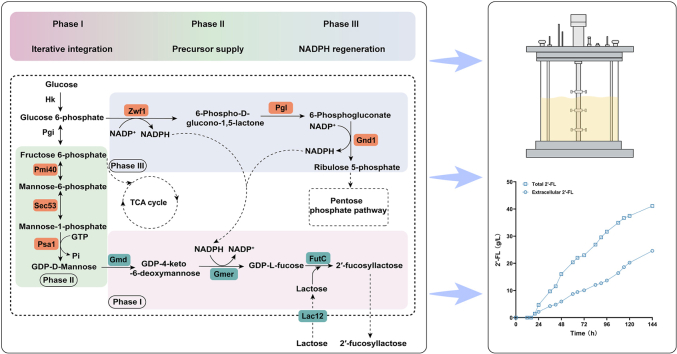
Abstract
2′-Fucosyllactose (2′-FL) has great application value as a nutritional component and the whole cell biosynthesis of 2′-FL has become the focus of current research. Yarrowia lipolytica has great potential in oligosaccharide synthesis and large-scale fermentation. In this study, systematic engineering of Y. lipolytica for efficient 2′-FL production was performed. By fusing different protein tags, the synthesis of 2′-FL was optimized and the ubiquitin tag was demonstrated to be the best choice to increase the 2′-FL production. By iterative integration of the related genes, increasing the precursor supply, and promoting NADPH regeneration, the 2′-FL synthesis was further improved. The final 2′-FL titer, 41.10 g/L, was obtained in the strain F5-1. Our work reports the highest 2′-FL production in Y. lipolytica, and demonstrates that Y. lipolytica is an efficient microbial chassis for the synthesis of oligosaccharides.
Keywords: 2′-Fucosyllactose, Yarrowia lipolytica, Iterative integration, Pathway enhancement, NADPH regeneration
Graphical abstract
1. Introduction
Human milk oligosaccharides (HMOs), a class of important nutritional components in human milk, are essential for the healthy growth of infants because of the regulatory and protective influence on the infant's immune system and gastrointestinal system [[1], [2], [3]]. 2′-Fucosyllactose (2′-FL), as the oligosaccharide with the highest content in HMOs, has been permitted for using in infant formula as nutritional additives by several authorities for its unique immune function [4,5]. Therefore, the production methods of 2′-FL have become the focus of current research.
Various 2′-FL production routes have been reported [6,7]. With the development of synthetic biology techniques, microbial synthesis shows significant potential for practical applications [8,9]. The salvage pathway and the de novo pathway are the main pathways for 2′-FL synthesis using microorganisms and have been achieved in a variety of microorganisms [[10], [11], [12], [13]]. Among these microorganisms, Escherichia coli is the most common chassis, and the remarkable production of 2′-FL, 112.56 g/L, was obtained by optimizing the source of the α-1, 2-fucosyltransferase [[14], [15], [16]]. Bacillus subtilis also showed certain advantages and 88.3 g/L 2′-FL was attained in the recombinant B. subtilis strain by pathway optimization [11]. In addition, yeasts with generally recognized as safe (GRAS) status are also potential cell factories for 2′-FL synthesis, such as Saccharomyces cerevisiae and Yarrowia lipolytica have been engineered to synthesize 2′-FL, and 32.05 g/L was obtained in S. cerevisiae, while 24 g/L 2′-FL was reported in Y. lipolytica [13,17,18].
Y. lipolytica has become a commonly used chassis for the biosynthesis of many types of target products, including oils, organic acids, terpenoids, flavones, and proteins [[19], [20], [21], [22], [23], [24], [25]]. The capacity to tolerate stress from the external environment and the ability to utilize different substrates to synthesize high-value compounds make it suitable for the industrial production of products of interest, particularly functional food ingredients [[26], [27], [28]]. Besides, Y. lipolytica has a high glycolytic flux, which facilitates the synthesis of the precursor GDP-d-mannose, making it suitable for 2′-FL production. There is no colanic acid synthesis pathway in Y. lipolytica cells, which reduces the consumption of GDP-l-fucose. However, the production in Y. lipolytica still has no advantage over that in other microorganisms; therefore, it is necessary to optimize the synthesis of 2′-FL to achieve large-scale industrial production using Y. lipolytica.
Many metabolic engineering strategies have been applied to improve 2′-FL production, including optimizing the expression of key enzyme genes, enhancing the supply of precursors, promoting cofactor regeneration, and so on. We present examples of the optimization of the 2′-FL synthesis pathway in various microorganisms using the above strategies, which have been shown to increase 2′-FL production (Table S1) [10,11,13,15,16,[29], [30], [31], [32], [33], [34]]. α-1, 2-fucosyltransferase (FutC) is a rate-limiting enzyme in the synthesis of 2′-FL. Screening for FutC from different sources or increasing the expression of FutC by adding protein tags has been proved to be an effective way to promote the production of 2′-FL [29,30]. GDP-l-fucose is a direct precursor for the synthesis of 2′-FL, while GDP-d-mannose is a key precursor for the synthesis of GDP-l-fucose. The GDP-d-mannose synthesis pathway is present in most microorganisms, which is catalyzed by three main enzymes, including mannose-6-phosphate isomerase (Pmi40), phosphomannomutase (Sec53), and mannose-1-phosphate guanylyltransferase (Psa1). Studies have shown that the synthesis of GDP-d-mannose can be enhanced by overexpression of the genes encoding the above enzymes, thus providing more precursors for 2′-FL synthesis and promoting the accumulation of 2′-FL [11,[32], [33], [34]]. NADPH also plays an important role in the synthesis of 2′-FL as an essential cofactor. The synthesis of GDP-l-fucose requires the participation of NADPH. Overexpression of genes involved in NADPH regeneration pathways could also facilitate the synthesis of 2′-FL [30,32]. Here, optimizing the 2′-FL synthesis pathway and the cofactor regeneration pathway, the final recombinant Y. lipolytica strain produced 41.10 g/L 2′-FL. Our work provides another alternative efficient production platform for 2′-FL synthesis.
2. Material and methods
2.1. Strains and culture conditions
The E. coli strains were used for plasmids construction and cultured in Luria-Bertani (LB) medium. Y. lipolytica strains used for 2′-FL synthesis (Table 1) were grown using YPD medium, or in SD minimal medium lacking leucine or tryptophan (TaKaRa, Beijing, China) for selection. 5 g/L lactose and 80 g/L glucose were used for shake-flask fermentation.
Table 1.
Y. lipolytica strains used in this study.
| Strains | Description | Source |
|---|---|---|
| PO1f | MatA, Leu2-270, URA3-302, xpr2-322, axp-2 | INRA |
| F0 | PO1f with episomal plasmid pLEP-hpFutC-GGL | This study |
| F0-SUMO | PO1f with episomal plasmid pLEP-SUMO-hpFutC-GGL | This study |
| F0-Ub | PO1f with episomal plasmid pLEP-Ub-hpFutC-GGL | This study |
| F0-DsbA | PO1f with episomal plasmid pLEP-DsbA-hpFutC-GGL | This study |
| F0-MBP | PO1f with episomal plasmid pLEP-MBP-hpFutC-GGL | This study |
| F0-TrxA | PO1f with episomal plasmid pLEP-TrxA-hpFutC-GGL | This study |
| F0-GST | PO1f with episomal plasmid pLEP-GST-hpFutC-GGL | This study |
| F1 | PO1f with integrated fragment of linearized plasmid pULUF-GG | This study |
| F1-4ΔUra3 | F1-4 recycling URA3 selection marker | This study |
| F2 | F1-4ΔUra3 with integrated fragment of linearized plasmid pULUF-GG | This study |
| F3 | F2-4 with integrated fragment of linearized plasmid pHLUF-GG | This study |
| F4 | F3-5 with integrated fragment of linearized plasmid pMAN | This study |
| F4-ZWF1 | F4with episomal plasmid pTrp-ZWF1 | This study |
| F4-PGL | F4 with episomal plasmid pTrp-PGL | This study |
| F4-GND1 | F4 with episomal plasmid pTrp-GND1 | This study |
| F5 | F4-2 with integrated fragment of linearized plasmid pZPG | This study |
For shake-flask fermentation, the 2′-FL-production strains were firstly activated on a solid YPD plate, then transferred into liquid YPD for a second activation, and finally transferred into 50 mL of YPD medium containing 80 g/L glucose at a concentration of 2 % and incubated for 144 h before measuring the 2′-FL titer. For the fed-batch fermentation, the 2′-FL-production strains were activated on solid YPD medium for 24 h; and a single colony was picked up to 50 mL of liquid YPD for the second activation, then transferred into 200 mL of liquid YPD medium for the third activation, and finally transferred into fermentation medium (2 % yeast extract, 4 % tryptone, 7 % initial glucose). 10 M NaOH was used to adjust the pH to about 5.5, and the pO2 was controlled at 20 % with the control of agitation speed, and the aeration was maintained at the rate of 2 v/v/min.
2.2. Construction of plasmids
To construct the 2′-FL-producing strains, the TEFin promoter, the EXP promoter and the GPD promoter were used to regulate the expression of klLAC12 (encoding lactose transporter, Lac12), maGMD (encoding GDP-d-mannose-4,6-dehydratase, Gmd), and hpGMER (encoding GDP-l-fucose synthase, Gmer), respectively. HpFutC (encoding ɑ-1,2-fucosyltransferase, FutC) was inserted into the plasmid pLEP-leu to construct the episomal plasmid pLEP-hpFutC [35]. Small ubiquitin-related modifier (SUMO), glutathione S-transferase (GST), thioredoxin A (TrxA), dithiol oxidase (DsbA), ubiquitin (Ub) and maltose binding protein (MBP) were codon optimized and synthesized (Table S2). The protein tags were fused to the N-terminus of hpFutC, and cloned into the plasmid pLEP-leu, respectively, using the methods described above. Finally, the klLAC12, maGMD and hpGMER gene expression cassettes were cloned into the NdeI restriction site of pLEP-hpFutC series plasmids to obtain the episomal plasmids pLEP-hpFutC-GGL, pLEP-Ub-hpFutC-GGL, pLEP-DsbA-hpFutC-GGL and so on.
For the construction of integrated plasmids, the expression fragment of Ub-hpFutC was amplified by PCR using pLEP-Ub-hpFutC-GGL as the template, and the expression fragments for klLAC12, maGMD and hpGMER were also amplified by PCR. The vector backbone containing the URA3 selection marker was amplified using plasmid 113-GPD-TEF as the template [36]. The gene expression fragments were assembled to obtain the plasmid pULUF-GG, and the linearized plasmid was obtained by digestion with the restriction enzyme PmlI. By inserting the hygromycin resistance gene (hyg) into the SmaI restriction site of pULF-GG, the plasmid pHLUF-GG was obtained, and linearization of the plasmid was acquired by PmlI digestion.
The genes PMI40 (encoding Pmi40, YALI0_B18348g), SEC53 (encoding Sec53, YALI0_D13112g), and PSA1 (encoding Psa1, YALI0_C06490g), were amplified from the genome of Y. lipolytica PO1f. PMI40, SEC53 and PSA1 were regulated by the TEF promoter, hp4d promoter and TEFin promoter as well as the VMA terminator, XPR terminator and LIP terminator, respectively. The vector backbone containing the Leu selection marker was amplified using plasmid 114-hp4d-EXP as the template. The plasmid 114-PMI40-SEC53-PSA1 was obtained by Gibson assembly method [37], and the linearized plasmid was obtained by XbaI digestion.
The plasmids of pTrp-ZWF1, pTrp-PGL, pTrp-GND1, and pZPG were constructed to enhance NADPH regeneration. ZWF1 (encoding glucose-6-phosphate dehydrogenase, Zwf1, YALI0_E22649g), PGL (encoding 6-phosphogluconolactonase, Pgl, YALI0_C19085g), and GND1 (encoding 6-phosphogluconate dehydrogenase, Gnd1, YALI0_B15598g) were regulated by the GPD promoter in the episomal plasmids pTrp-ZWF1, pTrp-PGL, and pTrp-GND1, and regulated by the TEFin promoter, GPD promoter, and TEF promoter in the plasmid pZPG, respectively. The vector backbone containing the Trp selection marker was obtained from the plasmid pCAS1yl-Trp and these above plasmids were constructed by Gibson assembly method [38]. The restriction enzyme XbaI was used to linearize the plasmid pZPG. All plasmids were listed in Table S3. Primers (Table S4) and the heterologous genes were synthesized by TsingKe (Beijing, China).
In some experiments, the URA3 selection marker was recycled through the Cre-loxP system which was achieved by transformation of the episomal plasmid pUB4-Cre; and the knockout of Trp gene was achieved by CRISPR/Cas9 system. The lithium acetate method was used for the transforming of Y. lipolytica [39].
2.3. Analytical methods
The concentration of 2′-FL, glucose, and lactose were detected by HPLC. The Shimadzu LC-20AT and a Rezex ROA Organic Acid H + (8 %) Column were used. The column temperature was set at 65 °C and the refractive index detector temperature was set at 40 °C. 5 mM H2SO4 was used as the mobile phase (0.6 mL/min). The extracellular 2′-FL and lactose were measured from the supernatant of the fermentation broth. To determine intracellular 2′-FL production, 1 mL of fermentation broth was centrifuged (12,000 g, 2 min), collected, washed, and the sediment was resuspended with distilled water, and crushed by high-speed grinding, then boiled for 10 min to lyse the cells, and then the supernatant was collected (12,000 g, 5 min) and detected by HPLC. The cells were collected, dried and weighed to get the dry cell weight (DCW) and the optical density at 600 nm (OD600) was used to evaluate the cell growth. The total 2′-FL concentration is the sum of the extracellular 2′-FL concentration and the intracellular 2′-FL concentration.
3. Results and discussion
3.1. Construction of the 2′-FL biosynthesis pathway and promoting its synthesis by fusion protein tags
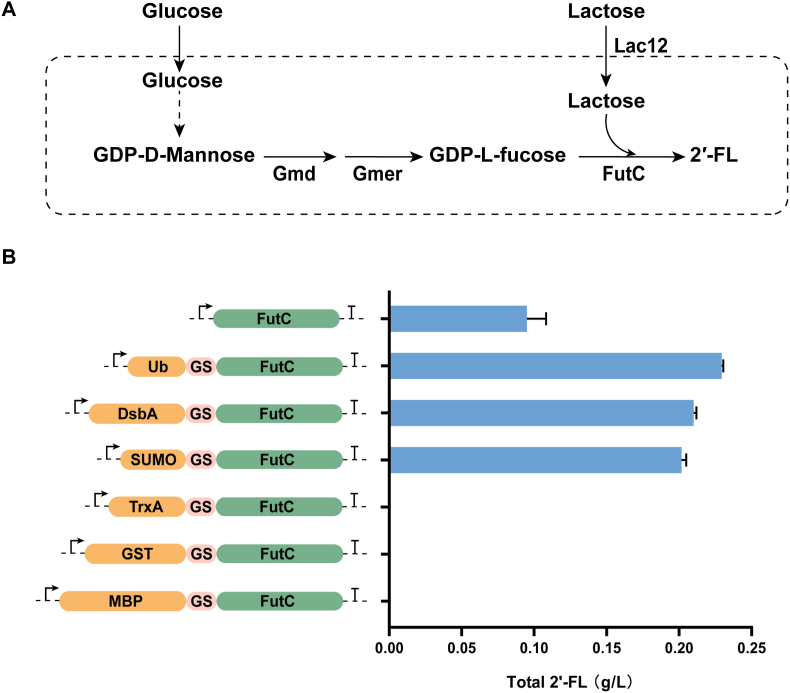
Four exogenous enzymes, Lac12 (XP_452193), Gmd (ADC54120), Gmer (WP_001002442), and FutC (ABO61750), were introduced into Yarrowia lipolytica PO1f (Fig. 1A) to obtain a 2′-FL-producing strain. Lac12 from Kluyveryomyces lactis was introduced into Y. lipolytica to enable it to transport lactose from the extracellular to the intracellular; Gmd from Mortierella alpina and Gmer from Helicobacter pylori were employed to synthesize the precursor GDP-l-fucose; FutC from Helicobacter pylori 26695 was selected to catalyze the last step reaction of 2′-FL synthesis because, in Y. lipolytica, it was proven to be more appropriate for the production of 2′-FL [10,11,13]. MaGMD, hpGMER, klLAC12, and hpFutC were overexpressed using strong promoter PEXP1, promoter PGPD, promoter PTEFin, and promoter Put8, respectively. The strain F0 was obtained after transformation with the episomal plasmid pLEP-hpFutC-GGL containing the above genes, in which 0.10 g/L 2′-FL was detected (Fig. 1B), whereas no 2′-FL was detected in PO1f.
Fig. 1.
Effect of expression of FutC fused with different protein tags on 2′-FL titer. (A) Schematic digram of the 2′-FL biosynthetic pathway. (B) Effect of different protein tags on 2′-FL production. The data was derived from the results of three repeated experiments.
It has been proved that suitable protein tags can improve the expression of target protein [40,41] and maintain the stability of protein [42]. Hence, we fused FutC with various protein tags including SUMO, TrxA, DsbA, GST, Ub and MBP to optimize its expression and ultimately achieve the purpose of enhancing 2′-FL synthesis. The protein tags were attached to FutC via a GS linker and transformed into yeast. The results showed that the strains expressing FutC fused with SUMO tag (F0-SUMO), Ub-tag (F0-Ub), and DsbA-tag (F0-DsbA) can produce 2′-FL, with 0.20 g/L, 0.23 g/L, and 0.21 g/L 2′-FL, respectively, while the strains expressing FutC fused with other protein tags did not accumulate 2′-FL (Fig. 1B).
In some reports on 2′-FL biosynthesis, protein tags have been used to optimize the expression of the key enzyme FutC and thus promote 2′-FL synthesis. Lin et al. tried several protein tags to increase the intensity of FutC expression in E. coli, including TrxA, MBP, SUMO, and NusA, and finally found that the strain expressing TrxA-futC had the highest 2′-FL titer, showing a 1.7-fold increase over the untagged FutC strain [10]; Protein tags have not only been shown to have a promotional effect on FutC expression in prokaryotes, but are equally effective in eukaryotes. Hollands et al. found that expression of FutC fused with SUMO tags increased the 2′-FL titer of S. cerevisiae by 30 %, and increased the 2′-FL titer of Y. lipolytica by about 10-fold [13]. In this study, the expression of FutC fused with Ub tag, SUMO tag and DsbA tag clearly promoted 2′-FL accumulation, especially for Ub tag. Our research demonstrated that fusing FutC with a protein tag could indeed facilitate the accumulation of more 2′-FL. This might be due to the fact that the introduction of protein tags has certain effects on the expression level or stability of FutC. However, no 2′-FL was detected in strains expressing FutC fused with TrxA, MBP, and GST tags, and we hypothesized that there could be several reasons for this. First, although TrxA, MBP, and GST can promote the expression of FutC in E. coli, and the effect of the TrxA tag is very significant, due to the differences between prokaryotes and eukaryotes in the process of translation and modification of proteins, these tags may not be suitable for promoting the accumulation of 2′-FL in Y. lipolytica. Second, the fusion of TrxA, GST, and MBP may result in decreased expression or stability of FutC, resulting in low production of 2′-FL that could not be detected by HPLC.
The 2′-FL accumulation in the strain F0-Ub was 1.15 times than that in the F0-SUMO strain, indicating that Ub tag is another protein tag besides SUMO tag that can promote the synthesis of 2′- FL in Y. lipolytica. Ubiquitin is a stable folded protein with a small molecular weight and has a variety of functions in cells [43]. In E. coli, the α-helical antimicrobial peptide A20L fused with the Ub tag had a higher expression level than of that fused with the SUMO tag [44]. In our research, the strains expressing Ub-FutC were able to produce more 2′-FL, which we speculated might be due to the fact that the fusion of the Ub tag could promote the functional expression of FutC or help maintain the stability of FutC. However, low copy episomal plasmids were used to keep the copy number of the genes encoding the different fusion proteins equal in the cells, resulting in low expression of the FutC, making it difficult to further verify the expression of FutC at the protein level. Our study found that the Ub tag fusion could promote 2′-FL accumulation, and even exhibited better effects than the SUMO tag. It provides a new optimization method for the expression of exogenous proteins in Y. lipolytica. Therefore, the Ub tag was chosen for optimizing FutC expression in Y. lipolytica.
3.2. Iterative integration of the 2′-FL synthesis pathway
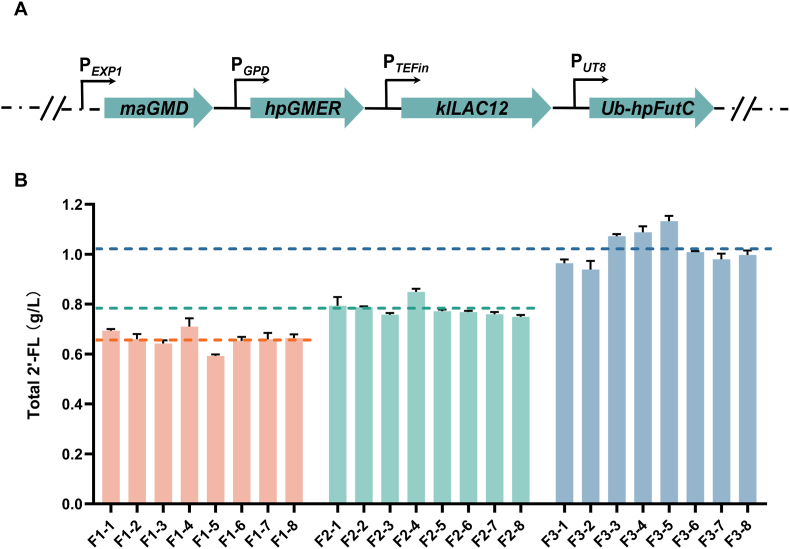
To obtain strains stably producing 2′-FL, the integration plasmid pULUF-GG was constructed and transformed into Y. lipolytica PO1f, and the best transformant F1-4 producing 0.71 g/L 2′-FL was obtained (Fig. 2A and B).
Fig. 2.
Enhanced 2′-FL production by iterative integration. (A) Plasmid construction for co-overexpression of Ub-hpFutC, klLAC12, maGMD and hpGMER. (B) 2′-FL production in strains obtained after the first, second, and third rounds of plasmid integration, respectively. The dashed line indicates the average level of 2′-FL production for each group of strains. The data was derived from the results of three repeated experiments.
It has been shown that multicopy integration of related genes can promote 2′-FL accumulation [10,45]. Iterative integration is an effective method for multicopy integration. For example, in Y. lipolytica, the synthesis of β-carotene has been greatly enhanced by the iterative integration of related enzymes [46]. Therefore, to promote the 2′-FL accumulation, we performed a second round of integration of 2′-FL synthesis-related genes. First, the URA3 selection marker of the strain F1-4 was recycled through the Cre-LoxP system to obtain the strain F1-4Δura3. Then, the linearized plasmid pULUF-GG was transformed into the strain F1-4Δura3 for the second round of integration, and 8 transformed strains were selected for shake-flask fermentation. Here, 0.85 g/L 2′-FL was produced by the strain F2-4 (Fig. 2B), and it was about 19.7 % higher than that of the strain F1-4. Compared with the F1-series strains obtained after the first round of integration, the F2-series strains obtained after the second round of integration showed an increase in average 2′-FL production (Fig. 2B), indicating that iterative integration did promote the 2′-FL accumulation. Then, a third round of integration was performed based on the strain F2-4.
For the third round of integration, the plasmid pHLUF-GG containing the selection marker of hyg was linearized and transformed into the strain F2-4 for random integration into the genome. The average 2′-FL production of the F3 series strains was significantly improved again. We obtained the strain F3-5 with 1.13 g/L 2′-FL (Fig. 2B). The titer of the strain F3-5 showed about 32.9 % higher than that of the strain F2-4.
The results showed that iterative integration significantly improved the titer of the product. Iterative integration can increase gene copy numbers and thus improve the expression of the related enzymes. Increased expression of Gmd and Gmer might drive more GDP-l-fucose synthesis, which, combined with high levels of FutC expression, leads to more 2′-FL accumulation.
3.3. Increasing the precursor GDP-d-mannose supply
As the direct precursor of 2′-FL, GDP-l-fucose was synthesized by Gmd and Gmer using GDP-d-mannose as a precursor. Studies have shown that increasing the supply of GDP-d-mannose could enhance 2′-FL production [33]. Thus, we overexpressed the GDP-d-mannose synthesis pathway in hopes of achieving the target of promoting 2′-FL synthesis.
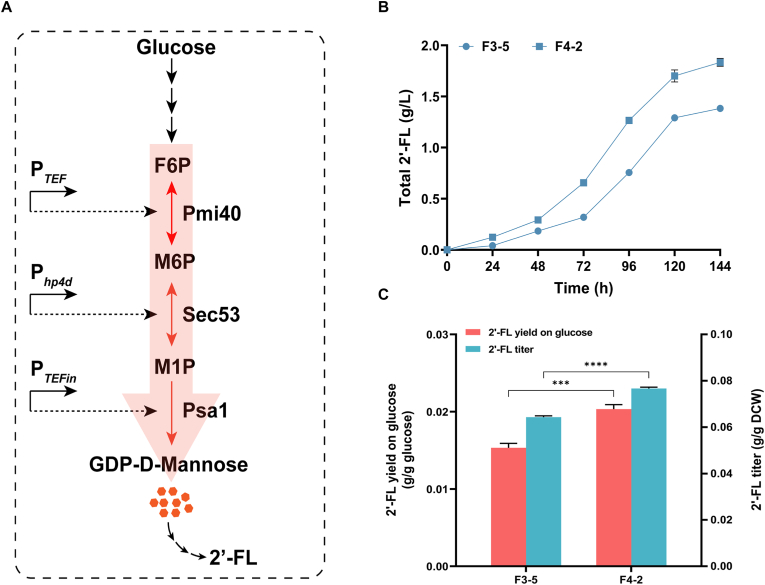
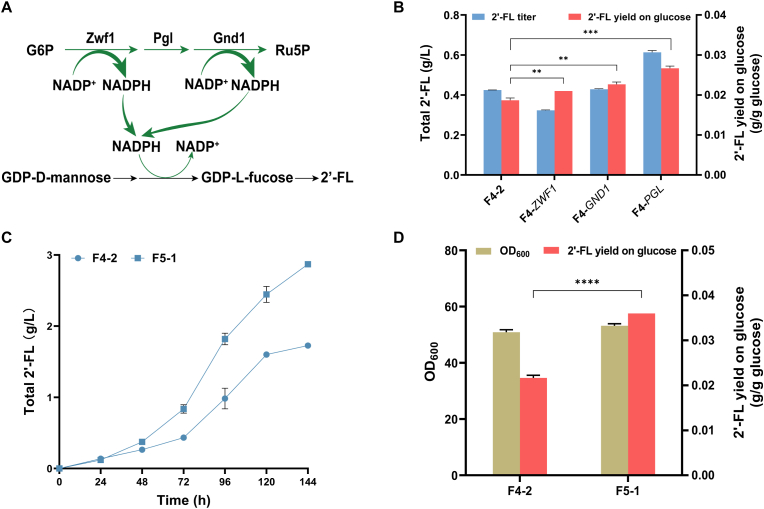
GDP-d-mannose can be produced from fructose-6-phosphate through the catalysis of three enzymes: Pmi40, Sec53, and Psa1, encoded by PMI40, SEC53, and PSA1, respectively. Therefore, the GDP-d-mannose synthesis pathway was enhanced by simultaneously overexpressing PMI40, SEC53, and PSA1 to facilitate 2′-FL synthesis. PMI40, SEC53, and PSA1 were controlled by PTEF, Php4d, and PTEFin, respectively (Fig. 3A) and the linearized plasmid pMAN was transformed into the strain F3-5, to generate strains F4-1 to F4-8, respectively. As shown in Fig. 3B, the strain F4-2 produced 1.84 g/L 2′-FL, which showed a significant increase compared with the strain F3-5. Compared with the strain F3-5 (0.064 g/g DCW), the 2′-FL yield of the strain F4-2 (0.077 g/g DCW) was increased by 20.3 %, and the 2′-FL yield on glucose of the strain F4-2 was also increased by 33.3 % (Fig. 3C), indicating that overexpression of PMI40, SEC53, and PSA1 under the control of strong promoters resulted in a greater carbon flux for the GDP-d-mannose synthesis, and that enhancing the precursor synthesis pathway is an effective approach to promoting 2′-FL synthesis. The GDP-d-mannose synthesis pathway is equivalent to the competition pathway of glycolysis pathway. The enhancement of the GDP-d-mannose synthesis pathway not only promoted the synthesis of downstream products, but also did not exert pressure on the growth of the strain, indicating that Y. lipolytica were able to maintain a balance between growth and production.
Fig. 3.
Enhanced 2′-FL production by enhancing GDP-d-mannose synthesis pathway. (A) Schematic view of the strategies to strengthen GDP-d-mannose synthesis by co-overexpressing of PMI40, SEC53, and PSA1. (B) 2′-FL production of the strains F3-5 and the strain F4-2. (C) 2′-FL yield on glucose and 2′-FL titer (g/g DCW) of the strain F3-5 and the strain F4-2. The data was derived from the results of three repeated experiments. T-test was performed to determine p values. ***p < 0.001; ****p < 0.0001.
3.4. Enhancement of NADPH regeneration pathway
It has been reported that 2′-FL production can be promoted by regulating the regeneration of NADPH [30]. Previous research has shown that overexpression of ZWF1 could facilitate NADPH regeneration and has a positive effect on 2′-FL accumulation [10]. Thus, to further enhance the synthesis of 2′-FL, we optimized the NADPH regeneration pathway based on the strain F4-2. For NADPH regeneration, ZWF1, PGL, and GND1 in the pentose phosphate pathway (PPP) were respectively overexpressed (Fig. 4A).
Fig. 4.
The enhancement of NADPH regeneration pathway promotes 2′-FL accumulation. (A) Schematic view of the enhanced NADPH regeneration pathway. (B) 2′-FL production and 2′-FL yield on glucose of the strains overexpressing ZWF1, PGL, and GND1, respectively. (C) 2′-FL titer of the strain F4-2 and the strain F5-1. (D) 2′-FL yield on glucose and OD600 of the strain F4-2 and the strain F5-1. The data was derived from the results of three repeated experiments. T-test was performed to determine p values. **p < 0.01; ***p < 0.001; ****p < 0.0001.
Overexpression of individual genes resulted in different levels of 2′-FL production. Overexpression of Pgl increased the titer of 2′-FL by 44.5 % compared to the control strain F4-2 (Fig. 4B). Pgl catalyzes the conversion of 6-Phospho-D-glucono-1,5-lactone to 6-phospho-d-gluconate, which is the second reaction in the PPP. There are two configurations of 6-Phospho-D-glucono-1,5-lactone, namely delta and gamma. The oxidation product of glucose 6-phosphate is delta, and delta can be converted to gamma through intramolecular rearrangement, which is the “dead end” of the branch, and Pgl accelerates the hydrolysis of the delta to produce 6-phospho-d-gluconate, thereby preventing it from converting to the gamma to ensure the normal operation of the PPP [47]. The overexpression of PGL in Y. lipolytica might facilitate the formation of 6-phospho-d-gluconate and prevent further formation of the gamma form, thereby promoting the regeneration of NADPH. This result also indicated the importance of Pgl for the PPP.
In most reports, NADPH regeneration was mainly achieved by overexpressing of ZWF1 and GND1 rather than PGL, because Zwf1 and Gnd1 can directly catalyze NADPH generation [48]. However, here, increasing ZWF1 and GND1 expression did not appear to further increase 2′-FL production (Fig. 4B). This may be because the activity of Zwf1 and Gnd1 was already relatively high, and the overexpression of these two genes put some pressure on cell metabolism, affecting cell growth (Fig. S1). However, we found that the 2′-FL yield on glucose was 0.021 and 0.023 g/g glucose for strains F4-ZWF1 and F4-GND1, respectively, while it was 0.019 g/g glucose for the strain F4-2 (Fig. 4B), indicating that the enhancement of ZWF1 and GND1 still had a positive effect on 2′-FL synthesis. Therefore, we concluded that the overexpression of ZWF1, PGL and GND1 indeed promoted the regeneration of NADPH.
To stably facilitate NADPH regeneration, the co-overexpression of ZWF1, PGL, and GND1 was performed in the strain F4-2 to obtain strains F5-1 to F5-8. 2.87 g/L 2′-FL was produced by the strain F5-1 (Fig. 4C). The maximum OD600 of the strain F5-1 was essentially identical as that to the strain F4-2, while the 2′-FL yield on glucose of the strain F5-1 (0.036 g/g glucose) was superior to that of the strain F4-2 (0.022 g/g glucose) (Fig. 4D), indicating that the co-overexpression of ZWF1, PGL, and GND1 might further enhance the NADPH supply and then promote more carbon flow to the synthesis of 2′-FL.
3.5. Efficient production of 2′-FL by fed-batch fermentation
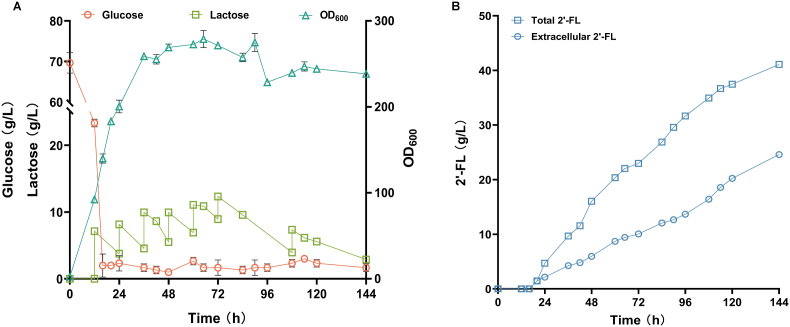
The capacity of the strain F5-1 for the production of 2′-FL was evaluated in a 5 L bioreactor. After 16 h, all of the initial glucose had been used up. To prevent the accumulation of by-products such as mannitol due to a hyperosmotic environment, glucose was added with the rate of ∼4 g/L/h to ensure that the glucose does not accumulate in the medium (Fig. 5A). The addition of lactose was started when the OD600 reached approximately 85, and the content of lactose in the medium was detected by HPLC (Fig. 5A). After the addition of lactose, 2′-FL production increased at a relatively steady rate. The maximum titer of 2′-FL reached 41.10 g/L (intracellular 16.52 g/L, extracellular 24.58 g/L) after 144 h (Fig. 5B). Although it did not reach the level of 2'-FL produced by E. coli and B. subtilis, it was the highest level of 2′-FL produced by Y. lipolytica to date. The 2′-FL yield on lactose of the strain F5-1 after 5 L fed-batch fermentation was about 0.98 mol/mol, which means that almost all of the absorbed lactose was used for 2′-FL synthesis. This also shows the advantage of 2′-FL production by Y. lipolytica, which is able to convert almost all of the lactose absorbed into 2′-FL instead of consuming it through other pathways. In addition, the 2′-FL content (g/g DCW) reached 0.47, which is higher than previously reported [13]. This indicates that the optimization strategy adopted in this study improves the efficiency of 2′-FL synthesis in Y. lipolytica. The OD600 was maintained between 230 and 270 after 36 h, indicating that the strain has good capability for continuous production, and is suitable for long-term and large-scale fermentation. These findings demonstrate that Y. lipolytica is a promising platform for 2′-FL synthesis and has good potential for industrial application.
Fig. 5.
Biosynthesis of 2’ -FL by fed-batch fermentation. (A) Cell density (OD600), glucose concentration and lactose concentration in the culture medium during fermentation. (B) 2’ -FL production during fermentation.
According to the fermentation results, the ratio of extracellular 2′-FL to intracellular 2'-FL was 1.49:1, and about 40 % of 2′-FL remained in the cells. This is different from previous reports that most of the 2′-FL accumulates intracellular in Y. lipolytica [13]. We hypothesized that there may be endogenous transporters that can help 2′-FL efflux in Y. lipolytica. The expression of appropriate transporters can effectively improve the efflux efficiency of 2′-FL [49]. By screening endogenous transporters and optimizing their expression, the production of 2′-FL may be further improved. In addition, the ratio of extracellular and intracellular 2′-FL was also greatly affected by different media. Lee et al. suggested that the activity of endogenous transporters may be different in different media [31]. Therefore, it is possible that the culture medium affected the activity of the endogenous 2′-FL transporter, which caused 40 % of the 2′-FL residues in intracellular. The findings suggest that Y. lipolytica has the potential to enhance 2′-FL production. By screening the endogenous 2′-FL transporter and regulating its expression, it will further promote the efflux of 2′-FL and possibly promote the synthesis of 2′-FL to obtain higher titer.
4. Conclusions
To further develop the application of Y. lipolytica in producing oligosaccharides, the strain F5-1 with highly efficient production of 2′-FL was constructed. By fusing Ub tag to α-1, 2-fucosyltransferase, the 2′-FL accumulation was promoted. Subsequently, the 2′-FL synthesis pathway was strengthened by the strategy of iterative integration. To increase the supply of precursors, the GDP-d-mannose synthesis pathway was overexpressed. Finally, the NADPH regeneration was facilitated by co-overexpressing of ZWF1, PGL, and GND1, further promoting the synthesis of 2′-FL. The 2′-FL titer reached 41.10 g/L by fed-batch fermentation, demonstrating that Y. lipolytica is a potential platform for oligosaccharide production, and thus laying the foundation for large-scale industrial production of 2′-FL.
CRediT authorship contribution statement
Yan Zhang: Conceptualization, Investigation, Formal analysis, Writing – original draft. Xuejing Zhang: Investigation, Writing – review & editing. Haiyan Liu: Investigation, Formal analysis. Jin Hou: Methodology, Writing – review & editing. Mengmeng Liu: Formal analysis, Writing – review & editing. Qingsheng Qi: Writing – review & editing, Conceptualization, Visualization.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Chengjia Zhang of the Core Facilities for Life and Environmental Sciences, State Key laboratory of Microbial Technology of Shandong University for his assistance in the performance of the fed-batch fermentation experiments. This work was supported by the Key R&D Program of Shandong Province (2020CXGC010602).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2023.11.002.
Contributor Information
Mengmeng Liu, Email: liummeng@sdu.edu.cn.
Qingsheng Qi, Email: qiqingsheng@sdu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eiwegger T., Stahl B., Schmitt J., Boehm G., Gerstmayr M., Pichler J., Dehlink E., Loibichler C., Urbanek R., Szepfalusi Z. Human milk--derived oligosaccharides and plant-derived oligosaccharides stimulate cytokine production of cord blood T-cells in vitro. Pediatr Res. 2004;56:536–540. doi: 10.1203/01.PDR.0000139411.35619.B4. [DOI] [PubMed] [Google Scholar]
- 3.Zuurveld M., van Witzenburg N.P., Garssen J., Folkerts G., Stahl B., Van't Land B., Willemsen L.E.M. Immunomodulation by human milk oligosaccharides: the potential role in prevention of allergic diseases. Front Immunol. 2020;11:801. doi: 10.3389/fimmu.2020.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobata A. Structures and application of oligosaccharides in human milk. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:731–747. doi: 10.2183/pjab.86.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandenplas Y., Berger B., Carnielli V.P., Ksiazyk J., Lagstrom H., Sanchez Luna M., Migacheva N., Mosselmans J.M., Picaud J.C., Possner M., Singhal A., Wabitsch M. Human milk oligosaccharides: 2'-fucosyllactose (2'-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients. 2018;10 doi: 10.3390/nu10091161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agoston K., Hederos M.J., Bajza I., Dekany G. Kilogram scale chemical synthesis of 2'-fucosyllactose. Carbohydr Res. 2019;476:71–77. doi: 10.1016/j.carres.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Faijes M., Castejon-Vilatersana M., Val-Cid C., Planas A. Enzymatic and cell factory approaches to the production of human milk oligosaccharides. Biotechnol Adv. 2019;37:667–697. doi: 10.1016/j.biotechadv.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Petschacher B., Nidetzky B. Biotechnological production of fucosylated human milk oligosaccharides: prokaryotic fucosyltransferases and their use in biocatalytic cascades or whole cell conversion systems. J Biotechnol. 2016;235:61–83. doi: 10.1016/j.jbiotec.2016.03.052. [DOI] [PubMed] [Google Scholar]
- 9.Liao L., Huang H., Wang Y., Du G., Kang Z. Yeast surface display of leech hyaluronidase for the industrial production of hyaluronic acid oligosaccharides. Engineering Microbiology. 2023;3 [Google Scholar]
- 10.Lin L., Gong M., Liu Y., Li J., Lv X., Du G., Liu L. Combinatorial metabolic engineering of Escherichia coli for de novo production of 2'-fucosyllactose. Bioresour Technol. 2022;351 doi: 10.1016/j.biortech.2022.126949. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q., Liu Z., Xia H., Huang Z., Zhu Y., Xu L., Liu Y., Li J., Du G., Lv X., Liu L. Engineered Bacillus subtilis for the de novo production of 2'-fucosyllactose. Microb Cell Factories. 2022;21:110. doi: 10.1186/s12934-022-01838-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S., Liu J.J., Yun E.J., Kwak S., Kim K.H., Jin Y.S. Production of a human milk oligosaccharide 2'-fucosyllactose by metabolically engineered Saccharomyces cerevisiae. Microb Cell Factories. 2018;17:101. doi: 10.1186/s12934-018-0947-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollands K., Baron C.M., Gibson K.J., Kelly K.J., Krasley E.A., Laffend L.A., Lauchli R.M., Maggio-Hall L.A., Nelson M.J., Prasad J.C., Ren Y., Rice B.A., Rice G.H., Rothman S.C. Engineering two species of yeast as cell factories for 2'-fucosyllactose. Metab Eng. 2019;52:232–242. doi: 10.1016/j.ymben.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.W., Kwak S., Liu J.J., Yun E.J., Jin Y.S. 2'-Fucosyllactose production in engineered Escherichia coli with deletion of waaF and wcaJ and overexpression of FucT2. J Biotechnol. 2021;340:30–38. doi: 10.1016/j.jbiotec.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Sun X., Peng Z., Li C., Zheng Y., Cheng Y., Zong J., Lu F., Li Y., Li Q. Combinatorial metabolic engineering and tolerance evolving of Escherichia coli for high production of 2'-fucosyllactose. Bioresour Technol. 2023;372 doi: 10.1016/j.biortech.2023.128667. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Zhu Y., Wang H., Chen R., Liu Y., Zhang W., Mu W. De novo biosynthesis of 2'-fucosyllactose in a metabolically engineered Escherichia coli using a novel a1,2-fucosyltransferase from Azospirillum lipoferum. Bioresour Technol. 2023;374 doi: 10.1016/j.biortech.2023.128818. [DOI] [PubMed] [Google Scholar]
- 17.Xu M., Sun M., Meng X., Zhang W., Shen Y., Liu W. Engineering pheromone-mediated quorum sensing with enhanced response output increases fucosyllactose production in Saccharomyces cerevisiae. ACS Synth Biol. 2023;12:238–248. doi: 10.1021/acssynbio.2c00507. [DOI] [PubMed] [Google Scholar]
- 18.Qiu Y., Wu M., Bao H., Liu W., Shen Y. Engineering of Saccharomyces cerevisiae for co-fermentation of glucose and xylose: current state and perspectives. Engineering Microbiology. 2023;3 [Google Scholar]
- 19.Cui Z., Gao C., Li J., Hou J., Lin C.S.K., Qi Q. Engineering of unconventional yeast Yarrowia lipolytica for efficient succinic acid production from glycerol at low pH. Metab Eng. 2017;42:126–133. doi: 10.1016/j.ymben.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Liu M., Zhang J., Ye J., Qi Q., Hou J. Morphological and metabolic engineering of Yarrowia lipolytica to increase beta-carotene production. ACS Synth Biol. 2021;10:3551–3560. doi: 10.1021/acssynbio.1c00480. [DOI] [PubMed] [Google Scholar]
- 21.Liu P., Zhang T., Zheng Y., Li Q., Su T., Qi Q. Potential one-step strategy for PET degradation and PHB biosynthesis through co-cultivation of two engineered microorganisms. Engineering Microbiology. 2021;1 [Google Scholar]
- 22.Yuzbasheva E.Y., Agrimi G., Yuzbashev T.V., Scarcia P., Vinogradova E.B., Palmieri L., Shutov A.V., Kosikhina I.M., Palmieri F., Sineoky S.P. The mitochondrial citrate carrier in Yarrowia lipolytica: its identification, characterization and functional significance for the production of citric acid. Metab Eng. 2019;54:264–274. doi: 10.1016/j.ymben.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Yu X., Wang K., Lin L., Liu H.H., Ledesma-Amaro R., Ji X.J. Reprogramming the fatty acid metabolism of Yarrowia lipolytica to produce the customized omega-6 polyunsaturated fatty acids. Bioresour Technol. 2023;383 doi: 10.1016/j.biortech.2023.129231. [DOI] [PubMed] [Google Scholar]
- 24.Wang G., Olofsson-Dolk M., Hansson F.G., Donati S., Li X., Chang H., Cheng J., Dahlin J., Borodina I. Engineering yeast Yarrowia lipolytica for methanol assimilation. ACS Synth Biol. 2021;10:3537–3550. doi: 10.1021/acssynbio.1c00464. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Liu X., Chen B., Liu W., Guo Z., Liu X., Zhu X., Liu J., Zhang J., Li J., Zhang L., Gao Y., Zhang G., Wang Y., Choudhary M.I., Yang S., Jiang H. Metabolic engineering of Yarrowia lipolytica for scutellarin production. Synthetic and Systems Biotechnology. 2022;7:958–964. doi: 10.1016/j.synbio.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobrowolski A., Drzymala K., Mitula P., Mironczuk A.M. Production of tailor-made fatty acids from crude glycerol at low pH by Yarrowia lipolytica. Bioresour Technol. 2020;314 doi: 10.1016/j.biortech.2020.123746. [DOI] [PubMed] [Google Scholar]
- 27.Duan X.Y., Tian Y., Song Z.Q., Song L.P., Lin W.B., Wang C., Yang H., Lu X.Y., Ji X.J., Liu H.H. High-level de novo biosynthesis of cordycepin by systems metabolic engineering in Yarrowia lipolytica. Bioresour Technol. 2022;363 doi: 10.1016/j.biortech.2022.127862. [DOI] [PubMed] [Google Scholar]
- 28.Bankar A.V., Kumar A.R., Zinjarde S.S. Environmental and industrial applications of Yarrowia lipolytica. Appl Microbiol Biotechnol. 2009;84:847–865. doi: 10.1007/s00253-009-2156-8. [DOI] [PubMed] [Google Scholar]
- 29.Chin Y.W., Kim J.Y., Lee W.H., Seo J.H. Enhanced production of 2'-fucosyllactose in engineered Escherichia coli BL21star(DE3) by modulation of lactose metabolism and fucosyltransferase. J Biotechnol. 2015;210:107–115. doi: 10.1016/j.jbiotec.2015.06.431. [DOI] [PubMed] [Google Scholar]
- 30.Huang D., Yang K., Liu J., Xu Y., Wang Y., Wang R., Liu B., Feng L. Metabolic engineering of Escherichia coli for the production of 2′-fucosyllactose and 3-fucosyllactose through modular pathway enhancement. Metab Eng. 2017;41:23–38. doi: 10.1016/j.ymben.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Lee J.W., Kwak S., Liu J.J., Yu S., Yun E.J., Kim D.H., Liu C., Kim K.H., Jin Y.S. Enhanced 2'-Fucosyllactose production by engineered Saccharomyces cerevisiae using xylose as a co-substrate. Metab Eng. 2020;62:322–329. doi: 10.1016/j.ymben.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Li M., Li C., Hu M., Zhang T. Metabolic engineering strategies of de novo pathway for enhancing 2'-fucosyllactose synthesis in Escherichia coli. Microb Biotechnol. 2022;15:1561–1573. doi: 10.1111/1751-7915.13977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M., Meng X., Zhang W., Shen Y., Liu W. Improved production of 2'-fucosyllactose in engineered Saccharomyces cerevisiae expressing a putative alpha-1, 2-fucosyltransferase from Bacillus cereus. Microb Cell Factories. 2021;20:165. doi: 10.1186/s12934-021-01657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Zhu Y., Wan L., Chen R., Zhang W., Mu W. High-level de novo biosynthesis of 2'-fucosyllactose by metabolically engineered Escherichia coli. J Agric Food Chem. 2022;70:9017–9025. doi: 10.1021/acs.jafc.2c02484. [DOI] [PubMed] [Google Scholar]
- 35.Cui Z.Z.H., Zhang J., Jiang Z., Zhu Z., Liu X., Qi Q., Hou J. A CRISPR/Cas9-Mediated, homology-independent tool developed for targeted genome integration in Yarrowia lipolytica. Appl Environ Microbiol. 2021;87:6. doi: 10.1128/AEM.02666-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui Z., Jiang X., Zheng H., Qi Q., Hou J. Homology-independent genome integration enables rapid library construction for enzyme expression and pathway optimization in Yarrowia lipolytica. Biotechnol Bioeng. 2019;116:354–363. doi: 10.1002/bit.26863. [DOI] [PubMed] [Google Scholar]
- 37.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 38.Gao S., Tong Y., Wen Z., Zhu L., Ge M., Chen D., Jiang Y., Yang S. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J Ind Microbiol Biotechnol. 2016;43:1085–1093. doi: 10.1007/s10295-016-1789-8. [DOI] [PubMed] [Google Scholar]
- 39.Chen D.C.B.J., Gaillardin C. One-step transformation of the dimorphic yeast Yarrowia lipolytica. Appl Microbiol Biotechnol. 1997;48:232–235. doi: 10.1007/s002530051043. [DOI] [PubMed] [Google Scholar]
- 40.Kuo D.N.M., Courey A.J. SUMO as a solubility tag and in vivo cleavage of SUMO fusion proteins with Ulp1. Methods Mol Biol. 2014;1177:71–80. doi: 10.1007/978-1-4939-1034-2_6. [DOI] [PubMed] [Google Scholar]
- 41.Xu M., Xie W., Luo Z., Li C.-X., Hua Q., Xu J.-H. Improving solubility and copy number of taxadiene synthase to enhance the titer of taxadiene in Yarrowia lipolytica. Synthetic and Systems Biotechnology. 2023;8:331–338. doi: 10.1016/j.synbio.2023.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malakhov M.P.M.M., Malakhova O.A., Drinker M., Weeks S.D., Butt T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genom. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 43.Akimoto G., Fernandes A.P., Bode J.W. Site-Specific protein ubiquitylation using an engineered, chimeric E1 activating enzyme and E2 SUMO conjugating enzyme Ubc9. ACS Cent Sci. 2022;8:275–281. doi: 10.1021/acscentsci.1c01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi T., Sun S., Huang Y., Chen Y. Prokaryotic expression and mechanism of action of alpha-helical antimicrobial peptide A20L using fusion tags. BMC Biotechnol. 2015;15:69. doi: 10.1186/s12896-015-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., Zhu Y., Wan L., Guang C., Mu W. Pathway optimization of 2'-fucosyllactose production in engineered Escherichia coli. J Agric Food Chem. 2021;69:1567–1577. doi: 10.1021/acs.jafc.0c07224. [DOI] [PubMed] [Google Scholar]
- 46.Gao S., Tong Y., Zhu L., Ge M., Zhang Y., Chen D., Jiang Y., Yang S. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous beta-carotene production. Metab Eng. 2017;41:192–201. doi: 10.1016/j.ymben.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Miclet E., Stoven V., Michels P.A., Opperdoes F.R., Lallemand J.Y., Duffieux F. NMR spectroscopic analysis of the first two steps of the pentose-phosphate pathway elucidates the role of 6-phosphogluconolactonase. J Biol Chem. 2001;276:34840–34846. doi: 10.1074/jbc.M105174200. [DOI] [PubMed] [Google Scholar]
- 48.Ahmad I., Shim W.Y., Jeon W.Y., Yoon B.H., Kim J.H. Enhancement of xylitol production in Candida tropicalis by co-expression of two genes involved in pentose phosphate pathway. Bioproc Biosyst Eng. 2012;35:199–204. doi: 10.1007/s00449-011-0641-9. [DOI] [PubMed] [Google Scholar]
- 49.Parschat K., Schreiber S., Wartenberg D., Engels B., Jennewein S. High-titer de novo biosynthesis of the predominant human milk oligosaccharide 2′-fucosyllactose from sucrose in Escherichia coli. ACS Synth Biol. 2020;9:2784–2796. doi: 10.1021/acssynbio.0c00304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.