Abstract
Chronic pulmonary infection with Pseudomonas aeruginosa is a common and serious problem in patients with cystic fibrosis (CF). The P. aeruginosa isolates from these patients typically have a mucoid colony morphology due to overproduction of the exopolysaccharide alginate, which contributes to the persistence of the organisms in the CF lung. Most of the alginate biosynthetic genes are clustered in the algD operon, located at 34 min on the chromosome. Alginate biosynthesis begins with the formation of an activated monomer, GDP-mannuronate, which is known to occur via the products of the algA, algC, and algD genes. Polymannuronate forms in the periplasm, but the gene products involved in mannuronate translocation across the inner membrane and its polymerization are not known. One locus of the operon which remained uncharacterized was a new gene called algK between alg44 and algE. We sequenced algK from the mucoid CF isolate FRD1 and expressed it in Escherichia coli, which revealed a polypeptide of the predicted size (52 kDa). The sequence of AlgK showed an apparent signal peptide characteristic of a lipoprotein. AlgK-PhoA fusion proteins were constructed and shown to be active, indicating that AlgK has a periplasmic subcellular localization. To test the phenotype of an AlgK− mutant, the algK coding sequence was replaced with a nonpolar gentamicin resistance cassette to avoid polar effects on genes downstream of algK that are essential for polymer formation. The algKΔ mutant was nonmucoid, demonstrating that AlgK was required for alginate production. Also, AlgK− mutants demonstrated a small-colony phenotype on L agar, suggesting that the loss of AlgK also caused a growth defect. The mutant phenotypes were complemented by a plasmid expressing algK in trans. When the algKΔ mutation was placed in an algJ::Tn501 background, where algA was not expressed due to polar transposon effects, the growth defect was not observed. AlgK− mutants appeared to accumulate a toxic extracellular product, and we hypothesized that this could be an unpolymerized alginate precursor. High levels of low-molecular-weight uronic acid were produced by the AlgK− mutant. When AlgK− culture supernatants were subjected to dialysis, high levels of uronic acids diffused out of the dialysis sac, and no uronic acids were detectable after extensive dialysis. In contrast, the mucoid wild-type strain produced only polymerized uronic acids (i.e., alginate), whereas the algKΔ algJ::Tn501 mutant produced no uronic acids. Thus, the alginate pathway in an AlgK− mutant was blocked after transport but at a step before polymerization, suggesting that AlgK plays an important role in the polymerization of mannuronate to alginate.
Chronic pulmonary disease in patients with cystic fibrosis is commonly caused by mucoid strains of Pseudomonas aeruginosa that overproduce an exopolysaccharide called alginate. Conversion to the mucoid phenotype occurs as the result of mutations in alginate regulatory loci that may encode a stress response system (14, 16, 24, 25, 29). Alginate has been implicated in facilitating the persistence of P. aeruginosa in the lungs by conferring an antiphagocytic barrier (3, 37), frustrating phagocytic cells (42), obstructing the bronchioles by its high viscosity (17), and providing a mechanism for adherence (23, 32) leading to the formation of biofilms (21). Additionally, the high resistance of P. aeruginosa to antibiotics makes these strains intractable, and they can overwhelm the cystic fibrosis patient by their sheer numbers (16, 41).
Alginate is a linear polymer of high molecular weight composed of the uronic acids β-d-mannuronate (M) and its C-5 epimer, α-L-guluronate (G), which are linked by β-1,4 glycosidic bonds (9). Analysis of P. aeruginosa alginate by nuclear magnetic resonance shows that the two moieties are present in blocks of poly-M and poly-MG but not poly-G (6). Most of the genes required for alginate biosynthesis are in the algD biosynthetic operon at 34 min on the 75-min P. aeruginosa chromosome (6, 8). The only known exception is algC, at 10 min on the chromosome, which encodes phosphomannose mutase, an enzyme that is also involved in lipopolysaccharide biosynthesis (15, 43).
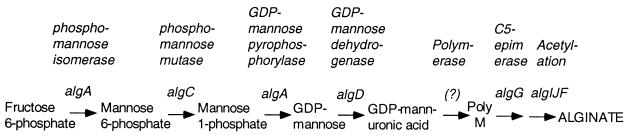
The early steps in the biosynthesis of alginate to form GDP-mannuronic acid (Fig. 1) require the products of algA and algC to convert fructose-6-phosphate to GDP-mannose and then require the product of algD to convert this to the uronic acid form, GDP-mannuronate (for a review, see reference 26). However, the mechanisms by which this activated monomer is utilized to form the next intermediate, and how it is transported across the inner membrane and polymerized, are still largely unknown. The genes called alg8 and alg44, immediately downstream of algD (22), are required for alginate production (8). Sequence comparison studies suggest that the alg8 product may be a glycosyl transferase (34). The algG gene encodes a periplasmic C-5-epimerase that introduces Gs into alginate by epimerization of Ms at the polymer level in the periplasm (5, 11). Alginate is also modified by acetylation of M moieties in the periplasm at positions O-2 and/or O-3, and this requires the products of algI, algJ, and algF (12, 13). The algE gene encodes a protein that may form a porin in the outer membrane through which the polymer is excreted out of the cell (7, 33). The algL gene encodes an alginate lyase whose role in alginate biosynthesis is unclear, and algX is a gene of unknown function (36).
FIG. 1.
Pathway leading to the synthesis of alginate in P. aeruginosa. Known genes required at each step and the products they encode are indicated. The transport and polymerization enzymes are unknown.
A region of approximately 1.2 kb between alg44 and algE remained uncharacterized, although it may encode a potentially important component of the alginate biosynthetic machinery. A recent report showed that in strain 8873 this region contains an open reading frame, which was termed algK (1). Here we extend this initial observation and describe a genetic characterization of algK, which includes the effects of nonpolar mutations in FRD1, a cystic fibrosis isolate and mucoid strain. Our results suggest that AlgK plays a critical role in the formation of poly-M from activated monomers in the pathway leading to alginate secretion.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are shown in Table 1. P. aeruginosa and Escherichia coli were routinely cultured in L broth (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl [per liter]). Triparental matings were used to mobilize plasmids from E. coli to P. aeruginosa with the conjugative helper plasmid pRK2013 (10). A 1:1 mixture of Pseudomonas Isolation Agar (Difco) and L agar (Difco) was used to select for P. aeruginosa following matings with E. coli. MAP, a defined medium that promotes alginate production by P. aeruginosa, was previously described (13). When used, antibiotics were at the following concentrations (micrograms per milliliter): ampicillin, 100; carbenicillin, 300; gentamicin, 15 for E. coli and 250 for P. aeruginosa; and kanamycin, 40.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotypea | Source or reference |
|---|---|---|
| E. coli | ||
| HB101 | proA2 leuB6 thi-1 lacY1 hsdR hsdM recA13 supE44 rpsL20 | This laboratory |
| JM109(DE3) | F′ traD36 lacIq Δ(lacZ)M15 proA+ B+/e14− (McrA−) Δ(lac-proAB) thi gyrA96 endA1 hsdR17 (rK− mK+) relA1 supE44 recA1 with DE3, a λ prophage carrying the T7 RNA polymerase gene under control of PlacUV5 | |
| P. aeruginosa | ||
| FRD1 | Cystic fibrosis isolate, Alg+ | 28 |
| FRD1003 | algJ::Tn501-3 Hgr Alg− | 13 |
| FRD1100 | algKΔ::Gmr Alg− | This study |
| FRD1105 | algKΔ::GmralgJ::Tn501-3 Hgr Alg− | This study |
| PAO1 | Prototrophic wild-type strain (nonmucoid) | 19 |
| Plasmids | ||
| pALG2 | pBR322 cos oriT Tn5 Apr Kmr with 35 kb of P. aeruginosa DNA containing the alginate biosynthetic operon and argF | 5 |
| M13mp18 | Derivative of M13 | New England Biolabs |
| pBluescript II KS(−) | ColE1 oriV lacZa Apr | Stratagene |
| pCC75 | pMMB22 (IncQ) with Ptac control of algA on a 2.0-kb fragment | 6 |
| pEX100T | bla (Cb/Apr) sacB oriT | 40 |
| pPHO7 | P15A oriV (Tcr) phoA (promoterless) | 18 |
| pMF54 | Ptrc ColE1 replicon Apr with oriV-SF oriT(RK2) lacIq | 11 |
| pRK2013 | ColE1-Tra(RK2)+ Kmr | 10 |
| pSJ5 | M13mp18 with 1.2-kb KpnI fragment from pALG2 | This study |
| pMF100 | pBluescript II KS(−) with PT7 control of alg44 algK on a 3.8-kb EcoRI fragment from pALG2 | This study |
| pSJ10 | pEX100T with alg44 algK on a 3.8-kb EcoRI fragment from pMF100 | This study |
| pSJ12 | pBluescript II KS(−) with 0.7-kb SmaI fragment containing a nonpolar Gmr cartridge | This study |
| pSJ15 | pSJ10 with algKΔ::Gmr (i.e., 1.2-kb KpnI fragment replaced by SmaI fragment from pSJ12) | This study |
| pSJ42 | pMF54 with the N-terminal 360-bp NcoI-XbaI fragment of algK | This study |
| pSJ43 | pMF54 with the N-terminal 1,020-bp NcoI-XbaI fragment of algK | This study |
| pSJ44 | pMF54 with alg44 algK on a 3.8-kb EcoRI fragment | This study |
| pSJ46 | pSJ42 with an algK (1-120)-phoA protein fusion by insertion of a 2.3-kb XbaI-XhoI fragment from pPHO7 | This study |
| pSJ47 | pSJ43 with an algK (1-340)-phoA protein fusion by insertion of a 2.3-kb XbaI-XhoI fragment from pPHO7 | This study |
| pSJ137 | pBluescript II KS(−) with PT7 control of alg44 algKΔ::Gmr | This study |
| pSJ151 | pMF54 with Ptac control of alg44 algKΔ::Gmr | This study |
Apr, ampicillin resistance; Cbr, carbenicillin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Hgr, mercury resistance; Gmr, gentamicin resistance; Alg+, alginate overproduction.
DNA manipulations.
Restriction endonucleases were purchased from Boehringer Mannheim or New England Biolabs. Klenow polymerase was used to blunt the ends of restriction fragments. Large-scale preparations of plasmid DNA were made by using the QIAprep Plasmid Midi system (Qiagen). DNA fragments for cloning were purified from agarose gels by using the QIAEX II Gel Extraction system (Qiagen). Oligonucleotide primers were synthesized on an Applied Biosystems 380B synthesizer. DNA for sequence analysis was obtained from pALG2 (5), which contains the entire alginate biosynthetic operon from strain FRD1; a 1.2-kb KpnI fragment from pALG2 containing DNA between alg44 and algE was cloned into the single-stranded phage vector M13mp18 in both orientations, to generate pSJ5-1 and pSJ5-2. DNA sequences were determined on pSJ5-1 and pSJ5-2 with the Taq DyeDeoxy Terminator Cycle Sequencing system and an automated sequencer (model 373A) from Applied Biosystems. Primers used for sequencing were the M13mp18 universal forward and reverse primers as well as primers designed from the sequences obtained (a total of 32) to determine the sequence on both DNA strands. Adjacent sequences were determined with pMF100 as a template. Sequence data were analyzed with Lasergene software (DNA-Star) on a Macintosh (Apple) computer. Sequences were aligned with published sequences upstream and downstream of this region. Homology searches and alignments were performed with the Basic Local Alignment Search Tool Network Service at the National Center for Biotechnology Information, National Institutes of Health (2).
Gene expression under control of the T7 promoter.
E. coli JM109(DE3), which carries the gene encoding T7 RNA polymerase under control of the inducible lacUV5 promoter, was used for expression of plasmid-encoded genes transcribed under control of the T7 promoter. Following the addition of isopropylthiogalactopyranoside (IPTG) for induction of rifampin (300 μg/ml) to inactivate the host RNA polymerase, the cells were labeled with 10 μCi of [35S]methionine, and whole-cell proteins were subjected to sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis as previously described (11).
Gene fusions with phoA.
To construct algK-phoA translational fusions, algK DNA was amplified by PCR with pMF100 as the template. One primer was specific to a region 240 bp upstream of the algK start codon that contained an NcoI site at the 5′ end (5′-TGGCCCATGGCACCCGGGTGAACTTCCAGGT-3′). The other primer used corresponded to oligonucleotides encoding either AlgK residues 120 or 340 (5′-AGGCTCTAGAGCCTCGCGGTGCTCGGCGTCG-3′ and 5′-CTGGTCTAGAGCCTTGAGCAGGTGCCGCTCG-3′, respectively), and each had an XbaI site downstream from the coding region. The PCR products were cut with NcoI and XbaI and cloned into the NcoI-XbaI sites of the expression vector pMF54 (11) to give rise to pSJ42 and pSJ43, respectively. An XbaI-XhoI fragment from pPHO7 (18), encoding alkaline phosphatase without a signal sequence, was ligated into the XbaI-XhoI sites of pSJ42 and pSJ43 to give rise to pSJ46 and pSJ47, respectively, encoding AlgK-PhoA with fusion joints at residue 120 or 340. Protein fusions with PhoA were verified by Western blot analysis, which was performed as previously described (27). The primary antibody used was rabbit anti-bacterial alkaline phosphatase (5 Prime→3 Prime, Inc), and the secondary antibody was a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G. The detection system consisted of a hydrogen peroxide substrate-based colorimetric assay. Positive activity of the phoA fusions (PhoA+) in E. coli and P. aeruginosa was tested by screening colonies for blue color on L agar containing 5-bromo-4-chloro-3-indolylphosphate (X-P) at a concentration of 40 μg/ml.
Replacement of chromosomal algK with a nonpolar gentamicin cassette.
Plasmid pUCGmΩ (39) contains a cassette encoding a polar gentamicin resistance (Gmr) gene between transcriptional stop sequences. This was used as a template to amplify by PCR a DNA fragment encoding a nonpolar Gmr gene, along with its promoter sequence but without the transcriptional stop sequences. The upstream primer used for this matched sequences 115 bp upstream of the start codon: 5′-CGCGCCCGGGTTGACATAAGCCTGTTCGGTTCGTAA-3′. The other oligonucleotide primer was complementary to sequences 12 bp downstream of the stop codon: 5′-CGCGCCCGGGAAGCCGATCTCGGCT-3′. Both primers were designed to produce SmaI sites at both ends of the PCR product. The 0.7-kb SmaI-digested PCR product was then ligated into the SmaI site of pBluescript II KS(−) to obtain pSJ12, which conferred Gmr. A 3.8-kb EcoRI fragment from pMF100 containing algK was cloned into the EcoRI site of the gene replacement vector pEX100T (40) to obtain pSJ10; this was digested with KpnI (to delete most of algK and a few nucleotides upstream), blunt ended, and ligated to the Gmr SmaI fragment from pSJ12 to obtain pSJ15. Triparental matings were used to mobilize pSJ15 from E. coli HB101 to P. aeruginosa strains with selection for Gmr. The plasmid had a narrow host range, and Gmr colonies obtained were due to homologous recombination with the chromosome. Colonies that had undergone double crossovers, leading to gene replacement, were identified by plating on L agar containing 7.5% sucrose, indicating loss of the sacB (sucrose sensitivity) gene on the plasmid, and also by screening for loss of the plasmid-encoded bla (carbenicillin resistance) marker.
PCR was used to verify insertion of the Gmr gene in the chromosome as follows. Oligonucleotides used for this purpose included primer a, a primer specific to the region 48 bp upstream of algK, which was 5′-TGAACAAGGCCGTGACCCTGGCCACCG-3′; primer b, a primer in the reverse orientation specific to a region within algK 987 bp downstream of the start codon, which was 5′-AGGTGCCGCTCGGCCTTGCGCGG-3′; and primer c, a primer in the reverse orientation specific to the 3′ end of the Gmr gene, which was the same one used to obtain a nonpolar Gmr cassette. Primers a-b and a-c were then used to amplify genomic DNAs of the mutants and wild-type FRD1. Primers specific either to the algK gene or to the Gmr-conferring gene were used in combination with a primer specific to the DNA upstream of algK (common in both the wild type and mutants) to generate amplified PCR products by using the genomic DNA of the wild-type FRD1 or the algKΔ mutant as a template. The PCR products were examined on an agarose gel, and FRD1 gave bands of the desired size with only the algK and upstream-specific primers. The mutant gave bands of the expected size only with the Gmr gene and upstream-specific primers.
Uronic acid assay.
P. aeruginosa cultures were grown in 10 ml of MAP defined medium for 24 h, and cells were removed by centrifugation (10,000 × g for 1.5 h). The supernatants were placed in dialysis bags (Spectra/Por membrane) (molecular weight cutoff, 10,000; 1.8 ml/cm) and dialyzed against an equal volume of 10 mM Tris-HCl (pH 7.6) at 4°C overnight. The uronic acid concentrations in the dialyzed and dialysate fractions were determined by the carbazole method of Knutson and Jeanes (20). Briefly, 30 μl of the fraction was mixed with 1.0 ml of borate-sulfuric acid reagent (100 mM H3BO3 in concentrated H2SO4), and 30 μl of carbazole reagent (0.1% in ethanol) was added. The mixture was heated to 55°C for 30 min, and the absorbance at 530 nm was determined. Uronic acid concentrations were determined from a plot with Macrocystis pyrifera alginate (Sigma) as a standard.
Nucleotide sequence accession number.
The nucleotide sequence data for the fragment containing algK from strain FRD1, a sputum isolate from a cystic fibrosis patient (28), have been deposited in the GenBank database under accession no. AF039535.
RESULTS
Nucleotide sequence of algK from P. aeruginosa FRD1 and its expression in E. coli.
We determined the sequence of the DNA in a region of the alginate biosynthetic operon between alg44 and algE by using DNA derived from the alginate-overproducing (Alg+) cystic fibrosis isolate FRD1. Upon aligning the sequences at each end with those already known, an open reading frame (algK) of 1,428 bp (accession no. AF039535) was observed. This open reading frame predicted the synthesis of a 52-kDa polypeptide with an isoelectric point of 5.8 and a charge of −4.3. The stop codon of algK overlapped with the start codon of the downstream gene, algE, suggesting that these two genes may be translationally coupled. The AlgK sequence from P. aeruginosa 8873 was recently reported (1), and it differed at Arg-78 (AGG) in FRD1, which was a Ser (AGT) in strain 8873, and Phe-177 (TTC) in FRD1, which was a Leu (CTC) in 8873. Homology searches did not reveal any obvious similarity to a protein of known function that might suggest a function for AlgK.
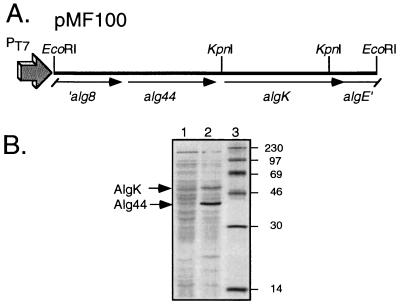
To verify the 52-kDa size of AlgK predicted from the sequence analysis, algK was manipulated to permit expression in E. coli. A 3.8-kb EcoRI fragment from pALG2 that contained algK and alg44 was cloned in the correct orientation relative to a vector’s T7 promoter to form plasmid pMF100 (Fig. 2A). E. coli JM109(DE3) carries the gene encoding T7 RNA polymerase, which is inducible from the lacUV5 promoter, and this was used as the expression host for pMF100. Following induction by the addition of IPTG and addition of rifampin to inactivate host RNA polymerase, the cells were labeled with [35S]methionine. Whole-cell proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiographed. The results (Fig. 2B) revealed two proteins specifically encoded by the clone that were approximately 52 and 42 kDa. These sizes corresponded to the predicted molecular masses of the polypeptides encoded by the two plasmid-encoded genes, algK and alg44, respectively.
FIG. 2.
Expression of algK in E. coli. (A) Partial restriction map of P. aeruginosa FRD1 DNA in pMF100, showing a 3.8-kb EcoRI fragment containing algK and alg44 that was used in this study for expression in E. coli under control of the T7 promoter (PT7). (B) Autoradiogram of [35S]methionine-labeled proteins expressed in E. coli JM109(DE3) from the vector pBluescript II KS(−) (lane 1) or the clone pMF100 (lane 2) in the presence of IPTG and rifampin. Sizes of molecular weight markers (lane 3), in thousands, are shown. The positions of the AlgK and Alg44 proteins expressed from pMF100 are indicated with arrows.
Periplasmic localization of AlgK.
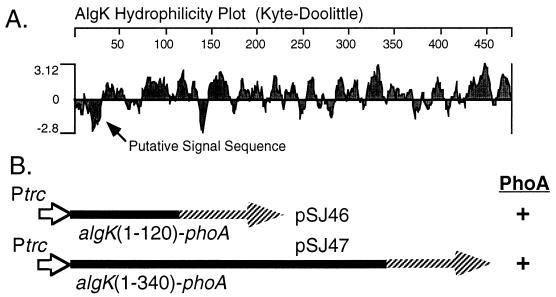
The hydrophilicity plot of the AlgK sequence predicted a generally hydrophilic protein, except for its amino terminus (approximately 25 residues), which displayed considerable hydrophobic character (Fig. 3A). This was followed by two potential signal peptidase cleavage (Ala-X-Ala) sites (35), suggesting that AlgK may possess a signal sequence for transport across the inner membrane. To determine if AlgK localized to the periplasm, algK DNA was manipulated to encode AlgK-PhoA fusion proteins containing the first 120 or 340 residues of AlgK (Fig. 3B). The 5′ algK sequences were ligated upstream and in frame to a phoA gene lacking its own signal sequence. Expression of the resulting algK-phoA fusions in the broad-host-range expression vector pMF54 was examined in E. coli HB101. AlgK-PhoA fusion proteins of the appropriate sizes were observed in a Western blot analysis utilizing alkaline phosphatase antibodies (data not shown). When grown on X-P indicator plates, the E. coli strains producing both AlgK-PhoA fusions were blue (PhoA+). This indicated that the AlgK-PhoA proteins had localized to the periplasm, utilizing the AlgK signal sequence, followed by cleavage of the phosphate group on the chromogenic X-P substrate. When these plasmids were mobilized to P. aeruginosa PAO1, the colonies also turned blue on the X-P agar medium. Colonies of HB101 and PAO1 containing the vector were not blue. These studies indicated that AlgK normally resides in the periplasm. In addition, a cysteine residue was observed immediately after the presumptive signal sequence in AlgK (Cys-28), which suggests that the protein may undergo lipid modification (31); the other two cysteines in AlgK (Cys-195 and Cys-205) probably form a disulfide bond.
FIG. 3.
Localization of AlgK to the periplasm. (A) A hydrophilicity plot of the predicted AlgK polypeptide shows a generally hydrophilic protein except for the amino terminus, which represents a putative signal sequence. (B) Aligned to the hydrophilicity plot are shown DNA fragments encoding 5′ portions of algK (solid lines) that were used to construct algK-phoA translational fusions. These algK-phoA fusions are under control of the Ptrc promoter (open arrow) in a broad-host-range expression vector (pMF54), and both showed alkaline phosphatase activity (PhoA+) in E. coli and P. aeruginosa as shown by blue colony formation on an X-P-containing medium.
Construction of a nonpolar algK deletion mutant.
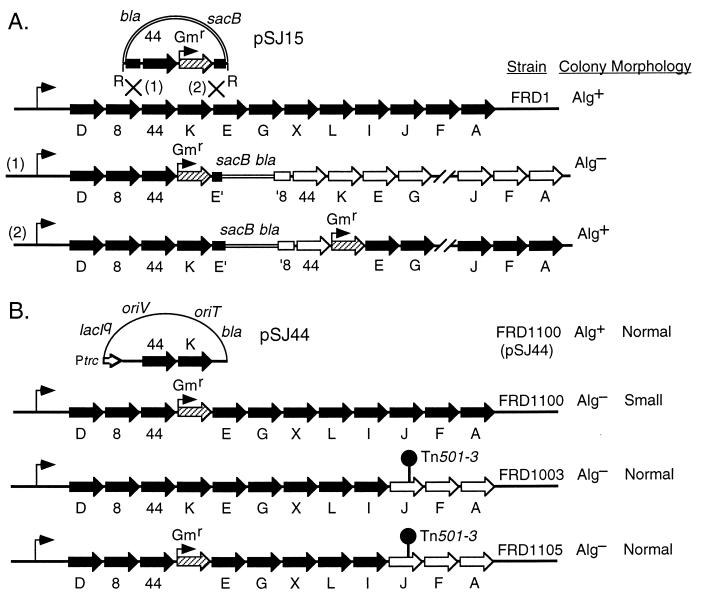
A defined algK deletion (algKΔ) mutant was constructed in the mucoid FRD1 strain background to evaluate the potential role of this gene in alginate production. Our strategy was to replace algK in the chromosome with a marker that would not affect expression of any downstream genes in the operon that are essential for alginate precursor formation (e.g., algA). This was accomplished by first cloning the 3.8-kb EcoRI fragment (Fig. 2A) containing algK and adjacent sequences into pEX100T, a gene replacement vector that contains sacB, which confers sucrose sensitivity in P. aeruginosa (38). A nonpolar Gmr cartridge was synthesized by PCR and used to replace a KpnI fragment in the clone to delete most of algK but none of the adjacent alg44 or algE sequences, forming pSJ15. When this EcoRI fragment containing alg44 algKΔ::Gmr was placed under control of the T7 promoter (pSJ137), AlgK expression was lost but Alg44 expression was not affected (data not shown). The algKΔ::Gmr construct in pEX100T (pSJ15) was introduced into mucoid FRD1 by triparental mating with selection for the vector-encoded bla (β-lactam resistance) gene by plating on L agar with carbenicillin (Fig. 4A). Due to the narrow host range of the plasmid, the numerous colonies that formed were merodiploids resulting from homologous recombination with the chromosome by a single crossover. Approximately three-fourths of the colonies obtained were nonmucoid, and one-fourth were mucoid. This was as predicted and depended on whether the crossover had occurred in the 2.0 kb upstream of algKΔ::Gmr or in the downstream 0.5-kb region (Fig. 4A). The former would result in the integrated vector having a polar effect on the downstream biosynthetic genes, whereas the latter would allow the promoter of the Gmr gene to drive transcription of downstream genes in the algD operon. The presence of colonies with the mucoid phenotype also verified that the Gmr cassette developed here was nonpolar on downstream gene transcription. To remove the vector sequences and obtain the algKΔ::Gmr mutants (i.e., gene replacement), mucoid merodiploid colonies were grown in L broth (without antibiotic selection) to permit spontaneous recombination, followed by selection for gentamicin and sucrose resistances. The colonies obtained also demonstrated carbenicillin sensitivity, as expected, due to loss of the vector. All the algKΔ::Gmr mutants constructed in the mucoid FRD1 background were nonmucoid, indicating that AlgK was essential for alginate production.
FIG. 4.
Construction of algKΔ mutants of P. aeruginosa. (A) Plasmid pSJ15 contains a 3.8-kb EcoRI (R) fragment of the algD operon in FRD1 cloned into a gene replacement vector (pEX100T), with a nonpolar Gmr cartridge (hatched arrow) replacing algK. Shown are the predicted genotypes of merodiploids that formed when pSJ15 integrated into the chromosome of mucoid FRD1 to form mucoid (Alg+) and nonmucoid (Alg−) colonies. When a crossover occurs at site 1, the integrated vector has a polar effect on the transcription of downstream genes (open arrows), and the strain should be Alg−. When a crossover occurs at site 2, the promoter of the Gmr gene should drive transcription of downstream genes in the algD operon so that all genes in the operon are expressed (black arrows), and this strain should be Alg+. (B) Diagrams of pSJ44, which expresses algK in trans, and the algD operons in the mutants used in this study along with their colony morphologies are shown. Note that in a strain overproducing alginate (FRD1), a nonpolar algKΔ::Gmr mutation resulted in a small-colony (i.e., sick) phenotype (strain FRD1100) and that this phenotype could be suppressed (strain FRD1105) with an algJ::Tn501-3 mutation (shown as a pin) which was polar on algA. Open arrows indicate genes that are not expressed. The genes of the algD operon are not drawn to scale.
algKΔ mutants display a small-colony phenotype that is suppressed by a mutation polar on algA.
In addition to the loss of alginate production, another interesting phenotype of the algKΔ mutants was their small-colony phenotype. One of the algKΔ mutants, called FRD1100, which displayed the typical small-colony phenotype, was chosen for further study; its chromosomal replacement of algK by the Gmr cartridge was verified by PCR analysis (see Materials and Methods). We also tested the effect of algK in trans in FRD1100. For this, the P. aeruginosa expression vector pMF54 (11) was used to clone the 3.8-kb EcoRI fragment that contained algK in the proper orientation relative to the plasmid’s trc promoter, which formed pSJ44 (Fig. 4B). A similar plasmid (pSJ151) that contained the alg44 and algKΔ::Gmr alleles was constructed as a control. When pSJ44 and pSJ151 were conjugally transferred to FRD1100 (as well as to the other algKΔ mutants) with selection for carbenicillin resistance, only the transconjugants with pSJ44 (i.e., algK+) exhibited both the wild-type mucoid (Alg+) and normal growth phenotypes. This also indicated that the chromosomal algKΔ::Gmr mutation was nonpolar.
FRD1 is a strain that normally overproduces alginate. However, the algKΔ mutation in strain FRD1100 appeared to have a deleterious affect on its growth. Given that the algKΔ mutation should have no effect on the production of AlgA, AlgC, or AlgD, it was likely that GDP-mannuronic acid was still being produced at high levels in FRD1100 (Fig. 1). We next asked whether a precursor of alginate being synthesized in this strain might be toxic to the cells. Genetic evidence for this was obtained by using FRD1003 (Fig. 4B), a derivative of FRD1 with an algJ::Tn501 mutation that was polar on algA transcription (13). AlgA (phosphomannose isomerase) is required for the initial step in alginate biosynthesis, so FRD1003 should contain no alginate precursors. It also formed normal-sized Alg− colonies. An algKΔ mutant was constructed in the FRD1003 AlgA− background and called FRD1105 (Fig. 4B). The colony morphology of this double mutant (algKΔ algJ::Tn501) was indistinguishable (Fig. 5B) from that of the parental Alg− strain, FRD1003 (not shown), which is in sharp contrast to the sickly, small-colony phenotype of the algKΔ mutant FRD1100 (Fig. 5A). When FRD1003 contained pCC75, providing algA in trans, the Alg+ phenotype was restored because the production of alginate precursors was restored. However, we did not obtain any transconjugants when we attempted to introduce algA in trans (via pCC75) into the algKΔ mutant (FRD1105), suggesting that the formation of unpolymerized alginate precursors was deleterious to the cells.
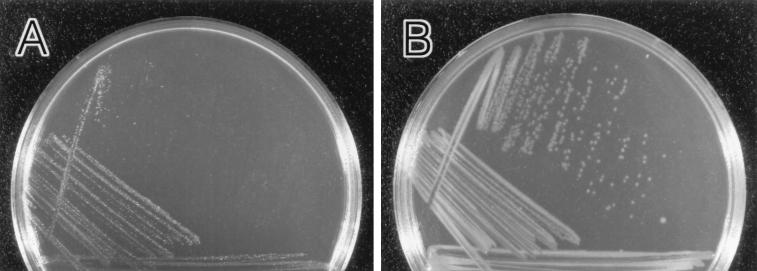
FIG. 5.
Photographs of L-agar plates containing colonies of P. aeruginosa strains that formed after 24 h at 37°C. Note that in an Alg+ strain (FRD1) background, a nonpolar algKΔ::Gmr mutation (strain FRD1100) resulted in Alg− and small-colony (i.e., sick) phenotypes. These phenotypes could be suppressed with a polar algJ::Tn501-3 mutation (strain FRD1105). (A) algKΔ mutant FRD1100. (B) algKΔ algJ::Tn501-3 mutant FRD1105.
The algKΔ mutant secretes unpolymerized uronic acids.
The studies described above suggested that the small-colony (i.e., slow-growth) phenotype of algKΔ mutants was due to the production of toxic alginate precursors. Given that AlgK localized to the periplasm, we hypothesized that a mannuronate precursor might still be transported to the periplasm and, if unpolymerized, may permeate through the outer membrane and into the medium. Initial tests showed that culture supernatants of strains grown in L broth (22 h, 37°C, with aeration) were lower in pH with FRD1100 than with FRD1105, and this was consistent with our hypothesis (data not shown). To test culture supernatants for unpolymerized uronic acids and to avoid the background interference that arises with L-broth cultures, the P. aeruginosa strains were grown in the defined medium MAP (11), which promotes alginate production. Uronic acids in the supernatants of Alg+ FRD1 cultures (grown for 22 h at 37°C with aeration) were compared to those of the algKΔ mutant (FRD1100); the algKΔ algJ::Tn501 double mutant (FRD1105) served as the background negative control. At first, each sample was dialyzed against just an equal volume (7 ml) of buffer, and then the uronic acid contents of both the dialyzed fraction and the dialysate were assayed (Table 2). As expected, the Alg+ FRD1 sample had uronic acids almost exclusively in the dialyzed fraction, because the uronic acids (i.e., mannuronate and guluronate) were in the polymeric (i.e., alginate) form, which is too large to pass through the dialysis membrane. This was also true of the Alg+ strain FRD1100(pSJ44) (data not shown). In contrast, the algKΔ mutant FRD1100 showed approximately equal amounts of uronic acids in both fractions. The amount of uronic acid produced by the algKΔ mutant, determined by adding the values observed in the dialyzed and dialysate fractions, was similar to that of the polymerized product of parent strain FRD1. When each fraction was then exhaustively dialyzed, all of the FRD1 uronic acids remained in the dialysis sac (Table 2). However, none of the FRD1100 uronic acids was detectable following this, because they had all dialyzed away. This suggests that algKΔ mutants produced large amounts of a low-molecular-weight precursor of alginate that may have been properly localized but not polymerized. This high concentration of unpolymerized uronic acids in the environment of FRD1100 was apparently what was detrimental to its growth and led to the small-colony morphology observed.
TABLE 2.
Amounts and relative sizes of uronic acid-containing material secreted by Alg+ P. aeruginosa FRD1 and its Alg− algKΔ derivative FRD1100
| Strain | Relevant genotype | Uronic acid concn (μg/ml) in treated samplesa
|
||
|---|---|---|---|---|
| Dialyzed against same vol | Dialysate of same vol | Dialyzed exhaustively | ||
| FRD1 | algK+ | 1,800 | 70 | 1,800 |
| FRD1100 | algKΔ | 700 | 700 | 0 |
MAP culture supernatants of FRD1 and FRD1100 were dialyzed against an equal volume (7 ml) of Tris-HCl buffer. Uronic acids in the bag and those small enough to diffuse into the dialysate were determined by the carbazole method. Both supernatants contained high concentrations of uronic acids, but only the algKΔ mutant (FRD1100) produced dialyzable uronic acids. The low values obtained with negative control strain FRD1105 were subtracted as background values to produce the values shown. Following extensive dialysis against buffer, all uronic acids produced by FRD1 remained in the sac, but all those produced by FRD1100 had diffused out of the dialysis sac and became undetectable.
DISCUSSION
Alginate biosynthesis is comprehensively regulated by a number of factors that ultimately converge on the promoter of the algD biosynthetic operon to limit the production of enzymes for alginate biosynthesis (for a review, see reference 29). However, mucoid strains from cystic fibrosis patients appear to undergo deregulation, probably due to adaptive mutations occurring in vivo, which result in the copious overproduction of the viscous polymer. This conversion to the Alg+ phenotype is regarded as an important pathogenic mechanism. Alginate biosynthesis in P. aeruginosa also provides an attractive model system to better understand the secretion of exopolysaccharides in gram-negative bacteria. The steps in the pathway leading to the formation of an activated monomeric unit (GDP-mannuronate) have been well characterized (26). This involves the participation of two genes, algD and algA, which are the first and last genes of the large algD operon. Recently, the late steps involved in postpolymerization modifications, such as epimerization and acetylation, are beginning to be understood. These processes require the gene products of algG, algI, algJ, and algF in the operon (5, 11–13). However, the middle process in the pathway of alginate biosynthesis, in which the inner membrane is crossed by the next intermediate (monomer or oligomer) and the polymerization of these moieties to poly-mannuronic acid occurs, is still unknown.
A complete understanding of the alginate biosynthetic pathway was not possible until all of the components involved had been identified. There was one segment (∼1.2 kb) of the algD operon, located between alg44 and algE, that was still uncharacterized. Our sequence analysis of this region from FRD1 revealed a potential open reading frame (1,428 kb), and we showed here that it encoded the predicted 52-kDa protein. During the final stages of this work, Aarons et al. (1) also sequenced this region from strain 8873 and called it algK; we have adopted their nomenclature. The amino terminus of the predicted protein resembled a signal sequence recognized by type II signal peptidase, suggesting that the algK gene may encode a lipoprotein (31). The hydrophilicity plot predicted a hydrophilic protein, suggesting its localization to the periplasm. We confirmed this by constructing algK-phoA protein fusions, which were positive for alkaline phosphatase activity and strongly suggested that the protein is indeed periplasmic. These studies complement those of Aarons et al. (1), who also predicted such localization by using β-lactamase (bla) fusions.
A better understanding of AlgK function was obtained through the construction and analysis of an algK deletion mutant in the Alg+ FRD1 background. The mucoid or nonmucoid phenotype of such a mutant would show whether AlgK played a role either in polymer production or in modification of the polymer. The genetic manipulations involved in this construction were simplest if algK in the chromosome was replaced with a selectable marker. This laboratory has previously used Tn501 insertions to study the algD operon, but their polar nature requires that the downstream genes, which are essential for polymer formation, be provided in trans (12). Thus, it was more desirable here to have a selectable marker that would be nonpolar on downstream gene expression so that even the last gene (algA) would still be expressed, as this is required for the formation of all alginate precursors. Using PCR amplification, we constructed a Gmr cassette that contained no transcriptional stop sequences, and thus it was predicted to be nonpolar. By replacing algK with this nonpolar Gmr cassette (which still retained its promoter sequence), we obtained algKΔ mutants. These demonstrated a nonmucoid phenotype, indicating that AlgK was required for alginate production rather than its modification. By providing algK in trans under control of a trc promoter, alginate production was restored, which indicated that the algKΔ::Gmr mutation was nonpolar on all downstream genes.
An interesting and useful phenotype of the algKΔ mutants was their small-colony morphology, suggesting that they were sick as a result of the mutation. We hypothesized that such algKΔ mutants produced an alginate precursor that was potentially toxic to the cells. We constructed an algKΔ mutation in an AlgA− (i.e., algJ::Tn501) strain background, and these double mutants grew as well as the parent strain, which was in contrast to the small colony size of the algKΔ mutant. This implied that AlgK acted at a step subsequent to precursor formation. If there was no precursor available in the double mutant, then loss of algK did not have an effect on cell growth. Bringing algA in trans should then give the double mutant a growth defect. However, we were unsuccessful in attempts to transfer algA on a high-copy-number plasmid to such strains, suggesting that an algKΔ mutation was deleterious when alginate precursors were produced. If algK was involved in the assembly and/or transport of alginate, then a deletion mutation of the gene could potentially result in accumulation of toxic levels of either GDP-mannose or a precursor polymer within the cells. In the case of K1 capsule formation in E. coli, mutations that block transport across the inner membrane lead to the accumulation of polymer within the cell, and this can be observed by electron microscopy (4, 30). However, our electron micrographs of FRD1 and the algK mutants constructed here showed no evidence of polymer accumulation in the cells (data not shown). Efforts to determine whether the algKΔ mutation causes the accumulation of GDP-mannuronate within the cytoplasm are in progress.
However, it seemed reasonable that if AlgK was a periplasmic component involved in alginate production, then it may be a part of a polymer assembly apparatus in the periplasm. If AlgK− did not affect precursor formation or transport across the membrane, then this might lead to accumulation of a monomeric or oligomeric precursor in the periplasm, which could diffuse through the outer membrane. The first suggestion that the algKΔ mutation affects polymerization was the observation that the AlgK− mutant supernatants were more acidic than those of an AlgA− mutant, suggesting that uronic acids were being released by the cells. Indeed, the AlgK− mutant was shown to produce large quantities of unpolymerized uronic acids. This material was readily dialyzable, indicating that it was of low molecular weight. This uronic acid material is currently being purified for further analysis. However, we would predict that it is some form of d-mannuronic acid, either monomer, short oligomer, or sugar nucleotide. This analysis should provide further insight toward our understanding of the immediate precursor of poly-M in the pathway of alginate biosynthesis and the process involved in polymer formation.
ACKNOWLEDGMENTS
We thank Michael J. Franklin for providing clones and strains and for many insightful discussions concerning this project. We gratefully acknowledge the Molecular Resources Center of the University of Tennessee, Memphis, for assistance in oligonucleotide synthesis and the sequence analysis described here.
This work was supported by Veterans Administration Medical Research Funds (to D.E.O.) and by Public Health Service grant AI-19146 from the National Institute of Allergy and Infectious Diseases (to D.E.O.).
REFERENCES
- 1.Aarons S J, Sutherland I W, Chakrabarty A M, Gallagher M P. A novel gene, algK, from the alginate biosynthetic cluster of Pseudomonas aeruginosa. Microbiology. 1997;143:641–652. doi: 10.1099/00221287-143-2-641. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore R S, Mitchell M. Immunologic investigations of mucoid strains of Pseudomonas aeruginosa: comparison of susceptibility to opsonic antibody in mucoid and nonmucoid strains. J Infect Dis. 1982;141:238–247. doi: 10.1093/infdis/141.2.238. [DOI] [PubMed] [Google Scholar]
- 4.Boulnois G J, Roberts I S. Genetics of capsular polysaccharide production in bacteria. Curr Top Microbiol Immunol. 1990;150:1–18. doi: 10.1007/978-3-642-74694-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Chitnis C E, Ohman D E. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J Bacteriol. 1990;172:2894–2900. doi: 10.1128/jb.172.6.2894-2900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 7.Chu L, May T B, Chakrabarty A M, Misra T K. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene. 1991;107:1–10. doi: 10.1016/0378-1119(91)90290-r. [DOI] [PubMed] [Google Scholar]
- 8.Darzins A, Wang S-K, Vanags R I, Chakrabarty A M. Clustering of mutations affecting alginic acid biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1985;164:516–524. doi: 10.1128/jb.164.2.516-524.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans L R, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin M J, Chitnis C E, Gacesa P, Sonesson A, White D C, Ohman D E. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J Bacteriol. 1994;176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin M J, Ohman D E. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993;175:5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin M J, Ohman D E. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J Bacteriol. 1996;178:2186–2195. doi: 10.1128/jb.178.8.2186-2195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg J B, Gorman W L, Flynn J L, Ohman D E. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol. 1993;175:1303–1308. doi: 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldberg J B, Hatano K, Pier G B. Synthesis of lipopolysaccharide O side chains by Pseudomonas aeruginosa PAO1 requires the enzyme phosphomannomutase. J Bacteriol. 1993;175:1605–1611. doi: 10.1128/jb.175.6.1605-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govan J R W, Harris G S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986;3:302–308. [PubMed] [Google Scholar]
- 18.Gutierrez C, Devedjian J C. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holloway B W, Morgan A F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- 20.Knutson C A, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 21.Lam J, Chan R, Lam K, Costerton J R W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maharaj R, May T B, Wang S K, Chakrabarty A M. Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene. 1993;136:267–269. doi: 10.1016/0378-1119(93)90477-k. [DOI] [PubMed] [Google Scholar]
- 23.Marcus H, Baker N R. Quantitation of adherence of mucoid and nonmucoid Pseudomonas aeruginosa to hamster tracheal epithelium. Infect Immun. 1985;47:723–729. doi: 10.1128/iai.47.3.723-729.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin D W, Schurr M J, Mudd M H, Govan J R W, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathee K, McPherson C J, Ohman D E. Posttranslational control of the algT (algU)-encoded ς22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May T B, Shinabarger D, Maharaj R, Kato J, Chu L, DeVault J D, Roychoudhury S, Zielinski N A, Berry A, Rothmel R K, Misra T K, Chakrabarty A M. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev. 1991;4:191–206. doi: 10.1128/cmr.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIver K S, Kessler E, Ohman D E. Substitution of active site His-223 in Pseudomonas aeruginosa elastase and expression of the mutated lasB alleles in Escherichia coli show evidence for autoproteolytic processing of proelastase. J Bacteriol. 1991;173:7781–7789. doi: 10.1128/jb.173.24.7781-7789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohman D E, Chakrabarty A M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohman D E, Mathee K, McPherson C J, DeVries C A, Ma S, Wozniak D J, Franklin M J. Regulation of the alginate (algD) operon in Pseudomonas aeruginosa. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 472–483. [Google Scholar]
- 30.Pavelka J, M S, Wright L F, Silver R P. Identification of two genes, kpsM and kpsT, in region 3 of the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1991;173:4603–4610. doi: 10.1128/jb.173.15.4603-4610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramphal R, Guay C, Pier G B. Pseudomonas aeruginosa adhesins for tracheobronchial mucin. Infect Immun. 1987;55:600–603. doi: 10.1128/iai.55.3.600-603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rehm B H, Boheim G, Tommassen J, Winkler U K. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J Bacteriol. 1994;176:5639–5647. doi: 10.1128/jb.176.18.5639-5647.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxena I M, Brown R M, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: Implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatz P A, Beckwith J. Genetic analysis of protein export in Escherichia coli. Annu Rev Genet. 1990;24:215–248. doi: 10.1146/annurev.ge.24.120190.001243. [DOI] [PubMed] [Google Scholar]
- 36.Schiller N L, Monday S R, Boyd C M, Keen N T, Ohman D E. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J Bacteriol. 1993;175:4780–4789. doi: 10.1128/jb.175.15.4780-4789.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarzmann S, Boring J R., III Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect Immun. 1971;3:762–767. doi: 10.1128/iai.3.6.762-767.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 39.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 40.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 41.Seale T W, Thirkhill H, Tarpay M, Flux M, Rennert O M. Serotypes and antibiotic susceptibilities of Pseudomonas aeruginosa isolates from single sputa of cystic fibrosis patients. J Clin Microbiol. 1979;9:72–78. doi: 10.1128/jcm.9.1.72-78.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson J A, Smith S E, Dean R T. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J Gen Microbiol. 1988;134:29–36. doi: 10.1099/00221287-134-1-29. [DOI] [PubMed] [Google Scholar]
- 43.Zielinski N A, Chakrabarty A M, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]