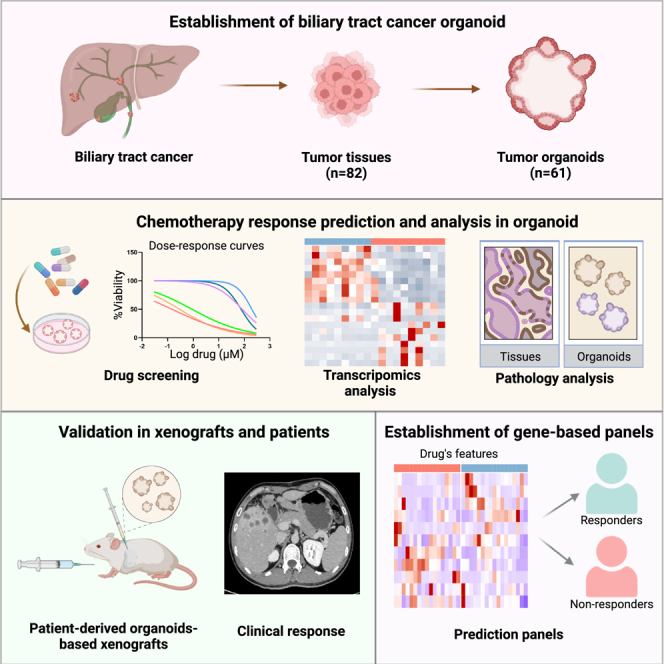
Summary
Patients with biliary tract cancer (BTC) show different responses to chemotherapy, and there is no effective way to predict chemotherapeutic response. We have generated 61 BTC patient-derived organoids (PDOs) from 82 tumors (74.4%) that show similar histological and genetic characteristics to the corresponding primary BTC tissues. BTC tumor tissues with enhanced stemness- and proliferation-related gene expression by RNA sequencing can more easily form organoids. As expected, BTC PDOs show different responses to the chemotherapies of gemcitabine, cisplatin, 5-fluoruracil, oxaliplatin, etc. The drug screening results in PDOs are further validated in PDO-based xenografts and confirmed in 92.3% (12/13) of BTC patients with actual clinical response. Moreover, we have identified gene expression signatures of BTC PDOs with different drug responses and established gene expression panels to predict chemotherapy response in BTC patients. In conclusion, BTC PDO is a promising precision medicine tool for anti-cancer therapy in BTC patients.
Keywords: biliary tract cancer, organoids, chemotherapy, personalized drug screening
Graphical abstract
Highlights
-
•
A living biobank of biliary tract cancer (BTC) organoids is derived from patients
-
•
Organoids allow effective chemotherapeutic drug screening that is validated in vivo
-
•
Identification of gene expression signatures of BTC with different drug responses
-
•
Establishment of gene-based panels to predict chemotherapy response in BTC patients
Ren et al. establish patient-derived BTC organoids with characteristics of originated tumors that could be applied for personalized drug screening of BTC patients. Moreover, they identify gene expression signatures of BTC organoids with different drug responses and establish gene expression panels to predict chemotherapy response in BTC patients.
Introduction
Biliary tract cancer (BTC) ranks among the most lethal human malignancies, and it refers to a spectrum of invasive adenocarcinomas, including intrahepatic cholangiocarcinoma (ICC), extrahepatic cholangiocarcinoma (ECC), and gallbladder carcinoma (GBC).1 The incidence and mortality of BTC, especially ICC, are increasing rapidly.2 Surgical resection combined with postoperative chemotherapy is the only potentially curative therapy for BTC patients with 65% relapse rate in 5 years.3,4,5 The majority of BTC patients are diagnosed at an unresectable stage, and the median overall survival is reported to be 7.4–13.8 months,6 for whom only systemic therapies are available.7 Although immune checkpoint inhibitors and targeted therapies, such as anti-PD-L1 antibody, mutant-IDH1 inhibitor, and FGFR inhibitor, were reported to modestly improve patients’ outcome recently, systemic chemotherapy represents the backbone of palliative treatment.8,9,10,11 The most common chemotherapeutic drugs for BTC include gemcitabine, cisplatin, 5-fluoruracil (5-FU), oxaliplatin, irinotecan, mitomycin C, and paclitaxel. However, the efficacy of chemotherapy is highly varied in patients with BTC.
The development of effective treatments for BTC has been hindered by the shortage of effective ways to predict chemotherapeutic response. Patient-derived organoid (PDO), which is a culture system of a three-dimensional (3D) structure isolated and cultured from cancer cells, was reported to accurately recapitulate tissue architecture in solid tumors such as colon, pancreas, and liver cancers. Some researchers have reported the establishment of PDOs derived from BTC patients.12,13,14 However, the sample sizes of previous studies of drug screening on BTC PDOs were relatively small, and gene expression signatures related to chemosensitivity based on PDOs’ drug sensitivity remain largely unknown. Therefore, personalized drug screening based on a large biobank of BTC organoids with analyses about gene signatures is needed to identify marker genes related to chemosensitivity.
In this study, we established a living BTC organoid library including 61 human BTC organoid cultures that represented each patient’s tumor characteristics. We analyzed the molecular characteristics of BTC tissues with successful or failed organoid cultures. Personalized drug screening was performed on PDOs being treated with clinically used drugs. The responses to drugs in PDOs were consistent with the treatment effects in PDO-based xenografts (PDOXs) in mice and in actual treatment response in individual BTC patients. Finally, we identified gene expression features of BTC tissues with different drug sensitivities in PDOs and established gene expression-based panels to provide novel clinical decision-making support tools for BTC patients.
Results
Establishment of a living BTC organoid biobank
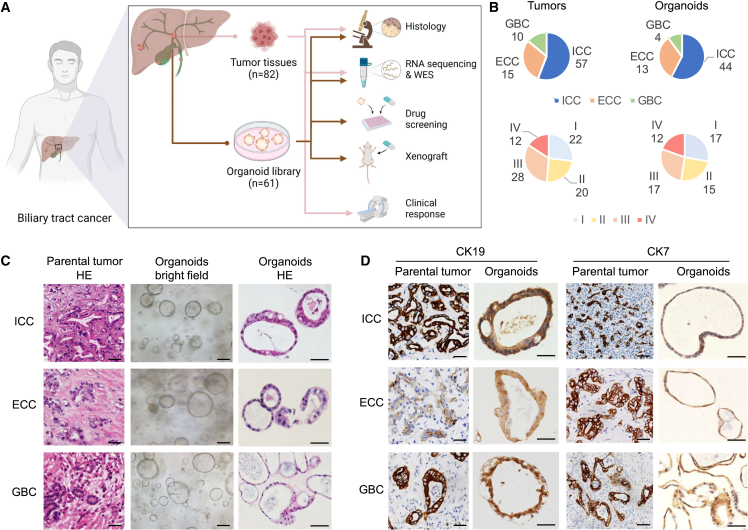
To establish a living BTC organoid biobank, we obtained 82 primary tumor and malignant ascite samples from 72 patients with BTC (Figure 1A), including 57 ICCs, 15 ECCs, and 10 GBCs (Figure 1B). The detailed clinicopathological information is shown in Table S1. We established 61 organoids from dissociated BTC tissues with a success rate of 74.4% (61/82) (Table S1), including 44 ICCs, 13 ECCs, and 4 GBCs (Figure 1B). BTC organoid was defined as the multicellular in vitro structures derived from fresh BTC tissues that resemble features of their original tissues and that have the ability to self-organize and self-renew. Each PDO was established with the irregularly shaped cyst-like structure via primary culturing in Matrigel and then passaged one time to confirm viability (Figure 1C).15,16,17 Among the successfully established organoids, 77% (47/61) exhibited long-term growth and were used to perform a drug sensitivity test with seven drugs (Table S3), while 23% (14/61) exhibited short-term growth and were tested by one to two drugs (Table S3). This organoid biobank encompassed different anatomical subtypes of BTC, as well as different clinical tumor stages (Figure 1B), indicating that organoids can be established from different subtypes of BTCs.
Figure 1.
PDOs established from BTC patients
(A) Experimental design. Fresh BTC samples were obtained from patients (patient information is detailed in Table S1) and processed as described in STAR Methods.
(B) Pie charts comparing the stratification of all tumors with that of all derived organoids based on histological type (top) and tumor-node-metastasis (TNM) stage (bottom).
(C) Representative H&E and bright-field microscopy images of patient-derived BTC organoids and matched primary tumors from three main histological subtypes. Scale bars, 50 μm.
(D) IHC analysis for the BTC markers (CK19 and CK7) on parental tumors and organoids. Scale bars, 50 μm.
To evaluate the histological features of BTC organoids, hematoxylin and eosin (H&E) staining (Figure 1C) and immunohistochemistry (IHC) (Figure 1D) were performed. BTC organoids exhibited irregularly shaped cyst-like structures and glandular shapes, similar to the H&E staining results of tumor tissues from which the organoids were originally derived (Figure 1C). The protein expression pattern of cytokeratin 19 (CK19) and cytokeratin 7 (CK7) was matched with PDOs and tissues (Figure 1D), even for those derived from biopsies or ascites (Figure S1A) of BTC patients. Furthermore, the organoids and their corresponding parental tumors shared similar mutational profiles with a concordance rate greater than 51% (51%–81%) (Figure S1B) as determined by whole-exome sequencing. This is consistent with previous reports.18,19 Importantly, both the recurrent mutations (Figure S1C) and copy number alterations (Figure S1D) of the organoids recapitulated the genomic alterations in human BTC. We further established an orthotopic model and subcutaneous model, respectively. The xenografted organoids grew robustly in vivo (Figure S1E). H&E staining and IHC of CK19 staining showed that tumors from xenografted organoids recapitulated the histopathological features of the primary tumors obtained from BTC patients (Figure S1F). Taken together, we established a living BTC organoid biobank in which these BTC organoids recapitulated and retained the histological and genetic characteristics of their original tumor tissues.
Clinical and molecular characterizations of BTCs that could establish PDOs
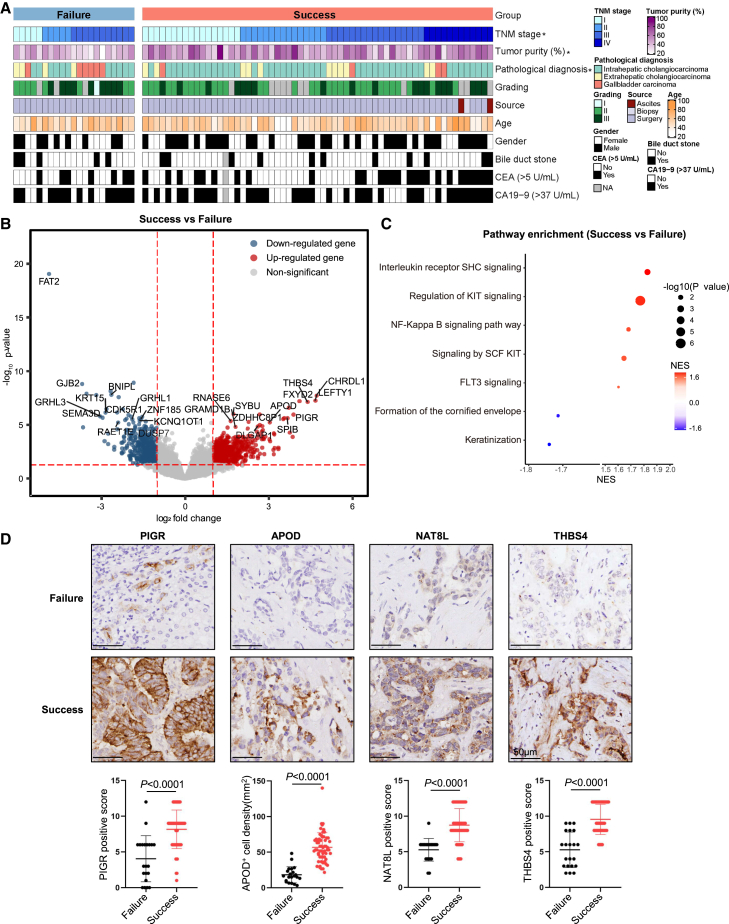
To characterize signatures of tumor tissues with successful organoid cultures, we compared their clinical features and expression profiles to the tumor tissues that failed to establish organoid cultures. All advanced BTC tissues (TNM stage IV) could successfully culture organoids (Figure 2A; Tables S1 and S2). The tumor content of the original tissues evaluated by H&E staining was also positively associated with PDO derivation (Figures 2A and S2A). In addition, it was easier for ICCs and more difficult for GBCs to establish PDOs (Figure 2A; Table S2). Except for TNM stage, tumor content, and pathological diagnosis, other clinicopathologic characteristics did not show significant correlation with the success of BTC organoid generation (Figure 2A; Table S2).
Figure 2.
Features of the successful organoid culture tissues
(A) Clinical features of successful organoid culture tissues. Clinical features are indicated with a color code. ∗p < 0.05.
(B) Volcano plots showing differentially expressed genes of successful organoid culture tissues and failed organoid culture tissues. Wilcoxon rank-sum test. Benjamini-Hochberg (BH) adjusted p value < 0.05.
(C) Pathways enriched in successful and failed organoid culture tissues. Permutation test. BH adjusted p value < 0.05. NES, normalized enrichment scores.
(D) Representative IHC images of formalin-fixed paraffin-embedded BTC samples belonging to the two groups and stained with the indicated specific antibodies. Quantification of staining intensities for the indicated markers was performed in 80 BTC tissue samples. Scale bar, 50 μm.
RNA sequencing was conducted to compare the expression profiles between BTC tissues with successful or failed organoid cultures. Typical stemness and proliferation genes including GRAMD1B, DLGAP1, PIGR, and GNMT were highly expressed in tumor tissues with successful organoid cultures, whereas tumor suppressor genes and anti-proliferation genes such as CRYM-AS1, KCNQ1OT1, PLAT, and DHRS9 were upregulated in tumor tissues with failed organoid cultures (Figure S2B). We found that stem cell-related genes ANPEP, PIGR, and APOD and proliferation genes CHRDL1, FXYD2, THBS4, LEFTY1, and NAT8L were highly expressed, whereas genes that inhibit tumor growth such as FAT2, GJB2, BNIPL, GRHL3, and KRT15 were significantly downregulated in the successful group (Figures 2B and S2C). Consistently, gene set enrichment analysis (GSEA) of differentially expressed genes (DEGs) confirmed that proliferation- and stemness-related pathways were significantly enriched in tumor tissues with successful organoid cultures (Figure 2C). The protein expression levels of stem cell-related genes PIGR and APOD as well as proliferation-related genes NAT8L and THBS4 were significantly higher in successful organoid cultures (Figure 2D) as determined by IHC. These results indicated that BTCs with the features of TNM stage IV, high tumor purity, and enhanced gene expression of stemness and proliferation were more likely to generate organoid successfully.
Drug screening of conventional chemotherapeutics in established BTC PDOs
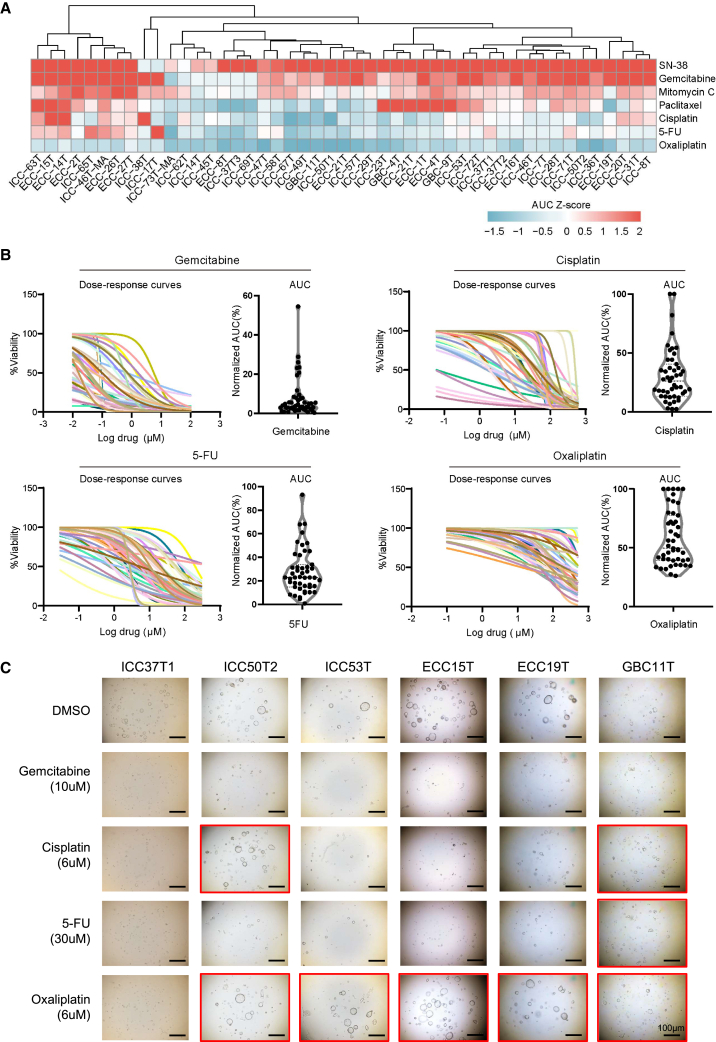
To explore the responses of BTC patient-derived tumor organoids to different chemotherapeutics, seven commonly used chemotherapeutic drugs including gemcitabine, cisplatin, capecitabine or 5-fluorouracil (5-FU), SN-38 (active metabolite of irinotecan), oxaliplatin, mitomycin C, and paclitaxel were treated in 47 BTC organoids. Each drug was screened at five different concentrations. Specifically, two different passages of organoid lines were treated with the above drugs in different concentrations to evaluate their responses. The number of viable cells was quantified and analyzed at the end of treatment in each experiment by dose-response curves and area under the curve (AUC). We tested the AUC values of two organoid passages from the same organoid line, and the AUC values of early-passage and late-passage organoids were highly consistent (Pearson correlation R2 = 0.927) (Figure S3A). AUC values of the same organoid line tested by different operators also showed high consistency (Pearson correlation R2 = 0.98) (Figure S3B). In addition, we tested AUC values of the same organoid line before freezing and after thawing, and the results were highly consistent (Pearson correlation R2 = 0.9293) (Figure S3C). These results indicated that our culture system is stable and almost unaffected by organoid passages, different operators, freezing, and thawing.
We observed that the responses to chemotherapeutic drugs varied among organoids generated from different BTC patients (Figure 3A). For each chemotherapeutic agent, the BTC PDOs were divided into three subgroups: resistant (top 33% AUC), sensitive (lowest 33% AUC), and intermediate response (middle 34% AUC). The detailed AUC values and sensitivities are shown in Table S3. Drug sensitivities of BTC PDOs showed substantial interpatient variability in response to single chemotherapy agents as evaluated using dose-response curves and the corresponding AUC, including gemcitabine, cisplatin, 5-FU, oxaliplatin (Figure 3B), SN-38, mitomycin C, and paclitaxel (Figure S3B). Some PDOs were resistant and some partially sensitive or sensitive to gemcitabine, cisplatin, 5-FU, oxaliplatin (Figure 3C), SN-38, mitomycin C, and paclitaxel (Figure S3C). For instance, 5-FU, as a guideline recommend adjuvant agent following curative resection of BTC, showed heterogeneous responses in organoid drug testing (Figure 3C). Thus, BTC PDOs are potentially effective tools for drug screening with varied responses to different conventional chemotherapeutics.
Figure 3.
Clinically relevant responses to chemotherapy in BTC organoids ex vivo
(A) The heatmap consists of 47 BTC organoids’ chemosensitivity responses, which included gemcitabine, cisplatin, 5-FU, SN-38, oxaliplatin, mitomycin C, and paclitaxel (n = 3). Chemosensitivity responses were awarded by AUC Z scores. Colors range from blue (low scores, meaning resistant) to red (high sores, meaning sensitive).
(B) Summary of chemosensitivity responses for 47 BTC organoids ex vivo to gemcitabine, cisplatin, 5-FU, and oxaliplatin, and results are displayed in the form of dose-response curves (n = 3). AUC was calculated from the raw dose-response data and is displayed as a violin plot.
(C) Effects on the viability of chemotherapy drug using an organoid-formation assay. Red square, resistant; no square, no viable cells. Scale bars, 100 μm.
Drug response status observed in BTC PDOs were validated in PDOX mice and in corresponding follow-up patients
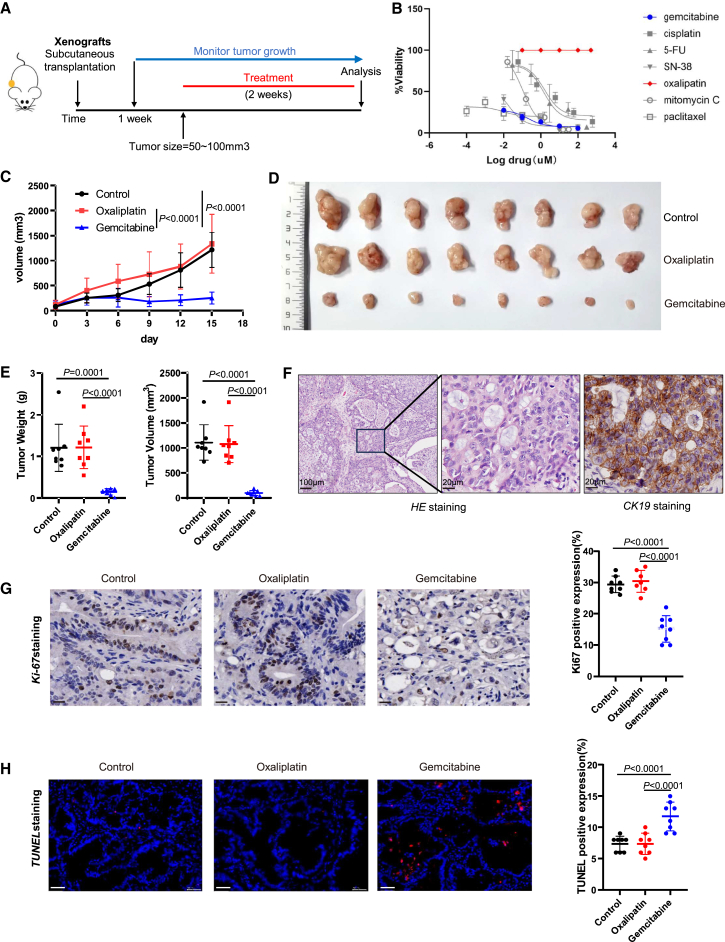
To validate the drug screening results in BTC PDOs, we established a PDOX mice model. Either lines of BTC organoids ICC47T-PDO, ICC46T-PDO, or ICC62T-PDO were transplanted subcutaneously into NOD-Prkdcem26Cd52Il2rgem26Cd22/Nju (NCG) mice. Once tumors reached 50–100 mm3, the mice were injected intraperitoneally with either chemotherapeutic drugs or the vehicle twice a week (Figure 4A). According to the drug-testing results of ICC47T-PDO (Figure 4B), ICC46T-PDO (Figure S4A), and ICC62T-PDO (Figure S4E) in vitro, we chose gemcitabine as the sensitive drug and oxaliplatin as the resistant drug. Similar conclusions of ICC46T-PDO and ICC62T-PDO were reached by drug screening (Figure S4A and Figure S4E). As expected, we observed significant inhibition of tumor growth in the gemcitabine treatment group, while the mice showed no response in the oxaliplatin treatment group in ICC47T-PDOXs (Figure 4C), ICC46T-PDOXs (Figure S4B), and ICC62T-PDOXs (Figure S4F). The tumor burdens, as indicated by tumor weight and tumor volume, were significantly decreased in the gemcitabine group compared with the oxaliplatin group and the control group (Figures 4D and 4E). Both H&E staining and positive staining of CK19 confirmed the pathological diagnosis of ICC (Figure 4F). Similar results were observed in ICC46T-PDOXs (Figures S4C and S4D) and ICC62T-PDOXs (Figures S4G and S4H). In keeping with this, cell proliferation was significantly decreased (p < 0.001) (Figure 4G), while cell apoptosis was significantly increased (p < 0.001) (Figure 4H) in tumor tissues from PDOXs treated with gemcitabine, compared with untreated PDOXs, whereas no difference was observed in the oxaliplatin treatment PDOXs. Taken together, these results demonstrated the drug screening results identified in BTC PDOs were verified in PDOX mice.
Figure 4.
Clinically relevant responses to chemotherapy in BTC organoids in vivo
(A) Experimental design. BTC organoids were transplanted subcutaneously in immunocompromised NCG mice and treated with either chemotherapeutic drugs or the vehicle twice a week when tumor sizes grow to 50–100 mm3.
(B) PDO sensitivity profiles. Dose-response curves of BTC organoids treated with the chemotherapeutic drugs, as measured using the Cell Titer-Glo cell viability assay (n = 3).
(C) Growth curves of PDO subcutaneous xenografts in NCG mice. Mice were treated with oxaliplatin, gemcitabine, or vehicle twice a week for 14 days (n = 8). Results are shown as the tumor volume (mean ± SD).
(D) A representative image of tumor xenografts harvested 14 days after treatment with oxaliplatin, gemcitabine, or vehicle (n = 8).
(E) The tumor weight and volume of the drugs/vehicle mice at the experimental endpoint.
(F) Histological analysis of the origin on mice’s xenografts was assessed by H&E and CK19. Representative H&E (left) and CK19 IHC (right). Scale bars from left to right magnification are as follows: 100 μm, 20 μm (H&E), 20 μm (CK19).
(G) Histological analysis of the tumor proliferation on PDO xenografts was assessed after the chemotherapy treatment. Representative Ki-67 IHC for oxaliplatin, gemcitabine, and vehicle group and quantification of the relative intensity for Ki-67-positive nuclear immunostaining in xenografts (n = 8). No difference was observed between vehicle- and oxaliplatin-treated groups. Scale bar, 20 μm (Ki-67).
(H) Histological analysis of the antitumor efficacy on PDO xenografts was assessed after the chemotherapy treatment. Representative TUNEL staining for each quantification of the relative intensity for TUNEL-positive in xenografts (n = 8). Scale bar, 50 μm (TUNEL).
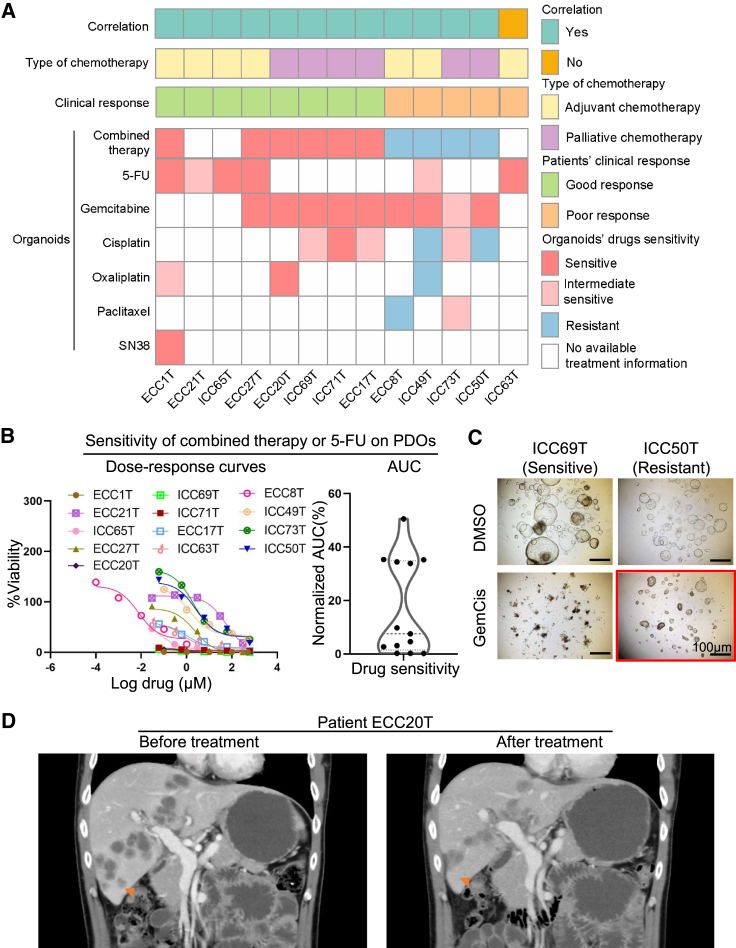
We further validated the drug screening results from PDOs in BTC patients with actual clinical treatment and outcomes (Figure 5A; Table S4). In terms of drug screening results from PDOs, we defined the chemotherapy regimen as resistant when it contained a resistant chemotherapeutic agent.20 In addition to single chemotherapy agents, we tested the response of PDOs to combined treatments of different drugs, the same as the corresponding BTC patients’ regimen (Figure 5B). Consistently, the combined therapy showed heterogeneous responses in organoid drug testing as the response observed in BTC patients (Figure 5C).
Figure 5.
Comparison of drug response between patients on drug treatment and their corresponding cancer organoids
(A) The heatmap summarizes the results for the 13 PDOs and corresponding patient drug responses.
(B) Ex vivo chemosensitivity of 13 BTC organoids to combined chemotherapy or 5-FU in the form of dose-response curves are displayed for each organoid (n = 3 independent experiments for each). AUC was calculated from the raw dose-response data and is displayed as a violin plot.
(C) Effects on the viability of treatment with gemcitabine plus cisplatin (GemCis) using an organoid-formation assay. Red square, resistant; no square, no viable cells. Scale bar, 100 μm.
(D) CT scan of patient ECC20T before and 3 months after treatment with gemcitabine plus oxaliplatin. The liver metastasis disappeared or showed shrinkage in size.
Thirteen BTC patients who underwent chemotherapy and had the drug screening in their derived organoids were followed up for 4 to 26 months (Table S4). According to the BILCAP trial,5 a patient who had recurrence within 17.5 months after surgery was defined as a clinically poor responder to adjuvant treatment. Seven patients underwent adjuvant treatment after surgery. Among these patients, cases ECC1T, ICC65T, and ECC27T were clinically sensitive responders, which was consistent with the drug responses in their derived PDOs (Figure 5A; Table S4). Patient ECC21T remained disease-free for 22 months after surgery and thus was a clinically sensitive responder to 5-FU, whose PDOs showed an intermediate response to 5-FU. The remaining three patients were resistant to adjuvant chemotherapy, two of whom (ECC8T and ICC49T) were consistent with drug responses in their PDOs (Figure 5A; Table S4). Moreover, six advanced BTC patients received palliative chemotherapy. Based on Response Evaluation Criteria in Solid Tumors (version 1.1),6 four patients (ECC20T, ICC69T, ICC71T, and ECC17T) showed partial response or stable disease. For example, the metastasis nodules in liver or retroperitoneal lymph nodes in patient ECC20T shrunk significantly. Accordingly, PDO drug screening showed sensitive chemotherapy (Figure 5D). On the other hand, patient ICC50T progressed rapidly under treatment with gemcitabine plus cisplatin (Figure S5A), whose PDO was markedly resistant to cisplatin. Patient ICC73T, with a large tumor (52 × 36 mm) in liver and many metastatic nodules in abdomen, was clinically resistant to combination therapy, whose PDOs showed intermediate response to cisplatin-gemcitabine-nab-paclitaxel. All six patients who received palliative chemotherapy were consistent with the matched PDOs’ drug sensitivity testing (Figure 5A). In total, 12/13 (92.3%) BTC patients showed consistent results of drug response as observed in their derived PDOs (Table S4). These results indicated that PDOs are useful tools for drug screening and prediction of drug response in BTC patients.
Transcriptional characteristics of BTC PDOs are associated with the responses to chemotherapy
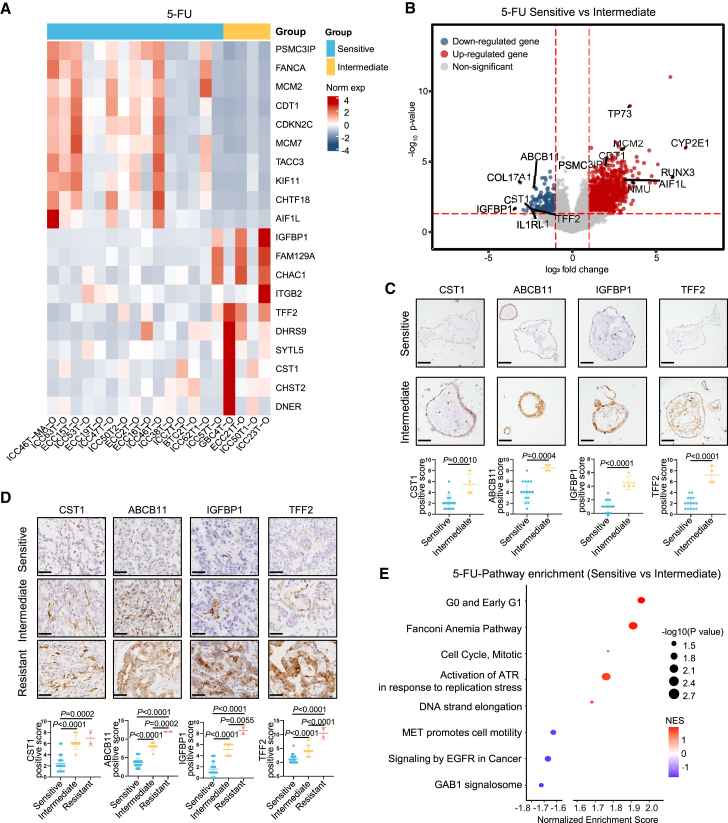
To investigate whether PDOs’ transcriptional characteristics could predict treatment responses of BTC patients to chemotherapy, 19 BTC organoids were used for transcriptome sequencing and divided into three subgroups based on their responses to each drug: resistant (top-tier AUC), sensitive (low-tier AUC), and intermediate (middle-tier AUC). We further explored the gene expression characteristics of PDOs from each drug group. 5-FU is the active metabolite of capecitabine, which is the first-line adjuvant treatment for BTC after surgery. We identified 1,174 DEGs between the 5-FU-sensitive group and 5-FU-intermediate group (Figures 6A and 6B). Significantly upregulated genes related to tumor apoptosis and sensitivity to chemotherapy were identified in the 5-FU-sensitive group compared to 5-FU-intermediate organoids, including PSMC3IP, FANCA, MCM2, CDT1, and CDKN2C (Figure 6A). Meanwhile, some drug resistance-related genes, such as IGFBP1, FAM129A, TFF2, and DHRS9, were significantly upregulated in the 5-FU-intermediate group (Figure 6A). Specifically, PDOs from the 5-FU-sensitive group showed high expression of apoptosis regulation factor TP7321 and CDT1,22 pro-chemotherapy sensitive factor MCM2,23 and antitumor factor AIF1L22 (Figure 6B). The protein expression of these genes was confirmed immunohistochemically in derived organoids (Figure S6A) and tumor specimens (Figure S6B) by IHC.
Figure 6.
Transcriptional characteristics of 5-FU-sensitive group
(A) Heatmap of the expression levels of differentially expressed genes between 5-FU-sensitive group and 5-FU-intermediate group.
(B) Volcano plots showing differentially expressed genes of 5-FU-sensitive group and 5-FU-intermediate group. Wilcoxon rank-sum test. BH adjusted p value < 0.05.
(C) IHC staining of 5-FU-resistant markers in the derived organoids. Quantification of staining intensities for the indicated markers was performed in 19 organoid samples. Scale bars, 20 μm.
(D) IHC staining of 5-FU-resistant markers in tumor specimens of the whole cohort. Quantification of staining intensities for the indicated markers was performed in 45 BTC tissue samples. Scale bars, 50 μm.
(E) Pathways enriched in the 5-FU-sensitive group and 5-FU-intermediate group. Permutation test. BH adjusted p value < 0.05. NES, normalized enrichment scores.
On the other hand, protein expression of pro-resistance factors CST124 and ABCB1125 and pro-stemness factors IGFBP126 and TFF227 was significantly upregulated in PDOs from the 5-FU-intermediate group (Figure 6B). The protein expression features of these drug resistance-related genes were verified in organoids (Figure 6C) and BTC patients (Figure 6D) by IHC. We next performed GSEA to identify potential regulated pathways. Fanconi anemia pathway, which could regulate the tumors’ hypersensitive to chemotherapy, was upregulated in the 5-FU-sensitive group (Figure 6E). ATR pathway was also activated in response to stalled DNA replication forks to promote antitumor activity in the 5-FU-sensitive group (Figure 6E).
Administration of gemcitabine plus cisplatin (GemCis) is the current standard therapy for advanced BTC. We further compared different PDOs’ transcriptional characteristics in response to cisplatin. We identified feature genes from the cisplatin-sensitive group, cisplatin-intermediate group, and cisplatin-resistant group (Figure S7A). Dozens of genes were affected in cisplatin-resistant PDOs, such as DNAJC12, HTR7, IFRD1, and DLL4 (Figure S7A). The protein expression of drug resistance-related genes was verified in PDOs (Figure S7B) and tumor tissues (Figure S7C) by IHC. Sensitive biomarkers for cancer chemotherapy response such as CEACAM5 and ZC3H12D were upregulated in the cisplatin-sensitive group (Figure S7A). Besides, the protein expression levels of cisplatin-sensitive-related genes were consistent with RNA expression through IHC staining in organoids (Figure S7D) and BTC patients (Figure S7E). Moreover, programmed cell death and TP53 regulation pathways were enriched in the cisplatin-sensitive group (Figure S7F). Overall, these results indicated that the PDOs’ transcriptional characteristics helped to explain a potential mechanism leading to diverse responses of BTC PDOs to chemotherapy.
Prediction model of drug response based on drug sensitivity of BTC organoids
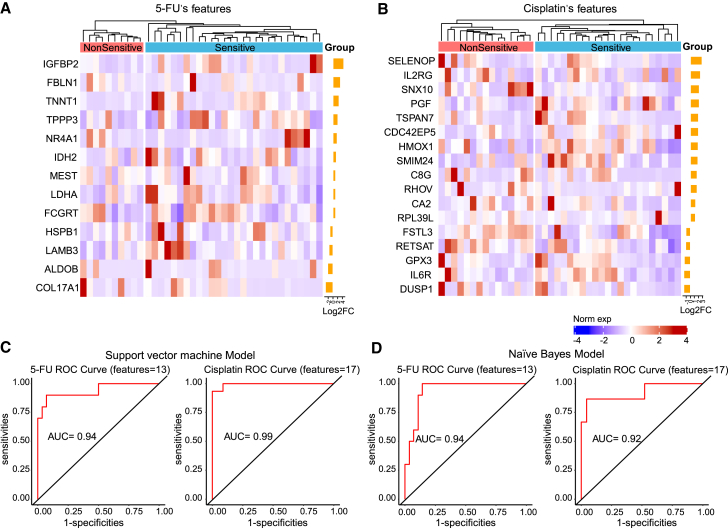
5-FU is a guideline recommend adjuvant agent following curative resection of BTC, and GemCis is the standard first-line treatment of advanced BTC. We thus explored gene expression biomarkers of tumor tissues to discriminate sensitive PDOs from non-sensitive (including intermediate and resistant) PDOs to 5-FU (Figure 7A) and cisplatin (Figure 7B), respectively.
Figure 7.
Prediction model based on drug sensitivity of BTC organoids
(A and B) Heatmap shows drug response signatures were selected from DEGs between sensitive group and non-sensitive group for 5-FU (A) and cisplatin (B).
(C) ROC curve of SVM model based on drug sensitivity of 5-FU and cisplatin.
(D) ROC curve of NB prediction model based on drug sensitivity of 5-FU and cisplatin.
A panel containing 13 genes was identified in 5-FU-sensitive PDOs compared to 5-FU-non-sensitive PDOs (Figure 7A). These 13 genes were utilized to construct support vector machine (SVM) and naive Bayes (NB) models. Receiver operating characteristic (ROC) curves and AUC were calculated to evaluate model quality. This gene expression biomarker panel could discriminate 5-FU-sensitive patients and non-sensitive patients at AUCs of 93.9% (95% confidence interval (CI): 83.9%–99.9%) for SVM model (Figure 7C) and 93.9% (95% CI: 86.7%–99.9%) for NB model (Figure 7D). A similar analysis was performed to identify the gene expression biomarkers to discriminate sensitive PDOs from non-sensitive PDOs to cisplatin treatment (Figure 7B). The expression biomarker panel of 17 genes could discriminate cisplatin-sensitive patients and non-sensitive patients at AUCs of 99.4% (95% CI: 98.0%–99.9%) for SVM model (Figure 7C) and 92.2% (95% CI: 82.4%–99.9%) for NB model (Figure 7D). These findings suggested that gene expression biomarker panels from BTC organoids could distinguish sensitive patients from non-sensitive patients to chemotherapies of 5-FU and cisplatin.
Discussion
In this study, we successfully generated a BTC PDO library and demonstrated that PDOs can predict chemotherapy response for patients with BTC. Our BTC organoid library included 61 human BTC organoid cultures that represented each patient’s tumor characteristics, with a success rate of 74.4% for organoid cultures. We assessed the phenotypic and genetic characteristics of BTC PDOs and confirmed the consistency between organoids and their corresponding tumors tissues. In particular, our results showed that BTC PDOs exhibited similar histological features and protein expression as the subtypes of their corresponding primary tumors. Hence, BTC organoids were suitable for cancer research and drug response assessment in vitro.
To identify the gene expression features associated with the establishment of organoid culture, we compared the transcriptome differences between the tumor tissues that successfully established organoid and those failed to form organoids. Stemness- and proliferation-related pathways were found to be significantly activated in the successful organoid culture tissues, which is supported by a recent study from Nuciforo et al.,19 and a report showed a positive correlation between the derivation success rate and the proliferation index of the original tumor.12 Thus, BTC tissues with stronger stemness and proliferative capacity are more likely to succeed in establishing organoids.
We established a robust PDO drug screening system and found substantial interpatient heterogeneity of responses to different chemotherapeutic agents through drug sensitivity assays using PDOs. Our results demonstrated that the responses to chemotherapeutic agents varied among organoids generated from different BTC patients. For each drug, the BTC PDOs also present diverse responses. The response to compounds in PDO cultures was consistent with the subcutaneous xenograft tumors in mice. In particular, we testified to the drug screening results of PDOs in BTC patients. Eight BTC patients retained no evidence of disease status or stable disease, and their organoids showed sensitivity to chemotherapy in vitro, while four BTC patients whose organoids showed resistance to chemotherapy in vitro had recurrence or metastasis after chemotherapy in our cohort. These patients’ clinical outcomes further supported that BTC organoids are effective tools and can predict response to chemotherapy.
We further explored the potential mechanisms of sensitivity to chemotherapy in BTC organoids through RNA sequencing, which were verified in derived organoids and tumor tissues. In organoids sensitive to chemotherapeutics, multiple well-acknowledged genes that sensitized tumor cells to cytotoxic drugs and to apoptosis were upregulated including RUNX328 and CDT1,22 which were previously reported to inhibit tumor cells and to facilitate drug transport. In organoids resistant to chemotherapeutics, several drug resistance-related genes and invasion-related genes were upregulated, including IGFBP1 in 5-FU-intermediate-response PDOs and DNAJC12 in cisplatin-resistant PDOs. Overexpression of IGFBP1 induced resistance in first-line treatment by activation of the extracellular signal-regulated kinases 1/2 (ERK1/2).26 DNAJC12 could enhance aerobic glycolysis and drug resistance to chemotherapy through regulating β-catenin expression.29 We revealed that Fanconi anemia pathway, activation of ATR in response to replication stress pathway, and TP53 regulation pathways were significantly activated in the 5-FU and cisplatin chemotherapy-sensitive group. On the other hand, signaling by epidermal growth factor receptor (EGFR) in the cancer pathway and MET-related pathway was enriched in the 5-FU chemotherapy-resistance group. Alteration of the above pathways are classic mechanisms of drug response.30,31 With the PDO model, not only patients can benefit from precision medicine based on individual drug screening results; we can also explore the potential causes of drug resistance, which might help overcome drug resistance in future studies.
Based on individual drug sensitivities in organoids and DEGs found in PDOs’ transcriptional characteristics, we derived the transcriptional signatures from corresponding tumor tissues and established prediction models of drug response to 5-FU and cisplatin, with AUC of 0.92–0.99. Since approximately 30% of BTC patients failed to establish PDOs, organoid culture system has not been widely established, and because some patients could not afford the high cost of a drug test using organoids, it is hard to use PDOs to test drug sensitivities for every patient. Thus, prediction models could help identify responders to chemotherapy agents and enable stratification of BTC patients so that treatments could be tailored for each individual patient. However, these models need to be refined and prospectively evaluated on larger cohorts of BTC patients.
BTC organoids are promising to be used to help clinical decision-making and to optimize patient outcome. Tumor organoids also have great potential in preclinical research such as disease modeling, drug discovery, and genetic engineering. The current culture system of tumor organoids did not contain the tumor microenvironment such as fibroblasts or immune cells. In the future, it is essential to establish the co-culture system of organoid with autologous fibroblasts or immune cells so as to predict the treatment response to immunotherapy. In addition, the patients’ sample size is still limited.
In conclusion, we successfully established a patient-derived BTC organoid library. The PDOs represented the original tumor histological characteristics of original BTC tissues from patients. The established PDOs were effective tools for personalized chemotherapeutic drug screening. A chemotherapy response prediction gene panel was established based on the drug sensitivities of BTC organoids for helping in the selection of effective drugs for individual BTC patients.
Limitations of the study
Among the limitations of the study, our current culture system of tumor organoids did not contain the tumor microenvironment such as vascular structures, fibroblasts, or immune cells. This remains an obstacle in predicting the treatment response to immunotherapy and target therapy. In addition, the prospective study of BTC organoids with a large cohort is needed before implementation in clinical cancer care can be considered. Lastly, the clinical implications and translational potentials of our findings require further mechanistic studies and validation in larger patient cohorts.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Cytokeratin 7 | Abcam | Cat#ab68459; RRID: AB_1139824 |
| Anti-Cytokeratin 19 | Abcam | Cat#ab76539; RRID: AB_1523469 |
| Anti-Ki67 | Abcam | Cat#ab15580; RRID: AB_443209 |
| Anti-PIGR | Abcam | Cat#ab275020; RRID: N/A |
| Anti-NAT8L | Abcam | Cat#ab76842; RRID: AB_1523625 |
| Anti-APOD | Abcam | Cat#ab256496; RRID: N/A |
| Anti-THBS4 | Abcam | Cat#ab263898; RRID: AB_2922811 |
| Anti-IGFBP1 | CST | Cat#31025S; RRID: AB_2798998 |
| Anti-CST1 | Proteintech | Cat#16025-1-AP; RRID: AB_10916385 |
| Anti-TFF2 | Abcam | Cat#ab267474; RRID: N/A |
| Anti-ABCB11 | Proteintech | Cat#18990-1-AP; RRID: AB_2878571 |
| Anti-TP73 | Abcam | Cat#ab189896; RRID: AB_2941918 |
| Anti-MCM2 | Abcam | Cat#ab108935; RRID: AB_10859977 |
| Anti-CDT1 | Proteintech | Cat#14382-1-AP; RRID: AB_2076871 |
| Anti-AIF1L | Abcam | Cat#ab204493; RRID: N/A |
| Anti-DNAJC12 | Proteintech | Cat#12338-1-AP; RRID: AB_2246188 |
| Anti-HTR7 | Proteintech | Cat#13830-1-AP; RRID: AB_2122706 |
| Anti-IFRD1 | Santa cruz | Cat#sc-515012; RRID: N/A |
| Anti-DLL4 | Proteintech | Cat#21584-1-AP; RRID: AB_2878888 |
| Anti-CEACAM5 | Origene | Cat#TA803413; RRID: AB_2626814 |
| Anti-ZC3H12D | Proteintech | Cat#24991-1-AP; RRID: AB_2879834 |
| Anti-CLDN18 | Proteintech | Cat#21126-1-AP; RRID: AB_10733638 |
| Anti-ARID3A | Proteintech | Cat#14068-1-AP; RRID: AB_2060390 |
| Biological samples | ||
| Human biliary tract tumor sample | First Affiliated Hospital of Sun Yat-sen University | N/A |
| Human biliary tract tumor sample | Shunde Hospital of Southern Medical University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Cisplatin | Selleck | Cat#S1166 |
| Oxaliplatin | Selleck | Cat#S1224 |
| Gemcitabine | Selleck | Cat#S1714 |
| 5-Fluorouracil | Selleck | Cat#S1209 |
| SN-38 | Selleck | Cat#S4908 |
| Paclitaxel | Selleck | Cat#S1150 |
| Mitomycin C | Selleck | Cat#S8146 |
| B-27™ Supplement, serum free | Thermo Fisher Scientific | Cat#17504044 |
| N-2 Supplement | Thermo Fisher Scientific | Cat#17502048 |
| Nicotinamide | Sigma | Cat#N0636 |
| GlutaMAX | Gibco | Cat#35050061 |
| HEPES | Gibco | Cat#15630080 |
| N-acetyl-L-cysteine | Sigma | Cat#A9165-5G |
| Gastrin-1, human | Genscript | Cat#PR12740 |
| Recombinant Human EGF | Peprotech | Cat#AF-100-15 |
| Human HGF | Novoprotein | Cat#CJ72-50 |
| Recombinant Human FGF-10 | Peprotech | Cat#100-26 |
| A 83-01 | TOCRIS | Cat#2939 |
| Y-27632 | Selleck | Cat#S1049 |
| Recombinant Human Noggin | Peprotech | Cat#120-10C |
| Matrigel | Corning | Cat#356231 |
| Advanced DMEM/F-12 | Gibco | Cat#12634028 |
| Forskolin | Sigma | Cat#F6886-10MG |
| Pen/Strep | Gibco | Cat#15140122 |
| Collagenase type IV | Sigma | Cat#C5138 |
| dispase type II | Invitrogen | Cat#17101015 |
| Critical commercial assays | ||
| DeadEnd™ Fluorometric TUNEL System | Promega | Cat#G3250 |
| REAL™ EnVision™ Detection System | Dako | Cat#K500711-2 |
| CellTiter-Glo assay | Promega | Cat#G7570 |
| PureLink™ RNA | Thermo Fisher | Cat#12183020 |
| Deposited data | ||
| Raw and analyzed data | This paper | GSA: HRA005081 |
| Experimental models: Organisms/strains | ||
| NCG (NOD-Prkdcem26Cd52Il2rgem26Cd22/Nju) mice | GemPharmatech, Nanjing, China | Cat#T001475 |
| Software and algorithms | ||
| GraphPad 9.0 | GraphPad Software | N/A |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and inquiries regarding resources and reagents should be directed to the lead contact, Prof. Xu (xulixia@mail.sysu.edu.cn), who will fulfill these requests.
Materials availability
This study did not generate new unique reagents.
Data and code availability
The sequencing data reported in this paper are deposited in Genome Sequence Archive (GSA) with project number HRA005081 and publicly available as of the date of publication. The software and algorithms for data analyses used in this study are published and referenced throughout the STAR Methods section. No original code is featured in this paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Human specimens
Human biliary tract tumor specimen (1–4 cm3) were obtained through liver resection performed at the department of hepatobiliary surgery in the First Affiliated Hospital of Sun Yat-sen University and Shunde Hospital of Southern Medical University. Samples were confirmed to be biliary tract tumor on the basis of histopathological assessment. The diagnosis of each case was confirmed on routine combining H&E-stained slides and IHC-stained slides by two independent histopathologists. For each tumor specimen, samples were split into three parts and processed for histology, RNA isolation, or dissociated and processed for organoid culture. For three tumor specimens, samples were also processed for DNA isolation. This study was centrally approved by the Ethical committee of the First Affiliated Hospital of Sun Yat-sen University (No. [2020]052) and complied with all relevant ethical regulations.
PDOX models and chemotherapy in NCG mice
All mouse experiments have been regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by Sun Yat-sen University Animal Welfare and Ethical Review Body and have been performed in accordance with the license. For orthotopic grafts, 1,000,000 organoids in 20 μL Matrigel were injected under the hepatic capsule of 4-6-week-old male NCG (NOD-Prkdcem26Cd52Il2rgem26Cd22/Nju) mice (GemPharmatech, Nanjing, China). For subcutaneous grafts, 1,000,000 organoids and 100 μL Matrigel were mixed and injected into right axilla of 4-6-week-old male NCG. Visible tumors developed in 2–4 weeks approximately. Mice were sacrificed and tumors were culled when tumor size reached 2000mm3. We cut the tumors into 10 mm3 pieces and transplanted them into right axilla of NCG mice. After 2 weeks observation, mice with established subcutaneous tumors in approximate size were randomly divided into three groups (n = 8), which consists of drug-resistant group (oxaliplatin), drug-sensitive group (gemcitabine) and negative control group (physiological saline). Mice were injected with oxaliplatin (10 mg/kg) or gemcitabine (50 mg/kg) twice a week, and the tumors were measured three times a week using a caliper. Volumes were calculated by applying the formula v = 0.5 × L × w2 in vivo and v = 0.5 × L × w × h in vitro, where v is volume, L is length, w is width and h is height. Investigators performing tumor measurements were blinded to treatment groups. Mice were sacrificed after taking two weeks drug treatment. Tumor tissues were collected by applying to RNA and histological analyses.
Method details
Isolation and culture of tumoral organoids
Tumor-derived organoids were isolated by adapting this method as follows. Briefly, the patient-derived specimen was confirmed to be obtain from tissue less than 30 min and the sample was reserved in tissue storage solution (Miltenyi Biotec, Bergisch Gladbach, Germany) at 4°C. Then, specimen was minced and incubated at 37°C for 150 rpm with the digestion solution. The digestion medium involves advanced DMEM/F12 (Gibco, California, USA), Pen/Strep (Gibco), 100 U ml−1 collagenase type IV (Sigma Aldrich, Madison, USA), and 125 μg/mL dispase type II (Invitrogen, California, USA). Tissue digestion lasted for 1.5–2 h according to the content of fibrous tissue, which was evaluated on the resistance of the tissue to be minced. If a significant part of the original tissue was still underdigested, using 5mL trypsin-EDTA in the digestion solution at 37°C for 5 min could get a good yield of tumoral cells. After specimen digestion, the mixture was then filtered through a 70-μm nylon cell strainer and spun for 5 min at 2000 rpm. Then cell pellet was disposed by 1x RBC lysis buffer for 5 min at 4°C in order to remove the red blood cells and then spun for 5 min at 2000rpm. Isolated cells were embedded in Matrigel (Corning, NY, USA) and then seeded in a 48-multiwell plate with 35ul per well. After Matrigel had solidified, each well obtained 200ul organoid-specific isolation medium. The organoid medium contained: Advanced DMEM/F12 supplemented with 1% Pen/Strep, 1% glutamax (Gibco), 10-mM HEPES (Gibco), 1:50 B27 supplement without vitamin A (Thermo Fisher Scientific, Massachusetts, USA), 1:100 N2 supplement (Thermo Fisher Scientific), 1.25-mM N-acetyl-L-cysteine (Sigma), 10-mM nicotinamide(Sigma), 10-nM recombinant human (Leu15)-gastrin I (Genscript, Nanjing, China), 50 ng/mL recombinant human EGF (Gibco), 100 ng/mL recombinant human FGF10 (Novoprotein, Suzhou, China), 25 ng/mL recombinant human HGF (Novoprotein), 10 μM forskolin (Sigma), 5 μM A83-01 (TOCRIS, Bristol, England) and 10 μM Y27632(Selleck, Houston, USA). BTC organoids were passaged after incubation in TrypLE (GIBCO) for 15 min.
Histological evaluation of tissues and organoids under H&E and IHC staining
H&E and IHC were carried out using 5μm paraffin embedded tumor tissues or organoids. All of slides were dewaxed and hydrated. For H&E, the slides were stained with hematoxylin and eosin. For IHC, the slides were subjected to antigen retrieval using Ethylene Diamine Tetraacetic Acid (EDTA) sodium and blocked in 20% goat serum for 30 min to reduce background. The primary antibody, including anti-CK7 antibody (1:1000, Abcam, London, England), anti-CK19 antibody (1:2000, Abcam), anti-Ki67 antibody (1:500, Abcam), Anti-PIGR (1:200, Abcam), Anti-APOD (1:1000, Abcam), Anti-NAT8L (1:500, Abcam), Anti-THBS4 (1:300, Abcam), Anti-IGFBP1 (1:200, CST), Anti-CST1 (1:1000, Proteintech), Anti-TFF2 (1:500, Abcam), Anti-ABCB11 (1:2000, Proteintech), Anti-TP73 (1:1000, Abcam), Anti-MCM2 (1:500, Abcam), Anti-CDT1 (1:1000, Proteintech), Anti-AIF1L (1:200, Abcam), Anti-DNAJC12 (1:200, Proteintech), Anti-HTR7 (1:500, Proteintech), Anti-IFRD1 (1:400, Santa cruz), Anti-DLL4 (1:400, Proteintech), Anti-CEACAM5 (1:1000, Origene), Anti-ZC3H12D (1:400, Proteintech), Anti-CLDN18 (1:400, Proteintech) and Anti-ARID3A (1:500, Proteintech) was used for overnight at 4°C. After inoculating with HRP conjugated secondary antibody (Dako REAL EnVision Detection System, Peroxidase/DAB+, Rabbit/Mouse, Danmark) for 30 min, the slides were disposed of 3,3′-diaminobenzidine (DAB) for chromogenic detection. Pictures were grasped by a microscope (Leica, Weztlar, Germany) and camera. To quantify staining, five random areas per section were captured under a bright field microscope with 5× magnification, and the positive score or density of positively-stained cells was determined by two independent experienced pathologists.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining
For TUNEL assay, DeadEnd Fluorometric TUNEL System (Promega, Madison,USA) was used in the instructions’ guidance. Paraffin embedded tissue sections were deparaffinized and rehydrated. After fixing the sections with 4% methanol-free formaldehyde solution, the tissue sections were covered with 20 μg/mL Proteinase K to permeabilize tissues easily for 10 min. Slides then were incubated with rTdT incubation buffer and stained by propidium iodide solution. To quantify staining, five random areas per section were captured under a fluorescence microscope with 5× magnification, and the percentage of positively-stained cells was determined by two independent experienced pathologists.
Drug screening and cell viability assay
Organoids were resuspended in Matrigel and embedded in suspension in a 96 well plate (500 cells per 10μL Matrigel per well). The cells were allowed to culture for 1 day. Medium was replaced with fresh culture medium with varying concentrations of the drugs for another 5 days. For chemotherapy drugs treatments, organoids were cultured with drugs at 10-fold gradient dilution. Assay was done in triplicate wells, and each experiment was repeated two times at least. The chemotherapy drugs include 5-FU (Selleck), cisplatin (Selleck), gemcitabine (Selleck), oxaliplatin (Selleck), paclitaxel (Selleck), SN-38 (Selleck) and mitomycin C (Selleck). At the end of treatment in each experiment, the number of viable cells was determined by CellTiter-Glo assay (Promega) according to the kit protocol. Dose-response curves were fit to the data using the three-parameter logistic regression with variable slope and constraints at 100% and 0% viability using GraphPad Prism 9.0. AUC was calculated using the raw experimental data and normalized uniformly by dividing the AUC by the total maximum area a curve could occupy from 0 to 100% viability over the range of drug concentrations analyzed.
DNA isolation and DNA data generation
Genomic DNA was extracted from tumor and normal snap-liver frozen tissues using QIAGEN DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). To generate whole exome sequencing (WES) libraries for exome capture, the qualified DNA of tissue and organoid samples were sheared into 200–300 bp by Covaris technology. Ligation-mediated PCR (LM-PCR) was used to amplify extracted DNA and then hybridized to the Agilent human exome array. After washing out the non-hybridized fragments, the post-hybridization amplification products were obtained with 2 × 150-bp paired-end reads. The HiSeq X TEN platform (Illumina, San Diego, California, USA) was used to sequence those products. The data has been deposited in Genome Sequence Archive (GSA) with project number HRA005081.
Exome sequencing alignment and quality control
Two matched tumor/normal pairs tissue and their paired organoids were included in our dataset. Fastq files were filtered adapter sequence and low-quality reads with fastp32 and then gapped aligned to the hg38 using BWA-mem33 with default parameters. Before mutation calling, an addition step was applied for marking duplicates reads in the resulting BAM files using sambamba.34
Mutation calling
To identify all the variants in the samples, we used mutect2 in GATK Toolkit35 for single nucleotide variants (SNVs) and indels. Somatic mutation calls were annotated with ANNOVAR36 based on comparison to other variant databases, including gnomAD, Exome Aggregation Consortium (ExAC) and Catalog Of Somatic Mutations In Cancer (COSMIC).
Copy-number analyses
For WES data, FACETS (v0.5.0) was applied for calling copy number alterations.37 The paired tumor and normal tissue sorted bam files containing SNP locations, were used to calculate the counts of the reference nucleotide, alternative nucleotide, errors, and deletions of each SNP. Then the result files were used in facets as input to estimate cellular fraction and copy numbers.
RNA isolation and RNA-seq data generation
Organoids were released from Matrigel by using RNA cracking liquid. Tissues and organoids’ RNA were extracted using PureLink RNA Microextraction Kit (Thermo Fisher) according to the manufacturer’s procedure. Total isolated mRNA was cut into short fragments from fresh frozen tissue and tumoroids. For cDNA synthesis, buffer, random hexamer-primer, RNase H, dNTPs and DNA polymerase I was used respectively. The cDNA was purified with QIAQuick PCR extraction kit (Qiagen, Hilden, Germany) and resolved with EB buffer for end-repair, A-base addition and ligation of the sequencing adapters. Suitable fragments were then selected as templates for PCR amplification. Finally, paired-end libraries were sequenced with a sequence coverage of 150bp paired-end reads using HiSeq X TEN platform. The data have been deposited in Genome Sequence Archive (GSA) with project number HRA005081.
RNA-seq data processing
Following filtering out bad reads and cutting adapters with the fastp tools, fastq files of 79 samples (60 from tumor tissues, 19 from cultured organoids) were mapped to a human reference genome hg19).32 The hisat2-RNASeQC2 pipeline38,39,40 was used to obtain the Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) values and counts value.
Differential expression analysis and gene set enrichment analysis
To identify differentially expressed genes between two groups, we performed differential expression analysis using DESeq241 (v 1.28.1) R package with count data from RNA-seq data processing. Significance was set at p ≤ 0.05 and | log2 fold change | ≥ 1. Gene Set Enrichment Analysis were performed using the Hallmark gene sets, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome gene sets in Molecular Signatures Database42 (MSigDB) with GSEA43,44 (v 4.1.0) local software. ComplexHeatmap45 (v 2.4.3), ggplot2 (v 2.0.0) and EnhancedVolcano (v 1.9.5, https://github.com/kevinblighe/EnhancedVolcano) were used to visualize the result.
Ki67 staining
Each slide was manually scanned with film scanner, then got a score for the area of greatest Ki67 positivity randomly selected 5 view a slide at 10X objective by two independent experienced pathologists. Ki67-positive cells were counted using ImageJ ‘cell counter’ plug-in. According to counting the percentage of Ki67-positive cells for each slide, we estimated whether it is existing a difference between different groups by using GraphPad Prism 9.0.
Drug screening prediction model
We divided 38 patient tissue bulk RNA-seq samples into sensitive group (lowest 33% AUC) and non-sensitive group (top 67% AUC, combining the intermediate and resistant groups) and were used as train dataset. The gene expression panels were established using the RNA-seq data from both PDOs and patients. In organoid bulk RNA-seq samples, the differentially expression genes (DEGs) between the sensitive group and the non-sensitive group (including resistant and intermediate response groups) were used as input for constructing prediction models of drug sensitivity. We used the Support Vector Machine (SVM)46 and the Naive Bayes (NB)47 algorithm as classifiers in this study. The RFE method48 was then performed to select the optimal gene panel that can achieve the best prediction performance in both SVM and NB model. The gene panels were validated in two different machine learning models and demonstrated good performance. For prediction of unseen data, a score can be calculated by the constructed SVM or NB model using the expression values of our selected gene panels. Codes for computing such score have been uploaded in GitHub, and can be publicly accessed via https://github.com/huangmingle314/refactored-octo-system/. Finally, the prediction performance of two models was visualized with receiver operating characteristic (ROC) curve. All programs were executed with default parameters.
Quantification and statistical analysis
All summary data are presented as means ± S.D. or representative images of at least two independent experiments. All statistical analyses were performed in R (v 4.0.2), python (v 3.9.7) and GraphPad Prism software (GraphPad 9.0). Sample size (n) values used for statistical analyses are provided in the relevant figures and supplementary figures. Individual data point is graphed or can be found in Source Data. Tests for differences between two groups were performed using Mann–Whitney’s two-tailed test, Student’s two-tailed unpaired t-test or log-rank test, as specified in the figure legends. When using t-test we assumed normality and equal distribution of variance between the different groups. No data points were excluded from the statistical analyses. Significance was set at p ≤ 0.05.
Acknowledgments
This work was supported by National Natural Science Foundation of China (82173191), Guangdong Basic and Applied Basic Research Foundation (2019B151502009), and Kelin Outstanding Young Scientist of the First Affiliated Hospital of Sun Yat-sen University (Y12002).
Author contributions
X.X.R., W.X.W., and Y.F.W. performed experiments and drafted the manuscript. M.L.H. and Y.B.X. performed bioinformatics analysis. S.H.Z. helped perform animal experiments. Y.Z., D.M.L., J.M.J., S.L.S., J.L., and M.K. collected human samples and commented on the study. L.X.X., J.Y., and X.X.L. designed and supervised the study and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 8, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101277.
Contributor Information
Xiaoxing Li, Email: lixiaox23@mail.sysu.edu.cn.
Jun Yu, Email: junyu@cuhk.edu.hk.
Lixia Xu, Email: xulixia@mail.sysu.edu.cn.
Supplemental information
References
- 1.Sia D., Hoshida Y., Villanueva A., Roayaie S., Ferrer J., Tabak B., Peix J., Sole M., Tovar V., Alsinet C., et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valle J.W., Kelley R.K., Nervi B., Oh D.Y., Zhu A.X. Biliary tract cancer. Lancet. 2021;397:428–444. doi: 10.1016/S0140-6736(21)00153-7. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Li J., Xia Y., Gong R., Wang K., Yan Z., Wan X., Liu G., Wu D., Shi L., et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J. Clin. Oncol. 2013;31:1188–1195. doi: 10.1200/JCO.2012.41.5984. [DOI] [PubMed] [Google Scholar]
- 4.Wang S.J., Lemieux A., Kalpathy-Cramer J., Ord C.B., Walker G.V., Fuller C.D., Kim J.S., Thomas C.R., Jr. Nomogram for predicting the benefit of adjuvant chemoradiotherapy for resected gallbladder cancer. J. Clin. Oncol. 2011;29:4627–4632. doi: 10.1200/JCO.2010.33.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Primrose J.N., Fox R.P., Palmer D.H., Malik H.Z., Prasad R., Mirza D., Anthony A., Corrie P., Falk S., Finch-Jones M., et al. Capecitabine compared with observation in resected biliary tract cancer (BILCAP): a randomised, controlled, multicentre, phase 3 study. Lancet Oncol. 2019;20:663–673. doi: 10.1016/S1470-2045(18)30915-X. [DOI] [PubMed] [Google Scholar]
- 6.Phelip J.M., Desrame J., Edeline J., Barbier E., Terrebonne E., Michel P., Perrier H., Dahan L., Bourgeois V., Akouz F.K., et al. Modified FOLFIRINOX Versus CISGEM Chemotherapy for Patients With Advanced Biliary Tract Cancer (PRODIGE 38 AMEBICA): A Randomized Phase II Study. J. Clin. Oncol. 2022;40:262–271. doi: 10.1200/JCO.21.00679. [DOI] [PubMed] [Google Scholar]
- 7.Valle J.W. Advances in the treatment of metastatic or unresectable biliary tract cancer. Ann. Oncol. 2010;21 doi: 10.1093/annonc/mdq420. vii345-348. [DOI] [PubMed] [Google Scholar]
- 8.Rizzo A., Ricci A.D., Brandi G. Durvalumab: an investigational anti-PD-L1 antibody for the treatment of biliary tract cancer. Expert Opin. Investig. Drugs. 2021;30:343–350. doi: 10.1080/13543784.2021.1897102. [DOI] [PubMed] [Google Scholar]
- 9.Ricci A.D., Rizzo A., Brandi G. The DNA damage repair (DDR) pathway in biliary tract cancer (BTC): a new Pandora's box? ESMO Open. 2020;5 doi: 10.1136/esmoopen-2020-001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh D.Y., Lee K.H., Lee D.W., Yoon J., Kim T.Y., Bang J.H., Nam A.R., Oh K.S., Kim J.M., Lee Y., et al. Gemcitabine and cisplatin plus durvalumab with or without tremelimumab in chemotherapy-naive patients with advanced biliary tract cancer: an open-label, single-centre, phase 2 study. Lancet. Gastroenterol. Hepatol. 2022;7:522–532. doi: 10.1016/S2468-1253(22)00043-7. [DOI] [PubMed] [Google Scholar]
- 11.Vogel A., Bridgewater J., Edeline J., Kelley R.K., Klümpen H.J., Malka D., Primrose J.N., Rimassa L., Stenzinger A., Valle J.W., et al. Biliary tract cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2023;34:127–140. doi: 10.1016/j.annonc.2022.10.506. [DOI] [PubMed] [Google Scholar]
- 12.Broutier L., Mastrogiovanni G., Verstegen M.M., Francies H.E., Gavarró L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., Guo Y., Jin Y., Zhang X., Geng H., Xie G., Ye D., Yu Y., Liu D., Zhou D., et al. Establishment and drug screening of patient-derived extrahepatic biliary tract carcinoma organoids. Cancer Cell Int. 2021;21:519. doi: 10.1186/s12935-021-02219-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z., Jin Y., Guo Y., Tan Z., Zhang X., Ye D., Yu Y., Peng S., Zheng L., Li J. Conversion Therapy of Intrahepatic Cholangiocarcinoma Is Associated with Improved Prognosis and Verified by a Case of Patient-Derived Organoid. Cancers. 2021;13 doi: 10.3390/cancers13051179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J., Liu I.H., Chiou S.H., Salahudeen A.A., Smith A.R., et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018;175:1972–1988.e16. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macias R.I.R., Cardinale V., Kendall T.J., Avila M.A., Guido M., Coulouarn C., Braconi C., Frampton A.E., Bridgewater J., Overi D., et al. Clinical relevance of biomarkers in cholangiocarcinoma: critical revision and future directions. Gut. 2022;71:1669–1683. doi: 10.1136/gutjnl-2022-327099. [DOI] [PubMed] [Google Scholar]
- 17.Veninga V., Voest E.E. Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell. 2021;39:1190–1201. doi: 10.1016/j.ccell.2021.07.020. [DOI] [PubMed] [Google Scholar]
- 18.Lee S.H., Hu W., Matulay J.T., Silva M.V., Owczarek T.B., Kim K., Chua C.W., Barlow L.J., Kandoth C., Williams A.B., et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell. 2018;173:515–528.e17. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuciforo S., Fofana I., Matter M.S., Blumer T., Calabrese D., Boldanova T., Piscuoglio S., Wieland S., Ringnalda F., Schwank G., et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018;24:1363–1376. doi: 10.1016/j.celrep.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiriac H., Belleau P., Engle D.D., Plenker D., Deschênes A., Somerville T.D.D., Froeling F.E.M., Burkhart R.A., Denroche R.E., Jang G.H., et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller M., Schleithoff E.S., Stremmel W., Melino G., Krammer P.H., Schilling T. One, two, three--p53, p63, p73 and chemosensitivity. Drug Resist. Updat. 2006;9:288–306. doi: 10.1016/j.drup.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Wu K., Huynh K.Q., Lu I., Moustakim M., Miao H., Yu C., Haeusgen M.J., Hopkins B.D., Huang L., Zheng N., et al. Inhibitors of cullin-RING E3 ubiquitin ligase 4 with antitumor potential. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2007328118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y., Cheng Z., Liu S. MCM2 in human cancer: functions, mechanisms, and clinical significance. Mol. Med. 2022;28:128. doi: 10.1186/s10020-022-00555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Shi Q., Yang Z., Wang K., Zhang Z., Huang Z., Cui X., Li F. An Extracellular Matrix-Based Signature Associated With Immune Microenvironment Predicts the Prognosis and Therapeutic Responses of Patients With Oesophageal Squamous Cell Carcinoma. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.598427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasui K., Mihara S., Zhao C., Okamoto H., Saito-Ohara F., Tomida A., Funato T., Yokomizo A., Naito S., Imoto I., et al. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res. 2004;64:1403–1410. doi: 10.1158/0008-5472.can-3263-2. [DOI] [PubMed] [Google Scholar]
- 26.Berberich A., Kessler T., Thomé C.M., Pusch S., Hielscher T., Sahm F., Oezen I., Schmitt L.M., Ciprut S., Hucke N., et al. Targeting Resistance against the MDM2 Inhibitor RG7388 in Glioblastoma Cells by the MEK Inhibitor Trametinib. Clin. Cancer Res. 2019;25:253–265. doi: 10.1158/1078-0432.CCR-18-1580. [DOI] [PubMed] [Google Scholar]
- 27.Wuputra K., Ku C.C., Pan J.B., Liu C.J., Liu Y.C., Saito S., Kato K., Lin Y.C., Kuo K.K., Chan T.F., et al. Stem Cell Biomarkers and Tumorigenesis in Gastric Cancer. J. Pers. Med. 2022;12 doi: 10.3390/jpm12060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Zhang Y., Zhang Y., Qiao T., Wu K., Ding J., Liu J., Fan D. RUNX3 inhibits growth of HCC cells and HCC xenografts in mice in combination with adriamycin. Cancer Biol. Ther. 2008;7:669–676. doi: 10.4161/cbt.7.5.5664. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Li M., Jin F., Liu J., Chen M., Yin J. DNAJC12 promotes lung cancer growth by regulating the activation of beta-catenin. Int. J. Mol. Med. 2021;47 doi: 10.3892/ijmm.2021.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haider T., Pandey V., Banjare N., Gupta P.N., Soni V. Drug resistance in cancer: mechanisms and tackling strategies. Pharmacol. Rep. 2020;72:1125–1151. doi: 10.1007/s43440-020-00138-7. [DOI] [PubMed] [Google Scholar]
- 31.Zamame Ramirez J.A., Romagnoli G.G., Kaneno R. Inhibiting autophagy to prevent drug resistance and improve anti-tumor therapy. Life Sci. 2021;265 doi: 10.1016/j.lfs.2020.118745. [DOI] [PubMed] [Google Scholar]
- 32.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen R., Seshan V.E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graubert A., Aguet F., Ravi A., Ardlie K.G., Getz G. RNA-SeQC 2: Efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics. 2021;37:3048–3050. doi: 10.1093/bioinformatics/btab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu L.X., He M.H., Dai Z.H., Yu J., Wang J.G., Li X.C., Jiang B.B., Ke Z.F., Su T.H., Peng Z.W., et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann. Oncol. 2019;30:990–997. doi: 10.1093/annonc/mdz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas B.J., Dobin A., Li B., Stransky N., Pochet N., Regev A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20:213. doi: 10.1186/s13059-019-1842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davidson N.M., Majewski I.J., Oshlack A. JAFFA: High sensitivity transcriptome-focused fusion gene detection. Genome Med. 2015;7:43. doi: 10.1186/s13073-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 46.Bennett K.P., Campbell C.J.A.S.E.N. Support Vector Machines: Hype or Hallelujah? Acm Sigkdd Explorations Newsletter. 2000;2:1–13. doi: 10.1145/380995.380999. [DOI] [Google Scholar]
- 47.Zhang H. The Optimality of Naïve Bayes. Florida AI Research Society. 2004 https://aaai.org/papers/flairs-2004-097/ [Google Scholar]
- 48.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V.J.J.o. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011 https://dl.acm.org/doi/10.5555/1953048.2078195 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported in this paper are deposited in Genome Sequence Archive (GSA) with project number HRA005081 and publicly available as of the date of publication. The software and algorithms for data analyses used in this study are published and referenced throughout the STAR Methods section. No original code is featured in this paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.