Abstract
Objective:
Natural language processing (NLP) combined with machine learning (ML) techniques are increasingly used to process unstructured/free-text patient-reported outcome (PRO) data available in electronic health records (EHRs). This systematic review summarizes the literature reporting NLP/ML systems/toolkits for analyzing PROs in clinical narratives of EHRs and discusses the future directions for the application of this modality in clinical care.
Methods:
We searched PubMed, Scopus, and Web of Science for studies written in English between 1/1/2000 and 12/31/2020. Seventy-nine studies meeting the eligibility criteria were included. We abstracted and summarized information related to the study purpose, patient population, type/source/amount of unstructured PRO data, linguistic features, and NLP systems/toolkits for processing unstructured PROs in EHRs.
Results:
Most of the studies used NLP/ML techniques to extract PROs from clinical narratives (n=74) and mapped the extracted PROs into specific PRO domains for phenotyping or clustering purposes (n=26). Some studies used NLP/ML to process PROs for predicting disease progression or onset of adverse events (n=22) or developing/validating NLP/ML pipelines for analyzing unstructured PROs (n=19). Studies used different linguistic features, including lexical, syntactic, semantic, and contextual features, to process unstructured PROs. Among the 26 NLP systems/toolkits we identified, 14 used rule-based NLP, 8 used hybrid NLP, and 4 used ML algorithms embedded in NLP.
Conclusions:
This study supports the potential utility of different NLP/ML techniques in processing unstructured PROs available in EHRs for clinical care. Though using annotation rules for NLP/ML to analyze unstructured PROs is dominant, deploying novel neural ML-based methods is warranted.
Keywords: Natural Language Processing, Machine Learning, Patient-reported Outcome, Electronic Health Record, Electronic Medical Record, Unstructured Clinical Narrative
INTRODUCTION
Patient-reported outcomes (PROs) provide information about a patient’s physical, psychological, somatic symptoms, daily functional status, health-related quality-of-life (HRQOL), and satisfaction with healthcare services that facilitate clinical decision-making and outcome evaluation [1]. PROs are conventionally assessed through validated questionnaires or semi-structured interviews. However, these methods may not capture the full patient experience as their responses are bound by predetermined items. Additionally, it is challenging to collect PRO data from patients during time-limited clinical encounters [2]. Therefore, finding effective approaches to assess, retrieve, and extract already available, unstructured PRO data from alternative sources, e.g., medical notes in electronic health records (EHRs), is important [3].
PRO data in EHRs are often stored in an unstructured and free-text format (e.g., symptom narratives in a physician note) and cannot be directly used in clinical tasks (e.g., disease prediction, classification). Natural language processing (NLP) and machine learning (ML) can turn unstructured PROs into a quantitative or structured format for clinical use [4]. The application of NLP/ML techniques in clinical settings includes text pre-processing (e.g., tokenization, lemmatization, or stemming from the corpus), linguistic feature extraction for clinical narratives (e.g., encoding, affirmed/negated expressions), and clinical applications [5]. Given the significant expansion of EHR ecosystems and the development of novel NLP/ML techniques over the past decade, the interest in processing unstructured PROs from clinical narratives through automatic or semi-automatic NLP/ML pipelines of free-text-based PROs is emerging [6, 7].
Previous reviews have described the application of NLP/ML methods for automatic extraction of non-PRO clinical narratives (e.g., disease progression, adverse drug reactions, medications, and treatments) from EHRs [5, 8], yet review studies of NLP/ML techniques on unstructured PROs are limited. One review article investigated text-mining NLP techniques to extract symptom information through patient-authored data collected from social media (e.g., Twitter, WebMD, and Reddit) [9]. Another review article collected the applications of NLP in analyzing symptom-only data documented in EHRs [10]. However, these studies mainly focus on the traditional rule-based NLP methods to process symptom data, followed by non-neural ML-based classifiers (e.g., support vector machine, logistic regression classifier), rather than novel neural network (e.g., CNN, RNN) or large language models (e.g., GPT, BERT) to analyze associations of PROs and clinical outcomes.
With the recent advances in NLP/ML techniques, this study aimed to summarize research applying NLP/ML for processing and analyzing unstructured PRO data collected in EHRs. Specifically, we evaluated studies that analyzed unstructured PROs for clinical care or research, the type of unstructured PRO data, and the uniqueness of NLP/ML systems/toolkits and techniques to process unstructured PROs. In contrast to previous review studies [9, 10], the findings from this study will improve our clinical insights of using NLP/ML techniques to process EHR-based unstructured PROs in a broader category (including symptom, functioning, and quality-of-life), together with other clinical parameters for clinical application.
METHODS
Data Retrieval
We searched studies written in English between January 1st, 2000 and December 31st, 2020 through PubMed, Scopus, and Web of Science. Following the guideline of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses, we identified 690 studies from PubMed, 739 from Scopus, and 886 from Web of Science (Figure 1). The search strategies are reported in Supplementary Table S1.
Figure 1.
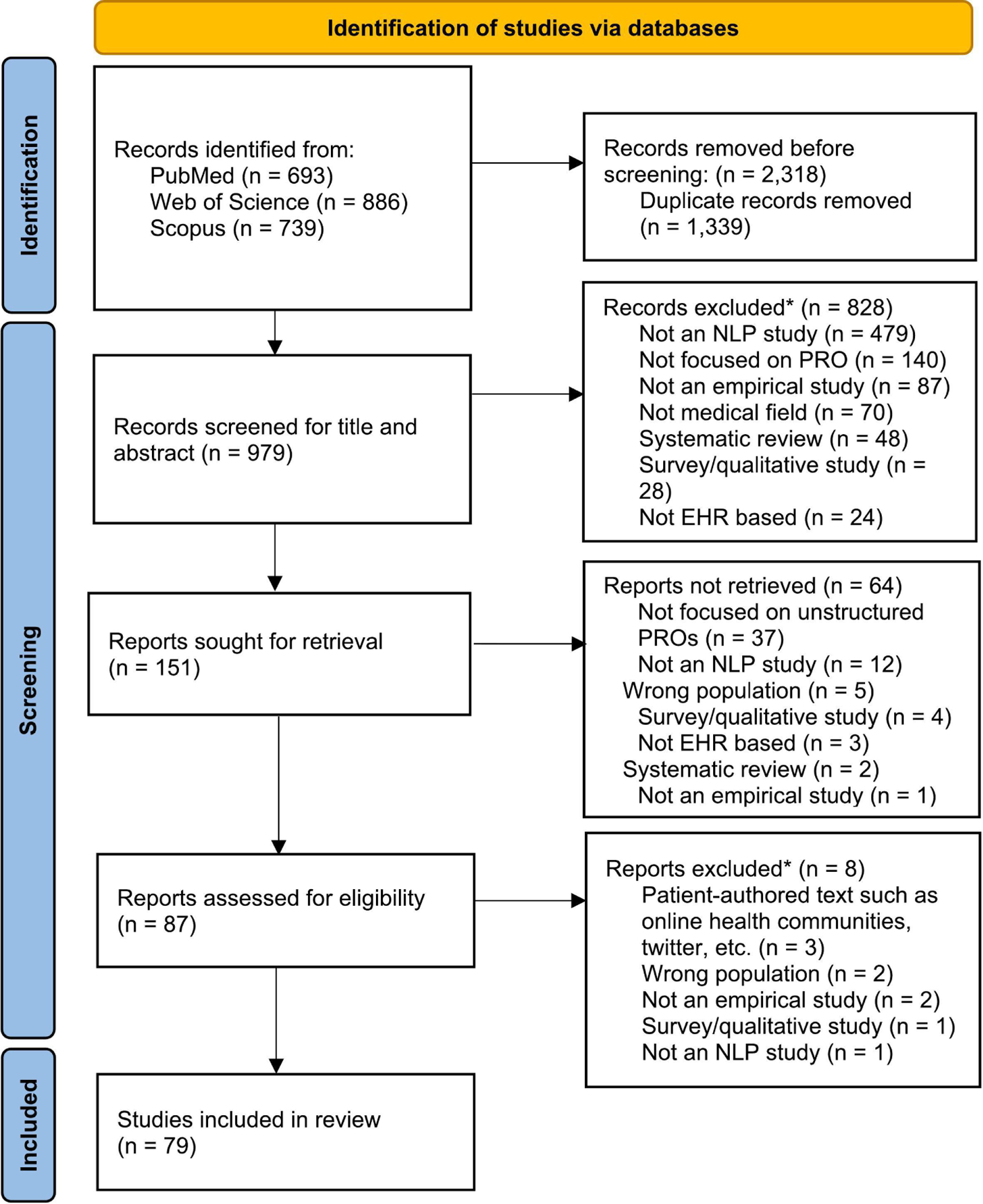
Flow diagram of included articles
Article Selection
Among the 2,318 studies identified from these three databases, 979 non-duplicate studies were retained for the title and abstract screening. Studies were considered for inclusion if they 1) focused on unstructured PRO data in EHRs or medical notes, and 2) used NLP techniques or applications accompanied by ML algorithms to extract or process unstructured PRO data. Studies were excluded if they 1) did not apply NLP/ML techniques, 2) were non-empirical studies (e.g., case reports, commentary), 3) were non-EHR-based studies (e.g., patient-authored data collected from social media), 4) were survey-based studies containing quantitative PRO data, 5) were previous systematic review studies or E-pub ahead of print, and/or 6) focused on non-chronic disease topics (e.g., infectious disease/vaccination). Based on these criteria, the first author (JAS) and the senior author (ICH) independently reviewed the title and abstract of all 979 studies retrieved from the literature search and retained 151 studies. Subsequently, the same two authors reviewed the full‐text articles, resolved any discrepancies, and selected 79 studies for inclusion in this study (Figure 1).
Data Extraction and Summary
We implemented the following steps to collect data from the original articles: 1) the first author (JAS) manually extracted data from the 79 studies and documented information in a database, 2) the senior author (ICH) reviewed all of the extracted information in the database, 3) both co-authors met to confirm the extracted data through reviewing each of the original articles included in this study, and 4) the second author (XH) adjudicated the discrepancy raised by the first and senior authors, and reviewed information summarized in the tables for identifying any errors. For each selected study, the characteristics of the study sample, the objective of the study, the type/format, size, and unit of unstructured PRO data, specific PRO content, and NLP/ML systems/toolkits and linguistic features used to process PRO data were abstracted and reported.
RESULTS
Study Characteristics
Supplementary Table S2 displays the characteristics of 79 studies selected for inclusion in this systematic review. These studies included various participant sample sizes, ranging from 22 to 267,855, and different disease diagnoses, including any type (n=24) [7, 11–33], chronic disease (n=17) [6, 34–50], mental illness (n=23) [51–73], cancer (n=8) [74–81], and others (n=6). The majority of studies (n=73) focused on adults, six on pediatrics [19, 25, 38, 54, 82, 83], and one on both adult and pediatric patient populations [7]. Most of the PRO narrative data were primarily obtained from inpatient or outpatient EHRs (n=75), while several studies relied on the open data repositories/resources (e.g., MIMIC-III; n=4) [24, 33, 35, 50] and the national database (e.g., Taiwan’s National Health Insurance Research Database; n=1) [72]. Narrative data from both inpatient settings (e.g., admission notes, discharge summaries, nursing narratives, emergency department documents, intensive care unit reports; n=57) and outpatient settings (e.g., primary care documents, psychiatric evaluation notes; n=35) were generated by healthcare professionals. The number of PRO narrative documents/grammatical units for NLP/ML analysis ranged from 100 to 4.2 million. Approximately 85% of the studies were based on English-based EHRs (USA: 66%, UK: 15%, and Canada, Island, and Australia: 4%), 10% from Chinese-based EHRs (China: 7% and Taiwan: 3%), and 5% from EHRs with other languages (Danish: 3%, Swedish: 1%, and Egyptian Arabic: 1%).
The structure of the free-text PRO documents included keywords/phrases, sentences, paragraphs, or the entire document/medical note. Different vocabulary systems representing standardized clinical terminologies or nomenclatures (e.g., SNOMED-CT (n=10) [35, 42–44, 47, 56, 57, 60, 73, 78], UMLS (n=17) [17, 25, 26, 31, 33, 36, 42–45, 47, 59, 64, 74, 76, 82], LOINC (n=1) [18], DSM (n=3) [59, 60, 73], ICD-codes (n=26) [11, 15, 17–20, 23, 28, 34, 36, 38, 40, 42, 43, 50, 53, 54, 61–63, 67, 68, 83–85]) were used to extract the representation of unstructured PROs. Among 37 studies that used a rule-based NLP approach, 14 adopted an extant rule-based NLP system, and 21 created study-specific/custom rules to extract the representation of unstructured PROs. Additionally, 20 studies used clinical/medical dictionaries to incorporate the standard clinical terminologies (e.g., LOINC, UMLS, SNOMED-CT) or created custom or study-specific terms/concepts [12, 14, 16, 28–31, 37–40, 55, 60, 64, 69, 72] for extracting unstructured PROs.
Main Objectives of the NLP/ML Application in Analyzing PRO Data
Table 1 summarizes the features of studies that used NLP/ML techniques to process unstructured PRO data. Most of the studies used NLP/ML algorithms to identify or extract unstructured PROs from clinical narratives (n=74 studies), map or classify extracted PRO information into specific PRO domains (n=16), phenotype unstructured PROs to capture specific PRO features (n=9), and classify co-occurrence of multiple PRO problems (e.g., symptom clustering; n=5). Some studies used NLP/ML techniques to analyze unstructured PRO to predict the risk of disease progression or adverse medical events (n=22), develop/validate NLP/ML pipelines for analyzing unstructured PROs (n=19), test associations with clinical outcomes (n=5), and stratify or group patients for delivering tailored healthcare services per distinct patterns of PRO problems (n=3).
Table 1:
Purpose of NLP/ML applications for analyzing PRO data among the 79 studies included in the systematic review*
| Classifications | Specific roles of NLP/ML tasks | Task description | N | % |
|---|---|---|---|---|
| Information extraction/text identification | 74 | 93.7 | ||
| PRO content detection, identification, extraction | Detect or identify PRO keywords or terminologies from free text | 47 | 59.5 | |
| PRO annotation | Perform semi-automated or manual annotation for PROs in free text | 37 | 46.8 | |
| PRO affirmation/ negation | Declare whether symptoms or symptom-related outcomes exist or equivalent expression or negative statement for having symptoms | 38 | 48.1 | |
| Vocabulary mapping | Map or assign PROs or PRO-related vocabulary words to appropriate indexes or labels | 13 | 16.5 | |
| Classification/phenotyping/clustering | 26 | 32.9 | ||
| PRO classification | Assign or classify extracted PROs into specific categories | 16 | 20.3 | |
| PRO phenotyping | Indicate specific characteristics of single or multiple PROs features | 9 | 11.4 | |
| PRO clustering | Identify two or more PROs that are related to each other or co-occur | 5 | 6.3 | |
| Develop or validate NLP/ML pipelines | 19 | 24.1 | ||
| Development of NLP/ML pipelines | Develop new NLP/ML pipelines or build NLP software | 10 | 12.7 | |
| Evaluation/validation | Evaluate and validate the performances of NLP system/pipeline | 12 | 15.2 | |
| Risk prediction or stratification for clinical outcomes | 25 | 31.6 | ||
| Risk prediction | Predict the risk of outcomes using extracted PROs based on unstructured narratives | 22 | 27.8 | |
| Risk stratification | Identify the right level of care and services for distinctive subgroups of patients. | 3 | 3.8 | |
| Investigate associations between PROs and clinical outcomes | 5 | 6.3 | ||
| Relationship detection | Detect semantic associations or relationships between unstructured PROs | 5 | 6.3 |
Each study may include multiple study purposes and NLP/ML tasks.
Domains of Unstructured PROs Included in EHRs
Supplementary Table S3 displays the domains of unstructured PROs documented in EHRs that were extracted by NLP/ML approaches. The most popular documented PRO domains were psychological symptoms (e.g., anxiety, depression, stress; 57.0% of 79 studies), followed by general symptoms (e.g., fatigue, pain, insomnia; 48.8%), metabolic/hormonal symptoms (e.g., obese, weight loss; 28.8%), and physical symptoms in the digestive/gastrointestinal (e.g., bloating, constipation, diarrhea; 42.5%), nervous (e.g., arousal, attention problem, dysarthria; 41.3%), respiratory (e.g., cough, sneezing, dyspnea; 37.5%), cardiovascular (e.g., cardiac problem, angina, swelling of arms and legs; 28.8%), and dermatologic (e.g., itching, edema, rashes; 20.0%) organ systems. Several PRO domains (e.g., symptoms in the head/neck, musculoskeletal, urinary, sexual/reproductive organ systems; physical and social functioning) were documented by <20% of the studies.
Types and Methods for Linguistic Features
Table 2 provides examples of linguistic feature types derived from unstructured PROs: 1) lexical, 2) syntactic, 3) semantic, and 4) contextual features. Lexical features address the word-level characteristics of clinical narratives primarily using n-grams (e.g., uni- and bi-grams) representing the concept of unstructured PROs [51, 57, 61, 74]. Syntactic features address the phrase, clause, sentences, and part-of-speech tagging [12, 31, 45, 56, 82]. While syntactic features represent grammatical patterns (e.g., noun and adjective/adverb phrases) [31, 45], semantic features focus on the meaning of words and terms, typically defined in a custom dictionary, controlled medical terminology, or lexicon (e.g., LOINC, SNOMED-CT, UMLS) [31, 39, 51, 59, 62]. Representing the relevance of specific words to individual texts, as opposed to their prevalence in a corpus, is another method used to process semantic features and semantic keywords for clustering the symptoms [62]. Contextual features address the relative or absolute characteristics of unstructured PROs by considering the linguistic components (i.e., words, phrases, and sentences) neighboring around words or phrases of interest rather than searching keywords of unstructured PROs [6, 7, 22, 51, 76]. One study shows that subject terms (e.g., “mother”, “patient”), negation terms (e.g., “does not”), hypothetical terms (e.g., “if”), temporal terms (e.g., “previously”, “when”), and termination terms (e.g., “however”) were examples of contextual analytic features [39]. Another study labeled the contextual feature as “if-clause” for the text “I recommended nitroglycerin if he should develop chest pain”[22].
Table 2:
Examples of frequently used linguistic features
| Levels of linguistic features | Description | Methods | Some example methods explained in selected studies |
|---|---|---|---|
| Lexical features | Numerical characteristics of tokens in text documents, such as token count and length. | N-gram (e.g., uni- and bi-grams), capitalization (uppercase, title case), stemming, lemmatization, stopwords removal, lexicon, word embeddings | “Stop-words removal (e.g., “is”, “an”, “the”, etc.), stemming, and number to string conversation.” [Banerjee, 2019] “Lexical variances in the extraction rules [i.e., misspellings (e.g., obese* instead of obsessive)].” [Chandran, 2019] “N-grams represent concepts of serious mental illness symptomatology.” [Jackson, 2018] “Text processing included lower casing; removal of punctuation, stop words, and numbers; word stemming; and tokenization.” [Obeid, 2020] |
| Syntactic features | Patterns of sentence structures defined by language grammar. | Part-of-speech (POS) tags, constituency grammar, dependency grammar | “POS tagger and multi-word term identification to identify symptoms and non-symptoms were used.” [Divita, 2017] “POS tags in conjunction with knowledge engineering features generated to build a sentence classifier.” [Jackson, 2017] “Syntactic phrases representative of patients’ functional status including noun phrases (e.g. “patient”), prepositional phrases (e.g. “with pain”), and adjective/adverb phrases (e.g. “very tired”) using two reference standards.” [Pakhomov, 2011] “Syntactic patterns of concept phrases were mined from continuous, non-permuted forms of synonyms, and these patterns were used to detect discontinuous and/or permuted concept phrases.” [Torii, 2018] |
| Semantic features | Linguistic units of meaning-holding components that represent word meaning, such as lexicon definitions, dependency between tokens, and semantic networks. | Semantic definitions from lexicons (LOINC, SNOMEDCT, UMLS, etc.), relative temporal words (next, later, until etc.), absolute temporal expressions (a.m., p.m., etc.), meaning of the numbers (doses, levels), deidentification, topics of the section | “Semantic variances in terms of obsessive and compulsive in the extraction (alternative meanings beyond their definition in the context of Obsessive Compulsive Symptoms (OCS).” [Chandran, 2019] “Semantic keywords identifying the Altered mental status cluster of symptoms in the context of pulmonary embolism.” [Obeid, 2019] “UMLS semantic networks which are relevant to clinical findings were used” [Torii, 2018] “The symptom dictionary was based on UMLS, which includes a semantic network.” [Le, 2018] |
| Contextual features | Linguistic neighboring components (e.g., word, phrase, or sentence) of tokens or sentences that represent similar semantic meanings. | Negation/affirmation, complex temporal relations, discourse structure, line position, order of sections, implicit context dependent information, feature representations from pre-trained neural embeddings | “Distinguishing between instances where a patient is described as experiencing a particular symptom from instances where the texts state that the patient is not experiencing that symptom, or where it is someone else (e.g. a friend or relative) who is experiencing that specific symptom.” [Chandran, 2019] “The “conditional” context label is considered when the term is mentioned in the following context (e.g., “I recommended nitroglycerin if he should develop chest pain”).” [Pakhomov, 2008] “Depending on the context, weight gain could indicate either fluid accumulation because of worsening heart failure or an improvement in appetite because of decreased gut edema associated with a higher dose of diuretics.” [Leiter, 2020] “Subject terms (e.g., ‘mother’, ‘patient’), negation terms (e.g., ‘does not’), hypothetical terms (e.g., ‘if’), temporal terms (e.g., ‘previously’) and termination terms (e.g., ‘however’).” [Iqbal, 2017] |
NLP Systems/Toolkits for Processing Unstructured PROs
Table 3 displays the NLP systems/toolkits used to process unstructured PROs in EHRs, inclusive of 15 generic (e.g., ConText, FMA, GATE, Hiedel Time, MALLET, NegEX, SUTime) and 11 clinical (e.g., MEDLEE, TextHunter, v3NLP, cTAKES, ClinREAD, Clamp, MetaMap, RedCap, V3NLP) systems/toolkits. Techniques utilized under each system/toolkit were mostly rule-based NLP (n=14 techniques), followed by hybrid NLP (n=8 techniques) and ML algorithms embedded in NLP (n=4 techniques). The rule-based approach uses rules created by experts to categorize or label unstructured PRO data [71, 86], and some studies validated the rules with unstructured data from different patient samples and subsequently applied the rules to the new samples [87]. ML-based approaches often train established classification algorithms with statistical inference techniques through previously annotated text-corpus [13]. Once ML algorithms learn the unstructured data of a new system, they can be applied to other lexical, semantic, and contextual meanings without referring to any rules [88]. In contrast, the hybrid approach adopts both rule- and ML-based methods, and integrates expert-generated, ruled-based systems to guide ML algorithms to perform the analysis [16, 45, 58]. Fifteen NLP systems/toolkits were rule-based (e.g., generic toolkits: ConText, FMA, Heidel Time, NegEX, Protégé, SUTime, and Wordnet; clinical toolkits: CliX NLP, ClinREAD, MedLEE, MedEx, NLP-PAC, and V3NLP). Specific ML-embedded systems/toolkits included MALLET and GENSIM with R, and hybrid systems/toolkits included GATE, UIMA, Clamp, cTAKES, MetaMap, and MedTagger. A full list of references is in Supplementary Table S4.
Table 3:
The reported NLP systems or toolkits*
| Systems/toolkits | Full Names | Purposes** | Techniques*** |
|---|---|---|---|
| Generic toolkits | |||
| ConText | N/A | Feature extraction and representation: sentence classification | Rule-based NLP |
| FMA | Freetext Matching Algorithm (FMA) | Preprocessing: annotation Feature extraction and representation: information extraction | Rule-based NLP |
| GATE | General Architecture for Text Engineering | Preprocessing: tokenization, sentence splitting, POS tagging, annotation Feature extraction and representation: name entity recognition, information extraction Non-neural ML: sentence classification | Hybrid NLP: rule-based, non-neural ML (support vector machine; SVM, WEKA ML) |
| GENSIM (R) | GENSIM | Feature extraction and representation: topic modeling and word embedding | Non-neural ML |
| Heidel Time | High quality rule-based extraction and normalization of temporal expressions | Preprocessing: tagging, normalization Feature extraction and representation: information extraction | Rule-based NLP |
| MALLET | MAchine Learning for LanguagE Toolkit | Feature extraction and representation: document classification, clustering, topic modeling, information extraction | Non-neural ML |
| NegEX | N/A | Feature extraction and representation: negation, affirmation | Rule-based NLP |
| Punkt Sentence Tokenizer | nltk.tokenize.punkt module | Preprocessing: tokenization | Non-neural ML |
| Protégé | Feature extraction and representation: ontology editor or framework | Rule-based NLP | |
| NLTK (Python) | Natural Language Toolkit | Preprocessing: text tokenization, stemming, stop word removal, classification, clustering, POS tagging, parsing, and semantic reasoning | Non-neural ML |
| SUTime | Stanford NLP annotator | Preprocessing: annotation, recognizing and normalizing time expressions (TIMEx) | Rule-based NLP |
| Tagger_Date Normalizer plugin | Not available | Preprocessing: tagging, normalization | Rule-based NLP |
| TextHunter | N/A | Preprocessing: tokenization, stemming, POS tagging Feature extraction and representation: information extraction Non-neural ML: automated concept identification | Hybrid NLP: rule-based, non-neural ML (SVM based “batch learning”) |
| WordNet | N/A | Lexical database for NLP (ontology) | Rule-based NLP |
| Clinical NLP toolkits | |||
| Clamp | Clinical Language Annotation, Modeling, and Processing Toolkit | Preprocessing: tokenization, POS tagging, annotation, sentence boundary detection, section header identification Feature extraction and representation: assertion, negation, name entity recognition, UMLS encoder Non-neural ML: sentence boundary detetion, section header identification, classification | Hybrid NLP: rule-based NLP, non-neural ML (conditional random fields, CRF). |
| cTAKES | clinical Text Analysis and Knowledge Extraction System | Preprocessing: sentence boundary detection, tokenization, parsing, dictionary lookup annotation, normalization, POS tagging Feature extraction and representation: affirmation, negationname d section identification, name entity recognition, information extraction Non-neural ML: classification of medical information | Hybrid NLP: rule-based NLP, non-neural ML (CRF, SVM) |
| CliX NLP | Clinical NLP tools for SNOMED CT | Feature extraction and representation: processing system based on SNOMEDCT | Rule-based NLP |
| ClinREAD | Rapid Clinical Note Mining for New Languages | Preprocessing: tokenization, POS tagging, vocabulary mapping Feature extraction and representation: name entity recognition | Rule-based NLP |
| MedLEE | Medical Language Extraction and Encoding System Processing System | Preprocessing: parsing Feature extraction: clinical entities extraction, assertions Non-neural ML: word disambiguation, classification of medical information, generate rules for classifying medical conditions | Hybrid NLP: rule-based NLP, non-neural ML (CRF, SVM) |
| MedTagger | N/A | Preprocessing: tokenization, POS tagging Feature extraction and representation: information extraction, assertion, negation Non-neural ML: sentence detection, concept identification, patient level risk factor classification | Hybrid NLP: rule-based NLP, non-neural ML (WEKA ML) |
| MetaMap | N/A | Preprocessing: vocabulary mapping, parsing, tokenization, POS tagging, sentence boundary determination Feature extraction and representation: lexical lookup of input words in the SPECIALIST lexicon – an information extraction system based on UMLS | Rule-based NLP |
| MTERMS | Medical Text Extraction, Reasoning and Mapping System | Preprocessing: parsing, tokenization, POS tagging, vocabulary mapping, information extraction Feature extraction and representation: affirmation, negation | Rule-based NLP |
| MedEx | Medication Information Extraction System for Clinical Narratives | Preprocessing: tokenizer, tagging, semantic tagger, parsing, encoding Feature extraction and representation: information extraction | Rule-based NLP |
| NLP-PAC | NLP algorithms for Predetermined Asthma Criteria | Feature extraction and representation: information extraction, affirmation, negation | Rule-based NLP |
| V3NLP | Not available | Preprocessing: annotation Feature extraction and representation: information extraction | Rule-based NLP |
See Supplementary Table S4 for a list of references;
The purpose of NLP systems/toolkits used to process unstructured PROs;
Hybrid NLP approach uses both rule-based and ML-based methods
NLP/ML Techniques for Processing Unstructured PROs
Table 4 reports the 3-step NLP/ML methods to process unstructured PROs from EHR clinical narratives: Step 1 is data pre-processing (n=60 studies), Step 2 is feature extraction and representations (n=69 studies), and Step 3 is data analysis (n=61 studies, including 39 using non-neural NLP/ML methods and 22 using neural NLP/ML methods). The step-by-step process was also summarized in Figure 2. The most common techniques for data pre-processing were annotation and text tokenization. The most common techniques for feature extraction and representations were rule-based NLP, affirmation/negation, and word2vec/bag-of-words (BOW). Regarding NLP/ML analytic methods, the most common non-neural ML methods were Support Vector Machine (SVM), decision tree, and Conditional Random Field (CRF). In contrast, commonly used neural NLP/ML methods included Convolutional Neural Network (CNN), Recurrent Neural Network (RNN; e.g., LSTM), and Artificial Neural Network (ANN or feed forward network). As an example of the way neural NLP/ML methods are used in clinical settings, the novel contextual embeddings of the BERT model use a context-based representation of chief complaints to predict specific signs/symptoms (e.g., pain, cognitive confusion) labeled by the experts and map semantically similar chief complaints to nearby points of vector space [7]. A full list of references was provided in Supplementary Table S5.
Table 4:
The 3-step NLP/ML application and corresponding techniques among the 79 studies included in the systematic review*
| Steps and techniques | N | % |
|---|---|---|
| Step 1: Preprocessing | 60 | 75.9 |
| Annotation | 38 | 48.1 |
| Text tokenization | 36 | 45.6 |
| Remove stop-words | 18 | 22.8 |
| Part-of-speech (POS) tagging | 16 | 20.3 |
| Normalization | 14 | 17.7 |
| Lemmatization/ stemming | 12 | 15.2 |
| Step 2: Feature extraction and representations | 69 | 87.3 |
| Rule-based NLP | 37 | 46.8 |
| Affirmation/ negation | 33 | 41.8 |
| Word2vec/ bag-of-words (BOW) | 23 | 29.1 |
| Name entity recognition (NER) | 16 | 20.3 |
| N-gram (Term Frequency–Inverse Document Frequency [TF-IDF], Document-Term Matrix [DTM], Term-Document Matrix [TDM]) | 15 | 19.0 |
| Latent Dirichlet Allocation (LDA) for topic modeling | 5 | 6.3 |
| Latent semantic indexing (LSI) | 1 | 1.3 |
| Knowledge graph | 1 | 1.3 |
| Step 3: Data analysis (non-neural ML) | 39 | 49.4 |
| Support vector machine (SVM) | 18 | 22.8 |
| Decision tree (DT) | 6 | 7.6 |
| Conditional random fields (CRF) | 9 | 11.4 |
| Logistic regression classifier | 8 | 10.1 |
| Naïve Bayesian | 6 | 7.6 |
| Random forest (RF) | 6 | 7.6 |
| K-means clustering | 3 | 3.8 |
| K-nearest neighborhood (KNN) | 3 | 3.8 |
| Boosting (e.g., Light Gradient Boosting Machine [LightGBM], eXtreme Gradient Boosting [XGBoost]) | 2 | 2.5 |
| Linear regression classifier | 2 | 2.5 |
| Bagging | 1 | 1.3 |
| Step 3: Data analysis (neural ML) | 22 | 27.8 |
| Convolutional neural network (CNN) | 10 | 12.7 |
| Recurrent Neural Network (RNN) (e.g., Bi-LSTM, GRU, Glove) | 10 | 12.7 |
| Artificial neural network (ANN) Feed forward network (FFN) | 7 | 8.9 |
| Transformer (e.g, Bidirectional Encoder Representations from Transformers [BERT], Bio-BERT) | 3 | 3.8 |
| Auto encoder | 3 | 3.8 |
| Embeddings from Language Model (ELMo) | 1 | 1.3 |
| Others | 2 | 2.5 |
Abbreviations: Bi-LSTM, Bi-Long Short-Term Memory; BERT, Bidirectional Encoder Representations from Transformers.
See Supplementary Table S5 for a list of references
Figure 2:
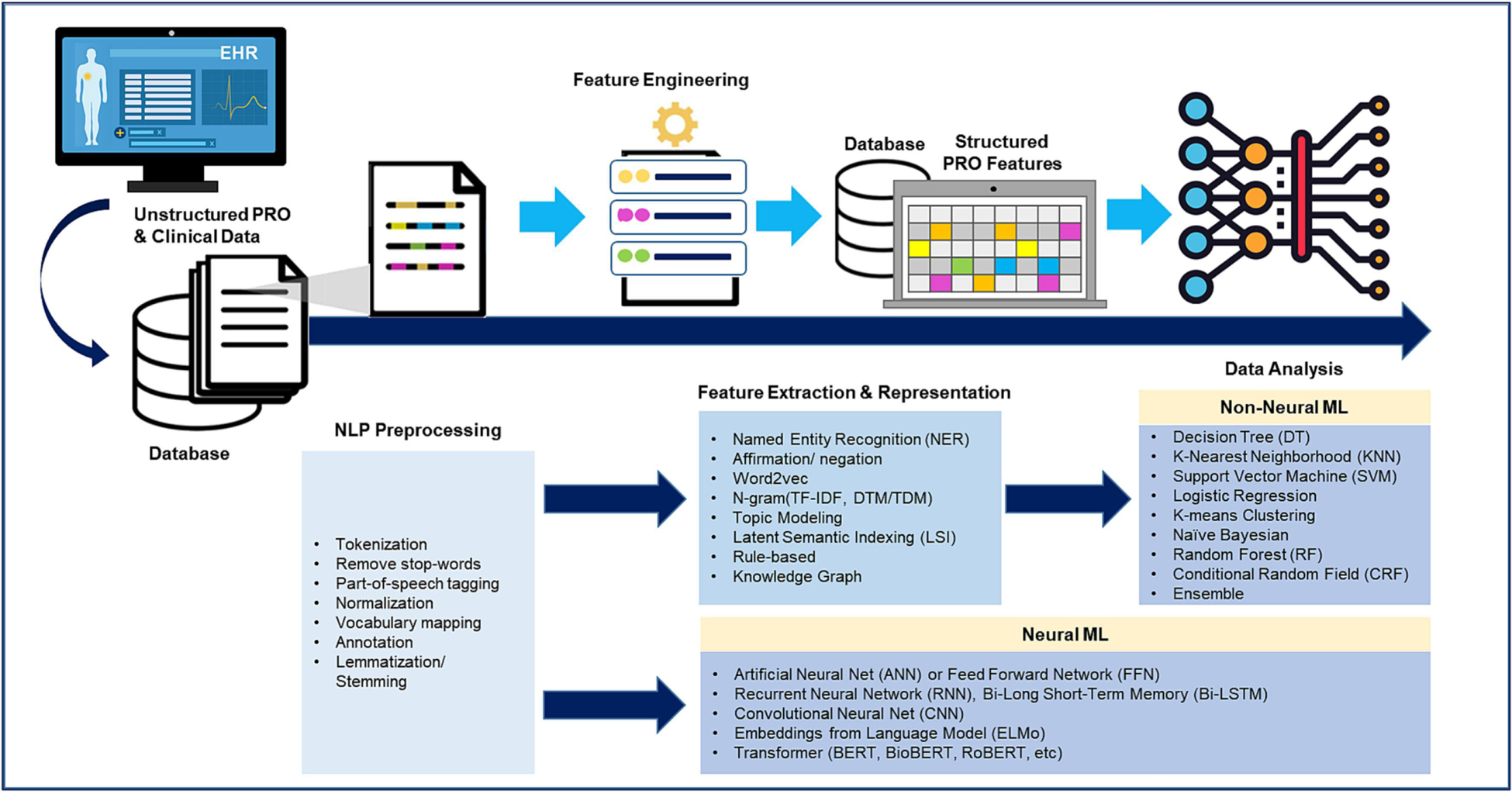
NLP/ ML pipeline for processing unstructured PRO data in EHRs
DISCUSSION
This systematic review study focuses on PRO-related studies and the findings have a significant contribution to the literature by summarizing the NLP/ML applications in PROs (e.g., classification, phenotyping, and clustering; PRO-based risk prediction and stratification for clinical outcomes), the available NLP systems or toolkits, and NLP/ML pipelines (e.g., preprocessing, feature extraction and representations, and data analysis using non-neural or neural ML methods). Among 79 selected studies, most studies (>90%) used NLP/ML techniques for extracting free-text PRO data, followed by predicting the risk of adverse events (30%), classifying, phenotyping or clustering PROs (20%), and testing associations between PROs and clinical outcomes (8%). Given the challenges of using standard surveys to assess PROs in busy clinics, as well as well-described barriers to PRO instrument application into routine clinical care [2], NLP/ML application provides a convenient mechanism to integrate PROs available in EHRs into clinical workflows for clinical decision-making [89].
We found that different types/units of unstructured PROs (e.g., keyword/phrase, sentence, paragraph, entire document/note) were used in NLP/ML analyses. Vocabularies from standard clinical terminologies or nomenclatures (e.g., SNOMED-CT, UMLS, LOINC, DSM-5, and ICD-codes) were commonly used to process unstructured PROs. These rule-based systems (e.g., ontologies, medical terminologies) were typically used to identify the meaning of the words and terms from free-text PROs [90]. Systematized Nomenclature of Medicine–Clinical Terms (SNOMED–CT) is deemed the most comprehensive computer collection of medical terms and medical relationships [91]. The feasibility of mapping other medical terminologies (e.g., ICD-9 or ICD-10 codes) to the SNOMED–CT makes the translation between different terminology systems feasible. To achieve semantic interoperability [90]. many studies used medical vocabulary systems to map PRO words or terminologies [13, 17, 25, 28, 36, 37, 40, 44, 53, 57, 62, 76, 78]. Additionally, to account for disease-specific content, some studies created dictionaries for incorporating existing clinical terminologies and/or added new terminologies to complement the functionality of extant rule-based or vocabulary systems [12, 14, 16, 28–31, 37–40, 55, 60, 64, 69, 72]. In the clinical setting, PRO data are often created by clinicians through handwriting or typing in an unstructured format and stored in EHRs, and then coders follow medical terminologies (e.g., ICD diagnosis code, SNOMED-CT, UMLS) or vocabulary systems to map or annotate the unstructured data into the structured format. Our review study details the implementation of different NLP/ML techniques for transforming unstructured PROs into vectorized structured formats, together with the ML techniques for clinical use (e.g., disease prediction).
The studies included in our review used 15 generic and 11 clinical NLP systems/toolkits to process unstructured PRO data and transformed information into structured PROs. NLP systems/toolkits typically transfer unstructured PROs into numerically computable information through pre-processing free-text PRO and clinical data, extracting specific features of the data, and then data normalization (i.e., converting a token into its base form). The systems/toolkits for free-text PRO processing included rule-based NLP, ML-based, or hybrid approaches. Practically, NLP/ML techniques were commonly used for feature extractions and representation (n=70), and nearly half of these studies (n=37) relied on rule-based methods [6, 16, 17, 20–22, 25–28, 30, 31, 33–36, 39, 45, 49–51, 55, 56, 58–60, 64, 70–72, 74, 76, 80, 81, 83–85]. However, the rule-based method is not efficient because it requires manual extraction of knowledge that may involve trial- and-error [92]. Furthermore, the accuracy of the rule-based model depends on appropriate and available rules and domain expertise. Few studies included in our present review found comparable or superior performance of ML-based or hybrid approaches (e.g., accuracy, precision, recall, F1) to that of the rule-based NLP approach [51, 53, 74, 75]. Technically, ML-based or hybrid approaches learn the patterns between phrases and sentences, which improves accuracy and generalizability in representing clinical narratives beyond rule-based approaches [16, 45, 58].
Modern neural network-based ML algorithms (e.g., RNN, transformer-style model) are a breakthrough for processing unstructured clinical narratives. Several studies in our review reported better performance of newer neural network-based ML algorithms (e.g., BERT, GPT-3) compared to traditional NLP or other neural network-based ML algorithms (e.g., ELMO, LSTM) [7, 24, 53]. The superior performance of the BERT model to other NLP/ML methods is because the BERT model uses effective pre-training methods (i.e., masked language modeling and next sentence prediction) and deploys multiple transformer layers to account for contextual information of natural language [93]. The BERT model has been shown to effectively capture linguistic features (i.e., syntactic, semantic, contextual features) [94]. In comparison, the traditional feature extraction and representation methods (e.g., bag-of-words, NER, n-gram) require domain knowledge to design complex feature engineering with less generalizability [95]. As a result, the semantic features derived from the BERT model may incorporate more contextualized meanings of words than traditional NLP models when contexts of words vary. Recently, clinical BERT models (e.g., Bio-BERT, Cancer-BERT, and BlueBERT) have been developed, which continue fine-tune BERT models through the corpus of PubMed, MIMIC, or other clinical sources, to meet different purposes [96]. These findings suggest the usefulness of domain-specific neural network-based ML methods for processing unstructured PROs in the future.
While this review collects studies prior to 2021, we have covered the state-of-the-art NLP models (e.g., RNN, Bio-BERT, and Cancer-BERT) and those techniques have not been changed in PRO-related studies since 2020. The recent rise of deep learning approaches typically brings novel embedding techniques that can jointly integrate lexical, syntactic, semantic, and contextual patterns into unified vectors, which significantly reduces labor expenses and domain knowledge requirements of feature engineering and design. The core idea of developing embedding techniques is to use context to define language and therefore train embedding models (i.e., neural networks). Co-occurrence, word prediction, and neighboring sentence matching are useful approaches [94, 97] to obtain the embedding models for PRO research. Studies building deep learning models [6, 14, 50, 53, 55, 62] for PRO assessment benefit from the power and flexibility of embedding techniques to represent features from token to document levels [7, 24, 50, 53, 96]. Those studies have demonstrated their success in embedding techniques to learn representative features of free-text data in PRO research, such as phenotype inference [50], information extraction [14], and diagnosis [53]. Nevertheless, ML models trained on word embeddings output tables of weights can make debugging missed predictions more challenging compared to the models trained on simpler linguistic features.
Though various NLP systems/toolkits used to analyze unstructured PROs from clinical narratives have been reported [22, 27, 45, 48, 67, 81], the validity of using unstructured PROs derived from clinical narratives versus standard PRO surveys remains unclear. The standard PRO surveys may likely measure generic PRO concepts across all patients, whereas unstructured PROs from clinical narratives can capture patient-unique PROs that may not be included in the standard PRO surveys. Several studies in our review used PRO data collected from the survey to evaluate the validity of NLP/ML pipelines for unstructured PROs [22, 43, 45, 67]. Agreement of PRO data between data collected from self-reports and EHR-based unstructured PROs analyzed with NLP/ML ranged from a range of 67–82% [45]. A similar finding (63–75% agreement) was found in the patient responses to standard surveys versus unstructured symptom data (e.g., chest pain, dyspnea, cough) documented in the physicians’ medical notes [22]. Several studies have noted the value of unstructured PROs as anchors in predicting or correlating various clinical outcomes (e.g., disease onset, suicidal ideation, readmission, mortality) [20, 30, 33, 59, 61, 68, 79, 83]. These findings suggest that unstructured PROs may be a surrogate for the standard PRO surveys [93].
The findings of this review study have important implications for future clinical research and care. Although NLP/ML techniques are evolving, successful NLP/ML application requires the implementation of integrated platforms that seamlessly interconnect EHR functionality and NLP/ML algorithms to facilitate clinical interpretation. Evidence shows that integrating unstructured PROs into EHRs likely improves cancer survivorship care by predicting late effects based on worsening symptoms and other clinical data [7, 21, 30, 33, 44, 61, 63, 75, 79, 84]. However, there is a critical need to find novel platforms/systems for integrating unstructured PROs into EHRs and using NLP/ML techniques to annotate unstructured PROs. Further effort is warranted to create meaningful PRO scores from free-text narratives based on the features derived from NLP/ML to be comparably interpretable to scores from standard PRO surveys.
There are implementation barriers to collecting and integrating unstructured PROs from EHRs for NLP/ML applications. The major barriers include technical complexity, system interoperability, and concerns about the quality of unstructured clinical narratives (e.g., fragmented or incomplete words or sentences to represent a patient’s health status or symptoms) in EHRs [98]. Adopting the Common Data Model or Common Data Warehouse to enhance model portability [99] may improve the collection of standard and high-quality unstructured PRO data from clinical narratives. Instead of relying on text-based PRO data, alternative methods include the use of speech or voice recognition systems for collecting unstructured PROs and developing audio-based NLP/ML pipelines to automatically analyze PRO information from patient-doctor conversations in clinics. There are different ways to manage conversation-based PROs: 1) using NLP/ML algorithms to automatically annotate and analyze the conversation-based PROs and 2) using software to transcribe voice data, followed by NLP/ML to annotate and analyze the transcribed data. If systems were set up appropriately, acoustic recordings of clinical interactions would involve less paperwork and be less prone to data collection artifacts. However, compared to text, audio contains richer information (e.g., prosody) that humans process together with lexical meaning to alter language’s propositional content. For example, an ironic phrasing of “yeah, that medication worked” captured in audio would be interpreted as a declaration of efficacy if it were simply converted into text. Once the voice-based PRO data are annotated, the NLP/ML pipelines could be applied for data analysis and clinical application. Few-shot learning, or prompting, could also be used to improve parts of PRO prediction by significantly augmenting the amount of relevant ground truth training data [100].
This review study contains some limitations. First, several selected studies did not report specific information to meet our inclusion criteria (e.g., frequently used linguistic features); therefore, the results may not fully reflect the status of NLP/ML applications in unstructured PROs. Second, we did not evaluate the quality of the selected studies because the standards or guidelines for evaluating NLP/ML applications in unstructured PROs have yet been established. Finally, we included studies that were published by the end of 2020, while NLP/ML techniques evolve day by day. However, to our best knowledge, our study has covered the latest trends and techniques of applying NLP/ML techniques (e.g., BERT) for researching unstructured PROs. We are aware of large generative language models (e.g., ChatGPT), however, those newer generative AI techniques have not been applied in recent PRO-related studies.
While the NLP revolution, specifically generative AI, has taken place in the past 2–3 years, our literature search until 2023 found no instances of generative AI being applied to unstructured PROs. Large language models become the promising direction for future PROs studies, and contemporary NLP techniques (e.g., BERT or GPT) discussed in this review study can provide a solid background for future PROs research. One notable example of generative AI is ChatGPT (e.g., GPT-4 developed by OpenAI) which aims to generate a patient summary using information provided by clinicians or patients and to facilitate interactive text-based communication with users. It is important to note that although ChatGPT can address potential bias in the training data, it has not undergone a comprehensive fine-tuning and validation process for PROs and other medical data. Moreover, its integration into EHRs for annotating and analyzing vast amounts of unstructured PROs in medical notes remains uncertain [101–103]. Currently, ChatGPT is valuable for providing a PRO summary based on the data provided by clinicians or patients, but it should not be relied upon for clinical interpretation or decision-making until empirical studies establish its validity. We believe the methods and resources of traditional NLP/ML reported in this review will be valuable to inform the generative NLP approaches (e.g., GPT) of how to incorporate domain knowledge and promote evidence-based PRO inferences from unstructured medical data. Our present recommendation is to employ established, validated NLP/ML approaches for feature extraction, including rule-based or neural machine/deep learning algorithms, and utilize NLP systems/toolkits for the analysis of unstructured PROs within EHRs.
CONCLUSION
This systematic review study reports the usefulness of NLP/ML techniques in processing unstructured PRO data. Currently, using the established rules to annotate unstructured PROs through NLP/ML systems/toolkits is the dominant method, though the use of novel neural ML-based methods is increasing. Transformer NLP/ML models (e.g., BERT) are the most cutting-edge and dominating techniques to process unstructured PROs in EHRs. Although we did not come across any studies utilizing generative large language models like ChatGPT for extracting and analyzing unstructured PROs and integrated into EHRs, the rapid advancements in generative AI offer a potential opportunity for future exploration and evaluation in this area.
Supplementary Material
Highlights.
Most of the studies used NLP/ML techniques to extract PROs from clinical narratives (81%), map the extracted PROs into specific PRO domains for phenotyping or clustering purposes (33%), process PROs for predicting disease progression or onset of adverse events (28%) or develop/validate NLP/ML pipelines for analyzing unstructured PROs (24%).
Using annotation rules for NLP/ML to analyze unstructured PROs is dominant, yet deploying novel neural ML-based methods is warranted.
FUNDING STATEMENT
The research reported in this manuscript was supported by the U.S. National Cancer Institute under award numbers U01CA195547 (Hudson/Ness), R01CA238368 (Huang/Baker), and R01CA258193 (Huang/Yasui). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST STATEMENT
All co-authors declare no conflict of interest.
DATA AVAILABILITY
Extracted data for this systematic review is available upon request.
REFERENCES
- [1].Wilson IB, Cleary PD. Linking clinical variables with health-related quality of life. A conceptual model of patient outcomes. JAMA. 1995;273:59–65. [PubMed] [Google Scholar]
- [2].Foster A, Croot L, Brazier J, Harris J, O’Cathain A. The facilitators and barriers to implementing patient reported outcome measures in organisations delivering health related services: a systematic review of reviews. J Patient Rep Outcomes. 2018;2:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alzu’bi AA, Watzlaf VJM, Sheridan P. Electronic Health Record (EHR) Abstraction. Perspect Health Inf Manag. 2021;18:1g. [PMC free article] [PubMed] [Google Scholar]
- [4].Kong HJ. Managing Unstructured Big Data in Healthcare System. Healthc Inform Res. 2019;25:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gonzalez-Hernandez G, Sarker A, O’Connor K, Savova G. Capturing the Patient’s Perspective: a Review of Advances in Natural Language Processing of Health-Related Text. Yearb Med Inform. 2017;26:214–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Leiter RE, Santus E, Jin Z, Lee KC, Yusufov M, Chien I, et al. Deep Natural Language Processing to Identify Symptom Documentation in Clinical Notes for Patients With Heart Failure Undergoing Cardiac Resynchronization Therapy. J Pain Symptom Manage. 2020;60:948–58 e3. [DOI] [PubMed] [Google Scholar]
- [7].Chang D, Hong WS, Taylor RA. Generating contextual embeddings for emergency department chief complaints. JAMIA Open. 2020;3:160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kreimeyer K, Foster M, Pandey A, Arya N, Halford G, Jones SF, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. J Biomed Inform. 2017;73:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dreisbach C, Koleck TA, Bourne PE, Bakken S. A systematic review of natural language processing and text mining of symptoms from electronic patient-authored text data. Int J Med Inform. 2019;125:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Koleck TA, Dreisbach C, Bourne PE, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc 2019;26:364–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chapman AB, Peterson KS, Alba PR, DuVall SL, Patterson OV. Detecting Adverse Drug Events with Rapidly Trained Classification Models. Drug Saf. 2019;42:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Divita G, Luo G, Tran LT, Workman TE, Gundlapalli AV, Samore MH. General Symptom Extraction from VA Electronic Medical Notes. Stud Health Technol Inform. 2017;245:356–60. [PubMed] [Google Scholar]
- [13].Fodeh SJ, Finch D, Bouayad L, Luther SL, Ling H, Kerns RD, et al. Classifying clinical notes with pain assessment using machine learning. Med Biol Eng Comput. 2018;56:1285–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gong L, Zhang Z, Chen S. Clinical Named Entity Recognition from Chinese Electronic Medical Records Based on Deep Learning Pretraining. J Healthc Eng. 2020;2020:8829219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu BT, Bajracharya A, Yu H. Generating Medical Assessments Using a Neural Network Model: Algorithm Development and Validation. JMIR Med Inf. 2020;8:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ji B, Liu R, Li S, Yu J, Wu Q, Tan Y, et al. A hybrid approach for named entity recognition in Chinese electronic medical record. BMC Med Inform Decis Mak. 2019;19:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karagounis S, Sarkar IN, Chen ES. Coding Free-Text Chief Complaints from a Health Information Exchange: A Preliminary Study. AMIA Annu Symp Proc. 2020;2020:638–47. [PMC free article] [PubMed] [Google Scholar]
- [18].Landi I, Glicksberg BS, Lee HC, Cherng S, Landi G, Danieletto M, et al. Deep representation learning of electronic health records to unlock patient stratification at scale. Npj Digit Med. 2020;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McCoy TH, Wiste AK, Doyle AE, Pellegrini AM, Perlis RH. Association between child psychiatric emergency room outcomes and dimensions of psychopathology. Gen Hosp Psychiat. 2019;59:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McCoy TH, Castro VM, Rosenfield HR, Cagan A, Kohane IS, Perlis RH. A clinical perspective on the relevance of research domain criteria in electronic health records. Am J Psychiatry. 2015;172:316–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Owlia M, Dodson JA, King JB, Derington CG, Herrick JS, Sedlis SP, et al. Angina Severity, Mortality, and Healthcare Utilization Among Veterans With Stable Angina. J Am Heart Assoc. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pakhomov S, Jacobsen SJ, Chute CG, Roger VL. Agreement between patient-reported symptoms and their documentation in the medical record. Am J Manag Care. 2008;14:530. [PMC free article] [PubMed] [Google Scholar]
- [23].Shao YJ, Zeng QT, Chen KK, Shutes-David A, Thielke SM, Tsuang DW. Detection of probable dementia cases in undiagnosed patients using structured and unstructured electronic health records. BMC Med Inform Decis. 2019;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Steinkamp JM, Bala W, Sharma A, Kantrowitz JJ. Task definition, annotated dataset, and supervised natural language processing models for symptom extraction from unstructured clinical notes. J Biomed Inform. 2020;102:103354. [DOI] [PubMed] [Google Scholar]
- [25].Tang H, Solti I, Kirkendall E, Zhai H, Lingren T, Meller J, et al. Leveraging Food and Drug Administration Adverse Event Reports for the Automated Monitoring of Electronic Health Records in a Pediatric Hospital. Biomed Inform Insights. 2017;9:1178222617713018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang X, Chused A, Elhadad N, Friedman C, Markatou M. Automated knowledge acquisition from clinical narrative reports. AMIA Annu Symp Proc. 2008:783–7. [PMC free article] [PubMed] [Google Scholar]
- [27].Wang L, Wang Q, Bai H, Liu C, Liu W, Zhang Y, et al. EHR2Vec: Representation Learning of Medical Concepts From Temporal Patterns of Clinical Notes Based on Self-Attention Mechanism. Front Genet. 2020;11:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yehia E, Boshnak H, AbdelGaber S, Abdo A, Elzanfaly DS. Ontology-based clinical information extraction from physician’s free-text notes. J Biomed Inform. 2019;98:103276. [DOI] [PubMed] [Google Scholar]
- [29].Zhang ZC, Zhang Y, Zhou T, Pang YL. Medical assertion classification in Chinese EMRs using attention enhanced neural network. Math Biosci Eng. 2019;16:1966–77. [DOI] [PubMed] [Google Scholar]
- [30].Zhang H, Ni W, Li J, Zhang J. Artificial Intelligence-Based Traditional Chinese Medicine Assistive Diagnostic System: Validation Study. JMIR Med Inform. 2020;8:e17608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Torii M, Yang, Elly W, Doan, Son. A preliminary study of clinical concept detection using syntactic relations. AMIA Annu Symp Proc. 2018;2018:1028. [PMC free article] [PubMed] [Google Scholar]
- [32].Wang Y, Yu Z, Chen L, Chen Y, Liu Y, Hu X, et al. Supervised methods for symptom name recognition in free-text clinical records of traditional Chinese medicine: an empirical study. J Biomed Inform. 2014;47:91–104. [DOI] [PubMed] [Google Scholar]
- [33].Ye JC, Yao L, Shen JH, Janarthanam R, Luo Y. Predicting mortality in critically ill patients with diabetes using machine learning and clinical notes. BMC Med Inform Decis. 2020;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Byrd RJ, Steinhubl SR, Sun J, Ebadollahi S, Stewart WF. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. Int J Med Inform. 2014;83:983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chan LL, Beers K, Yau AA, Chauhan K, Duffy A, Chaudhary K, et al. Natural language processing of electronic health records is superior to billing codes to identify symptom burden in hemodialysis patients. Kidney Int. 2020;97:383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chase HS, Mitrani LR, Lu GG, Fulgieri DJ. Early recognition of multiple sclerosis using natural language processing of the electronic health record. BMC Med Inform Decis. 2017;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ford E, Carroll J, Smith H, Davies K, Koeling R, Petersen I, et al. What evidence is there for a delay in diagnostic coding of RA in UK general practice records? An observational study of free text. BMJ open. 2016;6:e010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Geva A, Abman SH, Manzi SF, Ivy DD, Mullen MP, Griffin J, et al. Adverse drug event rates in pediatric pulmonary hypertension: a comparison of real-world data sources. J Am Med Inform Assoc. 2020;27:294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Iqbal E, Mallah R, Rhodes D, Wu H, Romero A, Chang N, et al. ADEPt, a semantically-enriched pipeline for extracting adverse drug events from free-text electronic health records. PLoS One. 2017;12:e0187121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kirk IK, Simon C, Banasik K, Holm PC, Haue AD, Jensen PB, et al. Linking glycemic dysregulation in diabetes to symptoms, comorbidities, and genetics through EHR data mining. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McCoy TH, Han L, Pellegrini AM, Tanzi RE, Berretta S, Perlis RH. Stratifying risk for dementia onset using large-scale electronic health record data: A retrospective cohort study. Alzheimers Dement. 2020;16:531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nagamine T, Gillette B, Pakhomov A, Kahoun J, Mayer H, Burghaus R, et al. Multiscale classification of heart failure phenotypes by unsupervised clustering of unstructured electronic medical record data. Sci Rep. 2020;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pakhomov SS, Hemingway H, Weston SA, Jacobsen SJ, Rodeheffer R, Roger VL. Epidemiology of angina pectoris: role of natural language processing of the medical record. Am Heart J. 2007;153:666–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pakhomov S, Shah N, Hanson P, Balasubramaniam S, Smith SA. Automatic quality of life prediction using electronic medical records. AMIA Annu Symp Proc. 2008;2008:545. [PMC free article] [PubMed] [Google Scholar]
- [45].Pakhomov SV, Shah ND, Van Houten HK, Hanson PL, Smith SA. The role of the electronic medical record in the assessment of health related quality of life. AMIA Annu Symp Proc. 2011;2011:1080–8. [PMC free article] [PubMed] [Google Scholar]
- [46].Park SY, Camilleri M, Packer D, Monahan K. Upper gastrointestinal complications following ablation therapy for atrial fibrillation. Neurogastroenterol Motil. 2017;29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Topaz M, Adams V, Wilson P, Woo K, Ryvicker M. Free-Text Documentation of Dementia Symptoms in Home Healthcare: A Natural Language Processing Study. Gerontol Geriatr Med. 2020;6:2333721420959861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vijayakrishnan R, Steinhubl SR, Ng K, Sun J, Byrd RJ, Daar Z, et al. Prevalence of heart failure signs and symptoms in a large primary care population identified through the use of text and data mining of the electronic health record. J Card Fail. 2014;20:459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wi CI, Sohn S, Rolfes MC, Seabright A, Ryu E, Voge G, et al. Application of a Natural Language Processing Algorithm to Asthma Ascertainment. An Automated Chart Review. Am J Respir Crit Care Med. 2017;196:430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yang Z, Dehmer M, Yli-Harja O, Emmert-Streib F. Combining deep learning with token selection for patient phenotyping from electronic health records. Sci Rep. 2020;10:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chandran D, Robbins DA, Chang CK, Shetty H, Sanyal J, Downs J, et al. Use of natural language processing to identify obsessive compulsive symptoms in patients with schizophrenia, schizoaffective disorder or bipolar disorder. Sci Rep. 2019;9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Colling C, Khondoker M, Patel R, Fok M, Harland R, Broadbent M, et al. Predicting high-cost care in a mental health setting. BJPsych Open. 2020;6:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dai HJ, Su CH, Lee YQ, Zhang YC, Wang CK, Kuo CJ, et al. Deep Learning-Based Natural Language Processing for Screening Psychiatric Patients. Front Psychiatry. 2020;11:533949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Downs J, Dean H, Lechler S, Sears N, Patel R, Shetty H, et al. Negative Symptoms in Early-Onset Psychosis and Their Association With Antipsychotic Treatment Failure. Schizophr Bull. 2019;45:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Geraci J, Wilansky P, de Luca V, Roy A, Kennedy JL, Strauss J. Applying deep neural networks to unstructured text notes in electronic medical records for phenotyping youth depression. Evid Based Ment Health. 2017;20:83–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jackson RG, Patel R, Jayatilleke N, Kolliakou A, Ball M, Gorrell G, et al. Natural language processing to extract symptoms of severe mental illness from clinical text: the Clinical Record Interactive Search Comprehensive Data Extraction (CRIS-CODE) project. BMJ Open. 2017;7:e012012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jackson R, Patel R, Velupillai S, Gkotsis G, Hoyle D, Stewart R. Knowledge discovery for Deep Phenotyping serious mental illness from Electronic Mental Health records. F1000Research. 2018;7:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Karystianis G, Nevado AJ, Kim CH, Dehghan A, Keane JA, Nenadic G. Automatic mining of symptom severity from psychiatric evaluation notes. Int J Methods Psychiatr Res. 2018;27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Le DV, Montgomery J, Kirkby KC, Scanlan J. Risk prediction using natural language processing of electronic mental health records in an inpatient forensic psychiatry setting. J Biomed Inform. 2018;86:49–58. [DOI] [PubMed] [Google Scholar]
- [60].Liu Q, Woo M, Zou X, Champaneria A, Lau C, Mubbashar MI, et al. Symptom-based patient stratification in mental illness using clinical notes. J Biomed Inform. 2019;98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].McCoy TH Jr., Yu S, Hart KL, Castro VM, Brown HE, Rosenquist JN, et al. High Throughput Phenotyping for Dimensional Psychopathology in Electronic Health Records. Biol Psychiatry. 2018;83:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Obeid JS, Weeda ER, Matuskowitz AJ, Gagnon K, Crawford T, Carr CM, et al. Automated detection of altered mental status in emergency department clinical notes: a deep learning approach. BMC Med Inform Decis Mak. 2019;19:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Obeid JS, Dahne J, Christensen S, Howard S, Crawford T, Frey LJ, et al. Identifying and Predicting Intentional Self-Harm in Electronic Health Record Clinical Notes: Deep Learning Approach. JMIR Med Inform. 2020;8:e17784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Parthipan A, Banerjee I, Humphreys K, Asch SM, Curtin C, Carroll I, et al. Predicting inadequate postoperative pain management in depressed patients: A machine learning approach. PLoS One. 2019;14:e0210575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Patel R, Lloyd T, Jackson R, Ball M, Shetty H, Broadbent M, et al. Mood instability is a common feature of mental health disorders and is associated with poor clinical outcomes. BMJ Open. 2015;5:e007504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Patel R, Jayatilleke N, Broadbent M, Chang C-K, Foskett N, Gorrell G, et al. Negative symptoms in schizophrenia: a study in a large clinical sample of patients using a novel automated method. BMJ Open. 2015;5:e007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Perlis R, Iosifescu D, Castro V, Murphy S, Gainer V, Minnier J, et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med. 2012;42:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rumshisky A, Ghassemi M, Naumann T, Szolovits P, Castro VM, McCoy TH, et al. Predicting early psychiatric readmission with natural language processing of narrative discharge summaries. Transl Psychiatry. 2016;6:e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sorup FKH, Eriksson R, Westergaard D, Hallas J, Brunak S, Ejdrup Andersen S. Sex differences in text-mined possible adverse drug events associated with drugs for psychosis. J Psychopharmacol. 2020;34:532–9. [DOI] [PubMed] [Google Scholar]
- [70].Viani N, Kam J, Yin L, Verma S, Stewart R, Patel R, et al. Annotating Temporal Relations to Determine the Onset of Psychosis Symptoms. Stud Health Technol Inform. 2019;264:418–22. [DOI] [PubMed] [Google Scholar]
- [71].Viani N, Kam J, Yin L, Bittar A, Dutta R, Patel R, et al. Temporal information extraction from mental health records to identify duration of untreated psychosis. J Biomed Semantics. 2020;11:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wu CS, Kuo CJ, Su CH, Wang SH, Dai HJ. Using text mining to extract depressive symptoms and to validate the diagnosis of major depressive disorder from electronic health records. J Affect Disorders. 2020;260:617–23. [DOI] [PubMed] [Google Scholar]
- [73].Zhou L, Baughman AW, Lei VJ, Lai KH, Navathe AS, Chang F, et al. Identifying Patients with Depression Using Free-text Clinical Documents. Stud Health Technol. 2015;216:629–33. [PubMed] [Google Scholar]
- [74].Banerjee I, Li K, Seneviratne M, Ferrari M, Seto T, Brooks JD, et al. Weakly supervised natural language processing for assessing patient-centered outcome following prostate cancer treatment. JAMIA Open. 2019;2:150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Forsyth AW, Barzilay R, Hughes KS, Lui D, Lorenz KA, Enzinger A, et al. Machine Learning Methods to Extract Documentation of Breast Cancer Symptoms From Electronic Health Records. J Pain Symptom Manage. 2018;55:1492–9. [DOI] [PubMed] [Google Scholar]
- [76].Heintzelman NH, Taylor RJ, Simonsen L, Lustig R, Anderko D, Haythornthwaite JA, et al. Longitudinal analysis of pain in patients with metastatic prostate cancer using natural language processing of medical record text. J Am Med Inform Assoc. 2013;20:898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Hyun S, Johnson SB, Bakken S. Exploring the ability of natural language processing to extract data from nursing narratives. Comput Inform Nurs. 2009;27:215–23; quiz 24–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hong JC, Fairchild AT, Tanksley JP, Palta M, Tenenbaum JD. Natural language processing for abstraction of cancer treatment toxicities: accuracy versus human experts. JAMIA Open. 2020;3:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jensen K, Soguero-Ruiz C, Oyvind Mikalsen K, Lindsetmo RO, Kouskoumvekaki I, Girolami M, et al. Analysis of free text in electronic health records for identification of cancer patient trajectories. Sci Rep. 2017;7:46226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tamang S, Patel MI, Blayney DW, Kuznetsov J, Finlayson SG, Vetteth Y, et al. Detecting unplanned care from clinician notes in electronic health records. J Oncol Pract. 2015;11:e313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Weegar R, Kvist M, Sundström K, Brunak S, Dalianis H. Finding cervical cancer symptoms in Swedish clinical text using a machine learning approach and NegEx. AMIA Annu Symp Proc. 2015;2015:1296. [PMC free article] [PubMed] [Google Scholar]
- [82].Deleger L, Brodzinski H, Zhai H, Li Q, Lingren T, Kirkendall ES, et al. Developing and evaluating an automated appendicitis risk stratification algorithm for pediatric patients in the emergency department. J Am Med Inform Assoc. 2013;20:e212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].McCoy TH, Pellegrini AM, Perlis RH. Research Domain Criteria scores estimated through natural language processing are associated with risk for suicide and accidental death. Depress Anxiety. 2019;36:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hane CA, Nori VS, Crown WH, Sanghavi DM, Bleicher P. Predicting Onset of Dementia Using Clinical Notes and Machine Learning: Case-Control Study. JMIR Med Inf. 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shah AD, Bailey E, Williams T, Denaxas S, Dobson R, Hemingway H. Natural language processing for disease phenotyping in UK primary care records for research: a pilot study in myocardial infarction and death. J Biomed Semantics. 2019;10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Liu H, Gegov A, Cocea M. Rule-based systems: a granular computing perspective. Granular Computing. 2016;1:259–74. [Google Scholar]
- [87].Aubaid AM, Mishra A. A Rule-Based Approach to Embedding Techniques for Text Document Classification. Appl Sci. 2020;10:4009. [Google Scholar]
- [88].Khurana D, Koli A, Khatter K, Singh S. Natural language processing: state of the art, current trends and challenges. Multimed Tools Appl. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Velupillai S, Suominen H, Liakata M, Roberts A, Shah AD, Morley K, et al. Using clinical Natural Language Processing for health outcomes research: Overview and actionable suggestions for future advances. J Biomed Inform. 2018;88:11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Tayefi M, Ngo P, Chomutare T, Dalianis H, Salvi E, Budrionis A, et al. Challenges and opportunities beyond structured data in analysis of electronic health records. WIREs Comput Stat. 2021;13:e1549. [Google Scholar]
- [91].Gaudet-Blavignac C, Foufi V, Bjelogrlic M, Lovis C. Use of the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) for Processing Free Text in Health Care: Systematic Scoping Review. J Med Internet Res. 2021;23:e24594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cronin R, Fabbri D, Denny J, Rosenbloom S, Jackson G. A comparison of rule-based and machine learning approaches for classifying patient portal messages. Int J Med Inform. 2017;105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lu Z, Sim JA, Wang JX, Forrest CB, Krull KR, Srivastava D, et al. Natural Language Processing and Machine Learning Methods to Characterize Unstructured Patient-Reported Outcomes: Validation Study. J Med Internet Res. 2021;23:e26777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv:181004805. 2018. [Google Scholar]
- [95].Gasparetto A, Marcuzzo M, Zangari A, Albarelli A. A Survey on Text Classification Algorithms: From Text to Predictions. Information. 2022;13:83. [Google Scholar]
- [96].Zhou S, Wang N, Wang L, Liu H, Zhang R. CancerBERT: a cancer domain-specific language model for extracting breast cancer phenotypes from electronic health records. J Am Med Inform Assoc. 2022;29:1208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst. 2013;26. [Google Scholar]
- [98].Ajami S, Arab-Chadegani R. Barriers to implement Electronic Health Records (EHRs). Mater Sociomed. 2013;25:213–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Schneeweiss S, Brown JS, Bate A, Trifirò G, Bartels DB. Choosing Among Common Data Models for Real-World Data Analyses Fit for Making Decisions About the Effectiveness of Medical Products. Clin Pharmacol Ther. 2020;107:827–33. [DOI] [PubMed] [Google Scholar]
- [100].Pereg D, Villiger M, Bouma B, Golland P. Less is More: Rethinking Few-Shot Learning and Recurrent Neural Nets. arXiv:220914267. 2022. [Google Scholar]
- [101].Cascella M, Montomoli J, Bellini V, Bignami E. Evaluating the Feasibility of ChatGPT in Healthcare: An Analysis of Multiple Clinical and Research Scenarios. J Med Syst. 2023;47:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Baumgartner C The potential impact of ChatGPT in clinical and translational medicine. Clin Transl Med. 2023;13:e1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Thirunavukarasu AJ, Ting DSJ, Elangovan K, Gutierrez L, Tan TF, Ting DSW. Large language models in medicine. Nat Med. 2023;29:1930–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Extracted data for this systematic review is available upon request.


