Abstract
SacY antiterminates transcription of the sacB gene in Bacillus subtilis in response to the presence of sucrose in the growth medium. We have found that it can substitute for BglG, a homologous protein, in antiterminating transcription of the bgl operon in Escherichia coli. We therefore sought to determine whether, similarly to BglG, SacY is regulated by reversible phosphorylation in response to the availability of the inducing sugar. We show here that two forms of SacY, phosphorylated and nonphosphorylated, exist in B. subtilis cells and that the ratio between them depends on the external level of sucrose. Addition of sucrose to the growth medium after SacY phosphorylation in the cell resulted in its rapid dephosphorylation. The extent of SacY phosphorylation was found to be proportional to the cellular levels of SacX, a putative sucrose permease which was previously shown to have a negative effect on SacY activity. Thus, the mechanism by which the sac sensory system modulates sacB expression in response to sucrose involves reversible phosphorylation of the regulator SacY, and this process appears to depend on the SacX sucrose sensor. The sac system is therefore a member of the novel family of sensory systems represented by bgl.
Sucrose induces the expression of two loci in Bacillus subtilis: the sacB gene, encoding levansucrase, and the sacPA operon, encoding a sucrose permease and a phosphosucrase (32, 48). Expression of these loci is controlled by transcriptional antitermination. Thus, in the absence of sucrose, transcription initiates constitutively and terminates at ρ-independent transcriptional terminators located between the respective promoters and the first structural genes; in the presence of sucrose, transcription proceeds through the putative terminators. The transcriptional antiterminators SacY and SacT are required for the sucrose-dependent readthrough of sacB and sacPA, respectively (7, 17, 18). Expression of sacB is negatively regulated by SacX, a sugar phosphotransferase-like protein (17). The sacX and sacY genes are contiguous and probably constitute an operon (55).
The B. subtilis antiterminators SacY and SacT highly resemble the BglG protein from Escherichia coli, both in sequence and in the mechanism of action (18, 47, 55). BglG prevents transcription termination at two terminators within the bgl operon by binding to the RNA chain and preventing the formation of the terminator structure (27). The RNA sequence recognized by BglG is highly conserved, and similar motifs which are found in the leader of both sacB and sacPA were suggested to be recognized by SacY and SacT, respectively (10). The activity of BglG is regulated by reversible phosphorylation (2, 3, 44) which, in turn, modulates the protein activity by controlling its dimeric state (4). Thus, BglG exists in the cell in two forms: an inactive, monomeric phosphorylated form and an active, dimeric nonphosphorylated form. Phosphorylation of BglG was recently shown to occur on a histidine residue (6) and was localized to His 208 (15). The state of BglG phosphorylation depends on the availability of β-glucosides; their addition to the growth medium leads to BglG dephosphorylation (3). The protein which functions as BglG kinase and phosphatase is BglF, an enzyme II of the phosphoenolpyruvate-dependent phosphotransferase system (PTS), which is in charge of the transport and phosphorylation of β-glucosides (2, 3, 44). The way β-glucosides lead to bgl operon induction is by stimulating BglF to dephosphorylate BglG, thus allowing it to function as an antiterminator. In the absence of sugar, BglF inactivates BglG by phosphorylating it. BglG and BglF represent a novel family of systems which utilize a sensor and a regulator to transduce a signal from the cell surface to the transcription machinery (5).
Based on indirect evidence, regulation of sacB expression in B. subtilis seems to resemble bgl regulation in E. coli. Controlled readthrough of both systems is induced by sugars, sucrose and β-glucosides, respectively, and is positively regulated by homologous transcriptional antiterminators SacY and BglG, respectively. Moreover, while bgl expression is negatively regulated by BglF, the β-glucoside PTS permease, sacB, is negatively regulated by SacX, a sucrose PTS permease homolog (9). Based on genetic studies, Crutz et al. (17) concluded that SacX suppresses sacB transcription in the absence of sucrose by inhibiting the antitermination activity of SacY. The general PTS proteins were also shown to negatively control sacB induction, most likely by inhibiting the function of SacY (17, 47). Based on these observations and on the striking similarity to the bgl system, a model was proposed which suggests that SacX is a sucrose sensor which is phosphorylated by the PTS general proteins and regulates SacY activity by phosphorylation according to sucrose availability in the medium (17).
To test this idea, we tested whether different forms of SacY, phosphorylated and nonphosphorylated, exist in B. subtilis cells. Two protein isoforms which differ in their charges were indeed precipitated by anti-SacY antibodies from extracts of cells induced for SacY expression. The isoforms were separated on two-dimensional (2-D) gels (isoelectric focusing in the first dimension and sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] in the second). Due to its sensitivity to phosphatase, the more acidic form was identified as the phosphorylated form of the protein. Our results indicate that SacY is regulated in vivo by reversible phosphorylation. The phosphorylation state of SacY depends on the availability of sucrose, the inducer of the sac system, and on the cellular level of SacX. Based on experiments which tested the stability of the bond between SacY and the phosphoryl group, we suggest that SacY, like BglG, is phosphorylated on a histidine residue. The similarity between the two systems, sac and bgl, was manifested in the ability of SacY to antiterminate transcription of the bgl operon when it was expressed in E. coli. Thus, the sac system is a member of the bgl family of sensory systems.
MATERIALS AND METHODS
Bacterial strains.
E. coli AG1688 (MC1061 F′128 lacIq lacZ::Tn5 [4]) and TG1 [supE thi hsdD5 Δ(lac-proAB) (F′ traD36 proAB lacIqZ ΔM15) (24)] were used for plasmid construction. E. coli MA152, which is Δbgl and carries a bgl′-lacZ fusion on its chromosome (λ bglR7 bglG′ lacZ+ lacY+ [34]), was used to measure antitermination. The B. subtilis strain used throughout this study is IS58 (trpC2 lys [46]).
Plasmids.
Plasmids pT712 and pT713 (used for overexpression of genes in E. coli), containing the phage T7 late promoter, were obtained from Bethesda Research Laboratories. Plasmid pDG148 (used for overexpression of genes in B. subtilis [50]) contains the Pspac promoter (composed of the SPO-1 phage promoter and the lac operator [54]) and also the lacI gene (specifying for the lac repressor). Plasmid pSL85 carries the entire sacX and sacY genes (17). Plasmid pSL85-ΔX1 carries the entire sacY gene but contains a deletion within the SacX coding sequence (17). Plasmid pSL165ATG, containing a sacX allele in which the first codon, TTG, was replaced by an ATG, was obtained from M. Steinmetz. Plasmid pMN25 carries the entire bglG gene cloned in pBR322 (34).
The following plasmids were constructed. pOACY is a derivative of pMN25. The PpuMI-EcoRI fragment of pMN25 which carries the entire bglG gene was replaced by a 930-bp fragment carrying the entire sacY gene, which was prepared by PCR amplification with pSL85 as a template. pT7OAC-Y contains a 1,042-bp AvaI-NruI fragment from pSL85, carrying the entire sacY gene, cloned into pT713, which was digested with AvaI and HincII. pT7OAC-XY contains a 2,518-bp SalI-NruI fragment from pSL85, carrying the entire sacX and sacY genes, cloned into pT712, which was digested with SalI and SmaI. pT7OAC-XATG contains a 1,741-bp SalI-BamHI fragment from pSL165ATG, carrying a mutant sacX allele that contains an ATG translation initiation codon, cloned into pT712, which was digested with SalI and BamHI. pMI1 is the same as pT7OAC-Y, except that the EcoRI site was replaced by an HindIII site. This was achieved by digesting pT7OAC-Y with EcoRI, incubating it with calf intestinal phosphatase (CIP), and ligating it to an oligonucleotide which contains an HindIII site and complements the sticky ends generated after EcoRI digestion. pMI3 contains a 1,071-bp HindIII fragment from pMI1, carrying the entire sacY gene, cloned into the HindIII site on pDG148. pMI4 is the same as pT7OAC-XY, except that the EcoRI site was replaced by an HindIII site. It was constructed in the same way as pMI1. pMI5 contains a 2,550-bp HindIII fragment, from pMI4, carrying the entire sacX and sacY genes, cloned into the HindIII site on pDG148. pMI4XATG was constructed by ligating a 2,158-bp NcoI-PvuII fragment from pMI4, carrying the C-terminal part of the sacX gene and the entire sacY gene, to the 3,154-bp NcoI-PvuII fragment from pT7OAC-XATG, carrying the N-terminal part of the sacX gene, which contains an ATG translation initiation codon. pMI5XATG contains a 2,550-bp HindIII fragment from pMI4XATG, carrying the sacX allele with the ATG translation initiation codon and the sacY gene, cloned into the HindIII site of pDG148.
Media.
For [35S]methionine labeling of proteins in B. subtilis, Spizizen minimal medium (25) was used with the following modifications. Lysine (100 μg/ml) and sodium glutamate (10 mg/ml) were also included, and the tryptophan concentration was 100 μg/ml. The medium used for growing B. subtilis for DNA transformation was as described previously (25). For all other purposes, E. coli and B. subtilis were grown in Luria broth. Ampicillin (30 to 200 μg/ml) and kanamycin (10 to 30 μg/ml) were included in the media when strains which contain plasmids that confer resistance to either one of these antibiotics were being grown.
Genetic and cloning techniques.
All manipulations with recombinant DNA were carried out by standard procedures (43). Competent B. subtilis cells were prepared and transformed with plasmid DNA following the Groningen method (described in reference 25). Plasmid DNA was isolated from B. subtilis cells by a modification of the alkaline lysis method (11) which was described previously (25). Assays for β-galactosidase activity were carried out as described by Miller (37). The cells used for these assays were grown in minimal medium which was supplied with 0.4% succinate as a carbon source.
Induction of expression and [35S]methionine labeling of proteins.
B. subtilis cells, containing plasmids carrying the sacY gene with or without the different sacX alleles under the control of the Pspac promoter, were grown in Spizizen minimal medium. Expression of the cloned genes was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to cells that reached the exponential growth phase (optical density at 600 nm, 0.2 to 0.3) at a final concentration of 1 mM. Cells were grown for an additional 1 to 3 h for 2-D gel analysis or 6 h for SDS-PAGE, followed by Western blot analysis. Following this growth period, [35S]methionine (1,200 Ci/mmol; Du Pont) was added for 15 to 25 min at a final concentration of 30 μCi/ml. Cells were either precipitated and washed immediately or incubated with 500 μg of methionine per ml for 5 to 15 min (chase).
Preparation of B. subtilis cell extracts.
B. subtilis cells, harvested and washed with 10 mM Tris-HCl (pH 8.0) and PI (phosphatase inhibitor mixture: 100 mM sodium pyrophosphate, 100 mM NaF, 4 mM EDTA, 4 mM sodium vanadate, adjusted to pH 7.6 with HCl), were lysed by one of two methods. By the first method, cells were resuspended in lysis buffer I (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.6 mg of lysozyme per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], and PI) and incubated for 30 min at 37°C. Following the incubation, SDS was added at a final concentration of 1% and the mixture was boiled for 2 min and centrifuged for 5 min in an Eppendorf centrifuge. The supernatant was used for further analyses. By the second method, cells were resuspended in lysis buffer II (30 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mg of lysozyme per ml, 1 mM PMSF, and PI) and incubated for 30 min at 4°C and 30 min at 30°C. Following the incubation, five cycles of freeze and thaw were performed, the mixture was spun as described above, and the supernatant was collected.
Immunoprecipitation and dephosphorylation of proteins.
Protein A-Sepharose beads (Pharmacia) were preincubated for 2 h at 4°C with 5 μl of rabbit nonimmune serum and then washed three times with Triton buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.25% Triton X-100, and 0.5 mM PMSF). The beads were subsequently incubated for 2 h at 4°C in a solution containing 30 to 40 μl of extract of B. subtilis cells induced for SacY overproduction and 4 to 5 mg of bovine serum albumin, which was brought to a final volume of 400 μl with Triton buffer containing 0.5% Triton X-100. After centrifugation, the supernatant was incubated with 5 μl of anti-SacY antiserum for 15 h at 4°C. Protein A-Sepharose was added, and incubation in the cold was continued for an additional 2 h. After the beads were washed, proteins were eluted from them by boiling in a solution of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1% SDS. For dephosphorylation, extracts of cells induced for SacY overproduction were incubated for 10 min at 30°C with CIP (20 U/μl; Boehringer). The reaction was stopped by adding isoelectric focusing sample buffer.
Hydrolysis of proteins with hydroxylamine.
The effect of hydroxylamine on phosphorylated SacY was tested essentially as described by Hokin et al. (26). Hydroxylamine was prepared in the cold just before use by adding 2 parts of 8 N NaOH to 5 parts of 4 M hydroxylamine hydrochloride. Extracts of cells, which were induced for SacY overproduction, were incubated in 0.1 M acetate buffer (pH 5.2) and 0.8 M hydroxylamine for 10 min at 30°C. Controls contained cell extracts incubated with 0.1 M sodium acetate (pH 5.2) alone.
Electrophoresis, immunoblotting, autoradiography, and densitometry.
Two-dimensional gel electrophoresis was performed essentially as described by O’Farrell (38) with the modification described by Messika et al. (36), except that the proteins were solubilized in Garrels’ sample buffer (23) and the ampholytes (pIs, 5 to 7, 6 to 8, and 3.5 to 10) in the first dimension (isoelectric focusing) were mixed at a ratio of 2:2:1. For regular separation of proteins according to molecular mass, SDS-PAGE was performed (30). After electrophoresis, gels with 35S-labeled proteins were dried and exposed either to Kodak XAR-5 X-ray film or Fujix imaging plate and detected by the Bio-imaging analyzer BAS1000. The relative amounts of radioactivity in the two forms of SacY were quantitated by densitomery performed with the Bio-imaging analyzer BAS1000 after the scanning. For immunoblotting, unlabeled proteins were transferred from an SDS-polyacrylamide gel to a nitrocellulose filter as previously described (52). The filter was incubated for 1 h at room temperature in blocking buffer (5% dried milk [1% fat] in phosphate-buffered saline) and then for 12 to 16 h at 4°C with anti-SacY antibodies diluted 1:500 in blocking buffer. After three washes in phosphate-buffered saline containing 0.1% Tween 20, the filter was incubated for 1 h with horseradish peroxidase goat anti-rabbit antibody (Jackson Immunoresearch Laboratories) diluted 1:20,000 in blocking buffer. Binding of antibodies to the membrane was probed with the enhanced chemiluminescence light-based detection kit (Amersham). Antibody-bound proteins were detected by exposure to Kodak XAR-5 X-ray film.
RESULTS
SacY from B. subtilis can replace BglG in antiterminating transcription of the bgl operon in E. coli.
The high degree of homology between the two antiterminators, BglG from E. coli and SacY from B. subtilis, and their RNA target sites stimulated us to try to determine whether SacY can replace BglG in antiterminating transcription of the bgl operon. We therefore introduced SacY-expressing plasmids into the E. coli strain MA152. This strain is deleted for the bgl operon and carries a chromosomal bgl′-lacZ fusion (34). The lacZ gene is not expressed in MA152, because transcription terminates at the bgl terminator, which is located upstream to the lacZ gene. Expression of plasmid-encoded BglG, the bgl antiterminator, renders the lacZ expression in this strain constitutive (34) (Table 1). The ability of plasmid-encoded SacY to antiterminate transcription at the bgl terminator and enable lacZ expression in MA152 was tested by observing the color of the colonies containing these plasmids on MacConkey lactose plates and by measuring the β-galactosidase levels produced by the cells expressing them. As shown in Table 1, SacY behaved like BglG in its ability to allow for lacZ expression. This result indicates that not only are SacY and BglG similar in their sequences and activities but they also function in an identical manner.
TABLE 1.
SacY can antiterminate transcription of a chromosomal bgl-lacZ fusion in E. colia
| Plasmid | Plasmid-encoded protein | Phenotype on MacConkey-lactose mediumb | β-Galatosidase activity (U)c |
|---|---|---|---|
| pMN25 | BglG | + | 47 |
| pSL85-XΔ1 | SacY | + | 39 |
| pOACY | SacY | + | 51 |
| pBR322 | − | 3 |
The experiment was carried out with MA152, an E. coli strain which is Δbgl and carries a bgl-lacZ transcriptional fusion (35).
Expression of the bgl-lacZ fusion was partly determined by colony color on MacConkey-lactose plates: +, red colonies; −, white colonies.
The values, in Miller units, represent the averages of four independent measurements.
SacY is phosphorylated in the B. subtilis cell.
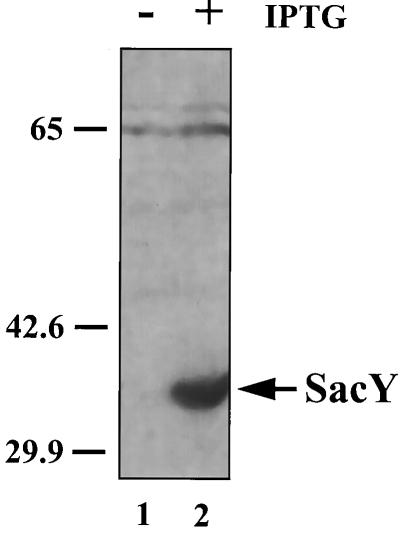
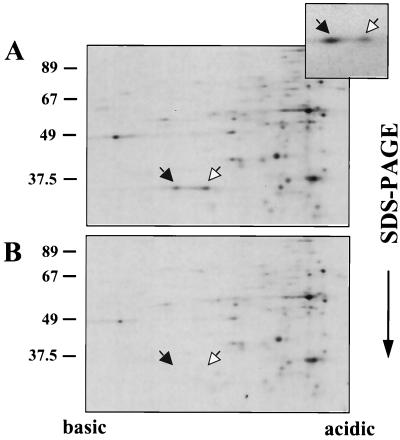
Based on the similarity between the sac system from B. subtilis and the bgl system from E. coli and on the ability of SacY to replace BglG in antiterminating transcription in E. coli, we speculated that sac is regulated similarly to bgl, i.e., that the signal transduction mechanism involves protein phosphorylation. It was previously suggested that SacY activity might be regulated by phosphorylation (17). To examine this hypothesis, we sought to determine whether SacY forms, which differ in their charges, are present in growing B. subtilis cells. Such forms can be separated from one another and from other proteins in the cell by the use of 2-D gels. Phosphorylation of BglG in vivo was demonstrated in this way (3). To allow detection of SacY, we expressed it from plasmid pMI3, which harbors sacY under control of the inducible Pspac promoter, and labeled the cellular proteins with [35S]methionine. Efficient expression of sacY in B. subtilis cells containing pMI3, depending exclusively upon the addition of IPTG to the growth medium, was demonstrated by SDS-PAGE followed by Western blot analysis with anti-SacY antibodies (Fig. 1, compare lanes 1 and 2). We then labeled B. subtilis cells metabolically with [35S]methionine in the presence and absence of IPTG and analyzed the labeled proteins on 2-D gels. Two labeled spots, with a molecular size expected for SacY, were detected only in cells treated with IPTG (Fig. 2, compare A and B). These spots were immunoprecipitated by anti-SacY antibodies (Fig. 2A, insert). We could thus conclude that two forms of SacY are present in B. subtilis cells.
FIG. 1.
Induction of sacY expression from the Pspac promoter in B. subtilis. Proteins were extracted from B. subtilis cells, containing the sacY gene cloned under Pspac promoter control on plasmid pMI3, which were grown either without (lane 1) or with (lane 2) IPTG. Samples were analyzed by SDS-PAGE, followed by Western blot analysis with anti-SacY antibodies. Molecular masses of protein standards are given in kilodaltons.
FIG. 2.
SacY protein is present in two forms in vivo. (A) B. subtilis cells containing the sacY gene cloned under Pspac promoter control on plasmid pMI3 were induced for SacY production by adding IPTG. The cellular proteins were extracted, after being labeled with [35S]methionine, and analyzed by 2-D gel electrophoresis (isoelectric focusing in one dimension and SDS-PAGE in the second dimension), followed by autoradiography. The insert (top, right) shows fractionation of the same proteins, which were immunoprecipitated with anti-SacY antibodies prior to the 2-D gel analysis. (B) The same cells as described for panel A except that IPTG was not added to the cells. Arrows indicate the positions of the two forms of SacY. Molecular masses of protein standards are given in kilodaltons.
To test whether the two forms of SacY represent phosphorylated and nonphosphorylated forms of the protein, we treated the 35S-labeled cellular proteins with alkaline phosphatase before gel analysis. This treatment led to the loss of the more acidic form of SacY (compare Fig. 3A, nontreated, to 3B, phosphatase treated), indicating that this indeed is the phosphorylated form (as expected from the charge conferred by a phosphoryl group). This result shows that SacY is phosphorylated in vivo. The fraction of the phosphorylated form of SacY detected by us in this strain varied between 30 to 60% in different experiments, depending on the conditions of the induction and the length of the pulse-labeling and whether a short chase period was included in the experiment. Therefore, in each of the experiments described below, we repeated all the controls rather than comparing different experiments.
FIG. 3.
SacY is phosphorylated in vivo. (A) B. subtilis cells containing the sacY gene cloned in plasmid pMI3 were induced for SacY production and labeled with [35S]methionine, and proteins were extracted and fractionated as described in the legend for Fig. 2A. (B) The same as described for panel A, except that the extracted proteins were treated with CIP prior to the 2-D gel analysis. Closed arrows indicate nonphosphorylated SacY; open arrows indicate phosphorylated SacY.
Sucrose affects the state of SacY phosphorylation.
SacY antiterminates transcription of the sacB gene in response to the presence of sucrose in the growth medium. Its E. coli homolog, BglG, antiterminates transcription of the bgl operon, depending on the presence of β-glucosides in the medium. Therefore, we expected sucrose to influence the state of SacY phosphorylation analogously to the known effect of β-glucosides on the extent of BglG phosphorylation (2, 3). To test this hypothesis, sucrose was added to the growth medium of B. subtilis cells during SacY production at a final concentration of 30 mM (the concentration which leads to SacY-dependent sacB expression). Cells overproducing SacY in the absence of sucrose served as a control. Whereas the fraction of the phosphorylated form of SacY in the absence of sucrose was approximately 40% (Fig. 4A), this form could hardly be detected when sucrose was included in the medium (Fig. 4B). Thus, in the presence of sucrose, SacY is present mainly in its nonphosphorylated form, and we can therefore conclude that the extent of SacY phosphorylation in vivo is influenced by the presence of sucrose in the growth medium.
FIG. 4.
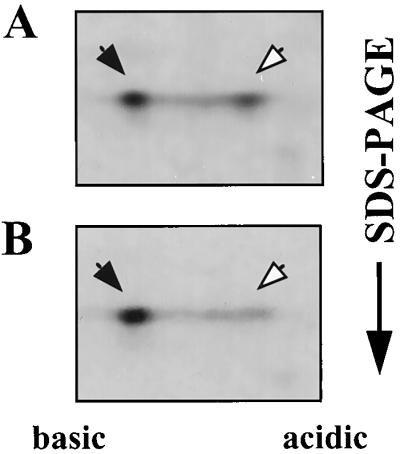
Influence of sucrose on the state of SacY phosphorylation in vivo. (A) B. subtilis cells containing the sacY gene cloned in plasmid pMI3 were induced for SacY production and labeled with [35S]methionine, and proteins were extracted and fractionated as described in the legend for Fig. 2A. (B) The same as described for panel A, except that 30 mM sucrose was added to the growth medium together with the IPTG. Closed arrows indicate nonphosphorylated SacY; open arrows indicate phosphorylated SacY.
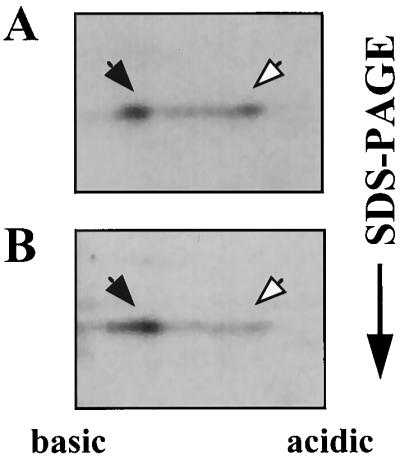
In order to determine whether sucrose prevents SacY phosphorylation or can actually lead to dephosphorylation of this protein, similarly to the ability of β-glucosides to lead to BglG dephosphorylation (2, 3), we carried out pulse-chase experiments in which sucrose was added only after SacY had been phosphorylated in the cell. Addition of unlabeled methionine to cells which were pulse-labeled with [35S]methionine did not reduce the relative amount of phosphorylated SacY (compare Fig. 5A and B). However, when sucrose was added to the cells together with the unlabeled methionine, all of the phosphorylated SacY was converted to the nonphosphorylated form of this protein (Fig. 5C). Thus, addition of sucrose to the growth medium leads to active dephosphorylation of SacY in vivo, and not merely to the inhibition of SacY phosphorylation.
FIG. 5.
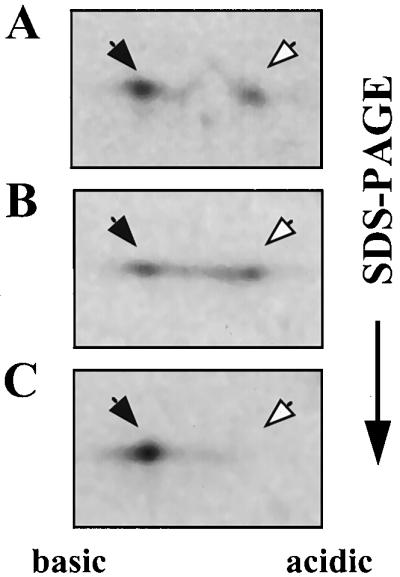
Sucrose stimulates dephosphorylation of SacY in vivo. (A) B. subtilis cells containing the sacY gene cloned under Pspac promoter control on plasmid pMI3 were induced for SacY production by adding IPTG and were then labeled by incubation with [35S]methionine for 15 min. Cells were further incubated with excess unlabeled methionine for 5 (A) or 15 (B) min or as in panel B except that sucrose was added during the last 10 min at a final concentration of 60 mM (C). Proteins were extracted and fractionated as described in the legend for Fig. 2A. Closed arrows indicate nonphosphorylated SacY; open arrows indicate phosphorylated SacY.
The effect of SacX on the state of SacY phosphorylation.
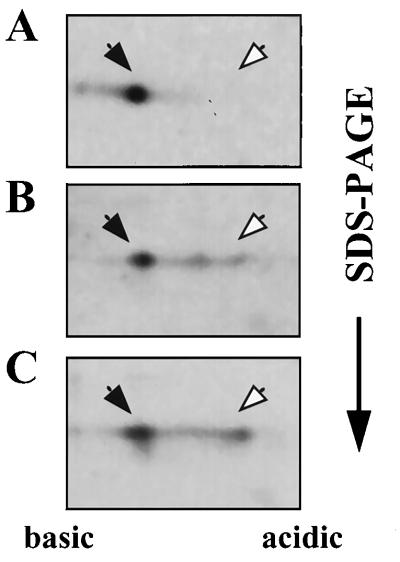
Expression of sacB, which is positively regulated by SacY, is repressed by SacX, a PTS enzyme II-like protein (9). Analogously to the negative regulation of BglG due to its phosphorylation by BglF, an enzyme II of PTS (2, 3), it was suggested that SacX negatively regulates SacY activity by phosphorylating it (17). To determine whether SacX is involved in SacY phosphorylation, we asked whether the extent of SacY phosphorylation correlates with the level of SacX produced in the cell. We therefore analyzed the extent of SacY phosphorylation in cells producing different levels of SacX. To this end, we constructed plasmids pMI5 and pMI5XATG, both carrying sacX and sacY cloned after Pspac, but whereas the first carries the wild-type sacX gene, which starts with a TTG codon, the second carries a sacX allele, which starts with ATG, a more efficient initiation codon than TTG (53). SDS-PAGE analysis of labeled proteins from B. subtilis strains which carry a chromosomal copy of sacX and contain either pMI3 (carrying sacY alone), pMI5, or pMI5XATG revealed a protein with the molecular size expected for SacX only when SacX was overproduced from the plasmids, as expected (data not shown). It is worth mentioning that the level of overexpressed SacX, even from the more efficiently translated allele, was lower than the level of overexpressed SacY, similar to the case of BglF and BglG (3). Analysis by 2-D gel electrophoresis of SacY produced in the three cell backgrounds, in the absence of sucrose, indicated that the fraction of phosphorylated SacY increased with increased SacX production (Fig. 6). The approximate fraction of SacY present in the phosphorylated form was 30% in the first background (Fig. 6A), 50% in the second (Fig. 6B), and more than 70% in the third (Fig. 6C). This experiment was carried out under conditions that result in a relatively low extent of SacY phosphorylation in the absence of SacX overproduction (short induction period, low level of inducer, relatively short pulse, and no chase). However, a similar dependence of SacY phosphorylation on the level of SacX production was also observed under other experimental conditions. Based on these results, we suggest that the extent of SacY phosphorylation is proportional to the level of SacX in the cell and thus that SacX appears to be involved in SacY phosphorylation.
FIG. 6.
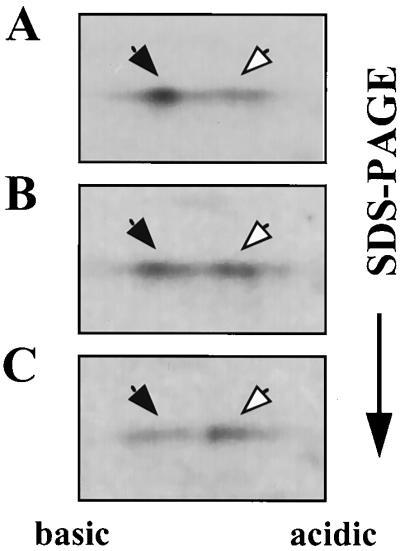
Influence of SacX on the state of SacY phosphorylation in vivo. B. subtilis cells were treated with IPTG and labeled with [35S]methionine, and proteins were extracted and fractionated as described in the legend for Fig. 2A. The cells contained the following plasmids: pMI3, which expresses sacY from Pspac (A); pMI5, which expresses both sacY and sacX from Pspac (B); and pMI5ATG, which is the same as pMI5 except that it carries a sacX allele which starts with an ATG codon (C). Closed arrows indicate nonphosphorylated SacY; open arrows indicate phosphorylated SacY.
Characterization of the bond between SacY and the phosphoryl group.
Phosphorylation on histidine and aspartate residues (which form phosphoramidates and acyl phosphates, respectively) is widespread among prokaryotes, although it also occurs on other residues (42). BglG is phosphorylated on a histidine (6, 15), while regulators of the two-component family are phosphorylated on an aspartate (see Discussion). To study the nature of the bond between SacY and the phosphoryl group, we tested the stability of P∼SacY under conditions which destabilize acyl phosphates and phosphoramidates but not phosphate esters. Only the former two are susceptible to rapid aminolysis at a pH of <5.5 in the presence of hydroxylamine (12, 26). We therefore incubated an extract of cells overproducing SacY with hydroxylamine in acetate buffer, pH 5.2. This led to the complete disappearance of P∼SacY (Fig. 7A). Incubation in acetate buffer (pH 5.2) without hydroxylamine did not affect the ratio between phosphorylated and nonphosphorylated SacY, and the result was as in the control presented in Fig. 7C (data not shown). This result indicates that SacY-phosphate is present either as an N-phosphate or as an acyl phosphate. Because both compounds are known for their relative sensitivity to elevated temperatures (1), we tested the stability of P∼SacY to heat by boiling the proteins extracted from cells overproducing SacY. This treatment also resulted in a sharp decrease in the intensity of the phosphorylated form of SacY (Fig. 7B). A new spot migrating between the two forms of SacY, which are routinely observed, appeared after the boiling. The fraction of the protein transferred to this intermediate location did not decrease upon prolonged heating. An explanation for the stability of this new form to heat is suggested in the Discussion.
FIG. 7.
Sensitivity of the bond between SacY and the phosphoryl group to hydroxylamine and heat. B. subtilis cells containing the sacY gene cloned in plasmid pMI3 were induced for SacY production and labeled with [35S]methionine. Proteins were extracted and were either subjected to treatment with hydroxylamine as described in Materials and Methods (A) or boiled for 10 min (B). Treated samples and an untreated control (C) were analyzed by 2-D gel electrophoresis, followed by autoradiography. Closed arrows indicate nonphosphorylated SacY; open arrows indicate phosphorylated SacY.
DISCUSSION
The bgl system in E. coli, composed of a membrane-bound sensor, BglF, and a cytoplasmic regulator, BglG, is not a member of the known family of two-component sensory systems (reviewed in references 40, 41, and 49). BglF and BglG have no homology with the sensors and regulators of the two-component systems, respectively. Moreover, it was recently shown that BglG is phosphorylated on a histidine residue, unlike response regulators of the two-component family, which are phosphorylated on an aspartate (6). Thus, bgl represents a novel family of systems involved in processing sensory data (reviewed in reference 5). Based on sequence homology and mechanistic similarity, other systems, responding to the presence of various sugars, were suggested to affiliate to this family (see below). Recent findings, showing that the two-component family and histidine phosphorylation are not confined to prokaryotes (14, 33, 39), raise the possibility that eukaryotic systems of the bgl family will be found in the future. To elucidate the general features of the mechanism that governs signal transduction in the bgl family, it is important to study other members of this family. Does the communication between sensors and regulators of the new family involve reversible phosphorylation, which depends on the presence of the respective sugars, as a general theme?
BglG and BglF have considerable homology with pairs of regulatory proteins, including the B. subtilis pairs SacY and SacX (48, 55), SacT and SacP (18, 21), LicT and BglP (29, 31, 45), and LevR and Lev-PTS (a complex of three proteins) (19, 35), and the Erwinia chrysanthemi pair ArbG and ArbF (20). Detailed analysis of the two sac systems in B. subtilis, which are induced by different concentrations of sucrose (48), indicates that SacY and SacT, which antiterminate transcription of the sacB gene and sacPA operon, respectively, function in a manner analogous to that of BglG (7, 8, 10, 17) by binding to ribonucleic acid antiterminator sequences highly homologous to the target site of BglG on the bgl transcript (7, 10). The homology between BglG and SacY is striking (above 35% identity and 65% similarity [55]), especially in light of the evolutionary distance between the two organisms. Moreover, as we show here, SacY expressed in E. coli can replace BglG in antiterminating transcription of the bgl operon. Thus, the two proteins act identically and are therefore expected to be regulated similarly. Indeed, as shown in this paper, like BglG, SacY exists in vivo in two forms, phosphorylated and nonphosphorylated. The ratio between the SacY forms depends on the presence of sucrose in the growth medium, similarly to the dependence of the BglG phosphorylation state on β-glucosides. Therefore, the same model which was deduced for the regulation of BglG by reversible phosphorylation, depending on the availability of the inducing sugar, holds for SacY regulation. Moreover, our results suggest that the negative effect of SacX, a putative PTS sucrose permease, on SacY activity (9, 17, 55) is due to its involvement in SacY phosphorylation, similarly to the negative effect of BglF, the PTS β-glucoside permease, on the activity of BglG (2, 3, 44). We reached this conclusion by comparing the relative amounts of phosphorylated and nonphosphorylated SacY in strains that produce different levels of SacX. It was difficult to accurately quantitate the amounts of SacX produced by these strains, due to the unavailability of anti-SacX antibodies. However, one finding that emerged from these experiments is that SacX was produced at lower levels than SacY in all the strains (even when both proteins were overproduced from the same plasmid). Thus, SacX, like BglF, acts in a catalytic rather than a stoichiometric way. In light of the catalytic mode of action of BglF (a ratio of less than 1:200 between BglF and BglG was enough to phosphorylate 50 to 60% of the overproduced BglG [3]), the observed phosphorylation of overproduced SacY by SacX expressed from a chromosomal gene is expected. Our results suggest that SacX is involved in SacY phosphorylation but do not rule out the possibility that the effect of SacX on SacY phosphorylation is indirect. It is hoped that future in vitro studies of SacY phosphorylation will answer the question of whether SacX is the SacY kinase.
Genetic studies have demonstrated that the general PTS proteins, enzyme I and HPr, negatively regulate SacY activity, and it was suggested that they exert this effect through SacX (17). Interestingly, unlike BglG and SacY, the BglG-like proteins SacT and LevR are positively regulated by the general PTS proteins and were shown to be phosphorylated by them in vitro (7, 8, 51). Thus, the general PTS proteins repress the activity of some antiterminators from the bgl family and activate others. The ability of SacY to antiterminate transcription of the bgl operon in E. coli at a level comparable to BglG (Table 1) rules out the possibility that the E. coli general PTS proteins can negatively regulate SacY activity by phosphorylation. Nevertheless, the possibility that the B. subtilis enzyme I and HPr exert a negative effect on SacY activity by directly phosphorylating it, in addition to their indirect effect via SacX, cannot be ruled out.
Another question is whether the phosphorylation of BglG on a histidine residue is a general theme in the bgl family of sensory systems, similarly to the phosphorylation of the response regulators of the two-component systems on an aspartate. The sensitivity of SacY-P to hydroxylamine and heat suggests that it is either a phosphoramidate or an acyl phosphate. Based on the types of amino acids that are known to form these types of bonds in proteins, phosphorylation is suggested to occur either on a histidine or an aspartate. One approach to decide between these possibilities is to determine the stability of SacY-P to acid and base, as was done with BglG-P (6). While phosphoramidates are stable in basic conditions but sensitive to acidic conditions (22), acyl phosphates are labile at either pH extreme (28). We tried to incubate extracts of cells overproducing SacY in HCl and NaOH prior to their analysis, but the 2-D gel technique turned out to be sensitive to such harsh treatments of the analyzed proteins, and the samples precipitated and smeared in a way that made the interpretation of the results difficult. However, the appearance, after boiling, of an additional spot on the 2-D gel, which migrates between the two forms of SacY, provides a hint about the type of amino acid which is phosphorylated in this protein. The most plausible explanation for this phenomenon is the occurrence of a phosphotransfer, i.e., the phosphoryl is transferred to a nearby residue (not necessarily adjacent on the primary sequence) to form a phosphoester which is heat stable. The magnitude of the mobility shift caused by phosphorylation on an aspartate in isoelectric focusing is smaller than that of any other shift which is caused by phosphorylation of other amino acids (13, 16). Therefore, a phosphotransfer from an aspartate to a serine, threonine, or tyrosine should not lead to the appearance of a spot with intermediary migration behavior but, rather, to a shift to the other direction, i.e., to a more acidic position. Phosphotransfer from a histidine to the phosphoester-forming residues will generate the observed shift, both in direction and in magnitude. Thus, this result supports the notion that SacY is phosphorylated on a histidine rather than an aspartate. Theoretically, phosphorylation on a lysine or an arginine, though not discovered yet, should yield the same result and cannot be ruled out. An alternative explanation to this result is phosphorylation of SacY on two histidines or aspartates, combined with a higher sensitivity of one of the two to heat, due to a difference in their immediate surroundings. This explanation is unfavorable because, contrary to what we have observed, prolonged heating in this case should lead to a decrease in the intensity of the intermediate spot.
ACKNOWLEDGMENTS
We gratefully acknowledge M. Baniash and O. Avni for their advice and help with the 2-D gel technique. We especially acknowledge M. Banish for fruitful discussions. We warmly thank M. Steinmetz and S. Aymerich for their support, openness, and willingness to exchange information. We acknowledge them also for the gift of plasmids pSL85 and pSL165ATG. We thank A. M. Crutz for the gift of the anti-SacY antiserum. We thank G. Glaser, L. Sonenshein, R. Rudner, and P. Stragier for advice on B. subtilis techniques and for the gifts of plasmid pDG148 and strain IS58. We thank O. Pines for critically reading the manuscript. Investigation of the ability of SacY to act in E. coli was initiated when O.A.-C. was a postdoctoral fellow in A. Wright’s lab.
This research was supported by the Israel Science Foundation administered by the Israel Academy of Sciences and Humanities.
Footnotes
This work is dedicated to the memory of Michel Steinmetz.
REFERENCES
- 1.Amir-Zaltsman Y, Salomon Y. Phosphorylation of proteins in rat ovarian plasma membranes by [γ-32P]GTP: evidence for the formation of a high energy phosphoprotein. Mol Cell Endocrinol. 1989;63:175–187. doi: 10.1016/0303-7207(89)90094-4. [DOI] [PubMed] [Google Scholar]
- 2.Amster-Choder O, Houman F, Wright A. Protein phosphorylation regulates transcription of the β-glucoside utilization operon in E. coli. Cell. 1989;58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 3.Amster-Choder O, Wright A. Regulation of activity of a transcriptional antiterminator in E. coli by phosphorylation in vivo. Science. 1990;249:540–542. doi: 10.1126/science.2200123. [DOI] [PubMed] [Google Scholar]
- 4.Amster-Choder O, Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science. 1992;257:1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- 5.Amster-Choder O, Wright A. Transcriptional regulation of the bgl operon of Escherichia coli involves phosphotransferase system-mediated phosphorylation of a transcriptional antiterminator. J Cell Biochem. 1993;51:83–90. doi: 10.1002/jcb.240510115. [DOI] [PubMed] [Google Scholar]
- 6.Amster-Choder O, Wright A. BglG, the response regulator of the Escherichia coli bgl operon, is phosphorylated on a histidine residue. J Bacteriol. 1997;179:5621–5624. doi: 10.1128/jb.179.17.5621-5624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnaud M, Débarbouillé M, Rapoport G, Saier M H, Jr, Reizer J. In vitro reconstitution of transcriptional antitermination by the SacT and SacY proteins of Bacillus subtilis. J Biol Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- 8.Arnaud M, Vary P, Zagorec M, Klier A, Debarbouille M, Postma P, Rapoport G. Regulation of the sacPA operon of Bacillus subtilis: identification of phosphotransferase system components involved in SacT activity. J Bacteriol. 1992;174:3161–3170. doi: 10.1128/jb.174.10.3161-3170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aymerich S, Steinmetz M. Cloning and preliminary characterization of the sacS locus from Bacillus subtilis which controls the regulation of the exoenzyme levansucrase. Mol Gen Genet. 1987;208:114–120. doi: 10.1007/BF00330431. [DOI] [PubMed] [Google Scholar]
- 10.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birnboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitte L, Kabat D. Isotopic labeling and analysis of phosphoproteins from mammalian ribosomes. Methods Enzymol. 1974;30:563–590. doi: 10.1016/0076-6879(74)30056-0. [DOI] [PubMed] [Google Scholar]
- 13.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 14.Chang C, Kwok S F, Bleecker A B, Meyerovitz E M. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Engelberg-Kulka H, Amster-Choder O. The localization of the phosphorylation site of BglG, the response-regulator of the E. coli bgl sensory system. J Biol Chem. 1997;272:17263–17268. doi: 10.1074/jbc.272.28.17263. [DOI] [PubMed] [Google Scholar]
- 16.Cooper J A. Estimation of phosphorylation stoichiometry by separation of phosphorylated isoforms. Methods Enzymol. 1991;201:251–261. doi: 10.1016/0076-6879(91)01023-u. [DOI] [PubMed] [Google Scholar]
- 17.Crutz A-M, Steinmetz M, Aymerich S, Richter R, Le Coq D. Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system. J Bacteriol. 1990;172:1043–1050. doi: 10.1128/jb.172.2.1043-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debarbouille M, Arnaud M, Fouet A, Klier A, Rapoport G. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J Bacteriol. 1990;172:3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Débarbouillé M, Martin-Verstraete I, Klier A, Rapoport G. The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both ς54- and phosphotransferase system-dependent regulators. Proc Natl Acad Sci USA. 1991;88:2212–2216. doi: 10.1073/pnas.88.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Hassouni M, Henrissat B, Chippaux M, Barras F. Nucleotide sequences of the arb genes, which control β-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new β-glycohydrolase family including enzymes from eubacteria, archaebacteria, and humans. J Bacteriol. 1992;174:765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouet A, Arnaud M, Klier A, Rapoport G. Bacillus subtilis sucrose-specific enzyme II of the phosphotransferase system: expression in Escherichia coli and homology to enzymes II from enteric bacteria. Proc Natl Acad Sci USA. 1987;84:8773–8777. doi: 10.1073/pnas.84.24.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujitaki J M, Smith R A. Techniques in the detection and characterization of phosphoramidate-containing proteins. Methods Enzymol. 1984;107:23–36. doi: 10.1016/0076-6879(84)07004-x. [DOI] [PubMed] [Google Scholar]
- 23.Garrels J I. Two-dimensional gel electrophoresis and computer analysis of protein synthesized by clonal cell lines. J Biol Chem. 1979;254:7961–7977. [PubMed] [Google Scholar]
- 24.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, United Kingdom: Cambridge University; 1984. [Google Scholar]
- 25.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons; 1990. [Google Scholar]
- 26.Hokin L E, Sastry P S, Galsworthy P R, Yoda A. Evidence that a phosphorylated intermediate in brain transport adenosine triphosphatase is an acyl phosphate. Proc Natl Acad Sci USA. 1965;54:177–184. doi: 10.1073/pnas.54.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houman F, Diaz-Torres M R, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 28.Koshland D E., Jr Effects of catalysts on the hydrolysis of acetyl phosphate. Nucleophilic displacement mechanisms in enzymatic reactions. J Am Chem Soc. 1952;74:2286–2292. [Google Scholar]
- 29.Krüger S, Hecker M. Regulation of the putative bglPH operon for aryl-β-glucoside utilization in Bacillus subtilis. J Bacteriol. 1995;177:5590–5597. doi: 10.1128/jb.177.19.5590-5597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of the structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Le Coq D, Lindner C, Krüger S, Steinmetz M, Stülke J. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lepesant J-A, Kunst F, Pascal M, Lepesant-Kejzlarova J, Steinmetz M, Dedonder R. Specific and pleiotropic regulatory mechanisms in the sucrose system of Bacillus subtilis 168. In: Schlessinger D, editor. Microbiology—1976. Washington, D.C: American Society for Microbiology; 1976. pp. 58–69. [Google Scholar]
- 33.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 34.Mahadevan S, Reynolds A E, Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990;214:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- 36.Messika E J, Avni O, Gallily R, Yefenof E, Baniyash M. Identification and characterization of a novel protein associated with macrophage complement receptor 3. J Immunol. 1995;154:6563–6570. [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 38.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 39.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 41.Russo F D, Silhavy T J. The essential tension: opposed reactions in bacterial two-component regulatory systems. Trends Microbiol. 1993;1:306–310. doi: 10.1016/0966-842x(93)90007-e. [DOI] [PubMed] [Google Scholar]
- 42.Saier M H., Jr Protein phosphorylation and signal transduction in bacteria. J Cell Biochem. 1993;51:1–6. doi: 10.1002/jcb.240510102. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Schnetz K, Rak B. β-Glucoside permease represses the bgl operon of E. coli by phosphorylation of the antiterminator protein and also interacts with glucose-specific enzyme II, the key element in catabolic control. Proc Natl Acad Sci USA. 1990;87:5074–5078. doi: 10.1073/pnas.87.13.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnetz K, Stülke J, Gertz S, Krüger S, Krieg M, Hecker M, Rak B. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J Bacteriol. 1996;178:1971–1979. doi: 10.1128/jb.178.7.1971-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith I, Paress P, Cabane K, Dubnau E. Genetics and physiology of the rel system of Bacillus subtilis. Mol Gen Genet. 1980;178:271–279. doi: 10.1007/BF00270472. [DOI] [PubMed] [Google Scholar]
- 47.Steinmetz M, Aymerich S, Le Coq D, Gonzy-Tréboul G. Levansucrase induction by sucrose in Bacillus subtilis involves an antiterminator. Homology with the Escherichia coli bgl operon. In: Ganesan A T, Hoch J A, editors. Fourth International Conference on Genetics and Biotechnology of Bacilli. Vol. 2. New York, N.Y: Academic Press, Inc.; 1988. pp. 11–16. [Google Scholar]
- 48.Steinmetz M, Le Coq D, Aymerich S. Induction of saccharolytic enzymes by sucrose in Bacillus subtilis: evidence for two partially interchangeable regulatory pathways. J Bacteriol. 1989;171:1519–1523. doi: 10.1128/jb.171.3.1519-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stock J B, Stock A M, Mottonen J M. Signal transduction in bacteria. Nature. 1990;334:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 50.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 51.Stülke J, Martin-Verstraete I, Charrier V, Klier A, Deutscher J, Rapoport G. The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6928–6936. doi: 10.1128/jb.177.23.6928-6936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vellanoweth R L, Rabinowitz J C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 54.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zukowski M M, Miller L, Cosgwell P, Chen K, Aymerich S, Steinmetz M. Nucleotide sequence of the sacS locus of Bacillus subtilis reveals the presence of two regulatory genes. Gene. 1990;90:153–155. doi: 10.1016/0378-1119(90)90453-x. [DOI] [PubMed] [Google Scholar]