Abstract
Background & Aims
Altered plasma acylcarnitine levels are well-known biomarkers for a variety of mitochondrial fatty acid oxidation disorders and can be used as an alternative energy source for the intestinal epithelium when short-chain fatty acids are low. These membrane-permeable fatty acid intermediates are excreted into the gut lumen via bile and are increased in the feces of patients with inflammatory bowel disease (IBD).
Methods
Herein, based on studies in human subjects, animal models, and bacterial cultures, we show a strong positive correlation between fecal carnitine and acylcarnitines and the abundance of Enterobacteriaceae in IBD where they can be consumed by bacteria both in vitro and in vivo.
Results
Carnitine metabolism promotes the growth of Escherichia coli via anaerobic respiration dependent on the cai operon, and acetylcarnitine dietary supplementation increases fecal carnitine levels with enhanced intestinal colonization of the enteric pathogen Citrobacter rodentium.
Conclusions
In total, these results indicate that the increased luminal concentrations of carnitine and acylcarnitines in patients with IBD may promote the expansion of pathobionts belonging to the Enterobacteriaceae family, thereby contributing to disease pathogenesis.
Keywords: Microbiota, IBD, Metabolism, Carnitine
Graphical abstract
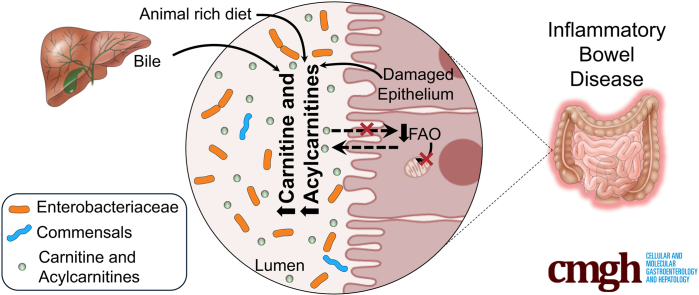
Summary.
The gut microbiota in inflammatory bowel disease is dysbiotic with increased Enterobacteriaceae as well as metabolites such as carnitine and acylcarnitines. The gut microbiota can consume these metabolites, thereby promoting the growth of specific Enterobacteriaceae species both in vitro and in vivo.
Alteration of gut microbiota composition associated with chronic diseases, sometimes referred to as dysbiosis, has been best characterized for intestinal inflammatory disease states that are mediated immunologically, such as in inflammatory bowel diseases (IBDs), as well as in infections such as Clostridioides difficile.1, 2, 3, 4 Not only do these alterations serve as biomarkers of disease, studies in rodent models provide evidence that the dysbiotic microbiota plays a role in disease pathogenesis,5 a notion that also may be the case in human beings based on the results of studies showing efficacy of fecal microbiota transplantation in the treatment of certain diseases such as C difficile infection.6, 7, 8 Dysbiosis typically features a decrease in bacterial diversity and an increase in the proportion of taxa belonging to the Proteobacteria phylum, particularly Enterobacteriaceae.9
The etiology of the dysbiotic microbiota is partially dependent on enhanced anaerobic respiration and tolerance of oxidative stress by Enterobacteriaceae.10,11 In turn, dysbiosis has been associated consistently with alterations in the fecal metabolome including the reduction in both short-chain fatty acid (SCFA) production and conversion of primary to secondary bile acids.12 Recently, we and others have shown that fecal acylcarnitines are increased in patients with IBD, especially during dysbiosis.12,13
The function of acylcarnitines has been well characterized in mammalian physiology. Fatty acids are esterified to coenzyme A (CoA) to form acyl-CoAs upon cellular uptake. Long-chain acyl-CoAs (>C12) are converted to acylcarnitines at the outer mitochondrial membrane via the activity of carnitine palmitoyltransferase in the presence of the quaternary amine, carnitine, which allows for their transport into the mitochondrial matrix where β-oxidation occurs. Hepatically produced acylcarnitines can be either transported into the plasma for systemic circulation or secreted into bile for excretion. Increased levels in blood and urine serve as biomarkers for inborn errors of fatty acid metabolism and are monitored in newborn screening panels.14,15 In addition, increased plasma acylcarnitines are associated with diabetes, sepsis, cancer, nonalcoholic liver disease, and heart failure.16, 17, 18, 19 Plasma levels of acylcarnitines also are increased during fasting and exercise, and are thought to represent incomplete fatty acid oxidation.4,20,21 In addition, these molecules serve as an energy source in brown adipose tissue during thermogenesis and in the intestinal epithelium in the absence of SCFAs.13,22 However, the mechanism(s) responsible for the increase of fecal acylcarnitines in IBD and their biological function have not been elucidated.
IBD is associated with alterations in intestinal fatty acid metabolism.23 Rats treated rectally with a fatty acid oxidation inhibitor developed acute colitis and perturbations that decrease the cellular concentrations of coenzyme A and carnitine, essential co-factors in fatty acid oxidation, increase susceptibility to IBD.24, 25, 26 The primary energy substrate for the colonic epithelium, SCFAs, are reduced in the feces of patients with dysbiosis in intestinal inflammatory disease states.12,27 Decreased SCFA availability in the gut has been hypothesized to play a role in the pathogenesis of IBD via reduced colonic barrier function.23 Recently, we showed that acylcarnitines serve as an alternative energy source for the colonic epithelium when mitochondrial dysfunction reduces both SCFAs and longer-chain acylcarnitine oxidation during intestinal inflammation.13
Herein, we provide a detailed characterization of both carnitine and acylcarnitines associated with pediatric IBD. Levels were correlated positively with both dysbiosis and disease activity, as quantified by fecal calprotectin levels. Using mouse models, we provide evidence that the increase of carnitine and acylcarnitines in the gut lumen are the result of host-dependent factors, including an increase in biliary secretions and possibly the release of host cellular material into the gut lumen resulting from intestinal damage. A comparative analysis of conventionally housed and germ-free mice, as well as in a human subject intervention study, suggests that carnitine and acylcarnitines are consumed by the gut microbiota. A bacterial screen of several facultative and obligate anaerobic bacterial strains showed species-specific patterns in carnitine and acylcarnitine consumption. In particular, carnitine, which was consumed by Escherichia coli in vitro, also was correlated positively with both E coli abundance in fecal samples from patients with IBD and healthy human subjects consuming an omnivorous diet, as well as abundance of genes in the cai operon, which regulates bacterial carnitine metabolism under anaerobic conditions. We show that cai operon–dependent metabolism of carnitine promotes anaerobic growth of E coli on an alternative carbon source. Furthermore, acetylcarnitine dietary supplementation increases carnitine levels in the gut and promotes the growth of Citrobacter rodentium, a rodent model of enteropathogenic E coli infection. In total, these results provide evidence for the impact of host-derived fatty acid oxidation intermediates on the gut microbiota in IBD.
Results
Carnitine and Acylcarnitines Are Correlated With Dysbiosis in Pediatric IBD
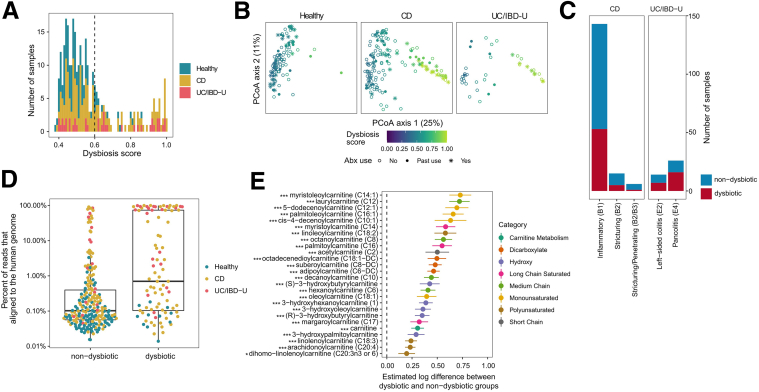
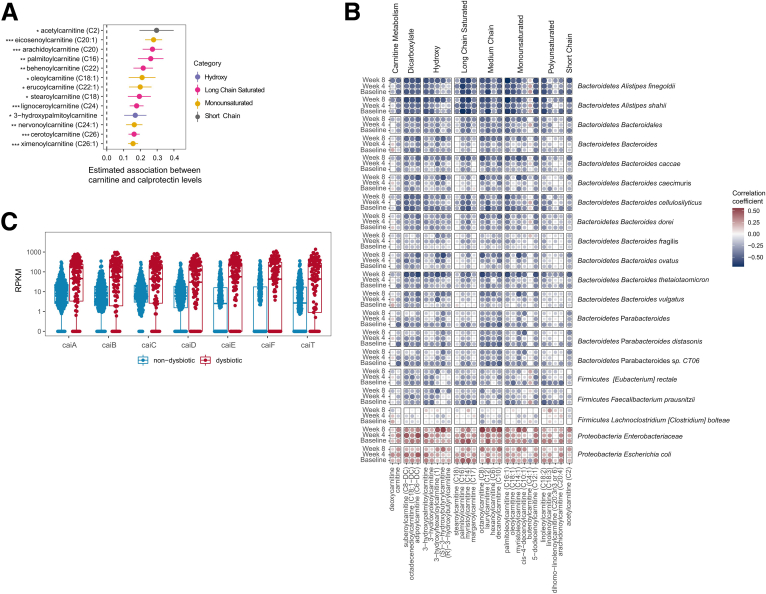
Fecal acylcarnitine levels recently were shown to be a robust biomarker for IBD in which strong associations were observed consistently between numerous acylcarnitine species and dysbiosis, the exception being C20:4 carnitine, which was associated negatively.12,13 To determine whether similar associations could be observed in a prospective cohort of patients with pediatric ulcerative colitis (UC)/IBD unclassified (IBD-U)28 and Crohn’s disease (CD), as well as identify correlations with disease phenotype and activity, we analyzed fecal acylcarnitines in the DYsbiosis, iNflammatory bowel disease, and Antibiotics in the MICrobiome (DYNAMIC) pediatric IBD study.2 The number of patients with UC, IBD-U, CD, and healthy controls, as well as their clinical characteristics, are provided in Tables 1 and 2. Fecal samples at baseline, 4 weeks, and 8 weeks were collected and analyzed by both shotgun metagenomic sequencing and liquid chromatography/mass spectrometry. Patients with a dysbiotic microbiota were identified using a previously described dysbiosis scoring method12 at baseline (Figure 1A). A principal component analysis showed that dysbiosis was observed in both CD and UC, where it was associated with antibiotic use (Figure 1B). Although the greatest number of patients in the DYNAMIC cohort used for this study had the inflammatory B1 phenotype in CD, there was no statistically significant correlation between the dysbiotic microbiota and clinical phenotypes for either CD or UC/IBD-U (Figure 1C). Dysbiosis also was associated with the abundance of human fecal DNA consistent with the notion that greater levels of human cellular material are released into the gut lumen in patients with dysbiosis, similar to our previously described observations29 (Figure 1D). Linear mixed-effects modeling was used to estimate the difference in log acylcarnitine levels across study groups with antibiotic use included as a covariate. Carnitine as well as all 25 acylcarnitines measured were associated with a dysbiotic microbiota across all 3 time points (Figure 1E). Linear-effects modeling also showed that acylcarnitines also were highly correlated with disease activity as quantified by fecal calprotectin levels (Figure 2A). Finally, carnitine and many acylcarnitines were correlated negatively with many bacterial species consistently across all 3 time points (Figure 2B). By contrast, the family Enterobacteriaceae, including the species E coli, were correlated positively with carnitine and these acylcarnitines. Interestingly, these correlations were not observed for the longest chain acylcarnitines (>C20). Although information about the metabolism of acylcarnitines by bacterial species is limited,30 anaerobic metabolism of carnitine by the cai operon in Enterobacteriaceae has been described previously.30,31 Consistent with this, the abundance of various genes in the cai operon were correlated positively with dysbiosis in IBD (Figure 2C and Table 3). These results provide evidence that fecal levels of carnitine and acylcarnitines are correlated with the dysbiotic microbiota in pediatric patients with IBD.
Table 1.
Demographics of Patient Population in the DYNAMIC Cohort
| Healthy | IBD | |
|---|---|---|
| Pediatric patients, n | 37 | 69 |
| Median age at enrollment, y (range) | 14.08 (5.60–19.74) | 14.81 (7.53–19.89) |
| Female, n (%) | 27 (72.97) | 29 (52.83) |
| Ethnicity: not Hispanic, n (%) | 33 (89.19) | 65 (94.20) |
| Race, n (%) | ||
| Caucasian | 27 (72.97) | 55 (79.71) |
| Asian | 2 (5.41) | 1 (1.45) |
| Black/African American | 7 (18.92) | 10 (14.49) |
| Mixed race | 0 (0.00) | 3 (4.35) |
| Decline to answer | 1 (2.70) | 0 (0.00) |
| Current antibiotic use at enrollment, n (%) | 12 (32.43) | 14 (20.29) |
| Antibiotic use within 90 days before enrollment, n (%) | 8 (21.62) | 12 (17.39) |
Table 2.
Clinical Characteristics of Pediatric Patients with Inflammatory Bowel Disease in the DYNAMIC Cohort
| Diagnosis, n (%) | |
| Crohn’s disease | 55 (79.71) |
| Ulcerative colitis | 8 (11.59) |
| IBD-U | 6 (8.70) |
| Median age at diagnosis, y (range) | 12.97 (7.52–17.74) |
| Years since diagnosis at enrollment, median (range) | 0.02 (0.00–9.66) |
| Crohn’s disease–Paris Classification lower GI tract location, n (%) | |
| Ileal L1 | 12 (22.64) |
| Colonic L2 | 9 (16.98) |
| IIeocolonic L3 | 30 (56.60) |
| None | 2 (3.77) |
| Crohn’s disease–Paris Classification phenotype, n (%) | |
| Inflammatory B1 | 48 (87.27) |
| Stricturing B2 | 5 (9.09) |
| Stricturing/penetrating B2/B3 | 2 (3.64) |
| Ulcerative colitis extent/IBD-U, n (%) | |
| E2 left-sided colitis | 5 (41.67) |
| E4 pancolitis | 7 (58.33) |
| Patients initiating therapy at enrollment, n (%) | 31 (44.93) |
| Current medication for IBD, n (%) | |
| Mesalamine | 26 (37.68) |
| Biologic | 44 (63.77) |
| Steroids | 8 (11.59) |
| Immunomodulators | 15 (21.74) |
| Calprotectin level, ug/dL, median (IQR) | |
| Baseline | 689 (180–1702.5) |
| Week 4 | 421 (94–998) |
| Week 8 | 300 (48–809) |
| Disease activity index, median (range) | |
| Pediatric Crohn’s Disease Activity Index | |
| Baseline | 30 (0–50) |
| Week 4 | 7.5 (0–37.5) |
| Week 8 | 7.5 (0–55) |
| Pediatric Ulcerative Colitis Activity Index | |
| Baseline | 45 (0–75) |
| Week 4 | 10 (0–40) |
| Week 8 | 10 (0–45) |
GI, gastrointestinal; IQR, interquartile range.
Figure 1.
Correlation analysis of dysbiosis in IBD and fecal acylcarnitines levels. (A) Distribution of microbial dysbiosis scores among all patient samples by diagnosis: CD, UC, and IBD-U. Dotted line denotes the threshold above which samples are classified as dysbiotic. (B) Principal component analysis (PCoA) plots based on the Bray–Curtis dissimilarity of the gut microbiome. All samples were used to make a common plot, then samples by diagnosis are each displayed separately. Dysbiosis scores are color coded, and symbols reflect antibiotic use. (C) Association between dysbiosis and IBD clinical phenotypes. (D) Association between the percentage sequencing reads aligned to the human genome and the presence of dysbiosis. (E) The 26 metabolites increased significantly in dysbiotic samples as compared with nondysbiotic samples. The x-axis is the estimated log difference of metabolite levels between dysbiotic and nondysbiotic groups. Various structural categories of acylcarnitines have been color coded. False discovery rate: ∗P < .05 and ∗∗∗P < .001. Abx, antibiotics.
Figure 2.
Acylcarnitine levels are correlated with fecal calprotectin and bacterial taxa. (A) Association between acylcarnitines and calprotectin in pediatric patients with IBD with acylcarnitines. Various structural categories of acylcarnitines have been color coded. False discovery rate: ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. (B) Spearman correlation between fecal acylcarnitines with bacterial taxa at individual time points among all pediatric subjects. The size of each circle indicates the strength of correlation and the correlations have been color coded as shown in the panel. Blue indicates a negative correlation and red indicates a positive correlation. (C) Reads per kilobase million (RPKM) of genes in the cai operon across samples labeled as nondysbiotic and dysbiotic.
Table 3.
Linear Mixed-Effects Modeling of Time-Dependent Alterations in the FARMM Study Comparing Alterations in the Relative Abundance of Genes in the cai Operon Across the 3 Diets Before and After Antibiotic Treatment
| KEGG Orthology Identifier | Gene name | Term | Value | SE | Degrees of Freedom | t value | P value | FDR | |
|---|---|---|---|---|---|---|---|---|---|
| Antibiotic treatment | K05245 | caiT | Study day | -0.70 | 0.26 | 41 | -2.72 | .01 | 0.022 |
| K08299 | caiD | Study day | -1.13 | 0.25 | 41 | -4.43 | .00 | 0.00018 | |
| Postantibiotics | K02182 | caiC | Study day | 0.43 | 0.08 | 157 | 5.32 | .00 | 1.1e-06 |
| K05245 | caiT | Study day | 0.56 | 0.08 | 157 | 7.19 | .00 | 1.9e-10 | |
| K05245 | caiT | Study day | -0.36 | 0.11 | 157 | -3.19 | .00 | 0.036 | |
| K08277 | caiF | Study day | 0.60 | 0.08 | 157 | 7.18 | .00 | 1.9e-10 | |
| K08279 | caiE | Study day | 0.54 | 0.08 | 157 | 6.96 | .00 | 4.6e-10 | |
| K08297 | caiA | Study day | 0.50 | 0.08 | 157 | 6.12 | .00 | 2.6e-08 | |
| K08298 | caiB | Study day | 0.58 | 0.08 | 157 | 7.18 | .00 | 1.9e-10 | |
| K08299 | caiD | Study day | 0.51 | 0.08 | 157 | 6.63 | .00 | 2.1e-09 |
FDR, false discovery rate. See Figure 3C.
Microbiome Depletion Is Associated With Higher Luminal Concentrations of Carnitine and Acylcarnitines
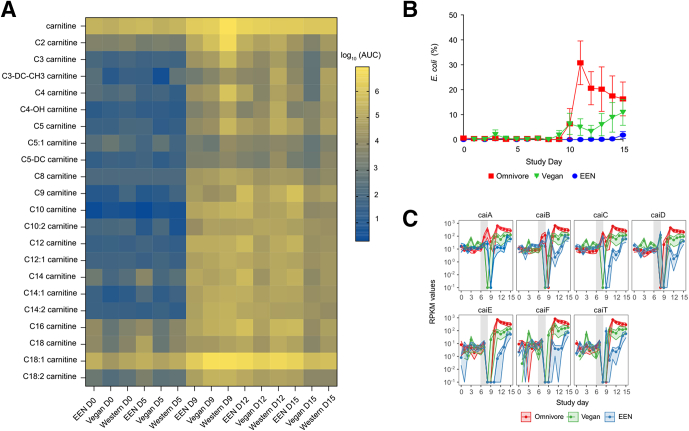
We previously provided evidence in batch culture that a specific acylcarnitine, palmitoyl carnitine (C16), can be consumed by a complex gut microbial community.13 To determine if the gut microbiota consumes carnitine and acylcarnitines in human beings, we analyzed fecal samples collected from subjects in the Food and Resulting Microbiota and Metabolome (FARMM) study.32 Subjects in this study consumed either a Western (omnivore), vegan, or exclusive enteral nutrition (EEN) diet that lacked fiber over a 15-day period. The dietary phase (days 1–5) was followed by a gut purge with polyethylene glycol and treatment with 2 oral antibiotics (days 6–9), after which the recovery of the microbiota was monitored from days 9 to 15. The reduction in gut microbiota biomass, by approximately 5 logs,32 by the gut purge at the end of day 9 led to a significant increase in fecal carnitine and nearly all acylcarnitines measured (Figure 3A) (all 21 acylcarnitines measured were significantly different in the vegan and omnivore groups between days 5 and 9, and 18 acylcarnitines were significantly different in the EEN group; paired Wilcoxon tests with false discovery rate corrections, adjusted P < .05) (Table 4). This effect occurred across all 3 diets, although the levels of carnitine as well as C2, C3, C4, and C5 acylcarnitines were higher on a Western diet after the purge (all higher in the omnivore group relative to the vegan and EEN groups; adjusted P < .05 using the Dunn multiple comparison test). Carnitine is particularly abundant in red meat and other animal products, and previous studies have shown that circulating levels are higher in omnivores compared with vegans or vegetarians.33 By day 15, there also were lower levels of numerous acylcarnitines on a Western and vegan diet relative to EEN (Dunn multiple comparisons test, adjusted P < .05) (Table 4), which is consistent with the delayed recovery of the gut microbiota in the absence of dietary fiber in EEN.32
Figure 3.
Fecal acylcarnitines over the course of a human dietary intervention study and corresponding changes in E coli abundance. (A) Heatmap of log-transformed average acylcarnitine levels in the FARMM study. Values below the limit of detection were imputed as one half of the lowest detected level for each carnitine species. (B) Average relative abundance of E coli over the course of the FARMM study. Data are expressed as means ± SEM. (C) Reads per kilobase million (RPKM) of genes in the cai operon throughout the course of the FARMM study in which the 3 diets consumed have been color coded as indicated. AUC, area under the curve.
Table 4.
Statistical Analyses for Time- and Diet-Dependent Alterations in Fecal Carnitine Acylcarnitine Levels in the FARMM Study
| Paired Wilcoxon tests with FDR corrections | EEN | Vegan | Omnivore |
|---|---|---|---|
| Day 5 vs 9, within group | |||
| Carnitine | 0.021 | 0.000 | 0.002 |
| C2 carnitine | 0.012 | 0.000 | 0.002 |
| C3 carnitine | 0.005 | 0.000 | 0.002 |
| C3–DC–CH3 carnitine | 0.033 | 0.000 | 0.002 |
| C4 carnitine | 0.016 | 0.000 | 0.002 |
| C4–OH carnitine | 0.005 | 0.000 | 0.002 |
| C5 carnitine | 0.005 | 0.000 | 0.002 |
| C5:1 carnitine | 0.008 | 0.023 | 0.002 |
| C5–DC carnitine | 0.008 | 0.000 | 0.002 |
| C8 carnitine | 0.005 | 0.000 | 0.002 |
| C9 carnitine | 0.005 | 0.000 | 0.002 |
| C10 carnitine | 0.005 | 0.000 | 0.002 |
| C10:2 carnitine | 0.005 | 0.000 | 0.002 |
| C12 carnitine | 0.005 | 0.000 | 0.002 |
| C12:1 carnitine | 0.005 | 0.000 | 0.002 |
| C14 carnitine | 0.041 | 0.000 | 0.002 |
| C14:1 carnitine | 0.009 | 0.000 | 0.002 |
| C14:2 carnitine | 0.005 | 0.000 | 0.002 |
| C16 carnitine | 0.271 | 0.000 | 0.002 |
| C18 carnitine | 0.092 | 0.000 | 0.002 |
| C18:1 carnitine | 0.064 | 0.000 | 0.002 |
| C18:2 carnitine | 0.033 | 0.000 | 0.002 |
| Dunn multiple comparison tests | EEN vs vegan | EEN vs omnivore | Vegan vs omnivore |
|---|---|---|---|
| Day 15, between groups | |||
| Carnitine | >0.9999 | 0.7912 | >0.9999 |
| C2 carnitine | 0.1426 | >0.9999 | 0.3824 |
| C3 carnitine | 0.0752 | >0.9999 | 0.6092 |
| C3–DC–CH3 carnitine | 0.5854 | 0.0089 | <0.0001 |
| C4 carnitine | 0.1512 | 0.8924 | 0.0081 |
| C4–OH carnitine | 0.8753 | 0.001 | 0.0332 |
| C5 carnitine | 0.0564 | >0.9999 | 0.0045 |
| C5:1 carnitine | 0.0582 | >0.9999 | 0.0168 |
| C5–DC carnitine | >0.9999 | 0.2252 | 0.0274 |
| C8 carnitine | 0.0356 | >0.9999 | 0.3282 |
| C9 carnitine | 0.0002 | 0.0144 | 0.6683 |
| C10 carnitine | 0.001 | 0.015 | >0.9999 |
| C10:2 carnitine | >0.9999 | >0.9999 | >0.9999 |
| C12 carnitine | 0.0002 | 0.0033 | >0.9999 |
| C12:1 carnitine | 0.0645 | >0.9999 | 0.3637 |
| C14 carnitine | <0.0001 | 0.0082 | 0.3121 |
| C14:1 carnitine | 0.0287 | 0.0287 | >0.9999 |
| C14:2 carnitine | >0.9999 | 0.147 | 0.7911 |
| C16 carnitine | <0.0001 | 0.0024 | >0.9999 |
| C18 carnitine | 0.0001 | 0.0384 | 0.2961 |
| C18:1 carnitine | 0.0357 | 0.0069 | >0.9999 |
| C18:2 carnitine | 0.0925 | 0.0034 | 0.8241 |
DC, decarboxylated; FDR, false discovery rate; OH, hydroxylated. For additional details see Figure 3A.
Given the positive association of Enterobacteriaceae and E coli with fecal carnitine and acylcarnitines in the DYNAMIC cohort (Figure 2B), and the association of dysbiosis with the cai operon (Figure 2C), we examined the abundance of E coli across the 3 diets in the FARMM study. We observed high levels of E coli after day 9, predominantly in subjects consuming a Western diet (Figure 3B). Mixed-effects linear modeling showed a significant time (P = .0051) and group effect (P = .0042) between the omnivore and the other 2 diets. Given that E coli is capable of metabolizing carnitine via the cai operon, we investigated the abundance of the cai operon genes throughout the course of the study.34 Although the relative abundance of these genes were similar for all diets before microbiome depletion, gene abundance increased in all dietary groups during the recovery phase (Figure 3C and Table 3). However, from day 11 onward, the abundance of genes in the cai operon were significantly greater in the fecal microbiomes of omnivore study participants compared with the vegan and EEN groups (Figure 3C and Table 5).
Table 5.
Linear Mixed-Effects Modeling of Diet-Dependent Alterations in the FARMM Study Comparing Omnivore With Either EEN or Vegan Diets in the Relative Abundance of Genes in the cai Operon
| KEGG Orthology Identifier | Gene name | Term | Value | SE | Degrees of Freedom | t value | P value | FDR |
|---|---|---|---|---|---|---|---|---|
| K02182 | caiC | Study group vegan | -6.64 | 2.05 | 15 | -3.24 | .00 | 0.012 |
| K02182 | caiC | Study group EEN | -9.09 | 2.04 | 15 | -4.45 | .00 | 0.00036 |
| K02182 | caiC | Study group vegan: study day | 0.43 | 0.15 | 101 | 2.87 | .00 | 0.017 |
| K02182 | caiC | Study group EEN: study day | 0.52 | 0.15 | 101 | 3.48 | .00 | 0.0019 |
| K05245 | caiT | Study group vegan | -5.55 | 1.93 | 25 | -2.87 | .01 | 0.014 |
| K05245 | caiT | Study group EEN | -8.04 | 1.93 | 25 | -4.17 | .00 | 0.00056 |
| K05245 | caiT | Study group vegan: study day | 0.34 | 0.14 | 101 | 2.42 | .02 | 0.03 |
| K05245 | caiT | Study group EEN: study day | 0.40 | 0.14 | 101 | 2.80 | .01 | 0.011 |
| K08277 | caiF | Study group vegan | -6.26 | 2.10 | 25 | -2.97 | .01 | 0.014 |
| K08277 | caiF | Study group EEN | -8.00 | 2.10 | 25 | -3.82 | .00 | 0.0011 |
| K08277 | caiF | Study group vegan: study day | 0.39 | 0.16 | 101 | 2.50 | .01 | 0.03 |
| K08277 | caiF | Study group EEN: study day | 0.39 | 0.15 | 101 | 2.54 | .01 | 0.018 |
| K08279 | caiE | Study group vegan | -6.98 | 1.95 | 25 | -3.58 | .00 | 0.01 |
| K08279 | caiE | Study group EEN | -9.43 | 1.94 | 25 | -4.85 | .00 | 0.00028 |
| K08279 | caiE | Study group vegan: study day | 0.45 | 0.14 | 101 | 3.13 | .00 | 0.016 |
| K08279 | caiE | Study group EEN: study day | 0.49 | 0.14 | 101 | 3.46 | .00 | 0.0019 |
| K08297 | caiA | Study group vegan | -5.59 | 2.29 | 25 | -2.44 | .02 | 0.022 |
| K08297 | caiA | Study group EEN | -6.50 | 2.28 | 25 | -2.86 | .01 | 0.0085 |
| K08297 | caiA | Study group vegan: study day | 0.35 | 0.17 | 101 | 2.02 | .05 | 0.048 |
| K08298 | caiB | Study group vegan | -5.26 | 1.95 | 25 | -2.69 | .01 | 0.017 |
| K08298 | caiB | Study group EEN | -9.17 | 1.95 | 25 | -4.71 | .00 | 0.00028 |
| K08298 | caiB | Study group vegan: study day | 0.33 | 0.14 | 101 | 2.28 | .02 | 0.034 |
| K08298 | caiB | Study group EEN: study day | 0.53 | 0.14 | 101 | 3.65 | .00 | 0.0019 |
| K08299 | caiD | Study group vegan | -5.10 | 2.05 | 25 | -2.49 | .02 | 0.022 |
| K08299 | caiD | Study group EEN | -6.30 | 2.04 | 25 | -3.09 | .00 | 0.0057 |
| K08299 | caiD | Study group vegan: study day | 0.30 | 0.15 | 101 | 2.01 | .05 | 0.048 |
FDR, false discovery rate. For additional details see Figure 3C.
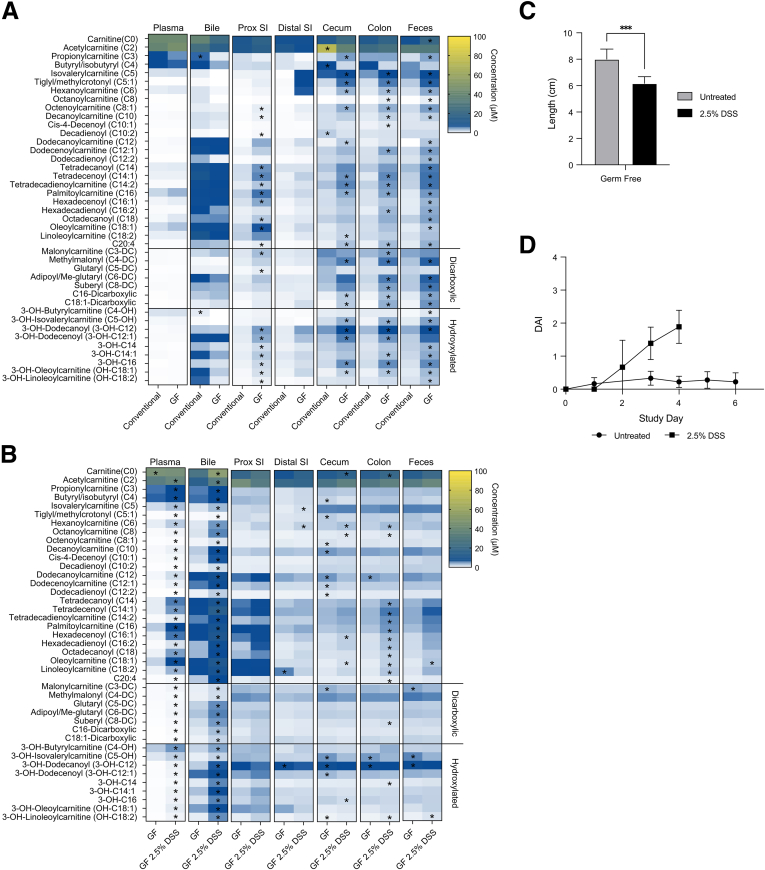
To characterize the effect of the gut microbiota on carnitine and acylcarnitine availability throughout the length of the intestinal tract, as well as in bile and plasma, we quantified their abundance in conventionally housed and germ-free mice (Figure 4A). We previously proposed that the high levels of acylcarnitines in bile is a major contributor to fecal levels in mice.13 Despite somewhat lower levels of carnitine and acylcarnitines in the bile of germ-free mice relative to conventionally housed mice, germ-free mice had significantly higher levels of both throughout the length of their intestinal tract, with the most robust differences observed in the cecum, colon, and feces (Figure 4A). Together with the results of the FARMM study, this provides evidence that the gut microbiota consumes both carnitine and acylcarnitines.
Figure 4.
Quantification of acylcarnitine levels in conventional and germ-free mice. (A) Heatmap of carnitine and acylcarnitine concentrations in various biospecimens from conventionally raised and germ-free (GF) mice, n = 4–5 for conventional mice and n = 5 for germ-free mice. (B) Heatmap of carnitine and acylcarnitine concentrations in various biospecimens from GF mice with and without treatment with 2.5% DSS to induce colitis, n = 4–6 for GF mice and n = 5–6 for GF mice with 2.5% DSS. ∗Significant P value using unpaired t tests with false discovery rate corrections. (C) Colonic length of germ-free mice treated with 2.5% DSS in drinking water to induce colitis as well as germ-free controls. n = 6. ∗∗∗P < .001. (D) Time-dependent Disease Activity Index (DAI) in germ-free mice treated with 2.5% DSS in drinking water to induce colitis as well as germ-free controls. n = 6 in each group. DSS-treated mice met the threshold for euthanasia by day 4 of the study. Prox, proximal; SI, small intestine.
Colitis Leads to an Increase in Luminal Carnitine and Acylcarnitines in the Bile and Plasma as Well as in the Intestinal Tract
The mechanism(s) leading to increased fecal levels of carnitine and acylcarnitines in patients with IBD (Figures 1 and 2) have not been defined and could be influenced by both host- and gut microbiota–dependent factors. Because the microbiota has an impact on the levels of carnitine and acylcarnitines in the intestinal lumen (Figure 4A), we examined whether intestinal inflammation could increase luminal levels independent of the microbiota. Because dextran sodium sulfate (DSS)-induced colitis is one of the only described models of intestinal inflammation in the absence of a microbiota, we evaluated the effect of DSS on carnitine and acylcarnitine levels in germ-free mice.35 DSS (2.5%) in drinking water induced colitis, leading to a shortening of the colon as well as a significant increase in disease quantified by a previously described disease activity index36 (Figure 4C and D). Germ-free mice are particularly sensitive to DSS treatment and reached humane end point criteria before the predetermined end of the experiment. We found that colitis led to a significant increase in carnitine and acylcarnitine levels in the bile and plasma of DSS-treated compared with untreated germ-free mice (Figure 4B). Colitis also led to a more modest increase in levels of medium-chain acylcarnitines in the colon, which may reflect host cellular material, such as colonic epithelium, inflammatory cells, and blood, being released into gut lumen. This would be consistent with the higher levels of human DNA associated with dysbiosis in patients with IBD (Figure 1D).29 The mechanism by which colitis leads to higher levels of acylcarnitines in both bile and plasma, presumably a reflection of enhanced hepatic synthesis,22,37 remains to be characterized. These larger pools may serve as an important source of energy for both brown adipose tissue to maintain thermal regulation22,37, 38, 39 and the intestinal epithelium for epithelial regeneration in the setting of colitis.13,40 Because the gut microbiota consumes both carnitine and acylcarnitines (Figures 3A and 4A), these results are consistent with the notion that the increase of these metabolites in the feces of patients with IBD is driven by increased biliary delivery of acylcarnitines to the lumen combined with the reduced number and function of mitochondria in the colonic epithelium as previously reported.13
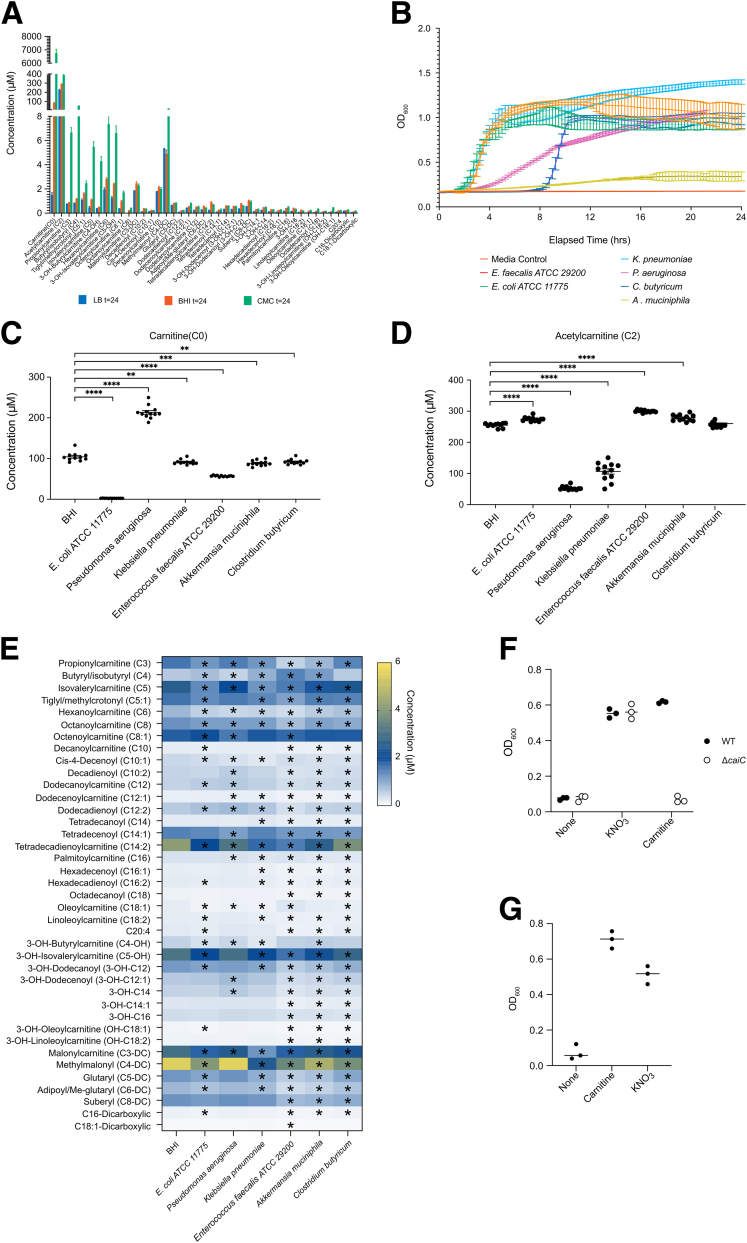
Multiple Bacterial Species Consume Carnitine and Acylcarnitines in a Taxon-Specific Pattern
Consumption of carnitine and acylcarnitines by the gut microbiota (Figures 3A and 4A) suggests that enhanced delivery of these molecules by the host during colitis might have a functional effect on the gut microbiota. To identify the capability of several bacterial species to consume carnitine and acylcarnitines, we tested if routine bacterial culture media contains quantifiable levels of these molecules because most contain cellular components derived from eukaryotic organisms. Indeed, lysogeny broth (LB), brain heart infusion (BHI), and chopped meat carbohydrate broth all contain carnitine and acylcarnitines (Figure 5A) at levels similar to those observed in fecal material (Figure 4A).13 After anaerobic growth of 6 different enteric bacterial species individually in BHI media for 24 hours (Figure 5B), carnitine and acylcarnitine levels were quantified. E coli and Pseudomonas aeruginosa were included as positive controls because previous literature has reported their ability to consume carnitine and short-chain acylcarnitines, respectively.41,42 Carnitine indeed was consumed by E coli whereas growth of P aeruginosa led to higher levels of carnitine in the medium, likely owing to consumption and hydrolysis of acetylcarnitine resulting in carnitine excretion by the hydrolase of O-acylcarnitine, short chains (HocS) esterase (Figure 5C and D).42 Novel patterns of carnitine and acylcarnitine consumption also were observed. For example, unlike P aeruginosa, the consumption of acetylcarnitine by Klebsiella pneumoniae did not lead to an increase in carnitine in BHI medium, possibly owing to an endogenous ability to consume carnitine. By profiling the abundance of a variety of acylcarnitines, we provide evidence that individual bacterial species may consume numerous acylcarnitines in distinct patterns within the context of a rich bacterial culture medium (Figure 5E).
Figure 5.
Consumption of carnitine and acylcarnitines by various bacterial species and the effect of the cai operon on anaerobic respiration. (A) Concentration of acylcarnitines naturally present in rich bacterial culture media, LB, BHI, and chopped meat carbohydrate (CMC) broth. (B) Growth curves for bacterial strains under anaerobic conditions at 37°C in BHI, n = 12. Concentrations of (C) carnitine, (D) acetylcarnitine, and (E) longer-chain acylcarnitines in BHI media after a 24-hour incubation with or without bacteria under anaerobic conditions at 37°C. n = 12. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001 after unpaired t test with false discovery rate correction. (F) Anaerobic growth of wild-type (WT) E coli MP1 and a mutant of the cai operon (ΔcaiC) in M9 minimal medium with glycerol as the sole carbon source in the presence of either nitrate or carnitine. (G) Growth of the adherent invasive E coli strain NRG 857C in M9 minimal medium with glycerol as the sole carbon source in the presence of either nitrate or carnitine. OD600, optical density at 600 nm.
The positive correlation between Enterobacteriaceae and fecal carnitine and acylcarnitines in IBD (Figure 1E) is consistent with previous studies showing that anaerobic supplementation of Enterobacteriaceae with carnitine or its metabolite, crotonobetaine, induces the expression of the caiTABCDE and fixABCX operons and stimulates growth.31,43 The expression of these proteins drives the conversion of carnitine to γ-butyrobetaine via crotonobetaine. Enterobacteriaceae thus may utilize carnitine metabolism to serve as an electron acceptor to enhance their growth.44 Indeed, the cai operon genes are correlated positively with dysbiosis in IBD (Figure 2C), which suggests their importance within the dysbiotic community. To directly demonstrate the role that the cai operon plays in anaerobic growth, we examined the ability of both a wild-type and an isogenic caiC mutant strain (ΔcaiC) of E coli to use glycerol for growth as a carbon source under anaerobic conditions, a process that that requires an electron acceptor (Figure 5F). Anaerobic growth on glycerol was observed in both strains in the presence of nitrate as an electron acceptor, but is observed in the wild-type but not the ΔcaiC mutant strain in the presence of L-carnitine as an electron acceptor. Similar results were obtained using the well-characterized adherent invasive E coli strain NRG 857C associated with CD (Figure 5G).45 Indeed, in a study of roughly 7500 high-quality E coli genomes, caiA and caiB were present in 98% of the strains, suggesting that the utilization of carnitine in this manner may be a general characteristic of E coli.46
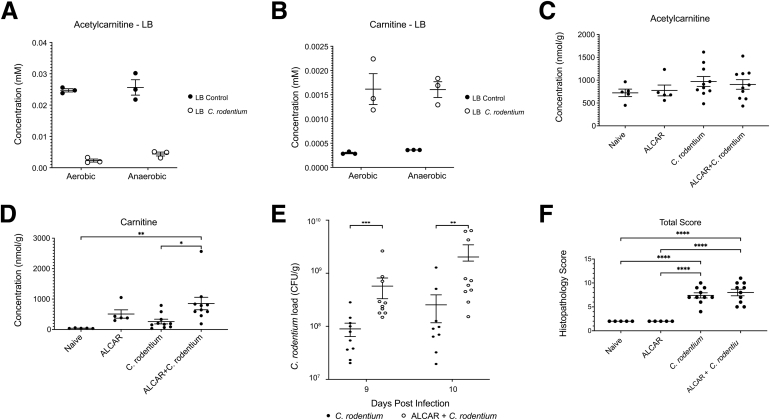
Carnitine and Acetylcarnitine Promote the Growth of Enterobacteriaceae Both In Vitro and in the Murine Gut
Because acetylcarnitine is the most abundant acylcarnitine in the cecum and colon of mice (Figure 4A), we examined the ability of C rodentium, an Enterobacteriaceae model of human enteropathogenic and enterohemorrhagic E coli infection,47 to consume acetylcarnitine and determined its effect on the development of murine colitis. Acetylcarnitine is depleted in LB by the growth of C rodentium overnight in culture (Figure 6A), with an associated increase in carnitine (Figure 6B). Although dietary supplementation of acetylcarnitine in drinking water did not alter fecal levels of acetylcarnitine (Figure 6C), similar to the effects observed in LB there was an increase in fecal carnitine levels both without and with C rodentium infection (Figure 6D). Dietary acetylcarnitine supplementation also led to a significant increase in C rodentium bacterial load (Figure 6E), although there was only a trend toward worsened disease by histologic scoring of colonic inflammation (Figure 6F). In total, these results support the notion that carnitine and acylcarnitines may play a role in IBD disease pathogenesis by altering the metabolic physiology of Enterobacteriaceae.
Figure 6.
The effect of carnitine and acetylcarnitine metabolism on the growth of Enterobacteriaceae in vitro and in vivo. (A) Acetylcarnitine levels before and after overnight aerobic and anaerobic culture of C rodentium in LB broth. (B) Carnitine levels before and after overnight aerobic and anaerobic culture of C rodentium in LB broth. (C) Cecal acetylcarnitine levels in naïve and C rodentium–infected mice without and with acetylcarnitine dietary supplementation (P = .43, 1-way analysis of variance [ANOVA]). (D) Cecal carnitine levels in naïve and C rodentium–infected mice without and with acetylcarnitine dietary supplementation (P = .006, 1-way ANOVA; P = .02 and P = .009, ALCAR + C rodentium vs C rodentium alone and naïve, respectively; Tukey multiple comparisons tests). (E) C rodentium load on days 9 and 10 postinfection in mice with and without acetylcarnitine dietary supplementation, n = 10 per condition. ∗∗P < .01, ∗∗∗P < .001 by unpaired t test. (F) Effect of C rodentium infection and 15 mmol/L acetylcarnitine supplementation in drinking water on histologic disease activity, n = 5 for naïve and ALCAR, n = 10 for C rodentium and ALCAR + C rodentium. ∗∗∗∗P < .0001 by unpaired t test. ALCAR, acetylcarnitine; CFU, colony-forming unit.
Discussion
There are numerous examples in which host interactions with its gut microbiota are altered in the setting of intestinal inflammation via the metabolome. In many cases, alterations in the composition of the gut microbiota in the disease state, that we describe as dysbiosis, provide insights into the mechanistic underpinnings responsible for these alterations in fecal metabolites. Examples include a reduction in fecal secondary bile acids and SCFAs associated with a dysbiotic microbiota in patients with IBD owing in part to a loss in Clostridium species capable of producing these 2 classes of metabolites.12 Although we and others recently reported that the dysbiotic microbiota in patients with IBD is associated with increased fecal levels of carnitine and acylcarnitines,12,13 the responsible mechanism(s) have not been identified. Herein, we provide evidence that intestinal inflammation leads to the increased delivery of carnitine and acylcarnitines into the gut lumen, where they are consumed by the bacterial gut microbiota.
By characterizing the relationship between fecal levels of carnitine and acylcarnitine in pediatric IBD, we show that previous results12 are relevant not only to pediatric CD29 but also to UC in a prospective cohort. In addition to showing a robust association between dysbiosis and increased fecal carnitine and most acylcarnitines, we also show that this pattern is stable temporally. Increased fecal acylcarnitines were observed in CD patients who have an inflammatory phenotype and is associated with both higher levels of human fecal DNA and disease activity. We have shown a clear inverse relationship between Enterobacteriaceae, particularly E coli, which correlates positively with carnitine and acylcarnitines, and many taxa belonging to the Firmicutes and Bacteroidetes phyla, which are correlated negatively with carnitine and acylcarnitines. It is interesting that these correlations have been observed for carnitine as well as short-chain (C2–C5), medium-chain (C6–C11), and long-chain (C12–C20) acylcarnitines of varying degrees of saturation and hydroxylation. By contrast, limited correlations were observed with very long chain acylcarnitines (>C20).
The mechanism(s) by which IBD leads to increased levels of fecal carnitine and acylcarnitines have not been characterized. Because very few bacterial species are capable of de novo carnitine synthesis and the generation of acylcarnitines via carnitine palmitoyltransferase is a mitochondrial function not observed in prokaryotic organisms, these metabolites must be delivered into the gut lumen by the mammalian host, including through biliary secretions.13,30 By characterizing levels of carnitine and acylcarnitines in germ-free mice, we show that colitis induced by DSS increases luminal concentrations of some acylcarnitines in the colon concurrent with increased levels in both bile and plasma.
Although SCFAs are the preferred substrate used by the colonic epithelium for fatty acid oxidation, we have shown that acylcarnitines can be an alternative source of energy for oxidative metabolism.13 Although an intact epithelium can use acylcarnitines, the colonic mitochondria in IBD patients are fewer and dysfunctional, thus limiting the tissue’s capacity for fatty acid oxidation.13 This, combined with increased delivery of cellular material from the host into the gut lumen in the setting of intestinal inflammation, would result in higher levels of unused acylcarnitines in the stool of these patients. This signature of increased acylcarnitines, be it in stool or plasma, often is thought to represent incomplete fatty acid oxidation and also is seen in other diseases whose etiology often is attributed to dysfunctional mitochondria.4,18,19 Often, it is not known whether increased acylcarnitines are a symptom or cause of the pathology,48 but in the case of IBD, in which the gut microbiota is also a factor, they are likely to be both.
Although additional investigation will be needed to characterize the mechanism(s) associating DSS colitis with increased plasma and biliary levels of acylcarnitines, an unanticipated finding, possible explanations include direct enterohepatic signaling similar to the regulation of hepatic bile acid synthesis via the activation of intestinal Farnesoid X receptor.49 Indeed, bile acid absorption is impaired in animal models of intestinal inflammation, including DSS colitis, as well as in IBD, and Farnesoid X receptor signaling has been shown to regulate hepatic fatty acid oxidation and acylcarnitine levels.50,51 Alternatively, the effect of colitis on energy homeostasis as described in patients with IBD38,52 may lead to the induction of hepatic acylcarnitine synthesis to increase plasma delivery, as has been shown in mice in which plasma acylcarnitines fuel thermogenesis.22,37 Indeed, increases in liver-derived acylcarnitines may help explain other comorbidities with IBD such as nonalcoholic fatty liver disease53 and cardiovascular risk,54 both of which correlate with altered acylcarnitine levels and aberrant lipid metabolism.18,19
Our goal was to examine whether these increased levels have a functional effect on the gut microbiota. Indeed, by studying fecal samples collected from healthy human subjects in whom the gut microbiota has been depleted, as well as comparing levels in germ-free and conventionally housed mice, we provide evidence that carnitine and acylcarnitines are consumed by the gut microbiota. Particularly notable are the high levels of fecal carnitine and acetylcarnitine in the feces of human subjects on a Western diet after a gut purge compared with subjects consuming a vegan or EEN diet in our FARMM study. This agrees with the previously proposed suggestion that diet regulates the luminal concentrations of these metabolites within the context of cardiovascular disease pathogenesis.33
The association between fecal levels of carnitine on a Western diet and the robust induction of E coli is particularly notable given the positive correlation between fecal carnitine levels and E coli in pediatric IBD. Indeed, we also show that the cai operon, needed for anaerobic bacterial metabolism of carnitine, also is correlated positively with E coli levels in both of these populations. This might be relevant to the relationship between diet and IBD, for which previous studies have reported that consumption of red meat, a rich source of dietary carnitine, is both epidemiologically associated with the prevalence of IBD and exacerbates colitis in an animal model.55 Furthermore, red meat consumption is specifically prohibited in the Crohn’s Disease Exclusion Diet, which has shown promise in the treatment of CD.56 By contrast, epidemiologic studies have suggested that a plant-based diet reduces the risk for IBD, potentially because plants have levels of acylcarnitines 2–3 logs lower than animal products.57
Carnitine is a quaternary amine that is known to be used by a variety of bacteria as a carbon and/or nitrogen source, osmoprotectant, or electron acceptor.30,41,58,59 Much less is known about the ability of bacteria to consume acylcarnitines, with only a few examples existing in the literature.30,60 To more broadly characterize the consumption of these metabolites by specific bacterial taxa, we developed a novel method to monitor the change in carnitine and a variety of acylcarnitines normally abundant in bacterial culture media, such as BHI, after growth under anaerobic conditions in batch cultures. The results verify the ability of E coli to consume carnitine and P aeruginosa to metabolize acetylcarnitine.41,42 Using this screen, we identified unique patterns of acylcarnitine metabolism by a select number of additional bacterial species that we have shown to correlate in abundance with fecal acylcarnitines in IBD, such as Akkermansia muciniphila, Clostridium butyricum, Enterococcus faecalis, and K pneumonia, indicating that acylcarnitines may be a previously underappreciated modality by which the host influences the biology of the gut microbiota. Based on current evidence, it is reasonable to hypothesize that the release of carnitine and its cognate fatty acid through the hydrolysis of acylcarnitines might provide either carbon- or nitrogen-based substrates for growth, facilitate energy production through β-oxidation of fatty acids, and/or enhance cellular protection by fortifying bacterial membranes or serving as an osmoprotectant.30 Additional studies are needed to determine if the effect of carnitine and acylcarnitine metabolism by different bacterial taxa leads to a competitive growth advantage either in vitro or in vivo.
Facultative anaerobes such as Enterobacteriaceae can use oxygen or alternative electron acceptors, such as nitrate, to fuel their electron transport chains and generate energy more quickly than via fermentation alone. In a healthy gut, these molecules are not abundant but have been shown to increase in intestinal inflammation.1,61 Although inflammation is an oxidative process, the lumen of the gut likely remains mostly anaerobic due to the ability of facultative anaerobes to consume oxygen rapidly.62,63 We speculate that carnitine, which is readily available from the host and through the diet, also may serve as an electron acceptor for Enterobacteriaceae under these conditions. Indeed, we show that the metabolism of carnitine by E coli can facilitate anaerobic growth using glycerol as a carbon source, suggesting that the increased levels of carnitine in the gut lumen associated with IBD might enhance the development of dysbiosis by expanding the range of substrates that Enterobacteriaceae can consume anaerobically. Similar effects have been shown for other genera belonging to the Enterobacteriaceae family including Salmonella and Proteus species.30
Using C rodentium as a model disease-inducing member of the Enterobacteriaceae family, we provide evidence that it can consume acetylcarnitine (C2) in LB, thereby leading to the release of carnitine into the culture medium. Focusing on acetylcarnitine (C2), the most abundant acylcarnitine throughout the luminal tract of mice, we show that dietary acetylcarnitine supplementation not only leads to greater levels of fecal carnitine, similar to the in vitro culture, but also enhances growth of C rodentium. Future studies are needed to determine whether or not C rodentium has an acetylcarnitine hydrolase, as has been described for P aeruginosa,42 and whether its increased growth in the murine gut is owing to carnitine-dependent metabolism as an electron acceptor to promote carbon-based growth, as we have shown for E coli. It also is interesting that the higher levels of C rodentium burden induced by acetylcarnitine dietary supplementation was not associated with a significant increase in disease activity, a finding that deserves additional study. It is possible that carnitine levels in normal mouse chow may have led to a reduced difference in C rodentium burden between the control and dietary supplement groups because engraftment of this pathogen has been shown to be dependent on both inoculum size and diet.64 We also cannot exclude the possibility that acetylcarnitine may have direct effects on host biology congruent with our observation that acylcarnitines can be used as a nutrient source by the colonic epithelium13 and that acetylcarnitine has been shown to have anti-inflammatory and anti-oxidant effects.65 Nevertheless, the enhanced growth of C rodentium through the dietary supplementation of acetylcarnitine, a commonly used dietary supplement,66 is consistent with epidemiologic studies showing that the consumption of animal products, which are rich in carnitine and acylcarnitines, are associated with an increased risk for IBD, whereas the consumption of plants and vegetables are associated with a decreased risk.67
In total, the enhanced delivery of carnitine and acylcarnitines from the host into the luminal environment of the gut in IBD serves as metabolic substrates for a wide range of bacterial taxa in the gut microbiota where their utilization is complex and taxon-specific. As a high-dimensional analytic feature, the pattern of fecal acylcarnitines, perhaps together with bacterial taxonomy, may have utility as a biomarker for the presence or prognosis of IBD. In addition, based on currently available information about the impact of carnitine on the biology of Enterobacteriaceae, acylcarnitines also may have an important functional effect on the biology of the gut microbiota that is relevant to the pathogenesis or course of disease in patients with IBD.
Methods
Human Studies
The pediatric IBD fecal metabolomic analysis was performed as part of the DYNAMIC study that has been described previously.2 Healthy children and children with IBD were recruited at the Children’s Hospital of Philadelphia between September 2015 and August 2018 (Institutional Review Board #15-011817). Exclusion criteria for healthy children were diarrhea, antibiotic use within 90 days of enrollment, a family member with C difficile infection in the study, or a chronic diagnosis. The demographic characteristics of the healthy children and children with IBD are provided in Table 1 and the clinical characteristics of children with IBD are provided in Table 2. Fecal microbiome analysis was performed.2 In brief, genomic DNA was extracted using the PowerSoil htp kit (MO BIO Laboratories) and sequencing libraries were prepared with the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA). Libraries were sequenced on an Illumina HiSeq 2500 using the paired-end 125–base pair sequencing protocol. Sequence reads were processed to remove adapter sequences and low-quality reads with Trimmomatic v. 0.33.68 Taxonomic assignments were generated with Kraken2.69 Untargeted metabolomics via ultra-high-performance liquid chromatography/tandem high-resolution and accurate mass spectrometry was performed by Metabolon, Inc (North Carolina) using reverse-phase and hydrophilic interaction chromatography. Additional analysis of acylcarnitines for this study was performed using R.70
Acylcarnitine levels from a dietary intervention study in healthy adults were obtained from the FARMM study.32 The University of Pennsylvania Institutional Review Board approved the research protocol and considered it exempt from clinical trial registration requirements based on the protocol’s stated objectives. Metabolomic analysis of fecal samples were performed using liquid chromatography–tandem mass spectrometry on a Nexera X2 ultra-high-performance liquid chromatography (Shimadzu Scientific Instruments) with a 150 × 2 mm Atlantic hydrophilic interaction chromatography column (Waters, Milford, MA) and a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The column was eluted isocratically at a flow rate of 250 mL/min with 5% mobile phase A (10 mmol/L ammonium formate and 0.1% formic acid in water) for 1 minute followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 minutes. Analyses were performed using electrospray ionization in the positive ion mode using full-scan analysis over m/z 70–800 at 70,000 resolution and a 3-Hz data acquisition rate. Additional settings were as follows: ion spray voltage, 3.5 kV; capillary temperature, 350°C; probe heater temperature, 300°C; sheath gas, 40; auxiliary gas, 15; and S-lens radio frequency level 40. Raw data were processed using Progenesis QI software (NonLinear Dynamics) for feature alignment, nontargeted signal detection, and signal integration. Targeted processing of a subset of known metabolites was conducted using TraceFinder 3.3 software (Thermo Fisher Scientific).
Animals: DSS Colitis, C rodentium Colitis, Fecal and Luminal Sample Collection
All experiments were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania Perelman School of Medicine. C57BL/6J mice were obtained from the Jackson Laboratory. Germ-free mice were obtained from the University of Pennsylvania Gnotobiotic Mouse Facility. All animals used in experiments were 6- to 12-week-old female mice. Animals were kept on 12-hour light/12-hour dark cycles. In the DSS colitis model, mice were fed an irradiated AIN-76 diet (D10001i; Research Diets, Inc, New Brunswick, NJ) and treated with 2.5% DSS (J63606.22; Thermo Fisher Scientific) in their drinking water. Blood was collected from the submandibular vein and then mice were killed by carbon dioxide asphyxiation followed by cervical dislocation when mice reached humane end point criteria based on body condition scores.71 Biospecimens were collected and the intestinal weights and lengths were recorded. For studies using the C rodentium model of colitis, mice were kept on an irradiated AIN-76 diet (D10001i) and were pretreated with or without 15 mmol/L acetylcarnitine (Fisher Scientific) in drinking water for 1 week. Mice then were inoculated by oral gavage with 7 × 107 colony-forming units of C rodentium (strain DBS100, a gift from David Artis, Weill Cornell Medicine at Cornell University, New York, NY). Fecal samples were collected daily and the C rodentium load was calculated by counting colonies on MacConkey agar plates. Mice were killed by carbon dioxide asphyxiation followed by cervical dislocation at the peak of infection on day 10.
Colonic Histology in C rodentium Colitis
Swiss-rolled portions of mouse cecum and colon were fixed with 4% paraformaldehyde and then paraffin-embedded, sectioned, and stained with H&E. Slides were analyzed and scored by a blinded pathologist for disease activity according to the multiparametric semiquantitative scoring system developed for C rodentium colitis.47
Bacterial Culture
Amuciniphila ATCC BAA-835, C butyricum ATCC 19398, E coli ATCC 11775, E faecalis ATCC 29200, K pneumoniae ATCC 13883, and P aeruginosa ATCC BAA-47 were obtained from the American Type Culture Collection (Manassas, VA). All strains were grown on brain heart infusion broth at 37°C in either an anaerobic glove box (Coy Laboratory Products, Inc, Grass Lake, MI) or an aerobic incubator (Fisher Scientific). Growth curves were inoculated from standing overnight cultures and performed in an Epoch2 automated microplate spectrophotometer (Agilent, Santa Clara, CA) with 12 replicates.
For studies on anaerobic growth of E coli in glycerol, E coli strains were MP13 (wild type), a tetracycline-resistant derivative of MP1,72 MP377, an MP13 derivative containing a caiC deletion and NRG 857C. MP377 was constructed from the ΔcaiC::kan Keio knockout strain JW003673 by transduction with P1vir.74 For all of the experiments described here, M9 minimal medium consisted of M9 base medium74 (made in house) supplemented with 2 mmol/L FeSO4 (Fisher Scientific) and 0.4% glycerol (Fisher Scientific). Triplicate cultures of each strain were grown overnight aerobically at 37°C in M9. These saturated cultures were introduced into the anaerobic chamber and diluted 1/200 in M9 that had been pre-equilibrated for several weeks in the anaerobic chamber. Electron acceptors were added to the cultures to final concentrations of 30 mmol/L for KNO3 (Fisher Scientific) and 20 mmol/L for carnitine. The latter was added from a stock solution of 200 mmol/L carnitine hydrochloride (Fisher Scientific) in M9 salts and with the pH adjusted to 7.2 with NaOH (Fisher Scientific). Cultures were incubated in the anaerobic chamber for 48 hours at 37°C. They then were removed from the anaerobic chamber, and streptomycin (Fisher Scientific) immediately was added to a final concentration of 250 μg/mL to stop growth. Cell density was assayed by measuring optical density at 600 nm. Statistical analysis of results of bacterial culture studies were performed in GraphPad Prism 9 (GraphPad Software, San Diego, CA).
Acylcarnitine Analysis in Animal and In Vitro Studies by Mass Spectrometry
To analyze acylcarnitines in intestinal contents and fecal pellets they were first taken up in ultrapure water at a concentration of 100 mg/mL. Samples then were subjected to 3 freeze/thaw cycles and 1 round of sonication to fully liberate the acylcarnitines from the solid material. Then, as with plasma, bile, and bacterial samples, acylcarnitines were extracted with ethanol and added to tubes containing stable isotope-labeled internal standards (NSK-B; Cambridge Isotope Laboratories). Ethanol extractions were dried down under a stream of nitrogen and acylcarnitines were derivatized to their butyl ester forms by incubating with butanolic hydrochloric acid (Regis Technologies) at 65°C for 15 minutes. The dried samples were reconstituted with acetonitrile/water (80:20) and 5 μL sample was delivered to a Xevo TQ-XS tandem mass spectrometer (Waters Corporation) through direct flow injection. Data were acquired in multiple reaction monitoring and focused on collecting the parent compounds of fragment mass m/z 85. Quantitation was against the nearest chain-length stable isotope–labeled internal standard. The mobile phase was acetonitrile/water (80:20, v/v). The flow rate was 0.1 mL/min. The source temperature was 150°C and the desolvation temperature was 200°C, with gas flow of 550 L/h. The cone and capillary voltage were 50 V and 3.2 kV, respectively. The mass spectrometer was operated in positive ion mode with resolving power of 140,000 at m/z 200, with a mass scanning range of m/z 140–600. Unless otherwise stated, statistical analysis of acylcarnitine levels was performed in GraphPad Prism 9 (GraphPad Software, San Diego, CA).
Analysis of Clinical Data Sets
Shotgun metagenomic data were analyzed using Sunbeam version 2.1.1.75 Adapters and low-quality regions were trimmed using default parameters in Trimmomatic.68 Reads that aligned to the human and PhiX genomes using bwa76 were discarded. The abundance of bacteria was estimated using Kraken.77 Reads also were mapped to the Kyoto Encyclopedia of Genes and Genomes database78 to estimate the abundance of bacterial gene orthologs. Sample similarity was assessed by Bray–Curtis distance.
The dysbiosis score was calculated for each sample as the median Bray–Curtis distance to the samples from healthy subjects as previously described.12 A dysbiosis threshold was calculated as the 90% quantile of the healthy subjects’ dysbiosis score.
Untargeted metabolomics identified 37 metabolites that belong to acylcarnitine metabolism. Linear mixed-effects models were used to estimate the difference in log10 transformed metabolite levels between nondysbiotic and dysbiotic samples. Antibiotic use was added as a covariate and subject identification was added as a random effect.
Spearman correlation was calculated between log10-transformed bacterial abundances and log10-transformed metabolite levels for each time point. Only the bacteria that have a mean relative abundance of 1% across all samples have been tested.
The P values from multiple tests were corrected for false discovery rate using the Benjamini–Hochberg method.
Acknowledgments
Johanna M. S. Lemons and Maire Conrad contributed equally to this manuscript.
CRediT Authorship Contributions
Daria Krzikalla (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Software: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Equal)
Alena Laschtowitz (Formal analysis: Equal; Investigation: Equal; Writing – review & editing: Supporting)
Lisa Leypoldt (Formal analysis: Equal; Investigation: Equal; Writing – review & editing: Supporting)
Cornelia Gottwick (Investigation: Supporting; Writing – review & editing: Supporting)
Pia Averhoff (Investigation: Supporting; Writing – review & editing: Supporting)
Sören Weidemann (Methodology: Supporting; Writing – review & editing: Supporting)
Ansgar W. Lohse (Conceptualization: Supporting; Funding acquisition: Supporting; Writing – review & editing: Supporting)
Samuel Huber (Methodology: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Christoph Schramm (Conceptualization: Supporting; Writing – review & editing: Supporting)
Dorothee Schwinge (Conceptualization: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Johannes Herkel (Conceptualization: Lead; Data curation: Lead; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Supporting; Methodology: Equal; Project administration: Equal; Resources: Lead; Supervision: Lead; Writing – original draft: Lead)
Antonella Carambia (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Investigation: Supporting; Methodology: Lead; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the Crohn's and Colitis Foundation (G.D.W.), the PennCHOP Microbiome Program (C.T., E.S.F., R.N.B., K.B., and G.D.W.), the Penn Center for Nutritional Science and Medicine (L.C., E.S.F., and G.D.W.), the Sherman Prize, the Center for Molecular Studies in Digestive and Liver Disease grant P30DK050306 (L.C., E.S.F., K.B., and G.D.W.), and the Commonwealth Universal Research Enhancement (C.U.R.E.) program (SAP #4100068710).
Data Availability Data are available upon request.
Contributor Information
LinShu Liu, Email: linshu.liu@usda.gov.
Gary D. Wu, Email: gdwu@pennmedicine.upenn.edu.
References
- 1.Winter S.E., Lopez C.A., Baumler A.J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushman F.D., Conrad M., Ren Y., et al. Multi-omic analysis of the interaction between Clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe. 2020;28:422–433 e427. doi: 10.1016/j.chom.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrello C., Garavaglia F., Cribiu F.M., et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018;9:5184. doi: 10.1038/s41467-018-07359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koves T.R., Ussher J.R., Noland R.C., et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Zhu W., Winter M.G., Byndloss M.X., et al. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018;553:208–211. doi: 10.1038/nature25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton M.J., Weingarden A.R., Sadowsky M.J., et al. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol. 2012;107:761–767. doi: 10.1038/ajg.2011.482. [DOI] [PubMed] [Google Scholar]
- 7.Hvas C.L., Dahl Jorgensen S.M., Jorgensen S.P., et al. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent Clostridium difficile infection. Gastroenterology. 2019;156:1324–1332 e1323. doi: 10.1053/j.gastro.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 8.van Nood E., Vrieze A., Nieuwdorp M., et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 9.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Winter S.E., Winter M.G., Xavier M.N., et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera-Chavez F., Lopez C.A., Baumler A.J. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017;105:93–101. doi: 10.1016/j.freeradbiomed.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd-Price J., Arze C., Ananthakrishnan A.N., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith S.A., Ogawa S.A., Chau L., et al. Mitochondrial dysfunction in inflammatory bowel disease alters intestinal epithelial metabolism of hepatic acylcarnitines. J Clin Invest. 2021;131 doi: 10.1172/JCI133371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knottnerus S.J.G., Bleeker J.C., Wust R.C.I., et al. Disorders of mitochondrial long-chain fatty acid oxidation and the carnitine shuttle. Rev Endocr Metab Disord. 2018;19:93–106. doi: 10.1007/s11154-018-9448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuda F., Narayan S.B., Squires R.H., Jr., et al. Bile acylcarnitine profiles in pediatric liver disease do not interfere with the diagnosis of long-chain fatty acid oxidation defects. Clin Chim Acta. 2006;367:185–188. doi: 10.1016/j.cca.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 16.Li S., Gao D., Jiang Y. Function, detection and alteration of acylcarnitine metabolism in hepatocellular carcinoma. Metabolites. 2019;9:36. doi: 10.3390/metabo9020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCann M.R., George De la Rosa M.V., Rosania G.R., et al. L-carnitine and acylcarnitines: mitochondrial biomarkers for precision medicine. Metabolites. 2021;11:51. doi: 10.3390/metabo11010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enooku K., Nakagawa H., Fujiwara N., et al. Altered serum acylcarnitine profile is associated with the status of nonalcoholic fatty liver disease (NAFLD) and NAFLD-related hepatocellular carcinoma. Sci Rep. 2019;9 doi: 10.1038/s41598-019-47216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz M., Labarthe F., Fortier A., et al. Circulating acylcarnitine profile in human heart failure: a surrogate of fatty acid metabolic dysregulation in mitochondria and beyond. Am J Physiol Heart Circ Physiol. 2017;313:H768–H781. doi: 10.1152/ajpheart.00820.2016. [DOI] [PubMed] [Google Scholar]
- 20.Costa C.C., de Almeida I.T., Jakobs C., et al. Dynamic changes of plasma acylcarnitine levels induced by fasting and sunflower oil challenge test in children. Pediatr Res. 1999;46:440–444. doi: 10.1203/00006450-199910000-00013. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguti K., Kuratsune H., Watanabe Y., et al. Acylcarnitine metabolism during fasting and after refeeding. Biochem Biophys Res Commun. 1996;225:740–746. doi: 10.1006/bbrc.1996.1244. [DOI] [PubMed] [Google Scholar]
- 22.Simcox J., Geoghegan G., Maschek J.A., et al. Global analysis of plasma lipids identifies liver-derived acylcarnitines as a fuel source for brown fat thermogenesis. Cell Metab. 2017;26:509–522 e506. doi: 10.1016/j.cmet.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roediger W.E. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet. 1980;2:712–715. doi: 10.1016/s0140-6736(80)91934-0. [DOI] [PubMed] [Google Scholar]
- 24.Roediger W.E., Nance S. Metabolic induction of experimental ulcerative colitis by inhibition of fatty acid oxidation. Br J Exp Pathol. 1986;67:773–782. [PMC free article] [PubMed] [Google Scholar]
- 25.Roediger W.E.W. Causation of human ulcerative colitis: a lead from an animal model that mirrors human disease. JGH Open. 2019;3:277–280. doi: 10.1002/jgh3.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peltekova V.D., Wintle R.F., Rubin L.A., et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 27.Aldars-Garcia L., Gisbert J.P., Chaparro M. Metabolomics insights into inflammatory bowel disease: a comprehensive review. Pharmaceuticals (Basel) 2021;14:1190. doi: 10.3390/ph14111190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine A., Griffiths A., Markowitz J., et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–1321. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J.D., Chen E.Z., Baldassano R.N., et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meadows J.A., Wargo M.J. Carnitine in bacterial physiology and metabolism. Microbiology (Reading) 2015;161:1161–1174. doi: 10.1099/mic.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichler K., Bourgis F., Buchet A., et al. Molecular characterization of the cai operon necessary for carnitine metabolism in Escherichia coli. Mol Microbiol. 1994;13:775–786. doi: 10.1111/j.1365-2958.1994.tb00470.x. [DOI] [PubMed] [Google Scholar]
- 32.Tanes C., Bittinger K., Gao Y., et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe. 2021;29:394–407 e395. doi: 10.1016/j.chom.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koeth R.A., Lam-Galvez B.R., Kirsop J., et al. l-Carnitine in omnivorous diets induces an atherogenic gut microbial pathway in humans. J Clin Invest. 2019;129:373–387. doi: 10.1172/JCI94601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buchet A., Eichler K., Mandrand-Berthelot M.A. Regulation of the carnitine pathway in Escherichia coli: investigation of the cai-fix divergent promoter region. J Bacteriol. 1998;180:2599–2608. doi: 10.1128/jb.180.10.2599-2608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., et al. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Cooper H.S., Murthy S.N., Shah R.S., et al. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 37.Jain R., Wade G., Ong I., et al. Determination of tissue contributions to the circulating lipid pool in cold exposure via systematic assessment of lipid profiles. J Lipid Res. 2022;63 doi: 10.1016/j.jlr.2022.100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mingrone G., Capristo E., Greco A.V., et al. Elevated diet-induced thermogenesis and lipid oxidation rate in Crohn disease. Am J Clin Nutr. 1999;69:325–330. doi: 10.1093/ajcn/69.2.325. [DOI] [PubMed] [Google Scholar]
- 39.Man K., Bowman C., Braverman K.N., et al. A thermogenic fat-epithelium cell axis regulates intestinal disease tolerance. Proc Natl Acad Sci U S A. 2020;117:32029–32037. doi: 10.1073/pnas.2012003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiko G.E., Ryu S.H., Koues O.I., et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleber H.P. Bacterial carnitine metabolism. FEMS Microbiol Lett. 1997;147:1–9. doi: 10.1111/j.1574-6968.1997.tb10212.x. [DOI] [PubMed] [Google Scholar]
- 42.Meadows J.A., Wargo M.J. Characterization of Pseudomonas aeruginosa growth on O-acylcarnitines and identification of a short-chain acylcarnitine hydrolase. Appl Environ Microbiol. 2013;79:3355–3363. doi: 10.1128/AEM.03943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchet A., Nasser W., Eichler K., et al. Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol Microbiol. 1999;34:562–575. doi: 10.1046/j.1365-2958.1999.01622.x. [DOI] [PubMed] [Google Scholar]
- 44.Walt A., Kahn M.L. The fixA and fixB genes are necessary for anaerobic carnitine reduction in Escherichia coli. J Bacteriol. 2002;184:4044–4047. doi: 10.1128/JB.184.14.4044-4047.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitz W., Montero D.A., Pardo M., et al. Characterization of adherent-invasive Escherichia coli (AIEC) outer membrane proteins provides potential molecular markers to screen putative AIEC strains. Int J Mol Sci. 2022;23:9005. doi: 10.3390/ijms23169005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horesh G., Blackwell G.A., Tonkin-Hill G., et al. A comprehensive and high-quality collection of Escherichia coli genomes and their genes. Microb Genom. 2021;7:499. doi: 10.1099/mgen.0.000499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouladoux N., Harrison O.J., Belkaid Y. The mouse model of infection with Citrobacter rodentium. Curr Protoc Immunol. 2017;119:191511–191525. doi: 10.1002/cpim.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schooneman M.G., Vaz F.M., Houten S.M., et al. Acylcarnitines: reflecting or inflicting insulin resistance? Diabetes. 2013;62:1–8. doi: 10.2337/db12-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kliewer S.A., Mangelsdorf D.J. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig Dis. 2015;33:327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitzpatrick L.R., Jenabzadeh P. IBD and bile acid absorption: focus on pre-clinical and clinical observations. Front Physiol. 2020;11:564. doi: 10.3389/fphys.2020.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu D., Liu Y., Luo Y., et al. Intestinal farnesoid X receptor signaling controls hepatic fatty acid oxidation. Biochim Biophys Acta Mol Cell Biol Lipids. 2022;1867 doi: 10.1016/j.bbalip.2021.159089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Jaouni R., Hebuterne X., Pouget I., et al. Energy metabolism and substrate oxidation in patients with Crohn's disease. Nutrition. 2000;16:173–178. doi: 10.1016/s0899-9007(99)00281-6. [DOI] [PubMed] [Google Scholar]
- 53.Papaefthymiou A., Potamianos S., Goulas A., et al. Inflammatory bowel disease-associated fatty liver disease: the potential effect of biologic agents. J Crohns Colitis. 2022;16:852–862. doi: 10.1093/ecco-jcc/jjab212. [DOI] [PubMed] [Google Scholar]
- 54.Bigeh A., Sanchez A., Maestas C., et al. Inflammatory bowel disease and the risk for cardiovascular disease: does all inflammation lead to heart disease? Trends Cardiovasc Med. 2020;30:463–469. doi: 10.1016/j.tcm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Li D.P., Cui M., Tan F., et al. High red meat intake exacerbates dextran sulfate-induced colitis by altering gut microbiota in mice. Front Nutr. 2021;8 doi: 10.3389/fnut.2021.646819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine A., Wine E., Assa A., et al. Crohn's disease exclusion diet plus partial enteral nutrition induces sustained remission in a randomized controlled trial. Gastroenterology. 2019;157:440–450 e448. doi: 10.1053/j.gastro.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 57.Bourdin B., Adenier H., Perrin Y. Carnitine is associated with fatty acid metabolism in plants. Plant Physiol Biochem. 2007;45:926–931. doi: 10.1016/j.plaphy.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Rajakovich L.J., Fu B., Bollenbach M., et al. Elucidation of an anaerobic pathway for metabolism of l-carnitine-derived gamma-butyrobetaine to trimethylamine in human gut bacteria. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2101498118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jameson E., Quareshy M., Chen Y. Methodological considerations for the identification of choline and carnitine-degrading bacteria in the gut. Methods. 2018;149:42–48. doi: 10.1016/j.ymeth.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aragozzini F., Manzoni M., Cavazzoni V., et al. L-carnitine resolution by Fusarium oxysporum. Biotechnol Lett. 1986;8:95–97. [Google Scholar]
- 61.Litvak Y., Byndloss M.X., Baumler A.J. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362 doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Riedel T.E., Berelson W.M., Nealson K.H., et al. Oxygen consumption rates of bacteria under nutrient-limited conditions. Appl Environ Microbiol. 2013;79:4921–4931. doi: 10.1128/AEM.00756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman E.S., Bittinger K., Esipova T.V., et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci U S A. 2018;115:4170–4175. doi: 10.1073/pnas.1718635115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caballero-Flores G., Pickard J.M., Fukuda S., et al. An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe. 2020;28:526–533 e525. doi: 10.1016/j.chom.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jamali-Raeufy N., Alizadeh F., Mehrabi Z., et al. Acetyl-L-carnitine confers neuroprotection against lipopolysaccharide (LPS)-induced neuroinflammation by targeting TLR4/NFkappaB, autophagy, inflammation and oxidative stress. Metab Brain Dis. 2021;36:1391–1401. doi: 10.1007/s11011-021-00715-6. [DOI] [PubMed] [Google Scholar]
- 66.Dambrova M., Makrecka-Kuka M., Kuka J., et al. Acylcarnitines: nomenclature, biomarkers, therapeutic potential, drug targets, and clinical trials. Pharmacol Rev. 2022;74:506–551. doi: 10.1124/pharmrev.121.000408. [DOI] [PubMed] [Google Scholar]
- 67.Lo C.H., Lochhead P., Khalili H., et al. Dietary inflammatory potential and risk of Crohn's disease and ulcerative colitis. Gastroenterology. 2020;159:873–883 e871. doi: 10.1053/j.gastro.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team (2022) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- 71.Ullman-Cullere M.H., Foltz C.J. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab Anim Sci. 1999;49:319–323. [PubMed] [Google Scholar]
- 72.Lasaro M., Liu Z., Bishar R., et al. Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J Bacteriol. 2014;196:1723–1732. doi: 10.1128/JB.01296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baba T., Ara T., Hasegawa M., et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller J.H. Cold Spring Harbor Laboratory Press; 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. [Google Scholar]
- 75.Clarke E.L., Taylor L.J., Zhao C., et al. Sunbeam: an extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome. 2019;7:46. doi: 10.1186/s40168-019-0658-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ogata H., Goto S., Sato K., et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]