Summary
Quantifying cellular characteristics from a large heterogeneous population is essential to identify rare, disease-driving cells. A recent development in the combination of high-throughput screening microscopy with single-cell profiling provides an unprecedented opportunity to decipher disease-driving phenotypes. Accurately and instantly processing large amounts of image data, however, remains a technical challenge when an analysis output is required minutes after data acquisition. Here, we present fast and accurate real-time cell tracking (FACT). FACT can segment ∼20,000 cells in an average of 2.5 s (1.9–93.5 times faster than the state of the art). It can export quantifiable features minutes after data acquisition (independent of the number of acquired image frames) with an average of 90%–96% precision. We apply FACT to identify directionally migrating glioblastoma cells with 96% precision and irregular cell lineages from a 24 h movie with an average F1 score of 0.91.
Keywords: machine-learning-based cell segmentation, real-time cell tracking, cell tracking correction, high-throughput imaging, live-cell imaging, lineage tracking
Graphical abstract

Highlights
-
•
FACT combines GPU-based GTWeka segmentation and real-time cell-to-cell linking
-
•
FACT exports quantifiable features within minutes after image acquisition
-
•
Automatic cell track correction of FACT improves tracking accuracy
-
•
FACT and accurately identifies rare and abnormal cell subpopulations
Motivation
The evolution of advanced volumetric fluorescence and high-throughput screening microscopy techniques has ushered in a new era of data-intensive cellular imaging. These advancements have triggered the development of computational algorithms for extracting biological insights from multidimensional image datasets. Various computational methods have been proposed to deal with object segmenting/tracking problems emerging from high-throughput data (>20,000 objects per field of view). However, challenges remain in capturing intricate cellular behaviors from densely populated cultures: cells that are prone to overlap make high-throughput cell segmentation difficult, resulting in a low cell tracking accuracy; large datasets are hard to process in real time, meaning existing computational pipelines can typically not provide instant (<10 min) tracking outcomes right after image acquisition. To bridge the technology gap, we present the FACT (fast and accurate real-time cell tracking) algorithm, which combines GPU-based, ground-truth-assisted trainable Weka segmentation (GTWeka) and real-time Gaussian-mixture-model-based cell linking.
Chou et al. develop a real-time cell segmentation and tracking algorithm, FACT, to enable instant processing of large-scale image data within minutes after image acquisition. The authors apply it to identify rare and abnormal cell subpopulations from longitudinal time-lapse movies with tens of thousands of cells/image frames.
Introduction
The advent of modern, large-scale volumetric fluorescence microscopy and high-throughput screening microscopy have driven rapid development in computational methods.1,2 Several advanced, automated computational methods have been recently introduced to accurately and rapidly process the big multidimensional image data generated from these new microscopes1,2,3,4 and to extract meaningfully biological information: cell migration, division, morphological changes, differentiation, or cell-cell interactions.1,2,3,5,6
Prior to these newly developed computational methods, terabytes of image data have required months of processing instead of a number of days.7,8,9 Real-time accurate cell-shape extractor (RACE),1 for example, can segment cells from 3,836 time points within 1.4 days compared to 100–700 days used by other state-of-the-art cell segmentation methods.7,8,9 To accurately and rapidly process cellular dynamics and lineages, the tracking Gaussian mixture model (TGMM) cell tracking algorithm,2 for another example, can process 26,000 cells/min (including cell segmentation + cell linking) compared to other state-of-the-art cell tracking algorithms, which can only process a number of thousands of cells/min.10,11
Inspired by TGMM, our recently developed cell tracking algorithm, mTGMM (modified TGMM),3 can process ∼42,000 cells/min. mTGMM has been successfully applied to process and extract quantifiable characteristics and phenotypic information of individual cells (i.e., cells with fast migration or spindle-shaped morphology)3 imaged through our high-throughput screening microscope, the ultrawide field-of-view optical (UFO) microscope.3 mTGMM can efficiently process UFO-acquired big image data, for instance, processing a 2 h time-lapse movie (less than 30 frames) with 20,000 cells/frame within 10 min. This processing time is sufficient for subsequent selective isolation of cells of interest from the imaging dish (for cells migrate 1 m/min), as those target cells remain close to the coordinates of the last acquired image frame. The capability to rapidly process and isolate subpopulations of cells from a large pool of heterogeneous cells opens an unprecedented possibility to dissect molecular mechanisms of phenotypes of interest, as the isolated cells with desired phenotypes can be directly subjected to downstream (single cell) sequencing and profiling. The profiled genomes, transcriptomes, or proteomes can be straightforwardly linked to their corresponding cellular phenotypes, namely genotype-to-phenotype linking.3,12,13,14
Despite the remarkable advances in fast cell tracking methods (cell segmentation + cell linking), these methods decrease tracking accuracy when cells have complicated cellular behaviors (like cells migrating across each other) or when cells are densely populated ( ∼100,000 cells/cm2); such a density often occurs when tracking cells or studying cell lineages over days or is required when searching for rare biological events (0%–5%). Most importantly, the processing speed of these new computational methods cannot satisfy the demand of instantly processing hundreds or thousands of image frames within a number of minutes, which is critical when isolating cells of interest from the imaging dish is required. These constraints stop us from directly profiling cells displaying, among others, abnormal, longitudinal migratory behaviors, anomalous cell lineages (both require longitudinal imaging), or irregular cell divisions (rare biological events), which are important in cancer research and treatment development. Deciphering the underlying mechanisms of these abnormal phenotypes is important, as they are linearly correlated with poor prognosis.15,16,17
To have superior cell linking and tracking performance, high accuracy in cell segmentation is extremely critical. Deep-neuronal-network-based cell segmentation have been demonstrated to address this challenge,18 but the amount of labeled data required for training to reach high performance often limits the widespread applications of these methods; the amount of training data is intensified when applying to different types of cells or samples as new training is often required.
To address the abovementioned challenges, here, we present FACT (fast and accurate real-time cell tracking algorithm). FACT is a real-time, instant cell segmentation and cell linking algorithm combining GPU-based, machine-learning-assisted cell segmentation (Figure 1) and a GMM-based cell tracking algorithm. FACT also incorporates automatic cell track correction to accurately and instantly process big image data (i.e., 20,000–30,000 cells/frame for more than hundreds of image frames) and can export quantitative characteristics of individual cells minutes after data acquisition. The algorithm can easily be adapted to segment different cell types with 30–60 min of human annotation to achieve peak performance for unseen sample types and has higher segmentation and tracking performance than the state of the art.
Figure 1.
Ground-truth-assisted trainable Weka segmentation (GTWeka) pipeline
The original TWS pipeline is highlighted in blue.
(A) Image features, generated by applying all default image filters on an input image.
(B) Data annotation (or labeling). Manual assignment of pixels to three classes: nucleus (blue), background (green), and edge (red).
(C) Training data, a combination of annotated pixels and their corresponding values from image features.
(D) Random forest classifier, which is trained with the input training data.
(E) We then use the well-trained model to perform the semantic segmentation. Connected component analysis is applied to generate the instance segmentation from the semantic segmentation. If needed, users can improve the segmentation performance by adding more annotations and re-training the classifier (steps A–D) until no further improvement is observed.
The processes from (A)–(E) take ∼30–60 min. Our GTWeka (highlighted in red) improves the segmentation speed via GPU-based operation and key feature selection and increases the segmentation accuracy by incorporating ground-truth reference data (F), which only requires 30–60 min of preparation. The reference data are used to optimize the random forest classifier. Reference data are generated from a number of cropped regions of the original input image and labeled as individual nuclei and background.
Results
Highly accurate cell segmentation with sparse annotation
As mentioned, deep-learning-based models18 (i.e., U-NET,19 mask R-CNN,20 StarDist,21 CellSeg,22 or Cellpose23) have been shown to perform well when used to tackle difficult problems in cellular image analysis and image segmentation.18 However, creating or (re-)training models that are customized for each sample type or experiment is often required to reach high performance, resulting in hours to days of data annotation, ground-truth preparation, and model training. Low-annotation machine learning methods, such as trainable Weka segmentation (TWS)24 or ilastik,25 on the other hand, can finish data annotation and training for each sample type within 30–60 min and therefore are easier to implement when model creation or (re-)training is required. Despite the great potential and promising results of TWS or ilastik, a better cell segmentation performance (i.e., 0.95 F1 score at an intersection over union [IoU] threshold of 0.5) is required to have higher accuracy in cell tracking and lineage construction. The necessary performance improvement seems difficult to reach using TWS, ilastik, or even deep learning methods18 (see below). In addition, a fast processing time is often required when screening and identifying rare biological events from a large quantity of cells, especially when immediate isolation of those rare cells is needed.
To improve segmentation performance and processing time, here, we introduce a GPU-based, ground-truth-assisted TWS method called GTWeka (Figure 1). GTWeka improves cell segmentation accuracy (0.95 F1 score at an IoU threshold of 0.5; Figure S1A) by optimizing random forest classification using sparse ground-truth reference data, which consist of foreground and background labeled images and which can be prepared within 30–60 min (Figures 1, S2, and S3; STAR Methods). The ground-truth data act as a reference during the process of selecting the best random forest parameter option (Figure S2B), i.e., the depth of the tree and the number of leaves, etc., by checking the segmentation performance (i.e., F1 score) (Figure 1; see Figure S2B for detail). The minimal effort needed for ground-truth data preparation makes the method easily adaptable to other sample types without compromising its performance. Furthermore, GTWeka improves the processing time by (1) computing the segmentation via GPU instead of CPU (STAR Methods; Figure 1) and by (2) selecting the most relevant image features for classification (instead of running all 57 image features)24,25 (Figures S1A and S2C). Image features are the images processed with a number of different filters (Gaussian filer, Sobel filter, difference of Gaussians filter, Hessian matrix, membrane projections, and median filter). They hold major information per image, e.g., a Sobel filter can find pixels that represent the edge of a nucleus while suppressing the edge-irrelevant pixels. The ground-truth reference data, used in the selection of the best random forest model, is also applied to the selection of the most relevant image features (see Figure S2C for details). By default, TWS generates 57 image features (all features) and uses them to construct a random forest model. With our GTWeka, we observed that a random forest model constructed using a subset of image features (selected features), selected by the forward selection method26 (see STAR Methods for details), achieves similar performance to one constructed using all features (Figure S1A) but is ∼8 times faster (Figure S1B). With this processing speed (2.5 s/frame), our GTWeka method makes it possible to screen, segment, and identify rare cell subpopulations from a population of close to a million cells in a matter of minutes. High-speed identifications of cellular subtypes in populations of this size are, for instance, essential for genotype-to-phenotype linking experiments,3,13 where 100 cells per subtype need to be identified and analyses of smaller populations are not sufficient. We can, for example, identify extremely rare tripolar dividing cells at a rate sufficient for follow-up sequencing: we have applied GTWeka to identify 50 tripolar division events from a population of ∼0.8 million MCF10A epithelial cells in ∼5 min (∼100 s of cell segmentation + ∼200 s of tripolar division detection) (Figure S4).
Ilastik,25 arguably the most commonly used, state-of-the-art Weka-based cell segmentation method,24 also allows users to select key features instead of using its all-feature method (32 image features are used in ilastik; STAR Methods). ilastik offers three different types of key feature selection methods, namely filter method, Gini importance, and wrapper method (Figure S1A); none of these feature selection methods outperform either our GTWeka feature selection method or ilastik’s all-feature method (Figure S1A).
To properly assess the performance, three different types of cell images that are often seen in biological data were further used in benchmarking the performance of GTWeka against other segmentation methods (Weka-based and deep-learning-based methods): (1) low signal-to-noise ratio images from densely packed cells (here, we chose MCF10A epithelial cells); (2) smaller nuclei size (here, we chose glioblastoma multiforme [GBM] cells); and (3) large nuclei size and decent signal-to-noise ratio images (here, we chose HeLa cancer cells). The indicated performance mentioned below is calculated from the average of these three cell image types.
When comparing our GTWeka method with state-of-the-art Weka-based methods (TWS and ilastik), GTWeka outperforms these methods, both in segmentation performance (average F1 score at an IoU of 0.5 or 0.7) and computational time (32.8 times faster than TWS and 2 times faster than ilastik; STAR Methods; Figure 2). Please note that the segmentation performance was evaluated based on the comparison of ground-truth data (unseen images) and the segmented results (Figure 2). The high performance on segmenting unseen data indicates that our model was not overfitted.
Figure 2.
Benchmarking of cell segmentation methods (GTWeka, TWS, ilastik, StarDist, CellSeg, and Cellpose)
(A) Cell segmentation using different Weka-based segmentation methods with different cell image datasets (MCF10A, GBM, and HeLa). Images from top to bottom: raw image, images processed by GTWeka_all features, GTWeka_selected features (3 features), TWS, ilastik_all features, and ilastik_selected features (3 features by the ilastik_filter method).
(B) Comparison of performance (average F1 score at IoU = 0.5 or 0.7) and processing time (s) of all the methods mentioned in (A). Fast processing, as much as possible to the degree of seconds, is crucial for applications of identifying rare cells out of a large population; see Figure S4.
(C) Cell segmentation using GTWeka and other deep-learning-based methods (StarDist, CellSeg, and Cellpose) with different cell image datasets (MCF10A, GBM, and HeLa). Images from top to bottom: raw image, images processed by GTWeka_selected features (3 features), StarDist, CellSeg, and Cellpose.
(D) Comparison of performance (average F1 score at IoU = 0.5 or 0.7) and processing time (s) of all the methods mentioned in (C).
As deep-learning-based segmentation methods have been shown to yield high performance in cell/nuclei segmentation, we then also compare our GTWeka method with state-of-the-art deep-learning-based segmentation methods, StarDist,21 CellSeg,22 and Cellpose.23 The average F1 scores of GTWeka_selected features, StarDist, CellSeg, and Cellpose are 0.95, 0.94, 0.93, and 0.85, respectively, for an IoU of 0.5 and are 0.81, 0.74, 0.78, and 0.53, respectively, for an IoU of 0.7; the average processing times of GTWeka_selected features, StarDist, CellSeg, and Cellpose are 2.5, 4.8, 233.8, and 39.3 s, respectively. Furthermore, we benchmarked our GTWeka against other segmentation methods using images consisting of cell nuclei with heterogeneous fluorescent intensities (Figures S5A and S5B), different shapes or non-convex shapes (Figure S5C). Our method either reached a similar performance to or, in most cases, outperformed the tested methods.
As shown, our GTWeka cell/nuclei segmentation method can handle different types of input image data well and outperforms both state-of-the-art Weka-based and deep-learning-based methods with a faster processing time (1.9–93.5 times faster).
Instant cell linking and cell track correction
Highly accurate cell segmentation is required and crucial for accurate cell tracking. With the high performance of our GTWeka cell segmentation, we then apply it to link and track cells from frame to frame. The ability to accurately link positions of individual cells frame to frame and to reconstruct the movement and divisions of cells is crucial for dissecting cell lineage and their correlation with cell functions and cell-fate decisions.17,27 TGMM2 and mTGMM3 are fast state-of-the-art cell linking and tracking algorithms, with ∼26,000 and ∼42,000 cells being processed per minute, respectively, compared to other cell tracking algorithms, which can only process a number of thousands of cells/min.10,11 Tracking cell lineages often requires over a day of image acquisition, resulting in hundreds to thousands of image frames. Even with mTGMM, the algorithm cannot sufficiently process this amount of image frames within a number of minutes, which is required for downstream phenotype-to-genotype linking experiments. We therefore develop an instant, GMM-based cell linking algorithm.
Similar to TGMM2 and mTGMM,3 individual cells are modeled as a 2D Gaussian, , with being the 2D coordinates, being the mean location, and being the covariance (i.e., representing the shape of a Gaussian blob). All Gaussians (representing all individual cells) of the entire image constitute a Gaussian mixture per image frame. Calculating the expected properties per cell (i.e., intensity, size, shape) over time is performed by a full Bayesian approach,2,28 with the assumption that cell properties between two consecutive frames are correlated. If correlated, the same cells of the current frame and the previous frame will be linked (see details in STAR Methods).
To instantly complete image analysis (independent of the number of frames) after the acquisition of the last image frame, we implemented a real-time cell segmentation and linking method: once a new image is generated, it is directly processed by our GTWeka cell segmentation; when two consecutive images are segmented, cells of these two image frames are linked and tracked; and lastly, when the tracked frames are 3, the algorithm reviews all the connected tracks (from the current frame and the past few frames, pre-defined by users) and performs cell track corrections if needed (see below). With this instant processing of freshly generated image frames, FACT can complete the cell segmentation and linking and export quantifiable features in a matter of minutes, independent of the number of image frames. In our case, processing a 24 h time-lapse movie (361 frames with a 4 min interval, ∼10,000 cells/frame) took 2.88 h after data acquisition to export quantifiable results by mTGMM, while FACT required 0.17 h to obtain the results. FACT is built based on mTGMM3 with further development of real-time cell linking and cell track correction. FACT’s computational speed maintains ∼30,000 cells/min, which tends to be slower than mTGMM, as we add the function of cell track correction. Accurate cell lineage reconstruction highly relies on precise detection of cell divisions and differentiation of true divisions from false divisions. False cell divisions often arise from either mis-segmented cells or cells crossing each other’s trajectory; methods to correct the former cases have been reported27,29,30 but not for the latter scenarios. When two cells bump into each other and share a fraction of their boundaries, this causes the disappearance of one cell track (Figure 3). We call this phenomenon cell merging. A false cell division occurs when the two cells demerge, meaning the two cells move apart from each other after merging (Figure 3). Figure 3B shows cell merging-demerging found in the GBM image data. The cell merging-demerging problem is exacerbated when cell density is high or cell behavior is complicated, such as the directionally walking pattern employed by GBM cells (Figure 3B). To address this challenge, FACT implemented an automatic, real-time cell track correction function, which includes three steps as follows: (1) when a cell division is detected, we search for any incomplete cell tracks nearby (Figure 4A, top). Should the division occur next to an incomplete cell track within a pre-defined (Euclidean) distance (i.e., 10 pixels) and time window (i.e., 5 image frames), the division is deemed false division. The pre-defined distance and time window are based on users’ prior knowledge for each cell type. (2) When a false division is detected, the link between the mother and daughter cells will be disconnected, and the falsely generated cell link will be removed accordingly (Figure 4A, middle). (3) Two incomplete tracks (after step 2) will be re-connected by generating “fake” cell(s) in any position between the two incomplete tracks (Figure 4A, bottom).
Figure 3.
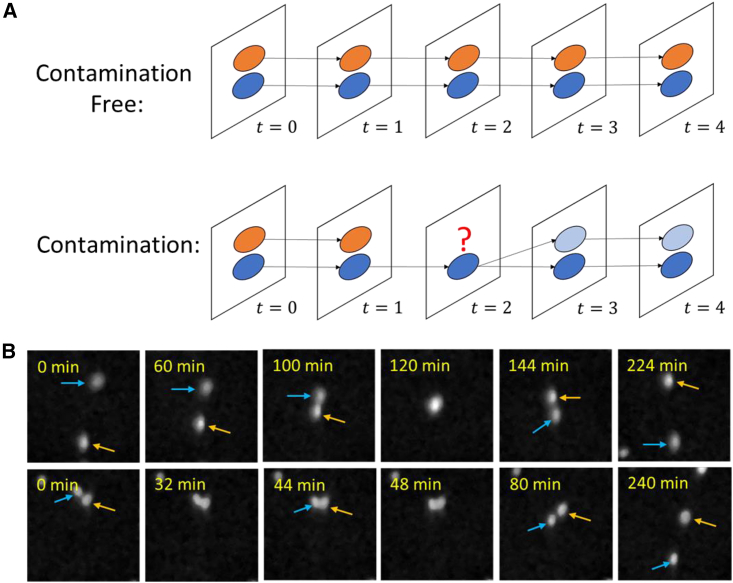
Cell merging and demerging
(A) Graphical illustration of two complete tracks (top) and incomplete tracks (bottom). For the contaminated case, two cells merge at frame , giving rise to (1) a false division at frame and (2) an incomplete track of the orange cell that ends at frame .
(B) Two examples of the merging-demerging problem shown in the raw image data of GBM cells. Top: two cells (blue and orange arrows) merge for over 30 min and then deviate. A snapshot at time “120 min” shows the merging event. Bottom: two cells (blue and orange arrows) merge twice at times “32 min” and “48 min.” This is caused by both complicated cell behaviors as well as false segmentation.
Figure 4.
Cell track correction
(A) Graphic illustration of cell track correction. (Top) Step 1: examine if a division is valid. We see that a merging at causes a cell track (orange) to disappear at location and a demerging at causes a division at location . The false cell division is detected when the event is next to an incomplete track within a pre-defined (Euclidean) distance ( between and ) and within a pre-defined time window (time difference, , between the merging and demerging events). (Middle) Step 2: remove the link of a false division. Compared to step 1, the frames with changes are highlighted in yellow. We disconnect the link (green) from to (in step 1), as this daughter cell (green) is closer to than the other daughter cell (blue). The link removal generates an incomplete track (green) from to . (Bottom) Step 3: close the gap between the incomplete tracks. Compared to step 2, the frames with changes are highlighted in yellow. The two incomplete tracks (orange and green) are to be connected. There is a gap between them, which refers to the disappeared cell at . We then construct a fake cell (gray) at this frame at any location of between and . We update the reconstructed cell to the subsequent frames . At , we obtain two complete tracks.
(B) Merging-demerging caused a false division (top). The tracks are corrected with our cell track correction method (bottom).
(C) Merging-demerging caused three false divisions (top) (merging happened at “76 min,” “120 min,” and “264 min”), and tracks were corrected with our cell track correction method (bottom).
We verified our cell track correction method on GBM image data (Figures 4B and 4C), as these cells often travel directionally from point to point31 and the cell merging-demerging issue is exacerbated in this cell type. For example, when tracking ∼3,000 cells over 5 h, ∼450 merging-demerging events were detected. With our cell track correction method, we have corrected those (∼450) falsely detected cell divisions; with correction, we reached 91% precision, while without correction, the precision is 12% due to heavy contamination from merging-demerging.
FACT identifies abnormal cancer cells
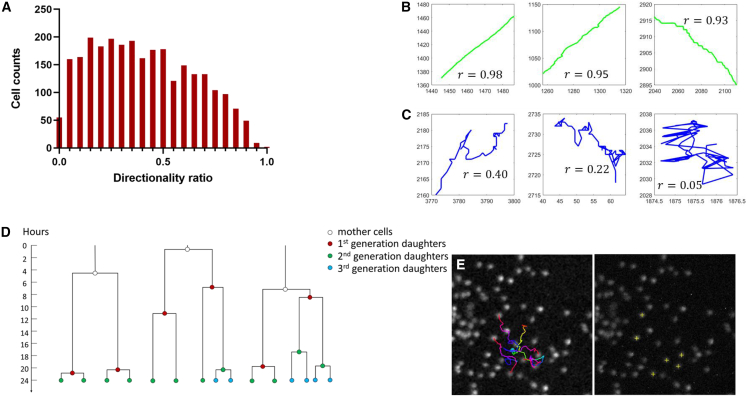
We then applied FACT to identify abnormally migrating GBM cells, namely the most directionally migrating cells, as this behavior is linearly correlated with cancer cell aggressiveness.31,32,33 We used the metric of directionality ratio () to measure the deviation between the cumulative distance (total distance traveled over all time points) and the linear distance (the distance from the start to the end time point). When is close to 1, a cell’s walk is close to directional walk. From a 5 h time-lapse movie, ∼3,000 GBM cells were tracked and analyzed via FACT. Please note that manual checking and validation of all the tracked trajectories are required, and therefore a relatively small number of cells were used in this particular application. The directionality ratios (r) of all the tracked cells were processed immediately and exported in <2 min after data acquisition. Based on the density distribution of all directionality ratios (Figure 5A), the mean ratio (0.40) and the standard deviation (0.24) were calculated. We set the ratio as the threshold to define a directional trajectory. With FACT, we found 28 single cells (∼1.0% of the whole population) displaying a directional walking pattern, and we have validated the results manually with a precision of 96% (Figures 5B, 5C, and S6).
Figure 5.
Application of FACT to identify abnormal cancer cells
(A–C) GBM cell tracking.
(A) Distribution of directionality ratio () over 2,724 cells. Over all cells, we looked for the ones giving a ratio r 0.90.
(B) Example trajectories of cells with directional walk; directionality ratio () is indicated per cell.
(C) Example trajectories of cells without directional walk; directionality ratio () is indicated per cell.
(D and E) Cell lineage tracking of MCF10A cells.
(D) Topology of lineages from 3 groups, “tree-3-div” (left), “tree-4-div” (middle), and “tree-5-div” (right). In each plot, time is represented as the vertical axis, going from 0 to 24 h. Daughter cells generated in different generations are color-coded. Videos of these cases are included in Video S1.
(E) Migratory trajectories of the lineage of interest (tree-5-div) (left image), and the final coordinates of the cells (yellow cross, right image).
Next, we applied FACT to segment and track MCF10A cells (∼10,000 cells/frame) from a 24 h time-lapse movie in real time (361 frames, 4 min/frame), from which cell lineages of individual cells will be extracted. The precision of cell tracking is, on average, 96% (see STAR Methods). We aim to identify abnormal cell lineages that deviate from the rest of lineages. MCF10A cells are epithelial cells with a tendency to densely pack and connect with each other and are therefore used as the sample model here.
After the FACT analysis, we grouped the lineage trees by the number of cell divisions. “tree-1-div,” “tree-2-div,” or “tree-5-div” indicates a lineage tree with 1, 2, or 5 cell divisions, respectively. For example, a lineage tree in the “tree-5-div” group includes 5 divisions within 24 h (Figure 5D).
From the assayed ∼10,000 MCF10A cells, FACT exported the lineage information of each cell less than ∼10 min after the data acquisition with an average F1 score of 0.91. We identified 52.8%, 12.2%, 6.9%, 0.5%, and 0.1% of cells dividing once, twice, three, four, and five times, respectively.
Figure 5D shows 3 representative lineage trees of fast-dividing cells (“tree-5-div”) identified from the assayed cells (see the original movies of these 3 lineages in Video S1), from which the “tree-5-div” case is shown in Figure 5E. The coordinates of those 5 daughter cells of this irregular lineage tree were immediately exported within 10 min after the image acquisition.
Discussion
We have developed a real-time cell segmentation and tracking algorithm, called FACT, to instantly process large-scale image data (104−6 cells/frame for 102−3 image frames) and export quantitative cellular characteristics within minutes after data acquisition. FACT implements ground-truth-assisted Weka-based cell segmentation (GTWeka), which outperforms state-of-the-art methods and only requires 30–60 min of human annotation to achieve peak performance for unseen sample types. High segmentation performance ( 0.95 F1 score at an IoU threshold of 0.5), in combination with real-time cell track correction, results in high tracking performance, as the program can correct wrongly detected cell divisions and falsely linked cell tracks in real time. As shown, FACT can correctly track GBM cells, which tend to migrate across each other, and track densely packed MCF10A epithelial cells, with an average of 90%–96% precision.
When combined with high-throughput screening microscopy, FACT can be used to instantly identify rare subpopulations of cells during large-scale image data acquisition, enabling immediate isolation of target cells for downstream assays, like single-cell sequencing or proteomic profiling.3,13 For example, FACT can instantly identify sparse, tripolar dividing cells, aggressively migrating cells, and fast-dividing cells; these are examples of extremely low occurrence events. Linking rare, abnormal cellular phenotypes (i.e., metastatic cells or abnormally dividing cells) to genotypes, transcriptomes, or proteomes is possible with FACT, as instant export of cellular characteristics is required for target cell identification and isolation. This creates an unprecedented opportunity to decipher the underlying mechanisms of the observed irregular or disease-driving phenotypes.
Limitations of the study
Potential limitations of the methods described here are as follows.
-
•
The instant segmentation of GTWeka comes from both a powerful GPU as well as a low number of selected features (i.e., 3 features). If a less powerful GPU or no GPU is implemented, and/or more selected features (>>3 features) are needed for a dataset, one may not achieve the processing time presented in this study.
-
•
Regarding tracking and track correction, FACT can correct merging of two cells but not more. If more than two cells are merging, FACT will not be able to correct tracks and close gaps correctly. If a dataset contains cells prone to overlapping, correction may take more time, as there will be more “incomplete tracks” to be fixed via FACT track correction.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| SPY650-DNA | Spirochrome | Cat#SC501 |
| Deposited data | ||
| Cell images (.tif files) used in this paper to identify abnormal cancer cells | This paper | Figshare https://doi.org/10.6084/m9.figshare.24187455 |
| Experimental models: Cell lines | ||
| Human: MCF10A | Laboratory of Dr. Agami (Netherlands Cancer Institute) | N/A |
| Human: Glioblastoma | Erasmus University Medical Center, Molecular Genetics | N/A |
| Human: HeLa | Erasmus University Medical Center, Molecular Genetics | N/A |
| Software and algorithms | ||
| All source codes used in this paper | This paper | Zenodo https://doi.org/10.5281/zenodo.8372760 |
| Automatic cell tracking analysis | You et al.3 | https://sourceforge.net/projects/funseq/ |
| Python | Python Software Foundation | https://www.python.org/ |
| Trainable Weka Segmentation | Arganda-Carrera et al.24 | https://ImageJ.net/plugins/tws/ |
| ilastik | Berg et al.25 | https://www.ilastik.org/ |
| Stardist | Schmidt et al.21 | https://github.com/stardist/stardist |
| CellSeg | Lee et al.22 | https://michaellee1.github.io/CellSegSite/index.html |
| Cellpose | Stringer et al.23 | https://www.cellpose.org/ |
| ImageJ | Schneider et al.34 | https://ImageJ.nih.gov/ij/ |
| MATLAB | MathWorks | https://nl.mathworks.com/products/MATLAB.html |
| TrackMate | Ershov et al.11 | https://ImageJ.net/plugins/trackmate/ |
Resource availability
Lead contact
-
•
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Miao-Ping Chien (m.p.chien@erasmusmc.nl).
Materials availability
-
•
This study did not generate new unique reagents.
Data and code availability
-
•
Cell images (.tif files) used in the application to identify abnormal cancer cells are available at Figshare https://doi.org/10.6084/m9.figshare.24187455.
-
•
The original code has been deposited in Zenodo https://doi.org/10.5281/zenodo.8372760.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
MCF10A cells
MCF10A epithelial cells, a gift from Dr. Reuven Agami (Dutch National Cancer Institute, NKI), were grown in DMEM (Dulbecco’s modified Eagle’s medium)/F-12 (ThermoFisher) supplemented with 5% horse serum, 1% penicillin/streptomycin, Epidermal growth factor (10 ng mL−1; ThermoFisher), Hydrocortisone (500 ng mL−1; Stem Cell), cholera toxin (100 ng mL−1; Sigma) and insulin (10 μg mL−1; ThermoFisher) in a 37°C incubator under 5% CO2.
Before conducting experiments, ∼50,000 cells were seeded on a fibronectin (0.1 mg/mL)- coated 35 mm-glass bottom dish with a 20 mm-microwell (Cellvis) in the MCF10A culture medium (described above) without phenol red and were stained with a SPY650-DNA nuclear dye (Spirochrome) (1:2000) (λex: 652, λem: 674nm). Experiments were performed ∼24 h after plating on the glass-bottom dishes.
Glioblastoma multiforme (GBM) cells
GBM cells were maintained on reduced growth factor basement membrane extract (Cultrex, Bio-techne R&D systems) coated petri dishes in serum-free Dulbecco’s Modified Eagle Medium (DMEM)/F12 supplemented with 1% penicillin/streptomycin, 2% B27 without vitamin A (Gibco, ThermoFisher), 20 ng/mL basic fibroblast growth factor (Gibco, ThermoFisher), 20 ng/mL epidermal growth factor (Sigma Aldrich), and 5 μg/mL heparin (Alfa Aesar, ThermoFisher). Cells were maintained in a humidified incubator (37°C, 5% CO2) and culture medium was refreshed every 3–4 days). Passaging of the cells was performed around 80–90% confluency using enzymatic dissociation (Accutase, Invitrogen). The use of GBM samples and study was approved by Erasmus University Medical Center ethics committee (MEC-2013-090).
Before conducting experiments, ∼19,000 cells were seeded on Cultrex (0.1 mg/mL)- coated 35 mm-glass bottom dish with a 20 mm-microwell (Cellvis) in the GBM culture medium (described above) without phenol red and were stained with a SPY650-DNA nuclear dye (Spirochrome) (1:2000) (λex: 652, λem: 674nm). Experiments were performed ∼24 h after plating on the glass-bottom dishes.
HeLa cells
HeLa human cervical carcinoma cells, a gift from Dr. Roland Kanaar (Erasmus University Medical Center), were cultured in DMEM medium (Thermo Fisher) with 10% Fetal Bovine Serum (Thermo Fisher), 1% penicillin/streptomycin at a 37°C incubator under 5% CO2.
Before conducting experiments, ∼200,000 cells were seeded on a fibronectin (0.1 mg/mL)- coated 35 mm-glass bottom dish with a 20 mm-microwell (Cellvis) in the HeLa culture medium (described above) without phenol red and were stained with a SPY650-DNA nuclear dye (Spirochrome) (1:2000) (λex: 652, λem: 674nm). Experiments were performed ∼24 h after plating on the glass-bottom dishes.
Method details
Image data preparation
The images used in this paper are either MCF10A epithelial cells, glioblastoma multiforme (GBM) primary culture cells or HeLa cells (see Supplementary Methods for the detail of cell culture). The images were taken by a custom-built microscope, ultrawide field-of-view optical (UFO) microscope,3 which incorporates a large chip-size camera (CMOS Point Gray, GS3-U3-123S6M-C (4,096 × 3,000 pixels, 3.45 μm per pixel) for MCF10A and GBM cell images; pco.panda 26 DS (5,120 × 5,120 pixels, 2.5 μm) for HeLa cell images) and a large-field-of-view (FOV) objective (Olympus MVP Plan Apochromat, ×0.63)3 with comparatively high numerical aperture (NA = 0.25).
Workstation and software information
All segmentation work was done via Ubuntu 18.04 environment with Intel(R) i9-9980XE CPU, 128 GB ram, two of Nvidia QUADRO RTX 8000 GPUs. GTWeka was implemented in Python 3 using open-source packages: NumPy,35 OpenCV,36 CuPy,37 cuML,38 scikit-image,39 SciPy40 and Matplotlib.41
Real-time cell linking was performed on a single workstation, Dell Precision 7920, with the following hardware components: dual Intel(R) Xeon(R) Gold 6130 CPUs, 12 × 32GB ram, a Nvidia GeForce GTX 1080 GPU.
Details of ground truth-assisted Trainable Weka Segmentation (GTWeka)
GTWeka is inspired by and modified from original Trainable Weka Segmentation (TWS).24 We first introduce the method, and we introduce and explain our improvements on this method.
Trainable Weka segmentation
Different image filters are used to generate image features as follows: Gaussian filer, Sobel filter, difference of Gaussians filter, Hessian matrix, Membrane projections and Median filter. TWS starts with semantic segmentation, which classifies each pixel into one of the three classes: background, foreground or edge. The class of foreground is of our interest. TWS uses a random forest classifier42 to determine a label for each pixel, i.e., a collection of decision trees where each tree represents a ‘test’ if a given pixel contributes enough or not to the class (i.e., background, foreground or edge) prediction.
After we obtained all the foreground pixels, we added a step of instance segmentation to identify each single cell, such as giving each object an id. We used connected component analysis39,43 on the semantic segmentation to generate the indices.
Trainable Weka segmentation on GPU
Processing a single image, e.g., 4096 3000 pixels, takes ∼55 s when using TWS from Fiji44 (TWS provides a Fiji plugin for users without specific programming skills). This version runs on CPU. We use GPU to accelerate the whole process, and it takes ∼16 s (for segmenting an image of 4096 3000 pixels). We observed significant improvement in processing speed by implementing it on GPU. We implemented GPUs (two NVIDIA QUADRO RTX 8000) to generate key image features by the RAPIDS cuCIM library.45
Reference dataset preparation (see Figure S3 for detail)
The reference image preparation was done with the Fiji image processing program.44 We randomly selected 3 cropped images from the raw images and use the LabKit plugin46 (Weka-base segmentation) to generate the labeling of foreground, background and edge (edge labeling is optional) (Step 1–2, Figure S3). After that, the (foreground) segmentation result can be generated (Step 3, Figure S3) and converted to regions of interest (Step 4, Figure S3). The wrongly segmented objects can be manually corrected (Step 5, Figure S3) and the final segmentation result can be saved as reference ground truth data (Step 6, Figure S3). The whole process takes ∼30 min.
Random forest and its structure
Implementing TWS on GPU is one of our improvements to achieve fast and accurate high-throughput cell segmentation. The other improvement lies on the random forest optimization, which is introduced subsequently.
The nature of a random forest classifier indicates that how the forest ‘look alike’ and is critical for pixel-classifications. Variation (i.e., the parameters) includes the number of trees, the depth of each tree, and the number of nodes per tree, etc. If parameters of the model change, the prediction might also change, yielding the question ‘what are the best model parameters for our data of interest’?
To find the best random forest model per dataset, we applied grid search (and cross validation) to find the best parameters for the model, including the number of estimators, the maximum number of features, the maximum depth of the tree and the minimum number of sample leaf. To do this, a ground truth reference dataset is required, which, in our case, is a foreground/background labeled imaging dataset. This ground truth reference data acts as a reference during the process of selecting the best random forest parameter option (i.e., the depth of the tree and the number of sample leaf etc.) by checking the segmentation performance (i.e., accuracy or F1 score) (Figure 1, see Figure S2 for detail).
Key feature selection
We call the features that are relevant to our pixel classification the ‘key features’. Now the question comes to how to figure out the features that are the key ones. We applied forward selection26 to find out the key features - Recall that we have in total 57 default features , and we want to find a subset as key features:
-
1.
With respect to each a classifier is trained, and corresponding segmentation accuracy is obtained via comparing to reference data.
-
2.
The feature, say , that contributes to the highest accuracy will be selected as the first key feature . There are hence 56 features left (), out of which we will choose the second key feature.
-
3.
We let where for every A classifier is trained with every feature subset , and the second key feature, say , is determined as together contributing to the highest accuracy.
If we want to continue the selection, we shall repeat Step 2 by adding a third key feature where for every . The forward selection continues till the number of elements from reaches a pre-defined number of features. For example, if the pre-defined number of features is 3, then the selection stops once we have .
In our case, using 3 key features provides us with the fastest and most accurate segmentation (Figures 2 and S1).
Training dataset preparation
For all the cell image datasets, we selected 3 frames at different timepoints (frame 0, 360 and 720) from the entire time lapse movie (1080 frames in total, 4 min/frame). For annotation, we focused only at a partial region of a FOV, and we manually annotated the chosen region with 3 classes, i.e., nuclei, background, and edge (Figure 1). All the annotation work was done with Fiji.
Benchmarking
Input data
We randomly selected cropped images from all the different cell image datasets as our test images (5 cropped images from the MCF10A 3-day image data, 6 cropped images from the GBM 2-day image data and 3 cropped images from the HeLa 1-day image data), and the ground truth on these test images were annotated manually. We compared our segmentation performance and computational time to other state-of-the-art Weka-based and neural network-based cell segmentation methods. The methods that are compared to are listed as follows.
-
•
TWS. This is the default setup of TWS, where there are 57 features generated for the random forest classifier (to determine if a pixel is of the Class foreground, background or edge).
-
•
GTWeka_all features. Unlike TWS which uses a fixed structure of random forest (by default) for any input dataset, we search for the structure, out of many combinations, that can give the highest accuracy per input dataset. Optimization is bounded to the reference data, which helps to optimize the best parameters of the random forest classifier. In this case the all features (57 features) are used. We aim to compare this method with TWS when the all features are included during classification.
-
•
GTWeka_selected features. Instead of using all features, we apply only the key features (described above) to the classifier. Optimization of the random forest classifier, including key feature selection, is restricted to the reference data. We aim to compare this method to TWS when a subset of features is chosen.
-
•
StarDist. StarDist,21 a cell segmentation method for microscopy images with star-convex shape priors. The ‘2D_versatile_fluo’ pre-trained model was used in this paper.
-
•
CellSeg. CellSeg22 is an open-source cell segmentation method using pre-trained models. The method is based on a Mask region-convolutional neural network (R-CNN) architecture.
-
•
Cellpose. Cellpose23 is an open-source cell segmentation method using various pre-trained models. The ‘Nucleus’ pre-trained model was used in this paper.
The comparison results can be found in Figure 2.
To compare only the feature selection methods, we examine:
-
•
Ilastik. Ilastik is a TWS-based cell segmentation method with an option of selecting key features. ilastik provides three kinds of feature selection methods25: filter method, GINI importance and wrapper method. According to the all features used for classifier, feature selection method helps users to choose a subset of it , on the classifier. We followed the filters which are provided by ilastik with sigma settings as 1, 2, 4, and 8. The used image filters are Gaussian filter, Laplace filter, gradient magnitude, difference of Gaussians, structure tensor eigen values and Hessian matrix eigen values. The total amount of image features provided in ilastik is 32. Please note that the image filters mentioned here are different than the filter setting used in the segmentation comparison analysis (Figure 2) as ilastik only offers the abovementioned filters (total 32 image features, compared to TWS or our GTWeka methods, 57 image features). Also, for ilastik’s wrapper method, we set the parameter of size penalty to 0.01, 0.04, 0.1 and this method only allows for up to 6 features.
-
•
GTWeka_selected features. Here we use the same training dataset and image feature setting. We compare our GTWeka methods with selected 3 key features with ilastik’s feature selection methods; our method outperforms ilastik’s feature selection methods.
The comparison results can be found in Figure S1a, where we selected 3 to 9 features from the all features.
Single cell tracking via FACT
The segmented cells are linked over time via the Gaussian Mixture Model-based method.2,47 FACT tracking steps are summarized in Figure 6. Details of the approach per step are explained afterward.
Figure 6.
FACT tracking steps
(A) Nuclear-mask images as input for tracking. The input images are the pixel-wise dot product of raw and GTWeka segmented images.
(B) Transforming each input into a mixture of Gaussian models (GMs) , where each cell is considered as one Gaussian . The properties of a Gaussian (i.e., mean, variance) can describe a cell’s location and shape. And a Gaussian distribution itself denotes a cell’s intensity. Frame-to-frame linking per cell is performed by “finding nearest neighbors of Gaussians between adjacent frames” if we say a cell's position in one frame is the nearest neighboring location to its position in the next frame. The nearest neighbors are searched in multidimensions, including changes of location, shape, and intensity. The algorithm behind is Bayesian inference, a probabilistic model that calculates expected changes (e.g., location, intensity, shape) of all cells (per frame) over time. Similar cells are linked over time. Division can be followed by examining if one Gaussian is splittable.
(C) Forwarding GMs gives initial tracking outcomes, which might be prone to contamination. The outcomes are saved in XML files. Here, we visualize them as a table. Cells that are tracked at each frame generate “complete tracks.” Cells that are missed for at least one frame generate “incomplete tracks.”
(D) Correction of tracks that are contaminated. The correction goes sequentially as follows: (1) looking for false divisions, with the information of detected divisions and incomplete tracks. (2) Breaking the mother-daughter link if a false division is confirmed. The breakup also generates new incomplete tracks. (3) Updating the incomplete tracks with new ones. (4) Bridging the incomplete tracks as one complete track (a way of closing gaps). A gap refers to the spatial and temporal distance between two (or more) track segments; meanwhile, these track segments are parts of one same cell track.
Nuclear-mask images as input for tracking
After a microscope generates raw images and GTWeka gives segmented nuclei-masks (Figure 6A), we obtain input images (Figure 6B) for mTGMM tracking. These are the dot product of raw images and nuclei-masks.
Cell intensity as a Gaussian
Connected pixels (from GTWeka) are considered as one single cell, and its intensity profile is modeled as a 2D Gaussian: with being the 2D coordinates, being the mean location and being the covariance (i.e., representing the shape of a Gaussian blob). Each cell/Gaussian is independent from each other. Hereby we treat an object as a probabilistic model: Using the information of a cell at , we could predict, for this cell, a ‘bounding box’ at (), which in our case is a probabilistic distribution of (). In other words, if we know the moving object distribution in the previous frame(s), we can locate the object in the next frame by tracking the expected changes. Tracking one cell means forwarding its Gaussian by calculating its expected changes, in the dimensions of location, shape and intensity.
Image as a Gaussian mixture
When dealing with an entire image, we want to forward all Gaussians from that image simultaneously over time. Then we move to the next step, modeling an image (of many objects) at frame as a Gaussian Mixture:
where is the number of nuclei at frame , are the 2D coordinates for the -th nucleus. The parameters , , define the -th Gaussian, i.e., respectively, they describe the contribution of the -th nucleus to the image, estimated mean location and shape. Calculating the expectation per cell over time is performed by a full Bayesian approach,2,28 with the assumption that cell properties between two consecutive frames are correlated.
Cell division
We handled cell divisions separately in this process: First we shall determine if a cell is going to divide, then we include the newly generated cell into a Gaussian mixture. For each nucleus we perform Otsu thresholding on only the foreground pixels, and we see if two unconnected regions are generated. If not, we do not find a case of dividing and we do not proceed with splitting one Gaussian into two components. If two unconnected regions are found, we further assess if the size per region is larger than a pre-defined value (as a region can be too small to represent a daughter cell).
Once we have detected a cell division at frame , we accordingly need to update the Gaussian mixture at , by letting .
Cell track correction
Solution to merging-demerging problem
We propose to correct the corrupted tracks by three steps (see Figure 4A).
-
•
Step 1: We examine if a cell division is valid. A cell division that is next to one or more incomplete tracks in both space and time is the consequence of a merging-demerging. In Figure 4A, a merging at causes a cell track (orange) disappearing at location , a demerging at causes a division at location . We call also the location of the incomplete track (orange). The cell division is next to the incomplete track in space if the (Euclidian) distance between and is smaller than a pre-defined value (such as 10 pixels). The cell division is next to the incomplete track in time if the time difference between the merging and demerging events is smaller than a pre-defined value (such as 5 frames). In this example, the cell division is found invalid, and we move to the next step.
-
•
Step 2: For an identified false division, we break the mother-daughter. The daughter cell that is closer to and forming smaller angle with respect to the incomplete track is excluded. We remove the link (green) from to , as this daughter (green) is closer to than the other daughter (blue) is, while angle angle (over ). The link removal generates an incomplete track (green) from to .
-
•
Step 3: We connect two incomplete tracks, and close the gap between the two incomplete tracks. The gap closing is done via linear assignment problem (LAP),48 which searches if (at each frame) any track end can be connected to a track start, and this connection is a 1-to-1 relationship. The cost to close a gap is constrained to where and when the tracks exist. For example, if the end of incomplete track is 10 frames (>30 min, a pre-defined temporal threshold) away from the start of incomplete track , the cost to connect will be too high to perform. In Figure 4A, the two incomplete tracks (orange and green) are to be connected (assuming the cost function agrees). There is a gap between them, which refers to the disappeared cell at . We then construct a fake cell (gray) at this frame at a random location between and . This fake cell resembles the two cells at and : its mean intensity is a random value between the two cells’ mean intensity, and its size (i.e., number of pixels) is also a random value tween the two cells’ sizes. Once the intensity, size and location of this fake cell are known, a Gaussian is constructed for this cell. This Gaussian is representing the ‘look’ of this fake cell. We update the reconstructed cell to the subsequent frames . At we obtain two complete tracks.
Quantification and statistical analysis
Segmentation performance
We examined the segmentation performance (average F1 score) over different methods: TWS, ilastik, StarDist, CellSeg, Cellpose and our GTWeka method with all features or with selected features (main text Figure 2). Cell segmentation images and results are shown in Figure 2. These outcomes were achieved using the same input image. Ground truth (GT) segmentation was prepared independently. We compared the segmentation outcome per method to the GT by calculating the IoU (intersection-over-union) value. We quantified the segmentation performance (see Figure 2) by looking at the segmentation accuracy per method at the IoU threshold of 0.5 or 0.7. We obtained a true positive (TP) when the calculated IoU is IoU threshold, a false positive (FP) if the calculated IoU is between 0.1 - IoU threshold, a false negative (FN) if the calculated IoU is <0.1. The computational time was an average of fourteen FOVs.
Tracking accuracy and computational time
Tracking accuracy
We assess the accuracy of cell linking by examining how many cell tracks, out of all, are accurately followed. We call a cell is accurately tracked if its centroids between any two adjacent time points are accurately linked: Let () represent the centroid coordinates of cell at with be the length of a time lapse movie. Cell track is accurate if () is accurately linked to ( +1) for every .
We selected two regions of time lapse movies (MCF10A cells) and manually checked the tracking accuracy: Region 1 contains 31 frames with a dimension of 1170 by 724 pixels, Region 2 contains 31 frames with a dimension of 1628 by 978 pixels. We compared the tracking results of FACT to the ground truth data and obtained an average precision of 96% per region (95% for Region 1 and 97% for Region 2).
The tracked images are shown in Supplementary Information: Video S2, left for Region 1 and Video S2, right for Region 2.
Tracking efficiency
For real-time experiment, the tracking efficiency is crucial. A real-time tracking method shall be able to catch up the speed of image acquisition. When tracking two image frames of MCF10A cells with ∼20,000 cells/frame, FACT requires ∼100 s (an average of three runs: 101.37, 98.50, and 99.96 s).
Cell lineage accuracy
We calculate, for each lineage tree, its F1 score: we check manually if each detected division from this tree is a TP or FP. We also check any missed divisions (compared to the raw images) and treat it as an FN. We define that a detected division is a TP if 1) it is truly a division from this lineage, as well as 2) the detected splitting time is +/− 5 frames compared to the true division time. For example, if a detected lineage tree has 5 divisions, with 2 divisions being FPs, 3 divisions being TPs, and 1 missed division as 1 FN, then the F1 score of this detected tree is 0.67 (=).
In Table S1 the performance of lineage tracking is shown. On average we can reach 0.91 F1 score per lineage.
In addition, we compared our cell tracking method with TrackMate,11 which also uses LAP to link cells and correct wrong cell tracks. The main difference of our approach from the existing LAP is that we only applied LAP on incomplete tracks, instead of all tracks (including the completed ones). Our approach not only shortens the time of wrong-track correction, but also improves linking accuracy, as correcting cell tracks by incorporating completed tracks could potentially wire incomplete tracks to complete ones, thereby creating false cell-to-cell linking and false cell divisions (see the comparison table below). Averaging over two regions, FACT gave 0.92 precision and 0.92 F1 score; TrackMate gave 0.80 precision and 0.83 F1 score. The detailed differences of our cell track correction method versus the current LAP method can be found in Figure 7.
Figure 7.
Comparing FACT tracking to other popular tracking approaches such as linear assignment problem (LAP)
When cells are prone to move toward each other, often causing overlapping, FACT gives higher precision and F1 score regarding cell division estimation. Details of cell track correction used by LAP47 and our FACT method are summarized in this figure.
Acknowledgments
We thank Dr. D. Brinks for critical reading of the manuscript. We thank the Josephine Nefkens Cancer Program for infrastructural support. M.-P.C. acknowledges support from the Oncode Institute, Cancer GenomiCs.nl (CGC), NWO (the Netherlands Organization for Scientific Research) Veni and Vidi Grants, Stichting Ammodo, a Dutch Cancer Society (KWF) grant, and a Flagship convergence grant.
Author contributions
T.-C.C. scripted the cell segmentation algorithm and analyzed the assayed data. L.Y. scripted the real-time cell tracking algorithm and analyzed the assayed data. C.B. contributed to the cell culture preparation for the experiments. K.J.F. contributed to the GBM cell preparation. J.S. provided the technical support regarding the UFO microscope. T.-C.C., L.Y., and M.-P.C. designed all the experiments. M.-P.C., L.Y., and T.-C.C. wrote the paper with input from all authors. M.-P.C. initiated the project and contributed to and supervised all aspects of the project.
Declaration of interests
The authors declare no competing interests.
Published: November 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100636.
Supplemental information
References
- 1.Stegmaier J., Amat F., Lemon W.C., McDole K., Wan Y., Teodoro G., Mikut R., Keller P.J. Real-Time Three-Dimensional Cell Segmentation in Large-Scale Microscopy Data of Developing Embryos. Dev. Cell. 2016;36:225–240. doi: 10.1016/j.devcel.2015.12.028. [DOI] [PubMed] [Google Scholar]
- 2.Amat F., Lemon W., Mossing D.P., McDole K., Wan Y., Branson K., Myers E.W., Keller P.J. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods. 2014;11:951–958. doi: 10.1038/nmeth.3036. [DOI] [PubMed] [Google Scholar]
- 3.You L., Su P.-R., Betjes M., Rad R.G., Chou T.-C., Beerens C., van Oosten E., Leufkens F., Gasecka P., Muraro M., et al. Linking the genotypes and phenotypes of cancer cells in heterogenous populations via real-time optical tagging and image analysis. Nat. Biomed. Eng. 2022;6:667–675. doi: 10.1038/s41551-022-00853-x. [DOI] [PubMed] [Google Scholar]
- 4.Pantazis P., Supatto W. Advances in whole-embryo imaging: a quantitative transition is underway. Nat. Rev. Mol. Cell Biol. 2014;15:327–339. doi: 10.1038/nrm3786. [DOI] [PubMed] [Google Scholar]
- 5.Masuzzo P., Van Troys M., Ampe C., Martens L. Taking Aim at Moving Targets in Computational Cell Migration. Trends Cell Biol. 2016;26:88–110. doi: 10.1016/j.tcb.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Oates A.C., Gorfinkiel N., González-Gaitán M., Heisenberg C.P. Quantitative approaches in developmental biology. Nat. Rev. Genet. 2009;10:517–530. doi: 10.1038/nrg2548. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez R., Das P., Mirabet V., Moscardi E., Traas J., Verdeil J.L., Malandain G., Godin C. Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat. Methods. 2010;7:547–553. doi: 10.1038/nmeth.1472. [DOI] [PubMed] [Google Scholar]
- 8.Khan Z., Wang Y.C., Wieschaus E.F., Kaschube M. Quantitative 4D analyses of epithelial folding during Drosophila gastrulation. Development. 2014;141:2895–2900. doi: 10.1242/dev.107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosaliganti K.R., Noche R.R., Xiong F., Swinburne I.A., Megason S.G. ACME: Automated Cell Morphology Extractor for Comprehensive Reconstruction of Cell Membranes. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ershov D., Phan M.-S., Pylvänäinen J.W., Rigaud S.U., Le Blanc L., Charles-Orszag A., Conway J.R.W., Laine R.F., Roy N.H., Bonazzi D., et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods. 2022;19:829–832. doi: 10.1038/s41592-022-01507-1. [DOI] [PubMed] [Google Scholar]
- 12.Smit M.M., Feller K.J., You L., Storteboom J., Begce Y., Beerens C., Chien M.P. Spatially Annotated Single Cell Sequencing for Unraveling Intratumor Heterogeneity. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.829509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su P.-R., You L., Beerens C., Bezstarosti K., Demmers J., Pabst M., Kanaar R., Hsu C.-C., Chien M.-P. Microscopy-based single-cell proteomic profiling reveals heterogeneity in DNA damage response dynamics. Cell Rep. Methods. 2022;2 doi: 10.1016/j.crmeth.2022.100237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smit M.M., Feller K.J., You L., Chien M.-P. Protocol for profiling in vitro intratumor heterogeneity using spatially annotated single-cell sequencing. STAR Protoc. 2023;4 doi: 10.1016/j.xpro.2023.102447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine M.S., Holland A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018;32:620–638. doi: 10.1101/gad.314351.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bravo-Cordero J.J., Hodgson L., Condeelis J. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Zhu H.H., Chu M., Liu Y., Zhang C., Liu G., Yang X., Yang R., Gao W.-Q. Symmetrical and asymmetrical division analysis provides evidence for a hierarchy of prostate epithelial cell lineages. Nat. Commun. 2014;5:4758. doi: 10.1038/ncomms5758. [DOI] [PubMed] [Google Scholar]
- 18.Caicedo J.C., Goodman A., Karhohs K.W., Cimini B.A., Ackerman J., Haghighi M., Heng C., Becker T., Doan M., McQuin C., et al. Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat. Methods. 2019;16:1247–1253. doi: 10.1038/s41592-019-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long F. Microscopy cell nuclei segmentation with enhanced U-Net. BMC Bioinf. 2020;21:8. doi: 10.1186/s12859-019-3332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai H.-F., Gajda J., Sloan T.F., Rares A., Shen A.Q. Usiigaci: Instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning. SoftwareX. 2019;9:230–237. doi: 10.1016/j.softx.2019.02.007. [DOI] [Google Scholar]
- 21.Schmidt U., Weigert M., Broaddus C., Myers G. In: Cell Detection with Star-Convex Polygons. held in Cham, 2018// Frangi A.F., Schnabel J.A., Davatzikos C., Alberola-López C., Fichtinger G., editors. Springer International Publishing); 2018. pp. 265–273. [Google Scholar]
- 22.Lee M.Y., Bedia J.S., Bhate S.S., Barlow G.L., Phillips D., Fantl W.J., Nolan G.P., Schürch C.M. CellSeg: a robust, pre-trained nucleus segmentation and pixel quantification software for highly multiplexed fluorescence images. BMC Bioinf. 2022;23:46. doi: 10.1186/s12859-022-04570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 24.Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K.W., Schindelin J., Cardona A., Sebastian Seung H. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 25.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 26.Ferri F.J., Pudil P., Hatef M., Kittler J. In: Machine Intelligence and Pattern Recognition. Gelsema E.S., Kanal L.S., editors. North-Holland); 1994. Comparative study of techniques for large-scale feature selection∗; pp. 403–413. ∗This work was suported by a SERC grant GR/E 97549. The first author was also supported by a FPI grant from the Spanish MEC, PF92 73546684. [DOI] [Google Scholar]
- 27.Tian C., Yang C., Spencer S.L. EllipTrack: A Global-Local Cell-Tracking Pipeline for 2D Fluorescence Time-Lapse Microscopy. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107984. [DOI] [PubMed] [Google Scholar]
- 28.Smal I., Meijering E., Draegestein K., Galjart N., Grigoriev I., Akhmanova A., van Royen M.E., Houtsmuller A.B., Niessen W. Multiple object tracking in molecular bioimaging by Rao-Blackwellized marginal particle filtering. Med. Image Anal. 2008;12:764–777. doi: 10.1016/j.media.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Al-Zaben N., Medyukhina A., Dietrich S., Marolda A., Hünniger K., Kurzai O., Figge M.T. Automated tracking of label-free cells with enhanced recognition of whole tracks. Sci. Rep. 2019;9:3317. doi: 10.1038/s41598-019-39725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panteli A., Gupta D.K., Bruijn N., Gavves E. In: Proceedings of the Third Conference on Medical Imaging with Deep Learning. Tal A., Ismail Ben A., Marleen de B., Maxime D., Herve L., Christopher P., editors. PMLR; 2020. Siamese Tracking of Cell Behaviour Patterns. [Google Scholar]
- 31.Nousi A., Søgaard M.T., Audoin M., Jauffred L. Single-cell tracking reveals super-spreading brain cancer cells with high persistence. Biochem. Biophys. Rep. 2021;28 doi: 10.1016/j.bbrep.2021.101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J., Verkman A.S. Increased migration and metastatic potential of tumor cells expressing aquaporin water channels. FASEB J. 2006;20:1892–1894. doi: 10.1096/fj.06-5930fje. [DOI] [PubMed] [Google Scholar]
- 33.Huda S., Weigelin B., Wolf K., Tretiakov K.V., Polev K., Wilk G., Iwasa M., Emami F.S., Narojczyk J.W., Banaszak M., et al. Lévy-like movement patterns of metastatic cancer cells revealed in microfabricated systems and implicated in vivo. Nat. Commun. 2018;9:4539. doi: 10.1038/s41467-018-06563-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji - an Open Source platform for biological image analysis. Nat Methods. 2012;9 doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradski G. The OpenCV Library; 2000. https://github.com/opencv/opencv/wiki/CiteOpenCV [Google Scholar]
- 37.Okuta R.a.U., Yuya, Nishino D., Hido S., Loomis C. Proceedings of Workshop on Machine Learning Systems (LearningSys) in the Thirty-First Annual Conference on Neural Information Processing Systems (NIPS) 2017. CuPy: A NumPy-Compatible Library for NVIDIA GPU Calculations. [Google Scholar]
- 38.Raschka S.a.P., Joshua, Nolet C. Machine Learning in Python: Main Developments and Technology Trends in Data Science, Machine Learning, and Artificial Intelligence. arXiv. 2020 doi: 10.3390/info11040193. Preprint at. [DOI] [Google Scholar]
- 39.Stéfan van der Walt J.L.S., Schönberger J.L., Boulogne F., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., Tony Yu and the scikit-image contributors scikit-image: Image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 42.Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 43.Wu K.O., Ekow, Shoshani A. 2005. Optimizing Connected Component Labeling Algorithms.https://escholarship.org/uc/item/7jg5d1zn#main [Google Scholar]
- 44.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Team, R.D. 2018. RAPIDS: Collection of Libraries for End to End GPU Data Science.https://rapids.ai [Google Scholar]
- 46.Arzt M., Deschamps J., Schmied C., Pietzsch T., Schmidt D., Tomancak P., Haase R., Jug F. LABKIT: Labeling and Segmentation Toolkit for Big Image Data. Front. Comput. Sci. 2022;4 doi: 10.3389/fcomp.2022.777728. [DOI] [Google Scholar]
- 47.Ban Z., Liu J., Cao L. Superpixel Segmentation Using Gaussian Mixture Model. IEEE Trans. Image Process. 2018;27:4105–4117. doi: 10.1109/TIP.2018.2836306. [DOI] [PubMed] [Google Scholar]
- 48.Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Cell images (.tif files) used in the application to identify abnormal cancer cells are available at Figshare https://doi.org/10.6084/m9.figshare.24187455.
-
•
The original code has been deposited in Zenodo https://doi.org/10.5281/zenodo.8372760.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.