Abstract
Paracoccidioides spp. is the etiologic agent of Paracoccidioidomycosis (PCM), a systemic disease with wide distribution in Latin America. Macrophages are very important cells during the response to infection by P. brasiliensis. In this study, we performed a proteomic analysis to evaluate the consequences of P. brasiliensis yeast cells on the human THP-1 macrophage proteome. We have identified 443 and 2247 upregulated or downregulated proteins, respectively, in macrophages co-cultured with yeast cells of P. brasiliensis in comparison to control macrophages unexposed to the fungus. Proteomic analysis revealed that interaction with P. brasiliensis caused metabolic changes in macrophages that drastically affected energy production pathways. In addition, these macrophages presented regulated many factors related to epigenetic modifications and gene transcription as well as a decrease of many proteins associated to the immune system activity. This is the first human macrophage proteome derived from interactions with P. brasiliensis, which contributes to elucidating the changes that occur during the host response to this fungus. Furthermore, it highlights proteins that may be targets for the development of new therapeutic approaches to PCM.
Keywords: Paracoccidioides brasiliensis, proteomic analysis, metabolic reprogramming, immunometabolism, macrophages
1. Introduction
Paracoccidioidomycosis (PCM) is a systemic mycosis widely distributed in Latin America; Brazil is responsible for the largest number of reported cases. The etiological agent of PCM consists of thermodimorphic fungus of the genus Paracoccidioides, which presents the mycelium form in the soil (25°C) and the yeast form in the host (37°C). Paracoccidioides brasiliensis has a wide distribution in Latin America and is the most studied species. PCM affects mainly rural workers, and infection occurs by inhalation of the infectious propagules present in the soil that, when reaching the lungs, differentiate into yeast cells. The lung is the main organ affected during the disease; however, yeast cells can proliferate in the lung and spread through the host organism affecting organs such as lymph nodes, liver, and skin (Shikanai-Yasuda et al., 2006; Mendes et al., 2017; Chaves et al., 2021).
Individuals infected with Paracoccidioides spp. may present different clinical manifestations depending on both pathogen and host factors. The infection mainly affects children, adolescents, and young adults, who usually develop a T helper (Th) lymphocyte response with a Th2/Th9 profile. This profile is associated with susceptibility and characterizes the acute form of PCM. On the other hand, the chronic form of the disease is characterized by a slow progression and predominantly affects adults over 30 years of age, who usually develop a Th17/Th22 type response, and a substantial participation of Th1 lymphocytes. This mixed immune response is crucial for infection control. Asymptomatic individuals usually develop a Th1-type immune response, which can contain the spread of the fungus and prevent the progression of the disease (Oliveira et al., 2002; De Castro et al., 2013; Martinez, 2017).
The innate immune mechanisms responsible for pathogen elimination are induced by pathogen-associated molecular patterns (PAMPs) of the fungus that activate pattern-recognizing receptors (PRRs) on the host cells. Therefore, fungus cell wall composition is a critical factor for the pathogen-host interaction and, consequently, the outcome of the infection (Gow et al., 2017). The hypha to yeast transition of Paracoccidioides spp. involves changes in the composition and structure of the cell wall, which are necessary for the establishment of the infection. Yeast cells present a thicker cell wall as well as a higher amount of chitin and α-glucans than hyphae (Chaves et al., 2021). As β-glucan is a relevant PAMP to activate the innate immune response against Paracoccidioides spp. (Loures et al., 2014), the masking of β-glucan by α-glucan is considered an immune evasion mechanism (Puccia et al., 2011). In addition, P. brasiliensis has other virulence factors such as proteases and adhesins, which facilitate the invasion and colonization of tissue by the fungus (Vicentini et al., 1994; Hernández et al., 2010; Bailão et al., 2012; Lacerda Pigosso et al., 2017).
Macrophage are a crucial cell in the immune response against fungal infections. The recognition of fungal PAMPs and/or activation by cytokines triggers macrophage microbicidal mechanisms. These responses include the production of reactive oxygen (ROS) or nitrogen (RNS) species, which are responsible by the killing of the pathogen (Bogdan et al., 2000). However, even though living inside macrophages is a big challenge Paracoccidioides spp. may evade of the immune response and multiply inside these cells (Brown et al., 2009). Therefore, efficient infection control depends on how macrophages are activated. As described above, type 1 cytokines, such as interferon-gamma (IFNγ) and tumor necrosis factor (TNF), activate macrophages, contributing to a pro-inflammatory phenotype and increased microbicidal activity. On the other hand, in the presence of type 2 or regulatory cytokines (IL-4, IL-13, IL-10) macrophages are alternatively activated, showing an anti-inflammatory profile, and contribute to tissue repair (Ferrante and Leibovich, 2012). Recently, our group demonstrated that murine alveolar macrophages activated with IFNγ are less permissive to P. brasiliensis survival and proliferation (Chaves et al., 2019). Other studies have also demonstrated its relevance for P. brasiliensis infection control (Brummer et al., 1988; Brummer et al., 1989; Moscardi-Bacchi et al., 1994; Gonzalez et al., 2000).
It has been shown that during the infections, the host cells and fungi can undergo reprogramming in their gene expression and metabolism. The fungus changes to establish infection and evade the immune system while the host cell adopts mechanisms to eradicate the infection (Silva et al., 2008; De Arruda Grossklaus et al., 2013; Parente-Rocha et al., 2015; Lacerda Pigosso et al., 2017; Weerasinghe and Traven, 2020). Our group has studied the proteome of Paracoccidioides spp. after interaction with macrophages. Members of the Paracoccidioides complex have been observed to respond to host cell effector mechanisms through an oxidative stress response characterized by increase of several antioxidant enzymes and virulence factors. In addition, fungus presents metabolic reprogramming changing from glycolysis to gluconeogenesis, beta-oxidation, and catabolism of amino acids (De Arruda Grossklaus et al., 2013; Parente-Rocha et al., 2015; Lacerda Pigosso et al., 2017; Chaves et al., 2019). Despite these descriptions, so far there is only one study that performed a proteomic analysis focusing on the host response to fungal infection whose objective was to identify serum biomarkers of severe pulmonary sequelae and relevant signaling pathways in different clinical stages of patients with PCM (Santos et al., 2021). Santos et al. (2021) have identified 72 proteins differentially present in the serum of patients with severe sequelae compared to the serum of patients with mild/moderate sequelae. In addition, the study showed that the development of severe sequelae is associated with an increase in glycolytic pathway proteins and proteins involved in oxygen exchange, suggesting high cellular activity and deregulation of wound healing. Understanding the host-pathogen interface is an important tool that can help the development of new therapeutic strategies for diseases. THP-1 cells, a human leukemia monocytic cell line, have been one of the widely used models for studies on monocytes and macrophages. Among several advantages, THP-1 cells are easily derived into macrophages and are advantageous for studies that require a large number of cells. Furthermore, these cells can be stored for several years, and as they are immortalized, they can be cultured for a long time (Chanput et al., 2014). Thus, THP-1 macrophages are a good experimental in vitro model to study the human host response to P. brasiliensis infection. The present study aimed to evaluate the effect of P. brasiliensis on the THP-1 macrophage proteome, highlighting the proteins from the host cell are produced in response to the first 24 h of this interaction.
2. Materials and methods
2.1. Culture of Paracoccidioides brasiliensis yeast cells
Yeast cells of P. brasiliensis (Pb18 - ATCC 32069) were grown in complete solid Fava-Netto medium [1% (w/v) peptone (Kasvi, São José dos Pinhais, PR, Brazil), 0.5% (w/w) yeast extract (BD, Franklin Lakes, NJ, USA), 0.3% (w/v) proteose peptone (Sigma-Aldrich, Saint Louis, MO, USA), 0.5% (w/v) meat extract (BD), 0.5% (w/v) NaCl (Sigma-Aldrich), 4% (w/w) glucose (Sigma-Aldrich), 1% (w/v) brain and heart infusion (BD) and 1.2% (w/v) agar (BD), pH 7.2]. The medium was supplemented with 5% (v/v) fetal bovine serum (FBS, Cripion, Andradina, SP, Brazil) and 10 μg/mL gentamicin (Sigma-Aldrich). The culture was maintained at 36°C and sub cultivations were made every seven days (Bastos et al., 2007). To obtain yeast cells of P. brasiliensis, fungi were cultured in brain and heart infusion (BD) medium, supplemented with 4% (w/w) glucose, 5% (v/v) FBS, and 10 μg/mL gentamicin (Complete BHI medium). The culture was maintained at 36 °C during three days under agitation, centrifuged (1,400 ɡ, 5 min), washed three times with phosphate-buffered saline solution (PBS), and the supernatant was discarded. Yeast cell quantification was performed on a hematocytometer, and vital staining with 0.1% (w/v) trypan blue in PBS assessed cell viability.
2.2. Obtaining THP-1 macrophage and interaction assays
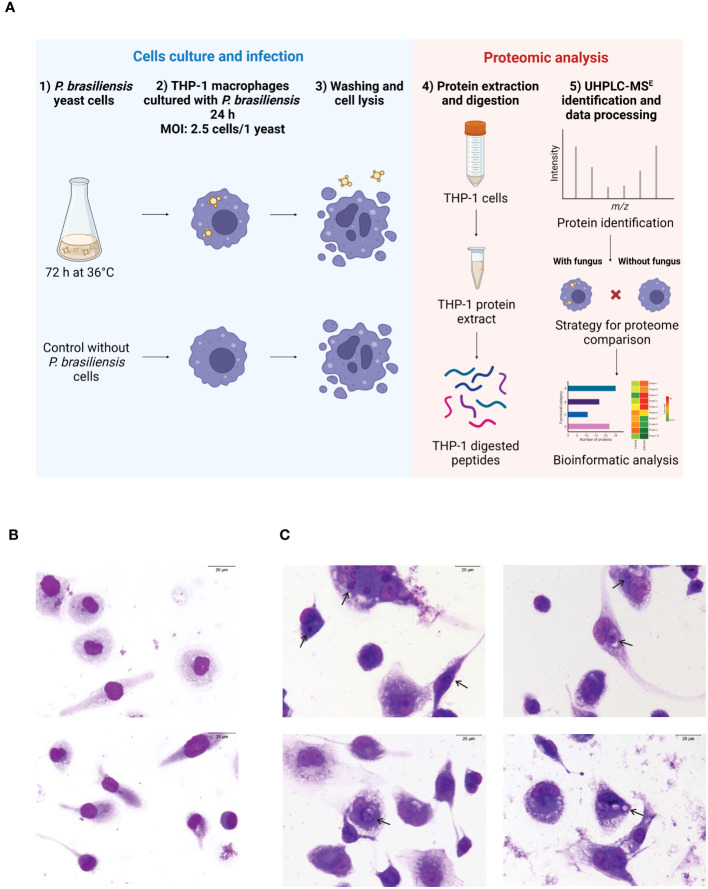
The human monocyte cell line THP-1 was cultured in RPMI 1640 medium (Sigma), supplemented with 10% inactivated FBS (Gibco), 2 mM L-glutamine (Sigma), 11 mM sodium bicarbonate, 100 U/mL of penicillin (Sigma), and 100 µg/mL of streptomycin (Sigma), referred as complete medium. Cells were maintained in a humidified atmosphere with at 5% CO2, at 37°C. To obtain macrophages, THP-1 cells were cultivated in petri dishes for cell culture (TPP, Switzerland) (106 cells/mL) and incubated with 100 ng/mL of phorbol myristate acetate (PMA) for 48 h. After incubation, the cells were washed 5x with PBS at 37°C. After washing, the complete medium was replaced, and the cells were re-incubated for 24 h (Lund et al., 2016). Then, the THP-1 macrophages were incubated with yeast cells of P. brasiliensis (MOI: 2.5 cells/1 yeast) for 24 h. To obtain photomicrographs of THP-1 macrophages co-cultured with the fungi, glass coverslips (Knittel Glass, SP, São Paulo) were added to a 24-well plate (Corning-Costar - Kennebunk, ME, USA), and the same experimental procedure for cell derivation and macrophage co-cultured with the fungus was performed as described above. Coverslips were harvested after 24 h, and cells were fixed with absolute methanol, and stained with Giemsa (Merck Millipore, Damstadt, Germany). Photomicrographs were taken under a light microscope (400x magnification). The percentage of infected cells and the mean of fungi per cell were obtained by counting a total of 300 cells per coverslips (experimental triplicate). The infection index was calculated as % x mean of fungi per cell. A schematic presentation of the cell and fungus interaction conditions is presented in Figure 1A .
Figure 1.
THP-1 macrophages and Paracoccidioides brasiliensis yeast cells were co-cultured for 24 h, and proteomic analysis was done by UHPL-MSE. (A) Flowchart of the methodology (Created with BioRender.com). (B) Photomicrographs of macrophages in the absence of fungus. (C) Photomicrographs of macrophages co-cultured with the fungi. Arrows indicate cells with internalized fungus (400 times magnification). Scale bar is 20 μm. Cells were stained with Giemsa.
2.3. Preparation and digestion of protein extracts
After 24h infection, the macrophages were rinsed to remove the unphagocytosed yeast cells. THP-1 macrophages co-cultured with P. brasiliensis or not (control macrophages) were removed from the plates with ice-cold PBS using a cell scraper (Corning), washed twice by centrifugation (600 g for 10 min, at 10 °C) and the pellet was resuspended in ammonium bicarbonate buffer (NH4HCO3) 50 mM, pH 8.5. Then, the cells were centrifuged at 600 g for 10 min at 4°C, and the supernatant was discarded; NH4HCO3 50 mM, pH 8.5, and glass beads were added to the pellet. Cells were mechanically lysed in bead beater equipment (BioSpec Products Inc., Bartlesville, OK, USA) for 5 cycles of 30 s. The lysate was centrifuged three times (10,000 ɡ, 10 min, at 4°C); for protein quantification, the Bradford reagent (Sigma Aldrich) was used. The enzymatic digestion of proteins was performed based on Murad et al. (2011). In brief, to microcentrifuge tubes (Axygen, Tewksbury, MA) were added 150 μg of protein sample, 10 μL of NH4HCO3 at 50 mM, pH 8.5 and 75 μL of RapiGest SF solution (0.2% v/v) (Waters, Milford, MA, USA). The samples were vortexed and incubated in a thermoblock at 80°C for 15 min. To reduce disulfide bonds, 2.5 μL of 100 mM dithiothreitol (DTT) (GE Healthcare, Little Chalfont, UK) was added and the tubes were incubated at 60°C for 30 min. Then, for cysteine alkylation, 2.5 μL of 300 mM iodocetamide (GE Healthcare, Piscataway, NJ, USA) was added, followed by vortexing and incubation at room temperature in the dark, for 30 min. Proteins were digested with 30 μL of trypsin solution (Promega, Madison, WI, USA) 0.05 mg/mL in 50 mM ammonium bicarbonate, then vortexed and incubated in a thermoblock at 37°C for 16 h. Stopping trypsin digestion and RapiGEST precipitation were performed by acidifying samples with 30 μL of 5% (v/v) trifluoroacetic acid (TFA) (Sigma-Aldrich). The samples were vortexed, incubated in a thermoblock at 37°C for 90 min and centrifuged at 18,000 ɡ, at 4°C for 30 min. Then supernatant was transferred to another tube and dried in a speed vacuum device (Eppendorf, Hamburg, Germany). The peptides were resuspended in 80 μL of ammonium formate (20 mM NH4HCO3, pH 10.0).
2.4. Data processing, protein identification and bioinformatics analysis
We used the ACQUITY UPLC® M-Class system (Waters Corporation, Manchester, UK) coupled to Synapt G1 HDMS™ mass spectrometer (Waters Micromass, Manchester, UK) to perform Ultra High-Performance Liquid Chromatography (Baeza et al., 2017). Oryctolagus cuniculus glycogen phosphorylase_muscle form (MassPREP™ Digestion Standard) was used as internal standard (200 fmol/mL.) and [GLU1]-Fibrinopeptide B (GFB) for calibration. ProteinLynx Global Server software (PLGS) version 3.0.2 (Waters, Manchester, UK) loaded with a specific database for Homo sapiens was used to process the mass spectra according to Geromanos et al. (2009). So, the protein IDs were obtained by using a one-randomized Homo sapiens databank in the PLGS software. In summary, the PLGS software uses fourteen parameters to identify a protein in the provided database. According to Moraes et al. (2023), the following protein identification parameters were used: the detection of at least two fragment ions per peptide; five fragment per protein; the determination of at least one unique peptide per protein; carbamidomethylation of cysteine as a fixed modification; variable modifications were considered to be serine, threonine and tyrosine phosphorylations and methionine oxidation. In addition, a maximum protein mass (600 kDa); a lost cleavage site was allowed for trypsin; and 4% of the maximum false positive ratio were considered. Proteins identified with differences in abundance of minimum fold-change of 50% were considered as regulated, that is, fold-change ≤ 0.67 (ratio 1/1.5) to downregulated proteins and fold change ≥ 1.5 (ratio 1.5/1) to upregulated proteins). The PLGS score, the result of different mathematical-statistical models for predicting the assignment of peptides and fragments, was presented for each protein. The number of protein identifications in the three replicates can be verified in column B of Supplementary Spreadsheet 1 , with the number one indicating that the protein was identified in two replicates and the number two indicating that it was identified in three replicates. Proteins identified in only one replicate were excluded from the analysis. To classify and separate identified proteins according to their function in different biological pathways were used online tools: Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (https://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/#browse). The proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022; Deutsch et al., 2023), partner repository with the dataset identifier PXD043727.
2.5. Assessment of macrophage mitochondrial activity
To verify whether P. brasiliensis yeasts altered the mitochondrial membrane integrity and mitochondrial activity of THP-1 macrophages co-cultured or not with the fungi, cells were stained with Rhodamine 123 (Sigma-Aldrich) and MitoTracker Green FM (Sigma-Aldrich). After 24 h of interaction with the fungi, macrophages were collected and simultaneously stained with Rhodamine 123 (2.4 μM) and MitoTracker Green FM (400 μM) and incubated for 45 min in the dark, at room temperature. Cells were centrifuged and washed three times with PBS. Fluorescence was analyzed using a fluorescence microscope (Zeiss Axiocam MRc-Scope A1) using a wavelength of 515–575 nm for Rhodamine and 450–490 nm for MitoTracker Green. The images were captured at 400x magnification. The AxioVision software (Carl Zeiss) evaluated the pixel density. The quantification of pixels was performed in triplicate and all macrophage cells that showed fluorescence, delimited borders and without overlapping were evaluated (according to Rocha et al. (2022). Data represent the mean and standard deviation of three biological replicates. The log of 2.7 was used to normalize the data for statistical analyses. For statistical analysis, the Student’s t test was used, and the established significance level was p ≤ 0.05.
2.6. Evaluation of the production of reactive oxygen species by THP-1 macrophages
To evaluate the production of ROS by THP-1 macrophages co-cultured or not with fungi, cells were stained with 2’,7’-dichlorodihydrofluorescein diacetate (DCFH-DA) (Sigma-Aldrich) at 25 µM in the dark, for 25 min. After this period, the macrophages were washed three times with PBS and analyzed in a fluorescence microscope (Axiocam MRc-Scope A1) using the 450-490 nm filter. Pixel density was determined using the AxioVision software (Carl Zeiss). The images were captured at 400x magnification. The quantification of pixels was performed in triplicate and all macrophage cells that showed fluorescence, delimited borders and without overlapping were evaluated. Data represent the mean and standard deviation of three biological replicates. The log of 2.7 was used to normalize the data for statistical analyses. Data was analyzed by Student’s t test and the established significance level was p ≤ 0.05.
3. Results
3.1. P. brasiliensis infects THP-1 macrophages
A flowchart of the methodology is shown in Figure 1A . Macrophages derived from THP-1 monocytes were obtained as described in Section 2.2. To investigate the effect of P. brasiliensis on THP-1 macrophage proteome, these cells were incubated in the absence ( Figure 1B ) or presence of yeast cells of P. brasiliensis for 24 h ( Figure 1C ). This incubation time was selected based on experiments with THP-1-derived macrophages, showing 24 has adequate time to observe production of cytokines, enzymes such as inducible nitric oxide synthase (iNOS) and other proteins following activation by microorganisms or PAMPs (He et al., 2012; Meijer et al., 2015; Dos Santos et al., 2017). Macrophages were efficiently infected by the fungi, as depicted in Figure 1C . After 24 h of co-cultures, the percentage of infected macrophages was 24% with a mean of 1.5 yeasts per cell, leading to an infection index 36.
3.2. Differentially regulated proteins in TH1-P macrophages co-cultured with P. brasiliensis
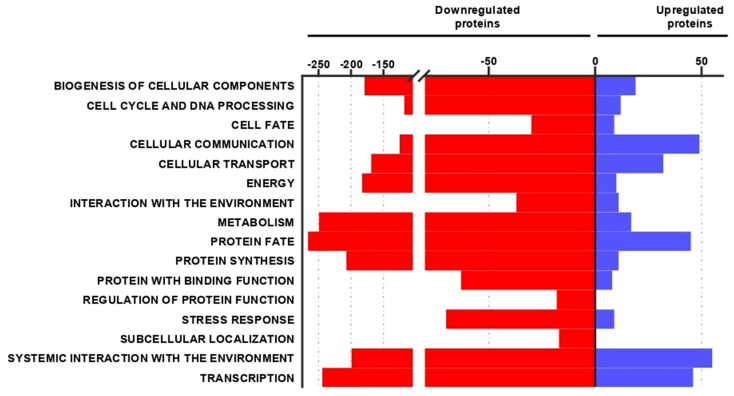
To study the effects of P. brasiliensis on human macrophage proteomes, these host cells were cultured in the absence or presence of the fungal yeasts. In this large-scale analysis, macrophages were collected at 24 h of incubation. Samples were analyzed in biological triplicates and mass spectrometry analysis provided 2,905 identified proteins ( Supplementary Spreadsheet 1 ). Of all proteins, 65% were identified in three replicates and 35% were identified in two replicates. Considering a 1.5-fold change value, the proteomic analysis identified 2,690 differentially expressed proteins ( Supplementary Spreadsheet 1 ), among them 2,247 (83.53%) proteins decreased ( Supplementary Table 1 ) and 443 (16.47%) proteins increased ( Supplementary Table 2 ). These proteins are organized in different functional categories so that we can synthesize information and evaluate the main differences between the protein profile of macrophages cultured without or with P. brasiliensis yeasts. The regulated proteins are related to various biological processes, as shown by Figure 2 . Down-regulated proteins are mainly related to: protein fate (12%), metabolism (11%), transcription (11%), protein synthesis (10%), systemic interaction with the environment, mainly immune response (9%), and energy (8%). Most of P. brasiliensis-induced proteins in macrophages play a role in systemic interaction with the environment (16%), cellular communication (15%), transcription (14%), protein fate (13%) and cellular transport (10%). These functional categories are subdivided into smaller groups, for example: protein fate (folding, modification and destination); metabolism (carbohydrates, proteins, lipids, vitamins, etc.); transcription (RNA synthesis, and RNA processing); energy (glycolysis and gluconeogenesis, tricarboxylic acid cycle (TCA), electron transport and membrane-associated energy conservation, etc.).
Figure 2.
Functional classification of decreased and increased proteins in macrophages co-cultured with Paracoccidioides brasiliensis. The top part of the graph shows the number of proteins regulated in each biological process. Red bars and blue bars correspond to the number of proteins repressed or increased, respectively.
3.3. P. brasiliensis induces changes in THP-1 macrophages decreasing enzymes of energy metabolic processes and affecting mitochondrial activity
P. brasiliensis decreased proteins involved in macrophage energy production, impairing glycolysis, the TCA and oxidative phosphorylation ( Tables 1 , 2 ). Noteworthy, 24 enzymes of glycolysis, including their isoforms, were decreased in macrophages co-cultured with the fungus as depicted in Table 1 . In addition, the isoforms of lactate dehydrogenase and alcohol dehydrogenase were decreased in macrophages exposed to P. brasiliensis indicating an inhibition of fermentation. Also, the downregulation of glycogen phosphorylase suggests a reduction in glycogen degradation, consequently leading to decreased glucose consumption by macrophages ( Table 1 ). Additionally, 14 TCA enzymes and 12 proteins of the oxidative phosphorylation (OXPHOS) were decreased in macrophages co-cultured with P. brasiliensis ( Table 2 ).
Table 1.
Decreased proteins in macrophages infected with Paracoccidioides brasiliensis yeast cells involved in glucose metabolism.
| Accession number1 | Protein description and biological process2 | Score3 | Fold change4 |
|---|---|---|---|
| Glycolysis and Gluconeogenesis | |||
| A0A2U3TZU2 | Glucose-6-phosphate isomerase | 3765.44 | 0.394 |
| A0A7I2V3Z0 | ATP-dependent 6-phosphofructokinase | 6534.87 | * |
| P17858 | ATP-dependent 6-phosphofructokinase_ liver type | 670.40 | * |
| F8VZI0 | ATP-dependent 6-phosphofructokinase_ muscle type | 416.68 | * |
| Q01813 | ATP-dependent 6-phosphofructokinase_ platelet type | 6601.00 | * |
| J3KPS3 | Fructose-bisphosphate aldolase | 31785.80 | * |
| P05062 | Fructose-bisphosphate aldolase B | 1668.83 | * |
| P09972 | Fructose-bisphosphate aldolase C | 12130.18 | * |
| P60174 | Triosephosphate isomerase | 35540.98 | 0.379 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 21535.55 | * |
| O14556 | Glyceraldehyde-3-phosphate dehydrogenase_ testis-specific | 399.52 | * |
| P00558 | Phosphoglycerate kinase 1 | 23595.51 | * |
| P07205 | Phosphoglycerate kinase 2 | 1115.35 | * |
| P18669 | Phosphoglycerate mutase 1 | 26342.06 | * |
| P15259 | Phosphoglycerate mutase 2 | 17441.32 | * |
| Q8N0Y7 | Probable phosphoglycerate mutase 4 | 13757.52 | * |
| P06733 | Alpha-enolase | 63037.82 | * |
| P13929 | Beta-enolase | 5425.63 | * |
| P09104 | Gamma-enolase | 5319.81 | * |
| E5RGZ4 | 2-phospho-D-glycerate hydro-lyase | 5340.21 | * |
| F5H0C8 | 2-phospho-D-glycerate hydro-lyase | 5297.14 | * |
| A0A2R8Y6G6 | 2-phospho-D-glycerate hydro-lyase | 23270.41 | * |
| P30613 | Pyruvate kinase PKLR | 1599.20 | * |
| P14618 | Pyruvate kinase PKM | 47876.48 | 0.132 |
| Fermentation | |||
| P11766 | Alcohol dehydrogenase class-3 | 5335.49 | * |
| P00338 | L-lactate dehydrogenase A chain | 13796.04 | * |
| Q6ZMR3 | L-lactate dehydrogenase A-like 6A | 703.09 | * |
| Q9BYZ2 | L-lactate dehydrogenase A-like 6B | 914.21 | * |
| P07195 | L-lactate dehydrogenase B chain | 41051.41 | * |
| P07864 | L-lactate dehydrogenase C chain | 748.34 | * |
| Glycogen metabolism | |||
| E9PK47 | Alpha-1_4 glucan phosphorylase | 1890.55 | * |
| P11216 | Glycogen phosphorylase_ brain form | 2313.76 | 0.051 |
| P06737 | Glycogen phosphorylase_ liver form | 1890.55 | * |
| P11217 | Glycogen phosphorylase_ muscle form | 8381.66 | 0.477 |
1 Accession number of matched protein from Homo sapiens’ macrophages Uniprot database (https://www.uniprot.org/).
2 Proteins annotation from Homo sapiens’ database or by homology in NCBI database (http://www.ncbi.nlm.nih.gov/) and biological process according to the classification of KEGG (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (http://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/).
3 PLGS score is the result of different mathematical models for peptide and fragment assign prediction.
4 Fold-change values were obtained by dividing the values of protein abundance (in fmol) from macrophages during infection by live PB by the abundance in the uninfected macrophages. Proteins with a minimum fold-change of 50% (≤ 0.67) were considered to be dowregulated.
* Proteins detected only in uninfected macrophages.
A fold equal to or less than 0.67 was considered.
Table 2.
Decreased proteins in macrophages infected with Paracoccidioides brasiliensis yeast cells involved in energy production.
| Accession number1 | Protein description and biological process2 | Score3 | Fold change4 |
|---|---|---|---|
| Tricarboxylic-acid cycle | |||
| P53396 | ATP-citrate synthase | 1520.56 | * |
| O75390 | Citrate synthase_ mitochondrial | 12200.29 | * |
| A2A274 | Aconitate hydratase_ mitochondrial | 916.17 | * |
| P50213 | Isocitrate dehydrogenase [NAD] subunit alpha_ mitochondrial | 1918.82 | * |
| O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | 975.87 | * |
| P09622 | Dihydrolipoyl dehydrogenase_ mitochondrial | 1686.78 | * |
| P53597 | Succinate–CoA ligase [ADP/GDP-forming] subunit alpha_ mitochondrial | 651.86 | * |
| A0A5K1VW95 | Malate dehydrogenase | 15614.68 | * |
| C9JRL4 | Cytosolic malate dehydrogenase (Fragment) | 13862.17 | * |
| P07954 | Fumarate hydratase_ mitochondrial | 2669.02 | * |
| Oxidative phosphorylation | |||
| P13804 | Electron transfer flavoprotein subunit alpha_ mitochondrial | 1834.68 | * |
| C9J8T6 | Cytochrome c oxidase copper chaperone | 7210.97 | * |
| Q5SQT6 | Inorganic diphosphatase | 403.07 | * |
| H0Y9D8 | Inorganic diphosphatase (Fragment) | 1522.39 | * |
| Q15181 | Inorganic pyrophosphatase | 721.87 | * |
| Q9H2U2 | Inorganic pyrophosphatase 2_ mitochondrial | 1668.87 | * |
| K7EJP1 | ATP synthase subunit alpha_ mitochondrial (Fragment) | 17166.89 | * |
| P06576 | ATP synthase subunit beta_ mitochondrial | 18491.35 | * |
| P36542 | ATP synthase subunit gamma_ mitochondrial | 1169.11 | * |
| A8MUH2 | ATP synthase-coupling factor 6_ mitochondrial | 1566.69 | * |
| Q93050 | V-type proton ATPase 116 kDa subunit a1 | 2050.49 | * |
| O75323 | Protein NipSnap homolog 2 | 2351.76 | * |
1 Accession number of matched protein from Homo sapiens’ macrophages Uniprot database (https://www.uniprot.org/).
2 Proteins annotation from Homo sapiens’ database or by homology in NCBI database (http://www.ncbi.nlm.nih.gov/) and biological process according to the classification of KEGG (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (http://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/).
3 PLGS score is the result of different mathematical models for peptide and fragment assign prediction.
4 Fold-change values were obtained by dividing the values of protein abundance (in fmol) from macrophages during infection by live PB by the abundance in the uninfected macrophages. Proteins with a minimum fold-change of 50% (≤ 0.67) were considered to be dowregulated.
* Proteins detected only in uninfected macrophages.
A fold equal to or less than 0.67 was considered.
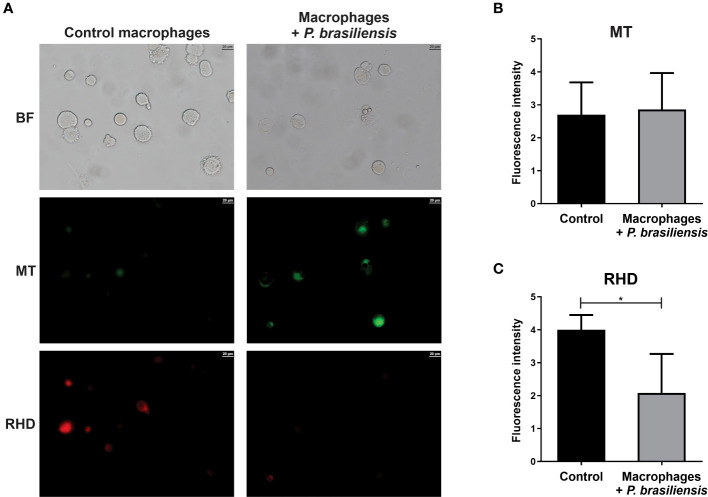
As enzymes related to the electron transport chain were decreased in P. brasiliensis-exposed macrophages, we assessed whether the integrity and activity of mitochondria in macrophages were impaired. There was no difference in mitochondrial membrane integrity in macrophages co-cultured or not with P. brasiliensis yeasts ( Figure 3 ). Photomicrographs presented in Figure 3A show macrophages with fluorescence of MitoTracker Green FM whereas in Figure 3B , the results of pixels of fluorescence quantification.We observed a significant decrease in mitochondrial activity in macrophages co-cultured with P. brasiliensis compared to the controls ( Figure 3 , p < 0.05). In Figure 3A the photomicrographs of cells and in 3C the quantification of pixels of fluorescence of Rhodamine 123 in macrophages exposed to the fungi was lower than in control macrophages. Therefore, the data indicate that P. brasiliensis infection affects mitochondrial activity what is probably due to the repression of enzymes in energy production pathways.
Figure 3.
Mitochondrial membrane activity in THP-1 macrophages co-cultured with Paracoccidioides brasiliensis yeast cells (for 24 h). Mitochondria integrity and activity was analyzed by fluorescence microscopy using MitoTracker Green FM and Rhodamin 123 dyes, respectively. (A) Macrophages in absence or presence of P. brasiliensis were photographed using an Axio-Scope A1 microscope at 400x magnification. (B) Fluorescence intensity (in pixels) of cells stained with MT (C) Fluorescence intensity (in pixels) of cells stained with RDH. Data represent the mean and standard deviation of three biological replicates. Brightfield (BF), Rhodamine 123 (RDH) and MitoTracker Green (MT). *p ≤ 0.05 by using Student’s t-test.
3.4. P. brasiliensis regulated detoxification proteins and increased ROS production in THP-1 macrophages
P. brasiliensis induced changes in the proteins that play a role in the stress response of THP-1 macrophages. Among the repressed proteins related to the stress response, three proteins of the heat shock family were identified as well as catalase, glutathione reductase_mitochondrial, glutathione S-transferase omega-1, peroxiredoxin-1, 4, 5 and 6, superoxide dismutase [Cu-Zn] (SOD1), and five proteins of the thioredoxin system ( Table 3 ). On the other hand, among the P. brasiliensis-induced proteins, glutathione peroxidase 1(GPX1), glutathione S-transferase kappa (GSTK1), glutathione S-transferase P (GSTP1), and peroxiredoxin-2 (PRDX2) ( Table 4 ), all very important in the response to oxidative stress, were identified.
Table 3.
Decreased proteins in macrophages infected with live Paracoccidioides brasiliensis yeast cells involved in stress response.
| Accession number1 | Protein description and biological process2 | Score3 | Fold change4 |
|---|---|---|---|
| Stress response | |||
| A0A7I2V2S7 | 75 kDa glucose-regulated protein | 7343.21 | 0.677 |
| P04040 | Catalase | 6229.48 | * |
| P00390 | Glutathione reductase_ mitochondrial | 3504.57 | * |
| P78417 | Glutathione S-transferase omega-1 | 1469.15 | * |
| Q5TA02 | Glutathione-dependent dehydroascorbate reductase (Fragment) | 1469.15 | * |
| A8MX94 | GST class-pi | 21612.53 | 0.386 |
| A0A6Q8PGK1 | Heat shock 27 kDa protein | 30162.08 | * |
| H0Y8K0 | Heat shock 70kDa protein 9B (Mortalin-2)_ isoform CRA_a | 5920.49 | * |
| P04792 | Heat shock protein beta-1 | 30168.58 | * |
| H3BSU0 | Liver carboxylesterase 1 (Fragment) | 407.80 | * |
| P55145 | Mesencephalic astrocyte-derived neurotrophic factor | 12025.95 | * |
| G5E977 | Nicotinate phosphoribosyltransferase | 2537.61 | * |
| A1KZ92 | Peroxidasin-like protein | 344.36 | * |
| A0A0A0MRQ5 | Peroxiredoxin-1 | 6482.96 | * |
| Q13162 | Peroxiredoxin-4 | 8387.69 | * |
| P30044 | Peroxiredoxin-5_ mitochondrial | 1369.47 | * |
| P30041 | Peroxiredoxin-6 | 13986.24 | * |
| P10768 | S-formylglutathione hydrolase | 1667.33 | * |
| P38646 | Stress-70 protein_ mitochondrial | 7419.77 | 0.670 |
| P31948 | Stress-induced-phosphoprotein 1 | 10056.83 | * |
| P00441 | Superoxide dismutase [Cu-Zn] | 8491.53 | * |
| P10599 | Thioredoxin | 7245.89 | * |
| O95881 | Thioredoxin domain-containing protein 12 | 1161.27 | * |
| Q16881 | Thioredoxin reductase 1_ cytoplasmic | 4239.45 | * |
| P30048 | Thioredoxin-dependent peroxide reductase_ mitochondrial | 5641.69 | * |
| A0A182DWI3 | Thioredoxin-disulfide reductase | 4235.96 | * |
1 Accession number of matched protein from Homo sapiens’ macrophages Uniprot database (https://www.uniprot.org/).
2 Proteins annotation from Homo sapiens’ database or by homology in NCBI database (http://www.ncbi.nlm.nih.gov/) and biological process according to the classification of KEGG (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (http://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/).
3 PLGS score is the result of different mathematical models for peptide and fragment assign prediction.
4 Fold-change values were obtained by dividing the values of protein abundance (in fmol) from macrophages during infection by live PB by the abundance in the uninfected macrophages. Proteins with a minimum fold-change of 50% (≤ 0.67) were considered to be dowregulated.
* Proteins detected only in uninfected macrophages.
A fold equal to or less than 0.67 was considered.
Table 4.
Increased proteins in macrophages infected with Paracoccidioides brasiliensis yeast cells involved in stress response.
| Accession number1 | Protein description and biological process2 | Score3 | Fold change4 |
|---|---|---|---|
| Stress response | |||
| P07203 | Glutathione peroxidase 1 | 4743.71 | 2.225 |
| Q9Y2Q3 | Glutathione S-transferase kappa 1 | 843.45 | * |
| P09211 | Glutathione S-transferase P | 22608.57 | 1.682 |
| K7EN27 | Maillard deglycase (Fragment) | 13395.05 | 2.117 |
| K7ELW0 | Parkinson disease protein 7 | 17649.65 | 3.706 |
| P32119 | Peroxiredoxin-2 | 1915.38 | 1.896 |
1 Accession number of matched protein from Homo sapiens’ macrophages Uniprot database (https://www.uniprot.org/).
2 Proteins annotation from Homo sapiens’ database or by homology in NCBI database (http://www.ncbi.nlm.nih.gov/) and biological process according to the classification of KEGG (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (http://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/).
3 PLGS score is the result of different mathematical models for peptide and fragment assign prediction.
4 Fold-change values were obtained by dividing the values of protein abundance (in fmol) from macrophages during infection by live PB by the abundance in the uninfected macrophages. Proteins with a minimum fold-change of 50% (≥ 1.5) were considered to be upregulated.
* Proteins detected only in infected macrophages.
A fold equal to or higher than 1.5 was considered.
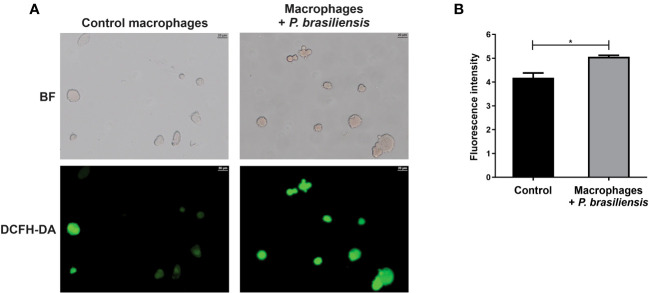
The ROS are generated during oxidative phosphorylation, but the mitochondrial dysfunction can lead to an exacerbated ROS production, which can cause tissue damage (Peoples et al., 2019; Rizwan et al., 2020). Since we have observed a decrease in mitochondrial activity in macrophages exposed to P. brasiliensis as well as a regulation of proteins relative to oxidative stress response, we investigated whether those macrophages could produce ROS. In fact, we observed that macrophages co-cultured with yeasts showed higher ROS production than control cells (an increase of 18%), as shown in Figure 4 .
Figure 4.
Production of reactive oxygen species in THP-1 macrophages co-cultured with P. brasiliensis yeast cells (for 24 h). Macrophages were stained with 2’,7’-dichlorodihydrofluorescein diacetate (DCFH-DA). (A) Macrophages were photographed using an Axio-Scope A1 microscope at 400× magnification. (B) Fluorescence intensities (in pixels) of cells stained with DCFH. Brightfield (BF) and 2’,7’-dichlorodihydrofluorescein diacetate (DCFH-DA). Data represent the mean and standard deviation of three biological replicates. *p ≤ 0.05 by using Student’s t-test.
3.5. Proteins of DNA processing, cell cycle, and transcription are regulated in THP-1 macrophages co-cultured with P. brasiliensis
Co-culture with P. brasiliensis and macrophages caused a repression of 21 and 17 proteins that play a role in DNA processing and cell cycle in macrophages, respectively. Some of these proteins are important in the process of DNA replication and repair such as ATP-dependent DNA helicase 2 subunit 1, DNA damage-binding protein 1 and proliferating cell nuclear antigen. Other proteins, which play a role in chromatin remodeling such as histones H1.2, H1.3, H1.4, H2A type 3, and HIST2H3PS2 were reduced ( Supplementary Table 1 ). Among the proteins increased in macrophages co-cultured with P. brasiliensis involved in DNA processing ( Supplementary Table 2 ) there are two subunits of the DNA polymerase enzyme, which plays a key role in DNA replication, and three other proteins involved in chromatin remodeling [Lethal(3)malignant brain tumor-like protein 3 (L3MBTL3), Lysine-specific demethylase 5A (KDM5A) and SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 (SMARCAD1)], which means that access to chromatin is altered and, consequently, there are changes in gene expression. Regarding the cell cycle, the decreased proteins are those ones involved with the organization and maintenance of microtubules, formation of the mitotic spindle, progression and regulation of the cell cycle, and histone transport ( Supplementary Table 1 ). Therefore, the repression of these proteins can deregulate the cell cycle, causing damage to the cell or decreasing its ability to proliferate.
It is also relevant to highlight that 61 proteins involved in gene transcription control were negatively ( Supplementary Table 1 ) whereas 22 proteins were positively regulated ( Supplementary Table 2 ). Among the repressed proteins, the Forkhead box protein M1 transcription factor (FOXM1), Hox family proteins, and several zinc finger proteins were identified ( Supplementary Table 1 ). The presence of the fungus also induced transcription elongation factor SPT6 (SPT6), some zinc finger proteins and the FOX transcription factor family proteins (FOX) C1, C2, G1, and L1 ( Supplementary Table 2 ).
3.6. P. brasiliensis regulated many proteins of the immune responses in host macrophages
Regarding the immune response, 88 proteins were negatively regulated in macrophages exposed to P. brasiliensis ( Table 5 ), among them: protein subunits of the Arp2/3 complex, Actin-like protein 3, Arf-GAP with SH3 domain _ ANK repeat and PH domain-containing protein 2 and Cofilin-2, which are relevant for cell adhesion and phagocytosis. Protein as Toll-like receptor (TLR)-9 that recognizes pathogen DNA was also decreased in the proteoma. Proteins of antigen processing were also identified such as Cathepsin B and S, Endoplasmic reticulum chaperone BiP, Heat shock-related 70 kDa protein 2, and Ras-related protein Rab-3C. Thirty-five histones participating in the formation of extracellular traps (ETs) were identified. Other proteins are related to the inflammatory process (Annexin, Macrophage migration inhibitory factor), the complement system (Complement component 1 Q subcomponent-binding protein_mitochondrial), leukocyte adhesion, and transmigration (Integrin beta-2, Lymphocyte-specific protein 1), differentiation of lymphocytes [Chitinase-3-like protein 1, Transforming growth factor beta (TGFβ-1), Retinoic acid receptor alpha, beta, and gamma], among other processes ( Table 5 ).
Table 5.
Some proteins decreased in macrophages infected vwith Paracoccidioides brasiliensis yeast cells involved in immune response.
| Accession number1 | Protein description and biological process2 | Score3 | Fold change4 |
|---|---|---|---|
| Immune response | |||
| O15143 | Actin-related protein 2/3 complex subunit 1B | 8740.81 | 0.631 |
| O15144 | Actin-related protein 2/3 complex subunit 2 | 3617.00 | * |
| O15145 | Actin-related protein 2/3 complex subunit 3 | 17431.37 | * |
| R4GN08 | Actin-related protein 2/3 complex subunit 4 (Fragment) | 1106.88 | * |
| O15511 | Actin-related protein 2/3 complex subunit 5 | 3627.45 | * |
| F8WE84 | Actin-related protein 3 | 9712.19 | * |
| D6RBL5 | Annexin | 13850.32 | 0.650 |
| P04083 | Annexin A1 | 18072.62 | * |
| P08758 | Annexin A5 | 30602.12 | 0.644 |
| O43150 | Arf-GAP with SH3 domain_ ANK repeat and PH domain-containing protein 2 | 594.14 | * |
| G5E9J0 | Arp2/3 complex 34 kDa subunit | 475.38 | * |
| P61769 | Beta-2-microglobulin | 3926.66 | * |
| A0A7I2V440 | Cathepsin B | 6042.11 | * |
| P25774 | Cathepsin S | 1155.57 | * |
| P36222 | Chitinase-3-like protein 1 | 1597.86 | * |
| Q07021 | Complement component 1 Q subcomponent-binding protein_ mitochondrial | 7230.48 | * |
| A0A7P0TB36 | Endoplasmic reticulum chaperone BiP | 21012.03 | 0.582 |
| P54652 | Heat shock-related 70 kDa protein 2 | 11903.81 | 0.001 |
| P09429 | High mobility group protein B1 | 1668.83 | * |
| C9J0D1 | Histone H2A | 7828.83 | * |
| P0C0S8 | Histone H2A type 1 | 8902.71 | * |
| Q96QV6 | Histone H2A type 1-A | 3271.32 | * |
| P04908 | Histone H2A type 1-B/E | 8902.71 | * |
| Q93077 | Histone H2A type 1-C | 8902.71 | * |
| P20671 | Histone H2A type 1-D | 8902.71 | * |
| Q96KK5 | Histone H2A type 1-H | 8902.71 | * |
| Q99878 | Histone H2A type 1-J | 8902.71 | * |
| Q6FI13 | Histone H2A type 2-A | 8902.71 | * |
| Q8IUE6 | Histone H2A type 2-B | 2747.75 | * |
| Q16777 | Histone H2A type 2-C | 8902.71 | * |
| Q9BTM1 | Histone H2A.J | 8902.71 | * |
| Q71UI9 | Histone H2A.V | 8676.91 | * |
| P0C0S5 | Histone H2A.Z | 8676.91 | * |
| P16104 | Histone H2AX | 3273.08 | * |
| U3KQK0 | Histone H2B | 16807.41 | * |
| Q96A08 | Histone H2B type 1-A | 7130.46 | * |
| P33778 | Histone H2B type 1-B | 17593.68 | * |
| P62807 | Histone H2B type 1-C/E/F/G/I | 16807.41 | * |
| P58876 | Histone H2B type 1-D | 16807.41 | * |
| Q93079 | Histone H2B type 1-H | 16807.41 | * |
| P06899 | Histone H2B type 1-J | 17593.68 | * |
| O60814 | Histone H2B type 1-K | 16807.41 | * |
| Q99880 | Histone H2B type 1-L | 16807.41 | * |
| Q99879 | Histone H2B type 1-M | 16807.41 | * |
| Q99877 | Histone H2B type 1-N | 16807.41 | * |
| P23527 | Histone H2B type 1-O | 17593.68 | * |
| Q16778 | Histone H2B type 2-E | 17593.68 | * |
| A0A2R8Y619 | Histone H2B type 2-E1 | 311.58 | * |
| Q5QNW6 | Histone H2B type 2-F | 16807.41 | * |
| Q8N257 | Histone H2B type 3-B | 17593.68 | * |
| P57053 | Histone H2B type F-S | 16800.03 | * |
| Q6NXT2 | Histone H3.3C | 3853.37 | * |
| P62805 | Histone H4 | 7844.59 | * |
| P05107 | Integrin beta-2 | 2518.87 | * |
| P33241 | Lymphocyte-specific protein 1 | 820.58 | * |
| P14174 | Macrophage migration inhibitory factor | 4042.46 | * |
| Q96E17 | Ras-related protein Rab-3C | 1115.57 | * |
| P19793 | Retinoic acid receptor RXR-alpha | 654.98 | * |
| P28702 | Retinoic acid receptor RXR-beta | 1177.33 | * |
| P48443 | Retinoic acid receptor RXR-gamma | 630.23 | * |
| H0Y858 | Toll-like receptor 9 (Fragment) | 624.89 | * |
| A0A499FJK2 | Transforming growth factor beta | 618.77 | * |
1 Accession number of matched protein from Homo sapiens’ macrophages Uniprot database (https://www.uniprot.org/).
2 Proteins annotation from Homo sapiens’ database or by homology in NCBI database (http://www.ncbi.nlm.nih.gov/) and biological process according to the classification of KEGG (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (http://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/).
3 PLGS score is the result of different mathematical models for peptide and fragment assign prediction.
4 Fold-change values were obtained by dividing the values of protein abundance (in fmol) from macrophages during infection by live PB by the abundance in the uninfected macrophages. Proteins with a minimum fold-change of 50% (≤ 0.67) were considered to be dowregulated.
* Proteins detected only in uninfected macrophages.
A fold equal to or less than 0.67 was considered.
Our analysis identified 14 proteins related to immune response that were increased in macrophages exposed to P. brasiliensis ( Table 6 ). Among them we highlight proteins associated to inflammation (Caspase recruitment domain family_ member 14_ isoform CRA_d), proteins involved in the adhesion process, related to leukocyte transmigration and phagocytosis (Actin_cytoplasmic 2, Ezrin, Moesin, Stabilin-2). Protein involved in the antigen processing (Heat shock cognate 71 kDa protein, Heat shock 70 kDa protein 1A, Heat shock 70 kDa protein 1B, Heat shock 70 kDa protein 6) and respiratory burst (Neutrophil cytosol factor 2 - NCF2), were also increased in the macrophage during infection with P. brasiliensis. Figure 5 shows an overview of the proteome changes observed in THP-1 macrophages exposed to P. brasiliensis.
Table 6.
Increased proteins in macrophages infected with Paracoccidioides brasiliensis yeast cells involved in immune response.
| Accession number1 | Protein description and biological process2 | Score3 | Fold change4 |
|---|---|---|---|
| Immune response | |||
| A0A7P0TBL1 | Actin_ cytoplasmic 2 | 1198.38 | 314.190 |
| P61160 | Actin-related protein 2 | 5478.23 | 2.013 |
| Q7Z6M3 | Allergin-1 | 326.88 | * |
| I3L414 | Caspase recruitment domain family_ member 14_ isoform CRA_d | 406.41 | 37.712 |
| P15311 | Ezrin | 1969.29 | 2.293 |
| V9GZ37 | Heat shock 70 kDa protein 1A | 129.76 | * |
| A0A0G2JIW1 | Heat shock 70 kDa protein 1B | 2588.57 | 5.365 |
| P17066 | Heat shock 70 kDa protein 6 | 2359.95 | 196.369 |
| E9PQQ4 | Heat shock cognate 71 kDa protein (Fragment) | 21826.13 | 5.312 |
| A0A1B0GU45 | Inositol 1_4_5-triphosphate receptor-associated 2 | 514.40 | * |
| P26038 | Moesin | 11172.13 | 2.534 |
| P19878 | Neutrophil cytosol factor 2 | 334.05 | * |
| Q06330 | Recombining binding protein suppressor of hairless | 338.53 | * |
| Q8WWQ8 | Stabilin-2 | 238.15 | * |
1 Accession number of matched protein from Homo sapiens’ macrophages Uniprot database (https://www.uniprot.org/).
2 Proteins annotation from Homo sapiens’ database or by homology in NCBI database (http://www.ncbi.nlm.nih.gov/) and biological process according to the classification of KEGG (https://www.genome.jp/kegg/), UniProt database (https://www.uniprot.org/), NCBI database (http://www.ncbi.nlm.nih.gov/) and CORUM database (http://mips.helmholtz-muenchen.de/corum/).
3 PLGS score is the result of different mathematical models for peptide and fragment assign prediction.
4 Fold-change values were obtained by dividing the values of protein abundance (in fmol) from macrophages during infection by live PB by the abundance in the uninfected macrophages. Proteins with a minimum fold-change of 50% (≥ 1.5) were considered to be upregulated.
* Proteins detected only in infected macrophages.
A fold equal to or higher than 1.5 was considered.
Figure 5.
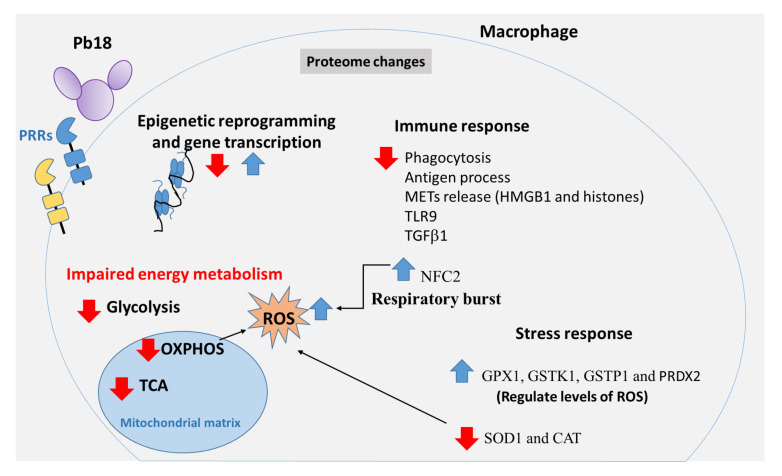
Graphical summary of proteomic changes in human THP-1 macrophages co-cultured with Paracoccidioides brasiliensis. Pb18 yeast cells induce changes in the macrophage proteome. The interactions between Pb18 yeast cells and macrophages include recognition of the fungal PAMPs by macrophage PRRs as well as other fungal and host factors which induce changes in the proteome of macrophages. Co-cultures of THP-1 macrophages and P. brasiliensis yeasts caused increase and decrease of macrophage proteins participating in epigenetic reprogramming, and gene transcription processes such as several histones, L3MBTL3, KDM5A, SMARCAD1, FOXM1, SPT6, FOXC1 and ZN140. P. brasiliensis-exposed macrophages showed impaired glycolysis, TCA and OXPHOS pathways, and increased ROS production. In addition, P. brasiliensis repressed several proteins of the immune system, relevant for phagocytosis, antigen processing and presentation, besides components of METs, innate immune receptors, cytokines, among others. Macrophage interaction with P. brasiliensis also increases NCF2, a functional subunit of NADPH oxidase complex, which may contribute to the ROS production. Some enzymes involved with oxidative stress were also increased (GPX1, GSTK1, GSTP1, and PRDX2). Other proteins related to the reaction to oxidative stress were repressed: SOD1 and catalase CAT. Pattern-recognizing receptors (PRRs); Tricarboxylic acid cycle (TCA); Phosphorylative oxidation (OXPHOS); Reactive oxygen species (ROS); Extracellular traps of macrophages (METs); Neutrophil cytosol factor 2 (NCF2); Glutathione peroxidase 1 (GPX1); Glutathione S-transferase kappa (GSTK1); Glutathione S-transferase P (GSTP1); Peroxiredoxin-2 (PRDX2); Superoxide dismutase (SOD1); Catalase (CAT).
4. Discussion
In the present study, we have performed a proteomic analysis of the THP-1 macrophage in response to interaction with P. brasiliensis yeast cells. Since cells and fungi were co-cultured, the macrophage proteome was a result of the fungus effects on infected and bystander non-infected macrophages for 24 h of incubation. Although a survival assay was not performed to evaluate fungal replication within 24 h, we have shown that macrophages were efficiently infected by yeast cells. There is no information in the literature about the growth of P. brasiliensis in THP-1 macrophages; however, we have demonstrated that the number of viable fungal cells (assessed by the number of colony-forming units) increases during murine alveolar macrophages infection (AMJ2-C11) from 6 hours and continues to increase up to 12 hours after interaction (Chaves et al., 2019). Thus, probably the fungi proliferated during the incubation time (24 h) in THP-1 macrophages.
We showed that P. brasiliensis infection regulates proteins of a variety of biological processes. To respond to an infection, immune cells undergo a metabolic adaptation, which is coupled to the effector mechanisms of the immune response, which require energy and/or specific metabolites (Domínguez-Andrés et al., 2017; Tucey et al., 2018). On the other hand, the pathogen also undergoes changes and tries to escape the immune system (De Arruda Grossklaus et al., 2013; Parente-Rocha et al., 2015; Lacerda Pigosso et al., 2017; Weerasinghe and Traven, 2020). In recent years, some studies in the field of immunometabolism have been carried out, with the aim of understanding the role of metabolic events in regulating the response of immune cells to pathogens (O’Neill et al., 2016; Domínguez-Andrés et al., 2017; Tucey et al., 2018; Weerasinghe and Traven, 2020).
Of relevance, in this study, proteins of the cellular processes for energy production decreased in macrophages interacting with P. brasiliensis. In these cells, it was detected a decreased glycolysis, TCA, and oxidative phosphorylation. Proteomic data were confirmed by assessing the mitochondrial activity, which was decreased as tested by staining with Rhodamine 123. Studies have shown that recognition of some PAMPs by monocytes and macrophages induces aerobic glycolysis, also called as Warburg effect. This effect consists in increased glucose consumption, repression of the TCA and OXPHOS as well as increased lactate production. These metabolic effect are triggered, for example, in monocytes and macrophages after recognition of β-glucans from Candida albicans (Cheng et al., 2014; Domínguez-Andrés et al., 2017) or melanin in Aspergillus fumigatus (Gonçalves et al., 2020). However, this change in metabolism is not the rule for all pathogens and is probably due to differences in PRRs triggered by PAMPs and other intrinsic factors produced by each microorganism. Similar to our result, Shin et al. (2005) observed in a proteomic analysis a decrease in energy metabolism enzymes in murine macrophages infected with C. albicans hyphae and this contributes along with other factors to host cell death. However, in murine alveolar epithelial cells infected with A. fumigatus, enrichment of phosphorylative oxidation proteins was observed, indicating increased energy production (Seddigh et al., 2017). Also according to our findings, it has already been demonstrated that Salmonella typhimurium compromises glycolysis, impairing phagosome acidification and, consequently, decreasing bacterial clearance in murine macrophages (Gutierrez et al., 2020). Additionally, McGettrick et al. (2016) showed that the metabolite indolepyruvate from Trypanosoma brucei decreases glycolysis and hypoxia-inducible transcription factor-1α (HIF-1α) in murine macrophages as an evasion mechanism. Therefore, we can suggest that P. brasiliensis somehow affects energy production pathways in the host cells in an attempt to benefit itself.
In the present study, proteins of cellular stress response have been identified. Among the increased proteins are three enzymes of glutathione metabolism (GPX1, GSTK1 and GSTP1) and PRDX2, important cellular antioxidants, which acting to regulate necessary levels of ROS (Hayes and McLellan, 1999; Rhee, 2016). On the contrary, the enzymes SOD1, and catalase were decreased. SOD1 catalyzes the dismutation of superoxide anion (O2·-) into oxygen and hydrogen peroxide (H2O2) whereas catalase does the decomposition of H2O2 into water and oxygen. It has been demonstrated that H2O2 and O2 ·- are important effector molecules to kill P. brasiliensis in human macrophages and neutrophils activated by cytokines (Carmo et al., 2006; Rodrigues et al., 2007). The lack of these enzymes can lead to a significant increase in ROS and, consequently, cause exacerbated tissue damage. Nevertheless, in some cases, it can be important to increase ROS at toxic levels to eliminate pathogens (Gebicka and Krych-Madej, 2019). In our study, we demonstrated that there is an increase in ROS production by macrophages interacting with P. brasiliensis, thus suggesting that P. brasiliensis-infected macrophages can be regulated by ROS. We need to check whether the levels of ROS produced during the first 24 h of macrophage and fungus interaction are able to directly kill the fungus or are used to regulate immune responses, for example by activating inflammasome (Tavares et al., 2013).
Proteomic analysis identified regulation of several proteins that play important roles in DNA replication and transcription processes. Some histones, enzymes and transcription factors that contribute to epigenetic modifications have changed in macrophages after interaction with P. brasiliensis. This has implications for chromatin accessibility by the transcription machinery. Epigenetic remodeling is related to metabolic changes, since some metabolites act as cofactors for enzymes that regulate chromatin modifications and, consequently, alter gene transcription and cell proteome (Netea et al., 2016; Sun et al., 2022). The increased proteins L3MBTL3 and KDM5A, participate in demethylation of lysine 4 of histone 3 (H3K4) (Christensen et al., 2007; Xu et al., 2017), thus causing gene repression, since H3K4 methylation is associated with decondensed chromatin (Ciccone and Chen, 2009). Another increased protein was SMARCAD1, which mediates the deacetylation of histones 3 and 4, which can be related to gene repression. Deletion of SMARCAD1 in mice with colitis leads to increased expression of several genes, including those related to innate immune response (Kazakevych et al., 2020). Nevertheless, the SPT6 also increased here and participates in the demethylation of lysine 27 of histone 3 (H3K27) (Wang et al., 2013) and the methylation of lysine 36 of the same histone (Gopalakrishnan et al., 2019). Thus, SPT6 helps the opening of chromatin and activation of transcription. Therefore, the increase of the SMARCAD1 protein as well as L3MBTL3 and KDM5A in the present study, probably contributed to repress several genes. The induction of SPT6 could have induced the expression of other genes, which is consistent with changes found in the proteome of macrophages interacting with P. brasiliensis.
Among the increased proteins that have functions in transcription process, Forkhead box protein C1 (FOXC1), and Zinc finger protein 140 (ZN140) also called our attention. Zhang et al. (2019) have demonstrated that FOXC1 increases the expression of TLR3, and TLR4, and is related to pro-inflammatory effects in a murine model of cardiac ischemia. One study suggested that TLR3 is a negative regulator of host response against P. brasiliensis, what was associated with a decrease of the proinflammatory response, NO production, and activation of CD8+ T lymphocytes (Jannuzzi et al., 2019). It has also been demonstrated that TLR4 is relevant to the production of IFNγ in human neutrophils infected with P. brasiliensis (Rodrigues et al., 2014). Besides, TLR4 cooperates with dectin-1 (for β-glucan) and mannose receptor in the differentiation of Th17 cells stimulated by murine dendritic cells (Loures et al., 2015). Although the cytokine IFNγ and Th17 cells are important for fungal control, studies have associated TLR4 with the pathogenesis of PCM (Acorci-Valério et al., 2010; Loures et al., 2010). We cannot identify TLR4 in this study, but we can speculate that by contributing to the production of IFNγ and the differentiation of Th17 cells, TLR4 may lead to a mix of Th1/Th17 response that is associated with the chronic form of PCM, and crucial for the control of the infection (De Castro et al., 2013). The ZN140 protein was associated with gene repression of the Fc gamma (γ) RIIB receptor (Nishimura et al., 2001). This receptor inhibits phagocytosis and release of some pro-inflammatory cytokines in macrophages, and is important for a balance between mechanisms of pathogen elimination and exacerbated inflammation (Clynes et al., 1999; Clatworthy and Smith, 2004). There are no studies with this receptor in PCM, however, in a murine model, FcγRIIB deficiency was associated with severe cryptococcosis, increased phagocytosis, and cytokine production (Surawut et al., 2017). Although FcγRIIB, TLR3 and TLR4 were not identified in our proteomic analysis, it can be suggested that ZN140 increase leads to inhibition of FcγRIIB expression, and that FOXC1 increases the expression of these TLRs in the context of PCM and that this has some relevance for the control of P. brasiliensis.
Among the repressed proteins, it is relevant to mention the nuclear phosphoprotein nucleolin, which is important to the processes of synthesis and maturation of ribosomes (Ginisty et al., 1999). Therefore, repression of this protein must have impaired ribosome formation and protein synthesis in macrophages co-cultured with P. brasiliensis. In line with this, our analysis of the proteome from those macrophages identified 83 ribosomal proteins that were decreased ( Supplementary Table 1 ). Another repressed protein was Forkhead box protein M1 (FOXM1). Inhibition of this transcription factor has already been shown to increase production of ROS in diabetic wounds (Sawaya et al., 2022). Similarly, in our study we have also observed an increase of ROS in macrophages exposed to P. brasiliensis.
Recognition of the pathogen initiates changes in defense cells that work for controlling the infection; however, these changes may also provide opportunities for the fungus to evade the immune system (Weerasinghe and Traven, 2020). Here, proteomic analysis of host defense cells co-cultured with P. brasiliensis showed that the fungus regulates the expression of different proteins of the immune system. Previously, Silva et al. (2008) evaluated a group of genes in murine macrophages infected with opsonized yeasts of P. brasiliensis. They described alterations in the transcription of several genes of the immune response. After 24 h of infection, 59 genes were differentially expressed, 54 of which were induced and five were repressed. Genes related to inflammation, regulation of transcription, and signal transduction pathways, phagocytosis and the process of apoptosis were increased (Silva et al., 2008). Unlike the results by Silva et al. (2008), our proteomic analysis showed that human THP-1 macrophages co-cultured with P. brasiliensis showed predominantly the repression of several proteins for PAMP recognition in endosome (TLR9), adhesion, and phagocytosis as well as a decrease of some proteins that participate in the inflammatory process. The differences are probably due to the models and experimental procedures used in each study as well as to different levels of gene expression and protein production. It is important to consider that not everything that is transcribed is translated into protein.
We also observed a decrease of protein processing and antigen presentation pathways. A result consistent with this is the positive regulation of the KDM5A enzyme mentioned above. A study has shown that this demethylase silences genes in the antigen processing pathway during antitumor immune responses (Liu et al., 2022). In the current study, the analysis also identified repression of 34 histones related to the formation of ETs. It has already been demonstrated that P. brasiliensis induces the release of neutrophil ETs (NETs) that contribute to the fungus control in human neutrophils (Bachiega et al., 2016). To our knowledge up to now, there are no studies showing the release of ETs by macrophages (METs) infected with P. brasiliensis; however the release of METs by THP-1 macrophages infected with Trichomonas vaginalis has already been demonstrated (Fei et al., 2019). Another repressed protein, high mobility group protein B1 (HMGB1) is also associated with the NET formation. HMGB1 is a nuclear protein that acts as a molecular pattern associated with damage (DAMP) and its interaction with the TLR4 triggers increased NET release by murine neutrophils (Tadie et al., 2013). Therefore, our data suggest that P. brasiliensis may impair the release of ETs by THP-1 macrophages in a HMGB1-dependent manner.
Among the decreased proteins in macrophages co-cultured with P. brasiliensis, it is also relevant to discuss TLR9 in detail. The TLR9 is an endosome receptor mainly present in innate immunity cells that recognizes unmethylated CpG dinucleotides. Studies demonstrated that TLR9 is important for the survival of mice during the first 48 hours of infection by P. brasiliensis (Menino et al., 2013). Additionally, Morais et al. (2016) confirmed the role of TLR9 in the initial protection against P. brasiliensis, showing that immunization of mice with the recombinant protein rPb27 associated with CPG oligodeoxynucleotide motifs provides high protection against the fungus, 30 days after infection. Another important protein, which was decreased in the macrophage proteome of the present study is TGFβ-1, a multifunctional cytokine that regulates several processes of the immune responses including the differentiation of Th17 cells (Mangan et al., 2006), and the regulation of the healing process (Lodyga and Hinz, 2020). Restrepo et al. (1998) showed, in a murine model, that pulmonary fibrosis induced by P. brasiliensis conidia was associated with an increased TGFβ-1 production. Another study showed that peripheral blood monocytes from patients with PCM, evaluated before and 20 days after treatment, produced elevated endogenous levels of TGFβ-1 compared to healthy controls, and this cytokine was present in fibrotic areas of the patients’ lesions (Parise-Fortes et al., 2006). Therefore, it appears that this cytokine plays an important role in tissue repair as well as in fibrosis during PCM. In the current study, TGFβ-1 was repressed during the first 24 h of macrophage-P. brasiliensis interaction probably to avoid early suppression of inflammatory process or innate immunity.
Among the increased proteins, neutrophil cytosol factor 2 (NFC2) was identified; this protein is a functional subunit of NADPH oxidase complex, which is very important for the production of ROS during the process of respiratory burst in phagocytes (Nauseef, 2004). Besides the role of NADPH oxidase complex to ROS production, which are important microbicidal molecules, this enzyme complex may play other roles in the immune response (Hogan and Wheeler, 2014) such as induction of NETs, and inflammasome activation (Tavares et al., 2013; Mejía et al., 2015). Consistent with the increased NFC2 level, we have observed an increase in ROS production by macrophages co-cultured with P. brasiliensis.
In conclusion, we demonstrated that interaction of human macrophages with P. brasiliensis yeast cells resulted in modification of the macrophage proteome, which presented impaired production of proteins participating in energy metabolism, regulation of epigenetic modifications, and gene transcription. Most importantly, data showed repression of many proteins crucial for antifungal immune responses. Although some conclusions are limited and experimental validations are necessary to elucidate the role of the main regulated proteins in macrophages exposed to P. brasiliensis, our study contributes to the understanding of the host-pathogen interface, and may provide important information for future studies aiming to develop new therapeutic approaches for PCM.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material . Data is available at: http://www.ebi.ac.uk/pride/archive/projects/PXD043727.
Author contributions
AF: Conceptualization, Writing – review & editing, Formal Analysis, Methodology, Validation, Writing – original draft. DM: Methodology, Writing – original draft, Data curation, Software. AB: Conceptualization, Supervision, Writing – review & editing. OR: Writing – review & editing, Methodology. LS: Writing – original draft. FR-D: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. CS: Conceptualization, Supervision, Writing – review & editing, Funding acquisition, Project administration, Resources.
Acknowledgments
The authors thank Mariana V. Tomazett and Juliana S. de Curcio for their experimental support. The authors are grateful to CAPES, CNPq and FAPEG for the financial support provided to this study.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was done at Universidade Federal de Goiás with financial support of Fundação de Apoio a Pesquisa de Goiás (FAPEG) and in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES)/Ministry of Education, Brazil-Finance Code 001.; AF is a fellow of CAPES; FR-D and CS are fellow researchers of National Council for Scientific and Technological Development (CNPq), Brazil; FR-D and CS are members of National Institute of Science and Technology of the strategies in host-pathogen interaction (INCT-IPH), supported by FAPEG (grant n. 465771/2014-9), Brazil.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1275954/full#supplementary-material
All proteins identified in the proteomic analysis.
References
- Acorci-Valério M. J., Bordon-Graciani A. P., Dias-Melicio L. A., De Assis Golim M., Nakaira-Takahagi E., De Campos Soares Â.M.V. (2010). Role of TLR2 and TLR4 in human neutrophil functions against Paracoccidioides brasiliensis . Scandinavian J. Immunol. 71, 99–108. doi: 10.1111/j.1365-3083.2009.02351.x [DOI] [PubMed] [Google Scholar]
- Bachiega T. F., Dias-Melicio L. A., Fernandes R. K., De Almeida Balderramas H., Rodrigues D. R., Ximenes V. F., et al. (2016). Participation of dectin-1 receptor on NETs release against Paracoccidioides brasiliensis: Role on extracellular killing. Immunobiology 221, 228–235. doi: 10.1016/j.imbio.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Baeza L. C., Da Mata F. R., Pigosso L. L., Pereira M., De Souza G. H. M. F., Coelho A. S. G., et al. (2017). Differential metabolism of a two-carbon substrate by members of the Paracoccidioides genus. Front. Microbiol. 8, 2308. doi: 10.3389/fmicb.2017.02308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailão A. M., Nogueira S. V., Rondon Caixeta Bonfim S. M., De Castro K. P., De Fátima Da Silva J., Mendes Giannini M. J. S., et al. (2012). Comparative transcriptome analysis of Paracoccidioides brasiliensis during in vitro adhesion to type I collagen and fibronectin: identification of potential adhesins. Res. Microbiol. 163, 182–191. doi: 10.1016/j.resmic.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Bastos K. P., Bailão A. M., Borges C. L., Faria F. P., Felipe M. S., Silva M. G., et al. (2007). The transcriptome analysis of early morphogenesis in Paracoccidioides brasiliensis mycelium reveals novel and induced genes potentially associated to the dimorphic process. BMC Microbiol. 7, 29. doi: 10.1186/1471-2180-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C., Röllinghoff M., Diefenbach A. (2000). Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr. Opin. Immunol. 12, 64–76. doi: 10.1016/S0952-7915(99)00052-7 [DOI] [PubMed] [Google Scholar]
- Brown A. J., Haynes K., Quinn J. (2009). Nitrosative and oxidative stress responses in fungal pathogenicity. Curr. Opin. Microbiol. 12, 384–391. doi: 10.1016/j.mib.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer E., Hanson L. H., Restrepo A., Stevens D. A. (1989). Intracellular multiplication of Paracoccidioides brasiliensis in macrophages: killing and restriction of multiplication by activated macrophages. Infect. Immun. 57, 2289–2294. doi: 10.1128/iai.57.8.2289-2294.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummer E., Hanson L. H., Stevens D. A. (1988). Gamma-interferon activation of macrophages for killing of Paracoccidioides brasiliensis and evidence for nonoxidative mechanisms. Int. J. Immunopharmacol. 10, 945–952. doi: 10.1016/0192-0561(88)90041-0 [DOI] [PubMed] [Google Scholar]
- Carmo J. P. M., Dias-Melicio L. A., Calvi S. A., Peraçoli M. T. S., Soares A. M. V. C. (2006). TNF-α activates human monocytes for Paracoccidioides brasiliensis killing by an H2O2-dependent mechanism. Med. Mycol 44, 363–368. doi: 10.1080/13693780500536885 [DOI] [PubMed] [Google Scholar]
- Chanput W., Mes J. J., Wichers H. J. (2014). THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 23, 37–45. doi: 10.1016/j.intimp.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Chaves A. F. A., Navarro M. V., de Barros Y. N., Silva R. S., Xander P., Batista W. L. (2021). Updates in Paracoccidioides biology and genetic advances in fungus manipulation. J. Fungi 7, 116. doi: 10.3390/jof7020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves E. G. A., Parente-Rocha J. A., Baeza L. C., Araújo D. S., Borges C. L., Oliveira M. A. P. D., et al. (2019). Proteomic analysis of Paracoccidioides brasiliensis during infection of alveolar macrophages primed or not by interferon-gamma. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.-C., Quintin J., Cramer R. A., Shepardson K. M., Saeed S., Kumar V., et al. (2014). mTOR- and HIF-1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684. doi: 10.1126/science.1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P. A. C., Pasini D., Rose S., Sennels L., et al. (2007). RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076. doi: 10.1016/j.cell.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Ciccone D. N., Chen T. (2009). Histone lysine methylation in genomic imprinting. Epigenetics 4, 216–220. doi: 10.4161/epi.8974 [DOI] [PubMed] [Google Scholar]
- Clatworthy M. R., Smith K. G. C. (2004). FcγRIIb balances efficient pathogen clearance and the cytokine-mediated consequences of sepsis. J. Exp. Med. 199, 717–723. doi: 10.1084/jem.20032197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes R., Maizes J. S., Guinamard R., Ono M., Takai T., Ravetch J. V. (1999). Modulation of immune complex–induced inflammation in vivo by the coordinate expression of activation and inhibitory fc receptors. J. Exp. Med. 189, 179–186. doi: 10.1084/jem.189.1.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arruda Grossklaus D., Bailão A. M., Vieira Rezende T. C., Borges C. L., De Oliveira M. A. P., Parente J. A., et al. (2013). Response to oxidative stress in Paracoccidioides yeast cells as determined by proteomic analysis. Microbes Infection 15, 347–364. doi: 10.1016/j.micinf.2012.12.002 [DOI] [PubMed] [Google Scholar]
- De Castro L. F., Ferreira M. C., Da Silva R. M., Blotta M. H. D. S. L., Longhi L. N. A., Mamoni R. L. (2013). Characterization of the immune response in human paracoccidioidomycosis. J. Infection 67, 470–485. doi: 10.1016/j.jinf.2013.07.019 [DOI] [PubMed] [Google Scholar]
- Deutsch E. W., Bandeira N., Perez-Riverol Y., Sharma V., Carver J. J., Mendoza L., et al. (2023). The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 51, D1539–D1548. doi: 10.1093/nar/gkac1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Andrés J., Arts R. J. W., Ter Horst R., Gresnigt M. S., Smeekens S. P., Ratter J. M., et al. (2017). Rewiring monocyte glucose metabolism via C-type lectin signaling protects against disseminated candidiasis. PloS Pathog. 13, e1006632. doi: 10.1371/journal.ppat.1006632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos J. C., Heinhuis B., Gomes R. S., Damen M. S. M. A., Real F., Mortara R. A., et al. (2017). Cytokines and microbicidal molecules regulated by IL-32 in THP-1-derived human macrophages infected with New World Leishmania species. PloS Negl. Trop. Dis. 11, e0005413. doi: 10.1371/journal.pntd.0005413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei L., Zhengkai W., Weina J., Lili C., Yuhang G., Zhengtao Y., et al. (2019). Trichomonas vaginalis triggers the release of THP-1 extracellular traps. Parasitol. Res. 118, 267–274. doi: 10.1007/s00436-018-6139-5 [DOI] [PubMed] [Google Scholar]
- Ferrante C. J., Leibovich S. J. (2012). Regulation of macrophage polarization and wound healing. Adv. Wound Care 1, 10–16. doi: 10.1089/wound.2011.0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebicka L., Krych-Madej J. (2019). The role of catalases in the prevention/promotion of oxidative stress. J. Inorganic Biochem. 197, 110699. doi: 10.1016/j.jinorgbio.2019.110699 [DOI] [PubMed] [Google Scholar]
- Geromanos S. J., Vissers J. P. C., Silva J. C., Dorschel C. A., Li G.-Z., Gorenstein M. V., et al. (2009). The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695. doi: 10.1002/pmic.200800562 [DOI] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B., Bouvet P. (1999). Structure and functions of nucleolin. J. Cell Sci. 112, 761–772. doi: 10.1242/jcs.112.6.761 [DOI] [PubMed] [Google Scholar]
- Gonçalves S. M., Duarte-Oliveira C., Campos C. F., Aimanianda V., Ter Horst R., Leite L., et al. (2020). Phagosomal removal of fungal melanin reprograms macrophage metabolism to promote antifungal immunity. Nat. Commun. 11, 2282. doi: 10.1038/s41467-020-16120-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., De Gregori W., Velez D., Restrepo A., Cano L. E. (2000). Nitric Oxide Participation in the Fungicidal Mechanism of Gamma Interferon-Activated Murine Macrophages against Paracoccidioides brasiliensis Conidia. Infect. Immun. 68, 2546–2552. doi: 10.1128/IAI.68.5.2546-2552.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan R., Marr S. K., Kingston R. E., Winston F. (2019). A conserved genetic interaction between Spt6 and Set2 regulates H3K36 methylation. Nucleic Acids Res. 47, 3888–3903. doi: 10.1093/nar/gkz119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N. A. R., Latge J.-P., Munro C. A. (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 5, 5.3.01. doi: 10.1128/microbiolspec.FUNK-0035-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez S., Fischer J., Ganesan R., Cildir G., Wolke M., Pessia A., et al. (2020). S. Typhimurium impairs glycolysis-mediated acidification of phagosomes to evade macrophage defense. SSRN J. 17, 1–26. doi: 10.2139/ssrn.3624438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. D., McLellan L. I. (1999). Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radical Res. 31, 273–300. doi: 10.1080/10715769900300851 [DOI] [PubMed] [Google Scholar]
- He X., Shu J., Xu L., Lu C., Lu A. (2012). Inhibitory effect of Astragalus Polysaccharides on lipopolysaccharide-induced TNF-a and IL-1β Production in THP-1 cells. Molecules 17, 3155–3164. doi: 10.3390/molecules17033155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández O., Almeida A. J., Gonzalez A., Garcia A. M., Tamayo D., Cano L. E., et al. (2010). A 32-kilodalton hydrolase plays an important role in Paracoccidioides brasiliensis adherence to host cells and influences pathogenicity. Infect. Immun. 78, 5280–5286. doi: 10.1128/IAI.00692-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D., Wheeler R. T. (2014). The complex roles of NADPH oxidases in fungal infection: NADPH oxidases in fungal infection. Cell Microbiol. 16, 1156–1167. doi: 10.1111/cmi.12320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannuzzi G. P., De Almeida J. R. F., Amarante-Mendes G. P., Romera L. M. D., Kaihami G. H., Vasconcelos J. R., et al. (2019). TLR3 is a negative regulator of immune responses against Paracoccidioides brasiliensis . Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakevych J., Denizot J., Liebert A., Portovedo M., Mosavie M., Jain P., et al. (2020). Smarcad1 mediates microbiota-induced inflammation in mouse and coordinates gene expression in the intestinal epithelium. Genome Biol. 21, 64. doi: 10.1186/s13059-020-01976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda Pigosso L., Baeza L. C., Vieira Tomazett M., Batista Rodrigues Faleiro M., Brianezi Dignani De Moura V. M., Melo Bailão A., et al. (2017). Paracoccidioides brasiliensis presents metabolic reprogramming and secretes a serine proteinase during murine infection. Virulence 8, 1417–1434. doi: 10.1080/21505594.2017.1355660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Lin J., Zhou W., Moses R., Dai Z., Kossenkov A. V., et al. (2022). KDM5A inhibits antitumor immune responses through downregulation of the antigen-presentation pathway in ovarian cancer. Cancer Immunol. Res. 10, 1028–1038. doi: 10.1158/2326-6066.CIR-22-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodyga M., Hinz B. (2020). TGF-β1 – A truly transforming growth factor in fibrosis and immunity. Semin. Cell Dev. Biol. 101, 123–139. doi: 10.1016/j.semcdb.2019.12.010 [DOI] [PubMed] [Google Scholar]
- Loures F. V., Araújo E. F., Feriotti C., Bazan S. B., Calich V. L. G. (2015). TLR-4 cooperates with Dectin-1 and mannose receptor to expand Th17 and Tc17 cells induced by Paracoccidioides brasiliensis stimulated dendritic cells. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loures F. V., Araújo E. F., Feriotti C., Bazan S. B., Costa T. A., Brown G. D., et al. (2014). Dectin-1 induces M1 macrophages and prominent expansion of CD8+IL-17+ Cells in pulmonary paracoccidioidomycosis. J. Infect. Dis. 210, 762–773. doi: 10.1093/infdis/jiu136 [DOI] [PubMed] [Google Scholar]
- Loures F. V., Pina A., Felonato M., Araújo E. F., Leite K. R. M., Calich V. L. G. (2010). Toll-like receptor 4 signaling leads to severe fungal infection associated with enhanced proinflammatory immunity and impaired expansion of regulatory T cells. Infect. Immun. 78, 1078–1088. doi: 10.1128/IAI.01198-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M. E., To J., O’Brien B. A., Donnelly S. (2016). The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J. Immunol. Methods 430, 64–70. doi: 10.1016/j.jim.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Mangan P. R., Harrington L. E., O’Quinn D. B., Helms W. S., Bullard D. C., Elson C. O., et al. (2006). Transforming growth factor-β induces development of the TH17 lineage. Nature 441, 231–234. doi: 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Martinez R. (2017). New trends in paracoccidioidomycosis epidemiology. JoF 3, 1. doi: 10.3390/jof3010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGettrick A. F., Corcoran S. E., Barry P. J. G., McFarland J., Crès C., Curtis A. M., et al. (2016). Trypanosoma brucei metabolite indolepyruvate decreases HIF-1α and glycolysis in macrophages as a mechanism of innate immune evasion. Proc. Natl. Acad. Sci. U.S.A. 113, E7778–E7787. doi: 10.1073/pnas.1608221113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer K., Weening D., De Vries M. P., Priebe M. G., Vonk R. J., Roelofsen H. (2015). Quantitative proteomics analyses of activation states of human THP-1 macrophages. J. Proteomics 128, 164–172. doi: 10.1016/j.jprot.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Mejía S. P., Cano L. E., López J. A., Hernandez O., González Á. (2015). Human neutrophils produce extracellular traps against Paracoccidioides brasiliensis . Microbiology 161, 1008–1017. doi: 10.1099/mic.0.000059 [DOI] [PubMed] [Google Scholar]
- Mendes R. P., Cavalcante R. D. S., Marques S. A., Marques M. E. A., Venturini J., Sylvestre T. F., et al. (2017). Paracoccidioidomycosis: current perspectives from Brazil. TOMICROJ 11, 224–282. doi: 10.2174/1874285801711010224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menino J. F., Saraiva M., Gomes-Alves A. G., Lobo-Silva D., Sturme M., Gomes-Rezende J., et al. (2013). TLR9 activation dampens the early inflammatory response to Paracoccidioides brasiliensis, impacting host survival. PloS Negl. Trop. Dis. 7, e2317. doi: 10.1371/journal.pntd.0002317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes D., Assunção L. D. P., Silva K. L. P. D., Soares C. M. D. A., Silva-Bailão M. G., Bailão A. M. (2023). High copper promotes cell wall remodeling and oxidative stress in Histoplasma capsulatum, as revealed by proteomics. Fungal Biol. S1878614623000582, 1–15. doi: 10.1016/j.funbio.2023.05.004 [DOI] [PubMed] [Google Scholar]
- Morais E. A., Chame D. F., Melo E. M., De Carvalho Oliveira J. A., De Paula A. C. C., Peixoto A. C., et al. (2016). TLR 9 involvement in early protection induced by immunization with rPb27 against Paracoccidioidomycosis. Microbes Infection 18, 137–147. doi: 10.1016/j.micinf.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Moscardi-Bacchi M., Brummer E., Stevens D. A. (1994). Support of Paracoccidioides brasiliensis multiplication by human monocytes or macrophages: inhibition by activated phagocytes. J. Med. Microbiol. 40, 159–164. doi: 10.1099/00222615-40-3-159 [DOI] [PubMed] [Google Scholar]
- Murad A. M., Souza G. H. M. F., Garcia J. S., Rech E. L. (2011). Detection and expression analysis of recombinant proteins in plant-derived complex mixtures using nanoUPLC-MSE. J. Sep. Sci. 34, 2618–2630. doi: 10.1002/jssc.201100238 [DOI] [PubMed] [Google Scholar]
- Nauseef W. M. (2004). Assembly of the phagocyte NADPH oxidase. Histochem Cell Biol. 122, 277–291. doi: 10.1007/s00418-004-0679-8 [DOI] [PubMed] [Google Scholar]
- Netea M. G., Joosten L. A. B., Latz E., Mills K. H. G., Natoli G., Stunnenberg H. G., et al. (2016). Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098. doi: 10.1126/science.aaf1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Narita T., Miyazaki E., Ito T., Nishimoto N., Yoshizaki K., et al. (2001). Characterization of the human FcγRIIB gene promoter: human zinc-finger proteins (ZNF140 and ZNF91) that bind to different regions function as transcription repressors. Int. Immunol. 13, 1075–1084. doi: 10.1093/intimm/13.8.1075 [DOI] [PubMed] [Google Scholar]
- Oliveira S. J., Mamoni R. L., Musatti C. C., Papaiordanou P. M. O., Blotta M. H. S. L. (2002). Cytokines and lymphocyte proliferation in juvenile and adult forms of paracoccidioidomycosis: comparison with infected and non-infected controls. Microbes Infection 4, 139–144. doi: 10.1016/S1286-4579(01)01521-0 [DOI] [PubMed] [Google Scholar]
- O’Neill L. A. J., Kishton R. J., Rathmell J. (2016). A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565. doi: 10.1038/nri.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente-Rocha J. A., Parente A. F. A., Baeza L. C., Bonfim S. M. R. C., Hernandez O., McEwen J. G., et al. (2015). Macrophage interaction with Paracoccidioides brasiliensis yeast cells modulates fungal metabolism and generates a response to oxidative stress. PloS One 10, e0137619. doi: 10.1371/journal.pone.0137619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise-Fortes M. R., Marques S. A., Soares A. M. V. C., Kurokawa C. S., Marques M. E. A., Peracoli M. T. S. (2006). Cytokines released from blood monocytes and expressed in mucocutaneous lesions of patients with paracoccidioidomycosis evaluated before and during trimethoprim-sulfamethoxazole treatment: Cytokines in lesions of paracoccidioidomycosis. Br. J. Dermatol. 154, 643–650. doi: 10.1111/j.1365-2133.2006.07161.x [DOI] [PubMed] [Google Scholar]
- Peoples J. N., Saraf A., Ghazal N., Pham T. T., Kwong J. Q. (2019). Mitochondrial dysfunction and oxidative stress in heart disease. Exp. Mol. Med. 51, 1–13. doi: 10.1038/s12276-019-0355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., KamatChinathan S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccia R., Vallejo M. C., Matsuo A. L., Longo L. V. G. (2011). The Paracoccidioides cell wall: past and present layers toward understanding interaction with the host. Front. Microbio. 2. doi: 10.3389/fmicb.2011.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A., Restrepo S., Najvar L., Gomez B. L., Graybill J. R., Franco L. (1998). Experimental pulmonary fibrosis induced by Paracoccidioides brasiliensis conidia: measurement of local host responses. Am. J. Trop. Med. Hygiene 58, 424–430. doi: 10.4269/ajtmh.1998.58.424 [DOI] [PubMed] [Google Scholar]
- Rhee S. G. (2016). Overview on peroxiredoxin. Molecules Cells 39, 1–5. doi: 10.14348/molcells.2016.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwan H., Pal S., Sabnam S., Pal A. (2020). High glucose augments ROS generation regulates mitochondrial dysfunction and apoptosis via stress signalling cascades in keratinocytes. Life Sci. 241, 117148. doi: 10.1016/j.lfs.2019.117148 [DOI] [PubMed] [Google Scholar]
- Rocha O. B., Freitas E Silva K. S., De Carvalho Junior M. A. B., Moraes D., Alonso A., Alonso L., et al. (2022). Proteomic alterations in Paracoccidioides brasiliensis caused by exposure to curcumin. J. Proteomics 266, 104683. doi: 10.1016/j.jprot.2022.104683 [DOI] [PubMed] [Google Scholar]
- Rodrigues D. R., Dias-Melicio L. A., Calvi S. A., Peraçoli M. T. S., Soares A. M. V. C. (2007). Paracoccidioides brasiliensis killing by IFN-γ, TNF-α and GM-CSF activated human neutrophils: role for oxygen metabolites. Med. Mycol 45, 27–33. doi: 10.1080/13693780600981676 [DOI] [PubMed] [Google Scholar]
- Rodrigues D. R., Fernandes R. K., Balderramas H. D. A., Penitenti M., Bachiega T. F., Calvi S. A., et al. (2014). Interferon-gamma production by human neutrophils upon stimulation by IL-12, IL-15 and IL-18 and challenge with Paracoccidioides brasiliensis . Cytokine 69, 102–109. doi: 10.1016/j.cyto.2014.05.009 [DOI] [PubMed] [Google Scholar]
- Santos A. R. D., Dionizio A., Fernandes M. D. S., Buzalaf M. A. R., Pereira B., Donanzam D. D. F. A., et al. (2021). Proteomic analysis of serum samples of paracoccidioidomycosis patients with severe pulmonary sequel. PloS Negl. Trop. Dis. 15, e0009714. doi: 10.1371/journal.pntd.0009714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya A. P., Stone R. C., Mehdizadeh S., Pastar I., Worrell S., Balukoff N. C., et al. (2022). FOXM1 network in association with TREM1 suppression regulates NET formation in diabetic foot ulcers. EMBO Rep. 23, e54558. doi: 10.15252/embr.202154558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddigh P., Bracht T., Molinier-Frenkel V., Castellano F., Kniemeyer O., Schuster M., et al. (2017). Quantitative analysis of proteome modulations in alveolar epithelial type II cells in response to Pulmonary Aspergillus fumigatus infection. Mol. Cell. Proteomics 16, 2184–2198. doi: 10.1074/mcp.RA117.000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai-Yasuda M. A., Telles Filho F. D. Q., Mendes R. P., Colombo A. L., Moretti M. L. (2006). Consenso em paracoccidioidomicose. Rev. Soc Bras. Med. Trop. 39, 297–310. doi: 10.1590/S0037-86822006000300017 [DOI] [PubMed] [Google Scholar]
- Shin Y.-K., Kim K.-Y., Paik Y.-K. (2005). Alterations of protein expression in macrophages in response to Candida albicans infection. Mol. Cells 20, 271–279. [PubMed] [Google Scholar]
- Silva S. S., Tavares A. H. F. P., Passos-Silva D. G., Fachin A. L., Teixeira S. M. R., Soares C. M. A., et al. (2008). Transcriptional response of murine macrophages upon infection with opsonized Paracoccidioides brasiliensis yeast cells. Microbes Infection 10, 12–20. doi: 10.1016/j.micinf.2007.09.018 [DOI] [PubMed] [Google Scholar]
- Sun L., Zhang H., Gao P. (2022). Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 13, 877–919. doi: 10.1007/s13238-021-00846-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surawut S., Ondee T., Taratummarat S., Palaga T., Pisitkun P., Chindamporn A., et al. (2017). The role of macrophages in the susceptibility of Fc gamma receptor IIb deficient mice to Cryptococcus neoformans . Sci. Rep. 7, 40006. doi: 10.1038/srep40006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadie J.-M., Bae H.-B., Jiang S., Park D. W., Bell C. P., Yang H., et al. (2013). HMGB1 promotes neutrophil extracellular trap formation through interactions with Toll-like receptor 4. Am. J. Physiology-Lung Cell. Mol. Physiol. 304, L342–L349. doi: 10.1152/ajplung.00151.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A. H., Magalhães K. G., Almeida R. D. N., Correa R., Burgel P. H., Bocca A. L. (2013). NLRP3 inflammasome activation by Paracoccidioides brasiliensis . PloS Negl. Trop. Dis. 7, e2595. doi: 10.1371/journal.pntd.0002595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucey T. M., Verma J., Harrison P. F., Snelgrove S. L., Lo T. L., Scherer A. K., et al. (2018). Glucose homeostasis is important for immune cell viability during candida challenge and host survival of systemic fungal infection. Cell Metab. 27, 988–1006.e7. doi: 10.1016/j.cmet.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A. P., Gesztesi J. L., Franco M. F., De Souza W., De Moraes J. Z., Travassos L. R., et al. (1994). Binding of Paracoccidioides brasiliensis to laminin through surface glycoprotein gp43 leads to enhancement of fungal pathogenesis. Infect. Immun. 62, 1465–1469. doi: 10.1128/iai.62.4.1465-1469.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Zare H., Mousavi K., Wang C., Moravec C. E., Sirotkin H. I., et al. (2013). The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J. 32, 1075–1086. doi: 10.1038/emboj.2013.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerasinghe H., Traven A. (2020). Immunometabolism in fungal infections: the need to eat to compete. Curr. Opin. Microbiol. 58, 32–40. doi: 10.1016/j.mib.2020.07.001 [DOI] [PubMed] [Google Scholar]
- Xu T., Park S., Giaimo B. D., Hall D., Ferrante F., Ho D. M., et al. (2017). RBPJ/CBF1 interacts with L3MBTL3/MBT 1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD 1. EMBO J. 36, 3232–3249. doi: 10.15252/embj.201796525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Yang R., Shang J., Gao T., Wang R., Peng X., et al. (2019). FOXC1 up-regulates the expression of toll-like receptors in myocardial ischaemia. J. Cell. Mol. Medi 23, 7566–7580. doi: 10.1111/jcmm.14626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All proteins identified in the proteomic analysis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material . Data is available at: http://www.ebi.ac.uk/pride/archive/projects/PXD043727.