Abstract
Deamidation is a major age-related modification in the human lens that is highly prevalent in crystallins isolated from the insoluble fraction of cataractous lenses and also causes protein aggregation in vitro. However, the mechanism by which deamidation causes proteins to become insoluble is not known, because of only subtle structural changes observed in vitro. We have identified Asn14 and Asn76 of γS-crystallin as highly deamidated in insoluble proteins isolated from aged lenses. These sites are on the surface of the N-terminal domain and were mimicked by replacing the Asn with Asp residues in order to generate recombinant human γS and deamidated mutants. Both N14D and N76D had increased light scattering compared to wildtype-γS and increased aggregation during thermal-induced denaturation. Aggregation was enhanced by oxidized glutathione suggesting deamidation may increase susceptibility to disulfide bonding. These changes were correlated to changes in protein dynamics determined by NMR spectroscopy. Heteronuclear NMR spectroscopy was used to measure amide hydrogen exchange and 15N relaxation dynamics to identify regions with increased dynamics compared to wildtype-γS. Residue-specific changes in solvent accessibility and dynamics identified significant changes both near and distant from the sites of deamidation, suggesting that deamidation had both local and global effects on the protein structure at slow (ms to s) and fast (μs to ps) time scales. Thus, a potential mechanism for γS deamidation-induced insolubilization in cataractous lenses is altered dynamics due to local regions of unfolding and increased flexibility in both the N- and C-terminal domains particularly at surface helixes. This conformational flexibility increases the likelihood of aggregation which would be enhanced in the oxidizing cytoplasm of the aged and cataractous lens. The NMR findings combined with the in vivo insolubility and in vitro aggregation findings support a model that deamidation drives changes in protein dynamics that facilitate protein aggregation associated with cataracts.
Keywords: crystallin, cataracts, nuclear magnetic resonance, dynamics, deamidation
Graphical Abstract
INTRODUCTION
Cataracts are the leading cause of low vision over age 40 in the United States and the leading cause of blindness worldwide. The number of individuals affected continues to increase due to our aging population1. Lens transparency relies on the major structural proteins, called crystallins, to achieve close packing at the high concentrations found in the lens without causing crystallization, which would lead to a cataract2.
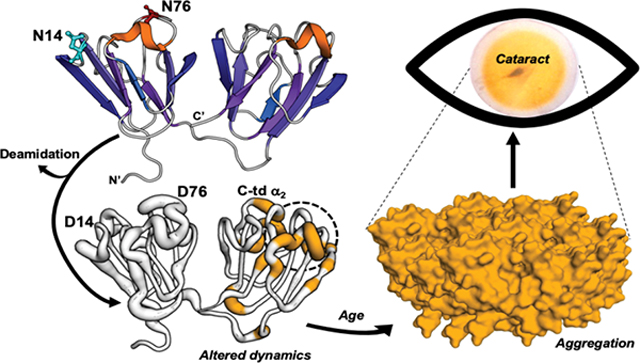
The γS-crystallin is in high abundance in human lenses, about 9% of the total crystallins in the young lens3, and shares a high degree of homology within the β/γ-crystallin family. The β/γ-crystallins contain about 30% β-strand content in extremely stable Greek key motifs necessary for their long life spans, several surface helical loops, and N-and C-terminal domains (N-td and C-td) along with their extensions4. The γS-crystallin shares these features, but has a short 5-residue N-td extension and no C-td extension. The γS crystallin is a ~21kDa globular protein consisting of four highly stable Greek key motifs joined by disordered loops, with two structurally mirrored halves connected by an unstructured linker (Fig. 1). This monomeric structure solved by NMR5, 6 is consistent with the known C-td7 and dimer8 crystal structures.
Figure 1. Location of labile Asn residues, N14 and N76, in human γS.
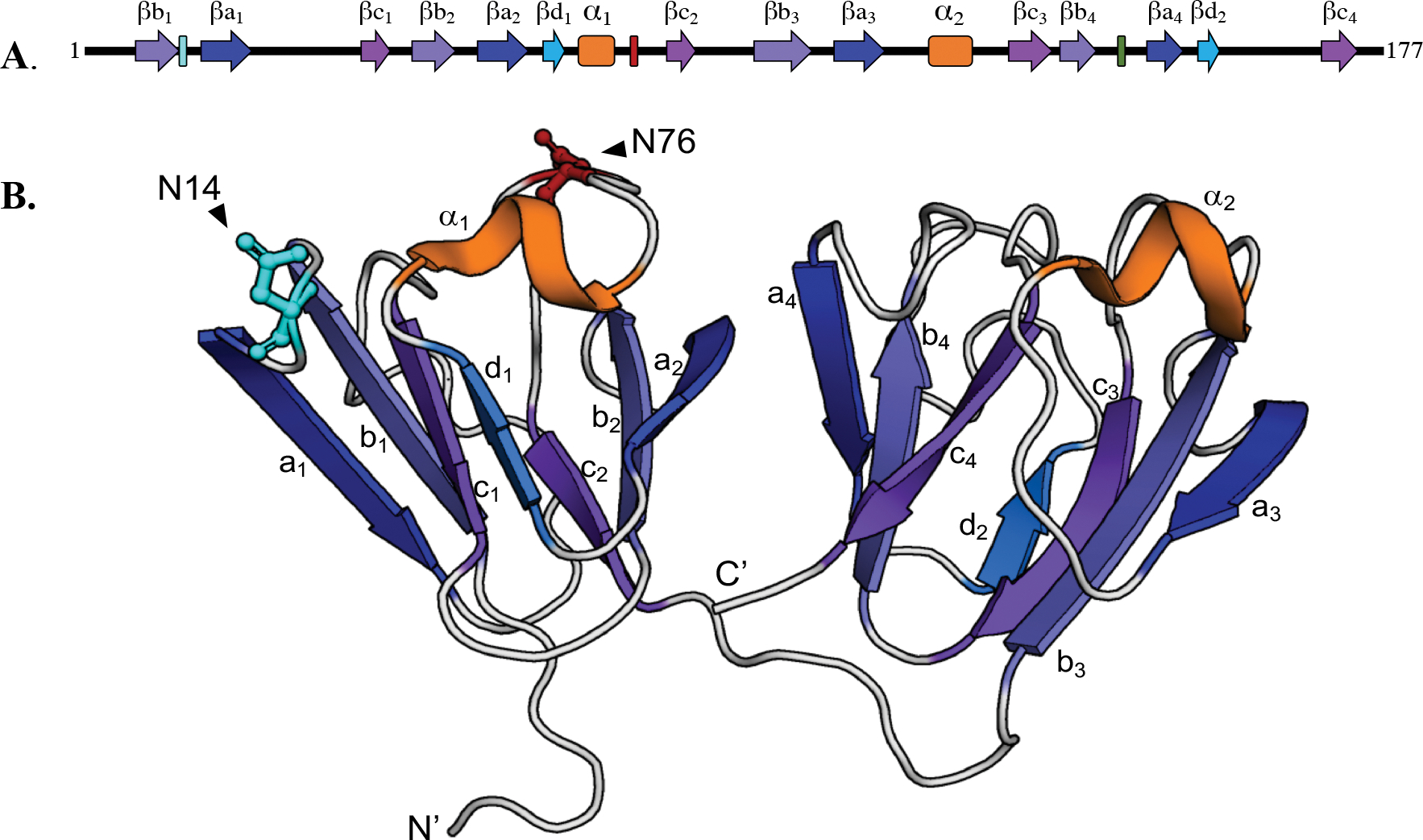
(A) N14 and N76 are in the N- terminal domain between β-strands b1 and a1 of Greek key motif 1 and between α-helix1 and β-strand c2 of Greek key motif 2, respectively. (B) Ribbon structure showing N14 and N76 located on the surface of the N- terminal domain (N-td) (PDB: 2M3T).
The major age-related modifications of lens crystallins are oxidation of Trp and Met residues, disulfide bond formation between Cys residues, truncation of the flexible N- and C-terminal extensions, and deamidation and isomerization of labile Asn and Gln residues9–15. More recently, crosslinking of succinimide-forming amino acids (Asn and Asp) to Lys16 and crosslinking of dehydroalanine and dehydrobutyrine (generated from Ser and Thr) with Cys17 have been identified. The accumulation of these modifications are thought to facilitate unfolding of lens crystallins during normal aging, facilitating further modifications, such as UV-induced oxidation, so that a threshold is reached where the protective α-crystallin chaperone is no longer able to maintain their solubility and both the chaperone and the unfolded crystallin aggregate, become insoluble, and cause light scatter18–20. Other work in the crystallin field has looked at the liquid-liquid phase separation properties of modified proteins as a cause of light scatter21. Given the extensive deamidation in aged and cataractous lenses and the prevalence of deamidated proteins in the isolated insoluble protein fraction, deamidation likely contributes to protein insolubilization and cataracts. Although the mechanism of how deamidation, particularly the abundant surface deamidations, disrupts proteostasis has not been established.
We have identified deamidation as a major age-related modification in the lens and determined it is associated with insolubilization of crystallins in vivo10, 12, 22. Deamidation can occur by nucleophilic attack of the amide nitrogen on the tertiary carbonyl group of an Asn (or Gln) residue resulting in an imide ring and release of the tertiary amino group. When the ring is cleaved in aqueous environments, the resulting Asp (or Glu) carboxylate group introduces a negative charge. Deamidation of Asn in vivo leads to isomerization (β-linkage) and epimerization (conversion of l to d stereoisomers) of resulting Asp residues9, 23–25. The b-Asp isomer has been suggested to be favored in the lens26 and this form may indeed be more closely associated with cataract. However, it is not possible to readily express these isoforms and data presented here represent baseline conformational changes at labile Asn residues upon which other modifications would only be expected to further modify the conformational dynamics.
Deamidation occurs at more surface accessible sites in both cataractous and aged clear human lenses with several sites more deamidated in cataractous lenses compared to age-matched controls26–30. We, along with others, have identified deamidation at Asn 14 (N14) and Asn 76 (N76) in γS as the most abundant in the water-insoluble proteins and that deamidation at N76 is also more prevalent in human cataractous lenses than normal lenses12, 27, 28, 30. Our findings reported here using high resolution mass spectrometry also support high levels of deamidation at these two sites. Deamidation at N76 has been shown to decrease protein stability while, deamidation at Asn143 (N143) and Gln92/Gln120 had less of an effect31, 32. The impact of deamidation at N14 remains unknown. Therefore, the effects of deamidation at N14 and N76 on γS structure were further investigated in this study using NMR.
We have mimicked deamidation in vitro by replacing Asn/Gln residues with Asp/Glu residues in human β-crystallins, which leads to subtle structural changes, but with significantly decreased stability4, 33, 34. During unfolding, deamidated crystallins rapidly aggregate and are not rescued by α-crystallin chaperone with both proteins precipitating18, 19. Similar effects of deamidation on γD and γS have been observed by others31, 32, 35, 36. In γS, deamidation at N76 leads to decreased stability during chemical unfolding and ineffective chaperoning by α-crystallin32. Deamidation at N76 and N143 together led to higher attractive interactions and a tendency towards aggregation with little structural perturbation31. However, none of these studies identified specific residues involved in the altered interactions that led to decreased stability or aggregation.
Here we report that mimicking deamidations at N14 and N76 located on the surface of γS in the N-td disrupt regions in the C-td distant to the deamidation site, exposing a helical region in the C-td and altering surface loop regions that are known protein interactions sites5, thus providing a potential mechanism for aberrant interactions of γS and its insolubilization in cataractous lenses.
MATERIALS and METHODS
Quantification of modifications at labile Asn in aged human lenses.
Eyes from donors of 82- and 85-years of age were procured from the Lions Eye Bank of Oregon (approved through the Institutional Review Board at Oregon Health & Science University). Lenses were dissected and imaged. Lens images were captured using Epson Expression 1600 scanner and RGB measurements of the central brunescent (yellowed) region of each lens determined using ImageJ (version 1.52a, National Institutes of Health). The attenuation of blue light was determined using the RGB Measure plug-in. Lenses were stored at −70 °C until used. The nuclear region of each frozen lens was isolated using a 4 mm corneal trephine to remove a core from the center of the lens. This core was then homogenized in 20 mM phosphate buffer (pH 7.0) containing 1 mm EDTA, centrifuged at 20,000 × g and the supernatant removed for the water-soluble fraction. The pellet was then washed once and resuspended by sonication for the water-insoluble fraction. The protein content in both fractions was assayed (BCA Assay, Thermo Scientific), lyophilized, and stored frozen at −70°C.
Protein samples were resuspended and solubilized in 8M deionized urea, 1.0 M Tris (pH 8.5), 8 mM CaCl2, and 0.2 M methylamine. Reduction and alkylation of proteins was then performed using dithiothreitol and iodoacetamide, and samples digested with trypsin (Sequencing grade modified trypsin, Promega, Madison, WI) overnight at 37°C after dilution of the urea to 2M concentration. Digestion was stopped by additional of formic acid, samples centrifuged and the supernatants removed. One 1 μg portions of the digests were injected onto a C18 trap column at 5 μl/min in mobile phase A containing water and 0.1% formic acid, washed for 5 min, then switched on-line to a 2 μm C18, 75 μm × 25 cm EasySpray column (Thermo Scientific) maintained a 40°C with a 300 nl/min flow rate. Peptides were separated by a 7.5–30% mobile phase B (acetonitrile, 0.1% formic acid) gradient over 205 min and analyzed using an EasySpray source and Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). Wide isolation single ion monitoring (WiSIM) scans at 500,000 resolution in the Orbitrap to produce high resolution accurate mass data for precursor ions were alternated with data-independent acquisition (DIA) MS/MS scans in the ion trap for sequence confirmation of detected peptides from observed fragment ions. Quantification of the relative amount of deamidation in peptides was determined using Skyline software37, using the 19 mDa mass defect between isotopic peaks of non-deamidated and deamidated peptide ions38. Precursor ion peaks for non-deamidated and deamidated forms of peptides 7–18 (ITFYEDKNFQGR) and 72–78 (WMGLNDR) of γS-crystallin were integrated and % deamidation in each peptide calculated. These calculations included the multiple peptide peaks resulting from isomerization and racemization of N and D residues, as well as the doubly deamidated form of peptide 7–18. The single peak resulting from deamidation at only Q16 was excluded from the calculation of % deamidation at N14, and occurred at a much lower rate than that at N14 as previously reported28. The chromatograms used for these calculations for deamidation at N14 are shown in Fig. S1.
Recombinant expression of human γS-crystallin and deamidated mutants.
Deamidation sites were chosen based on the high levels of deamidation and their association with γS-crystallin water-insolubilzation in aged brunescent lenses (Fig. 2). The plasmid containing the gene for human γS WT was engineered into the pE-SUMO (Small Ubiquitin-like Modifier) vector containing a N-terminal 6XHis tagged fusion protein (LifeSensors Inc., Malvern, PA). The γS N14D and γS N76D were generated via site-directed mutagenesis using QuikChangeXL Kit (Agilent Technologies, Santa Clara, CA). Escherichia coli cell stocks containing the plasmid of interest were grown at 37 °C, induced for protein expression with IPTG and their growth continued for 4 h. Following cell lysis, proteins were purified using HisPur Cobalt Resin according to the manufacturer’s protocol (Thermo Scientific, Rockford, IL.) and Ulp-1 protease (Ubl-specific protease 1) used to remove the SUMO-tag. Protein was separated from the SUMO-tag by a subsequent pass-through the HisPur Cobalt Resin. Pure protein was aliquoted and stored at −80 °C. In all purification steps, 2.5 mM DTT was present to keep the cysteines reduced. Typical yields of protein were several mg per 100 ml of E. coli culture. Purity and masses of all expressed proteins were confirmed by mass spectrometry to match predicted masses and be greater than 90% pure. Protein concentration was determined by measuring absorbance of folded protein at 280 nm and calculating concentrations using the protein extinction coefficient calculated using ExPasy ProtParam tool (γS WT 42.860 cm−1 M−1).
Figure 2. Deamidation at γS N14 and N76 is associated with insolubility in aged human lenses.
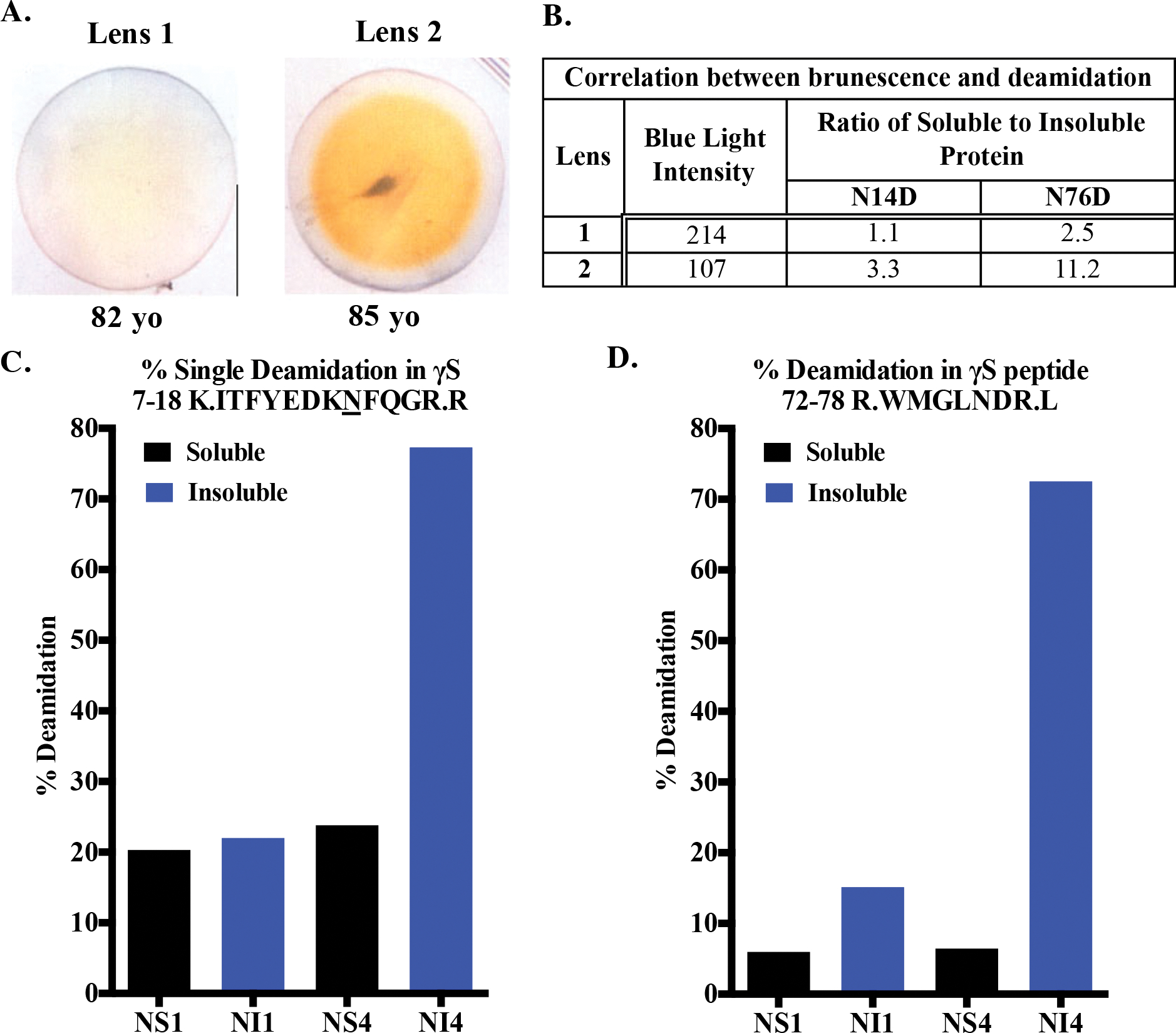
(A) Photographs of 82 and 85-year old lenses. (B) Brunescence estimated by measuring loss of intensity of blue light as yellowing lenses preferentially absorb blue light. (C and D) The percent deamidation at γS N14 and N76, respectively, were measured by mass spectrometry of tryptic peptides from the nuclear soluble (NS) and nuclear insoluble (NI) fractions of lenses 1 and 2 shown in (A).
Stable isotope labeled γS WT, N14D, and N76D were expressed in MJ9 minimal media at 37°C with 15NH4Cl as the sole nitrogen source39. Double-labeled WT γS-crystallin was expressed in MJ9 minimal media with 13C-Glucose and 15NH4Cl as the sole carbon and nitrogen sources, respectively. Recombinant protein expression was induced with 0.4 mM IPTG at a cell density OD600 of 0.6 and growth continued at 26 °C for 16 hours. Cells were harvested and recombinant protein purified using TALON His-Tag Purification protocol (Clontech Laboratories, Mountain View, CA.) The SUMO tag was cleaved by Ulp-1 as described above and the protein was further purified using anion-exchange chromatography (GE Healthcare HiPrep™ Q HP 16/10). In all purification steps, 2.5 mM DTT was present. The purity of the recombinant proteins, as assessed by SDS-polyacrylamide gels, was >95%.
Detection of light scattering of γS-crystallin and deamidated mimics.
The weighted-average molecular weights (Mw) of γS crystallins WT, N14D and N76D were determined by size-exclusion chromatography in-line with a multiangle laser scattering (MALS) detector (Dawn Heleos II, Wyatt Tech.) using measured Rayleigh light scattering and software provided by the manufacturer (Astra VII, Wyatt Tech.). Either 50 or 100 uL samples at 5 mg/mL in 58mM phosphate, 100 mM KCl, 1 mM EDTA and 1 mM DTT were injected. A dn/dc of 0.19331 and absorbance at 280 nm or a refractive index detector were used to determined concentration (Optilab Rex, Wyatt Tech.). The polydispersity was the Mw/Mn, where Mn is the weighted average number of molecules in each slice under the peak. All values were calculated in ASTRA VII software and variability is a percentage of the average value under the protein peak. Chromatography was repeated 2–4 times for each protein and included different concentrations (Fig. S2).
Heat-induced aggregation of γS-crystallin and deamidated mimics.
γS WT was compared to deamidated mimics by measuring changes in their turbidity during heating at 70° C using previously published methods40. This temperature was chosen based on the midpoint of thermal aggregation at 65° C previously reported32. A 100 μL aliquot of each protein was diluted to 24 μM using 100 mM sodium-phosphate (pH 7.4), 1 mM EDTA, 2.5 mM TCEP, and 100 mM KCl and heated to 70 °C in Peltier Thermal Cycler-200 (MJ Research, Hercules, CA). Turbidity of each solution was measured in 96-well plate as the change in optical density at 405 nm using Synergy-HT Microplate Reader (Bio-Tek, Winooski, VT). Data were fit to a nonlinear regression with PRISM 6 (GraphPad Software, CA) and initial rates of aggregate formation determined from the curves. Significance at each time point was determined via 2-way ANOVA with p < 0.05 (*), n=3 with errors bar=SEM. Experiments were repeated with purified γS-crystallins with a final concentration of 8.5 mM DTT and 15 mM GSSG to induce oxidation. Oxidized samples at 1.0 mg/ml were heated at 60 °C according to methods previously published with Student’s t-test performed on 3–4 independent replicates41–43.
Nuclear magnetic resonance of γS-crystallin and deamidated mimics.
Samples of 15N labeled γS crystallin were prepared in 10 mM sodium phosphate (pH 6.9), 50 mM sodium chloride, 2.5 mM DTT, a protease inhibitor mixture (Roche Applied Science, Madison, WI), and 2–2 dimethylsilapentane-5-sulfonic acid (DSS) for 1H chemical shift referencing. All NMR experiments were collected at 22° C, on a Bruker 800 MHz Avance III HD spectrometer equipped with a triple resonance cryogenic probe. NMR resonances were manually assigned using previously reported assignments (PDB: 2M3T) imported from BMRB (accession number 17576). All 2D spectra were processed using Topspin (Bruker Biosciences; RRID:SCR_014227) and analyzed in CcpNmr44. Spectra were referenced to DSS at 0 ppm and chemical shift differences were calculated as described previously39. A summary of methods used is presented in Fig. S3.
Changes in amide hydrogen solvent accessibility determined by H/H and H/D exchange with NMR.
CLEANEX-HSQC experiments for H/H measurements were collected with a mixing time of 100 ms using a recycle delay of 1.5 s. For H/D exchange, NMR samples were dehydrated at room temperature in a Savant SVC 100H Speed Vacuum Concentrator with RH 40–11 rotor and resuspended in an equivalent volume of D2O. We used a Best-TROSY HSQC experiment to assess H/D exchange. The experimental parameters for these experiments were similar to those used in the CLEANEX experiments, except that the recycle delay was reduced to 0.2 s, resulting in a total acquisition time of 13 minutes. The first Best-TROSY spectrum for each sample was taken ~30min (including the 13min collection time) after resuspension. Six sequential measurements were taken immediately to capture more rapid exchange. Additional measurements were taken over the course of 14 days for a total of 20 time points. Peak intensities were measured as heights determined through CcpNmr Analysis. CLEANEX and core residue peak intensities were normalized to respective TROSY spectra. For non-core resonances in the H/D exchange spectra, the measured intensities were normalized to an average of unaffected residue intensities. H/D exchange rates for non-core residues were calculated by fitting to an exponential decay as described previously45.
Relaxation measurements and analysis.
1H-15N Heteronuclear NOE experiments were collected with 16 scans, a recycle delay of 8 s, 2048 total points in the direct dimension and 256 total points in the indirect dimension. NOE values were obtained from the ratios of peak intensities in the presence and absence of proton saturation, and uncertainty was determined from the baseline noise as described elsewhere46. The delay times for the T1 experiment were 0.02, 0.06, 0.1, 0.2, 0.4, 0.6, 0.8, 1.2 s. The delay time for the T2 experiment were 16.9, 33.9, 50.9, 67.8, 84.8, 135.7, 169.6, 237.4 ms. All time points were collected in random order and two replicate points were included to assist with error estimation. T1 and T2 relaxation rates were determined using CcpNmr and error was estimated by the bootstrap method using 1000 samples46. The T1, T2 and heteronuclear NOE data were analyzed using five standard models of increasing complexity46. Model selection and model fitting were performed using FastModelFree and ModelFree447, 48.
RESULTS
Increased deamidation at Asn14 and Asn76 in human γS-crystallin.
To justify the selection of the Asn14 and Asn76 as sites of interest, we first measured the extent of deamidation at these sites in aged human lens. Human γS-crystallin contains 5 Asn and 9 Gln residues that are potential deamidation sites, of these, N14 and N76 are especially labile and are found on the surface of the N-td (Fig. 1). The N14 is between β-strands b1 and a1 of Greek key motif 1 and N76 is between α-helix1 and β-strand c2 of Greek key motif 2 (PDB:2M3T). Due to the presence of the most aged proteins in the nucleus of the lens, water-soluble and water-insoluble proteins were isolated from the center of the 82- and 85-year old lenses (Fig. 2a). Brunescence, an indicator of opacity49, 50, was estimated from the yellowing of the lenses, which was measured by the decrease in blue light pixal density (Fig. 2b). The level of deamidation at N14 and N76 was determined by liquid chromatography/mass spectrometry analysis of tryptic peptides 7-ITFYEDKNFQGR-18 and 72-WMGLNDR-78 (Fig. 2c and d). Deamidation of N14 and N76 was 77 and 72%, respectively, in the water-insoluble fraction of the nucleus of the more brunescent lens compared to 22 and 15%, respectively in the similar aged, but less brunescent lens. Both deamidations were associated with water-insolubility, especially for deamidation at N76, which was 11-fold more abundant in the water-insoluble fraction than the soluble fraction (Fig. 2b). Deamidation was also detected at Q16, but at only 24% and occurred most often in peptides that also contained deamidation at N14 (Fig. S1). The Asn to β-Asp modification has been reported to be favored over the α-Asp isomer26. Multiple chromatographic peaks were detected suggesting these isoforms were also present in the aged lenses (Fig. S1). These results confirm our previously reported levels using mass spectrometry spectra counting12 and those of others28, 51, 52 with levels ranging from 30–60% and 8–54% for N14 and N76, respectively. Variability reflects the different ages, stage of opacity, and total vs. nuclear insoluble proteins analyzed.
Increased light scattering and thermal-induced aggregation of deamidation mimics at Asn14 and Asn76 in γS-crystallin.
To determine potential changes in γS-crystallin stability and structure due to deamidation at N14 and N76, deamidation was mimicked by site-directed mutagenesis, replacing the Asn with Asp residues at these positions. The light scattering of each protein was determined at room temperature (Fig. 3a). Two light scattering peaks eluted during size exclusion chromatography- the main monomer peak and a high molecular weight (HMW) peak just after the void peak. Others have also reported a dimer due to disulfide bond formation, which would not have been observed in this study due to the presence of reducing agents in the buffers32. A molar mass could not be determined of the HMW peak due to the low UV signal. However, the peak is likely due to a small amount of a large oligomers of γS (Fig. 3b). The relative Rayleigh light scatter of the HMW peak was highest in the N76D and then the N14D compared to the WT control (Fig. 3c). Chromatography runs were repeated at different concentrations with similar increases in the HMW peaks from N14D and N76D compared to WT (Fig. S2).
Figure 3. Increased light scattering of deamidated human γS crystallin.
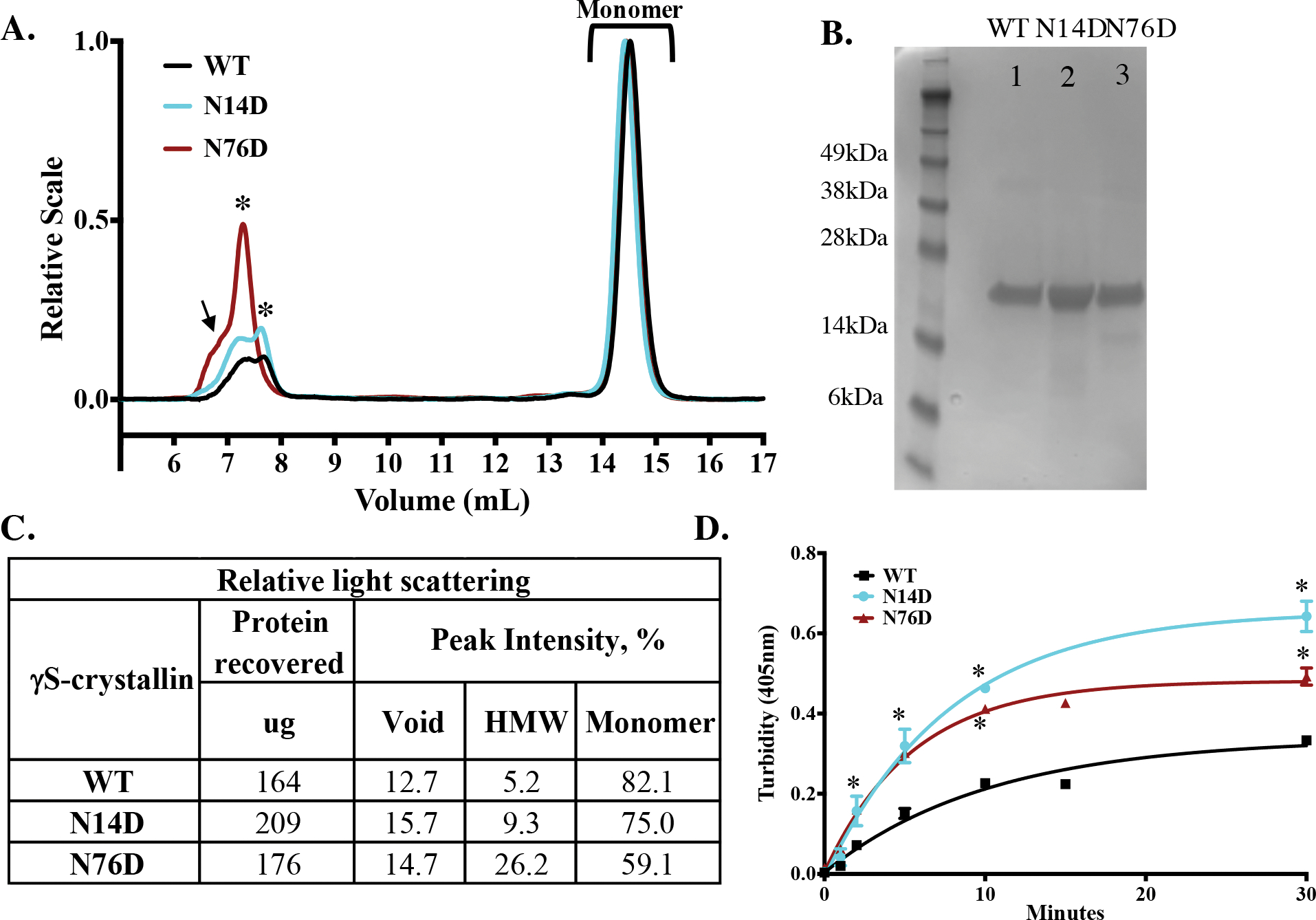
(A) Relative Rayleigh light scattering traces of γS WT (black), N14D, (cyan) and N76D (dark red) during size-exclusion chromatography in-line with multiangle laser scattering. A high molecular weight peak (HMW, starred) was detected just after the void peak (arrow). (B) SDS-PAGE of WT (Lane 1), N14D (Lane 2) and N76D (Lane 3) γS-crystallins. (C) Table of relative light scattering intensity of peaks from B. (D) Relative turbidity of γS-crystallins during thermal-induced denaturation at 70 °C of WT (black), γS N14D (cyan), and γS N76D (red). Turbidity measured as change in O.D. at 405 nm, n=3 and SEM calculated via 2-way ANOVA with p < 0.05 (*). Curve fits were generated using nonlinear regression with PRISM 6. Initial rates of increase in turbidity were 0.03 min−1 for WT and 0.06 min−1 for N14D and N76D.
The thermal aggregation of these mutants was determined by measuring their turbidity during denaturation at 70 °C (Fig. 3d). This temperature was chosen because it was above the midpoint of the thermal transition for WT γS32. The initial rates of aggregation were significantly greater in both deamidation mimics, 0.06 min−1, compared to WT, 0.03 min−1 (Student’s t-test, p = 0.04) suggesting deamidation decreased heat stability. Experiments were repeated at 60 °C with addition of the oxidizing agent, oxidized glutathione (GSSG)41,43. Oxidized N14D and N76D had significantly greater turbidity than oxidized WT and non-oxidized samples (Fig. 4).
Figure 4. Increased thermal-induced aggregation of oxidized, deamidated human γS.
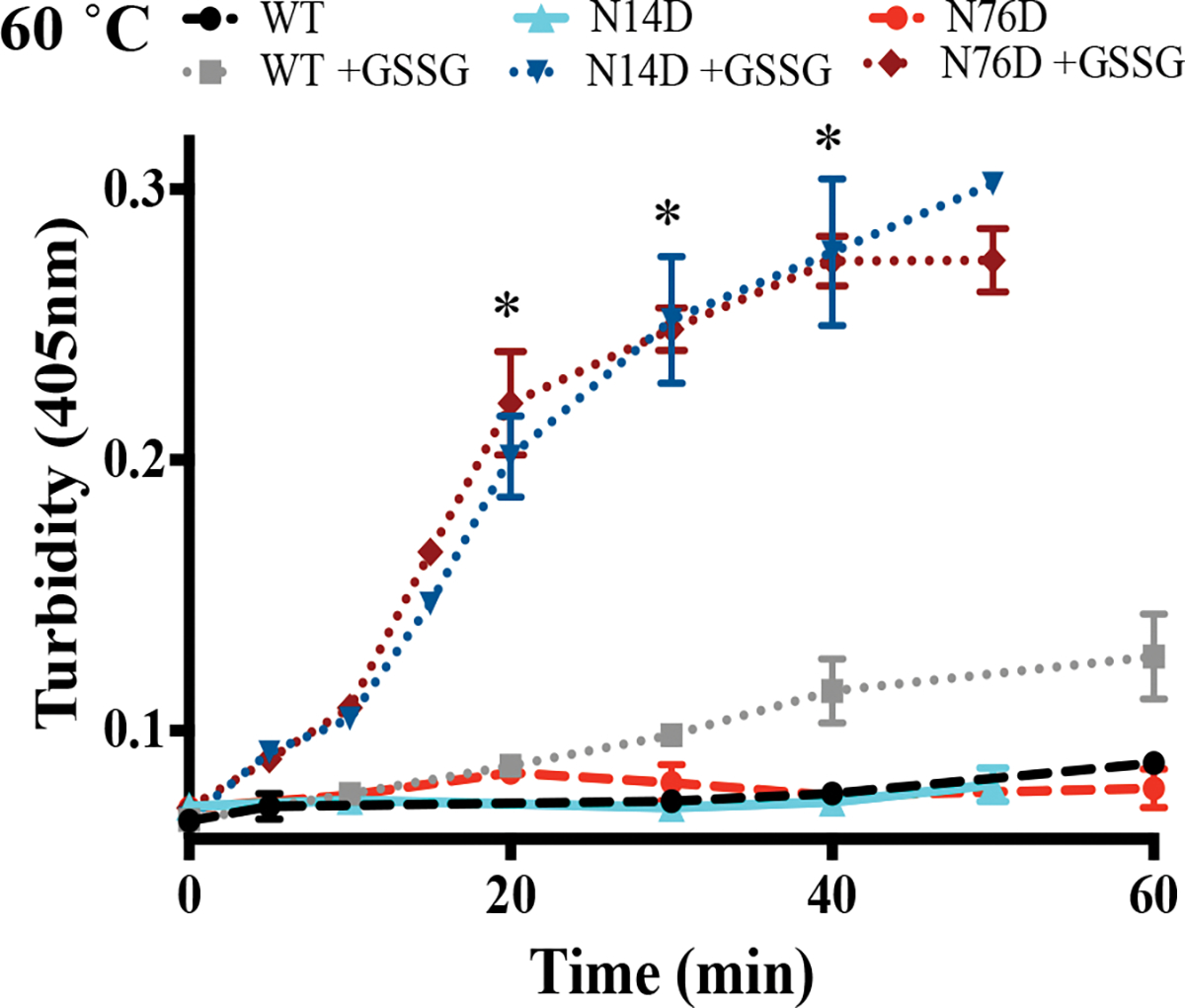
Thermal-denaturation at 60 °C was performed in the presence of oxidized glutathione, GSSG. Resulting turbidity was measured of WT, N14D, and N76D γS crystallins with (dotted lines) and without (dashed lines) GSSG. Each sample contained 1.0 mg/ml of protein. (*) denotes significant differences from oxidized WT using Student’s t-test, p < 0.0007.
Dynamics of WT human γS crystallin determined by NMR.
NMR spectra of γS-crystallin showed widely dispersed peaks indicative of a well-ordered protein and consistent with published NMR structures (Fig. 5)5, 6, 53. Resonance assignments of ~90% of backbone amides aided by previously assigned spectra53 were confirmed by 3D HNCACB experiments. The few missing peaks correspond to residues in βb3, βc3, and βd2, and the flanking loops, similar to what was reported earlier7. Missing peaks due to peak broadening could arise from internal backbone flexibility or from molecular aggregation. Spectra at 10-fold dilution did not show any additional peaks, suggesting conformational exchange at microseconds time scales.
Figure 5. 1H-15N spectra of WT human γS-crystallin.
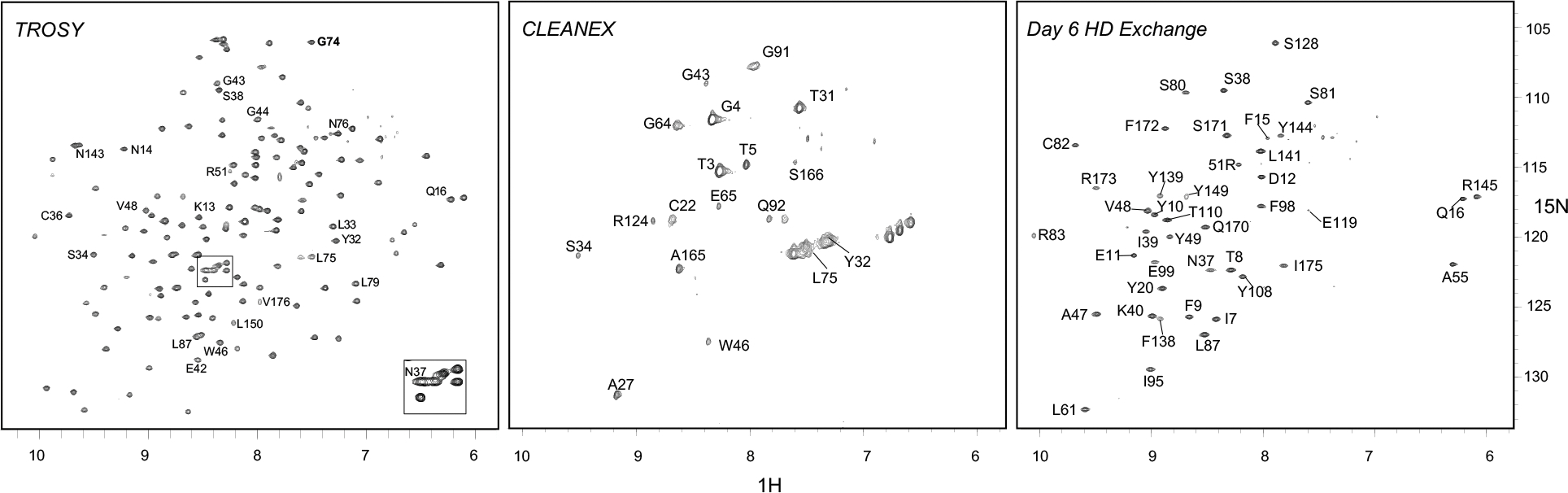
TROSY (left), CLEANEX (middle), and TROSY after 6 days H/D exchange (right). Peaks that shift significantly in N14D or N76D are labeled on WT TROSY and shifts are visualized in Fig. S4. Labeled peaks in CLEANEX and Day 6 H/D exchange spectra are colored pink or blue, respectively, in Fig. 6, and were plotted by normalized peak intensity in Fig. S5.
To identify amide protons exposed at the protein surface or in flexible loops, we used CLEANEX experiments that measure fast backbone amide proton exchange with water (H/H) (Fig. 6a, top row WT and Fig. 5). As expected, the most rapidly exchanging amide protons (the peaks with the highest intensities) were in the N-terminal extension (residues 3–5), loops (residues 27–32, 75, 165–166), and in the linker connecting the N-td and C-td (residues 91–92). These residues also showed multiple conformations in the NMR ensemble structure (PDB: 2M3T)5, 53. Not expected from the structure, was the relatively fast exchange observed for residues 64 and 65 within βd1 and to a lesser extent at the ends of β-strands a1, c1, a2, b2, a3, a4, and c4.
Figure 6. Hydrogen exchange of WT human γS-crystallin and N14D and N76D variants measured by NMR.
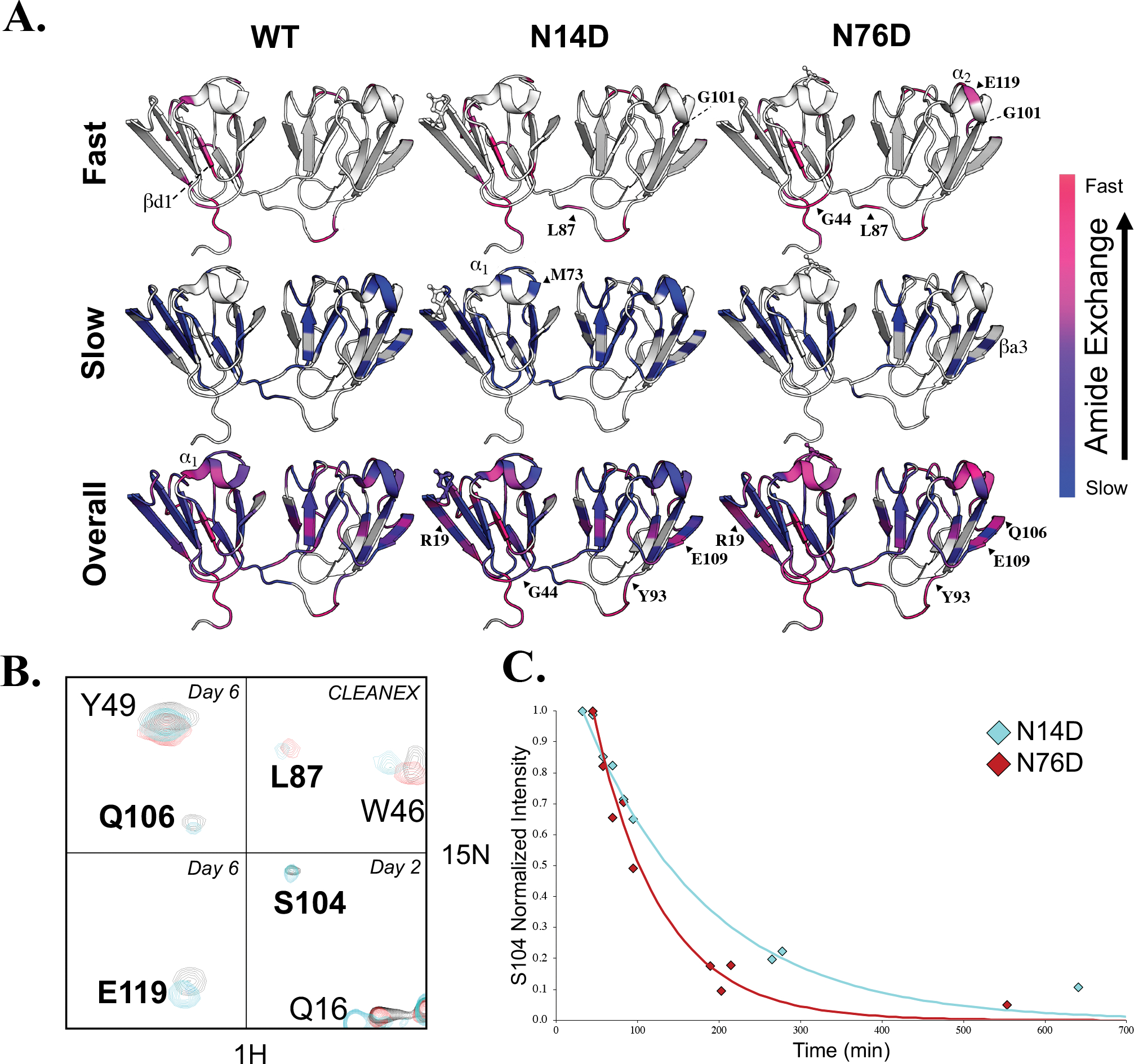
(A, Top) H/H exchange of WT (left), N14D (middle), and N76D (right) showed altered solvent accessibility compared to WT. High intensity peaks that appeared in CLEANEX spectra for amide protons that exchanged the fastest are mapped on the structure in hotpink and lower intensity CLEANEX peaks are mapped in magenta. (A, Middle) H/D exchange showing the slowest, core residues mapped in blue whose intensity did not significantly change after fourteen days in D2O. (A, Bottom) Intermediate exchange rates with purple denoting residues that exchanged before the first Best-TROSY spectrum was obtained (in D2O for ~30min) but weren’t visible in CLEANEX, followed by residues that exchanged after 1 day in D2O (pink), after 2–3 days (magenta-purple), and after 6 days in D2O (dark purple). Those residues that did not have NMR peak assignments available or that were too broad to confidently measure their exchange rates, were mapped on the structure in white. (B) NMR spectral overlay of WT (black), N14D (cyan) and N76D (red). Residues which showed significantly altered H/D exchange compared to WT were in bold. Day 6 H/D exchange spectra of Q106 and Y49 (top left), day 6 H/D exchange spectra of E119 (bottom left), CLEANEX spectra of L87 and W46 (top right), and day 2 H/D exchange spectra of S104, Q16, and R145 (bottom right). (C) H/D exchange rates of S104 are different for N14D (cyan) and N76D (red) and are reported in Table S1.
To identify the slowly exchanging amide protons that are buried and/or involved in H-bonding, we measured hydrogen deuterium (H/D) exchange over a period of two weeks (Fig. 6a, middle row WT). A large number of residues in the β-strands never exchanged significantly (blue) within this time window (Figs. 6a, middle row and 4). The interface between the N-td and C-td was not readily solvent accessible (Fig. 6a, magenta-to-purple, bottom row WT). This finding is similar to the slow exchange we have reported for the domain interface in β-crystallins54. Within the flexible connecting peptide, slowly exchanging amide protons were detected for residues 85–87 near the N-td suggesting they were buried within the interface (Fig. 6a, blue, middle row WT). On average, amide protons within the C-td were slower exchanging than those in the N-td. In particular, α2 exchanged more slowly than α1, as did the connecting loops in the C-td compared to those in the N-td. Exchange rates for amide protons measured in this time window of 1–6 days, and that are shown in Fig. 6a (pink to purple, bottom row WT), are listed in Table S1.
In summary, both H/H and H/D exchange data show that the WT N-td was more dynamic and heterogeneous relative to the C-td, most prominently at N-terminal extension residues, in α1 and linker residues connecting the domains.
Effects of mimicking deamidation on human γS structure and dynamics.
Comparison of WT spectra to those of Asn-to-Asp mutations showed chemical shift differences at the sites of mutation as expected, but also significant differences at unexpected sites distant from the mutation site (Fig. S4). These sites included residues in the region between βc1 and βb2, α1, residue L87 in the connecting peptide, C-terminal interface βa4, α2, and the most distant β-strand, βa3.
The same or nearby residues observed to have altered chemical shifts in mutant constructs also had significantly altered amide H/H and H/D exchange measurements (Figs. 6a–c and 7). For N76D, faster exchange rates were observed in general (Figs. 6a and S5). Several new peaks appeared in the CLEANEX spectra, including L87 and G101 in both mutants, as well as G44 and E119 in N76D (Fig. 6a, top row). Peaks for residues newly appearing in the CLEANEX experiment, such as L87, were of greater intensity in N76D than in N14D. Residues with faster exchange rates compared to WT in both mutants included those of βa1 (blue to magenta/pink), and E109 and S104 in βa3 (deep blue to purple) (Figs. 6a, bottom row and 6b, c and Table S1). In N76D, exchange rates were elevated throughout the protein, in particular α1 residues (purple to pink) and E119 in α2 (blue to pink). Furthermore, those residues with increased exchange rates in N14D such as S104 (cyan in overlaid spectra) were significantly more increased in N76D, disappearing by day 2 (red) (Figs. 6b, c). In both mutants, backbone NH S104 exchanged faster in N76D and N14D than in WT. Conversely, WT NH S104 is part of the slowly exchanging core (Fig. 6a, middle row, near G101 visible in structure).
Figure 7. 1H-15N chemical shifts and R2/R1 values for WT, N14D, and N76D.
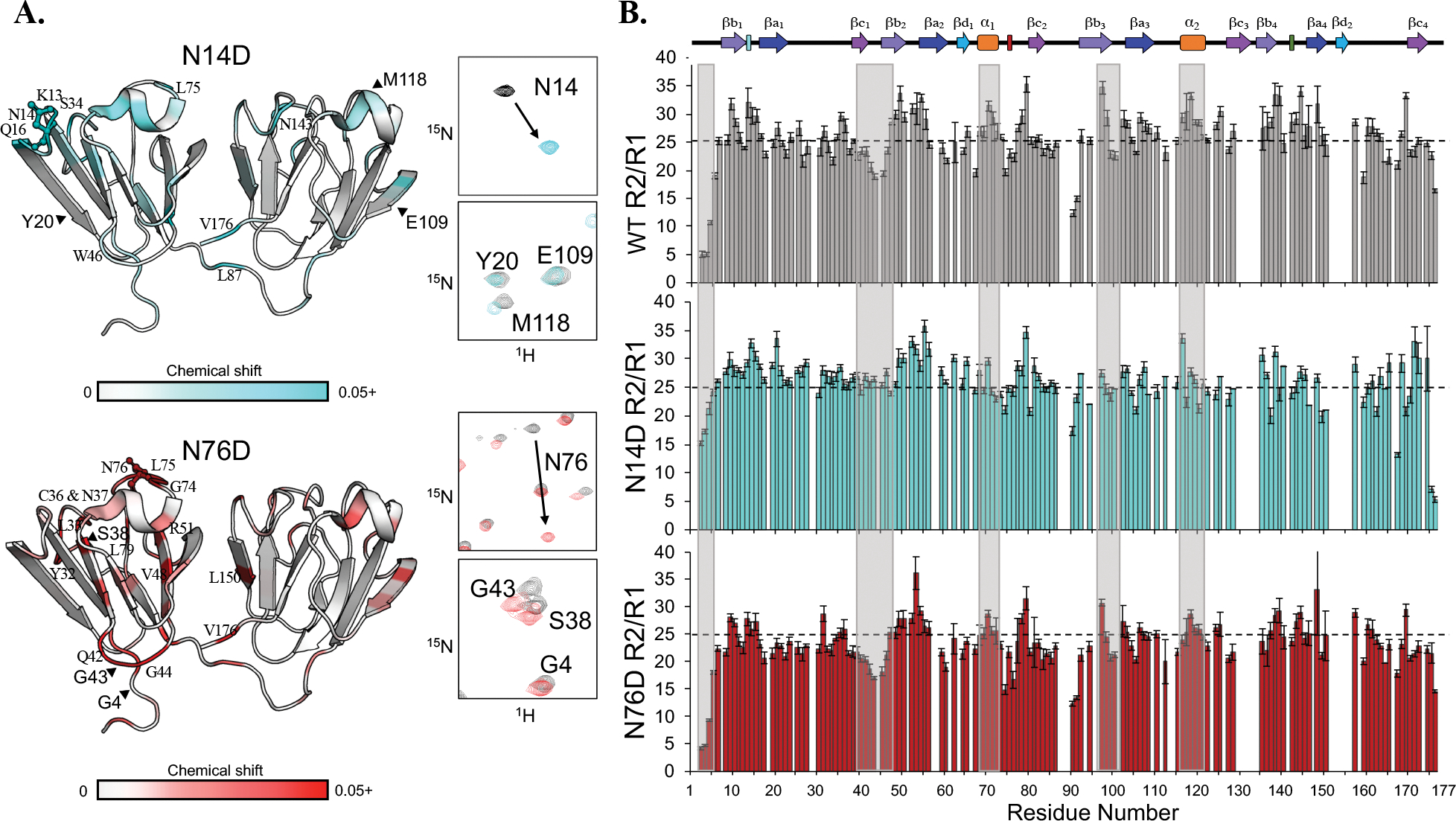
(A) Chemical shift differences mapped on the structure in N14D (top) and N76D (bottom). Shifts shown range from 0.02–0.8 to 0.02–0.15 ppm for N14D and N76D, respectively, and are colored on the structure here from 0–0.5 ppm to distinguish small changes. Full mutant spectra overlaid with WT are shown in Fig. S4. (B) R2/R1 plots of WT (grey), N14D (cyan), and N76D (red). Boxes highlight key differences. The dotted line was placed at the WT average of 25 for comparison.
Compared to WT γS, exchange rates were decreased for some residues in N14D, but not for N76D. Examples in N14D are residue M107 in the βa3 (Fig. 6a, magenta to purple, bottom row N14D, near E109) and G44 (Fig. 6a, magenta to purple, bottom row, N14D). Overall, there was a higher number of amide protons that exchanged within 1–6 days measured for N14D and N76D compared to WT (31 for N14D, and 24 for N76D, but only 18 for WT) (Table S1).
To compare dynamics between deamidation mimics and WT γS at the μs to ps time scale, we measured longitudinal and transverse relaxation rates, R1 and R2, and heteronuclear NOE (Figs. 7 and S5). Heteronuclear NOE measurements for both mutants were on average similar to WT with only modest decrease (Fig. S6). The R2/R1 values were most informative in identifying differences in ms to μs time scale. The R2/R1 values ranging from ~5–35 for WT γS identify regions of heterogeneous dynamics. Regions with the highest R2/R1 values included β-strands within the first two Greek key motifs, βa1–4, and βb1–4, and both α-helices, α1 and α2. Those with the lowest R2/R1 values included the unstructured N-terminal extension and the loop connecting the β-strands βc1, βb2 and βb3. Additionally, linker residues 90–92 had significantly decreased values compared to other residues in the linker. The R2/R1 values for N14D showed more homogeneous dynamics in the N-td compared to the C-td, which remained dynamically heterogeneous (Fig. 7b, middle row). For example, residues in the N-terminal extension and in the loop residues between βc1 and βb2 have higher values compared to WT. Conversely, N76D showed similar heterogeneity relative to WT, but with lower values overall (Fig. 7b, bottom row). From R2/R1, we determined rotational correlation times of 11.9, 11.8, and 11.3 ns for WT, N14D, and N76D, respectively. The lower correlation time estimated for N76D suggests that mimicking deamidation at this site causes faster tumbling than expected for a globular protein of ~21kDa53 suggesting a global increase in flexibility.
Local mobility parameters of backbone amide N-H vectors interpreted using the FAST-ModelFree analysis48 revealed differences in dynamics at both the ms to μs conformational exchange (intermediate exchange, measured as Rex for heterogenous dynamics specific for these regions) and motions faster than the overall tumbling correlation time on ns to ps timescale (measured as S2, where numbers closer to 1 imply restricted motion and those closer to zero imply fully disordered). While WT and N76D had 10 and 7 Rex terms respectively, N14D had 37, which was mostly localized to the N-terminus, suggesting deamidation at N14 resulted in multiple conformations specifically at the N-td (Fig. 8a). The C-td also showed regions of exchange but not to the same extent (29 residues with Rex terms in the N-td vs 8 in the C-td).
Figure 8. FastModelFree analysis of N14D and N76D.
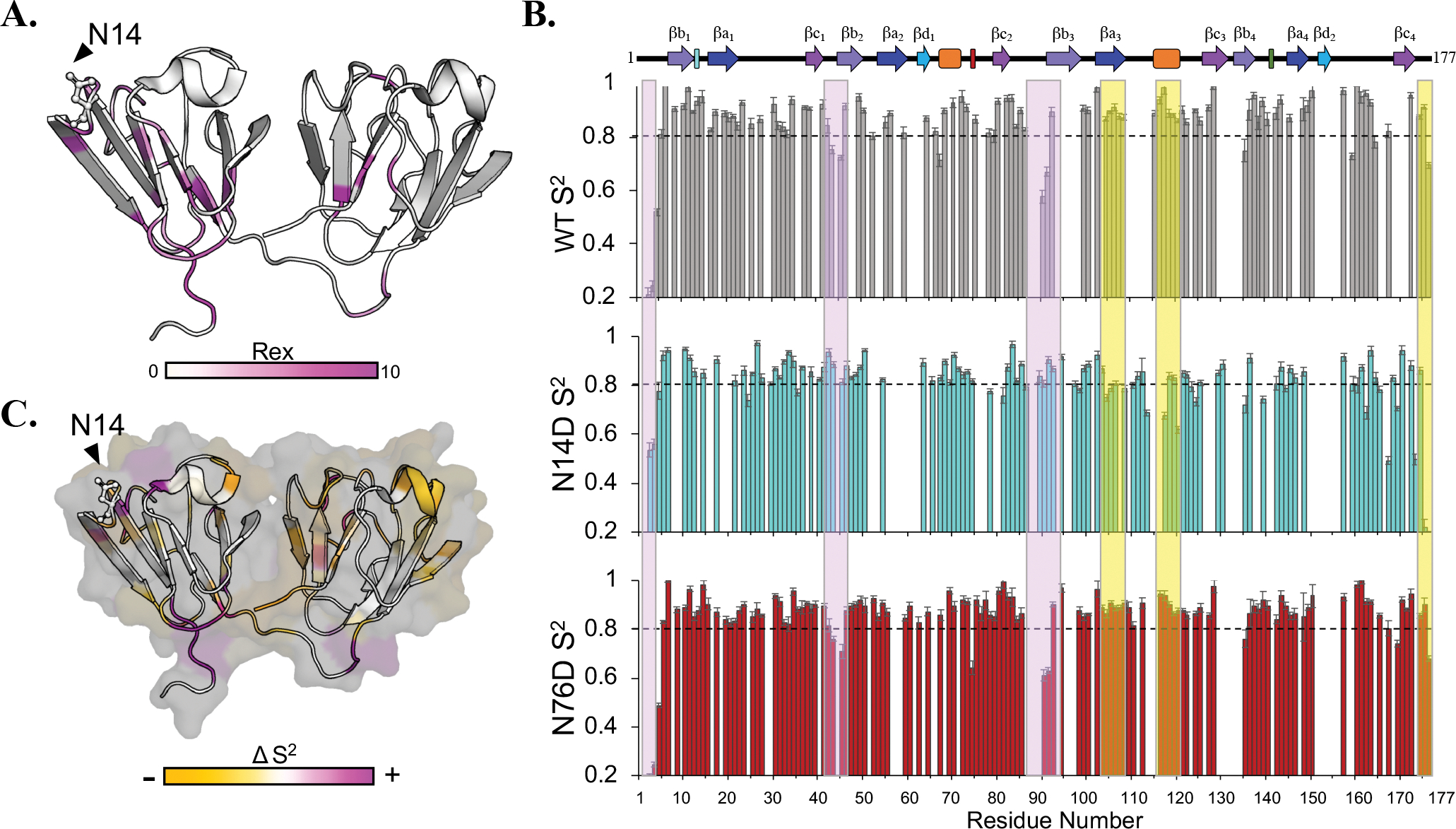
(A) N14D Rex values ranging from 0.38–9.01 were mapped on the 3D structure from light to bright magenta. Residues colored white have no Rex values determined from the model. (B) S2 plots of WT (grey), N14D (cyan), and N76D (red). Boxes highlight a decrease (yellow) or increase (purple) in S2 in N14D. The dotted line at 0.8 was provided for comparison. Few significant differences were observed for N76D at this timescale. (C) Changes in S2 for N14D were colored by residue onto the structure as a decrease (yellow) or increase in S2 (magenta).
Both mutants showed decreased S2 values overall, suggesting an increase in fast dynamics. N14D showed significant increases in S2 in segments highlighted in purple in the N-td, including in the N-terminal extension, the loop connecting βc1 and βb2, and the end of the linker, leading into the C-td. In contrast, the C-td of N14D showed a decrease in S2 specifically in βa3 and α2 highlighted in yellow (Fig. 8b). Although H/D exchange and chemical shift comparisons show more significant changes in N76D than in N14D, FAST-ModelFree48 analysis revealed more significant changes in N14D.
DISCUSSION
Age-related deamidation is thought to be associated with cataracts due to their increased levels in the insoluble proteins isolated from cataractous lenses. However, the mechanism for deamidation-induced insolubilization of lens crystallins is not known, because in vitro studies to date have focused on global changes to protein structure and stability. In this study, we confirmed that in the water-insoluble fraction of an aged, brunescent lens, extensive deamidation occurs in γS-crystallin at amino acid positions N14 and N76. Deamidation at N76 and N14 has been observed by us12 and others as the most prevalent deamidations in γS associated with insoluble proteins28, 52.
We mimicked these deamidation sites using mutagenesis. Thermal-induced turbidity and light scattering was increased in deamidated mutants, suggesting that deamidation increases propensity of γS to aggregate and form large soluble oligomers. Thorn et al. recently found increased aggregation of oxidized γS-dimer formed through an intermolecular disulfide bond at Cys24 in the N-td41. We report here increased aggregation of oxidized deamidated γS compared to oxidized WT γS, suggesting that deamidation increases aggregation at least partially due to disulfide bond formation. Although the specific Cys residues involved in disulfide bonding were not determined, Cys24 was among the fastest exchanging residues in WT and N14D, while Cys114 exchanged faster in N76D compared to WT and N14D, and Cys36 had conformational exchange in all three proteins (Table S1). The Cys residues involved in disulfide formation may differ between the proteins. Serebrynay et al. has shown similar aggregation of oxidized γD-crystallin at room temperature due to disulfide exchange between monomers involving Cys108 and Cys110 in the C-td43. In γS, there is not a homologous Cys at residue 110 as found in γD. The higher temperature required for oxidation-induced aggregation of γS may reflect differences between γS and γD in the environment of the Cys residues and their N- and C-td conformations.
The propensity of γS to aggregate was correlated to residue-specific allosteric effects of deamidation by measuring changes in solvent accessibility and protein dynamics at different time scales using NMR. The emerging theme is that insolubility and aggregation due to deamidation may be caused by allosteric conformational changes summarized below and in Table 1.
Table 1.
Summary of human γS dynamics parameters measured by NMR.
| WT | N14D | N76D | |
|---|---|---|---|
| Number of fast exchanging residues (kex ≥ 500 sec−1*10−6)a | 34 | 38 | 40 |
| Number of intermediate exchanging residues (0.1 ≤ kex ≤ 500 sec−1*10−6)b | 57 | 55 | 78 |
| Number of slow exchanging core residues (kex < 0.1 sec−1*10−6)c | 36 | 35 | 9 |
| (R2/R1)d | 25.6 ± 0.4 | 26.0 ± 0.9 | 23.3 ± 1.2 |
| Number of Rex terms e | 10 | 37 | 7 |
| (S2)f | 0.86 ± 0.02 | 0.82 ± 0.02 | 0.86 ± 0.02 |
| NOE g | 0.82 ± 0.01 | 0.79 ± 0.01 | 0.8 ± 0.03 |
In both N14D and N76D, chemical shifts were detected in the vicinity of the mutation site in the N-td as expected. In addition, they both showed significant changes in the homologous region of the opposite domain, particularly in the surface helix loop region of the C-td. Most notably, changes were observed in both the α2 helix and βa3 strand of the 3rd Greek key motif (Fig. 1). Dynamics measurements clearly demonstrated an increase in flexibility in the N-td, and also in the same region of the C-td that showed chemical shift changes. With N76D, considerably faster exchange was observed in βa3 and α2, suggesting local unfolding specific to this region of the C-td (Fig. 6a and Table 1). In contrast, with N14D, changes in this region of the C-td were not detected by hydrogen exchange, but were observed instead as a decrease in the order parameters obtained from relaxation measurements on the ns to ps motions (Fig. 8b and c), also primarily for βa3 and α2. Further changes in N14D dynamics were most notably detected as Rex terms for ms to μs fluctuations (Fig. 8a and Table 1), primarily in the N-td, reporting multiple conformations.
Large changes in dynamics of the linker connecting the N-td and C-td were also observed for both N14D and N76D by hydrogen exchange measurements. Residues L87 and Y93, for example, which were buried and slowly exchanging in the WT became considerably exposed and fast exchanging in both mutants. While it is not immediately clear how mutations introduced in the N-td disrupted dynamics in the C-td, it is possible that changes in both the N-td and connecting peptide led to the observed changes in the C-td. This is plausible due to the contribution of the domain interface to the stability to γ-crystallins55. In support of this, the N-td γS mutant, F9S, was reported to show evidence of an aggregation-prone intermediate with increased ms-μs dynamics, propagating from the N-td, through the domain interface, to the C-td, by CPMG T2-relaxation dispersion NMR and fluorescence experiments56. We have also previously reported that disrupting the dimer interface in βB2-crystallin by introducing deamidation sites led to changes in solvent accessibility distant from the mutation site54. Taken together, these results suggest that age-related deamidations on the surface of γS cause dynamic changes not only near the site of mutation, but also in the stabilizing domain interface and in distant sites in the C-td.
While, mimicking deamidations at N14 and N76 affected similar regions within γS, particularly the surface helixes in both the N- and C-td, the NMR parameter affected differed between the two mutants (Table 1). Mimicking deamidation at N76 in γS led to greater solvent accessibility predicting unfolding of the surface helix regions, while mimicking deamidation at N14 led to more heterogeneous dynamics in the N-td predicting multiple conformations. The unfolding of surface regions is likely a greater contributor to insolubilization, since, deamidation at N76 was more closely correlated with insolubilization in vivo (Fig. 2b) and with formation of a large light scattering oligomer in vitro (Fig. 3c).
A recent study has examined the effect of the congenital cataract variant, G57W mutation on the structure and dynamics of γS-crystallin6, 57, 58. The G57W mutation perturbed the structure around the mutation site and induced local conformational changes destabilizing the N-td but leaving the C-td structurally unperturbed. In contrast, our study examined the impact of the age-related modification of deamidation, which does not introduce a large change in side chain size, and measured perturbations at multiple time scales. Importantly, we observed only modest chemical shift perturbations outside of the mutated residues, suggesting that the changes in 3D structure induced by deamidation were small. Yet, dynamic perturbations in the ns to ps and slower motions probed by hydrogen exchange and relaxation measurements detected significant changes occurring not only in the N-td but also in the C-td, despite the mutations being in the N-td.
In summary, deamidation increases dynamics and induces local unfolding distant from the deamidation site. Residue-specific changes in solvent accessibility and dynamics indicate that surface deamidation sites result in changes both near and distant from the site of deamidation, suggesting that site specific deamidation, has both local and global effects on the protein structure at slow (ms to s) and fast (μs to ps) time scales. Altered protein dynamics at sites distant from the mutation site, particularly at exposed loop regions, provide a plausible mechanism for the insolubilization observed in vivo. These changes in conformation and dynamics could be magnified at the higher concentrations of 300–500 mg/ml found in vivo leading to disruption of protein-protein interactions. Further perturbation of the structure would be expected in vivo due to the associated isomerization and epimerization that could not be mimicked here with mutagenesis. In addition, the charged aspartic acids resulting from deamidation may increase susceptibility to further modifications such as disulfide bonds and/or cross-linking resulting from the succinimide intermediate16.
While, preventing age-related deamidation may not be possible, stabilizing the disrupted structural regions might be, and thus compounds designed to stabilize the disrupted interface between domains could be developed as potential therapeutics. The NMR findings combined with the in vivo insolubility and in vitro aggregation findings support a model that deamidation drives changes in protein dynamics that may more readily unfold the protein and expose regions buried in the WT protein increasing the propensity for aggregation and eventually leading to a cataract. These findings have relevance to other age-related diseases such as Alzheimer’s where the involvement of deamidation in amyloid formation has been implicated59.
Supplementary Material
Figure S1. Chromatograms of doubly charged γS peptide 7–18 (ITFYEDKNFQGR).
Figure S2. Multiangle laser scattering in-line with size exclusion chromatography of γS crystallins.
Figure S3. Summary of NMR methods.
Figure S4. TROSY overlay of WT with N14D and with N76D.
Figure S5. CLEANEX and day 6 core residue intensities of WT, N14D, and N76D.
Figure S6. NOE values for WT, N14D, and N76D.
Table S1. Measured H/D exchange rates for WT, N14D, and N76D.
ACKNOWLEDGEMENTS
General:
Authors wish to thank Mr. Samuel Wheeler for technical help with thermal-induced turbidity assays.
Funding:
This work was supported by NSF MCB 1617019 to (EB), NIH EY027012 (KJL), NIH EY027768 (KJL and LLD). Mass spectrometry support includes Ophthalmology Proteomics Core NIH P30 EY010572 and S10 OD012246 to Oregon Health and Science University. NMR support includes Oregon State University NMR Facility NIH HEI Grant 1S10OD018518 and the M. J. Murdock Charitable Trust grant # 2014162.
Footnotes
Competing interests: None
PROTEIN ACCESSION ID
The accession ID for human γS is P22914 (CRYGS_HUMAN).
REFERENCES
- [1].Congdon N, O’Colmain B, Klaver CC, Klein R, Munoz B, Friedman DS, Kempen J, Taylor HR, and Mitchell P (2004) Causes and prevalence of visual impairment among adults in the United States, Arch Ophthalmol 122, 477–485. [DOI] [PubMed] [Google Scholar]
- [2].Delaye M, and Tardieu A (1983) Short-range order of crystallin proteins accounts for eye lens transparency, Nature 302, 415–417. [DOI] [PubMed] [Google Scholar]
- [3].Robinson NE, Lampi KJ, Speir JP, Kruppa G, Easterling M, and Robinson AB (2006) Quantitative measurement of young human eye lens crystallins by direct injection Fourier transform ion cyclotron resonance mass spectrometry, Mol Vis 12, 704–711. [PubMed] [Google Scholar]
- [4].Lampi KJ, Oxford JT, Bachinger HP, Shearer TR, David LL, and Kapfer DM (2001) Deamidation of human beta B1 alters the elongated structure of the dimer, Exp Eye Res 72, 279–288. [DOI] [PubMed] [Google Scholar]
- [5].Kingsley CN, Brubaker WD, Markovic S, Diehl A, Brindley AJ, Oschkinat H, and Martin RW (2013) Preferential and specific binding of human alphaB-crystallin to a cataract-related variant of gammaScrystallin, Structure 21, 2221–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bari KJ, Sharma S, and Chary KVR (2018) Sequence specific (1)H, (13)C and (15)N resonance assignments of a cataract-related variant G57W of human gammaS-crystallin, Biomol NMR Assign 12, 51–55. [DOI] [PubMed] [Google Scholar]
- [7].Purkiss AG, Bateman OA, Goodfellow JM, Lubsen NH, and Slingsby C (2002) The X-ray crystal structure of human gamma S-crystallin C-terminal domain, J Biol Chem 277, 4199–4205. [DOI] [PubMed] [Google Scholar]
- [8].Sagar V, Chaturvedi SK, Schuck P, and Wistow G (2017) Crystal Structure of Chicken gammaS-Crystallin Reveals Lattice Contacts with Implications for Function in the Lens and the Evolution of the betagamma-Crystallins, Structure 25, 1068–1078.e1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fujii N, Takata T, Fujii N, and Aki K (2016) Isomerization of aspartyl residues in crystallins and its influence upon cataract, Biochim Biophys Acta 1860, 183–191. [DOI] [PubMed] [Google Scholar]
- [10].Lampi KJ, Ma Z, Hanson SR, Azuma M, Shih M, Shearer TR, Smith DL, Smith JB, and David LL (1998) Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry, Exp Eye Res 67, 31–43. [DOI] [PubMed] [Google Scholar]
- [11].Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, and David LL (1997) Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens, J Biol Chem 272, 2268–2275. [DOI] [PubMed] [Google Scholar]
- [12].Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, Pevzner PA, and David LL (2006) Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility?, J Proteome Res 5, 2554–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Z, Smith DL, and Smith JB (2003) Human beta-crystallins modified by backbone cleavage, deamidation and oxidation are prone to associate, Exp Eye Res 77, 259–272. [DOI] [PubMed] [Google Scholar]
- [14].Harrington V, McCall S, Huynh S, Srivastava K, and Srivastava OP (2004) Crystallins in water soluble-high molecular weight protein fractions and water insoluble protein fractions in aging and cataractous human lenses, Mol Vis 10, 476–489. [PubMed] [Google Scholar]
- [15].Harrington V, Srivastava OP, and Kirk M (2007) Proteomic analysis of water insoluble proteins from normal and cataractous human lenses, Mol Vis 13, 1680–1694. [PubMed] [Google Scholar]
- [16].Friedrich MG, Wang Z, Schey KL, and Truscott RJW (2018) Spontaneous cross-linking of proteins at aspartate and asparagine residues is mediated via a succinimide intermediate, Biochem J 475, 3189–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Z, Lyons B, Truscott RJ, and Schey KL (2014) Human protein aging: modification and crosslinking through dehydroalanine and dehydrobutyrine intermediates, Aging Cell 13, 226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lampi KJ, Fox CB, and David LL (2012) Changes in solvent accessibility of wild-type and deamidated betaB2-crystallin following complex formation with alphaA-crystallin, Exp Eye Res 104, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Michiel M, Duprat E, Skouri-Panet F, Lampi JA, Tardieu A, Lampi KJ, and Finet S (2010) Aggregation of deamidated human betaB2-crystallin and incomplete rescue by alpha-crystallin chaperone, Exp Eye Res 90, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mafia K, Gupta R, Kirk M, Wilson L, Srivastava OP, and Barnes S (2008) UV-A-induced structural and functional changes in human lens deamidated alphaB-crystallin, Mol Vis 14, 234–248. [PMC free article] [PubMed] [Google Scholar]
- [21].Annunziata O, Ogun O, and Benedek GB (2003) Observation of liquid-liquid phase separation for eye lens gammaS-crystallin, Proc Natl Acad Sci U S A 100, 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ma Z, Hanson SR, Lampi KJ, David LL, Smith DL, and Smith JB (1998) Age-related changes in human lens crystallins identified by HPLC and mass spectrometry, Exp Eye Res 67, 21–30. [DOI] [PubMed] [Google Scholar]
- [23].Fujii N, Matsumoto S, Hiroki K, and Takemoto L (2001) Inversion and isomerization of Asp-58 residue in human alphaA-crystallin from normal aged lenses and cataractous lenses, Biochim Biophys Acta 1549, 179–187. [DOI] [PubMed] [Google Scholar]
- [24].Fujii N, Takata T, and Fujii N (2015) Quantitative analysis of isomeric (l-alpha-, l-beta-, D-alpha-, D-beta-) aspartyl residues in proteins from elderly donors, J Pharm Biomed Anal 116, 25–33. [DOI] [PubMed] [Google Scholar]
- [25].Geiger T, and Clarke S (1987) Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation, J Biol Chem 262, 785–794. [PubMed] [Google Scholar]
- [26].Takemoto L, Fujii N, and Boyle D (2001) Mechanism of asparagine deamidation during human senile cataractogenesis, Exp Eye Res 72, 559–563. [DOI] [PubMed] [Google Scholar]
- [27].Hooi MY, Raftery MJ, and Truscott RJ (2012) Age-dependent deamidation of glutamine residues in human gammaS crystallin: deamidation and unstructured regions, Protein Sci 21, 1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lapko VN, Purkiss AG, Smith DL, and Smith JB (2002) Deamidation in human gamma S-crystallin from cataractous lenses is influenced by surface exposure, Biochemistry 41, 8638–8648. [DOI] [PubMed] [Google Scholar]
- [29].Takemoto L (2001) Deamidation of Asn-143 of gamma S crystallin from protein aggregates of the human lens, Curr Eye Res 22, 148–153. [DOI] [PubMed] [Google Scholar]
- [30].Takemoto L, and Boyle D (2000) Increased deamidation of asparagine during human senile cataractogenesis, Mol Vis 6, 164–168. [PubMed] [Google Scholar]
- [31].Pande A, Mokhor N, and Pande J (2015) Deamidation of Human gammaS-Crystallin Increases Attractive Protein Interactions: Implications for Cataract, Biochemistry 54, 4890–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ray NJ, Hall D, and Carver JA (2016) Deamidation of N76 in human gammaS-crystallin promotes dimer formation, Biochim Biophys Acta 1860, 315–324. [DOI] [PubMed] [Google Scholar]
- [33].Lampi KJ, Amyx KK, Ahmann P, and Steel EA (2006) Deamidation in human lens betaB2-crystallin destabilizes the dimer, Biochemistry 45, 3146–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Takata T, Oxford JT, Brandon TR, and Lampi KJ (2007) Deamidation alters the structure and decreases the stability of human lens betaA3-crystallin, Biochemistry 46, 8861–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Acosta-Sampson L, and King J (2010) Partially folded aggregation intermediates of human gammaD-, gammaC-, and gammaS-crystallin are recognized and bound by human alphaB-crystallin chaperone, Journal of molecular biology 401, 134–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Flaugh SL, Mills IA, and King J (2006) Glutamine deamidation destabilizes human gammaD-crystallin and lowers the kinetic barrier to unfolding, J Biol Chem 281, 30782–30793. [DOI] [PubMed] [Google Scholar]
- [37].MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments, Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Robinson NE, Lampi KJ, McIver RT, Williams RH, Muster WC, Kruppa G, and Robinson AB (2005) Quantitative measurement of deamidation in lens betaB2-crystallin and peptides by direct electrospray injection and fragmentation in a Fourier transform mass spectrometer, Mol Vis 11, 1211–1219. [PMC free article] [PubMed] [Google Scholar]
- [39].Hall J, Hall A, Pursifull N, and Barbar E (2008) Differences in dynamic structure of LC8 monomer, dimer, and dimer-peptide complexes, Biochemistry 47, 11940–11952. [DOI] [PubMed] [Google Scholar]
- [40].Lampi KJ, Kim YH, Bachinger HP, Boswell BA, Lindner RA, Carver JA, Shearer TR, David LL, and Kapfer DM (2002) Decreased heat stability and increased chaperone requirement of modified human betaB1-crystallins, Mol Vis 8, 359–366. [PubMed] [Google Scholar]
- [41].Thorn DC, Grosas AB, Mabbitt PD, Ray NJ, Jackson CJ, and Carver JA (2019) The Structure and Stability of the Disulfide-Linked gammaS-Crystallin Dimer Provide Insight into Oxidation Products Associated with Lens Cataract Formation, Journal of molecular biology 431, 483–497. [DOI] [PubMed] [Google Scholar]
- [42].Serebryany E, Woodard JC, Adkar BV, Shabab M, King JA, and Shakhnovich EI (2016) An Internal Disulfide Locks a Misfolded Aggregation-prone Intermediate in Cataract-linked Mutants of Human gammaD-Crystallin, J Biol Chem 291, 19172–19183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Serebryany E, Yu S, Trauger SA, Budnik B, and Shakhnovich EI (2018) Dynamic disulfide exchange in a crystallin protein in the human eye lens promotes cataract-associated aggregation, J Biol Chem 293, 17997–18009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kraulis J, Clore GM, Nilges M, Jones TA, Pettersson G, Knowles J, and Gronenborn AM (1989) Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing, Biochemistry 28, 7241–7257. [DOI] [PubMed] [Google Scholar]
- [45].Sarkar R, Comment A, Vasos PR, Jannin S, Gruetter R, Bodenhausen G, Hall H, Kirik D, and Denisov VP (2009) Proton NMR of (15)N-choline metabolites enhanced by dynamic nuclear polarization, J Am Chem Soc 131, 16014–16015. [DOI] [PubMed] [Google Scholar]
- [46].Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, and Kay LE (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation, Biochemistry 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- [47].Mandel AM, Akke M, and Palmer AG 3rd. (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme, Journal of molecular biology 246, 144–163. [DOI] [PubMed] [Google Scholar]
- [48].Cole R, and Loria JP (2003) FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data, J Biomol NMR 26, 203–213. [DOI] [PubMed] [Google Scholar]
- [49].Lou MF, Dickerson JE Jr., Tung WH, Wolfe JK, and Chylack LT Jr. (1999) Correlation of nuclear color and opalescence with protein S-thiolation in human lenses, Exp Eye Res 68, 547–552. [DOI] [PubMed] [Google Scholar]
- [50].Thompson JR, Deane JS, Hall AB, and Rosenthal AR (1997) Associations between lens features assessed in the Oxford Clinical Cataract Classification and Grading System, Ophthalmic Epidemiol 4, 207–212. [DOI] [PubMed] [Google Scholar]
- [51].Hains PG, and Truscott RJ (2007) Post-translational modifications in the nuclear region of young, aged, and cataract human lenses, J Proteome Res 6, 3935–3943. [DOI] [PubMed] [Google Scholar]
- [52].Hooi MY, Raftery MJ, and Truscott RJ (2012) Racemization of two proteins over our lifespan: deamidation of asparagine 76 in gammaS crystallin is greater in cataract than in normal lenses across the age range, Invest Ophthalmol Vis Sci 53, 3554–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Brubaker WD, and Martin RW (2012) (1)H, (1)(3)C, and (1)(5)N assignments of wild-type human gammaS-crystallin and its cataract-related variant gammaS-G18V, Biomol NMR Assign 6, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Takata T, Smith JP, Arbogast B, David LL, and Lampi KJ (2010) Solvent accessibility of betaB2-crystallin and local structural changes due to deamidation at the dimer interface, Exp Eye Res 91, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mills-Henry IA, Thol SL, Kosinski-Collins MS, Serebryany E, and King JA (2019) Kinetic Stability of Long-Lived Human Lens gamma-Crystallins and Their Isolated Double Greek Key Domains, Biophys J 117, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mahler B, Doddapaneni K, Kleckner I, Yuan C, Wistow G, and Wu Z (2011) Characterization of a transient unfolding intermediate in a core mutant of gammaS-crystallin, Journal of molecular biology 405, 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bari KJ, Sharma S, and Chary KVR (2019) Enhanced H/D exchange unravels sequential structural excursions in G57W variant of human gammaS-crystallin with pro-cataractogenic conformations, Biochem Biophys Res Commun 514, 901–906. [DOI] [PubMed] [Google Scholar]
- [58].Bari KJ, Sharma S, and Chary KVR (2019) Conformational dynamics study on human gammaScrystallin as an efficient route to childhood blindness, Biochem Biophys Res Commun 511, 679–684. [DOI] [PubMed] [Google Scholar]
- [59].Sadakane Y, and Kawahara M (2018) Implications of Metal Binding and Asparagine Deamidation for Amyloid Formation, Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Chromatograms of doubly charged γS peptide 7–18 (ITFYEDKNFQGR).
Figure S2. Multiangle laser scattering in-line with size exclusion chromatography of γS crystallins.
Figure S3. Summary of NMR methods.
Figure S4. TROSY overlay of WT with N14D and with N76D.
Figure S5. CLEANEX and day 6 core residue intensities of WT, N14D, and N76D.
Figure S6. NOE values for WT, N14D, and N76D.
Table S1. Measured H/D exchange rates for WT, N14D, and N76D.


