Summary
From the beginning of the COVID-19 pandemic, children have exhibited different susceptibility to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, reinfection, and disease compared with adults. Motivated by the established significance of SARS-CoV-2-neutralizing antibodies in adults, here we characterize SARS-CoV-2-specific antibody repertoires in a young cohort of individuals aged from 5 months to 18 years old. Our results show that neutralizing antibodies in children possess similar genetic features compared to antibodies identified in adults, with multiple antibodies from children belonging to previously established public antibody clonotypes in adults. Notably, antibodies from children show potent neutralization of circulating SARS-CoV-2 variants that have cumulatively resulted in resistance to virtually all approved monoclonal antibody therapeutics. Our results show that children can rely on similar SARS-CoV-2 antibody neutralization mechanisms compared to adults and are an underutilized source for the discovery of effective antibody therapeutics to counteract the ever-evolving pandemic.
Keywords: antibodies, B cells, public clonotype, SARS-CoV-2, neutralization, virus, infection, children
Graphical abstract
Highlights
-
•
Applied LIBRA-seq to isolate cross-reactive coronavirus (CoV) B cells from children
-
•
Identified a potent antibody against diverse SARS-CoV-2 variants, including XBB.1.5
-
•
CoV antibodies from children and adults exhibited similar/public sequence signatures
Wall et al. demonstrate that SARS-CoV-2-reactive antibodies isolated from children can potently neutralize a broad range of virus variants, including highly neutralization-resistant XBB1.5, and can exhibit similar (public) genetic signatures compared to antibodies isolated from adults.
Introduction
The human immune response to a pathogen depends on a wide variety of factors, including the history of prior exposure to the same or related pathogens, as well as the current state of an individual’s immune system. Compared with adults, children’s infection and vaccination histories are more limited, and their immune systems are not fully mature. As a result, substantial differences in disease phenotype can be observed for a variety of infectious diseases between children and adults.1,2,3 In particular, children tend to be more susceptible to severe disease phenotypes for several common infectious diseases, including influenza, respiratory syncytial virus, human metapneumovirus, and others.1,2,3 However, this trend appeared to be reversed in the case of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), with children experiencing significantly less severe disease phenotypes compared with adults.4,5 Adolescents also have less incidence of severe COVID-19, and those with severe disease exhibit lower rates of hospitalization, require shorter hospital stays, and have lower mortality than adults.6 While there have been some efforts aiming to understand these differences in how children and adults respond to SARS-CoV-2 infection, little is still known about the specific factors that are responsible for this phenomenon.
While a number of immune system components have been implicated in the response to SARS-CoV-2 infection and vaccination,7,8,9 antibodies that are capable of effectively neutralizing the virus are among the most critical factors.10 In fact, the elicitation of neutralizing antibodies to the spike glycoprotein of SARS-CoV-2 is the primary target for the majority of current vaccines on the market.11,12 The initial SARS-CoV-2 vaccine candidates were developed based on the knowledge that neutralizing antibodies previously had been shown to protect against infection with a variety of other pathogens,13 long before ultimately showing that such antibodies are a correlate of protection against SARS-CoV-2 as well.10,14 Antibody responses to SARS-CoV-2 infection and vaccination have been extensively studied in adults, resulting in a plethora of information about the genetic, molecular, and functional characteristics of SARS-CoV-2-specific antibodies.8,12,15,16 While antibodies in adults have been found to target a variety of different epitopes on the spike glycoprotein, several epitope clusters have been identified that are commonly targeted at the population level.17 Importantly, the ability of different individuals to target similar epitopes is in part attributed to the existence of “public” antibodies with highly similar genetic features that are found in many individuals.18,19 In practice, public antibodies in the context of SARS-CoV-2 have been defined based on the use of identical combinations of variable heavy (VH) and variable light (VL) genes,8,20 although more stringent requirements related to high sequence identity in the third complementarity-determining regions of the heavy and light chains (CDRH3 and CDRL3, respectively) can also be incorporated.18,19 Overall, the identification of a variety of public antibodies indicates that the immune system in adults can converge on a few common modes of SARS-CoV-2 spike recognition that can effectively counteract the virus.
Within the first few months of the pandemic, monoclonal antibodies (mAbs) isolated from adults were translated into the clinic as both preventive and therapeutic measures.21,22,23,24,25,26,27,28 The critical significance of translating multiple mAbs with different genetic, epitope, and functional characteristics into the clinic became especially evident once the effects of SARS-CoV-2 variant evolution came to the forefront. While the approved mAbs were initially effective against the wild-type virus, mutations within the spike protein soon enabled viral escape from some, and now virtually all, mAbs on the market.29,30,31,32,33,34,35,36 Further efforts toward the development of next-generation mAbs with an expanded breadth of SARS-CoV-2 variant reactivity is currently ongoing.37,38,39
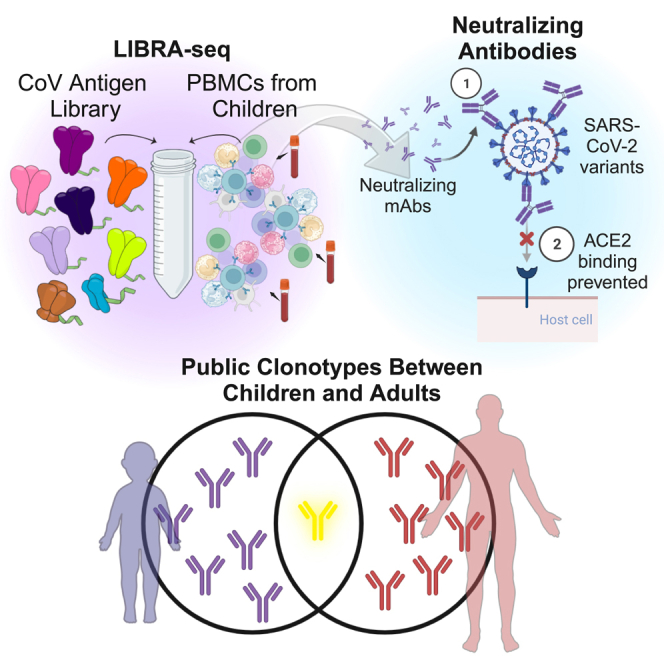
Unlike the expansive studies of antibody responses to SARS-CoV-2 in adults, there is little to no information about the types and specificities of neutralizing antibodies in children, with studies generally limited to the assessment of polyclonal responses.40,41,42 To address this, we employed LIBRA-seq (linking B cell receptor to antigen specificity through sequencing), a powerful tool that combines a DNA oligo barcoded antigen library, fluorescence-activated cell sorting (FACS), next-generation sequencing (NGS), and a computational pipeline to enable high-throughput mapping of antibody sequence to antigen specificity while maintaining single-cell resolution with high precision.43 Peripheral blood mononuclear cells (PBMCs) from children are generally challenging to acquire, and LIBRA-seq facilitated the utilization of limited samples to efficiently discover cross-reactive B cells predicted to bind the spike protein of SARS-CoV-2, its variants, and other betacoronaviruses. Due to the different disease phenotypes observed in children compared with adults and the established significance of the antibody response to SARS-CoV-2 in adults, we set out to explore the characteristics of SARS-CoV-2-specific antibodies in children. In this work, we aimed to delineate similarities and differences between SARS-CoV-2 antibodies in children and previously characterized antibody responses in adults, focusing on both genetic features and functional characteristics like epitope specificity and neutralization phenotypes. The results presented here highlight the properties of SARS-CoV-2-specific antibody responses in children and emphasize the potential of samples from children as an underutilized source for clinical mAb candidates and can help inform the design of next-generation pediatric vaccines.
Results
Cohort of samples from children
We obtained PBMCs from peripheral blood samples remaining after diagnostic tests were performed in the Clinical Hematology Laboratory at Vanderbilt University Medical Center (VUMC). Samples were collected for children up to 18 years of age between July and August 2021. Samples were divided into two groups: 71281, no known exposure to SARS-CoV-2 infection or vaccination, and 71282, known exposure to SARS-CoV-2 infection or vaccination, determined through self-reporting. Group 1 included a total of 44 samples, with ages ranging from 5 months to 17 years, and a median age of 9.5, while group 2 included a total of 12 samples, with ages ranging from 13 to 18 years, and a median age of 15.5 (Figure S1). Due to low sample volumes (ranging from 200 μL to 3 mL, average ∼1 mL), all samples within a group were pooled together for subsequent characterization.
Discovery of SARS-CoV-2 antibodies from children’s samples
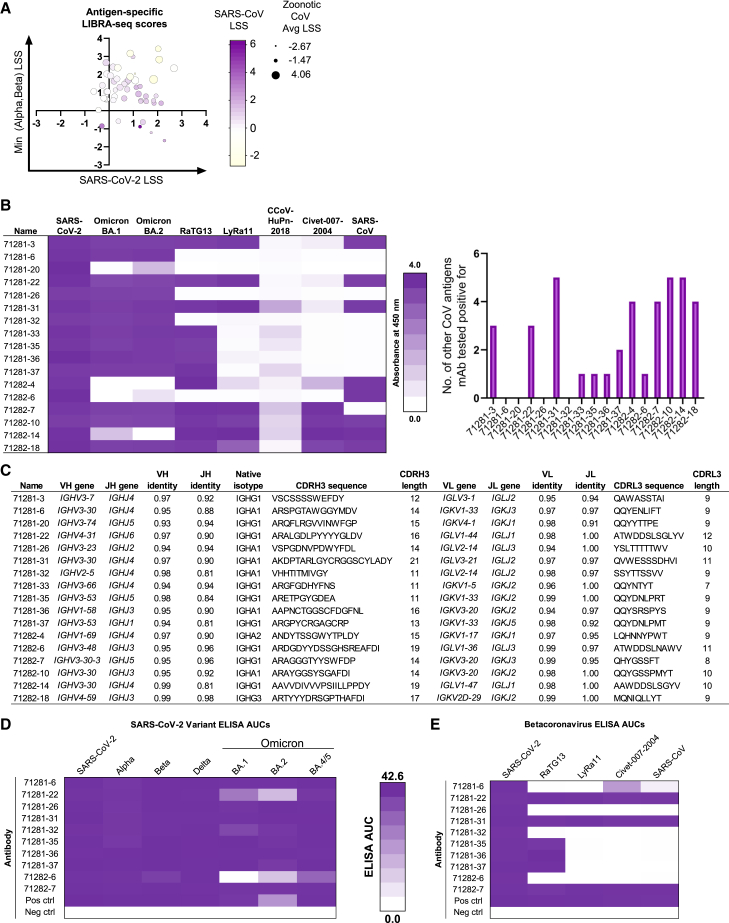
For each of the two sample groups, we performed a LIBRA-seq discovery experiment to determine the cross-reactivity patterns of SARS-CoV-2+ B cells from children. The antigen screening library incorporated a panel of stabilized recombinant spike antigens from SARS-CoV-2 variants (WA1, Alpha, and Beta) and from other sarbecoviruses with potential for zoonosis (SARS-CoV, RaTG13, LYRa11, and Civet-007-2004). Applying this antigen library to the two groups of samples described above resulted in the identification of a number of B cells with predicted cross-reactivity between multiple SARS-CoV-2 variants, multiple sarbecoviruses, or both (Figure 1A). Based on these LIBRA-seq results, a set of B cells were prioritized for further characterization as recombinant immunoglobulin G1 (IgG1) mAbs. Microscale expression of paired heavy-light chains (see STAR Methods), followed by antigenicity characterization by ELISA, revealed 17 antibodies with breadth within SARS-CoV-2 and/or other non-SARS-CoV-2 coronavirus recognition from each of the two groups of samples (Figure 1B). These antibodies derived from multiple different isotypes (IgG1, IgG3, and IgA1) and utilized diverse heavy- and light-chain variable genes with varying CDRH3 and CDRL3 lengths (CDRH3: 11–21 amino acids; CDRL3: 7–12 amino acids) and somatic hypermutation levels (heavy chain: 1%–7%; light chain: 1%–6%) (Figure 1C). Antibody leads were then expressed in large scale, purified, and tested for recognition of an expanded set of antigens. The antibodies showed diverse antigen specificities, with recognition of different subsets of SARS-CoV-2 variants, including the omicron lineage variants BA.1, BA.2, and BA.4/5, (Figures 1D and S2) or other sarbecoviruses (Figures 1E and S2). Together, these results revealed the existence of SARS-CoV-2 antibodies in samples from children with a wide range of breadth within SARS-CoV-2 and related sarbecoviruses.
Figure 1.
Using LIBRA-seq with a diverse CoV antigen panel to discover antibodies from children’s samples
(A) 59 IgG and IgA cells identified by LIBRA-seq are shown as circles, with their respective LIBRA-seq scores (LSSs) for SARS-CoV-2 (x axis), minimum score for the Alpha and Beta SARS-CoV-2 variants (y axis), and SARS-CoV (color heatmap) and average score for the potentially zoonotic CoV spikes used in the LIBRA-seq library (size of circle).
(B) Recombinant antibodies were produced by microexpression and screened by ELISA against a panel of recombinant CoV spike antigens. For the antibodies that were positive for SARS-CoV-2 spike binding, shown are (left) a binding heatmap for each antibody (rows) against a panel of SARS-CoV-2 variants and other coronaviruses (columns) and (right) the number of non-SARS-CoV-2 antigens (y axis) that each antibody (x axis) was able to recognize.
(C) For each antibody (rows), shown are the CDR amino acid sequences and lengths, V-gene and J-gene and percentage of nucleotide identities, and isotype (columns).
(D and E) ELISA area under the curve (AUC) values from purified monoclonal antibody binding to (D) SARS-CoV-2 variant spike proteins and (E) other betacoronavirus spike proteins are shown as heatmaps from minimum (white) to maximum (purple) binding. ELISA controls are described in Figure S2.
Antibodies from children neutralize recently emergent SARS-CoV-2 variants
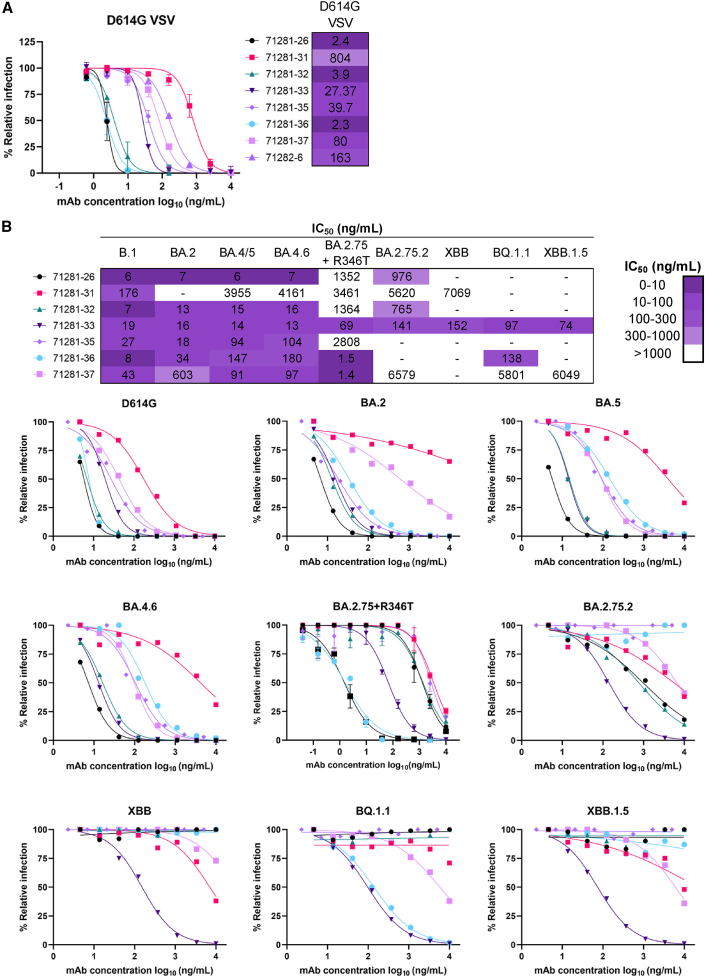
We next sought to explore whether the SARS-CoV-2 antibodies identified from children’s samples exhibit neutralization breadth and potency against diverse SARS-CoV-2 variants (Figure 2). Indeed, 8 of the 14 antibodies from children that showed binding to at least one SARS-CoV-2 variant or SARS-CoV, when tested in a vesicular stomatitis virus (VSV)-based neutralization assay,44,45 potently neutralized the SARS-CoV-2 variant harboring the D614G mutation,46 with a half maximal inhibitory concentration (IC50) of <10 ng/mL observed for 3 antibodies (Figures 2A and S3A). Similar neutralization patterns were also observed against the D614G variant in a pseudotyped-virus-based neutralization assay (Figure 2B). The pseudotyped virus assay was also used for evaluating the breadth of the antibodies against recently emergent Omicron variants that have been associated with resistance to virtually all available mAb therapies.33,34,36,47 Notably, 6 of the 7 antibodies from children exhibited neutralization breadth against 4 or more of the tested variants with IC50s <1 μg/mL, and all of the tested antibodies neutralized BA.4/5 (Figure 2B). In particular, antibody 71281-33 retained neutralization potency against all 9 SARS-CoV-2 pseudovirus variants tested: D614G, BA.2, BA.2.75+R346T, BA.4, BA.4.6, BA.2.75.2, XBB, BQ.1.1, and XBB.1.5, the last 5 of which have been implicated in resistance to mAb neutralization, including to bebtelovimab, Evusheld (a cocktail of tixagevimab and cilgavimab), and sotrovimab.33,34,36,47 Together, these data show that antibodies identified from children’s samples can display high levels of neutralization breadth and potency against SARS-CoV-2 variants even without prior vaccination or exposure to that variant. More generally, these results indicate that children’s samples can play an important role in the discovery of effective SARS-CoV-2 antibody therapeutics.
Figure 2.
Antibodies from children show diverse neutralization profiles of SARS-CoV-2 variants
(A) SARS-CoV-2 VSV-G neutralization was performed for a panel of antibodies against SARS-CoV-2 VSV-G with the D614G mutation.
(B) Additional lentiviral pseudovirus neutralization assays were performed against SARS-CoV-2 D614G and Omicron variants (BA.2, BA.4/5, BA.4.6, BA.2.75.2, BA.2.75+R346T, XBB, XBB.1.5, and BQ1.1). The IC50 values calculated in GraphPad Prism software by four-parameter best-fit analysis are shown on the bottom right as a color heatmap from least (white) to most (purple) potent. Data represent the percentage of neutralization or relative infection as mean ± SD; data are representative of at least two independent experiments performed in technical duplicate.
Antibodies from children target multiple diverse epitopes on the SARS-CoV-2 spike
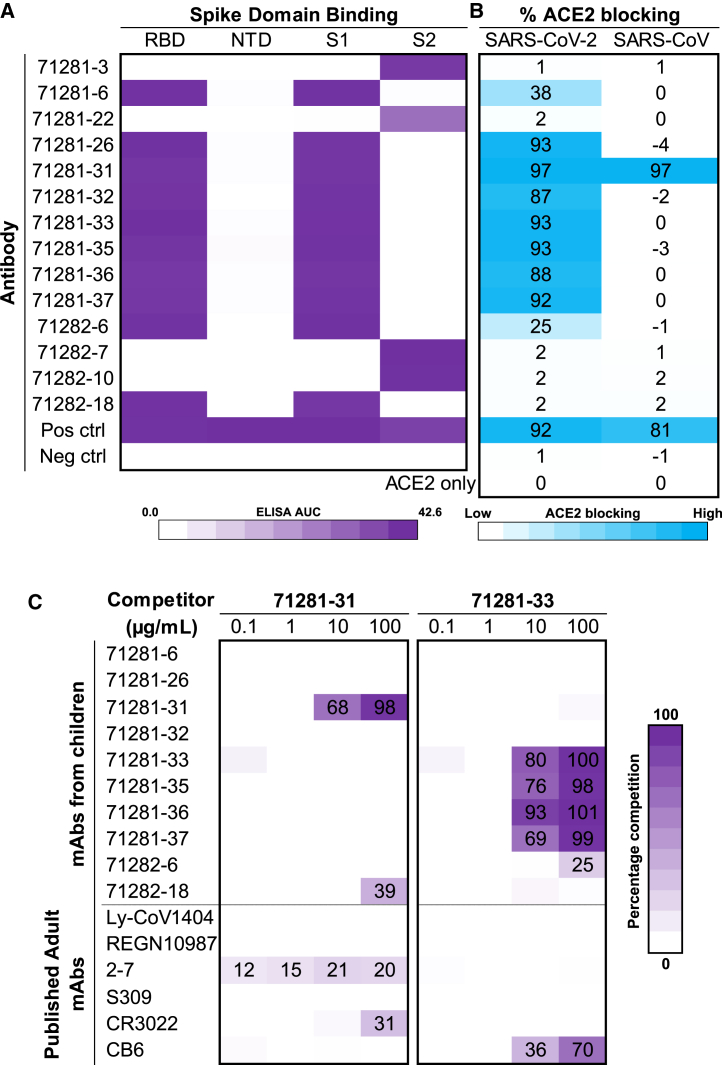
To evaluate the breadth of epitopes targeted by antibodies from children, we first tested binding against different spike subdomains (Figures 3A and S2B). Both receptor-binding domain (RBD) and S2 subdomain antibodies were identified. Further, antibody competition for spike-receptor ACE2 binding was tested in an ELISA, revealing high levels (>85%) of ACE2-blocking activity by 7/10 RBD-specific antibodies and medium levels (25%–38%) of blocking activity by 2 of the remaining RBD-specific antibodies (Figure 3B). Notably, antibody 71281-31 showed strong ACE2-blocking ability against both SARS-CoV-2 and SARS-CoV, in line with the SARS-CoV/SARS-CoV-2 cross-reactivity (Figure 1E) and RBD recognition (Figure 3A) observed for this antibody. Antibody 71281-31 was also able to neutralize both SARS-CoV-2 (Figure 2A) and SARS-CoV (Figure S3B), albeit at low potency, indicating a potential trade-off between high breadth of virus recognition and neutralization potency. As a next step, we selected the exceptionally broadly SARS-CoV-2-neutralizing antibody 71281-33 and the SARS-CoV/SARS-CoV-2 cross-reactive antibody 71281-31 to further delineate their epitope regions in antibody competition assays, both against other antibodies from children, as well as against published adult SARS-CoV-2 antibodies (Figure 3C). Antibody 71281-31 did not compete with any other antibodies, including the published S309 RBD SARS-CoV/SARS-CoV-2-reactive antibody,23 suggesting that it targets a unique cross-reactive epitope. In contrast, antibody 71281-33 competed with a subset of the other antibodies from children, and to some extent with the published class 1 RBD antibody CB6 (etesevimab),25 but no other antibodies in the set that was tested. Next, we performed negative-stain electron microscopy (nsEM) experiments in order to further delineate the epitope of neutralizing antibody 71281-33. Reconstructive 3D mapping of the SARS-CoV-2 spike in complex with Fab 71281-33 indicated that the epitope of that antibody is near the receptor-binding motif (RBM) of the RBD. Notably, the nsEM reconstruction showed Fab 71281-33 binding to the RBD in both the up and down states (Figures S3C–S3E). This epitope, along with the IGHV3-66 gene usage, short CDRH3, and mild competition with CB6, indicates that mAb 71281-33 is likely a class 1 RBD antibody.17 Together, these results suggest that the SARS-CoV-2 antibodies that we identified from children samples target a variety of different epitopes both on the spike in general and within the RBD subdomain.
Figure 3.
Epitope and ACE2-blocking analysis of antibodies from children
(A) ELISA AUC values for recombinant SARS-CoV-2 spike antigen domains are shown as heatmaps from minimum (white) to maximum (purple) binding. Controls are described in Figure S2B.
(B) Percentage of ACE2 blocking by ELISA is shown for SARS-CoV-2 and SARS-CoV spike proteins and is depicted as a heatmap from 0% (white) to 100% (blue).
(C) Antibody-antibody competition ELISAs were performed for antibodies 71281-31 and 71281-33 against other child antibodies identified here, as well as previously published adult antibodies. Percentage of competition is shown as a heatmap from 0% (white) to 100% (purple). Non-biotinylated competitor antibodies were coated first, and then biotinylated 71281-31 and 71281-33 were added to detect competition, as described in more detail in the STAR Methods.
Antibodies from children have sequence features that are associated with known public clonotypes in adults
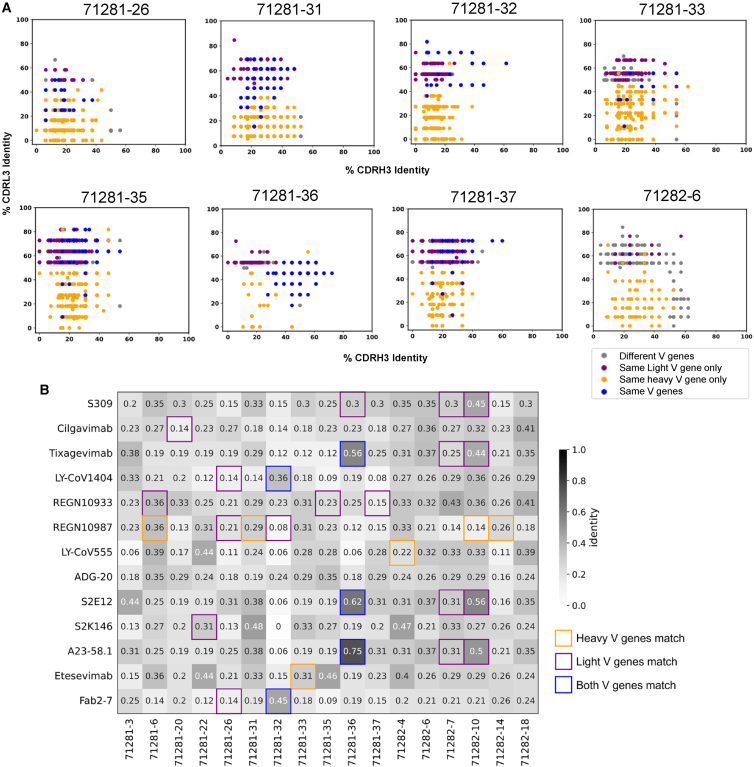
Characterization of the antibody responses to SARS-CoV-2 infection and vaccination in adults has led to the discovery of common genetic characteristics for SARS-CoV-2-reactive antibodies that are observed in multiple individuals.17,19,20,48,49 Such antibodies define public clonotypes and highlight a convergent mechanism of viral antigen recognition that can be observed at the population level.17,18,19,20,42 We therefore set out to explore how similar the antibodies discovered here are compared to known SARS-CoV-2-reactive antibodies in adults. To that end, utilizing the genetic features identified in Figure 1C, we compared each of the SARS-CoV-2-reactive antibodies identified here to a database of published SARS-CoV-2 antibody sequences.50 Notably, 16 of 17 antibodies that showed binding to SARS-CoV-2 spike utilized a combination of VH and VL chain genes that was identical to previously published adult SARS-CoV-2 antibodies (Figures 4A and S4). Of these, 11 antibodies also had CDRH3 identity of >50% to at least one published adult SARS-CoV-2 antibody, and 4 antibodies had CDRH3 identity of >70% (Figures 4A and S4). For all of the spike-binding antibodies from children, previously published SARS-CoV-2 antibodies were identified that utilized an identical VH chain gene, independent of the VL gene, with 15 of the antibodies having CDRH3 identity of >50% to at least one published adult SARS-CoV-2 antibody, and 5 antibodies had CDRH3 identity of >70% (Figures 4A and S4). Specifically focusing on the subset of 8 antibodies from children that were found to be neutralizing, all 8 utilized a combination of VH and VL genes that was identical to previously published adult SARS-CoV-2 antibodies, and 5 of the 8 also had CDRH3 identity of >50% to at least one published adult SARS-CoV-2 antibody (Figure 4A). Some of these combinations included well-documented public SARS-CoV-2 clonotypes that have been defined as public clonotypes specifically based on the variable gene usage, including IGHV1-58/IGKV3-2019,51,52,53 (used by antibody 71281-36) and IGHV2-5/IGLV2-1422,54,55 (utilized by 71281-32), as well as the IGHV3-53/3-66 public class of SARS-CoV-2 antibodies19,53,56 (utilized by 71281-33, 71281-35, and 71281-37).
Figure 4.
Public clonotype analysis of SARS-CoV-2 antibodies from children
(A) For each antibody from children (separate plot), shown are previously published adult antibodies (dots), with the respective CDRH3 (x axis) and CDRL3 (y axis) identity, colored according to V-gene usage: blue if both the VH and VL genes of the given child and adult antibodies match, orange if only the VH genes match, purple if only the VL genes match, and gray if neither match but at least one of the CDRH3 and CDRL3 have >50% sequence identity for the child vs. adult antibody.
(B) For each antibody from children (column), shown is the CDRH3 identity (values; heatmap) against a set of adult antibodies in clinical use or advanced preclinical development (rows). Cell border color corresponds to the V-gene usage described in (A).
In addition to the large-scale database comparison, we also evaluated the similarity of the antibodies identified here to a set of marketed, in-development, or well-characterized SARS-CoV-2 antibodies. Of note, several of the antibodies from children belonged to the same clonotypes as these published mAbs. In particular, 71281-32 shared identical VH and VL chain gene usage with LY-CoV1404 (bebtelovimab) and 2-7, and 71281-36 shared identical VH and VL chain genes with tixagevimab, S2E12, and A23-58.1, with CDRH3 identities of 56%, 62%, and 75%, respectively (Figure 4B). Together, these results suggest that the development of SARS-CoV-2 antigen specificity may use similar antibody sequence solutions in children to those used in adults.
Discussion
The state-of-the-art technologies that have revolutionized the field of antibody research in recent years have had limited applications in the context of children, likely due to the difficulty of obtaining and studying samples from children. Given the established importance of antibodies as a major component of immune responses to infection and vaccination, efforts aiming to fill this gap in knowledge can have significant implications for basic immunology, through shedding light on the fundamental rules that govern antigen recognition by antibodies in children. Such efforts can also be of clinical and translational significance by providing insights for the development of mAb therapies and vaccine modalities that are specifically tailored toward children.
Due to the significant differences in disease phenotype between children and adults, studying antigen-specific antibody repertoires in children in the context of SARS-CoV-2 is well motivated. To that end, using the LIBRA-seq technology for high-throughput antibody discovery, we identified and characterized a number of SARS-CoV-2-spike-specific antibodies from a cohort of children. Importantly, these antibodies targeted a variety of different spike epitopes and were associated with diverse genetic features, suggesting a broad portfolio of potential solutions for antigen targeting in children. Moreover, several of the antibodies from children belonged to known classes of public antibodies from adults, suggesting that there is a convergence of antigen-specific antibody responses for a wide range of ages. We note, however, that while child and adult antibodies can belong to overlapping public clonotypes based on common variable gene usage, there can be sufficient differences in the antibody sequence (e.g., in the CDRH3 region; Figure 4A) that could lead to major differences in the neutralization sensitivity phenotypes for these antibodies (Figure 2B) compared with adult antibodies from the same public clonotypes. This indicates that optimization of the antibody neutralization phenotype can be achieved through fine-tuning of antigen recognition even within individual public clonotypes.20,51,57,58
Notably, a set of antibodies from children showed potent neutralization of different currently circulating SARS-CoV-2 variants, including variants that have successfully escaped from the currently available mAb products. This result points to samples from children as a potential source for the development of mAb candidates with improved breadth of SARS-CoV-2 reactivity and further highlights the importance of deciphering the antigen-specific antibody repertoires in children for other infectious diseases with high biomedical significance.
Limitations of the study
This study investigates the genetic features, antigen cross-reactivity, and coronavirus neutralization breadth of antibodies discovered in a cohort of children aged 5 months to 18 years. Due to the limited number of samples that we were able to obtain in the lower end of the age range (0–5 years), samples were pooled, and group 71282 consisted of samples from only adolescent ages 12–17. Future studies that include stratified children’s age groups with retained donor source information for the identified B cells can better reflect the different characteristics of B cell repertoires in children. Additionally, while samples from group 71281 had no history of infection or vaccination, we were unable to experimentally confirm this, so we cannot rule out the possibility that donors in that cohort had been exposed at some point. Finally, public clonotype analysis was defined purely through sequence features. A comprehensive mapping of epitopes would need to be performed to confirm that adult and child antibodies that meet the sequence criteria to be defined within the same clonotype bind the spike protein in near-identical ways.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 Mouse Anti-Human CD14 | BD | Cat# 561709; RRID: AB_10893806 |
| FITC Anti-Human CD3 (OKT3) | Tonbo Biosciences | Cat# 35–0037; RRID: AB_2621662 |
| BV711 Mouse Anti-Human CD19 | BD | Cat# 563036; RRID: AB_2737968 |
| PE-Cy5 Mouse Anti-Human IgG | BD | Cat# 551497; RRID: AB_394220 |
| Goat anti-human IgG antibody (peroxidase) | Jackson ImmunoResearch | Cat#109-035-088, RRID:AB_2337584 |
| Anti-Mouse IgG HRP Conjugate | Promega | Cat#W4021 |
| Monoclonal anti-influenza 3602-1707 | Setliff et al.43 | N/A |
| Monoclonal anti-SARS-CoV-2 NTD 5317-9 | Shiakolas et al.37 | N/A |
| Monoclonal anti-SARS-CoV-2 71281-3 | This paper | GenBank: OR486977, OR486978 |
| Monoclonal anti-SARS-CoV-2 71281-6 | This paper | GenBank: OR486983, OR486984 |
| Monoclonal anti-SARS-CoV-2 71281-20 | This paper | GenBank: OR487011, OR487012 |
| Monoclonal anti-SARS-CoV-2 71281-22 | This paper | GenBank: OR487015, OR487016 |
| Monoclonal anti-SARS-CoV-2 71281-26 | This paper | GenBank: OR487023, OR487024 |
| Monoclonal anti-SARS-CoV-2 71281-31 | This paper | GenBank: OR487033, OR487034 |
| Monoclonal anti-SARS-CoV-2 71281-32 | This paper | GenBank: OR487035, OR487036 |
| Monoclonal anti-SARS-CoV-2 71281-33 | This paper | GenBank: OR487037, OR487038 |
| Monoclonal anti-SARS-CoV-2 71281-35 | This paper | GenBank: OR487041, OR487042 |
| Monoclonal anti-SARS-CoV-2 71281-36 | This paper | GenBank: OR487043, OR487044 |
| Monoclonal anti-SARS-CoV-2 71281-37 | This paper | GenBank: OR487045, OR487046 |
| Monoclonal anti-SARS-CoV-2 71282-4 | This paper | GenBank: OR487057, OR487058 |
| Monoclonal anti-SARS-CoV-2 71282-6 | This paper | GenBank: OR487061, OR487062 |
| Monoclonal anti-SARS-CoV-2 71282-7 | This paper | GenBank: OR487063, OR487064 |
| Monoclonal anti-SARS-CoV-2 71282-10 | This paper | GenBank: OR487069, OR487070 |
| Monoclonal anti-SARS-CoV-2 71282-14 | This paper | GenBank: OR487077, OR487078 |
| Monoclonal anti-SARS-CoV-2 71282-18 | This paper | GenBank: OR487085, OR487086 |
| LY-CoV1404 | Westendorf et al.54 | N/A |
| REGN10987 | Copin et al.59 | N/A |
| 2–7 | Liu et al.22 | N/A |
| CR3022 | Yuan et al.55 | N/A |
| CB6 | Shi et al.25 | N/A |
| DENV-2D22 | James Crowe Jr. | N/A |
| Bacterial and virus strains | ||
| VSV-SARS-CoV-2 | Case et al.44 | N/A |
| VSV-SARS-CoV | This paper | N/A |
| SARS-CoV-2 D614G pseudotyped lentivirus | Sheward et al.34 | N/A |
| SARS-CoV-2 BA.2 pseudotyped lentivirus | Daniel Sheward | N/A |
| SARS-CoV-2 BA.5 pseudotyped lentivirus | Sheward et al.34 | N/A |
| SARS-CoV-2 BA.4.6 pseudotyped lentivirus | Sheward et al.34 | N/A |
| SARS-CoV-2 BA.2.75.2 pseudotyped lentivirus | Sheward et al.34 | N/A |
| SARS-CoV-2 XBB pseudotyped lentivirus | Daniel Sheward | N/A |
| SARS-CoV-2 BQ.1.1 pseudotyped lentivirus | Daniel Sheward | N/A |
| SARS-CoV-2 XBB.1.5 pseudotyped lentivirus | Daniel Sheward | N/A |
| SARS-CoV-2 BA.2.75 + R346T pseudotyped lentivirus | This paper | N/A |
| Biological samples | ||
| PBMCs from donors | Mary Arildsen | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Ghost dye red 780 | Tonbo biosciences | Cat#13-0865 |
| Live/Dead fixable aqua dead cell stain kit | Thermo Fisher Scientific | Cat#L34957 |
| Streptavidin HRP | Thermo Fisher Scientific | Cat#ENN100 |
| Polyethyleniminine Linear MW 25000 | Polysciences | Cat#23966-1 |
| 1-Step Ultra TMB-ELISA Substrate Solution | Thermo Fisher Scientific | Cat#34029 |
| ACE2 with mouse Fc | Sino Biological | Cat#10108-H05H-100 |
| Streptavidin R-phycoerythrin (SA-PE) | Invitrogen | Cat#S866 |
| Galanthus nivalis lectin | Vector Laboratories | Cat#AL-1243-5 |
| Protein A resin | GenScript | Cat#L00210 |
| SARS-CoV-2 S1 | Sino Biological | Cat#40591-V08H |
| SARS-CoV-2 S2 | Sino Biological | Cat#40590-V08B |
| SARS-CoV-2 RBD | Sino Biological | Cat#40592-VNAH |
| SARS-CoV-2 NTD | Sino Biological | Cat#40591-V49H |
| HCoV-229E S | Sino Biological | Cat#40605-V08B |
| HCoV-NL63 S | Sino Biological | Cat#40604-V08B |
| SARS-CoV-2 S (Hexapro) | PMID: 32703906 | N/A |
| SARS-CoV S (S-2P) | PMID: 28807998 | N/A |
| HCoV-OC43 S (S-2P) | McLellan Lab | N/A |
| HCoV-HKU1 S (S-2P) | PMID: 28807998 | N/A |
| MERS-CoV S (S-2P) | PMID: 28807998 | N/A |
| A/New Caledonia/20/99 (H1N1) | Barney Graham | GenBank: ACF41878 |
| BG505.N332T.SOSIP.664 gp140 trimer | Ivelin Georgiev | N/A |
| SARS-CoV-2 S (Hexapro Alpha) | This paper | N/A |
| SARS-CoV-2 S (Hexapro Beta) | This paper | N/A |
| SARS-CoV-2 S (Hexapro Delta) | This paper | N/A |
| SARS-CoV-2 S (Hexapro Omicron BA.1) | This paper | N/A |
| SARS-CoV-2 S (Hexapro Omicron BA.2) | This paper | N/A |
| SARS-CoV-2 S (Omicron BA.4/5) | Sino Biological | Cat#40589-V08H32 |
| LYRa11 S (S-2P) | This paper | N/A |
| RaTG13 S (S-2P) | This paper | N/A |
| Civet-007-2004 S (S-2P) | This paper | N/A |
| Critical commercial assays | ||
| EZ link Sulfo-NHS-LC-biotin | Thermo Fisher Scientific | Cat#21327 |
| Solulink protein-oligonucleotide conjugation kit | TriLink Biotechnologies | Cat#S-9011 |
| B cell Single Cell V(D)J solution | 10X Genomics | N/A |
| ExpiFectamine™ 293 Transfection Kit | Thermo Fisher Scientific | Cat#A14526 |
| StrepTrap XT prepacked chromatography column | Cytiva | Cat#29401322 |
| Deposited data | ||
| 71281 heavy and light chain sequences | This paper | GenBank: OR486973-OR487050 |
| 71282 heavy and light chain sequences | This paper | GenBank: OR487051-OR487090 |
| Raw next-generation sequencing data | This paper | SRA: PRJNA1014462 |
| 71281-33 Fab nsEM map data | This paper | EMDB: EMD-41075, EMD-41076 |
| Experimental models: Cell lines | ||
| Human: Expi293F cells | Thermo Fisher Scientific | Cat#A14527 |
| ExpiCHO cells | Thermo Fisher Scientific | Cat#A29127 |
| Vero E6 cells | ATCC | Cat#CRL-1586 |
| HEK-293T-hACE2 cells | BEI | Cat#NR-52511 |
| HEK-293T/17 cells | ATCC | Cat#CRL-11268 |
| HEK-293T cells | ATCC | Cat#CRL-3216 |
| Oligonucleotides | ||
| Oligonucleotides for protein DNA-barcoding | Setliff et al.43 | N/A |
| Software and algorithms | ||
| Flowjo v10 | TreeStar | https://www.flowjo.com/ |
| GraphPad Prism 9.5.0 | https://www.graphpad.com:443/ | N/A |
| Cell Ranger | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| HighV-Quest | Alamyar et al.60 | http://www.imgt.org/IMGTindex/IMGTHighVQUEST.php |
| ChangeO | Gupta et al.61 | https://changeo.readthedocs.io/en/stable/ |
| cryoSPARC | Punjani et al.62 | https://cryosparc.com/updates |
| ChimeraX | Petterson et al.63 | https://www.cgl.ucsf.edu/chimerax/download.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Ivelin Georgiev (ivelin.georgiev@vanderbilt.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement. Please direct resource and reagent requests to the lead contact specified above, Ivelin Georgiev.
Experimental models and subject details
Human subjects
All research in this study complies with relevant ethical regulations. Vanderbilt University Medical Center Institutional Review Board approval was obtained (VUMC #210733). All donors were children between the ages of 5 months and 18 years old. Blood samples were collected with consent on June 24th and August 18th of 2021 and then PBMCs were isolated upon receipt. PBMCs were pooled into either group 71281 (no known history of SARS-CoV-2 infection or vaccination) or group 71282 (known history of infection or vaccination) based on self-reporting. Additional details on the age and gender distribution of both groups is further detailed in Figure S1. Additional information about these donors is unavailable.
Cell lines
A variety of cell lines were used for different assays in this study. Expi293F mammalian cells (ThermoFisher, A14527) were maintained in FreeStyle F17 expression medium supplemented with a final concentration of 0.1% Pluronic Acid F-68 and 4mM L-Glutamine. ExpiCHO cells (ThermoFisher, A29127) were maintained in ExpiCHO Expression medium(ThermoFisher, A2910002). Cells were cultured at 37 C with 8% CO2 saturation while shaking. Vero E6 cells (ATCC, CRL-1586) and all HEK293T cell lines (ATCC, CRL-3216; ATCC, CRL-11268; BEI, NR-52511) were maintained in Dulbecco’s minimal essential medium (DMEM) supplemented with 10mM HEPES pH 7.3, 1X non-essential amino acids, 1mM sodium pyruvate, 100U/mL of penicillin-streptomycin, and 10% fetal bovine serum and grown in 37 C with 5% CO2. Authentication analysis was not performed on the cell lines used.
Viruses
The generation of a replication-competent VSV expressing SARS-CoV-2 S protein with a 21 amino acid C-terminal deletion that replaces the VSV G protein (VSV-SARS-CoV-2) was described previously (Case et al., 2020b). The S protein-expressing VSV virus was propagated in MA104 cell culture monolayers (African green monkey, ATCC CRL-2378.1) as described previously (Case et al., 2020b), and viral stocks were titrated on Vero E6 cell monolayer cultures. VSV plaques were visualized using neutral red staining. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and A-BSL3 facilities at Washington University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Method details
Blood sample collection and preparation
Blood was collected for diagnostic testing into K2EDTA-coated Becton-Dickenson tubes and mixed to prevent coagulation. Blood was refrigerated after routine hematology testing was performed, until selection for the study (within 48 h). Blood samples were selected and delivered to the Georgiev lab and PBMCs were isolated immediately upon receipt. Use of the leftover diagnostic peripheral blood samples is covered by Vanderbilt University Medical Center IRB approval (#210733).
PBMC isolation
Upon receiving samples from VUMC Diagnostic Laboratories, whole blood was separated and pooled based on the parameters described in the results section. Pooled blood samples were diluted with 3 parts DPBS. 12.5 mL of Ficoll was added to 50 mL conical tubes and then the diluted blood (up to 30mL per 50 mL conical) was pipetted to mix before carefully layering it on top of the Ficoll. Without disrupting the Ficoll/Blood interface, tubes were centrifuged at 400 g for 30 min. The buffy coat was carefully removed and transferred to a new 50 mL conical up to a maximum of 25 mL per tube. Tubes were then topped to the 50 mL mark with DPBS and inverted to mix. Tubes were again centrifuged at 400 g. For each tube, the pellets were resuspended in ACK lysing buffer and incubated for 1 min at room temperature. Resuspended pellets from the same pooled samples were combined and then cells were counted. Tubes were filled up to 50 mL with DPBS and centrifuged at 200 g for 5 min at room temperature. Supernatants were decanted and pellets were resuspended in CryoStor Freeze Media in aliquots of 1 mL each between 10 and 100 million cells/mL. Isolated PBMCs were subject to slow freezing and then placed into liquid nitrogen for storage.
Some PBMCs were isolated using the EasySep Direct Human PBMC Isolation Kit (Stemcell) according to kit instructions. In short, EDTA was added to pooled blood samples, mixed with the isolation cocktail, and incubated. RapidSpheres were then added to the sample and mixed, followed by incubation of the tube inside of the “The Big Easy” magnet (Cat# 18001). The enriched cell suspension was then poured into a new tube, followed by mixing with more RapidSpheres. Cell mixture was incubated in the magnet for a 2nd time for additional separation. The enriched cell suspension was poured into a new tube and the process was repeated for a third time. Isolated cells were poured into a new tube and subsequently counted and frozen for use as detailed above.
Antigen expression and purification
An assortment of recombinant soluble protein antigens was used in the LIBRA-seq experiment and assays. All Expi293F cells were cultured at 8% CO2 saturation and 37°C with shaking in FreeStyle F17 expression media (Thermo Fisher) supplemented to a final concentration of 0.1% Pluronic Acid F-68 and 4 mM L-glutamine.
Plasmids were transiently transfected in Expi293F cells using polyethylenimine or ExpiFectamine transfection reagent (Thermo Fisher Scientific) and encoded the following: residues 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 S Hexapro (HP)); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations L18F, D80A, L242-244L del, R246I, K417N, E484K, N501Y, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 spike HP Beta); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations 69-70del, Y144del, N501Y, A570D, P681H, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 spike HP Alpha); residues 1–1190 of the SARS-CoV spike with proline substitutions at positions 968 and 969, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV S-2P); residues 1–1291 of the MERS-CoV spike with a mutated S1/S2 cleavage site, proline substitutions at positions 1060 and 1061, and a C-terminal T4-fibritin trimerization motif, an AviTag, an 8x HisTag, and a TwinStrepTag (MERS-CoV S-2P Avi); residues 1–1278 of the HCoV-OC43 spike with proline substitutions at positions 1070 and 1071, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (HCoV-OC43 S-2P); residues 1–1277 of the HCoV-HKU1 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 1067 and 1068, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (HCoV-HKU1 S-2P); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations T19R, del157/158, L452R, T478K, D614G, P681R, D950N, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (SARS-CoV-2 Delta S HP); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations A67V, del69/70, T95I, G142D, del143/145, del11, L212I, G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, Q496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (SARS-CoV-2 Omicron BA.1 S HP); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations T19I, Del24-26, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (SARS-CoV-2 Omicron BA.2 S HP); 1–1204 of the RaTG13 spike (ENA QHR63300.2) with proline substitutions at positions 982 and 983, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (RaTG13 S-2P); 1–1194 of the LYRa11 (ENA AHX37558.1) spike with proline substitutions at positions 972 and 973, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (LYRa11 S-2P); 1–1190 of the Civet-007/2004 spike (ENA AAU04646.1) with proline substitutions at positions 968 and 969, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (Civet-007-2004 S-2P); 1–1387 of the CCoV-HuPn-2018 spike64 with proline substitutions at positions 1140 and 1141, and a C-terminal T4-fibritin trimerization motif, Avitag, HRV3C, 8x HisTag, and a TwinStrepTag (CCoV-HuPn-2018 S-2P). All coronavirus spike supernatants were collected 5–7 days post transfection, sterile filtered, and purified over a StrepTrap XT column (Cytiva Life Sciences). Purified proteins were further purified using a size exclusion Superose6 Increase column (Cytiva Life Sciences). For LIBRA-seq, the purified antigens were then biotinylated with the EZ-Link Sulfo-NHS-Biotin (Thermo Fisher Scientific) using a 50:1 biotin to protein molar ratio for calculations.
Recombinant HIV-1 gp140 SOSIP BG505 N332T trimer65 was cultured in Expi293F cells and transfected in the same method as above. The clarified supernatant was run over an affinity column of agarose-bound Galanthus nivalis lectin (GNA, Snowdrop) slowly at 4°C. The column was washed with 1X PBS and bound protein was eluted with 1M methyl-a-D-mannopyranoside in PBS. The protein eluate was buffer exchanged into 1X PBS and then purified by size exclusion chromatography using a Superdex 200 Increase 10/300 GL Sizing column on the AKTA FPLC system (GE Life Sciences). The fractions of purified protein were analyzed by SDS-PAGE and binding was confirmed using ELISA with known antibodies.
The recombinant HA protein (A/New Caledonia/20/99 H1N1 GenBank ACF41878 (NC99)) was produced using Expi 293F cells and the Expifectamine 293 transfection reagent. The protein contains the HA ectodomain with a point mutation at the sialic acid-binding site (Y98F), T4 fibritin foldon trimerization domain, AviTag, and hexahistidine-tag. The cells were cultured for 4–5 days, then the supernatant was harvested and sterile filtered. The pH and NaCl concentration were adjusted by adding 1M Tris-HCl (pH 7.5) and 5M NaCl to 50 mM and 500 mM, respectively. The supernatant was then mixed with Ni Sepharose excel resin (GE Healthcare) to capture the hexahistidine tag. The resin was isolated in a column by gravity and the captured HA protein was eluted by a Tris-NaCl (pH 7.5) buffer containing 300 mM imidazole. The eluted protein was further purified by size exclusion chromatography using a HiLoad 16/60 Superdex 200 column (GE Healthcare). Fractions containing the appropriate sized HA protein were concentrated, analyzed by SDS-PAGE, and tested for antigenicity by ELISA using known antibodies. The proteins were then stored at −80C until use.
The SARS-CoV-2 S1, SARS-CoV-2 S2, SARS-CoV-2 RBD, and SARS-CoV-2 NTD subdomains as well as recombinant SARS-CoV-2 Omicron BA.4/5 S HP, and HCoV-229E S were purchased from Sino Biological.
Oligonucleotide barcodes
We used oligonucleotides composed of a 15 bp antigen barcode, a sequence designed for annealing to the template switch oligo on the 10X bead-delivered oligos, and contains truncated TruSeq small RNA read 1 sequences in the following structure: 5′-CCTTGGCACCCGAGAATTCCANNNNNNNNNNNNNCCCATATAAGA∗A∗A-3′, where Ns represent the antigen barcode. For each antigen we used the following barcode sequences: SARS-CoV-2 S HP (ACAATTTGTCTGCGA), SARS-CoV-2 Alpha S HP (GGTAGCCCTAGAGTA), SARS-CoV-2 Beta S HP (ATTCGCCTTACGCAA), SARS-CoV S-2P (GACCTCATTGTGAAT), LYRa11 S-2P (AGACTAATAGCTGAC), RaTG13 S-2P (TGTGTATTCCCTTGT), Civet-007-2004 S-2P (GACAAGTGATCTGCA), MERS S-2P (AACCTTCCGTCTAAG), HCoV-OC43 S-2P (TAACTCAGGGCCTAT), HCoV-HKU1-S-2P (CAGCCCACTGCAATA), HCoV-229E S (TACGCCTATAACTTG), BG505 gp140 SOSIP N332T (AACCCACCGTTGTTA), HA NC99 (GCTCCTTTACACGTA). Oligos were ordered from Sigma-Aldrich and IDT with a 5′ amino modification and HPLC purified.
Labeling antigens with DNA oligonucleotide barcodes
For each antigen described above, the unique DNA barcodes were directly conjugated to the antigen using a SoluLINK Protein-Oligonucleotide Conjugation kit (TriLink, S-9011) according to kit protocol. In short, we desalted the oligonucleotide and protein, modified the amino-oligonucleotide with the 4FB cross-linker, and modified the biotinylated antigen with S-HyNic. Afterward, the 4FB-oligonucleotide and the HyNic-antigen were mixed to form a stable bond between the protein and the oligonucleotide. The antigen-oligonucleotide concentrations were then determined using a bicinchoninic acid (BCA) assay, and the HyNic molar substitution ratios of each antigen-oligonucleotide conjugate was determined using a NanoDrop according to SoluLINK protocol instructions. Excess oligonucleotide was removed from the protein-oligonucleotide conjugates using an AKTA FPLC and were subsequently verified using SDS-PAGE and silver stain. The optimal amounts of antigen-oligonucleotide conjugates to be used in antigen-specific B cell sorting were then determined through flow cytometry titration experiments on cell lines expressing BCRs of known specificities.
Antigen specific B cell sorting
To start, PBMCs were thawed, washed, and counted. Viability was evaluated using Trypan Blue. The cells were then washed with a solution of DPBS supplemented with 0.1% Bovine serum albumin (BSA). Afterward, the cells were resuspended in DPBS-BSA and stained with cell markers: Ghost Red 780 for viability, CD14-APC-Cy7, CD3-FITC, CD19-BV711 and IgG-PE-Cy5. Additionally, antigen-oligo conjugates were added to the stain. After a 30-min incubation in the dark at room temperature, the cells were washed again three times with DPBS-BSA at 300 g for 5 min. Then, the cells were incubated for 15 min at room temperature with Streptavidin-PE to label cells with bound antigen. The cells were again washed three times with DPBS-BSA, resuspended in DPBS, and sorted by FACS. Antigen positive B cells were bulk sorted and delivered to the Vanderbilt Technologies for Advanced Genomics (VANTAGE) sequencing core at an appropriate target concentration for 10X Genomics library preparation and subsequent sequencing. FACS data were analyzed using FlowJo.
Sample and library preparation for sequencing
Single-cell suspensions were processed using the Chromium Controller microfluidics device (10X Genomics) and the B cell Single Cell V(D)J solution as per the manufacturer’s instructions. The aim was to capture 10,000 B cells per 1/8 10X cassette. Slight modifications were made to intercept, amplify, and purify the antigen barcode libraries, as previously described.43
Sequence processing and bioinformatics analysis
Our established pipeline was followed, which takes paired-end FASTQ files of oligonucleotide libraries as input, to process and annotate reads for cell barcodes, unique molecular identifiers (UMIs) and antigen barcodes, resulting in a cell barcode-antigen barcode UMI count matrix.43 B cell receptor contigs were processed using CellRanger (10x Genomics) and GRCh38 as reference, while the antigen barcode libraries were also processed using CellRanger (10x Genomics). The cell barcodes that overlapped between the two libraries formed the basis of the subsequent analysis. Cell barcodes that had only non-functional heavy chain sequences as well as cells with multiple functional heavy chain sequences and/or multiple functional light chain sequences, were eliminated, reasoning that these may be multiplets. We also aligned the B cell receptor contigs (filtered_contigs.fasta file output by CellRanger, 10x Genomics) to IMGT reference genes using HighV-Quest.60 The output of HighV-Quest was parsed using ChangeO,61 and combined with an antigen barcode UMI count matrix. Finally, we determined the LIBRA-seq score for each antigen in the library for every cell as previously described.43
High-throughput antibody microscale expression and purification
For high-throughput production of recombinant antibodies, a microscale approach was employed. For antibody expression, microscale transfection was performed (∼1 mL per antibody) with CHO cell cultures using the Gibco ExpiCHO Expression System and a protocol for deep 96-well blocks (Thermo Fisher Scientific). Briefly, synthesized antibody-encoding DNA (∼2 μg per transfection) was added to OptiPro serum-free medium (OptiPro SFM), incubated with ExpiFectamine CHO Reagent, and added to 800 μL of ExpiCHO cell cultures in deep 96-well blocks using a ViaFlo384 liquid handler (Integra Biosciences). The plates were incubated on an orbital shaker at 1,000 r.p.m. with an orbital diameter of 3 mm at 37°C in 8% CO2. The next day after transfection, ExpiFectamine CHO Enhancer and ExpiCHO Feed reagents (Thermo Fisher Scientific) were added to the cells, followed by 4 more days of incubation. Culture supernatants were collected after centrifuging the blocks at 450 g for 5 min and were stored at 4°C until use. For high-throughput microscale antibody purification, fritted deep-well plates were used containing 25 μL of settled Protein G resin (GE Healthcare Life Sciences) per well. Clarified culture supernatants were incubated with protein G resin for antibody capturing, washed with PBS using a 96-well plate manifold base (Qiagen) connected to the vacuum and eluted into 96-well PCR plates using 86 μL of 0.1 M glycine-HCl buffer, pH 2.7. Purified antibodies were then neutralized with 14 μL of 1 M Tris-HCl pH 8.0 and buffer exchanged into PBS using Zeba Spin Desalting plates (Thermo Fisher Scientific). Purified antibodies were then stored at 4°C until use.
Antibody expression and purification
Variable heavy and light genes were inserted into custom plasmids that encode the constant region for the human IgG1 heavy chain and respective lambda and kappa light chains (pTwist CMV BetaGlobin WPRE Neo vector, Twist Bioscience). The antibodies were expressed in Expi293F cells by co-transfecting heavy chain and light chain expressing plasmids using polyethylenimine or Expifectamine transfection reagent, and the cells were cultured for 4–5 days. These cells were maintained as previously described in the antigen purification methods. Cultures were harvested, centrifuged and supernatant was 0.45 μm filtered with Nalgene Rapid Flow Disposable Filter Units with PES membrane. The filtered supernatant was run over a column containing Protein A agarose resin that was equilibrated with PBS. The column was washed with PBS, and then the antibodies were eluted with 100mM Glycine HCl at 2.7 pH directly into a 1:10 volume of 1M Tris-HCl pH 8.0. Eluted antibodies were buffer exchanged into PBS using using Amicon Ultra centrifugal filter units, centrifuging and topping off three times with PBS, and finally concentrated. The antibodies were analyzed by SDS-PAGE. Antibody plasmids were sequenced to confirm the expected heavy and light chain match.
ELISA
To evaluate the binding of the expressed antibodies, soluble purified antigen was plated at a concentration of 2 μg/mL and incubated overnight at 4°C. The next day, the plates were washed three times with a PBS solution containing 0.05% Tween 20 (PBS-T) and then coated with 5% milk powder in PBS-T. The plates were incubated for 1 h at room temperature and then washed three times with PBS-T. The primary antibodies were diluted in 1% milk in PBS-T, starting at a concentration of 10 μg/mL with a serial 1:5 or 1:10 dilution and then added to the plate. The plates were incubated for an additional hour at room temperature and then washed three times with PBS-T. The secondary antibody, goat anti-human IgG conjugated to peroxidase, was added at a dilution of 1:10,000 in 1% milk in PBS-T to the plates, which were incubated for 1 h at room temperature. The plates were washed three times with PBS-T and then developed by adding TMB substrate to the plates. The plates were incubated for 10 min at room temperature, and the reaction was stopped with 1N sulfuric acid. Plates were read at 450 nm. The data is shown as one representative biological replicate with the mean ± SEM for one ELISA experiment. The ELISAs were repeated 2 or more times. The area under the curve (AUC) was calculated using GraphPad Prism 9.5.0.
ACE2 competition ELISA
96-well plates were coated with 2 μg/mL of purified recombinant SARS-CoV-2 spike protein, and left to incubate overnight at 4°C. The following day, the plates were washed three times with a PBS solution containing 0.05% Tween 20 (PBS-T) and then coated with 5% nonfat dry milk in PBS-T. The plates were incubated for 1 h at room temperature and then washed three times with PBS-T. Next, purified antibodies were diluted in 1% nonfat dry milk in PBS-T at a concentration of 10 μg/mL and added to the wells in triplicate, followed by incubation for an additional hour at room temperature. Without washing, recombinant human ACE2 protein with a mouse Fc tag was added to the wells for a final concentration of 0.4 μg/mL of ACE2 and incubated for 40 min at room temperature. The plates were washed three times with PBS-T and bound ACE2 was detected using HRP-conjugated anti-mouse Fc antibody and TMB substrate. The plates were incubated for 10 min at room temperature, and then the reaction was stopped with 1N sulfuric acid. Plates were read at 450 nm ACE2 binding without antibody served as a control. The experiment was done in biological replicate and technical triplicates.
Antibody competition ELISA
Antibody competition ELISAs were performed similarly to the regular binding ELISA protocol described above, with some alterations. After antigen coating and blocking, 25 μL of serially diluted non-biotinylated competitor antibody was added to each well at a concentration of 100, 10, 1, and 0.1 μg/mL, and incubated at 37°C for 10 min. Then, without washing, 75 μL of biotinylated antibody (final well concentration of 0.1 μg/mL) was added and incubated at 37°C for 1 h. After washing three times with PBS-T, streptavidin-HRP was added at a dilution of 1:10,000 in 1% milk in PBS-T and incubated for 1 h at room temperature. The plates were washed, and substrate and sulfuric acid were added as described earlier. ELISAs were repeated at least 2 times. Data is shown as the percentage decrease in binding relative to no competitor.
Real-time cell analysis (RTCA) neutralization assay
To determine neutralizing activity of IgG, Fab, or F(ab′)2 proteins, we used real-time cell analysis (RTCA) assay on an xCELLigence RTCA MP Analyzer (ACEA Biosciences Inc.) that measures virus-induced cytopathic effect (CPE).45 Briefly, 50 μL of cell culture medium (DMEM supplemented with 2% FBS) was added to each well of a 96-well E-plate using a ViaFlo384 liquid handler (Integra Biosciences) to obtain background reading. A suspension of 18,000 Vero-E6 cells in 50 μL of cell culture medium was seeded in each well, and the plate was placed on the analyzer. Measurements were taken automatically every 15 min, and the sensograms were visualized using RTCA software version 2.1.0 (ACEA Biosciences Inc). VSV-SARS-CoV-2 (0.01 MOI, ∼120 PFU per well) was mixed 1:1 with a dilution of mAb in a total volume of 100 μL using DMEM supplemented with 2% FBS as a diluent and incubated for 1 h at 37°C in 5% CO2. At 16 h after seeding the cells, the virus-mAb mixtures were added in replicates to the cells in 96-well E-plates. Triplicate wells containing virus only (maximal CPE in the absence of mAb) and wells containing only Vero cells in medium (no-CPE wells) were included as controls. Plates were measured continuously (every 15 min) for 48 h to assess virus neutralization. Normalized cellular index (CI) values at the endpoint (48 h after incubation with the virus) were determined using the RTCA software version 2.1.0 (ACEA Biosciences Inc.). Results are expressed as percent neutralization in a presence of respective mAb relative to control wells with no CPE minus CI values from control wells with maximum CPE. RTCA IC50 values were determined by nonlinear regression analysis using Prism software.
SARS-CoV-2 VSV-G virus production
The generation of a replication-competent VSV expressing SARS-CoV-2 S protein with a 21 amino acid C-terminal deletion that replaces the VSV G protein (VSV-SARS-CoV-2) was described previously.44 The S protein-expressing VSV virus was propagated in MA104 cell culture monolayers (African green monkey, ATCC CRL-2378.1) as described previously,44 and viral stocks were titrated on Vero E6 cell monolayer cultures. VSV plaques were visualized using neutral red staining. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and A-BSL3 facilities at Washington University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Pseudovirus production and neutralization assays
Lentiviral pseudovirus production and neutralizations were done in one of two different ways.
-
(1)
For the pseudoviruses D614G, BA.2, BA.5, BA.4.6, BA.2.75.2, XBB, BQ.1.1, and XBB.1.5: HEK293T cells and HEK293T-hACE2 (stably expressing human ACE2) cells were cultured in Dulbecco’s Modified Eagle Medium (high glucose, with sodium pyruvate) supplemented with 10% fetal bovine serum, 100 units/ml Penicillin, and 100 μg/mL Streptomycin. Cultures were maintained in a humidified 37C incubator (5% CO2). SARS-CoV-2 spike-pseudotyped lentiviruses were generated by the co-transfection of HEK293T cells with a spike-encoding plasmid (with a 19 amino acid C-terminal truncation), a lentiviral packaging plasmid (Addgene #8455), and a transfer plasmid encoding firefly luciferase (Addgene #170674) using polyethylenimine (PEI). Growth media was replaced 12–16 h after transfection, and pseudotyped viruses were harvested from the supernatant at 48- and 72-h post transfection, clarified by centrifugation, and stored at −80C. Pseudotyped viruses previously titrated to generate ∼100,000 relative light units (RLUs) were incubated with serial 3-fold dilutions of antibody for 60 min at 37C, and then ∼10,000 HEK293T-hACE2 cells were added to each well. Plates were incubated at 37C for 44–48 h and luminescence was then measured using Bright-Glo (Promega) on a GM-2000 luminometer (Promega) per the manufacturer’s instructions. Neutralization was calculated relative to the mean of eight control wells infected in the absence of antibody and fit using a four-parameter logistic curve in Prism v9 (GraphPad Software).
-
(2)
For the pseudovirus BA.2.75 + R346T: Lentiviral pseudoviruses bearing the S glycoproteins of SARS-CoV-2 variants were generated using a previously described protocol.66 Briefly, HEK-293T/17 cells (ATCC, cat. CRL-11268) were seeded in a culture flask at a density that would give approximately 50–70% confluency on the day of transfection. The morning of the transfection, media was changed to fresh DMEM supplemented with sodium pyruvate, penicillin/streptomycin 25 mM HEPES, and 10% (v/v) FBS. To generate lentiviral-based reporter pseudoviruses, cells were co-transfected with a pHAGE-CMV-Luc2-IRES-ZsGreen-W plasmid encoding the lentiviral genome (BEI Resources, cat. NR-52516), the lentiviral packaging plasmids HDM-Hgpm2 (BEI Resources, cat. NR-52517), HDM-tat1b (BEI Resources, cat. NR-52518), and pRC-CMV- Rev (BEI Resources, cat. NR-52519), and a plasmid encoding the SARS-CoV-2 S gene with a 21 amino acid deletion. Approximately 16–18 h later, media was removed and replaced with fresh DMEM supplemented with sodium pyruvate, penicillin/streptomycin 25 mM HEPES, and 2% (v/v) FBS. Approximately 48 h after transfection, the supernatant was collected from each flask, clarified by centrifugation, and filtered through a 0.2 μm filter. Aliquots of pseudovirus stocks were prepared and stored at −80°C.
Lentivirus-based pseudoviral neutralization assays were executed based on a previous described protocol.66 One day prior to the assay, HEK-293T cells transduced to stably express human ACE2 (293T-hACE2 cells, BEI Resources NR-52511) were seeded at a density of 1.25 × 104 cells per well in DMEM+10% FBS onto 96-well tissue culture plates that had previously been coated with poly-D-lysine (ThermoFisher Scientific, cat. A3890401) The day of the assay, monoclonal antibodies were diluted in DMEM+2% FBS using a 4-fold dilution series (with each antibody in technical duplicate) in a 96-well polypropylene microtiter plate and incubated with pseudovirus for 1 h at 37°C in the presence of a final concentration of 5 μg/mL polybrene (EMD Millipore). After the 1 h incubation, pseudovirus-mAb mixtures were added to 293T-hACE2 monolayers. Plates were incubated at 37°C for 48–60 h, at which point cells were lysed using the Bright-Glo Luciferase Assay System (Promega) and luciferase activity was quantified using a CLARIOStar plate reader (BMG LabTech). The luminescence signal from wells in each plate where no pseudovirus or antibody was added was averaged and subtracted from each value, after which the percent infection of each well was determined relative to the average of pseudovirus only control wells present in each plate. IC50 values were determined by nonlinear regression using Prism v.9.5 (GraphPad) using a four-parameter [inhibitor] vs. response curve fit with top and bottom values constrained to 100 and 0, respectively. Each neutralization assay was repeated twice.
Public antibody analysis
For generation of plots for each child antibody in comparison with previously published adult antibodies with respective to percent CDRH3 (x axis) and percent CDRL3 (y axis) identity, CovAbDab database was used.50 Each dot is colored according to V-gene usage: blue if both the VH and VL of the given child and adult antibodies match, orange if only the VH match, purple if only the VL match, and gray if neither match but at least either one of the CDRH3 or CDRL3 have ≥50% sequence identity for the child vs. adult antibody.
Sequences and gene annotations for antibodies in clinical use were manually curated from IMGT/3Dstructure-DB67 and compared to the SARS-CoV-2 antibodies from children using a custom Python script. Heavy chain CDR3 identity between each antibody pair was defined by calculating the Levenshtein distance between the amino acid sequences and dividing by the length of the longer CDR3 region.
Negative stain electron microscopy (nsEM) sample and grid preparation, imaging and processing of spike–Fab complexes
To perform electron microscopy imaging, Fabs were produced by digesting recombinant chromatography-purified IgGs using resin-immobilized cysteine protease enzyme (FabALACTICA, Genovis). The digestion occurred in 100 mM sodium phosphate and 150 mM NaCl pH 7.2 (PBS) for around 16 h at ambient temperature. To remove cleaved Fc from intact IgG, the digestion mix was incubated with CaptureSelect Fc resin (Genovis) for 30 min at ambient temperature in PBS buffer. Spike protein were incubated with the Fab at an Fab:S protein molar ratio of 4:1 for about 1 h at ambient temperature, and approximately 3 μL of the sample at concentrations of about 10–15 μg/mL was applied to a glow-discharged grid with continuous carbon film on 400 square mesh copper electron microscopy grids (Electron Microscopy Sciences). The grids were stained with 2% uranyl formate.68 Images were recorded on a Gatan US4000 4k×4k CCD camera using an TF20 transmission electron microscope (TFS) operated at 200 keV and control with Serial EM.69 All images were taken at 50,000× magnification with a pixel size of 2.18 Å/pixel in low-dose mode at a defocus of 1.5–1.8 μm. The total dose for the micrographs was around 30e− per Å2. Image processing was performed using the cryoSPARC software package.62 Images were imported, CTF-estimated and particles were picked. The particles were extracted with a box size of 256 pixels and binned to 128 pixels (4.36 Å/pixel). 2D class averages were performed and good classes selected for ab initio model and NU refinement without symmetry with final resolution of ∼20 Å. Model docking to the EM map was done in ChimeraX.63 For spike protein model (PDB:7XIW) was used and PDB:12E8 was used for the Fab. ChimeraX software was used to make all figures, and the EM map has been deposited into EMDB (EMD-41075, EMD-41076).
Quantification and statistical analysis
The ELISA error bars (± standard error of the mean) were calculated using Graphpad Prism 9.5.0. Mean ± SEM or mean ± SD were determined for continuous variables as noted. Technical and biological replicates are described in the figure legends. Details of the statistical analysis can be found in the main text and respective figure captions.
Acknowledgments
We thank A. Jones, L. Raju, and J. Roberson of VANTAGE for their expertise regarding NGS and library preparation; D. Flaherty and B. Matlock of the Vanderbilt Flow Cytometry Shared Resource for help with flow panel optimization; and members of the Georgiev laboratory for comments on the manuscript. The Vanderbilt VANTAGE Core provided technical assistance for this work. VANTAGE is supported, in part, by CTSA grant 5UL1 RR024975-03, the Vanderbilt-Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956). This work was conducted, in part, using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. Flow cytometry experiments were performed in the Vanderbilt University Medical Center (VUMC) Flow Cytometry Shared Resource. The VUMC Flow Cytometry Shared Resource is supported by the Vanderbilt-Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). EM data collections were conducted at the Center for Structural Biology Cryo-EM Facility at Vanderbilt University. The following reagent was obtained through BEI Resources, NIAID, NIH: human embryonic kidney cells (HEK-293T) expressing human angiotensin-converting enzyme 2, HEK-293T-hACE2 cell line, NR-52511. We thank the G2P-UK National Virology consortium funded by MRC/UKRI (MR/W005611/1) and the Barclay group at Imperial College for providing spike expression plasmids. For work described in this manuscript, I.S.G., S.C.W., A.R.S., C.M.H., P.T.W., and Y.P.S. were supported, in part, by NIH National Institute of Allergy and Infectious Diseases (NIAID) award R01 AI131722-S1, the Hays Foundation COVID-19 Research Fund, Fast Grants, and the G. Harold and Leila Y. Mathers Charitable Foundation. J.E.C., N.S., E.C.C., and R.H.C. were supported, in part, by Defense Advanced Research Projects Agency (DARPA) grant HR0011-18-2-0001, US NIH contract 75N93019C00074, NIH grant R01 AI157155, the Dolly Parton COVID-19 Research Fund at Vanderbilt, and a grant from Fast Grants, Mercatus Center, George Mason University. J.E.C. is a recipient of the 2019 Future Insight Prize from Merck KGaA, which supported this work with a grant. The graphical abstract was created using BioRender.com.
Author contributions
S.C.W., A.R.S., and I.S.G. developed the methodology. S.C.W., N.S., C.K., A.R.S., C.M.H., E.B.I., P.T.W., Y.P.S., S.J.Z., E.C.C., E.B., and D.J.S. performed the investigations. E.C. and M.A.T.-A. provided blood samples from children. S.C.W. performed validations. S.C.W. and I.S.G. wrote the original draft. All authors reviewed and edited the manuscript. I.S.G., J.E.C., and R.H.C. acquired funding. I.S.G., J.E.C., D.J.S., and R.H.C. provided resources. D.J.S., R.H.C., and I.S.G. supervised the work.
Declaration of interests
S.C.W. and I.S.G. are listed as inventors on antibodies described here. A.R.S. and I.S.G. are co-founders of AbSeek Bio. I.S.G. and A.R.S. are listed as inventors on patent applications for the LIBRA-seq technology. D.J.S. has served as a consultant for AstraZeneca AB. J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Board of Meissa Vaccines, and is founder of IDBiologics. The Crowe laboratory has received funding support in sponsored research agreements from AstraZeneca, IDBiologics, and Takeda. The Georgiev laboratory at VUMC has received unrelated funding from Takeda Pharmaceuticals.
Published: November 6, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101267.
Supplemental information
Data and code availability
-
•
Sequences for antibodies identified and characterized in this study have been deposited to GenBank under GenBank accession numbers Genbank: OR486973-OR487050 (for 71281 antibodies) and Genbank: OR487051-OR487090 (for 71282 antibodies). Raw sequencing data used in this study are available on the Sequence Read Archive under BioProject accession number SRA: PRJNA1014462. nsEM map data of Fab 71281–33 in complex with SARS-CoV-2 spike is available on the Electron Microscopy Databank under accession numbers EMDB: EMD-41075 and EMDB: EMD-41076.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ruf B.R., Knuf M. The burden of seasonal and pandemic influenza in infants and children. Eur. J. Pediatr. 2014;173:265–276. doi: 10.1007/s00431-013-2023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shafagati N., Williams J. Human metapneumovirus - what we know now. F1000Res. 2018;7:135. doi: 10.12688/f1000research.12625.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi T., McAllister D.A., O'Brien K.L., Simoes E.A.F., Madhi S.A., Gessner B.D., Polack F.P., Balsells E., Acacio S., Aguayo C., et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet. 2017;390:946–958. doi: 10.1016/s0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmermann P., Curtis N. Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Arch. Dis. Child. 2020;106:429–439. doi: 10.1136/archdischild-2020-320338. [DOI] [PubMed] [Google Scholar]
- 5.Mehta N.S., Mytton O.T., Mullins E.W.S., Fowler T.A., Falconer C.L., Murphy O.B., Langenberg C., Jayatunga W.J.P., Eddy D.H., Nguyen-Van-Tam J.S. SARS-CoV-2 (COVID-19): What Do We Know About Children? A Systematic Review. Clin. Infect. Dis. 2020;71:2469–2479. doi: 10.1093/cid/ciaa556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou J., Thomas P.G., Randolph A.G. Immunology of SARS-CoV-2 infection in children. Nat. Immunol. 2022;23:177–185. doi: 10.1038/s41590-021-01123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D'Andrea K., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369 doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramer K.J., Wilfong E.M., Voss K., Barone S.M., Shiakolas A.R., Raju N., Roe C.E., Suryadevara N., Walker L.M., Wall S.C., et al. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat. Commun. 2022;13:3466. doi: 10.1038/s41467-022-31142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond M.S., Kanneganti T.-D. Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol. 2022;23:165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cromer D., Steain M., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Kent S.J., Triccas J.A., Khoury D.S., Davenport M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet. Microbe. 2022;3:e52–e61. doi: 10.1016/s2666-5247(21)00267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., Dipiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 15.Collier D.A., Ferreira I.A.T.M., Kotagiri P., Datir R.P., Lim E.Y., Touizer E., Meng B., Abdullahi A., CITIID-NIHR BioResource COVID-19 Collaboration. Elmer A., et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596:417–422. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Setliff I., McDonnell W.J., Raju N., Bombardi R.G., Murji A.A., Scheepers C., Ziki R., Mynhardt C., Shepherd B.E., Mamchak A.A., et al. Multi-Donor Longitudinal Antibody Repertoire Sequencing Reveals the Existence of Public Antibody Clonotypes in HIV-1 Infection. Cell Host Microbe. 2018;23:845–854.e6. doi: 10.1016/j.chom.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen E.C., Gilchuk P., Zost S.J., Suryadevara N., Winkler E.S., Cabel C.R., Binshtein E., Chen R.E., Sutton R.E., Rodriguez J., et al. Convergent antibody responses to the SARS-CoV-2 spike protein in convalescent and vaccinated individuals. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan T.J.C., Yuan M., Kuzelka K., Padron G.C., Beal J.R., Chen X., Wang Y., Rivera-Cardona J., Zhu X., Stadtmueller B.M., et al. Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain. Nat. Commun. 2021;12:3815. doi: 10.1038/s41467-021-24123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 23.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 24.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 26.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.09.30.318972. Preprint at. [DOI] [Google Scholar]
- 27.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y., Ho M. Emerging antibody-based therapeutics against SARS-CoV-2 during the global pandemic. Antib. Ther. 2020;3:246–256. doi: 10.1093/abt/tbaa025.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisblum Y., Schmidt F., Zhang F., Dasilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/elife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., Vanblargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheward D.J., Kim C., Fischbach J., Sato K., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. Lancet Infect. Dis. 2022;22:1538–1540. doi: 10.1016/s1473-3099(22)00663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai M., Ito M., Kiso M., Yamayoshi S., Uraki R., Fukushi S., Watanabe S., Suzuki T., Maeda K., Sakai-Tagawa Y., et al. Efficacy of Antiviral Agents against Omicron Subvariants BQ.1.1 and XBB. N. Engl. J. Med. 2023;388:89–91. doi: 10.1056/nejmc2214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiakolas A.R., Kramer K.J., Johnson N.V., Wall S.C., Suryadevara N., Wrapp D., Periasamy S., Pilewski K.A., Raju N., Nargi R., et al. Efficient discovery of SARS-CoV-2-neutralizing antibodies via B cell receptor sequencing and ligand blocking. Nat. Biotechnol. 2022;40:1270–1275. doi: 10.1038/s41587-022-01232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y., Jian F., Zhang Z., Yisimayi A., Hao X., Bao L., Yuan F., Yu Y., Du S., Wang J., et al. Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Astrazeneca . 2022. AstraZeneca Data on File - REF-173560. [Google Scholar]
- 40.Dowell A.C., Butler M.S., Jinks E., Tut G., Lancaster T., Sylla P., Begum J., Bruton R., Pearce H., Verma K., et al. Children develop robust and sustained cross-reactive spike-specific immune responses to SARS-CoV-2 infection. Nat. Immunol. 2022;23:40–49. doi: 10.1038/s41590-021-01089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraley E., Lemaster C., Banerjee D., Khanal S., Selvarangan R., Bradley T. Cross-reactive antibody immunity against SARS-CoV-2 in children and adults. Cell. Mol. Immunol. 2021;18:1826–1828. doi: 10.1038/s41423-021-00700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang F., Nielsen S.C.A., Hoh R.A., Röltgen K., Wirz O.F., Haraguchi E., Jean G.H., Lee J.Y., Pham T.D., Jackson K.J.L., et al. Shared B cell memory to coronaviruses and other pathogens varies in human age groups and tissues. Science. 2021;372:738–741. doi: 10.1126/science.abf6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Setliff I., Shiakolas A.R., Pilewski K.A., Murji A.A., Mapengo R.E., Janowska K., Richardson S., Oosthuysen C., Raju N., Ronsard L., et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell. 2019;179:1636–1646.e15. doi: 10.1016/j.cell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suryadevara N., Gilchuk P., Zost S.J., Mittal N., Zhao L.L., Crowe J.E., Jr., Carnahan R.H. Real-time cell analysis: A high-throughput approach for testing SARS-CoV-2 antibody neutralization and escape. STAR Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11:6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., Lee H., Jung J., Kang C.K., Song K.H., et al. Stereotypic neutralizing V(H) antibodies against SARS-CoV-2 spike protein receptor binding domain in patients with COVID-19 and healthy individuals. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong J., Zost S.J., Greaney A.J., Starr T.N., Dingens A.S., Chen E.C., Chen R.E., Case J.B., Sutton R.E., Gilchuk P., et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat. Microbiol. 2021;6:1233–1244. doi: 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y., Yuan M., Lv H., Peng J., Wilson I.A., Wu N.C. A large-scale systematic survey reveals recurring molecular features of public antibody responses to SARS-CoV-2. Immunity. 2022;55:1105–1117.e4. doi: 10.1016/j.immuni.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., Van Der Lee R., Hendle J., Pustilnik A., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan M., Wang Y., Lv H., Tan T.J.C., Wilson I.A., Wu N.C. Molecular analysis of a public cross-neutralizing antibody response to SARS-CoV-2. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q., Ju B., Ge J., Chan J.F.-W., Cheng L., Wang R., Huang W., Fang M., Chen P., Zhou B., et al. Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2. Nat. Commun. 2021;12:4210. doi: 10.1038/s41467-021-24514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.-J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kramer K.J., Johnson N.V., Shiakolas A.R., Suryadevara N., Periasamy S., Raju N., Williams J.K., Wrapp D., Zost S.J., Walker L.M., et al. Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Copin R., Baum A., Wloga E., Pascal K.E., Giordano S., Fulton B.O., Zhou A., Negron N., Lanza K., Chan N., et al. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. 2021;184:3949–3961.e11. doi: 10.1016/j.cell.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alamyar E., Duroux P., Lefranc M.-P., Giudicelli V. Humana Press; 2012. IMGT® Tools for the Nucleotide Analysis of Immunoglobulin (IG) and T Cell Receptor (TR) V-(D)-J Repertoires, Polymorphisms, and IG Mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS; pp. 569–604. [DOI] [PubMed] [Google Scholar]
- 61.Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 63.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. scp>UCSF ChimeraX</scp> : Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlasova A.N., Diaz A., Damtie D., Xiu L., Toh T.H., Lee J.S.Y., Saif L.J., Gray G.C. Novel Canine Coronavirus Isolated from a Hospitalized Patient With Pneumonia in East Malaysia. Clin. Infect. Dis. 2022;74:446–454. doi: 10.1093/cid/ciab456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Georgiev I.S., Joyce M.G., Yang Y., Sastry M., Zhang B., Baxa U., Chen R.E., Druz A., Lees C.R., Narpala S., et al. Single-Chain Soluble BG505.SOSIP gp140 Trimers as Structural and Antigenic Mimics of Mature Closed HIV-1 Env. J. Virol. 2015;89:5318–5329. doi: 10.1128/jvi.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaas Q., Ruiz M., Lefranc M.P. IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res. 2004;32:D208–D210. doi: 10.1093/nar/gkh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohi M., Li Y., Cheng Y., Walz T. Negative staining and image classification — powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mastronarde D.N. SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microsc. Microanal. 2003;9:1182–1183. doi: 10.1017/s1431927603445911. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Sequences for antibodies identified and characterized in this study have been deposited to GenBank under GenBank accession numbers Genbank: OR486973-OR487050 (for 71281 antibodies) and Genbank: OR487051-OR487090 (for 71282 antibodies). Raw sequencing data used in this study are available on the Sequence Read Archive under BioProject accession number SRA: PRJNA1014462. nsEM map data of Fab 71281–33 in complex with SARS-CoV-2 spike is available on the Electron Microscopy Databank under accession numbers EMDB: EMD-41075 and EMDB: EMD-41076.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.