissue, thus providing a platform for personalized diagnosis and therapy. 15 , 16 ASC‐derived organoids are mainly generated from epithelial tissues (intestine, colon, stomach, liver, pancreas, lung, bladder and so forth) and need to isolate the tissue‐specific stem cell population, thus recapitulating only the epithelium of organs. 14 , 15 , 17 ASC‐derived organoids are typically cystic and highly polarized epithelium, lacking nerves, blood vessels and stromal elements. 18 , 19 , 20 Compared with ASC‐derived organoids, PSC‐derived organoids present higher complexity in structure. 20 PSC‐derived organoids can be established from non‐epithelial tissues, including blood vessels, the brain and the retina. 21 , 22 , 23 PSC‐derived embryoid bodies (EBs) can be guided to differentiate into three primary germ layers (ectoderm, mesoderm and endoderm), which recapitulates early embryonic development and finally generate a large variety of organoids. 15 , 24
Currently, various human organoids have been successfully established, such as brain, retinal, liver, kidney, stomach, intestinal, lung, cardiac and colon organoids and are widely applied for regenerative medicine and modelling human diseases, including infectious diseases. For instance, human brain organoid was utilized to understand Zika virus (ZIKV)‐induced adverse effects on the human brain. Qian's lab first generated brain organoids to elucidate ZIKV‐mediated neurodevelopmental disorder and model microcephaly. 25 ZIKV was found to hijack host cells to increase viral replication and preferentially target neural progenitor cells, induce cell death, proliferation and premature differentiation of neural progenitors in human brain organoids. 25 , 26 , 27 Furthermore, brain organoids were used for drug discovery against ZIKV infection. 28 The study from Xu's group showed that Emricasan (a pan‐caspase inhibitor) and Niclosamide (The United States Food and Drug Administration (FDA)‐approved anthelmintic drug) were found to reduce ZIKV replication and protect neural progenitor cells from ZIKV‐induced cell death in brain organoids. 29 Similarly, cholesterol‐25‐hydroxylase (CH25H) was observed to suppress viral infection and reduce tissue injuries in human ZIKV‐infected cortical organoids. 30 These data stimulated the field to apply human organoids for studying viral infectivity, vaccine development and drug discovery of SARS‐CoV‐2.
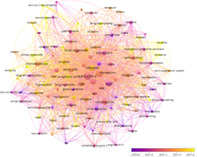
Over the last 3 years, growing worldwide concerns regarding the application of human organoids for SARS‐CoV‐2 infection during the COVID‐19 epidemic scenario. Data from the Web of Science (WOS) Core Collection showed that the publication numbers dramatically increased, especially from 2020 to 2021 (Figure 1A). The major fields of organoid‐based COVID‐19 study are involved in Cell Biology, Biochemistry Molecular Biology, Cell Tissue Engineering, Medicine Research Experimental, Multidisciplinary Sciences, Microbiology, Immunology, Virology and so on (Figure 1B). Given the rapid development of organoid applications in SARS‐CoV‐2 infection, the research status and development trend of this field are systematically and comprehensively reviewed in the present study. Bibliometric analysis could be applied to determine the development, hotspots and trend directions of organoid applications in SARS‐CoV‐2 infection. 31 , 32 , 33 , 34 In this review, bibliometric method was used to reveal the research trends regarding the most influential countries and institutes, contributing journals and hotspots of the research field in organoid‐based COVID‐19 studies. We further provide a detail discussion on the application of human organoids in studying viral infectivity, vaccine development and drug discovery of SARS‐CoV‐2. At last, existing challenges and future perspectives in this field will be discussed. We hope this work will provide an objective angle to identify the current trend and give novel insights for guiding the future development of human organoid applications during the COVID‐19 pandemic scenario.
FIGURE 1.
Characteristics of publications regarding human organoids applied in the COVID‐19 study. (A) Semi‐annual publications of human organoid application for SARS‐CoV‐2 infection. (B) Major fields of organoid application in COVID‐19 research. (C) The top 10 contributing countries/regions in organoid‐based COVID‐19 study.
2. BIBLIOMETRIC ANALYSIS
2.1. Data source and search strategy
The WOS Core Collection Advanced Search based on Science Citation Index Expanded (SCI‐EXPANDED) and Social Sciences Citation Index (SSCI) was applied to retrieve records on organoid applications in SARS‐CoV‐2 research. The data was obtained from the period between 1 December 2019 and 27 November 2022, due to the first case occurred in Wuhan, China on 1 December 2019. Subsequently, WOS analysis and VOSviewer (version 1.6.18, from the Centre of Science and Technology Studies at Leiden University) application were applied to analyse the general information, including growing trends of publication and citation, the most productive/influential countries/regions and organizations, journal distribution, co‐cited references and sources and key research areas. The retrieval strategy was as follows: TS = (‘COVID‐19’ OR ‘coronavirus disease 2019’ OR ‘2019‐nCov’ OR ‘2019 novel coronavirus’ OR ‘SARS‐CoV‐2’ OR ‘Severe acute respiratory syndrome coronavirus 2’ OR ‘novel coronavirus disease 19’ OR ‘novel coronavirus disease‐19’ OR ‘SARS2’ OR ‘SARS‐2’ OR ‘COVID‐2019’ OR ‘COVID19’) AND (‘organoid*’). The inclusion criteria were publications written in English.
2.2. Publication and citation characteristics
A total of 284 records were retrieved, which included 154 original articles, 90 reviews, 18 editorials, 10 early access, 10 letters and 2 others. The articles and reviews constituted the largest share (54.22% and 31.69%, respectively) of the full share of the publications and were analysed in our subsequent analysis. As seen in Figure 1A, both the publications and citations of papers regarding organoid‐based COVID‐19 have increased rapidly over the last 3 years. This data suggests a growing interest in organoid applications in COVID‐19 research.
2.3. The most contributing countries/regions and organizations
Up to 45 countries or regions with publications related to organoid applications for SARS‐CoV‐2 infection. Figure 1C presents the top 10 contributing countries or regions in this field. As indicated, the United States ranks first with 101 publications and has the highest H‐index (25), which was normally utilized to evaluate the publication number and citation performance. 35 , 36 China (54 articles, 16) and Germany (28 articles, 11) rank second and third place, respectively. England, the Netherlands and India also are influential countries/regions in this field. Interestingly, in terms of average citations (citations per publication), Canada ranks first with 126.55 average times. The Netherlands (78.33) and England (50.06) rank second and third place, respectively. Regarding the most contributing organizations, 667 institutes around the world were identified associated with organoid‐based COVID‐19 study. As shown in Table S1, the University of California System and the Chinese Academy of Science rank first and second in terms of publication numbers, with 21 and 15 publications, respectively. The H‐indexes of the University of California System and the University of California San Diego are also in the leading place, reaching 12 and 9, respectively. Although the publications from the Karolinska Institutet put it in ninth place, it reached the highest average citations of 165.22. These top 10 contributing organizations are mainly from the United States, China and European countries. Taken together, scientists from the United States, China and European countries/regions paid great efforts in the application of organoids for SARS‐CoV‐2 infection.
2.4. Co‐citation analysis of references and sources
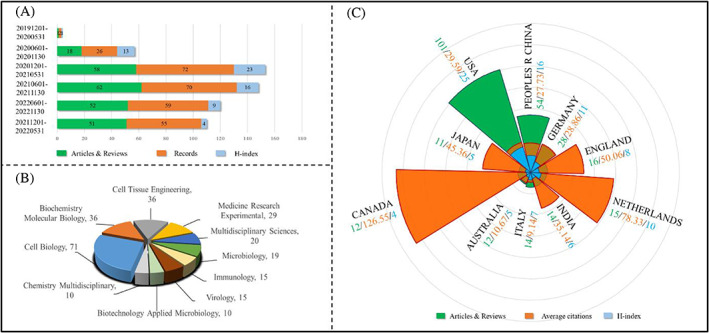
Co‐citation analysis will be helpful for the reader to know the frequency of co‐cited references and journals and can promote the development of the disciplines. 37 , 38 Hence, VOSviewer was applied to visualize the corresponding journals in the field of organoid applications in COVID‐19 research. A total of 69 journals were identified with more than 50 co‐cited papers, as shown in Figure 2A. The intensity of hotspots indicates the frequency of occurrences of that item. Warm colours represent hot items and cool colours represent cool items. The top 10 contributing journals based on citations are Nature, Cells, Science, Cell Stem Cell, New England Journal of Medicine, Proceedings of the National Academy of Sciences of the United States of America, Nature Communications, the Journal of Virology, Nature Methods and Lancet. In terms of co‐cited references, the top 5 references were listed in Figure 2B. The paper ‘SARS‐CoV‐2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor’ (https://doi.org/10.1016/j.cell.2020.02.052) was published in Cell in 2020 and received the highest co‐citation (119 times) in this field. The authors provided critical insights into the early stage of SARS‐CoV‐2 infection, which depends on the host cell factor ACE2 and co‐factor TMPRSS2 (transmembrane serine protease 2), thus identified potential targets for protection against the virus. 39 Although the study did not focus on organoids, this groundbreaking will help decipher the pathomechanism of viral infection and drug candidates screening for COVID‐19 therapeutics. The flowing four publications were closely associated with human intestinal, kidney, liver, lung and colonic organoids, respectively. 11 , 40 , 41 , 42 These data indicate that human organoids contribute to the COVID‐19 study.
FIGURE 2.
Co‐citation analysis of the publications regarding organoid application for COVID‐19 study. (A) The density visualized network of the contributing journals (more than 50 citations). The intensity of hotspots indicates the frequency of occurrences of that item. Warm colours represent hot terms and cool colours represent cool items. (B) The top 5 co‐cited references for organoid‐based COVID‐19 research.
2.5. The hotspots of in organoid‐based COVID‐19 field
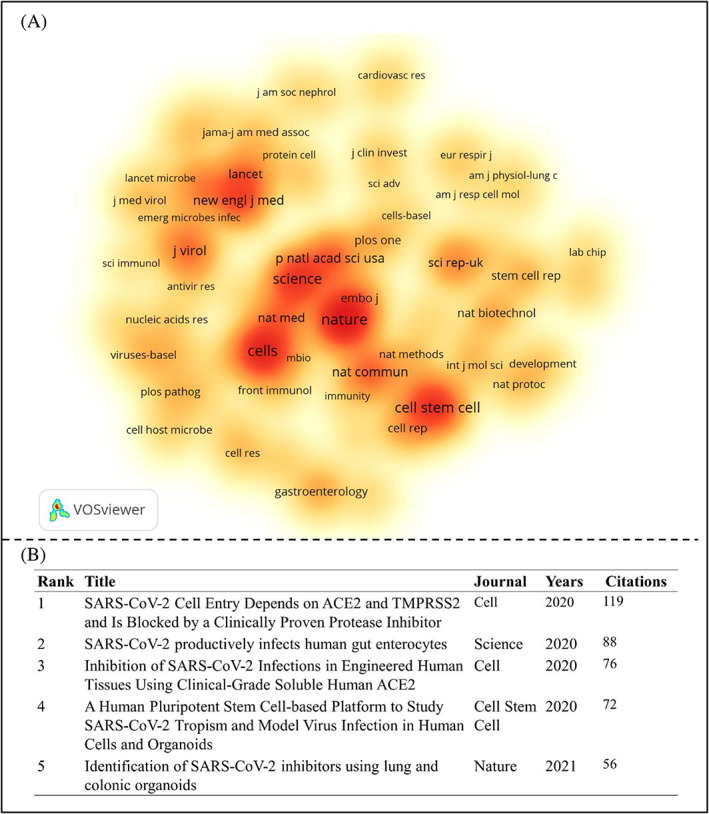
To better reveal the core topic in the organoid‐based COVID‐19 field, keyword co‐occurrence analysis was performed by the VOSviewer application. As shown in Figure 3, a total of 117 keywords were identified within the visualized network with a frequency of more than 3. The top 5 most frequently emerging keywords are ‘sarce‐cov‐2’ (110 co‐occurrence times), ‘covid‐19’ (99), ‘organoids’ (65), ‘ace2’ (50) and ‘infection’ (49). All 117 keywords were classified into eight clusters with distinct colours. The largest cluster, red colour, involved keywords associated with stem cells and disease models, such as ‘airway organoids’, ‘lung organoids’, ‘epithelial‐cells’, ‘transmission’, ‘disease’ and ‘virus‐infection’; Cluster 2 (green) involved in keywords associated with drug discovery and viral replication, such as ‘inhibitors’, ‘inhibition’, ‘resistance’ and ‘expression’; Cluster 3 (blue) involved in keywords associated with viral cell entry and virus–host interaction, such as ‘spike protein’, ‘entry’, ‘receptor’, ‘ace2’ and ‘tmprss2’; Cluster 4 (yellow) involved in keywords related to SARS‐CoV‐2 infection and immune response, such as ‘immunity’, ‘responses’, ‘t‐cells’ and ‘airway epithelium’; Cluster 5 (purple) involved in keywords associated with neural infection, such as ‘brain organoids’, ‘choroid‐plexus’, ‘neural progenitors’, ‘tau’ and ‘sars‐cov‐2 neurotropism’; and other clusters are associated with ‘stem cells’, ‘air–liquid interface’ and ‘intestinal organoids’.
FIGURE 3.
Keyword co‐occurrence (at least three times) network visualization of the research of organoids for SARS‐CoV‐2 infection. Node size indicates the occurrence frequency; node colour represents the cluster; cluster resolution = 1.00.
To obtain the emergence of keywords over time, the overlay visualization of keywords regarding organoid‐based COVID‐19 research was presented in Figure S1, a derivative of Figure 3. As indicated, the node colour stands for the average publication year (purple means that keywords occurred earlier and marked with yellow stands that keywords are vigorous recently). For example, keywords, such as ‘virus‐infection’, ‘sars‐cov‐2 infection’, ‘replication’, ‘airway’, ‘airway epithelium’, ‘acute respiratory syndrome’, ‘pneumonia’ and ‘ace2’ are active in the early era. This suggests more studies focused on pulmonary infection using organoid models at the early stages of the COVID‐19 pandemic. Recently, keywords included ‘drug repurposing’, ‘personalized medicine’, ‘repair’, ‘therapy’, ‘resistance’ and ‘therapeutics’ are more popular. Therefore, more strategies are urgently demanded for SARS‐CoV‐2 therapeutics based on organoid technology.
3. HUMAN ORGANOIDS FOR STUDYING SARS‐CoV‐2 INFECTIVITY
Human organoids are in vitro 3D tissues formed by the self‐assembly of human stem cells. Stem cell‐derived EBs can generate derivatives of three embryonic germ layers, including ectoderm, mesoderm and endoderm and thus various organoids are generated. Given human organoids can mimic the key features of tissues and organs in the human body, such as cell diversity, structural and functional properties and physiological microenvironment, organoid platforms have been applied for studying SARS‐CoV‐2 infectivity.
3.1. Respiratory organoids
The human airways comprise the nasal cavity, proximal and intermediate airways, respiratory bronchioles and alveoli. 43 The airway epithelium is the primary target of SARS‐CoV‐2, which causes severe cough, excessive mucous production, shortness of breath, chest tightness and wheezing. 2 , 44 Studies regarding viral lung infection appeared relatively earlier in the early stages of the COVID‐19 pandemic (Figure S1). ACE2 and TMPRSS2, the key host proteins that promote cellular entry of SARS‐CoV‐2, were highly expressed on the lung epithelium. 45 To date, nasal, bronchial, bronchioalveolar and alveolar organoids have been generated to investigate SARS‐CoV‐2 infectivity (Figure 4A–C). 2 , 48 , 50 , 51 , 52 , 53 , 54 , 55 The upper airway, particularly the nasopharyngeal epithelium, is the entry portal and primary site for SARS‐CoV‐2 replication and transmission. 51 , 56 , 57 Recently, nasal epithelial cells procured via a non‐invasive procedure was used to generate 3D differentiated nasal organoids that adequately simulate the native nasal epithelium, accommodating all airway epithelial cell types, such as ciliated, basal, goblet and club cells. 51 The differentiated nasal organoids accurately recapitulated the differential infectivety of emerging variants. Moreover, the study revealed the potentially viral pathogenesis was closely associated with ciliary damage and tight junction disruption. 51
FIGURE 4.
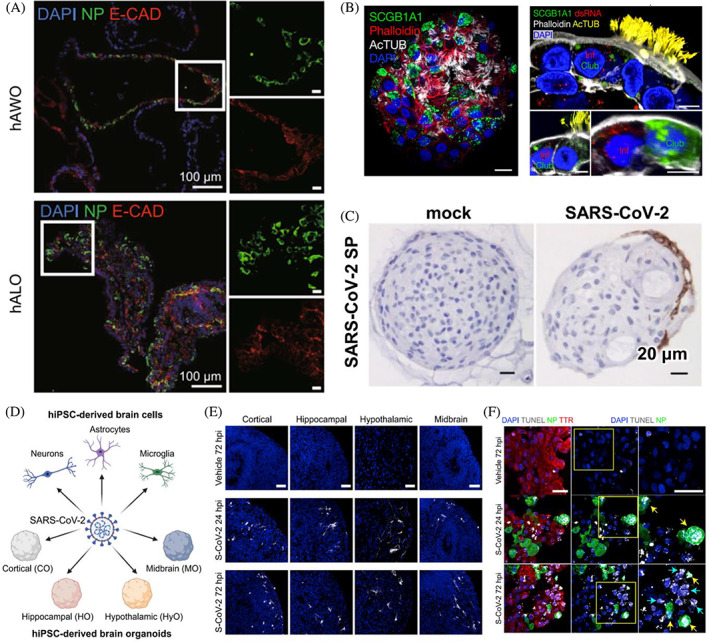
Human lung and brain organoids for modelling SARS‐CoV‐2 infection. (A) Generation of human airway organoids (hAWOs) and human alveolar organoids (hALOs) from hESCs for SARS‐CoV‐2 infection. Reproduced with permission. 46 Copyright 2020, Oxford University Press. (B) Generation of apical‐out distal lung organoids for modelling SARS‐CoV‐2 infection. Reproduced with permission. 47 Copyright 2020, Springer Nature. (C) Human bronchial organoids were infected by SARS‐CoV‐2. Reproduced with permission. 48 Copyright 2022, Springer Nature. (D–F) hiPSC‐derived cortical, hippocampal, hypothalamic, midbrain organoids (D,E) and choroid plexus organoids (F) could be infected by SARS‐CoV‐2. Reproduced with permission. 49 Copyright 2020, Elsevier.
hPSC‐derived airway organoids (AWOs), containing functional multi‐ciliated cells, basal cells, mucus‐producing secretory cells and CC10‐secreting club cells, were also used to monitor SARS‐CoV‐2 infection, showing ciliated and club cells were infected. 46 , 58 , 59 In the human lung, ciliated cells are decreased while club cells are increased from the proximal to the distal airway. 46 Therefore, it was speculated that SARS‐CoV‐2 sequentially infected ciliated and club cells along the upper airway down to the alveoli. Moreover, SARS‐CoV‐2 infection could cause metabolic disorders, as illustrated by downregulated lipid metabolism and upregulated glycolysis. 46 , 60 The data of RNA‐sequence (RNA‐seq) of infected AWOs showed the upregulation of cytokine/chemokine signalling, which was consistent with exuberant inflammatory cytokine production in human COVID‐19 pulmonary infections. 60 , 61 In this case, AWOs thus provide a promising platform for anti‐viral therapeutic drug discovery. Moreover, human bronchial organoids (BCOs), containing transient secretory, goblet and ciliated cells, were generated for SARS‐CoV‐2 research. 48 , 62 The study from Sano's group proved that SARS‐CoV‐2 efficiently infected ciliated cells in BCOs, whereas not basal cells. It is probably due to ciliated cells expressing ACE2, but not basal cells, or club cells could dedifferentiate into basal cells. 48 , 63 The differentially expressed genes caused by viral infection were closely associated with immune response and cytokines. 64
In addition to nasal organoids and AWOs, alveolar lung organoids (ALOs) were also used for SARS‐CoV‐2 studies. The ALO consists of type I cells (AT1) and alveolar type II cells (AT2), which functionally mimic the alveolar epithelium. 65 hPSC‐derived ALOs have been applied to reveal the alveolar infection of SARS‐CoV‐2, showing that AT2 cells were permissive to SARS‐CoV‐2 infection. 40 , 47 , 66 Given the complicated protocol and less reproducibility of PSC‐derived lung organoids, more ALOs have been established from purified primary alveolar stem cells. 67 , 68 For example, Salahudeen and colleagues established human distal lung organoids, including AT2 organoid and basal organoid, for exploring SARS‐CoV‐2 infection. 47 The organoids with apical‐out polarity presenting ACE2 promoted viral infection in AT2, basal and club cells. Similarly, Chiu et al. proved that two‐dimensional (2D) ALOs sustained productive SARS‐CoV‐2, but the virus replicative fitness was lower than that in 2D AWOs. 65 This could be due to more ACE2+ cells in AWOs compared to ALOs. This study suggests that higher viral transmissibility could occur in the lung airway than in the alveoli. More interestingly, it was reported that the Omicron variant only occurred in AWOs, not in ALOs. 65 Consistently, researchers from the LKS Faculty of Medicine at the University of Hong Kong (HKUMed) proved that Omicron SARS‐CoV‐2 could infect and spread faster than ancestral strains in human bronchus but with lower severity in the lung. 69 Another study from Chiu et al. showed that the differentiated nasal organoid monolayers accurately reproduced the highest transmissibility of the Omicron variant. 51 Moreover, the Omicron's replicative advantage was more remarkable in the nasal organoid monolayer than that in the AWOs. This evidence suggests that nasal organoids could more adequately recapitulate the variable replication capacity and transmissibility of emerging SARS‐CoV‐2 variants than AWOs.
3.2. Brain organoids
Besides respiratory insufficiency, existing studies have highlighted severe neurological complications in COVID‐19 patients, ranging from headache and loss of smell to confusion and disabling strokes. 70 , 71 Despite viral RNA being detected in patient‐derived brain samples, it remains unclear the neurotropism of SARS‐CoV‐2 and its potential pathogenesis. 72 , 73 , 74 Given the great potential to elucidate the relationship between ZIKV infection and microcephaly, brain organoids show great promise for understanding the neurotropism and neurotoxic effects of SARS‐CoV‐2. 25 , 66 At the early onset of the COVID‐19 pandemic, SARS‐CoV‐2 (NRW‐42) isolated from a nasopharyngeal and oropharyngeal swab specimen of infected patients was found to enter human organoids with 2‐day viral exposure but did not appear to actively replicate. 75 The study also showed that SARA‐CoV‐2 preferred relatively mature neurons in brain organoids. For instance, mature cortical neurons in the brain organoids were susceptible to SARA‐CoV‐2 exposure. 75 , 76 However, this is indeed markedly inconsistent with ZIKV infection, which was found to preferentially target neural progenitor cells and trigger them to premature differentiation. 25 , 26 , 77 The authors also proved that SARS‐CoV‐2 infection was related to aberrant Tau localization from axons to soma and hyperphosphorylation, which might lead to neuronal death. 75 High‐density viral regions have more apoptotic cells compared with the low‐density viral area. 76 Moreover, SARS‐CoV‐2 infection was accompanied by metabolic alteration in infected and neighbouring neurons, which may create a resource‐restricted environment for neural cells.
Recently, region‐specific brain organoids, including cortical, hippocampal, hypothalamic and midbrain organoids, as well as choroid‐plexus organoids (CPOs) have been established to systematically test SARS‐CoV‐2 neurotropism (Figure 4D–F). 49 , 78 The study from Jacob's team showed that the virus exhibited high infectivity of choroid plexus epithelial cells while limited affinity with neurons and astrocytes. 49 The transcriptional alteration in infected CPOs showed that SARS‐CoV‐2 caused inflammatory responses and damaged the choroid plexus, indicating that the choroid plexus epithelium might be a potential gateway for viral entry to the brain and contribute to neural infection. Consistently, Pellegrini et al. proved that SARS‐CoV‐2 could infect choroid plexus epithelial cells of the brain but not neurons, leading to a potential breakdown of blood‐cerebrospinal fluid barrier integrity and development of neurological complications. 79 This could be closely associated with neural progenitors and neurons do not specifically express SARS‐CoV‐2 entry factors or co‐factors, ACE2 and TMPRSS2. 71 , 79 , 80 Conversely, in line with ZIKV infection, Zhang and colleagues proved that ACE2 and TMPRSS2 expressed in neural progenitor cells and SARS‐CoV‐2 directly infected neural progenitor cells in brain organoids. 81 Moreover, Wang et al. demonstrated that SARS‐CoV‐2 infected hiPSC‐derived neurons, astrocytes and brain organoids, especially ApoE4 astrocytes exhibited high susceptibility to SARS‐CoV‐2. 82 These discrepancies could be caused by the difference in differentiation protocol and differential neuronal phenotypes regulating SARS‐CoV‐2 susceptibility. These findings unambiguously demonstrate that brain organoids provide great promising platforms for probing the neurotropic features and neurotoxic effects of SARS‐CoV‐2.
3.3. Gastrointestinal organoids
Multiple studies have reported that COVID‐19 patients developed gastrointestinal symptoms, such as diarrhoea, vomiting, or abdominal pain, 11 , 83 , 84 , 85 suggesting that gastrointestinal tracts are considered targets of SARS‐CoV‐2. Given live virus was detected in the faeces of COVID‐19 patients, SARS‐CoV‐2 could be transmitted through the faecal‐oral route. 84 , 86 , 87 Gastrointestinal organoids, such as intestinal organoids, colonic organoids and gastric organoids have been generated to unravel the susceptibility of gastrointestinal tracts to SARS‐CoV‐2 (Figure 5A). 40 , 87 , 88 , 93 , 94 , 95 Human and bat intestinal organoids were successfully generated by Zhou's group and first applied for modelling enteric SARS‐CoV‐2 infection. 96 Active viral replication was found in intestinal organoids, which contain enterocytes, goblet cells, Paneth cells and enteroendocrine cells. The study showed that enterocytes were the main targets of SARS‐CoV‐2 probably due to ACE2 and TMPRSS2 being highly expressed in differentiated enterocytes. This finding is consistent with the data from Zhang's study, showing active SARS‐CoV‐2 replication in ACE2+ human mature enterocytes. 97 The authors also proved that TMPRSS2 and TMPRSS4 facilitated SARS‐CoV‐2 entry into host cells by promoting viral spike fusogenic activity. Enterocytes in human colonic organoids were also sensitive to SARS‐CoV‐2 infection. 40 Promoting enterocyte differentiation in the organoid could increase ACE2 expression. 98 RNA‐seq analysis proved that viral infection in colonic organoids involved cytokines and chemokines signalling pathways. 40
FIGURE 5.
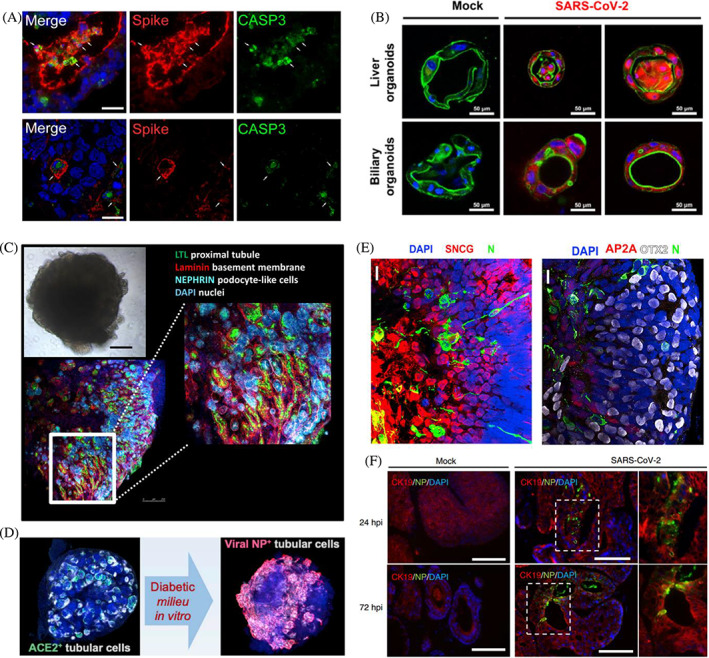
Human organoids for modelling SARS‐CoV‐2 infection. (A) Generation of human intestinal organoids for modelling SARS‐CoV‐2 infection. Reproduced with permission. 88 Copyright 2021, Elsevier. (B) SARS‐CoV‐2 infected liver organoids and biliary organoids. Reproduced with permission. 89 Copyright 2022, BMJ Publishing Group Ltd. (C) Generation of human kidney organoids for modelling SARS‐CoV‐2 infection. Reproduced with permission. 42 Copyright 2020, (D) Diabetic conditions enhanced SARS‐CoV‐2 infections in human kidney organoids. Reproduced with permission. 90 Copyright 2022, Elsevier. (E) SARS‐CoV‐2 infected and replicated in retinal organoids. Reproduced with permission. 91 Copyright 2022, Elsevier. (F) SARS‐CoV‐2 infected salivary gland organoids. Reproduced with permission. 92 Copyright 2022, Springer Nature.
Apart from intestinal and colonic organoids, gastric organoids are also susceptible to SARS‐CoV‐2 infection. Giobbe et al. generated gastric organoids from fetal, pediatric and adult biopsies for modelling SARS‐CoV‐2 infection and demonstrated that late fetal and pediatric organoids allowed higher viral replication. 93 This could be caused by the lower levels of ACE2 and TMPRSS2 in the undifferentiated organoids, including early fetal and adult organoids, compared to organoids following differentiation. Consistently, several studies showed that differentiated intestinal organoids exhibited higher susceptibility to viral infection than undifferentiated ones. 95 , 99 SARS‐CoV‐2 infection was also found to induce mild innate antiviral responses, particularly involved in the interferon family in late fetal and pediatric organoids as illustrated by the transcription analysis. Likewise, single‐cell RNA‐seq (scRNA‐seq) analysis of viral‐infected intestinal organoids highlighted infected cells activated strong proinflammatory events and induced interferon production. 100 These data indicate that human gastrointestinal organoids serve as a valuable platform for SARS‐CoV‐2 gastrointestinal infection and the development of therapeutic strategies.
3.4. Liver organoids
As epidemiologic studies reported that COVID‐19 patients had abnormal liver function mainly involved in hepatocytes and cholangiocytes, thus human hepatocyte and cholangiocyte organoids were used to validate these findings. 101 , 102 , 103 ACE2 and TMPRSS2 were expressed in hepatocytes and cholangiocytes, making cells highly vulnerable to SARS‐CoV‐2 infection. 11 , 104 Therefore, viral replication was observed in both hepatocyte and cholangiocyte organoids after viral infection. 11 Meanwhile, the SARS‐CoV‐2 pseudo‐entry virus was found to infect hepatocytes. The SARS‐CoV‐2 infection activated cellular cytokine–cytokine receptor interaction, IL‐17, chemokine, TNF (tumor necrosis factor) and NF‐κB signalling pathways and downregulated metabolism in infected organoids. 11 Similarly, Zhao et al. revealed activated TNF signalling and apoptosis pathways in the infected liver and biliary organoids, implying that SARS‐CoV‐2 infection might trigger cell death of hepatocytes and cholangiocytes (Figure 5B). 89 A subset of human‐specific ACE2+/TMPRSS2+ cholangiocytes was identified in human liver ductal organoids, thus the organoids were extremely susceptible to SARS‐CoV‐2, leading to robust viral replication was detected in organoids. 104 Furthermore, SARS‐CoV‐2 exposure caused the disruption of the barrier and bile acid transporting functions of cholangiocytes through the dysregulation of gene expression, which was closely associated with a tight junction formation and bile acid transportation. Therefore, SARS‐CoV‐2 infection might induce cholangiocyte injuries and consequent bile acid accumulation in COVID‐19 patients.
3.5. Kidney organoids
Kidney failure was also frequently observed in COVID‐19 patients. 105 Recent studies detected viral RNA in urine samples of COVID‐19 patients. 106 ACE2 was expressed in kidney proximal convoluted tubules, indicating that renal tubule cells could be potential targets of SARS‐CoV‐2. 107 , 108 Thus, 3D human kidney organoids containing proximal tubule epithelial cells were generated for SARS‐CoV‐2 infection (Figure 5C). 42 , 109 Compared with traditional 2D culture, ACE2 was higher expressed in organoids, indicating the organoids would be more permissive to SARS‐CoV‐2. 109 Apart from proximal tubular cells, podocytes and stromal cells were also infected by SARS‐CoV‐2 in iPSC‐derived kidney organoids, which was confirmed by the results from the scRNA‐seq analysis. 110 Activated profibrotic signalling was also found in infected kidney organoids and could be inhibited by a protease blocker. More recently, Garreta and colleagues established diabetic human kidney organoids that exhibited higher susceptibility to SARS‐CoV‐2 compared to non‐diabetic control (Figure 5D). 90 This was linked to diabetic‐induced upregulation of ACE2 and metabolic programming. Therefore, kidney organoids are useful tools for investigating kidney complications for SARS‐CoV‐2 infection and looking for therapeutic drugs.
3.6. Other organoids
SARS‐CoV‐2 infection can cause cardiac injury and dysfunction in patients and increase the risk of mortality. 111 , 112 Human cardiac organoids and vascular organoids have been established to explore SARS‐CoV‐2 infectivity in the cardiovascular system. 42 , 113 , 114 Mils et al. demonstrated that infected cardiac organoids could recapitulate key clinical features of diastolic malfunction in COVID‐19 patients. 115 The study also showed that cytokine‐induced cardiac dysfunction in cardiac organoids could be attenuated by using bromodomain and extraterminal family inhibitors (BETi).
To determine the ocular involvement and potential virus–host ocular interactions, Eriksen et al. generated a whole‐eye organoid, including the retina, retinal pigment epithelium, ciliary margin, iris, lens and cornea, for SARS‐CoV‐2 infection. 116 The study showed that higher viral replication was detected in limbus than in other ocular surface cells in eye organoids. Moreover, retinal organoids also were used to investigate retinal SARS‐CoV‐2 infection (Figure 5E). 91 , 117 Although relatively low expression of ACE2 in the retina, the study proved that SARS‐CoV‐2 infected retinal organoids, replicated in retinal cells, such as retinal ganglion cells and photoreceptors and induced the expression of inflammatory genes, such as interleukin 33. 91
It was recently reported that saliva could be a potential source of SARS‐CoV‐2 transmission due to the virus being identified in the salary glands of COVID‐19 patients. 60 Indeed, salivary glands express ACE2 and TMPRSS2 and serve as reservoirs of viruses. 60 , 92 , 118 Tanaka and colleagues generated salivary gland organoids (SGOs) from hiPSCs to model SARS‐CoV‐2 infection of salary glands (Figure 5F). 92 The human SGOs mimicked human embryonic salary gland characteristics and functions and expressed viral entry factors and co‐factors, including ACE2 and TMPRSS2. Thus, viral infection and replication were found in organoids after exposure to SARS‐CoV‐2. Although SGO is a prospective model for studying SARS‐CoV‐2 infection, it is not yet clear how SARS‐CoV‐2 replicated in salivary glands was secreted into saliva.
4. HUMAN ORGANOIDS FOR VACCINE DEVELOPMENT OF SARS‐COV‐2
Currently, COVID‐19 is still spreading and posing threats to widespread health, society and economy. 119 Although, as of 30 November 2022, approximately 13 billion vaccine doses have been administered, more effective vaccines are urgently demanded to prevent further spread and keep people from contracting the virus. 1 , 120 , 121 The development of a novel vaccine is still a lengthy process, including the initial vaccine design, preclinical testing in animal models and clinical trials (phases I–IV), which could take 15 years or more, great efforts have been paid in vaccine development responding to the COVID‐19 pandemic. 121 , 122 According to the WHO, as of 6 December 2022, 175 candidate vaccines are under clinical testing for treating COVID‐19 and 199 candidate vaccines are in pre‐clinical studying. 123 Of the 175 candidates, 49 and 11 are undergoing further validations of safety and efficacy in phase III and phase IV clinical trials with a large number of volunteers, respectively. The technology platforms can be divided into traditional whole virus vaccines (inactivated or live attenuated vaccines), recombinant protein‐based vaccines (protein subunit vaccines, virus‐like particles), viral vector vaccines and nucleic acid vaccines (DNA‐ and RNA‐based vaccines). Among them, protein subunits and RNA are mainly candidate vaccines. 123
As viral infections are the prototypic species‐specific diseases and pronounced species‐specific differences in adaptive immunity, many candidate vaccines that worked in animals while failing in human trails. 124 , 125 , 126 , 127 Therefore, novel animal‐free test methods may accelerate the testing programs for drug and vaccine development. 124 Although in vitro human organoids are devoid of the host immune system, organoid‐based assays could provide unique opportunities for studying the host–virus interaction and testing the efficacy and neutralizing antibodies of candidate vaccines. 40 Several studies have proved that antibodies were effective in neutralizing SARS‐CoV‐2. For instance, Pei and colleagues evaluated the inhibitory effect of a neutralizing antibody CB6 on SARS‐CoV‐2 infection in human lung organoids. 46 The study showed that CB6 notably suppressed viral replication in infected lung organoids. This was consistent with the previous study that reported CB6 repressed SARS‐CoV‐2 infection in rhesus monkeys. 128 Moreover, a new tetravalent neutralizing antibody (15033‐7) and a synthetic dipeptidyl peptidase‐4 (DPP4) peptide were observed to reduce SARS‐CoV‐2 entry in human lung organoids and modulate innate immunity and inflammatory response. 129 These data will further aid vaccine and drug development for COVID‐19. Advanced engineering approaches, such as bioreactor or chip‐based engineering devices, showing great promise to generate immune cell organoids, but they cannot recapitulate the complex of human adaptive immunity. 130 , 131 , 132 Importantly, the presence of immune elements, including B cells, T cells and microphages in organoid models might facilitate vaccine development. 40
Recently, Wagar et al. established human tonsil organoids (TOs) from reaggregating the dissociated primary human tonsils to evaluate human adaptive immune responses to the SARS‐CoV‐2 vaccine. 126 The specialized germinal centre (GC)‐like structure with distinct T cells and B cells was identified in human TOs, which is essential to produce antigen‐specific antibodies, affinity maturation, B cell differentiation and class‐switch recombination (Figure 6A). Increased B cell differentiation and influenza‐specific antibody production were observed in organoids after exposure to live attenuated influenza vaccine. To clear the responses of individual cells to influenza, the authors analysed the changes in plasmablast differentiation and antigen‐specific antibody production after antigen‐presenting cell (APC), T cell and B cell subsets depletion. The results showed that naive B cells, APCs, CD4+, or CD45− stromal cells were minimally needed to constantly maintain plasmablast differentiation and naive antibody responses. 126 Subsequently, TOs from different donors were used to investigate the response to the adenovirus‐based SARS‐CoV‐2 vaccine. Increased plasmablast differentiation, CD8+ T cell activation, and IgG and IgA antibody production were observed in TOs after 14‐day stimulation. Therefore, human TO application in COVID‐19 could help decipher immune responses to viruses and accelerate vaccine design. 134 Although this model was not lacking in immune components, it is worth noting that human organoids used for viral vaccine development are in isolation. The organoid model could not well mimic the immune cell migration from pathogen entry sites to the lymph nodes, tonsils, or spleen, where GCs form in humans. 126 Future studies are needed to incorporate more methods to capture features of human adaptive responses. Moreover, safety considerations are the most important things for vaccine development. 120 It remains to be seen whether new SARS‐CoV‐2 vaccines could trigger aberrant immune responses and induce potential multi‐organ injuries.
FIGURE 6.
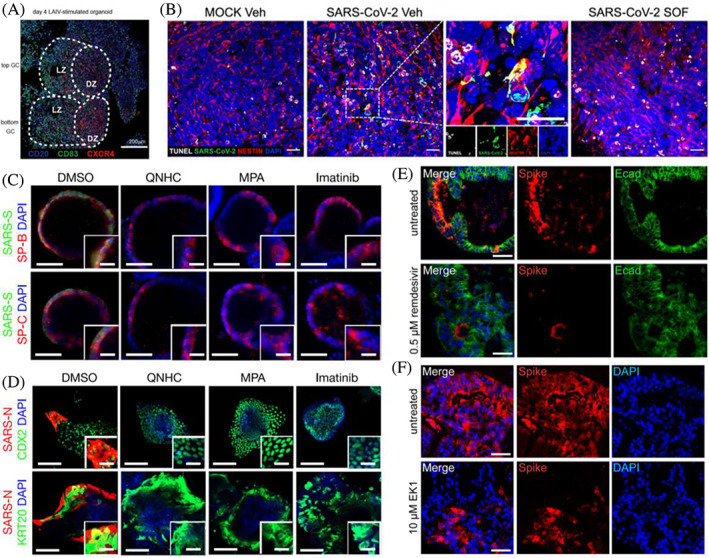
Human organoids for vaccine development and drug discovery of SARS‐CoV‐2. (A) Generation of human tonsil organoids for SARS‐CoV‐2 infection and vaccine development. Reproduced with permission. 126 Copyright 2021, Springer Nature. (B) SOF (sofosbuvir) could inhibit SARS‐CoV‐2 replication in brain cells and rescue neurological impairments in human brain organoids. Reproduced with permission. 133 Copyright 2022, Public Library of Science. (C,D) QNHC (quinacrine dihydrochloride), imatinib, and MPA (mycophenolic acid) blocked the entry of SARS‐CoV‐2 in both hPSC‐derived lung organoids (C) and colonic organoids (D). Reproduced with permission. 40 Copyright 2021, Springer Nature. (E,F) EK1 and remdesivir inhibited infection of human intestinal organoids with SARS‐CoV‐2. Reproduced with permission. 88 Copyright 2021, Elsevier.
5. HUMAN ORGANOIDS FOR DRUG DISCOVERY OF SARS‐COV‐2
Given traditional cell lines cannot recapitulate the physiologically relevant kinetics of in vivo SARS‐CoV‐2 infection, human organoids provide promising models for drug discovery of COVID‐19. The respiratory system is the initial viral target, thus human lung organoids were used to test the efficacy of drugs and compounds, focusing on viral entry and replication. 135 , 136 For instance, Xu et al. demonstrated that inhibition of receptor‐interacting serine/threonine‐protein kinase 1 (RIPK1 kinase) caused by RIPK1 inhibitor Nec‐1s reduced viral load and inflammatory responses in infected lung organoids. 137 The results suggest that the function of RIPK1 in SARS‐CoV‐2 propagation and suppression of RIPK1 might provide a strategy for COVID‐19 therapeutics and prevention. Since ACE2 serves as the entry receptor, a humanized decoy antibody (ACE2‐Fc fusion protein) has been designed to target the interaction between ACE2 and viral spike protein. 138 The study proved that ACE2‐Fc effectively blocked the entry of SARS‐CoV‐2 spike‐expressing pseudotyped virus into lung organoids, thus suppressing viral replication, suggesting that ACE2‐Fc provides a latent prospect for COVID‐19 treatment. Samuel and colleagues demonstrated that androgen (AR) signalling regulated the entry factors and co‐factors of SARS‐CoV‐2 directly, ACE2 in addition to TMPRSS2. 139 Thus, drugs inhibiting AR signalling were found to reduce viral infection in hESC‐derived lung organoids. The study underscores the critical roles of AR signalling in viral infection and lays the foundation for antiandrogenic drug application in COVID‐19 therapeutics.
Primary human lung epithelial infection models, consisting of differentiated air–liquid interface (ALI) cultures of proximal airway epithelium and alveosphere cultures of distal lung AT2 cells, were applied to validate the efficacy of remdesivir, hydroxychloroquine, and interferon beta 1 (IFN‐β1) in anti‐SARS‐CoV‐2. 140 The results confirmed that three molecules significantly suppressed the infection and replication of SARS‐CoV‐2. Remdesivir, an inhibitor of the viral RNA‐dependent, is the most promising FDA‐approved drug in clinical use for hospitalized COVID‐19 patients. 128 , 141 Treatment of remdesivir inhibited viral replication and alleviated alveolar‐capillary barrier disruption on bioengineered human alveolus chip. 142 Similarly, Pei et al. demonstrated remdesivir inhibited viral production in AWO and ALO models. 46 Since the role of interferon lambda (IFN‐λ) in the innate antiviral immunity in the respiratory tract, Lamers and colleagues proved that SARS‐CoV‐2 replication and dissemination were abrogated by low‐dose IFN‐λ1 treatment in the organoid‐derived bronchioalveolar model. 54 The study from Katsura's group also proved that pre‐treatment with a low dose of IFN alpha (IFN‐α) and IFN gamma (IFN‐γ) blocked SARS‐CoV‐2 replication in alveospheres. 67 Moreover, pre‐treatment with both IFN‐λ and IFN‐β1 was observed to significantly impaired SARS‐CoV‐2 infection in colonic organoids. 143 These data indicate interferons will be a viable therapeutic option for COVID‐19. 144 , 145
Human airway microfluidic devices, containing two parallel microchannels, lung bronchial‐airway basal stem cells located in an ‘airway channel’ exposed to air and lung endothelium cultured on the opposite side (‘vascular channel’) exposed to continuous fluid perfusion, have been established to test the efficacy of antiviral drug candidates (Figure 7A). 146 The airway chip faithfully recapitulated key features of influenza‐induced human lung responses and the anti‐influenza efficacy of Oseltamivir. Moreover, the ‘airway channel’ of the airway chip was exposed to SARS‐CoV‐2 pseudoparticles to simulate viral airborne infection, confirming that SARS‐CoV‐2 pseudoparticles could infect human airway epithelial cells efficiently. Subsequently, the ‘vascular channel’ was continuously perfused by candidate drugs to simulate circulatory distribution after oral administration, and the results showed that amodiaquine and its metabolite inhibited SARS‐CoV‐2 infection. 146 Although amodiaquine is used to prevent and treat malaria, this medicine showed great potential in antiviral therapeutics and prophylactics.
FIGURE 7.
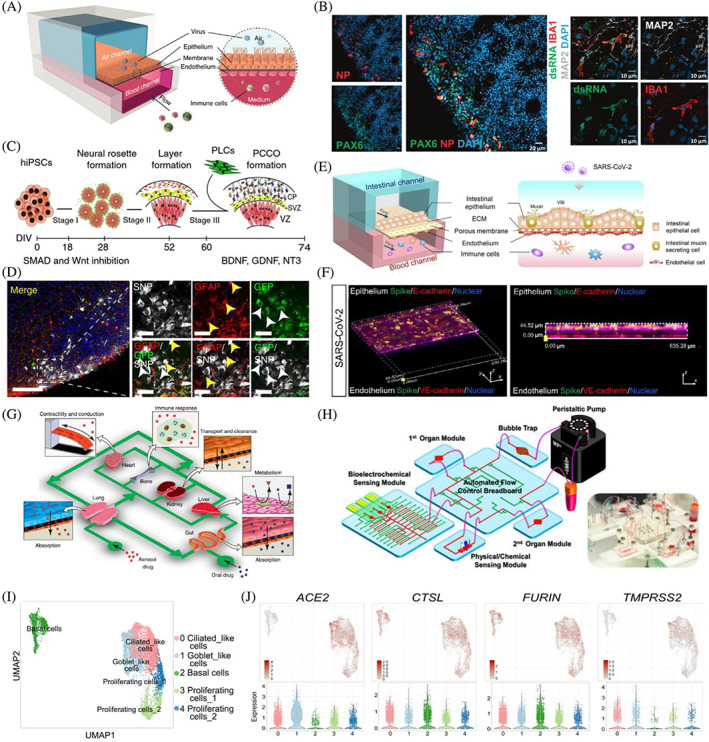
Building advanced human organoids by integrating novel strategies and technologies. (A) A human‐airway‐on‐a‐chip for studying viral infection. Reproduced with permission. 146 Copyright 2021, Springer Nature. (B) SARS‐CoV‐2 infection in brain organoids with innately developing microglia. Reproduced with permission. 147 Copyright 2022, Springer Nature. (C,D) Pericyte‐like cell‐containing cortical organoids were generated by the integration of pericyte‐like cells into cortical brain organoids and were prone to be infected by SARS‐CoV‐2. Reproduced with permission. 148 Copyright 2021, Springer Nature. (E,F) SARS‐CoV‐2 induced intestinal responses with a biomimetic human gut‐on‐chip. Reproduced with permission. 149 Copyright 2021, Science China Press. (G) The human multi‐organ‐on‐a‐chip concept. Reproduced with permission. 150 Copyright 2011, Elsevier. (H). Generation multisensor‐integrated organs‐on‐chips platform. Reproduced with permission. 151 Copyright 2017, National Academy of Sciences. (I,J). Single‐cell RNA‐seq analysis of hPSC‐derived airway organoids. Reproduced with permission. 60 Copyright 2021, Elsevier.
hPSC‐derived lung and colon organoids were also applied for high‐throughput screening FDA‐approved drugs, such as imatinib, mycophenolic acid, and quinacrine dihydrochloride (Figure 6C,D). 40 The study showed that all three drugs could inhibit SARS‐CoV‐2 infection in lung and colon organoids in a dose‐dependent manner. RNA‐seq data highlighted that imatinib‐treated infected organoids were closely associated with lipid metabolism. Tiwari et al. found that EK1 peptide (spike protein inhibitor) and camostat/nafamostat (TMPRSS2 inhibitors) could block SARS‐CoV‐2 entry into lung organoids. 66 This result was consistent with previous studies, showing the antiviral efficacy of EK1 and camostat mesylate. 39 , 152 Pei et al. also proved camostat exerted a slight suppressive effect on viral infection in AWOs, but not in ALOs. 46 A recent study reported that human lung organoids were also applied to explore the antiviral activity of CH25H. 153 Mechanistically, CH25H converted cholesterol to 25‐hydroxylase (25HC), triggering cholesterol depletion on the plasma membrane and affecting virus–cell membrane fusion, resulting in inhibiting viral entry. As 25HC is a natural product without toxicity at effective concentrations, it provides a potential antiviral agent to fight against SARS‐CoV‐2 infection.
As SARS‐CoV‐2‐infected intestinal organoids exhibited the disruption of intestinal integrity and organoid deterioration, mimicking SARS‐CoV‐2‐induced gastrointestinal damage, intestinal organoids were used to test drug efficacy in COVID‐19 patients. 88 A study has shown that remdesivir could effectively inhibit SARS‐CoV‐2 infection dose‐dependently at low micromolar concentrations. However, famotidine did not impact viral infection and spread in intestinal organoids, which was inconsistent with a previous retrospective cohort study that proved famotidine use would be related to alleviative clinical outcomes in hospitalized COVID‐19 patients. 154 Similar to lung organoids, EK1 peptide could inhibit viral infection of intestinal organoids (Figure 6E,F). 88 Interferon‐inducible transmembrane (IFITM) proteins are highly expressed in intestinal organoids and function as entry co‐factors for efficient SARS‐CoV‐2 infection. 155 Thus, IFITM‐derived peptides or antibodies targeting the N‐terminus of IFITMs were found to strongly reduce viral entry and replication in gut organoids.
A recent study showed that ACE2 expression was regulated by the farnesoid X receptor (FXR) signalling in the gastrointestinal and respiratory systems. 156 Therefore, suppression of the FXR signalling by the off‐patent drug ursodeoxycholic acid or the over‐the‐counter compound z‐guggulsterone downregulated the expression of ACE2, subsequently reducing susceptibility to SARS‐CoV‐2 infection in cholangiocyte, airway, and intestinal organoids. The study identified a new regulation of ACE2 through the FXR signalling and provided a new therapeutic target for COVID‐19. More recently, clinical‐grade human recombinant soluble ACE2 (hrsACE2) has been found to inhibit SARS‐CoV‐2 infections of human kidney and capillary organoids dose‐dependently. 42 Similarly, Wysocki et al. also proved that a novel soluble ACE2 variant, consisting of 618 amino acids fused with an albumin binding domain (ABD), could neutralize SARS‐CoV‐2 in human kidney organoids. 157 More intriguingly, Monteil and colleagues studied the additive effects of combination treatment with hrsACE2 and remdesivir on infected kidney organoids. 158 The data showed that combining both drugs at low doses caused a notably reduced viral production. This finding lays the foundations for combinatorial regimes in future COVID‐19 clinical trials. MEDS433, a new inhibitor of the human dihydroorotate dehydrogenase, also exhibited a high potency of anti‐SARS‐CoV‐2 activity in hESC‐derived kidney organoids. 159
A study conducted by Mesci's group showed that SARS‐CoV‐2 infected brain organoids and caused neuron death, which was accompanied by excitatory synapses in neurons. 133 SARS‐CoV‐2 regulated specific gene expression in neurons implicated in antigen presentation, viral entry via the endocytic pathway, neuronal projection development, oxidative stress, and the complement pathway. Treatment with sofosbuvir, a FDA‐approved antiviral drug, efficiently repressed SARS‐CoV‐2 replication and rescued viral‐induced neuronal damage in infected brain organoids (Figure 6B). 133 In addition to lung and intestinal organoids, remdesivir was also found to effectively suppress SARS‐CoV‐2 infection and rescue disease phenotypes in neurons and astrocytes. 82 Therefore, brain organoids also provide invaluable in vitro tools for COVID‐19 drug discovery and validation. Moreover, the therapeutic efficacy of remdesivir against SARS‐CoV‐2 was also validated in the tonsil organoid model. 160
6. CHALLENGES AND FUTURE CONSIDERATIONS
Although human organoids provide valuable platforms for deciphering the pathology of SRAS‐CoV‐2 infection and looking for effective vaccines and potential treatments, these models still have several problems that need to be addressed. One major remaining challenge is the absence of cellular complexity and cell–cell communications, thus organoid microenvironment cannot mimic in vivo organ situations because of lacking vascular, neural, and immune cells. COVID‐19 caused by SARS‐COV‐2 infection could induce multiple organ failure, whereas conventional organoid models are devoid of physiological relevance and organ‐organ interaction. 161 , 162 Therefore, advanced methods and technologies are urgently needed to improve organoid advancement.
6.1. Immune vascular organoids
The host immune system plays a pivotal role in anti‐SARS‐CoV‐2. 163 Given most organoids lack immune cells, human organoids co‐culturing with immune cells have been applied to investigate the pathophysiology of COVID‐19 involved with immune responses. Recently, Zhang's group established a biomimetic human alveolus chip to recapitulate the alveolar‐capillary barrier by coculture of the alveolar epithelial cells, microvascular endothelial cells, and circulating immune cells with perfusing media flow. 142 Notably, SARS‐CoV‐2 caused immune cell recruitment and endothelium detachment and promoted inflammation, indicating the critical roles of immune cells in SARS‐CoV‐2‐induced lung alveolar‐capillary barrier injury and inflammatory response. Another study generated an immune‐cardiac coculture system containing cardiomyocytes and macrophages to study macrophage‐mediated host cell responses to SARS‐CoV‐2 infection. 164 The results proved that macrophages induced reactive oxygen species production and apoptosis in cardiomyocytes after viral exposure. In this case, ranolazine and tofacitinib were then found to alleviate the macrophage‐induced cardiotoxicity. As astrocytes are permissive to SARS‐CoV‐2, brain organoids co‐culturing with astrocytes might boost the viral infection rate. 82 Microglia, the resident immune cells of the central nervous system, play vital roles in brain development, homeostasis, and diseases, incorporating microglia into neural organoids provides unprecedented opportunities to study infective neurological disorders. 165 , 166 , 167 , 168 , 169 For instance, upon ZIKV infection, microglia became activated and targeted neurons and excessively pruned synapses in microglia‐containing brain organoids. 168 Similarly, Samudyata et al. established a protocol for generating a brain organoid model with innately developing microglia for studying SARS‐CoV‐2 infection (Figure 7B). 147 The authors proved that viral infection induced neuronal cell death and increased microglial engulfment of postsynaptic termini in infected microglia‐containing brain organoids. ScRNA‐seq analysis showed that upregulation of interferon‐responsive genes could promote microglia‐mediated synapse elimination secondary to SARS‐CoV‐2 exposure, subsequently causing disruption in circuit integrity, leading to potential cognitive impairments. The data indicate that microglia contribute to SARS‐CoV‐2‐induced neural damage. 147 , 170 , 171 , 172 Thus, microglia‐containing brain organoid models may provide an opportunity for studying SARS‐CoV‐2‐induced neuronal damage and preventing COVID‐19 neurological sequelae.
Notably, vascular networks play critical roles in organogenesis and organ identity. 173 , 174 Currently, evidence is accumulating regarding how to generate vascularized human organoids. 175 , 176 , 177 Therefore, human organoid vascularization would promote their applications in studying SARS‐CoV‐2 infection. For instance, as brain pericytes are proposed as SARS‐CoV‐2 infection points, Wand et al. established pericyte‐like cell (PLC)‐containing cortical organoid (PCCO) ‘assembloid’ through the integration of PLCs into cortical organoids to model SARS‐CoV‐2‐induced neuropathology (Figure 7C,D). 148 PLCs were found to promote astrocytic maturation and capture the features of PLC–basement membrane–astrocyte structure, mimicking human brain pericyte functions in vivo. Compared with conventional cortical organoids, PCCOs exhibited higher susceptibility to SARS‐CoV‐2 due to PLCs within PCCOs serving as viral ‘replication hubs’, promoting virus cell‐to‐cell spread. 148 The study indicates that incorporating neurovascular unit components with brain organoids could provide a promising platform for modelling neural infection of SARS‐CoV‐2. As ACE2 is expressed throughout the vasculature of the body, virus–host interactions on the vascular barrier could contribute to virus access to multiple organ systems. 178 , 179 To date, 3D microfluidic chips modelling the human blood–brain barrier, 179 respiratory vascular barrier, 180 and intestinal epithelium–vascular endothelium barrier (Figure 7E, F) 149 have been established to investigate the effect of SARS‐CoV‐2 on the endothelial barrier. Consistently, SARS‐CoV‐2 was found to disrupt vascular endothelial adherent junctions.
6.2. Multi‐organoids on a chip
In recent years, the microphysiological system, named the ‘human‐on‐a‐chip’ platform, provides specific compartments for multiple organoid interconnections through microchannels (Figure 7G). 150 , 181 The fluidic flow in the chip is analogous to the vasculature through the organ and tissue residence time. 182 , 183 Compared with ‘single‐organoid‐on‐a‐chip’ (single‐OoC) models, ‘muti‐organoid‐on‐a‐chip’ (multi‐OoC) models present advantages in providing a systemic approach to decipher cross‐organ communications. 184 Multi‐OoC platforms could be classified into two groups, namely coupling of single‐OoC units or integration of multiple organoids into one dish. However, the latter offers higher throughput and is more valuable in identifying potential biomarkers and therapeutic targets when compared to the former. 184 Multi‐OoCs hold great promise for systemic assessment of SARS‐CoV‐2 infection, determining organ susceptibility, and looking for anti‐viral therapeutic drugs. Indeed, SARS‐CoV‐2 infection presents multi‐organ injuries and acts at a high level of organ‐organ interaction. 161 , 162 Therefore, a multi‐organoid system could help to study the inter‐organ communications within organs after SARS‐CoV‐2 infection and the secondary effects of the infected organ on another organ. Since multi‐OoCs have been widely applied for toxicity assessment, thus multi‐OoC platforms could be used for the safety evaluation of COVID‐19 vaccines and drugs. 24 , 185 Moreover, muti‐OoCs provide promising models to evaluate the complex process of drug absorption (intestine), organ distribution (along with blood circulation), drug metabolism (liver), and excretion (kidney), as well as potentially reduce the cost of preclinical animal models before conducting clinical trials.
Although the muti‐OoC system has shown enormous potential as an advanced organoid model, muti‐OoC technology is still in its infancy and maintains scientific and technical challenges. First, it is important to optimize the culture medium for co‐culturing different organoids due to tissue‐specific small molecule and growth factor requirements through the differentiation process in guided stem cell differentiation. The culture medium should be provided to meet the needs of each organoid. Noteworthy, the flow rate of the medium needs to be optimized to ensure that sufficient nutrients and oxygen meet the requirements of the next organoids. Moreover, it is necessary to consider that the metabolites generated by the organoid could be toxic to other organoids through the distribution of flow. 186 Therefore, the OoC platform needs a sophisticated programme for nutrient‐waste management. Second, highly integrated monitoring platforms could be used to real‐time monitor cellular activity and organoid behaviours, such as cell vitality, metabolites, inflammatory cytokines, and electrophysiology. 187 , 188 Multisensor‐integrated OoC platform will enhance their performance in drug screening by providing biomarker profiling (Figure 7H). 151 To overcome the high variability of organoids, multi‐organoids derived from the same hiPSC lines could resolve this problem and provide opportunities for an accurate model for personalized prediction and treatment. 181 To date, further improvements are still demanded until multi‐OoCs become a standard COVID‐19 evaluation tool.
6.3. Multi‐omics analysis
The increased resolution of scRNA‐seq technologies has led to great breakthroughs and improved our understanding of cellular antiviral responses, such as hepatitis B virus (HBV), 189 , 190 Ebola virus, 191 influenza virus, 192 , 193 rabies virus, 194 human immunodeficiency virus (HIV), 195 , 196 ZIKV, 197 , 198 , 199 and SARS‐CoV‐2. 189 , 200 , 201 , 202 , 203 , 204 , 205 , 206 , 207 scRNA‐seq enables transcriptome‐wide expression profile at a single‐cell resolution and provides a powerful method to investigate virus–host and host–host interactions and intercellular communication networks upon viral infection. 208 , 209 , 210 Previous evidence has shown that early scRNA‐seq technology has been applied to investigate the expression profiling of the SARS‐CoV‐2 entry factors and co‐factors, such as ACE2, TMPRSS2 and FURIN. 45 , 211 , 212 , 213 As human organoids contain multiple differentiated cell types and allow to explore cell–cell interactions, organoid platforms combined with scRNA‐seq can be used to identify the gene expression of entry factors and co‐factors in diverse cell phenotypes. 214 , 215 For instance, Duan and colleagues proved that scRNA‐seq analysis validated that ACE2, TMPRSS2, and FURIN were expressed in the ciliated‐like cells, allowing SARS‐CoV‐2 entry in AWOs (Figure 7IJ). 60 Similarly, the results of scRNA‐seq also showed that ACE2, TMPRSS2, and FURIN were enriched in the AT2‐like cell population in lung organoids. 40 It is worth noting that the data from scRNA‐seq require to be validated by immunostaining and flow cytometry analyses. Although scRNA‐seq has gained tremendous attention, challenges in high sample preparation cost, severe batch effects, time‐consuming process, numerous sample requirements, and relatively low‐throughout manners limited the widespread adoption and scope of scRNA‐seq. 210 , 216 The existing strategies for sample multiplexing for scRNA‐seq with DNA‐based barcoding will help address these limitations. 217 , 218 Multiplexing of samples in scRNA‐seq in combination with human organoids could be successfully applied to study viral pathogenesis and develop viral vaccines and antiviral therapeutics.
In addition to transcriptomic analysis, epigenomic, proteomic, and metabolic analyses are needed to be integrated into exploring the wide dysregulation of tissues or organs upon SARS‐CoV‐2 infection. 219 , 220 , 221 The paired analyses empowered the study's ability to comprehensively reveal potential mechanisms and regulatory networks that respond to SARS‐CoV‐2 infection. More recently, novel single‐cell multi‐omics have been used to investigate to better understand SARS‐CoV‐2‐induced immune responses. 222 , 223 , 224 , 225 , 226 For instance, scRNA‐seq, CITE‐seq (cellular indexing of transcriptomes and epitopes sequencing), TCR‐seq (T cell receptor sequencing) and BCR‐seq (B cell receptor sequencing) have been utilized to illustrate the dynamic responses of the innate and adaptive immune systems to SARS‐CoV‐2. 227 The high‐throughput immune profiling revealed progressive COVID‐19 was associated with dyssynchrony of the innate and adaptive immune responses. Similarly, Wilk's lab performed scRNA‐seq, scATAC‐seq (single‐cell assay for transposase‐accessible chromatin sequencing), and CyTOF (cytometry by time of flight) on the COVID‐19 patient‐derived peripheral immune cells. 221 The epigenomic, proteomic and transcriptomic profiling revealed the aberrant peripheral immune responses to SARS‐CoV‐2, including neutrophil and natural killer cell hyperactivation. Regarding these, we foresee that multi‐omics technology integrated with organoid models will provide large‐scale information and improve the understanding of virus–host interactions.
7. CONCLUSION REMARKS
The present review aims to reveal the status and research trend of organoid applications systematically and comprehensively for COVID‐19 research, including studying the infection mechanism, vaccine development and drug discovery of SARS‐CoV‐2. Although fruitful data have been obtained, it cannot be denied that the organoid‐based COVID‐19 study is still in its infancy. Human organoids are closely resembling embryonic or fetal organs rather than mature adult organs. It remains to be seen whether organoid maturation could cause diverse cellular responses and susceptibility to viral infections between immature and mature organs. Moreover, differences in different organoid protocols might induce diverse susceptibility to SARS‐CoV‐2 infection. Building advanced human organoids by integrating novel strategies and technologies could address the challenges and promote the development of COVID‐19 research. We hope this study could fully present the status and research trend of organoid‐based SARS‐CoV‐2 study and aid in understanding COVID‐19 pathogenesis and facilitating vaccine development and drug discovery.
AUTHOR CONTRIBUTIONS
Minghui Li: Conceptualization, supervision, data curation, writing review, and editing. Yuhan Yuan, Ting Zou, and Zongkun Hou: Data curation, review, and editing. Liang Jin and Bochu Wang: Conceptualization, supervision, review and editing.
CONFLICT OF INTEREST STATEMENT
The authors declare no potential conflicts of interest.
Supporting information
Appendix S1: Supporting information.
ACKNOWLEDGEMENTS
This work is supported by funding from the National Natural Science Foundation of China (grant number 82204083); the Natural Science Foundation of Chongqing (grant number cstc2021jcyjmsxmX0171).
Li M, Yuan Y, Zou T, Hou Z, Jin L, Wang B. Development trends of human organoid‐based COVID‐19 research based on bibliometric analysis. Cell Prolif. 2023;56(12):e13496. doi: 10.1111/cpr.13496
Contributor Information
Minghui Li, Email: mhli1988@outlook.com.
Liang Jin, Email: jinliang4002@126.com.
Bochu Wang, Email: wangbc@cqu.edu.cn.
DATA AVAILABILITY STATEMENT
The datasets analyzed during the current research are available from the corresponding author on reasonable request.
REFERENCES
- 1. WHO . Who coronavirus (COVID‐19) dashboard. 2022. Available from: https://covid19.who.int/
- 2. Han Y, Yang L, Lacko LA, Chen S. Human organoid models to study SARS‐CoV‐2 infection. Nat Methods. 2022;19:418‐428. [DOI] [PubMed] [Google Scholar]
- 3. Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. SARS‐CoV‐2 omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 2022;7:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thakur V, Ratho RK. Omicron (b.1.1.529): a new SARS‐CoV‐2 variant of concern mounting worldwide fear. J Med Virol. 2022;94:1821‐1824. [DOI] [PubMed] [Google Scholar]
- 5. Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (b.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62:412‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cui Z, Liu P, Wang N, et al. Structural and functional characterizations of infectivity and immune evasion of SARS‐CoV‐2 omicron. Cell. 2022;185:860‐871.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ai J, Zhang H, Zhang Y, et al. Omicron variant showed lower neutralizing sensitivity than other SARS‐CoV‐2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. 2022;11:337‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perez‐Then E, Lucas C, Monteiro VS, et al. Neutralizing antibodies against the SARS‐CoV‐2 delta and omicron variants following heterologous coronavac plus bnt162b2 booster vaccination. Nat Med. 2022;28:481‐485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu X, Wei D, Xu W, et al. Reduced sensitivity of SARS‐CoV‐2 omicron variant to antibody neutralization elicited by booster vaccination. Cell Discov. 2022;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takayama K. In vitro and animal models for SARS‐CoV‐2 research. Trends Pharmacol Sci. 2020;41:513‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang L, Han Y, Nilsson‐Payant BE, et al. A human pluripotent stem cell‐based platform to study SARS‐CoV‐2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125‐136.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu J, Li C, Wang S, Li T, Zhang H. Genetic variants are identified to increase risk of COVID‐19 related mortality from UK biobank data. Hum Genomics. 2021;15:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pairo‐Castineira E, Clohisey S, Klaric L, et al. Genetic mechanisms of critical illness in COVID‐19. Nature. 2021;591:92‐98. [DOI] [PubMed] [Google Scholar]
- 14. Drost J, Clevers H. Translational applications of adult stem cell‐derived organoids. Development. 2017;144:968‐975. [DOI] [PubMed] [Google Scholar]
- 15. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21:571‐584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rookmaaker MB, Schutgens F, Verhaar MC, Clevers H. Development and application of human adult stem or progenitor cell organoids. Nat Rev Nephrol. 2015;11:546‐554. [DOI] [PubMed] [Google Scholar]
- 17. Szabo L, Seubert AC, Kretzschmar K. Modelling adult stem cells and their niche in health and disease with epithelial organoids. Semin Cell Dev Biol. 2023;144:20‐30. [DOI] [PubMed] [Google Scholar]
- 18. Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586‐1597. [DOI] [PubMed] [Google Scholar]
- 19. Schutgens F, Rookmaaker MB, Margaritis T, et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol. 2019;37:303‐313. [DOI] [PubMed] [Google Scholar]
- 20. Schutgens F, Clevers H. Human organoids: tools for understanding biology and treating diseases. Annu Rev Pathol. 2020;15:211‐234. [DOI] [PubMed] [Google Scholar]
- 21. Eiraku M, Takata N, Ishibashi H, et al. Self‐organizing optic‐cup morphogenesis in three‐dimensional culture. Nature. 2011;472:51‐56. [DOI] [PubMed] [Google Scholar]
- 22. Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wimmer RA, Leopoldi A, Aichinger M, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M, Gong J, Gao L, Zou T, Kang J, Xu H. Advanced human developmental toxicity and teratogenicity assessment using human organoid models. Ecotoxicol Environ Saf. 2022;235:113429. [DOI] [PubMed] [Google Scholar]
- 25. Qian X, Nguyen HN, Song MM, et al. Brain‐region‐specific organoids using mini‐bioreactors for modeling zikv exposure. Cell. 2016;165:1238‐1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gabriel E, Ramani A, Karow U, et al. Recent zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell. 2017;20:397‐406.e5. [DOI] [PubMed] [Google Scholar]
- 27. Priyathilaka TT, Laaker CJ, Herbath M, Fabry Z, Sandor M. Modeling infectious diseases of the central nervous system with human brain organoids. Transl Res. 2022;250:18‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu YP, Qiu Y, Zhang B, et al. Zika virus infection induces RNAi‐mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019;29:265‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu M, Lee EM, Wen Z, et al. Identification of small‐molecule inhibitors of zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101‐1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li C, Deng YQ, Wang S, et al. 25‐hydroxycholesterol protects host against zika virus infection and its associated microcephaly in a mouse model. Immunity. 2017;46:446‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen C, Song M. Visualizing a field of research: a methodology of systematic scientometric reviews. PLoS One. 2019;14:e0223994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Y, Zhang X, Chen S, et al. Bibliometric analysis of mental health during the COVID‐19 pandemic. Asian J Psychiatr. 2021;65:102846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gu C, Wang Z, Pan Y, Zhu S, Gu Z. Tungsten‐based nanomaterials in the biomedical field: a bibliometric analysis of research progress and prospects. Adv Mater. 2022;35:2204397. [DOI] [PubMed] [Google Scholar]
- 34. Li C, Zhu X, Zhang Y, Zhang J, Jeon CO, Jia B. COVID‐19 influences both physical and mental health: lessons from bibliometric analysis. Travel Med Infect Dis. 2022;49:102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bornmann L, Daniel HD. The state of h index research. Is the h index the ideal way to measure research performance? EMBO Rep. 2009;10:2‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hirsch JE. Does the h index have predictive power? Proc Natl Acad Sci U S A. 2007;104:19193‐19198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boyack KW, Klavans R. Co‐citation analysis, bibliographic coupling, and direct citation: which citation approach represents the research front most accurately? J Am Soc Inform Sci Technol. 2010;61:2389‐2404. [Google Scholar]
- 38. Trujillo CM, Long TM. Document co‐citation analysis to enhance transdisciplinary research. Sci Adv. 2018;4:e1701130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han Y, Duan X, Yang L, et al. Identification of SARS‐CoV‐2 inhibitors using lung and colonic organoids. Nature. 2021;589:270‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lamers MM, Beumer J, van der Vaart J, et al. SARS‐CoV‐2 productively infects human gut enterocytes. Science. 2020;369:50‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ace2. Cell. 2020;181:905‐913.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Basil MC, Katzen J, Engler AE, et al. The cellular and physiological basis for lung repair and regeneration: past, present, and future. Cell Stem Cell. 2020;26:482‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Upadhya S, Rehman J, Malik AB, Chen S. Mechanisms of lung injury induced by SARS‐CoV‐2 infection. Physiology (Bethesda). 2022;37:88‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ace2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016‐1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pei R, Feng J, Zhang Y, et al. Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS‐CoV‐2 infection. Protein Cell. 2021;12:717‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salahudeen AA, Choi SS, Rustagi A, et al. Progenitor identification and SARS‐CoV‐2 infection in human distal lung organoids. Nature. 2020;588:670‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sano E, Suzuki T, Hashimoto R, et al. Cell response analysis in SARS‐CoV‐2 infected bronchial organoids. Commun Biol. 2022;5:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jacob F, Pather SR, Huang WK, et al. Human pluripotent stem cell‐derived neural cells and brain organoids reveal SARS‐CoV‐2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27:937‐950.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen KG, Park K, Spence JR. Studying SARS‐CoV‐2 infectivity and therapeutic responses with complex organoids. Nat Cell Biol. 2021;23:822‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chiu MC, Li C, Liu X, et al. Human nasal organoids model SARS‐CoV‐2 upper respiratory infection and recapitulate the differential infectivity of emerging variants. MBio. 2022;13:e0194422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Deng X, Garcia‐Knight MA, Khalid MM, et al. Transmission, infectivity, and neutralization of a spike l452r SARS‐CoV‐2 variant. Cell. 2021;184:3426‐3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Honzke K, Obermayer B, Mache C, et al. Human lungs show limited permissiveness for SARS‐CoV‐2 due to scarce ace2 levels but virus‐induced expansion of inflammatory macrophages. Eur Respir J. 2022;60:2102725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lamers MM, van der Vaart J, Knoops K, et al. An organoid‐derived bronchioalveolar model for SARS‐CoV‐2 infection of human alveolar type ii‐like cells. EMBO J. 2021;40:e105912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mlcochova P, Kemp SA, Dhar MS, et al. SARS‐CoV‐2 b.1.617.2 delta variant replication and immune evasion. Nature. 2021;599:114‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ahn JH, Kim J, Hong SP, et al. Nasal ciliated cells are primary targets for SARS‐CoV‐2 replication in the early stage of COVID‐19. J Clin Invest. 2021;131:e148517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu CT, Lidsky PV, Xiao Y, et al. SARS‐CoV‐2 replication in airway epithelia requires motile cilia and microvillar reprogramming. Cell. 2023;186:112‐130.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Elbadawi M, Efferth T. Organoids of human airways to study infectivity and cytopathy of SARS‐CoV‐2. Lancet Respir Med. 2020;8:e55‐e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sachs N, Papaspyropoulos A, Zomer‐van Ommen DD, et al. Long‐term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duan X, Tang X, Nair MS, et al. An airway organoid‐based screen identifies a role for the hif1alpha‐glycolysis axis in SARS‐CoV‐2 infection. Cell Rep. 2021;37:109920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181:1036‐1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Suzuki T, Itoh Y, Sakai Y, et al. Generation of human bronchial organoids for SARS‐CoV‐2 research. bioRxiv. 2020. [Google Scholar]
- 63. Chen F, Fine A. Stem cells in lung injury and repair. Am J Pathol. 2016;186:2544‐2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fang KY, Cao WC, Xie TA, et al. Exploration and validation of related hub gene expression during SARS‐CoV‐2 infection of human bronchial organoids. Hum Genomics. 2021;15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chiu MC, Li C, Liu X, et al. A bipotential organoid model of respiratory epithelium recapitulates high infectivity of SARS‐CoV‐2 omicron variant. Cell Discov. 2022;8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tiwari SK, Wang S, Smith D, Carlin AF, Rana TM. Revealing tissue‐specific SARS‐CoV‐2 infection and host responses using human stem cell‐derived lung and cerebral organoids. Stem Cell Rep. 2021;16:437‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Katsura H, Sontake V, Tata A, et al. Human lung stem cell‐based alveolospheres provide insights into SARS‐CoV‐2‐mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell. 2020;27:890‐904.e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Youk J, Kim T, Evans KV, et al. Three‐dimensional human alveolar stem cell culture models reveal infection response to SARS‐CoV‐2. Cell Stem Cell. 2020;27:905‐919.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chan M. Hkumed finds omicron SARS‐CoV‐2 can infect faster and better than delta in human bronchus but with less severe infection in lung. Braz J Implantol Health Sci. 2022;4:50‐54. [Google Scholar]
- 70. Ellul MA, Benjamin L, Singh B, et al. Neurological associations of COVID‐19. Lancet Neurol. 2020;19:767‐783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Iadecola C, Anrather J, Kamel H. Effects of COVID‐19 on the nervous system. Cell. 2020;183:16‐27.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kral AH, Lambdin BH, Wenger LD, Davidson PJ. Evaluation of an unsanctioned safe consumption site in the United States. N Engl J Med. 2020;383:589‐590. [DOI] [PubMed] [Google Scholar]
- 73. Vanderver A, Adang L, Gavazzi F, et al. Janus kinase inhibition in the aicardi‐goutieres syndrome. N Engl J Med. 2020;383:986‐989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhan M, Qin Y, Xue X, Zhu S. Death from COVID‐19 of 23 health care workers in China. N Engl J Med. 2020;382:2267‐2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ramani A, Muller L, Ostermann PN, et al. SARS‐CoV‐2 targets neurons of 3d human brain organoids. EMBO J. 2020;39:e106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Song E, Zhang C, Israelow B, et al. Neuroinvasion of SARS‐CoV‐2 in human and mouse brain. J Exp Med. 2021;218:e20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cugola FR, Fernandes IR, Russo FB, et al. The brazilian zika virus strain causes birth defects in experimental models. Nature. 2016;534:267‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hou Y, Li C, Yoon C, et al. Enhanced replication of SARS‐CoV‐2 omicron ba.2 in human forebrain and midbrain organoids. Signal Transduct Target Ther. 2022;7:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pellegrini L, Albecka A, Mallery DL, et al. SARS‐CoV‐2 infects the brain choroid plexus and disrupts the blood‐csf barrier in human brain organoids. Cell Stem Cell. 2020;27:951‐961.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen R, Wang K, Yu J, et al. The spatial and cell‐type distribution of SARS‐CoV‐2 receptor ace2 in the human and mouse brains. Front Neurol. 2020;11:573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang BZ, Chu H, Han S, et al. SARS‐CoV‐2 infects human neural progenitor cells and brain organoids. Cell Res. 2020;30:928‐931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang C, Zhang M, Garcia G Jr, et al. Apoe‐isoform‐dependent SARS‐CoV‐2 neurotropism and cellular response. Cell Stem Cell. 2021;28:331‐342.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ferm S, Fisher C, Pakala T, et al. Analysis of gastrointestinal and hepatic manifestations of SARS‐CoV‐2 infection in 892 patients in queens. Clin Gastroenterol Hepatol. 2020;18:2378‐2379.e2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Guo M, Tao W, Flavell RA, Zhu S. Potential intestinal infection and faecal‐oral transmission of SARS‐CoV‐2. Nat Rev Gastroenterol Hepatol. 2021;18:269‐283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pan L, Mu M, Yang P, et al. Clinical characteristics of COVID‐19 patients with digestive symptoms in Hubei, China: a descriptive, cross‐sectional, multicenter study. Am J Gastroenterol. 2020;115:766‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA. 2020;323:1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS‐CoV‐2. Gastroenterology. 2020;158:1831‐1833.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kruger J, Gross R, Conzelmann C, et al. Drug inhibition of SARS‐CoV‐2 replication in human pluripotent stem cell‐derived intestinal organoids. Cell Mol Gastroenterol Hepatol. 2021;11:935‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhao Y, Ren X, Lu J, et al. Mechanistic insight of SARS‐CoV‐2 infection using human hepatobiliary organoids. Gut. 2022;72:216‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Garreta E, Prado P, Stanifer ML, et al. A diabetic milieu increases ace2 expression and cellular susceptibility to SARS‐CoV‐2 infections in human kidney organoids and patient cells. Cell Metab. 2022;34:857‐873.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Menuchin‐Lasowski Y, Schreiber A, Lecanda A, et al. SARS‐CoV‐2 infects and replicates in photoreceptor and retinal ganglion cells of human retinal organoids. Stem Cell Rep. 2022;17:789‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tanaka J, Senpuku H, Ogawa M, et al. Human induced pluripotent stem cell‐derived salivary gland organoids model SARS‐CoV‐2 infection and replication. Nat Cell Biol. 2022;24:1595‐1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Giobbe GG, Bonfante F, Jones BC, et al. SARS‐CoV‐2 infection and replication in human gastric organoids. Nat Commun. 2021;12:6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jang KK, Kaczmarek ME, Dallari S, et al. Variable susceptibility of intestinal organoid‐derived monolayers to SARS‐CoV‐2 infection. PLoS Biol. 2022;20:e3001592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID‐19: a randomised, double‐blind, placebo‐controlled, multicentre trial. Lancet. 2020;395:1569‐1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhou J, Li C, Liu X, et al. Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nat Med. 2020;26:1077‐1083. [DOI] [PubMed] [Google Scholar]
- 97. Zang R, Gomez Castro MF, McCune BT, et al. Tmprss2 and tmprss4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5:eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Heuberger J, Trimpert J, Vladimirova D, et al. Epithelial response to IFN‐gamma promotes SARS‐CoV‐2 infection. EMBO Mol Med. 2021;13:e13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhao Z, Chen X, Dowbaj AM, et al. Organoids. Nat Rev Methods Prim. 2022;2:94. doi: 10.1038/s43586-022-00174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Triana S, Metz‐Zumaran C, Ramirez C, et al. Single‐cell analyses reveal SARS‐CoV‐2 interference with intrinsic immune response in the human gut. Mol Syst Biol. 2021;17:e10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fan Z, Chen L, Li J, et al. Clinical features of COVID‐19‐related liver functional abnormality. Clin Gastroenterol Hepatol. 2020;18:1561‐1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Richards A, Friesen M, Khalil A, Barrasa MI, Gehrke L, Jaenisch R. SARS‐CoV‐2 infection of human pluripotent stem cell‐derived liver organoids reveals potential mechanisms of liver pathology. iScience. 2022;25:105146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhao B, Ni C, Gao R, et al. Recapitulation of SARS‐CoV‐2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Soler MJ, Jacobs‐Cacha C. The COVID‐19 pandemic: progress in nephrology. Nat Rev Nephrol. 2022;18:80‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Li Z, Wu M, Yao J, Guo J, Liao X, Song S, et al. Caution on kidney dysfunctions of COVID‐19 patients. medRXiv. 2020. [Google Scholar]
- 108. Pan XW, Xu D, Zhang H, Zhou W, Wang LH, Cui XG. Identification of a potential mechanism of acute kidney injury during the COVID‐19 outbreak: a study based on single‐cell transcriptome analysis. Intensive Care Med. 2020;46:1114‐1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xia S, Wu M, Chen S, et al. Long term culture of human kidney proximal tubule epithelial cells maintains lineage functions and serves as an ex vivo model for coronavirus associated kidney injury. Virol Sin. 2020;35:311‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jansen J, Reimer KC, Nagai JS, et al. SARS‐CoV‐2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell. 2022;29:217‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guo T, Fan Y, Chen M, et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID‐19). JAMA Cardiol. 2020;5:811‐818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID‐19 in Wuhan, China. JAMA Cardiol. 2020;5:802‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Arhontoulis DC, Kerr CM, Richards D, et al. Human cardiac organoids to model COVID‐19 cytokine storm induced cardiac injuries. J Tissue Eng Regen Med. 2022;16:799‐811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Khan AO, Reyat JS, Hill H, et al. Preferential uptake of SARS‐CoV‐2 by pericytes potentiates vascular damage and permeability in an organoid model of the microvasculature. Cardiovasc Res. 2022;118:3085‐3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Mills RJ, Humphrey SJ, Fortuna PRJ, et al. Bet inhibition blocks inflammation‐induced cardiac dysfunction and SARS‐CoV‐2 infection. Cell. 2021;184:2167‐2182.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Eriksen AZ, Moller R, Makovoz B, Uhl SA, tenOever BR, Blenkinsop TA. SARS‐CoV‐2 infects human adult donor eyes and hesc‐derived ocular epithelium. Cell Stem Cell. 2021;28:1205‐1220.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ahmad Mulyadi Lai HI, Chou SJ, Chien Y, et al. Expression of endogenous angiotensin‐converting enzyme 2 in human induced pluripotent stem cell‐derived retinal organoids. Int J Mol Sci. 2021;22:1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xu J, Li Y, Gan F, Du Y, Yao Y. Salivary glands: potential reservoirs for COVID‐19 asymptomatic infection. J Dent Res. 2020;99:989. [DOI] [PubMed] [Google Scholar]
- 119. Li Y, Tenchov R, Smoot J, Liu C, Watkins S, Zhou Q. A comprehensive review of the global efforts on COVID‐19 vaccine development. ACS Cent Sci. 2021;7:512‐533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS‐CoV‐2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Krammer F. SARS‐CoV‐2 vaccines in development. Nature. 2020;586:516‐527. [DOI] [PubMed] [Google Scholar]
- 122. Thanh Le T, Andreadakis Z, Kumar A, et al. The COVID‐19 vaccine development landscape. Nat Rev Drug Discov. 2020;19:305‐306. [DOI] [PubMed] [Google Scholar]
- 123. WHO . COVID‐19 vaccine tracker and landscape. 2022. Available from: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 124. Busquet F, Hartung T, Pallocca G, Rovida C, Leist M. Harnessing the power of novel animal‐free test methods for the development of COVID‐19 drugs and vaccines. Arch Toxicol. 2020;94:2263‐2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Herati RS, Wherry EJ. What is the predictive value of animal models for vaccine efficacy in humans? Consideration of strategies to improve the value of animal models. Cold Spring Harb Perspect Biol. 2018;10:a031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wagar LE, Salahudeen A, Constantz CM, et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med. 2021;27:125‐135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the merck HIV‐1 vaccine in humans. Nat Med. 2008;14:617‐621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor‐binding site of SARS‐CoV‐2. Nature. 2020;584:120‐124. [DOI] [PubMed] [Google Scholar]
- 129. Spitalieri P, Centofanti F, Murdocca M, et al. Two different therapeutic approaches for SARS‐CoV‐2 in hiPSCs‐derived lung organoids. Cells. 2022;11:1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Giese C, Marx U. Human immunity in vitro—solving immunogenicity and more. Adv Drug Deliv Rev. 2014;69‐70:103‐122. [DOI] [PubMed] [Google Scholar]
- 131. Goyal G, Prabhala P, Mahajan G, et al. Ectopic lymphoid follicle formation and human seasonal influenza vaccination responses recapitulated in an organ‐on‐a‐chip. Adv Sci (Weinh). 2022;9:e2103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hammel JH, Zatorski JM, Cook SR, Pompano RR, Munson JM. Engineering in vitro immune‐competent tissue models for testing and evaluation of therapeutics. Adv Drug Deliv Rev. 2022;182:114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mesci P, de Souza JS, Martin‐Sancho L, et al. SARS‐CoV‐2 infects human brain organoids causing cell death and loss of synapses that can be rescued by treatment with sofosbuvir. PLoS Biol. 2022;20:e3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wagar L. Small centers of defense. Science. 2022;375:830. [DOI] [PubMed] [Google Scholar]
- 135. Duarte RRR, Copertino DC Jr, Iniguez LP, et al. Identifying fda‐approved drugs with multimodal properties against COVID‐19 using a data‐driven approach and a lung organoid model of SARS‐CoV‐2 entry. Mol Med. 2021;27:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Peng L, Gao L, Wu X, et al. Lung organoids as model to study SARS‐CoV‐2 infection. Cells. 2022;11:2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Xu G, Li Y, Zhang S, et al. SARS‐CoV‐2 promotes ripk1 activation to facilitate viral propagation. Cell Res. 2021;31:1230‐1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Huang KY, Lin MS, Kuo TC, et al. Humanized COVID‐19 decoy antibody effectively blocks viral entry and prevents SARS‐CoV‐2 infection. EMBO Mol Med. 2021;13:e12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Samuel RM, Majd H, Richter MN, et al. Androgen signaling regulates SARS‐CoV‐2 receptor levels and is associated with severe COVID‐19 symptoms in men. Cell Stem Cell. 2020;27:876‐889.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mulay A, Konda B, Garcia G Jr, et al. SARS‐CoV‐2 infection of primary human lung epithelium for COVID‐19 modeling and drug discovery. Cell Rep. 2021;35:109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of COVID‐19—final report. N Engl J Med. 2020;383:1813‐1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhang M, Wang P, Luo R, et al. Biomimetic human disease model of SARS‐CoV‐2‐induced lung injury and immune responses on organ chip system. Adv Sci (Weinh). 2021;8:2002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Stanifer ML, Kee C, Cortese M, et al. Critical role of type iii interferon in controlling SARS‐CoV‐2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Andreakos E, Tsiodras S. COVID‐19: lambda interferon against viral load and hyperinflammation. EMBO Mol Med. 2020;12:e12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Jhuti D, Rawat A, Guo CM, Wilson LA, Mills EJ, Forrest JI. Interferon treatments for SARS‐CoV‐2: challenges and opportunities. Infect Dis Ther. 2022;11:953‐972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Si L, Bai H, Rodas M, et al. A human‐airway‐on‐a‐chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng. 2021;5:815‐829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Samudyata OAO, Malwade S, Rufino de Sousa N, Goparaju SK, Gracias J, et al. SARS‐CoV‐2 promotes microglial synapse elimination in human brain organoids. Mol Psychiatry. 2022;27:3939‐3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wang L, Sievert D, Clark AE, et al. A human three‐dimensional neural‐perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS‐CoV‐2 neuropathology. Nat Med. 2021;27:1600‐1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Guo Y, Luo R, Wang Y, et al. SARS‐CoV‐2 induced intestinal responses with a biomimetic human gut‐on‐chip. Sci Bull (Beijing). 2021;66:783‐793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs‐on‐chips. Trends Cell Biol. 2011;21:745‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhang YS, Aleman J, Shin SR, et al. Multisensor‐integrated organs‐on‐chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A. 2017;114:E2293‐E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Xia S, Liu M, Wang C, et al. Inhibition of SARS‐CoV‐2 (previously 2019–nCoV) infection by a highly potent pan‐coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wang S, Li W, Hui H, et al. Cholesterol 25‐hydroxylase inhibits SARS‐CoV‐2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39:e106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Freedberg DE, Conigliaro J, Wang TC, et al. Famotidine use is associated with improved clinical outcomes in hospitalized COVID‐19 patients: a propensity score matched retrospective cohort study. Gastroenterology. 2020;159:1129‐1131.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Prelli Bozzo C, Nchioua R, Volcic M, et al. Ifitm proteins promote SARS‐CoV‐2 infection and are targets for virus inhibition in vitro. Nat Commun. 2021;12:4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Brevini T, Maes M, Webb GJ, et al. Fxr inhibition may protect from SARS‐CoV‐2 infection by reducing ace2. Nature. 2023;615:134‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wysocki J, Ye M, Hassler L, et al. A novel soluble ace2 variant with prolonged duration of action neutralizes SARS‐CoV‐2 infection in human kidney organoids. J Am Soc Nephrol. 2021;32:795‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Monteil V, Dyczynski M, Lauschke VM, et al. Human soluble ace2 improves the effect of remdesivir in SARS‐CoV‐2 infection. EMBO Mol Med. 2021;13:e13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Calistri A, Luganini A, Mognetti B, et al. The new generation hdhodh inhibitor meds433 hinders the in vitro replication of SARS‐CoV‐2 and other human coronaviruses. Microorganisms. 2021;9:1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kim HK, Kim H, Lee MK, et al. Generation of tonsil organoids as an ex vivo model for SARS‐CoV‐2 infection. Biomaterials. 2020;283:121460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gao CC, Li M, Deng W, et al. Differential transcriptomic landscapes of multiple organs from SARS‐CoV‐2 early infected rhesus macaques. Protein Cell. 2022;13:920‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Liu J, Li Y, Liu Q, et al. SARS‐CoV‐2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Vabret N, Britton GJ, Gruber C, et al. Immunology of COVID‐19: current state of the science. Immunity. 2020;52:910‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Yang L, Han Y, Jaffre F, et al. An immuno‐cardiac model for macrophage‐mediated inflammation in COVID‐19 hearts. Circ Res. 2021;129:33‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Muffat J, Li Y, Omer A, et al. Human induced pluripotent stem cell‐derived glial cells and neural progenitors display divergent responses to zika and dengue infections. Proc Natl Acad Sci U S A. 2018;115:7117‐7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Ormel PR, Vieira de Sa R, van Bodegraven EJ, et al. Microglia innately develop within cerebral organoids. Nat Commun. 2018;9:4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Popova G, Soliman SS, Kim CN, et al. Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell. 2021;28:2153‐2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Xu R, Boreland AJ, Li X, et al. Developing human pluripotent stem cell‐based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. Stem Cell Reports. 2021;16:1923‐1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Zhang W, Jiang J, Xu Z, et al. Microglia‐containing human brain organoids for the study of brain development and pathology. Mol Psychiatry. 2022;28:96‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Awogbindin IO, Ben‐Azu B, Olusola BA, et al. Microglial implications in SARS‐CoV‐2 infection and COVID‐19: lessons from viral RNA neurotropism and possible relevance to parkinson's disease. Front Cell Neurosci. 2021;15:670298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Frank MG, Nguyen KH, Ball JB, et al. SARS‐CoV‐2 spike s1 subunit induces neuroinflammatory, microglial and behavioral sickness responses: evidence of pamp‐like properties. Brain Behav Immun. 2022;100:267‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Poloni TE, Medici V, Moretti M, et al. COVID‐19‐related neuropathology and microglial activation in elderly with and without dementia. Brain Pathol. 2021;31:e12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. DeFalco T, Bhattacharya I, Williams AV, Sams DM, Capel B. Yolk‐sac‐derived macrophages regulate fetal testis vascularization and morphogenesis. Proc Natl Acad Sci U S A. 2014;111:E2384‐E2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ewald ML, Chen YH, Lee AP, Hughes CCW. The vascular niche in next generation microphysiological systems. Lab Chip. 2021;21:3244‐3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Grebenyuk S, Ranga A. Engineering organoid vascularization. Front Bioeng Biotechnol. 2019;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Li M, Gao L, Zhao L, Zou T, Xu H. Toward the next generation of vascularized human neural organoids. Med Res Rev. 2023;43:31‐54. [DOI] [PubMed] [Google Scholar]
- 177. Vargas‐Valderrama A, Messina A, Mitjavila‐Garcia MT, Guenou H. The endothelium, a key actor in organ development and hpsc‐derived organoid vascularization. J Biomed Sci. 2020;27:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Bargehr J, Rericha P, Petchey A, et al. Cardiovascular ace2 receptor expression in patients undergoing heart transplantation. ESC Heart Fail. 2021;8:4119‐4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Buzhdygan TP, DeOre BJ, Baldwin‐Leclair A, et al. The SARS‐CoV‐2 spike protein alters barrier function in 2d static and 3d microfluidic in‐vitro models of the human blood‐brain barrier. Neurobiol Dis. 2020;146:105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Hashimoto R, Takahashi J, Shirakura K, et al. SARS‐CoV‐2 disrupts respiratory vascular barriers by suppressing claudin‐5 expression. Sci Adv. 2022;8:eabo6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Lee SH, Sung JH. Organ‐on‐a‐chip technology for reproducing multiorgan physiology. Adv Healthc Mater. 2018;7:1700419. [DOI] [PubMed] [Google Scholar]
- 182. Ching T, Toh YC, Hashimoto M, Zhang YS. Bridging the academia‐to‐industry gap: organ‐on‐a‐chip platforms for safety and toxicology assessment. Trends Pharmacol Sci. 2021;42:715‐728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Truskey GA. Human microphysiological systems and organoids as in vitro models for toxicological studies. Front Public Health. 2018;6:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Picollet‐D'hahan N, Zuchowska A, Lemeunier I, Le Gac S. Multiorgan‐on‐a‐chip: a systemic approach to model and decipher inter‐organ communication. Trends Biotechnol. 2021;39:788‐810. [DOI] [PubMed] [Google Scholar]
- 185. Ramezankhani R, Solhi R, Chai YC, Vosough M, Verfaillie C. Organoid and microfluidics‐based platforms for drug screening in COVID‐19. Drug Discov Today. 2022;27:1062‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Goncalves IM, Carvalho V, Rodrigues RO, et al. Organ‐on‐a‐chip platforms for drug screening and delivery in tumor cells: a systematic review. Cancers (Basel). 2022;14:935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Clarke GA, Hartse BX, Niaraki Asli AE, et al. Advancement of sensor integrated organ‐on‐chip devices. Sensors (Basel). 2021;21:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Lopez‐Muñoz GA, Mughal S, Ramón‐Azcón J. Sensors and biosensors in organs‐on‐a‐chip platforms. Adv Exp Med Biol. 2022;1379:55‐80. [DOI] [PubMed] [Google Scholar]
- 189. Bost P, Giladi A, Liu Y, et al. Host–viral infection maps reveal signatures of severe COVID‐19 patients. Cell. 2020;181:1475‐1488.e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Ho DW, Tsui YM, Chan LK, et al. Single‐cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of hbv‐associated hepatocellular carcinoma. Nat Commun. 2021;12:3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kotliar D, Lin AE, Logue J, et al. Single‐cell profiling of ebola virus disease in vivo reveals viral and host dynamics. Cell. 2020;183:1383‐1401.e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Gao KM, Derr AG, Guo Z, et al. Human nasal wash RNA‐seq reveals distinct cell‐specific innate immune responses in influenza versus SARS‐CoV‐2. JCI Insight. 2021;6:e152288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Steuerman Y, Cohen M, Peshes‐Yaloz N, et al. Dissection of influenza infection in vivo by single‐cell RNA sequencing. Cell Syst. 2018;6:679‐691.e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Patiño M, Lagos WN, Patne NS, Tasic B, Zeng H, Callaway EM. Single‐cell transcriptomic classification of rabies‐infected cortical neurons. Proc Natl Acad Sci U S A. 2022;119:e2203677119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Kazer SW, Aicher TP, Muema DM, et al. Integrated single‐cell analysis of multicellular immune dynamics during hyperacute HIV‐1 infection. Nat Med. 2020;26:511‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Wang S, Zhang Q, Hui H, Agrawal K, Karris MAY, Rana TM. An atlas of immune cell exhaustion in HIV‐infected individuals revealed by single‐cell transcriptomics. Emerg Microbes Infect. 2020;9:2333‐2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Nowakowski TJ, Pollen AA, Di Lullo E, Sandoval‐Espinosa C, Bershteyn M, Kriegstein AR. Expression analysis highlights axl as a candidate zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016;18:591‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Onorati M, Li Z, Liu F, et al. Zika virus disrupts phospho‐tbk1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016;16:2576‐2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zanini F, Pu SY, Bekerman E, Einav S, Quake SR. Single‐cell transcriptional dynamics of flavivirus infection. Elife. 2018;7:e32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS‐CoV‐2 identified by high‐throughput single‐cell sequencing of convalescent patients' b cells. Cell. 2020;182:73‐84.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Grant RA, Morales‐Nebreda L, Markov NS, et al. Circuits between infected macrophages and t cells in SARS‐CoV‐2 pneumonia. Nature. 2021;590:635‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Liao M, Liu Y, Yuan J, et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020;26:842‐844. [DOI] [PubMed] [Google Scholar]
- 203. Melms JC, Biermann J, Huang H, et al. A molecular single‐cell lung atlas of lethal COVID‐19. Nature. 2021;595:114‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Ren X, Wen W, Fan X, et al. COVID‐19 immune features revealed by a large‐scale single‐cell transcriptome atlas. Cell. 2021;184:1895‐1913.e1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Sinha S, Rosin NL, Arora R, et al. Dexamethasone modulates immature neutrophils and interferon programming in severe COVID‐19. Nat Med. 2022;28:201‐211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Sungnak W, Huang N, Bécavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Tang X, Uhl S, Zhang T, et al. SARS‐CoV‐2 infection induces beta cell transdifferentiation. Cell Metab. 2021;33:1577‐1591.e1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Cristinelli S, Ciuffi A. The use of single‐cell RNA‐seq to understand virus–host interactions. Curr Opin Virol. 2018;29:39‐50. [DOI] [PubMed] [Google Scholar]
- 209. Justet A, Zhao AY, Kaminski N. From COVID to fibrosis: lessons from single‐cell analyses of the human lung. Hum Genomics. 2022;16:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Ratnasiri K, Wilk AJ, Lee MJ, Khatri P, Blish CA. Single‐cell RNA‐seq methods to interrogate virus–host interactions. Semin Immunopathol. 2022;45:71‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Song H, Seddighzadeh B, Cooperberg MR, Huang FW. Expression of ace2, the SARS‐CoV‐2 receptor, and tmprss2 in prostate epithelial cells. Eur Urol. 2020;78:296‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Zhao Y, Zhao Z, Wang Y, Zhou Y, Ma Y, Zuo W. Single‐cell RNA expression profiling of ace2, the receptor of SARS‐CoV‐2. Am J Respir Crit Care Med. 2020;202:756‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Zhou L, Niu Z, Jiang X, et al. SARS‐CoV‐2 targets by the pscrna profiling of ace2, tmprss2 and furin proteases. iScience. 2020;23:101744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Aguilar C, Alves da Silva M, Saraiva M, Neyazi M, Olsson IAS, Bartfeld S. Organoids as host models for infection biology—a review of methods. Exp Mol Med. 2021;53:1471‐1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Dutta D, Clevers H. Organoid culture systems to study host–pathogen interactions. Curr Opin Immunol. 2017;48:15‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Hicks SC, Townes FW, Teng M, Irizarry RA. Missing data and technical variability in single‐cell RNA‐sequencing experiments. Biostatistics (Oxford, England). 2018;19:562‐578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Mylka V, Matetovici I, Poovathingal S, et al. Comparative analysis of antibody‐ and lipid‐based multiplexing methods for single‐cell RNA‐seq. Genome Biol. 2022;23:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Shin D, Lee W, Lee JH, Bang D. Multiplexed single‐cell RNA‐seq via transient barcoding for simultaneous expression profiling of various drug perturbations. Sci Adv. 2019;5:eaav2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Byeon SK, Madugundu AK, Garapati K, et al. Development of a multiomics model for identification of predictive biomarkers for COVID‐19 severity: a retrospective cohort study. Lancet Digit Health. 2022;4:e632‐e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Du M, Cai G, Chen F, Christiani DC, Zhang Z, Wang M. Multiomics evaluation of gastrointestinal and other clinical characteristics of COVID‐19. Gastroenterology. 2020;158:2298‐2301.e2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Wilk AJ, Lee MJ, Wei B, et al. Multi‐omic profiling reveals widespread dysregulation of innate immunity and hematopoiesis in COVID‐19. J Exp Med. 2021;218:e20210582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Bernardes JP, Mishra N, Tran F, et al. Longitudinal multi‐omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID‐19. Immunity. 2020;53:1296‐1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Li CX, Gao J, Zhang Z, et al. Multiomics integration‐based molecular characterizations of COVID‐19. Brief Bioinform. 2022;23:bbab485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Tan HW, Xu YM, Lau ATY. Human bronchial‐pulmonary proteomics in coronavirus disease 2019 (COVID‐19) pandemic: applications and implications. Expert Rev Proteomics. 2021;18:925‐938. [DOI] [PubMed] [Google Scholar]
- 225. Tang Z, Fan W, Li Q, et al. Mvip: multi‐omics portal of viral infection. Nucleic Acids Res. 2022;50:D817‐D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Tian Y, Carpp LN, Miller HER, Zager M, Newell EW, Gottardo R. Single‐cell immunology of SARS‐CoV‐2 infection. Nat Biotechnol. 2022;40:30‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Unterman A, Sumida TS, Nouri N, et al. Single‐cell multi‐omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID‐19. Nat Commun. 2022;13:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information.
Data Availability Statement
The datasets analyzed during the current research are available from the corresponding author on reasonable request.


