Abstract
Inbreeding depression has become an urgent issue in cosmopolitan breeds where the massive genetic progress achieved in the latest generations is counterbalanced by a dramatic loss of genetic diversity causing increased health issues. Thus, the aim of this study was to estimate inbreeding depression on productive traits in Holstein dairy cattle. More precisely, we aimed to i) determine the level of inbreeding in 27,735 Italian Holstein dairy cows using pedigree and genotype data, ii) quantify the effect of inbreeding on 305-d in milk yield (MY; kg), fat yield (FY; kg), and protein yield (PY; kg) based on different statistical approaches, iii) determine if recent inbreeding has a more harmful impact than ancestral ones, and iv) quantify chromosomal homozygosity effect on productive traits. Quality control was performed on the autosomal chromosomes resulting in a final dataset of 84,443 single nucleotide polymorphisms. Four statistical models were used to evaluate the presence of inbreeding depression, which included linear regression analysis and division of FPED and FROH into percentile classes. Moreover, FROH was partitioned into i) length classes to assess the role of recent and ancestral inbreeding and ii) chromosome-specific contributions (FROH-CHR). Results evidenced that inbreeding negatively impacted the productive performance of Italian Holstein Friesian cows. However, differences between the estimated FPED and FROH coefficients resulted in different estimates of inbreeding depression. For instance, a 1% increase in FPED and FROH was associated with a decrease in MY of about 44 and 61 kg (P < 0.01). Further, when considering the extreme inbreeding percentile classes moving from the 5th lowest to the 95th highest, there was a reduction of −263 kg and −561 kg per lactation for FPED and FROH. Increased inbreeding, estimated by FPED and FROH, had also a negative effect on PY and FY, either fit as a regressor or percentile classes. When evaluating the impact of inbreeding based on runs of homozygosity (ROH) length classes, longer ROH (over 8 Mb) had a negative effect in all traits, indicating that recent inbreeding might be more harmful than the ancestral one. Finally, results within chromosome homozygosity highlighted specific chromosomes with a more deleterious effect on productive traits.
Keywords: dairy cattle, FPED, FROH, genomics, inbreeding depression, production
Inbreeding in Italian Holstein dairy cows is present, and it reduces milk, fat, and protein yields, with recent inbreeding having a stronger negative impact than the ancestral ones.
Avoiding inbreeding is important to sustain a highly productive dairy sector, and specific homozygous regions across the genome could be monitored to limit their detrimental effects in breeding programs.
Introduction
Inbreeding depression is the decrease in the biological fitness of individuals due to a reduction in heterozygosity. Inbreeding depression is related to the increased frequency of deleterious recessive homozygous genotypes which in turn has a negative effect on the survival of the affected animals (Charlesworth and Willis, 2009; Leroy, 2014). In dairy cattle, inbreeding depression can be observed in reduced performance of productive and reproductive traits, and resilience (Parland et al., 2007; Howard et al., 2017; Martikainen et al., 2017; Doekes et al., 2019). The genomic era has led to a massive increase in genetic gain especially in dairy cattle where the increase in genetic progress was recently estimated to range from 50% to 100% for yield traits and from 200% to 300% for fitness traits per year in American Holstein due to genomic selection (García-Ruiz et al., 2016). However, there is increasing evidence that the continuous increase in genetic gain in recent decades is counterbalanced by a reduction of genetic diversity within a population. It has been shown that genomics plays a crucial role in precisely evaluating the interplay between genetic improvement and inbreeding depression (Gutiérrez-Reinoso et al., 2022). Indeed, based on genotype data, it has been shown that the annual inbreeding rate (ΔF) has increased more after the implementation of genomic selection compared to what happened in the past in several dairy cattle populations (Stachowicz et al., 2011; Baes et al., 2019; Doublet et al., 2019; Makanjuola et al., 2020). The Italian Holstein breed exactly resembles this scenario with a significant increase in the ΔF since the introduction of genomic selection in the breed (Ablondi et al., 2022). Therefore, there is an urgent need to quantify the potential negative impact of increased inbreeding on dairy cows’ performance. Inbreeding coefficients (F) can be combined with phenotypic data to estimate the effect that increased F has on phenotypes. Traditionally, F is derived from pedigree data (FPED) routinely recorded by breeding associations. However, FPED has some limitations since it does not capture variation due to Mendelian sampling and linkage during gamete formation (Hill and Weir, 2011). Additionally, the accuracy of genetic diversity estimated by pedigree data highly relies on the quality and depth of the recorded genealogical data. Single nucleotide polymorphisms (SNPs) are genetic markers widely used nowadays in breeding programs to estimate genomic breeding values but also to estimate F (Howard et al., 2017). Various ways exist to estimate F from SNP data (Dadousis et al., 2022), with the most robust being the runs of homozygosity (FROH) across the genome (McQuillan et al., 2008). Specifically, runs of homozygosity (ROH) are long consecutive homozygous segments distributed across the genome, which can arise from identical-by-descendent haplotypes (McQuillan et al., 2008). These regions can provide information on the level of inbreeding of an individual (FROH), its ancestry, and the population it belongs to Aguilar et al. (2011). By combining the estimated inbreeding level from pedigree and genotype data with phenotype data, the presence of inbreeding depression can be quantified (Howard et al., 2017). Among several economically relevant traits in the dairy industry, fat, and protein are two of the most important milk components included in payment systems. For dairy cattle farmers, achieving high protein yield (PY) and fat yield (FY) is key for maximizing the economic value of their herds (Koutouzidou et al., 2022). This is especially prominent in the Italian context where roughly 80% of the milk is transformed into cheese (Ablondi et al., 2021). Milk with higher protein and fat yield can result in higher cheese and other dairy product yields. Therefore, dairy farmers often focus on breeding and management strategies that promote high PY and FY in their cows, such as selecting bulls with high genetic potential for milk quality and providing cows with balanced diets that meet their nutritional needs. Thus, knowledge of the reduction of fat and protein yields due to inbreeding depression is fundamental for the dairy industry. Therefore the objectives of this study were to i) estimate the level of inbreeding in 27,735 Italian Holstein dairy cows from pedigree and ~85k SNP imputed genotype data, ii) assess the effect of inbreeding on 305-d in milk (milk yield [MY]; kg), FY (kg), and PY (kg) based on different statistical methods, iii) compare the effect of recent and ancestral inbreeding on the aforementioned traits, and iv) quantify chromosome homozygosity-specific contribution (FROH-CHR) on inbreeding depression.
Material and Methods
Ethical statement
All the dairy cows involved in this study were reared in commercial private farms and were not subjected to any invasive procedures. Milk samples used for the analyses were collected during routine milking. Therefore, no ethics approvals were necessary for this study.
Animals and herds data
Pedigree, genomic, and phenotypic data were provided by the Italian National Association of Holstein, Brown Swiss, and Jersey Breeders (ANAFIBJ). The raw dataset included a total of 41,049 cows which was subjected to a series of filtering criteria to guarantee high-quality data. A filter on the number of cows available per herd was considered, excluding herds where dairy cows are occasionally genotyped (at least 5 animals per herd). This latter filter was applied to reduce the potential bias of the breeder’s choice to genotype only highly selected animals. Those cows had lactations during a period of 10 yr from 2011 to 2020 and were either at their first, second, or third parity.
The pedigree database included 393,607 cattle born between 1898 and 2020 with 26,226 bulls and 367,381 cows over 24 generations of pedigree depth. To assess pedigree completeness, the complete generation equivalent (CGE) (Maignel et al., 1996) was computed which averaged to 10.6 (SD = 1.02). The CGE was computed as the sum of (1/2)n of all known ancestors of each individual, with n as the number of generations from a given ancestor to the individual. To limit the effect of incomplete pedigree information on the inbreeding depression estimates, cows with a CGE lower than 5 were excluded since the correlation between FPED and FROH dropped when considering animals with less pedigree depth and completeness. Cows were genotyped with a variety of SNP panels, but all with at least medium-density panels (over 40K SNPs) to ensure less variability due to the SNP numbers that are included in the imputed pipeline (Dadousis et al., 2023). Genotypes were imputed to medium density (85K) using PedImpute (Nicolazzi et al., 2013). To guarantee high accuracy during the imputation pipeline, females retained for this study were only those with both sire and sire of the dam genotyped and used in the imputation pipeline. The reference dataset for imputation is made by over 500.000 Holstein genotypes, which passed Quality control (QC) for: known ancestry (at least sire, dam, and maternal grandsire), call rate, and known sex. The bulls included in the reference dataset are from the Intercontinental consortium: Italy, USA, Canada, UK, and Switzerland animals together with bulls from several other countries which send them in on voluntary bases. The imputation pipeline ANAFIBJ is currently running accommodates for 53 different genomic chips.
The bulls included in the reference dataset are from the Intercontinental consortium: Italy, USA, Canada, UK, and Switzerland animals together with bulls from several other countries which send them in on a voluntary bases. The imputation pipeline ANAFIBJ is currently running accommodates 53 different genomic chips. QC was performed on the 29 autosomal chromosomes in PLINK v1.90 (Purcell et al., 2007). The QC was based on the following criteria: call rate of <95%, parent-offspring SNP mismatch of <0.01, minor allele (<0.01) and genotype (<0.001) frequencies, and extreme deviation from the Hardy–Weinberg equilibrium (P < 0.005). The final dataset contained 84,443 autosomal SNPs.
In terms of phenotypic traits, the 305-d MY (kg), 305-d FY (kg), and 305-d PY (kg) were considered in this study. For each of the traits, values outside the range of ±3 SD from the mean were excluded from further analysis. In total, 27,735 cows from 939 herds were included in the study which resulted from the above-mentioned filtering criteria. Those animals were 14,679 primiparous, 9,036 secondiparous, and 4,023 tertiparous. Although by the application of those filters we have included 67% of the observations available in the raw dataset, high standards in terms of distribution of phenotypic data as well as imputed genotypes were guaranteed.
Statistical analysis
Inbreeding coefficients
Pedigree-based inbreeding coefficients (FPED) were defined as the probability that an individual has two identical alleles by descendant and was computed using the OptiSel R package (Wellmann, 2019). Genomic inbreeding coefficients were derived by means of ROH assessment. The ROH segments were detected using the DetectRUNS package (Biscarini et al., 2019) in R using a sliding window approach (15 SNPs) and defined as follows: at least 15 SNPs in a run, a minimum length of a run equal to 1 Mb, a maximum distance between consecutive SNPs in a window 500 kb, a lower density limit of 1 SNP per 100 kb and allowing for a maximum of one missing and one heterozygous SNP in a run. The genomic inbreeding coefficients (FROH) were calculated as the proportion of individual genome size covered by ROH as performed in previous studies (Ablondi et al., 2022; Dadousis et al., 2022).
Inbreeding depression
The presence of inbreeding depression on 305-d in MY, FY, and PY was evaluated by using several linear mixed models [ASReml 4.1(Gilmour et al., 2015)] where FPED and FROH were included in different ways in the statistical model. More specifically, firstly they were included as regressors; secondly as fixed effects by dividing FPED and FROH into seven percentile classes, where each percentile included the same number of animals, to further quantify the effect in terms of least square means (LSM). In addition, the FROH divided by length classes (ROH 1: 1 to 2 Mb, ROH 2: 2 to 4 Mb, ROH 3: 4 to 8 Mb, ROH 4: 8 to 16 Mb, ROH 5: 16 to 32 Mb) were modeled simultaneously following the method proposed by (Doekes et al., 2019) to evaluate the effect of ancestral and recent inbreeding. Finally, FROH was calculated per each of the 29 autosomal chromosomes separately, and each of them was included in the model as a regressor to determine the effect of chromosome-specific inbreeding. In the latest model, the FROH-CHR was computed as the ratio between the length of the chromosome covered by ROH and the total length of the chromosome. In all the tested models, the following terms were also included: herd-year of calving (2,242 classes), parity (3 classes), and month of calving (12 classes) as fixed effects while animal as random effect as reported below:
Where is the response variable being either 305-d MY, FY, and PY, is the mean of the population, is the combined effect of herd-year of calving, which is the parity class, is the month of calving, are the inbreeding coefficients included in the model either as a regressor or fixed factor as above mentioned for the lth cow, cowi is the random genetic effect for the lth cow, and is the random error term. The cow
effect was assumed to follow N (0, A ), where A is the numerator relationship matrix and the additive genetic variance.
Results and Discussion
Effect of inbreeding depression on productive performances
The Italian dairy industry is highly oriented in the production of high value and traditional cheeses, in particular protected designation of origin (PDO), which highlights the Italian deep-rooted tradition in cheese making. More than fifty cheeses produced in the Italian territory are registered at the European Commission as PDO since they have proven traditional land of origin and specific procedures for milk processing. Across the prosperous variety of dairy products made in this country, the most widely reared dairy cattle breed is the Italian Holstein, accounting 2022 for 1,148,844 herd-tested cows (AIA, 2023). Therefore, milk quality in this breed, which is mainly used for cheese production, is key in Italy and its composition, in particular fat and protein, is of particular interest. The descriptive statistics of productive traits (MY, FY, and PY) of the 27,735 Italian Holstein dairy cows included in this study are presented in Table 1. On average, MY was 10,836 kg, fat 412 kg, and protein 363 kg for 305-d in milk. These values are in line with those reported for the Canadian Holstein, and are largely superior compared to other specialized dairy and dual-purpose breeds (Brito et al., 2021).
Table 1.
Descriptive statistics of quantity of milk, fat, and protein, expressed in kg on 305-d in milk
| Trait | Mean | Min | Max | SD |
|---|---|---|---|---|
| Milk | 10,836 | 4,861 | 16,781 | 1,929 |
| Fat | 412.3 | 170 | 659 | 77.79 |
| Protein | 363.3 | 183 | 544 | 59.01 |
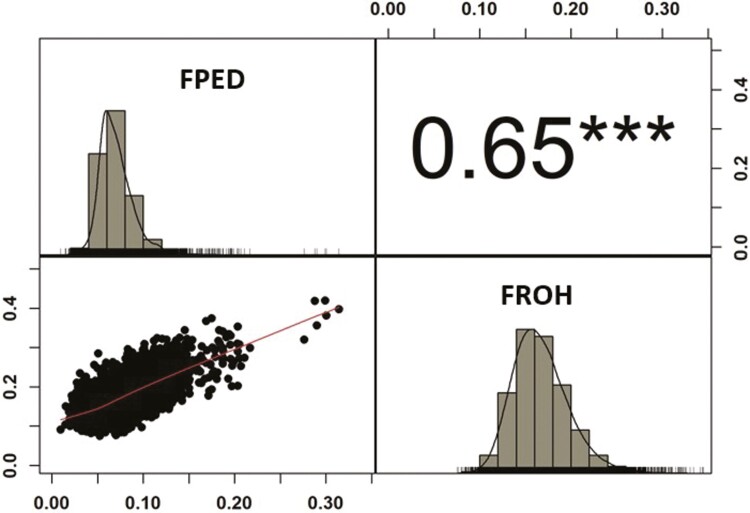
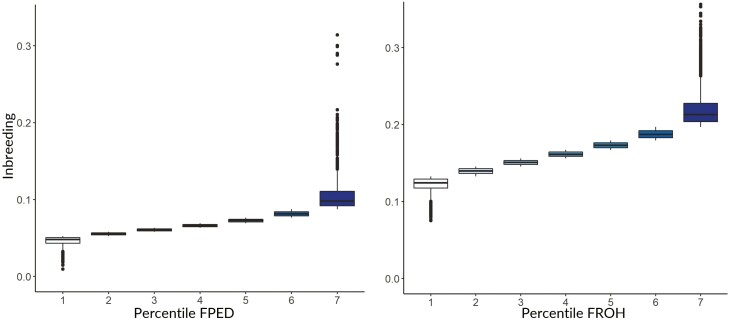
Overall, FROH (FROH = 0.16 ± 0.03) was higher than FPED (FPED = 0.07 ± 0.02, CGE = 10.59), evidencing that the observed homozygosity in the population (FROH) is greater than the expected homozygosity based on pedigree (FPED) (Figure 1). The moderate positive Pearson correlation of 0.65 indicates that as FPED increases, FROH also tends to increase. As previously shown in terms of average value, FPED fails to fully account for all the lost heterozygosity throughout generations. Nevertheless, a correlation of one is not expected between FPED and FROH, since the former does not account for the Mendelian sampling variation as the latter does, and FPED highly relies on pedigree completeness and correctness (Curik et al., 2014; Baes et al., 2019). Nevertheless, the obtained correlation is in line with the values found in previous studies (Rodríguez-Ramilo et al., 2015; Ablondi et al., 2022). To quantify the effect of different levels of FPED and FROH on milk and productive traits, they were divided into seven percentile classes and included as fixed factors in the model (Figure 2). Interesting to notice is that, although each percentile class contains the same number of observations, more variation was observed for the highest class (seventh percentile) both in terms of FPED and FROH. Overall, the variability of the individual percentile classes was slightly higher in the case of FROH than FPED.
Figure 1.
Pearson product-moment correlation (above diagonal) with the level of significance of the correlation (P-value ***≤0.001), and x-y plot (below diagonal) between FPED (pedigree-based) and FROH (runs of homozygosity-based) inbreeding coefficients. On diagonal the distributions of FPED and FROH inbreeding coefficients.
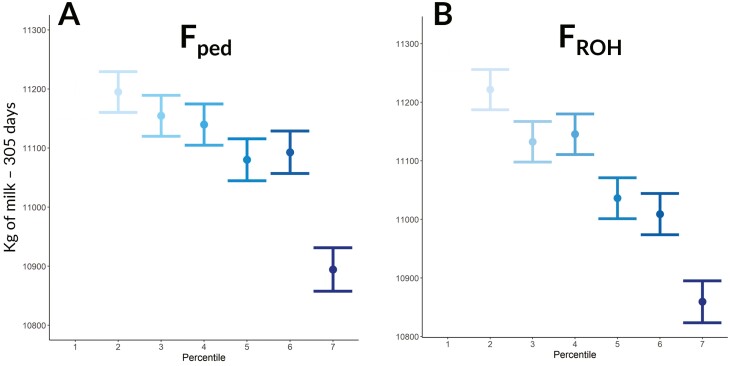
Figure 2.
Seven percentile classes based on FPED (pedigree-based inbreeding) (A) and FROH (runs of homozygosity-based inbreeding) (B).
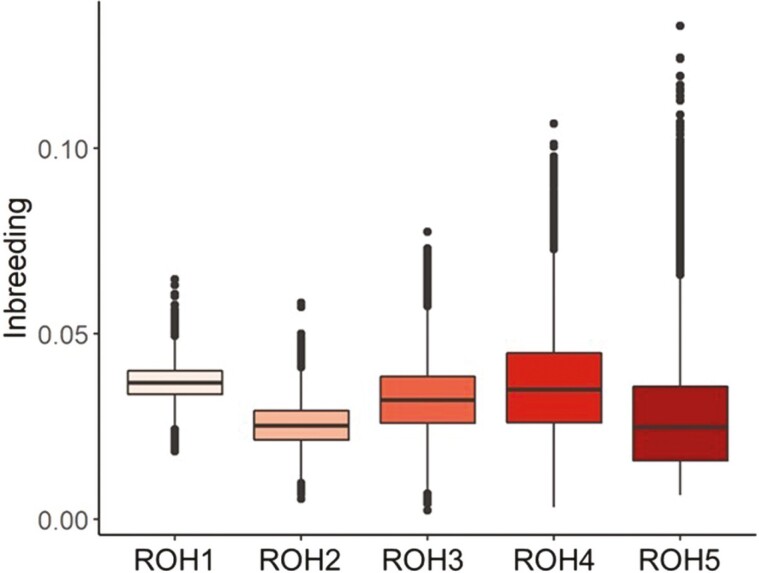
In Figure 3, the level of inbreeding based on FROH estimates divided by length classes (Mb). The rationale behind this classification was to determine if a recent inbreeding, described by longer ROH, could be more detrimental in terms of inbreeding depression compared to ancestral ones. The mean level of inbreeding was comparable among ROH classes starting from 0.026 in the 2 to 4 Mb to 0.037 in the 1 to 2 Mb class. Although extreme values (over the mean ± 3 SD as called hereafter outliers) were found for all the assessed ROH length classes, in the longest class, the highest number of outliers were observed (Figure 3). This could be mostly due to the presence of repeated mating among close relatives in the recent generations that has led to the formation of longer and more contiguous stretches of homozygous regions in the genome (Bosse et al., 2012; Zhang et al., 2015).
Figure 3.
Inbreeding estimates divided by the following 5 ROH (Runs of homozygosity-based) classes: ROH 1: 1 to 2 Mb, ROH 2: 2 to 4 Mb, ROH 3: 4 to 8 Mb, ROH 4: 8 to 16 Mb, ROH 5: 16 to 32 Mb.
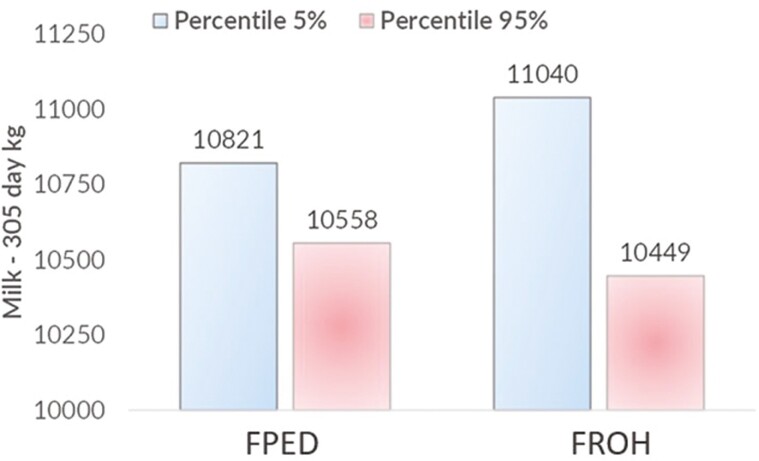
Inbreeding depression was observed for MY, FY, and PY for both inbreeding estimates (FPED and FROH) (P < 0.01) when included as regressors in the model. As an example, when both FPED and FROH increase by 1%, a reduction in MY of about 44 and 61 kg (P < 0.01) was observed. Thus, by evaluating the extreme in terms of inbreeding estimates as 5% lowest and highest (Figure 4), the effect on reduced milk production was equal to -263 kg and -561 kg per lactation for FPED and FROH. The latter result translates into a 2.4% and 5.2% decline compared to the average MY per lactation of 10,836 kg. The same negative trend was found for both FY and PY (P < 0.01), with a decrease of 1.31 and 2.45 kg per lactation, respectively, if based on FROH, and a reduction of 1.41 and 2.00 kg for FY and PY when based on FPED. These reductions in fat and PY can have significant implications for cheese production, as both are principal components of cheese (Fox et al., 2017). Lower fat and PYs may result in reduced cheese yield per lactation. This is because milk with a lower protein content may result in less recovery of fat in the cheese (Cipolat-Gotet et al., 2018), and lower milk fat content may lead to reduced firmness or texture (Martin et al., 2005). This could potentially impact the overall profitability and quality of cheese production from a dairy cattle population experiencing inbreeding depression. To give a practical example from the Italian dairy scenario, we could consider the context of Parmigiano Reggiano (PR) production, which is the second largest Italian PDO consortium, with 160,097 tons of hard cheese produced in 2022 and an overall production value of 2.7 billion euro (CLAL., 2023). When looking at losses due to inbreeding in terms of PR production and comparing the two most extreme 5% inbred cows, this translated into over half a wheel of PR cheese per FPED and over one per FROH, leading to an economic loss of roughly 310 about 600 euros, respectively for FPED and FROH per cow per lactation, if considering the price of a 24-mo-aged PR cheese (CLAL, 2022).
Figure 4.
Differences in milk production expressed in kg on 305-d in milk between the lowest and the highest fifth percentile based on FPED (pedigree-based) and FROH (runs of homozygosity-based) inbreeding coefficients.
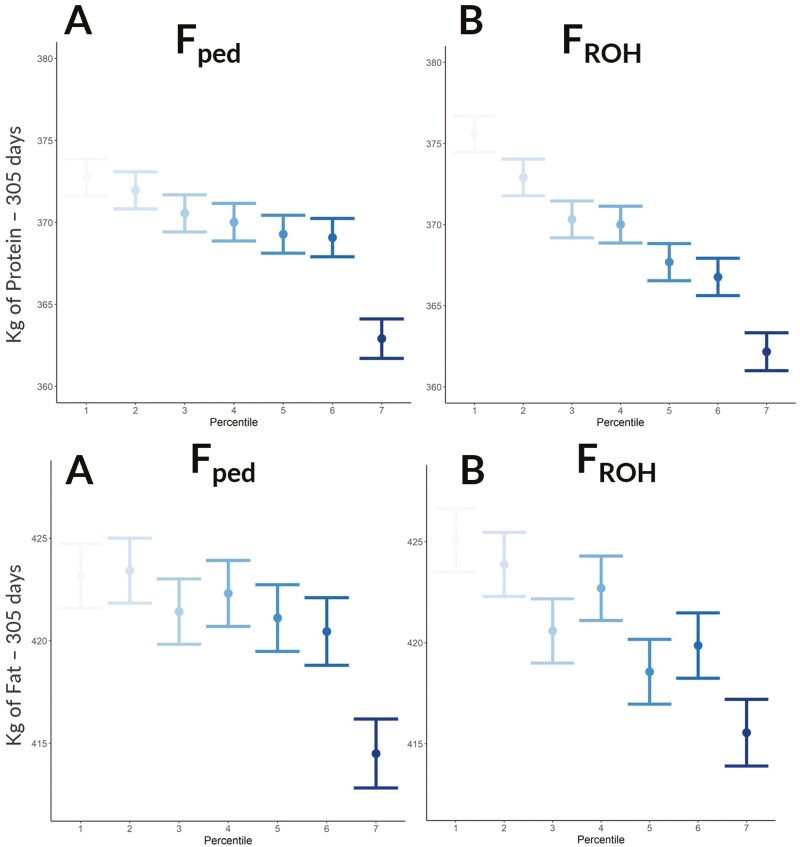
When the inbreeding was included as a fixed factor in the linear mixed model in percentile classes, the effect resulted significantly for both estimates (FPED and FROH) across all the tested productive traits (P < 0.05). The percentage of decline when comparing the two extreme percentile classes was 3.79% and 2.57% for MY, 2.35% and 2.13% for FY, and 3.46% and 2.69% for PY, respectively, for FROH and FPED. As expected, for all the evaluated traits the differences in terms of decrease of MY, FY, and PY per increase of percentile classes were more evident when including in the model the inbreeding as FROH compared to FPED. Indeed, the differences in terms of the increase of FPED percentile were only significant when comparing the highest class with the remaining ones (Figures 5 and 6), demonstrating that FPED captures less inbreeding depression compared to FROH. This could be explained since FROH captures the random nature of recombination and segregation, whereas with pedigree data this latter aspect cannot be assessed. In addition, as pointed out in a previous study, the recording errors that can happen in the pedigree might cause an attenuation of the slope towards zero, a statistical phenomenon known as regression dilution (Doekes et al., 2019). Figure 6 shows that FY displayed a more erratic trend across percentiles in both FPED and FROH compared to PY, and this is related to the high variability of the milk fat itself, as reported in the literature (Cipolat-Gotet et al., 2013).
Figure 5.
Milk production expressed in kg on 305-d in milk for FPED (pedigree-based) (A) and FROH (runs of homozygosity-based) (B) inbreeding coefficients across percentile classes.
Figure 6.
Milk protein and fat production expressed in kg on 305-d in milk for FPED (pedigree-based) (A) and FROH (runs of homozygosity-based) (B) across percentile classes.
It is key to contextualize the negative effect of inbreeding compared to what sthe gain thanks to the realized genetic progress in the breed to fully quantify the effect and the cost of inbreeding. For example, for a 305-d MY, a reduction of about 61 and 44 kg of milk per 1% increase in FROH and FPED was found. If we consider the last 5 yr evaluated in this study (2015-2020), the pedigree ΔF based on a previous work (Ablondi et al., 2022) was equal to + 2.35% and this means a loss of roughly 143 kg per lactation if estimated by FROH and of 103 kg per lactation if based on FPED. In the same 5-yr period, at the population level, the realized genetic progress for MY in the Italian Holstein breed was equal to + 415 kg (ANAFIBJ, 2023). Thus, the missed genetic progress due to inbreeding compared to the realized one can be quantified as 34.5% and 24.9% based on FROH and FPED, respectively. In other words, if expressed in potential genetic progress, it translates into 25.7% and 19.9% for FROH and FPED, respectively, if inbreeding would have not occurred. Thus, this result showed that the realized genetic progress has so far successfully counterbalanced the negative effect of inbreeding for MY; however, this might not be true for the overall sustainability of the breed over future time. This is because the overall costs of inbreeding should be quantified not only in terms of productive traits but also in terms of fitness-related traits as shown in previous studies (Leroy, 2014; Doekes et al., 2019) and by the expectation of rapidly increasing inbreeding levels in future generations. On top of that, as shown in the following analyses on the ROH length effect, recent inbreeding seems to be more harmful than ancestral ones. Thus, the current loss of genetic diversity both in terms of economic losses and the long-term adaptability of the breed in the long terms should be carefully considered.
Effect of recent and ancestral inbreeding and chromosomal homozygosity on productive performances
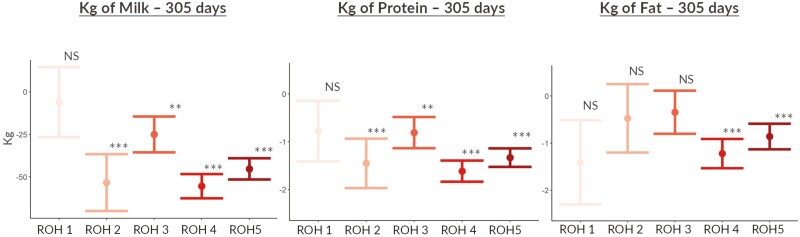
When the inbreeding was included as a fixed factor based on five different classes of ROH length, its effect was mainly significant for longer ROH which reflects more recent inbreeding compared to shorter ones (Figure 7). This result was expected and in line with a previous study in Holstein and Jersey where a stronger depression effect for 305-d MY was observed only when long ROH was included (Pryce et al., 2014). Long ROH segments are more likely to reflect recent inbreeding events, where the offspring of close relatives have inherited longer stretches of identical genomic regions which did not undergo recombination yet and might potentially carry recessive deleterious variants. Indeed, it has been shown that longer ROH segments are more likely to harbor deleterious or recessive alleles due to recent inbreeding, leading to a higher genetic load or burden (Forutan et al., 2018). The genetic load refers to the accumulation of harmful genetic variants in the genome, which can increase the risk of various health issues in the population. In contrast, shorter ROH segments as a signature of older genetic diversity reduction may carry fewer deleterious alleles or have a positive effect (Szpiech et al., 2013). Nevertheless, it is important to mention that the medium density panel used in this study (85K) might result in a false positive ROH shorter than 2 Mb as previously pointed out (Ferenčaković et al., 2013). Thus, the results from shorter ROH effects, especially for the ROH class 1: 1 to 2 Mb, should be interpreted with caution. The inability to detect short ROH precisely might be the reason behind the highest standard error found when estimating the effect of ROH class 1 on productive traits (Figure 7). Therefore, thanks to the decreasing cost of high-density panels as well as sequence data, we suggest further exploring the effect of ancestral and recent inbreeding with higher-density SNP panels. For the sake of completeness, it is important to mention that ROH of 2 to 4 Mb seemed to have the similar negative effect of longer ones (above 8 Mb) for MY and PY. However, since the standard error of the ROH 2 to 4 Mb effect was much higher, did not align with previously found in the literature, and for the above-mentioned lack of precision when detecting shorter ROH with medium density panel, we preferred to not make any inference on this result.
Figure 7.
Effect of inbreeding on milk, fat and protein yields, expressed in kg on 305-d in milk based on different ROH (Runs of homozygosity) length classes: ROH 1: 1 to 2 Mb, ROH 2: 2 to 4 Mb, ROH 3: 4 to 8 Mb, ROH 4: 8 to 16 Mb, ROH 5: 16 to 32 Mb (* = P ≤ 0.05, ** = P ≤ 0.01, and *** = P ≤ 0.001).
Finally, it is known that ROH varies across chromosomes and thus their contribution to the overall individual inbreeding, ranging in the Italian Holstein from 12.6% in Bos taurus autosome (BTA) 18 to 22.4% in BTA10 (Supplementary Figure S1). Accordingly, we aimed to quantify the effect on productive traits of each chromosomal homozygosity calculated as FROH per each chromosome. A total of 16 FROH-CHR had a negative effect on MY ranging between −1.54 and −3.53 kg of fat per lactation for BTA21 and BTA14, respectively (Table 2). Interestingly for FY, homozygosity on BTA2, which was mostly characterized by ROH limited in size (average length of 3.88 Mb) had a positive effect which highlights that homozygosity can also be signatures of positive selection for favorable traits. Moreover, a total of 19 FROH-CHR had a negative effect on PY which overall accounted for a reduction of −1.20 kg of protein per lactation for each 1% increase of inbreeding in those 19 chromosomes (Table 2). Those results on chromosome level might be used in mating schemes to constrain homozygosity at specific regions with a more deleterious effect on productive traits.
Table 2.
Estimates of the regression coefficients of FROH per significant chromosome (FROH-CHR) on quantity of milk, fat and protein, expressed in kg on 305-d in milk
| Trait | Chromosome | FROH-CHR | SE | P value |
|---|---|---|---|---|
| Milk | ||||
| BTA3 | −3.25 | 0.85 | *** | |
| BTA4 | −2.15 | 0.80 | ** | |
| BTA6 | −2.02 | 0.86 | ** | |
| BTA7 | −1.72 | 0.75 | ** | |
| BTA10 | −1.68 | 0.66 | ** | |
| BTA11 | −2.57 | 0.82 | *** | |
| BTA13 | −3.18 | 0.78 | *** | |
| BTA14 | −3.53 | 0.73 | *** | |
| BTA15 | −2.01 | 0.85 | ** | |
| BTA16 | −2.41 | 0.73 | ** | |
| BTA17 | −2.78 | 0.75 | *** | |
| BTA18 | −2.82 | 0.82 | ** | |
| BTA19 | −2.43 | 0.78 | ** | |
| BTA20 | −2.87 | 0.61 | *** | |
| BTA21 | −1.57 | 0.74 | * | |
| BTA24 | −2.29 | 0.69 | ** | |
| Fat | ||||
| BTA2 | 0.08 | 0.04 | ** | |
| BTA9 | −0.10 | 0.04 | ** | |
| BTA10 | −0.07 | 0.04 | ** | |
| BTA11 | −0.18 | 0.04 | *** | |
| BTA17 | −0.08 | 0.03 | ** | |
| BTA18 | −0.08 | 0.04 | ** | |
| BTA19 | −0.10 | 0.03 | ** | |
| BTA21 | −0.12 | 0.03 | *** | |
| BTA24 | −0.06 | 0.03 | * | |
| BTA27 | −0.07 | 0.31 | ** | |
| Protein | ||||
| BTA3 | −0.05 | 0.03 | * | |
| BTA4 | −0.05 | 0.03 | * | |
| BTA5 | −0.07 | 0.03 | ** | |
| BTA6 | −0.08 | 0.03 | ** | |
| BTA7 | −0.06 | 0.03 | ** | |
| BTA9 | −0.05 | 0.03 | * | |
| BTA10 | −0.06 | 0.02 | ** | |
| BTA11 | −0.10 | 0.02 | *** | |
| BTA13 | −0.08 | 0.02 | *** | |
| BTA14 | −0.10 | 0.03 | *** | |
| BTA16 | −0.05 | 0.02 | ** | |
| BTA17 | −0.07 | 0.02 | ** | |
| BTA18 | −0.07 | 0.02 | ** | |
| BTA19 | −0.06 | 0.03 | * | |
| BTA20 | −0.05 | 0.02 | ** | |
| BTA21 | −0.05 | 0.02 | ** | |
| BTA23 | −0.07 | 0.02 | *** | |
| BTA24 | −0.05 | 0.02 | ** | |
| BTA27 | −0.04 | 0.02 | * | |
* = P ≤ 0.05, ** = P ≤ 0.01 and *** = P ≤ 0.001; FROH: Inbreeding based on Runs of Homozygosity; BTA: Bos taurus autosome.
Implications of inbreeding depression in the dairy industry
Production traits such as milk, fat and protein yields are crucial components for cheese manufacture. They are fundamental to produce high-quality and labeled dairy foods, where a reduction in milk, fat, and protein yields can lead to lower-quality cheeses that may not meet the stringent standards required for their production. Moreover, since fat and protein are key for cheese yield, their reduction is associated with decreased profitability for the cheese factories. Over time, selection indexes have been improved by targeting specific market demands or tackling emerging issues related to the sustainability of the dairy industry, as well as by using genomic data. All those changes in the breeding programs have enhanced the productivity while reducing the genetic variability of the dairy cattle population due to intense use of few elite sires. Thus, the economic impact of inbreeding today translates into increased economic losses along the dairy chain due to inbreeding depression although so far counterbalanced by genetic progress. This study corroborated the negative effects of inbreeding on production traits and suggested that more awareness and understanding of inbreeding depression in dairy cattle are needed to adopt mitigation measurements for the future sustainability of the dairy sector. The results of this study are even more alarming when combined with what previously found at genetic diversity level in the breed throughout a period of 18 yr (2002 to 2020) analyzing 74,485 cows where a significant increase in the ΔF was found since the introduction of genomic selection in the breed (Ablondi et al., 2022).
Conclusions
This study compared the different levels of inbreeding based on pedigree (FPED) and SNP data (FROH), and the presence of inbreeding depression for productive performance in the Italian Holstein Friesian cows. In particular, the inbreeding included both as regressors as well as percentile classes was significant for both methods (FPED and FROH) and all the evaluated traits. Moreover, the differences in terms of decrease of MY, FY, and PY per increase of percentile classes were larger when included in the model inbreeding as FROH compared to FPED, suggesting that FPED captures less inbreeding depression compared to FROH. In addition, when including inbreeding based on classes of ROH length, the effect of inbreeding was mainly significant for longer ROH compared to shorter ones, meaning that the recent inbreeding has a more deleterious effect than the ancestral one. Finally, based on a chromosomal homozygosity evaluation, we could identify specific chromosomes which seem to have a more deleterious effect on productive traits. Thus, it is key to further implement strategies at a mating level to reduce inbreeding and thus the damage from the loss of genetic variation in the future generations.
Supplementary Material
Acknowledgments
This research was granted by University of Parma through the action Bando di Ateneo 2021 per la ricerca co-funded by MUR-Italian Ministry of Universities and Research—D.M. 737/2021—PNR—PNRR—NextGenerationEU. This study was also supported by “Latteco2 project, sottomisura 10.2 of the National Rural Development Program (PSRN) - Biodiversity 2020–2023” [Ministero delle politiche agricole, alimentari e forestali (MIPAAF). D.M. No. 465907 del 24/09/2021, project unique code J12C21004080005], National Breeders Association of Italian Holstein, Brown, and Jersey (ANAFIBJ, Cremona, Italy).
Glossary
Abbreviations
- CGE
complete generation equivalent
- F
inbreeding
- FPED
pedigree-based inbreeding coefficient
- FROH
SNP-based inbreeding coefficient
- FROH-CHR
chromosome homozygosity-specific contribution
- FY
fat yield
- MY
milk yield
- PDO
protected designation of origin
- PY
protein yield
- ROH
runs of homozygosity
- SNP
single nucleotide polymorphism
Contributor Information
Michela Ablondi, Department of Veterinary Science, University of Parma, 43126 Parma, Italy.
Andrea Summer, Department of Veterinary Science, University of Parma, 43126 Parma, Italy.
Giorgia Stocco, Department of Veterinary Science, University of Parma, 43126 Parma, Italy.
Raffaella Finocchiaro, Associazione Nazionale Allevatori della Razza Frisona Bruna e Jersey Italiana (ANAFIBJ), 26100 Cremona, Italy.
Jan-Thijs van Kaam, Associazione Nazionale Allevatori della Razza Frisona Bruna e Jersey Italiana (ANAFIBJ), 26100 Cremona, Italy.
Martino Cassandro, Associazione Nazionale Allevatori della Razza Frisona Bruna e Jersey Italiana (ANAFIBJ), 26100 Cremona, Italy; Department of Agronomy, Food, Natural resources, Animals and Environment, University of Padova, 35020 Legnaro, Italy.
Christos Dadousis, Department of Veterinary Science, University of Parma, 43126 Parma, Italy.
Alberto Sabbioni, Department of Veterinary Science, University of Parma, 43126 Parma, Italy.
Claudio Cipolat-Gotet, Department of Veterinary Science, University of Parma, 43126 Parma, Italy.
Conflict of interest statement. The authors declare no real or perceived conflicts of interest.
Literature Cited
- Ablondi, M., Malacarne M., Cipolat-Gotet C., van Kaam J.-T., Sabbioni A., and Summer A... 2021. Genome-wide scan reveals genetic divergence in Italian Holstein cows bred within PDO cheese production chains. Sci. Rep. 11:12601. doi: 10.1038/s41598-021-92168-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablondi, M., Sabbioni A., Stocco G., Cipolat-Gotet C., Dadousis C., van Kaam J.-T., Finocchiaro R., and Summer A... 2022. Genetic diversity in the italian holstein dairy cattle based on pedigree and SNP data prior and after genomic selection. Front. Vet. Sci 8:1–11. doi: 10.3389/fvets.2021.773985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar, I., Misztal I., Legarra A., and Tsuruta S... 2011. Efficient computation of the genomic relationship matrix and other matrices used in single-step evaluation. J. Anim. Breed. Genet. 128:422–428. doi: 10.1111/j.1439-0388.2010.00912.x [DOI] [PubMed] [Google Scholar]
- AIA. 2023. Italy: milk recording activity - Official Statistics - Year 2023. Roma. [Google Scholar]
- ANAFIBJ. 2023. ANAFIBJ Annual Evaluation. Accessed November 20, 2021. http://www.anafi.it/it/servizi/web-anafi-mate
- Baes, C. F., Makanjuola B. O., Miglior F., Marras G., Howard J. T., Fleming A., and Maltecca C... 2019. Symposium review: The genomic architecture of inbreeding: How homozygosity affects health and performance. J. Dairy Sci. 102:2807–2817. doi: 10.3168/jds.2018-15520 [DOI] [PubMed] [Google Scholar]
- Biscarini, F., Cozzi P., Gaspa G., and Marras G... 2019. detectRUNS: an R package to detect runs of homozygosity and heterozygosity in diploid genomes. CRAN (The Comprehensive R Archive Network); 2018. https://orca.cardiff.ac.uk/id/eprint/108906/.
- Bosse, M., Megens H.-J., Madsen O., Paudel Y., Frantz L. A. F., Schook L. B., Crooijmans R. P. M. A., and Groenen M. A. M... 2012. Regions of homozygosity in the porcine genome: consequence of demography and the recombination landscape. PLoS Genet. 8:e1003100. doi: 10.1371/journal.pgen.1003100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito, L. F., Bedere N., Douhard F., Oliveira H. R., Arnal M., Peñagaricano F., Schinckel A. P., Baes C. F., and Miglior F... 2021. Review: Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal. 15:100292. doi: 10.1016/j.animal.2021.100292 [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., and Willis J. H... 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10:783–796. doi: 10.1038/nrg2664 [DOI] [PubMed] [Google Scholar]
- Cipolat-Gotet, C., Cecchinato A., De Marchi M., and Bittante G... 2013. Factors affecting variation of different measures of cheese yield and milk nutrient recovery from an individual model cheese-manufacturing process. J. Dairy Sci. 96:7952–7965. doi: 10.3168/jds.2012-6516 [DOI] [PubMed] [Google Scholar]
- Cipolat-Gotet, C., Cecchinato A., Malacarne M., Bittante G., and Summer A... 2018. Variations in milk protein fractions affect the efficiency of the cheese-making process. J. Dairy Sci. 101:8788–8804. doi: 10.3168/jds.2018-14503 [DOI] [PubMed] [Google Scholar]
- CLAL. 2022. Parmigiano Reggiano Cheese Production. [Accessed May 12, 2023]. https://www.clal.it/en/?section=produzioni_parmigiano
- CLAL. 2023. Production Volumes of the Italian PDO Cheeses. [Accessed May 5, 2023]. https://www.clal.it/en/?section=formaggi_dop
- Curik, I., Ferenčaković M., and Sölkner J... 2014. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 166:26–34. doi: 10.1016/j.livsci.2014.05.034 [DOI] [Google Scholar]
- Dadousis, C., Ablondi M., Cipolat-Gotet C., van Kaam J.-T., Marusi M., Cassandro M., Sabbioni A., and Summer A... 2022. Genomic inbreeding coefficients using imputed genotypes: assessing different estimators in Holstein-Friesian dairy cows. J. Dairy Sci. 105:5926–5945. doi: 10.3168/jds.2021-21125 [DOI] [PubMed] [Google Scholar]
- Dadousis, C., Ablondi M., Cipolat-Gotet C., van Kaam J.-T., Finocchiaro R., Marusi M., Cassandro M., Sabbioni A., and Summer A... 2023. Genomic inbreeding coefficients using imputed genotypes: assessing differences among SNP panels in Holstein-Friesian dairy cows. Front. Vet. Sci. 10:1–10. doi: 10.3389/fvets.2023.1142476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doekes, H. P., Veerkamp R. F., Bijma P., de Jong G., Hiemstra S. J., and Windig J. J... 2019. Inbreeding depression due to recent and ancient inbreeding in Dutch Holstein–Friesian dairy cattle. Genet. Sel. Evol. 51:54. doi: 10.1186/s12711-019-0497-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet, A. C., Croiseau P., Fritz S., Michenet A., Hozé C., Danchin-Burge C., Laloë D., and Restoux G... 2019. The impact of genomic selection on genetic diversity and genetic gain in three French dairy cattle breeds. Genet. Sel. Evol. 51:1–13. doi: 10.1186/s12711-019-0495-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenčaković, M., Sölkner J., and Curik I... 2013. Estimating autozygosity from high-throughput information: effects of SNP density and genotyping errors. Genet. Sel. Evol. 45:42. doi: 10.1186/1297-9686-45-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forutan, M., Ansari Mahyari S., Baes C., Melzer N., Schenkel F. S., and Sargolzaei M... 2018. Inbreeding and runs of homozygosity before and after genomic selection in North American Holstein cattle. BMC Genomics. 19:98. doi: 10.1186/s12864-018-4453-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, P.F., Guinee T.P., Cogan T.M., and McSweeney P.L.H... 2017. Fundamentals of Cheese Science. Boston, MA: Springer US. [Google Scholar]
- García-Ruiz, A., Cole J. B., VanRaden P. M., Wiggans G. R., Ruiz-López F. J., and Van Tassell C. P... 2016. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. U.S.A. 113:E3995–E4004. doi: 10.1073/pnas.1519061113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, A.R., Gogel B.J., Cullis B.R., Welham S.J., and Thompson R... 2015. ASReml User Guide. [Google Scholar]
- Gutiérrez-Reinoso, M. A., Aponte P. M., and García-Herreros M... 2022. A review of inbreeding depression in dairy cattle: current status, emerging control strategies, and future prospects. J. Dairy Res. 89:3–12. doi: 10.1017/s0022029922000188 [DOI] [PubMed] [Google Scholar]
- Hill, W. G., and Weir B. S... 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. (Camb). 93:47–64. doi: 10.1017/S0016672310000480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. T., Pryce J. E., Baes C., and Maltecca C... 2017. Invited review: inbreeding in the genomics era: inbreeding, inbreeding depression, and management of genomic variability. J. Dairy Sci. 100:6009–6024. doi: 10.3168/jds.2017-12787 [DOI] [PubMed] [Google Scholar]
- Koutouzidou, G., Ragkos A., Theodoridis A., and Arsenos G... 2022. Entrepreneurship in dairy cattle sector: key features of successful administration and management. Land 11:1736. doi: 10.3390/land11101736 [DOI] [Google Scholar]
- Leroy, G. 2014. Inbreeding depression in livestock species: review and meta-analysis. Anim. Genet. 45:618–628. doi: 10.1111/age.12178 [DOI] [PubMed] [Google Scholar]
- Maignel, L., Boichard D., and Verrier E... 1996. Genetic variability of French dairy breeds estimated from pedigree information. Interbull Bull. 14:49–53. [Google Scholar]
- Makanjuola, B. O., Miglior F., Abdalla E. A., Maltecca C., Schenkel F. S., and Baes C. F... 2020. Effect of genomic selection on rate of inbreeding and coancestry and effective population size of Holstein and Jersey cattle populations. J. Dairy Sci. 103:5183–5199. doi: 10.3168/jds.2019-18013 [DOI] [PubMed] [Google Scholar]
- Martikainen, K., Tyrisevä A. M., Matilainen K., Pösö J., and Uimari P... 2017. Estimation of inbreeding depression on female fertility in the Finnish Ayrshire population. J. Anim. Breed. Genet. 134:383–392. doi: 10.1111/jbg.12285 [DOI] [PubMed] [Google Scholar]
- Martin, B., Verdier-Metz I., Buchin S., Hurtaud C., and Coulon J.-B... 2005. How do the nature of forages and pasture diversity influence the sensory quality of dairy livestock products? Anim. Sci. 81:205–212. doi: 10.1079/asc50800205 [DOI] [Google Scholar]
- McQuillan, R., Leutenegger A.-L., Abdel-Rahman R., Franklin C. S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A.,. et al. 2008. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83:359–372. doi: 10.1016/j.ajhg.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolazzi, E. L., Biffani S., and Jansen G... 2013. Short communication: imputing genotypes using PedImpute fast algorithm combining pedigree and population information. J. Dairy Sci. 96:2649–2653. doi: 10.3168/jds.2012-6062 [DOI] [PubMed] [Google Scholar]
- Parland, S. M., Kearney J. F., Rath M., and Berry D. P... 2007. Inbreeding effects on milk production, calving performance, fertility, and conformation in Irish Holstein-Friesians. J. Dairy Sci. 90:4411–4419. doi: 10.3168/jds.2007-0227 [DOI] [PubMed] [Google Scholar]
- Pryce, J. E., Haile-Mariam M., Goddard M. E., and Hayes B. J... 2014. Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle. Genet. Sel. Evol. 46:71. doi: 10.1186/s12711-014-0071-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R. R., Bender D., Maller J., Sklar P., de Bakker P. I. W. W., Daly M. J.,. et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ramilo, S. T., Fernández J., Toro M. A., Hernández D., and Villanueva B... 2015. Genome-Wide estimates of coancestry, inbreeding and effective population size in the Spanish Holstein population. PLoS One 10:e0124157–e0124111. doi: 10.1371/journal.pone.0124157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowicz, K., Sargolzaei M., Miglior F., and Schenkel F. S... 2011. Rates of inbreeding and genetic diversity in Canadian Holstein and Jersey cattle. J. Dairy Sci. 94:5160–5175. doi: 10.3168/jds.2010-3308 [DOI] [PubMed] [Google Scholar]
- Szpiech, Z. A., Xu J., Pemberton T. J., Peng W., Zöllner S., Rosenberg N. A., and Li J. Z... 2013. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 93:90–102. doi: 10.1016/j.ajhg.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmann, R. 2019. Optimum contribution selection for animal breeding and conservation: the R package optiSel. BMC Bioinf. 20:25. doi: 10.1186/s12859-018-2450-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Calus M. P. L., Guldbrandtsen B., Lund M. S., and Sahana G... 2015. Estimation of inbreeding using pedigree, 50k SNP chip genotypes and full sequence data in three cattle breeds. BMC Genet. 16:1–11. doi: 10.1186/s12863-015-0227-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.