Summary
Homologous recombination (HR)-mediated DNA repair is a prerequisite for maintaining genome stability. Cancer cells displaying HR deficiency (HRD) are selectively eliminated by poly(ADP-ribose) polymerase inhibitors (PARPis). To date, sequencing of HR-associated genes and analyzing genome instability have been used as clinical predictions for PARPi therapy. However, these genetic tests cannot reflect dynamic changes in the HR status. Here, we have developed a virus- and activity-based functional assay to quantify real-time HR activity directly. Instead of focusing on a few HR-associated genes, our functional assay detects endpoint HR activity and establishes an activity threshold for identifying HRD across cancer types, validated by PARPi sensitivity and BRCA status. Notably, this fluorescence-based assay can be applied to primary ovarian cancer cells from patients to reflect their level of HRD, which is associated with survival benefits. Thus, our work provides a functional test to predict the response of primary cancer cells to PARPis.
Keywords: BRCA, DNA repair activity, DNA repair deficiency, HRD, functional test, ovarian cancer, breast cancer, colorectal cancer, PARP inhibitor
Graphical abstract
Highlights
-
•
The fluorescence-based activity score reflects DNA repair status in real time
-
•
The activity score is correlated with BRCA status and PARPi sensitivity
-
•
A standard activity threshold identifies repair-deficient cells among cancer types
-
•
The activity score is correlated with the duration of progression-free survival
Lee et al. develop a virus-based assay to quantify DNA repair activity in cells and to identify different types of cancer cells that are susceptible to poly(ADP-ribose) polymerase inhibitors (PARPis). Notably, their functional test predicts the clinical response to platinum-based first-line treatment in patients with ovarian cancer, demonstrating clinical applicability.
Introduction
Homologous recombination (HR) is an error-free DNA double-strand break repair pathway. Genetic and epigenetic alteration of HR-related genes has been shown to elicit HR deficiency (HRD),1 a phenotype characterized by defective HR-mediated DNA repair activity in cells. For example, the tumor-suppressor genes BRCA1 and BRCA2 exert essential roles in maintaining genome integrity through HR-directed DNA repair.2,3 Germline mutation in BRCA1 or BRCA2 greatly increases the lifetime risk of developing breast cancer to 82%.4 Additionally, carriers of BRCA1 and BRCA2 mutations have lifetime risks of developing ovarian cancer of approximately 54% and 23%, respectively,4 highlighting a strong correlation between HRD and tumorigenesis. Cancer cells featuring HRD provide a unique opportunity for precision cancer treatment. It has been well demonstrated that poly(ADP-ribose) polymerase inhibitors (PARPis), which inhibit single-strand DNA break sensing and repair to induce DNA double-strand break formation, exhibit selective synthetic lethality preferentially in cancer cells defective in HR.5,6 Promisingly, targeting HRD in patients carrying BRCA mutations and diagnosed with ovarian, breast, pancreatic, or prostate cancers by means of PARPi significantly lowers the risk of tumor recurrence.7,8,9,10,11
Although BRCA mutations in somatic breast tumors are scarce and only account for ∼15% of ovarian cancer,12,13,14 other dysregulated mechanisms can lead to HRD phenotypes, including hypermethylation of the BRCA1 promoter, mutations in other HR-associated genes, and amplification of genes that attenuate BRCA2 transcription.1 Given the various mechanisms linked to HRD, many studies have suggested a high proportion of patients with breast (46%), ovarian (51%), pancreatic (14%), and prostate cancers (19%)15,16,17,18,19 displaying HRD by means of comprehensive molecular assessment, pathway analyses, and whole-genome sequencing. Thus, there is a pronounced clinical need to develop strategies for identifying patients with cancer with HRD beyond those linked to BRCA mutations.
Several studies have established genomic-scar-based approaches to quantify large-scale genomic aberrations related to HRD,20,21,22 including large-scale transitions (LSTs),23 telomeric allelic imbalance (TAI),20 and loss of heterozygosity (LOH) events.24 Notably, a previous investigation defined cells with a genomic-scar-based HRD score, calculated as the unweighted sum of LST, TAI, and LOH events, with a threshold of ≥42 as indicative of HRD.25 Ever since, this genomic-scar-based HRD score has been applied in clinical trials as a predictive test. Although the genomic-scar-based HRD scores correlated with clinical responses to platinum-containing neoadjuvant chemotherapy in triple-negative breast cancer in that study, the false positive rate (47%) was high.25 Moreover, assays identifying genomic instability cannot detect chromosomal abnormalities of <10 Mbp in length, implying a detection limit due to low sensitivity.21 Notably, the genetic test employed in the past to assess abnormal genomic structures does not precisely reflect the real-time HR status of tumor cells in patients receiving treatments. Thus, a major limitation of sequencing-based approaches is their inability to promptly reveal dynamic changes in HR status, including abrogation of the HRD phenotype, which is a key element contributing to clinical resistance.21,26 Moreover, recent phase 3 studies have shown that genome instability is not necessarily correlated with clinical responses arising from drugs targeting HRD.27,28 Thus, the European Society for Medical Oncology (ESMO) has highlighted that better real-time assays are urgently needed to identify HRD patients for first-line treatment of ovarian cancer.26,29
Although RAD51 foci-based functional assays can successfully detect HRD patients with ovarian or breast cancer,30,31 a standard HRD threshold has not yet been proposed. In addition, RAD51 foci cannot reflect HRD cells defective in steps downstream of RAD51 formation.32 Here, we have developed a real-time functional assay for directly assessing HR activity that may circumvent the aforementioned limitations. As measured by our functional test, HR activity scores strongly correlate with cellular sensitivity to PARPis. Notably, our work identifies an activity threshold for HRD among different cancer types. Furthermore, we show the clinical applicability of our activity-based assay in primary ovarian cancer cells. Apart from its correlation with ex vivo PARPi sensitivity, the HR status detected by our functional test is also associated with patients’ clinical responses to platinum-based first-line treatment. Patients with lower HR activity scores showed longer progression-free survival than those with higher activity scores. Additionally, we conducted a small-scale comparative analysis among the activity-based functional test, the sequencing-based test, and the RAD51 foci-based assay to support the promising potential for a further prospective biomarker study.
Results
The activity-based functional test detects HR status in real time
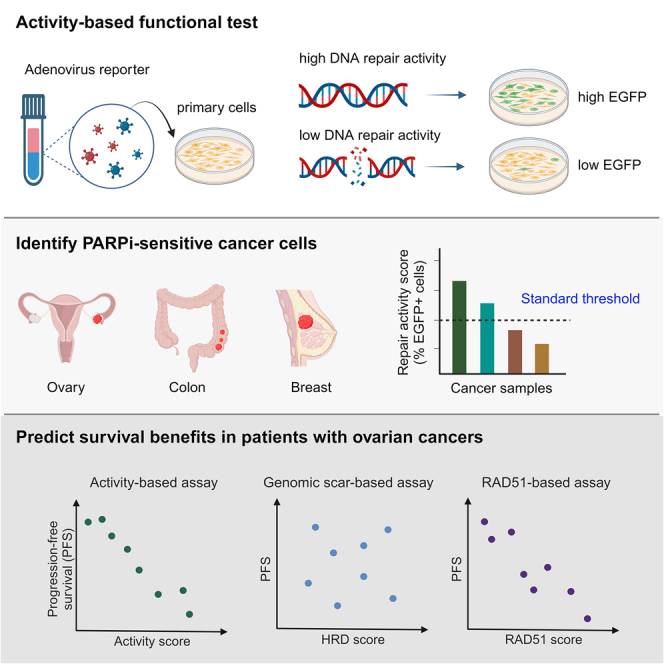
The DR-GFP (direct repeat-green fluorescent protein)-based reporter assay was first established over 20 years ago,33 and it is widely used to measure HR activity in laboratories. In this assay, the reporter cassette is composed of two copies of mutated enhanced GFP (EGFP) fragments: one copy harbors the recognition site of the endonuclease I-SceI, and the other, which serves as a repair template, contains a 5′ and 3′ truncation fragment. Thus, neither copy can express functional EGFP. Upon expression of I-SceI in cells, a double-strand break (DSB) is generated in the EGFP sequence, followed by homology-directed repair to restore and produce a functional EGFP (Figure S1A).33 Thus, resulting EGFP signals represent cellular HR activity. However, only a few available human cell lines (e.g., HEK293 and U2OS) harbor in their genome the integrated reporter cassette for studying HR.34,35 Consequently, simultaneous co-transfection of two plasmids encoding I-SceI and DR-GFP (5.6 and 8.6 kb, respectively) is necessary to detect HR activity in desired cell lines.36 Due to significantly low co-transfection efficiencies in primary cells, this type of HR reporter assay has not been successfully applied for clinical diagnostics of HRD patients. Moreover, comparing HR activity between different cell lines without normalizing co-transfection efficiency remains controversial.
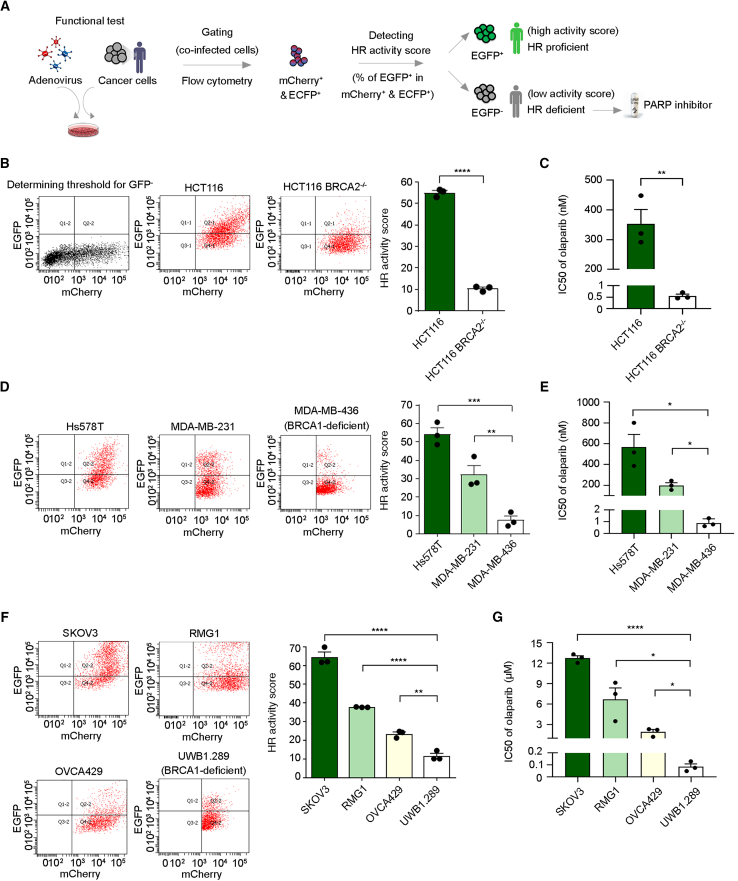
We have optimized this established DR-GFP assay into an adenovirus-based HR reporter system to detect HRD in clinical samples and act as an activity-based HR test (Figures 1A and S1A). The advantage of adenovirus is that it can infect a wide range of cell types, including primary cancer cells, with high efficiency.37 We chose adenovirus serotype 5, the most frequently used virus vector in clinical settings worldwide, which does not integrate into the host genome, unlike lentivirus. Two adenovirus constructs were designed to monitor HR rate: one construct harbors I-SceI and the red fluorescent protein mCherry, and the other contains the DR-GFP cassette and enhanced cyan fluorescent protein (ECFP) (Figure S1A). The mCherry and ECFP fluorescent proteins act as indicators in subsequent flow cytometry analysis to confirm that cells have been co-infected by two types of viruses harboring I-SceI and the DR-GFP reporter cassette, respectively. Only mCherry- and ECFP-positive cells were then used to measure the EGFP signal, which reflects cellular HR activity. The HR activity score is the percentage of EGFP-positive cells among the mCherry- and ECFP-positive population (Figures 1A, S1B, and S1C). The procedure for conducting our activity-based assay is straightforward. Target cells are incubated with the optimized virus-mediated HR reporter. Then, automatic fluorescence emission is analyzed by flow cytometers, which are generally available in laboratory medicine departments in medical centers or hospitals (Figure 1A).
Figure 1.
The activity-based assay predicts sensitivity to a PARPi
(A) Flowchart of the adenovirus-based detection method to assess cellular HR activity to accompany PARPi treatment as precision medicine.
(B) HR activity of parental or BRCA2-deficient colorectal carcinoma HCT116 cells was assessed 72 h after adenovirus infection.
(C) Viability of paired HCT116 cells was measured upon treatment with olaparib for 12 days to evaluate the half-maximal inhibitory concentration (IC50) of olaparib.
(D) HR activity of triple-negative breast cancer cell lines Hs578T, MDA-MB-231, and BRCA1-deficient MDA-MB-436 cells was analyzed 72 h after adenovirus infection.
(E) Cell sensitivity to olaparib was measured after treatment with olaparib for 12 days.
(F) HR activity of ovarian cancer cell lines SKOV3, RMG1, OVCA429, and BRCA1-deficient UWB1.289 cells was assessed.
(G) Ovarian cancer cells were treated with olaparib for 12 days, followed by measurement of the IC50 of olaparib.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Data are the mean ± SEM from three independent experiments (n = 3 biological replicates). Statistical analysis was performed by unpaired two-tailed Student’s t test in (B) and (C) and one-way ANOVA with Tukey’s post hoc test in (D)–(G).
See also Figure S1.
Next, to assess the accuracy of the DR-GFP assay in the adenovirus system, we investigated the impact of modulating RAD51 recombinase activity, a critical effector of HR. Consistent with a prior study employing the plasmid-based DR-GFP assay,38 small interfering RNA (siRNA)-mediated knockdown of RAD51 reduced HR activity by approximately 75% in the osteosarcoma U2OS cell line in both plasmid-based and adenovirus-based systems (Wan et al.38; Figure S1D), supporting the reliability of our optimized adenovirus-based test. Furthermore, our method successfully detected HR activity attenuation resulting from dose-dependent inhibition of RAD51 enzymatic activity using the small-molecule inhibitor B0239 (Figure S1E). These findings collectively demonstrate that virus-based functional assay effectively characterizes real-time cellular HR defects, manifesting as low HR activity scores.
The HR activity scores correlate with PARPi sensitivity across cancer types
The traditional plasmid-based DR-GFP assay has been employed in laboratories since 1999,33 but previous studies have not explored whether an HR activity value could be deployed as a threshold for distinguishing HRD samples from HR-proficient (HRP) ones due to varied co-transfection rates. To address this issue, we sought to identify an HR activity threshold that would differentiate HRD samples from HRP ones across different cell types. For this purpose, we conducted a comprehensive analysis of the correlation between HR activity scores obtained from our functional test and cell sensitivity to PARPis in diverse cell types.
We adopted a pair of colorectal carcinoma cell lines, HCT116, with one deficient in BRCA2 and the other harboring its wild-type form. It is important to note that the parental BRCA2-proficient HCT116 cell line carries a mixture of wild-type and mutant alleles of MRE11, leading to an impaired DNA damage response.40,41 Consequently, the parental HCT116 cells have been reported to exhibit mild sensitivity to PARPis compared with the PARPi-resistant SW620 cells, an MRE11-proficient colorectal cancer cell line.42 Nevertheless, despite this potential confounding factor, the parental BRCA2-proficient HCT116 cells demonstrated significantly higher HR activity scores compared with the BRCA2-deficient cells (Figure 1B). In addition, our activity-based assay detected significant difference in HR activity, which was highly correlated with cell sensitivity to the PARPi olaparib (Figures 1B, 1C, and S1F). Notably, our activity-based test could also be applied to triple-negative breast cancer cells (Figure 1D) and ovarian cancer cells (Figure 1F) to distinguish HRD cells attributable to BRCA1 deficiency and those that exhibited the most robust sensitivity to olaparib (Figures 1E, 1G, S1G, and S1H). It is worth noting that different BRCA1/2-deficient cell types consistently presented an HR activity score of ≤12 (10, 8, and 12 for Figures 1B, 1D, and 1F, respectively), suggesting that a standard threshold can be defined using our activity-based analysis for HRD detection across different cell types, including at least colorectal, triple-negative breast, and ovarian carcinoma cells. We elaborate further on this significant feature in the next section. Importantly, these results demonstrate the feasibility of applying our functional test to multiple cancer types.
The activity-based functional assay identifies primary ovarian cancer cells susceptible to a PARPi
Next, we evaluated the clinical applicability of our activity-based assay. To do so, we isolated primary epithelial cancer cells derived from tumor tissue or ascites of patients with ovarian cancer using negative selection to eliminate unwanted cells (Figure S2A). The tumor cell isolation kit we used (Miltenyi Biotec) contains a cocktail of monoclonal antibodies to magnetically label non-tumor cells in the samples, including lymphocytes, fibroblasts, and endothelial cells. Upon applying a magnetic field, unlabeled human tumor cells can then be harvested. Moreover, the identity of primary ovarian cancer cells was confirmed by staining for epithelial cytokeratin markers and ovarian-cancer-associated transcription factor PAX8 (Figure S2B). By analyzing the percentage of pan-cytokeratin-positive cells in the unlabeled tumor cell fraction, we found that the isolation kit exhibited an efficiency of ≥95% in purifying epithelial cells from other types of cells (Figure S2B). Furthermore, the percentage of PAX8-positive cells in the unlabeled tumor fraction ranged from 90% to 93%, confirming the high efficiency and accuracy of our negative selection procedure. In addition, a high prevalence of p53 deficiency (96%) is a unique characteristic of high-grade serous ovarian carcinoma (HGSOC), a subtype of ovarian cancer.15 Therefore, we subjected primary ovarian cancer cells derived from the ascites of HGSOC to immunoblotting analysis. As expected, all isolated HGSOC ascites samples harbored p53 deficiency, comparable to the p53-null SKOV3 ovarian cancer cell line (Figure S2C), supporting our validation of the primary ovarian cancer cells we had isolated. Notably, we observed a high co-infection rate (∼30%) of two adenoviruses in primary ovarian cancer cells (Figure S2D). Furthermore, the clinical utility of our activity-based assay can be extended to frozen samples. No statistical difference in HR activity scores was detected for primary ovarian cancer cells derived from fresh or frozen tumor tissue or ascites (Figure S2E).
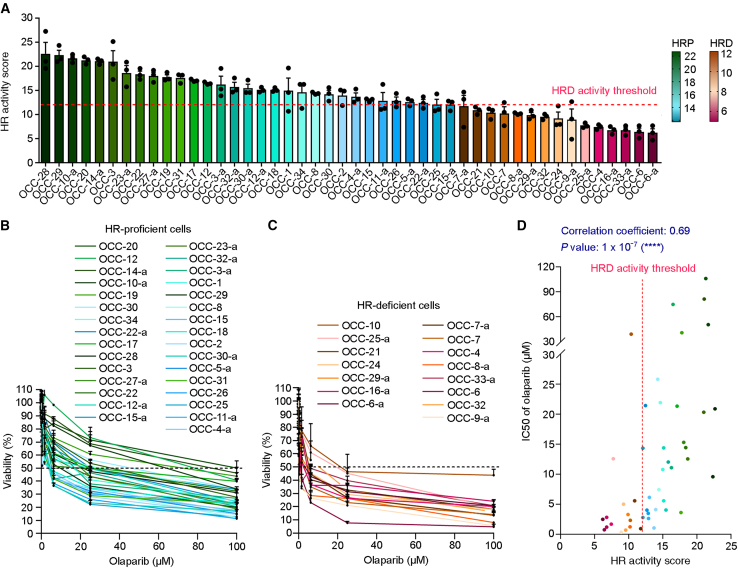
Importantly, our activity-based assay successfully determined the HR status of 46 primary cancer cells obtained from 34 patients with ovarian cancer. Among these, 25 primary cancer cells were isolated from tumor tissue, and the remaining 21 were obtained from ascites (Figure 2A). Given that the mean values for HR activity scores of BRCA1/2-deficient colorectal, triple-negative breast, and ovarian carcinoma cells were ≤12 (10, 8, and 12 for Figures 1B, 1D, and 1F, respectively), we set a score of 12 to classify the cellular HR activity of primary ovarian cancer cells, with an HR activity score >12 reflecting HRP cells, and those ≤12 defined as being HRD (Figure 2A). To further ascertain the association between HR activity and PARPi sensitivity, we treated all primary ovarian cancer cells with olaparib to determine their drug sensitivity. Overall, many cells in the HRD group displayed greater sensitivity to olaparib compared with the HRP group; specifically, 80% (12/15) of HRD cells and only 35.5% (11/31) of HRP cells exhibited <50% viability at 6.25 μM olaparib (Figures 2B, 2C, S3A, and S3B). Next, we tested whether HR activity scores quantified by the functional assay directly correlate with PARPi sensitivity by the Spearman correlation test. As shown in Figures 2D and S3C, the result further supported a significant correlation between HR activity scores and PARPi sensitivity of primary ovarian cancer cells (correlation coefficient = 0.69 and p = 1 × 10−7).
Figure 2.
The activity-based assay assesses HR deficiency in primary ovarian cancer cells
(A) Primary ovarian cancer cells were arranged in order of HR activity scores, representing high to low HR activity. The cells with HR activity higher than the HR deficiency (HRD) threshold, an HR activity score of 12 (dashed line), were classified as the HR-proficient (HRP) group and represented by a color palette ranging from green to cyan. Cells having HR activity ≤12 were classified as the HRD group and represented by a color palette ranging from brown to red-violet. The heatmaps for color palettes and associated HR activity scores are indicated at right. The primary ovarian cancer cells derived from an individual patient’s tumor tissue have been labeled in the form OCC-X, with respective ascites being labeled as OCC-X-a.
(B and C) Primary ovarian cancer cells were treated with olaparib for 4 days, followed by measurement of the IC50 of olaparib by alamarBlue cell viability assay. The dashed line represents 50% viability.
(B) The survival curves for HRP cells.
(C) The survival curves for HRD cells.
(D) HR activity and PARPi sensitivity of cancer cells derived from tumor tissue or ascites were analyzed separately and then subjected to Spearman correlation test, assessing the relationship between two variables.
∗∗∗∗p < 0.0001. Data are the mean ± SEM from three independent experiments (n = 3 biological replicates).
See also Figures S2 and S3.
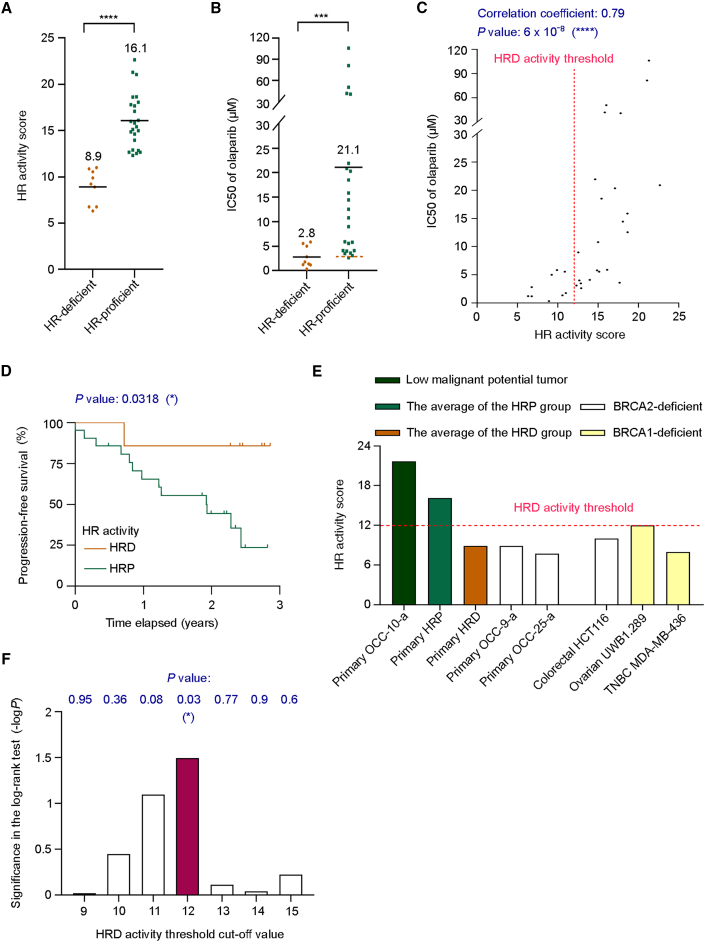
Intra-tumor heterogeneity in ovarian cancer is critical in determining clinical responses to treatments.43 To account for this heterogeneity, a previous RAD51 foci-based study reported that different biopsies from the same patient with ovarian cancer occasionally displayed varying HR statuses.44 Therefore, to alleviate the influence of tumor heterogeneity on determining HR status, it has been proposed that the HR score should be calculated as an average of multiple samples from patients.44 Thus, for patients whose tumor tissue and ascites were available simultaneously, we determined HR status by calculating the average activity score of primary cancer cells derived from tissue biopsies and ascites of the same patient. The mean HR activity of the HRP group was about 1.8-fold greater than that of the HRD group (16.1 vs. 8.9; Figure 3A). Notably, the difference in HR activity between the two groups was reflected in a 7.5-fold difference in half-maximal inhibitory concentration (IC50) value for olaparib (21.1 vs. 2.8 μM; Figure 3B). Furthermore, our results demonstrate that 96% of samples classified in the HRP group displayed higher IC50 values than the average IC50 value of the HRD group (Figure 3B). Notably, the Spearman correlation test revealed significant (p = 6 × 10−8) and strong correlation (correlation coefficient = 0.79)45 between the IC50 of olaparib and average HR activity scores (Figure 3C). Thus, these data support the notion arising from RAD51 foci-based assay,44 i.e., that the HR status of a patient should be interpreted as the average HR activity score from multiple biopsies. Altogether, our activity-based test can select a subset of cells susceptible to PARPis based on low HR activity scores.
Figure 3.
The HR activity of primary ovarian cancer cells strongly correlates with cell sensitivity to PARPis and clinical response
(A) Distributions of HR activity scores for HRP and HRD groups. The means of the two groups are shown.
(B) The difference in the means of IC50 of olaparib between the HRP and HRD groups. The brown dashed line represents the average IC50 value for the HRD group.
(C) A Spearman correlation test was used to measure the relationship between the HR activity score and PARPi sensitivity of primary ovarian cancer cells. For patients with multiple clinical samples, HR activity has been calculated as their average.
(D) Kaplan-Meier analysis for the progression-free survival in 28 patients with ovarian cancer. The brown line represents patients with HRD tumors, while the green line represents those with HRP tumors. All patients received platinum-based chemotherapy. Statistical analysis of survival benefits was performed by log-rank test. See also Table S1.
(E) Distribution of HR activity scores for primary ovarian cancer cells and BRCA-deficient cancer cell lines.
(F) The Kaplan-Meier analysis and log-rank test evaluated different HRD activity threshold cutoff values in analyzing progression-free survival benefits between the HRD and HRP groups.
Statistical significance is represented as the −log10 transformation of the p value in the log-rank test. Each data point shown in (A)–(C) is the mean from at least three independent experiments (n = 3, biological replicates); thirteen of them, which represent patients with multiple clinical samples, are the mean from six independent experiments (n = 6 biological replicates). ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Statistical analysis was performed by unpaired two-tailed Student’s t test in (A) and Mann-Whitney U test in (B).
See also Figure S3.
The HR activity status correlates with progression-free survival benefits
Next, to better assess the relationship between HR activity scores and the clinical response, the Kaplan-Meier estimate for 28 patients who received platinum-based chemotherapy was used for survival analysis. There was no difference in clinical and pathologic characteristics between patients with and without HRD (Table S1). Although our current study is limited in terms of patient sample size, the patients classified into the HRD group presented superior progression-free survival (PFS) to those in the HRP group (p = 0.0318; Figure 3D); in the HRD group, only one patient suffered disease progression compared with the 61.9% of patients in the HRP group that displayed tumor recurrence.
In the HR activity score range of 9–15, we observed some overlap for in vitro PARPi sensitivity between the HRD and HRP groups (Figures 3C and S3D). To thoroughly assess the accuracy of HR status within this range, we analyzed the clinical response in 14 patients whose tumors exhibited HR activity scores between 9 and 15 in the functional test. The Kaplan-Meier analysis and log-rank test yielded significant discrimination between HRD and HRP patients in this selected cohort (Figure S3E; p = 0.0051). Remarkably, approximately 89% of patients (8/9) in the HRP group experienced tumor progression, whereas none of the patients (0/5) in the HRD group did. These findings underscore the effectiveness of the activity-based functional test in identifying patients with a favorable clinical response to first-line treatment, even among primary cancer cells displaying similar in vitro PARPi sensitivity, as determined by short-term drug exposure.
To validate the HRD activity threshold (score of 12), we analyzed BRCA1/2 status in patients with available records. Remarkably, two patients exhibited BRCA2 pathogenic mutations (p.E1895X in OCC-9-a and p.L1227fs in OCC-25-a), and both primary ovarian tumor lines displayed an HR activity score lower than 12, aligning with the average of the HRD group and other BRCA-deficient cancer cell lines (Figure 3E). To further verify the reliability of our defined HRD activity threshold, we utilized a primary ovarian low malignant potential (LMP) tumor line, OCC-10-a, as an HRP control, representing an intermediate state between benign and malignant tumors. As expected, the functional HR activity score of the ovarian LMP tumor was 21.7, surpassing the HRD activity threshold we established (Figure 3E). Furthermore, to assess the performance of different HRD activity cutoff values ranging from 9 to 15, i.e., in the range where we had observed partial overlap in olaparib IC50 values between the HRD and HRP groups, we employed Kaplan-Meier analysis and the log-rank test to evaluate clinical response. The results (presented in Figure 3F) demonstrate that only the HRD cutoff value of 12 exhibited statistical significance in distinguishing the PFS of the HRD group from the HRP group (p = 0.03, represented as the −log10 transformation of the p value in the log-rank test). Taken together, these findings indicate that although some overlap in short-term drug response assays exists between HRD and HRP groups, patients with HRD, i.e., characterized by HR activity scores equal to or lower than 12 as determined by our functional test, displayed a longer survival benefit compared with those classified as HRP after long-term clinical follow-up.
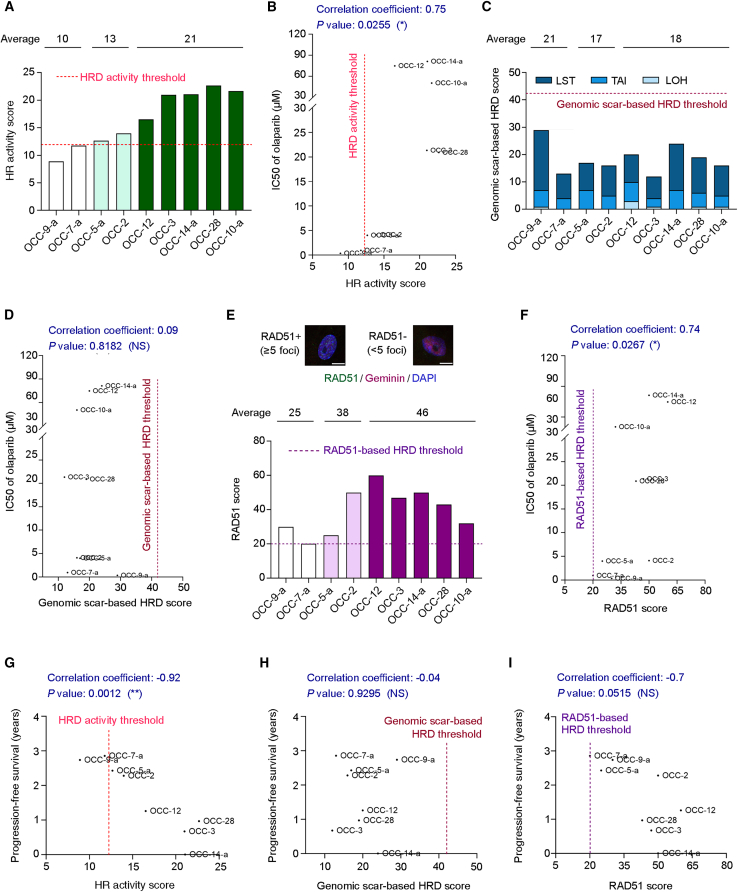
Next, we compared the activity-based functional test with other HRD detection approaches. Genomic sequencing-based HRD assays, including genomic-scar-based HRD scores22 and mutational signatures,46 place a lot of emphasis on training the algorithm to identify HRD and then verifying predictions according to BRCA status. However, the strength of the association between predicted results and cell sensitivity to PARPis needed to be clarified. Therefore, we examined if sequencing-based HRD detection is directly associated with olaparib sensitivity in primary ovarian cancer cells. We deployed three standard measures—LSTs,23 TAI,20 and LOH events24—to reflect genomic instability. Previous studies have used the unweighted sum of these measures as a genomic-scar-based HRD score and set a score of ≥42 as representing HRD25 to inform clinicians of possible HRD patients.21,25,26,27,28 To compare our activity-based functional assay with the genomic sequencing-based method for identifying HRD patients, nine primary ovarian cancer samples showing different cell sensitivities to PARPis (Figures 4A and 4B) were subjected to whole-exome sequencing (Figure 4C).
Figure 4.
HR activity scores correlate more strongly with PARPi sensitivity and clinical responses than the genomic-scar-based test and the RAD51 foci-based HRD assay
(A) The HR activity scores of nine primary ovarian cancer cell lines. The value of the HRD threshold for the activity-based functional test is 12.
(B) According to a Spearman correlation test, primary ovarian cancer cells can be classified as PARPi-susceptible cells (OCC-9-a and OCC-7-a; IC50 values: 0.35 and 0.94 μM, respectively), PARPi less sensitive cells (OCC-5-a and OCC-2; IC50 values: 4.01 and 4.11 μM, respectively), or PARPi-resistant cells (OCC-12, OCC-3, OCC-14-a, OCC-28, and OCC-10-a; IC50 values ranging from 20.9 to 81.3 μM). The correlation coefficient represents the strength of correlation between two variables.
(C) The genomic DNA of nine primary ovarian cancer cells mentioned in (A) was subjected to next-generation sequencing. Three indicators of HRD-associated genomic scar include large-scale transitions (LSTs), telomeric allelic imbalance (TAI), and loss of heterozygosity (LOH) events, which are summed and presented as genomic-scar-based HRD scores. The value of the HRD threshold for the genomic-scar-based analysis is 42.
(D) A Spearman correlation test was conducted to analyze the correlation between the genomic-scar-based HRD score and PARPi sensitivity of primary ovarian cancer cells.
(E) Primary ovarian cancer cells were co-stained for RAD51 and geminin, a marker of S/G2 cell phases, 2 h after 5 Gy radiation exposure. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The RAD51 scores, the percentage of geminin-positive cells with ≥5 RAD51 foci, of the nine primary ovarian cancer cells mentioned in (A) are shown. The value of the HRD threshold for the RAD51-based functional assay is 20. Scale bar, 10 μm. See also Figure S4.
(F) The relationship between the RAD51 scores and the PARPi sensitivity of primary ovarian cancer cells, as analyzed using the Spearman correlation test.
(G‒I) A Pearson correlation test was used to analyze the strength of the correlation between the duration of progression-free survival and (G) HR activity score, (H) genomic-scar-based HRD score, or (I) RAD51 score. A minus sign indicates an inverse relationship between two variables in the correlation test.
∗p < 0.05; ∗∗p < 0.01; NS, not significant (p > 0.05).
According to the PARPi sensitivity, nine primary ovarian cancer cells were classified into three groups (Figure 4B): PARPi-susceptible cells (white bars in Figure 4A; IC50 values: 0.35 and 0.94 μM), PARPi less-sensitive cells (light green bars in Figure 4A; IC50 values: 4.01 and 4.11 μM), and PARPi-resistant cells (dark green bars in Figure 4A; IC50 values ranged from 20.9 to 81.3 μM). Surprisingly, all nine samples could be categorized as HRP according to their sum of LST, TAI, and LOH being <42, i.e., the threshold defined in a previous study25; even two PARPi highly susceptible samples exhibited genomic-scar-based HRD scores lower than the threshold (Figure 4C; HRD scores: 29 and 13). Furthermore, although the PARPi-resistant group showed the highest average HR activity score among the three groups (Figure 4A; 21 vs. 10 and 13), it did not display the lowest average genomic-scar-based HRD score (Figures 4C and 4D; 18 vs. 21 and 17). Thus, the genomic-scar-based HRD scores presented a weaker relationship with cell sensitivity to PARPis (Figure 4D, correlation coefficient = 0.09, p = 0.8182) than did HR activity scores (Figure 4B, correlation coefficient = 0.75, p = 0.0255).
We further conducted a comparative analysis between the activity-based functional test and the RAD51 foci-based assay. The RAD51 foci-based assay serves as another functional HR method, enabling us to assess the initial step of RAD51-mediated DNA repair, which involves binding of recombinase RAD51 to damaged DNA. To perform the RAD51 foci-based assay, we utilized the same group of nine patients described above and employed two established protocols for patient-derived ovarian cancer samples, with or without ex vivo ionizing radiation (IR) exposure.44,47,48,49 In the absence of IR, detecting endogenous DNA DSBs is a critical requirement for the RAD51 assay. However, due to the low frequency of detectable endogenous DNA damage (as indicated by γH2AX signal) in primary ovarian cancer cells (Figure S4A), we adopted a protocol involving irradiating the cells with 5 Gy gamma radiation and then performing the RAD51 assay after a 2 h interval.47,50 This approach confirmed the presence of IR-induced DNA damage in most primary cells (Figure S4B). Additionally, given that the S and G2 phases of the cell cycle are when HR is most active, we co-stained the primary ovarian cancer cells for RAD51 and geminin (a marker of the S/G2 phases) to ensure that we detected RAD51 foci during the appropriate phases (Figure 4E), thereby adhering to the criteria established previously for both protocols.47,49
However, the main challenge in determining HR status using the RAD51 score, which represents the percentage of geminin-positive cells with ≥5 RAD51 foci, lies in the absence of a standardized HRD threshold. Various studies have defined HRD thresholds for ovarian tumor blocks using RAD51 scores of 6, 10, and 15 in the absence of IR48,49,51 and for fresh ovarian tumor tissue and primary ovarian cancer cells using a RAD51 score of 20 as the HRD cutoff value for the RAD51 test with IR.44,47 To classify the HR status of our primary ovarian cancer cells with IR, we used a RAD51 score of 20 (Figure 4E) as the HRD threshold, aligning with previous studies.44,47 In Figure S4C, we present the distribution of numbers of RAD51 foci per geminin-positive primary ovarian cancer cell. Among the nine samples, olaparib-sensitive OCC-7-a displayed a RAD51 score of 20 and was the only one classified as being an HRD sample (Figures 4E and 4F). However, although OCC-9-a was more susceptible to olaparib than OCC-7-a (IC50 values: 0.35 vs. 0.94 μM, respectively), it was classified as an HRP sample based on a RAD51 score of 30. This outcome may be explained by the fact that the RAD51 foci-based assay specifically assesses the initial step of RAD51-mediated DNA repair, which cannot reflect HRD cells defective in downstream steps. Nevertheless, given the correlation between RAD51 scores and cell sensitivity to PARPis, we found that the correlation coefficients are comparable between our activity-based functional test and the RAD51 foci assay (0.75 vs. 0.74, respectively; Figures 4B and 4F). Thus, both our activity-based test and the RAD51 assay, which are functionally based, appear to offer a more precise approach to identifying PARPi-sensitive cells than the genomic-scar-based approach, which exhibited a correlation coefficient of 0.09 (Figure 4D).
In addition to in vitro PARPi sensitivity, we analyzed the association between different HRD detection methods and clinical responses to first-line, platinum-based chemotherapy. Notably, a Pearson correlation test revealed a significantly strong inverse relationship between HR activity scores and the duration of PFS, with a correlation coefficient of −0.92 and a significant p value of 0.0012. Thus, the lower the HR activity score, the longer the PFS (Figure 4G). In contrast, we detected no significant link between genomic-scar-based HRD scores and patients’ clinical responses (Figures 4H, correlation coefficient = −0.04, p = 0.9295). Moreover, RAD51 scores presented a moderate correlation with patients’ PFS, with a correlation coefficient of −0.7 and a near significant p value of 0.0515 (Figure 4I). Accordingly, these findings imply that genomic-scar-based HRD scores may have a limited capacity to identify patients who may benefit from treatment targeting the DNA repair defect, in line with a previous phase 3 study.27 Although the comparative analysis is limited in sample size, these results support the clinical trial applicability of our activity-based functional test and reinforce the considerable utility of comparing it with other HRD detection approaches in a comprehensive prospective biomarker study.
Discussion
HRD cells display a striking sensitivity to PARPis that is 1,000 times greater than for HRP cells, implying a large therapeutic window (or index) both in vitro and in vivo.6 Importantly, PARPis have become the first clinically approved antitumor drug targeting DNA repair deficiency.52 Nevertheless, it has remained challenging to select HRD-positive patients beyond those that display germline or somatic BRCA1/2 mutations. Mutational signature profiling and genomic-scar-based detection methods have been applied to identify HRD cells. However, these detection methods analyze the accumulation of genomic instability over time, which may not accurately reflect HR status in real time, such as for cells experiencing HRD reversion mutation during tumorigenesis. Thus, an activity-based assay is necessary to serve as a real-time test. To meet this urgent need, we have developed an adenovirus- and activity-based functional test as a companion predictive assay (Figure 1A). Our results show that the HR activity scores detected by the functional test correlate with the BRCA status and PARPi sensitivity of colorectal, ovarian, and triple-negative breast cancers. Most importantly, we have evaluated our functional test against primary ovarian cancer cells derived from tumor tissue or ascites from patients, revealing a strong correlation between detected HR activity scores and cell sensitivity to the PARPi olaparib. Notably, we have established an activity threshold to identify HRD cancer cells susceptible to PARPi. In addition, PFS was longer in patients with HRD tumors characterized by the functional test than in those with HRP tumors. Therefore, these data support the capability of the functional test we have developed to select patients who will likely benefit from PARPi treatment.
It is worth noting that not only can our activity-based test identify HRD patients but also when they are HRD, given that HR activity of tumor cells can be revealed in real time by our approach. Moreover, unlike genetic tests that can only be applied to surgery-derived tumor tissue, we have successfully tested our activity-based assay on ovarian cancer ascites in liquid biopsies (N = 21). Thus, our activity-based functional assay can be applied in follow-up care for patients with cancer. For example, we tested one primary sample (OCC-5-a) derived from follow-up ascites fluid analysis after chemotherapy. Accordingly, our activity-based analysis can contribute to decision making for first-line HRD treatment and can determine whether PARPi is still a suitable therapy after a tumor has recurred. Although the PARPi sensitivity of primary cancer cells can be measured in vitro, our data show that the IC50 values of various cancer types differ significantly. We found that HRD colorectal and triple-negative breast cancer cells were highly sensitive to olaparib treatment, exhibiting IC50 values in the picomolar range (0.55 and 0.93 nM for Figures 1C and 1E, respectively). However, the IC50 value of HRD ovarian cancer cells ranged from nanomolar to micromolar: 0.08 μM in ovarian cancer cell line UWB1.289 (Figure 1G) and an average of 2.8 μM in primary ovarian cancer cells (Figure 3B). These outcomes indicate that it may be challenging to establish a standard reference to determine PARPi sensitivity among various cancer types. In contrast, the HRD activity threshold we identified herein can be applied to at least three cancer types to select cancer cells showing the highest PARPi sensitivity. Thus, we propose that an HRD activity threshold may be more applicable to tackling diverse cancer types than identifying a standard PARPi sensitivity.
Apart from ovarian cancers, malignant ascites is associated with pancreatic, liver, and colon cancers.53 Moreover, fluid biopsies of lung or breast cancer also encompass malignant pleural effusion.54 Our activity-based assay may be used together with liquid biopsy to evaluate HRD patients with these types of cancer.
Limitations of the study
Our functional-based assay may not encompass all factors influencing PARPi sensitivity, as HR activity alone may not be the sole determinant. Previous studies have demonstrated that PARPi sensitivity can also be observed in HRP cells under specific conditions.55,56,57 For instance, such cells can exhibit sensitivity to PARPi when they are deficient in ribonuclease H2-initiated ribonucleotide excision repair and fork protection.55,56,57 Another limitation of this study is that the primary ovarian cancer cells were not 100% of tumor origin, as the efficiency of isolating tumor cells from the clinical samples was ∼93%. Therefore, we cannot rule out the possibility that a few remaining non-tumor cells were analyzed together with primary cancer cells in our analyses. Additionally, the number of patients included in our clinical analysis is limited. The functional assay has been applied to 34 patients with ovarian cancers. Except for 9 patients who received concordant genomic-scar-based, HR-activity-based, and RAD51-based HRD tests, Kaplan-Meier analysis was performed to estimate the PFS benefit in 28 patients who received platinum-based chemotherapy. As shown in Figure 3D, only one patient with an HRD tumor suffered disease progression, whereas 61.9% of patients with HRP tumors displayed disease progression, indicating a better trend for PFS in patients with HRD tumors than those with HRP tumors, as classified by our activity-based functional test (p = 0.0318). Thus, we advocate for initiating extensive clinical trials on ovarian and other HRD-associated cancers to refine the HR activity threshold and comprehensively compare predictive powers among the activity-based functional test, the genomic-scar-based approach, and the RAD51 foci-based assay. Such endeavors hold great promise for optimizing HRD assessment and advancing personalized cancer therapies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-RAD51 (clone H-92) | Santa Cruz Biotechnology | Cat# sc-8349; RRID: AB_2253533 |
| anti-γH2AX (clone 20E3) | Cell Signaling Technology | Cat# 9718; RRID: AB_2118009 |
| anti-geminin (clone 1A8) | Abcam | Cat# ab104306; RRID: AB_10889692 |
| anti-α tubulin | GeneTex | Cat# GTX112141; RRID: AB_10722892 |
| anti-p53 | GeneTex | Cat# GTX102965; RRID: AB_1952339 |
| anti-GAPDH | GeneTex | Cat# GTX100118; RRID: AB_1080976 |
| anti-pan cytokeratin (clone AE1/AE3)-Alexa Fluor 488 conjugated | Invitrogen | Cat# 53-9003-82; RRID: AB_1834350 |
| anti-PAX8 | Proteintech | Cat# 10336-1-AP; RRID: AB_2236705 |
| Goat anti-rabbit IgG (HRP) | GeneTex | Cat# GTX213110–01; RRID: AB_10618573 |
| Goat anti-rabbit IgG-DyLight 488 conjugated | Thermo Fisher Scientific | Cat# 35553; RRID: AB_1965947 |
| Goat anti-mouse IgG-Alexa Fluor 647 conjugated | Thermo Fisher Scientific | Cat# A-21236; RRID: AB_2535805 |
| Bacterial and virus strains | ||
| AdenoX-pCMV-I-SceI-mCherry | This paper | N/A |
| AdenoX-pCMV-DR-GFP-pPGK-ECFP | This paper | N/A |
| Biological samples | ||
| Primary ovarian cancer cells derived from tumor tissue | This paper | N/A |
| Primary ovarian cancer cells derived from ascites | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Olaparib | Selleckchem | Cat# S1060 |
| B02 | Cayman Chemical | Cat# 22133 |
| Critical commercial assays | ||
| Adeno-X Adenoviral System 3 | Clontech | Cat# 6322666 |
| Adeno-X Maxi Purification Kit | Clontech | Cat# 631533 |
| alamarBlue Cell Viability Reagent | Thermo Fisher Scientific | Cat# DAL1025 |
| Tumor Dissociation Kit | Miltenyi Biotech | Cat# 130-095-929 |
| Ammonium Chloride Solution | STEMCELL Technologies | Cat# 07850 |
| Tumor Cell Isolation kit | Miltenyi Biotech | Cat# 130-108-339 |
| QIAamp DNA Mini kit | QIAGEN | Cat# 51304 |
| Deposited data | ||
| whole-exome sequencing datasets for OCC-2 | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX18246139 |
| whole-exome sequencing datasets for OCC-3 | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742482 |
| whole-exome sequencing datasets for OCC-5-a | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742483 |
| whole-exome sequencing datasets for OCC-7-a | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742484 |
| whole-exome sequencing datasets for OCC-9-a | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742485 |
| whole-exome sequencing datasets for OCC-10-a | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742477 |
| whole-exome sequencing datasets for OCC-12 | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742478 |
| whole-exome sequencing datasets for OCC-14-a | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX16742479 |
| whole-exome sequencing dataset for OCC-28 | This paper | https://www.ncbi.nlm.nih.gov/sra/SRX17907740 |
| Experimental models: Cell lines | ||
| HeLa cells | ATCC | Cat# CCL-2; RRID: CVCL_0030 |
| U2OS cells | ATCC | Cat# HTB-96 |
| Hs578T cells | ATCC | Cat# HTB-126 |
| MDA-MB-23 cells | ATCC | Cat# CRM-HTB-26 |
| MDA-MB-436 cells | ATCC | Cat# HTB-130 |
| SKOV3 cells | ATCC | Cat# HTB-77 |
| TOV-21G cells | ATCC | Cat# CRL-3577 |
| UWB1.289 cells | ATCC | Cat# CRL-2945 |
| OVCA429 cells | Obtained from Ruby Yun-Ju Huang (National Taiwan University, Taiwan) | N/A |
| RMG1 cells | Obtained from Wendy W. Hwang-Verslues (Academia Sinica, Taiwan) | N/A |
| HCT116 cells | ECACC | Cat# 91091005 |
| HCT116 BRCA2−/− cells | ECACC | Cat# 16071901 |
| AdenoX-293 cells | Clontech | Cat# 632271 |
| Oligonucleotides | ||
| siRNAs targeting human RAD51 | GE Dharmacon | Cat# J-003530-10-0020 |
| siRNA non-targeting pool control | GE Dharmacon | Cat# D-001810-10-20 |
| Recombinant DNA | ||
| pCMV6-AC-GFP | OriGene | Cat# PS100010 |
| pAdenoX-PRLS | Clontech | Cat# 632260 |
| pAdenoX-pCMV-GFP | This paper | N/A |
| pAdenoX-pCMV-I-SceI-mCherry | This paper | N/A |
| pAdenoX-pCMV-DR-GFP-pPGK-ECFP | This paper | N/A |
| Software and algorithms | ||
| Burrows-Wheeler Aligner v0.5.9 | https://bio-bwa.sourceforge.net/ | https://bio-bwa.sourceforge.net/ |
| SAM tools v0.1.18 | https://samtools.sourceforge.net/ | https://samtools.sourceforge.net/ |
| Picard v1.54 | https://github.com/broadinstitute/picard | https://github.com/broadinstitute/picard |
| Genome Analysis Toolkit v4 | https://gatk.broadinstitute.org/hc/en-us | https://gatk.broadinstitute.org/hc/en-us |
| GraphPad Prism v7 | https://www.graphpad.com/ | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Peter Chi (peterhchi@ntu.edu.tw).
Materials availability
All reagents generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Experimental model and subject details
Patients
This study was approved by the Institutional Review Board of National Taiwan University Hospital and complied with its ethical regulations (IRB: 200706002R). To evaluate the clinical applicability of our activity-based functional assay, we enrolled 34 female patients aged 29–74 years with high-grade serous ovarian carcinomas, low-grade serous ovarian carcinomas, ovarian clear cell carcinoma, ovarian endometrioid carcinoma, mucinous ovarian carcinoma, ovarian carcinosarcoma, ovarian fibrothecoma, or ovarian serous tumor of low malignant potential (borderline tumor). All of the patients provided signed informed consent and agreed to publication of the research results. Regarding deposition of the whole-exome sequencing dataset in the NCBI Sequence Read Archive (SRA) database, de-identification was conducted to comply with IRB regulations and to preserve patients' privacy.
Cell culture
HeLa, U2OS, Hs578T, OVCA429, and MDA-MB-436 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Biological Industries). MDA-MB-231, SKOV3, and TOV-21G cells were cultured in DMEM/Nutrient Mixture F-12 (DMEM/F-12, Gibco) supplemented with 10% FBS. The RMG1 cell line was cultured in F-12 (Gibco) supplemented with 10% FBS. HCT116 paired cells were cultured in McCoy’s 5A medium (Gibco) supplemented with 10% FBS. UWB1.289 cells were cultured in 48.5% RPMI 1640 medium (Gibco), 48.5% Mammary Epithelial Growth Medium (Lonza), and 3% FBS. All cells were cultured at 37°C and 5% CO2. All cell lines tested negative for mycoplasma contamination using PlasmoTest (InvivoGen) and were authenticated using short tandem repeat profiling.
Method details
Plasmids
The CMV enhancer, CMV promoter, TurboGFP flanked by two SwaI cutting sites, and hGH polyA signal (pCMV6-AC-GFP; OriGene) were all cloned into the linear double-stranded DNA (dsDNA) pAdenoX-PRLS (Clontech), forming a circular dsDNA plasmid named pAdenoX-pCMV-GFP. The adenovirus vector for downstream construction was prepared by cutting pAdenoX-pCMV-GFP with SwaI to release the pCMV-GFP fragment. To construct plasmids encoding mCherry-tagged I-SceI (pAdenoX-pCMV-I-SceI-mCherry) and the cyan fluorescent protein (ECFP)-tagged HR reporter (pAdenoX-pCMV-DR-GFP-pPGK-ECFP), a HiFi DNA assembly kit (NEB) was used to assemble the following fragments: (1) the CMV promoter, I-SceI, mCherry, and transcription terminator were assembled with the adenovirus vector; or (2) the CMV promoter, DR-GFP cassette, PGK promoter, ECFP, and terminator were cloned into the adenovirus vector.
Adenovirus production
AdenoX-293 cells (Adeno-X Adenoviral System 3; Clontech) were seeded in 60-mm dishes to 70% confluency at transfection with the recombinant PacI-digested pAdeno-X DNA using a CalPhos mammalian transfection kit (Clontech). One week later, cells were harvested and prepared for primary virus stock according to the manufacturer’s instructions (Clontech) when the cytopathic effect (CPE) had occurred. Then, AdenoX-293 cells were infected by the primary virus stock to amplify the secondary virus stock. Three days later, a secondary virus stock of pAdenoX-pCMV-I-SceI-mCherry was harvested. For pAdenoX-pCMV-DR-GFP-pPGK-ECFP, secondary virus stock was prepared eight days after transduction. Then, AdenoX-293 cells were seeded in 100-mm dishes to 70% confluency at transduction with secondary virus stock. Two days later, high-titer virus stocks were prepared and purified according to the manufacturer’s instructions (Adeno-X Maxi Purification Kit; Clontech). The adenoviruses used in this study are available from the corresponding author upon request.
Adenovirus-based HR reporter assay
Cells were seeded in 6-well plates to 60% confluency at transduction with recombinant adenoviruses at a multiplicity of infection (MOI) of 10–100. Cells were then harvested and analyzed using an LSRFortessa flow cytometer (BD Biosciences) three to four days later. A fluorescence filter for BV510 (525/50) was used to detect ECFP signal. Other bandpass filters employed were 610/20 for mCherry and 530/30 for EGFP. First, the fluorescence threshold for ECFP-negative and EGFP-negative cells was established by analyzing cells infected with pAdenoX-pCMV-I-SceI-mCherry alone. Then, the threshold for mCherry-negative cells was determined by analyzing cells infected with pAdenoX-pCMV-DR-GFP-pPGK-ECFP alone. Finally, cells co-infected with the two different viruses were gated for ECFP-positive and mCherry-positive profiles, representing cells successfully infected simultaneously by the two viruses. The proportion of EGFP-positive cells was then determined to represent HR activity.
siRNA transfection
For gene silencing, siRNA sequences were CUAAUCAGGUGGUAGCUCAUU (targeting human RAD51; GE Dharmacon), UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA (non-targeting pool control; GE Dharmacon). Briefly, cells were seeded in 60-mm dishes to 50% confluency on the day of transfection. Then, siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions for 24 h before infection with adenoviruses.
Clonogenic survival assay
Cell lines and their seeding number per well in 6-well plates were: HCT116 WT (2.5 × 102), HCT116 BRCA2−/− (1.5 × 104), Hs578T (2.5 × 102), MDA-MB-231 (2.5 × 102), MDA-MB-436 (4 × 104), SKOV3 (2 × 103), RMG1 (1.2 × 104), OVCA429 (8 × 102), and UWB1.289 (2 × 103). The next day, cells were treated with the indicated concentration of olaparib for 12 days. Fresh medium containing olaparib was replaced every 4 days. Cell survival was determined by fixing the cells with ice-cold methanol:acetone (1:1) fixative solution for 5 min on ice. The cells were then washed with PBS and stained with 0.5% w/v crystal violet in distilled water containing 25% v/v methanol at room temperature for 3 h before washing again with PBS and air-drying. Cells were imaged before adding 360 μL de-staining solution (15% v/v methanol and acetic acid each in distilled water) into each well and incubating for 20 min. Finally, 120 μL of the solution was loaded into a 96-well plate for optical density measurement at 570 nm.
Cell viability assay
Primary ovarian cancer cells were seeded in 96-well plates at a density ranging from 500–2000 cells per well. The next day, cells were treated with the indicated amount of olaparib for 96 h. Cell viability was determined using alamarBlue cell viability reagent (ThermoFisher). Data collection and analyses were performed by different laboratory members to achieve single-blind analyses.
Immunoblotting analysis
Cells were washed with PBS and incubated in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 10% glycerol, and 1 mM EDTA) containing 10 μg mL−1 aprotinin, 10 μg mL−1 chymostatin, 10 μg mL−1 leupeptin, 10 μg mL−1 pepstatin A, 1.5 mM PMSF, freshly prepared 2.5 mM sodium pyrophosphate, and 1 mM β-glycerolphosphate on ice for 20 min, and then subjected to sonication. The lysate was clarified by centrifugation, and protein concentration was determined using a BCA Protein Assay kit (Pierce). After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), proteins were transferred onto polyvinylidene fluoride (PVDF) membrane. The membrane was treated with 5% bovine serum albumin (BSA) in PBS containing 0.1% Tween 20 (PBST) for 1 h, and then incubated at 4°C overnight with the primary antibodies including anti-RAD51 (clone H-92, Santa Cruz, 1:1000 dilution), anti-α tubulin (GeneTex, 1:10,000 dilution), anti-p53 (GeneTex, 1:5000 dilution), and anti-GAPDH (GeneTex, 1:10,000 dilution). After washing three times with PBST, the membrane was incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (GeneTex, 1:5000 dilution) at room temperature for 1 h. Then, the membrane was washed three times again with PBST before being incubated with enhanced chemiluminescent horseradish peroxidase substrate (ThermoFisher) for 5 min. Immunoblot images were acquired using a BioSpectrum imaging system (UVP).
Immunofluorescence and RAD51-based HRD assay
Primary ovarian cancer cells were seeded in 24-well plates at a density of 5 × 102 cells per well for staining of pan-CK (Invitrogen, 1:100 dilution) or PAX8 (Proteintech, 1:100 dilution). The next day, cells were fixed with 4% paraformaldehyde at 25°C for 10 min and permeabilized in PBS containing 0.3% Triton X-100 for 10 min. After being washed twice with PBS, the cells were incubated in blocking buffer (5% BSA, 0.1% NP-40 in PBS) at room temperature for 30 min and then incubated with primary antibodies at 4°C overnight. Next, the cells were washed with PBS containing 0.1% NP-40 and then incubated with DyLight 488-conjugated goat anti-rabbit IgG antibody (ThermoFisher, 1:400 dilution) at 37°C for 1 h. Cell nuclei were revealed by staining with 1 μg mL−1 DAPI (4′,6-diamidino-2-phenylindole) at 25°C for 2 min.
To perform RAD51-based HRD assay, primary ovarian cancer cells were seeded in 12-well plates at a density of 1.1 × 104 cells per well before being exposed to 5 Gy ionizing radiation (XCELL140, Kubtec). After 2 h of recovery from irradiation, cells were fixed with 4% paraformaldehyde at 25°C for 10 min and permeabilized in 0.5% Triton X-100 for 10 min. Next, samples were washed and blocked with 5% BSA as described above. Then, cells were stained with anti-γH2AX (clone 20E3, Cell Signaling, 1:400 dilution) or anti-RAD51 (clone H-92, Santa Cruz, 1:200 dilution) antibody and co-stained with anti-geminin (clone 1A8, Abcam, 1:100 dilution) antibody at 4°C overnight. After washing three times with PBS containing 0.1% NP-40, the cells were incubated with DyLight 488-conjugated goat anti-rabbit IgG (ThermoFisher, 1:400 dilution) and Alexa Fluor 647-conjugated goat anti-mouse IgG (ThermoFisher; 1:300 dilution) antibodies at 37°C for 1 h. Finally, cells were counterstained with DAPI at 25°C for 2 min.
Immunofluorescence images were acquired using a TCS SP5 confocal microscope (Leica Microsystems). For the RAD51-based assay, at least 100 geminin-positive cells were analyzed per sample. To ensure sufficient DNA damage had been induced by the ionizing radiation, the samples were only eligible for assessment of RAD51 score when we detected >25% geminin-positive cells with 2 or more γH2AX foci. Next, the RAD51 scores were determined by quantifying the percentage of geminin-positive cells with 5 or more RAD51 foci. To achieve single-blind analysis, data collection and analyses were performed by different laboratory members.
Tumor tissue processing
Ovarian tumor tissues from ovarian cancer patients were placed into sterile 15-mL conical tubes containing ice-cold DMEM/F-12 (Gibco) supplemented with 10% FBS (Biological Industries), 100 units/mL penicillin (Gibco), 100 μg/mL streptomycin (Gibco), 2.5 μg/mL amphotericin B, and 50 μg/mL gentamicin sulfate. All samples underwent tissue processing within 20 min of collection. Tumor tissues were washed with phosphate-buffered saline (PBS) and cut using sterile fine-tip forceps and a scalpel blade into smaller pieces of approximately 2 mm3. The specimens were washed with PBS and digested with a tumor dissociation kit (Miltenyi Biotech). First, specimens were incubated in 4.7 mL RPMI 1640 medium (Gibco), 200 μL Enzyme H (Miltenyi Biotech), 100 μL Enzyme R (Miltenyi Biotech), 25 μL Enzyme A (Miltenyi Biotech), and 10 μM Y-27632 (inhibitor of Rho-associated, coiled-coil containing protein kinase, StemCell) in 6-well tissue culture plates for 90 min at 37°C. The plates were gently swirled every 30 min. After incubation, the digested tissue suspension was applied to a 70 μm cell strainer (Corning) placed on a 50 mL tube with a syringe plunger, and then washed with 20 mL RPMI 1640 medium. The cell filtrate was centrifuged at 300 × g for 5 min at room temperature. The cell pellet was resuspended in Ovarian Carcinoma Modified Ince (OCMI) culture medium,58 which induces primary ovarian cancer cells to phenocopy original ovarian tumors. OCMI medium comprises equal volumes of DMEM/F-12 and M199 medium (Gibco) supplemented with 2 mM glutamine (Gibco), 20 μg/mL insulin, 10 μg/mL transferrin, 0.2 pg/mL triiodothyronine, 5 μg/mL o-phosphoryl ethanolamine, 8 ng/mL selenous acid, 25 ng/mL all-trans retinoic acid, 500 ng/mL hydrocortisone, 25 ng/mL cholera toxin (all from Sigma-Aldrich), 10 ng/mL epidermal growth factor (PeproTech), 5 μg/mL linoleic acid (Cayman Chemicals), 100 nM 17β-Estradiol (Cayman Chemicals), 5% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, 2.5 μg/mL amphotericin B, and 50 μg/mL gentamicin sulfate. Cells were then cultured in 6-well tissue culture plates. To test the clinical applicability of our activity-based assay for frozen samples, tumor tissues from the same patients were divided into two portions; one was processed as described above (defined as ‘freshly processed clinical samples’), and the other was cut into smaller pieces and resuspended in OCMI medium containing 10% dimethyl sulfoxide (DMSO) in cryovials. These latter samples were placed in an isopropanol chamber and stored at −80°C overnight, before being transferred to liquid nitrogen. After thawing these frozen samples, they underwent the same tissue processing steps as described above for freshly processed clinical samples.
Ascites fluid processing
Ascites fluid from ovarian cancer patients was collected into sterile 50-mL conical tubes and centrifuged at 300 × g for 5 min at room temperature. The cell pellet was treated with red blood cell lysis buffer (ammonium chloride solution, StemCell) for 10 min at 4°C, before being centrifuged at 300 × g for 5 min at room temperature. The cell pellet was resuspended in PBS and applied to a 70-μm cell strainer placed on a 50 mL tube with a syringe plunger, followed by washing with PBS. The cell filtrate was centrifuged at 300 × g for 5 min at room temperature, and then the cell pellet was resuspended in OCMI medium and cultured in 6-well tissue culture plates or in OCMI medium containing 10% DMSO to freeze cells.
Tumor cell isolation and primary cell culture
A Tumor Cell Isolation kit (Miltenyi Biotech) was used to isolate tumor cells from a heterogeneous sample by means of a negative selection strategy. In brief, when cells reached 70% confluency they were trypsinized with 0.05% w/v trypsin/EDTA for 3 min at 37°C. After adding Ovarian Carcinoma Modified Ince (OCMI) medium58 and centrifugation at 300 × g for 5 min at room temperature, the cell pellet was washed with PBS and centrifuged again. The cell pellet containing up to 1×107 total cells was resuspended in 60 μL buffer I (0.5% bovine serum albumin in PBS), 20 μL non-tumor cell depletion cocktail A, and 20 μL cocktail B (Miltenyi Biotech). Cells were incubated for 15 min at 4°C, before adding 400 μL buffer I. A MACS LS column and MACS separator were used to magnetically separate target tumor cells. Cells were cultured in OCMI medium at 37°C and 5% CO2.
Genomic scar-based HRD test
Genomic DNA was purified from 1 × 106 cultured cells using a QIAamp DNA Mini kit (QIAGEN) according to the manufacturer’s instructions. The quality of purified genomic DNA was evaluated using a NanoDrop system, a Qubit dsDNA HS Assay Kit (Invitrogen), a Fragment Analyzer with DNF-930 Reagent kit (Agilent), and a Fragment Analyzer with DNF-464 HS Large Fragment kit (Agilent). The DNA libraries were then generated with Truseq DNA Library Prep kits (Illumina, USA), according to the manufacturer’s manual. The target gene library was then generated with NimblGen whole-exome capture kits (Roche NimblGen Inc.). The samples were sequenced using an Illumina NovaSeq system with paired-end reads of 300 nucleotides.
We used an analytical algorithm based on our previously published protocol.59 In brief, the raw sequencing data were aligned with a reference human genome (Dec. 2013, GRCh38) using Burrows-Wheeler Aligner software (version 0.5.9). SAM tools (version 0.1.18) and Picard (version 1.54) were used for data conversion, sorting, and indexing. Genome Analysis Toolkit (GATK; version 4) was employed for variant calling using Mutect2 parameters. After variant calling, ANNOVAR was used to annotate the genetic variants. ClinVar, dbSNP (version 150), Exome Sequencing Project 6500 (ESP6500), 1000 Genomes Variant Dataset (2014Sep), ExAC, and genomeAD databases were used for filtering common variants of sequencing results. Pathogenic/likely pathogenic variants were defined according to the interpretation guideline of the American College of Medical Genomics and Genetics.60
HRD was considered according to the two following conditions: (1) when a pathogenic/likely pathogenic variant of BRCA1 or BRCA2 was detected, those tumor cells were characterized as HRD; or (2) we used scarHRD software to calculate the HRD score (sum of LST, TAI, and LOH) and when it was ≥42, we defined those cells as being HRD.22
To achieve single-blind analysis, the samples were only identified by a number for downstream analysis.
Quantification and statistical analysis
GraphPad Prism 7 (GraphPad Software) was used to perform all statistical tests and analyze statistical significance. Data are presented as scatterplots with bars (means ± s.e.m). The normality of data was tested by the Shapiro-Wilk and the D’Agostino-Pearson test. Similar sample variance among multiple groups was first analyzed by the Brown-Forsythe test, followed by one-way ANOVA with Tukey’s post hoc test to compare the difference in means of multiple groups. For comparison between two groups, an F test and unpaired two-tailed Student’s t test were carried out to confirm the similar sample variance and the statistical difference between the means of the two groups, respectively. In the absence of a normal distribution, an unpaired two-tailed nonparametric Mann-Whitney U-test was performed to compare the medians of two groups. The strength of correlation between two variables was calculated using Pearson correlation test or Spearman correlation test (nonparametric data). Progression-free survival was analyzed with Kaplan-Meier analysis and the log rank test. Statistical significance was defined as p < 0.05.
Acknowledgments
We are grateful to the patients involved in this study. We would like to acknowledge the service provided by the Flow Cytometric Analyzing and Sorting Core of the First Core Laboratory at National Taiwan University College of Medicine, Core Facility of the Institute of Biochemical Sciences at National Taiwan University, and by the Laboratory Animal Center at Taipei Medical University for technical support. The graphical abstract was created with BioRender.com. The icons shown in Figure S2A were made using Freepik, Witdhawaty, and wanicon from www.flaticon.com. We thank Ching-Ying Kuo (National Taiwan University) for her comments on the manuscript. We appreciate Yen-Ju Chen’s effort to illustrate this study with diagrams. An illustration of MACS untouched cell isolation was derived from Miltenyi Biotec. This work was supported by Academia Sinica, National Taiwan University, and the National Science and Technology Council (MOST 111-2326-B-002-019).
Author contributions
C.-Y.L. and P.C. conceived the study. C.-Y.L., W.-F.C., P.-H.L., Y.-L.C., and P.C. designed the experiments. C.-Y.L., S.-H.H., K.-Y.C., and P.-H.L. analyzed primary ovarian cancer cells derived from tumor tissue and ascites. C.-Y.L., S.-H.H., K.-H.L., M.-Y.K., and K.-Y.C. performed cell-based experiments. C.-Y.L. provided statistical analysis. C.-Y.L. and P.C. wrote the paper. All authors discussed the results and contributed to the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101247.
Supplemental information
Data and code availability
-
•
De-identified whole-exome sequencing datasets produced in this study have been deposited at NCBI Sequence Read Archive (SRA) repository. They are publicly available as of the date of publication. DOIs are listed in the Key resources table.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Turner N., Tutt A., Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 2.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/S1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 3.Moynahan M.E., Pierce A.J., Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/S1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 4.King M.-C., Marks J.H., Mandell J.B., New York Breast Cancer Study Group Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 5.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H., McCabe N., Lord C.J., Tutt A.N.J., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Moore K., Colombo N., Scambia G., Kim B.-G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 8.González-Martín A., Pothuri B., Vergote I., DePont Christensen R., Graybill W., Mirza M.R., McCormick C., Lorusso D., Hoskins P., Freyer G., et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 9.Litton J.K., Rugo H.S., Ettl J., Hurvitz S.A., Gonçalves A., Lee K.-H., Fehrenbacher L., Yerushalmi R., Mina L.A., Martin M., et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018;379:753–763. doi: 10.1056/nejmoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golan T., Hammel P., Reni M., Van Cutsem E., Macarulla T., Hall M.J., Park J.-O., Hochhauser D., Arnold D., Oh D.-Y., et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/nejmoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bono J., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K.N., Sartor O., Agarwal N., Olmos D., et al. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 2020;382:2091–2102. doi: 10.1056/nejmoa1911440. [DOI] [PubMed] [Google Scholar]
- 12.Futreal P.A., Liu Q., Shattuck-Eidens D., Cochran C., Harshman K., Tavtigian S., Bennett L.M., Haugen-Strano A., Swensen J., Miki Y., et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 13.Lancaster J.M., Wooster R., Mangion J., Phelan C.M., Cochran C., Gumbs C., Seal S., Barfoot R., Collins N., Bignell G., et al. BRCA2 mutations in primary breast and ovarian cancers. Nat. Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]
- 14.Pal T., Permuth-Wey J., Betts J.A., Krischer J.P., Fiorica J., Arango H., LaPolla J., Hoffman M., Martino M.A., Wakeley K., et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104:2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 15.The Cancer Genome Atlas Research Network. Berchuck A., Birrer M., Chien J., Cramer D.W., Dao F., Dhir R., DiSaia P., Gabra H., Glenn P., et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abeshouse A., Ahn J., Akbani R., Ally A., Amin S., Andry C., Annala M., Aprikian A., Armenia J., Arora A., et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singhi A.D., George B., Greenbowe J.R., Chung J., Suh J., Maitra A., Klempner S.J., Hendifar A., Milind J.M., Golan T., et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019;156:2242–2253.e4. doi: 10.1053/j.gastro.2019.02.037. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Network. Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F., Fulton L.L., Dooling D.J., Ding L., Mardis E.R., et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manié E., Popova T., Battistella A., Tarabeux J., Caux-Moncoutier V., Golmard L., Smith N.K., Mueller C.R., Mariani O., Sigal-Zafrani B., et al. Genomic hallmarks of homologous recombination deficiency in invasive breast carcinomas. Int. J. Cancer. 2016;138:891–900. doi: 10.1002/ijc.29829. [DOI] [PubMed] [Google Scholar]
- 20.Birkbak N.J., Wang Z.C., Kim J.-Y., Eklund A.C., Li Q., Tian R., Bowman-Colin C., Li Y., Greene-Colozzi A., Iglehart J.D., et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins J.A., Irshad S., Grigoriadis A., Tutt A.N.J. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211. doi: 10.1186/2Fbcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sztupinszki Z., Diossy M., Krzystanek M., Reiniger L., Csabai I., Favero F., Birkbak N.J., Eklund A.C., Syed A., Szallasi Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. npj Breast Cancer. 2018;4:16. doi: 10.1038/s41523-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popova T., Manié E., Rieunier G., Caux-Moncoutier V., Tirapo C., Dubois T., Delattre O., Sigal-Zafrani B., Bollet M., Longy M., et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 24.Abkevich V., Timms K.M., Hennessy B.T., Potter J., Carey M.S., Meyer L.A., Smith-McCune K., Broaddus R., Lu K.H., Chen J., et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telli M.L., Timms K.M., Reid J., Hennessy B., Mills G.B., Jensen K.C., Szallasi Z., Barry W.T., Winer E.P., Tung N.M., et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller R.E., Leary A., Scott C.L., Serra V., Lord C.J., Bowtell D., Chang D.K., Garsed D.W., Jonkers J., Ledermann J.A., et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020;31:1606–1622. doi: 10.1016/j.annonc.2020.08.2102. [DOI] [PubMed] [Google Scholar]
- 27.Tutt A., Tovey H., Cheang M.C.U., Kernaghan S., Kilburn L., Gazinska P., Owen J., Abraham J., Barrett S., Barrett-Lee P., et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat. Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgson D., Lai Z., Dearden S., Barrett J.C., Harrington E.A., Timms K., Lanchbury J., Wu W., Allen A., Senkus E., et al. Analysis of mutation status and homologous recombination deficiency in tumors of patients with germline BRCA1 or BRCA2 mutations and metastatic breast cancer: OlympiAD Ann. Oncol. 2021;32:1582–1589. doi: 10.1016/j.annonc.2021.08.2154. [DOI] [PubMed] [Google Scholar]
- 29.Vergote I., González-Martín A., Ray-Coquard I., Harter P., Colombo N., Pujol P., Lorusso D., Mirza M.R., Brasiuniene B., Madry R., et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann. Oncol. 2022;33:276–287. doi: 10.1016/j.annonc.2021.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Meijer T.G., Verkaik N.S., Sieuwerts A.M., van Riet J., Naipal K.A.T., van Deurzen C.H.M., den Bakker M.A., Sleddens H.F.B.M., Dubbink H.-J., den Toom T.D., et al. Functional ex vivo assay reveals homologous recombination deficiency in breast cancer beyond BRCA gene defects. Clin. Cancer Res. 2018;24:6277–6287. doi: 10.1158/1078-0432.CCR-18-0063. [DOI] [PubMed] [Google Scholar]
- 31.Llop-Guevara A., Loibl S., Villacampa G., Vladimirova V., Schneeweiss A., Karn T., Zahm D.M., Herencia-Ropero A., Jank P., van Mackelenbergh M., et al. Association of RAD51 with homologous recombination deficiency (HRD) and clinical outcomes in untreated triple-negative breast cancer (TNBC): analysis of the GeparSixto randomized clinical trial. Ann. Oncol. 2021;32:1590–1596. doi: 10.1016/j.annonc.2021.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Modesti M., Budzowska M., Baldeyron C., Demmers J.A.A., Ghirlando R., Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 33.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/2Fgad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gunn A., Stark J.M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 35.Lee C.-Y., Su G.-C., Huang W.-Y., Ko M.-Y., Yeh H.-Y., Chang G.-D., Lin S.-J., Chi P. Promotion of homology-directed DNA repair by polyamines. Nat. Commun. 2019;10:65. doi: 10.1038/s41467-018-08011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng G., Chun-Jen Lin C., Mo W., Dai H., Park Y.-Y., Kim S.M., Peng Y., Mo Q., Siwko S., Hu R., et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat. Commun. 2014;5:3361. doi: 10.1038/ncomms4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X., et al. Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan L., Han J., Liu T., Dong S., Xie F., Chen H., Huang J. Scaffolding protein SPIDR/KIAA0146 connects the Bloom syndrome helicase with homologous recombination repair. Proc. Natl. Acad. Sci. USA. 2013;110:10646–10651. doi: 10.1073/pnas.1220921110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F., Mazina O.M., Zentner I.J., Cocklin S., Mazin A.V. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J. Med. Chem. 2012;55:3011–3020. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- 40.Giannini G., Ristori E., Cerignoli F., Rinaldi C., Zani M., Viel A., Ottini L., Crescenzi M., Martinotti S., Bignami M., et al. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248–254. doi: 10.1093/embo-reports/kvf044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen Q., Scorah J., Phear G., Rodgers G., Rodgers S., Meuth M. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol. Biol. Cell. 2008;19:1693–1705. doi: 10.1091/mbc.e07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oplustilova L., Wolanin K., Mistrik M., Korinkova G., Simkova D., Bouchal J., Lenobel R., Bartkova J., Lau A., O’Connor M.J., et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle. 2012;11:3837–3850. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz R.F., Ng C.K.Y., Cooke S.L., Newman S., Temple J., Piskorz A.M., Gale D., Sayal K., Murtaza M., Baldwin P.J., et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumiati M., Hietanen S., Hynninen J., Pietilä E., Färkkilä A., Kaipio K., Roering P., Huhtinen K., Alkodsi A., Li Y., et al. A functional homologous recombination assay predicts primary chemotherapy response and long-term survival in ovarian cancer patients. Clin. Cancer Res. 2018;24:4482–4493. doi: 10.1158/1078-0432.CCR-17-3770. [DOI] [PubMed] [Google Scholar]
- 45.Mukaka M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012;24:69–71. https://pubmed.ncbi.nlm.nih.gov/23638278 [PMC free article] [PubMed] [Google Scholar]
- 46.Davies H., Glodzik D., Morganella S., Yates L.R., Staaf J., Zou X., Ramakrishna M., Martin S., Boyault S., Sieuwerts A.M., et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Wijk L.M., Vermeulen S., Meijers M., van Diest M.F., ter Haar N.T., de Jonge M.M., Solleveld-Westerink N., van Wezel T., van Gent D.C., Kroep J.R., et al. The RECAP test rapidly and reliably identifies homologous recombination-deficient ovarian carcinomas. Cancers. 2020;12 doi: 10.3390/cancers12102805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Wijk L.M., Kramer C.J.H., Vermeulen S., ter Haar N.T., de Jonge M.M., Kroep J.R., de Kroon C.D., Gaarenstroom K.N., Vrieling H., Bosse T., Vreeswijk M.P.G. The RAD51-FFPE test; Calibration of a functional homologous recombination deficiency test on diagnostic endometrial and ovarian tumor blocks. Cancers. 2021;13 doi: 10.3390/cancers13122994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Compadre A.J., van Biljon L.N., Valentine M.C., Llop-Guevara A., Graham E., Fashemi B., Herencia-Ropero A., Kotnik E.N., Cooper I., Harrington S.P., et al. RAD51 foci as a biomarker predictive of platinum chemotherapy response in ovarian cancer. Clin. Cancer Res. 2023;29:2466–2479. doi: 10.1158/1078-0432.ccr-22-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naipal K.A.T., Verkaik N.S., Ameziane N., van Deurzen C.H.M., ter Brugge P., Meijers M., Sieuwerts A.M., Martens J.W., O'Connor M.J., Vrieling H., et al. Functional ex vivo assay to select homologous recombination–deficient breast tumors for PARP inhibitor treatment. Clin. Cancer Res. 2014;20:4816–4826. doi: 10.1158/1078-0432.ccr-14-0571. [DOI] [PubMed] [Google Scholar]
- 51.Pellegrino B., Herencia-Ropero A., Llop-Guevara A., Pedretti F., Moles-Fernández A., Viaplana C., Villacampa G., Guzmán M., Rodríguez O., Grueso J., et al. Preclinical in vivo validation of the RAD51 test for identification of homologous recombination-deficient tumors and patient stratification. Cancer Res. 2022;82:1646–1657. doi: 10.1158/0008-5472.can-21-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lord C.J., Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta A., Sedhom R., Beg M.S. Ascites, or fluid in the belly, in patients with cancer. JAMA Oncol. 2020;6:308. doi: 10.1001/jamaoncol.2019.5409. [DOI] [PubMed] [Google Scholar]
- 54.West H.J. Malignant pleural effusions. JAMA Oncol. 2015;1:260. doi: 10.1001/jamaoncol.2015.0671. [DOI] [PubMed] [Google Scholar]
- 55.Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., et al. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmermann M., Murina O., Reijns M.A.M., Agathanggelou A., Challis R., Tarnauskaitė Ž., Muir M., Fluteau A., Aregger M., McEwan A., et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 2018;559:285–289. doi: 10.1038/2Fs41586-018-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cong K., Peng M., Kousholt A.N., Lee W.T.C., Lee S., Nayak S., Krais J., VanderVere-Carozza P.S., Pawelczak K.S., Calvo J., et al. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell. 2021;81 doi: 10.1016/j.molcel.2021.06.011. 3227–3144.e3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ince T.A., Sousa A.D., Jones M.A., Harrell J.C., Agoston E.S., Krohn M., Selfors L.M., Liu W., Chen K., Yong M., et al. Characterization of twenty-five ovarian tumour cell lines that phenocopy primary tumours. Nat. Commun. 2015;6:7419. doi: 10.1038/ncomms8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin P.-H., Chen M., Tsai L.-W., Lo C., Yen T.-C., Huang T.Y., Chen C.-K., Fan S.-C., Kuo S.-H., Huang C.-S. Using next-generation sequencing to redefine BRCAness in triple-negative breast cancer. Cancer Sci. 2020;111:1375–1384. doi: 10.1111/cas.14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
De-identified whole-exome sequencing datasets produced in this study have been deposited at NCBI Sequence Read Archive (SRA) repository. They are publicly available as of the date of publication. DOIs are listed in the Key resources table.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.