Abstract
A citrate lyase (EC 4.1.3.6) was purified 25-fold from Leuconostoc mesenteroides and was shown to contain three subunits. The first 42 amino acids of the β subunit were identified, as well as an internal peptide sequence spanning some 20 amino acids into the α subunit. Using degenerated primers from these sequences, we amplified a 1.2-kb DNA fragment by PCR from Leuconostoc mesenteroides subsp. cremoris. This fragment was used as a probe for screening a Leuconostoc genomic bank to identify the structural genes. The 2.7-kb gene cluster encoding citrate lyase of L. mesenteroides is organized in three open reading frames, citD, citE, and citF, encoding, respectively, the three citrate lyase subunits γ (acyl carrier protein [ACP]), β (citryl-S-ACP lyase; EC 4.1.3.34), and α (citrate:acetyl-ACP transferase; EC 2.8.3.10). The gene (citC) encoding the citrate lyase ligase (EC 6.2.1.22) was localized in the region upstream of citD. Protein comparisons show similarities with the citrate lyase ligase and citrate lyase of Klebsiella pneumoniae and Haemophilus influenzae. Downstream of the citrate lyase cluster, a 1.4-kb open reading frame encoding a 52-kDa protein was found. The deduced protein is similar to CitG of the other bacteria, and its function remains unknown. Expression of the citCDEFG gene cluster in Escherichia coli led to the detection of a citrate lyase activity only in the presence of acetyl coenzyme A, which is a structural analog of the prosthetic group. This shows that the acetyl-ACP group of the citrate lyase form in E. coli is not complete or not linked to the protein.
Lactic acid bacteria of the genus Leuconostoc play important roles in the dairy industry because of their ability to produce carbon dioxide and C4 aroma compounds through lactose heterofermentation and citrate utilization. The carbon dioxide produced is responsible for eye formation in certain types of cheese. Citrate utilization by these bacteria leads to the production of diacetyl, which is considered a main flavor compound of a range of fermented dairy products such as cultured butter, buttermilk, and cottage cheese.
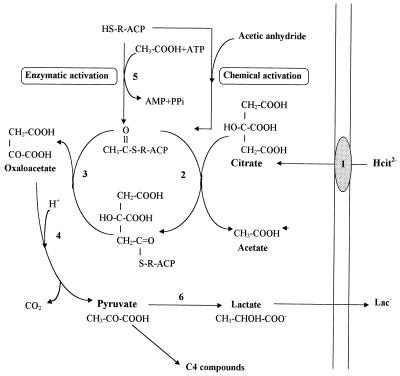
The citrate utilization by lactic acid bacteria requires specifically three enzymes involved in the conversion of citrate to pyruvate: a citrate permease, a citrate lyase, and an oxaloacetate decarboxylase. The energetic role of citrate metabolism in Leuconostoc mesenteroides has been recently described (24, 25). The citrate permease catalyzes an electrogenic exchange of divalent anionic citrate and monovalent lactate, resulting in the generation of a membrane potential (Fig. 1, reaction 1) (24, 25). The intracellular citrate is cleaved by a citrate lyase (EC 4.1.3.6), yielding acetate and oxaloacetate (Fig. 1, reactions 2 and 3). The oxaloacetate is decarboxylated into carbon dioxide and pyruvate in a reaction catalyzed by the enzyme oxaloacetate decarboxylase (Fig. 1, reaction 4).
FIG. 1.
Citrate fermentation pathway in L. mesenteroides and role of the different subunits in the reaction catalyzed by citrate lyase (EC 4.1.3.6). The proteins involved are citrate permease (1), citrate lyase α subunit citrate:acetyl-ACP transferase (EC 2.8.3.10) (2), citrate lyase β subunit citryl–S-ACP lyase (EC 4.1.3.34) (3) oxaloacetate decarboxylase (4), acetate:SH-CL ligase (EC 6.2.1.22) (5), and lactate dehydrogenase (6). ACP, γ subunit of ACP; R, prosthetic group. Acetic anhydride is used for chemical specific acetylation of the prosthetic group. Acetic anhydride is an analog of the mixed anhydride of citric and acetic acids which corresponds probably to an intermediate analog in the acyl-exchange reaction (7a, 14a).
Understanding of the molecular genetics of these lactic acid bacteria is not far advanced, and the genes encoding the enzymes citrate lyase and oxaloacetate decarboxylase are unknown.
On the basis of previous studies (22, 33), the citrate lyase of Lactococcus lactis subsp. lactis biovar diacetylactis and Leuconostoc can be considered a functional complex (Mr, 585,000) composed of three proteins: α, β, and γ subunits in a stoichiometric relationship of 6:6:6. The structure and the mechanism of action are similar to those of the citrate lyase of Klebsiella pneumoniae, which has been extensively studied (1, 15, 16, 34, 36). The citrate lyase is active only if the thioester residue of the prosthetic group linked to its acyl carrier protein (ACP) (γ subunit) is acetylated. This activation is catalyzed by an acetate:SH-citrate lyase ligase (CL ligase) (EC 6.2.1.22), which converts HS-ACP with ATP and acetate into the acetyl-S-ACP (Fig. 1, reaction 5) (32). The α subunit replaces the acyl group with a citryl group to form the citryl-S-ACP (Fig. 1, reaction 2) (16). At last, the β subunit cleaves citryl-S-ACP into oxaloacetate and regenerates the acyl-S-ACP (Fig. 1, reaction 3) (16).
Different mechanisms of regulation of citrate lyase have been reported, such as configurational changes, reversible covalent modification by acetylation-deacetylation, and phosphorylation-dephosphorylation (1, 2). In microorganisms like Klebsiella, in which the reactions of the tricarboxylic acid cycle are operative and therefore contain citrate synthase, a strict regulation of citrate lyase activity is necessary to avoid a futile cycle between citrate fermentation and the l-glutamate biosynthetic pathway. After citrate depletion from the growth medium or upon transfer from an anaerobic citrate medium to an aerobic glucose medium, the synthesis of l-glutamate from oxaloacetate and acetyl coenzyme A (CoA) via citrate can be ensured only if the citrate fermentation pathway is turned off. The intracellular l-glutamate concentration controls these pathways by modulating the activity of the citrate lyase complex (1, 2).
An induction of citrate lyase activity has been observed in Leuconostoc but never in all Lactococcus strains tested (21, 26). In L. mesenteroides, the citrate lyase activity is induced by citrate and rapidly repressed after the citrate consumption in the medium. However, the regulation mechanisms remain unknown. In this paper, we report the purification of L. mesenteroides citrate lyase and an approach based on reverse genetics that yielded the full-length sequence of CL ligase and citrate lyase genes encoding the α, β, and γ subunits. The citrate lyase and CL ligase genes were sequenced and expressed in Escherichia coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. L. mesenteroides strains were grown at 30°C without shaking in MRS broth (12). E. coli TG1 was used for cloning purposes. E. coli AR 1062 and E. coli BL21(DE3) were used as hosts for gene expression studies. E. coli strains were routinely grown at 37°C in Luria-Bertani broth or on Luria-Bertani agar (4). Antibiotics were used at concentrations as follows: ampicillin, 100 μg/ml, and erythromycin, 200 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | 20 |
| AR1062 | F−thr leu ara azi fhuA lacY tsx minA gal rpsL xyl mtl thi hsdR | 13 |
| BL21(DE3) | hsdS gal (λclts857 ind-1 sam-7 nin-5 lacUV5-T7 gene 1) | 35 |
| L. mesenteroides | ||
| 195 | Chr. Hansen (Milwaukee, Wis.) | |
| 18T | Our collection | |
| Plasmids | ||
| pJDC9 | EmrlacZ | 9 |
| pBluescript KS+ | AprlacZ | Stratagene |
| pCL9 | pJDC9 containing the 4.4-kb Sau3A DNA fragment from L. mesenteroides subsp. cremoris 195 with citDEFG gene cluster | This work |
| pNA2028 | pBluescript carrying the 4.4-kb SacI-XbaI insert of pCL9 under the control of the T7 promoter | This work |
| pNA2030 | pBluescript carrying the citC gene | This work |
Assay for citrate lyase activity and protein concentration.
Citrate lyase activity was determined at 25°C in a coupled spectrophotometric assay with malate and lactate dehydrogenases. The assay mixture contained, in a final volume of 1 ml, 100 mM phosphate buffer (pH 7.2), 5 mM trisodium citrate, 3 mM MgCl2, 0.23 mM NADH, 11 U of malate dehydrogenase, 16 U of lactate dehydrogenase (Boehringer Mannheim, Mannheim, Germany), and 20 to 100 μl of enzyme. The oxidation of NADH was measured in a spectrophotometer at 340 nm. The protein concentration was estimated by the method of Bradford (7) with bovine serum albumin as standard or by monitoring absorbance at 280 nm during purification procedures. One unit of enzyme activity is defined as 1 pmol of citrate converted to acetate and oxaloacetate per min under the conditions used. For chemical acetylation of citrate lyase, 100 μl of enzyme sample was incubated for 1 min at 25°C with 5 mM acetic anhydride and then used immediately for determination of citrate lyase activity.
Purification of citrate lyase.
All the purification steps were performed at 4°C, and all solutions contained 3 mM MgCl2. The bacterial cell pellet was obtained by centrifugation (4,000 × g for 15 min) from an exponential growth phase culture, washed twice, and then resuspended in buffer A (50 mM potassium phosphate, pH 7.2). The cells were broken by passage through a French pressure cell (14,500 lb/in2). The crude cell extract was cleared by centrifugation (15,000 × g for 30 min) to give the soluble extract. This extract was treated with protamine sulfate (10 to 50 μg per mg of protein) in order to eliminate nucleic acids. After each addition of protamine sulfate, an aliquot was centrifuged and the citrate lyase activity of the supernatant was determined. The suspension was stirred for 20 min, and the precipitate was collected by centrifugation (15,000 × g for 40 min). The proteins from the supernatant were precipitated by the addition of ammonium sulfate (30 to 65% [wt/vol] in 5% increments). The pellet of the fractions collected by centrifugation (15,000 × g for 15 min) was resuspended in buffer A. Citrate lyase activity precipitated between 40 and 60% ammonium sulfate. The fractions containing citrate lyase activity were pooled and then loaded on a Bio-Gel HT hydroxylapatite column (10 by 30 mm; Bio-Rad Laboratories, Ivry sur Seine, France) equilibrated in buffer A. Proteins were eluted from the column by using a linear gradient of 40 to 200 mM phosphate buffer. The enzyme was eluted between 50 and 60 mM. The active fractions were pooled in a dialysis tube and then concentrated by spraying them with flakes of polyethylene glycol 20,000. The concentrate was loaded onto a Macro-Prep 50Q Sepharose anion-exchange chromatography column (16 by 70 mm; Pharmacia, LKB Biotech, Uppsala, Sweden) equilibrated with buffer A and eluted with a 50 to 400 mM NaCl gradient. The enzyme was eluted at 300 mM salt. Fractions with citrate lyase activity were pooled, desalted by dialysis against buffer B (1 mM potassium phosphate [pH 7.2]), concentrated as described above, and stored at −20°C in 40% (vol/vol) glycerol.
SDS-PAGE and Western blotting.
Proteins were separated by sodium dodecyl sulfate (SDS)–14% polyacrylamide gel electrophoresis (PAGE) according to the method of Laemmli (23) and then either stained with Coomassie brilliant blue R-250 or electroblotted onto Hybond C nitrocellulose (Amersham, Les Ulis, France). A semidry electroblotting apparatus (Bio-Rad Laboratories) operating at an applied potential of 12 V for a period of 30 min was used. After preincubation for 30 min in phosphate-buffered saline containing 5% milk powder, the blots were washed in 0.2% (vol/vol) Triton X-100 in phosphate-buffered saline and then incubated (1 h at 25°C) with monoclonal antibodies raised against the α or β subunit of lactococcal citrate lyase (11). Antibodies bound to cross-reacting proteins were probed by using a rabbit anti-mouse immunoglobulin G peroxidase conjugate (Bio-Rad Laboratories) and visualized by the development of a brown color after addition of diaminobenzidine.
N-terminal sequencing.
For N-terminal sequence analysis, the Mono Q fraction (2 to 5 μg) was resolved on an SDS–14% polyacrylamide gel as described above. The protein was electroblotted onto a polyvinylidene difluoride membrane with a Bio-Rad semidry electroblotting apparatus operating at an applied potential of 12 V for a period of 40 min and then visualized by staining with Coomassie blue. The section of the membrane containing α or β subunit as revealed by Western blotting was excised and subjected to N-terminal Edman degradation. The analysis was performed in the blot cartridge on the model 476A pulsed liquid phase Sequenator with on-line phenylthiohydantoin analysis on a 120A analyzer (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Sequencing reagent and solvents were obtained from the same firm.
Enzymatic cleavage of membrane-bound α subunit.
The in situ digest was performed as described by Fernandez et al. (18). Briefly, the polyvinylidene difluoride pieces, containing approximately 6 μg of α subunit, were cut into small pieces and washed twice with 200 μl of 50% methanol in ultrapure water. Fifty microliters of digest buffer (1% Triton X-100, 10% acetonitrile, 100 mM Tris-HCl [pH 8.6]) were added to the sample together with 0.6 μg of endoproteinase LysC (Wako, Osaka, Japan) (enzyme/substrate ratio of 1/10 [wt/wt]). The digest was carried out at 37°C for 24 h. After digestion, peptides were extracted by sonication for 10 min and then centrifuged at 2,500 × g for 5 min followed by consecutive washes with 50 μl of digest buffer and 50 μl of 0.1% TFA-MQ water. The pooled fractions were freeze-dried in a Speed Vac concentrator (Savant, Hicksville, N.Y.) and redissolved in 100 μl of 0.05% trifluoroacetic acid in ultrapure water. The extracted peptides were separated on a reversed-phase high-pressure liquid chromatography column (C2-C18; 2.1 by 30 mm; 5 μm) installed on the SMART system (Pharmacia Biotechnology). A linear gradient was formed with two solvents, solvent A (0.05% trifluoroacetic acid in ultrapure water) and solvent B (0.04% TFA–70% ACN). The applied gradient was as follows: 0 to 60 min, 1 to 60% B; 60 to 75 min, 60 to 100% B; 75 to 85 min, 100% B; 85 to 90 min, 100 to 1% B.
DNA manipulations.
The methods described by Sambrook et al. (30) were used for construction and isolation of recombinant DNA. Restriction and ligation were performed with enzymes from Boehringer Mannheim under conditions recommended by the supplier. DNA fragments used as probes in Southern blotting and colony hybridizations were radiolabelled by random priming with [α-32P]dATP by the multiprime labelling system (Gibco-BRL, Cergy Pontoise, France). DNA was transferred to a nylon membrane (Hybond-N; Amersham) as recommended by the manufacturer. Oligodeoxynucleotides used as sequencing or PCR primers were synthesized by Eurogentec (Seraing, Belgium). Aerobically grown mid-logarithmic-phase E. coli cells were prepared for electroporation and transformed according to the Bio-Rad standard protocol.
Amplification of specific probes.
For the citrate lyase cloning, degenerated oligonucleotide primers designed by using amino acid sequence information for the α and β subunits were cLB1 (5′ACNATGATGTTYGTNCCNGG3′) and cLA (5′GTDATRTTNGTNACNACNCC3′). For the CL ligase cloning, the cLC degenerated oligonucleotide primer (5′ATGAAYGCNAAYCCNTTYAC3′), designed by using a conserved amino acid sequence of K. pneumoniae and Haemophilus influenzae CL ligases, and cLD (5′GGAGATGAAGCAGTATGGCA3′), a primer designed by using the N-terminal sequence of the ACP, were used (see Fig. 3). Each reaction mixture contained 10 μl of 10× Taq DNA polymerase buffer, 2 mM MgCl2, deoxynucleotide triphosphate at a concentration of 50 μM, 50 ng of genomic DNA, 20 pmol of each primer, and 2.5 U of Goldstar Taq polymerase (Eurogentec) in a final volume of 100 μl. DNA amplification was performed in a temperature cycler (Hybaid, Aubervilliers, France) for 35 cycles consisting of a denaturation step for 40 s at 92°C, an annealing step for 1 min at 55°C, and an elongation step for 1 min at 72°C. The PCR product was analyzed on a 1% agarose gel. The 1.2-kb DNA fragment was recovered from agarose with a Jetsorb kit (Genomed, Bad Oeynhausen, Germany) and then sequenced.
FIG. 3.
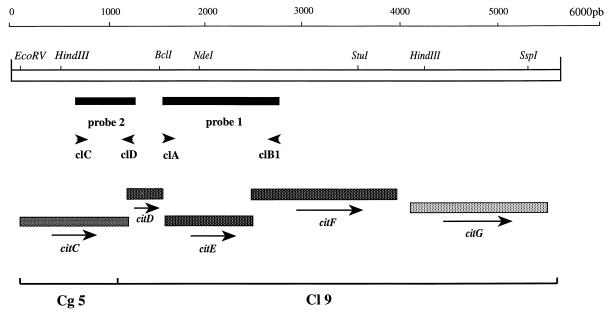
Restriction map of L. mesenteroides citCDEFG (citC, CL ligase; citD, γ subunit; citE, β subunit; citF, α subunit; citG, unknown function). The directions of transcription are indicated by arrows. The solid bars indicate the locations of the probes used to screen the L. mesenteroides subsp. cremoris genomic bank: probe 1 (1.2 kb) was used for citrate lyase cloning; probe 2 (0.5 kb) was used for CL ligase cloning. The locations of the oligonucleotides used to synthesize these probes are indicated by the convergent arrows.
DNA cloning, sequencing, and sequence analysis.
Double-stranded DNA from the recombinant plasmid was purified with a Qiagen plasmid kit (Qiagen) and sequenced by the dideoxynucleotide chain termination method (31) with a 370A sequencer and the Taq Dye-Deoxy TM terminator cycle sequencing kit (Perkin-Elmer, Applied Biosystems Division). DNA and deduced amino acid sequences were submitted to the National Center for Biotechnology Information (Bethesda, Md.) for similarity searches in the GenBank, Swissprot, and Pirprot protein sequence databases. This DNA probe was used for screening a genomic library of Leuconostoc mesenteroides subsp. cremoris. The library has been constructed by ligation into pJDC9 (9, 10) of fragments (5 to 7 kb) isolated from a Sau3A partial digestion of L. mesenteroides subsp. cremoris 195 chromosomal DNA. Recombinant plasmids were transformed in E. coli TG1 to give a library of 2,500 clones. The comparisons and alignments of nucleotides and amino acid sequences were performed with the Palign program (paired and clustal alignment programs) available in the PC/GENE package (IntelliGenetics, Mountain View, Calif.).
Identification of plasmid-encoded proteins.
The selective l-[35S]methionine (Isotopchim, Ganagobie Peyrius, France) labelling of encoded proteins was performed with minicells (3) or under the T7 RNA polymerase control as described by Studier et al. (35).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been submitted to the EMBL, GenBank, and DDBJ nucleotide databases under accession no. Y10621.
RESULTS
Purification of L. mesenteroides citrate lyase.
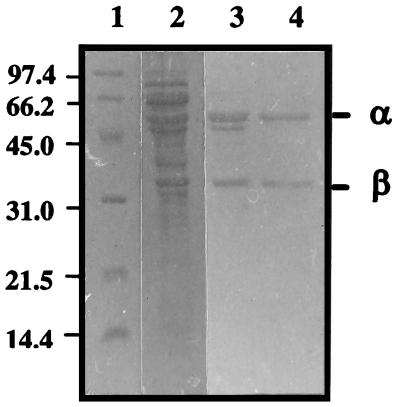
We purified citrate lyase from Leuconostoc mesenteroides subsp. dextranicum. The citrate lyase was found to be present primarily in the cell pellet with less than 5% of citrate lyase activity in the culture supernatant. Citrate lyase activity was always eluted as a single symmetrical peak from hydroxylapatite and Q-Sepharose ion-exchange chromatography. During the course of the purification, a significant loss of citrate lyase activity was found when the enzyme fractions were stored overnight at 4°C after the ammonium sulfate step. The acetic anhydride treatment allowed recovery of 60 to 90% of the initial activity, showing that inactivation of the enzyme results mainly from the loss of the essential acetyl group. The purified enzyme could not be reactivated by incubation with acetate and ATP, showing that acetate–SH-citrate lyase ligase, which catalyzes the acetylation, was not copurified with the citrate lyase subunits. The purification procedure gave a 25-fold enrichment of enzyme specific activity and an overall yield of purified enzyme of 8% (Table 2). The relatively low yield is due to the instability of the enzyme. The enzyme consisted of three subunits as determined by denaturing PAGE: two prominent bands of approximately 55,000 and 34,000 Da (Fig. 2) and a weakly staining band of approximately 14,000 Da. The homogeneity of the purified citrate lyase subunit was also confirmed by immunoblotting with monoclonal antibodies directed against lactococcal α or β subunit which cross-reacted with the Leuconostoc proteins (data not shown).
TABLE 2.
Purification steps of citrate lyase from L. mesenteroides
| Step | Protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Crude extract | 377 | 181 | 0.48 | 100 | 1 |
| Protamine sulfate | 182 | 165 | 0.91 | 91.3 | 1.8 |
| Ammonium sulfate | 62 | 130 | 2.10 | 72 | 4.3 |
| Hydroxylapatite | 4.3 | 25 | 5.70 | 13.5 | 11.8 |
| Ion exchange | 1.3 | 16 | 12.10 | 8.8 | 25.3 |
FIG. 2.
SDS-PAGE analysis of the citrate lyase subunits from L. mesenteroides at various steps of purification. Lane 1, molecular weight markers; lane 2, preparation after ammonium sulfate precipitation; lane 3, preparation after elution from hydroxylapatite column; lane 4, preparation after elution from Q-Sepharose column fractions of the citrate lyase. The numbers at the left indicate molecular weights (103). The band corresponding to the γ subunit, which does not appear in the photograph, was very weakly visible in some preparations.
Partial amino acid sequence of the α and β citrate lyase subunits.
Since an initial N-terminal analysis of the electroblotted α subunit indicated that this polypeptide was N-terminally blocked, we cleaved the membrane-bound protein enzymatically to obtain sequence information from internal peptide fragments. An in situ digest with endoproteinase LysC was performed, and the extracted peptides were separated on a reversed-phase liquid chromatography column (Materials and Methods). The resulting chromatogram revealed some peptides, of which three were subjected to sequence analysis. Fraction Kc 26 contained three peptides present in equimolar amounts (result not shown). Sequence analysis of the fraction Kc 33 yielded one stretch of about 20 amino acids, GVVTNITSSGMRGTLGDTVHQDA.
N-terminal sequence analysis of the blotted β subunit yielded an unambiguous sequence of 42 amino acids, ANTEERLRRTMMFVPGNNPAMIKDAGIYGADSIMFDLEDAVS.
Cloning and sequencing of the citrate lyase gene cluster.
In order to clone the L. mesenteroides subsp. cremoris 195 genes encoding citrate lyase, PCR was carried out to obtain a partial clone of the enzyme. Three degenerated 20-mer oligodeoxynucleotides were designed. One of them (cLA) was based on a segment of an α-subunit internal peptide sequence (GVVTNIT, residues 1 to 7); the two others were modeled after segments of the β-subunit N-terminal sequence (cLB1, TMMFVPG, residues 10 to 16; cLB2, MFDLEDA, residues 35 to 41). The localization and orientation of the primers used for PCR amplification are shown in Fig. 3.
PCR amplification of L. mesenteroides subsp. cremoris 195 genomic DNA, with the internal primer cLA in conjunction with the N-terminal primer cLB1, generated a 1.2-kb single major product. No PCR amplification was detected when cLB2 and cLA were used. This result suggested that the gene encoding the β subunit is located upstream of that encoding the α subunit. The 1.2-kb PCR product was used as template for a second round of PCR with the pair of β-subunit N-terminal primers. A second PCR product was generated, one which, as predicted from the amino acid sequence, was 60 bp in length (results not shown).
The 1.2-kb PCR product was sequenced with cLB1. Computer-assisted analysis of the deduced amino acid sequence revealed that the PCR fragment encoded a portion of the citrate lyase β subunit (data not shown).
The PCR product was then used as a probe to isolate a larger clone from a genomic library of L. mesenteroides as previously described. Restriction analysis of the 10 positive clones selected by colony hybridization indicated seven different profiles. PCR amplification with primers cLB1 and cLA, with plasmid DNA purified from these clones as a template, was performed. Only PCR amplification from one clone resulted in a 1.2-kb fragment, showing that the cloned DNA contained at least DNA coding for the β subunit and the beginning of the α subunit. The recombinant plasmid was designated pCL9.
The genomic origin of the pCL9 insert was confirmed by Southern blot analysis of L. mesenteroides subsp. cremoris 195 genomic DNA (data not shown).
Sequence analysis.
The 4.4-kb nucleotide sequence from the pCL9 insert contains four open reading frames (ORFs) (Fig. 4).
FIG. 4.
DNA nucleotide and deduced amino acid sequences of citC, citD, citE, citF, and citG and flanking DNA from L. mesenteroides subsp. cremoris. The deduced citCDEFG gene products are indicated below the nucleotide sequence, by the one-letter code. Potential RBSs are enclosed in boxes. The putative start codons are white letters in black boxes, and the stop codons are indicated by asterisks. The numbers on the right are the nucleotide positions. Convergent arrows indicate a putative terminator structure.
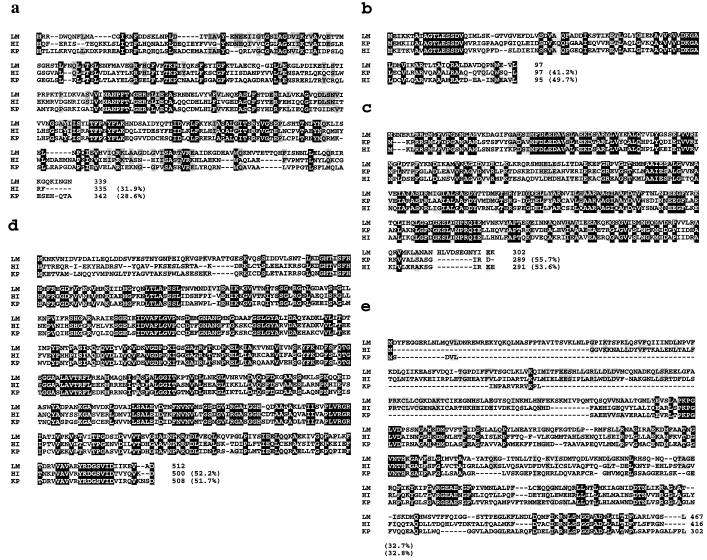
The first ORF (citD) was 327 nucleotides long, and a putative ATG start codon (nucleotide 1173) was identified. This ORF encodes a 97-amino-acid protein (calculated molecular mass, 10.472 Da) preceded by a putative ribosome binding site (RBS) (5′AAAGGG3′) which is complementary to the 3′ end of the L. mesenteroides 16S rRNA 5′CACCTCCTTT3′ sequence (complementary nucleotides are underlined) (37). The protein sequence is 41 and 49% identical to the γ subunit (ACP) of K. pneumoniae and H. influenzae, respectively (5, 19) (Fig. 5). The most conserved region of the γ subunits is located at the beginning of the sequence residues (9-AGTLESSDV-17). This region very likely corresponds to the attachment site of the prosthetic group (14, 27, 28, 34).
FIG. 5.
Amino acid sequence alignment of the predicted citC, citD, citE, citF, and citG gene products of L. mesenteroides subsp. cremoris (LM) with similar proteins. The alignments are of deduced gene products of citC (a), citD (b), citE (c), citF (d), and citG (e). HI, H. influenzae (19); KP, K. pneumoniae (5). Amino acids identical in all the sequences are black boxed; amino acids identical to those of the L. mesenteroides sequence are shaded. The numbering of the amino acids for the individual proteins is shown to the right of the sequence. The percentages on the right are the identities between the protein sequences deduced from L. mesenteroides subsp. cremoris genes and the sequences of the previously described proteins.
The putative start codon ATG of the second ORF (citE) was found at the same position as the TAA stop codon terminating citD, which overlapped the citE coding region by only 1 nucleotide (Fig. 4). A putative RBS (5′GGAGG3′) preceding the start codon was complementary to the 3′ end of L. mesenteroides 16S rRNA 5′CACCTCCTTT3′ (complementary nucleotides are underlined) (37). The citE gene codes for a 302-amino-acid polypeptide (Fig. 5) with a predicted molecular mass of 33,354 Da. This peptide exhibits 55 and 53% sequence identity with the citrate lyase β subunit of K. pneumoniae and H. influenzae, respectively. The deduced N-terminal protein sequence closely matches the corresponding segment previously obtained by Edman degradation of the purified protein, except for six substitutions, which could result from strain polymorphism. The amino acid N-terminal sequence of the β subunit was ANTEER from positions 1 to 6, while the nucleotide sequence predicted MNNERL. Direct amino acid sequence analysis had indicated the presence of an isoleucine at position 22 and a tyrosine at position 28, while the nucleotide sequence predicted a valine and a phenylalanine, respectively. In order to exclude cloning and sequencing artifacts, the corresponding region was amplified from genomic DNA of L. mesenteroides subsp. cremoris by PCR and sequenced. The DNA sequence was 100% identical to the one obtained originally. This indicates that the discrepancies described above may be due to the use of a strain for the Edman degradation different from the one used to clone the gene.
The citF gene overlapped the citE coding region by 11 nucleotides. The same putative RBS as found for citE is present upstream from a TTG start codon (nucleotide 1466) which has already been reported as an initiation codon for some genes of gram-positive bacteria (17). citF codes for a 512-amino-acid polypeptide (Fig. 5), with a predicted molecular mass of 55,015 Da, which is 52% identical to the sequence of citrate lyase α subunit of K. pneumoniae and H. influenzae.
Downstream of the TAA stop codon of citF, inspection of the noncoding region revealed an inverted repeat sequence with four unbound bases (Fig. 4). This sequence could form a stem and loop structure in mRNA with a calculated ΔGf of −9 kcal/mol (−37.6 kJ/mol). This structure probably functions as a rho-independent transcriptional terminator (29).
One more ORF (citG [Fig. 3]) was found on the 3′ side of citF. The deduced amino acid sequence of citG (Fig. 5) displays similarity with those of CitG proteins from other bacteria, except in the N-terminal region, which has an additional stretch of 169 amino acids compared with the corresponding sequence of K. pneumoniae (Fig. 5). However, no significant similarity to other genes in GenBank was found, so that citG encodes a protein whose functional role remains unknown.
Cloning and nucleotide sequence of the CL ligase gene.
Preliminary sequencing and analysis of the upstream region of the citD gene cloned in pCL9 revealed a truncated ORF. Computer-assisted analysis of the partial deduced amino acid sequence revealed similarities with the C terminus of the citC product (CL ligase) of K. pneumoniae (5) and of H. influenzae (19). A Leuconostoc probe for the gene encoding CL ligase was isolated by the following strategy. The amino acid sequences from K. pneumoniae (5) and H. influenzae (19) CL ligases were aligned, and a region of conserved sequence, MNANPFT, was selected for PCR primer design. The degenerated oligonucleotide (cLC) based on this sequence was synthesized and used as a PCR primer with oligonucleotide cLD. The localization and orientation of the primers used for PCR amplification are shown in Fig. 3. PCR amplification generated a 0.5-kb single major product, which was close to the expected size. The 0.5-kb PCR product was sequenced with the primer cLD. Computer-assisted analysis of the deduced amino acid sequence revealed that the PCR fragment encoded a portion of the CL ligase (data not shown). The PCR product was then used as a probe to isolate a clone from the genomic library as described above. One clone showing a restriction profile of plasmid DNA different from pCL9 was selected. The recombinant plasmid was designated pCg5 and was retained for further characterization. The nucleotide sequence of the upstream region of citD was determined by primer walking with pCg5 as a template. The sequence analysis revealed one ORF preceded by a putative RBS (5′AAAGGA3′). This ORF encoded the first 285 amino acids of CL ligase, and so the citC gene was not cloned entirely in pCg5. The complete citC gene was amplified from genomic DNA of L. mesenteroides subsp. cremoris by PCR and sequenced. The sequence analysis revealed that DNA fragments cloned in pCg5 and pCL9 were contiguous. The DNA sequence was 100% identical to those obtained from the 0.5-kb PCR product and pCg5. The citC gene overlapped the citD coding region by 10 nucleotides (Fig. 4). This ORF (citD) encodes a 348-amino-acid protein (calculated molecular mass, 38,246 Da) which exhibits 32 and 29% sequence identity with the CL ligases of H. influenzae and K. pneumoniae, respectively.
Expression of the citCDEFG gene cluster in E. coli.
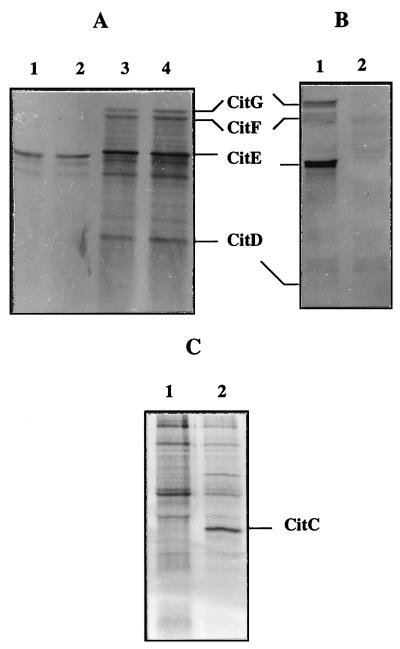
The SacI-XbaI fragment from pCL9 was subcloned behind the T7 promoter in the vector pBluescript KS+ (Stratagene, Heidelberg, Germany). The resulting plasmid (pNA2028) was then transformed in E. coli AR 1062. The AR 1062 strain was used to prepare minicells. Proteins were labelled with l-[35S]methionine during IPTG (isopropyl-β-d-thiogalactopyranoside) induction and were analyzed by SDS-PAGE (Fig. 6). The proteins were also present without IPTG induction, suggesting that the L. mesenteroides promoter or a related sequence from the E. coli promoter was recognized by the E. coli RNA polymerase. The complete citC gene amplified from genomic DNA of L. mesenteroides subsp. cremoris by PCR was subcloned behind the T7 promoter in the vector pBluescript KS+. The resulting plasmids pNA2030 and pNA2028 were transformed separately in E. coli BL21(DE3), which is a λ lysogen strain carrying a chromosomal integrated gene for T7 RNA polymerase under the control of a lacUV5 promoter. Expression of the citC and citDEFG gene products in E. coli BL21 was verified by protein labelling with l-[35S]methionine in the presence of rifampin (Fig. 6).
FIG. 6.
Detection of the citCDEFG gene product in E. coli. (A) SDS-PAGE (14% separating gel) of 35S-labelled proteins synthesized in AR 1062 minicells containing pBluescript KS+ (lanes 1 and 2) and pNA2028 (lanes 3 and 4). Minicells were labelled in the absence (lanes 1 and 3) or in the presence (lanes 2 and 4) of 5 mM IPTG. The positions of the subunits γ (ACP), β (citryl-S-ACP lyase), and α (citrate:acetyl-ACP transferase) and of the citG gene product (CitG) are indicated by lines. (B) SDS-PAGE (15% separating gel) of 35S-labelled proteins synthesized in BL21(DE3) containing pNA2028 (lane 1) and pBluescript KS+ (lane 2). (C) SDS-PAGE (14% separating gel) of 35S-labelled proteins synthesized in BL21(DE3) containing pBluescript KS+ (lane 1) and pNA2030 (lane 2).
Cell extracts from L. mesenteroides subsp. cremoris 195 and E. coli BL21 either enclosing or not enclosing pNA2028 were tested for citrate lyase activity. However, no significant citrate lyase activity was detected in E. coli (Table 3). We attempted to activate citrate lyase by in vitro acetylation. When the citDEFG genes were expressed in E. coli or when the cell extracts from BL21/pNA2028 and BL21/pNA2030 were mixed, the negligible citrate lyase activity of cell extract could not be stimulated in the presence of acetate and ATP, nor by treatment with acetic anhydride (Table 3). However, in the presence of acetyl-CoA (5 mM) and ATP a citrate lyase activity was found in cell extracts from E. coli BL21/pNA2028 but not in those from E. coli BL21 (Table 3).
TABLE 3.
Citrate lyase assays in E. coli/pBluescript and E. coli harboring the citDEFG cluster
| Strain | Citrate lyase activity (U/mg)
|
||
|---|---|---|---|
| Initial activity | After treatment with acetic anhydride | In the presence of acetyl-CoA (5 mM) | |
| BL21/pBluescript KS+ | 0.051 | 0.060 | 0.059 |
| BL21/pNA2028 | 0.073 | 0.085 | 0.162 |
DISCUSSION
In this study, the DNA fragment carrying the citCDEFG gene cluster of L. mesenteroides subsp. cremoris was subcloned, sequenced, and expressed in E. coli. This work represents the first genetic analysis of citrate lyase of a gram-positive bacterium.
We purified the enzyme 25-fold by a combination of anion-exchange and hydroxylapatite chromatography. The anion-exchange chromatography was consistently the most powerful one to separate citrate lyase from the other proteins present in the cellular extract. The citrate lyase showed similarity to the Klebsiella enzyme with regard to subunit composition. The three nonidentical α (55 kDa), β (34 kDa), and γ (14 kDa) subunits are similar in size to the three subunits of citrate lyase from K. pneumoniae. The cross-reaction between Leuconostoc proteins and monoclonal antibodies directed against lactococcal α and β subunits suggests that the two proteins are closely immunologically related. The purified protein exhibits an important loss in its initial activity. The ability to reactivate the purified Leuconostoc enzyme shows that the active form is an acetyl enzyme like all the known citrate lyases.
The deduced amino acid sequence of L. mesenteroides subsp. cremoris citrate lyase exhibits similarity to the sequences of other bacterial citrate lyases. The prosthetic group 5-phosphoribosyl-dephospho-CoA of K. pneumoniae citrate lyase is attached by its ribose-5-phosphate moiety via a phosphodiester linkage to serine 14 (14, 27, 28, 34). This residue and the neighboring residues (9-AGTLESSDV-17) were found to be conserved in all ACPs of citrate lyases. No similarity could be found with ACPs involved in acyl group activation and transfer reaction in the biogenesis of fatty acids, nor with any other protein sequence available in data banks. Our results suggest that the ACPs of citrate lyase belong to a family distinct from other ACPs.
The calculated molecular masses of the α and β subunits are in good agreement with those determined by SDS-PAGE. However, a difference between the calculated and estimated molecular masses of ACP was observed (Fig. 6). The difference between the predicted molecular mass (10,472 Da) and the apparent one (14,000 Da) may be due to the difference in the mass between apo-ACP and the ACP linked to the prosthetic group (holo-ACP) or to the migration conditions.
Upstream of citDEF, the citC gene encoding CL ligase was found. The citCDEF genes encode overlapping ORFs.
Downstream of the citCDEF genes, a fifth ORF, encoding a protein of yet unknown function, was found and called citG. Similar ORF-encoding proteins were found also in the two other citrate lyase gene clusters. On the basis that at least two enzymatic activities are required in the formation of the prosthetic group (5-phosphoribosyl-dephospho-CoA) of Klebsiella citrate lyase, CitG may be an enzyme involved in its formation (5). This enzyme may catalyze (i) the formation of the phosphodiester bond between the hydroxyl group of serine-14 and the 5-phosphoribosyl moiety or (ii) the formation of the α-1,2 glycosidic linkage with dephospho-CoA (5). The composition of the lactococcal and Leuconostoc prosthetic group is not clearly established, and the function of CitG remains to be elucidated.
Since a citrate lyase was synthesized in E. coli BL21, activity was determined by using this strain. No detectable activity was found in cells after chemical or enzymatic activation. However, citrate lyase activity was stimulated in the presence of acetyl-CoA. We conclude that the inactivity of the enzyme formed in E. coli is not due to the absence of acetylation of the prosthetic group. Similar results have been reported elsewhere for the expression of citrate lyase genes of K. pneumoniae in E. coli (5). It has been demonstrated previously that the citrate lyase of K. pneumoniae is active with acetyl-CoA as substrate because of the structural similarity of acetyl-CoA with ACP-R-acyl (8). The acetyl-CoA can be considered a free analog of the ACP. Our results suggest that the prosthetic group of the citrate lyase in E. coli was not complete or not linked to the protein, probably because of the lack of enzyme activities involved in its formation.
The molecular approach to studying citrate lyase in Leuconostoc could permit determination of the number of genes involved in the enzymatic activity. In this work, we have demonstrated that the citCDEFG genetic organization is identical to that found in K. pneumoniae. In this bacterium, the genes encoding the enzymes required for anaerobic catabolism of citrate, citS (Na+-dependent citrate carrier), citC (CL ligase), citDEF (citrate lyase), citG (protein of unknown function), and oadGAB (Na+ pump oxaloacetate decarboxylase) are clustered on the chromosome (5). The citCDEFG genes are located upstream and are divergent from the citS-oadGAB genes (6).
A study of the regulation of citrate lyase and CL ligase genes should aid in the understanding of mechanisms involved in the regulation of this multienzyme complex in Leuconostoc.
ACKNOWLEDGMENTS
S.B. was the recipient of a fellowship from the Programme Intergouvernemental Franco-Algérien. J.V.B. is indebted to the Flemish Government for the Concerted Research Action 12052293. This work was supported by a grant from the Conseil Régional de Bourgogne.
We thank Clair-Yves Bocquien for generously providing monoclonal antibodies directed against α and β subunits of lactococcal citrate lyase. We are grateful to Georges Corrieu, Corinne Joyeux, Jean Guzzo, and Jean-François Cavin for helpful discussion and expert help in protein purification and electrophoresis.
REFERENCES
- 1.Antranikian G, Giffhorn F. Citrate metabolism in anaerobic bacteria. FEMS Microbiol Rev. 1987;46:175–198. [Google Scholar]
- 2.Antranikian G, Gottschalk G. Phosphorylation of citrate lyase ligase in Clostridium sphenoides and regulation of anaerobic citrate metabolism in other bacteria. Biochemistry. 1989;71:1029–1037. doi: 10.1016/0300-9084(89)90107-7. [DOI] [PubMed] [Google Scholar]
- 3.Bally M, Wretlind B, Lazdunski A. Protein secretion in Pseudomonas aeruginosa: molecular cloning and characterization of the xcp-1 gene. J Bacteriol. 1989;171:4342–4348. doi: 10.1128/jb.171.8.4342-4348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bott M, Dimroth P. Klebsiella pneumoniae genes for citrate lyase and citrate lyase ligase: localization, sequencing, and expression. Mol Microbiol. 1994;14:347–356. doi: 10.1111/j.1365-2958.1994.tb01295.x. [DOI] [PubMed] [Google Scholar]
- 6.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7a.Buckel W. Acetic anhydride: an intermediate analogue in the acyl-exchange reaction of citramalate lyase. Eur J Biochem. 1976;64:263–267. doi: 10.1111/j.1432-1033.1976.tb10296.x. [DOI] [PubMed] [Google Scholar]
- 8.Buckel W, Ziegert K, Eggerer H. Acetyl-CoA-dependent cleavage of citrate on inactivated citrate lyase. Eur J Biochem. 1973;37:295–304. doi: 10.1111/j.1432-1033.1973.tb02988.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen J-D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 10.Dartois V, Phalip V, Schmitt P, Diviès C. Purification, properties and DNA sequence of the D-lactate dehydrogenase from Leuconostoc mesenteroides subsp. cremoris. Res Microbiol. 1995;146:291–302. doi: 10.1016/0923-2508(96)81052-7. [DOI] [PubMed] [Google Scholar]
- 11.David P. Suivi de la fermentation lactique par dosage immunochimique d’un marqueur de croissance bactérienne: la citrate lyase. Ph.D. thesis. Compiègne, France: Université Technologique de Compiègne; 1991. [Google Scholar]
- 12.De Man J C, Rogosa M, Sharpe M E. A medium for the cultivation of Lactobacilli. J Appl Bacteriol. 1960;23:130–135. [Google Scholar]
- 13.D’Enfert C, Ryter A, Plugsey A. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for the production, surface localisation and secretion of the lipoprotein pullulanase. EMBO J. 1987;6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimroth P. The prosthetic group of citrate-lyase acyl-carrier protein. Eur J Biochem. 1976;64:269–281. doi: 10.1111/j.1432-1033.1976.tb10297.x. [DOI] [PubMed] [Google Scholar]
- 14a.Dimroth P, Loyal R, Eggerer H. Characterization of the isolated transferase subunit of citrate lyase as a CoA-transferase. evidence against a covalent enzyme-substrate intermediate. Eur J Biochem. 1977;80:479–488. doi: 10.1111/j.1432-1033.1977.tb11903.x. [DOI] [PubMed] [Google Scholar]
- 15.Dimroth P. The role of vitamins and their carrier proteins in citrate fermentation. In: Kleinkauf H, Von Döhren H, Jaenicke L, editors. The roots of modern biochemistry. Berlin, Germany: Walter de Gruyter; 1988. pp. 191–204. [Google Scholar]
- 16.Dimroth P, Eggerer H. Isolation of subunits of citrate lyase and characterization of their function in the enzyme complex. Proc Natl Acad Sci USA. 1975;72:3458–3462. doi: 10.1073/pnas.72.9.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferain T, Garmyn D, Bernard N, Hols P, Delcour J. Lactobacillus plantarum ldhL gene: overexpression and deletion. J Bacteriol. 1994;176:596–601. doi: 10.1128/jb.176.3.596-601.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez J, Andrews L, Mische S M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 19.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, Mckenney K, Sutton G, Fitzhugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liul I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblome E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 20.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, England: Cambridge University; 1984. [Google Scholar]
- 21.Hugenholtz J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:165–178. [Google Scholar]
- 22.Kummel A, Behrens G, Gottschalk G. Citrate lyase from Streptococcus diacetylactis. Association with its acetylating enzyme. Arch Microbiol. 1975;102:111–116. doi: 10.1007/BF00428354. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Marty-Teysset C, Lolkema J S, Schmitt P, Divies C, Konings W N. Membrane potential-generating transport of citrate and malate catalyzed by CitP of Leuconostoc mesenteroides. J Biol Chem. 1995;270:25370–25376. doi: 10.1074/jbc.270.43.25370. [DOI] [PubMed] [Google Scholar]
- 25.Marty-Teysset C, Posthuma C, Lolkema J S, Schmitt P, Diviès C, Konings W N. Proton motive force generation by citrolactic fermentation in Leuconostoc mesenteroides. J Bacteriol. 1996;178:2178–2185. doi: 10.1128/jb.178.8.2178-2185.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellerick D, Cogan T M. Induction of some enzymes of citrate metabolism in Leuconostoc lactis and other heterofermentative lactic acid bacteria. J Dairy Res. 1981;48:497–502. [Google Scholar]
- 27.Oppenheimer J N, Singh M, Sweeley C C, Sung S J, Srere P A. The configuration and location of the ribosidic linkage in the prosthetic group of citrate lyase (Klebsiella aerogenes) J Biol Chem. 1979;254:1000–1002. [PubMed] [Google Scholar]
- 28.Robinson J B, Singh M, Srere P A. Structure of the prosthetic group of Klebsiella aerogenes citrate (pro-3S)-lyase. Proc Natl Acad Sci USA. 1976;73:1872–1876. doi: 10.1073/pnas.73.6.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmellenkamp B, Eggerer H. Mechanism of enzymic acetylation of desacetyl citrate lyase. Proc Natl Acad Sci USA. 1974;71:1987–1991. doi: 10.1073/pnas.71.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh M, Srere P. Purification and properties of citrate lyase from Streptococcus diacetylactis. J Biol Chem. 1975;250:5818–5825. [PubMed] [Google Scholar]
- 34.Singh M, Srere P A, Klapper D G, Capra J D. Subunit and chemical composition of citrate lyase from Klebsiella pneumoniae. Eur J Biochem. 1976;251:2911–2915. [PubMed] [Google Scholar]
- 35.Studier W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian C, Sivaraman C. Bacterial citrate lyase. J Biosci. 1984;6:379–401. [Google Scholar]
- 37.Yang D, Woese C R. Phylogenetic structure of the “Leuconostoc”: an interesting case of rapidly evolving organism. Syst Appl Microbiol. 1989;12:145–149. [Google Scholar]