Abstract
Mouse-virulent Salmonella typhimurium strains SR-11 and ATCC 14028-1s express curli fibers, thin aggregative fibers, at ambient temperature on plates as judged by Western blot analysis and electron microscopy. Concomitantly with curli expression, cells develop a rough and dry colony morphology and bind the dye Congo red (called the rdar morphotype). Cloning and characterization of the two divergently transcribed operons required for curli biogenesis, csgBA(C) and csgDEFG, from S. typhimurium SR-11 revealed the same gene order and flanking genes as in Escherichia coli. The divergence of the curli region between S. typhimurium and E. coli at the nucleotide level is above average (22.4%). However, a high level of conservation at the protein level, which ranged from 86% amino acid homology for the fiber subunit CsgA to 99% homology for the lipoprotein CsgG, implies functional constraints on the gene products. Consequently, S. typhimurium genes on low-copy-number plasmids were able to complement respective E. coli mutants, although not always to wild-type levels. rpoS and ompR are required for transcriptional activation of (at least) the csgD promoter. The high degree of conservation at the protein level and the identical regulation patterns in E. coli and S. typhimurium suggest similar roles of curli fibers in the same ecological niche in the two species.
Proteinaceous, filamentous appendices on bacterial surfaces, called fimbriae or pili, enable the bacterial cell to make contact with inanimate surfaces and eukaryotic or prokaryotic cells. Tight contact, called adherence, precedes, e.g., colonization of surfaces and invasion of eukaryotic cells by the bacteria. Fimbriae are best studied in the family Enterobacteriaceae, particularly in Escherichia coli and Salmonella enterica (49), in the context of pathogen-host interactions (44). Related species, subspecies, and even particular strains can have a specific set of fimbrial genes which are often located on pathogenicity islands on the chromosome or on plasmids (28, 31, 43, 45). The need for flexibility in the strategy of adhesion in order to overcome the host immune system, for example, has also led to a variability in fimbrial genes derived from a common ancestor. The immunogenic and adhesive properties of these fimbriae, which can be encoded either by the fimbrial subunit gene, as in the case of K88 fimbriae, or by separate genes, as in the case of the Pap pili, can be exchanged as gene cassettes in the context of a common frame (45). Therefore, fimbrial genes often do not appear to fit the phylogenetic classification of the bacterium but are shared by more distantly related organisms occupying the same ecological niche (45, 63).
Most of the fimbriae identified in Salmonella enterica subsp. enterica serotype Typhimurium (in this paper, referred to as S. typhimurium) have been described only phenotypically; the few whose genes have been cloned and sequenced (6, 15, 16, 28, 56) are unique to S. enterica or a subset of its subspecies (5, 56). However, curli fibers, thin aggregative fibers, seem to be present and expressed in almost all Salmonella spp. and E. coli (5, 17, 23) and maybe also in other Enterobacteriaceae, such as Shigella, Citrobacter, and Enterobacter spp. (23). So far, two nomenclature systems exist (20, 34). The genes for curli biogenesis (csg) in E. coli are called agf (thin aggregative fibers) in Salmonella enteritidis. For convenience, we use one nomenclature system in this communication.
Curli fibers detected, for example, on S. typhimurium strains causing acute salmonellosis in pigeons (32) and on E. coli isolates causing bovine mastitis (54) mediate binding to fibronectin (54), a variety of other human serum and tissue matrix proteins (7, 54, 62), and the dye Congo red (CR) (18). In E. coli MC4100, two divergently transcribed operons, csgDEFG and csgBA(C), which are separated by a 513-bp intergenic region are required for the biogenesis of curli fibers (34). Transposon insertions in the csgD gene, which encodes a transcriptional regulator belonging to the LuxR family as identified by the sequence similarity of the DNA binding helix-turn-helix motif, completely abolished transcription of the csgBA operon (34). Assembled by the extracellular nucleation-precipitation pathway, the secreted fiber subunit CsgA is polymerized on the surface-exposed nucleator CsgB (35), which, in addition, is present along the filament in minor amounts (8). CsgA and CsgB show 49% similarity and contain repeat regions whose interaction triggers polymerization of CsgA (8, 35). The outer-membrane-located lipoprotein CsgG is required to protect CsgA and CsgB from proteolysis (48). The roles of csgE and csgF are just beginning to be elucidated. csgE is required for the fibronectin and CR binding properties of curli fibers but does not significantly affect polymerization of the fiber subunit (36). The nucleation function is impaired in a csgF mutant, in which CsgA is released into the growth medium (37). Curli expression in E. coli MC4100 and YMel is highly regulated by environmental conditions; it is restricted to low temperature on plates containing medium with a low salt concentration.
The alternative sigma factor RpoS (ςS) is a global regulator controlling the expression of a large number of genes during starvation and other stress conditions in E. coli (52) and S. typhimurium (26). rpoS-deficient S. typhimurium strains are impaired in their virulence in the mouse model for typhoid fever (22, 26); the rpoS deficiency also seems to be the cause of attenuation of common laboratory derivatives of strain LT2 (65, 68). Transcription by the RNA polymerase containing ςS at different promoters can include complex interactions with additional regulators (25). The stationary-phase-induced transcription of the genes for curli biogenesis is dependent on ςS in E. coli. It has not been resolved whether rpoS is needed only for transcription from the E. coli csgD promoter or also affects the CsgD-dependent csgBA promoter (34). Absence of H-NS has been shown to make at least the csgD promoter independent of rpoS, suggesting an efficient repression of rpoD-dependent transcription by hns (2, 34).
Increasing osmolarity has been shown to shut off expression of curli genes at the transcriptional level (53) but to increase the levels of RpoS (42). Therefore, other regulators must also influence the transcription from the csgD and csgBA promoters. OmpR is a transcriptional regulator which was studied mainly for its role in regulating transcription of the outer membrane proteins OmpF and OmpC in response to surrounding osmolarity sensed by EnvZ in E. coli (55). Besides OmpF and OmpC, a tripeptide permease (TppB) is known to be regulated by OmpR in S. typhimurium (30). ompR mutants of S. typhimurium are attenuated in vivo (24) and unable to kill macrophages in vitro (47). ompR has been reported to be required for transcriptional activation of both the csgBA and the csgDEFG promoters in E. coli (40); however, no experimental data have been reported so far.
In this paper, we report the cloning and characterization of the two operons for curli biogenesis from S. typhimurium SR-11. The highly conserved genes displayed the same arrangement in the same chromosomal context as in E. coli. Consequently, S. typhimurium-derived genes on plasmids could functionally replace their E. coli counterparts, although not always to the wild-type levels. Regulation of curli biogenesis in S. typhimurium SR-11 and ATCC 14028-1s was reminiscent of E. coli MC4100 and YMel. Curli biogenesis was restricted to ambient temperature on plates, and transcription from the csgDEFG and csgBA promoters required rpoS and ompR. The conservation of genes and of the regulation pattern implies an important role of curli fibers in the lifestyle of E. coli and Salmonella spp. in the same ecological niche.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used and constructed in this study are given in Tables 1 and 2, respectively. In the beginning, S. typhimurium SR-11 was used for the analysis of curli biogenesis. However, the initial lack of tools for genetic analysis led to a subsequent shift to strain ATCC 14028-1s, a virulent derivative of the well-characterized LT2 strain. Curli expression in E. coli and S. typhimurium was monitored by growth on solid Yesca (35) and Luria-Bertani (LB) medium without salt, respectively. Media were supplemented with CR (40 μg/ml) and Coomassie brilliant blue (20 μg/ml) to judge colony morphology and color (34, 35). However, CR slightly inhibits the growth of S. typhimurium; therefore, the colony morphology develops later than on medium without CR. Recombinant clones were grown on LB medium supplemented with recommended concentrations of antibiotics (4), if required.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| S. typhimurium | ||
| ATCC 14028-1s | wt1a | American Type Culture Collection |
| SR-11 | wt1a | R. Curtiss III |
| LB5010 | metA22 metE551 ilv-452 leu-3121 trpD2 xyl-404 galE856 hsdLT6 hsdSA29 hsdSB121 rpsL120 | 12 |
| 146 | polA-2 zig214::Tn10 (Tetr) | M. Rhen |
| SF1005 | rpoS::pRR10(ΔtrfA)(Ampr) | D. Guiney |
| UMR1 | ATCC 14028-1s Nalr | This study |
| UMR3 | SR-11 Nalr | This study |
| MAE1 | UMR1 ΔcsgA101::Kmr | This study |
| MAE2 | UMR3 ΔcsgA101::Kmr | This study |
| MAE5 | UMR1 ΔcsgA101 | This study |
| INK1 | SR-11 zcg-101::Kmr | This study |
| MAE40 | SF1005 × UMR1; UMR1 rpoS::pRR10(ΔtrfA)(Ampr) | This study |
| MAE29 | SF1005 × SR-11; SR-11 rpoS::pRR10 (ΔtrfA)(Ampr) | This study |
| MAE46 | UMR1 ΔompR101::Ampr | This study |
| MAE34 | UMR3 ΔompR101::Ampr | This study |
| E. coli K-12 | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | Laboratory collection |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | Laboratory collection |
| XL1 Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| HLO7 | MC4100 Δ(csgG-csgC)::Kmr | 47a |
| MHR204 | MC4100 csgA2::Tn105 Cmr | 34 |
| MHR210 | MC4100 csgG1::Tn105 Cmr | 34 |
| MHR261 | MC4100 csgB2 | 35 |
| MHR426 | MC4100 csgF4 | 36 |
| MHR480 | MC4100 ΔcsgE3 | 37 |
| MHR503 | MC4100 csgD6 | This study |
wt1, wild type 1.
TABLE 2.
Plasmids used and constructed in this study
| Plasmid | Characterization | Reference |
|---|---|---|
| pLAFR3 | Tetr RK2 oriT | 64 |
| pMAK700 | Cmr temperature-sensitive replicon derived from pSU101 | 33 |
| pMAK705 | pMAK700 containing pUC19 polylinker | 33 |
| pRK2013 | Kmr ColE1 mob tra | 27 |
| pUC4K | Ampr Kmr cassette | Pharmacia |
| pWSK29 | Ampr pSC101 ori | 67 |
| pWKS30 | Ampr pSC101 ori | 67 |
| pUMR2b | pMAK700 ΔcsgA101; Cmr | This study |
| pUMR2c-1 | pMAK700 ΔcsgA101::Kmr; Cmr Kmr | This study |
| pUMR10-7 | pBS, NsiI-BamHI-cut CSGD1-CSGB3 from SR-11; Ampr | 56a |
| pMU1 | pUC18, PstI fragment of zcg-101; Ampr | This study |
| pMU2 | pUC18, HindIII-PstI fragment of zcg-101; Ampr | This study |
| pMU3a | pMAK705, HindIII-PstI fragment of zcg-101; Cmr | This study |
| pMU3b-1 | pMAK705, zcg-101::Km; Cmr Kmr | This study |
| pMU4 | pLAFR3 containing a 39-kb PstI fragment harboring the csg genes; Tetr Kmr | This study |
| pCSGA | pWKS30, PstI-BamHI-cut CSGB2-CSGC; Ampr | This study |
| pCSGB | pWKS30, HindIII-PstI-cut CSGBD-CSGA1; Ampr | This study |
| pCSGD | pWSK29 (SmaI/EcoRI), EcoRI-cut SP2-SP5; Ampr | This study |
| pCSGE | pWSK29 (SmaI/BamHI), BstYI-cut SP5-SP9; Ampr | This study |
| pCSGF | pWKS30 (EcoRI/SmaI), EcoRI-cut SP6-SP17; Ampr | This study |
| pCSGEFG | pWSK29 (PstI/EcoRI), NsiI-cut CSGD2-SP21; Ampr | This study |
| pUMR7b-2 | pMAK705 ΔompR101::Ampr; Cmr Ampr | This study |
DNA techniques.
Isolation of plasmid, cosmid, and chromosomal DNAs and all enzymatic manipulations (restriction digestion, ligation, phosphorylation, and PCR) were carried out by standard protocols (4, 58) with enzymes from Boehringer Mannheim or Biolabs. Southern blotting was done as described previously (57). Individual PCR fragments were purified with a Quiaquick PCR purification kit (Quiagen); otherwise, a Geneclean II kit (Bio 101, Inc.) was used after electrophoresis. After polyethylene glycol precipitation (1), plasmids were sequenced with a Cycle Sequencing Ready Reaction kit (Perkin-Elmer). All sequence analyses were performed with the Genetics Computer Group package, version 8 or 9 (GCG, University of Wisconsin).
Cloning of the genes for curli biogenesis from S. typhimurium SR-11.
The curli genes from S. typhimurium SR-11 were cloned in the following way. First, a DNA fragment downstream of the csgC gene was sought. Therefore, plasmid pCurli, containing a 3.1-kb HindIII fragment (analogous to the fragment described in reference 17) from the non-curli-producing strain S. typhimurium SL2965 in pUC18, was integrated into the chromosome of a polA2 derivative of strain LB5010. After determination of appropriate restriction sites by hybridization with the vector, genomic DNA isolated from an integrant was cut with PstI and ligated under conditions favoring the intramolecular reaction (pMU1). A HindIII/PstI digest with subsequent subcloning depleted pMU1 from any DNA fragments containing curli sequences (pMU2).
As the second step, a resistance marker was placed downstream of csgC. The 6.6-kb HindIII/PstI fragment of pMU2 was cloned into pMAK705 (pMU3a), and a Kmr cassette was introduced into a single NsiI site (pMU3b-1). The Kmr marker was placed on the chromosome of SR-11 by a procedure described below (see “Strain construction”), thereby creating the zcg-101::Kmr allele on the chromosome.
For cloning of the curli operons, after the position of the fragment containing the curli genes and the Kmr marker was checked by Southern hybridization of a PstI-digested chromosomal DNA, the DNA fragments of respective size were isolated from a gel by the Freeze-Squeeze method (66) and ligated with PstI-cut pLAFR3. The partial cosmid library was packed (Gigapack III Gold; Stratagene) and amplified in E. coli XL1 Blue MR in liquid culture with kanamycin resistance as an additional selection marker. The cells were plated for individual colonies to be examined further, and subsequently one cosmid clone (pMU4) which complemented HLO7 [Δ(csgG-csgC)] was chosen for sequencing. Sequence identity between SR-11 and ATCC 14028-1s was confirmed for csgD and the intergenic region.
Strain construction.
Phage P22 HT105/1 int-201 (60) was used for transduction of LT2 strains and SR-11 according to the recommended protocol (13, 50). In order to detect lytically infected cells, LT2 strains were streaked on green plates. Lysogens were detected by streaking LT2 derivatives against phage H5. Since SR-11 does not support the propagation of P22, the purification procedures were skipped. DNA translocation into bacteria was also done by using competent cells (41), electrocompetent cells (9), and conjugation by triparental mating. A deletion in csgA was constructed as follows. Primers CSGBD (dACGAAAGCTTGCACTGCTGTGGGTTG [HindIII restriction site underlined]), CSGA1 (dCGTCTGCAGGATTGCTGCGAATGCTGC [PstI site underlined]), CSGA2 (dCGTCTGCAGTGGAACGCTAAAAACTC [PstI site underlined]), and CSGC (dCGAGGATCCGGCCATTGTTGTGATAAA [BamHI site underlined]) were used to create fragments PCR1 (CSGBD↔CSGA1) and PCR2 (CSGA2↔CSGC). After restriction digestion, PCR1 and PCR2 were directly cloned into pMAK700 cut with HindIII and BamHI, which resulted in plasmid pUMR2b. Cloning of the Kmr cassette of pUC4K into the single PstI site of pUMR2b resulted in pUMR2c-1. The plasmids were passaged through LB5010 and finally electroporated into S. typhimurium wild-type strains. After propagation of the strains at 28°C, the temperature was shifted to 44°C in order to integrate the pMAK derivative into the chromosome. For pUMR2c-1, for which a selectable marker was available, individual colonies were streaked on chloramphenicol and kanamycin plates in order to screen for a double crossover event. With pUMR2b, 10 individual integrants were selected and incubated at 28°C in order to resolve the plasmid again. Strains harboring a deletion were selected by their white color on CR plates. All constructs were checked by Southern hybridization and/or PCR. A deletion in ompR was created in the same way. The ompR primers OMPR1 (dTGGAAGCTTTTGTTTGAGTGTTTCGT [HindIII site underlined]), OMPR2 (dCTTAGATCTCTCTTGCATTGTCTGT [BglII site underlined]), OMPR3 (dCCTGAGATCTGTCTTTGTACCGGAC [BglII site underlined]), and OMPR4 (dGGCTCTAGAACTTCTACCTGAAACCAG [XbaI site underlined]) were selected on the basis of sequence X12374 (EMBL database [46]). After restriction digestion, PCR fragments PCR3 (OMPR1↔OMPR2) and PCR4 (OMPR3↔OMPR4) were cloned into XbaI/HindIII-digested pMAK705, yielding pUMR7a. Cloning of the ampicillin resistance gene from pWKS30 (BglII/BamHI digested) into the single BglII site resulted in pUMR7b-2.
Plasmid construction.
Individual genes from the curli operon were cloned into low-copy-number vector pWSK29 or pWSK30. csgA was amplified by using primers CSGB2 (dCGTCTGCAGTGCAGAAACAGTCGCA [PstI site underlined]) and CSGC (dCGAGGATCCGGCCATTGTTGTGATAAA [BamHI site underlined]), and for csgB primers CSGBD (HindIII) and CSGA1 (PstI) were used. The restriction sites at the primer ends were used to clone the fragments into pWKS30, yielding plasmids pCSGA and pCSGB, respectively. csgD was amplified with primers SP2 (dTTTCTCTTTCTGGATAATGGG) and SP5 (dTGTTTAACACGCATGACAGC), csgE was amplified with primers SP5 and SP9 (dCCTGACGATTATCCCTACC), and csgF was amplified with primers SP6 (dGATTGTTAACCGACCATACC) and SP17 (dGCAGGTAAGTGCGTCAAATC). The ends of the PCR fragments were treated with Klenow polymerase. Primer pair SP2-SP5 was digested with EcoRI, SP5-SP9 was digested with BstYI, and SP6-SP17 was digested with EcoRI. SP2-SP5 and SP5-SP9 were cloned into pWSK29 digested with the respective restriction enzymes, yielding pCSGD and pCSGE, respectively, and SP6-SP17 was cloned into pWKS30 in order to create pCSGF. The gene sequences for csgEFG were also amplified by using primers CSGD2 (dTGGATGCATACCCAGGCAGTTTCATGG [NsiI site underlined]) and SP21 (dGCTTTGTCGTATTCATCAGG) and cloned into pWSK29, yielding pCSGEFG.
RNA techniques.
Total RNA was prepared from 10 mg of S. typhimurium cells by the hot-phenol method. Cells were resuspended in 300 μl of 0.3 M sucrose–0.01 M sodium acetate (pH 4.5) and the same amount of 0.01 M sodium acetate (pH 4.5)–2% sodium dodecyl sulfate (SDS). After being mixed with an equal amount of hot acidic phenol, the cells were incubated at 65°C for 5 min. The extraction was repeated once with hot acidic phenol and twice with cold acidic phenol. After precipitation, the remaining DNA was digested with 10 U of RQ1 DNase (Promega) for 30 min in 0.05 M Tris (pH 7.5)–0.05 M NaCl–0.01 M MgCl2, and phenol-CHCl3 extraction was carried out twice. The concentration of the RNA dissolved in water was determined spectroscopically. A 10-μg sample of RNA was loaded on a 1.2% morpholine propanesulfonic acid (MOPS)-formaldehyde gel (4) which was run for 4 h at 4 V/cm with a 0.24- to 9.5-kb RNA ladder (Life Technologies) as a standard. After the gel was soaked in H2O twice (20 min each), the RNA was transferred to an Amersham Hybond-N membrane overnight by capillary blotting with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (4). Single-stranded probes complementary to the RNA template on the blot were constructed by an asymmetric PCR with primers CSGB2 and SP2 on symmetric PCR templates spanning the region of the csgA (primers CSGB2 and SP8 [dCTAAATTAATACTGGTTGA]) and csgD (primers SP2 and SP24 [dTAACTCTGCTGCTACAATCC]) genes, respectively, and labeled with the RadPrime Labelling System (Life Technologies) using 30 μCi of [α-32P]dCTP (3,000 Ci/nmol; Amersham). Hybridization (using less than 6 ng of probe per ml) and washing of blots were carried out according to standard procedures (4). The quality of transfer to the membrane was checked by probing with part of the 16S RNA sequence from plasmid pKK3535 (11) cut by HindIII. Signals were analyzed with a radioisotope imaging system (PhosphorImager 445SI; Molecular Dynamics) and quantified by integration over all bands detected by a single probe.
For primer extension, 10 μg of RNA and 2 pmol of primer PEXD1 (dTGACAGATGTTGCACTGCTG) were diluted in 9 μl of 1× avian myeloblastosis virus (AMV) reverse transcriptase buffer (Boehringer Mannheim). A 10-pmol amount of PEXD1 had been labeled with 30 μCi of [γ-32P]ATP (3,000 Ci/nmol; Amersham) by use of 10 U of polynucleotide kinase (Boehringer Mannheim). After incubations at 95°C for 5 min, 67°C for 3 min, and 50°C for 5 min, 5 U of AMV reverse transcriptase (Boehringer Mannheim) and 5 mM deoxynucleoside triphosphates (Pharmacia) were added, and primer extension was performed at 50°C for 1 h. The reaction mixture, containing 5 μl of formamide loading buffer, was heated at 95°C for 5 min and cooled on ice, and 6 μl was analyzed on a 7% denaturing polyacrylamide gel (4). A sequencing ladder generated with a T7 Sequencing kit (Pharmacia) using the PEXD1 primer and pUMR10-7 as template DNA was run as a standard.
Western blotting.
Bacteria were grown for 48 to 60 h on plates at 28°C and for 17 to 24 h at 37°C. In order to depolymerize the curli fiber into subunits, the cells have to be treated with strong acids (19). After resuspension of the cells in 100 μl of 99% formic acid and incubation for 10 min on ice, the liquid was removed by evaporation in a Speed Vac. The pellet was resuspended in 200 μl of SDS-polyacrylamide gel electrophoresis sample buffer (4), while 3 μl was loaded on a gel (15% separating gel with a 4% stacking gel, with a double concentration of buffer used in the separation gel and the running buffer). Proteins were transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore). The membranes were blocked and incubated with a 1:4,000 dilution of an anti-E. coli CsgA antiserum (34); a secondary goat antibody against rabbit immunoglobulin G conjugated with horseradish peroxidase was used for detection according to the protocol of the manufacturer (Boehringer Mannheim).
Electron microscopy.
Bacteria were grown on plates under the same conditions used for Western blotting. A concentrated bacterial suspension in water was allowed to adhere to a carbon-coated copper grid for 2 min. The liquid was removed by blotting, and staining was carried out for 30 s with 0.7% ammonium molybdate–150 μg of bacitracin per ml. The samples were examined with a Zeiss microscope at 80 kV.
Nucleotide sequence accession number.
The nucleotide sequence of the genes for curli biogenesis has been submitted to the EMBL data library under accession no. AJ002301.
RESULTS
Detection of curli expression in S. typhimurium SR-11 and ATCC 14028-1s on plates.
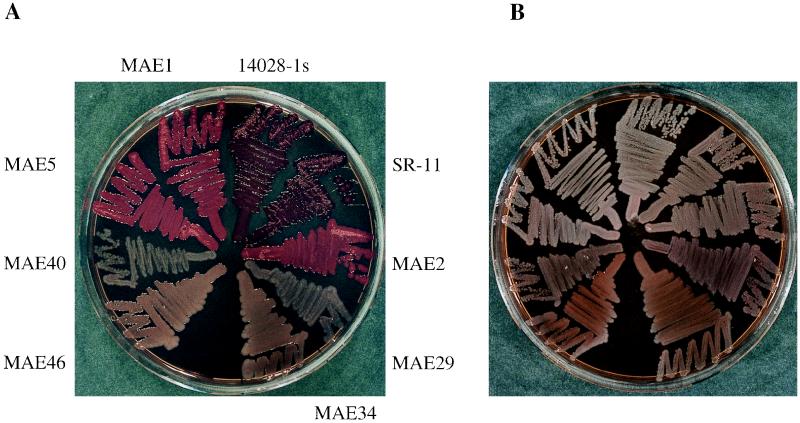
The mouse-virulent strains SR-11 and ATCC 14028-1s exhibited distinct colony morphologies when grown on CR plates at different temperatures; a white and smooth colony morphology was seen at 37°C, while a red, dry, and rough colony morphology was displayed at 28°C (Fig. 1). We called the two morphotypes saw37 and rdar28, respectively. Morphotypes similar to the rdar morphotype were previously described for E. coli MC4100 and YMel (34) and Salmonella enteritidis (18) and shown to be tightly linked to the expression of a thin aggregative fiber called curli fiber or SEF 17, respectively.
FIG. 1.
Colony morphology and color of S. typhimurium ATCC 14028-1s and SR-11 and their respective derivatives. (A) Cells grown at 28°C on LB medium plates without salt containing the dye CR. SR-11 and ATCC 14028-1s (here shown as the Nalr derivatives UMR3 and UMR1, respectively) developed a rough colony morphology with a dry surface and showed a deep red color by binding the dye CR, the rdar morphotype. The strains with deletions in csgA (MAE1, MAE2, and MAE5) were also rough but had a shinier surface. Binding of the dye CR led to a pinkish color of the colonies (the pdar morphotype). The strains with deletions in rpoS (MAE40 and MAE29) and ompR (MAE46 and MAE34) were white. (B) The same strains as in panel A but grown at 37°C. All cells were white and smooth, the saw morphotype.
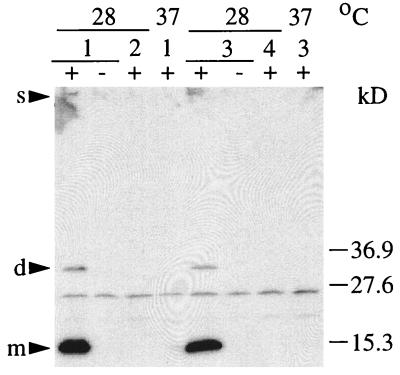
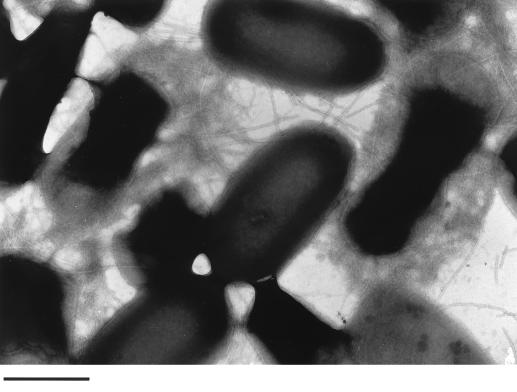
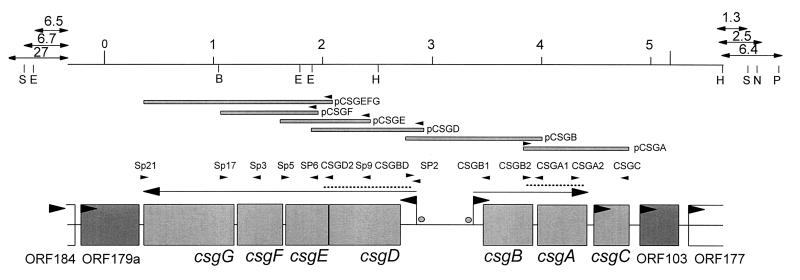
Both S. typhimurium strains were analyzed for expression of curli fibers by immunoblot analysis using a polyclonal antiserum against CsgA (34), detection of fibers on the surface of the cells by electron microscopy, and gene replacement of csgA, the fiber subunit gene, the sequence of which was taken from the one published for S. enteritidis (17). In accordance with the colony morphology, a signal for CsgA was detected only at 28°C in both strains (Fig. 2). By electron microscopy, an abundance of fibers was detected at 28°C (Fig. 3), while very few were occasionally seen at 37°C (data not shown). However, electron micrographs might not reflect the actual amount of curli fibers present on the cells at 28°C, since the cells clump together and only a fraction of them can be released from the tight extracellular matrix. Replacement of the curli subunit gene csgA with a Kmr cassette (MAE1 and MAE2) or an in-frame deletion of csgA (MAE5) abolished the rdar28 phenotype. Instead of rdar, mutant strains of SR-11 and ATCC 14028-1s, MAE2 and MAE1, respectively, and MAE5 displayed a pink colony which developed a delayed roughness, the pdar28 morphotype (Fig. 1). This phenotype seems to be more common in Salmonella spp., since it was also described for S. enteritidis 27655-3b after gene replacement of csgA (17), whereas E. coli MC4100 and YMel gave white colonies after knockout of the fiber subunit gene (34, 35). All these experiments showed that the curli fibers are expressed in SR-11 and ATCC 14028-1s in a temperature- and surface-dependent manner; therefore, the curli operon from SR-11 was cloned and characterized (see Materials and Methods). The organization of the two divergently transcribed csg operons, csgDEFG and csgBAC, is shown in Fig. 4.
FIG. 2.
Western blot analysis of fiber-derived CsgA from whole cells grown at 28 and 37°C. The cell pellets were immediately resuspended in SDS sample buffer (−) or treated with formic acid (+) as described in Materials and Methods. Minor signals for the fiber subunit which could vary in their intensities were regularly found in the slot (s) and in the gel corresponding to a dimer (d), while the major signal was consistent with the running behavior of a monomer (m). SR-11 and ATCC 14024-1s showed a signal for CsgA only at 28°C and not at 37°C. csgA knockouts did not show any signal at all. Lanes: 1, SR-11; 2, MAE2; 3, ATCC 14028-1s; 4, MAE1.
FIG. 3.
Electron micrograph of negatively stained ATCC 14028-1s cells grown at 28°C on LB agar without salt. The cells which are surrounded by a thick layer of curli fibers show a different cell morphology. Bar, 1 μm.
FIG. 4.
Organization of the csg region on the S. typhimurium SR-11 chromosome. A restriction map containing sites important for the cloning and localization of the csg region is shown. The positions of the genes required for curli biogenesis, csgDEFG and csgBAC (boxes), the position of the transcriptional start sites and the direction of polymerization (flags), and the position of the promoter (circle) are indicated. Experimentally confirmed RNA full-length transcripts (arrows above gene boxes) and open reading frames for which transcriptional analysis had not been carried out or for which no transcript had been detected so far (boxes with arrowheads inside) are also shown. The DNA fragments used as probes in RNA transcript analysis (dotted lines), primers used for the PCR amplification of DNA fragments (arrowheads above dotted lines), and subclones used to complement E. coli isolates with mutations in the csg genes (pCSGA, pCSGB, pCSGD, pCSGE, pCSGF, pCSGG, and pCSGEFG) (bars with small arrows indicating the transcription from the lacZ promoter) are indicated. B, BstYI; E, EcoRI; H, HindIII; N, NsiI; P, PstI; S, SacI.
Transcriptional analysis of the two curli operons.
Primer extension analysis of the csgD operon revealed the nucleotide G 174 bp upstream of the putative translation start of the csgD transcript as the transcription start site in S. typhimurium ATCC 14028-1s (Fig. 5). Transcription initiation of the E. coli csgD operon takes place 2 bp further upstream (34). A signal was obtained only when RNA isolated from cells grown on plates at 28°C, and not at 37°C, was used. Primer extension analysis of the csgBAC operon confirmed previous results (2, 17; also data not shown).
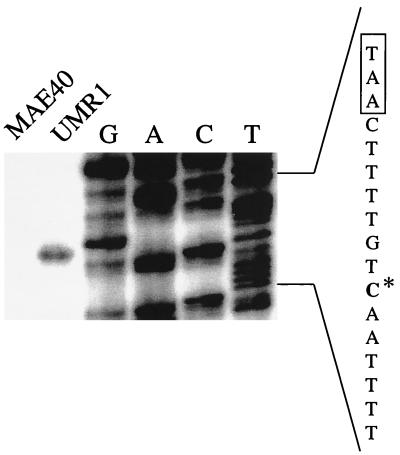
FIG. 5.
Primer extension analyses for the determination of the transcriptional start site of the csgDEFG operon. RNA was prepared from strains UMR1 and MAE40 (the rpoS derivative of ATCC 14028-1s) grown at 28°C on plates, and primer extension was carried out as described in Materials and Methods. An extension product is seen for UMR1 but not for MAE40. Primer PEXD1, located 58 bp downstream of the csgDEFG start codon, was used for the extension reaction as well as for the sequencing reaction on pUMR10-7 as a template. The sequence derived from the PEXD1 primer on pUMR10-7 is complementary to the RNA template, so the coding strand is automatically shown. The transcriptional start site (asterisk) and bases belonging to the putative promoter sequences (boxed) are indicated.
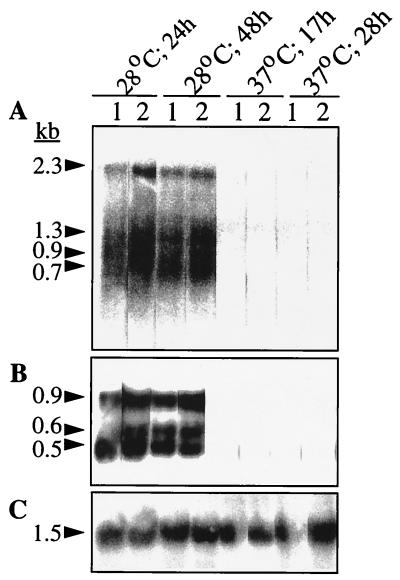
In order to determine the expression state of both csg operons, analysis of the steady-state levels of the RNA transcripts of csgD, the transcriptional regulator, and csgA, the fiber subunit gene, was carried out by Northern blot analysis (Fig. 6). By using a probe encompassing the whole csgD gene, four major bands of 2.3, 1.3, 0.9, and 0.7 kb which could correspond to the transcripts csgDEFG, csgDEF, csgDE, and csgD (with theoretical sizes of 2.5, 1.6, 1.2, and 0.8 kb, respectively) were detected; the csgA probe hybridized to three bands of 0.9, 0.6, and 0.5 kb, as in S. enteritidis (17). Signals of the same intensity were detected for ATCC 14028-1s and SR-11 after the cells were grown at 28°C for 24 or 48 h on plates but not for cells grown at 37°C for 17 or 28 h. Therefore, detection of csgD and csgA transcripts is concomitant with the expression of CsgA at 28°C, and transcripts are present long after entrance into the stationary phase.
FIG. 6.
Northern blot analysis of RNA transcripts of the csg region using S. typhimurium ATCC 14028-1s and SR-11 grown on LB medium plates without salt at 28 and 37°C for different periods. Probes covering the whole csgD and csgA genes were used; their locations are indicated in Fig. 4. Lanes: 1, UMR3; 2, UMR1. The sizes of the bands detected by the respective probes are shown on the left, calculated by using a 0.24- to 9.5-kb RNA ladder (Life Technologies) as a standard. (A) Hybridization with the csgD probe; (B) hybridization with the csgA probe; (C) control hybridization with 16S RNA as described in Materials and Methods.
Comparative sequence analysis of the curli region.
The csg genes are located at the same positions on the chromosomes of E. coli K-12 and S. typhimurium LT2 (26 centisomes [20]). In S. typhimurium, two divergently transcribed operons, csgBAC and csgDEFG, flank a 521-bp intergenic region (Fig. 4), a situation as in E. coli MC4100 (34). The nucleotide sequences of the two species showed an identity of 77.6%, a value which is below the average sequence conservation of 84.4% (61). The overall G+C contents in S. typhimurium and E. coli are similar; however, single genes show some variability in G+C content conservation (Table 3). The intergenic region between the transcriptional start sites has a low G+C content and is the least conserved (71%) (Table 3). However, the intergenic region has not homogeneously diverged but can be subdivided into four regions. Only four nucleotide substitutions occurred in the 60 bp upstream of the transcription start site of the csgD operon. The csgD promoter shares the characteristics of promoters transcribed by ςD both in S. typhimurium and E. coli, with the E. coli −35 box being closer to the consensus sequence. The region upstream of the csgD promoter has a very low G+C content (21.2%) which is identical in S. typhimurium and E. coli, and a conspicuous peak of curvature at position −147 (data not shown) was found by calculation with the DNase I-based parameters using the bend.it server (29). Sequence identity drops to 42.7% between 165 and 443 bp upstream of the transcriptional start site of csgD. The csgB promoter has a −10 box, which resembles more the proposed consensus sequence of ςS than that of ςD (38). Two alternative −35 boxes have been found which both suggest an unusually wide spacing of 19 or 21 bp between the −35 and −10 boxes. Some promoters whose transcription can be initiated in the absence of specific −35 hexamer contacts contain an upstream extension of the −10 element (10) by 5′-Tgn-3′, a sequence which was not found at the csgB promoter.
TABLE 3.
Properties of the csg genes and surrounding open reading frames
| Gene or region | % G+C content (S. typhimurium/ E. coli) | Sequence identity (%)a | Amino identity/ similarity (%)a | KS |
|---|---|---|---|---|
| ORF179ab | 52.2/54.1 | 75.7 | 86.6/92.5 | NDe |
| csg operon | 43.5/43.2 | 77.6 | ||
| csgG | 49.0/50.1 | 83.4 | 96.0/99.3 | 1.065 |
| csgF | 44.6/42.4 | 81.0 | 89.9/94.2 | 1.018 |
| csgE | 44.4/43.1 | 80.8 | 91.4/96.9 | 1.496 |
| csgD | 40.7/41.8 | 81.1 | 92.1/95.8 | 1.155 |
| Intergenic region | 33.7c/34.4 | 73.0c | ||
| 30.6d/32.2 | 70.6d | |||
| csgB | 43.4/41.8 | 82.9 | 82.1/90.7 | 0.564 |
| csgA | 51.5/50.1 | 73.2 | 74.8/86.1 | 1.021 |
| csgC | 45.0/42.6 | 71.9 | 73.1/87.0 | 1.263 |
| ORF103b | 47.7/43.6 | 45.2 | 39.4/57.6 | ND |
Between S. typhimurium SR-11 and E. coli MG1655.
Nomenclature and sequence from E. coli MG1655 and W3110; EMBL files with accession no. ecae205 and ecae206 and ecd740 to ecd742 were used.
Sequence between the translational start sites of csgD and csgB.
Sequence between the transcriptional start sites of csgD and csgB.
ND, not determined.
The putative proteins encoded by the genes of the two operons showed variability in sequence conservation. With the exception of CsgC, whose role has not been unambiguously determined, the proteins for the fiber subunit, CsgA, and the nucleator, CsgB, displayed the lowest amino acid identities (74.8 and 82.1%, respectively). These values lie at the lower end of gene conservation between S. typhimurium and E. coli, which ranges from 100 to 74.3% amino acid identity (61), but are surprisingly high when homologies among fimbriae, even of common origin within a species, are considered (45). In addition, the amino acid identities of CsgA, CsgB, and CsgC were 100% when the proteins of S. typhimurium SR-11 and S. enteritidis 27655-36 were compared (see reference 17 for further characterization of the genes).
The degree of conservation of the CsgD, CsgE, CsgF, and CsgG proteins between S. typhimurium and E. coli is high and lies between 89.5 and 96% amino acid identity (Table 3). The lipoprotein CsgG, which has a putative molecular mass of 30 kDa, was most conserved and showed only conservative amino acid exchanges. The helix-turn-helix DNA binding motif of CsgD, the putative transcriptional regulator of 25 kDa, is completely conserved between the two species. Classified according to the sequence similarity of the DNA binding motif, CsgD belongs to the LuxR family. The closest sequence homology is to regulators belonging to a two-component sensory transduction system, such as DegU of Bacillus subtilis and NarP of E. coli.
Complementation of E. coli csg mutants with S. typhimurium genes.
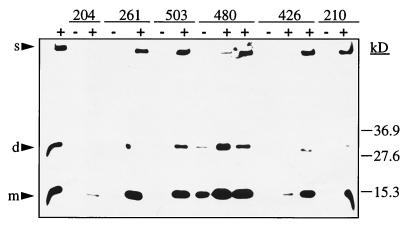
Considering the high homology of the Csg proteins between S. typhimurium and E. coli, functional complementation of gene products seemed likely. In order to test this hypothesis, PCR-generated gene fragments from S. typhimurium were cloned into the low-copy-number vectors pWSK29 and pWKS30 so that transcription occurred from the lacZ promoter (see Materials and Methods). All E. coli csg mutants were white and, with the exception of MHR480 (the csgE mutant), produced no curli fibers. MHR261 (csgB2) and MHR426 (csgF4) secrete CsgA in a soluble form (35, 36), but the soluble form of the protein was not detected in the whole-cell preparations used here (Fig. 7). Complementation was judged by the development of a red and rough colony morphology type and by Western blots detecting CsgA as an acid-sensitive polymer (Fig. 7 and Table 4). As seen in Table 4, all S. typhimurium genes complemented the respective E. coli csg mutants, although to different degrees. Single csgB, csgD, and csgE genes complemented the respective E. coli mutants to the wild-type phenotype. The csgA and csgF mutations could be only very poorly replaced by the copy on the plasmid. In the case of csgA, two signals of almost equal intensity were seen on an overexposed Western blot, one of which has a slightly higher molecular weight than the wild-type signal and is most likely a premature form of CsgA (data not shown). In addition, the very low signal intensity could be explained by a lower specificity of the anti-E. coli CsgA antiserum against CsgA from S. typhimurium. MHR426, the csgF mutant used in this study, has a polar effect on csgG expression, leading to decreased amounts of CsgG (36). The reduced amount of CsgG might limit the full complementation of MHR426 by pCSGF, as occurs with the respective E. coli gene (36). Neither the vector control nor the csgA, csgD, csgE, and csgF genes cloned in the direction which would allow transcription from the T7 promoter gave a change in the color of the colonies or their morphology with respect to the wild type or in signal intensity of CsgA on Western blots (data not shown). We conclude from the available data that interspecies complementation of the csg genes is possible in principle.
FIG. 7.
Western blot analysis of the complementation of the E. coli csg mutants. Results for whole-cell preparations treated with formic acid are shown. The mutant (−) and the mutant harboring the respective complementing plasmid (+) were run next to each other. Except for MHR480 (ΔcsgE3), none of the E. coli mutants showed a CsgA signal derived from polymerized fibers. MHR261 (csgB2) and MHR503 (csgD6) were complemented to wild-type levels by using plasmids with the single genes. MHR426 (csgF4) was complemented to wild type only when a plasmid carrying csgEFG was used. Faint signals of CsgA were detected when pCSGA was introduced into MHR204 (csgA::Tn105).
TABLE 4.
Complementation of E. coli csg mutants with S. typhimurium genes
| Mutation in E. coli | E. coli strain | Complement- ing plasmid | Colony morphology | Detection of CsgA derived from fibers |
|---|---|---|---|---|
| csgA2::Tn105 | MHR204 | pCSGA | Brown but not rough | Very weak signal |
| csgB2 | MHR261 | pCSGB | wta | wt levels |
| csgD6 | MHR503 | pCSGD | wt | wt levels |
| ΔcsgE3 | MHR480 | pCSGE | wt | wt levels |
| pCSGEFG | wt | wt levels | ||
| csgF4 | MHR426 | pCSGF | Reddish but not rough | Weak signal |
| pCSGEFG | wt | wt levels | ||
| csgG1::Tn105 | MHR210 | pCSGEFG | wt | wt levels |
wt, wild type.
Analysis of the effect of rpoS and ompR mutations on colony morphology and transcription from the csg promoters.
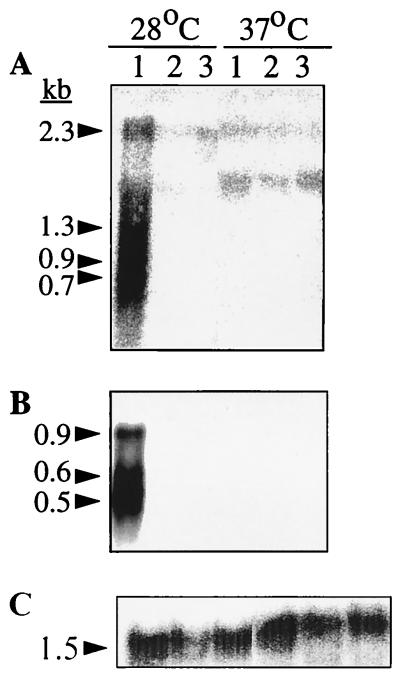
For E. coli, it was demonstrated that transcription of the csgD and csgBA operon requires ςS. Transduction of the mutant rpoS allele from SF1005 into ATCC 14028-1s and SR-11 gave white and smooth colonies, a saw28 morphotype (Fig. 1). Transcriptional analysis with the csgD and csgA probes on RNA extracted from the rpoS mutant of ATCC 14028-1s (MAE40) grown at 28 and 37°C detected no signal for either probe (Fig. 8). In addition, no extension product was seen in the latter strain by primer extension analysis (Fig. 5). Therefore, rpoS is also required for transcription of the csg operons in S. typhimurium strains.
FIG. 8.
Northern blot analysis of RNA transcripts from S. typhimurium ATCC 14028-1s and rpoS and ompR derivatives grown on LB medium plates without salt at 28 and 37°C. Probes covering the whole csgD and csgA genes were used; their locations are indicated in Fig. 4. Lanes: 1, UMR1; 2, MAE40 (rpoS); 3, MAE46 (ompR). The sizes of the bands detected by the respective probes are shown on the left, calculated by using a 0.24- to 9.5-kb RNA ladder (Life Technologies) as a standard. (A) Hybridization with the csgD probe; (B) hybridization with the csgA probe; (C) control hybridization with 16S RNA as described in Materials and Methods.
It is also known that ompR is necessary for transcription from both the csgBA and the csgD promoters in E. coli (40). An ompR mutant was constructed by introducing a deleted ompR gene carrying an Ampr cassette instead of the major part of its open reading frame into the chromosome of ATCC 14028-1s and SR-11 by double crossover, yielding strains MAE46 and MAE34, respectively (Table 1; see Materials and Methods). The ompR mutants were white at 28 and 37°C (Fig. 1). No CsgA signal was detected for MAE46 in Western blots (data not shown). Northern blot analysis of RNA extracted from strain MAE46 grown at both temperatures gave no signal for csgD or csgA (Fig. 8). Since ompR was shown to be necessary for the transcription of the csgD and csgBA promoters, the intergenic region was examined for putative ompR binding sites. One nucleotide sequence which has one mismatch base pair to the recently proposed consensus sequence for independent binding was found (39). This sequence is centered at position −50.5 relative to the transcriptional start site of the csgD promoter. If this sequence is used for OmpR binding, it can be imagined that transcriptional regulation takes place mainly at the csgD promoter. CsgD may then act upon the csgBA promoter to initiate transcription there.
DISCUSSION
Although many fimbrial gene clusters have been isolated from Salmonella spp. and E. coli, variation in operon structure and genes encoding regulatory control features of pili from the same structural class suggests frequent remodeling of DNA sequences due to the necessity to respond flexibly to changing environmental conditions and various host environments (49). In contrast to this behavior of fimbrial operons, the operons for curli biogenesis embedded in the same context on the chromosome (reference 20 and this study) are remarkably conserved between E. coli and S. typhimurium. In addition, the sequence similarities at the amino acid level of all proteins in the operon, ranging from the fiber subunit CsgA (86%) to the transcriptional regulator CsgD (96%) and the lipoprotein CsgG (99%), are higher than the homologies for most functionally and sequentially related fimbrial gene products, even within a species (49, 51, 56). It is therefore suggested that the thin aggregative fibers in Salmonella be named curli fibers, as for the products of the homologous E. coli genes (csg) (59).
The sequence similarity at the amino acid level is reflected by the successful complementation of individual csg gene mutants of E. coli by S. typhimurium genes. The S. typhimurium csgB gene could complement the homologous E. coli gene to wild-type levels, showing that the four-repeat structure consisting of 22-amino-acid-long putative β-strand–turn–β-strand–turn units (35) which are all required for organelle assembly (8) functions between the species. It can be imagined that interspecies exchange of organelle subunits can occur in a natural environment where bacteria are tightly packed, such as the gut flora. The consensus sequence for the repeat structure of CsgB was determined to be NLA-I-Q-GS-N-A-I-Q-G--, where the underlined amino acids show 100% conservation between S. typhimurium and E. coli. The consensus motif for CsgA, which has five 23-amino-acid-long units, is NS--T-TQYG-GN-AT-DQTAA-. There may be several reasons for the lack of complementation to a full wild type for some genes, such as functional restriction of the proteins, imbalance of protein ratios, or instability of the RNA message created from the plasmid, which remain to be elucidated. Alternatively, polar effects of the csg mutations on downstream genes may prevent the full complementation to the wild-type phenotype.
The high degree of conservation at the protein level contradicts the low conservation at the nucleotide level. The nucleotide sequences encoding csgDEFG diverged more than average, and so does KS (0.94 on the average), a value for the estimation of the number of synonymous substitutions per site (Table 3). csgE has a KS value of 1.5 and is therefore almost saturated with substitutions. The high rate of nucleotide substitutions could reflect the chromosomal location of the csg operons proximal to the terminus of replication, where more nucleotide changes take place, and/or the low expression state of the genes from the csgDEFG operon (61). The high conservation of proteins could indicate a lack of selective pressures, such as the immune response in a host, on the fibers or functional constraints on the macromolecules and their specificities, which do not tolerate an evolution which develops too far from a common ancestor (45).
The two csg operons are flanked by the same open reading frames as in the fully sequenced E. coli MG1655 strain (EMBL accession no. ecae205, ecae206, and ecd740 to ecd742). The homologous ORF179a showed the same ambivalent conservation scheme as the curli genes, low conservation on the nucleotide level and high conservation of the amino acid sequence (Table 3); the intergenic region between csgG and ORF179a showed no homology. Downstream of csgC, the nucleotide homology already starts to decline at the end of csgC (the stop codon has changed with respect to E. coli) to a level of 60%, which continues within ORF103.
Another fiber, type 1 fimbria, which seems to be present in all Salmonella species, shares morphological and adhesive properties with E. coli type 1 fimbria. However, the different location on the chromosome in the two species and the high degree of sequence diversity and gene rearrangements (14), together with different regulation schemes having no overlapping regulating factors, suggest that these genes were acquired independently by S. typhimurium and E. coli and subject to evolutionary pressures (45). Therefore, curli genes are the only fimbrial genes detected which were already present in a common ancestor of E. coli and Salmonella spp.
The regulation of the curli genes in S. typhimurium SR-11 and ATCC 14028-1s by temperature, rpoS, and ompR is identical to that in E. coli MC4100 and YMel (2, 3, 34, 40, 53), implying that fiber expression responds to the same environmental cues in general. The complementation to wild-type levels of the E. coli csgD gene by the respective S. typhimurium gene shows that the recognition sites for the transcriptional activator are also conserved between the species. The correspondence in regulation pattern in addition to gene conservation points to a similar or identical function of the curli fibers in the two species in the same ecological niche. The expression pattern on plates at low temperature and low osmolarity suggests a role for curli fibers primarily outside a host, on surfaces. A participation of curli fibers in a bacterial network such as a biofilm (21) is possible, considering the adhesive nature of the fibers and the colony morphology of cells. However, more environmental studies need to be performed before a firm role of curli fibers in biofilm formation can be concluded.
ACKNOWLEDGMENTS
We thank the following people for providing plasmids, strains and antibodies: D. G. Guiney for SF1005, S. R. Kushner for pWSK29 and pWKS30, A. Lazdunski for pLAFR3, H. Loferer for HLO7, E. Morfeldt and S. Arvidson for anti-OmpA antiserum, and M. Rhen for 146, LB5010, and pCurli. We are grateful to H. Loferer for helpful technical advice.
U.R. was the recipient of a fellowship from the program “Infektionsbiologie” from the German Bundesministerium für Forschung und Technologie (BMFT). This work was supported by grants from the Swedish Medical Research Council (B96-16X-10843-03A) and the Swedish Natural Science Research Council and by an unrestricted grant for infectious disease research from the Bristol-Myers Squibb Company to S. Normark.
REFERENCES
- 1.Applied Biosystems, Inc. User bulletin 18. Foster City, Calif: Applied Biosystems, Inc.; 1991. [Google Scholar]
- 2.Arnqvist A, Olsén A, Normark S. ςS-dependent growth-phase induction of the csgBA promoter can be achieved in vivo by ς70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 3.Arnqvist A, Olsén A, Pfeifer J, Russell D G, Normark S. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol. 1992;6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 5.Bäumler A J, Gilde A J, Tsolis R M, van der Velden A W M, Ahmer B M M, Heffron F. Contribution of horizontal gene transfer and deletion events to development of distinctive patterns of fimbrial operons during evolution of Salmonella serotypes. J Bacteriol. 1997;179:317–322. doi: 10.1128/jb.179.2.317-322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bäumler A J, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben Nasr A, Olsen A, Sjöbring U, Müller-Esterl W, Björck L. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol Microbiol. 1996;20:927–935. doi: 10.1111/j.1365-2958.1996.tb02534.x. [DOI] [PubMed] [Google Scholar]
- 8.Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bio-Rad Laboratories. Gene pulser electroprotocol. Hercules, Calif: Bio-Rad Laboratories; 1993. [Google Scholar]
- 10.Bown J, Belyaeva T, Busby S, Minchin S. Extended −10 promoters. Nucleic Acids Mol Biol. 1997;11:41–52. [Google Scholar]
- 11.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 12.Bullas L R, Ryu J-I. Salmonella typhimurium LT2 strains which are r−m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 14.Clegg S, Swenson D L. Salmonella fimbriae. In: Klemm P, editor. Fimbriae, adhesion, genetics, biogenesis and vaccine. Boca Raton, Fla: CRC Press; 1994. pp. 105–113. [Google Scholar]
- 15.Clouthier S C, Collinson S K, Kay W W. Unique fimbriae-like structures encoded by sefD on the SEF14 fimbrial gene cluster of Salmonella enteritidis. Mol Microbiol. 1994;12:893–903. doi: 10.1111/j.1365-2958.1994.tb01077.x. [DOI] [PubMed] [Google Scholar]
- 16.Clouthier S C, Müller K-H, Doran J L, Collinson S K, Kay W W. Characterization of three fimbrial genes, sefABC, of Salmonella enteritidis. J Bacteriol. 1993;175:2523–2533. doi: 10.1128/jb.175.9.2523-2533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collinson S K, Clouthier S C, Doran J L, Banser P A, Kay W W. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collinson S K, Doig P C, Doran J L, Clouthier S, Trust T J, Kay W W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collinson S K, Emödy L, Müller K-H, Trust T J, Kay W W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collinson S K, Liu S-L, Clouthier S C, Banser P A, Doran J L, Sanderson K E, Kay W W. The location of four fimbrin-encoding genes, agfA, fimA, sefA and sefD, on the Salmonella enteritidis and/or S. typhimurium XbaI-BlnI genomic restriction maps. Gene. 1996;169:75–80. doi: 10.1016/0378-1119(95)00763-6. [DOI] [PubMed] [Google Scholar]
- 21.Costerton J W, Cheng K-J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 22.Coynault C, Robbe-Saule V, Norel F. Virulence and vaccine potential of Salmonella typhimurium mutants deficient in the expression of the RpoS (ςS) regulon. Mol Microbiol. 1996;22:149–160. doi: 10.1111/j.1365-2958.1996.tb02664.x. [DOI] [PubMed] [Google Scholar]
- 23.Doran J L, Collinson S K, Burian J, Sarlos G, Todd E C D, Munro C K, Kay C M, Banser P A, Peterkin P I, Kay W W. DNA-based diagnostic tests for Salmonella species targeting agfA, the structural gene for thin, aggregative fimbriae. J Clin Microbiol. 1993;31:2263–2273. doi: 10.1128/jcm.31.9.2263-2273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorman C J, Chatfield S, Higgins C F, Hayward C, Dougan G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect Immun. 1989;57:2136–2140. doi: 10.1128/iai.57.7.2136-2140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenstark A, Calcutt J M, Backer-Hapak M, Ivanova A. Role of Escherichia coli rpoS and associated genes in defense against oxidative damage. Free Radical Biol Med. 1996;21:975–993. doi: 10.1016/s0891-5849(96)00154-2. [DOI] [PubMed] [Google Scholar]
- 26.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative ς factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 29.Gabrielian A, Simoncsits A, Pongor S. Distribution of bending propensity in DNA sequences. FEBS Lett. 1996;393:124–130. doi: 10.1016/0014-5793(96)00837-x. [DOI] [PubMed] [Google Scholar]
- 30.Gibson M M, Ellis E M, Graeme-Cook K A, Higgins C F. OmpR and EnvZ are pleiotropic regulatory proteins: positive regulation of the tripeptide permease (tppB) of Salmonella typhimurium. Mol Gen Genet. 1987;207:120–129. doi: 10.1007/BF00331499. [DOI] [PubMed] [Google Scholar]
- 31.Giron J A, Livine M M, Kaper J B. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol Microbiol. 1994;12:71–82. doi: 10.1111/j.1365-2958.1994.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 32.Grund S, Weber A. A new type of fimbriae on Salmonella typhimurium. J Vet Med B. 1988;35:779–782. doi: 10.1111/j.1439-0450.1988.tb00560.x. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 35.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammar, M., H. Loferer, A. Olsén, and S. Normark. Nucleation dependent extracellular formation of curli; requirement of the CsgF protein. Submitted for publication.
- 37.Hammar, M., A. Olsén, H. Loferer, and S. Normark. The binding property of adhesive curli organelles in Escherichia coli requires nagA, an N-acetylglucosamine-6-phosphate deacetylase, and CsgE. Submitted for publication.
- 38.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 39.Huang K-J, Igo M M. Identification of the bases in the ompF regulatory region which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 40.Hultgren S J, Hal Jones C, Normark S. Bacterial adhesions and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 2730–2756. [Google Scholar]
- 41.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 42.Jishage M, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of ς70 and ς38. J Bacteriol. 1995;177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knapp S, Hacker J, Jarchau T, Goebel W. Large, unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli O6 strain 536. J Bacteriol. 1994;168:22–30. doi: 10.1128/jb.168.1.22-30.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogfelt K A. Bacterial adhesion: genetics, biogenesis, and role in pathogenesis of fimbrial adhesions of Escherichia coli. Rev Infect Dis. 1991;13:721–735. doi: 10.1093/clinids/13.4.721. [DOI] [PubMed] [Google Scholar]
- 45.Kusters J G, Gaastra W. Fimbrial operons and evolution. In: Klemm P, editor. Fimbriae, adhesion, genetics, biogenesis and vaccine. Boca Raton, Fla: CRC Press; 1994. pp. 179–196. [Google Scholar]
- 46.Liljeström P, Laamanen I, Palva E T. Structure and expression of the ompB operon, the regulatory locus for the outer membrane porin regulon in Salmonella typhimurium LT2. J Mol Biol. 1988;201:663–673. doi: 10.1016/0022-2836(88)90465-2. [DOI] [PubMed] [Google Scholar]
- 47.Lindgren S W, Stojikovic I, Heffron F. Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:4197–4201. doi: 10.1073/pnas.93.9.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Loferer, H. Unpublished data.
- 48.Loferer H, Hammar M, Normark S. Availability of the fiber subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 49.Low D, Braaten B, van der Woude M. Fimbriae. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 146–157. [Google Scholar]
- 50.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 51.Marc D, Dho-Moulin M. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J Med Microbiol. 1996;44:444–452. doi: 10.1099/00222615-44-6-444. [DOI] [PubMed] [Google Scholar]
- 52.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 54.Olsén A, Jansson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 55.Pratt L A, Silhavy T J. Porin regulon of Escherichia coli. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 105–127. [Google Scholar]
- 56.Purcell B K, Pruckler J, Clegg S. Nucleotide sequences of the genes encoding type 1 fimbrial subunits of Klebsiella pneumoniae and Salmonella typhimurium. J Bacteriol. 1987;169:5831–5834. doi: 10.1128/jb.169.12.5831-5834.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Römling, U. Unpublished data.
- 57.Römling U, Heuer T, Tümmler B. Bacterial genome analysis by pulsed field gel electrophoresis techniques. In: Chrambach A, Dunn M J, Radola B J, editors. Advances in electrophoresis. Weinheim, Germany: VCH; 1994. pp. 355–406. [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sanderson K E, Hessel A, Liu S L, Rudd K E. The genetic map of Salmonella typhimurium, edition VIII. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1903–1999. [Google Scholar]
- 60.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 61.Sharp P M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 62.Sjöbring U, Pohl G, Olsen A. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA) Mol Microbiol. 1994;14:443–452. doi: 10.1111/j.1365-2958.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 63.Spangenberg C, Fislage R, Römling U, Tümmler B. Disrespectful type IV pilins. Mol Microbiol. 1997;25:203–204. [PubMed] [Google Scholar]
- 64.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swords W E, Cannon B M, Benjamin W H., Jr Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun. 1997;65:2451–2453. doi: 10.1128/iai.65.6.2451-2453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tautz D, Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983;132:14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- 67.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 68.Wilmes-Riesenberg M R, Foster J W, Curtiss R., III An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]