Summary
Background
HIV-1-associated immune activation drives CD4+ T cell depletion and the development of acquired immunodeficiency syndrome. We aimed to determine the role of nicotinamide mononucleotide (NMN), the direct precursor of nicotinamide adenine dinucleotide (NAD) co-enzyme, in CD4+ T cell modulation during HIV-1 infection.
Methods
We examined HIV-1 integrated DNA or transcribed RNA, intracellular p24 protein, and T cell activation markers in CD4+ T cells including in vitro HIV-1-infected cells, reactivated patient-derived cells, and in HIV-1-infected humanized mice, under NMN treatment. RNA-seq and CyTOF analyses were used for investigating the effect of NMN on CD4+ T cells.
Findings
We found that NMN increased the intracellular NAD amount, resulting in suppressed HIV-1 p24 production and proliferation in infected CD4+ T cells, especially in activated CD25+CD4+ T cells. NMN also inhibited CD25 expression on reactivated resting CD4+ T cells derived from cART-treated people living with HIV-1 (PLWH). In HIV-1-infected humanized mice, the frequency of CD4+ T cells was reconstituted significantly by combined cART and NMN treatment as compared with cART or NMN alone, which correlated with suppressed hyperactivation of CD4+ T cells.
Interpretation
Our results highlight the suppressive role of NMN in CD4+ T cell activation during HIV-1 infection. It warrants future clinical investigation of NMN as a potential treatment in combination with cART in PLWH.
Funding
This work was supported by the Hong Kong Research Grants Council Theme-Based Research Scheme (T11-706/18-N), University Research Committee of The University of Hong Kong, the Collaborative Research with GeneHarbor (Hong Kong) Biotechnologies Limited and National Key R&D Program of China (Grant2021YFC2301900).
Keywords: HIV-1, AIDS, Nicotinamide mononucleotide, CD4 T cell, T cell activation
Research in context.
Evidence before this study
NAD is an essential co-enzyme for both energy metabolism and cellular function in host cells, which may affect various physiological behaviours and systems of the body. Previous studies reported that HIV-1 infection decreased the intracellular NAD level in peripheral blood cells and that nicotinamide treatment might inhibit the infection in vitro by maintaining the amount of intracellular NAD. We, therefore, sought to determine the role of NAD precursors in regulating host immune cells against HIV-1 infection. Notably, HIV-1-induced immune activation associates not only with the progression of AIDS but also with the failure of CD4+ T cell recovery after cART treatment. It remains unclear if NAD precursors would be useful for modulating immune activation, assisting the recovery of CD4+ T cells in cART-treated PLWH. To date, the impact of NMN, the direct precursor of NAD, in modulating CD4+ T cell activation has not been investigated.
Added value of this study
Using multiple infection models, we found that NMN treatment suppressed viral production in HIV-1-infected ACH-2 and primary CD4+ T cells at the post-transcription stage. On the one hand, NMN treatment preferentially decreased the intracellular HIV-1 p24 production in CD25+CD4+ T cells. On the other hand, NMN inhibited the expression of late T cell activation markers including CD25 and HLA-DR consistently using in vitro, ex vivo, and in vivo models. Furthermore, NMN improved the therapeutic efficacy of cART in HIV-1-infected humanized mice by improving the CD4+ T cell reconstitution.
Implications of all the available evidence
Our findings suggest that NMN may be further tested as a potential drug in combination with cART for PLWH to improve CD4+ T cell recovery, especially for those cART-treated PLWH with persistently low CD4+ T cell count.
Introduction
During human immunodeficiency virus type one (HIV-1) infection, although the reduction of viremia and mortality has been achieved by combination antiretroviral therapy (cART), up to 30% of individuals failed to reconstitute CD4+ T cell count regardless of successfully suppressed viral load.1, 2, 3 People living with HIV (PLWH) with poor CD4+ T cell recovery are considered as immunological non-responders (INRs) and may have higher clinical risks of developing acquired immune deficiency syndromes (AIDS) and other diseases.4 Some researchers found that Vitamin D and Vitamin B3 (niacin, also known as a NAD booster5), rather than Vitamin A, Vitamin B6 or Vitamin B12, could affect immune activation and CD4+ T cell reconstitution in INRs.6, 7, 8 Promoting CD4+ T cell recovery through suppressing immune activation by Vitamin D, Vitamin B3 or any other useful nutrients becomes a potential strategy to reduce clinical risks and disease progression of HIV-INRs.
Nicotinamide adenine dinucleotide (NAD) is an essential co-enzyme for energy metabolism and function in host cells.9 NAD depletion with ageing associates with disease conditions in nerve, liver, kidney, intestine, muscle, haematopoiesis and cardiovascular systems.9 Thus, boosting the NAD amount in patients using NAD precursors or NAD-consuming enzyme inhibitors is a promising therapeutic method for maintaining NAD homeostasis and alleviating disorders related to the NAD depletion.9,10 Majority of NAD boosters under clinical trials are NAD precursors including nicotinamide ribose (NR), nicotinamide (NAM) and nicotinamide mononucleotide (NMN).10,11 Recently, besides ageing-associated diseases, researchers have investigated the role of NAD in modulating immune system against infections or cancer through activating some NAD-consuming factors such as sirtuins, poly(ADP-ribose) polymerases (PARPs) and CD38.12,13 The anti-inflammatory role of NAD has also been documented during immune responses.14 Enhanced sirtuin 1 activity through boosted NAD could deacetylate nuclear factor-kappa B (NF-κB), downregulate NF-κB-regulated pro-inflammatory cytokines, and modulate CD4+ T cell differentiation.14 Exogenous NAD boosted by NMN supplementation could restore LPS-induced PARP-activated inflammation.15 The NAD-consumer CD38 could control intracellular NAD, modulating NAD-dependent immunometabolic events, despite its controversial role in pro-inflammatory cytokine release.16 Critical factors modulated by NAD or NAD precursors remain to be investigated in the context of host immunoregulation against pathogens.
For NAD biosynthesis (Supplementary Figure S1), NMN is the direct precursor of NAD in the salvage pathway, whereas both NAM and NR are indirect precursors. The synthesis of NAD from NAM and NR requires enzymes including nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenylyltransferases (NMNATs).17 Niacin (Vitamin B3) can be catalysed by nicotinate phosphoribosyltransferase (NAPRT) in the Preiss-Handler pathway before entering the salvage pathway.17 In terms of in vivo activities, undesirable adverse effects have been documented with high-dose niacin, NR, and NAM in preclinical and clinical trials.18, 19, 20 Similar adverse effects have not been reported with high-dose NMN. It is, therefore, conceivable that NMN would be a better candidate for clinical use. However, the impact of NMN on HIV/AIDS has not been investigated. To test the hypothesis on whether NMN may modulate immune activation, we sought to determine the effects of NMN on human CD4+ T cells upon in vitro and in vivo HIV-1 infection.
Methods
Cell isolation
Peripheral blood from HIV-1-uninfected donors (HUD) and PLWH was used for isolating peripheral blood mononuclear cells (PBMCs) by density gradient centrifugation with Lymphoprep™ (Axis-Shield). Fresh PBMCs were used for purifying CD4+ T cells using RosetteSep™ Human CD4+ T Cell Enrichment Cocktail (Stemcell™ Technologies) by negative selection to avoid unnecessary activation. Human CD4+ T cell Isolation kit (Miltenyi Biotec) was used for purifying CD4+ T cells from frozen PBMCs. Resting CD69−CD25−HLA-DR-CD4+ T cells were purified using CD69 MicroBead Kit II (human, Miltenyi Biotec), CD25 MicroBeads II (human, Miltenyi Biotec) and Anti-HLA-DR MicroBeads (human, Miltenyi Biotec) together with Human CD4+ T cell Isolation kit by negative selection. Freshly isolated PBMCs or purified primary CD4+ T cells were cultured in R10 medium [RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mM l-glutamine (Gibco) and 100 U/mL penicillin/100 μg/mL streptomycin (Gibco)] with 5% CO2 at 37 °C. HEPES (10 mM, Gibco) would be added in the cell culture experiment involving NMN treatment. Freezing medium was prepared using 90% FBS and 10% dimethylsulfoxide (DMSO; Sigma Aldrich) for cell storage at −150 °C. Frozen cells were recovered in pre-warmed R10 Medium with 5% CO2 at 37 °C overnight.
Virus preparation and viral infection
MOLT-4 CCR5+ cell line (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: MOLT-4 CCR5+ Cells, ARP-4984, contributed by Dr. Masanori Baba, Dr. Hiroshi Miyake, and Dr. Yuji Iizaw) was used for propagating the HIV-1JRFL virus (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Human Immunodeficiency Virus-1 JR-FL, ARP-395, contributed by Dr. Irvin Chen) in the presence of interleukin-2 (IL-2, 10 U/mL; R&D systems) for 14 days as we previously described.21 HEK293T cell line (ATCC, RRID:CVCL_0063) was used for packaging the non-replicable HIVJRFL-nLuc pseudovirus by co-transfection with a vector containing the envelope of tier-2 JRFL and a pNL4-3. Luc.R-E- backbone vector containing a defective Nef, Env, and Vpr as well as a nanoluciferase reporter (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Human Immunodeficiency Virus 1 (HIV-1) NL4-3 ΔEnv Vpr Luciferase Reporter Vector (pNL4-3·Luc.R-E-), ARP-3418, contributed by Dr. Nathaniel Landau).22,23 Around 4 ng p24 of HIV-1JRFL virus or 4000 TCID of HIVJRFL-nLuc pseudotyped virus was inoculated into around 0.2 million purified primary CD4+ T cells or MOLT-4 CCR5+ cell line on 96-well V-bottom or U-bottom plates by spinoculation at 1200 g × 90 min without brake. After being incubated for at least 2 h or overnight, cells were washed with PBS (Gibco) for at least three times before being cultured with R10 medium supplemented with cytokines. For HIV-1JRFL virus infection, 10 U/mL of IL-2 was used as the supplementary cytokine; for HIVJRFL-nLuc pseudotyped virus infection, 10 ng/mL IL-2 and 200 ng/mL IL-15 (Miltenyi Biotech) were used as supplementary cytokines. An appropriate concentration of NMN or 10 mM NMN was used as experimental drug treatment. In some experiments, efavirenz (EFV, 10 nM) was used for preventing secondary HIV infection. Mock infection or pre-treatment of Maraviroc (MAR, 1 μM, for at least 30 min; Sigma) before infection served as the negative control.
Cell activation
For viral reactivation in the ex vivo primary cell model, purified resting CD4+ T cells were treated with PMA (50–500 ng/mL) plus Ionomycin (1 μg/mL) for 4 or 7 days. For viral reactivation in the ACH-2 cell line, cells were treated with PMA (50 ng/mL) for 24 h.
Cell transfection
MOLT-4 CCR5+ cells were transiently transfected with 10 nM of siHCG27 (Thermo) or control siRNA (Thermo) using the Lipofectamine™ RNAiMAX Transfection Reagent (Thermo) in Opti-MEM (Gibco). At 48 h after transfection, cells were treated without or with 10 mM NMN for additional 24 h before RNA extraction.
Cell lines
Cell lines used in this study, including HEK293T cell line, MOLT-4 CCR5+ cell line and ACH-2 cell line, were maintained in the appropriate medium according to the manufacturers’ instructions. These three cell lines were confirmed free of mycoplasma contamination by real-time PCR.
Anti-p24 ELISA
Supernatant from cell culture was collected and lysed for detecting supernatant p24 level by Human Immunodeficiency Virus type 1 (HIV-1) p24/Capsid Protein p24 ELISA Pair Set (#SEK11695-15, Sino Biological).
Luciferase assay
For the detection of intracellular NAD level, around 0.1 million CD4+ T cells were harvested and lysed for measuring relative luciferase units (RLU) with NAD/NADH-Glo™ Assay (Promega). For the detection of cell viability, cell lysate was used for measuring relative luciferase units (RLU) with CellTiter-Glo® Luminescent Cell Viability Assay (Promega). For the detection of supernatant luciferase responses from the HIVJRFL-nLuc pseudotyped virus infected cell culture, 30 μL of supernatant was used for measuring relative luciferase units (RLU) using Nano-Glo® Luciferase Assay System (Promega).
DNA extraction and RNA extraction
In some experiments, CD4+ T cells were harvested at 48 h after NMN treatment or 24 h after HIV-1 infection for DNA and RNA extraction using AllPrep DNA/RNA Mini Kit (Qiagen). In other experiments, total RNA was extracted using Trizol reagent (Takara), while DNA was harvested using QIAamp DNA Mini Kit (Qiagen). Viral RNA was extracted from mouse plasma or cell supernatant according to the manufacturer's instruction of QIAamp Viral RNA Mini Kit (Qiagen).
Real-time PCR
PrimeScript™ II 1st strand cDNA Synthesis Kit was used for synthesizing cDNA from extracted RNA samples before performing quantitative real-time PCR assay on ViiA 7 Real-Time PCR System. In Taqman-based real-time PCR assay, HIV-1 primers and probe that targets to LTR/gag region of HIV-1 [(5′→3′) forward: TACTGACGCTCTCGCACC; reverse: TCTCGACGCAGGACTCG; probe: FAM-CTCTCTCCTTCTAGCCTC-MGB] and/or CCR5 primers and probe [(5′→3′) forward: ATGATTCCTGGGAGAGACGC; reverse: AGCCAGGACGGTCACCTT; probe: VIC-AACACAGCCACCACCCAAGTGATCA-MGB] were used with TaqMan™ Universal PCR Master Mix (no AmpErase™ UNG; Applied Biosystems™), under the following PCR conditions: 50 °C for 2 min, 95 °C for 2 min, 45 cycles of 95 °C for 15 s and 60 °C for 1 min.24 CCR5 copies were used for calculated cell numbers with a CCR5-encoding plasmid standard.25 In SYBR green-based real-time PCR assay, TB Green® Premix Ex Taq™ II (Tli RNase H Plus, Takara) was applied under the following PCR conditions: 95 °C for 2 min; 45 cycles of 95 °C for 10 s and 55 °C for 30 s; 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s. Primers used in the SYBR green-based real-time PCR assay were listed in Supplemental Table S1.
Flow cytometry
For cell surface staining, cells were stained with antibodies (seen in Supplemental Table S2) at 1:50 dilution in staining buffer (2% FBS in PBS) at 4 °C for 30 min. Alternatively, Annexin V PE/Cy7 (Biolegend) was stained using Annexin V binding buffer (Biolegend) according to the manufacturer's instruction. If not proceeding with intracellular staining in the next step, cells were fixed with 2% paraformaldehyde before FACS analysis. If proceeding with intracellular staining in the next step, cells were fixed and permeabilized using Fixation/Permeabilization Solution (BD Biosciences) at 4 °C for 20 min, followed by washing with Perm/Wash™ buffer (BD Biosciences), and stained with antibodies at 1:50 dilution in Perm/Wash™ buffer at 4 °C overnight. Cells were washed twice before FACS analysis. Propidium Iodide (Sigma) or Zombie Aqua™ Kit (BioLegend) was used for distinguishing live or dead cells. Data were analysed using FlowJo v10.0.7.
Humanized mice experiments
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were bred at a density of 4–6 mice per cage under an AAALAC International accredited program at the Centre for Comparative Medicine Research, HKU under Specific Pathogen Free (SPF) conditions. Human peripheral blood leukocyte (huPBL) isolated from buffy coat blood from Red Cross was intraperitoneally (i.p.) injected into six-to-eight weeks old female NSG mice (1 × 107 huPBL per mouse), in order to generate humanized NSG-huPBL mouse model compatible for in vivo HIV challenge experiment, as we previously described.26,27 Female mice were used because they exhibited lower basal NAD level than male ones28 and they were reported as preferred recipients for human cells.29, 30, 31 Around three weeks after PBL injection, the frequency of human CD45+CD3+ cells was measured using peripheral blood from mice, in order to confirm whether huPBL mouse model was constructed successfully. Those mice that did not acquire detectable human CD45+CD3+ cells in peripheral blood would not be proceeded to the further steps. Twenty-three successfully constructed huPBL mice in two batches (n > 4 per group, sample size calculation based on α = 0.05 and power = 80% (β = 0.2), single-blind) were then performed HIV-1JRFL infection (10 ng p24 per mouse) via i. p. route, as we previously described.26,27 The daily cART treatment for mice contains 61.5 mg/kg lamivudine (3TC), 10.25 mg/kg dolutegravir (DTG), and 61.5 mg/kg tenofovir disoproxil fumarates (TDF), equivalent to the combination of 300 mg 3TC/50 mg DTG/300 mg TDF for human.32, 33, 34 The dose of NMN was 300 mg/kg/day for mice, which is equivalent to 25 mg/kg/day for human.34,35 HIV-infected huPBL mice were used for negative control. Mice were monitored daily after infection, and humane endpoint scoring would be recorded if mice showed signs of being unwell, until the humane endpoint was reached (for example, weight loss is ≥20%, inability to rise or ambulate, and labored respiration) or the experimental endpoint reached. Enrofloxacin (10 mg/kg) would be orally administrated if mice suffering diarrhea due to graft-versus-host disease. Plasma viral load and peripheral CD4/CD8 ratio, as well as the percentage of CD4+ T cells and CCR5+CD4+ T cells, were monitored weekly. On Day 28 post-infection, mice were sacrificed and spleen samples were collected for splenocyte isolation and tissue section. The percentage of CD4+ T cells, CCR5+ CD4+ T cells, p24+ CD4+ T cells, Annexin V+ CD4+ T cells, CD25+ CD4+ T cells, HLA-DR+CD38+ CD4+ T cells, and proliferating (ki67+) CD4+ T cells, as well as CD4/CD8 ratio, in splenocytes was assessed by flow cytometry. Protein levels of CD25 and p24 in spleen sections were assessed by IHC staining. Splenocyte proviral DNA load was detected via real-time PCR. Plasma NAD level on Day 28 post-infection was detected by NAD/NADH-Glo™ Assay.
CyTOF
Purified total CD4+ T cells were treated without or with 10 mM NMN for 4 days before CyTOF analysis. Maxpar® Cell Staining Buffer (Fluidigm) was used for cell washing and antibody incubation. The antibodies used for CyTOF analysis were purchased from Fluidigm (seen in Supplemental Table S2). Cell-ID™ intercalator-Ir (#201192A, Fluidigm) was diluted in Maxpar® Fix and Perm Buffer (Fluidigm) and used for distinguishing dead/live cells and singlets/doublets. EQ four-element calibration beads (Fluidigm) in Maxpar® Cell Acquisition Solution (Fluidigm) were used for data calibration. CyTOF data were acquired by Mass Cytometer (Helios) and analysed using FlowJo v10.7.1. After gating CD45+CD3+CD4+ singlets, the t-SNE plot was generated using opt-SNE configuration using gradient algorithm FFT-interpolation with the perplexity at 30,36,37 whereas the FlowSOM algorithm v3.0.16 was used for cell clustering.38
IHC staining
Paraffin-embedded sectioning slides from spleen tissues in mouse experiments were deparaffinized and unmasked for IHC staining. After being incubated at 0.1% Sudan black b (Sigma) at room temperature for 20 min followed by being subsequently rinsed with PBS, PBS-FSG-TritonX100 [0.5 mL Triton™ X-100 (Sigma) + 1 mL fish skin gelatin (FSG, Sigma) in 500 mL PBS] and PBS-FSG (1 mL FSG in 500 mL PBS). Slides were then incubated with 10% goat serum (NGS, Gibco) in PBS-FSG at room temperature for 1 h, and later incubated with primary antibodies [anti-p24 (Santa Cruz, #sc-65918, 1:50, mIgG2a), anti-CD25 (R&D systems, #MAB623, 1:50, mIgG 1), and anti-CD4 (Biolegend, #396202, 1:50, mIgG2b) in PBS-FSG] at room temperature for 1 h. Next, slides were rinsed with PBS-FSG-TritonX100 and PBS-FSG and incubated with secondary antibodies (goat anti-mouse IgG1 AF568, goat anti-mouse IgG2b AF488, goat anti-mouse IgG2a AF647; 1:1000) in PBS-FSG at room temperature for 1 h. Hoechst 33342 buffer (1:1000 in PBS, #62249, Thermo) was used for staining the nucleus. After being mounted with Fluorescence Mounting Medium (S302380), slides were scanned using PerkinElmer Vectra Polaris™ Automated Quantitative Pathology Imaging System and analysed using Inform Software (v2.4 and v2.6).
RNA-seq
Primary CD4+ T cells (n = 4) were treated with or without NMN (10 mM) for 24 h, followed by 24-h HIV-1 infection in the absence or presence of NMN (10 mM). Cellular RNA was extracted using AllPrep DNA/RNA Mini Kit (Qiagen) and tested by Bioanalyzer. One sample from the NMN treatment group failed QC with RIN below eight and was excluded for further sequencing and analysis. Sample QC, library preparation, and Illumina sequencing (Pair-End sequencing of 151bp) were done at Centre for PanorOmic Sciences (CPOS), Genomics Core, LKS Faculty of Medicine, The University of Hong Kong. Quality control of raw fastq data was carried out by FastQC v0.11.7 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and fastp.39 Then clean reads were aligned to the UCSC GRCh38 reference and the HIV-1 genome (GenBank: U63632.1) using Hisat2 v2.2.1.40 HTSeq v0.6.1 was used to generate the raw read count for each gene.41 Differential expression analysis was performed using DESeq2.42 Genes with |log2 fold change| >1 and adjusted p value <0.05 (FDR) were considered significantly different.42 The volcano plot was generated using the EnhancedVolcano R package (https://github.com/kevinblighe/EnhancedVolcano). Gene set enrichment analysis (GSEA) was performed using fgsea R package (https://github.com/ctlab/fgsea).
Statistics
All statistical analyses were performed using the GraphPad Prism 7 software. Shapiro–Wilk normality test was used for normality test before group comparison analysis, whereas D'Agostino & Pearson normality test was used for normality test before correlation analysis. For data passing normality test, comparison tests were analysed using a One-way ANOVA test or a Student's t-test. For data not passing normality test, comparison tests were analysed using a Mann–Whitney test or a Friedman test; correlation analysis was based on Spearman correlation method. For the one-way ANOVA or Friedman test, a post-hoc test can be corrected by Tukey's test or two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with alpha = 0.05 used for multiple comparison analysis. A multiple comparison post-hoc test was used when comparison among more than two groups was tested, and multiple t-tests with no multiple comparison correction were used when comparison between two groups was tested. Data represent mean with 95% CIs or mean with SDs of at least three independent experiments unless indicated. Significant differences were calculated with an appropriate α at 0.05 and power at 80%. p < 0.05 was considered statistically significant. For RNA-Seq data, DESeq2 was used for data analysis; the p values were calculated by the Wald test and corrected for multiple comparison tests using the Benjamini and Hochberg method by default along with defaulted statistical power, and adjusted p value <0.05 (FDR) were considered significantly different.
Ethics
The studies involving PLWH were approved by the Ethics Review Committee of Shenzhen third People's Hospital (2018–080) and the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (2018.445). Written informed consent was received before participation. The use of healthy human PBMCs was approved by the Institutional Review Board of the University of Hong Kong/Healthy Authority Hong Kong West Cluster (UW19-482, UW18-286 and UW19-833). Research only proceeded following review and approval from the HKU Committee on the Use of Live Animals in Teaching and Research (CULATR# 5340-20) and under licence from the Hong Kong SAR Government's Department of Health, complying with ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments, seen in Supplementary Table S3).
Role of funders
Funders had no roles in the study design, data collection, data analyses, data interpretation, or writing of the manuscript.
Results
NMN suppresses live HIV-1JRFL infection in primary CD4+ T cells
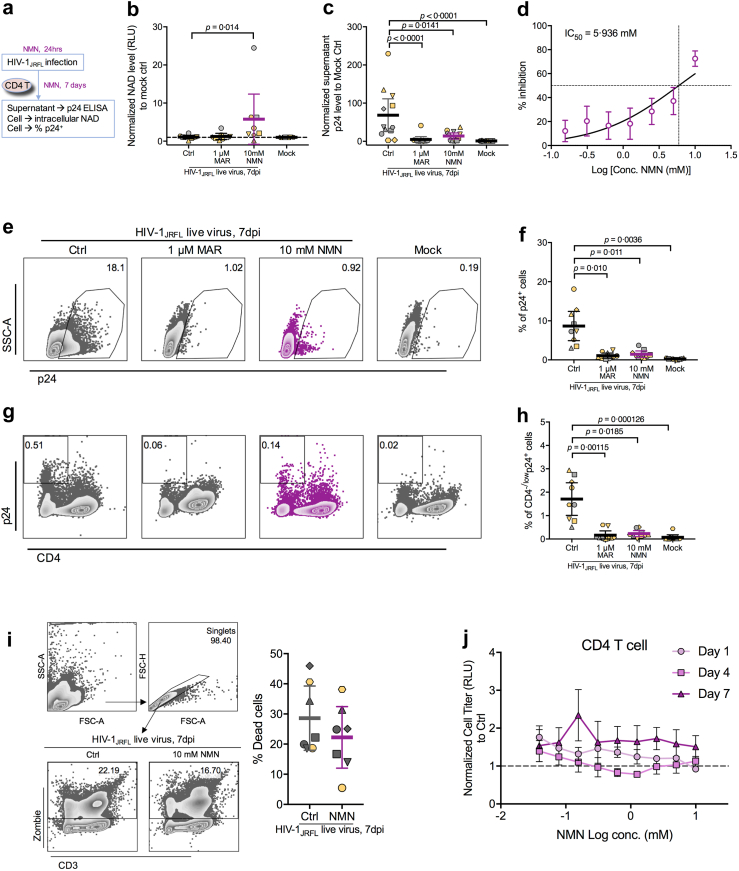
Previous studies reported that HIV-1 infection decreased the intracellular NAD level in peripheral blood cells and that NAM could inhibit the infection probably by maintaining the intracellular NAD.43,44 Considering that NMN is the direct NAD precursor, we sought to investigate its impact on HIV-1 infection in primary CD4+ T cells. First, we performed an in vitro live HIV-1JRFL infection experiment on NMN-treated primary CD4+ T cells (Fig. 1a). We did not find a significant decrease of intracellular NAD level in purified primary CD4+ T cells after live HIV-1JRFL infection, but a higher amount of intracellular NAD was detected in purified CD4+ T cells during treatment with 10 mM NMN (Fig. 1b). By detecting supernatant viral p24 protein from primary CD4+ T cells after the HIV-1JRFL infection for 7 days, we found that viral replication was suppressed by NMN with an IC50 value of 5.936 mM (Fig. 1c and d). Consistently, the intracellular p24 in primary CD4+ T cells was also dramatically decreased by NMN treatment on day 7 after the HIV-1JRFL infection (Fig. 1e and f); whereas the percentage of productively infected cells (typically as CD4−/lowp24+ cells) was also decreased (Fig. 1g and h). Notably, NMN treatment did not cause significant cell death during the experiments (Fig. 1i), suggesting that the reduced p24 production was not due to the cytotoxicity of NMN. Consistently, we found that in vitro 1-day, 4-day or 7-day NMN treatment did not show cytotoxicity to primary CD4+ T cells by assessing ATP-dependent cell viability (Fig. 1j). In addition, neither NMN treatment nor HIV-1JRFL infection altered the mRNA levels of NMN/NAD-related metabolic enzymes such as NAPRT, NAMPT and NMNATs (Supplementary Figure S2).
Fig. 1.
Live HIV-1JRFLinfection can be suppressed by NMN treatment in vitro. Primary CD4+ T cells were isolated from PBMCs and pre-treated with 1 μM Maraviroc (MAR), different concentrations of NMN (with serial dilution starting from 10 mM at 1:2 ratio) or 10 mM of NMN for 24 h before infection with HIV-1JRFL virus (2 ng p24 per 0.1 million cells). At 2 h post-infection, cells were washed and treated with or without NMN in the presence of IL-2 (10 U/mL) for seven days. (a) The experimental flowchart was displayed. On Day 7 post-infection, (b) the intracellular NAD level in CD4+ T cells (n = 8) was detected using NAD/NADH-Glo™ Assay. (c) Normalized supernatant p24 level (to infection control, n = 12) was detected using anti-p24 ELISA assay. (d) The IC50 value was calculated using supernatant p24 level (n = 12). Cells were also harvested for intracellular p24 staining and FACS analysis. (e) Representative plots of p24+ cells from CD4+ T cells were displayed. (f) The percentage of p24+ cells (n = 9) was analysed among groups. (g) Representative plots of CD4−/lowp24+ cells were displayed. (h) The percentage of CD4−/lowp24+ cells (n = 9) was analysed among groups. To detect cell death after infection, cells with or without NMN treatment (10 mM) were harvested for FACS analysis on dead cell analysis. (i) The percentage of dead (Zombie+) cells (n = 7) was analysed, along with representative plots showing the gating strategy. (j) Primary CD4 T cells (n = 3) treated with different concentrations of NMN (with serial dilution starting from 10 mM at 1:2 ratio) for 1 day, 4 days and 7 days were assessed by CellTiter-Glo® Luminescent Cell Viability Assay in one experiment. For (b), (c), (f), (h) and (i), data represent Mean ± 95% CI; for (d) and (j), data represent Mean ± SD. For (b) and (h), data did not pass normality test, and statistics were calculated using Friedman test with post-hoc multiple comparison tests; for (c) and (f), data passed normality test, and statistics were calculated using a One-way ANOVA test with a post-hoc test corrected by Tukey's test.
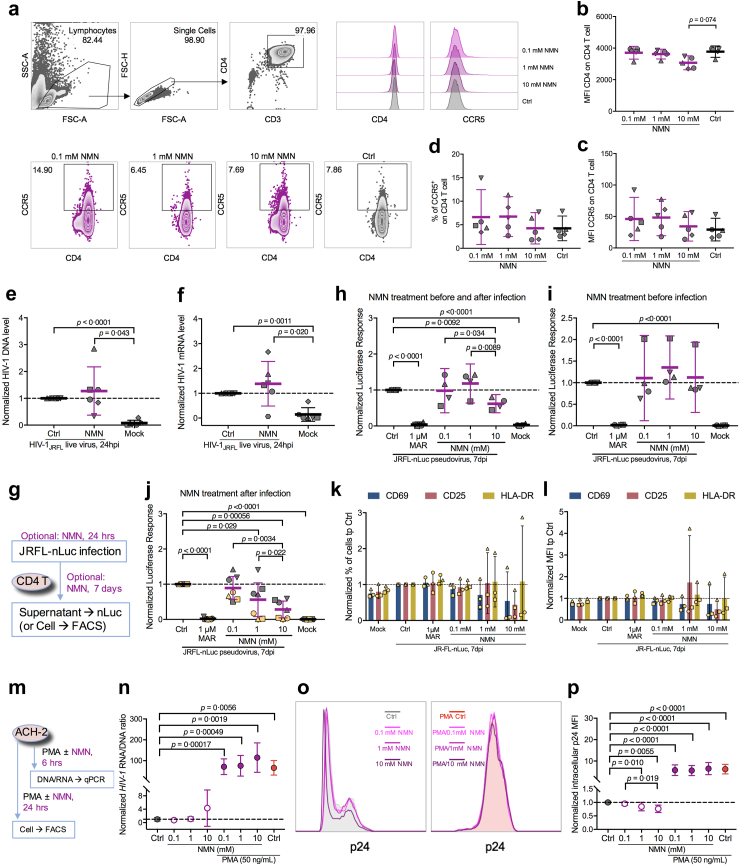
Next, we sought to determine how NMN affected HIV-1 replication. First, we investigated whether NMN treatment would alter the expression of HIV-1 receptor CD4 and co-receptor CCR5. No significant changes were found with MFI of CD4/CCR5 or with the frequency of CCR5+ cells in the NMN treatment groups as compared with the vehicle controls (Fig. 2a–d). In contrast, significant increases of both frequency and MFI of CXCR4 were found in NMN-treated CD4+ T cells (Supplementary Figure S3). Second, we measured the amount of HIV-1 integrated DNA and transcribed mRNA at 24 h post-infection (hpi) in infected primary CD4+ T cells with and without NMN. No significant differences were found between the NMN-treated cells and untreated controls (Fig. 2e and f). Consistently, NMN did not significantly decrease transcribed HIV-1 mRNA levels in infected MOLT-4 CCR5+ cell line or primary CD4+ T cells at 24 hpi by quantitative real-time PCR assay or RNA-seq analysis, respectively (Supplementary Figure S2a and S4). Based on the HIV-1 life cycle in infected T cells, the complete process of viral entry, reverse transcription, integration and transcription took place within 24 h after infection.45 Our results, therefore, indicated that NMN treatment might not affect the process of viral entry, reverse transcription, integration and transcription after the HIV-1JRFL infection. Next, we measured the effect of NMN treatment on post-transcription stages. We used the single-cycle HIVJRFL-nLuc pseudovirus to infect purified primary CD4+ T cells with or without NMN treatment before and/or after spinoculation (Fig. 2g). Interestingly, NMN treatment after the HIVJRFL-nLuc infection, rather than before infection, significantly reduced luciferase responses (Fig. 2h–j). Meantime, comparable frequencies and MFIs of CD69, CD25 or HLA-DR on T cells of 3 independent donors were detected in the NMN-treated groups as compared with the controls (Fig. 2k and l and Supplementary Figure S5). In addition, we found that NMN treatment could reduce p24 production but not HIV-1 RNA/DNA ratio dose-dependently in ACH-2 cell line (Fig. 2m–p), a T cell line that constitutively produces a low amount of HIV-1 p24 protein.46,47 NMN treatment, however, could not alleviate HIV-1 reactivation in ACH-2 cells treated with the phorbol 12-myristate 13-acetate (PMA) (Fig. 2m–p). These results demonstrated that NMN probably suppressed HIV-1 replication at a post-transcriptional stage during both live and pseudotyped viral infections.
Fig. 2.
In vitro NMN treatment suppresses HIV infection at the post-transcriptional stage. Purified human CD4+ T Cells were treated with indicated compounds [NMN (0.1, 1, 10 mM) or PBS control] for five days and harvested for FACS analysis. (a) Representative histogram plots were displayed. The MFI of CD4 (b) and CCR5 (c), as well as the percentage of CCR5+ cells (d), on CD4+ T cells were compared among groups (n = 5). Purified human CD4+ T cells were pre-treated with 10 mM of NMN for 24 h before infection with live HIV-1JRFL virus (2 ng p24 per 0.1 million cells). At 3 h post-infection, cells were washed and treated with or without NMN in the presence of IL-2 (10 U/mL) for 24 h. At 24 hpi, cellular DNA and mRNA were extracted for HIV-1 real-time PCR assay. Normalized HIV-1 DNA level (e) and HIV-1 mRNA level (f) to infection control were compared (n = 6). Purified human CD4+ T Cells were pre-treated without or with 1 μM Maraviroc (MAR) for 30 min or 0.1, 1, 10 mM of NMN for 24 h. Cells were mocked infected or infected with HIVJRFL-nLuc. At 24 h post-infection, cells were washed and treated without or with 0.1, 1, 10 mM of NMN in the presence of IL-2 (10 ng/mL)/IL-15 (200 ng/mL) for 7 days. Vehicle-treated infected cells served as control (Ctrl), whereas mock cells serve as mock control. (g) The experimental flowchart was displayed. The normalized supernatant luciferase responses to infection control (n = 4) were compared for treatment during the period of pre-infection and post-infection (h) and treatment during the period of pre-infection (i). The normalized supernatant luciferase responses to infection control (n = 7) were compared for treatment during the period of post-infection (j), with monitoring the frequencies (k) and expression (MFI, l) of CD69, CD25, and HLA-DR by flow cytometry. ACH-2 cell line was reactivated by PMA (50 ng/mL) in the presence or absence of NMN treatment (0.1, 1 or 10 mM). (m) The experimental flowchart was displayed. (n) The normalized HIV-RNA/HIV-DNA ratios to the unactivated control group at 6 h after treatment were compared among groups. For intracellular p24 expression at 24 h after treatment, (o) representative histogram plots were displayed. (p) Normalized intracellular p24 expression levels were compared among groups. For (b–f), (h–j), (n) and (p), data represent Mean ± 95% CI; for (k–l), data represent Mean ± SD. For (b–f) and (h–j), data passed normality test, and statistics were calculated using a One-way ANOVA test with an appropriate post-hoc test; for (n) and (p), data did not pass normality test, and statistics were calculated using a Mann–Whitney test.
NMN reduces CD25+CD4+ T cells and suppresses HIV-1 by modulating the proliferation of infected CD25+HLA-DR+CD4+ T cell subsets
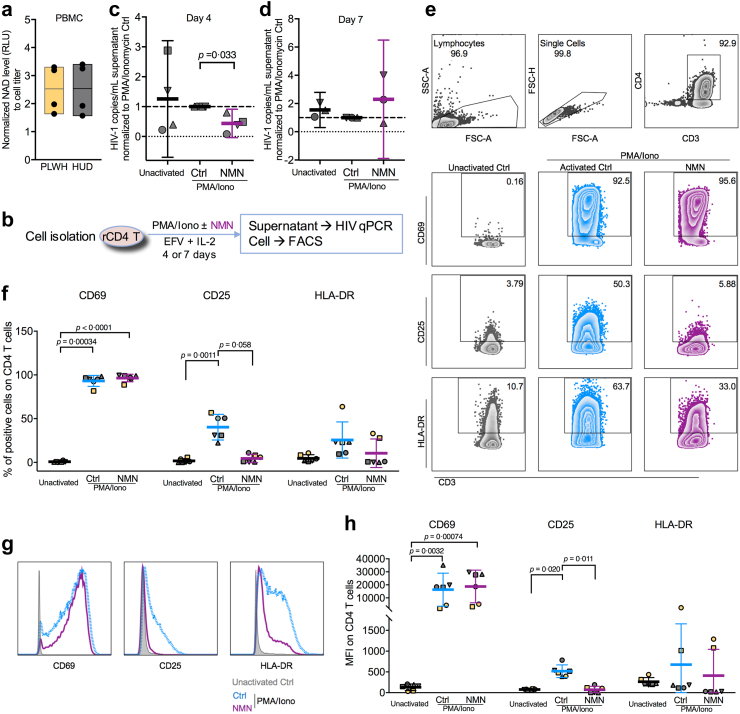
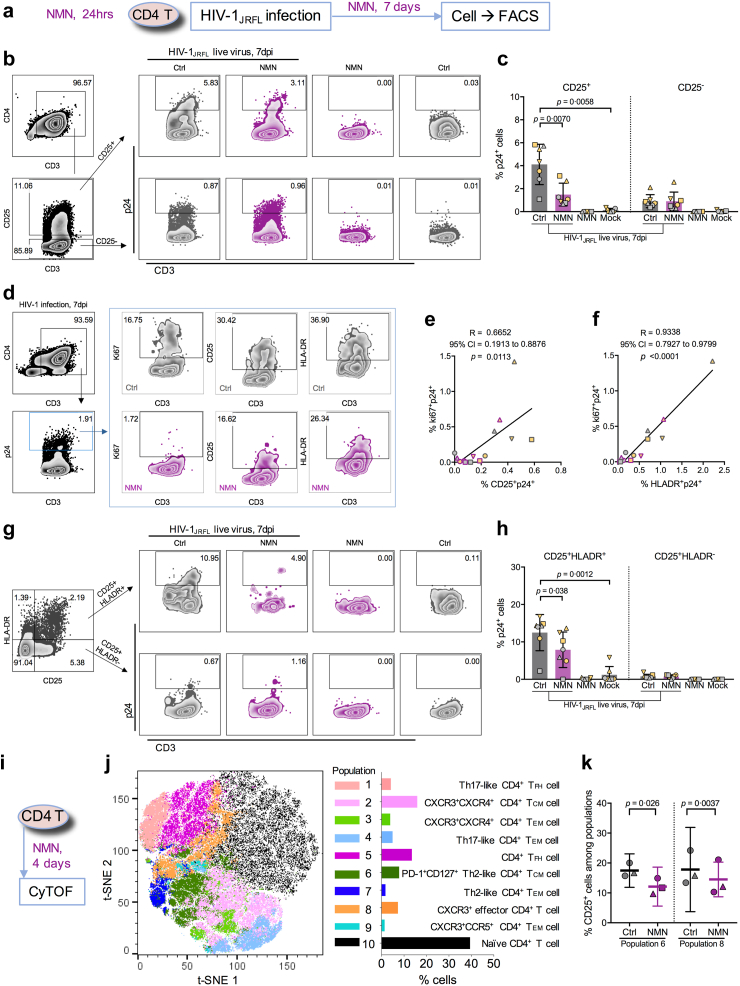
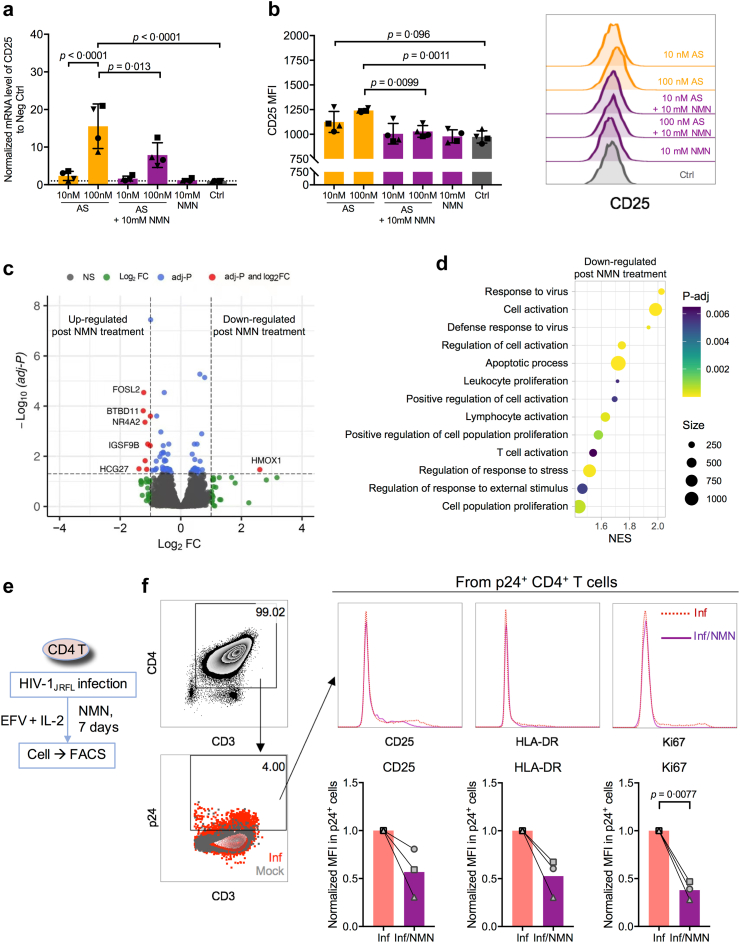
We then sought to investigate the ex vivo effect of NMN treatment on the reactivation of CD4+ T cells derived from cART-treated PLWH with undetectable viral load (clinical information seen in Supplementary Table S4). First, we measured the amount of intracellular NAD in PBMCs of cART-treated PLWH and HIV-1-uninfected donors (HUDs). We found comparable intracellular NAD amounts between PLWH and HUDs (Fig. 3a). The normalized NAD amount to cell titer in HUD was 2.536 ± 0.9264, while that in PLWH was 2.526 ± 0.8428. Second, we used PMA/Ionomycin to reactivate resting CD4+ T cells of cART-treated PLWH (Fig. 3b). Lower viral reactivation by NMN treatment was found on day 4, although not on day 7, after reactivation (Fig. 3c and d). We measured T cell early activation marker CD69 as well as T cell late activation markers CD25 and HLA-DR.48 We found that NMN treatment resulted in around 9-fold decreased frequencies of CD25+ and around 1.5-fold decreased frequencies of HLA-DR+ cells in spite of the lack of statistical difference, as well as significantly decreased CD25 MFI (Fig. 3e–h). In contrast, neither the frequency of CD69+ T cells nor CD69 MFI were affected. Similarly, NMN treatment reduced CD25+ and HLA-DR+ but not CD69+ on CD4+ T cells derived from HUDs under the same experimental condition (Supplementary Figure S6). Third, we investigated the effect of NMN treatment on HIV-1 expression in CD25+ or HLA-DR+ CD4+ T cells by measuring intracellular p24 on day 7 after viral infection (Fig. 4a). We found that NMN treatment suppressed intracellular p24 significantly in CD25+ CD4+ T cells, but neither in CD25− nor in HLA-DR+/− T cell subsets (Fig. 4b–c and Supplementary Figure S7). The frequency of ki67+p24+ CD4+ T cells was positively correlated with those of CD25+p24+ and HLA-DR+p24+ CD4+ T cells, suggesting that NMN might affect the proliferation of infected cells (Fig. 4d–f). We then measured the p24+ cell frequency among T cells co-expressing CD25 and HLA-DR. We found that NMN reduced the frequency of p24+ cells in CD25+HLA-DR+ but not in CD25+HLA-DR- CD4+ T cells (Fig. 4g–h).
Fig. 3.
Ex vivo NMN treatment suppresses resting CD4+T cell reactivation in cART-treated people living with HIV by reducing CD25+cells. Frozen PBMCs from four independent cART-treated people living with HIV (PLWH) and four independent HIV-uninfected donors (HUD) were used for NAD level detection via NAD/NADH-Glo™ Assay. (a) The intracellular NAD level in PBMCs between PLWH and HUDs was displayed. Data represent Mean with Mix to Max in floating bars. Fresh or frozen PBMCs from six independent cART-treated PLWH were used for resting CD4+ T cell isolation. Purified resting CD4+ T cells were treated with PMA (50–500 ng/mL) plus Ionomycin (1 μg/mL) (in short as PMA/Iono), PMA/Iono plus 10 mM NMN or mock, under treatment of 10 nM EFV and 10 U/ml IL-2. On day 4 or 7 after treatment, cells were collected for FACS analysis, whereas viral RNA in the supernatant was extracted for HIV-1 real-time PCR assay. (b) The experimental flowchart was displayed. Normalized HIV-1 mRNA levels to PMA/Iono activation control were compared on day 4 (c; n = 4, frozen PBMCs) and day 7 (d; n = 3, frozen PBMCs) after reactivation. For FACS analysis on activation markers, representative FACS plots (e) and histogram plots (g) were displayed. The percentage (f) and MFI (h) of CD69+, CD25+, and HLA-DR+ cells on CD4+ T cells was compared (n = 6). For (c and d), (f) and (h), data represent Mean ± 95% CI. For (c–d), data passed normality test, and statistics were calculated using a paired Student's t-test; for (f) and (h), data did not pass normality test, and statistics were calculated using a Friedman test with post-hoc multiple comparison tests.
Fig. 4.
NMN treatment predominantly reduces intracellular p24 level in CD25+CD4+T cells, correlating to proliferating p24-expressing CD4+T cells. Primary CD4+ T cells were isolated from PBMCs (n = 7) and pre-treated with 10 mM of NMN for 24 h before infection with live HIV-1JRFL virus (2 ng p24 per 0.1 million cells) or mock. After infection, cells were treated with 10 mM NMN for seven days. On Day 7 post-infection, cells were harvested for intracellular p24 staining and FACS analysis on CD25, HLA-DR, and ki67 expression. (a) The experimental flowchart was displayed. The gating strategies on p24+ cells in CD25+/−CD4+ T cells (b) and p24+ cells in CD25+HLA-DR+/−CD4+ T cells (g) among groups were displayed with representative plots. The percentage of p24+ cells in CD25+/−CD4+ T cells (c) and CD25+HLA-DR+/−CD4+ T cells (h) was compared. (d) The gating strategy on ki67+, CD25+ or HLA-DR+ cells in p24+CD4+ T cells from HIV-infected groups was displayed with representative plots. Correlation between the percentage of CD25+p24+CD4+ T cells (e) or of HLA-DR+p24+CD4+ T cells (f) and the percentage of ki67+p24+CD4+ T cells was calculated based on Spearman correlation. Purified CD4+ T cells were isolated from fresh PBMCs from 3 independent healthy donors. Cells were treated with 10 mM NMN or vehicle for 4 days before being collected for CyTOF analysis on CD25+ cells among various CD4+ T cell subsets in one experiment. (i) The experimental flowchart was displayed. The t-SNE plot of CyTOF data was generated by opt-SNE using the KNN algorithm and the Fit-SNE gradient algorithm. Clustering was processed using the FlowSOM algorithm in order to obtain 10 populations (meta clusters). (j) The combined CyTOF data (2 groups, 3 donors) were analysed by FlowSOM and visualized with 10 populations (meta clusters) in a two-dimensional grid. (k) The percentage of CD25+ cells in Populations 6 and 8 of CD4+ T cells were compared between the NMN group and the control group. For (c) and (h), data represent Mean ± SD. For (k), data represent Mean ± 95% CI. For (c) and (h), data did not pass normality test, and statistics were calculated using a Mann–Whitney test; for (k), data passed normality test, and statistics were calculated using a paired Student's t-test. Each dot represents one independent individual.
CD25 is also known as IL-2 receptor.49 To further investigate if NMN could impact IL-2-induced T cell proliferation and HIV-1 replication, we measured CD25+ and ki67+ cell frequencies among NMN-treated HIV-1-infected CD4+ T cells in the presence of dose-escalating IL-2 on day 4 and day 7 after infection (Supplementary Figure S8a). We found that the frequencies of CD25+ and Ki67+ CD4 T cells did not change significantly by IL-2, but that decreased CD25 MFI and increased ki67 MFI were found on day 7 when compared to day 4 (Supplementary Figure S8b). To determine the impact of NMN, we found that NMN reduced the frequency of CD25+ki67+ CD4+ T cells on both day 4 and 7 (Supplementary Figure S8c–e). By measuring the percentage of p24+ cells, NMN significantly reduced p24+ cells in CD25+ki67+ CD4+ T cells on day 7 rather than day 4 after infection (Supplementary Figure S8f and g). Besides, NMN significantly reduced the IL2-dose-dependently induced supernatant p24 amount on day 7 (Supplementary Figure S8h and i). These results demonstrated that NMN suppressed HIV-1 production likely by modulating the proliferation of infected CD25+ki67+ CD4+ T cells.
To determine the effect of NMN on primary CD4+ T cell subsets, we subsequently performed the CyTOF analysis. We treated primary CD4+ T cells consistently with 10 mM NMN for 4 days before the CyTOF analysis (Fig. 4i). We found 10 CD4+ T cell populations using the FlowSOM plot38 (Fig. 4j and Supplementary Figure S9a). Populations 6, 7 and 8 showed relatively higher frequencies of CD25; however, significantly reduced frequencies of CD25 were found primarily in populations 6 and 8 but not 7 after the NMN treatment (Fig. 4j and k and Supplementary Figure S9b–d). Population 6 displayed the phenotype of CD45RO+CCR7+PD-1+CD127+CCR4+, representing PD-1+CD127+ Th2-cell like central memory CD4+ T cells50 (Fig. 4j and Supplementary Figure S9a). Population 8 displayed the phenotype of CD127−CD45RA+CCR7+CXCR3+, representing CXCR3+ effector CD4+ T cells51 (Fig. 4j and Supplementary Figure S9a). These results demonstrated that NMN could modulate CD25 expression in certain CD25+CD4+ T cell subsets.
NMN reduces the proliferation of primary p24+CD4+ T cells significantly via CD25 downregulation
To further determine the role of NMN in modulating CD4+ T cell activation and HIV-1 suppression, we investigated whether NMN treatment would regulate CD25 (gene name: IL2RA) related gene expression in vitro. Using the Foxp3 inducer AS2863619 (shortly as AS) on MOLT-4 CCR5+ cell line to upregulate CD25 expression,52 AS could upregulate both mRNA and protein amounts of CD25. The induction of CD25 by AS, however, was reduced by 10 mM NMN (Fig. 5a and b). This result supported our aforementioned findings on NMN-suppressed CD25 expression on primary CD4+ T cells (Fig. 3e–h and Supplementary Figure S6b). To understand the effect of NMN treatment on transcriptomics in CD4+ T cells, we performed RNA-seq analysis using primary CD4+ T cells with or without 10 mM NMN treatment for 2 days. At the transcriptomic level, only a few genes including FOSL2, BTBD11, NR4A2, IGSF9B, HCG27 and HMOX1 were found being altered by the NMN treatment (Fig. 5c). After we validated these genes in MOLT-4 CCR5+ cells, only HCG27 was found to be upregulated significantly after the NMN treatment (Supplementary Figure S10a). However, long non-coding RNA HCG27 did not directly regulate CD25 (IL2RA) expression (Supplementary Figure S10b and c). By Gene Set Enrichment Analysis (GSEA) on gene ontology (GO), several GO pathways related to CD25 gene were found to be downregulated by the NMN treatment. These downregulated pathways include responses to virus, cell activation, cell proliferation, and cell apoptosis (Fig. 5d and Supplementary Figure S11). Foxp3 gene was particularly noted in some of the aforementioned CD25-related GO pathways for cell activation and proliferation (Supplemental File). Most genes in these pathways, however, were not found to be significantly changed by NMN, except for the downregulated HMOX1 gene, which is partially involved in cell activation, cell proliferation, and apoptotic process (Fig. 5c–d and Supplemental File). To understand whether NMN would decrease the proliferation of HIV-expressing cells, we detected the MFI values of CD25, HLA-DR, and ki67 in p24+CD4+ T cells in the presence of 10 nM efavirenz (EFV, for preventing secondary infection, similar to Fig. 3b) on day 7 after HIV-1 infection (Fig. 5e). Consistently, NMN treatment decreased the MFI and frequency of ki67 in p24+ CD4+ T cells, and moderately decreased the MFI and frequencies of CD25 and HLA-DR in p24+ CD4+ T cells (Fig. 5f and Supplementary Figure S12), suggesting reduced proliferation of p24+CD4+ T cells.
Fig. 5.
CD25(IL2RA) level associates with proliferating p24-expressing cells. MOLT-4 CCR5+ cells were treated with AS2863619 (shortly as AS; 10 nM or 100 nM) and/or 10 mM NMN for 24 h, followed by RNA extraction for real-time PCR assay and cell staining for flow cytometry assay. (a) Normalized mRNA level of CD25 to control group (ctrl) and (b) the MFI of CD25 from four replicates was compared among groups, along with the representative flow cytometry plot. Purified primary CD4+ T cells were treated with (n = 3) or without (n = 4) 10 mM of NMN for 48 h before harvesting for bulk RNA-seq analysis. (c) Volcano plot displays the Log2 FC and –Log10 (adj-P) of each gene comparing the untreated group and the NMN-treated group. Log2 FC of the control group to the NMN-treated group was calculated (Log2 FC > 0 represents gene upregulation in the control group as compared to the NMN-treated group). (d) CD25-related Gene Ontology (GO) of Gene Set Enrichment Analysis (GSEA) was displayed in a bubble chart. The normalized enrichment score (NES) of the control group to the NMN-treated group was calculated (NES >0 represents gene upregulation in the control group as compared to the NMN-treated group). The size of each bubble represents the number of genes involved in the corresponding pathways. Primary CD4+ T cells were isolated from PBMCs of 3 independent healthy donors and infected with live HIV-1JRFL virus (2 ng p24 per 0.1 million cells) or mock in one experiment. At 24 h post-infection, cells were treated without or with 10 mM NMN in the presence of 10 nM EFV and 10 U/mL IL-2. On Day 7 post-infection, cells were harvested for intracellular p24 staining and FACS analysis on CD25, HLA-DR, and ki67 expression. (e) The experimental flowchart was displayed. (f) The expression level of CD25, HLA-DR, and ki67 in p24+ cells were displayed in representative histogram plots, and the normalized MFI levels of CD25, HLA-DR, and ki67 to infected control were compared. For (a–b), data represent Mean ± SD; data passed normality test, and statistics were calculated using a One-way ANOVA test with a post-hoc test corrected by Tukey's test. Each dot represents one replicate. For (f), data represent Mean within line chart; data passed normality test, and statistics were calculated using a paired Student's t-test. Each dot represents one independent individual.
Combined NMN and cART treatment reconstitutes CD4+ T cells in HIV-1-infected humanized mice
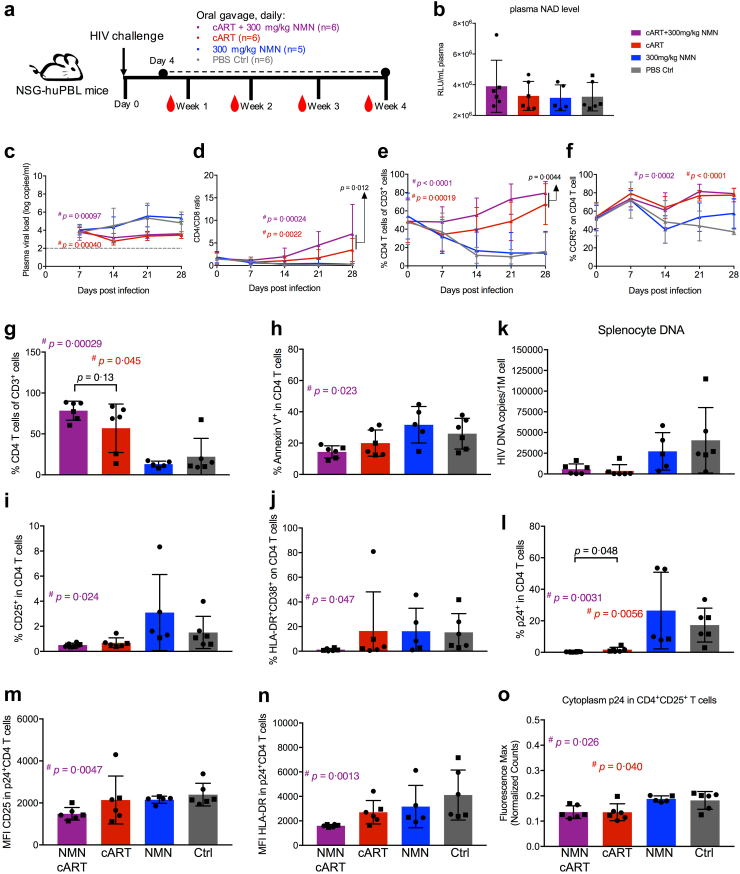
To determine the effect of NMN in vivo, we sought to investigate whether NMN treatment with or without cART would show any difference in the HIV-1-infected humanized mouse model. NSG-huPBL mice challenged with HIV-1JRFL live virus were orally administrated with NMN and/or cART daily starting from day 4 post-infection (Fig. 6a). The dose of NMN was 300 mg/kg per mouse as previously described by others.35 We measured the amount of plasma NAD on day 24 after NMN treatment. No significant increase in plasma NAD was found in the NMN treatment group as compared to the PBS control group (Fig. 6b). Similar plasma NAD amount was found in the cART-plus-NMN treatment group as compared with other groups due to lack of statistical significance (Fig. 6b). By monitoring plasma viral loads weekly, NMN alone was not able to suppress viral replication significantly as compared with the PBS control group likely due to insufficient concentration (<10 mM) (Fig. 6c). As expected, plasma viral loads were significantly suppressed in both cART-plus-NMN and cART treatment groups (Fig. 6c). Importantly, cART-plus-NMN treatment post-infection resulted in better CD4+ T cell reconstitution with significantly higher CD4/CD8 ratio and frequency of CD4+ T cells than the cART group over time (Fig. 6d and e and Supplementary Figure S13). However, the frequencies of CCR5+CD4+ T cells were restored similarly in both cART-plus-NMN and cART treatment groups (Fig. 6f and Supplementary Figure S13).
Fig. 6.
NMN treatment plus cART significantly increases CD4+T cell percentage in HIV-infected huPBL mouse model in vivo. (a) Schematic diagram of humanized mice experiment using human PBL reconstituted NSG mice (HuPBL). (b) Plasma NAD level was detected via luciferase assay on Day 28 post-infection. Plasma viral load (c), CD4/CD8 ratio (d), the percentage of CD4+ T cells in CD3+ cells (e), and the percentage of CCR5+ on CD4+ T cells (f), were monitored weekly before or post-infection. Among splenocytes collected on Day 28 post-infection, the percentage of CD4+ T cells in CD3+ cells (g), the percentage of Annexin V+ cells in CD4+ T cells (h), the percentage of CD25+ cells in CD4+ T cells (i), the percentage of HLA-DR+CD38+ cells in CD4+ T cells (j), and the percentage of p24+ cells in CD4+ T cells (l) from splenocytes were analysed by flow cytometry. The MFI values of CD25 (m) and HLA-DR (n) in p24+CD4+ T cells were also analysed by flow cytometry. (k) Splenocyte p24 DNA level was detected via real-time PCR. By immunohistochemistry (IHC) analysis on spleen tissue sections, IHC slides were scanned via PerkinElmer Vectra Polaris™ Automated Quantitative Pathology Imaging System and analysed by Inform Software. (o) The cytoplasm p24 max intensity (normalized counts) in CD25+CD4+ cells was calculated using Inform Software and compared among groups. Data represent Mean ± SD. For (c-f), the area under curve (AUC) was calculated and used for statistical analysis by an unpaired Student's t-test, and the p values reported in black text are the AUC analysis comparing ART alone to ART-plus-NMN; for (b) and (i), data did not pass normality test, and statistics were calculated using Mann–Whitney test; for (g–h) and (j–o), data passed normality test, and statistics were calculated using unpaired Student's t-tests. # represents the significant difference between experimental groups and the control group, and the corresponding p value was reported in the corresponding group colour (cART group in red, while cART-plus-NMN group in purple); ∗ represents the significant difference between cART group and cART-plus-NMN group, and the corresponding p value was reported in black colour. Two batches of mice were conducted in this experiment. Each dot represents one individual mouse. Circles show mice from Batch 1, whereas rectangles show mice from Batch 2.
On day 28 post-infection, mice were sacrificed, and their splenocytes were isolated for further analysis. Consistently, the most effective splenic CD4+ T cell reconstitution occurred in the cART-plus-NMN treatment group as compared to the PBS control group but not to the cART alone group (Fig. 6g and Supplementary Figure S14). Splenic CD4+ T cells in the cART-plus-NMN treatment group also displayed significantly lower frequencies of apoptotic (Annexin V+), hyperactivated (HLA-DR+CD38+) and CD25+ activated CD4+ T cells, as compared to the PBS control group but not to the cART alone group (Fig. 6h–j and Supplementary Figure S14). These findings, however, were not shown in the cART alone group when compared to the PBS control group (Fig. 6h–j and Supplementary Figure S14). Moreover, both CD25+ and CD25− CD4+ T cells from the cART-plus-NMN treatment group showed a dramatically decreased proportion of apoptotic cells, when compared to the PBS control but not to the cART alone group (Supplementary Figure S15). Regarding the proviral load in splenocytes, both the cART and cART-plus-NMN treatment groups exhibited similarly lower proviral loads as compared to the PBS control group (Fig. 6k). Importantly, the cART-plus-NMN treatment group showed significantly lower frequencies of both p24+CD4+ T cells and proliferating ki67+CD4+ T cells when compared with both the PBS control and the cART alone groups (Fig. 6l and Supplementary Figure S16a). Significantly decreased MFI values of both CD25 and HLA-DR in p24+CD4+ T cells were found in the cART-plus-NMN treatment group as compared to the PBS control group but not to the cART alone group (Fig. 6m and n). Comparable amounts of p24 MFI and ki67 MFI were noted in ki67+CD4+ and p24+CD4+ T cells, respectively, when compared the cART-plus-NMN with the cART alone (Supplementary Figure S16b and c). The immunohistochemistry (IHC) analysis of spleen tissues revealed decreased cytoplasm amounts of p24 in CD25+CD4+ cells in the cART-plus-NMN treatment group when compared to the PBS control but not to the cART alone group (Fig. 6o and Supplementary Figure S17). Therefore, the cART-plus-NMN but not cART alone showed a significant in vivo suppressive effect on T cell hyperactivation compared with the PBS control group, reducing frequencies of both p24+CD4+ and ki67+CD4+ T cells. Moreover, there were significant statistical differences between the ART alone and cART-plus-NMN groups for the frequencies of p24+CD4+ cells and proliferating ki67+CD4+ T cells. Notably, there were no significant differences between the NMN alone and the PBS control groups in the experiments, indicating the need of cART for combined in vivo effects with NMN. Results from the humanized mouse study demonstrated that NMN treatment could improve the therapeutic effect of cART in vivo on reconstituting CD4+ T cells and improving CD4/CD8 ratio significantly over time.
Discussion
In this study, we report that 10 mM NMN suppresses HIV-1 production in infected primary CD4+ T cells at the post translational level in vitro. The NMN-mediated HIV-1 suppression was found primarily in CD25+CD4+ T cells rather than in CD25−CD4+ T cells. Critically, 10 mM NMN also suppresses HIV-1 reactivation in primary CD4+ T cells derived from cART-treated PLWH. CyTOF analysis revealed consistently that the NMN treatment decreased the amounts of CD25 on both CXCR3+ effector and CCR4+ central memory CD4+ T cells. RNA-seq analysis then showed some upregulated genes (e.g. NR4A2) and the downregulated CD25-related GO pathways for cell proliferation, activation and apoptosis. Importantly, NMN improved the therapeutic effect of cART in HIV-1-infected humanized mice by improving the CD4+ T cell reconstitution, likely through reducing the frequencies of p24+CD4+ and ki67+CD4+ T cells. Our findings have implications to future investigation of NMN as a supplemental treatment to cART for improving CD4+ T cell reconstitution, especially for PLWH with persistently low CD4+ T cell count.
NMN acts on primary CD4+ T cells during HIV-1 replication. NMN suppressed HIV-1 with an IC50 value of 5.936 mM, which is much higher than those of existing anti-HIV drugs such as 3TC, DTG and TDF (e.g. mostly below 50 nM).53, 54, 55 It is, therefore, not surprising that NMN alone could not suppress plasma viral load or splenic proviral load significantly in HIV-1-infected humanized mice using the dose of 300 mg/kg. In support of this notion, we did not detect any significant increase of NAD in vivo at this dose. Nevertheless, a positive effect of NMN was observed when it was combined with cART for treatment in HIV-1-infected humanized mice. Such effect led to significantly improved CD4+ T cell reconstitution by reducing the frequencies of both p24+CD4+ and ki67+CD4+ T cells significantly as compared with the cART alone. Mechanistically, NMN displayed potent immunomodulating activity on primary CD4+ T cells. We found that 10 mM NMN suppressed late activation (CD25 and HLA-DR) instead of early activation (CD69) of patient-derived CD4+ T cells.48 When comparing the intracellular p24 amount in CD25+/− or HLA-DR+/− HIV-1-infected CD4+ T cells, we found a significant p24 reduction in HIV-1-infected CD25+CD4+ T cells after the NMN treatment. Our findings pointed to a selective effect of NMN on CD25+CD4+ T cells.49 By CyTOF analysis, CXCR3+ effector and CCR4+ central memory CD4+ T cells were found to be two distinguishable subsets affected by the NMN treatment in terms of the CD25 expression. These CXCR3+ effector and CCR4+ central memory CD4+ T cell subsets were of low expression of CCR6. A previous study indicated that CXCR3+/CCR4+ but CCR6− CD4+ T cells were relatively less permissive to R5-tropic HIV-1 infection than CXCR3+/CCR4+ but CCR6+ ones.56 However, the role of CD25 in modulating the permissive CCR5-tropic HIV-1 infection, as well as whether NMN-affected CD25+ cells are regulatory T cells or cells highly expressing NMN transporters, remains to be determined in future studies. Furthermore, by RNA-Seq analysis, we showed that the NMN treatment could downregulate several CD25-related GO pathways including cell proliferation, cell activation and cell apoptosis. Foxp3/CD25 was addressed in the GO pathways associated with cell activation and proliferation. Although Foxp3 is well known as a phenotypic marker of regulatory T cells, it may serve as a transient transcriptional regulator in activated CD4+ T cells upon T cell receptor activation.57,58 CD25 but not CD69 downregulation by the NMN treatment in CD4+ T cells of HUDs and PLWH in our study may explain why NMN suppressed late but not early T cell activation. These transcriptomic results are consistent with suppressed proliferating p24-expressing T cells after the in vitro NMN treatment and suppressed immune activation by the in vivo cART-plus-NMN treatment. Therefore, NMN/NAD may modulate CD4+ T cell activation and proliferation during HIV-1 infection.
NMN exhibited in vitro inhibitory effects on HIV-1 replication at 10 mM consistently. From our experimental finding, NMN may act both at the translation of HIV-1 p24 and at the proliferation of primary CD4 T cells. For action on the translation of HIV-1 p24, our results suggested that NMN did not inhibit HIV-1 infection at entry, integration, and transcription stages but at a post-transcription one. The single-cycle HIV-1 infection and the latent ACH-2 cell models were used to validate this finding. Moreover, the anti-HIV-1 activity of NMN was consistently obtained when PLWH-derived and in vitro infected primary CD4+ T cells were tested. For action on the proliferation of primary CD4 T cells, NMN may reduce IL-2-induced frequency of p24-expressing and proliferating CD4 T cells, due to the downregulation of CD25 (IL-2 receptor). Mechanistically, our RNA-seq results indicated that the upregulation of NR4A2 and the downregulation of CD25 by NMN treatment were likely related the modulation of transcriptional factor Foxp3. This finding was determined by using the inducer of Foxp3 namely AS2863619. Since only a very small proportion of cells might contain integrated HIV-1 in the PLWH-derived ex vivo model,59 the inhibitory activity of NMN could also be contributed by NMN-mediated suppression of T cell activation and proliferation. Therefore, NMN may suppress both cellular activation and viral reactivation in the ex vivo model. For the potential of NMN in the “block and lock” strategy, future studies are needed to determine whether NMN would be useful as a latency-locking agent60 and whether cellular NAD metabolism would modulate HIV-1 expression by testing NAD-consuming factors such as sirtuins, PARPs and CD38.12,13
NMN is likely a useful suppressive agent against immune hyperactivation in vivo. During HIV-1 chronic infection, persistent immune activation may lead to incomplete CD4+ T cell recovery despite cART in HIV-INRs.4,61 Various factors may contribute to the persistent immune activation in HIV-1 infection.62 These factors may include toll-like receptor activation by HIV-1 ssRNA, NF-κB activation triggered by viral protein, interferon-mediated stimulation, and caspase-induced cell apoptosis/pyroptosis and so on.62 To reduce the risk of AIDS progression, an intervention of reducing persistent immune activation in HIV-INRs is needed.4,61 Given the promising in vitro results of NMN in modulating CD4+ T cell activation in HIV-1 infection, we also investigated the potential of NMN as an intervention against immune activation in humanized mice. Interestingly, NMN alone was insufficient in suppressing HIV-1 replication in vivo at a dose of 300 mg/kg although this amount is much higher than 150 mg daily use as a food supplement in humans. HIV-infected huPBL mice treated with cART-plus-NMN, however, demonstrated significantly improved reconstitution of CD4+ T cells correlated with moderately alleviated hyperactivation by measuring the HLA-DR+CD38+ biomarkers and relatively decreased apoptotic cells by measuring Annexin V+ biomarker. One previous report provided in vitro evidence of 0.05–5 mM nicotinamide treatment on reducing apoptotic cells derived from PLWH, which supports our findings on decreased apoptosis by in vivo cART-plus-NMN treatment.63 Although cART-plus-NMN treatment showed an in vivo suppressive effect on immune hyperactivation compared to PBS control, it did not show statistical difference when compared to the cART treatment alone. Importantly, however, the cART-plus-NMN treatment resulted in better CD4+ T cell reconstitution with a higher CD4/CD8 ratio and a significantly higher frequency of CD4+ T cells than the cART group over time. The cART-plus-NMN treatment led to reduced frequencies of both p24+CD4+ and ki67+CD4+ T cells significantly as compared with the cART alone. Our findings, therefore, have implications to future clinical testing of daily high dose of NMN treatment to improve CD4+ T cell reconstitution in cART-treated PLWH, especially in INRs with persistently low CD4 count, by modulating immune activation.1,61
Contributors
Z.C. supervised the team, coordinated the collaboration and gained the research grants for this study. Y.M. and Z.C. designed the experiments, performed the experiments, collected the data, analysed the data, interpreted the data and wrote the article. M.Y. and Y.S. analysed and interpreted the RNA-seq data, and wrote the RNA-seq part of the article. Y.M., L.-Y.Y., Y.Z. and H.S. participated in experimental design, experimental performance, data analysis and data interpretation for CyTOF experiment. Y.M. and R.Z. participated in the experimental design and data interpretation for flow cytometry. Y.M., C.Y. and C.-Y.H.L. participated in in vitro experiments, and L.-Y.Y. participated in the experimental design and data interpretation for HIVJRFL-nLuc pseudotyped virus infection. Y.M., Q.P., T.-Y.L. and H.H. participated in weekly bleeding procedures and/or sacrificing procedures for animal experiments. L.L. participated in data analysis and data interpretation for the IHC experiment. Y.M., L.-Y.Y. and T.-Y.L. prepared PBMC samples from PLWH and/or healthy donors. Q.P., G.C.-Y.L. and H.W. collected the clinical samples and clinical data from PLWH. J.W. generated the NMN powder (purity ≥99%) for research use.
All authors read and approved the final version of the manuscript. Y.M., M.Y., C.Y. and Z.C. have verified the data.
Data sharing statement
RNA-seq data generated in this study were shared in NCBI (accession ID: GSE234308).
Declaration of interests
J.W. is an employee and shareholder of GeneHarbor (Hong Kong) Biotechnologies.
Acknowledgements
This work was supported by Theme-Based Research Scheme (T11-706/18-N to ZC) of the Hong Kong Research Grants Council, Seed Fund for Translational and Applied Research of University Research Committee (URC) of HKU, and the Collaborative Research with GeneHarbor (Hong Kong) Biotechnologies Limited; University Development Fund and Li Ka Shing Faculty of Medicine Matching Fund from HKU to AIDS Institute; and National Key R&D Program of China (Grant 2021YFC2301900).
We thank Prof Shui Shan Lee for recruiting some cART-treated PLWH in Hong Kong. We acknowledge Centre for PanorOmic Sciences (CPOS, Centre Manager: Dr. Agnes Chan) of Li Ka Shing Faculty of Medicine in The University of Hong Kong.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104877.
Appendix A. Supplementary data
References
- 1.Maartens G., Celum C., Lewin S.R. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014;384(9939):258–271. doi: 10.1016/S0140-6736(14)60164-1. [DOI] [PubMed] [Google Scholar]
- 2.Gazzola L., Tincati C., Bellistrì G.M., Monforte A., Marchetti G. The absence of CD4+ T cell count recovery despite receipt of virologically suppressive highly active antiretroviral therapy: clinical risk, immunological gaps, and therapeutic options. Clin Infect Dis. 2009;48(3):328–337. doi: 10.1086/595851. [DOI] [PubMed] [Google Scholar]
- 3.Nies-Kraske E., Schacker T.W., Condoluci D., et al. Evaluation of the pathogenesis of decreasing CD4(+) T cell counts in human immunodeficiency virus type 1-infected patients receiving successfully suppressive antiretroviral therapy. J Infect Dis. 2009;199(11):1648–1656. doi: 10.1086/598980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bono V., Augello M., Tincati C., Marchetti G. Failure of CD4+ T-cell recovery upon Virally-effective cART: an enduring gap in the understanding of HIV+ immunological non-responders. New Microbiol. 2022;45(3):155–172. [PubMed] [Google Scholar]
- 5.Pirinen E., Auranen M., Khan N.A., et al. Niacin cures systemic NAD(+) deficiency and improves muscle performance in adult-onset mitochondrial myopathy. Cell Metabol. 2020;31(6):1078–1090.e5. doi: 10.1016/j.cmet.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Eckard A.R., O'Riordan M.A., Rosebush J.C., et al. Vitamin D supplementation decreases immune activation and exhaustion in HIV-1-infected youth. Antivir Ther. 2018;23(4):315–324. doi: 10.3851/IMP3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shivakoti R., Ewald E.R., Gupte N., et al. Effect of baseline micronutrient and inflammation status on CD4 recovery post-cART initiation in the multinational PEARLS trial. Clin Nutr. 2019;38(3):1303–1309. doi: 10.1016/j.clnu.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebouché B., Yero A., Shi T., et al. Impact of extended-release niacin on immune activation in HIV-infected immunological non-responders on effective antiretroviral therapy. HIV Res Clin Pract. 2020;21(6):182–190. doi: 10.1080/25787489.2020.1866846. [DOI] [PubMed] [Google Scholar]
- 9.Katsyuba E., Romani M., Hofer D., Auwerx J. NAD(+) homeostasis in health and disease. Nat Metab. 2020;2(1):9–31. doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- 10.Kang B.E., Choi J.Y., Stein S., Ryu D. Implications of NAD(+) boosters in translational medicine. Eur J Clin Invest. 2020;50(10) doi: 10.1111/eci.13334. [DOI] [PubMed] [Google Scholar]
- 11.Reiten O.K., Wilvang M.A., Mitchell S.J., Hu Z., Fang E.F. Preclinical and clinical evidence of NAD(+) precursors in health, disease, and ageing. Mech Ageing Dev. 2021;199 doi: 10.1016/j.mad.2021.111567. [DOI] [PubMed] [Google Scholar]
- 12.Zheng M., Schultz M.B., Sinclair D.A. NAD(+) in COVID-19 and viral infections. Trends Immunol. 2022;43(4):283–295. doi: 10.1016/j.it.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navas L.E., Carnero A. NAD(+) metabolism, stemness, the immune response, and cancer. Signal Transduct Targeted Ther. 2021;6(1):2. doi: 10.1038/s41392-020-00354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Q., Dong L., Li Y., Liu G. SIRT1 and HIF1α signaling in metabolism and immune responses. Cancer Lett. 2018;418:20–26. doi: 10.1016/j.canlet.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Navarro M.N., Gómez de Las Heras M.M., Mittelbrunn M. Nicotinamide adenine dinucleotide metabolism in the immune response, autoimmunity and inflammageing. Br J Pharmacol. 2022;179(9):1839–1856. doi: 10.1111/bph.15477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piedra-Quintero Z.L., Wilson Z., Nava P., Guerau-de-Arellano M. CD38: an immunomodulatory molecule in inflammation and autoimmunity. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.597959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie N., Zhang L., Gao W., et al. NAD(+) metabolism: pathophysiologic mechanisms and therapeutic potential. Signal Transduct Targeted Ther. 2020;5(1):227. doi: 10.1038/s41392-020-00311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davidson M.H. Niacin use and cutaneous flushing: mechanisms and strategies for prevention. Am J Cardiol. 2008;101(8a):14b–19b. doi: 10.1016/j.amjcard.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 19.Sauve A.A. NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther. 2008;324(3):883–893. doi: 10.1124/jpet.107.120758. [DOI] [PubMed] [Google Scholar]
- 20.Shi W., Hegeman M.A., Doncheva A., Bekkenkamp-Grovenstein M., de Boer V.C.J., Keijer J. High dose of dietary nicotinamide riboside induces glucose intolerance and white adipose tissue dysfunction in mice fed a mildly obesogenic diet. Nutrients. 2019;11(10):2439. doi: 10.3390/nu11102439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung K.W., Wu T., Ho S.F., et al. α(4)β(7)(+) CD4(+) effector/effector memory T cells differentiate into productively and latently infected central memory T cells by transforming growth factor β1 during HIV-1 infection. J Virol. 2018;92(8) doi: 10.1128/JVI.01510-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 23.He J., Choe S., Walker R., Di Marzio P., Morgan D.O., Landau N.R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69(11):6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malnati M.S., Scarlatti G., Gatto F., et al. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc. 2008;3(7):1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 25.Cheung A.K.L., Huang Y., Kwok H.Y., Chen M., Chen Z. Latent human cytomegalovirus enhances HIV-1 infection in CD34(+) progenitor cells. Blood Adv. 2017;1(5):306–318. doi: 10.1182/bloodadvances.2016000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X., Guo J., Niu M., et al. Tandem bispecific neutralizing antibody eliminates HIV-1 infection in humanized mice. J Clin Invest. 2018;128(6):2239–2251. doi: 10.1172/JCI96764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X., Liu L., Cheung K.W., et al. Brain invasion by CD4(+) T cells infected with a transmitted/founder HIV-1BJZS7 during acute stage in humanized mice. J Neuroimmune Pharmacol. 2016;11(3):572–583. doi: 10.1007/s11481-016-9654-0. [DOI] [PubMed] [Google Scholar]
- 28.Siegel C.S., McCullough L.D. NAD+ and nicotinamide: sex differences in cerebral ischemia. Neuroscience. 2013;237:223–231. doi: 10.1016/j.neuroscience.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volk V., Schneider A., Spineli L.M., Grosshennig A., Stripecke R. The gender gap: discrepant human T-cell reconstitution after cord blood stem cell transplantation in humanized female and male mice. Bone Marrow Transplant. 2016;51(4):596–597. doi: 10.1038/bmt.2015.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drake A.C., Chen Q., Chen J. Engineering humanized mice for improved hematopoietic reconstitution. Cell Mol Immunol. 2012;9(3):215–224. doi: 10.1038/cmi.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Padura I., Agliano A., Marighetti P., Porretti L., Bertolini F. Sex-related efficiency in NSG mouse engraftment. Blood. 2010;116(14):2616–2617. doi: 10.1182/blood-2010-07-295584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO Policy brief: update of recommendations on first- and second-line antiretroviral regimens: World Health Organization. 2019. https://apps.who.int/iris/handle/10665/325892 Available from:
- 33.Nie J., Sun F., He X., et al. Tolerability and adherence of antiretroviral regimens containing long-acting fusion inhibitor albuvirtide for HIV post-exposure prophylaxis: a cohort study in China. Infect Dis Ther. 2021;10(4):2611–2623. doi: 10.1007/s40121-021-00540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair A.B., Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills K.F., Yoshida S., Stein L.R., et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metabol. 2016;24(6):795–806. doi: 10.1016/j.cmet.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belkina A.C., Ciccolella C.O., Anno R., Halpert R., Spidlen J., Snyder-Cappione J.E. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun. 2019;10(1):5415. doi: 10.1038/s41467-019-13055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linderman G.C., Rachh M., Hoskins J.G., Steinerberger S., Kluger Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods. 2019;16(3):243–245. doi: 10.1038/s41592-018-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Gassen S., Callebaut B., Van Helden M.J., et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87(7):636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 39.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray M.F., Nghiem M., Srinivasan A. HIV infection decreases intracellular nicotinamide adenine dinucleotide [NAD] Biochem Biophys Res Commun. 1995;212(1):126–131. doi: 10.1006/bbrc.1995.1945. [DOI] [PubMed] [Google Scholar]
- 44.Murray M.F., Srinivasan A. Nicotinamide inhibits HIV-1 in both acute and chronic in vitro infection. Biochem Biophys Res Commun. 1995;210(3):954–959. doi: 10.1006/bbrc.1995.1749. [DOI] [PubMed] [Google Scholar]
- 45.Mohammadi P., Desfarges S., Bartha I., et al. 24 hours in the life of HIV-1 in a T cell line. PLoS Pathog. 2013;9(1) doi: 10.1371/journal.ppat.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telwatte S., Morón-López S., Aran D., et al. Heterogeneity in HIV and cellular transcription profiles in cell line models of latent and productive infection: implications for HIV latency. Retrovirology. 2019;16(1):32. doi: 10.1186/s12977-019-0494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clouse K.A., Powell D., Washington I., et al. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J Immunol. 1989;142(2):431–438. [PubMed] [Google Scholar]
- 48.Reddy M., Eirikis E., Davis C., Davis H.M., Prabhakar U. Comparative analysis of lymphocyte activation marker expression and cytokine secretion profile in stimulated human peripheral blood mononuclear cell cultures: an in vitro model to monitor cellular immune function. J Immunol Methods. 2004;293(1-2):127–142. doi: 10.1016/j.jim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Ramilo O., Bell K.D., Uhr J.W., Vitetta E.S. Role of CD25+ and CD25-T cells in acute HIV infection in vitro. J Immunol. 1993;150(11):5202–5208. [PubMed] [Google Scholar]
- 50.Nakatani T., Kaburagi Y., Shimada Y., et al. CCR4 memory CD4+ T lymphocytes are increased in peripheral blood and lesional skin from patients with atopic dermatitis. J Allergy Clin Immunol. 2001;107(2):353–358. doi: 10.1067/mai.2001.112601. [DOI] [PubMed] [Google Scholar]
- 51.Lim H.W., Kim C.H. Loss of IL-7 receptor alpha on CD4+ T cells defines terminally differentiated B cell-helping effector T cells in a B cell-rich lymphoid tissue. J Immunol. 2007;179(11):7448–7456. doi: 10.4049/jimmunol.179.11.7448. [DOI] [PubMed] [Google Scholar]
- 52.Akamatsu M., Mikami N., Ohkura N., et al. Conversion of antigen-specific effector/memory T cells into Foxp3-expressing T(reg) cells by inhibition of CDK8/19. Sci Immunol. 2019;4(40) doi: 10.1126/sciimmunol.aaw2707. [DOI] [PubMed] [Google Scholar]
- 53.Rosenblum L.L., Patton G., Grigg A.R., et al. Differential susceptibility of retroviruses to nucleoside analogues. Antivir Chem Chemother. 2001;12(2):91–97. doi: 10.1177/095632020101200202. [DOI] [PubMed] [Google Scholar]
- 54.Alessandri-Gradt E., Collin G., Tourneroche A., et al. HIV-1 non-group M phenotypic susceptibility to integrase strand transfer inhibitors. J Antimicrob Chemother. 2017;72(9):2431–2437. doi: 10.1093/jac/dkx190. [DOI] [PubMed] [Google Scholar]
- 55.Li G., Wang Y., De Clercq E. Approved HIV reverse transcriptase inhibitors in the past decade. Acta Pharm Sin B. 2022;12(4):1567–1590. doi: 10.1016/j.apsb.2021.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gosselin A., Monteiro P., Chomont N., et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184(3):1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bacchetta R., Gambineri E., Roncarolo M.G. Role of regulatory T cells and FOXP3 in human diseases. J Allergy Clin Immunol. 2007;120(2):227–235. doi: 10.1016/j.jaci.2007.06.023. quiz 36-7. [DOI] [PubMed] [Google Scholar]
- 58.Wang J., Ioan-Facsinay A., van der Voort E.I., Huizinga T.W., Toes R.E. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37(1):129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 59.Wu V.H., Nordin J.M.L., Nguyen S., et al. Profound phenotypic and epigenetic heterogeneity of the HIV-1-infected CD4(+) T cell reservoir. Nat Immunol. 2023;24(2):359–370. doi: 10.1038/s41590-022-01371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeh Y.J., Ho Y.C. Shock-and-kill versus block-and-lock: targeting the fluctuating and heterogeneous HIV-1 gene expression. Proc Natl Acad Sci U S A. 2021;118(16) doi: 10.1073/pnas.2103692118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anthony K.B., Yoder C., Metcalf J.A., et al. Incomplete CD4 T cell recovery in HIV-1 infection after 12 months of highly active antiretroviral therapy is associated with ongoing increased CD4 T cell activation and turnover. J Acquir Immune Defic Syndr. 2003;33(2):125–133. doi: 10.1097/00126334-200306010-00002. [DOI] [PubMed] [Google Scholar]
- 62.Lv T., Cao W., Li T. HIV-related immune activation and inflammation: current understanding and strategies. J Immunol Res. 2021;2021 doi: 10.1155/2021/7316456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savarino A., Martini C., Orofino G.C., et al. Apoptotic DNA fragmentation, and its in vitro prevention by nicotinamide, in lymphocytes from HIV-1-seropositive patients and in HIV-1-infected MT-4 cells. Cell Biochem Funct. 1997;15(3):171–179. doi: 10.1002/(SICI)1099-0844(199709)15:3<171::AID-CBF736>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.