Abstract
Swarming motility of Serratia liquefaciens MG1 requires the expression of two genetic loci, flhDC and swrI. Here we demonstrate that the products of the flhDC operon (the flagellar master regulator) and the swrI gene (the extracellular signal molecule N-butanoyl-l-homoserine lactone) are global regulators which control two separate regulons.
Serratia liquefaciens is capable of two forms of flagellum-driven motility, swimming and swarming, depending on whether the growth medium is liquid or solid (1, 5). As in Escherichia coli and Salmonella typhimurium, control of flagellar expression is governed by the flhDC master operon, which encodes a positive transcription factor (10). In E. coli, this master regulatory system controls the expression of more than 40 genes that are organized in at least 15 operons (11, 12). The flhDC operon itself is subject to control by several regulatory circuits that are responsive to changes in environmental and nutritive conditions. Transcription is regulated by OmpR and requires cyclic AMP receptor protein-cyclic AMP complex (17, 21, 22). As a consequence, expression of the entire regulon is sensitive to fluctuations in the metabolite acetyl phosphate, medium osmolarity, and catabolite repression exerted by glucose. Furthermore, expression of flhDC is also dependent on the presence of the proteins DnaK, DnaJ, and GrpE (20), and it has been suggested that inhibition of motility at 42°C is caused by induction of the heat shock response. In S. liquefaciens, artificial transcriptional stimulation of the flhDC operon not only promotes swarm colony formation on an agar surface but also allows initiation of swarm cell differentiation in liquid culture without the surface contact that is otherwise obligatory (1). It was therefore suggested that the flhDC operon encodes a regulator whose concentration or activity status determines whether cells swim or swarm (1).
Recently, workers from our laboratory identified an additional gene, swrI, which is required for swarming motility of S. liquefaciens (2). This gene encodes a putative N-acyl-l-homoserine lactone (AHL) synthase which is involved in the synthesis of the extracellular signal molecules N-butanoyl-l-homoserine lactone (BHL) and N-hexanoyl-l-homoserine lactone (2). These two pheromones, which are produced in a molar ratio of approximately 10 to 1, accumulate in a growing culture until a certain threshold concentration (i.e., a concentration at which expression of target genes is triggered) is reached (2). This autoinduction circuit is therefore activated only when a certain culture density has been attained. Hence, swarming motility of S. liquefaciens is dependent on an AHL-mediated signaling mechanism that is used by a number of bacteria to monitor population size (3, 4, 19).
Except for one gene, swrA (9), the AHL-controlled genes are not known, nor are the roles of their respective gene products in the formation of a swarm colony understood. However, the requirement of fine-tuned expression of the flhDC master operon for swarming behavior led us to hypothesize that the AHL-dependent autoinduction circuit could be directly involved in the expression of flhDC. If this hypothesis is correct, then expression of the flhDC operon from an inducible promoter would be expected to overcome the need for the autoinduction circuit. Likewise, interference with the autoinduction circuit (by stimulation or destruction) should have marked effects on the expression of flagellar genes. In this study, we employed genetic approaches combined with an analysis of the pattern of protein synthesis to test our hypothesis.
Genetic analysis of swarming motility.
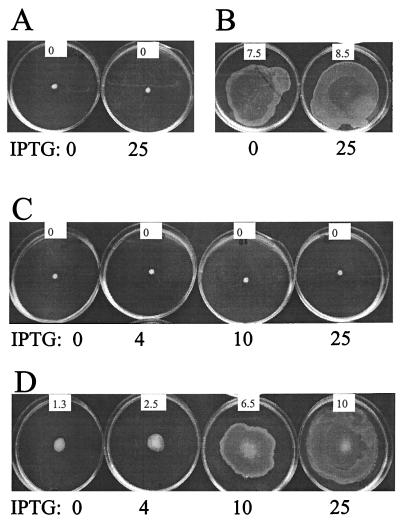
The AHL-dependent autoinducer circuit is indispensable for expression of swarming motility on Casamino Acid-supplemented minimal AB medium (2). On this medium, swarming motility of the swrI mutant can be restored to the level of the wild-type (wt) strain by the external addition of 0.2 μM BHL (2). Modulation of flhDC transcription and thus control over expression of the flagellar regulon can be achieved by introduction of the plasmid pMG600, which contains the entire flhDC operon under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptac promoter (1, 5). When pMG600 was introduced into the S. liquefaciens flhD strain MG3 (5), the addition of IPTG greatly stimulated colony expansion by means of swarming motility (2). However, if pMG600 was introduced into MG44 (which carries swrI) (2), the addition of increasing concentrations of IPTG had no effect, i.e., the colony was unable to expand on the surface (Fig. 1A). When the medium was supplemented with BHL, swarming was restored (Fig. 1B). MG44 is flhD+ flhC+. In order to investigate the effect of IPTG-controlled flhDC expression from pMG600, we constructed an flhD swrI double mutant which was designated MG50. This mutant carries, in addition to the inactivating mutation in swrI, the luxAB transposon originating from pJMS10 inserted into the flhD gene (8). The insertion into flhD was confirmed by DNA sequencing (data not shown). When pMG600 was introduced into MG50, the addition of increasing concentrations of IPTG had no effect on expansion (Fig. 1C). However, when the medium was supplemented with BHL, the stimulatory effect of IPTG on colony expansion was restored and the rate of colony expansion was found to correlate with increasing inducer concentrations (Fig. 1D).
FIG. 1.
Swarming behavior of S. liquefaciens strains harboring plasmid pMG600. (A and B) MG44 (swrI); (C and D) MG50 (swrI flhD). BHL (0.5 μM) was added to the medium in panels B and D. Micromolar concentrations of IPTG were added as indicated. Swarming motility was assayed in AB medium supplemented with 0.5% Casamino Acids and 0.5% glucose and containing 0.6% agar. Plates were photographed after 20 h of incubation at 30°C. The approximate rate (in millimeters per hour at which each colony expanded is indicated in the white inset on each plate.
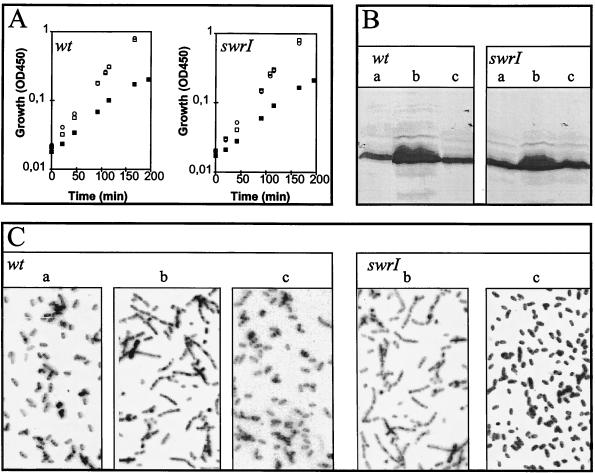
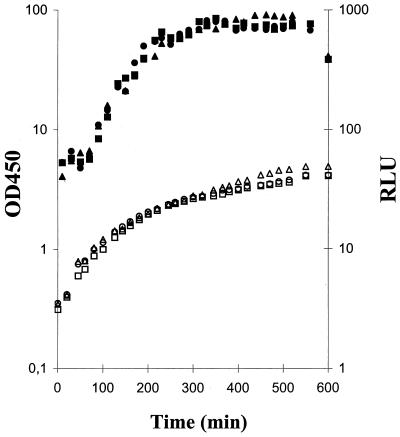
The failure of swrI cultures to expand in the absence of added BHL could be caused by the abolishment of FlhDC-controlled developmental processes leading to the formation of elongated, hyperflagellated swarm cells (1). A more detailed examination of wt and swrI cells containing pMG600 revealed that upon addition of IPTG, cells grew with a reduced doubling time, developed extensive flagellation, and elongated to become filamentous (Fig. 2). Cells harboring the vector pVLT33 neither elongated nor showed a reduced growth rate in the presence of IPTG (data not shown). These results indicate that swrI cells, as a result of flhDC stimulation, are able to differentiate into a filamentous, hyperflagellated form which is indistinguishable from cells isolated from the swarm of the wt cultures. The presence of externally added BHL (up to 20 μM) did not reduce the growth rate, nor did it induce cell elongation or flagellation (Fig. 2). Furthermore, we monitored the transcriptional activity of the flhDC operon by measuring the expression of bioluminescence from the flhD-luxAB fusion present in the swrI flhD double mutant MG50 (Fig. 3). First, transcription of flhDC was growth phase dependent, which in turn accounts for the growth phase-dependent expression of phospholipase and flagella as previously reported (5); second, externally added BHL (up to 20 μM) had no effect on the level of flhD-luxAB transcription. This is consistent with the data presented in Fig. 2B regarding the flagellar content of growing wt and swrI cells and in accordance with previously presented data regarding swarming wt cells and cells from a nonswarming swrI colony (6) which demonstrated that the flagellar protein content per unit of cell mass did not differ significantly between the two types of cells.
FIG. 2.
Analysis of S. liquefaciens strains harboring plasmid pMG600. (A) Growth; (B) flagellar content; (C) cell shape. Cells were grown in liquid cultures with no addition (□ and a), 0.5 mM IPTG (▪ and b), and 20 μM BHL (○ and c). Cells were harvested from the growing cultures at an optical density at 450 nm (OD450) of 0.25 for analysis in panels B and C. (B) Aliquots (50 μl) were heat denatured in sodium dodecyl sulfate-containing sample buffer, and the proteins were then separated by means of a standard sodium dodecyl sulfate-PAGE procedure, transferred to an Immobilon-P membrane (Millipore), and subjected to Western blotting analysis with rabbit antibodies directed against S. liquefaciens flagellar protein. Following binding of secondary alkaline phosphatase-labeled anti-rabbit immunoglobulin G, detection was performed with p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate. (C) Microscopic inspection of cell shape with a 50× long-working-distance objective.
FIG. 3.
Transcriptional activity of the flhDC operon. Liquid cultures of MG50 (flhD-luxAB swrI) were grown in the absence (squares) and presence (circles) of 2 μM BHL and in 20 μM BHL (triangles). Growth (open symbols) was monitored, and the optical density at 450 nm (OD450) was determined. For quantitation of bioluminescence (closed symbols), 0.1-ml samples were taken and added to 0.9 ml of fresh medium and 1 μl of n-decanal, and light emission, in relative light units (RLU), was determined in a TD-20e luminometer (Turner Designs, Sunnyvale, Calif.).
Global analysis of gene expression.
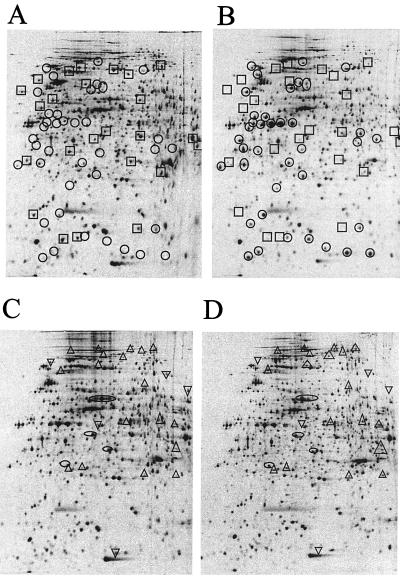
For further analysis, we employed the wt strain MG1, the swrI strain MG44, the flhD strain MG3, and finally MG3 harboring pMG600 for a two dimensional (2D) polyacrylamide gel electrophoresis (PAGE) (O’Farrell gel-based) global analysis of radioactively labeled synthesized proteins. Cultures of MG3 harboring pMG600 were incubated in the presence or absence of 0.2 mM IPTG; cultures of MG44 were incubated in the presence or absence of 2 μM BHL. The cultures of strains MG1, MG3, and MG44 were grown without any addition. Based on the comparison of gels obtained with MG1 (not shown), MG3, and MG3 harboring pMG600 in the absence (not shown) and presence of IPTG, the positions of FlhD- and FlhC-controlled proteins were identified (Fig. 4A and B). Based on the comparison of gels obtained with MG44 grown in the presence and absence of externally added BHL, the positions of BHL-controlled proteins were identified (Fig. 4C and D). This analysis strongly suggests that the products of the flhDC operon and the swrI gene are global regulators of gene expression. Syntheses of 62 and 28 proteins could be identified as controlled by the flhDC operon and by the BHL-dependent regulatory system, respectively. A comparison of Fig. 4B and D revealed that the positions of the FlhDC- and BHL-controlled proteins did not overlap. Furthermore, syntheses of many proteins are actually turned off in response to increased FlhD and FlhC expression. In addition to controlling flagellar expression, FlhD is involved in exoenzyme production and cell division (5, 17, 18). flhDC overexpression may cause inhibition of cell division and therefore indirect effects on the level of gene expression. Addition of BHL also leads to switch-off of gene expression. In light of a similar analysis of the pathogenic bacterium Yersinia enterocolitica and its derivative in which the swrI-analogous yenI gene had been disrupted (23), this may indicate that quorum-sensing systems are involved in both positive and negative control of gene expression. Interestingly, four spots in Fig. 4C and D show the presence of proteins that, in response to the addition of BHL, seem to have moved to the left in the first dimension of the 2D gels, indicating that they may have been modified rather than synthesized de novo (14). It is tempting to speculate that protein modification could be involved in quorum sensing in S. liquefaciens, although it has not been proven.
FIG. 4.
2D PAGE analysis of synthesized proteins. The growing liquid cultures (optical density at 450 nm = 1.5) were split in two, and inducers were added to one of the cultures. Twenty minutes later, cells were labeled with 30 μCi of 35[S]methionine per ml for 5 min. Cells were harvested and subjected to 2D PAGE analysis as described previously (16). Strains tested were MG3 (flhD) (A) and MG3/pMG600 (B) in the presence of 0.2 mM IPTG and MG44 (swrI) in the absence (C) and presence (D) of 2 μM BHL. ○ and □, proteins induced and repressed, respectively, by expression of flhDC. ▵ and ▿, proteins induced and repressed, respectively, by addition of BHL. ○, proteins that moved in a horizontal direction in response to BHL addition. The left and right sides correspond to pIs of 4 and 7, respectively, for proteins immobilized by isoelectric focusing (Pharmacia Immobiline strips). The top and bottom of the gel correspond to molecular masses of approximately 200 and 10 kDa, respectively.
Conclusions.
Neither the genetic nor the global analysis of the regulatory systems that control swarming motility of S. liquefaciens supported the hypothesis that the autoinducer circuit is involved in the regulation of the flhDC-encoded master regulator. None of the individual FlhD- or FlhC-controlled genes was found to be affected by the addition of BHL, nor did stimulation of the flhDC operon result in increased expression of BHL-controlled genes. Instead, our studies suggest that the flhDC operon and the BHL-dependent autoinducer circuit control separate regulons. Although the resolution of the second dimension of the gels does not allow for the detection of all proteins expressed under a given condition, the result of the global analysis is entirely consistent with that of the genetic and phenotypic analysis, which also suggests that the two regulatory systems work independently of each other. A model is emerging in which cell elongation and hyperflagellation, i.e., swarm cell differentiation, is controlled by the flhDC operon while the autoinducer circuit controls the expansion of the swarm colony, i.e., facilitates the outward movement of already-differentiated swarm cells (7). Work in progress by us demonstrates that the autoinduction circuit system controls the production of an extracellular biosurfactant which enables swarm cells to travel on top of surfaces (9). Consistent with our recent regulatory model, production of the surfactant is flhDC independent (7, 9). The requirement of biosurfactants for swarming motility of Serratia marcescens has been demonstrated previously (13). Interestingly, rhamnolipid biosurfactant production in Pseudomonas aeruginosa is controlled by BHL and a protein, RhlR, that belongs to the LuxR family of transcriptional regulators, suggesting regulatory similarities (15).
With S. liquefaciens, culture growth and expansion on top of a surface are accomplished by means of swarming motility. This trait is dispensable for cells that are maintained in liquid cultures. In nature, however, microbial activity is often associated with surfaces, and it appears that one of the most remarkable characteristics of bacteria is their ability to form structured and cooperative consortia. Formation of a swarming culture is an example of coordinated behavior that can be viewed as a primitive form of multicellularity. With S. liquefaciens, this requires the integration of diverse environmental signals involving metabolic potential (medium composition), quorum sensing, and contact with a surface. The present analysis strongly suggests that the tasks of signal integration and interpretation of all the information from the various sensory transducing pathways require the participation of at least two key regulatory systems, the cooperative action of which is required for the formation of a swarming colony.
Acknowledgments
This work was supported by grants from The Plasmid Foundation, The Nordic Research Network, and The Danish Biotechnology Program. L.E. is currently supported by the Research Training Program of the EU (contract BIO-4CT965025).
We are grateful to Linda Stabell for excellent technical assistance.
REFERENCES
- 1.Eberl L, Christiansen G, Molin S, Givskov M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J Bacteriol. 1996;178:554–559. doi: 10.1128/jb.178.2.554-559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eberl L, Winson M K, Sternberg C, Stewart G S A B, Christiansen G, Chhabra S R, Bycroft B, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behavior of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua W C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 4.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Givskov M, Eberl L, Christiansen G, Benedik M J, Molin S. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhDC. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 6.Givskov M, de Nys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg P D, Kjelleberg S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J Bacteriol. 1996;178:6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Givskov M, Eberl L, Molin S. Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol Lett. 1997;148:115–122. [Google Scholar]
- 8.Kristensen C S, Eberl L, Sanchez-Romero J M, Givskov M, Molin S, De Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindum, P. W., U. Anthoni, C. Christoffersen, L. Eberl, S. Molin, and M. Givskov. 1997. Unpublished data.
- 10.Liu X, Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Matsumura P. Differential regulation of multiple overlapping promoters in flagellar class II operons in Escherichia coli. Mol Microbiol. 1996;21:613–620. doi: 10.1111/j.1365-2958.1996.tb02569.x. [DOI] [PubMed] [Google Scholar]
- 12.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 13.Matsuyama T, Bhasin A, Harshey R M. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol. 1995;177:987–991. doi: 10.1128/jb.177.4.987-991.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nyström T, Neidhardt F C. Effects of overproducing the universal stress protein, UspA, in Escherichia coli K-12. J Bacteriol. 1996;178:927–930. doi: 10.1128/jb.178.3.927-930.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Östling J, Holmquist L, Kjelleberg S. Global analysis of the carbon starvation response of a marine Vibrio species with disruptions in genes homologous to relA and spoT. J Bacteriol. 1996;178:4901–4908. doi: 10.1128/jb.178.16.4901-4908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prüß B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 18.Prüß B M, Markovic D, Matsumura P. The Escherichia coli flagellar transcriptional activator flhD regulates cell division through induction of the acid response gene cadA. J Bacteriol. 1997;179:3818–3821. doi: 10.1128/jb.179.11.3818-3821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial ’enigma‘: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 20.Shi W, Zhou Y, Wild J, Adler J, Gross C A. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992;174:6256–6263. doi: 10.1128/jb.174.19.6256-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–4702. doi: 10.1128/jb.177.16.4696-4702.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman M, Simon M. Bacterial flagella. Annu Rev Microbiol. 1977;31:397–419. doi: 10.1146/annurev.mi.31.100177.002145. [DOI] [PubMed] [Google Scholar]
- 23.Throup J P, Camara M, Briggs G F, Winson M K, Chhabra S R, Bycroft B W, Williams P, Stewart G. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]