Abstract
The n-alkane-assimilating diploid yeast Candida tropicalis possesses three thiolase isozymes encoded by two pairs of alleles: cytosolic and peroxisomal acetoacetyl-coenzyme A (CoA) thiolases, encoded by CT-T1A and CT-T1B, and peroxisomal 3-ketoacyl-CoA thiolase, encoded by CT-T3A and CT-T3B. The physiological functions of these thiolases have been examined by gene disruption. The homozygous ct-t1aΔ/t1bΔ null mutation abolished the activity of acetoacetyl-CoA thiolase and resulted in mevalonate auxotrophy. The homozygous ct-t3aΔ/t3bΔ null mutation abolished the activity of 3-ketoacyl-CoA thiolase and resulted in growth deficiency on n-alkanes (C10 to C13). All thiolase activities in this yeast disappeared with the ct-t1aΔ/t1bΔ and ct-t3aΔ/t3bΔ null mutations. To further clarify the function of peroxisomal acetoacetyl-CoA thiolases, the site-directed mutation leading acetoacetyl-CoA thiolase without a putative C-terminal peroxisomal targeting signal was introduced on the CT-T1A locus in the ct-t1bΔ null mutant. The truncated acetoacetyl-CoA thiolase was solely present in cytoplasm, and the absence of acetoacetyl-CoA thiolase in peroxisomes had no effect on growth on all carbon sources employed. Growth on butyrate was not affected by a lack of peroxisomal acetoacetyl-CoA thiolase, while a retardation of growth by a lack of peroxisomal 3-ketoacyl-CoA thiolase was observed. A defect of both peroxisomal isozymes completely inhibited growth on butyrate. These results demonstrated that cytosolic acetoacetyl-CoA thiolase was indispensable for the mevalonate pathway and that both peroxisomal acetoacetyl-CoA thiolase and 3-ketoacyl-CoA thiolase could participate in peroxisomal β-oxidation. In addition to its essential contribution to the β-oxidation of longer-chain fatty acids, 3-ketoacyl-CoA thiolase contributed greatly even to the β-oxidation of a C4 substrate butyrate.
Candida tropicalis is an asporogenic diploid yeast which can utilize n-alkanes as a carbon source. The most striking feature of this yeast is profound proliferation of peroxisomes, ubiquitous organelles in eukaryotic cells, in growth on specific carbon sources such as n-alkanes and fatty acids (37). Peroxisomal proteins, including fatty-acid β-oxidation enzymes, are induced, as well as proliferation of peroxisomes (19, 28).
Thiolase catalyzes the thiolytic cleavage of 3-ketoacyl-coenzyme A (CoA) to acetyl-CoA and acyl-CoA, and this enzyme is classified into two types by substrate specificity. One type is acetoacetyl-CoA thiolase (EC 2.3.1.9), which catalyzes the thiolytic cleavage of acetoacetyl-CoA and the reverse condensation of acetyl-CoA. The other is 3-ketoacyl-CoA thiolase (EC 2.3.1.16), which has broad substrate specificity for 3-ketoacyl-CoAs in carbon length (≥C4). In bacterial cells, 3-ketoacyl-CoA thiolase takes part in fatty-acid β-oxidation (7) and acetoacetyl-CoA thiolase takes part in poly-β-hydroxybutyrate metabolism (36, 42). In eukaryotic cells, especially in mammalian cells, thiolases exhibit diversity in intracellular localization related to their metabolic functions as well as in substrate specificity. For example, they contribute to fatty-acid β-oxidation in peroxisomes and mitochondria (31, 34, 46, 49), ketone body metabolism in mitochondria (31), and the early steps of mevalonate pathway in peroxisomes and cytoplasm (11, 31, 48). In addition to biochemical investigations, analyses of genetic disorders have made clear the basis of their functions (33, 41). Genetic studies have also started to disclose the physiological functions of thiolases in the yeast Saccharomyces cerevisiae (10, 12).
In C. tropicalis pK233, there are at least three thiolase isozymes, cytosolic acetoacetyl-CoA thiolase (Cs-Thiolase I), peroxisomal acetoacetyl-CoA thiolase (Ps-Thiolase I), and peroxisomal 3-ketoacyl-CoA thiolase (Thiolase III) (23, 24, 26). Ps-Thiolase I and Thiolase III are each encoded by two extremely similar genes (CT-T1A and CT-T1B for Ps-Thiolase I, and CT-T3A and CT-T3B for Thiolase III) (17, 27). Recently, we have reported that Cs-Thiolase I and Ps-Thiolase I are derived from the same genes (18). As for individual roles of these isozymes in the metabolism of fatty acids, the exclusive localization of β-oxidation in peroxisomes and the inductive expression of peroxisomal isozymes led us to presume that Ps-Thiolase I and Thiolase III participate in peroxisomal β-oxidation, whereas the constitutive localization of Cs-Thiolase I in cytoplasm suggests that Cs-Thiolase I has a role in the mevalonate pathway (19, 25, 26, 51). Information about the physiological roles of thiolase isozymes will be a clue to understanding the evolution of the β-oxidation system and the regulation of peroxisomal β-oxidation. The mechanism of sorting of Thiolase I to two intracellular locations is also important with respect to the metabolic functions of Thiolase I.
In the present study, in order to genetically evaluate the physiological functions of these thiolase isozymes in C. tropicalis, we disrupted their genes and altered the localization of Thiolase I by the deletion of its putative peroxisomal targeting signal sequence. The growth phenotype of strains carrying various combinations of mutations on thiolase genes enabled us to understand the functions of thiolase isozymes.
MATERIALS AND METHODS
Strains and growth conditions.
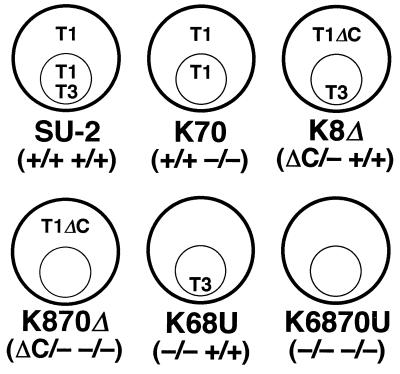
C. tropicalis strains used in this study are classified into representative and intermediate strains and are listed in Table 1 (see also Fig. 1). C. tropicalis SU-2 (ATCC 20913) (ura3a/ura3b) (9), derived from C. tropicalis pK233 (ATCC 20336), was used as a wild-type strain and as a host strain for transformation. Escherichia coli DH5α (3) was used for gene manipulation. S. cerevisiae MT8-1 (MATa ade his3 leu2 trp1 ura3) (47) was used as a host strain for the cloning of C. tropicalis URA3. Media for genetic experiments with C. tropicalis were as follows: YPD (10 g of yeast extract [Difco]/liter, 20 g of peptone [Difco]/liter, and 20 g of glucose/liter), SD (6.7 g of yeast nitrogen base without amino acid [Difco]/liter and 20 g of glucose/liter), SD+U (SD supplemented with 0.1 g of uracil/liter, 0.1 g of uridine/liter, and 0.1 g of UMP/liter), and SD+S (SD containing 1 M sorbitol). l-Mevalonolactone [(R)-(−)-3-hydroxy-3-methyl-5-pentanolide; Wako, Osaka, Japan] (5 g/liter) was used to supplement YPD, SD+U, and SD+S, if necessary.
TABLE 1.
List of C. tropicalis strains used in this study
| Strain | Genotypea | Presence or absence of thiolase genes (T1A/T1B T3A/T3B) |
|---|---|---|
| Representative | ||
| SU-2b | ura3a/ura3b T1A/T1B T3A/T3B | +/+ +/+ |
| K6 | ura3a/ura3b t1aΔ::lacZ/T1B T3A/T3B | −/+ +/+ |
| K8 | ura3a/ura3b T1A/t1bΔ::lacZ T3A/T3B | +/− +/+ |
| K68U | ura3a/ura3b t1aΔ::URA3/t1bΔ::lacZ T3A/T3B | −/− +/+ |
| K7 | ura3a/ura3b T1A/T1B t3aΔ::lacZ/T3B | +/+ −/+ |
| K0 | ura3a/ura3b T1A/T1B T3A/t3bΔ::lacZ | +/+ +/− |
| K70 | ura3a/ura3b T1A/T1B t3aΔ::lacZ/t3bΔ::lacZ | +/+ −/− |
| K870 | ura3a/ura3b T1A/t1bΔ::lacZ t3aΔ::lacZ/t3bΔ::lacZ | +/− −/− |
| K6870U | ura3a/ura3b t1aΔ::URA3/t1bΔ::lacZ t3aΔ::lacZ/t3bΔ::lacZ | −/− −/− |
| K8Δ | ura3a/ura3b T1AΔC6/t1bΔ::lacZ T3A/T3B | ΔC/− +/+ |
| K870Δ | ura3a/ura3b T1AΔC6/t1bΔ::lacZ t3aΔ::lacZ/t3bΔ::lacZ | ΔC/− −/− |
| Intermediate | ||
| K6ZUZ | ura3a/ura3b t1aΔ::ZUZ/T1B T3A/T3B | |
| K8ZUZ | ura3a/ura3b T1A/t1bΔ::ZUZ T3A/T3B | |
| K7ZUZ | ura3a/ura3b T1A/T1B t3aΔ::ZUZ/T3B | |
| K0ZUZ | ura3a/ura3b T1A/T1B T3A/t3bΔ::ZUZ | |
| K70ZUZ | ura3a/ura3b T1A/T1B t3aΔ::lacZ/t3bΔ::ZUZ | |
| K870ZUZ | ura3a/ura3b T1A/t1bΔ::ZUZ t3aΔ::lacZ/t3bΔ::lacZ | |
| K8ΔU | ura3a/ura3b t1aΔ::(T1AΔC6::URA3)/t1bΔ::lacZ T3A/T3Bc | |
| K870ΔU | ura3a/ura3b t1aΔ::(T1AΔC6::URA3)/t1bΔ::lacZ t3aΔ::lacZ/t3bΔ::lacZ |
Abbreviations: T1A, CT-T1A; T1B, CT-T1B; T3A, CT-T3A; T3B, CT-T3B; T1AΔC6, CT-T1AΔC6; ZUZ, lacZ-URA3-lacZ.
Haas et al. (1990) (9).
T1AΔC6 represents the gene encoding the mutant Thiolase I, of which the C-terminal 6 amino acid residues were deleted. t1aΔ::(T1AΔC6::URA3) indicates that the linearized plasmid pUT1AΔ containing CT-T1AΔC6 and URA3 is integrated on the CT-T1A locus.
FIG. 1.
Illustration of subcellular distribution of thiolase isozymes in wild-type and representative mutant strains prepared in this study. The presence or absence of each of the four thiolase genes (T1A/T1B T3A/T3B) in each strain is shown by plus or minus signs in parentheses. Outer and inner circles represent the yeast cell and peroxisome, respectively. Abbreviations: T1, Ps- or Cs-Thiolase I; T3, Thiolase III; T1ΔC, C-terminal-truncated Thiolase I.
C. tropicalis was cultivated aerobically at 30°C in a medium containing glucose (16.5 g/liter), glycerol (20 g/liter), ethanol (20 ml/liter), sodium propionate (10 g/liter), sodium butyrate (11 g/liter), or n-alkane mixture (C10 to C13) (10 ml/liter) as a sole carbon source (25, 50). The pH was adjusted to 5.2 for glucose-, glycerol-, ethanol-, or n-alkane-containing medium and to 6.0 for propionate- or butyrate-containing medium. Tween 80 (0.5 g/liter) was added to n-alkane medium used for preparation of cell extracts and for subcellular fractionation. Supplemental nutrients were added as described above, if necessary. Cell growth was monitored by measuring light scattering at 570 nm.
Cloning and sequencing of C. tropicalis URA3.
To make a minimal genomic DNA library of C. tropicalis (5, 35), genomic DNA of C. tropicalis was digested with NcoI and fractionated in size by 0.5% agarose gel electrophoresis. A fraction containing 6- to 9-kbp DNA fragments was cloned into the NcoI site of the E. coli-S. cerevisiae shuttle vector pMW1 containing the TRP1 selectable marker (16), which was modified to have an NcoI site in multicloning sites. This genomic DNA library was introduced into a uracil-auxotrophic (Ura−) strain, S. cerevisiae MT8-1, by the electroporation method (30). Six uracil prototrophs (Ura+) were obtained from 4.5 × 104 tryptophan-prototrophic (Trp+) transformants. Plasmids were recovered from Ura+ Trp+ candidates. The plasmids contained a 7-kbp fragment, in which a 1.7-kbp BglII fragment was enough to complement ura3 of S. cerevisiae MT8-1. Sequence analysis of the BglII fragment indicated that this fragment contained an 801-base open reading frame, and the deduced amino acid sequence exhibited high similarity to Ura3p’s from various organisms (data not shown). Sequence analysis was carried out with a PRISM DyeDeoxy Terminator Cycle Sequencing Kit and a DNA sequencer (model 373A; Applied Biosystems). The 1.7-kb C. tropicalis URA3 was subcloned into the BamHI site of pUC19 and into the BglII site of the modified pUC19, where a BglII linker was inserted in the SmaI site (the subclones were named pUC-URA3 and pUC-URA3Bg, respectively), and was used for the construction of disruption cassettes as described below.
Construction of disruption cassettes of thiolase isozyme genes.
To disrupt multiple thiolase genes by using the URA3 selectable marker in C. tropicalis SU-2 (uracil auxotrophy), the Ura-blasting procedure was applied (2). In this procedure, URA3 was placed between two directly repeated sequences in a disruption cassette (see Fig. 2). The 1.9-kb part of lacZ (EcoRV-EcoRI fragment) was used as a repeated sequence. Two lacZ fragments were inserted stepwise into pUC-URA3, one in the SmaI site and one in the XbaI site, and all the ends of the fragments were filled with T4 DNA polymerase. Thus a plasmid, pZUZ, containing the lacZ-URA3-lacZ module was constructed.
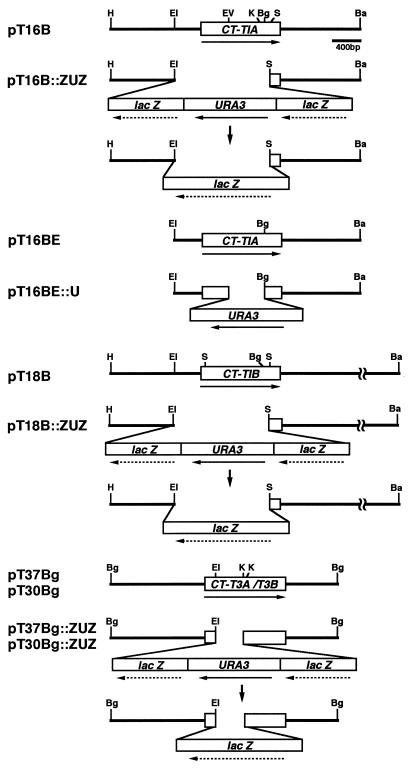
FIG. 2.
Physical maps of thiolase isozyme genes and disruption cassettes, and disruption strategies. Horizontal arrows indicate the orientation of transcription. Vertical arrows indicate the popping out of the lacZ-URA3-lacZ cassette. A boxed region of each thiolase gene shows the open reading frame. Restriction sites: Ba, BamHI; Bg, BglII; EI, EcoRI; EV, EcoRV; H, HindIII; K, KpnI; S, SalI.
pT16BE and pT16B contain the coding and flanking regions of CT-T1A, and pT18B contains those of CT-T1B (18, 27). pT37Bg, carrying the coding and flanking regions of CT-T3A, and pT30Bg, carrying those of CT-T3B, were constructed by insertion of the BglII fragments of λCT-KCT-A and λCT-KCT-B (17) into the modified pUC19 where a BglII linker was inserted into the HincII site and where the EcoRI site was deleted, respectively. EcoRI-SalI fragments (1,400 bp) of pT16B and pT18B were replaced with the lacZ-URA3-lacZ fragment (5,500 bp), which was excised from pZUZ by EcoRI and SalI. The EcoRI-KpnI (600 bp) fragments of pT37Bg and pT30Bg were replaced with the lacZ-URA3-lacZ fragment after the KpnI sites of pT37Bg and pT30Bg had been filled and ligated with a SalI linker, respectively. After a BglII linker was inserted into the EcoRV site of pT16BE, the EcoRV-BglII fragment (500 bp) of pT16BE was replaced with the BglII fragment (1,700 bp) of URA3 excised from pUC-URA3Bg. These constructs were named pT16B::ZUZ, pT18B::ZUZ, pT37Bg::ZUZ, pT30Bg::ZUZ, and pT16BE::U, respectively (see Fig. 2). Before these disruption cassettes were used for transformation, pT16BE::U was linearized with BamHI and EcoRI, pT16B::ZUZ and pT18B::ZUZ were linearized with BamHI, and pT37Bg::ZUZ and pT30Bg::ZUZ were linearized with BglII.
Transformation of C. tropicalis by the spheroplast method.
The spheroplast method developed for S. cerevisiae (4) was applied to the transformation of C. tropicalis with a slight modification: cells were lysed in 20 ml of KPE (1 M sorbitol, 10 mM potassium phosphate buffer [pH 7.2], and 10 mM EDTA) containing 40 μl of mercaptoethanol and 150 μl of Zymolyase 20T solution (10 mg in 1 ml of KPE) at 30°C for 15 min. After 2 to 4 days of incubation of cells transformed by this modified method at 30°C, Ura+ cells formed colonies on selective media at a frequency of approximately 103 colonies/μg of DNA. In order to pop out URA3, Ura+ cells in which the disruption cassette containing lacZ-URA3-lacZ had been integrated were inoculated on a minimal medium containing 5-fluoroorotic acid (5FOA) (SD+U containing 0.75 g of 5FOA/liter) at 30°C for 3 to 4 days. 5FOA-resistant colonies were used as host cells for the next round of transformation. These cells were subjected to Southern blot analysis and PCR at each stage of the process.
Construction of a Thiolase IA mutant with the C terminus deleted.
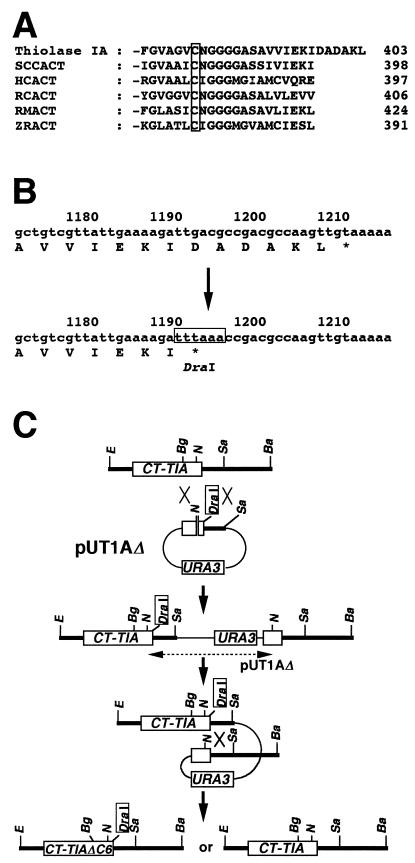
A site-directed mutation on CT-T1A was generated by PCR (13). PCR conditions were as follows: template, 50 pmol of primers, 0.2 mM deoxynucleoside triphosphates, 1× reaction buffer (supplied by the vendor), and 5 U of Pfu DNA polymerase (Stratagene, La Jolla, Calif.). Primers used were as follows: PRT1AN1, 5′-AACCATGGACGACGTCGTTATCG-3′; PRT1AD1, 5′-CTTGGCGTCGGTTTAAATCTTTTCA-3′; PRT1Cl, 5′-GTGCCGAATCGATGTCTAACA-3′; and M13 reverse, 5′-CAGGAAACAGCTATGAC-3′.
First, two sets of PCR were carried out with two primers each, one set with PRT1AN and PRT1AD1 and the other with PRT1Cl and M13 reverse. pT16B (18) was used as a template. After the heteroduplex of two PCR products was formed, the second round of PCR was carried out with PRT1AN1 and M13 reverse. The amplified fragment was digested with BglII and SacI and was cloned into the BamHI and SacI sites of pUC-URA3, with a BamHI linker inserted in the SmaI site. The plasmid was named pUT1AΔ. The inserted fragment was sequenced to check whether mutagenesis and PCR had been correctly performed.
The scheme for introducing the mutation on a chromosome by homologous recombination (40) is shown below (see Fig. 6C). pUT1AΔ was linearized with NcoI prior to its use in transformation. After pUT1AΔ was integrated into CT-T1A on the chromosome, URA3 and vector parts were popped out by 5FOA selection as described above. Subsequently, the desired strain carrying the mutation on CT-T1A was selected from Ura− candidates by Southern blot analysis.
FIG. 6.
(A) Comparison of C-terminal domains of acetoacetyl-CoA thiolases from various sources. The boxed residue is the catalytically important Cys in this domain. Abbreviations: Thiolase IA, C. tropicalis acetoacetyl-CoA thiolase encoded by CT-T1A; SCCACT, S. cerevisiae cytosolic acetoacetyl-CoA thiolase; HCACT, human cytosolic acetoacetyl-CoA thiolase; RCACT, radish cytosolic acetoacetyl-CoA thiolase; RMACT, rat mitochondrial acetoacetyl-CoA thiolase; ZRACT, Zooglea ramigera acetoacetyl-CoA thiolase. Amino acid sequences were retrieved from GenBank/EMBL/DDBJ as accession no. D13470 for Thiolase IA, L20428 for SCCACT, S70154 for HCACT, X78116 for RCACT, D00511 for RMACT, and J02631 for ZRACT. (B) Strategy for deletion of a putative peroxisomal targeting signal of Thiolase I by site-directed mutagenesis. Open box, DraI site. (C) Strategy for the introduction of the site-directed mutation on the CT-T1A locus. Restriction sites: Ba, BamHI; Bg, BglII; E, EcoRI; N, NcoI; Sa, SacI.
Preparation of cell extracts.
Yeast cells were cultivated in 10 ml of each medium and harvested at mid-logarithmic phase. Cells were suspended in 500 μl of 50 mM potassium phosphate buffer (pH 7.2) containing 10% (wt/vol) glycerol, 1 mM dithiothreitol, and protease inhibitors (0.2 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 0.05 g of pepstatin A/liter, 0.05 g of leupeptin/liter, 0.05 g of antipain/liter, and 0.05 g of chymostatin/liter) and were disintegrated by vortexing with 0.3 g of glass beads (diameter, 0.4 to 0.45 mm) in a microtube. Cell extracts were obtained by centrifugation at 14,000 × g for 20 min at 0°C.
Other methods.
Thiolase activity was assayed by monitoring acetoacetyl-CoA or 3-ketooctanoyl-CoA degradation as reported by Kurihara et al. (23). The protein concentration was determined by the Lowry method (29). Subcellular fractionation (51), Western blot analysis (52), and Southern blot analysis (27) were carried out as described previously. General methods for gene manipulation and yeast genetics were used as described in general protocols (3, 14).
Nucleotide sequence accession number.
The nucleotide sequence of C. tropicalis URA3 has been assigned GenBank/EMBL/DDBJ accession no. AB006207.
RESULTS
Development of disruptants of thiolase isozymes.
In order to genetically evaluate the physiological functions of thiolase isozymes in C. tropicalis, two pairs of genes for thiolase isozymes were individually disrupted. C. tropicalis URA3 was cloned to construct disruption cassettes for thiolase isozyme genes with the Ura-blasting procedure (Fig. 1) (see also Materials and Methods) (2).
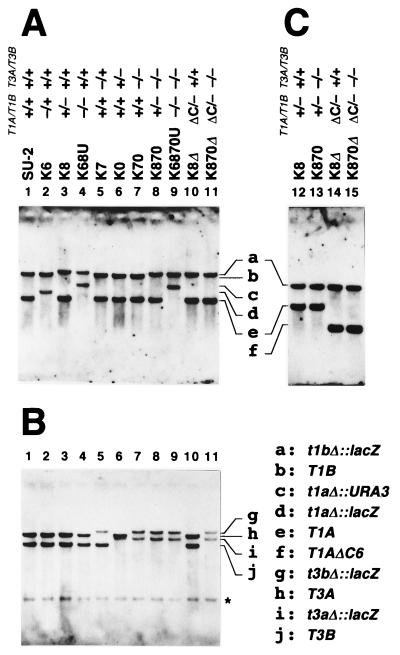
First, we disrupted a single gene among the four thiolase genes. Strains K6ZUZ, K8ZUZ, K7ZUZ, and K0ZUZ were obtained as Ura+ transformants from the wild-type strain C. tropicalis SU-2 (9, 38) by using disruption cassettes pT16B::ZUZ, pT18::ZUZ, pT37Bg::ZUZ, and pT30Bg::ZUZ (Fig. 2), respectively. Following selection of Ura− segregants on the basis of resistance to 5FOA, strains K6, K8, K7, and K0 were obtained. Southern blot analysis indicated that the desired chromosomal regions were correctly replaced with lacZ-URA3-lacZ in K6ZUZ, K8ZUZ, K7ZUZ, and K0ZUZ and that URA3 was eliminated in K6, K8, K7, and K0 by the 5FOA treatment (data not shown). Compared with thiolase genes in the wild-type strain, SU-2, the increased size of each disrupted thiolase gene on a Southern blot also showed that the first round of transformation was successful (Fig. 3). The shift of each band, or the disappearance of a band at the position seen in the SU-2 lane, revealed that each gene was present as a single copy, suggesting that the almost identical A and B genes were allelic in the diploid yeast C. tropicalis. Therefore, we can regard K6 and K8 as the hemizygous CT-T1A/T1B null mutants and K7 and K0 as the hemizygous CT-T3A/T3B null mutants.
FIG. 3.
Southern blot analysis of mutant strains derived from C. tropicalis SU-2. Genomic DNA was digested with EcoRI and BamHI (A and B) and with EcoRI, BamHI, and BanIII (C). The blots were probed with biotin-labeled cDNA of Ps-Thiolase I (28) (A and C) or Thiolase III (17) (B). Panels A and B are the same blot in each lane. The presence or absence of each thiolase gene is indicated as explained for Fig. 1. The genotype corresponding to each band is given in the key. Asterisk, nonspecific signal.
Second, the homozygous ct-t1aΔ/t1bΔ null mutant and the homozygous ct-t3aΔ/t3bΔ null mutant were developed from K8 and K7, respectively. Cs-Thiolase I is expected to have a role in the mevalonate pathway. The ct-t1aΔ/t1bΔ mutant, therefore, was expected to show mevalonate auxotrophy. Thus, in the selection of this mutant, l-mevalonolactone was included in the selective medium. However, the disruption using pT16::ZUZ was not successful despite the use of the medium containing l-mevalonolactone. Consequently, an improved disruption cassette for CT-T1A, pT16BE::U, was constructed (Fig. 2), in which one of two regions homologous to CT-T1A was exchanged with the region eliminated in the CT-T1B locus in K8 after the first round of transformation. Using this vector, we successfully disrupted CT-T1A to obtain the ct-t1aΔ/t1bΔ mutant K68U as shown on a Southern blot (Fig. 3). The ct-t3aΔ/t3bΔ mutant K70 was developed from K7 by using pT30::ZUZ, followed by the elimination of URA3 (Fig. 3).
Third, to obtain the homozygous ct-t1aΔ/t1bΔ ct-t3aΔ/t3bΔ null mutant K6870U, K70 was transformed by the same method that was applied to develop K68U from SU-2. K870ZUZ and K870 were obtained as intermediate strains. In K6870U, all bands of the genes encoding thiolase isozymes shifted in size (Fig. 3), revealing the correct disruption of all four genes.
Expression of thiolase isozymes in mutant strains.
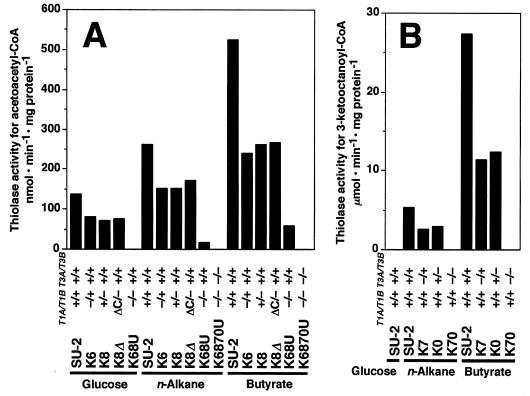
Development of a series of disruptants enabled us to examine the expression level of each isozyme and its contribution to thiolase activity in the cells. The expression of Thiolase I’s and Thiolase III in disruptants was monitored by thiolase activity and Western blot analysis (Fig. 4 and 5). According to substrate specificity, the activity of Thiolase I’s was represented mainly by the degradation of acetoacetyl-CoA and the activity of Thiolase III was represented exactly by the degradation of 3-ketooctanoyl-CoA (23, 24).
FIG. 4.
Thiolase activities of mutants for acetoacetyl-CoA (A) and 3-ketooctanoyl-CoA (B). The activities for acetoacetyl-CoA and 3-ketooctanoyl-CoA represent the activities of Thiolase I and Thiolase III, respectively. The presence or absence of each thiolase gene is indicated as explained for Fig. 1. Carbon sources for growth are displayed.
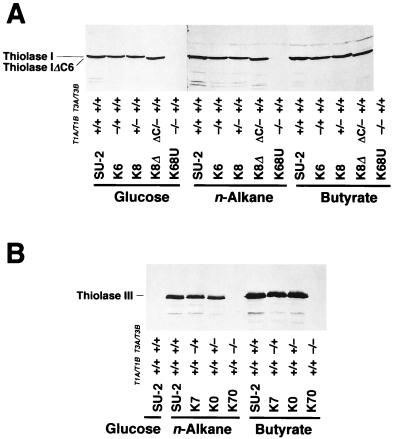
FIG. 5.
Western blot analysis of various Thiolase I (A) and Thiolase III (B) mutant strains. Aliquots of 50 μg (glucose) and 25 μg (n-alkane and butyrate) (A) and 20 μg (B) of cell extracts were run on gels. Thiolase I and Thiolase III were detected with anti-Ps-Thiolase I and anti-Thiolase III antiserum, respectively. The presence or absence of each thiolase gene is indicated as explained for Fig. 1. Carbon sources for growth are displayed. Thiolase IΔC6, C-terminus-truncated mutant of Thiolase I.
In the hemizygous CT-T1A/T1B null mutants, K6 and K8, the thiolase activity for acetoacetyl-CoA was half that of the wild-type strain, SU-2, on all carbon sources tested (Fig. 4A). Also, in the hemizygous CT-T3A/T3B null mutants, K7 and K0, the activity for 3-ketooctanoyl-CoA was half of that in SU-2 when these strains were grown on n-alkanes and butyrate (Fig. 4B). The band intensities of Thiolase I and Thiolase III in Western blot analysis paralleled the levels of thiolase activity in the wild-type and disruptant strains (Fig. 5). These results indicated that the expression of the A and B genes of Thiolase I and Thiolase III contributed equally to total intracellular thiolase activity and that their regulation in response to the carbon source was identical. These results confirmed that the A and B genes of both Thiolase I and Thiolase III were allelic. As for the homozygous null mutants, no thiolase activity for acetoacetyl-CoA was detected in the ct-t1aΔ/1bΔ mutant K68U grown on glucose (Fig. 4A). Residual activity for acetoacetyl-CoA was detected in K68U cells grown on n-alkanes and butyrate, but it was supposed to be the contribution of Thiolase III, which shows broad-chain-length specificity. This is strongly supported by the fact that this residual activity was abolished in the ct-t1aΔ/t1bΔ ct-t3aΔ/t3bΔ mutant K6870U (Fig. 4A). There was no protein detected by anti-Thiolase I antiserum in K68U grown on any of the carbon sources tested (Fig. 5A). No thiolase activity for 3-ketooctanoyl-CoA was detected in the ct-t3aΔ/t3bΔ mutant K70 grown on n-alkanes and butyrate (Fig. 4B), and no band was detected by anti-Thiolase III antiserum in K70 (Fig. 5B). Furthermore, in K6870U, no thiolase activity was found in the cells (Fig. 4A) and no protein was recognized by either anti-Thiolase I or anti-Thiolase III antiserum (data not shown) (see Fig. 5), indicating that in C. tropicalis there are only three thiolase isozymes encoded by the two pairs of alleles, as we have previously suggested (17, 27).
Development of the strains expressing the C-terminus-truncated protein of Thiolase I.
Compared with the amino acid sequences of acetoacetyl-CoA thiolases of other organisms, only Thiolase I of C. tropicalis has an additional 6 amino acid residues at the C terminus: DADAKL for Thiolase IA encoded by CT-T1A and DSDAKL for Thiolase IB encoded by CT-T1B, in which there is a putative motif of peroxisomal targeting signal type I (PTS1) (8, 27) (Fig. 6A). In order to investigate whether this sequence functions as a PTS1 and, furthermore, to distinguish the physiological roles of Cs- and Ps-Thiolase I’s, we developed two further strains, K8Δ (CT-T1AΔC6/ct-t1bΔ CT-T3A/CT-T3B) and K870Δ (CT-T1AΔC6/ct-t1bΔ ct-t3aΔ/ct-t3bΔ). These strains express only the C-terminus-truncated Thiolase I with and without Thiolase III, respectively. A nonsense codon was introduced into CT-T1A to delete the C-terminal 6 amino acids of Thiolase I by site-directed mutagenesis (Fig. 6B). A DraI restriction site was also introduced as a marker for this mutation, and then a mutation cassette, pUT1AΔ, was constructed. By using this cassette, these mutations were incorporated on the CT-T1A locus in K8 and K870 (Fig. 6C). Southern blot analysis showed that the CT-T1A locus in both K8Δ and K870Δ could be digested by DraI although BamHI-EcoRI fragments were identical to that of the wild type in size, indicating that the mutation was correctly introduced onto the CT-T1A locus in these strains (Fig. 3).
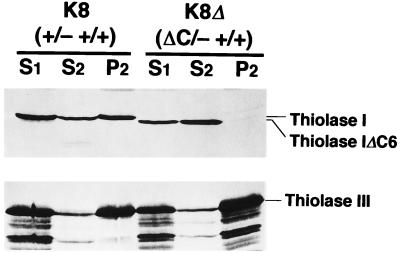
The expression and subcellular localization of the C-terminus-truncated Thiolase I in K8Δ were examined. The truncated Thiolase I was expressed as a slightly smaller protein than the wild-type Thiolase I (Fig. 5A). The thiolase activity for acetoacetyl-CoA and the band intensity of Thiolase I in K8Δ were essentially identical to those in the parent strain, K8, on all carbon sources tested (Fig. 4A and 5A). These results revealed that the C-terminal 6 amino acid residues of Thiolase I did not have any function for the enzymatic activity of Thiolase I and that this truncated protein was present in a completely active form. The truncated protein was also expressed in K870Δ (data not shown). The postnuclear supernatant fractions (S1) of strains K8 and K8Δ grown on n-alkanes were separated to cytoplasm/microsome fractions (S2) and organelle fractions (P2) at 20,000 × g (Fig. 7). Thiolase I was present only in the S2 fraction in K8Δ, whereas it was present in both the S2 and P2 fractions in K8. Proper subcellular fractionation was confirmed by the presence of the majority of Thiolase III in the P2 fraction. These results demonstrated that the C-terminal residues of Thiolase I functioned as a PTS1 in C. tropicalis and that the localization of Thiolase I was successfully restricted to the cytoplasm.
FIG. 7.
Subcellular distribution of the wild-type Thiolase I [K8 (+/− +/+)] and C-terminus-deleted Thiolase I [K8Δ (ΔC/− +/+)]. Cells grown on n-alkanes were harvested at mid-logarithmic phase, lysed to protoplast, homogenized, and separated to nuclear and postnuclear fractions (51). S1, S2, and P2 represent postnuclear supernatant, cytoplasm/microsome, and organelle fractions, respectively. Protein (20 μg) from each fraction was run on gels. Thiolases were detected with anti-Ps-Thiolase I (upper panel) and anti-Thiolase III (lower panel) antisera. Thiolase IΔC6, C-terminus-truncated Thiolase I.
Mevalonate requirement of mutant strains.
The ct-t1aΔ/t1bΔ mutants K68U and K6870U could be obtained in an SD+S medium containing l-mevalonolactone. In the yeast S. cerevisiae, cytosolic acetoacetyl-CoA thiolase encoded by ERG10 has been genetically shown to be essential for the mevalonate pathway (10). If a thiolase isozyme in C. tropicalis catalyzes the initial step of this pathway, its deficiency would result in mevalonate auxotrophy. K68U and K6870U, both of which lacked Thiolase I, could grow on YPD medium only when it was supplemented with l-mevalonolactone. K70, which lacks Thiolase III, K8Δ, which lacks Ps-Thiolase I, and K870Δ, which lacks Ps-Thiolase I and Thiolase III, did not require mevalonate, as was the case with the wild-type strain, SU-2. These results suggest that Cs-Thiolase I has an indispensable role in the mevalonate pathway.
Cell growth of mutant strains on various carbon sources.
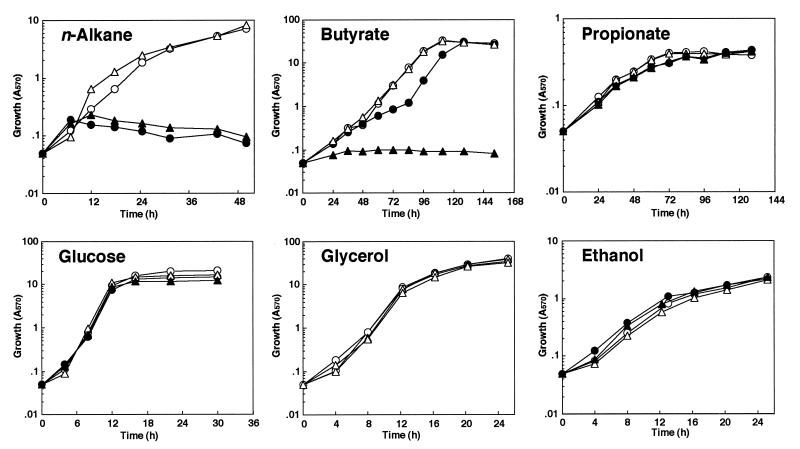
Previously, we reported that C. tropicalis could utilize a short-chain fatty acid, butyrate, as well as n-alkanes and longer-chain fatty acids (25). In butyrate-grown cells, peroxisomes and the enzymes of the peroxisomal β-oxidation system were induced. Therefore, the contributions of thiolase isozymes to fatty-acid β-oxidation and/or to other metabolic processes were examined by observation of the cell growth of thiolase disruptants on various carbon sources (Fig. 8).
FIG. 8.
Growth kinetics of wild-type and mutant strains on various carbon sources. Open circle, solid circle, open triangle, and solid triangle represent strains SU-2 (+/+ +/+), K70 (+/+ −/−), K8Δ (ΔC/− +/+), and K870Δ (ΔC/− −/−), respectively.
Significant differences in growth were not observed among the wild-type strain, SU-2, and the hemizygous thiolase gene null mutants K6, K8, K7, and K0 on glucose, n-alkane, and butyrate (data not shown), indicating that neither the A nor the B gene has an independent physiological role in cell growth. K70, which lacks Thiolase III, could not grow on n-alkanes (C10 to C13), whereas K8Δ, which lacks Ps-Thiolase I, exhibited growth on n-alkanes. K70 could not grow on oleic acid either (data not shown). These results demonstrated that Thiolase III was indispensable for β-oxidation of long-chain fatty acids. On butyrate, however, K8Δ also showed good growth, while the growth of K70 was retarded, but the growth of both strains reached almost the same level as that of the wild-type strain, SU-2, in the stationary-growth phase. No growth on butyrate was observed for K870Δ, which lacks both Ps-Thiolase I and Thiolase III, suggesting that butyrate was utilized solely through peroxisomal β-oxidation. This fact supports the induction of β-oxidation enzymes in butyrate-utilizing cells (25). There was no significant difference among strains SU-2, K70, and K8Δ in cell growth on another short-chain fatty acid, propionate, on glucose, or on the nonfermentable carbon sources, glycerol and ethanol (Fig. 8); these results also indicate the indispensable participation of peroxisomal thiolase isozymes in β-oxidation.
DISCUSSION
In the diploid yeast C. tropicalis, we disrupted the thiolase isozyme genes and altered the distribution of Thiolase I in order to elucidate the physiological functions of the thiolase isozymes. Intracellular thiolase activity was completely abolished in the double homozygous mutant lacking the Thiolase I and Thiolase III genes, indicating that there is no thiolase in C. tropicalis other than Cs-Thiolase I, Ps-Thiolase I, and Thiolase III, which are encoded by two pairs of alleles. For S. cerevisiae, it has been reported that there are peroxisomal 3-ketoacyl-CoA thiolase (Pot1p/Fox3p), cytosolic acetoacetyl-CoA thiolase (Erg10p), and mitochondrial acetoacetyl-CoA thiolase (10, 12, 22), although the gene encoding the mitochondrial enzyme has not been cloned yet. In mammalian cells, there are at least five thiolase isozymes, and they are encoded by distinct genes. Compared with these systems, C. tropicalis has a simple set of thiolase isozymes encoded by two pairs of allelic genes.
Experiments with deletions of the C-terminal 6 amino acid residues of Thiolase I, DADAKL, revealed the necessity of the sequence for targeting of the enzyme to peroxisomes in C. tropicalis. The last 3 residues, AKL, are in good agreement with one motif of PTS1 (8). The transport of peroxisomal proteins of C. tropicalis to peroxisomes has been examined for acyl-CoA oxidase and the multifunctional protein, but these studies were performed in in vitro systems or in heterologous in vivo systems (1, 15, 43). Therefore, the present result marks the first case for C. tropicalis in which a peroxisomal targeting signal was identified in a homologous in vivo system.
The expression of Thiolase I (Ps-Thiolase I and Cs-Thiolase I) genes was totally induced in response to n-alkane utilization (Fig. 5A) (17, 26), but Cs-Thiolase I is present constitutively (26) and contributes indispensably to the mevalonate pathway. Therefore, it is important that Thiolase I is sorted into peroxisomes and cytoplasm in a regulated manner. Many mechanisms for the sorting of a single protein to dual compartments have been proposed (6). Recently, the 4th residue of C-terminal PTS1 of human catalase has been shown to be important in determining the efficiency of transportation of catalase to peroxisomes, which was attributed to the binding affinity of PTS1 for PTS1 receptor (39). Further detailed analysis of the C-terminal 6 amino acids will be necessary to reveal their relation to the dual sorting mechanism of Thiolase I.
The present results demonstrated that Cs-Thiolase I was essential for the mevalonate pathway, the early steps of sterol synthesis, in C. tropicalis. This physiological function is consistent with the enzymatic properties of Thiolase I, i.e., the activity for condensation reaction of acetyl-CoA units (18). It has also been shown that Thiolase I which can catalyze the condensation reaction was present both in cytoplasm and in peroxisomes of C. tropicalis (18). In mammalian cells also, the condensation reaction of thiolase is detected in these two compartments (11, 32, 48), and additionally, 3-hydroxy-3-methylglutaryl-CoA reductase, which is the rate-limiting enzyme in the mevalonate pathway, is colocalized with the condensation reaction (20, 21), suggesting two pathways in the early steps of sterol synthesis. However, this reductase has not been detected in peroxisomes of C. tropicalis (26), and lack of Ps-Thiolase I had no significant effect on growth (Fig. 8). Therefore, it is suggested that the early steps of sterol synthesis occur only in the cytoplasm of this yeast.
The fatty-acid β-oxidation system is present only in peroxisomes in C. tropicalis (19). The present results for thiolase isozymes clarified this observation that peroxisomal β-oxidation exclusively contributes to fatty-acid degradation. On the other hand, unlike lack of Ps-Thiolase I, lack of Thiolase III resulted in growth retardation on butyrate (Fig. 8), suggesting that Thiolase III degraded acetoacetyl-CoA more efficiently than Ps-Thiolase I did in peroxisomal β-oxidation. The results were not consistent with the biochemical observations that Thiolase I has much higher specific activity for a C4 substrate, acetoacetyl-CoA, than Thiolase III (23, 24). Furthermore, most of the thiolase activity for acetoacetyl-CoA was due to Thiolase I (Fig. 4A). Therefore, we presume that the reasons for the growth retardation brought about by lack of Thiolase III are as follows. First, the contribution of each isozyme to β-oxidation might be determined by its quantity. In C. tropicalis peroxisomes, the amount of Thiolase III is much higher than that of Ps-Thiolase I. The molar ratio of the two isozymes (Thiolase III/Ps-Thiolase I) can be estimated as approximately 16 for the native enzyme and 5.2 for the subunit from specific activities of the peroxisomal fraction and the purified proteins (23, 24). Second, Thiolase III might be one component of a β-oxidation multienzyme complex in C. tropicalis. There is a “metabolon” hypothesis which suggests that enzymes included in a metabolic pathway form a multienzyme complex to bring about an efficient metabolic flux (44, 45). If Thiolase III belongs to a multienzyme complex, lack of Thiolase III would result in the inhibition of β-oxidation despite the presence of Ps-Thiolase I.
In C. tropicalis peroxisomes, either Ps-Thiolase I or Thiolase III allows this yeast to utilize butyrate through peroxisomal β-oxidation. This is the first genetic demonstration that acetoacetyl-CoA thiolase participates in peroxisomal β-oxidation. This system can be taken advantage of to alter the flux of peroxisomal β-oxidation in growing conditions and will give us insight into the relation between the control of flux and the regulation of gene expression in peroxisomal β-oxidation.
ACKNOWLEDGMENTS
We thank Takahito Suzuki and Shin-ichi Iwaguchi, Nara Women’s University, for their technical advice and suggestions on the transformation of C. tropicalis. We thank Akihiro Hara, Japan Energy Corp., for helpful discussions.
N.K. is a research fellow of the Japan Society for the Promotion of Science (JSPS). This study was supported in part by a grant-in-aid for JSPS fellows from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Aitchison J D, Murray W W, Rachubinski R A. The carboxyl-terminal tripeptide Ala-Lys-Ile is essential for targeting Candida tropicalis trifunctional enzyme to yeast peroxisomes. J Biol Chem. 1991;266:23197–23203. [PubMed] [Google Scholar]
- 2.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Burgers P M J, Percival K J. Transformation of yeast spheroplasts without cell fusion. Anal Biochem. 1987;163:391–397. doi: 10.1016/0003-2697(87)90240-5. [DOI] [PubMed] [Google Scholar]
- 5.Cregg J M, Barringer K J, Hessler A Y, Madden K R. Pichia pastoris as a host system for transformation. Mol Cell Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danpure C J. How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol. 1995;5:230–238. doi: 10.1016/s0962-8924(00)89016-9. [DOI] [PubMed] [Google Scholar]
- 7.Feigenbaum J, Schulz H. Thiolases of Escherichia coli: purification and chain length specificities. J Bacteriol. 1975;122:407–411. doi: 10.1128/jb.122.2.407-411.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gould S J, Keller G A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas L O C, Cregg J M, Gleeson M A G. Development of an integrative DNA transformation system for the yeast Candida tropicalis. J Bacteriol. 1990;172:4571–4577. doi: 10.1128/jb.172.8.4571-4577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiser L, Basson M E, Rine J. ERG10 from Saccharomyces cerevisiae encodes acetoacetyl-CoA thiolase. J Biol Chem. 1994;269:31383–31389. [PubMed] [Google Scholar]
- 11.Hovik R, Brodal B, Bartlett K, Osmundsen H. Metabolism of acetyl-CoA by isolated peroxisomal fractions: formation of acetate and acetoacetyl-CoA. J Lipid Res. 1991;32:993–999. [PubMed] [Google Scholar]
- 12.Igual J C, Matallana E, Gonzalez-Bosch C, Franco L, Pérez-Ortin J E. A new glucose-repressible gene identified from the analysis of chromatin structure in deletion mutants of yeast SUC2 locus. Yeast. 1991;7:379–389. doi: 10.1002/yea.320070408. [DOI] [PubMed] [Google Scholar]
- 13.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 15.Kamiryo T, Sakasegawa Y, Tan H. Expression and transport of Candida tropicalis peroxisomal acyl-coenzyme A oxidase in the yeast Candida maltosa. Agric Biol Chem. 1989;53:179–186. [Google Scholar]
- 16.Kanai T, Atomi H, Umemura K, Ueno H, Teranishi Y, Ueda M, Tanaka A. A novel heterologous gene expression system in Saccharomyces cerevisiae using the isocitrate lyase gene promoter from Candida tropicalis. Appl Microbiol Biotechnol. 1996;44:759–765. doi: 10.1007/BF00178615. [DOI] [PubMed] [Google Scholar]
- 17.Kanayama N, Ueda M, Atomi H, Kurihara T, Kondo J, Teranishi Y, Tanaka A. Comparison of molecular structures and regulation of biosynthesis of unique thiolase isozymes localized only in peroxisomes of n-alkane-utilizing yeast, Candida tropicalis. J Ferment Bioeng. 1994;78:273–278. [Google Scholar]
- 18.Kanayama N, Himeda Y, Atomi H, Ueda M, Tanaka A. Expression of acetoacetyl-CoA thiolase isozyme genes of n-alkane-assimilating yeast, Candida tropicalis: isozymes in two intracellular compartments are derived from the same genes. J Biochem (Tokyo) 1997;122:616–621. doi: 10.1093/oxfordjournals.jbchem.a021797. [DOI] [PubMed] [Google Scholar]
- 19.Kawamoto S, Nozaki C, Tanaka A, Fukui S. Fatty acid β-oxidation system in microbodies of n-alkane-grown Candida tropicalis. Eur J Biochem. 1978;83:609–613. doi: 10.1111/j.1432-1033.1978.tb12130.x. [DOI] [PubMed] [Google Scholar]
- 20.Keller G A, Barton M C, Shapiro D J, Singer S J. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase is present in peroxisomes in normal rat liver cells. Proc Natl Acad Sci USA. 1985;82:770–774. doi: 10.1073/pnas.82.3.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller G A, Pazirandeh M, Krisans S. 3-Hydroxy-3-methylglutaryl coenzyme A reductase localization in rat liver peroxisomes and microsomes of control and cholestyramine-treated animals: quantitative biochemical and immunoelectron microscopical analyses. J Cell Biol. 1986;103:875–886. doi: 10.1083/jcb.103.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblatt J A, Rudney H. Two forms of acetoacetyl coenzyme A thiolase in yeast. II. Intracellular location and relationship to growth. J Biol Chem. 1971;246:4424–4430. [PubMed] [Google Scholar]
- 23.Kurihara T, Ueda M, Tanaka A. Occurrence and possible roles of acetoacetyl-CoA thiolase and 3-ketoacyl-CoA thiolase in peroxisomes of an n-alkane-grown yeast, Candida tropicalis. FEBS Lett. 1988;229:215–218. doi: 10.1016/0014-5793(88)80830-5. [DOI] [PubMed] [Google Scholar]
- 24.Kurihara T, Ueda M, Tanaka A. Peroxisomal acetoacetyl-CoA thiolase and 3-ketoacyl-CoA thiolase from an n-alkane-utilizing yeast, Candida tropicalis: purification and characterization. J Biochem (Tokyo) 1989;106:474–478. doi: 10.1093/oxfordjournals.jbchem.a122876. [DOI] [PubMed] [Google Scholar]
- 25.Kurihara T, Ueda M, Okada H, Kamasawa N, Naito N, Osumi M, Tanaka A. β-Oxidation of butyrate, the short-chain-length fatty acid, occurs in peroxisomes in the yeast Candida tropicalis. J Biochem (Tokyo) 1992;111:783–787. doi: 10.1093/oxfordjournals.jbchem.a123836. [DOI] [PubMed] [Google Scholar]
- 26.Kurihara T, Ueda M, Okada H, Kamasawa N, Osumi M, Tanaka A. Physiological roles of acetoacetyl-CoA thiolase in n-alkane-utilizable yeast, Candida tropicalis: possible contribution to alkane degradation and sterol biosynthesis. J Biochem (Tokyo) 1992;112:845–848. doi: 10.1093/oxfordjournals.jbchem.a123987. [DOI] [PubMed] [Google Scholar]
- 27.Kurihara T, Ueda M, Kanayama N, Kondo J, Teranishi Y, Tanaka A. Peroxisomal acetoacetyl-CoA thiolase of an n-alkane-utilizing yeast, Candida tropicalis. Eur J Biochem. 1992;210:999–1005. doi: 10.1111/j.1432-1033.1992.tb17505.x. [DOI] [PubMed] [Google Scholar]
- 28.Lazarow P B, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 29.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Meilhoc E, Masson J M, Teissié J. High efficiency transformation of intact yeast cells by electric field pulses. Bio/Technology. 1990;8:223–227. doi: 10.1038/nbt0390-223. [DOI] [PubMed] [Google Scholar]
- 31.Middleton B. The oxoacyl-coenzyme A thiolases of animal tissues. Biochem J. 1973;132:717–730. doi: 10.1042/bj1320717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middleton B. The kinetic mechanism and properties of the cytoplasmic acetoacetyl-coenzyme A thiolase from rat liver. Biochem J. 1974;139:109–121. doi: 10.1042/bj1390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Middleton B, Bartlett K. The synthesis and characterization of 2-methylacetoacetyl coenzyme A and its use in the identification of the site of the defect in 2-methylacetoacetic and 2-methyl-3-hydroxybutyric aciduria. Clin Chim Acta. 1983;128:291–305. doi: 10.1016/0009-8981(83)90329-7. [DOI] [PubMed] [Google Scholar]
- 34.Miyazawa S, Osumi T, Hashimoto T. The presence of a new 3-oxoacyl-CoA thiolase in rat liver peroxisomes. Eur J Biochem. 1980;103:589–596. doi: 10.1111/j.1432-1033.1980.tb05984.x. [DOI] [PubMed] [Google Scholar]
- 35.Nicholls R D, Hill A V S, Clegg J B, Higgs D R. Direct cloning of specific genomic DNA sequences in plasmid libraries following fragment enrichment. Nucleic Acids Res. 1985;13:7569–7578. doi: 10.1093/nar/13.21.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oeding V, Schlegel H G. β-Ketothiolase from Hydrogenomonas eutropha H16 and its significance in the regulation of poly-β-hydroxybutyrate metabolism. Biochem J. 1973;134:239–248. doi: 10.1042/bj1340239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osumi M, Miwa N, Teranishi Y, Tanaka A, Fukui S. Ultrastructure of Candida yeasts grown on n-alkanes. Appearance of microbodies and its relationship to high catalase activity. Arch Microbiol. 1974;99:181–201. doi: 10.1007/BF00696234. [DOI] [PubMed] [Google Scholar]
- 38.Picataggio S, Deanda K, Mielenz J. Determination of Candida tropicalis acyl coenzyme A oxidase isozyme function by sequential gene disruption. Mol Cell Biol. 1991;11:4333–4339. doi: 10.1128/mcb.11.9.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purdue P E, Lazarow P B. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J Cell Biol. 1996;134:849–862. doi: 10.1083/jcb.134.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schram A W, Goldfischer S, van Roermund C W T, Brouwer-Kelder E M, Collins J, Hashimoto T, Heymans H S A, van den Bosch H, Schutgens R B H, Tager J M, Wanders R J A. Human peroxisomal 3-oxoacyl-coenzyme A thiolase deficiency. Proc Natl Acad Sci USA. 1987;84:2494–2496. doi: 10.1073/pnas.84.8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senior P J, Dawes E A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–238. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small G M, Szabo L J, Lazarow P B. Acyl-CoA oxidase contains two targeting sequences each of which can mediate protein import into peroxisomes. EMBO J. 1988;7:1167–1173. doi: 10.1002/j.1460-2075.1988.tb02927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srere P A. Complexes of sequential metabolic enzymes. Annu Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 45.Srere P A, Sumegi B. Processivity and fatty acid oxidation. Biochem Soc Trans. 1994;22:446–450. doi: 10.1042/bst0220446. [DOI] [PubMed] [Google Scholar]
- 46.Staack H, Binstock J F, Schulz H. Purification and properties of a pig heart thiolase with broad chain length specificity and comparison of thiolases from pig heart and Escherichia coli. J Biol Chem. 1978;253:1827–1831. [PubMed] [Google Scholar]
- 47.Tajima M, Nogi Y, Fukasawa T. Primary structure of the Saccharomyces cerevisiae GAL7 gene. Yeast. 1985;1:67–77. doi: 10.1002/yea.320010108. [DOI] [PubMed] [Google Scholar]
- 48.Thompson S L, Krisans S K. Rat liver peroxisomes catalyze the initial step in cholesterol synthesis. The condensation of acetyl-CoA units into acetoacetyl-CoA. J Biol Chem. 1990;265:5731–5735. [PubMed] [Google Scholar]
- 49.Uchida Y, Izai K, Orii T, Hashimoto T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. II. Purification and properties of enoyl-coenzyme A (CoA) hydratase/3-hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase trifunctional protein. J Biol Chem. 1992;267:1034–1041. [PubMed] [Google Scholar]
- 50.Ueda M, Okada H, Tanaka A, Osumi M, Fukui S. Induction and subcellular localization of enzymes participating in propionate metabolism in Candida tropicalis. Arch Microbiol. 1983;136:169–176. doi: 10.1007/BF00409839. [DOI] [PubMed] [Google Scholar]
- 51.Ueda M, Yamanoi K, Morikawa T, Okada H, Tanaka A. Peroxisomal localization of enzymes related to fatty acid β-oxidation in an n-alkane-grown yeast, Candida tropicalis. Agric Biol Chem. 1985;49:1821–1828. [Google Scholar]
- 52.Ueda M, Mozaffar S, Atomi H, Osumi M, Tanaka A. Characterization of peroxisomes in n-alkane-utilizable yeast, Candida tropicalis, grown on glucose and propionate. J Ferment Bioeng. 1989;68:411–416. [Google Scholar]