Summary
Background
Autochthonous transmission of Zika virus (ZIKV) has been reported in 87 countries since 2015. Although most infections are mild, there is risk of Guillain-Barré syndrome and adverse pregnancy outcomes. Vaccines are urgently needed to prevent Zika, but sufficient understanding of humoral responses and tools to assess ZIKV-specific immunity are lacking.
Methods
We developed a blockade-of-binding (BOB) ELISA using A9E and G9E, two strongly neutralising ZIKV-specific monoclonal antibodies, which do not react with dengue virus. Receiver operating characteristic curve analysis assessed A9E and G9E BOB serodiagnostic performance. BOB was then applied to samples from a surveillance cohort in Risaralda, Colombia, and phase 1 ZIKV vaccine trial samples, comparing results against traditional serologic tests.
Findings
In the validation sample set (n = 120), A9E BOB has a sensitivity of 93.5% (95% CI: 79.3, 98.9) and specificity 97.8 (95% CI: 92.2, 99.6). G9E BOB had a sensitivity of 100% (95% CI: 89.0, 100.0) and specificity 100% (95% CI: 95.9, 100). Serum from natural infections consistently tested positive in these assays for up to one year, and reactivity tracks well with ZIKV infection status among sera from endemic areas with complicated flavivirus exposures. Interestingly, a leading ZIKV vaccine candidate elicited minimal BOB reactivity despite generating neutralising antibody responses.
Interpretation
In conclusion, A9E and G9E BOB assays are sensitive and specific assays for detecting antibodies elicited by recent or remote ZIKV infections. Given the additional ability of these BOB assays to detect immune responses that target different epitopes, further development of these assays is well justified for applications including flavivirus surveillance, translational vaccinology research and as potential serologic correlates of protective immunity against Zika.
Funding
R21 AI129532 (PI: S. Becker-Dreps), CDCBAA 2017-N-18041 (PI: A. M. de Silva), Thrasher Fund (PI: M. H. Collins), K22 AI137306 (PI: M. H. Collins).
Keywords: Zika, Vaccinology, Virus neutralisation, Type-specific antibody, Humoral immunity
Research in context.
Evidence before this study
We searched PubMed with key words “Zika” and “antibody” for articles in English and Spanish published through November 2021 and accessed relevant cited articles. Neutralizing antibodies are a prominent component of the human immune response to flavivirus infections and known to mediate protection for several flaviviruses. Effective vaccines that generate neutralizing antibodies are available for yellow fever and other flaviviruses but are lacking for important global pathogens like dengue and Zika, which caused a pandemic in 2015–2016. While many strongly neutralizing antibodies against Zika have been identified, it is not known which epitopes are targeted by immunodominant antibody responses in natural infection or which epitopes may be most important for vaccine development. Over 40 Zika vaccine candidates are underdevelopment, with some demonstrating immunogenicity and safety sufficient to support Phase 2 clinical trials.
Added value of this study
Building on our prior work that identified two potent Zika-specific neutralizing monoclonal antibodies (mAbs, A9E and G9E) and found that Zika-immune sera from Nicaragua competed with both mAbs for Zika virion binding, we further developed blockade-of-binding (BOB) assays for each mAb. We find that the BOB assays have high sensitivity and specificity for identifying prior Zika infection via testing against a validation set of sera that included many non-Zika flavivirus-immune samples and that %BOB significantly but weakly correlates with neutralizing antibody titre. Interestingly, sera from a DNA vaccine study did not contain A9E or G9E competing antibodies despite having moderate levels of Zika-binding and neutralizing antibodies.
Implications of all the available evidence
A better understanding of Zika-specific antibody responses and serologic tools to assess these responses is needed to support public health activities such as surveillance and vaccine development. The data presented here justifies expanded use of BOB assays to track Zika immunity in diverse populations and to assess vaccine-elicited immunity. Recent work indicates that narrow reliance on neutralizing antibody titres determined by traditional assays may not account for all relevant properties of antibody responses that mediate protective immunity to Zika. Whether BOBs with A9E, G9E or other mAbs can contribute to optimal assessment of Zika immunity and correlate with protection against Zika requires further research.
Introduction
The 2015–2016 Zika virus (ZIKV) epidemic that spread throughout the Americas caused over one million infection and revealed unexpected phenotypes, most notably teratogenicity and the ability to be sexually transmitted.1,2 The outbreak resulted in up to 4700 cases of microcephaly in Brazil, the most notorious manifestation of congenital Zika syndrome (CZS).3 Since 2016, global incidence of ZIKV infection has fallen dramatically.4 However, it is estimated that billions of people remain at risk for ZIKV infection based on lack of immunity and geographical distribution and environmental suitability of the primary mosquito vector Aedes aegypti.5,6 Although most cases are inapparent, the risk of Guillain-Barré syndrome and the extensive risk to pregnant women and their developing foetuses remain a pressing global public health threat. Despite substantial effort, there are no approved ZIKV vaccines or antivirals.7,8
Clinically, approximately 20% of ZIKV-infected individuals will present with a self-limited illness characterized by rash and a constellation of other symptoms including fever, myalgia, arthralgia, malaise, and conjunctivitis. Molecular diagnostics are effective when patients present early during symptomatic ZIKV infection; Serodiagnostics can play important roles outside of that setting, including utility in both clinical and epidemiologic purposes.8 However, due to cross-reactivity observed for antibodies elicited by ZIKV and other flaviviruses [most notably the four dengue virus (DENV) serotypes], traditional serologic diagnostics often fail to differentiate between these viruses, limiting assay utility, particularly where DENV and ZIKV co-circulate.8,9 Neutralising antibody (NAb) testing may enable discrimination between infection by closely related flaviviruses, but widespread use of this testing modality is limited as it is resource intensive and requires handling of infectious virus. NAbs are also likely a key determinant of durable immunity against ZIKV infection. An increased understanding of NAb epitopes and the relationship of NAb quality and quantity with protective immunity is important for vaccine development.10,11
There were over 50 ZIKV vaccine candidates by 2018, but progress stagnated when low transmission precluded efficacy trials.12 Alternative approaches for evaluating and approving ZIKV vaccines, such as human challenge studies, have not been met with unified support. Comparing antibody responses elicited by vaccines to that from natural infection is often informative and important in vaccine development, particularly for viruses that appear to induce long term protective immunity following an acute and cleared infection. Thus, robust serologic tools are crucial for vaccine development, and the lack of standard correlates of protection also remains a significant limitation to further ZIKV vaccine development.10,13 One study in non-human primates did define a NAb threshold that correlated with protection from infection, explaining differential durability of protection elicited by distinct vaccine types, but such thresholds have not been defined for humans.11 Additionally, others have stressed the value of new or improved complimentary serologic assays to assess immunogenicity of candidate ZIKV vaccines, particularly alluding to competition assays based on well-characterized ZIKV-neutralising monoclonal antibodies (mAb) as an attractive approach.14
Our previous work identified A9E and G9E, two potently neutralising mAbs isolated from a person following primary ZIKV infection acquired in Bahia, Brazil in 2015.15 Both mAbs are ZIKV-specific and recognize distinct epitopes on the ZIKV envelope (E) protein. G9E binds across the E dimer16 similar to other described mAbs (Z20, Z-117 and ZIKV-195).17, 18, 19 A9E competes with ZKA190 for binding to ZIKV virions, indicating an epitope in or near to the lateral ridge of E domain III (EDIII), or the EDI/III linker region.15 The lateral ridge is also a known target of NAb on the DENV E protein.20 Both A9E and G9E exhibited protection in challenge experiments in an immunocompromised mouse model.15 We hypothesize that A9E and G9E define immunodominant NAb responses to ZIKV, which will correlate with protective immunity. If confirmed, simple serologic assays based on A9E and G9E competition could be highly valuable in assessing population immunity and guiding vaccine development. Here we expand upon our prior work, using a well-defined sample set with known flavivirus immune profiles to assess the performance of A9E and G9E BOB assays in specifically detecting ZIKV immunity. The advantage of the two mAbs studied here is that they strongly neutralize ZIKV by targeting two distinct epitopes on ZIKV E, which may be critical for mediating protective immunity. We further studied A9E and G9E BOB reactivity among additional sample sets to understand the potential of implementing these assays as tools for surveillance and for evaluating immunogenicity (and eventually protective immunity) induced by candidate ZIKV vaccines.
Methods
Ethics
All human subject research activities were conducted with review and approval from the appropriate institutional review boards and/or bioethics committees: ArboTrav, UNC IRB# 08–0895; ZIKV-TS, UNAN-León Acta 37, 2016 and UNC IRB# 16–054121; ZIKV Pilot, Emory IRB# 110,68322; YFV Vax, Emory IRB# 00045,98223; TWS, Emory IRB# 103,363; AVE, Emory IRB# 110,683; AIP, UTP Bioethics Committee, September 10, 20,21824 and Comité de Ética de Investigación, Institución Universitaria Visión de las Américas, Acta 88, June 21, 2021; VRC320, NCT0299641.25 Informed consent was obtained in the participant's native language. Many samples analysed in this study were archived serologic specimens previously collected under Institutional Review Board (IRB)-approved studies led by collaborators. These deidentified samples were made available in accordance with an Emory IRB determination letter (September 18, 2018). Study descriptions, sample sizes and reference to relevant IRB approvals are summarized in Table 1.
Table 1.
Samples used in analysis.
| Study source | Number of subjects | Total number of specimens | Characteristics | IRB if applicable | Reference |
|---|---|---|---|---|---|
| Used for ROC curve analysis | |||||
| ArboTrav | 34 | 34 | 9 ZIKV+, 9 primary DENV, 11 multitypic DENV, and 5 naïve with and without flavivirus vaccine exposure. Serum was collected from North Carolina residents with suspicion for flavivirus infection based on symptoms and recent travel history. Specimen were serologically characterized by virus-capture ELISA and FRNT. | UNC IRB# 08–0895 | N/a |
| ZIKV TS | 14 | 14 | Samples 28 DPSO and 6 months DPSO of subjects with PCR confirmed ZIKV infection in Nicaragua. Serial serum specimens were collected in León, Nicaragua during the 2016 Zika epidemic from people presenting to local health centres with fever or rash illness as part of a prospective cohort study. Serum samples were collected during acute illness and 2, 3, 4, 8, 12, and 24 weeks post symptom onset. Subjects were tested for ZIKV RT-PCR performed on the presentation sample (acute) and by paired acute and convalescent serology testing for ZIKV and DENV IgM and IgG. | UNAN-León Acta 37, 2016 and UNC IRB#16-0541 | 21 |
| ZIKV Pilot | 8 | 29a | Participants in the Atlanta area with confirmed travel-related ZIKV infection were recruited to donate longitudinal convalescent samples at the Emory Hope Clinic. ZIKV positive status was determined by RT-PCR and or serologic assessment. | Emory IRB# 110,683 | 22 |
| YFV vax | 44 | 44 | Serum was collected from participants 18–40 in a randomized, double blind clinical trial comparing the efficacy of the yellow fever vaccine with the vaccine administered with human immunoglobulin. Participants were recruited from the metro Atlanta area with no history of travel to yellow fever endemic areas. Serum was collected at days 5, 11, 30 and 91 days post vaccine administration.22 | Clinical Trial: NCT00254826 Emory IRB# 00045,982 |
23 |
| TWS | 18 | 18 | Serum from adults seen in pre-travel consultation at the Emory TravelWell Clinic prior to international travel that were determined to be flavi-naïve based on serologic testing. | Emory IRB# 103,363 | N/a |
| AVE | 2 | 2 | Serum samples from adults in the Atlanta area with a suspected past or acute emerging infection. Both these participants tested negative for DENV and ZIKV and the average OD readings for these samples were used as plate controls to calculate % BOB. | Emory IRB# 110,683 | N/a |
| Surveillance and vaccine cohorts | |||||
| Colombia AIP | 92 | 92 | Serum was collected from mothers age 18–43 in Risaralda, Colombia between November 2017 and June 2019 on the day they gave birth. Samples were tested for DENV and ZIKV immunity by antigen capture IgG ELISA and eFRNT. | UTP Bioethics Committee September 10, 2018; Comité de Ética de Investigación, Institución Universitaria Visión de las Américas, Acta 88, June 21, 2021. |
24 |
| VRC320 | 43 | 43 | The samples were collected under the Vaccine Research Center's (VRC), National Institutes of Allergy and Infectious Diseases (NIAID), National Institutes of Health protocol: NCT02996461) in compliance with the NIH Institutional Review Board (IRB) approved protocol and procedures. All subjects met protocol eligibility criteria and agreed to participate in the study by signing the NIH IRB approved informed consent. Research studies with these samples were conducted by protecting the rights and privacy of the study participants. Participants were healthy adults 18–50 and were recruited at the NIH Clinical Center. They received 3 doses of vrc5283 on weeks 0,4 and 8. The participants were split into three groups, each with a different vaccine delivery scheme.23 | Clinical Trial: NCT02996461 | 25 |
Description of sample sets used for ROC analysis and cohorts to which the assays were applied.
n/a, not applicable, no single publication available.
Only 1 timepoint was used for each participant in the ZIKV pilot study.
Samples from VRC320
Serum from this Phase I study were provided via a material transfer agreement for secondary use following a standard specimen request process in the Vaccine Research Center. Samples from three study groups were obtained: Group 1 received 4 mg vrc5283 in a single syringe injection; Group 2 received 4 mg vrc5283 split into two syringe doses (one in each arm); Group 3 received 4 mg vrc5283 as a split dose via needle-free injection (one dose in each arm).
ZIKV status determination for specimens included in receiver operating characteristics (ROC) curve analysis
ZIKV pilot samples were from participants with RT-PCR confirmed infections.22 ZIKV TS samples (Nicaragua) were confirmed by RT-PCR testing of individuals presenting for medical care of an acute fever or rash illness and also had IgG and/or IgM reactivity compatible with an acute flavivirus infection.21 Flavivirus infection status for ArboTrav specimens was determined by comparing relative NAb titres for ZIKV and DENV1-4 as previously described.26 Clinical testing data were available for some participants with confirmed infection of DENV (DT203) and ZIKV (DT206, 211,212, 213, and 244). YLV VAX samples were from a study that administered the yellow fever virus (YFV) 17-D vaccine.23
Viruses and cells
All in vitro assays were conducted with the DENV World Health Organization reference strains: DENV-1 West Pac 74, DENV-2 S-16803, DENV-3 CH53489, and DENV-4 TVP-360 (initially obtained from R. Putnak, Walter Reed Army Institute of Research, Silver Spring, MD, USA). ZIKV strain H/PF/2013 (originally obtained from the Centers for Disease Control) and DENV1-4 isolates were propagated in C6/36 cells (Obtained from ATCC cat# CRL-1660; RRID:CVCL_Z230) and titrated on Vero cells (Obtained from ATCC, cat # CCL-81–RRID:CVCL_0059) and titrated on Vero cells as previously described.15
Production and labelling of monoclonal probes
A9E and G9E were isolated from PBMCs of a participant in the UNC Dengue Travellers cohort using a process previously described15 and later produced in a mammalian expression platform (LakePharma, San Carlos, CA). Purified A9E and G9E mAbs were labelled with alkaline phosphatase (AP) for use in the BOB ELISA assay using the LYNX Rapid Alkaline Phosphatase Antibody Conjugation Kit® (BioRad, Cat# LNK012AP).
Antigen capture IgG ELISA
ZIKV-binding IgG was measured using antigen-capture ELISA. ZIKV was captured with plates coated with 100 ng of mouse 4G2 monoclonal antibody. 4G2 was produced in our lab from mouse hybridoma cells D1-4G2-4-15 (ATCC; RRID:CVCL_J890). Plates were blocked with 3% non-fat milk-PBS-T. IgG was detected using an anti-human IgG antibody linked to AP (Sigma Aldrich, cat#: A9544-1 ML) and measuring optical density (OD) at 405 nm after incubation with a p-nitrophenyl phosphate (p-NPP) (SIGMAFAST(TM) Sigma Aldrich, cat#: N1891-50SET). This protocol was modified to use biotinylated Zika-EDIII as antigen instead of the whole Zika virus, to detect Zika EDIII-specific IgG.27 Plates were coated with streptavidin (Fisher Scientific, cat# 434,301) to capture biotinylated Zika-EDIII and unspecific binding blocked with 3% non-fat milk-TBS-T (10X TBS from Teknova, cat#: T9545).
A9E and G9E Blockade-of-Binding (BOB) assay
The ZIKV BOB assay performed has been previously described.15 ZIKV was captured using antibody 4G2. Plates were blocked with 3% non-fat milk in PBS-T. Different dilutions of antigen, serum and labelled monoclonal probe were optimized prior to testing the validation sample set (Fig. S1). Plates were coated with 100ng/well of 4G2 and ZIKV antigen (virus culture supernatant) was captured by incubating antigen diluted 1:2 with blocking buffer on plates for 1 h at 37 °C. Sera were plated in duplicate at a 1:10 dilution and incubated at 4 °C overnight. 10ng/well of alkaline phosphate conjugated G9E or A9E was added to each well and incubated shaking for 1 h at room temperature. After washing unbound Ab, p-NPP was added, and OD was measured at 405 nm. Percentage of blockade-of-binding was calculated using the following equation (1 − [OD of sample/optical density of control]) × 100.
Focus forming neutralisation testing (FRNT)
In brief, 15,000 Vero cells (ATCC CCL-81) were seeded per well in 96 well plates and incubated for 24 h. The sera were serial diluted in Opti-MEM I Reduced Serum Media (Gibco™ Life Technologies Corporation, cat#: 11,058,021) and mixed with appropriate virus, also diluted in Opti-MEM. The virus-antibody mixtures were incubated for 1 h at 37 °C and transferred to a monolayer of Vero cells for infection. A 1.5% methyl cellulose/2% FBS-Opti-MEM overlay was then added after an hour of incubation at 37 °C and then plates incubated for 48 h (ZIKV and DENV4) or 72 h (DENV1-3). After incubation, cells were fixed and permeabilized with 100 μL methanol/acetone for 30 min. Then plates were blocked with 5% Milk in PBS buffer for 30 min, followed by 100 μL of monoclonal antibody (D1-4G2-4-15, ATCC) at 1 ng/μL in blocking buffer and incubated for 2 h at 37 °C. The plates were then washed and 100 μL of a 1:3000 solution of HRP-conjugated goat anti-mouse IgG antibody (Cell Signaling technology, cat#: 7076 V) was added and incubated for 1 h at 37 °C. Plates were visualized by addition of 100 μL of TrueBlue™ Peroxidase Substrate (KPL Sera Care, cat# 5510-0030) and a CTL ImmunoSpot analyzer (Cellular Technology Limited, OH, USA).
Estimated FRNT (eFRNT)
The FRNT assay was abbreviated to increase testing throughput and has been previously described.28,29 Four 4-fold serial dilution (1:20, 1:80, 1:320, 1:1280) were assessed. FRNT50 values were assigned to the most dilute sample with neutralisation greater than 50% compared to the negative control.
Single dilution FRNT (sdFRNT), The FRNT assay was abbreviated to increase testing throughput to provide a screen for previous DENV/ZIKV using a single dilution (1:50) of serum. Positivity was assigned if the sample had greater than 50% neutralisation at this dilution.
Validation of key reagents is provided in a Supplemental File.
Statistical analyses
ROC curve analysis: The A9E and G9E BOB ELISAs were validated by ROC curve analysis using Prism GraphPad 8.4.3. Positive sera came from ZIKV-confirmed participants from studies conducted in ZIKV endemic countries as well as from travellers in the United States. The negative sample set came from participants in studies that were designated as negative based on immune profile and/or travel history.
Linear regression: Because the VRC 320 data sets failed to meet normality assumptions required for simple linear regression a non-parametric test, Spearman's rank linear correlation was used to evaluate linear relationships between A9E and G9E BOB with other ZIKV immune markers of interest. GraphPad 8.4.3. was used to conduct the analysis. Either the r2 value or the Spearman's correlation coefficient (rs) and associated p value were reported when appropriate.
T-test: An unpaired t-test was used to assess if there was a difference in A9E and G9E responses in DENV-naïve and DENV immune individuals. The two-tailed p-value was reported.
ANOVA: Brown-Forsythe and Welch ANOVA tests were used to test the differences between A9E and G9E BOB activity in the three vaccine delivery groups in the VRC320 trial. Dunnett's multiple comparison tests were used to do pairwise comparisons between groups.
Role of funders
The funders of this work had no role in the study design, data collection, data analyses, interpretation or writing of this manuscript.
Results
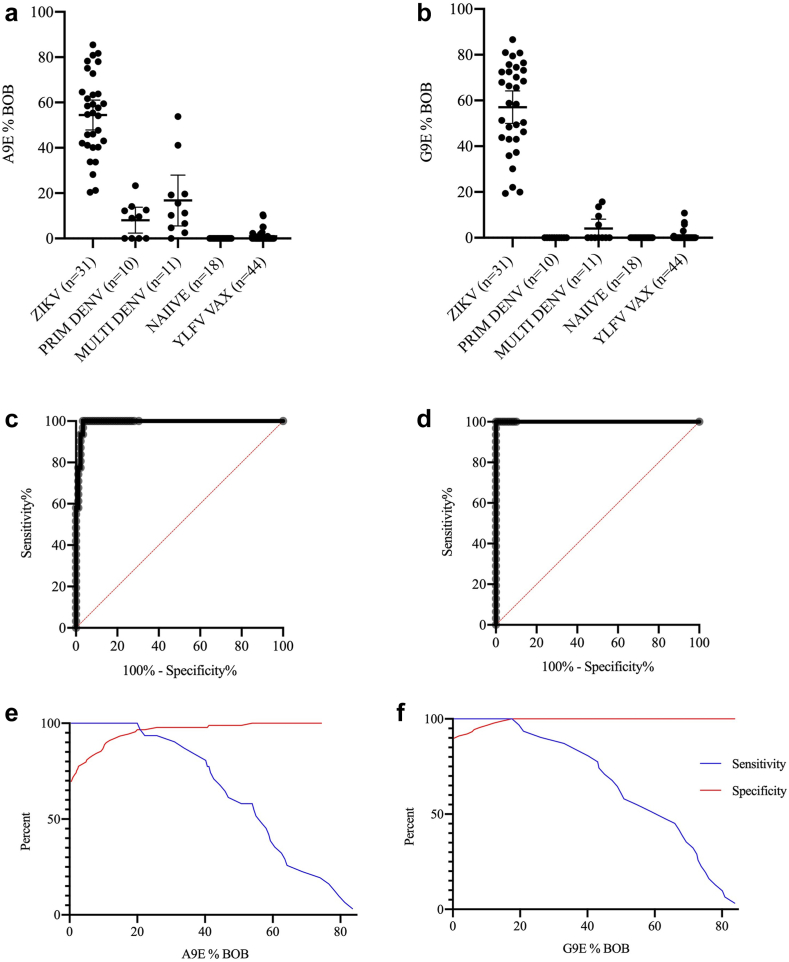
A9E and G9E BOB assays were validated using serum from persons with confirmed prior ZIKV infection, including those with (n = 14) and without (n = 17) prior DENV infection. Negative controls came from healthy travellers (n = 24) or travellers with a history of monotypic (n = 9) or multitypic DENV infection (n = 12). Additionally, sera from a cohort of flavivirus-naïve participants who had received the YFV 17-D vaccine (n = 44) as part of a randomized control trial were included in the validation set of negative controls.23 ZIKV positive samples consistently showed greater reactivity than all other subgroups (Fig. 1a and b). The average mean BOB difference between the ZIKV positive controls and ZIKV negative controls for the A9E BOB ELISA was 51.1% and was 56.3% for the G9E BOB ELISA. Two multitypic DENV-positive samples showed cross-reactivity in A9E BOB. Both reported having no symptoms associated with dengue in the last 20 years. Naïve and YFV-vaccinated specimen showed very little BOB activity. These data were used in ROC curve analyses to determine the optimal positivity threshold for the two BOB assays (Fig. 1c–f) (AUC A9E BOB = 0.992, AUC G9E BOB = 1.00). The first timepoint after 30 days post symptom onset (DPSO) was selected for ROC curve analysis if a participant donated samples at multiple timepoints. The optimal cut-offs were 25.75% and 17.58% blockade for the A9E and G9E BOB. At the optimal cut-off, the A9E BOB has a sensitivity of 93.5% (95% CI: 79.3, 98.9) and specificity 97.8 (95% CI: 92.2, 99.6). The G9E BOB has a sensitivity and specificity of 100% (95% CI: 89.0, 100.0) and 100% (95% CI: 95.9, 100.0). These cut-offs were used in all subsequent analyses.
Fig. 1.
A9E and G9E BOB reactivity in validation sample set. (a and b) Percent A9E and G9E blockade grouped by flavivirus-immune status. See Methods for details for flavivirus immune status determination. (c and d) ROC curves depicting results from validation samples (AUC = 0.992 for A9E BOB and AUC = 1.0 for G9E BOB). (e and f) Sensitivity and specificity plotted according to %BOB (x-axis) to visualize cut-off determination.
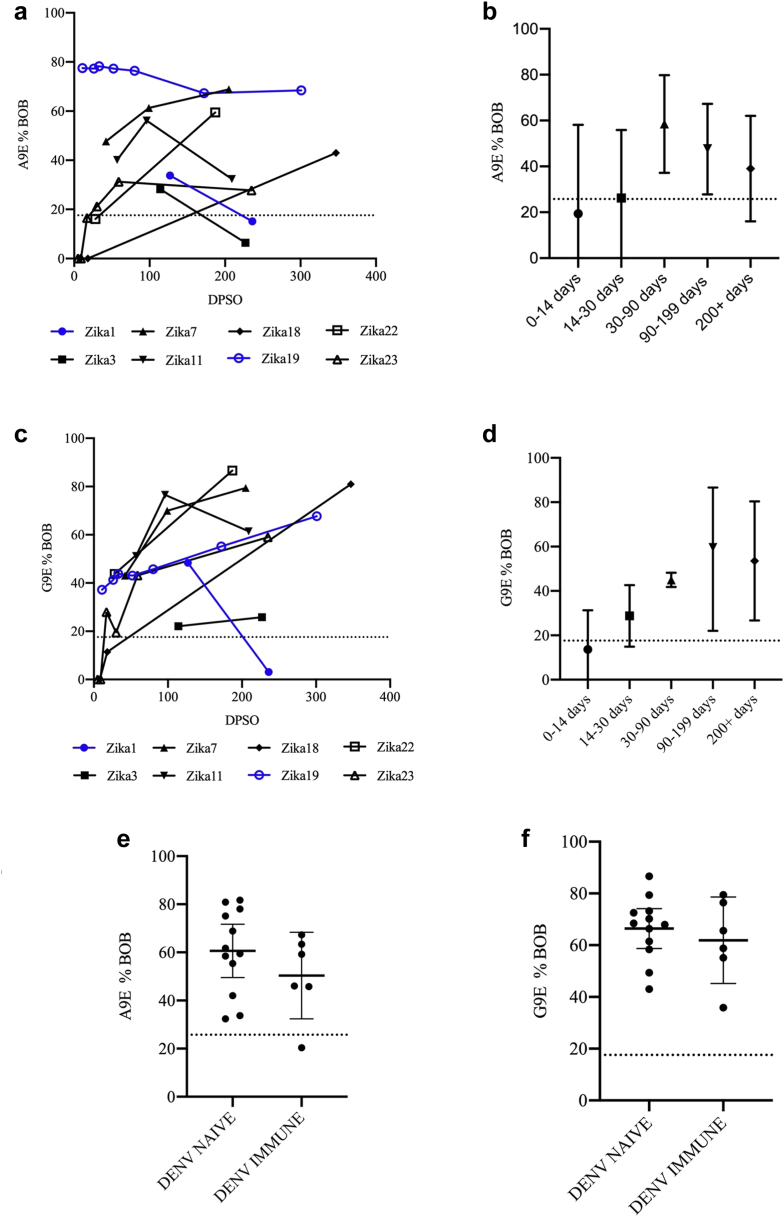
We examined the durability of the A9E and G9E blockade response using a subset of longitudinal samples from travellers with confirmed ZIKV infection (Table 1). Although there is individual variability in the magnitude of A9E and G9E BOB signal, BOB activity for A9E or G9E is detectable, and peaked in the majority (7/8) of people in the range of 30–90 days DPSO (Fig. 2a–d). Signal is increasing or stable for 4/8 samples in A9E BOB, with the other 4 exhibiting waning signals, to the point of seroreversion in 2 samples. For G9E BOB, 6/8 samples exhibit increased to stable signal and only 1 seroreversion of the two samples with waning signal.
Fig. 2.
Longitudinal analysis of A9E BOB and G9E BOB reactivity. (a and c) A9E %BOB and G9E %BOB reported by days post symptom onset (DPSO, x-axis) for ZIKV Pilot samples. Samples represented in blue were obtained from patients with a history of DENV infection, all others were flavivirus-naïve prior to ZIKV infection. (b and d) Summary longitudinal data grouped by DPSO for A9E %BOB and G9E %BOB. Error bars represent ±1 standard deviation around the mean. (e and f) A9E %BOB and G9E %BOB for DENV-naïve vs DENV-immune participants among those with prior ZIKV infection among samples from Zika Pilot and ZIKV TS samples. A single sample per participant was selected at a time point between 7 and 9 months post infection. Error bars represent the 95% CI of the mean. An unpaired t-test comparing the DENV naïve group to the DENV immune group yielded p-values of 0.25 and 0.50 for a A9E and G9E respectively.
Next, we asked if A9E- and G9E-like responses differed in DENV-naïve individuals who acquired ZIKV as a primary flavivirus infection compared to DENV-immune individuals experiencing Zika as a secondary flavivirus (Fig. 2e and f). Samples from the ZIKV Pilot Cohort (n = 4) and in the ZIKV TS Nicaragua cohort (n = 14) that were collected between five- and seven-months post-symptom onset were used for this analysis. There was no significant difference detected in the A9E (p = 0.25 [t-test]) and G9E (p = 0.51 [t-test]) response between the two groups.
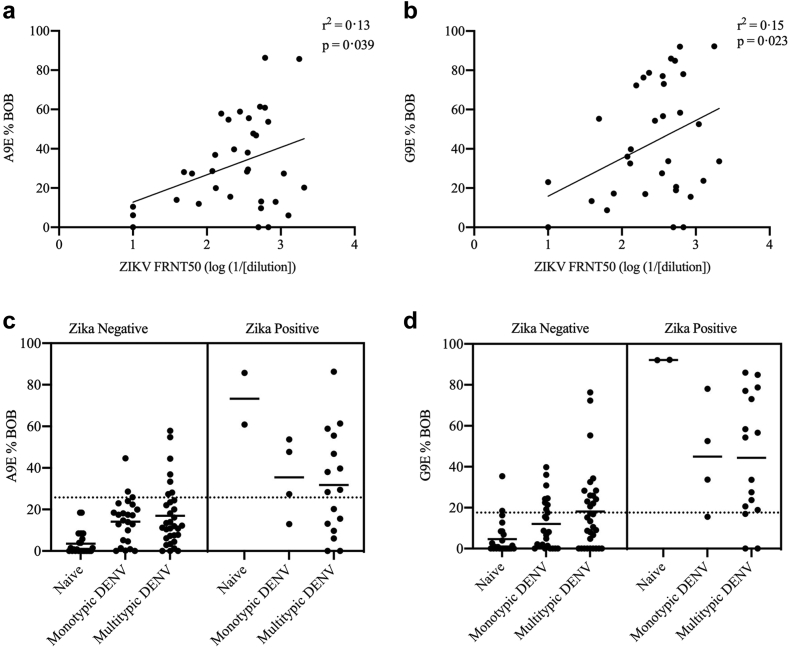
For flaviviruses such as ZIKV and DENV that share conserved epitopes leading to cross-reactive binding antibodies, understanding rates of prior infection by each virus has typically required neutralisation testing of multiple viruses for each sample in question. This is a labour-intensive process that is costly in time and expense. A simpler, rapid and cost-effective serological assay that distinguishes past ZIKV infection from other flaviviruses would substantially improve efficiency and feasibility of broad surveillance efforts. A9E and G9E BOB assays were applied to samples from a cross-sectional surveillance cohort study based in Risaralda, Colombia. The flavivirus immune profile for these sera was initially assessed by eFRNT,24 and a precise FRNT50 value against ZIKV was later determined. Samples with a ZIKV FRNT50 ≥ 1:200 were considered to likely have had previous ZIKV infection. DENV immune status was designated as monotypic or multitypic. Monotypic was defined by DENV eFRNT50 values for three serotypes that were at least 4-fold less than that of the serotype with the highest eFRNT50. Otherwise, a DENV + sample was assigned as multitypic. There was a significant, although weak association between the log of ZIKV FRNT and both A9E and G9E BOB responses using simple linear regression (A9E: r2 = 0.13, p = 0.039, G9E: r2 = 0.15, p = 0.023, Fig. 3a and b). The A9E and G9E responses were assessed between ZIKV positive and negative samples stratified by DENV status (Fig. 3c and d, Table 2). The A9E and G9E BOB false positivity rates were 14% and 31% respectively when using binary FRNT IC50 > 200 as the reference positivity standard (Table 2). Specificity was higher when requiring positivity in both A9E and G9E BOB for designating a sample as positive for prior ZIKV infection (Specificity = 92.5%).
Fig. 3.
A9E and G9E reactivity in a flavivirus endemic population. (a and b) Correlation between ZIKV eFRNT50 and A9E %BOB and G9E %BOB. For c and d, FRNT50 titres were available for n = 34 samples. The correlation coefficient and the p value from the simple linear regression are displayed in the upper right of each panel. (c and d) A9E and G9E %BOB grouped by flavivirus immune status as determined by FRNT and eFRNT testing for the Colombia AIP samples, a cross-sectional surveillance set of pregnant women in Risaralda, Colombia (n = 92). Each panel is divided into two adjoined panes corresponding to ZIKV immune status, using a cut-off of FRNT50 = 200. Dotted lines represent positivity threshold for A9E and G9E determined by ROC analysis.
Table 2.
Application of BOB assay to Colombia AIP samples.
| ZIKV status | DENV screen status | % A9E BOB positive | % G9EBOB positive | % A9E AND G9E BOB positive | % A9E OR G9E BOB positive |
|---|---|---|---|---|---|
| ZIKV Negative | Naïve | 0% (0/13) | 7.7% (1/13) | 0% (0/13) | 7.7% (1/13) |
| Monotypic DENV | 12% (3/25) | 32% (8/25) | 8% (2/25) | 36% (9/25) | |
| Multitypic DENV | 22% (7/32) | 40% (13/32) | 13% (4/32) | 50% (16/33) | |
| Total | 14% (10/70) | 31% (22/70) | 7.5% (6/70) | 37% (26/70) | |
| ZIKV Positive | Naïve | 100% (2/2) | 100% (2/2) | 100% (2/2) | 100% (2/2) |
| Monotypic DENV | 75% (3/4) | 75% (3/4) | 75% (3/4) | 75% (3/4) | |
| Multitypic DENV | 56% (9/16) | 81% (13/16) | 56% (9/16) | 81% (13/16) | |
| Total | 71% (14/22) | 82% (18/22) | 64% (14/22) | 82% (18/22) |
% A9E and G9E BOB ELISA positivity rate by ZIKV and DENV classification based on thresholds set by previous ROC analysis.
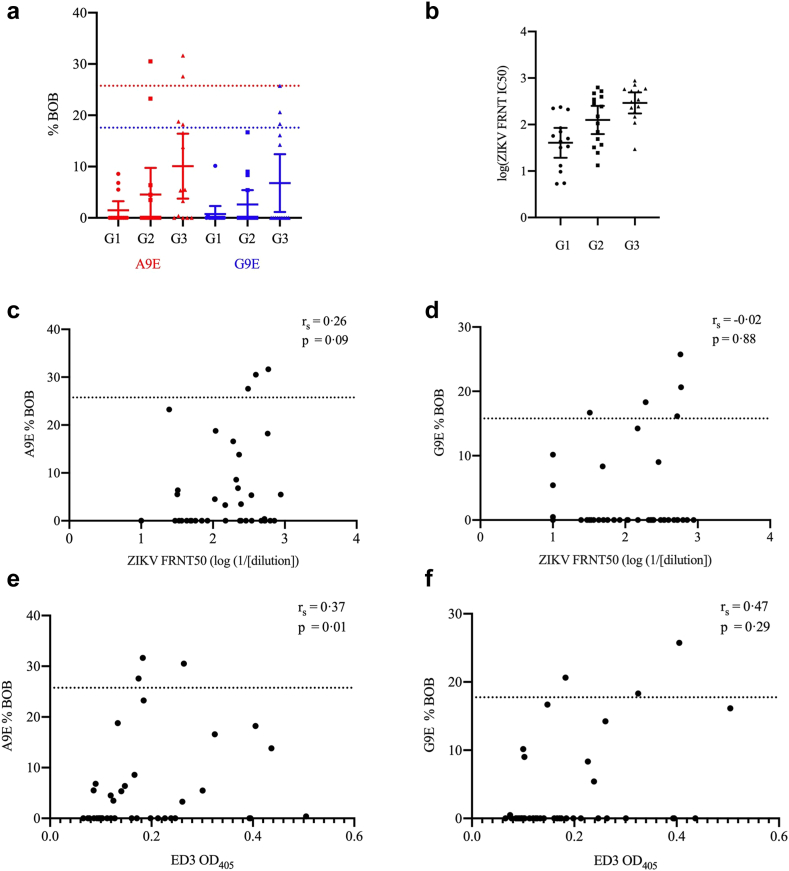
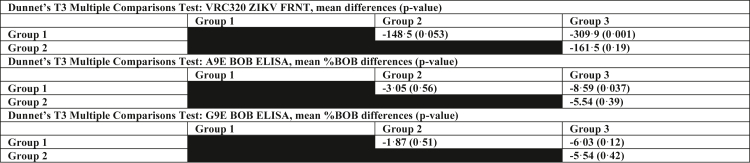
To determine whether vaccination with a PrM/E-encoding ZIKV DNA vaccine elicited A9E or G9E-like Abs as we observed in most cases of natural infection, we tested sera from the VRC320 Phase I vaccine trial.25 The vaccine candidate tested was the vrc5283 ZIKV plasmid, which was delivered in a schedule of three 4 mg-doses over 8 weeks. Subjects were assigned to three study groups corresponding to variation in vaccine delivery method. Samples from 8 weeks post-vaccination had only modest A9E or G9E BOB activity (Fig. 4a). Only six samples had A9E or G9E BOB activity above the positivity threshold and only one specimen was positive for both A9E and G9E BOB (Fig. 4a). Of these, one sample came from a participant who screened positive for DENV neutralising antibodies at a serum dilution of 1:50 (Tables 1 and 2, Fig. S2). In contrast, NAb activity was readily detected in all groups, with Group 3 exhibiting the highest titers (Fig. 4b), essentially replicating the data published in the Phase I immunogenicity study.25 When grouped according to trial study groups, group 3 exhibited greater A9E and G9E BOB reactivity than group 2 or 1, similar to the pattern of mean NAb responses, which progressively increased from group 1 to group 3 (Fig. 4a and b).25 A Dunnet's D3 multiple comparisons test found there to only be a significant FRNT50 titre difference between G1 and G3 (p = 0.0001) (Table 3). Likewise, there was only a statistically significant difference in A9E BOB activity between G1 and G3 (p = 0.04) (Table 3). The groups did not have significantly different G9E BOB activity (Table 3). Furthermore, A9E and G9E BOB results were compared to ZIKV FRNT50 results (Table 3). A sample's %BOB for A9E or G9E showed no association with neutralisation activity (A9E: rs = 0.26, p = 0.09, G9E: rs = −0.02, p = 0.88 [Spearman correlation], Fig. 4c and d). Although A9E and G9E responses among the vaccine sera were generally much lower compared to those of natural infection, there is evidence of a low degree of BOB activity in several samples, including seroconversion after vaccination (Fig. S3). A ZIKV ED3 antigen ELISA was also conducted to compare A9E or G9E BOB reactivity with binding to this antigenic region of ZIKV E [Spearman correlation]. There was a weak but significant correlation between A9E BOB and ED3 binding IgG (rs = 0.37, p = 0.01, Fig. 4e). ED3 OD405 was not significantly correlated with the G9E BOB response (rs = 0.47, p = 0.29, Fig. 4f).
Fig. 4.
vrc5283 DNA vaccination elicits NAb but not A9E- or G9E-like Ab responses. (a) A9E %BOB and G9E %BOB shown for the three VRC320 ZIKV DNA vaccine trial groups. Cut-off for positivity and data points are color coded for each mAb probe (Red, A9E; Blue, G9E). Error bars represent ±1 standard deviation around the mean. (b) NAb titre (FRNT50) shown for the same three VRC320 study groups. (c and d) Correlation between A9E %BOB and G9E %BOB and FRNT50. Dotted linen indicates positivity threshold as determined by ROC analysis. (e and f) Correlation between ZIKV EDIII ELISA OD405 and A9E and G9E %BOB. Group 1 received 4 mg vrc5283 in a single syringe injection; Group 2 received 4 mg vrc5283 split into two syringe doses (one in each arm); Group 3 received 4 mg vrc5283 as a split dose via needle-free injection (one dose in each arm).
Table 3.
Dunnet's multiple comparison tests for VRC320 ZIKV candidate vaccine groups.
Mean difference between groups and the adjusted p-value are reported for each comparison. Comparisons were done between groups for FRNT IC50, %A9E BOB and % G9E BOB.
Discussion
The ZIKV epidemic of 2015–2016 exposed profound public health shortcomings, particularly in the ability to protect pregnant women and children from emerging infectious diseases. Years later, the threat of future ZIKV outbreaks looms large, and there are still no licensed antivirals, vaccines, or other proven methods of preventing ZIKV infection, beyond vector avoidance. Vaccines and robust surveillance systems represent two of the greatest unmet needs that have persisted since the ZIKV epidemic. In this paper, we employed a innovative tool to analyse serologic features of samples following natural infection and vaccination. We found that A9E and G9E identify antigenic regions of ZIKV that are consistently targeted by Ab elicited by natural infection.15 When applying the optimized A9E and G9E BOB assays to samples from travellers or people living in endemic areas, we find A9E- and G9E-like responses are present in the vast majority of ZIKV-immune samples tested and these responses peak and persist in the period 3–12 months post infection.
A central hypothesis driving this work is that the mAbs A9E and G9E bind crucial neutralising epitopes frequently targeted in natural human infections that result in durable protective immunity. Thus, A9E and G9E BOB assays may provide a benchmark for predicting protective efficacy of vaccine-elicited Ab responses. Our data are consistent with this hypothesis, but additional study will be required to rigorously support or reject it. It is not yet known if A9E or G9E-like responses are necessary or sufficient for protective immunity to ZIKV. Moreover, this idea cannot be fully tested in the absence of an accepted correlate of protection for ZIKV infection and/or a vaccine efficacy trial in which vaccinated subjects experience a range of outcomes (protection, infection, disease). The degree of positivity (%BOB) in A9E and G9E BOB assays significantly correlated with NAb titre but the relationship was weaker than expected. Although NAb titres are commonly measured and thought to mediate immune protection, it is increasingly appreciated that in vitro detection of NAb may not correlate with protection. Recent evidence for both ZIKV and DENV indicates that commonly used neutralisation assays may not account for important properties of neutralising antibodies that impact protective efficacy.10,30 For ZIKV, assessing for neutralisation of mature virions accounted for the discrepancy in predicting protection in traditional neutralisation assays in one recent study.10 For DENV, it may be important to elicit serotype-specific NAb rather than cross-neutralising NAb detected by in vitro assays to achieve durable broad protection against DENV infection.31 We found that ample ZIKV- neutralising Abs were elicited by the DNA vaccine studied in VRC320, but A9E and G9E-competing Ab responses were absent or markedly diminished when compared to natural infection. One explanation for our findings is that the structural epitopes targeted by A9E and G9E are not well represented on the subviral particles (SVP) generated in vrc5283 recipients (VRC320 participants). During development of this and the sister ZIKV DNA construct (vrc5288), SVP were visualized by EM for vrc5288 but not vrc5283. Antigen production by the two constructs was assessed by an ELISA that utilizes mAb targeting simple cross-reactive epitopes on the fusion loop. SVP isolated from cell culture can be effectively used as antigen in ELISA assays that detect ZIKV-specific IgG following vaccination with the corresponding DNA construct32; however, the epitopes targeted by NAb in VRC320 vaccinees is unknown, and how closely the structure of vrc5283 SVPs reflects that of native ZIKV virions is not well defined. It would be interesting to determine whether A9E and G9E BOB reactivity is similar in specimens from VRC319 or other ZIKV human vaccine trials, particularly those utilising a live attenuated (NCT03611946) or purified inactivated ZIKV (NCT02963909) platforms. It should be noted that a lack of A9E BOB reactivity, with our current state of understanding, does not mean a vaccine will be ineffective. Safe and effective vaccines could certainly elicit robust and durable immunity that is qualitatively distinct from immunity elicited by natural infection. Additionally, A9E and G9E appear to identify two of a few or several important epitopes of ZIKV NAb but testing an expanded set of sera and ZIKV-neutralising mAbs will give greater resolution to our understanding of NAb response and frequency with which different epitopes are targeted. This information could further clarify the most promising antigens for ZIKV vaccine development and create a better set of reagents with which to comprehensively measure Ab immunity to ZIKV during vaccine development.19,33,34 Furthermore, we and others have shown that T cell responses and non-neutralising functional antibody responses like ADCC are likely components of protective immunity against ZIKV derived from either vaccines or infection.22,35 These additional immunologic parameters will likely need to be considered to fully assess the quality and protective potential of vaccine-elicited immunity to ZIKV.
Beyond vaccine development, sensitive, specific, and deployable serologic assays in DENV-endemic populations are central to meeting critical public health goals for ZIKV. Moreover, with multiple vaccine candidates to be tested and other intervention trials ongoing that include serologically determined endpoints for DENV, it is even more important to determine the flavivirus infection status of individuals with high specificity.13 Our ROC curve analysis for both A9E and G9E BOB in a well-defined sample set indicated that both assays were sensitive and specific, with AUC values above 0.99. This is encouraging given that traditional serologic diagnostic tools for flaviviruses are often complicated by cross-reactivity and ZIKV and DENV are widely co-endemic.8,36 Of note, a BOB assay using ZIKV NS1 antigen also performs well in this context.37 However, the picture in the flavivirus-endemic setting is somewhat more complex. In a cross-sectional surveillance cohort in Risaralda, Colombia, we found that the sensitivities and specificities of the A9E and G9E BOB assay were less than what was observed during assay development, using classification according to immune profiling by neutralization testing as the reference standard (Table 2). This is likely due to high levels of multitypic DENV infection in the study site that were not fully represented in our validation sample set. Alternatively, this could illustrate limitations of the eFRNT approach for precisely classifying flavivirus exposure history. Multitypic DENV infection may lead to such high levels of flavivirus cross-reactive Ab that stoichiometry increases steric hinderance of ZIKV-specific mAb binding by serum Ab binding ZIKV E within the same antigenic region. For example, though A9E is ZIKV-specific and nonreactive to any DENV serotype, a strongly neutralising mAb (MZ4) has been identified whose footprint has extensive overlap with A9E and that is cross-reactive with DENV, particularly DENV2. Sensitivity was also lower; however, this may be because of specimens being misclassified as ZIKV positive by eFRNT because of multitypic DENV that can lead to ZIKV FRNT50 ≥ 1:200 via cross-reactive Abs. In other words, if our abbreviated neutralisation screen for prior ZIKV is imperfectly specific, the sensitivity of A9E and G9E BOBs will be underestimated. Access to a set of samples prospectively collected, with flavivirus infection status defined by lab confirmation of acute illness could address this limitation in our study, though such ideal sample sets are not widely available. The specificity of A9E and G9E BOB assays for ZIKV was increased to 90% by requiring both to be positive, which could be sufficient for some public health purposes such as surveillance. Confirmation by a different test would likely be required, as would a large sample size since sensitivity would be modest if dual positivity were stipulated. Sensitivity could also decrease overtime due to viral evolution, but ZIKV has been remarkably antigenically stable over time,38 lowering the concern for this potential limitation, particularly if assays include mAb probes that target multiple distinct epitopes. For certain populations such as travellers, who have transient exposures and are more likely to have a primary flavivirus infection, the assays described here may perform much better, potentially reaching a sensitivity and specificity that could permit clinical use.
Future work is being pursued to address the main limitations of this study, which include 1) the need to further establish A9E and G9E BOB performance with larger, prospective samples sets that included confirmed flavivirus infections followed by longitudinal sampling. Because samples used here were collected by convenience sampling and/or under different study protocols, we are unable to group samples for the analyses presented here. This approach also precluded any effort to balance participant recruitment based on numerous factors (such as sex, demographic variables, and medical comorbidities) or perform meaningful multivariate analyses. Large longitudinal cohort studies would also support a more robust effort to resolve the immune interactions involved with sequential heterotypic flavivirus infection and vaccination. 2) There is a need to assess a larger array of epitopes on the ZIKV surface. 3) Finally, it would be interesting to consider epitope specificity of non-neutralising Ab effector function such as Ab-dependent cellular cytotoxicity in protective immunity to ZIKV.
In summary, this work demonstrates the diverse utility of a valuable serologic tool measuring ZIKV-specific antibody responses and provides further insight into the quality of humoral immunity elicited by ZIKV infection and vaccination. The findings and approaches in this study serve as a proof-of-principle of a conceptual, technical, and analytic framework that paves the way for future work including the further mapping of ZIKV Ab responses in previous infection and vaccination using additional ZIKV-specific neutralising mAbs. This approach will also be useful in studying other infectious diseases since the need for accurate diagnostics, efficient surveillance tools and effective vaccines is generally shared by many emerging viral infections.
Contributors
TS, study design, data generation and analysis, drafted the manuscript; DOE, data generation, edited manuscript; YZ data generation, edited manuscript; JACO, study concept, PI of cohort studies contributing samples, edited manuscript; NMB, PI of cohort studies contributing samples, edited manuscript; SBD, PI of cohort studies contributing samples, edited manuscript; NR, PI of cohort studies contributing samples, edited manuscript; AJRM, study concept, cohort study design, edited manuscript; FB, cohort study design, PI of cohort studies contributing samples, edited manuscript; SE; PI of clinical trial contributing samples, edited manuscript; LP, experimental design, data interpretation, provided key reagents, edited manuscript; MJM, study concept, PI of cohort studies contributing samples, edited manuscript; AMdS, study concept, provided key reagents, edited manuscript; MHC, study concept and oversight, data analysis, drafting and editing of manuscript. TS and MHC verified all data. All authors have read and approved the final version of the manuscript.
Data sharing statement
Samples were derived from several published studies, which has been indicated in Table 1 and in citations so readers may go to the primary reports as well as clinicaltrials.gov. Any primary data not included in the main text or supplement is freely available upon request: matthew.collins@emory.edu.
Declaration of interests
MHC and AMdS are co-inventors on US patent application No. 16/765,509. MJM reported potential competing interests: laboratory research and clinical trials funding from Lilly, Pfizer, and Sanofi; personal fees for Scientific Advisory Board service from Merck, Meissa Vaccines, Inc. and Pfizer.
SE and MHC reported potential competing interests: clinical trials funding from Sanofi. AJRM reported potential competing interests: consultant/speaker for Sanofi (2016–2017), Valneva (2021–2023), and Takeda (2021–2023). All other authors have no interests to declare.
Acknowledgements
We thank the many that participated in clinical research studies and consented to future use of samples to make this study possible. We thank Sci-Help, S.A.S. and the field team in Risaralda, Colombia managing cohort studies. We thank the Vaccine Research Center of the NIH for sharing samples from VRC320: Grace Chen, Principal Investigator; Ro Rothwell, Protocol Specialist; Laura Novik, VRC Study Coordinator. We thank Dr. Josephine Cox for assistance procuring samples from VRC320 and for critical feedback on the manuscript. We thank Dr. Lilin Lai for assistance procuring samples from travellers. We thank Drs. Christina Mehta and Bidemi Yusuf for thoughtful input on biostatistical approaches and analyses used in this study. We acknowledge several funding sources: R21 AI129532 (PI: S. Becker-Dreps), CDC BAA 2017-N-18041 (PI: A. M. de Silva), Minciencias, Convocatoria 897 de 2021, Project 85982 (PI: J. A. Cardona-Ospina), Thrasher Fund (PI: M. H. Collins), K22 AI137306 (PI: M. H. Collins).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104875.
Appendix A. Supplementary data
References
- 1.Lazear H.M., Diamond M.S. Zika virus: new clinical syndromes and its emergence in the western hemisphere. J Virol. 2016;90(10):4864–4875. doi: 10.1128/JVI.00252-16. http://www.ncbi.nlm.nih.gov/pubmed/26962217 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyne C.B., Lazear H.M. Zika virus - reigniting the TORCH. Nat Rev Microbiol. 2016;14(11):707–715. doi: 10.1038/nrmicro.2016.125. http://www.ncbi.nlm.nih.gov/pubmed/27573577 Available from: [DOI] [PubMed] [Google Scholar]
- 3.Victora C.G., Schuler-Faccini L., Matijasevich A., Ribeiro E., Pessoa A., Barros F.C. Microcephaly in Brazil: how to interpret reported numbers? Lancet. 2016;387(10019):621–624. doi: 10.1016/S0140-6736(16)00273-7. https://pubmed.ncbi.nlm.nih.gov/26864961/ [cited 2023 Jun 15] Available from: [DOI] [PubMed] [Google Scholar]
- 4.Siedner M.J., Ryan E.T., Bogoch Gone or forgotten? The rise and fall of Zika virus. Lancet Public Heal. 2018;3(3):e109–e110. doi: 10.1016/S2468-2667(18)30029-X. https://pubmed.ncbi.nlm.nih.gov/29519697/ [cited 2023 Jun 15] Available from: [DOI] [PubMed] [Google Scholar]
- 5.Zika: the continuing threat. Bull World Health Organ. 2019;97(1):6–7. doi: 10.2471/BLT.19.020119. https://pubmed.ncbi.nlm.nih.gov/30618459/ [cited 2023 Jun 15] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musso D., Nilles E.J., Cao-Lormeau V.-M. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect. 2014;20(10):O595–O596. doi: 10.1111/1469-0691.12707. http://www.ncbi.nlm.nih.gov/pubmed/24909208 Available from: [DOI] [PubMed] [Google Scholar]
- 7.Fauci A.S., Morens D.M. Zika virus in the Americas - yet another arbovirus threat. N Engl J Med. 2016;374(7):601–604. doi: 10.1056/NEJMp1600297. http://www.ncbi.nlm.nih.gov/pubmed/26761185 Available from: [DOI] [PubMed] [Google Scholar]
- 8.Petersen L.R., Jamieson D.J., Powers A.M., Honein M.A. Zika virus. Baden LR, editor. N Engl J Med. 2016;374(16):1552–1563. doi: 10.1056/NEJMra1602113. http://www.nejm.org/doi/10.1056/NEJMra1602113 Available from: [DOI] [PubMed] [Google Scholar]
- 9.Priyamvada L., Quicke K.M., Hudson W.H., et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016;113(28):7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maciejewski S., Ruckwardt T.J., Morabito K.M., et al. Distinct neutralizing antibody correlates of protection among related Zika virus vaccines identify a role for antibody quality. Sci Transl Med. 2020;12(547) doi: 10.1126/scitranslmed.aaw9066. https://pubmed.ncbi.nlm.nih.gov/32522807/ [cited 2021 Feb 3] Available from: [DOI] [PubMed] [Google Scholar]
- 11.Abbink P., Larocca R.A., Visitsunthorn K., et al. Durability and correlates of vaccine protection against Zika virus in rhesus monkeys. Sci Transl Med. 2017;9(420) doi: 10.1126/scitranslmed.aao4163. pmc/articles/PMC5747972/ [cited 2022 Mar 9] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin H.H., Yip B.S., Huang L.M., Wu S.C. Zika virus structural biology and progress in vaccine development. Biotechnol Adv. 2018;36(1):47–53. doi: 10.1016/j.biotechadv.2017.09.004. https://pubmed.ncbi.nlm.nih.gov/28916391/ [cited 2023 Jun 15] Available from: [DOI] [PubMed] [Google Scholar]
- 13.Collins M.H. Serologic tools and strategies to support intervention trials to combat zika virus infection and disease. Trop Med Infect Dis. 2019;4(2):68. doi: 10.3390/tropicalmed4020068. http://www.ncbi.nlm.nih.gov/pubmed/31010134 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckels K.H., De R.A., Barrera L., Putnak J.R. Immunological assays used to support efficacy of zika virus vaccines. Trop Med Infect Dis. 2019;4(3):97. doi: 10.3390/tropicalmed4030097. www.mdpi.com/journal/tropicalmed [cited 2021 Feb 4] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins M.H., Tu H.A., Gimblet-Ochieng C., et al. Human antibody response to Zika targets type-specific quaternary structure epitopes. JCI Insight. 2019;4(8) doi: 10.1172/jci.insight.124588. http://www.ncbi.nlm.nih.gov/pubmed/30996133 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams C., Carbaugh D.L., Shu B., et al. Structure and neutralization mechanism of a human antibody targeting a complex Epitope on Zika virus. PLoS Pathog. 2023;19(1) doi: 10.1371/journal.ppat.1010814. https://pubmed.ncbi.nlm.nih.gov/36626401/ [cited 2023 Jun 16] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasan S.S., Miller A., Sapparapu G., et al. A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat Commun. 2017;8 doi: 10.1038/ncomms14722. http://www.nature.com/doifinder/10.1038/ncomms14722 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long F., Doyle M., Fernandez E., et al. Structural basis of a potent human monoclonal antibody against Zika virus targeting a quaternary epitope. Proc Natl Acad Sci U S A. 2019;116(5):1591–1596. doi: 10.1073/pnas.1815432116. https://www.pnas.org/content/116/5/1591 [cited 2021 May 3] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., Yang H., Liu X., et al. Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus. Sci Transl Med. 2016;8(369):369ra179. doi: 10.1126/scitranslmed.aai8336. http://www.ncbi.nlm.nih.gov/pubmed/27974667 Available from: [DOI] [PubMed] [Google Scholar]
- 20.de Alwis R., Smith S.A., Olivarez N.P., et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A. 2012;109(19):7439–7444. doi: 10.1073/pnas.1200566109. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3358852&tool=pmcentrez&rendertype=abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman N.M., Bucardo F., Collins M.H., et al. Clinical and epidemiological features of acute zika virus infections in León, Nicaragua. Am J Trop Med Hyg. 2021;105(4):924–930. doi: 10.4269/ajtmh.20-1191. https://pubmed.ncbi.nlm.nih.gov/34370700/ [cited 2021 Oct 10] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai L., Rouphael N., Xu Y., et al. Innate, T-, and B-cell responses in acute human zika patients. Clin Infect Dis. 2018;66(1):1–10. doi: 10.1093/cid/cix732. https://academic.oup.com/cid/article/66/1/1/4083516 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed R., Keyserling H., Barrett A., et al. A randomized, double-blind, controlled trial of the 17D yellow fever virus vaccine given in combination with immune globulin or placebo: comparative viremia and immunogenicity. Am J Trop Med Hyg. 2013;88(1):172–177. doi: 10.4269/ajtmh.2012.12-0179. http://www.ncbi.nlm.nih.gov/pubmed/23208880 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardona-Ospina J.A., Trujillo A.M., Jiménez-Posada E.V., et al. Susceptibility to endemic Aedes-borne viruses among pregnant women in Risaralda, Colombia. Int J Infect Dis. 2022;122:832–840. doi: 10.1016/j.ijid.2022.07.017. https://pubmed.ncbi.nlm.nih.gov/35817285/ [cited 2022 Sep 24] Available from: [DOI] [PubMed] [Google Scholar]
- 25.Gaudinski M.R., Houser K.V., Morabito K.M., et al. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet. 2018;391(10120):552–562. doi: 10.1016/S0140-6736(17)33105-7. https://linkinghub.elsevier.com/retrieve/pii/S0140673617331057 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins M.H., McGowan E., Jadi R., et al. Lack of durable cross-neutralizing antibodies against zika virus from dengue virus infection. Emerg Infect Dis. 2017;23(5):773–781. doi: 10.3201/eid2305.161630. http://wwwnc.cdc.gov/eid/article/23/5/16-1630_article.htm Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Premkumar L., Collins M., Graham S., et al. Development of envelope protein antigens to serologically differentiate Zika from dengue virus infection. J Clin Microbiol. 2017;56(3):e01504–e01517. doi: 10.1128/JCM.01504-17. http://www.ncbi.nlm.nih.gov/pubmed/29263206 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willcox A.C., Collins M.H., Jadi R., et al. Seroepidemiology of dengue, zika, and yellow fever viruses among children in the democratic republic of the Congo. Am J Trop Med Hyg. 2018;99(3):756–763. doi: 10.4269/ajtmh.18-0156. http://www.ajtmh.org Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins M.H., Zepeda O., Blette B., et al. Serologic surveillance of maternal Zika infection in a prospective cohort in Leon, Nicaragua during the peak of the Zika epidemic. PLoS One. 2020;15(4) doi: 10.1371/journal.pone.0230692. https://dx.plos.org/10.1371/journal.pone.0230692 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadinegoro S.R., Arredondo-García J.L., Capeding M.R., et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med. 2015;373(13):1195–1206. doi: 10.1056/NEJMoa1506223. http://www.ncbi.nlm.nih.gov/pubmed/26214039 Available from: [DOI] [PubMed] [Google Scholar]
- 31.Henein S., Adams C., Bonaparte M., et al. Dengue vaccine breakthrough infections reveal properties of neutralizing antibodies linked to protection. J Clin Invest. 2021;131(13) doi: 10.1172/JCI147066. pmc/articles/PMC8245170/ [cited 2021 Oct 11] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ka D., Sy K., Km M., et al. Rapid development of a DNA vaccine for Zika virus. Science. 2016;354(6309):237–240. doi: 10.1126/science.aai9137. https://pubmed.ncbi.nlm.nih.gov/27708058/ [cited 2021 Aug 29] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stettler K., Beltramello M., Espinosa D.A., et al. Specificity, cross-reactivity and function of antibodies elicited by Zika virus infection. Science. 2016;353(6301):823–826. doi: 10.1126/science.aaf8505. http://www.ncbi.nlm.nih.gov/pubmed/27417494 Available from: [DOI] [PubMed] [Google Scholar]
- 34.Heinz F.X., Stiasny K. The antigenic structure of zika virus and its relation to other flaviviruses: implications for infection and immunoprophylaxis. Microbiol Mol Biol Rev. 2017;81(1) doi: 10.1128/MMBR.00055-16. http://www.ncbi.nlm.nih.gov/pubmed/28179396 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X., Anderson L.J., Rostad C.A., et al. Development and optimization of a Zika virus antibody-dependent cell-mediated cytotoxicity (ADCC) assay. J Immunol Methods. 2021;488 doi: 10.1016/j.jim.2020.112900. https://pubmed.ncbi.nlm.nih.gov/33075363/ [cited 2022 Oct 31] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierson T.C., Diamond M.S. The continued threat of emerging flaviviruses. Nat Microbiol. 2020;5(6):796–812. doi: 10.1038/s41564-020-0714-0. https://pubmed.ncbi.nlm.nih.gov/32367055/ [cited 2023 Jun 16] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balmaseda A., Stettler K., Medialdea-Carrera R., et al. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci U S A. 2017;114(31):8384–8389. doi: 10.1073/pnas.1704984114. http://www.ncbi.nlm.nih.gov/pubmed/28716913 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowd K.A., DeMaso C.R., Pelc R.S., et al. Broadly neutralizing activity of zika virus-immune sera identifies a single viral serotype. Cell Rep. 2016;16(6):1485–1491. doi: 10.1016/j.celrep.2016.07.049. http://linkinghub.elsevier.com/retrieve/pii/S2211124716309809 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.