Summary
Amphitropic proteins and peptides reversibly partition from solution to membrane, a key process that regulates their functions. Experimental approaches classically used to measure protein partitioning into lipid bilayers, such as fluorescence and circular dichroism, are hardly usable when the peptides or proteins do not exhibit significant polarity and/or conformational changes upon membrane binding. Here, we describe binding to lipid vesicles (B2LiVe), a simple, robust, and widely applicable nuclear magnetic resonance (NMR) method to determine the solution-to-membrane partitioning of unlabeled proteins or peptides. B2LiVe relies on previously described proton 1D-NMR fast-pulsing techniques. Membrane partitioning induces a large line broadening, leading to a loss of protein signals; therefore, the decrease of the NMR signal directly measures the fraction of membrane-bound protein. The method uses low polypeptide concentrations and has been validated on several membrane-interacting polypeptides, ranging from 3 to 54 kDa, with membrane vesicles of different sizes and various lipid compositions.
Keywords: amphitropic proteins, peptides, solution to membrane partitioning, protein membrane binding, affinity, partition coefficient, nuclear magnetic resonance, proton 1D-NMR fast-pulsing techniques, selective adiabatic pulses
Graphical abstract

Highlights
-
•
B2LiVe is a label-free 1D-1H-NMR method to quantify affinity of proteins for membranes
-
•
B2LiVe is based on selective excitation of amide resonances
-
•
B2LiVe is validated on several membrane-interacting peptides and proteins
-
•
B2LiVe can detect protein regions not embedded in the membrane
Motivation
Characterization of the interaction of peptides and proteins with lipid membranes is relevant for the study of various biological processes and is often challenging for polypeptides, which do not possess intrinsic fluorophores and do not exhibit significant structural content changes, as well as for those characterized by low membrane affinities. To meet these challenges, we have developed a label-free 1D-1H-NMR-based experimental approach, named B2LiVe, to measure the binding of polypeptides to lipid vesicles. B2LiVe complements the arsenal of label-free biophysical assays available to characterize protein-membrane interactions.
Sadi et al. describe a robust, label-free NMR-based experimental approach (B2LiVe) to measure the interactions of peptides or proteins with membranes. They validate the approach using several polypeptides and on various membrane vesicles. B2LiVe is an efficient and straightforward biophysical technique, complementing other label-free methods to characterize protein-membrane interactions.
Introduction
Characterization of the interaction of peptides and proteins with lipid membranes often begins with the determination of the affinity, or more appropriately, the partition coefficient, Kx.1,2 A common preliminary study consists in the identification of the lipid species as well as the lipid properties, such as lipid polymorphism, charge, and acyl chain fluidity, favoring the partitioning of proteins and peptides from solution to membrane. Moreover, knowledge of membrane affinity can be important to decipher molecular mechanisms or mutational analyses to identify regions or amino acid residues critical for membrane binding.
Several experimental approaches, such as surface plasmon resonance (SPR),3,4,5,6,7,8,9,10,11 interferometry,11,12,13,14,15 fluorescence,16,17,18,19,20,21,22,23,24 centrifugation,25,26,27 isothermal titration calorimetry (ITC),28,29,30 and circular dichroism31,32,33,34,35 are commonly used to measure the partitioning of soluble peptides and proteins into lipid bilayers. However, some peptides and proteins do not possess intrinsic fluorophores, or their secondary or tertiary structural contents do not change significantly enough to be used as a probe of their partitioning into membranes. Alternatively, they may exhibit a propensity to aggregate, complicating, if not precluding, the use of SPR, ITC, or centrifugation-based approaches. Besides, techniques based on phase separation (e.g., centrifugation or membrane flotation assays) are hardly applicable for peptides/proteins showing a low affinity for membranes.
In this article, we describe binding to lipid vesicles (B2LiVe), a simple and robust nuclear magnetic resonance (NMR)-based experimental approach to determine the solution-to-membrane partition coefficient, , of unlabeled peptides and proteins. The experimental strategy relies on proton 1D-NMR fast-pulsing techniques with selective adiabatic pulses to excite the amide resonances (HET-SOFAST) developed by Schanda et al.36 Under the low exchange regime on the chemical shift timescale (free-bound exchange rate much slower than the difference in frequency between the two states), membrane partitioning induces a large line broadening leading to a loss of peptide/protein NMR signals. The decrease of the NMR signal thus directly reports the fraction of membrane-bound peptide/protein. The validity of this approach has been established on several membrane-interacting peptides and proteins, ranging from 3 to 54 kDa, and on membrane vesicles of various sizes and different lipidic compositions. The method does not require any labeling and requires only low amounts of peptide/protein when using high-field spectrometers and sensitive probes. Our results show that the B2LiVe NMR-based method compares well with traditional fluorescence and circular dichroism (CD)-based techniques. Overall, B2LiVe should efficiently complement the arsenal of label-free biophysical techniques to characterize protein-membrane interactions.
Results
Protein binding to unilamellar vesicles of lipid bilayers induces a large line broadening in NMR signals, because of the slow tumbling rate of the particles, leading to a loss of protein signals. This observation has been exploited for characterization of the association of some 15N-labeled proteins with membranes.37,38,39 Here, we explored the possibility of quantifying membrane partitioning of unlabeled proteins by using proton 1D-NMR fast-pulsing techniques with selective excitation of amide protons using adiabatic pulses: in the slow exchange regime on the chemical shift time scale, the relative amount of protein or peptide remaining in solution at each lipid concentration should be proportional to the integral of the amide proton resonances. Hence, the decrease of the NMR signal should directly report on the fraction of protein bound to membranes.
To prove the general applicability of this approach, we investigated membrane partitioning of several proteins and peptides. Titrations were performed at constant protein and peptide concentrations and the lipid concentration was increased up to 10 mM lipids. Large and small unilamellar vesicles (LUV and SUV, respectively) of lipid bilayers were used. Protein partitioning into membranes was monitored by selective amide band 1D NMR (proton HET-SOFAST36,40) (see STAR Methods), by recording the loss of intensity of the integral of the amide proton resonances. In parallel, protein binding to membranes was measured by intrinsic tryptophan fluorescence changes (following the ratio of fluorescence intensities emitted at 320 and 370 nm as a proxy for polarity change upon membrane binding), and far-UV circular dichroism (for peptides). The partition coefficient, , which is directly related to the affinity constant, (Equation 7), is determined by fitting Equation 8 to the experimental data (see STAR Methods).
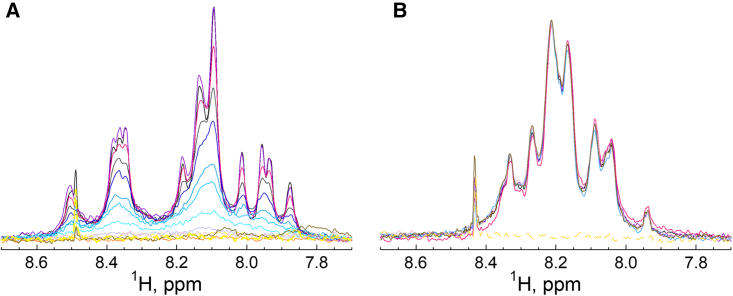
We first analyzed the partitioning of three amphitropic proteins into LUVs: apo-myoglobin (apo-Mb, 16.9 kDa),41,42 the diphtheria toxin translocation domain (T, 22 kDa),43,44 and anthrolysin O (ALO, 54 kDa).45 Figure 1 shows the 1D-NMR spectra of apo-Mb (Figure 1A), T (Figure 1B), and ALO (Figures 1C and 1D) with increasing concentrations of lipids.
Figure 1.
Membrane partitioning of amphitropic proteins monitored by 1D-1H amide-selective NMR spectroscopy
(A–D) The concentration of lipids ranges from 0 mM (black) to 2 mM (dark yellow). The spectrum of lipid vesicles in the absence of protein at the highest lipid concentration used is indicated as a black dashed line. Each protein is present at a concentration of 3 μM. Membrane partitioning and corresponding NMR spectra upon titration of apo-Mb with POPC:POPG 9:1 LUV (A), T with POPC:POPG 9:1 LUV (B), ALO with POPC:cholesterol 6:4 LUV (C), and ALO with POPC LUV (D). Membrane partitioning is followed by the disappearance of the amide envelope signal (see main text for details). The sharp signal at 8.43 ppm arises from a trace impurity of formic acid contained in D2O.
The intensity of the 1D-NMR spectra of apo-Mb and T (Figures 1A and 1B) decreases with the addition of LUV. ALO is a cholesterol-dependent cytolysin that requires the presence of cholesterol in lipid bilayers to partition from solution to membranes. Whereas the 1D-NMR spectra of ALO titrated by membranes composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and cholesterol show a loss of intensity (Figure 1C), the intensities of the 1D-NMR spectra do not significantly change for ALO titrated by LUV composed of POPC only (Figure 1D). This clearly establishes that the loss of 1D-NMR signal intensity is a straightforward indicator of ALO binding to membranes containing cholesterol.
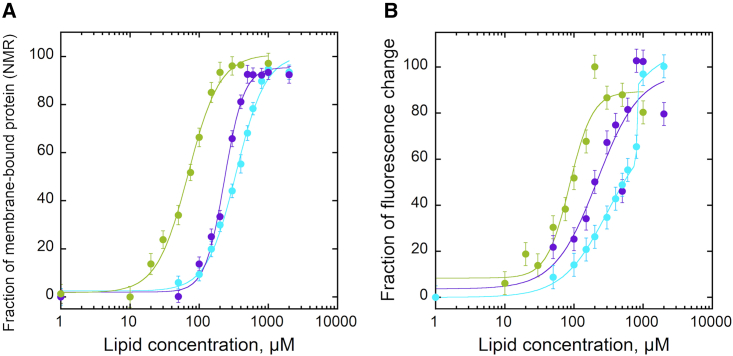
To estimate the partition coefficients of the different proteins, the decrease of the NMR signal intensity (i.e., integral of the amide proton envelope) of apo-Mb, T and ALO were plotted as a function of lipid concentrations as reported in Figure 2A. In parallel, membrane partitioning was monitored by recording the ratio of tryptophan fluorescence intensity for the same proteins as a function of lipid concentration (Figure 2B). The fluorescence data indicate that the three amphitropic proteins interact with membranes, as expected from the literature.42,45,46 Furthermore, fitting Equation 8 to 1D NMR and fluorescence experimental data provided similar partition coefficients for each protein (Table 1). To assess the reproducibility of each method, we performed independent experiments of membrane partitioning using ALO and POPC:cholesterol LUV (Figure S2). Fluorescence and NMR results show that the independent experiments provide reproducible results (see also Table S2 and Limitations of the study section below for the experimental reproducibility). Hence, as NMR and fluorescence provide similar quantitative results, we conclude that 1D-1H NMR can reliably be used to report protein partitioning into membranes.
Figure 2.
Membrane partitioning of amphitropic proteins
(A and B) Solution-to-membrane partitioning of apo-Mb (cyan), T (violet), and ALO (green) at a concentration of 3 μM each, in the presence of increasing lipid concentrations monitored by (A) The 1D-1H NMR (signal loss of the amide envelope) and (B) fluorescence (ratio of tryptophan fluorescence intensities at 320 nm and 370 nm). POPC:POPG 9:1 LUV were used for apo-Mb and T, POPC:Chol 6:4 LUV for ALO (green, closed circles) and 100% POPC LUV for ALO (green, open circles). The values and error bars in (A) are calculated from the integral of the amide envelope (8.4–7.7 ppm) and from the spectral noise standard deviation, respectively. Data have been normalized. Data in (B) are mean ± SEM. In NMR experiments, while for ALO and apo-Mb the signal completely disappears at high lipid concentrations, for the T domain at pH 4.2, circa 20% of the signal remains at the highest lipid concentration. As shown below with diffusion experiments performed at pH 4.5 with the T domain (Figure 5), this signal pertains to flexible regions of the membrane-bound proteins that are not embedded into the membrane.
Table 1.
Thermodynamics of solution-to-membrane partitioning of the amphitropic proteins and peptides determined by NMR, tryptophan fluorescence, and far-UV circular dichroism
| Proteins | Membranes | Methods | Membrane partition coefficient, Kx | Dissociation constants, KD, μM | nH | ΔGKx, kcal/mol |
|---|---|---|---|---|---|---|
| apo-Mb | POPC:POPG 9:1 LUV |
NMR | 170,000 ± 10,000 | 330 ± 10 | 2 ± 1 | −7.1 ± 0.1 |
| fluorescence | 180,000 ± 59,000 | 310 ± 50 (810 ± 5) | 2 ± 1 (9 ± 3) | −7.2 ± 0.1 | ||
| T | POPC:POPG 9:1 LUV |
NMR | 240,000 ± 11,000 | 230 ± 20 | 3 ± 1 | −7.4 ± 0.1 |
| fluorescence | 250,000 ± 100,000 | 220 ± 130 | 2 ± 1 | −7.4 ± 0.3 | ||
| ALO | POPC:chol 6:4 LUV |
NMR | 800,000 ± 54,000 | 70 ± 5 | 2 ± 1 | −8.1 ± 0.1 |
| fluorescence | 640,000 ± 94,000 | 90 ± 20 | 3 ± 1 | −7.9 ± 0.1 | ||
| P233 | POPC:POPG:chol 7:2:1 SUV |
NMR | 52,000 ± 1,000 | 1,000 ± 60 | 3 ± 1 | −6.4 ± 0.01 |
| fluorescence | 26,000 ± 10,000 | 2,100 ± 600 (4,100 ± 30) | 2 ± 1 (18 ± 3) | −6.0 ± 0.2 | ||
| Far-UV CD | 25,000 ± 2,000 | 2,200 ± 200 | 3 ± 1 | −6 ± 0.1 |
The table reports the membrane partition coefficient (Kx), dissociation constant (KD), Hill number (nH), and free energy (ΔGKx) values.
The thermodynamic values corresponding with the second transition of peptide/protein reorganization in membranes are reported in brackets because they do not report solution to membrane partitioning. For membrane partitioning, nH values have no biological meaning because protein membrane interaction corresponds to a change of environment (i.e., from solution to membrane) and not to a bimolecular interaction. Here, nH values help to provide a better fitting of the equation to the experimental data and to improve the estimation of the KD values.
To probe whether protein partitioning into SUVs, LUVs, and multilamellar vesicles (MLVs) can be quantified by NMR, we monitored membrane partitioning by tryptophan fluorescence and NMR using apo-Mb as standard amphitropic protein. Figure S3 shows that B2LiVe is appropriate for various lipid bilayer vesicles, including SUVs, LUVs, and MLVs.
As additional controls, we similarly characterized proteins that are not expected to interact with membranes of various lipid compositions: calmodulin (16.7 kDa), bovine holo-alpha-lactalbumin (14.2 kDa), and bovine serum albumin (66.5 kDa) (Figure S4). These proteins were chosen to cover a similar range of molecular masses as apo-Mb, T, and ALO. The addition of LUV does not affect the NMR signal or the tryptophan fluorescence (the fluorescence experiment could not be done for CaM, as it does not contain tryptophan), indicating that, as expected, these proteins do not interact with membranes under the experimental conditions used.47
A benefit of selective excitation of amides is that longitudinal relaxation is very fast, allowing one to apply short repetition delays such as the 150-ms delay used throughout this work and thereby increase sensitivity and reduce acquisition time. However, given that the membrane-bound protein is expected to show slower longitudinal relaxation than the protein in solution, we investigated whether using such a short delay could impact the affinity measurements. We, thus, compared experiments at 150 ms and a much longer delay of 500 ms with apo-Mb with SUV, LUV, and MLV and effectively obtained the same KD values within experimental error (Table S3).
Finally, the complementarity of the experimental approaches can be illustrated by the NMR and fluorescence results obtained with apo-Mb (Figure S5). NMR and fluorescence data indicate that the partition from solution to membrane is observed with a KD at circa 120 ± 10 μM. Indeed, the unique transition measured by NMR unambiguously indicates that the partition occurs during the first transition observed by fluorescence (common NMR and fluorescence KD values at circa 120 ± 10 μM). Then, a second transition monitored by tryptophan fluorescence is observed at circa 500 ± 50 μM, corresponding with a pH-dependent and lipid concentration-dependent conformational change of apo-Mb in membranes, as previously reported.41,42
Taken together, these results indicate that proton 1D-NMR fast-pulsing techniques with selective adiabatic pulses is a sensitive approach to quantitatively monitor protein partitioning into membranes. The decrease in the NMR signal directly reports on the fraction of membrane-bound proteins and can be easily recorded for unlabeled samples.
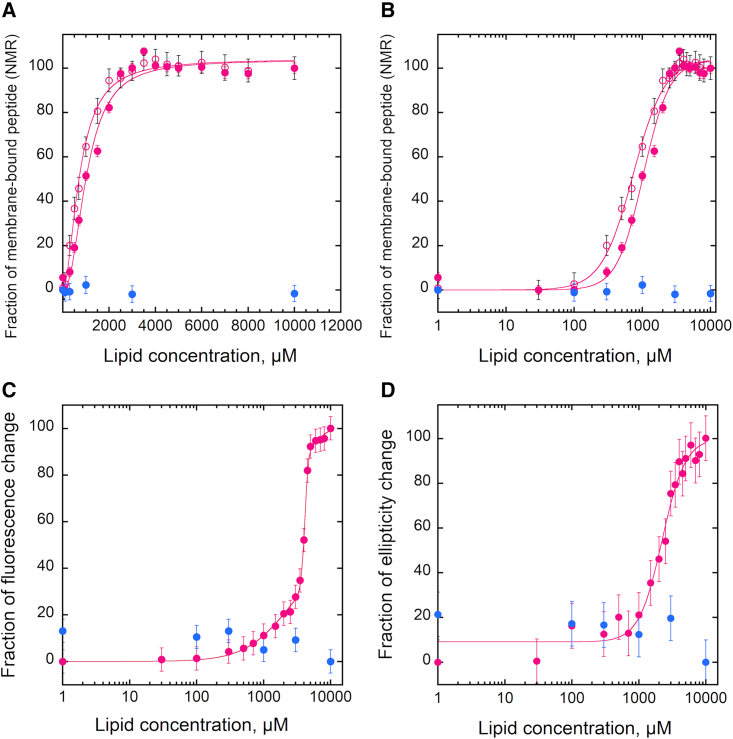
We then extended the study to peptide:membrane interactions. For this, we selected two peptides from the Bordetella pertussis adenylate cyclase toxin,48,49 which were previously characterized in our laboratory: P233 that was shown to interact with membranes and P414 that has no membrane binding activity.23 The NMR data shown in Figure 3 indicate that P233 interacts with POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol) [POPG]/cholesterol membranes with a complete loss of NMR signal at high lipid/peptide ratios, while in the same conditions the P414 peptide does not show any signal decrease and, therefore, does not bind to membranes. Because P233 was used at a high lipid/peptide molar ratio, which could induce membrane disruption, we analyzed the morphology of the peptide:SUV system at different concentrations of peptides by cryo-electron microscopy (EM) (Figure S6). Cryo-EM data provide direct evidence that the peptide does not disintegrate the lipid bilayer of the vesicles used at 100 μM and 1 mM lipids (lipid/protein ratio range at which peptide-induced vesicle disintegration might have occurred).
Figure 3.
Membrane interaction of peptides followed by 1D-1H-NMR spectroscopy
(A and B) NMR amide spectra upon titration of the P233 (A) and P414 (B) peptides with POPC:POPG:cholesterol 7:2:1 SUV. The lipid concentrations range from 0 (black) to 10 mM (dark yellow). The concentrations of the P233 and P414 peptides are 15 μM and 5 μM, respectively. The spectrum of lipid vesicles in the absence of peptides at the highest lipid concentration used is represented as a dashed line (yellow).
Membrane partitioning of P233 was monitored in parallel by NMR, tryptophan fluorescence, and far-UV circular dichroism (Figure 4). Overall, the experimental data revealed an excellent correspondence between the three different techniques that provide similar quantitative parameters (2-fold difference at most on KD, or 0.4 kCal/mol in free energy, roughly representing one-third of the energy of a hydrogen bond) for the peptide membrane partitioning process (Table 1). Hence, our data indicate that 1D-1H NMR is also readily applicable to quantify the membrane partitioning of standard, unlabeled peptides.
Figure 4.
Membrane partitioning of the P233 and P414 peptides followed by 1D-1H NMR, tryptophan fluorescence, and far-UV CD
(A–D) The experiments were performed in the presence of POPC:POPG:chol 7:2:1 SUV. Data for P233 and P414 as a function of lipid concentration are displayed in magenta and blue, respectively. Membrane partitioning followed by 1D-1H NMR is represented on a linear scale (A) and on a logarithmic scale (B). The ratio of tryptophan fluorescence intensity (320 nm/370 nm) (C) and the ellipticity at 222 ± 2 nm in the far-UV region (D) as a function of lipid concentration are shown. Error bars in (A and B) are calculated from the standard deviation of noise in 1D-NMR spectra. The signal loss in (A) and (B) of the tryptophan indol proton (open circles) followed by NMR is similar to that of the amide envelope (closed circles). Data in (C and D) are mean ± SEM.
Furthermore, the P233 peptide is unstructured in solution and can therefore be considered as a short intrinsically disordered protein (IDP). As IDPs show in general very intense and sharp NMR signals, B2LiVe should also be applicable to IDPs. Nevertheless, in unstructured peptides or IDPs, amides are solvent exposed in solution and exchange fast with water at high pH and temperature, but these might be protected against exchange in the bound form. We performed HETSOFAST amide saturation transfer experiments with water (see Materials and methods) with an unstructured 31-residue-long peptide (P454) at pH 7.4°C and 25°C to analyze whether, under these conditions, protection in the bound form could influence the apparent exchange of the visible solution state and bias the affinity measurement. Under the later conditions, the peptide shows relatively low affinity for membranes (KD = 890 ± 90 μM as determined with B2LiVe) and high intrinsic rates of exchange with water (mean = 47 s−1, half-time = 21 ms) (see legend of Figure S7) and most of its amide protons will exchange during the 100-ms exchange time of the experiment. As shown in Figure S7, the apparent exchange with water of the visible form (λwat value) is independent of the lipid concentration between 0 and 2 mM, discarding any effect of protection on the bound form.
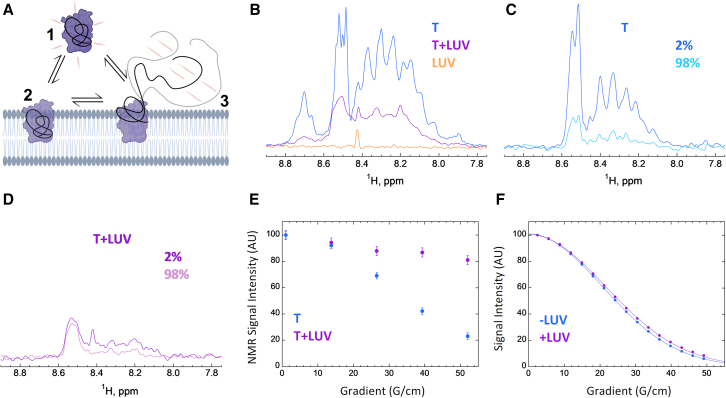
Interestingly, we noticed that, while in many cases (e.g., apo-Mb, ALO or peptide P233) no residual NMR signal is observed at saturating lipid concentrations (i.e., after reaching a plateau), a significant NMR signal remains for the T domain at the highest lipid concentrations tested. This signal might be due to either a fraction of protein unable to bind to membranes and remaining in solution, or it might arise from fully membrane-attached polypeptides containing flexible regions not directly bound to the lipids and floating above the membrane (disordered regions or ordered regions linked to the membrane-bound region[s] by a flexible linker) (Figure 5A). Of note, the T domain interaction with membrane is pH dependent: at pH 4 and below, T fully inserts into the membrane, while above pH 4, some regions remain in solution.46 To discriminate between these two possibilities, we performed NMR self-diffusion experiments as diffusion is expected to be different for proteins in solution or bound to the lipid vesicles.
Figure 5.
B2LiVe allows discrimination from partially and fully membrane-inserted proteins
For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.crmeth.2023.100624.
(A) Schematic model of three potential conformational states of an amphitropic protein exposed to membranes. The experimental results of the B2LiVe method report on the fraction of amide signal (red traces) that is lost upon partition of the protein from solution to membranes (arrows). The total loss of the NMR signal upon protein titration by membranes indicates that all proteins populate state #2 at the expense of state #1. If the NMR signal does not fully disappear and reaches an intermediate plateau, this indicates that a fraction of the proteins remains in solution. The remaining amide signal may arise either from a fraction of the population of proteins remaining in solution (state # 1) or from membrane-bound proteins that contain several segments remaining in solution (state #3). The DOSY experiment allows discrimination between these possibilities.
(B–F) Self-diffusion of the T domain in the absence and presence of LUV monitored by 1D-1H amide-selective diffusion NMR experiments.
(B) The 1D-1H amide-selective spectra of domain T (15 μM) at pH 4.5°C and 25°C in the absence (blue) or presence (violet) of LUV (15 mM concentration), and of LUV (POPC:POPG 9:1) without protein (orange). The envelope integral decays to 39% ± 3% in the presence of LUV.
(C–E) Diffusion experiments recorded with a 140 ms diffusion delay. The 1D spectra at 2% and 98% of the maximum gradient available (53.5 G/cm) of T alone (C) or in the presence of LUV (D) displayed on the same scale. (E) Diffusion rate-dependent decay of the intensity (integral) of the signal at 8.55 ppm of T with (violet) or without (blue) LUV. The much lower decrease in relative intensity of the signal in the presence of LUV indicates a slower diffusion rate.
(F) Diffusion rate-dependent decays of the HEPES signal (3.86 ppm) in samples (used in A–D) with (violet) or without (blue) lipid vesicles. Experiments in (F) were recorded with a 60-ms diffusion delay; data in (F) were fitted to a Gaussian decay of the intensity (I) from an Io value (without gradients) as a function of the gradient strength G and an apparent diffusion coefficient d (). The similarity of the diffusion decays in (F) indicates that the viscosity of the sample is not significantly increased by the LUV and is not at the origin of the slower diffusion of T in the presence of lipid vesicles shown in C, D and E. The sharp signal at 8.43 ppm arises from formic acid, a trace impurity contained in D2O.
Error bars in (E and F) are calculated from the standard deviation of noise in NMR spectra.
Diffusion experiments were performed with the T domain (15 μM) alone or in the presence of a 1,000-fold excess of lipids in LUV at pH 4.5, at which pH a substantial fraction (four among the nine helices) of the T domain remains in solution.46,50 Under these conditions, in amide-selective 1D spectra, we observed that in the presence of LUV, the signal was ∼40% (39% ± 3%) that of the protein without lipid vesicles (Figure 5B). We then submitted the samples to amide-selective diffusion experiments. In diffusion experiments in which all delays (diffusion delay, gradient pulses) are kept constant and only the gradient strength is varied, the signals show a Gaussian decay that depends on the diffusion coefficient and the applied gradient. As can be observed in Figures 5C and 5D, the relative decay on intensity of the spectra at high gradient strength is much higher for the T domain alone than that of the T domain in the presence of LUV. This can also be visualized on the diffusion curves (Figure 5E), which show the signal intensity as a function of the gradient strength. These data indicate that the diffusion coefficient of the species giving rise to the residual signal at a high lipid concentration are bound to lipid vesicles. To rule out the possibility that the slower diffusion of the T domain at a high lipid concentration could be caused by a higher viscosity resulting from the presence of LUV rather than a consequence of membrane binding, we used a buffer signal (HEPES resonance) as a control and showed that HEPES displays very similar diffusion decays with or without lipid vesicles in samples containing the T domain (Figure 5F). Hence, the residual signal pertains to membrane-bound protein, in agreement with previous results.46 In summary, in addition to quantifying membrane partitioning, the B2LiVe method as shown with diffusion-ordered spectroscopy (DOSY) can pinpoint toward the presence, within a membrane-bound protein, of polypeptide segments that are not embedded into membrane, but remain in the aqueous phase.
High concentrations of lipids occupy modest volumes and will not lead to crowding effects that could influence NMR measurements. For instance, 20 mM lipid SUV with a 30-nm diameter occupy 2% of the volume, while 200-nm LUV occupy only 1% of the volume. However, a high increase of viscosity with lipids would produce higher T1 and lower T2 relaxation times and possibly bias the NMR measurements. As observed for the T domain with 1.5 mM lipids in LUV, DOSY experiments for apo-Mb with SUV, LUV, and MLV (4 mM lipids) and ALO with LUV (1 mM lipids), high lipid concentrations with different compositions displayed a small increase in viscosity, from approximately 2% to approximately 11% (Table S4).
Range of affinities that can be evaluated by B2LiVe
Different considerations will limit the lower and higher KD values that can be determined by this method. For high-affinity systems, the limit will be given by the sensitivity. Indeed, as for any other biophysical direct method not relying on competition experiments, the protein/peptide concentration should be in the range of the KD or lower and hence a good signal to noise will be required: this will mainly depend on the spectrometer field, the probe sensitivity, the shape of the spectra (sharp or broad signals), and the size of the peptide or protein. In an 800 MHz equipped with a cryogenically cooled probe, using ALO at 0.25 μM, we were able to obtain a spectrum with a good signal to noise ratio in only 12 min (as used for 3 μM proteins herein) (Figure S8). By a rule of thumb, this means that a spectrum with similar quality could be obtained at 0.1 μM in 1 h, and that KDs of 10–50 nM could be determined, and even lower if a higher field were available. For short peptides the limit will be higher but could be micromolar or sub-micromolar.
Establishing the higher limit of measurable KDs is more complicated: it will be imposed by the slow exchange regime on the chemical shift timescale condition, which depends on many factors and is related to kinetics and not to thermodynamics (KD). Indeed, the method relies on integration of signals to determine the population of protein in solution and hence is only valid for systems in slow exchange, i.e., in which the sum (kex) of the on (kon × [lipids]) and off (koff) rates (kex = kon × [lipids] + koff) is much slower than the difference of resonance frequencies (expressed in Hertz) of the visible and bound forms. Thus, the kex rate will depend on the spectrometer field, on the lipid concentration dependent kinetics of the interaction, and on the difference of the bound and free environment of each amide. Nevertheless, we have shown here that B2LiVe can successfully measure KDs in the millimolar range. Moreover, knowing whether the system is in slow exchange or in the faster intermediate exchange regime can be readily detected on the variation of the shape of the spectra (line broadening) upon addition of lipids.
Discussion
We have shown that 1D-1H NMR with selective excitation of the amide region is a simple and robust approach to quantify membrane partitioning of unlabeled peptides or proteins. Selective excitation of the amide region allows one to efficiently filter-out the proton signals of water (111 M), buffers (tenths of millimolar range), and lipid vesicles (from 0 to 10 mM in this study), as well as most of the impurities that are usually observed at lower frequencies, without the need for 15N-labeling the proteins. In addition, the fast relaxation of the amide protons in the HET-SOFAST experiment36 warrants the use of short inter-scan repetition delays leading to high sensitivity. With high-field spectrometers and high-sensitivity, cryogenically cooled probes, data acquisition on low protein concentration samples can indeed be very short. In the experiments presented here, run on an 800-MHz spectrometer equipped with a cold probe, 1 data point was recorded with 2,048 scans in approximately 12 min for 3 μM protein in 180 μL samples.
The B2LiVe method requires low concentrations and low amounts of unlabeled proteins, is independent of the protein/peptide composition, and can be applied to a large range of protein sizes, which can go well above the range of the protein sizes shown in this study (>54 kDa, unpublished data). Importantly, the capacity to perform the experiments at low protein concentrations and high lipid/protein ratios, ensures the conditions of protein infinite dilution1,51 required to determine the thermodynamic partitioning constants (Equation 2). In contrast, given that the determination of the membrane-bound fraction is based on the disappearance of the solution protein signals, which is due to the large size of the lipid vesicles, the method is independent of the lipid bilayer particle size (SUV, LUV, or MLV) and is not negatively affected by high lipid concentration, which can cause light scattering problems in optical techniques like fluorescence and CD. This tolerance to particle size and high concentration allows the technique to quantify the solution-to-membrane partitioning for systems with low affinity. Although the range of KD values that can be accessed will depend on particular systems, NMR spectrometers, and the sensitivity of the probes available, we have shown here that the method might be applied for systems with KD values ranging from millimolar to micromolar/sub-micromolar for short peptides and tenths of nanomolar for proteins with good quality spectra and high NMR fields.
Furthermore, as we showed here for the T domain, and in contrast with other techniques, NMR can directly indicate if certain regions from a membrane-bound protein remain in the aqueous phase. In its molten globule state at acidic pH, the T domain is able to penetrate membranes on a pH dependent manner.43 Here, we showed that at pH 4.5, approximately 40% of the membrane-bound protein residues (39% ± 3% residual amide signal) are not directly attached to the membranes and remain flexible enough in solution so that their NMR signals can be observed. Previous works on the T domain, as well as with peptides encompassing its four N-terminal helices (TH1–TH4), indicate that under the conditions used in this study (pH 4.5, POPG:POPC 9:1 LUV), the N-terminal region does not interact with membranes46,50; membrane insertion of the N-terminal helices of T is only observed for more acidic pH (pH ≤ 4).43,50 Remarkably, this N-terminal amphiphilic region corresponds with approximately 40% of the residues of the T domain, in close agreement with our NMR results (39% ± 3% residual amide signal).
In conclusion, the proposed NMR-based B2LiVe method should be valuable to quantify the affinity of unlabeled peptides and proteins including IDPs for lipid bilayers in the context of structural biology and biophysical studies.
Limitations of the study
Protein/peptide aggregation
The B2LiVe method may be difficult to apply for proteins or peptides that have a strong propensity to aggregate. Nevertheless, for such peptides/proteins, other classical approaches such as SPR, ITC, and reflectometry-based or centrifugation-based partitioning assays will also be severely impeded and only membrane flotation assays might potentially be applicable to demonstrate their membrane-binding capacity. It should be stressed, however, that because B2LiVe can be carried out at high lipid:polypeptide molar ratios (i.e., in the low micromolar range for proteins and in the tens of millimolar range for lipids), the aggregation propensity of most proteins should be rather limited in such conditions.
Intermediate exchange regime
As stated, the quantification of populations by signal integration is only valid for systems in slow exchange on the chemical shift timescale. For systems with very low affinity, kex will be higher and, if it is of the same order of magnitude as the difference of frequencies between the solution and membrane-bound forms (approximately μs to ms time range), the system might be in intermediate exchange that will result in line broadening and possibly chemical shift variations of the visible form. Importantly, line broadening can be easily detected by visual inspection of the NMR spectra. We verified line broadening for all the titrations performed in this work, and we only noticed significant broadening for peptide P233 at concentrations of 800 μM lipid and higher and NMR measurements showed good agreement with fluorescence (Table 1).
If line broadening is present, especially at low lipid concentrations, the method will overestimate the affinity. In such cases, the B2LiVe method will still provide a good estimation of the affinity range (i.e., order of magnitude of KD). Moreover, it will still be useful for performing most comparative studies (e.g., effects of mutations, different lipid compositions), as line broadening will be associated with systems with higher exchange rates and in principle lower affinity. Also, if available, lower fields might switch the system from intermediate to slow exchange.
Amide proton exchange with water
An advantage of the HETSOFAST pulse program is that water is not suppressed or manipulated, and, if care is taken to not excite water, reproducible results are obtained. However, exchange with water at a high temperature (37°C) and pH in unstructured peptides and IDPs will likely result in broad lines (and absent signals) especially for basic, serine, and threonine residues. If the quality of the spectrum (broad lines, low intensity) precludes experiments at 37°C, these can be performed at a lower temperature. Indeed, we showed here for P233 and P454 that experiments are feasible at pH 7.4°C and 25°C and for P454 we further demonstrated that no effect of differential water accessibility in the soluble and bound forms affected the signal of the soluble form.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | Cat#850457C |
| 1-palmitoyl- 2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] | Avanti Polar Lipids | Cat#840457C |
| cholesterol | Avanti Polar Lipids | Cat#700000P |
| Deuterium oxide (D2O) | Eurisotop | Cat#D214 |
| The P233 peptide | Genosphere Biotechnologies | P233 |
| The P414 peptide | Genosphere Biotechnologies | P414 |
| The P454 peptide | Genosphere Biotechnologies | P454 |
| The P454FGR peptide | Genosphere Biotechnologies | P454FGR |
| The bovine serum albumin protein | Sigma-Aldrich | Cat#A0281 |
| The bovine α-lactalbumin | Sigma-Aldrich | Cat#L-6010 |
| The apo-myoglobin protein | Sigma-Aldrich | Cat#A8673-5X1VL |
| The Anthrolysin O protein | Bourdeau et al.45 | N/A |
| The Diphtheria Toxin Translocation Domain | Chenal et al.46 | N/A |
| Calmodulin | Karst et al.13 | N/A |
| Software and Algorithms | ||
| KaleidaGraph v5.01 | Synergy Software | https://www.synergy.com/ |
| NMRLIB 2.0 package | Vallet et al.52 | https://www.ibs.fr |
| Topspin 4.0.7 | Bruker | https://www.bruker.com/fr/products-and-solutions/mr/nmr-software/topspin.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, J. Iñaki Guijarro (inaki.guijarro@pasteur.fr).
Materials availability
This study did not generate new unique reagents.
Method details
Reagents
The lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, ref. 850457C), 1-palmitoyl- 2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG, ref. 840457C) and cholesterol (Chol, ref. 700000P) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). HEPES-Na (reference H9897), NaCl (reference S5150) and CaCl2 (ref. 21115) were purchased from Sigma Aldrich, USA. The D2O (reference D214) was purchased from Eurisotop, England.
Peptides
The synthetic peptides were purchased as powder from Genosphere Biotech (France) and their purity (95%) and composition were controlled by reverse-phase HPLC and MALDI-mass spectrometry, respectively. The peptides are capped on the N terminus with an acetyl group and on the C terminus with an amide group. The P233 and P414 peptides used in this study were derived from the CyaA protein.48,49,53 The P233 peptide23,54 corresponds to the H-helix of ACD (adenyl-cyclase domain), i.e., residues 233–254 of the CyaA toxin.50,54,55 The P233 peptide has the following sequence: LDRERIDLLWKIARAGARSAVG. The P414 peptide is derived from the segment 414–440 of CyaA23 and contains a F415W mutation. The P414 peptide has the following sequence: SWSLGEVSDMAAVEAAELEMTRQVLHA. The P454 peptide is derived from the segment 454–484 of CyaA.23,45,46,50,56 The P454 peptide has the following sequence: ASAHWGQRALQGAQAVAAAQRLVHAIALMTQ. The P454FGR peptide is derived from the segment 454–487 of CyaA.23 The FGR residues correspond to a native C-terminal extension of the CyaA sequence added on the previously characterized P454 peptide.23,45,46,50,56 The P454FGR peptide has the following sequence: ASAHWGQRALQGAQAVAAAQRLVHAIALMTQFGR. All peptides are synthetized with a C-terminal amide. Peptide powders were resuspended in 20 mM HEPES-Na, 150 mM NaCl, pH 7.4. Peptide concentrations were calculated from the absorbance at 280 nm subtracted by the absorbance at 320 nm of absorbance spectra recorded with a Jasco V630 spectrophotometer using Hellma cuvettes. Aliquots of the dissolved peptides were stored at −20°C.
Proteins
Bovine serum albumin (BSA, reference A0281), bovine α-lactalbumin (BLA, reference L-6010) and apo-myoglobin (apo-Mb, reference A8673-5X1VL) were purchased as powders from Sigma-Aldrich, USA. BSA was resuspended in H2O, BLA was resuspended in 20 mM HEPES-Na, 20 mM NaCl, 2 mM EDTA, pH 7.5 and apo-Mb was resuspended in 20 mM HEPES-Na, 20 mM NaCl, pH 7.5. After resuspension, BSA, BLA and apo-Mb were filtered through 0.2 μm syringe filters (reference F2504-8, Thermofisher Scientific). Anthrolysin O (ALO) production and purification were performed as described in.45 The diphtheria toxin translocation domain (T)56 was produced and purified as described in.46,50 The calcium-free apo-state of calmodulin, apo-CaM, was produced and purified as described elsewhere.13,55 The physical-chemical parameters of the proteins and peptides used in this study are reported in Table S1.
Lipid vesicle preparation
Multilamellar vesicles (MLV), large unilamellar vesicles (LUV) and small unilamellar vesicles (SUV) were used in this study. Lipid vesicles were prepared by reverse phase evaporation as described elsewhere57,58,59,60 at a lipid concentration of 40 mM. MLV produced by reverse phase evaporation were submitted to extrusion through 1.2 μm polycarbonate filters. LUV were prepared by further extrusion through 0.4 and 0.2 μm polycarbonate filters. The SUV were obtained by sonication of filtered MLV. The hydrodynamic diameters and dispersity were checked by dynamic light scattering (DLS) using a NanoZS instrument (Malvern Instruments, Orsay, France). Lipid vesicles were aliquoted and stored under argon at 4°C. Various lipid compositions were used: POPC:POPG at a 8:2 M ratio; POPC:POPG at a 9:1 M ratio, both in 20 mM HEPES-Na, 20 mM NaCl, pH 4; POPC:cholesterol at a 6:4 M ratio in 20 mM HEPES-Na, 20 mM NaCl, pH 7; POPC:POPG:cholesterol at a 7:2:1 ratio in 20 mM HEPES-Na, 150 mM NaCl, pH 7.
Sample preparation
The samples used for NMR, tryptophan fluorescence and far-UV CD were derived from a common stock solution to ensure the comparability of the three methods, when applicable. A typical titration series comprises 19 samples. Protein (1–3 μM) and peptide (5–15 μM) concentrations are kept constants for all samples, while the lipid concentration increases from 0 up to 10 mM. Apo-Mb and T titrations were performed at 25°C in the presence of LUV composed of POPC:POPG at a molar ratio of 9:1 in 20 mM HEPES-Na, 20 mM NaCl, 5% D2O, pH 4.2. The titrations of Apo-Mb by SUV, LUV and MLV composed of POPC:POPG at a molar ratio of 8:2 were performed at 25°C in 20 mM HEPES-Na, 20 mM NaCl, 5% D2O, pH 4.2. The titrations of ALO were performed in the presence of POPC:cholesterol 6:4 LUV in 20 mM HEPES-Na, 20 mM NaCl, 2 mM CaCl2, 5% D2O, pH 7.5 at 37°C. The titrations of the P233 and P414 peptides were performed at 25°C in the presence of POPC:POPG:cholesterol 7:2:1 SUV in 20 mM HEPES-Na, 150 mM NaCl, 2 mM CaCl2, 5% D2O, pH 7.5.
Nuclear magnetic resonance
NMR experiments were performed on an Avance Neo 800 MHz spectrometer (Bruker, Billerica, MA, USA) equipped with a triple resonance cryogenically cooled probe. Mono-dimensional (1D) proton experiments with selective excitation of the amide region40 of peptide (5–15 μM) and protein (1–3 μM) samples in the presence of varying lipid concentrations were recorded at 25°C or 37°C as indicated. Experiments were based on the proton HET-SOFAST pulse sequence36 implemented in the NMRLIB 2.0 package.52 Selective adiabatic polychromatic PC9 pulses with a 120° pulse centered at 9.5 ppm with a 3–4 ppm bandwidth were used to excite amide resonances. The recycling time between scans was 0.15 ms. Spectra were processed and analyzed with Topspin 4.0.7 (Bruker). The relative amount of peptide/protein remaining in solution at each lipid concentration was evaluated from the integral of the amide proton resonances, after subtraction of the spectrum of lipids at the highest concentration used, considered as the baseline.
Self-diffusion experiments of the T domain (15 μM) at pH 4.5 were performed in the presence or absence of 15 mM LUV (POPC:POPG 9:1) at 25°C by means of the amide selective self-diffusion experiment 1D_DOSY (Diffusion Ordered SpectroscopY) implemented in the NMRLIB 2.0 package.52 We used a diffusion delay of 140 ms, bipolar shaped gradients applied during 3.7 ms and 5 gradient strengths that varied from 2 to 98% of the probe maximum gradient (53.5 G/cm). The selective pulses were centered at 9.5 and had a 3.1-ppm width. Water was suppressed with an excitation sculpting scheme incorporated in the 1D_DOSY scheme. To evaluate the effect of the different viscosity of T samples ±15 mM LUV on diffusion, we followed the signal of HEPES at 3.89 ppm contained in the buffer in non-selective experiments. We recorded 16 stimulated-echo experiments with a diffusion delay of 60 ms, bipolar gradients applied during 1.6 ms at different gradient strength and excitation sculpting water suppression. The same strategy was followed to evaluate the viscosity of protein samples with different size, lipid composition and concentration (highest concentration used in the titrations), using samples of apo-Mb ± 4 mM SUV, LUV or MLV and ALO ±1 mM LUV.
Apparent solvent exposure of amide protons of the population in solution at different lipid concentrations was evaluated using the proton HETSOFAST experiment for p454 as described by Schanda et col.36 The λwat ratio, defined as the intensity ratio between a spectrum with water inversion (Iwat) and the reference spectrum (Iref), was determined using a 100 ms selective water inversion pulse. The mean of the per residue intrinsic exchange rate with water of P454 fully exposed amide protons at pH 7.4°C and 25°C was calculated according to Bai et al..61
Intrinsic tryptophan fluorescence
Measurements were performed at 25 or 37°C with an FP750 spectrofluorimeter (Jasco, Japan) equipped with a Peltier-thermostated cell holder, using a quartz cell (ref. 105.251-QS, 3 × 3 mm pathlength) from Hellma (France). The peptide tryptophan emission spectra were recorded from 300 to 400 nm (excitation at 280 nm at a scan rate of 100 nm/min). A bandwidth of 5 nm was used for both, excitation and emission beams. The ratio of fluorescence intensities at 320 and 370 nm extracted from the fluorescence emission spectra was used to monitor solution to membrane partitioning. Fluorescence spectra are shown in Figure S1 to assess the quality of the fluorescence experimental data.
Far-UV circular dichroism
Far-UV CD measurements were performed with a J-1500 circular dichroism spectrophotometer (Jasco, Japan) equipped with a Peltier-thermostated cell holder, using a quartz cell (ref. 101-QS, 1 mm pathlength) from Hellma (France). Spectra were acquired from 190 to 260 nm and subtracted from blank measurements. Far-UV CD spectra are shown in Figure S1 to assess the quality of the CD experimental data.
Determination of the partition coefficient Kx and of the free energy of partitioning ΔGKx
The solution to membrane partitioning, , was determined by analysis of the fluorescence intensity ratios at 320/370 nm, of 1D-NMR data and when applicable of CD data. The value of is defined as the ratio of peptide/protein concentrations in membrane and solution phases,1,23,51,58 given by Equation 1:
| (Equation 1) |
(protein in aqueous phase) and (protein in lipid phase) denote the concentration of soluble and membrane-bound protein, respectively; stands for total protein ( = + ); W is the concentration of water (55.55 M); L is the concentration of lipid. Since L ≫ and W ≫ , Equation 1 can be written as follows:
| (Equation 2) |
Equation 2 can, in turn, be expressed as follows:
| (Equation 3) |
The peptide fraction f(PL) partitioned into membrane is described by the following equation:
| (Equation 4) |
Based on Equations 3 and 4, Equation 5 can be expressed as follows:
| (Equation 5) |
Adding the experimental data offset (A), the amplitude of the experimental signal (B) and the Hill coefficient (n) for the interaction to Equation 5, leads to Equation 6, which is used to fit all parameters to the experimental data:
| (Equation 6) |
Equation 6 was fitted to the experimental data with KaleidaGraph (Synergy Software, Reading, USA). The constant can be expressed in terms of the dissociation constant of the peptide-membrane interaction and the water concentration:
| (Equation 7) |
From Equation 7, Equation 6 rewrites:
| (Equation 8) |
Finally, the free energy of solution to membrane partitioning was determined according to Equation 9:
| (Equation 9) |
Quantification and statistical analysis
Data in NMR experiments were measured from the integrals of the spectral regions of interest. Integral errors were estimated from the spectral noise standard deviation summed over an equivalent region. To calculate the error (Δλwat) in λwat, integral errors (ΔI) were further propagated assuming a normal distribution of noise [(Δλwat/λwat)2 = (ΔIref/Iref)2 + (ΔIwat/Iwat)2].
Membrane partitioning data in fluorescence experiments were obtained from fluorescence intensity ratios at 320/370 nm or for comparison purpose in selected experiments from intensities at fixed emission wavelengths. For CD, the mean residual ellipticity at 222 nm in the far-UV region was used.
Thermodynamic parameters (Kx, KD and nH) from NMR, fluorescence and CD data were obtained from fits of Equation 6 (Kx) and 8 (KD and nH) to experimental data using KaleidaGraph 5.01 (Synergy Software, Reading, USA). The free energy of membrane partitioning ΔGKx was obtained from Equation 9 using the fitted parameters. Errors in thermodynamic parameters correspond to the error of the fit.
Reproducibility of NMR and fluorescence partitioning experiments were assessed with independent repetition experiments with the protein ALO (see Figure S2). Values and errors in thermodynamic parameters were estimated from the mean and standard deviations, respectively.
Acknowledgments
M.S. was supported by the Pasteur - Paris University (PPU) International PhD Program. N.C. was supported by Institut Pasteur (DARRI-Emergence S-PI15006-12B). The 800-MHz NMR spectrometer of the Institut Pasteur was partially funded by the Région Ile de France (SESAME 2014 NMRCHR grant no. 4014526). We also acknowledge funding from the Agence Nationale de la Recherche (ANR 21-CE11-0014-01-TransCyaA), the CNRS (UMR 3528), and the Institut Pasteur (PTR 166-19, DARRI-Emergence S-PI15006-12B, PPUIP program). We thank W.J. Tang (University of Chicago) for the kind gift of purified ALO and Bruno Baron and the PFBMI platform at Institut Pasteur for assistance in the performance of the far-UV CD experiments. The funders have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization, J.I.G. and A.C.; methodology, J.I.G. and A.C.; software, B.V. and J.I.G.; calidation, B.V., D.L., J.I.G., and A.C.; formal analysis, M.S., N.C., C.L., B.V., J.I.G., and A.C.; investigation, M.S., N.C., C.L., B.V., and J.I.G.; resources, D.L., J.I.G., and A.C.; writing – original draft, M.S., D.L., J.I.G., and A.C.; visualization, M.S., N.C., C.L., J.I.G., and A.C.; supervision, J.I.G. and A.C.; project administration, J.I.G. and A.C.; funding acquisition, J.I.G. and A.C.
Declaration of interests
The authors declare no competing interests.
Published: October 30, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100624.
Contributor Information
J. Iñaki Guijarro, Email: inaki.guijarro@pasteur.fr.
Alexandre Chenal, Email: alexandre.chenal@pasteur.fr.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.White S.H., Wimley W.C., Ladokhin A.S., Hristova K. Protein folding in membranes: determining energetics of peptide-bilayer interactions. Methods Enzymol. 1998;295:62–87. doi: 10.1016/s0076-6879(98)95035-2. [DOI] [PubMed] [Google Scholar]
- 2.Wieprecht T., Apostolov O., Beyermann M., Seelig J. Membrane binding and pore formation of the antibacterial peptide PGLa: thermodynamic and mechanistic aspects. Biochemistry. 2000;39:442–452. doi: 10.1021/bi992146k. [DOI] [PubMed] [Google Scholar]
- 3.Salamon Z., Macleod H.A., Tollin G. Surface plasmon resonance spectroscopy as a tool for investigating the biochemical and biophysical of membrane protein systems. I: theoretical principles. Biochim. Biophys. Acta. 1997;1331:117–129. doi: 10.1016/s0304-4157(97)00004-x. [DOI] [PubMed] [Google Scholar]
- 4.Reimhult E., Larsson C., Kasemo B., Höök F. Simultaneous surface plasmon resonance and quartz crystal microbalance with dissipation monitoring measurements of biomolecular adsorption events involving structural transformations and variations in coupled water. Anal. Chem. 2004;76:7211–7220. doi: 10.1021/ac0492970. [DOI] [PubMed] [Google Scholar]
- 5.Besenicar M., Macek P., Lakey J.H., Anderluh G. Surface plasmon resonance in protein-membrane interactions. Chem. Phys. Lipids. 2006;141:169–178. doi: 10.1016/j.chemphyslip.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Wiltschi B., Knoll W., Sinner E.K. Binding assays with artificial tethered membranes using surface plasmon resonance. Methods. 2006;39:134–146. doi: 10.1016/j.ymeth.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Lambertz C., Martos A., Henkel A., Neiser A., Kliesch T.T., Janshoff A., Schwille P., Sönnichsen C. Single Particle Plasmon Sensors as Label-Free Technique To Monitor MinDE Protein Wave Propagation on Membranes. Nano Lett. 2016;16:3540–3544. doi: 10.1021/acs.nanolett.6b00507. [DOI] [PubMed] [Google Scholar]
- 8.Ahijado-Guzmán R., Menten J., Prasad J., Lambertz C., Rivas G., Sönnichsen C. Plasmonic Nanosensors for the Determination of Drug Effectiveness on Membrane Receptors. ACS Appl. Mater. Interfaces. 2017;9:218–223. doi: 10.1021/acsami.6b14013. [DOI] [PubMed] [Google Scholar]
- 9.Rascol E., Villette S., Harté E., Alves I.D. Plasmon Waveguide Resonance: Principles, Applications and Historical Perspectives on Instrument Development. Molecules. 2021;26:6442. doi: 10.3390/molecules26216442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson J., Bilotto P., Mears L.L.E., Fossati S., Ramach U., Köper I., Valtiner M., Knoll W. Solid-supported lipid bilayers - A versatile tool for the structural and functional characterization of membrane proteins. Methods. 2020;180:56–68. doi: 10.1016/j.ymeth.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Lee T.-H., Hirst D.J., Kulkarni K., Del Borgo M.P., Aguilar M.-I. Exploring Molecular-Biomembrane Interactions with Surface Plasmon Resonance and Dual Polarization Interferometry Technology: Expanding the Spotlight onto Biomembrane Structure. Chem. Rev. 2018;118:5392–5487. doi: 10.1021/acs.chemrev.7b00729. [DOI] [PubMed] [Google Scholar]
- 12.Cross G.H., Freeman N.J., Swann M.J. Handbook of biosensors and biochips. John Wiley and sons; 2007. Dual polarization interferometry: a real-time optical technique for measuring (bio) molecular orientation, structure and function at the solid/liquid interface. [Google Scholar]
- 13.Karst J.C., Barker R., Devi U., Swann M.J., Davi M., Roser S.J., Ladant D., Chenal A. Identification of a region that assists membrane insertion and translocation of the catalytic domain of Bordetella pertussis CyaA toxin. J. Biol. Chem. 2012;287:9200–9212. doi: 10.1074/jbc.M111.316166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallner J., Lhota G., Jeschek D., Mader A., Vorauer-Uhl K. Application of Bio-Layer Interferometry for the analysis of protein/liposome interactions. J. Pharm. Biomed. Anal. 2013;72:150–154. doi: 10.1016/j.jpba.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Wallner J., Lhota G., Schosserer M., Vorauer-Uhl K. An approach for liposome immobilization using sterically stabilized micelles (SSMs) as a precursor for bio-layer interferometry-based interaction studies. Colloids Surf. B Biointerfaces. 2017;154:186–194. doi: 10.1016/j.colsurfb.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Sinner E.K., Reuning U., Kök F.N., Saccà B., Moroder L., Knoll W., Oesterhelt D. Incorporation of integrins into artificial planar lipid membranes: characterization by plasmon-enhanced fluorescence spectroscopy. Anal. Biochem. 2004;333:216–224. doi: 10.1016/j.ab.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Caesar C.E.B., Esbjörner E.K., Lincoln P., Nordén B. Membrane interactions of cell-penetrating peptides probed by tryptophan fluorescence and dichroism techniques: correlations of structure to cellular uptake. Biochemistry. 2006;45:7682–7692. doi: 10.1021/bi052095t. [DOI] [PubMed] [Google Scholar]
- 18.Tiriveedhi V., Butko P. A fluorescence spectroscopy study on the interactions of the TAT-PTD peptide with model lipid membranes. Biochemistry. 2007;46:3888–3895. doi: 10.1021/bi602527t. [DOI] [PubMed] [Google Scholar]
- 19.Song H., Ritz S., Knoll W., Sinner E.K. Conformation and topology of amyloid beta-protein adsorbed on a tethered artificial membrane probed by surface plasmon field-enhanced fluorescence spectroscopy. J. Struct. Biol. 2009;168:117–124. doi: 10.1016/j.jsb.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Bocchinfuso G., Bobone S., Mazzuca C., Palleschi A., Stella L. Fluorescence spectroscopy and molecular dynamics simulations in studies on the mechanism of membrane destabilization by antimicrobial peptides. Cell. Mol. Life Sci. 2011;68:2281–2301. doi: 10.1007/s00018-011-0719-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiantia S., Klymchenko A.S., London E. A novel leaflet-selective fluorescence labeling technique reveals differences between inner and outer leaflets at high bilayer curvature. Biochim. Biophys. Acta. 2012;1818:1284–1290. doi: 10.1016/j.bbamem.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Cherry M.A., Higgins S.K., Melroy H., Lee H.-S., Pokorny A. Peptides with the same composition, hydrophobicity, and hydrophobic moment bind to phospholipid bilayers with different affinities. J. Phys. Chem. B. 2014;118:12462–12470. doi: 10.1021/jp507289w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voegele A., Subrini O., Sapay N., Ladant D., Chenal A. Membrane-Active Properties of an Amphitropic Peptide from the CyaA Toxin Translocation Region. Toxins. 2017;9:369. doi: 10.3390/toxins9110369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasquez-Montes V., Goldberg A.F.X., Thévenin D., Ladokhin A.S. Ca2+ and Mg2+ Influence the Thermodynamics of Peptide-Membrane Interactions. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2022.167826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tortorella D., Ulbrandt N.D., London E. Simple centrifugation method for efficient pelleting of both small and large unilamellar vesicles that allows convenient measurement of protein binding. Biochemistry. 1993;32:9181–9188. doi: 10.1021/bi00086a025. [DOI] [PubMed] [Google Scholar]
- 26.Lundqvist A., Lundahl P. Biomembrane-affinity centrifugal analyses of solute interactions with membrane proteins. J. Chromatogr. A. 1999;852:93–96. doi: 10.1016/s0021-9673(99)00261-7. [DOI] [PubMed] [Google Scholar]
- 27.Xi A., Bothun G.D. Centrifugation-based assay for examining nanoparticle–lipid membrane binding and disruption. Analyst. 2014;139:973–981. doi: 10.1039/c3an01601c. [DOI] [PubMed] [Google Scholar]
- 28.Situ A.J., Schmidt T., Mazumder P., Ulmer T.S. Characterization of membrane protein interactions by isothermal titration calorimetry. J. Mol. Biol. 2014;426:3670–3680. doi: 10.1016/j.jmb.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 29.Housden N.G., Kleanthous C. Thermodynamic dissection of colicin interactions. Methods Enzymol. 2011;488:123–145. doi: 10.1016/B978-0-12-381268-1.00006-9. [DOI] [PubMed] [Google Scholar]
- 30.Al-Kaddah S., Reder-Christ K., Klocek G., Wiedemann I., Brunschweiger M., Bendas G. Analysis of membrane interactions of antibiotic peptides using ITC and biosensor measurements. Biophys. Chem. 2010;152:145–152. doi: 10.1016/j.bpc.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Errico S., Ramshini H., Capitini C., Canale C., Spaziano M., Barbut D., Calamai M., Zasloff M., Oropesa-Nuñez R., Vendruscolo M., Chiti F. Quantitative Measurement of the Affinity of Toxic and Nontoxic Misfolded Protein Oligomers for Lipid Bilayers and of its Modulation by Lipid Composition and Trodusquemine. ACS Chem. Neurosci. 2021;12:3189–3202. doi: 10.1021/acschemneuro.1c00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klocek G., Schulthess T., Shai Y., Seelig J. Thermodynamics of melittin binding to lipid bilayers. Aggregation and pore formation. Biochemistry. 2009;48:2586–2596. doi: 10.1021/bi802127h. [DOI] [PubMed] [Google Scholar]
- 33.Tang T.-X., Jo A., Deng J., Ellena J.F., Lazar I.M., Davis R.M., Capelluto D.G.S. Structural, thermodynamic, and phosphatidylinositol 3-phosphate binding properties of Phafin2. Protein Sci. 2017;26:814–823. doi: 10.1002/pro.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohgita T., Takechi-Haraya Y., Okada K., Matsui S., Takeuchi M., Saito C., Nishitsuji K., Uchimura K., Kawano R., Hasegawa K., et al. Enhancement of direct membrane penetration of arginine-rich peptides by polyproline II helix structure. Biochim. Biophys. Acta Biomembr. 2020;1862 doi: 10.1016/j.bbamem.2020.183403. [DOI] [PubMed] [Google Scholar]
- 35.Sparks R.P., Lawless W., Arango A.S., Tajkhorshid E., Fratti R.A. Use of Microscale Thermophoresis to Measure Protein-Lipid Interactions. J. Vis. Exp. 2022 doi: 10.3791/60607. [DOI] [PubMed] [Google Scholar]
- 36.Schanda P., Forge V., Brutscher B. HET-SOFAST NMR for fast detection of structural compactness and heterogeneity along polypeptide chains. Magn. Reson. Chem. 2006;44:S177–S184. doi: 10.1002/mrc.1825. [DOI] [PubMed] [Google Scholar]
- 37.Ceccon A., D’Onofrio M., Zanzoni S., Longo D.L., Aime S., Molinari H., Assfalg M. NMR investigation of the equilibrium partitioning of a water-soluble bile salt protein carrier to phospholipid vesicles: NMR Study of BABP Binding to Lipid Vesicles. Proteins. 2013;81:1776–1791. doi: 10.1002/prot.24329. [DOI] [PubMed] [Google Scholar]
- 38.Sandin S.I., Gravano D.M., Randolph C.J., Sharma M., de Alba E. Engineering of Saposin C Protein Chimeras for Enhanced Cytotoxicity and Optimized Liposome Binding Capability. Pharmaceutics. 2021;13:583. doi: 10.3390/pharmaceutics13040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandin S.I., de Alba E. Quantitative Studies on the Interaction between Saposin-like Proteins and Synthetic Lipid Membranes. MPs. 2022;5:19. doi: 10.3390/mps5010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schanda P., Brutscher B. Very Fast Two-Dimensional NMR Spectroscopy for Real-Time Investigation of Dynamic Events in Proteins on the Time Scale of Seconds. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 41.Man P., Montagner C., Vernier G., Dublet B., Chenal A., Forest E., Forge V. Defining the interacting regions between apomyoglobin and lipid membrane by hydrogen/deuterium exchange coupled to mass spectrometry. J. Mol. Biol. 2007;368:464–472. doi: 10.1016/j.jmb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 42.Vernier G., Chenal A., Vitrac H., Barumandzadhe R., Montagner C., Forge V. Interactions of apomyoglobin with membranes: mechanisms and effects on heme uptake. Protein Sci. : a publication of the Protein Society. 2007;16:391–400. doi: 10.1110/ps.062531207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chenal A., Savarin P., Nizard P., Guillain F., Gillet D., Forge V. Membrane protein insertion regulated by bringing electrostatic and hydrophobic interactions into play. A case study with the translocation domain of diphtheria toxin. J. Biol. Chem. 2002;277:43425–43432. doi: 10.1074/jbc.M204148200. [DOI] [PubMed] [Google Scholar]
- 44.Perier A., Chassaing A., Raffestin S., Pichard S., Masella M., Ménez A., Forge V., Chenal A., Gillet D. Concerted protonation of key histidines triggers membrane interaction of the diphtheria toxin T domain. J. Biol. Chem. 2007;282:24239–24245. doi: 10.1074/jbc.M703392200. [DOI] [PubMed] [Google Scholar]
- 45.Bourdeau R.W., Malito E., Chenal A., Bishop B.L., Musch M.W., Villereal M.L., Chang E.B., Mosser E.M., Rest R.F., Tang W.J. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J. Biol. Chem. 2009;284:14645–14656. doi: 10.1074/jbc.M807631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chenal A., Prongidi-Fix L., Perier A., Aisenbrey C., Vernier G., Lambotte S., Haertlein M., Dauvergne M.T., Fragneto G., Bechinger B., et al. Deciphering membrane insertion of the diphtheria toxin T domain by specular neutron reflectometry and solid-state NMR spectroscopy. J. Mol. Biol. 2009;391:872–883. doi: 10.1016/j.jmb.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 47.Chenal A., Vernier G., Savarin P., Bushmarina N.A., Gèze A., Guillain F., Gillet D., Forge V. Conformational states and thermodynamics of alpha-lactalbumin bound to membranes: a case study of the effects of pH, calcium, lipid membrane curvature and charge. J. Mol. Biol. 2005;349:890–905. doi: 10.1016/j.jmb.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 48.Karst J.C., Ntsogo Enguéné V.Y., Cannella S.E., Subrini O., Hessel A., Debard S., Ladant D., Chenal A. Calcium, Acylation, and Molecular Confinement Favor Folding of Bordetella pertussis Adenylate Cyclase CyaA Toxin into a Monomeric and Cytotoxic Form. J. Biol. Chem. 2014;289:30702–30716. doi: 10.1074/jbc.M114.580852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Brien D.P., Cannella S.E., Voegele A., Raoux-Barbot D., Davi M., Douché T., Matondo M., Brier S., Ladant D., Chenal A. Post-translational acylation controls the folding and functions of the CyaA RTX toxin. Faseb. J. 2019;33:10065–10076. doi: 10.1096/fj.201802442RR. [DOI] [PubMed] [Google Scholar]
- 50.Montagner C., Perier A., Pichard S., Vernier G., Ménez A., Gillet D., Forge V., Chenal A. Behavior of the N-terminal helices of the diphtheria toxin T domain during the successive steps of membrane interaction. Biochemistry. 2007;46:1878–1887. doi: 10.1021/bi602381z. [DOI] [PubMed] [Google Scholar]
- 51.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 52.Favier A., Brutscher B. NMRlib: user-friendly pulse sequence tools for Bruker NMR spectrometers. J. Biomol. NMR. 2019;73:199–211. doi: 10.1007/s10858-019-00249-1. [DOI] [PubMed] [Google Scholar]
- 53.Veneziano R., Rossi C., Chenal A., Devoisselle J.M., Ladant D., Chopineau J. Bordetella pertussis adenylate cyclase toxin translocation across a tethered lipid bilayer. Proc. Natl. Acad. Sci. USA. 2013;110:20473–20478. doi: 10.1073/pnas.1312975110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karst J.C., Sotomayor Pérez A.C., Guijarro J.I., Raynal B., Chenal A., Ladant D. Calmodulin-induced conformational and hydrodynamic changes in the catalytic domain of Bordetella pertussis adenylate cyclase toxin. Biochemistry. 2010;49:318–328. doi: 10.1021/bi9016389. [DOI] [PubMed] [Google Scholar]
- 55.O’Brien D.P., Durand D., Voegele A., Hourdel V., Davi M., Chamot-Rooke J., Vachette P., Brier S., Ladant D., Chenal A. Calmodulin fishing with a structurally disordered bait triggers CyaA catalysis. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chenal A., Nizard P., Gillet D. Structure and function of diphtheria toxin: from pathology to engineering. J. Tox.-Tox. Rev. 2002;21:321–359. [Google Scholar]
- 57.Rigaud J.L., Lévy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 58.Subrini O., Sotomayor-Pérez A.C., Hessel A., Spiaczka-Karst J., Selwa E., Sapay N., Veneziano R., Pansieri J., Chopineau J., Ladant D., Chenal A. Characterization of a membrane-active peptide from the Bordetella pertussis CyaA toxin. J. Biol. Chem. 2013;288:32585–32598. doi: 10.1074/jbc.M113.508838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cannella S.E., Ntsogo Enguéné V.Y., Davi M., Malosse C., Sotomayor Pérez A.C., Chamot-Rooke J., Vachette P., Durand D., Ladant D., Chenal A. Stability, structural and functional properties of a monomeric, calcium-loaded adenylate cyclase toxin, CyaA, from Bordetella pertussis. Sci. Rep. 2017;7 doi: 10.1038/srep42065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voegele A., Sadi M., O’Brien D.P., Gehan P., Raoux-Barbot D., Davi M., Hoos S., Brûlé S., Raynal B., Weber P., et al. A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells. Adv. Sci. 2021;8 doi: 10.1002/advs.202003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai Y., Milne J.S., Mayne L., Englander S.W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.