Abstract
This study reports the cellular self-organization of primary human renal proximal tubule epithelial cells (RPTECs) around a minimal Matrigel scaffold to produce basal-in and apical-out proximal tubule organoids (tubuloids). These tubuloids are produced and maintained in hanging drop cultures for 90+ days, the longest such culture of any kind reported to date. The tubuloids upregulate maturity markers, such as aquaporin-1 (AQP1) and megalin (LRP2), and exhibit less mesenchymal and proliferation markers, such as vimentin and Ki67, compared to 2D cultures. They also experience changes over time as revealed by a comparison of gene expression patterns of cells in 2D culture and in day 31 and day 67 tubuloids. Gene expression analysis and immunohistochemistry reveal an increase in the expression of megalin, an endocytic receptor that can directly bind and uptake protein or potentially assist protein uptake. The tubuloids, including day 90 tubuloids, uptake fluorescent albumin and reveal punctate fluorescent patterns, suggesting functional endocytic uptake through these receptors. Furthermore, the tubuloids release kidney injury molecule-1 (KIM-1), a common biomarker for kidney injury, when exposed to albumin in both dose- and time-dependent manners. While this study focuses on potential applications for modeling proteinuric kidney disease, the tubuloids may have broad utility for studies where apical proximal tubule cell access is required.
Keywords: Tubuloid, kidney, basal-in organoid, proteinuria
1. Introduction
While animal experiments are effective for studying kidney function and disease, there are various species differences, ethical issues, and cost concerns [1,2]. Many have turned to in vitro platforms such as 2D cell cultures, Transwells, organ-on-a-chip systems, and organoids to complement animal models [3,4]. While 2D cultures are high-throughput and well standardized, they are difficult to maintain long-term (typically a week or two before overcrowding or detachment), and growth on stiff substrates induces epithelial-to-mesenchymal transition (EMT) [5]. Kidney-on-a-chip systems facilitate apical exposure of fluid shear stress (FSS) to promote enhanced polarization, barrier function, and overall cell maturity [6], but are generally lower-throughput [3,4,6] (Supplementary Figure 1).
Others have turned to organoids, which are self-organizing 3D structures that are derived from induced pluripotent stem cells (iPSCs), embryonic stem cells (ESCs) [7–16], or adult stem cells (ASCs) [17,18]. To date, most kidney organoid models have been derived from iPSCs or ESCs and have been used for studying healthy kidney function, diseased states, and nephrotoxicity testing. One major advantage of these 3D models is long-term culture.
Specific to the proximal tubule, there are 3D organotypic structures grown in scaffolds [19–21], bioprinted 3D perfusable proximal tubules [22], the OrganoPlate by Mimetas, and microfluidic chips by Nortis [23,24]; the latter two technologies combine hydrogels and plastic structures to grow tubular cultures [25–28]. While the techniques have varied widely, most of these models have been utilized to study drug-induced nephrotoxicity.
Recently, the kidney community has referred to tubular organoids as “tubuloids,” which are defined as 3D multicellular structures are derived from primary cells or ASCs. Tubuloids (representing proximal tubule, loop of Henle, distal tubule, or collecting duct) generally exclude endothelial cells, podocytes, parietal cells, and other glomerular components. In the past, kidney tubuloids have been utilized in applications involving viral infection, cystic fibrosis, and nephrotoxicity [17,25]. For some applications, the tubuloids are dissociated and re-seeded for apical access in microfluidic microphysiological systems [25].
One major limitation of many organoids and 3D organotypic cultures is the difficulty of apical access. An interesting emerging solution is the production of geometrically-inverted organoids, in which the basolateral and apical sides are reversed [29–32]. Production of such inverted organoids relies on mutations that alter cell behavior to have inverted polarity [33–36], rare chance occurrences that are unexplained [37–39], eversion of initially normally polarized organoids [31,32,40], and production of stably inverted organoids through biomaterials manipulation [29,30]. Our lab established eversion-free, stably basal-in cultures for breast organoids grown from MCF10A cells [29,30] in a one-drop, one-organoid hanging drop culture or in ultra-low attachment plates (ULA), which rely on specific biomaterials manipulations. Of interest, many have studied kidney cysts made of distal tubule cells (MDCK) that have reversed polarity [35–39]. To our knowledge, there have been no attempts to generate proximal tubule organoids with reversed polarity, in which the basolateral side faces inward, and the apical side is outward-facing. This paper describes methods for Matrigel manipulation and media optimization for the formation and long-term (>90 days) culture of the inverted tubuloids, molecular characterization, and functional demonstration focusing on albumin uptake and albumin-induced injury as it may relate to proteinuria (Figure 1).
Figure 1. Establishment of geometrically-inverted organoids.
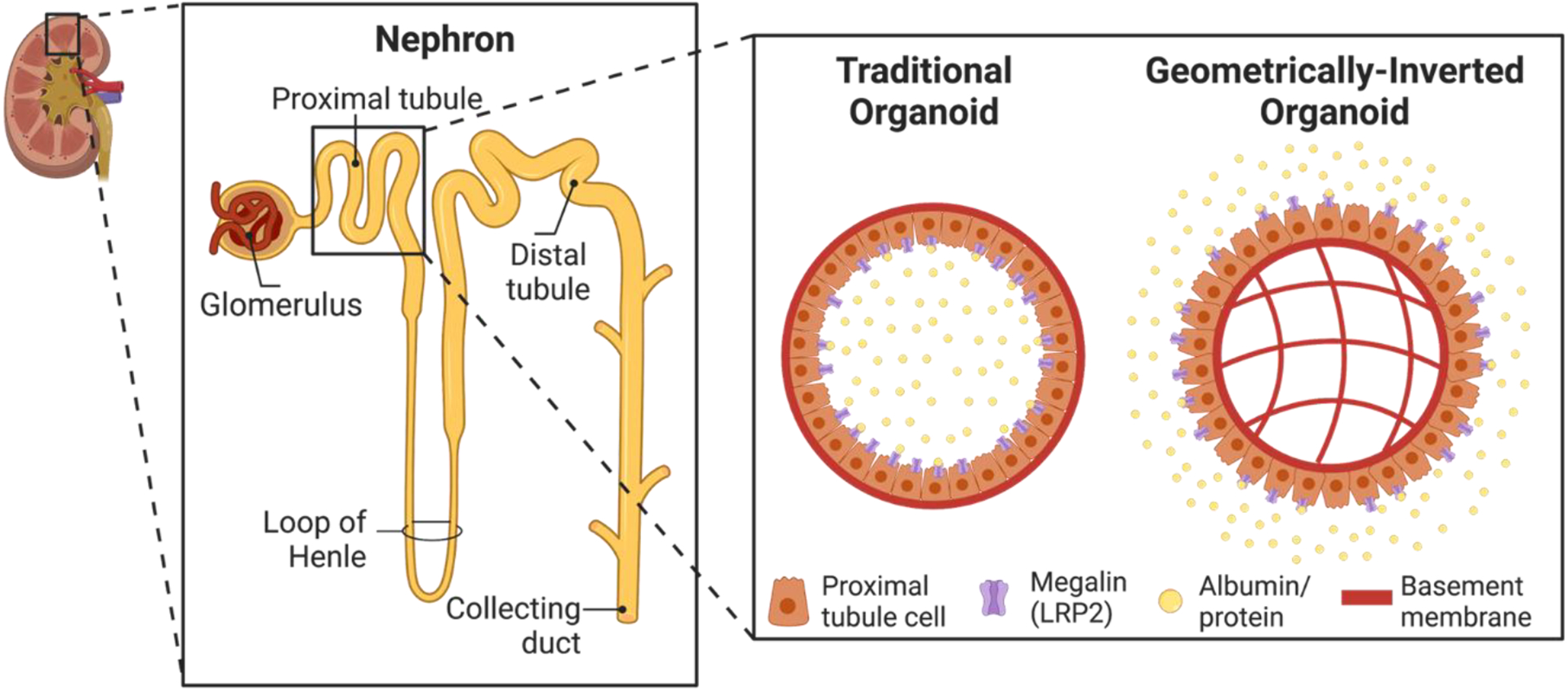
Schematic of a nephron, the functional unit of the kidney. Inset shows a comparison of typical organoids, and geometrically-inverted organoids. Since megalin is located on the apical surface of proximal tubule cells, apical-out organoids allow for more ease of access of megalin and therefore proteinuria studies.
2. Materials and methods
2.1. 2D culture
Human renal proximal tubule epithelial cells (RPTECs) were grown in renal epithelial cell basal media (ATCC) supplemented with 0.5% fetal bovine serum (FBS), 10 nM triiodothyronine, 10 ng/mL epidermal growth factor (EGF), 100 ng/mL hydrocortisone hemisuccinate, 5 ug/mL insulin, 1.0 μM epinephrine, 5 μg/mL transferrin, 2.4 mM L-glutamine, and 1% penicillin-streptomycin (pen-strep). RPTECs were maintained in T75 flasks and passaged at 95% confluency. Primary RPTECs were used until passage 6. All experiments were performed with lot 70001234, in which the donor was a 56-year-old Caucasian male.
2.2. 3D organoid culture
RPTEC organoids were maintained in hanging drop culture as previously described [30,41]. Briefly, custom hanging drop plates were soaked overnight in 0.1% Pluronic solution (Sigma, #542342). Plates were rinsed with water and sterilized using a UVP crosslinker (Analytik Jena). Hanging drop plates were sandwiched between a 96-well round bottom plate. To prevent droplet evaporation, a gauze pad and sterile water with 1% pen-strep were added to the side troughs, and 150 μL deionized water + 1% pen-strep was added to each well of the 96-well plate.
The RPTEC seeding solution consisted of 120,000 RPTEC cells/mL, 120 μg/mL Matrigel (Corning, #356231), 0.24% methylcellulose (Methocel), and 2.4% FBS. To ensure a basal-in and apical-out phenotype and proper encapsulation of Matrigel in the organoid core, cold Matrigel was added to prewarmed media, Methocel (A4M, Sigma), and FBS (Gemini). Upon the addition of Matrigel, the solution was mixed thoroughly, cells were immediately added to the solution, and 25 μL (corresponding to 3000 cells/droplet) was added to each droplet of the plate using a multi-channel repeater pipette. Hanging drop plates were incubated at 37°C and 5% CO2. After 72 hours, 3 consecutive media changes were performed using the Cybio FeliX liquid handler (Analytik Jena), each removing 9 μL and adding 10 μL. Media changes were then performed every 2–3 days, with 2 consecutive rounds of 9 μL removal and 9 μL addition.
Because the plates were maintained for 90+ days in culture, evaporation of the droplets needed to be carefully considered. To account for this, 5 μL of additional media was added, particularly to the outer droplets. The deionized water was replenished in the bottom receiver plate as needed. During routine media changes, the amount added or removed was adjusted accordingly based on the size of the droplets; if they looked too big, a little media was removed, and if they looked too small, additional media was added.
2.3. Live-organoid imaging and morphology quantification
RPTEC organoids were imaged every 2–3 days with an EVOS FL 2 Auto microscope (Thermo Fisher) with a 4x objective. To quantify the area, diameter, and roundness of the organoids, raw image files were imported into ImageJ [42]. The images were converted to binary, inverted, and holes were filled. The “Analyze Particles” feature was used to calculate the area and roundness. To determine the mean diameter, the organoid was assumed as a perfect circle, and the diameter was calculated.
2.4. Histology, immunofluorescence staining, and microscopy
RPTEC organoid histology, staining, and imaging were performed based on our previously described protocols [29,30]. Organoids were harvested from the hanging drop plate, washed with phosphate-buffered saline (PBS), and fixed for 30 minutes at room temperature in 4% paraformaldehyde (Alfa Aesar). Organoids were washed again with PBS and stained for 10 minutes in 0.5% methylene blue (Ricca Chemical Company) in PBS to aid in visualization during the sectioning process. Excess dye was then washed out, and the samples were transferred to a block and filled with optimal cutting temperature (OCT, Tissue-Tek). Isopentane (Sigma) was then cooled in liquid nitrogen, and samples were flash frozen and stored at −80°C until use.
A CryoStar NX70 cryostat (Thermo Fisher) was used to obtain 20 μm thick sections. For hematoxylin and eosin (H&E) staining, organoids were brought to room temperature and stained with an Autostainer XL (Leica). Slides were coverslipped with Xylene and Cytoseal 60 (Richard-Allan Scientific), and imaged in color with a DMi1 microscope (Leica) and 10x air objective. For immunostaining, slides were brought to room temperature, and a hydrophobic pen was used to highlight the region containing the organoid section. Samples were washed with PBS, permeabilized with 0.2% Triton X-100 (Sigma) in PBS 100 for 5 minutes, and washed again with PBS. Samples were incubated for 1 hour at room temperature with 4% BSA (Millipore Sigma, #82–067) in PBS. The following primary antibodies were prepared in 1% BSA in PBS solutions at the following dilutions: rabbit anti-megalin (Abcam #ab76969, 1:500 dilution), mouse anti-aquaporin-1 (Santa Cruz #sc-32737, 1:200 dilution), mouse anti-ezrin/villin-2 (BD #610602, 1:200 dilution), rabbit anti-Na+K+ATPase (Abcam #ab76020, 1:250 dilution), and rabbit anti-laminin-5 (Abcam #ab14509, 1:200 dilution), rabbit anti-laminin-1,2 (Thermo Fisher #PA1–16730, 1:1000 dilution), tubulin (Abcam #ab24610, 1:250 dilution), and anti-rat integrin α6 (Santa Cruz #sc-19622, dilution 1:200). Primary antibodies were added to the samples and incubated at 4°C overnight. Upon removal of the primary antibodies, samples were washed with 1% BSA in PBS, and the following secondary antibody solutions were prepared in 1% BSA: Goat anti-mouse Alexa Fluor 488 (Invitrogen #A-11001, 1:2000 dilution), goat anti-rabbit Alexa Fluor 594 (Invitrogen #A-11012, 1:1000 dilution), and donkey anti-rat Alexa Fluor 488 (Invitrogen #A21208, 1:2000 dilution). Secondary antibodies were incubated at room temperature for 2 hours along with Alexa Fluor 488 phalloidin (Invitrogen #A12379, 1:40 dilution). Secondary antibodies were removed, samples were washed with PBS, and incubated with DAPI (Thermo Fisher, 1.4 × 10−6 uM) for 15 minutes at room temperature. Samples were given a final rinse in PBS and sealed with ProLong Diamond Antifade (Thermo Fisher, #P36965) mounting media. The organoids were imaged using a DMi8 epifluorescence microscope (Leica) equipped with 10x, 20x, and 40x air objectives.
2.5. Quantification of organoid cell coverage and percentage of aquaporin-1 (AQP1) and megalin (LRP2) positive cells
To quantify the extent of organoid cell coverage (cells/perimeter), the images from H&E histology sections were utilized. The number of nuclei (only at the organoid periphery), represented by the dark purple spots, were manually counted in ImageJ. The projected cell area was quantified using the methods discussed in section 2.3. The number of cells was divided by the perimeter for each image. Five images were analyzed for days 8, 16, 30, and 90. For the AQP1 and LRP2 quantification, immunofluorescence images were used. For each image, the total number of nuclei were manually counted, and then the number of cells expressing AQP1 and LRP2 were counted. For each marker, the number of positive cells was divided by the total number of cells in the image to calculate the percentage of positive cells. Five images were analyzed for day 16, 30, and 90 organoids.
2.6. Bulk RNA sequencing
5–10 organoids per replicate were pooled from hanging drop plates to RNase-free tubes. Samples were prepared for 2D RPTECs (~106 cells) and day 31 and 67 organoids. Organoid media was then removed and 300 μl of RLT lysis buffer with 1% beta-mercaptoethanol was added to each tube. The lysate was vortexed for one minute and then immediately flash frozen with liquid nitrogen. Frozen lysates were kept at −80°C until further processing. The samples were shipped to the Emory Integrated Genomics Core (EIGC) for RNA extraction, sequencing library preparation, and sequencing. Illumina sequencers and kits were chosen based on the number of samples, availability, and read depths (>20M per sample).
2.7. Differential gene expression analysis
FASTQ files provided by EIGC were downloaded to Georgia Tech’s high-performance computing cluster called Partnership for an Advanced Computing Environment (PACE). Quality control was first performed using FastQC. Upon passing quality requirements, the reads were aligned using STAR or mapped using Salmon. A custom Python script was written to generate a gene count matrix. For Salmon transcripts, a gene count matrix was generated using R. Genes with less than 0.5 CPM (counts per million) in at least 3 samples (out of the total 9 samples) were filtered out. Differential gene expression analysis was conducted using the EdgeR package. PCA analysis was performed to examine the samples for possible outliers prior to further analysis. The counts were normalized based on library size using the default Trimmed mean of M-values (TMM) method from EdgeR. A comparison was run between the two groups through a generalized linear model (GLM), and likelihood-ratio test (LRT) was used to identify statistically significant differential expressions. The cutoff for significance was set at under an alpha value of 0.05 for the adjusted p-value, and the log2 fold change (LFC) of ±1. Volcano plots comparing 2D versus day 31 and day 31 vs day 67 organoid groups were then generated. The same alpha and LFC cutoffs were used to label upregulated, downregulated, and statistically insignificant genes.
2.8. Heatmap generation
A custom R script was written to plot heatmaps containing genes of interest. Briefly, the script takes in a spreadsheet with gene symbols and their categories. The R package pheatmap was used to scale the DESeq2-normalized gene counts transformed by variance stabilization to compute a Z-score matrix, which was then presented in a heatmap.
2.9. Protein uptake studies
For albumin uptake studies, we utilized all components of the RPTEC growth media, except 0.5% FBS. Fluorescein isothiocyanate-labeled bovine serum albumin (FITC-BSA) was added to the serum-free growth media at a concentration of 50 μg/mL. 200 μL of media was added to each well of the 96-well ultra-low attachment (ULA) plate (Corning, #7007). Organoids were then transferred from the hanging drop plate to an individual well of the ULA plate. Care was taken to introduce as little media from the hanging drop plate to the ULA plate as possible. The ULA plate was incubated at 37°C and 5% CO2 for 2 hours. After the incubation, the extracellular FITC-BSA was washed out with PBS or serum-free growth media. To prevent aspiration of the organoids from the bottom of the wells, we performed 10 consecutive half media changes (remove 100 μL, add 100 μL) to remove the excess FITC-BSA. Organoids were then imaged using an in-incubator microscope (Incucyte S3, Sartorius) using brightfield, phase contrast, and a GFP filter.
2.10. HSA toxicity and KIM-1 studies
As with the albumin uptake studies, serum-free growth media was utilized, as the serum contains some albumin that could interfere with the experiment. Human serum albumin (HSA, Sigma, #A4327) was added to the serum-free growth media at the desired concentration, and the solution was sterile-filtered. The stock solution was serially diluted using a 1:2 dilution ratio; concentrations ranged from 0.078 mg/mL to 2.5 mg/mL. 200 μL of each solution was transferred to wells in a 96-well ULA plate. Organoids were then transferred from the hanging drop plate to the HSA solutions in the ULA plates. The organoids were incubated for the desired timepoint at 37°C and 5% CO2. Organoids were incubated for 48 hours for dose-dependent experiments, and 3, 6, 12, 24, 48, and 72 hours for the time series experiments. At the appropriate timepoint, the supernatant from the wells was collected, flash frozen in liquid nitrogen, and stored at −80°C until needed. To quantify the level of kidney injury molecule-1 (KIM-1) production, an ELISA kit (Enzo, #ADI-900-226-0001) was utilized. The kit was used according to the manufacturer’s instructions, and a BioTek Synergy H4 microplate reader was used for absorbance readings at 450 nm.
2.11. Statistical analysis
To quantify cell area, diameter, and roundness, the same 16 organoids were tracked for 67 days. For the quantification of cell coverage, 5 samples were used per condition, and a one-way ANOVA with Tukey’s multiple comparisons was used to analyze the data. To identify differentially expressed genes from the RNAseq data, LRT was performed on each pre-filtered gene. Unadjusted p-values computed from LRT were corrected with the Benjamini-Hochberg procedure. For the quantification of percentages of AQP1 and LRP2 positive cells, 5 samples were used for each timepoint. A one-way ANOVA with Tukey’s multiple comparisons was used to analyze the data. For proteinuria experiments, analysis for days 35 and 90 organoids had n=4 samples per condition. A one-way ANOVA with Tukey’s multiple comparisons was applied to the data. For the KIM-1 time course data, n=4 samples were used per condition, and a two-way ANOVA with Šīdák’s multiple comparison tests was utilized. In all graphs, error bars represent the standard deviation. For all statistical tests, * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001.
3. Results and Discussion
3.1. Minimal Matrigel scaffolding allows for proximal tubule organoid formation
We previously described a biomaterials-driven, eversion-free method for the production of stably inverted mammary organoids using an immortalized epithelial cell line (MCF10A) [30]. Here we test the ability to adapt this method to commercially-available primary human renal proximal tubule epithelial cells (RPTECs). We screened different ranges of cell numbers, Matrigel concentrations, temperature conditions, and serum concentrations (Supplementary Figure 2) and found that new conditions were needed to adapt the method to RPTECs. While the cell numbers (3000 cells/drop), Matrigel concentrations (110 – 130 μg/mL), and 0.24% methylcellulose conditions were similar to our prior report [30], the FBS concentration was reduced from 10% to 2% in line with previous culture optimizations [30,41,43,44]. Finally, as with our previous report [30], the addition of cold Matrigel into thoroughly warmed cell suspension was critical (Supplementary Figure 2).
Upon the successful formation of RPTEC organoids and characterization of the minimal Matrigel scaffold, we maintained them in hanging drop culture for 90+ days, the longest hanging drop culture of any kind, including our previous reports [30]. Live/dead staining revealed that the organoids remained viable at days 65 and 90 of their culture (Supplementary Figure 3). We imaged and tracked organoids every 2–3 days, and representative images of organoids at day 8, 16, 30, and 90 are shown in Figure 2c. Histology and H&E staining was also performed at these timepoints (Figure 2d). Figures 2e and 2f show the morphological features of the particular organoid shown in Figure 2c. Figure 2g shows data from n=16 organoids, and demonstrates a decrease in area/diameter over time, an observation that can also be seen qualitatively in Figures 2c and 2d. Figure 2h shows that their roundness increases over time. These findings were consistent for multiple independent experiments (Supplementary Figure 4).
Figure 2. Optimization of geometrically-inverted proximal tubule organoid formation.
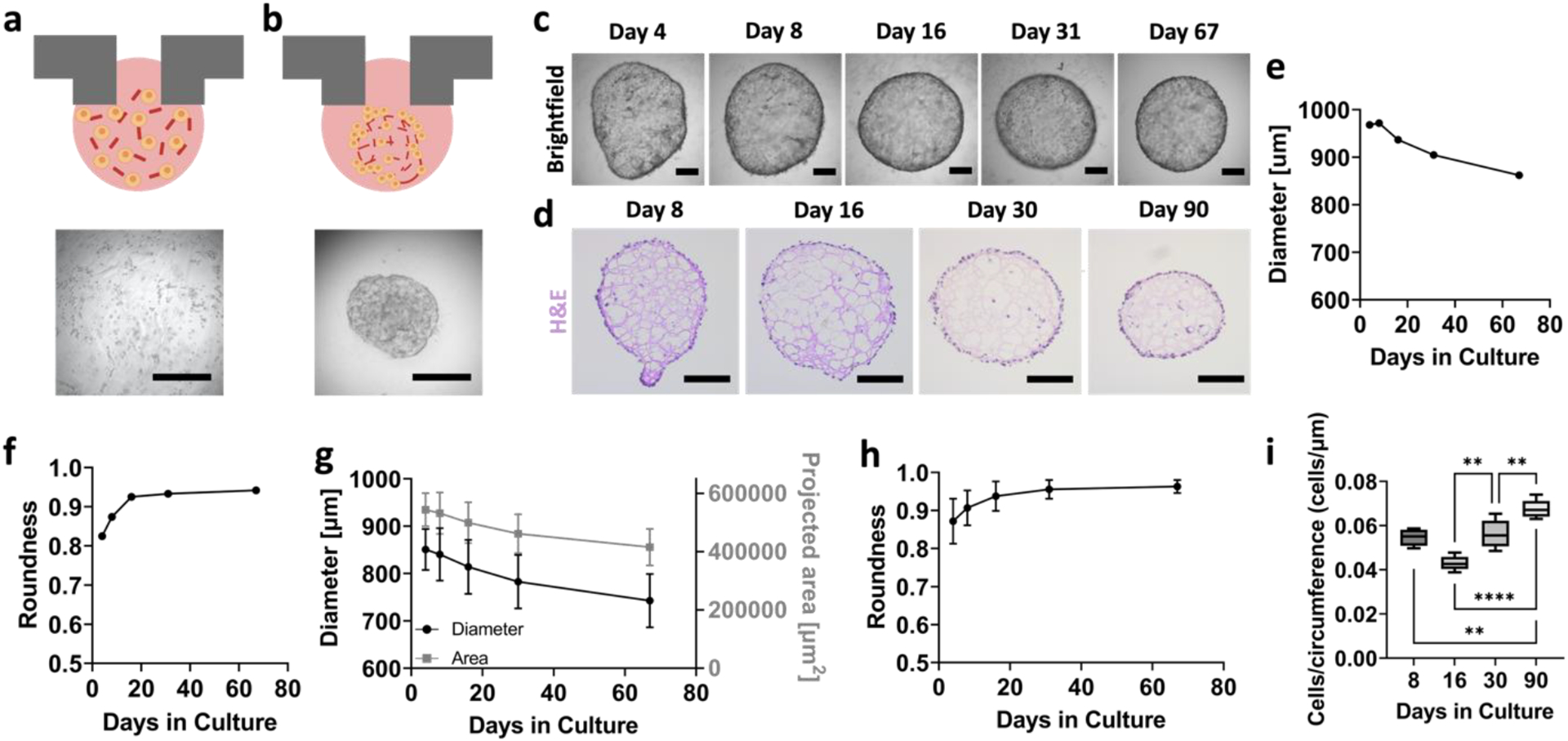
(a) Schematic and brightfield image of RPTECs at time of seeding, where cells are homogeneously mixed among partially gelled Matrigel. (b) Schematic and brightfield image of organoid formation within 24–48 hours. Scalebars represent 500 μm. (c) Time course brightfield of whole organoids, with the same organoid tracked over time. Scalebars represent 200 μm. (d) H&E images of representative organoid sections at days 8, 16, 30, and 90 of their growth. Scalebars represent 200 μm. (e) Diameter and (f) roundness values for the particular organoid shown in panel (c). (g) Area, diameter, and (h) roundness values for organoids for days 4–67 of their growth. (i) Quantification of cell coverage per circumference length of RPTEC organoids at days 8, 16, 30, and 90 of their growth. For panels (g) and (h), n = 16 per timepoint. For panel (i), n=5 per timepoint. One-way ANOVA was performed using Tukey’s multiple comparisons. ** denotes p ≤ 0.01 and **** denotes p ≤ 0.0001.
To quantify cell coverage of the organoid, we manually counted the number of cell nuclei from H&E images, and divided it by the organoid circumference. The number of cells per circumference length on the organoids increased between days 16, 30, and 90 of their culture. Although cell numbers were comparable between the conditions, the area decreased over time, especially by day 90 (Figure 2i). The organoids increase cell coverage, as is apparent also from the H&E images (Figure 2d). An additional description of extended cultures beyond 90 days is presented in Supplementary Figure 4.
From a materials perspective, the role of Matrigel in organoid formation and maintenance is important. Figure 2d shows a network of pink hues arising from Matrigel that is entrapped inside the organoid core. Interestingly, this pink hue fades over time, suggesting the degradation of Matrigel. Although the presence and maintenance of Matrigel in the core is the first indication of a basal-in phenotype, we aimed to further explore how this would affect the polarization of RPTEC organoids.
3.2. RPTEC tubuloids exhibit a basal-in phenotype
We utilized immunofluorescence staining of relevant apical and basolateral markers to assess tubuloid polarity. Staining at day 30 showed basolateral polarization of laminin-1,2 (Figure 3a) and integrin α6 (Figure 3b). Counterstaining with aquaporin-1 (AQP1), a transporter involved in water transport, showed physiologically consistent apical polarization. We further assessed changes over time. Staining of laminin-5 (red), a cell-secreted basement membrane marker, was localized to the interior side of the organoids at all timepoints. Counterstaining with DAPI revealed that the red is more localized to the interior of the organoid for days 16, 30, and 90 of culture (Figure 3c and Supplementary Figure 5). Generally, laminin-5 exhibited development, maturation, and thickening over time. Co-staining with phalloidin (apically located in proximal tubule cells) showed localization of the laminin-5 on the day 90 organoid interior and phalloidin to the organoid exterior (Figure 3c and Supplementary Figure 5). A similar trend can be observed when examining the co-staining of Na+K+ATPase, a basolateral marker, and ezrin/villin-2, an apical brush border marker, for days 16, 30, and 90, though not as pronounced as the laminin-5 staining (Figure 3d and Supplementary Figure 6). Day 90 co-staining of Na+K+ATPase, a basolateral marker, and ezrin/villin-2, an apical brush border marker showed localization of Na+K+ATPase to the organoid interior (basolateral side), and localization of ezrin/villin-2 to the organoid exterior (apical side) (Figure 3d). Finally, Figure 3e demonstrates the partial localization of tubulin to the apical side of the organoid (organoid exterior).
Figure 3. RPTEC organoids exhibit basal-in and apical-out phenotype.
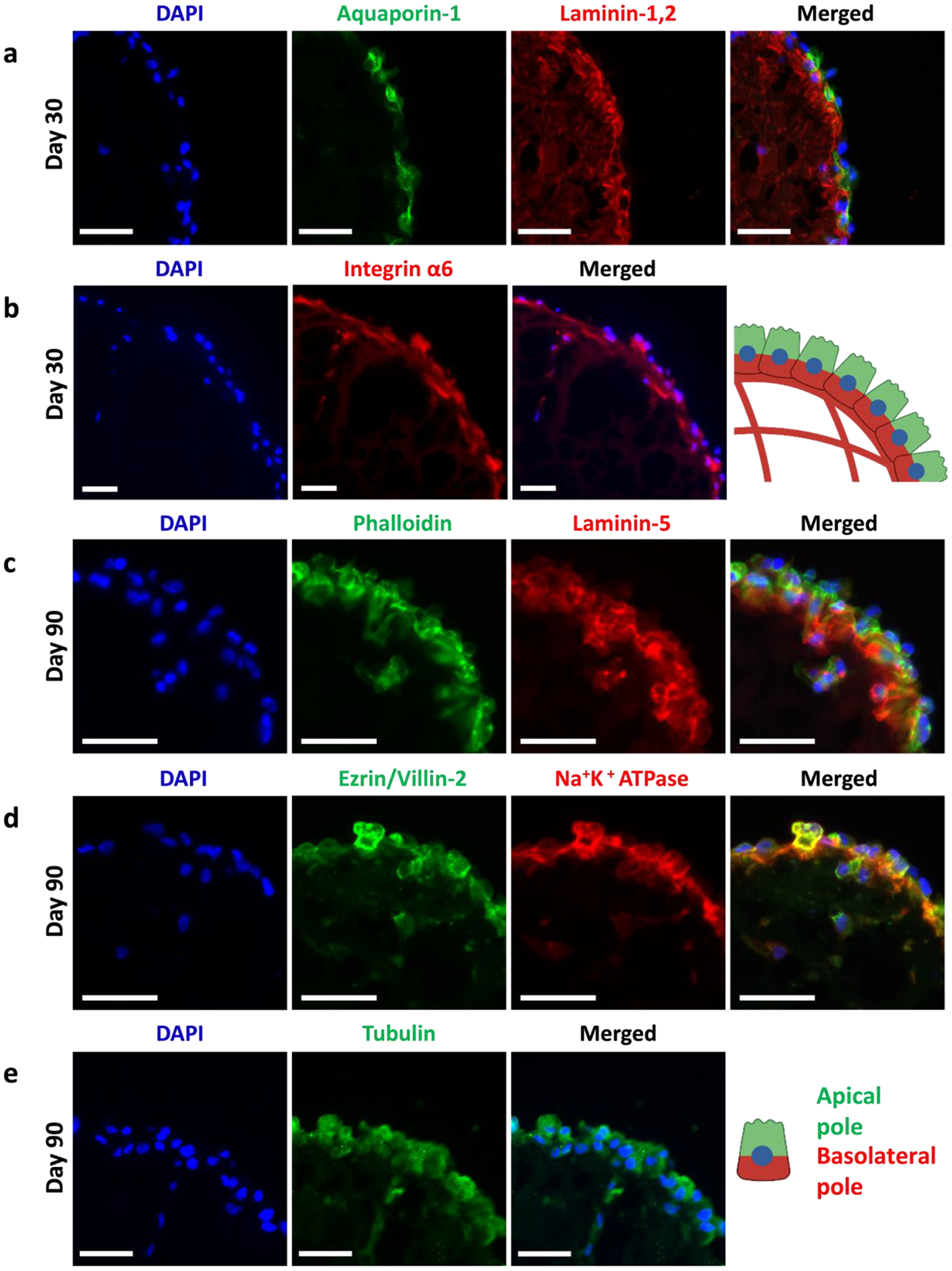
(a) Immunofluorescence staining images of: (a) DAPI (blue), aquaporin-1 (green), laminin-1,2 (red), and merged; (c) DAPI (blue), phalloidin (green), laminin-5 (red), and merged; (d) DAPI (blue), ezrin/villin-2 (green), and Na+K+ATPase (red), and merged; (e) DAPI (blue), tubulin (green), and merged. Schematic shows basal-in (red) and apical-out (green) phenotype. All scalebars represent 50 μm.
Overall, the process for formation of the inverted structures in RPTEC organoids, wherein the addition of cold Matrigel to warm media with serum promotes partial gelling and orientation of the basal pole towards the organoid interior, was consistent with our previous reports of forming geometrically-inverted organoids [29,30]. The presence of the Matrigel core was confirmed by laminin-1,2 immunostaining shown in Figure 3a. This interaction with Matrigel is the premise of the polarity reversal, and the basis for providing a soft scaffold to generate a 3D structure; rearrangement, cell movement, and rotation around the Matrigel ball could not be made possible in 2D culture. Such a mechanism highlights the importance of cell-material interactions for 3D culture to promote successful formation.
In our previous publication, we demonstrated the applicability of the basal-in organoid platform with immortalized cells (MCF10A) [30]. This is the first reported use of primary cells using our minimal Matrigel scaffolding method, and the longest reported hanging drop culture (90+ days) to date, suggesting broad applicability of the method. In comparison to immortalized MCF10A organoids, primary RPTEC organoids exhibited much more static growth patterns; in fact, RPTEC organoids decreased in the size over time, compared to the increase in size of MCF10A organoids. Despite some morphological differences, both geometrically-inverted organoids showed good basal-in polarization, and maturation of the basement membrane over time, as indicated by laminin-5 immunostaining.
While the organoids appeared to be polarized, as with MCF10A organoids, the basolateral polarization was stronger than apical polarization. Notably, the tubulin staining did not reveal strong apical polarization and demonstrated a lack of primary ciliation. As will be described in more detail later, however, the inverted tubuloids demonstrate functional protein uptake and injury marker release from the exterior-facing apical surface.
3.3. RPTEC organoid transcriptional profile
We performed bulk RNAseq for the RPTEC organoids at day 31 and 67 of culture. For each of the organoid timepoints, they were derived from distinct experimental batches; the organoids came from different hanging drop plates that were seeded at different times. RPTECs grown to confluency in 2D well-plates, and day 31 and 67 organoid timepoints were compared to each other. All samples were run in triplicate. A volcano plot comparing day 31 organoids versus 2D RPTEC cells (Figure 4a) showed 5177 genes upregulated and 4751 genes downregulated, and a volcano plot comparing day 67 and day 31 organoids (Figure 4b) showed 2683 genes upregulated and 2983 genes downregulated. A heatmap (Figure 4c) of representative gene expression markers suggested increased transporter expression, reduced EMT, increased maturity, and low proliferation in the day 31 tubuloids compared to 2D RPTECs. The lack of a stiff substrate may account for at least some of these differences [5,45]. The gene expression patterns were consistent with our tubuloids being sustainable for long-term cultures (90+ days), significantly different from 2D well-plate cultures. Furthermore, it is interesting to note that the organoids had higher E-cadherin and N-cadherin (Supplementary Figure 7); proximal tubule cells are one of the only epithelial cell types that express N-cadherin without undergoing EMT [46]. Finally, we assessed more specific markers of a mature transcriptional profile [12,47]. Compared to 2D well-plate culture, day 31 organoids had higher expression of NPAS2, CDH16, AQP1, LRP2, and PAX2 (Figure 4c and Supplementary Figure 7). Comparison of transcriptomics data between day 31 organoids to day 67 organoids showed further maturation and a decrease in EMT markers over extended periods (Figure 4c). Expression of cubilin (CUBN), a receptor that endocytoses proteins with assistance of LRP2 or amnionless (AMN) [48] was expressed but at similar levels in cells in 2D culture and day 31 and 67 tubuloids (Supplementary Figure 7).
Figure 4. RPTEC organoid transcriptomics.
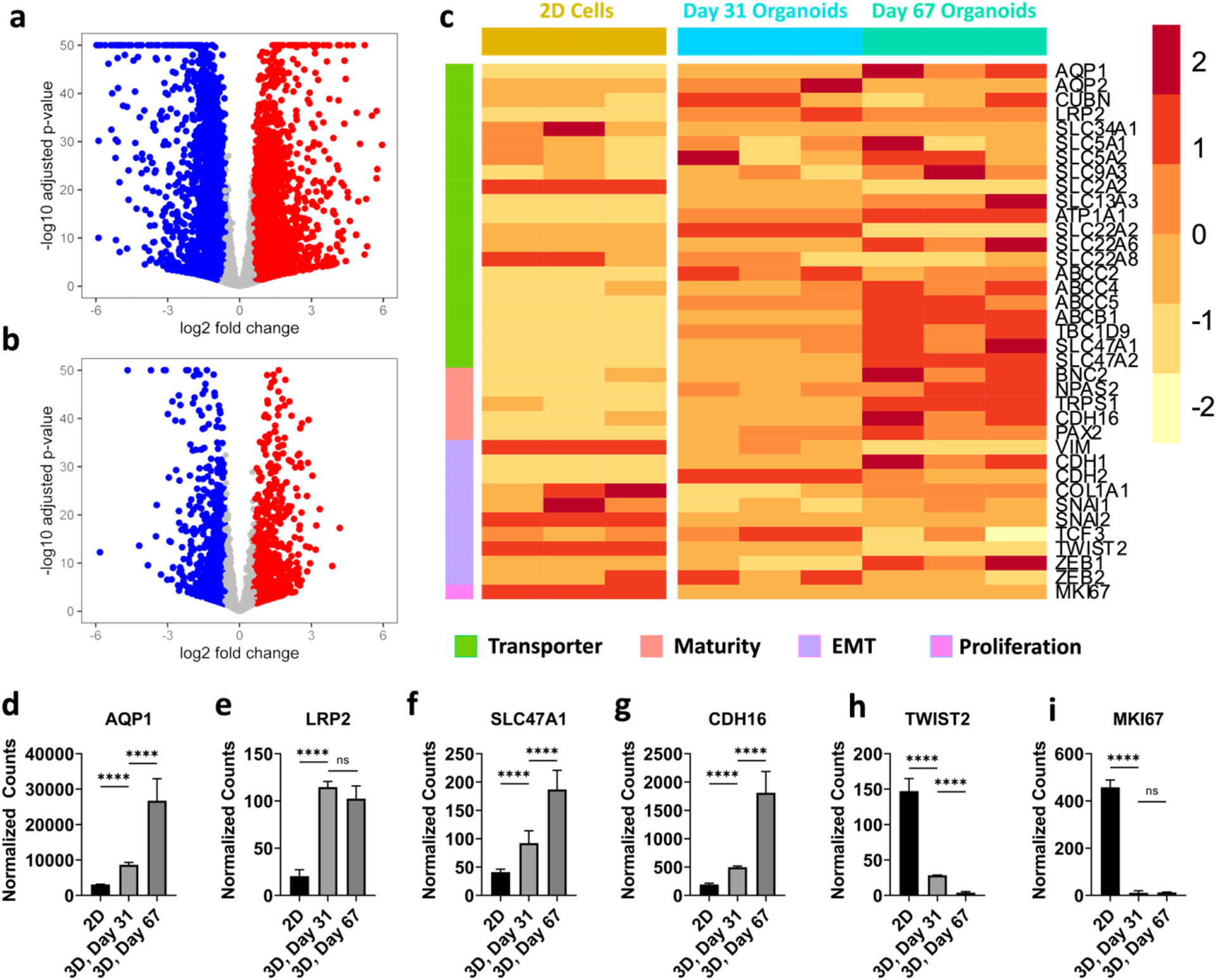
(a) Volcano plot from bulk RNAseq comparing day 31 organoids with 2D RPTECs, and (b) day 67 with day 31 organoids. (c) Heatmap from bulk RNAseq comparing day 67 organoids, day 31 organoids, and 2D RPTECs. The colors in the heatmaps represent Z-scores computed from normalized gene counts. Normalized counts of genes of day 67 organoids, day 31 organoids, and 2D RPTECs for (d) AQP1, (e) LRP2, (f) SLC47A1, (g) CDH16, (h) TWIST2, and (i) MKI67. n = 3 for both 2D and organoid samples. These genes were significantly different based on the LRT with the Benjamini-Hochberg correction. **** denotes p ≤ 0.0001.
We plotted individual genes found to be significantly different between conditions, based on the likelihood ratio test (LRT) with Benjamini-Hochberg multiple comparisons corrections (Figures 4d–4i). A more extensive list of significantly different genes, along with comparisons between various timepoints of organoids and 2D cultures, can be found in Supplementary Figure 7.
Taken together, these transcriptional results suggest that the organoids are in a mature and differentiated state, as they have downregulated proliferation compared to 2D cells, express increased maturity markers, and experience less EMT compared to 2D RPTECs. Using two different timepoints, we also see the advantages of growing the cells in 3D over extended periods. While these tubuloids are experiencing maturation as seen by upregulation of maturity markers over time, such as CDH16, it is possible that they are also selecting for a specific phenotype; it is likely a combination of both phenotype selection and maturation over time.
3.4. Megalin (LRP2) and aquaporin-1 (AQP1) increase expression over time
We also performed immunofluorescence staining initially focused on aquaporin-1 (AQP1) and megalin (LRP2), which are key markers of proximal tubule cell maturation [5,19,20]. At day 16 of organoid culture (Figure 5a), AQP1 and LRP2 staining are only present in certain cells. Not only does this indicate the lack of full maturity, but also that the organoids exhibit some extent of cellular heterogeneity. As they were grown to day 90 (Figure 5b), qualitative and quantitative (Figure 5c–5d) increase in both the percentage of AQP1 and LRP2 positive cells and signal strength is observed, although the change in the percentage of LRP2-positive cells between day 31 and 90 is not statistically significant. These observations match the transcriptomics data (Figures 4d–4e), supporting the notion that the tubuloids mature over the extended culture period. We note that megalin expression is reported to decrease on stiff substrates [49,50]. While it is challenging to unravel the exact complexities, our results and previous findings in the literature suggest that substrate stiffness, cell-matrix interactions, and the 3D nature of our cultures may contribute to enhanced megalin expression.
Figure 5. Organoids show maturity over 90 day growth period.
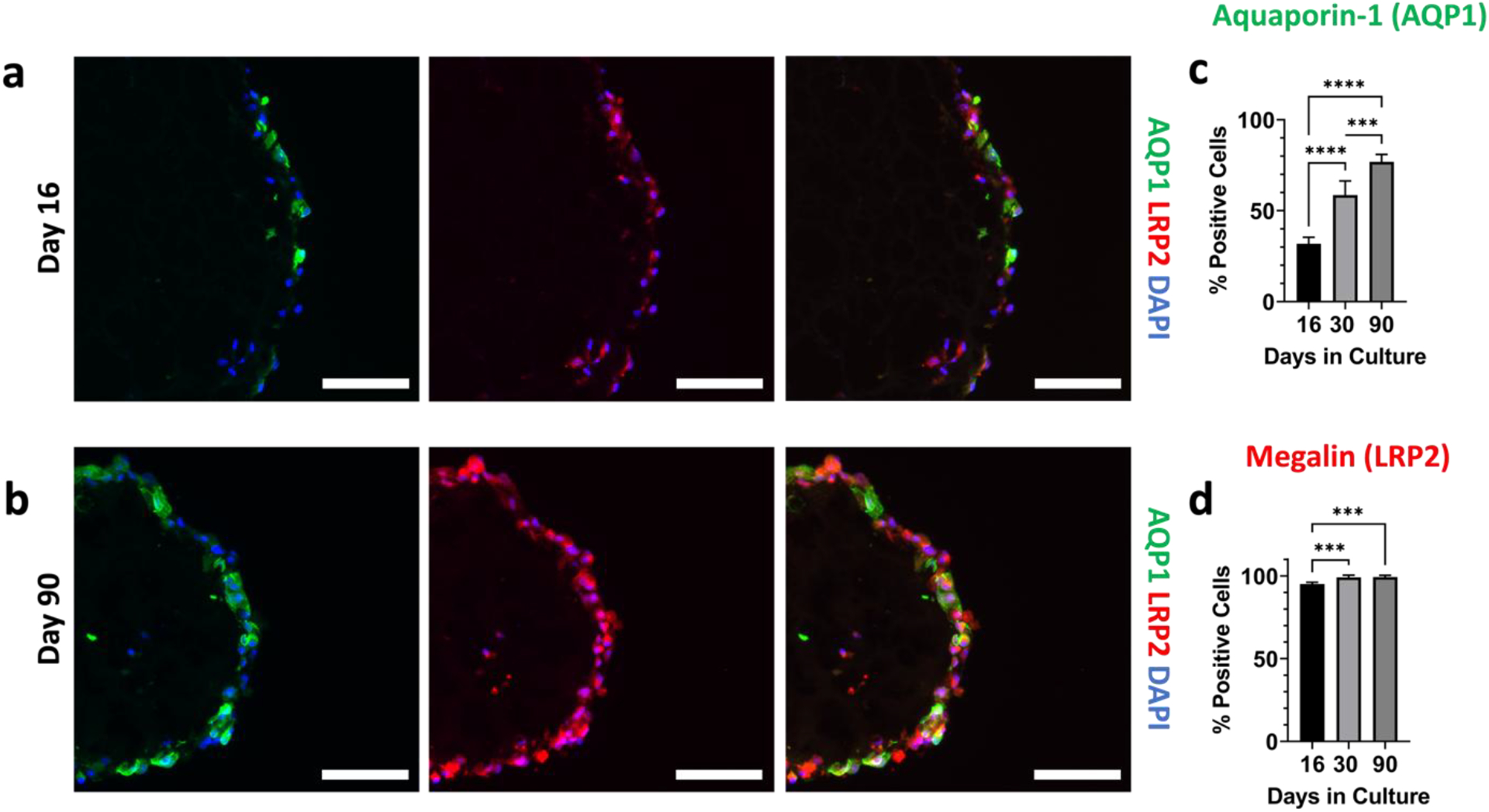
Immunofluorescence images of aquaporin-1 (green), megalin (red), DAPI (blue), and the channels merged at (a) day 16, and (b) day 90 of their culture. All scalebars represent 100 μm. (c) Quantification of the percentage of cells that were positive for aquaporin-1. (d) Quantification of the percentage of cells that were positive for megalin. For panels (c) and (d), n=5 per timepoint. One-way ANOVA was performed using Tukey’s multiple comparisons. *** denotes p ≤ 0.001 and **** denotes p ≤ 0.0001.
3.5. Tubuloids exhibit time- and dose-dependent responses to proteinuric conditions
Because our proximal tubule organoid model: 1) has 3D architecture, 2) can be grown long term, 3) exhibits reversed polarization, 4) possesses consistently high megalin expression, and 5) increases megalin expression over time, we were motivated to use it for proteinuria studies. We were interested in such an application because proteinuria studies require apical access, and surface expression of megalin is required to maintain apical endocytic pathway integrity; it is also essential for protein uptake in both normal and nephrotic conditions (Figure 1) [51,52].
First, we incubated RPTEC organoids with FITC-BSA for 2 hours at 50 μg/mL, washed out excess FITC-BSA, and imaged the organoids using an in-incubator microscope (Incucyte S3, Sartorius). We observed FITC-BSA uptake for RPTEC organoids at day 16, 30, and 90 (Figure 6a and Supplementary Figure 8). Punctate fluorescence patterns, as also observed in the literature [53], suggest successful uptake of the protein rather than simple diffusion of protein into the tubuloid interior.
Figure 6. RPTEC organoids uptake albumin, and show dose- and time-dependent KIM-1 production to HSA stimulus.
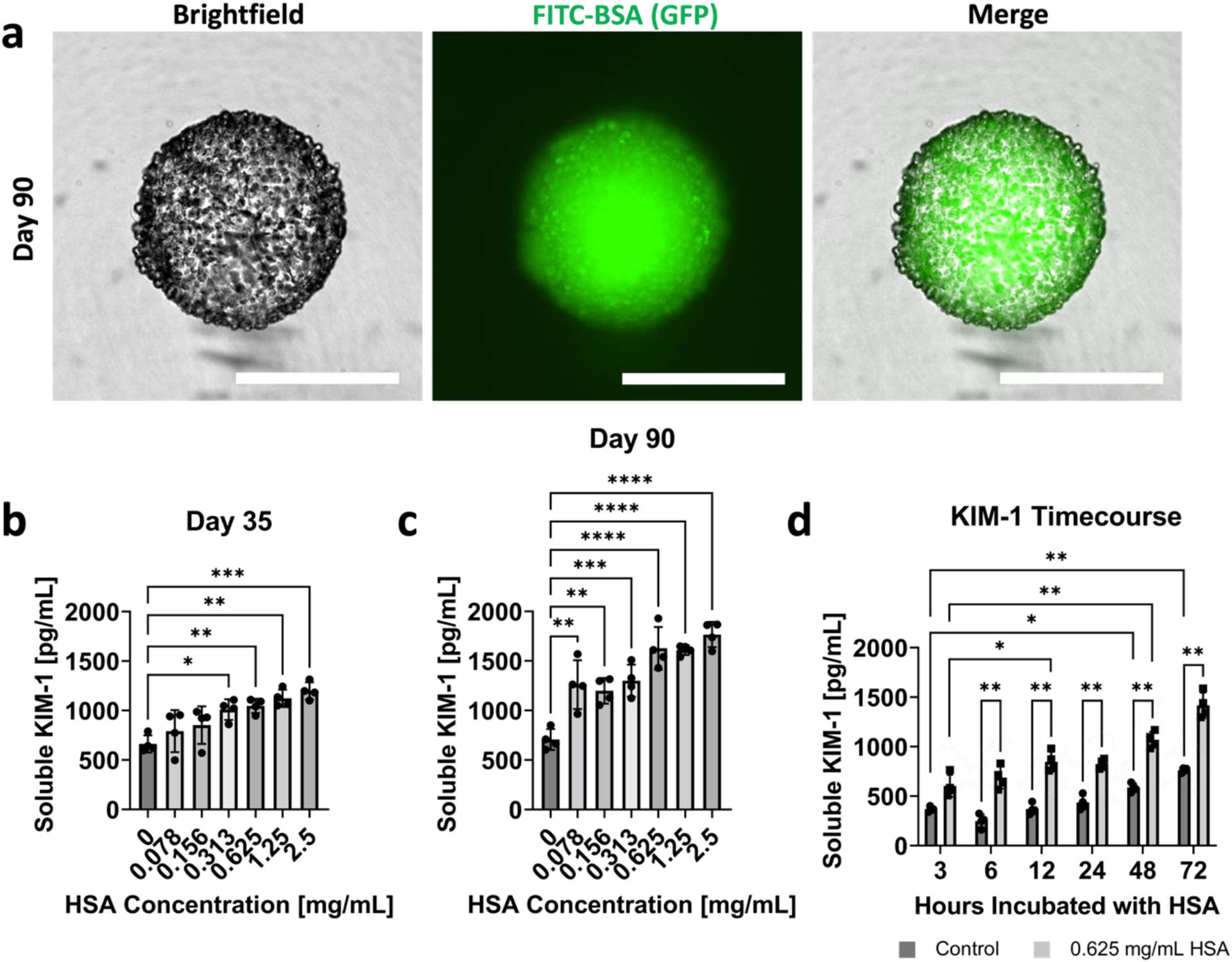
(a) Brightfield, GFP, and merged images of FITC-BSA uptake in day 90 RPTEC organoids. All scalebars represent 500 μm. Dose dependent KIM-1 production for organoids exposed to HSA at (b) day 35 and (c) day 90 of their culture. (d) Time-dependent KIM-1 production for control organoids and organoids exposed to 0.625 mg/mL HSA. For all plots, n = 4 was used for each dose or time point. One-way ANOVA with Tukey’s multiple comparisons was used for panels (b) and (c). Two-way ANOVA with Šīdák’s multiple comparison tests was utilized for panel (d). For all statistical tests, * indicates p ≤ 0.05, ** indicates p ≤ 0.01, *** indicates p ≤ 0.001, and **** indicates p ≤ 0.0001.
We next assessed whether high protein levels can cause injury in the proximal tubule cells. We chose kidney injury molecule 1 (KIM-1) as a readout, as it is an early non-invasive biomarker of proximal tubule injury and is apically released into the culture media or urine. KIM-1 is a specific proximal tubule injury marker that has been widely established for nephrotoxicity and other kidney injury studies, both in vitro and in vivo [4,54–56]. Conveniently, a portion of KIM-1 is secreted in the supernatant, making it easily detectable through an enzyme-linked immunoassay (ELISA). Consistent with the literature [57], we chose concentrations of HSA ranging from 0.078 mg/mL to 2.5 mg/mL in factors of 2, plus a control with no HSA present. First, we validated the ELISA kit with many acellular supernatant controls to ensure that no media components or growth factors caused a false signal KIM-1 signal (data not shown). Organoids were then incubated with HSA in ULA plates for 48 hours, and the supernatant was collected at the timepoint. Our results show that the KIM-1 was produced in a dose-dependent manner for organoids exposed to HSA at day 35 (Figure 6b) and day 90 (Figure 6c) of culture. The KIM-1 data suggest that there is a more substantial dose-dependent response with more mature organoids (day 90).
Since 0.625 mg/mL of HSA consistently showed a high response, we chose this condition to determine the time dependence of KIM-1 production in day 35 organoids. Supernatant was collected from both control samples and 0.625 mg/mL HSA samples at 3, 6, 12, 24, 48, and 72 hour timepoints. The results indicated a time-dependent increase in KIM-1 cleavage for both the control and proteinuric conditions (Figure 6d). For a given condition, although each timepoint is not significantly different from one another, there is an overall significance of the increase in KIM-1 production. Furthermore, at each of the timepoints, the KIM-1 production was significantly higher for the proteinuric condition compared to the control. Because of the dose- and time-dependent responses from HSA exposure seen in KIM-1 production, this geometrically-inverted proximal tubule organoid shows promise for future proteinuria studies. It also opens the possibility of studying nephrotoxicity of drugs such as gentamycin and cisplatin, both of which have been implicated in apical uptake via megalin [58,59].
4. Conclusions
This study establishes the first geometrically-inverted (basal-in, apical-out) proximal tubule organoid model, to our knowledge, using normal human primary RPTECs. These results contribute to the emerging sub-field, within the field of organoids, of engineering and using inverted organoids with reversed epithelial polarity. Our results expand the range of cell types amenable to such inverted organoid engineering as well as demonstrate the broader utility of our minimal Matrigel scaffolding technique. Some of the differentiating features of this method of inverted organoid production include being eversion-free, allowing the use of normal human cells now including primary RPTECs, and enabling stably inverted structures to be maintained for over 90+ days rather than obtaining temporarily- or partially-inverted structures, as is often the case in other methods. The method does have room for improvement, particularly for a better mechanistic understanding of epithelial polarization reversal and further enhancement of apical polarization. From a technological perspective, this extends the demonstrated limit of culture duration of continuous hanging drop culture by two months. Focusing on proteinuria as a proof-of-concept application enabled by geometrically-inverted organoids, we analyzed protein exposure to the exterior-facing apical tubuloid surface and subsequent cell injury response as monitored via an apically released injury marker. Although this paper focuses on the development of geometrically-inverted proximal tubule organoids with a proof-of-concept assay for albumin-uptake and injury, the ease of apical access together with the generally high levels of transporter and endocytic receptor expression suggests our model’s broader utility. Potential applications include certain types of drug-induced nephrotoxicity, where apical exposure and endocytic receptor uptake are involved.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health grant number SC1DK112151 and the Price Gilbert Jr. Chair Fund. This study was supported in part by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. We thank Professor Greg Gibson and the Georgia Tech Center for Integrative Genomics for advice on bioinformatics analysis. This research was supported in part through research cyberinfrastructure resources and services provided by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology, Atlanta, Georgia, USA. This material is also based upon work supported by the National Science Foundation Graduate Research Fellowship Program to EP (Grant Number: DGE-1650044). All schematics in this paper were generated using Biorender.
Footnotes
Credit author statement
EP, JHL, and AYL: Conceptualization, investigation, methodology, writing, formal analysis, and visualization. XZ and ST: Conceptualization, methodology, investigation, writing, and supervision.
Declaration of competing interests
The authors declare no competing interests.
Data availability
All data is available from the corresponding author upon reasonable request. Our RNAseq data have been deposited on Gene Expression Omnibus (GEO) of NCBI under accession number GSE198857.
References
- [1].Soo JYC, Jansen J, Masereeuw R, Little MH, Advances in predictive in vitro models of drug-induced nephrotoxicity, Nat. Rev. Nephrol 14 (2018) 378–393. 10.1038/s41581-018-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Knight A, Systematic Reviews of Animal Experiments Demonstrate Poor Contributions Toward Human Healthcare, Rev. Recent Clin. Trials 3 (2008) 89–96. 10.2174/157488708784223844. [DOI] [PubMed] [Google Scholar]
- [3].Nieskens TTG, Wilmer MJ, Kidney-on-a-chip technology for renal proximal tubule tissue reconstruction, Eur. J. Pharmacol 790 (2016) 46–56. 10.1016/j.ejphar.2016.07.018. [DOI] [PubMed] [Google Scholar]
- [4].Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R, Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening, Trends Biotechnol. 34 (2016) 156–170. 10.1016/j.tibtech.2015.11.001. [DOI] [PubMed] [Google Scholar]
- [5].King SM, Higgins JW, Nino CR, Smith TR, Paffenroth EH, Fairbairn CE, Docuyanan A, Shah VD, Chen AE, Presnell SC, Nguyen DG, 3D proximal tubule tissues recapitulate key aspects of renal physiology to enable nephrotoxicity testing, Front. Physiol 8 (2017) 1–18. 10.3389/fphys.2017.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kim S, LesherPerez SC, choul B Kim C, Yamanishi C, Labuz JM, Leung B, Takayama S, Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip, Biofabrication. 8 (2016) 15021. 10.1088/1758-5090/8/1/015021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Taguchi A, Nishinakamura R, Higher-Order Kidney Organogenesis from Pluripotent Stem Cells, Cell Stem Cell. 21 (2017) 730–746.e6. 10.1016/j.stem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- [8].Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, Nishinakamura R, Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells, Cell Stem Cell. 14 (2014) 53–67. 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- [9].Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Vander Werff R, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV, Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids, Nat. Commun 6 (2015). 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV, Nephron organoids derived from human pluripotent stem cells model kidney development and injury, Nat. Biotechnol 33 (2015) 1193–1200. 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, De Sousa Lopes SMC, Little MH, Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis, Nature. 526 (2015) 564–568. 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- [12].Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Lewis JA, Morizane R, Flow-enhanced vascularization and maturation of kidney organoids in vitro, Nat. Methods (2019). 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Miyoshi T, Hiratsuka K, Saiz EG, Morizane R, Kidney organoids in translational medicine: Disease modeling and regenerative medicine, Dev. Dyn 249 (2020) 34–45. 10.1002/dvdy.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tian P, Lennon R, The myriad possibility of kidney organoids, Curr. Opin. Nephrol. Hypertens 28 (2019) 211–218. 10.1097/MNH.0000000000000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Geuens T, van Blitterswijk CA, LaPointe VLS, Overcoming kidney organoid challenges for regenerative medicine, Npj Regen. Med 5 (2020). 10.1038/s41536-020-0093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nishinakamura R, Human kidney organoids: progress and remaining challenges, Nat. Rev. Nephrol 15 (2019) 613–624. 10.1038/s41581-019-0176-x. [DOI] [PubMed] [Google Scholar]
- [17].Yousef Yengej FA, Jansen J, Rookmaaker MB, Verhaar MC, Clevers H, Kidney Organoids and Tubuloids, Cells. 9 (2020) 1–20. 10.3390/cells9061326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rookmaaker MB, Schutgens F, Verhaar MC, Clevers H, Development and application of human adult stem or progenitor cell organoids, Nat. Rev. Nephrol 11 (2015) 546–554. 10.1038/nrneph.2015.118. [DOI] [PubMed] [Google Scholar]
- [19].Astashkina AI, Mann BK, Prestwich GD, Grainger DW, A 3-D organoid kidney culture model engineered for high-throughput nephrotoxicity assays, Biomaterials. 33 (2012) 4700–4711. 10.1016/j.biomaterials.2012.02.063. [DOI] [PubMed] [Google Scholar]
- [20].DesRochers TM, Suter L, Roth A, Kaplan DL, Bioengineered 3D Human Kidney Tissue, a Platform for the Determination of Nephrotoxicity, PLoS One. 8 (2013). 10.1371/journal.pone.0059219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X, Guo C, Chen Y, Tozzi L, Szymkowiak S, Li C, Kaplan DL, Developing a self-organized tubulogenesis model of human renal proximal tubular epithelial cells in vitro, J. Biomed. Mater. Res. - Part A 108 (2020) 795–804. 10.1002/jbm.a.36858. [DOI] [PubMed] [Google Scholar]
- [22].Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, Lewis JA, Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips, Sci. Rep 6 (2016) 1–13. 10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Weber EJ, Lidberg KA, Wang L, Bammler TK, MacDonald JW, Li MJ, Redhair M, Atkins WM, Tran C, Hines KM, Herron J, Xu L, Monteiro MB, Ramm S, Vaidya V, Vaara M, Vaara T, Himmelfarb J, Kelly EJ, Human kidney on a chip assessment of polymyxin antibiotic nephrotoxicity, JCI Insight. 3 (2018) 0–17. 10.1172/jci.insight.123673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nieskens TTG, Persson M, Kelly EJ, Sjögren AK, A multicompartment human kidney proximal tubule-on-a-chip replicates cell polarization–dependent cisplatin toxicity, Drug Metab. Dispos 48 (2020) 1303–1311. 10.1124/DMD.120.000098. [DOI] [PubMed] [Google Scholar]
- [25].Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M, Yengej FY, Derakhshan S, de Winter-de Groot KM, Artegiani B, van Boxtel R, Cuppen E, Hendrickx APA, van den Heuvel-Eibrink MM, Heitzer E, Lanz H, Beekman J, Murk JL, Masereeuw R, Holstege F, Drost J, Verhaar MC, Clevers H, Tubuloids derived from human adult kidney and urine for personalized disease modeling, Nat. Biotechnol (2019). 10.1038/s41587-019-0048-8. [DOI] [PubMed] [Google Scholar]
- [26].Vormann MK, Gijzen L, Hutter S, Boot L, Nicolas A, van den Heuvel A, Vriend J, Ng CP, Nieskens TTG, van Duinen V, de Wagenaar B, Masereeuw R, Suter-Dick L, Trietsch SJ, Wilmer M, Joore J, Vulto P, Lanz HL, Nephrotoxicity and Kidney Transport Assessment on 3D Perfused Proximal Tubules, AAPS J. 20 (2018) 1–11. 10.1208/s12248-018-0248-z. [DOI] [PubMed] [Google Scholar]
- [27].Vormann MK, Vriend J, Lanz HL, Gijzen L, van den Heuvel A, Hutter S, Joore J, Trietsch SJ, Stuut C, Nieskens TTG, Peters JGP, Ramp D, Caj M, Russel FGM, Jacobsen B, Roth A, Lu S, Polli JW, Naidoo AA, Vulto P, Masereeuw R, Wilmer MJ, Suter-Dick L, Implementation of a Human Renal Proximal Tubule on a Chip for Nephrotoxicity and Drug Interaction Studies, J. Pharm. Sci 110 (2021) 1601–1614. 10.1016/j.xphs.2021.01.028. [DOI] [PubMed] [Google Scholar]
- [28].Vriend J, Peters JGP, Nieskens TTG, Škovroňová R, Blaimschein N, Schmidts M, Roepman R, Schirris TJJ, Russel FGM, Masereeuw R, Wilmer MJ, Flow stimulates drug transport in a human kidney proximal tubule-on-a-chip independent of primary cilia, Biochim. Biophys. Acta - Gen. Subj 1864 (2020). 10.1016/j.bbagen.2019.129433. [DOI] [PubMed] [Google Scholar]
- [29].Lee S, Chang J, Kang SM, Parigoris E, Lee JH, High - throughput formation and image - based analysis of basal - in mammary organoids in 384 - well plates, Sci. Rep (2022) 1–9. 10.1038/s41598-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Parigoris E, Lee S, Mertz D, Turner M, Liu AY, Sentosa J, Djomehri S, Chang HC, Luker K, Luker G, Kleer CG, Takayama S, Cancer Cell Invasion of Mammary Organoids with Basal-In Phenotype, Adv. Healthc. Mater 10 (2021) e2000810. 10.1002/adhm.202000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Co JY, Margalef-Català M, Li X, Mah AT, Kuo CJ, Monack DM, Amieva MR, Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions, Cell Rep. 26 (2019) 2509–2520.e4. 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM, Flynn RA, Margalef-Català M, Santos AJM, Ju J, Batish A, Usui T, Zheng GXY, Edwards CE, Wagar LE, Luca V, Anchang B, Nagendran M, Nguyen K, Hart DJ, Terry JM, Belgrader P, Ziraldo SB, Mikkelsen TS, Harbury PB, Glenn JS, Garcia KC, Davis MM, Baric RS, Sabatti C, Amieva MR, Blish CA, Desai TJ, Kuo CJ, Progenitor identification and SARS-CoV-2 infection in human distal lung organoids, Nature. 588 (2020) 670–675. 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Subramanian B, Rudym D, Cannizzaro C, Perrone R, Zhou J, Kaplan DL, Tissue-engineered three-dimensional in vitro models for normal and diseased kidney, Tissue Eng. - Part A 16 (2010) 2821–2831. 10.1089/ten.tea.2009.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Akhtar N, Streuli CH, An integrin-ILK-microtubule network orients cell polarity and lumen formation in glandular epithelium, Nat. Cell Biol 15 (2013) 17–27. 10.1038/ncb2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yu W, Datta A, Leroy P, O’Brien LE, Mak G, Jou TS, Matlin KS, Mostov KE, Zegers MMP, β1-integrin orients epithelial polarity via Rac1 and laminin, Mol. Biol. Cell 16 (2005) 433–445. 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].O’Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, Mostov KE, Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly, Nat. Cell Biol 3 (2001) 831–838. 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- [37].Shen Y, Hou Y, Yao S, Huang P, Yobas L, In Vitro Epithelial Organoid Generation Induced by Substrate Nanotopography, Sci. Rep 5 (2015) 9293. 10.1038/srep09293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yonemura S, Differential sensitivity of epithelial cells to extracellular matrix in polarity establishment, PLoS One. 9 (2014) 1–10. 10.1371/journal.pone.0112922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang AZ, Ojakian GK, Nelson WJ, Steps in the morphogenesis of a polarized epithelium: II. Disassembly and assembly of plasma membrane domains during reversal of epithelial cell polarity in multicellular epithelial (MDCK) cysts, J. Cell Sci 95 (1990) 153–165. 10.1242/jcs.95.1.153. [DOI] [PubMed] [Google Scholar]
- [40].Wosen JE, Ilstad-Minnihan A, Co JY, Jiang W, Mukhopadhyay D, Fernandez-Becker NQ, Kuo CJ, Amieva MR, Mellins ED, Human intestinal enteroids model MHC-II in the gut epithelium, Front. Immunol 10 (2019) 1–8. 10.3389/fimmu.2019.01970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Djomehri SI, Burman B, Gonzalez ME, Takayama S, Kleer CG, A reproducible scaffold-free 3D organoid model to study neoplastic progression in breast cancer, J. Cell Commun. Signal 13 (2019) 129–143. 10.1007/s12079-018-0498-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods 9 (2012) 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tung Y-CC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S, High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array, Analyst. 136 (2011) 473–478. 10.1039/C0AN00609B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Leung BM, Lesher-Perez SC, Matsuoka T, Moraes C, Takayama S, Media additives to promote spheroid circularity and compactness in hanging drop platform, Biomater. Sci 3 (2015) 336–344. 10.1039/c4bm00319e. [DOI] [PubMed] [Google Scholar]
- [45].Ruiter FAA, Morgan FLC, Roumans N, Schumacher A, Slaats GG, Moroni L, LaPointe VLS, Baker MB, Soft, Dynamic Hydrogel Confinement Improves Kidney Organoid Lumen Morphology and Reduces Epithelial–Mesenchymal Transition in Culture, Adv. Sci (2022). 10.1002/advs.202200543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Keller C, Kroening S, Zuehlke J, Kunath F, Krueger B, Goppelt-Struebe M, Distinct mesenchymal alterations in N-cadherin and E-cadherin positive primary renal epithelial cells, PLoS One. 7 (2012). 10.1371/journal.pone.0043584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hiratsuka K, Monkawa T, Akiyama T, Nakatake Y, Oda M, Goparaju SK, Kimura H, Chikazawa-Nohtomi N, Sato S, Ishiguro K, Yamaguchi S, Suzuki S, Morizane R, Ko SBH, Itoh H, Ko MSH, Induction of human pluripotent stem cells into kidney tissues by synthetic mRNAs encoding transcription factors, Sci. Rep 9 (2019) 1–13. 10.1038/s41598-018-37485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nielsen R, Christensen EI, Birn H, Megalin and cubilin in proximal tubule protein reabsorption: From experimental models to human disease, Kidney Int. 89 (2016) 58–67. 10.1016/j.kint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- [49].Bosch-Fortea M, Rodriguez-Fraticelli AE, Herranz G, Hachimi M, Barea MD, Young J, Ladoux B, Martin-Belmonte F, Micropattern-based platform as a physiologically relevant model to study epithelial morphogenesis and nephrotoxicity, Biomaterials. 218 (2019) 119339. 10.1016/j.biomaterials.2019.119339. [DOI] [PubMed] [Google Scholar]
- [50].Love HD, Ao M, Jorgensen S, Swearingen L, Ferrell N, Evans R, Gewin L, Harris RC, Zent R, Roy S, Fissell WH, Substrate Elasticity Governs Differentiation of Renal Tubule Cells in Prolonged Culture, Tissue Eng. - Part A 25 (2019) 1013–1022. 10.1089/ten.tea.2018.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Weisz OA, Endocytic adaptation to functional demand by the kidney proximal tubule, 14 (2021) 3437–3446. 10.1113/JP281599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Silva-aguiar RP, Peruchetti DB, Florentino LS, Takiya CM, Marzolo M, Dias WB, Pinheiro AAS, Caruso-neves C, Albumin Expands Albumin Reabsorption Capacity in Proximal Tubule Epithelial Cells through a Positive Feedback Loop between AKT and Megalin, (2022). [DOI] [PMC free article] [PubMed]
- [53].Ren Q, Weyer K, Rbaibi Y, Long KR, Tan RJ, Nielsen R, Christensen EI, Baty CJ, Kashlan OB, Weisz OA, Distinct functions of megalin and cubilin receptors in recovery of normal and nephrotic levels of filtered albumin, Am. J. Physiol. - Ren. Physiol 318 (2020) F1284–F1294. 10.1152/ajprenal.00030.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhao X, Jiang C, Olufade R, Liu D, Emmett N, Kidney Injury Molecule-1 Enhances Endocytosis of Albumin in Renal Proximal Tubular Cells, J. Cell. Physiol 231 (2016) 896–907. 10.1002/jcp.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zhao X, Chen X, Zhang Y, George J, Cobbs A, Wang G, Li L, Emmett N, Kidney injury molecule-1 is upregulated in renal lipotoxicity and mediates palmitate-induced tubular cell injury and inflammatory response, Int. J. Mol. Sci 20 (2019). 10.3390/ijms20143406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV, Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury, Kidney Int. 62 (2002) 237–244. 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- [57].Zhao X, Chen X, Chima A, Zhang Y, George J, Cobbs A, Emmett N, Albumin induces CD44 expression in glomerular parietal epithelial cells by activating extracellular signal-regulated kinase 1/2 pathway, J. Cell. Physiol 234 (2019) 7224–7235. 10.1002/jcp.27477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hori Y, Aoki N, Kuwahara S, Hosojima M, Kaseda R, Goto S, Iida T, De S, Kabasawa H, Kaneko R, Aoki H, Tanabe Y, Kagamu H, Narita I, Kikuchi T, Saito A, Megalin blockade with cilastatin suppresses drug-induced nephrotoxicity, J. Am. Soc. Nephrol 28 (2017) 1783–1791. 10.1681/ASN.2016060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nagai J, Tanaka H, Nakanishi N, Murakami T, Takano M, Role of megalin in renal handling of aminoglycosides, Am. J. Physiol. - Ren. Physiol 281 (2001) 1–2. 10.1152/ajprenal.2001.281.2.f337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available from the corresponding author upon reasonable request. Our RNAseq data have been deposited on Gene Expression Omnibus (GEO) of NCBI under accession number GSE198857.


