Summary
Here, we present a protocol to study and describe immune cells that surround or infiltrate tumor cells or get through the body of a melanoma syngeneic mice model. We describe steps for creating and establishing the syngeneic mouse model, euthanasia, and tumor or organ harvest. We then detail procedures to rapidly achieve a single-cell suspension from different tissue samples to further quantify and analyze the phenotype of the immune cell population (lymphocytes T and B, tumor-associated macrophages, and myeloid-derived suppressor cells) by flow cytometry.
Subject areas: Cell Isolation, Single Cell, Flow Cytometry, Cancer, Immunology
Graphical abstract

Highlights
-
•
Single-cell suspension from different tissues samples without enzymatic approach
-
•
Isolated cells are suitable for flow cytometry analysis
-
•
Specific antibody panel to analyze different immune cell populations by flow cytometry
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol to study and describe immune cells that surround or infiltrate tumor cells or get through the body of a melanoma syngeneic mice model. We describe steps for creating and establishing the syngeneic mouse model, euthanasia, and tumor or organ harvest. We then detail procedures to rapidly achieve a single-cell suspension from different tissue samples to further quantify and analyze the phenotype of the immune cell population (lymphocytes T and B, tumor-associated macrophages, and myeloid-derived suppressor cells) by flow cytometry.
Before you begin
This protocol describes a process in syngeneic melanoma murine C57BL/6J mice model (murine 5555 melanoma cell lines). However, this protocol could be adapted to different oncogenic mice models to obtain a single cell preparation. In addition, this protocol could be adapted to other specific antibody panel to detect new immune cells that infiltrate or surround the tumor tissue.
Institutional permission
This research complies with all relevant ethical regulations. C57BL/6 mice were maintained by brother-sister mating under specific pathogen-free conditions at the University of Lleida. This study was carried out in accordance with the principles of the Basel Declaration and recommendations of the Catalan Government (Generalitat de Catalunya, 1201/2005) concerning the protection of animals for experimentation. The protocol was approved by the Committee on the Ethics of Research in Animal Experimentation of the University of Lleida, Spain.
Cell lines
Mouse melanoma cell line 55551,2 was used. The cell line should be regularly checked to ensure the original identity and for mycoplasma cell free infection.
When handling the chemical and biological materials used in this protocol, always wear suitable personal protective equipment, including a lab coat, gloves, safety goggles and, where indicated, a face shield. For any reagent listed in this protocol, appropriate institutional and governmental safety guidelines must be followed. Please refer to the appropriate materials safety data sheets.
Flow cytometer
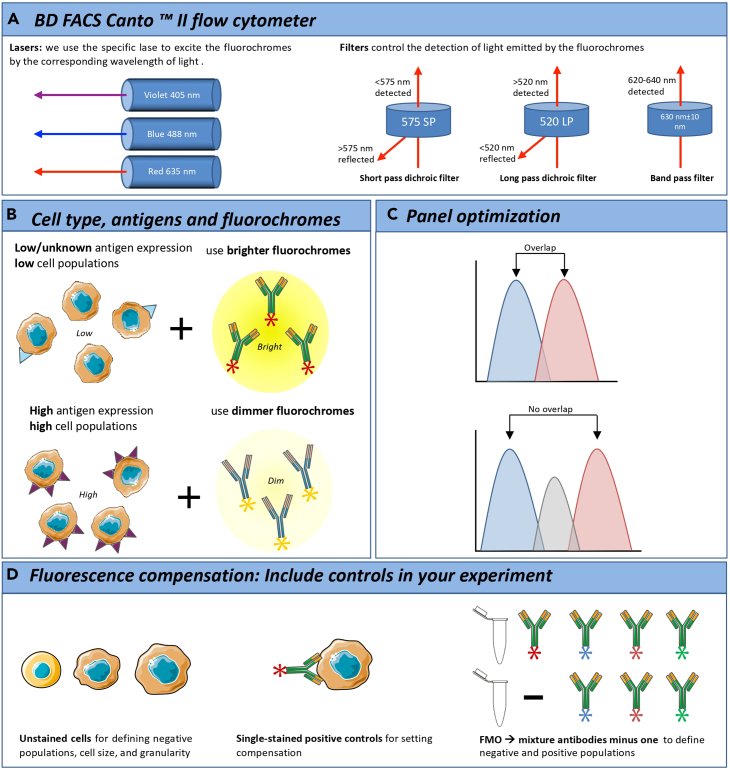
The panels used in this protocol were designed for a BD FACSCanto II flow cytometer equipped with the following three lasers 4-2-2 configuration: 4 channels for the blue 488 nm laser (with the following filters in front of the detectors: 530/30, 585/42, 670LP and 780/60), 2 channels for the red 635 nm laser (with 660/20 and 780/60 filters) and 2 channels for the violet 405 nm laser (with 450/50 and 510/50 filters) (Figure 2A).
Figure 2.
Optimization of immune antibody panel
(A) BD FACSCanto II flow cytometer.
(B) Cell type, antigens and fluorochromes.
(C) Panel optimization.
(D) Fluorescence compensation: Include controls in your experiment.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-F4/80 SB436 (clone BM8) | eBioscience | Cat # 62-4801-82 |
| Mouse monoclonal anti-Ly6G/Ly6C eFluor 506 (clone RB6-8C5) | eBioscience | Cat # 69-5931-82 |
| Mouse monoclonal anti-CD4 FITC (RM4-5) | eBioscience | Cat # 11-0042-82 |
| Mouse monoclonal anti-CD8 APC-eFluor 710 (clone 53-6.7) | eBioscience | Cat # 47-0081-82 |
| Mouse monoclonal anti-CD11b PerCP-eFluor 710 (clone M1/70) | eBioscience | Cat # 46-0112-82 |
| Mouse monoclonal anti-CD206 PE (clone MR6F3) | eBioscience | Cat # 12-2061-80 |
| Mouse monoclonal anti-MHC-II APC (clone M5/114.15.2) | eBioscience | Cat # 17-5321-81 |
| Mouse monoclonal anti-FoxP3 PE-Cyanine7 (clone FJK-16s) | eBioscience | Cat # 25-5773-82 |
| Mouse monoclonal anti-iNOS PE (clone CXNFT) | eBioscience | Cat # 12-5920-82 |
| Mouse monoclonal anti-arginase 1 APC (clone A1exF5) | eBioscience | Cat # 17-3697-82 |
| Mouse monoclonal anti-Ly6C APC-eFluor 780 (clone HK1.4) | eBioscience | Cat # 47-5932-80 |
| Mouse monoclonal anti-Ly6G FTIC (clone 1A8-Ly6g) | eBioscience | Cat # 11-9668-80 |
| Mouse monoclonal anti-galectin 3 eFluor 660 (clone M3/38) | eBioscience | Cat # 50-5301-82 |
| Mouse monoclonal anti-CD19 BV421 (clone 6D5) | BioLegend | Cat # 115537 |
| Mouse monoclonal anti-CD138 PE (clone 300506) | eBioscience | Cat # MA5-23527 |
| Mouse monoclonal anti-CD45 PerCP (clone EM-05) | eBioscience | Cat # MA1-10234 |
| Mouse monoclonal anti-CD38 APC-eFluor 780 (clone 90) | eBioscience | Cat # 47-0381-80 |
| Mouse monoclonal anti-B220 Alexa Fluor 647 (clone RA3-6B2) | BioLegend | Cat # 103226 |
| Chemicals, peptides and recombinant proteins | ||
| Dulbecco’s modified Eagle’s medium (DMEM) | Gibco | Cat # 41965-039 |
| Penicillin-streptomycin | Gibco | Cat # 15140-122 |
| Fetal bovine serum (FBS) | Corning | Cat # 35089001 |
| ANFO Fungizone | Gibco | Cat # 15290-018 |
| Trypsin-EDTA, 0.25% | Gibco | Cat # 25200-056 |
| PBS (phosphate-buffered saline) (pH 7.4) | Gibco | Cat # 10010023 |
| Paraformaldehyde (PFA) | Sigma-Aldrich | Cat # 158127 |
| Milli-Q water | N/A | N/A |
| RBC lysis buffer 10× | N/A | N/A |
| FACSFlow | BD | Cat # 12756528 |
| Isoflurane | Zoetis | Cat # 571398 |
| Heparin/EDTA tubes | Sarstedt | Cat # 16.434 |
| Experimental models: Organisms/strains | ||
| C57BL/6 mouse | Charles River, UK | RRID: IMSR_JAX:000664 |
| Mice melanoma cell lines (5555) | Richard Marais | Bibliography1,2 |
| Software and algorithms | ||
| BD FACSDiva software | BD Biosciences | N/A |
| GraphPad Prism 8 | GraphPad Software | N/A |
| Other | ||
| Syringes (23G) | N/A | N/A |
| Plungers (1 mL) | N/A | N/A |
| Caliper | N/A | N/A |
| 100 mm culture dishes | Sterilin | Cat # 101VR20 |
| P10 micropipette | VWR | Cat # 89079-962 |
| P20 micropipette | VWR | Cat # 89079-964 |
| P200 micropipette | VWR | Cat # 89079-970 |
| P1000 micropipette | VWR | Cat # 89079-974 |
| Pipette tips | ClearLine | Cat # 713118 |
| Pipette controller | Corning | Cat # 357469 |
| Tissue culture pipettes | Corning | Cat # CORN4101 |
| Sterile conical (micro) centrifuge tubes | Corning | Cat # 352070 |
| Sterile cell strainer 40 μm nylon mesh | Fisherbrand | Cat # 11587522 |
| Hemocytometer | Sigma-Aldrich | Cat # Z359629 |
| FACSCanto II | BD Biosciences | N/A |
Materials and equipment
Cell culture media
| Reagent | Final concentration | Amount |
| DMEM Dulbecco’s modified Eagle’s medium (DMEM), high glucose (4.5 g/L), L-glutamine (2 mM) | N/A | 450 mL |
| FBS (Fetal Bovine Serum) | 10% | 50 mL |
| P/S (Penicillin/Streptomycin) | 1% | 5 mL |
| Total | 555 mL |
Note: Store at 4°C. Stability for more than 6 months. Pre-warm the culture medium at 37°C before use.
PBS 10×
| Reagent | Final concentration | Amount |
|---|---|---|
| ClNa | N/A | 80 g |
| ClK | N/A | 2 g |
| KH2PO4 | N/A | 2 g |
| Na2HPO4 x 7H2O | N/A | 11.5 g |
| H2O Milli-Q | N/A | 1000 mL |
| Total | 1000 mL |
Note: Store at 4°C. Stability for more than 6 months. Adjust the pH to 7.4.
PBS 1×
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS 10× | 1% | 100 mL |
| H2O Milli-Q | N/A | 900 mL |
| Total | 1000 mL |
Note: Store at 4°C. Stability for more than 6 months. Adjust the pH to 7.4.
PFA 4%
| Reagent | Final concentration | Amount |
|---|---|---|
| PFA | 4% | 4 g |
| H2O Milli-Q | N/A | 90 mL |
| Total | 100 mL |
Note: To dissolve the PFA it has to be carried out with warm water (60°C–70°C). Adjust the pH to 7.5 and filter. Store at –20°C. Stability for more than 1 year.
Staining medium
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS 1× | N/A | 47.5 mL |
| FBS | 5% | 2.5 mL |
| Total | 50 mL |
Note: Store at 4°C. Make fresh buffer in each experiment.
RBC lysis buffer 10×
| Reagent | Final concentration | Amount |
|---|---|---|
| NH4Cl | N/A | 8.26 g |
| NaHCO3 | N/A | 1.19 g |
| EDTA 0.5 M pH 8 | 1 mM | 200 μL |
| H2O Milli-Q | Until 100 mL | |
| Total | 100 mL |
Note: Adjust the pH to 7.3 and filter. Store at 4°C. Stability for more than 6 months.
RBC lysis buffer 1×
| Reagent | Final concentration | Amount |
|---|---|---|
| RBC Lysis Buffer 10× | 1% | 50 mL |
| H2O Milli-Q | N/A | 450 mL |
| Total | 500 mL |
Note: Prepare fresh on the day of harvest and store at 21°C–23°C.
Step-by-step method details
Performing syngeneic tumor mice model
Timing: 2 h 30 min
This procedure details how to generate syngeneic xenograft mice model.
-
1.
Seed tumoral cells (3·106 cells if the doubling time is one day) into 100 mm culture dishes, and grow them until they reach 90% of confluence (at 37°C and 5% CO2) in appropriate culture medium (e.g., 5555 cells, use DMEM with 10% (vol/vol) FBS (See materials and equipment).
-
2.Remove medium from culture dish by gentle aspiration.
-
a.Wash cells twice with sterile 1× PBS, add trypsin-EDTA enough to completely cover the cells (1 mL is enough).
-
b.Place them at 37°C for 2 min.
-
c.Check under the microscope if the cells have detached from the plate.
-
a.
-
3.
After incubation, add 4 mL of complete medium to stop the trypsin-EDTA reaction and collect all the liquid in a sterile tube.
-
4.Centrifuge the cell suspension for 5 min at 400–600 g at RT.
-
a.Remove the trypsin-EDTA solution by aspiration.
-
b.Mix the cells with fresh medium containing serum (you can resuspend cells in 1 mL of fresh medium).
-
a.
-
5.
Determine cell concentration using a hemocytometer or Burker camera.
Note: In this step you can check cell viability, cell number, and concentration.
Note: C1–C3 represent the number of cells counted in each of the 3 sets of 16 squares, that is expected to be similar among them if there is a good resuspension of the sample. 3 is the number of sets that have been counted. 10.000 cells/mL is the equivalent of cell concentration of the surface of each set counted. 10 is the dilution factors used to count the cells.
-
6.
Calculate the number of total cells in your sample following the formula:
-
7.Wash cells with sterile 1× PBS (See materials and equipment).
-
a.Add 1× PBS to tube, and again spin down cells at 400–600 g for 5 min.
-
b.Aspirate PBS, resuspend cells in fresh PBS to a concentration of 1 × 106 cells/100 μL.
-
c.Transfer cells to a sterile 1.5 mL tube.
-
a.
Note: The cell number required depends upon the aggressiveness of the tumor cells and can vary by an order of magnitude.
Optional: Add an equal volume (μL) of thawed Matrigel to cells and mix carefully with pipette.
CRITICAL: Matrigel should be thawed on ice because it will solidify at room temperature.
Note: Matrigel provides a favorable environment for less aggressive cells to grow and it is often used to generate syngeneic melanoma mouse models.
-
8.
Slowly pull up 100 μL of cells alone or 200 μL of cell/Matrigel mixture using an insulin syringe.
Note: Cells can be damaged by the small gauge of the insulin needle; however, insulin syringes provide a more accurate volume measurement. Keep syringes with cells on ice to avoid polymerization of the Matrigel.
-
9.
Anesthetizing the mice using isoflurane.
Note: use isoflurane before the injection process should be less stressful for both, the mice and the researcher.
-
10.
Shave the skin working area before starting to inject the cell into the left or right flank of the mice.
-
11.
Once a small spot appears at the injection site of the mice measurements can be started.
Note: about 10 days after cell injection depending on the number of cells injected.
-
12.Measurements should be carried out three times a week using a caliper.
-
a.In order to determine tumor volume, the greatest longitudinal diameter (length, D) and the greatest transverse diameter (width, d) were determined.
-
b.Tumor size was calculated using the formula:
-
a.
Sacrifice mice and organs collection
Timing: 30 min each animal
The following steps detail the essential information required to sacrifice and obtain all the collected organs. This procedure details how to obtain blood, xenograft tumor, bone marrow and spleen organ.
Note: We recommend a direct cardiac puncture without opening the thorax for a quick and safe process.
CRITICAL: It requires practice and some prefer directly taking the blood from the visible heart, which particularly suits for smaller species.
Note: Blood should be taken as soon as possible after the death of the animal as blood can coagulate rapidly.
Note: Fix the needle to the 21G syringe and carefully loosen the needle cover. Expel the air from the syringe.
-
13.
Place the animal on its back, in a horizontal position. Clean the thoracic fur with ethanol.
-
14.
Locate the thoracic cage and xiphoid process (the lower part of the sternum) with one hand.
Note: making sure that the animal is well positioned (i.e., the xiphoid process is well centered).
-
15.With the other hand, insert the needle under the xiphoid process at about 30 degrees to the left.
-
a.While inserting the needle, gently pull the plunger to aspirate until the heart is perforated and blood begins to flow.
-
b.As soon as blood is flowing, continue pulling the plunger without moving the needle to avoid any more perforations in the heart, which will stop the flow.
 CRITICAL: If no blood appears in the needle, the action should be repeated. If the flow stops rapidly, the heart may have been perforated on the back: slowly withdraw and adjust the needle to restore the flow.
CRITICAL: If no blood appears in the needle, the action should be repeated. If the flow stops rapidly, the heart may have been perforated on the back: slowly withdraw and adjust the needle to restore the flow. -
c.As soon as enough blood is collected, release pressure on the plunger and withdraw the needle.
-
a.
-
16.
Slowly expel the blood from the syringe into the Heparin/EDTA tubes.
Note: To avoid damaging blood cells, we recommend disposing of the needle before expelling blood.
Optional: A syringe previously soaked with heparin can be used. Then transfer the blood to a 1.5 mL tube and centrifuge to obtain the plasma.
-
17.To carry out the dissection and collection of the organs of interest, put the animal ventral side up on the clean dissection board.
-
a.Clean the ventrum with ethanol to avoid introducing hairs in the body.
-
b.Pinch and raise the skin with dissecting forceps.
-
c.Cut through the body wall muscles just anterior to the genital opening and continue just to one side of the midline on the ventral side until the thoracic cavity.Note: Use blunt-end scissors to avoid damaging the organs.
-
d.Collect the organs of interest and process them accordingly to generate single cell suspension. (To see the sample processing read the generation of single cell suspension section).
-
a.
Generation of single cell suspension
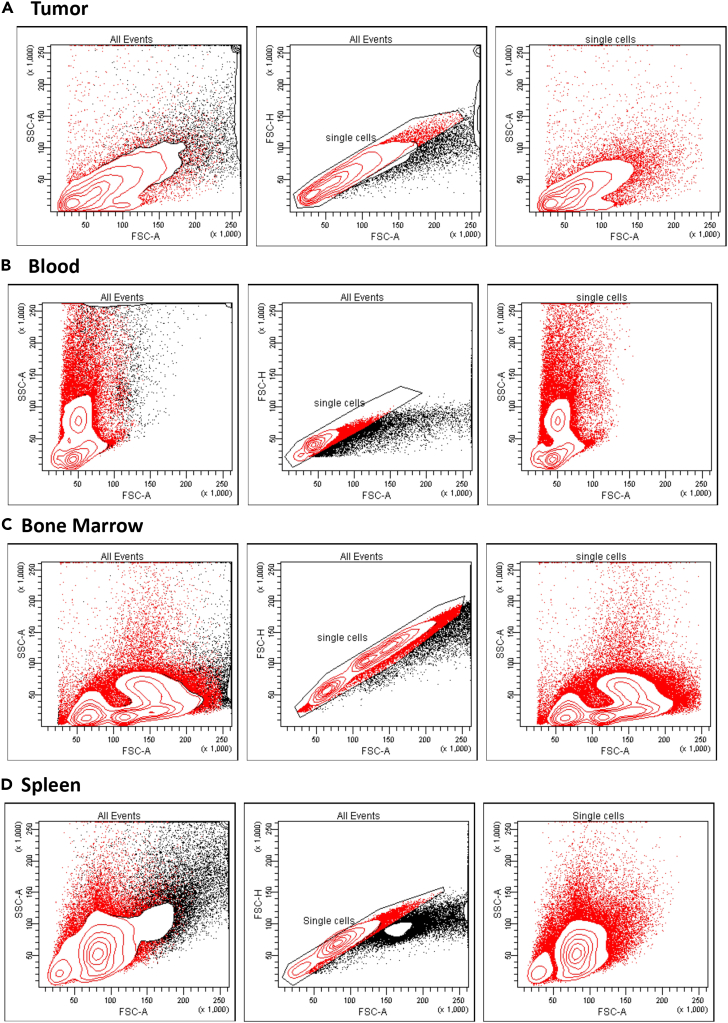
Single-cell suspension is the first step required for flow cytometry assays. Thus, the tissue samples require processing into single-cell suspension before flow cytometry analysis. This procedure details how to obtain a single cell suspension depending on the organ to be processed (Figure 1).
Figure 1.
Single-cell suspension from organs
Preparation and staining of single cell suspension from (A) tumor, (B) Blood, (C) Bone Marrow and (D) Spleen.
Tumor tissue
Timing: 30 min each tumor
-
18.
Take out the tumor tissue from the mice at the bench (Figure 1A) and wash tissue in 10 mL of PBS 1× one time.
-
19.Remove the skin around the tumor tissue.
-
a.Cut a small piece (≈0.5 cm) with autoclaved surgical instruments.
-
b.Put in a new plate (60 mm or 100 mm) with staining medium (See materials and equipment for its preparation).
-
a.
-
20.
Disaggregate tumor tissue mechanically with the rough side of a microscope slide (Figure 1A).
CRITICAL: It is well disaggregated when a turbid solution with few aggregates is obtained.
-
21.Filter the disaggregated tissue with a 40 μm strainer.
-
a.Wash the filter with staining medium to make sure it is well cleaned (Figure 1A).
 CRITICAL: Use 50 mL sterile conical centrifuge tubes to fit the filter properly.
CRITICAL: Use 50 mL sterile conical centrifuge tubes to fit the filter properly. -
b.Put the 50 mL tube on ice to maintain the cells alive at 4°C.Optional: A plunger can be used to improve cell filtration through the filter.
-
a.
-
22.
Centrifuge the cell suspension at 400–600 g for 5 min. After the centrifugation, decant off or aspirate the supernatant.
-
23.
Resuspend the cells with 1 mL of staining medium on ice (4οC).
-
24.
Count total single cells with the counting chamber (Burker Camera) or using an alternative method (hemocytometer).
CRITICAL: It is needed around 600.000 to 1 × 106 tumor cells per panel.
-
25.
Put the counted tumor cells in a microcentrifuge tub (1.5 mL) and centrifuge at 400–600 g for 5 min plus a fast spin of 10 s at maximum speed to recovery all the tumor cells.
-
26.
Incubate tumor cells with 100 μL of the mixture antibodies during 1 h at RT in the dark (Tables 1 and 2) (See immune cell staining section).
Table 1.
List of antibodies used to characterize the TIME (membrane staining)
| Antibody | Clone | Fluorochrome | Concentration; dilution | Company and catalog # |
|---|---|---|---|---|
| Macrophages panel | ||||
| CD11b | M1/70 | PerCP-eFluor 710 | 0.2 mg/mL; 0.1 μL/100 μL | eBioscience # 46-0112-82 |
| Ly6G/Ly6C (Gr) | RB6-8C5 | eFluor 506 | 0.2 mg/mL; 0.1 μL/100 μL | eBioscience # 69-5931-82 |
| F4/80 | BM8 | SB436 | 0.2 mg/mL; 0.8 μL/100 μL | eBioscience # 62-4801-82 |
| CD206 | MR6F3 | PE | 0.2 mg/mL; 0.1 μL/100 μL | eBioscience # 12-2061-82 |
| MHC-II | M5/114.15.2 | APC | 0.2 mg/mL; 0.05 μL/100 μL | eBioscience # 17-5321-82 |
| Lymphocytes T panel | ||||
| CD4 | RM4-5 | FITC | 0.5 mg/mL; 0.08 μL/100 μL | eBioscience # 11-0042-82 |
| CD8 | 53–6.7 | APC-eFluor 780 | 0.2 mg/mL; 0.42 μL/100 μL | eBioscience # 47-0081-82 |
| Myeloid Derived Suppressor Cells panel | ||||
| CD11b | M1/70 | PerCP-eFluor 710 | 0.2 mg/mL; 0.05 μL/100 μL | eBioscience # 46-0112-82 |
| Ly6C | HK1.4 | APC-eFluor 780 | 0.2 mg/mL; 0.65 μL/100 μL | eBioscience # 47-5932-82 |
| Ly6G | 1A8-Ly6g | FTIC | 0.5 mg/mL; 0.65 μL/100 μL | eBioscience # 11-9668-82 |
| CD11c | N418 | SB436 | 0.2 mg/mL; 0.65 μL/100 μL | eBioscience # 62-0114-82 |
| Galectin 3 | M3/38 | eFluor 660 | 0.2 mg/mL; 0.25 μL/100 μL | eBioscience # 50-5301-82 |
| Lymphocytes B panel | ||||
| CD45 | EM-05 | PerCP | 0.5 mg/mL; 0.2 μL/100 μL | eBioscience # MA1-10234 |
| CD19 | 6D5 | BV421 | 0.25 mg/mL; 1μL/100 μL | BioLegend # 115537 |
| B220 | RA3-6B2 | Alexa Fluor 647 | 0.5 mg/mL; 0.1 μL/100 μL | BioLegend # 103226 |
| CD38 | 90 | APC-eFluor 780 | 0.2 mg/mL; 0.1 μL/100 μL | eBioscience # 47-0381-82 |
| CD138 | 300506 | PE | 0.2 mg/mL; 0.2 μL/100 μL | eBioscience # MA5-23527 |
Table 2.
List of antibodies used to characterize the TIME (intracellular staining)
| Antibody | Clone | Fluorochrome | Concentration; Dilution | Company and catalog # |
|---|---|---|---|---|
| Macrophages panel | ||||
| CD11b | M1/70 | PerCP-eFluor 710 | 0.2 mg/mL; 0.1 μL/100 μL | eBioscience # 46-0112-82 |
| Ly6G/Ly6C | RB6-8C5 | eFluor 506 | 0.2 mg/mL; 0.1 μL/100 μL | eBioscience # 69-5931-82 |
| F4/80 | BM8 | SB436 | 0.2 mg/mL; 0.8 μL/100 μL | eBioscience # 62-4801-82 |
| iNOS | CXNFT | PE | 0.2 mg/mL; 0.05 μL/100 μL | eBioscience # 12-5920-82 |
| Arginase 1 | A1exF5 | APC | 0.2 mg/mL; 0.5 μL/100 μL | eBioscience # 17-3697-82 |
| Lymphocytes Treg panel | ||||
| CD4 | RM4-5 | FITC | 0.5 mg/mL; 0.08 μL/100 μL | eBioscience # 11-0042-82 |
| CD8 | 53–6.7 | APC-eFluor 710 | 0.2 mg/mL; 0.42 μL/100 μL | eBioscience # 47-0081-82 |
| FoxP3 | FJK-16s | PE-Cyanine7 | 0.2 mg/mL; 0.25 μL/100 μL | eBioscience # 25-5773-82 |
Blood
Timing: 30 min
-
27.
Blood collection by intracardiac puncture (See sacrifice mice and organs collection section).
CRITICAL: As much as blood you collected better should be the immune detection.
Note: 500 μL to 1 mL of blood is needed to perform the analysis.
-
28.Place blood in heparin tubes or EDTA tubes (≈300 μL of blood in each tube) to prevent coagulation.
-
a.Let the blood stand for 10 min at RT (Figure 1B).
-
b.Centrifuge at 400–600 g for 5 min to obtain plasma.
-
c.Aliquot the plasma in 1.5 mL tubs and store at −20°C or −80°C if it is for a long time.
-
a.
-
29.Transfer the blood pellet into a 50 mL tube.
-
a.Add 4 mL of Milli-Q water per heparin tube.Note: if you have collected 3 tubes of heparin, you will need to add 12 mL of Milli-Q water.
-
b.Shake gentle up and down the 50 mL tube on ice for 30 s (Figure 1B).Note: The objective of steps 5 and 6 is to lysate erythrocytes.
-
c.Stop the lysis reaction by adding 1× PBS up to 50 mL.
 CRITICAL: First add up to 20 mL, discard coagulates and then filed up to 50 mL.Optional: Erythrocyte lysis can be done using RBC 1× lysis buffer. 1 mL is added and let sit for 3 min in ice. (See materials and equipment for its preparation). To obtain 1× solution, dilute lysis buffer 10× with Milli-Q water.
CRITICAL: First add up to 20 mL, discard coagulates and then filed up to 50 mL.Optional: Erythrocyte lysis can be done using RBC 1× lysis buffer. 1 mL is added and let sit for 3 min in ice. (See materials and equipment for its preparation). To obtain 1× solution, dilute lysis buffer 10× with Milli-Q water.
-
a.
-
30.Centrifuge 50 mL tube at 400–600 g for 5 min at 4°C.
-
a.Decant the supernatant taking care not to lose the pellet.Note: Decanting can be done, but better if aspiration is available.
 CRITICAL: if the pellet is still very red, repeat steps 6–8 to improve erythrocyte lysis.
CRITICAL: if the pellet is still very red, repeat steps 6–8 to improve erythrocyte lysis. -
b.Resuspend the pellet with 1 mL of staining medium and transfer the contents into a 1.5 mL tube at 4οC.
-
a.
-
31.
Count total single cells.
Note: Cells can be counted with the counting chamber or using an alternative method.
CRITICAL: It is needed around 300.000–600.000 blood cells per panel.
-
32.
Incubate blood cells with 100 μL of mixture antibodies during 1 h at RT (See immune cell staining section).
Bone marrow
Timing: 45 min
-
33.Remove the femur and repel the skin of syngeneic mice (Figure 1C).
-
a.Cut both epiphyses a little to be able to pass either an insulin or a 23G syringe.
-
b.Flush 3 mL of staining medium through the femoral shaft to obtain bone marrow cells.
 CRITICAL: Pool the two femurs of the mouse in order to have enough bone marrow cells.
CRITICAL: Pool the two femurs of the mouse in order to have enough bone marrow cells. -
c.Disaggregate using a pipette the groups of cells that are stuck together (up and down).
-
a.
-
34.Centrifuge the cell suspension at 400–600 g for 5 min.
-
a.After centrifugation, decant off or aspirate the supernatant.
-
b.Add 900 μL of Milli-Q water in the pellet to lysate erythrocytes.
-
c.Once added, pipette or shake gently up and down during 30 s on ice.
-
d.Stop the reaction with 100 μL of 10× PBS.
-
a.
Optional: Erythrocyte lysis can be done using RCB 1× lysis buffer. 1 mL is added and let sit for 3 min in ice (See materials and equipment for its preparation). To obtain 1× solution, dilute lysis buffer 10× with Milli-Q water.
-
35.Centrifuge the cell suspension at 400–600 g for 5 min.
-
a.After the centrifugation, decant off or aspirate the supernatant and resuspend the pellet with 1 mL of staining medium.
-
b.Pass the cell suspension through a 40 μm strainer.
 CRITICAL: A plunger from a 50 mL syringe can be used to improve filtration.
CRITICAL: A plunger from a 50 mL syringe can be used to improve filtration. -
c.Pass through the strainer a bit of staining medium to clean it and recover as much as cells you can.
-
a.
-
36.Centrifuge the 50 mL tube at 400–600 g for 5 min at 4οC and spin at the end.
-
a.Resuspend the pellet with 1 mL of staining medium on ice.
-
b.Count total single cells. Cells can be counted with the counting chamber or using an alternative method.
-
a.
CRITICAL: It is needed around 300.000–600.000 bone marrow cells per panel.
-
37.
Incubate bone marrow cells with 100 μL of mixture antibodies during 1 h at RT (See immune cell staining section).
Spleen organ
Timing: 30 min
-
38.Disaggregate spleen tissue (half spleen; ≈1 cm) mechanically with the rough side of a microscope slide (Figure 1D).
 CRITICAL: It is well disaggregated when a red turbid solution with few aggregates is obtained.
CRITICAL: It is well disaggregated when a red turbid solution with few aggregates is obtained.-
a.Put the disaggregated mixture in a 50 mL tube.
-
b.Centrifuge the cell suspension at 400–600 g for 5 min.
-
c.After the centrifugation, decant off or vacuum the supernatant.
-
d.Lyse the erythrocytes from the cell suspension with 900 μL of Milli-Q water.
-
e.Pipette up and down almost 5 times to resuspend the pellet.Optional: Erythrocyte lysis can be done using RBC 1× lysis buffer. 1 mL is added and let sit for 3 min in ice (See materials and equipment for its preparation). To obtain 1× solution, dilute lysis buffer 10× with Milli-Q water.
-
f.Add 100 μL of 10× PBS to stop the lysis reaction.
-
a.
-
39.Centrifuge the cell suspension at 400–600 g for 5 min.
-
a.After the centrifugation spin and decant off or vacuum the supernatant.
-
b.Resuspend with 1 mL of staining medium on ice.
-
a.
-
40.
Filter through 40 μm strainer put on a 50 mL tube on ice.
CRITICAL: A plunger can be used to improve filtration.
-
41.Centrifuge the cell suspension at 400–600 g for 5 min.
-
a.After the centrifugation, decant off or vacuum the supernatant.
-
b.Count total single cells. Cells can be counted with the counting chamber or using an alternative method.
-
a.
CRITICAL: It is needed around 600.000 spleen cells per panel.
-
42.
Incubate spleen cells with 100 μL of mixture antibodies during 1 h at RT (See immune cell staining section).
Characterization and optimization of immune antibody panel
Timing: 1–2 h
Know your flow cytometer
Knowing which cytometer is available in your Flow Cytometry Facility is of outmost interest in order to properly set up a correct immune antibody panel.
Note: There are several parts that must be taken into account to help you choose your final antibody-fluorochrome combinations: number of lasers, number of detectors for each laser and type of filter in front of the detector (Figure 2A).
-
43.
You need to know which lasers are available in your cytometer.
Note: A cytometer with 5 lasers will probably let you choose at least one fluorochrome for each laser, so a combination of 5 colors would be quite easy to achieve, while a 3-laser cytometer would let you choose just only 3 different fluorochromes minimum.
-
44.
Final number of different fluorochromes will be determined by the number of detectors available for each laser.
-
45.
Final number of different fluorochromes will be determined by the filter that sits in front of the detector.
Note: All commercial cytometers have a tunable configuration that is usually set at the moment the cytometer is bought.
Note: Sometimes some filters might be interchanged afterwards.
-
46.You need to be aware of these considerations and ask for this information to your flow cytometry technician to know exactly:
-
a.How many lasers the cytometer has.
-
b.How many detectors the cytometer has.
-
c.Which range of wavelengths is able to detect by cytometer.
-
a.
Note: Knowing all this information will allow you to be sure that the fluorochromes you have chosen will be excited and detected in your flow cytometer.
Know your cell type, antigens and fluorochromes
Everything starts with understanding what the biological hypothesis is and which experiments need to be performed.
Note: This will dictate what populations need to be identified, and what information needs to be extracted from the data.
-
47.
Rank your antibodies based on cellular expression level and importance in answering the biological hypothesis (Figure 2B).
Note: For example, CD11b is a highly expressed antigen on monocyte population and is important in making primary gating decisions, while CD206 is a faintly expressed marker on cells undergoing activation and may be critical to answer the biological hypothesis
-
48.
Search for the appropriate fluorochromes for each of the populations you want to determine.
Note: This can be done by using a chart from different companies where fluorochromes are ranked from brightest to most dim.
Panel optimization
Once the fluorochromes have been selected for each of the study populations, the panel must be optimized and validated. Proper titration of antibodies, correct voltages and correcting for the spectral overlap are all part of a lengthy but very critical process to ensure that the panel works properly (Figure 2C). To correct for this spectral overlap, a process of fluorescence compensation is used. This ensures that the fluorescence detected in a particular detector derives from the fluorochrome that is being measured.
Here are some general steps for compensation and panel optimization (Figure 2D).
-
51.
Determine which fluorophores are involved in spectral overlap and select compensation controls.
Note: Include an unstained sample to correct for autofluorescence inherent to cells.
CRITICAL: Compensation controls are samples stained with single-color antibodies that are conjugated to the same fluorophores as those used in the experiment. If sample availability is critical, consider using compensation beads instead of cells.
-
52.
Collect compensation controls and experimental samples on the flow cytometer using identical instrument settings.
Note: Make sure that positive signal is as brighter as possible, at least as bright as your experimental sample positive signal.
-
53.
Use software to generate compensation matrices that describe the spectral overlap between the fluorophores.
Note: These matrices are used to mathematically adjust the fluorescence signals in each channel to correct for the spectral overlap.
-
54.
Apply the compensation matrices to the experimental data to correct for spectral overlap.
-
55.
Analyze the data to identify the different cell populations.
CRITICAL: It is important to optimize the compensation settings to minimize compensation errors and to validate the compensation strategy using appropriate positive and negative controls. It is also important to note that over-compensation can lead to inaccurate measurements and should be avoided.
Controls and Fluorescent minus one (FMOs)
Setting up the right controls is critical to determining how your cells are responding to treatment. It’s also important for correctly interpreting your data and drawing correct conclusions.
Note: Without the proper controls, you would not be able to neither compensate your flow cytometry experiments nor to identify your cells of interest correctly (Figure 2D).
Note: The Fluorescence Minus One Control (FMO) is a type of control used to properly interpret flow cytometry data. It is used to:
-
56.
Identify and gate cells in the context of data spread due to the multiple fluorochromes in a given panel.
-
57.
Use a FMO control that contains all the fluorochromes in a panel, except the one that is being measured (Figure 2D).
Note: The FMO control ensures that the any spread of the fluorochromes into the channel of interest is properly identified.
-
58.
Each day that an experiment is carried out, an unstained sample has to be run to determine the autofluorescence of the samples.
Immune cell staining
The aim of this part is to accomplish identification of immune cells that infiltrate or surround the tumor cells.
Membrane staining
Timing: 1 h 30 min
This section details the procedure of antibody staining of membrane antigen in single cell suspension.
-
59.
Prepare a single-cell suspension using the appropriate protocol (see Generation of single cell suspension protocol) and adjust the cell amount from 600.000 to 106 cells in staining medium in each tube.
-
60.
Centrifuge the single cell suspension 400–600 g during 5 min.
Note: Prepare as many staining tubes as needed: unstained control, and individually sample for each antibody mixture panel.
-
61.
Add the optimized dilution mixture of antibodies (Table 1) to the respective tubes and incubate them 1 h at RT in the dark.
-
62.Centrifuge the 1.5 mL tube at 400–600 g for 5 min.
-
a.Wash the cells once with ice-cold staining medium at 400–600 g for 5 min.
-
b.Resuspend with 100–200 μL FACS buffer.
-
a.
Note: You can repeat the washing step to better remove the antibodies before analyzing the cells in the cytometer.
Optional: You can fix the cells with PFA 1%. If the samples are fixed, they can be stored at 4οC during 24 h before analysis, otherwise they must be analyzed the same day.
-
63.
Analyze the stained cell preparation in the cytometer (See flow cytometry acquisition section).
Intracellular staining
Timing: 2 h–2 h 30 min
This section delineates the procedure of antibody staining of intracellular antigen in single cell suspension.
-
64.
Prepare a single cell suspension using a specific protocol depending on the tissue (See generation of single cell suspension section).
-
65.
First, we have to perform the cell membrane staining protocol or cell surface markers (See membrane staining section).
-
66.After this staining, centrifuge de 1.5 mL tube at 400–600 g for 5 min.
-
a.Discard the supernatant and add 100 μL of staining medium to wash the cells.
-
b.After the last wash, discard the supernatant and pulse vortex the sample to completely dissociate the pellet.
-
a.
CRITICAL: You should obtain a good suspension of individual cells in order to have a good preparation. You can also pipette up and down the pellet to homogenize better.
-
67.Fix the cells by adding 200 μL of Fixation/Permeabilization Buffer (Invitrogen, cat. no. 00-5123-43) (1:3 dilution with Fixation/Permeabilization Diluent (Invitrogen, cat. no. 00-5223-56) to each 1.5 mL tube.
-
a.Pipette up and down the pellet to homogenize better.
-
b.Incubate 30–60 min at RT.Note: Protect from light.Optional: Samples can be incubated for up to 18 h at 2°C–8°C in the dark.
-
c.Centrifuge samples at 400–600 g at room temperature for 5 min.
-
d.Discard the supernatant.
-
a.
-
68.Add 200 μL 1× Permeabilization Buffer (Invitrogen, cat. no. 00-8333-56) (1 part of 10× Permeabilization Buffer with 9 parts of Milli-Q water) to each tube.
-
a.Centrifuge samples 400–600 g at room temperature for 5 min.
-
b.Discard the supernatant.
-
a.
-
69.
Repeat Step 69.
-
70.
Resuspend pellet in residual volume.
Optional: Block with 2% normal mouse/rat serum by adding 2 μL directly to the cells. Incubate at room temperature for 15 min.
-
71.Without washing, add the recommended amount of directly conjugated antibody for the detection of intracellular antigen(s)/markers (See Table 2).
-
a.Incubate for at least 30 min at RT in the dark.
-
b.Centrifuge samples at 400–600 g at room temperature for 5 min.
-
c.Discard the supernatant.
-
a.
-
72.Add 200 μL of 1× Permeabilization Buffer to each 1.5 mL tube.
-
a.Centrifuge samples at 400–600 g at RT for 5 min.
-
b.Discard the supernatant.
-
a.
-
73.
Repeat Step 73.
-
74.
Resuspend stained cells in an appropriate volume of Flow Cytometry Buffer (100–200 μL of FACSFlow).
-
75.
Analyze by flow cytometry (See flow cytometry acquisition and phenotyping section).
Flow cytometry acquisition
Flow cytometer setup and quality control
Timing: 1 h
Setting up a flow cytometer involves several steps. Here are some general steps that may vary slightly depending on the specific flow cytometer model.
-
76.
Power on the flow cytometer and the computer that controls it.
-
77.
Check that the lasers are working correctly and calibrated according to the manufacturer’s specifications.
-
78.
Set up the appropriate filters and detectors for the fluorophores used in the experiment.
-
79.
Adjust the gain and voltage settings for the detectors to optimize the signal-to-noise ratio for each fluorescence channel.
-
80.
Perform a quality control check with calibration beads to ensure the flow cytometer is working correctly.
-
81.
Run compensation tubes to perform compensation and correct for the spectral overlap. Once voltage settings are properly set and compensation done, make sure that your experimental samples are recorded using the same settings.
-
82.
Prepare the sample and load it into the sample injection port or chamber.
-
83.
Run the sample through the flow cytometer and collect data.
-
84.
Analyze the data using appropriate software such as FlowJo 8.7 (FlowJo Software, BD) or BD FACSDiva Software to generate scatter plots and histograms to identify the different cell populations.
Note: It is important to follow the manufacturer's instructions for specific flow cytometer models and to maintain and clean the instrument regularly to ensure optimal performance.
Collecting samples
Timing: 1–4 h depending on the tubes
This step details the acquisition of our samples using the BD FACSCanto II flow cytometer from our institution.
Note: You might have to modify this step to accommodate to your institute’s instruments and guidelines (Figure 2A).
-
85.
Transfer the samples to individual FACS tubes.
Note: If your instrument has a plate reader attached follow your institute’s core facility’s guidelines.
-
86.
Use your unstained control to set the voltages of your FSC-A, SSC-A plot depending on the cell type (Blood, Bone Marrow, Spleen or Tumor cells (Figures 3A–3D).
-
87.
In the FSC-A and SSC-A plot, draw a gate around all cell events profile (Figure 3).
-
88.
In the FSC-A and FSC-H plot, draw a gate around the diagonal to select only the single cells (Figure 3).
-
89.
Draw other dot plots showing the different fluorochromes combinations depending on each immune panel.
Note: Use your FMO controls to set the proper gates in each dot plot.
Note: These controls will allow you to identify data spread in your fluorochrome combinations (see controls and fluorescent minus one (FMOs) section).
Note: Once spread has been identified, the gate will be correctly set to show real positivity/negativity for a given marker.
-
90.
Acquire all samples and ensure that record at least 10.000 events of your selected population.
-
91.
Perform the acquisition of the stained tubes using the BD FACSCanto II flow cytometer.
-
92.
Once acquisition is completed, export your data according to your institute’s core facility’s guidelines.
Note: This procedure will be explained in detail in the next section.
-
93.
Using BD FACSDiva (or the available acquisition software in your institute’s core facility) create a new data acquisition matrix.
Figure 3.
Gating strategy of the single-cell suspension
Flow cytometry gating strategy to select single cell from (A) melanoma tumor, (B) Blood, (C) Bone Marrow and (D) spleen.
Expected outcomes
After the acquisition of sample by flow cytometry panels (See flow cytometry acquisition) single cells were discriminated using the following gaiting strategies depending on the cell type (Figure 3).
In Lymphocyte T gating strategy, we identify CD4+ T helper cells (CD4+/CD8-), CD8+ cytotoxic T cells (CD4−/CD8+) and regulatory T-cells (Treg) (CD4+/FoxP3+/CD8-) (Figure 4A).
Figure 4.
Flow cytometry gating strategy to identify T lymphocytes and macrophages
Flow cytometry analysis of (A) Lymphocyte CD4+ T helper cells, CD8+ cytotoxic T cells and regulatory T-cells.
(B) Tumor associated macrophages (TAMs) and (C) TAMs M1-type or M2-type gating panel.
In Macrophages gating panel we could characterize monocytes (CD11b+/Ly6C + Ly6G–), Tumor associated macrophages (TAMs) (CD11b+/Ly6G−/F4/80+), TAM M1-type (CD11b+/Ly6G−/F4/80+/MHCII+/CD206- or CD11b+/Ly6G−/F4/80+/iNOS+) and TAM M2-type (CD11b+/Ly6G−/F4/80+/MHCII-/CD206+ or CD11b+/Ly6G−/F4/80+/Arg+) (Figures 4B and 4C).
In Myeloid Derived Suppressor Cells (MDSCs) we could identify monocytic MDSCs (M-MDSC (CD11b+/Ly6C+/Ly6G-/Galectin+)) polymorphonucleate MDSC (PM-MDSC (CD11b+/Ly6C+/mild, Ly6G+/Galectin low)) (Figure 5).
Figure 5.
Flow cytometry gating strategy to identify Myeloid Derived Suppressor Cells (MDSCs)
In Lymphocyte B cells we could classify B cells (CD45+/CD19+/B220+), plasmatic B cells (CD45+/CD19+/B220+/CD138+/CD38-) and memory B cells (CD45+/CD19+/B220+/CD138-/CD38+) (Figure 6).
Figure 6.
Flow cytometry gating strategy to identify plasmatic B cells and memory B cells
In this gating strategy, the researcher will be able to determine the percentage (%) of the specific immune population according to lymphocytic or monocytic lineage. Further, the researcher will be able to obtain the mean fluorescence intensity (MFI) of the fluorochrome, which determines the amount of antibody that specifically bound to the specific antigen used in each immune panel.
In addition, if you could use a cell sorter flow cytometer you should sort the specific cell population of interest for targeting downstream analysis such us proteomics or genomics profile.
Limitations
Here we describe an approach to study and describe the immune cells that surround or infiltrate the tumor, in this approach you obtain a higher amount of single cell population to perform all the specific panels at the same time. However, the melanoma tumor is poorly infiltrated with regulatory T cells (Treg) and lymphocyte B cells. Therefore, when a minority population has to be identified, a greater number of starting single cell (around 2–3 × 106 cells) is required to ensure the acquisition of adequate events of interest.
Depending on the cytometer available at your research center, panels can be made with more or less markers. This should be taken into account when panels are designed.
In the case of tumors, consider collecting the infiltrating part of the tumor. If the samples are examined and there is no staining, it means that a necrotic area or tumor body has been taken.
In case of blood sample, a proper blood collection is important in order to have enough cells for the subsequent steps. A minimum of 300.000 cells is required for each of the study panels. If this minimum amount is not obtained, there will not be enough events to perform good analysis.
In case of bone marrow sample, pool the two femurs to have enough cells for the subsequent steps. A minimum of 300,000 cells is required for each of the study panels. If this minimum amount is not obtained, there will not be enough events to perform good analysis.
In spleen, blood and bone marrow samples, it is important to perform a good erythrocytes’ lysis. If there is too much debris in the SSC-A vs. FSC-A gate it will not be possible to determine the population of the study.
Troubleshooting
Problem 1
High number of cell death in the single cell suspension.
Potential solution
The possible reason is because you do not perform rapidly the protocol or you do not put the single cell suspension on ice (4°C) before doing the antibody detection panel (Step 22–26). Also, the possibility of a long lysis period with Milli-Q water or the lysis buffer must also be ruled out (Step 30–31). If this is the case, reduce the incubation time.
Problem 2
You do not detect as much positive population as you want to analyze.
Potential solution
You could add a higher number of total single cells to get through the flow cytometer to detect as much as positive single cells of interests (Step 60). Another solution is to put higher amount of antibody in the mixture panel (Tables 1 and 2).
Problem 3
The disaggregated cells are clustered in the cell strainer and the liquid and cells do not pass through.
Potential solution
If the suspension is highly concentrated the cell strainer may be obstructed. To avoid obstruction, you can dilute the sample before filtration or help to get through it with a plunge syringe (Step 22 or 41).
Problem 4
Low yield of cells in the single cell staining suspension before flow cytometry acquisition.
Potential solution
The possible reason could be an error in centrifugation steps or during discarding the supernatant (Step 24–25 or 31 or 36–37 or 42). Maintain the correct protocol, take care about the visible pellet during all the steps and resuspend them in a correct buffer volume.
Problem 5
No positive staining of markers detected by flow cytometry.
Potential solution
You could repeat compensation settings to increase staining voltage or use different fluorophore that might work better in your combination panel (See panel optimization, Step 52–55). Make sure that your antibodies are tested by flow cytometry. Re-titration of your antibodies may be a good choice to enhance signal-to-noise ratio of your staining.
Problem 6
Intracellular stain panel cannot be detected.
Potential solution
You can increase Fixation/Permeabilization solution incubation (Step 68–69) and increase the amount of specific antibody to detect this epitope in your single cell suspension (See Tables 1 and 2).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anna Macià (amacia@irblleida.cat) and Pol Sisó (polsiso95@gmail.com).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This study has been funded by the Instituto de Salud Carlos III (ISCIII) through the project "PI18/00573 and PI21/294" and co-funded by the European Union. This work was also supported by a grant from Ministerio de Ciencia e Innovación (PID2019-109302RB-I00 to J.V.). P.S. holds a predoctoral fellowship from Universitat de Lleida (UdL)-IRBLleida. I.d.l.R. holds a predoctoral fellowship from AECC. C.R. holds a predoctoral fellowship from UdL. The cell culture experiments were performed in the Cell Culture Scientific and Technical Service from UdL, Lleida, Spain. The flow cytometry analysis was performed in the Flow Cytometry Core Facility from IRBLleida.
Author contributions
P.S., A.P., and A.M. designed the experiments and performed data analysis; P.S., I.d.l.R., and C.R. performed the experiments; J.V., R.M.M., and A.M. supervised the manuscript; P.S., A.P., and A.M. wrote and edited the manuscript; and J.V., R.M.M., and A.M. provided funding acquisition.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Pol Sisó, Email: polsiso95@gmail.com.
Anna Macià, Email: amacia@irblleida.cat.
Data and code availability
This study did not generate new datasets or code.
References
- 1.Dhomen N., Reis-filho J.S., Dias R., Hayward R., Savage K., Delmas V., Laure L., Pritchard C., Marais R. Oncogenic Braf Induces Melanocyte Senescence and Melanoma in Mice. Cancer Cell. 2009:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Hirata E., Girotti M.R., Viros A., Hopper S., Spence-Dene B., Matsuda M., Larkin J., Marais R., Sahai E. Intravital Imaging Reveals How BRAF Inhibition Generates Drug-Tolerant Microenvironments with High Integrin β1/FAK Signaling. Cancer Cell. 2015:574–588. doi: 10.1016/j.ccell.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new datasets or code.