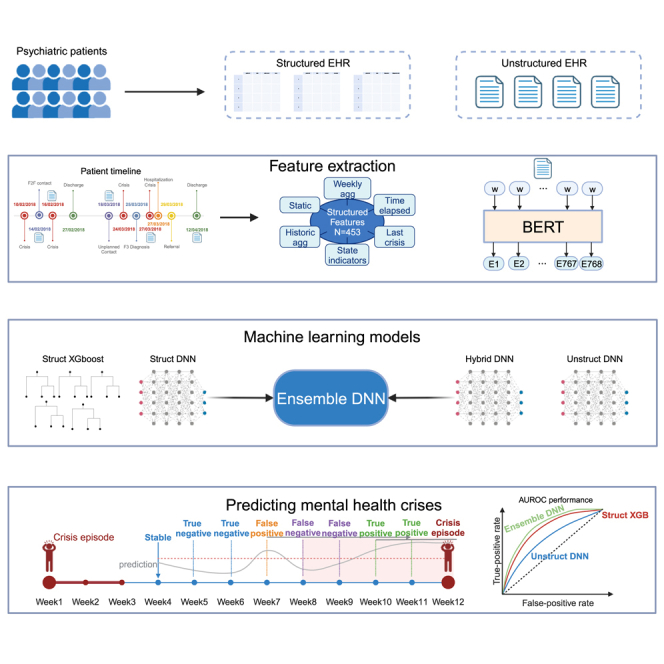
Summary
An automatic prediction of mental health crises can improve caseload prioritization and enable preventative interventions, improving patient outcomes and reducing costs. We combine structured electronic health records (EHRs) with clinical notes from 59,750 de-identified patients to predict the risk of mental health crisis relapse within the next 28 days. The results suggest that an ensemble machine learning model that relies on structured EHRs and clinical notes when available, and relying solely on structured data when the notes are unavailable, offers superior performance over models trained with either of the two data streams alone. Furthermore, the study provides key takeaways related to the required amount of clinical notes to add value in predictive analytics. This study sheds light on the untapped potential of clinical notes in the prediction of mental health crises and highlights the importance of choosing an appropriate machine learning method to combine structured and unstructured EHRs.
Keywords: natural language processing, mental health, electronic health records, machine learning, predictive analytics, AI
Graphical abstract
Highlights
-
•
Machine learning applied to clinical notes is able to predict mental health crises
-
•
An ensemble leveraging structured and unstructured EHRs improves model performance
-
•
The method used to combine both data sources is key to enhance predictive power
-
•
A minimum of 10% of weeks with notes is required to extract optimal value
Garriga et al. demonstrate the potential of clinical notes to predict mental health crises. An ensemble of machine learning models that effectively combines unstructured and structured electronic health records offers superior performance over single data source models. A sufficient volume of clinical notes is required to enhance predictive power.
Introduction
Mental disorders represent one of the leading causes of disease burden and disability worldwide.1 This situation is unlikely to change (at least not positively) in the foreseeable future because the demand for mental health services have been steadily increasing2,3 while the resources have long been limited.4,5 A considerable demand for mental healthcare resources is attributed to mental health crises, defined as situations where patients are unable to function effectively in the community or when there is a risk of hurting themselves or others.6 Such crisis episodes may include emotional or psychotic breakdowns, substance abuse, and suicide attempts, and they often require emergency care and hospitalization. Research has shown that intervening during early stages of a crisis can mitigate the risk of escalation or even prevent the crisis.7,8 However, the lack of crisis prediction tools combined with the fact that patients are usually attended to through emergency pathways at the peak of a crisis make healthcare systems ill equipped to anticipate demand and to enable preventative interventions.
The advances in machine learning, computational power, and data collection promised a better understanding of a variety of disorders, improvements in early detection, and better long-term management and outcomes—the approach frequently referred to as precision medicine.9 A particularly promising direction is to use electronic health records (EHRs) and predictive algorithms to inform clinical decision-making. Leveraging EHRs to predict important mental health events and to better deploy healthcare resources has been mainly limited to prediction of suicide attempts,10,11,12,13,14,15 self-harm, or the first episode of psychosis,16,17,18 which constitute only a small fraction of mental health demand. More recently, Garriga et al.19 demonstrated the feasibility to continuously predict a full breadth of mental health crises, and importantly, the authors showed the added value of such predictions in clinical practice. Although this pioneering study provided a proof of concept for computational analysis of structured EHRs, the predictive power of clinical notes has remained unexplored. For chronic disorders, clinical notes are particularly relevant for practical implementation of predictive analysis because the clinical notes typically dominate over structured data.20
EHRs have become the norm for collecting and storing records of patients’ medical history21 in structured (i.e., discrete variables, such as demographics, diagnoses, hospitalization events, etc.) and unstructured (i.e., narrative text, such as clinical notes, discharge reports, etc.) forms. Whereas structured data are easier to process, they do not provide a complete clinical context of the patient.21 Research has shown that complementing structured with unstructured data can improve cohort identification22 as well as the prediction accuracy of hospital readmission23,24,25 and suicide attempts.26,27 Furthermore, unlike in cardiovascular disorders, where objective measurements of blood pressure or electrocardiogram signals can be stored in structured EHRs, mental health assessments are highly subjective, and a wealth of information about patients’ mental health status is stored as observations in clinical notes. For this reason, understanding the untapped potential of unstructured data in predicting critical events in mental healthcare becomes essential to fully leverage the breadth and depth of information available in EHRs. However, the reality of clinical practice presents a considerable challenge; namely, the inconsistency of the availability of clinical notes across different patients. The volume of these notes is typically related to the severity and frequency of the patient’s mental health crises, resulting in a greater accumulation of notes for patients with more severe or recurrent episodes. This variability highlights not only the necessity to determine the minimum quantity of unstructured data that contribute to the accurate prediction of mental health crises but also to explore the development of models that remain effective across a spectrum of patient records, regardless of the volume of available clinical notes. This exploration is of significant interest to predictive analytics because it will elucidate the practicability of using clinical notes across diverse clinical scenarios, not just those featuring more severe or frequent mental health crises.
In this study, we analyzed anonymized unstructured and structured data from 59,750 de-identified patients collected over 8 years. Building on our previous study,19 in this work we develop an algorithm to predict mental health crisis within the next 28 days following weekly algorithm prediction and extend the state of the art in three key ways. First, we explore the predictive power of unstructured data alone, and we compare it with structured data. Second, with the objective to improve the performance of the prediction model, we develop and compare different methods for combining structured and unstructured data. Last, given that healthcare systems provide no strict requirements for the collection of unstructured data, we investigate the minimal availability of unstructured data that brings an added value to the prediction model.
Results
Cohort description
This study relied on a dataset containing structured and unstructured data from 59,750 de-identified patients. Structured data refers to information stored in a database with a predefined format and range of values. Unstructured data refers to the clinical notes captured by the hospital staff in a narrative format during interactions with patients or their caregivers. The dataset included a total of 2,709,626 crisis events that occurred from September 2012 until July 2020. These crisis events correspond to a total of 110,978 crisis episodes (that is, an average of 336 crisis episodes per week). Approximately 99% of the patients in our dataset had at least one written note, and over 81% had two notes or more. On average, the unstructured data yielded one note for each patient at an interval of every 10 weeks, with the average note consisting of around 110 words. The number of notes per patient, number of notes per patient-week, and the average number of weeks between notes per patient follow long-tailed distributions (Figure S1). The demographics as well as other patient characteristics are summarized in Table 1.
Table 1.
Demographics and patient characteristics over the training set together with the validation set and the test set
| Patient group | Number of patients | Number of crisis episodes |
|
|---|---|---|---|
| Training and validation | Test | ||
| All patients | 59,750 | 93,809 | 17,644 |
| Age (%) | |||
| <18 | 481 (0.8) | 3,437 (3.7) | 586 (3.3) |
| ≥18 and <34 | 22,000 (36.8) | 37,447 (39.9) | 7,268 (41.1) |
| ≥ 34 and <65 | 29,555 (49.5) | 45,124 (48.1) | 8,545 (48.4) |
| ≥65 | 7,714 (12.9) | 7,864 (8.4) | 1,262 (7.1) |
| Sex (%) | |||
| Female | 29,714 (49.7) | 46,221 (49.2) | 8,644 (48.9) |
| Male | 30,001 (50.2) | 47,588 (50.7) | 9,000 (51.0) |
| Ethnic group (%) | |||
| White | 38,677 (64.7) | 62,480 (66.6) | 11,418 (64.6) |
| Black | 4,173 (7.0) | 7,375 (7.9) | 1,304 (7.4) |
| Asian | 8,221 (13.8) | 13,245 (14.1) | 2,408 (13.6) |
| Mixed | 1,645 (2.8) | 3,128 (3.3) | 620 (3.5) |
| Not known | 1,657 (2.8) | 4,740 (5.0) | 1,251 (7.1) |
| Other | 4,916 (8.2) | 2,205 (2.3) | 477 (2.7) |
| Marital status (%) | |||
| Married | 7,015 (11.70) | 11,063 (11.8) | 1,616 (9.1) |
| Cohabit | 1,258 (2.1) | 2,133 (2.3) | 376 (2.1) |
| Single | 20,617 (34.5) | 42,162 (44.9) | 6,153 (34.8) |
| Divorced, separated, or widowed | 4,276 (7.2) | 7,686 (8.0) | 1,017 (6.0) |
| Unknown or not disclosed | 22,729 (38.0) | 24,421 (26.0) | 7,290 (41.0) |
| (ICD-10) Diagnosed disorder type (%) | |||
| (F0) Organic symptomatic mental disorders | 1,769 (3.0) | 2,156 (2.3) | 231 (1.3) |
| (F1) Substance use | 1,364 (2.3) | 4,336 (4.6) | 596 (3.4) |
| (F2) Schizophrenia schizotypal and delusional | 5,733 (9.6) | 13,547 (14.4) | 1,895 (10.7) |
| (F3) Mood affective disorders | 7,000 (11.7) | 13,547 (14.4) | 2,166 (12.3) |
| (F4) Neurotic stress related and somatoform | 3,454 (5.8) | 6,187 (6.6) | 1,084 (6.1) |
| (F6) Adult personality and behavior | 2,448 (4.1) | 8,616 (9.2) | 1,355 (7.7) |
| Other diagnosis | 1,461 (2.4) | 2,988 (3.2) | 498 (2.8) |
| No diagnosis | 36,521 (61.1) | 42,745 (45.5) | 9,837 (55.7) |
Prediction of mental health crises with structured and unstructured data
In total, 450 features were computed from the structured data along three broad categories; namely, static features (such as demographic information), variables that described the latest interaction with the hospital, and variables that quantified the time elapsed since a specific type of event (e.g., since the crisis episode). The complete list of features is shown in Table S1. From the unstructured data (i.e., clinical notes written by healthcare practitioners), we computed semantic features over 768 dimensions using a BERT model.28 The full process of computing the structured data features and the semantic features is detailed in the STAR Methods.
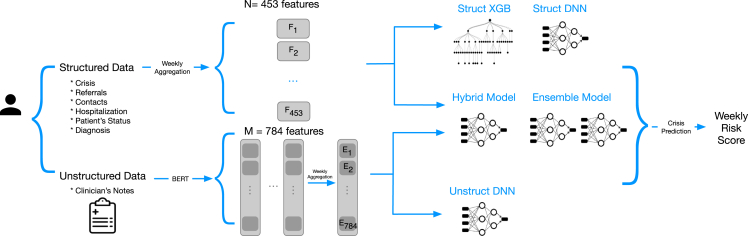
Relying on these features, we trained four models to predict the risk of relapse within the next 28 days as a binary classification problem (relapse versus no relapse) based on structured data only (Struct XGB and Struct DNN), unstructured data only (Unstruct DNN), and both data types (Hybrid DNN). Additionally, we created an ensemble model that uses the predictions from a version of the Hybrid DNN model when there were unstructured data available and the predictions from the Struct DNN model otherwise (Ensemble DNN) (Figures 1 and S2). Since relapses occur infrequently, with a prevalence of 1.3%, the models were tuned to maximize the area under the precision recall curve (AUPRC)29 on the validation set, which is a preferred metric to evaluate the performance of binary classification tasks with an unbalanced distribution.30 The model that performed the best using only structured data was an XGBoost model, a tree-based classifier that implements gradient boosting.31 For the dataset with only unstructured data and the dataset that combined structured and unstructured data, the best performing model was a feedforward deep neural network (DNN). We defined two baseline models: the first one (LogReg5) as a 5-factor logistic regression model inspired by the important variables suggested by the literature32 (see Table S1 for the list of features used) and the second one (OneYearTotalCrisis) as a heuristic model that ranks patients based on total number of crises experienced during the past year (last 53 weeks).
Figure 1.
Diagram of the five trained models and the data types used as input
Struct XGB is an XGBoost model, and the rest are feedforward neural networks, with Ensemble DNN combining the results of a neural network trained on structured data only and a neural network trained on structured and unstructured data.
The best-performing model overall was Ensemble DNN, which achieved a mean AUPRC of 0.133 and a mean area under the receiving operator curve (AUROC) of 0.865 (see Table 2 for the complete set of results). This model performed significantly (p < 0.001) better than the two baseline models with AUPRC of 0.064, AUROC of 0.772 for the LogReg5 and AUPRC of 0.040, AUROC of 0.729 for OneYearTotalCrisis and the other proposed models (namely, Unstruct DNN, Struct DNN, Hybrid DNN, and Struct XGB). While the Hybrid DNN model, which makes use of structured and unstructured data, demonstrated less remarkable performance than the Struct XGB, it would be incorrect to conclude that semantic features add no value. It is vital to consider that the Hybrid DNN is configured to rely on semantic features, even in the absence of unstructured data, which can negatively affect its performance. On the other hand, the Ensemble DNN adopts a flexible approach; it applies the Hybrid DNN model when unstructured data are present and reverts to the Struct DNN model when it is not. This adaptability enables it to draw additional insight from semantic features when available, resulting in higher overall performance. This superior predictive power of the Ensemble DNN in comparison with the other models is validated by the net reclassification improvement analysis. The Ensemble DNN achieved a value exceeding 0.160 compared with the remaining models (refer to Table S2). Despite minor variations, the predictive power of the Ensemble DNN model remained consistent across different gender and ethnic groups (Tables S6 and S7).
Table 2.
Performance of each model in terms of mean AUPRC and AUROC
| Mean AUPRC (SD) | Mean AUROC (SD) | |
|---|---|---|
| Baseline 1 (LogReg 5) | 0.064 (0.006) | 0.772 (0.010) |
| Baseline 2 (OneYearTotalCrisis) | 0.040 (0.004) | 0.729 (0.011) |
| Unstruct DNN | 0.080 (0.019) | 0.809 (0.077) |
| Struct DNN | 0.110 (0.010) | 0.823 (0.008) |
| Hybrid DNN | 0.129 (0.010) | 0.823 (0.008) |
| Struct XGB | 0.130 (0.012) | 0.831 (0.008) |
| Ensemble DNN | 0.133 (0.013) | 0.865 (0.011) |
SD, standard deviation.
Clinical notes available for at least 10% of weeks improve the prediction accuracy
We constructed six cohorts based on the percentage of weeks with available unstructured data, from patients with less than 10% of weeks with notes to patients with at least 50% of weeks with available notes, in 10% splits. Overall, 69.3% of patients had up to 10% of weeks with at least one note, while 3.6% of patients had 50% or more weeks with a note. The number of crisis episodes as well as the modeling target prevalence (the percentage of observations with a relapse of mental health crises) was higher for patients with a higher percentage of notes. This was expected, given that a considerable portion of data records come from hospital visits, which are more frequent for more severe patients (see Table S3 for more details).
We explored the prediction of patients’ 4-week risk of relapse using structured and unstructured EHRs in subgroups of patients selected according to the percentage of weeks with at least one clinical note recorded. We trained three models based on the different data inputs (structured only, unstructured only, and both) for each of the patient subgroups. We compared the model performance across input data types in each subgroup with AUPRC and AUROC.
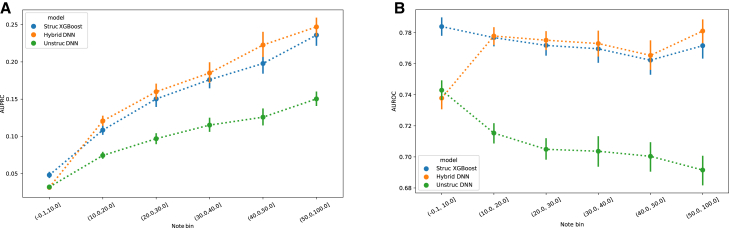
For the three evaluated models, AUPRC increases with the percentage of available notes (Figure 2A). This mainly stems from the fact that patients with a higher number of weeks with available notes have a higher prevalence of relapse (Table S3). The AUROC, on the other hand, shows a different trend (Figure 2B). The performance of the Unstruct DNN model decreases with the percentage of available notes, whereas the Hybrid DNN shows a considerable increase in performance for the patients with more than 20% of weeks with available notes, followed by an approximately steady performance until the point of having 50% or more available notes. Finally, the Struct XGB model shows a pattern similar to the Hybrid DNN, except for the category of patients with fewer than 10% of notes, where it outperforms the other models. The decreasing AUROC of the Unstruct DNN model as the percentage of available notes increases may initially appear counterintuitive. This highlights the complexity of predictive modeling in the context of mental health crises. Even though the quantity of available data per patient increases with the rise in the percentage of notes, the total number of patients conversely decreases, thereby affecting the performance of the neural network. This can be attributed to the nature of neural networks based on textual input, which tend to perform less effectively when the number of training instances is low.
Figure 2.
Clinical notes available for at least 10% of weeks improve the model’s performance
(A) AUPRC of the structured only model (Struct XGBoost), unstructured only model (Unstruct DNN), and structured and unstructured (combined) model (Hybrid DNN). Points and lines indicate mean and ± standard deviation values computed in the 52 weeks of the test set.
(B) AUROC of Struct XGBoost, Unstruct DNN, and Hybrid DNN. Points and lines indicate mean and ± standard deviation values computed in the 52 weeks of the test set.
The Struct XGBoost model is statistically significantly better (p < 0.001) than the other two models in both metrics for the group of patients with less than 10% of weeks with available notes. In contrast, the Hybrid DNN is statistically significantly better (p < 0.001) than the other two models in both metrics for the group of patients with more than 50% of the weeks with available notes. For patients who had between 10% and 50% of weeks with available notes, the Hybrid DNN model shows statistically significantly better (p < 0.01) AUPRC than Struct XGB, but the difference in AUROC is not statistically significant (p values ranging from 0.04–0.34) (see Tables S4 and S5 for detailed results).
Predictors and model interpretation
We extracted the SHAP (Shapley additive explanation) from the records from the dataset containing the full list of patients to (1) understand the predictive power of different types of data, (2) interpret the model predictions on the test data, and (3) infer the most important variables that impacted the algorithm. To understand how the amount of available notes impacts the model and the most predictive features, we analyzed the SHAP values for unstructured and structured data in different cohorts of patients with different ranges of the available clinical notes. SHAP values are based on the Shapley value from coalitional game theory and describe the relationship between features and the model output.33 Specifically, SHAP values provide a comprehensive overview of how each feature and the range of their values impact the prediction output. They account for feature interactions and allow instance-specific explanations as well as cohort-level exploration.
Predictors: Combined structured and unstructured data-related features
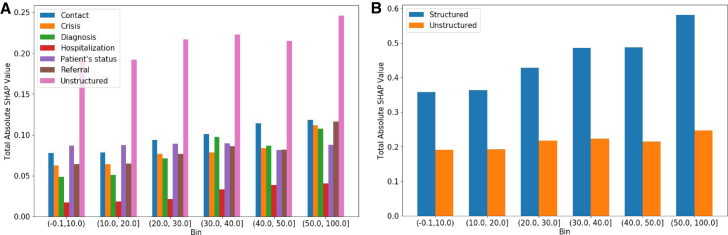
We present the total absolute SHAP values across different subgroups based on the note availability (Figure 3B). The structured data features are broken down into different categories reflecting the type of the data used for the features extraction (Table S1). Figure 3A presents the total absolute SHAP values across structured and unstructured categories. The total per category was computed as the sum of all SHAP values for each individual feature belonging to the corresponding category. As the percentage of notes increases, so do the total absolute SHAP values (Figures 3A and 3B). Moreover, the total SHAP values across all categories also increase, except in case of the patient status-related features, likely because of the dominance of event-based features. The category “unstructured” has the highest total SHAP; however, when grouping together all structured data categories, the structured data related features prevail in the SHAP values (Figure 3B). Finally, we did not observe significant changes across the top predictive features, where the structured features dominated the top 20 predictive features across all datasets. One reason behind the dominance of the structured data-related features (in the top 20 predictive features) can be related to the number of features available in each category; namely, the total number of unstructured features is almost two times higher than the number of the structured data-related features. We also observed that the cumulative total SHAP values of the unstructured data-related features dominates each bin. This suggests that each unstructured data-related feature does not independently carry a high predictive power (in the top 20), but the unstructured data-related features do so when combined, and they even surpass the total absolute SHAP of the single categories of the structured data features.
Figure 3.
Overall contribution of the different data categories per subgroup of patients based on the percentage of weeks with unstructured data
(A) The total absolute SHAP values for the Hybrid DNN extracted on the test set across the different datasets obtained based on the percentage of notes available from the patients.
(B) The total absolute SHAP values for the Hybrid DNN for structured and unstructured feature categories extracted on the test set across the different datasets obtained based on the percentage of notes available from the patients.
Predictors: Structured data features
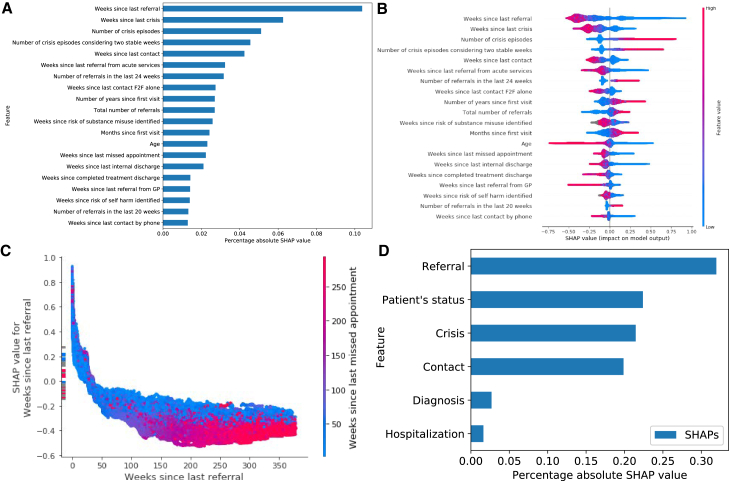
We use the Struct XGB model to analyze the SHAP values. We refrain from unpacking the other models because the unstructured data-related features cannot be directly interpreted because of their design (STAR Methods). In Figure 4, we provide the most important predictors of the Struct XGB model trained on the data from all patients by extracting the SHAP values from the test data. Exploration of different feature categories suggests that features extracted from the records related to referrals had the highest impact on the model’s predictions (Figure 4D). The top predictors (Figures 4A and 4B) also highlight the predictive power of this feature category; most features belong to the features related to referrals (namely, 6 of 20 features). We further explored the impact of the most predictive feature “weeks since last referral,” combined with the feature “weeks since last missed appointment,” which has the highest interaction effect with. We observe that the referral has a positive effect on the predicted risk score (PRS) during the first weeks and that its effect diminishes in time. The dependence plot in Figure 4C suggests that the feature decreases its effect on the PRS as the number of weeks since the last missed appointment increases. Similar trends were observed from the dependence plots of “weeks since last crisis.” The SHAP values in the bar summary depicted in Figure 4B provide a more comprehensive view of the top overall predictors and indicate features that contribute positively or negatively to the PRS depending on their value. Some features have a clear threshold of such separation (e.g., the lower the age, the higher the positive effect on the model risk prediction and vice versa).
Figure 4.
Most influential predictors derived from structured data
(A) The most impactful features on prediction based on the absolute SHAP values (ranked from the most to the least important).
(B) The distribution of the impact of each feature on the model output. The colors reflect the numerical value of the features: red represents larger values, while blue represents smaller values. The line is made of individual dots representing each crisis, and the thickness of the line is determined by the number of examples at a given value (for example, most patients have a low number of severe crises). A positive SHAP value (extending to the right) indicates an increased probability of a crisis prediction; symmetrically, a negative SHAP value (extending to the left) indicates a reduced probability.
(C) Shows the dependence plot of the top predictive feature in terms of SHAP values. The plot represents a scatterplot that shows the effect a single feature has on the predictions made by the model, where the x axis is the value of the feature, the y axis is the SHAP value for that feature, and the color corresponds to a second feature that may have an interaction effect with the feature we are plotting. The longer the time since the last referral and the longer the time since the last missed appointment, the lower the probability that the model will predict a crisis.
(D) The most impactful categories of features based on the total absolute SHAP values per category.
We delve deeper into the top 3 feature categories: “referrals,” “patient status,” and “crisis.” First, the analysis of the “referrals”-related features shows a positive effect on the model’s predictions with the decrease in the values of “weeks since last referral,” “weeks since last referral from acute services,” and “weeks since last referral from GP.” The opposite can be observed for “total number of referrals”; as the number of referrals increases, its positive effect on the prediction increases as well. Second, when it comes to the “patient’s status” category, positive effect on the predictions occurs as the age of the patient decreases. “Weeks since risk of substance misuse identified” is unique in not contributing positively to the predicted risk (highlighted in gray in Figure 4B); rather, it contributes negatively to the model’s predictions. “Weeks since risk of self harm identified” follows a similar pattern. Last, exploration of the “crisis” category indicates a positive effect on the model’s predictions with the decrease of “weeks since last crisis.” In contrast, the increase in “number of crisis episodes” and “number of crisis episodes considering two stable weeks” is associated with a positive impact on the model’s predictions.
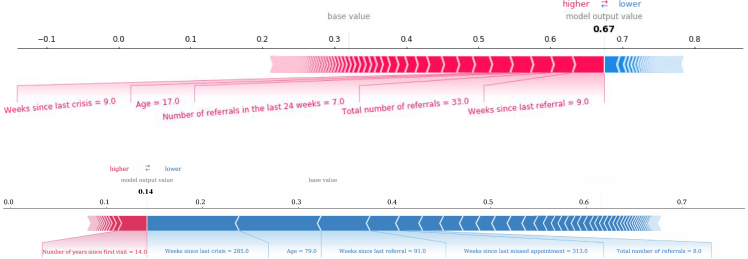
As an illustration of prediction interpretations at the individual level, we generated force plots for two selected crisis prediction cases (Figure 5); an example of a positive crisis prediction is shown in Figure 5A, whereas Figure 5B shows a negative crisis prediction example. The former example shows how the combination of “total number of referrals” with a value of 33 and “weeks since last crisis” with a value of 9 has a positive influence on the PRS, resulting in a positive prediction of 0.67. In the latter example, the feature “weeks since last crisis” with a value of 285 and a total number of referrals of 8 shows a negative effect on the model’s predictions, resulting in a negative prediction of 0.14. This example illustrates the complexity of the model and how the influence of each feature on the PRS varies depending on its value.
Figure 5.
Composition of individualized predictions for two patients
The coloring displays whether the feature contributed positively (red) or negatively (blue) to the probability computed by the model.
(A) Example of predicting a high risk for a crisis, mainly driven by the values of the weeks since last crisis and last referral, age and number of total referrals, as well as number of referrals in the last 24 weeks.
(B) Example of a low risk to have a crisis, driven mainly by a high number of weeks since the last referral, high number of weeks since the last crisis, and high number of weeks since the last contact.
Discussion
This study demonstrates the predictive power of clinical notes (i.e., unstructured EHRs) to evaluate the risk of the upcoming mental health crisis relapse within a 4-week period. When compared, the models trained solely on structured EHRs showed higher predictive capabilities than those built only on unstructured EHRs. However, the combination of structured and unstructured EHR can offer a better performance than either data type alone. We also developed and compared the models that combine the two data types by exploring different machine learning methods. The ensemble model that combines the predictions from two neural networks trained with different data streams (Ensemble DNN) achieved the highest performance: an AUPRC of 0.133 and an AUROC of 0.865. The ensemble model relies on the model trained only with structured data for cases with no available notes, whereas it relies on the model trained with structured and unstructured data in cases with available notes prior to the week related to the crisis prediction. Interestingly, this model achieves a significantly higher performance (p < 0.001) than the model trained on the entire data using both data categories (Hybrid DNN) and surpasses the performance of any of the models trained on a single data stream. This highlights a strong complementary value of the clinical notes when adding them to the models trained on the structured data. However, the method used to build a combined model is key to effectively extract value from both data streams.
The results also highlight the importance of the availability of unstructured data and its impact on the best-performing model. While the Struct XGBoost model outperformed the other models for the group of patients with less than 10% of weeks with available notes, the Hybrid DNN was the best for patients with more than 10% of weeks with at least one note (Figure 2). This suggests that the amount of available notes per patient needs to be considered when deciding whether to incorporate unstructured data into a mental health crisis prediction model, either for a specific category of patients or across an entire sample. When the overall quantity of unstructured data is limited, it becomes challenging to harness this data source and to enhance the predictive capabilities of the model. Conversely, when the quantity of clinical notes per patient is adequately large, we underscored potential challenges and demonstrated that integrating the two types of data, structured and unstructured, can improve the performance of the model. This finding is validated by the enhanced metrics of the Hybrid DNN for patients with more than 10% of weeks with notes and the overall metrics of the Ensemble DNN. This type of scenario may also hold in other predictive modeling use cases in the medical domain, so further research needs to be conducted for specific modeling tasks. Importantly, we refrain from establishing a specific threshold that defines the minimally required percentage of available notes because this threshold can depend not only on the availability of notes but also on the availability of the various sources of structured data and the nature of the modeling task. Rather, we argue for the importance of the analysis (such as the one conducted in this study) before deploying predictive analytics systems to determine which model and which data categories are more appropriate for each group of patients. Our approach provides a flexible methodology for blending both types of models depending on the available data sources.
An essential element of our system is the language model employed to process unstructured data. For this study, we applied a BERT model, one of the pioneer language models that uses the transformer architecture, a technique that has since become ubiquitous in the realm of natural language processing.28 Given the rapid advancements in the field of language models, particularly in clinical applications but also beyond, it is important to choose a state-of-the-art language model when deploying a system akin to the one we presented. Moreover, staying abreast with the latest developments in natural language processing is essential to ensure that the system performs at its optimum.
As the predictive models promise to power decision-making in healthcare, the explainability of the artificial intelligence (AI)-based models in such a high-stakes domain is often an important requirement. In particular, providing a case-specific explanation about how the model makes a certain decision alongside the model’s prediction is of paramount importance to gain the clinicians’ trust in the algorithm.34,35,36,37 In this regard, we used SHAP values to determine which features have the highest impact on the models’ predictions and aggregated them based on the type of information each feature brings to understand which data categories carry the most predictive power. A higher absolute SHAP value indicates a greater impact of the feature on the outcome of the model. Overall, we observed that the highest total absolute SHAP value from a single data source was obtained from unstructured data (Figure 3B), but the structured features dominated the top 20 predictive features in all subsets of patients. This suggests that each individual unstructured feature does not have a strong predictive power, but when combined, they surpass the total absolute SHAP of the structured features categories. In other words, individual semantic representations do not bring a considerable value to the model, but when combined, they carry a substantial weight in the model’s predictions. Furthermore, the total SHAP values within most categories, including unstructured, increase with the percentage of available notes (Figure 3A). The increase in absolute SHAP values per category across different bins suggests that the notes become more important as we restrict the sample to patients with more available notes. The categories related to referral, diagnosis, crisis, contact, and hospitalization show a similar pattern. Interestingly, the SHAP values of the structured features overall show a more significant increase compared with unstructured features. It is worth noting that the only category for which the SHAP values decrease with the increase in percentage of notes was the one of “patient’s status.” This may stem from the fact that the model leverages more event-based features for predicting mental health crises as the percentage of available notes increases; note that the increase in available notes is associated with the number of crisis episodes and that such patients are attended to more often at the hospital. One limitation concerning the explainability of our modeling framework is our inability to identify the specific text segments that play the most significant role in predicting a crisis. This is due to the unstructured features being precomputed as weekly aggregates from the clinical notes before performing the process of model training. As a result, we cannot trace back the predictions to individual words or sentences within the text. However, this limitation opens up opportunities for future improvements of our system. Performing end-to-end modeling, from the raw clinical notes to the final prediction, could enable more detailed explanations of the predictions, although it may result in increased complexity and demands on computational resources.
Previous research has also shown that incorporating both structured and unstructured EHR data for prediction of critical events in healthcare settings yields better results than using either of the two data streams independently. Zhang et al.23 demonstrated that combining structured data with clinical notes significantly improved the performance of the models overusing a single data category in three different modeling predictive tasks (in-hospital mortality, 30-day hospital readmission, and long length of stay prediction). Two different studies26,27 presented the benefits of using unstructured data together with structured EHR to predict suicide. Specifically, they shed light on the complementary value of clinical notes that is not embedded in the structured data (such as the well-being of patients). Our initial mental health crisis prediction model (a gradient boosting machine, XGBoost) relied solely on structured EHR data19 and achieved an AUROC of 0.797 for detecting crises within the cohort of patients with a history of relapse. In this study, we broadened the scope by including patients who have not had any crisis relapse registered in EHRs. Moreover, we expanded the analysis by adding unstructured data as well as new features extracted from structured data. In line with the previous study,19 the XGBoost model was the best-performing model based only on structured data, now with a higher AUROC of 0.831. When we incorporated unstructured data, the best-performing model was based on neural networks (Ensemble DNN) and achieved an AUROC of 0.865. Although we are unable to directly compare the performance of different models in this and the previous study because of an extended cohort of patients, the data used in both studies originated from the same hospital, and thus they represent a reasonable baseline for this study.
Because the rapid adoption of EHRs leads to massive amounts of computable clinical data, electronic phenotyping and predictive modeling for mental health crises presents the opportunity to improve clinical decision-making, management of resources, and, ultimately, health outcomes. The amount of digital clinical notes in mental healthcare has dramatically increased over the past years, opening new research avenues to develop game-changing preventative care systems. Our study is the first one to develop and evaluate a machine learning model by combining structured and unstructured EHRs to predict the risk of mental health crises. The lack of standardization in clinical note taking prompted us to also explore what the amount of available notes that brings an added value to the prediction of mental health crises, and we provided takeaways in identifying the required availability of clinical notes to enhance the performance of a structured data-based model. With the structured EHRs becoming a de facto standard and clinical notes remaining pervasive (and in most healthcare systems, a dominating source of data), this study represents an important milestone and a call to arms to leverage both sources of data to enable long-awaited preventative interventions.
Limitations of the study
This study is limited by the single-center data—findings and insights from our analysis may be biased by the cohort of patients that belongs to one hospital and the UK healthcare system, and hence, it may limit their generalizability. In addition, the models developed in this study were tested in a retrospective manner and not evaluated in clinical practice. However, our previous study showed the added value of mental health crisis predictions in clinical settings provided by a machine learning model based on structured data,19 and the results of the present work demonstrated a higher predictive power when incorporating unstructured data. Another limitation stems from the fact that the process of taking notes in hospitals is not standardized; each clinician may take notes in their own way depending on their subjective judgment or time availability. We expect that our approach should largely remain impartial to individual differences in notetaking, given the diverse range of note lengths, styles, and content included in our dataset. However, this study did not take into account other potential sources of variation, such as differences in local clinical practice protocols. Finally, selecting subgroups of patients based on the amount of available notes inevitably selects patients according to other characteristics correlated with the amount of notes (such as an individual frequency of crises). This is an unavoidable consequence of grouping patients based on certain conditions when these conditions are not independent from all other patient attributes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Processed data to generate results | This paper | https://doi.org/10.17632/rg5cssgmwg.2 |
| Software and algorithms | ||
| Crisis detection algorithm | This paper | https://doi.org/10.17632/rg5cssgmwg.2 |
| Python version 3.6.7 | Python Software Foundation | https://www.python.org/ |
| pandas = = 1.1.5 | The pandas Contributors | https://pandas.pydata.org |
| xgboost = = 1.0.2 | The XGBoost Contributors | https://xgboost.ai |
| keras = = 2.2.4 | Google LLC | https://keras.io |
| tensorflow = = 2.1.0 | Google LLC | https://www.tensorflow.org |
| scikit-learn = = 0.24.0 | The scikit-learn Contributors | https://scikit-learn.org/stable/ |
| transformers = = 2.3.0 | Hugging Face | https://huggingface.co/docs/transformers/index |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Roger Garriga Calleja (roger.garriga@bse.eu).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
The study was entirely computational and did not obtain data through intervention or interaction with human subjects. The dataset is constituted of anonymized electronic health records (EHR) collected between September 2012 and July 2020. The EHR belong to Birmingham and Solihull Mental Health Foundation Trust (BSMHFT), which operates over 40 sites and serves a culturally and socially diverse population of over a million people of the surrounding area of Birmingham and Solihull, United Kingdom. BSMHFT allowed access to the EHR via a Data Processing Agreement, but no patient identifiable data was accessed at any stage during the study. The available data included demographics, crisis events, hospitalizations, contacts with the hospital, referrals, risk and wellbeing assessments, diagnosis, as well as notes taken by healthcare professionals and social workers from 59,750 psychiatric patients. For this study, we included the data from patients that are alive with a history of mental health crisis episodes, i.e., those that had suffered at least one mental health crisis. We refrained from predicting the first crisis episode. Our primary focus primarily lies on the patients who mainly contribute to the demand for mental health services. Therefore, the predictive model was designed to be implemented in clinical settings, specifically targeting patients who have already experienced their first crisis. Patients with less than three months of records in the system were also excluded to make sure all patients have sufficient historical data for the algorithm to learn. Finally, no exclusion criteria based on age or diagnosed disorder was applied. The demographics of the cohort are provided at Table 1. From the aforementioned dataset we distinguished between patients from whom healthcare providers take frequent notes (e.g., patients that have 50% of weeks with at least one note) to those that have 0 to 10% of weeks with a note, as well as the ranges in between (10%–20%, 20%–30%, 30%–40%, and 40%–50%).
The Project Steering Group (of which, at the time was an NHS England and NHS Improvement commissioned program hosted by Birmingham and Solihull Mental Health Foundation Trust) approved the study following consultation with Information Governance specialist advisors. Due to the retrospective nature of the study and the use of anonymized data, it was concluded that data protection legislation did not require explicit patient consent, ethics committee review or HRA approvals to be obtained. Furthermore, the Trust’s Information Governance Steering Group maintained oversight of the project’s proposals, progress and approvals in respect to the use of anonymized data.
Method details
Crisis prediction algorithm
We predict patients’ four-week risk of mental health crisis relapse on a weekly basis for each living patient. In a given week w, the four-week risk of relapse for a given patient p, denoted by , is predicted by using a previously trained model φw for week w, which takes as inputs the structured and/or unstructured features computed from the patient’s EHR between the first interaction of the patient until week w and outputs an estimate of the probability of four-week relapse. To train the model, we
generated for each patient and each week the target-features tuple (), where is the binary target for patient
p at week w that takes a value of 1 if the patient had a crisis during the next four weeks and 0 otherwise, and Xs and Xu
p,w
p,w
are the structured and unstructured features for patient p at week w, respectively. We detail each of the steps in the following subsections.
Target generation
There is a wide range of approaches to defining a mental health crisis in the literature: self-defined (i.e., the user define the experience and recovery), risk-focused (i.e., risk of suicide, or harming themselves or others), negotiated definitions (i.e., a decision reached collaboratively between service user, carer or professional), and a pragmatic service-oriented approach (“crisis brings the service user to the attention of crisis services such as through the relapse of an existing mental health condition”).38 All in all, these definitions describe an event that substantially affects the life of a patient and the load on healthcare services regardless of the diagnosed disorder of the patient. The dataset included crisis events, registered hourly, every time a patient had an urgent need of mental health crisis services e.g., emergency assessment, inpatient admission, home treatment assessment, or hospitalization. Frequently, there are multiple crisis events registered when a patient is undergoing a mental health crisis. For that reason, we trained the machine learning models to detect the onset of a crisis episode, which contains one or more crisis events preceded by at least one full stable week without any crisis event. Specifically, we constructed the prediction targets yp,w for all patient as follows: Patient p at week w was assigned a value of 1 whenever there was an onset of crisis episode within the following 28 days and a 0 otherwise. This time window was selected according to clinical practice in our clinical setting as a reasonable time frame that allows the hospital to conduct a timely intervention and prevent the next crisis episode. The weeks that correspond to a crisis episode were excluded from the training process and no predictions were made for those weeks. This decision was based on the assumption that patients receive close attention and care during a crisis episode and cannot relapse until they are deemed stable.
Features generation
We processed separately the structured EHR and the unstructured EHR to build the features to be used in the models. The structured data contained 10 data sources and the unstructured data one. We handled the structured data sources independently and generated a total of ds = 450 features. These features are computed at a weekly basis using the information available up to the week. The features extracted can be categorized in 8 different types depending on the process applied to the data.
-
•
Static or semi-static. These features are derived from data that does not change over the history of the patient, such as date of birth, gender, or the date of the first visit to the hospital. Gender is kept as a static feature, while the features based on date of birth and date of the first visit vary over time in a monthly or yearly basis. Examples: months since first visit, current age, gender male.
-
•
Weekly aggregations. These features are computed from the interactions that the patient had with the hospital, which are aggregated to weekly level. Some examples of interactions are contacts (visits), crises, or referrals. For each week and type of interaction we counted the number of times the interaction occurred, whether all the days of the week had that type of interaction and whether there was at least one day with that type of interaction. Examples: number of crises during week, at least one contact during week, all days with crisis during week.
-
•
One hot encoded. Some interaction types had subcategories associated to it, these subcategories were one hot encoded by assigning a 1 to those weeks in which the subcategory occurred and a 0 otherwise. Examples: source of referral from carer, not attended contact, crisis allocation method hospitalization.
-
•
Time elapsed features. These features are constructed for each type of interaction and subcategory. At each week, the feature counts the number of weeks passed since the last time the type of interaction or subcategory occurred. NaN values were used to indicate that the patient has never had a certain type of interaction or subcategory. Examples: weeks since last crisis, weeks since last referral source self, time since last unplanned contact.
-
•
Lagged aggregations. For some important interactions that showed up as important in the previous literature,19 we aggregated a subset of the previous weekly aggregations to construct these features. Examples: crisis sum in the last 4 weeks, contact sum in the last 8 weeks, referrals sum in the last 12 weeks.
-
•
State indicators. There are a number of EHR that had a start and end date to define the period of time in which the patient was assigned a certain team, assessed with some risk or wellbeing indicators, or diagnosed with a certain disorder among others. In these cases, we constructed a feature that assigned a 1 to those weeks within the period and a 0 otherwise, or the value that was assigned during that period. For the risk and wellbeing indicators, NaN values were used to indicate that the indicators were unknown due to a lack of assessment. Examples: currently diagnosed with mood disorder, wellbeing assessment emotional score, referral to mental health community team.
-
•
History aggregations. At each week, for some important interactions or state indicators, we aggregated the whole history of the patient up to that week. These features capture the total number of some interactions or whether the patient has ever had some state indicator. Examples: number of crisis episodes, ever diagnosed with personality disorder, ever hospitalized with acute assessment.
-
•
Last crisis episode. Each crisis episode is summarized through a set of aggregated interactions that describe the severity and length of the episode. Then, at each week, the feature is constructed with the values of the aggregations from the last crisis episode the patient experienced. Examples: number of days in crisis during last episode, maximum length of hospitalization during last episode, maximum severity of crisis during last episode.
Note that diagnosis data was captured as either a state indicator or a history aggregation and it was missing for over 60% of the patients. This can be explained by the fact that in many instances the clinicians did not possess enough observational data to confidently ascertain a diagnosis, as a significant proportion of patients had either no relapses or only one relapse following their initial crisis episode. Moreover, it is vital to recognize that mental health disorders often carry a societal stigma. As a result, clinicians take great care and aim for accuracy when diagnosing, often opting to monitor the patient across multiple visits before reaching a definitive diagnosis.
We processed the unstructured data to generate a set of weekly features that represent the information gathered by clinicians through the following procedures.
-
•
Semantic representation of most recent notes. For each patient and each week all those weeks’ notes were parsed using a pretrained BERT language model, which produced a 768-dimensional vectors containing the semantic representation of each note. Each such vector is an aggregate of the vectors (of same dimension) describing the semantic meaning and position in the sentence of each relevant sub-word. Those vectors were then averaged, per patient and per week, to obtain a vector describing all the notes of a given week. Since most patients do not have notes on their EHR every week, we used as a feature the vector with the most recent representation of a patient’s notes. If none were available prior to a given week, then the zero vector with dimension 768 was used instead. For instance, if a patient had three notes during a certain week, the pretrained BERT model was applied to each of the notes independently generating three 768-dimensional vectors. Then, those three vectors were averaged dimension-wise resulting in a single 768-dimensional vector for the given week. Finally, if the patient had no notes in the subsequent four weeks, the same 768-dimensional vector will be used for those four weeks.
-
•
Time since last doctor’s note. In tandem with the feature described in the previous point, we defined a feature with the number of weeks since the most recently recorded note. This feature was built in a similar way to the time elapse features described in the structured data procedure.
In this manuscript, the features generated from unstructured data are referred as "unstructured features" and the features generated from structured data are referred as "structured features". Note that NaN values were used when generating many of the features and the use of NaN carry a specific and important meaning, denoting when an event has never happened in the history of the patient.
Machine learning prediction models
A trained model φw for week w takes as inputs the structured and unstructured features Xs and Xu for week w to generate
p,w
p,w
the prediction .
We call a model "structured" when we restrict the prediction to be done only using the structured features of patient p at w, and similarly, we call it "unstructured" when only is used for the prediction. Moreover, we built two models ("mixed" models) that make use of both types of features, and . In total, we present 5 models and 2 baselines:
-
•
Struct XGBoost. A structured model based on XGBoost,31 an implementation of Gradient Boosting Machines (GBM).39.
-
•
Struct DNN. A structured model based on a multi layer perceptron.
-
•
Unstruct DNN. An unstructured model based on a multi layer perceptron.
-
•
Hybrid DNN. A mixed model based on a multi layer perceptron.
-
•
Ensemble DNN. A mixed model built as an ensemble of Struct DNN and Hybrid∗ DNN: at inference time, if no notes exist on or prior to week w (meaning is a zero vector) then the Structured DNN model is called, otherwise, Hybrid∗ DNN is called, a version of Hybrid DNN trained on the subset of the data with nonzero u.
-
•
Baseline 1 LogReg5. A structured model based on a logistic regression trained using 5 features inspired by those important variables identified in the literature.32 Table S1 shows the list of features used in this baseline.
-
•Baseline 2 OneYearTotalCrisis. A heuristic model that ranks the patients based on total number of crisis experienced during the past year (last 53 weeks).
- Additionally, we explored other machine learning techniques that had lower performance to those presented, including the use of XGBoost as an unstructured and hybrid model, and the use of Logistic Regression and Random Forest.
- Note that for the DNN classifiers and the logistic regression-based baseline, we performed standard scaling and imputation of missing values. In particular, the "time since last event" type of features were log normalized and the NaN values were imputed with a constant value, larger than all the values of that feature in the training set; the rest of the features that contained missing values were imputed with 0. This was not necessary for the XGBoost model as it accepts NaN values and tree-based models do not benefit from scaling.
Training and hyperparameter tuning
We partitioned the dataset into training, validation, and testing segments according to temporal slices, mirroring the model’s potential application in routine practice. The training set comprised all data between September 2012 and December 2017, the validation set comprised data from January 2018 to December 2018 whereas the test set included data from January 2019 to December 2019. Data pertaining to the period between January 2020 and July 2020 was excluded from analysis due to the COVID-19 pandemic that significantly impacted the typical healthcare routines and therefore the EHRs. We performed hyperparameter tuning and model selection by training our models in the training set and evaluating in the validation set; for each of the models we run 100 rounds of hyperparameter tuning using Hyperopt,40 which applies a Bayesian Optimization algorithm called TreeParzen Estimator41 in a sequential manner. The best hyperparameters of each model were selected by optimizing the area under the precision recall curve (AUPRC) and then used to train the model in the training and validation sets.
Explainability of predictions
We measured the contribution of each feature to the models built using the SHAP values,33 which is a state of the art technique taken from game theory that is used for local interpretability of Machine learning models. We used the TreeExplainer algorithm to compute the SHAP values of the XGBoost model and the DeepExplainer algorithm to compute those for the neural network model. The TreeExplainer measures local feature interaction effects through an additive feature attribution method and allows to understand the global model structure by combining the local effects in a consistent way.42 The DeepExplainer algorithm represents a variation of the SHAP algorithm that is specifically optimized for explaining DNN models, where the conditional expectations of SHAP values are approximated using a selection of background samples.43 The SHAP values were computed for each prediction of test set and provide a numerical score to every feature that quantifies the influence (positive or negative) that the feature had to the predicted risk. In addition to the SHAP values for each feature, we grouped the features by categories and computed the total SHAP value per category as the sum of the SHAP values of each feature belonging to that category.
Quantification and statistical analysis
The metrics considered to evaluate the model’s performance were area under the receiver operating curve (AUROC) and area under the precision recall curve (AUPRC). We computed the percentile based Net Reclassification Improvement44 with 1000 categories to directly compare reclassification changes between models. All evaluation was done on a weekly basis to emulate the real case scenario. All the results reported throughout the manuscript were computed on the testing set except those that are specified otherwise. In particular, the metrics of the test set (comprising 52 weeks) were computed by making predictions week by week and then aggregated to generate the mean and standard deviation. Confidence intervals of the reported performance metrics were computed using the n = 52 temporal splits. Statistical analysis for model comparison was done through a paired sample T-test on the AUPRC weekly results and we used the DeLong test45 to compare ROC curves and the AUROC metrics.
Acknowledgments
The authors would like to thank P. Presland, J. Nolan, L. Hudson, E. Patterson, and R. Russell for their support and help throughout the course of this study. This work was supported by a Health Foundation grant (Call: Round 1 Advancing Applied Analytics Program, award reference number 709246; project title: Using predictive analysis to prevent mental health crisis), the Birmingham and Solihull Mental Health NHS Foundation Trust, and Koa Health (formerly Telefonica Alpha). This work was completed prior to T.S.B., J.O.I, and I.E.A. joining Meta, Telefonica, and Amazon, respectively.
Author contributions
All authors participated in the conceptualization of the study and the design of the machine learning approach. A.M. served as the principal supervisor. R.G. and T.S.B. pre-processed and cleaned the structured data, engineered the structured features, and implemented the structured-based models and their interpretation. T.S.B. prepared the reports of the models’ interpretation. J.O.I. described and summarized the data cohort. J.G. pre-processed and cleaned the clinical notes and implemented the unstructured-based and hybrid models with the support of J.O.I. and T.S.B. interpreted the unstructured-based models. R.G. designed and implemented the statistical analysis and the ensemble model. All authors contributed to the first draft of the manuscript and revised and approved the final version.
Declaration of interests
Koa Health (formerly Telefonica Innovation Alpha) has provided financial resources to support the realization of this project. All authors were employees of Telefonica Innovation Alpha (now, R.G., J.G., and A.M. are employees of Koa Health S.L.), and they received salary support during the realization of the study. The funders of the study had no role in the design, data analysis and model development, interpretation of the results, writing, and reviewing of the manuscript.
Published: October 31, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101260.
Supplemental information
Data and code availability
-
•
The anonymized electronic health records used in this study cannot be publicly available because they contain highly sensitive information about vulnerable populations. This data can only be accessed within the hospital infrastructure following a Data Processing Agreement (DPA) underpinned by rigorous information governance and data sharing legislation. To request access, contact Birmingham and Solihull Mental Health NHS Foundation Trust’s Information Governance Committee. The processed datasets derived from these data to produce the presented results have been deposited at Mendeley Data and are publicly available as of date of publication with the following https://doi.org/10.17632/rg5cssgmwg.2.
-
•
All original code has been deposited at Mendeley Data and is publicly available as of date of publication with the following https://doi.org/10.17632/rg5cssgmwg.2.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Patel V., Saxena S., Lund C., Thornicroft G., Baingana F., Bolton P., Chisholm D., Collins P.Y., Cooper J.L., Eaton J., et al. The Lancet Commission on global mental health and sustainable development. Lancet. 2018;392:1553–1598. doi: 10.1016/S0140-6736(18)31612-X. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2019 Mental Disorders Collaborators National burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019 (2022) Lancet Psychiatr. 2022;9:137–150. doi: 10.1016/S2215-0366(21)00395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiens K., Bhattarai A., Pedram P., Dores A., Williams J., Bulloch A., Patten S. A growing need for youth mental health services in Canada: examining trends in youth mental health from 2011 to 2018. Epidemiol. Psychiatr. Sci. 2020;29:e115. doi: 10.1017/S2045796020000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keynejad R., Spagnolo J., Thornicroft G. WHO mental health gap action programme (mhGAP) intervention guide: updated systematic review on evidence and impact. Evid. Base Ment. Health. 2021;24:124–130. doi: 10.1136/ebmental-2021-300254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olfson M. Building The Mental Health Workforce Capacity Needed To Treat Adults With Serious Mental Illnesses. Health Aff. 2016;35:983–990. doi: 10.1377/hlthaff.2015.1619. [DOI] [PubMed] [Google Scholar]
- 6.National Alliance on Mental Illness; 2018. Navigating a Mental Health Crisis: A NAMI Resource Guide for Those Experiencing a Mental Health Emergency. [Google Scholar]
- 7.Heyland M., Johnson M. Evaluating an Alternative to the Emergency Department for Adults in Mental Health Crisis. Issues Ment. Health Nurs. 2017;38:557–561. doi: 10.1080/01612840.2017.1300841. [DOI] [PubMed] [Google Scholar]
- 8.Miller V., Robertson S. A Role for Occupational Therapy in Crisis Intervention and Prevention. Aust. Occup. Ther. J. 2010;38:143–146. doi: 10.1111/j.1440-1630.1991.tb01710.x. [DOI] [Google Scholar]
- 9.Shandhi M.M.H., Dunn J.P. AI in medicine: Where are we now and where are we going? Cell Rep. Med. 2022;3:100861. doi: 10.1016/j.xcrm.2022.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook B.L., Progovac A.M., Chen P., Mullin B., Hou S., Baca-Garcia E. Novel Use of Natural Language Processing (NLP) to Predict Suicidal Ideation and Psychiatric Symptoms in a Text-Based Mental Health Intervention in Madrid. Comput. Math. Methods Med. 2016;2016:8708434–8708438. doi: 10.1155/2016/8708434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon G.E., Johnson E., Lawrence J.M., Rossom R.C., Ahmedani B., Lynch F.L., Beck A., Waitzfelder B., Ziebell R., Penfold R.B., Shortreed S.M. Predicting Suicide Attempts and Suicide Deaths Following Outpatient Visits Using Electronic Health Records. Aust. J. Pharm. 2018;175:951–960. doi: 10.1176/appi.ajp.2018.17101167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barak-Corren Y., Castro V.M., Javitt S., Hoffnagle A.G., Dai Y., Perlis R.H., Nock M.K., Smoller J.W., Reis B.Y. Predicting Suicidal Behavior From Longitudinal Electronic Health Records. Aust. J. Pharm. 2017;174:154–162. doi: 10.1176/appi.ajp.2016.16010077. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q., Zhang-James Y., Barnett E.J., Lichtenstein P., Jokinen J., D’Onofrio B.M., Faraone S.V., Larsson H., Fazel S. Predicting suicide attempt or suicide death following a visit to psychiatric specialty care: A machine learning study using Swedish national registry data. PLoS Med. 2020;17:e1003416. doi: 10.1371/journal.pmed.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulin C., Shiner B., Thompson P., Vepstas L., Young-Xu Y., Goertzel B., Watts B., Flashman L., McAllister T. Predicting the Risk of Suicide by Analyzing the Text of Clinical Notes. PLoS One. 2014;9:e85733. doi: 10.1371/journal.pone.0085733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes A.C., Dutta R., Velupillai S., Sanyal J., Stewart R., Chandran D. Identifying Suicide Ideation and Suicidal Attempts in a Psychiatric Clinical Research Database using Natural Language Processing. Sci. Rep. 2018;8:7426. doi: 10.1038/s41598-018-25773-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olfson M., Marcus S.C., Bridge J.A. Emergency Department Recognition of Mental Disorders and Short-Term Outcome of Deliberate Self-Harm. Aust. J. Pharm. 2013;170:1442–1450. doi: 10.1176/appi.ajp.2013.12121506. [DOI] [PubMed] [Google Scholar]
- 17.Raket L.L., Jaskolowski J., Kinon B.J., Brasen J.C., Jönsson L., Wehnert A., Fusar-Poli P. Dynamic ElecTronic hEalth reCord deTection (DETECT) of individuals at risk of a first episode of psychosis: a case-control development and validation study. Lancet. Digit. Health. 2020;2:e229–e239. doi: 10.1016/S2589-7500(20)30024-8. [DOI] [PubMed] [Google Scholar]
- 18.Irving J., Patel R., Oliver D., Colling C., Pritchard M., Broadbent M., Baldwin H., Stahl D., Stewart R., Fusar-Poli P. Using Natural Language Processing on Electronic Health Records to Enhance Detection and Prediction of Psychosis Risk. Schizophr. Bull. 2021;47:405–414. doi: 10.1093/schbul/sbaa126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garriga R., Mas J., Abraha S., Nolan J., Harrison O., Tadros G., Matic A. Machine learning model to predict mental health crises from electronic health records. Nat. Med. 2022;28:1240–1248. doi: 10.1038/s41591-022-01811-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheikhalishahi S., Miotto R., Dudley J.T., Lavelli A., Rinaldi F., Osmani V. Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review. JMIR Med. Inform. 2019;7:e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tayefi M., Ngo P., Chomutare T., Dalianis H., Salvi E., Budrionis A., Godtliebsen F. Challenges and opportunities beyond structured data in analysis of electronic health records. WIREs Computational Stats. 2021;13:e1549. doi: 10.1002/wics.1549. [DOI] [Google Scholar]
- 22.Edgcomb J.B., Zima B. Machine Learning, Natural Language Processing, and the Electronic Health Record: Innovations in Mental Health Services Research. PSIC. 2019;70:346–349. doi: 10.1176/appi.ps.201800401. [DOI] [PubMed] [Google Scholar]
- 23.Zhang D., Yin C., Zeng J., Yuan X., Zhang P. Combining structured and unstructured data for predictive models: a deep learning approach. BMC Med. Inform. Decis. Mak. 2020;20:280. doi: 10.1186/s12911-020-01297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golas S.B., Shibahara T., Agboola S., Otaki H., Sato J., Nakae T., Hisamitsu T., Kojima G., Felsted J., Kakarmath S., et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: a retrospective analysis of electronic medical records data. BMC Med. Inform. Decis. Mak. 2018;18:44. doi: 10.1186/s12911-018-0620-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahajan S.M., Ghani R. Combining Structured and Unstructured Data for Predicting Risk of Readmission for Heart Failure Patients. Stud. Health Technol. Inf. 2019;264:238–242. doi: 10.3233/SHTI190219. [DOI] [PubMed] [Google Scholar]
- 26.Tsui F.R., Shi L., Ruiz V., Ryan N.D., Biernesser C., Iyengar S., Walsh C.G., Brent D.A. Natural language processing and machine learning of electronic health records for prediction of first-time suicide attempts. JAMIA Open. 2021;4:ooab011. doi: 10.1093/jamiaopen/ooab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayramli I., Castro V., Barak-Corren Y., Madsen E.M., Nock M.K., Smoller J.W., Reis B.Y. Predictive structured–unstructured interactions in EHR models: A case study of suicide prediction. npj Digit. Med. 2022;5:15. doi: 10.1038/s41746-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devlin J., Chang M.-W., Lee K., Toutanova K. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Vol. 1. Association for Computational Linguistics; 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding; pp. 4171–4186. (Long and Short Papers) [DOI] [Google Scholar]
- 29.Boyd K., Eng K.H., Page C.D. In: Advanced Information Systems Engineering Lecture Notes in Computer Science. Salinesi C., Norrie M.C., Pastor Ó., editors. Springer Berlin Heidelberg; 2013. Area under the Precision-Recall Curve: Point Estimates and Confidence Intervals; pp. 451–466. [DOI] [Google Scholar]
- 30.Ozenne B., Subtil F., Maucort-Boulch D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015;68:855–859. doi: 10.1016/j.jclinepi.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM); 2016. XGBoost: A Scalable Tree Boosting System; pp. 785–794. [DOI] [Google Scholar]
- 32.Futoma J., Morris J., Lucas J. A comparison of models for predicting early hospital readmissions. J. Biomed. Inf. 2015;56:229–238. doi: 10.1016/j.jbi.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Lundberg S.M., Lee S.-I. Proceedings of the 31st international conference on neural information processing systems. 2017. A unified approach to interpreting model predictions; pp. 4768–4777. [Google Scholar]
- 34.Yoon C.H., Torrance R., Scheinerman N. Machine learning in medicine: should the pursuit of enhanced interpretability be abandoned? J. Med. Ethics. 2022;48:581–585. doi: 10.1136/medethics-2020-107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjoa E., Guan C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Transact. Neural Networks Learn. Syst. 2021;32:4793–4813. doi: 10.1109/TNNLS.2020.3027314. [DOI] [PubMed] [Google Scholar]
- 36.ElShawi R., Sherif Y., Al-Mallah M., Sakr S. Interpretability in healthcare: A comparative study of local machine learning interpretability techniques. Comput. Intell. 2021;37:1633–1650. doi: 10.1111/coin.12410. [DOI] [Google Scholar]
- 37.Diprose W.K., Buist N., Hua N., Thurier Q., Shand G., Robinson R. Physician understanding, explainability, and trust in a hypothetical machine learning risk calculator. J. Am. Med. Inf. Assoc. 2020;27:592–600. doi: 10.1093/jamia/ocz229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paton F., Wright K., Ayre N., Dare C., Johnson S., Lloyd-Evans B., Simpson A., Webber M., Meader N. Improving outcomes for people in mental health crisis: a rapid synthesis of the evidence for available models of care. Health Technol. Assess. 2016;20:1–162. doi: 10.3310/hta20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001;29 doi: 10.1214/aos/1013203451. [DOI] [Google Scholar]
- 40.Bergstra J., Yamins D., Cox D. In: Proceedings of the 30th International Conference on Machine Learning Proceedings of Machine Learning Research. Dasgupta S., McAllester D., editors. PMLR); 2013. Making a Science of Model Search: Hyperparameter Optimization in Hundreds of Dimensions for Vision Architectures; pp. 115–123. [Google Scholar]
- 41.Bergstra J., Bardenet R., Bengio Y., Kégl B. In: Advances in Neural Information Processing Systems. Shawe-Taylor J., Zemel R., Bartlett P., Pereira F., Weinberger K.Q., editors. Curran Associates, Inc.; 2011. Algorithms for Hyper-Parameter Optimization. [Google Scholar]
- 42.Lundberg S.M., Erion G., Chen H., DeGrave A., Prutkin J.M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S.-I. 2019. Explainable AI for Trees: From Local Explanations to Global Understanding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrikumar A., Greenside P., Kundaje A. 2017. Learning Important Features through Propagating Activation Differences. [DOI] [Google Scholar]
- 44.McKearnan S.B., Wolfson J., Vock D.M., Vazquez-Benitez G., O’Connor P.J. Performance of the Net Reclassification Improvement for Nonnested Models and a Novel Percentile-Based Alternative. Am. J. Epidemiol. 2018;187:1327–1335. doi: 10.1093/aje/kwx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The anonymized electronic health records used in this study cannot be publicly available because they contain highly sensitive information about vulnerable populations. This data can only be accessed within the hospital infrastructure following a Data Processing Agreement (DPA) underpinned by rigorous information governance and data sharing legislation. To request access, contact Birmingham and Solihull Mental Health NHS Foundation Trust’s Information Governance Committee. The processed datasets derived from these data to produce the presented results have been deposited at Mendeley Data and are publicly available as of date of publication with the following https://doi.org/10.17632/rg5cssgmwg.2.
-
•
All original code has been deposited at Mendeley Data and is publicly available as of date of publication with the following https://doi.org/10.17632/rg5cssgmwg.2.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.