Abstract
Lysinuric protein intolerance (LPI) is a rare, inherited aminoaciduria caused by biallelic pathogenic variants in the amino acid transporter gene SLC7A7 (OMIM *603593). Individuals with LPI show extreme variability in their clinical presentation, and LPI is included in the differential diagnosis of several disorders such as urea cycle disorders, lysosomal storage diseases, malabsorption diseases, autoimmune disorders, hemochromatosis, and osteoporosis. The phenotypic variability of LPI and the lack of a specific clinical presentation have caused various misdiagnoses. Here, we report two siblings diagnosed in their 4th decade of life with LPI, manifesting rare hyperferritinemia. Additionally, they presented with short stature, multiple bone fractures due to osteoporosis, and they showed an aversion to protein-rich food. Using a combination of exome sequencing, microarray analysis and qPCR, we identified a novel homozygous deletion in SLC7A7 encompassing exons 3 to 10, which is predicted to lead to disruption of SLC7A7 function. This is the first report of lysinuric protein intolerance in a Turkish family associated with this so far unknown deletion in SLC7A7.
Keywords: Lysinuric protein intolerance (LPI), SLC7A7 deletion, Hyperferritinemia, Multiple bone fractures
1. Background
Lysinuric protein intolerance (LPI) is an autosomal recessive disease caused by defective cationic amino acid (CAA; arginine, lysine, ornithine) transport in intestinal and renal tubular cells. Impairment of both, intestinal absorption and renal reabsorption of CAA, causes decreased circulating dibasic amino acid levels resulting in urea cycle disorder and hyperammonemia [1,2].
The first symptoms of LPI typically start after an infant is weaned from breast milk or formula, and the first signs include recurrent vomiting and episodes of diarrhea, especially after protein-rich meals, resulting in aversion to protein-rich food and failure to thrive. Over time, a multitude of health issues can arise, including osteoporosis, various problems involving the lungs such as progressive interstitial changes and pulmonary alveolar proteinosis or the kidneys as, e.g., progressive glomerular and proximal tubular disease, as well as hematologic disorders such as normochromic or hypochromic anemia, leukopenia, thrombocytopenia, and erythroblastophagocytosis observed in bone marrow aspirates. Hyperferritinemia, hypercholesterolemia, hypertriglyceridemia, and acute pancreatitis might also be observed [1]. The diagnosis of LPI is often complicated due to uncertain clinical presentation and a variety of clinical differential diagnoses presenting with similar biochemical anomalies. Classic symptoms of protein intolerance may remain unnoticed during the first and second decades of life due to unconscious avoidance of dietary protein. The presence of hyperammonemia in individuals with LPI often leads to a misdiagnosis of urea cycle disorder, though increased orotic aciduria associated with hyperexcretion of CAA is highly suggestive of LPI. Unsurprisingly, LPI is mainly diagnosed in countries such as Finland, Italy, and Japan, where the disorder is more prevalent and clinicians are familiar with it3.
LPI is a genetic disorder caused by homozygous or compound heterozygous pathogenic variants in the SLC7A7 gene, which is located on chromosome 14q11.2. SLC7A7(NM_003982.4) has 10 exons; transcription can be initiated by two promoter regions [4]. SLC7A7 encodes the Y + L amino acid transporter 1 (y + LAT-1) protein, which together with the 4F2hc cell-surface antigen heavy chain (also known as SLC3A2) forms a heteromeric amino acid transporter complex that transports CAA through the basolateral membrane of epithelial cells in the small intestine and kidney [3,5]. So far, 78 pathogenic variants have been identified in SLC7A7 in individuals with LPI including missense and nonsense variants as well as deletions, insertions, splicing variants, and large genomic rearrangements, all leading to impairment or loss of SLC7A7 protein function (listed by HGMD 24.04.2023) [6].
2. Case reports
Individual 1, a female patient and the second child of unrelated healthy parents of Turkish descent, presented with elevated ferritin levels (Table 1) at the age of 45 and was referred to the gastroenterology department. She received genetic counselling at the human genetics department of Umraniye Training and Research Hospital regarding the pre-diagnosis of hemochromatosis.
Table 1.
Clinical presentation and ferritin follow-ups by years.
| Clinical presentation | Individual 1 | Individual 2 |
|---|---|---|
| Failure to thrive | + | + |
| Protein-rich food aversion | + | + |
| Osteoporosis | + | + |
| Hyperferritinemia | + | + |
| Anemia | + | − |
| Coagulopathy | − | − |
| Recurrent bacterial or viral infections⁎ | − | + |
| Neurologic symptoms | − | − |
| Muscular hypotonia | + | + |
| Pulmonary alveolar proteinosis | − | − |
| Pneumonia | − | − |
| Ferritin levels⁎⁎ (ng/ml) | ||
| 2022 | 1750 ng /ml | 1257 ng /ml |
| 2021 | 1298 ng /ml | 918 ng /ml |
| 2020 | 672 ng /ml | − |
| 2019 | 1486 ng /ml | − |
| 2018 | 842 ng /ml | − |
| 2017 | 1004 ng /ml | − |
Varicella infections.
Ferritin threshold: 5–204 ng /ml.
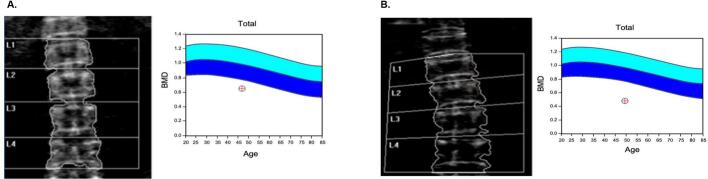
Upon admission, her body weight was 48 kg, and her height was 145 cm (BMI 22 kg/m2). She presented with short stature and thin extremities with muscle hypoplasia. She had loose skin that lost its turgor tone. She suffered from muscle weakness and osteoporosis (Fig. 1A). Her mental capacity was average, she had no known neurological or psychological disease.
Fig. 1.
Bone densitometry. (A) Individual 1. Z-score: −3 (B) Individual 2. Z-score: −4,5.
In 2017, hyperferritinemia was initially identified. Subsequent measurements consistently showed high levels of ferritin. Additionally, serum lactate hydrogenase was also elevated. Although the aspartate aminotransferase level and triglyceride levels were slightly raised, all other liver function tests were within the normal range for all measurements. Upon initial examination, the patient's ferritin level measured 1004 ng/ml, iron was at 102 μg/dl, and total iron binding capacity (TIBC) was at 210 μg/dl. Transferrin saturation (TS) was calculated at 48.5%. Based on the presence of mild hepatosplenomegaly, thrombocytopenia, and elevated prothrombin time, the possibility of chronic liver disease (CLD) was considered in the differential diagnosis. However, tests conducted for common etiologies showed normal results. During the gastroscopy, there were no signs of esophageal varices. Genetic analysis was conducted to exclude hereditary hemochromatosis (ferritin >200 ng/ml (premenopausal women), TS >48.5%) as a rare cause of CLD, and no genetic variant was detected in a hemochromatosis panel based on the expected pathogenesis of hyperferritinemia. Beta-glucosidase and chitotriosidase enzyme levels were analyzed to rule out Gaucher's disease, which is another rare etiology of chronic liver disease that can present only with hyperferritinemia. Enzyme levels were also normal (beta-glucosidase: 2.30 μmol/ml/h (>1.30), chitotriosidase: 24.8 μmol/L/h (<290)) and there was no hyperammonemia. Upon thorough examination of the biochemical data from the last five years, it has been observed that the LDH levels fall within the range of 419–827 U/L, and the triglyceride levels were between 187 and 215 mg/dl. The transaminase levels were slightly elevated, but there was no significant increase (AST: 24–44 U/L, ALT: 13–26 U/L). She had mild anemia. In the quantitative urine amino acid analysis, significant increases were observed in the levels of glutamine (1954 μmol/gkrea), lysine (1069 μmol/gkrea) and alanine (999 μmol/gkrea).
Individual 2 is the older sister of individual 1. At first admission, she was 48 years old. In 2021, after her sister had been initially diagnosed, the first ferritin measurement was conducted for her. She had also hyperferritinemia (Table 1). Furthermore, her serum lactate hydrogenase and triglyceride levels were elevated. She has been suffering from gastrointestinal problems since her childhood including vomiting and diarrhea, especially after eating protein-rich food. She had a failure to thrive and bone fractures starting at the age of two. She presented with severe osteoporosis (Fig. 1B) and had a history of frequent fractures of multiple bones until the age of eight years. She had loose skin and thin hair that massively fell out. Additionally, she had recurrent bullous vesicles in her childhood. Biochemical data at the first admission was quite similar to her sister's. In addition to an increased ferritin level, LDH was also found to be highly elevated, measuring 1089 U/L and there was no hyperammonemia overall. Along with other laboratory parameters, AST was monitored at 38 U/L, ALT: 17 U/L and ALP: 146 U/L. Her triglyceride level was 276 mg/dL; her cholesterol was 285 U/L. In the quantitative urine amino acid analysis, the increase in the levels of glutamine (2900 μmol/gkrea), lysine (5495 μmol/gkrea), serin (1591 μmol/gkrea) and alanine (2338 μmol/gkrea) was remarkable.
3. Genetic analyses
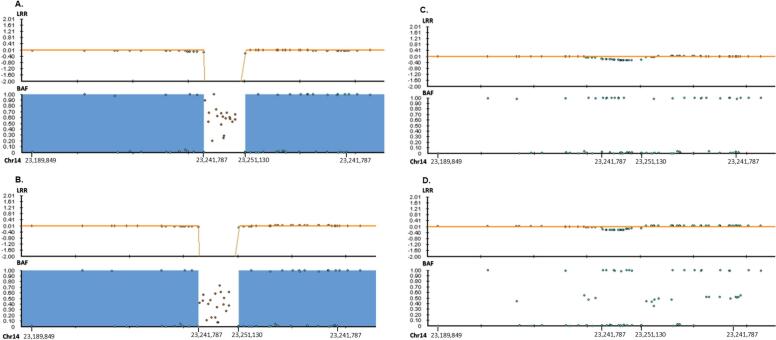
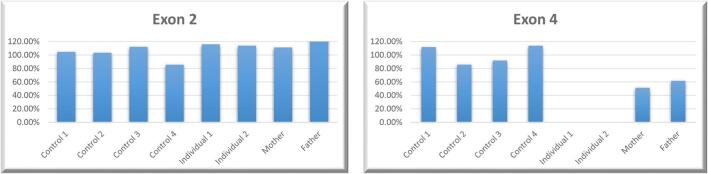
Based on the initial diagnosis of hemochromatosis, individual 1 was first screened with a virtual hemochromatosis panel including six genes (BMP2, HAMP, HFE, HJV, SLC40A1, TFR2). Next-generation sequencing (NGS) was performed using Illumina V2 chemicals and SOPHIA Clinical Exome Solution on the Illumina NextSeq 500 platform, and bioinformatics analyses and variant calling were performed using the Sophia-DDM-V5 bioinformatics analysis program. No pathogenic variants were detected in the selected six genes causative of hemochromatosis. For this reason, we performed a copy number variation (CNV) analysis based on the generated clinical exome data by Sophia Genetics' MUSKAT® software. This led to the detection of a homozygous deletion located from exon 3 to exon 10 within the SLC7A7 gene. Based on the clinical features that included hyperferritinemia, SLC7A7 was a good candidate gene for the observed phenotype. Infinium CytoSNP-850 K v1.2 microarray analysis was performed to confirm the large deletion extending from exon 3 to exon 10. The homozygous deletion was detected at chr14:23241787–23,251,130. Subsequently, microarray analysis via Illumina Infinium HumanCytoSNP-12 v2.1 for both parents and the affected older sister was carried out, and we were able to confirm that individual 2 carried the identical homozygous deletion within the SLC7A7 gene. However, no heterozygous deletion was detected in either of the parents at the BlueFuse Multi v4.5 Software Guide. After analyzing the target region, a decrease in LogR value was observed (Fig. 2). Therefore, we performed qPCR with four primer pairs for the targeted region (SLC7A7 exons 2, 4, 9 and OXAL1 exon 3). It was confirmed that both parents carry the heterozygous deletion at the same location in the SLC7A7 gene (Fig. 3).
Fig. 2.
Microarray analysis. (A) Individual 1 and (B) individual 2 have homozygous, (C) mother and (D) father have heterozygous deletions. Blue areas on individual 1 and 2 show the LOH regions that span between chr14:23,254,148-32,942,539. (X-axis: Chromosomal positions. Y-axis: LRR (LogR Ratio), BAF (B allele Frequency)).
Fig. 3.
Quantitative PCR analysis of SLC7A7. (A) There is a homozygous deletion in exon 4 on individual 1 and 2; both parents are heterozygous for the deletion. (B) There is no deletion in any individual in exon 2.
Following the genetic analysis, urinary amino acid analysis was performed for both patients to confirm the LPI diagnosis. In both patients, urinary lysine excretion was remarkably high (Individual 1:5495 μmol/gkrea, Individual 2: 1069 μmol/gkrea) however urinary arginine and ornithine excretion were normal. Based on the genetic findings, a low protein diet, l-carnitine, and calcium replacement were initiated for both individuals.
4. Discussion
LPI is a rare inherited metabolic disorder of dibasic amino acid transport. Currently, the incidence of LPI in Turkey is unknown. LPI is prevalent in Finland (1:50,000) and Northern Japan (1:60,000) [1,3]. The SLC7A7 gene encodes the light subunit of a cationic amino acid transporter. Together with the 4F2hc cell-surface antigen heavy chain (also known as SLC3A2); SLC7A7 forms a heteromeric amino acid transporter complex that is involved in renal tubular and intestinal transport of cationic amino acids. Disruption of SLC7A7 function as, e.g., observed in patients with LPI results in decreased circulating dibasic amino acid levels and high urinary excretion of cationic amino acids. The most commonly affected organs are the liver and the kidneys. Due to aversion to protein-rich food and mild intestinal malabsorption, affected individuals generally show physical retardation [7]. Patients with LPI are generally (52%) admitted to hospital based on their failure to thrive during childhood [8]. Interestingly, individual 1 was referred to the gastroenterology clinic with hyperferritinemia in adulthood, and she was diagnosed with LPI at 46 years of age. It is thought that in LPI patients, hyperammonemia and lysine-protein malnutrition cause fatty degeneration in the liver and injury to hepatocytes.
The main clinical features of the two individuals presented here were compatible with those of LPI, with typical routine laboratory indicators for LPI such as hyperammonemia after an oral protein load and high urinary excretion of lysine, ornithine, and arginine. Moreover, LDH and ferritin levels were constantly elevated. Both individuals had osteoporosis, which started in their childhood and resulted in frequent bone fractures. The developmental delay and protein aversion were also remarkable in childhood. Osteoporosis in LPI may result from defective matrix protein synthesis due to protein deprivation and lack of cationic amino acids, as well as increased collagen turnover [9]. Patients with LPI also have an increased risk for generalized varicella infections. The older sister's skin problems referred to as bullous vesicles might be varicella rash, which is an extremely rare presentation in LPI [10].
While in gastroenterology clinics, hyperferritinemia is often considered to be caused by hemochromatosis LPI is a rare cause of hyperferritinemia. Elevated ferritin levels in LPI patients, may result from impaired elimination of ferritin from plasma by injured hepatocytes [11]. Besides, this condition may be a result of hemophagocytic lymphohistiocytosis (HLH), which often accompanies LPI. HLH is a severe clinical condition characterized by fever, cytopenia, hypertriglyceridemia, hyperferritinemia, coagulopathy, splenomegaly, and neurological problems [12]. However, our patients did not meet full of the clinical diagnostic criteria. When an additional infectious disease accompanies LPI, intermittent hemophagocytic lymphohistiocytosis should be considered during follow-up. Nonetheless, HLH does not explain the elevated ferritin levels in this case. The expected decrease in ferritin level was not observed despite a protein-restricted diet and food supplements (ornithine, citrulline, lysine) for these cases. Serum ferritin concentration appears to be a valuable index of the control of this disease [13].
The diagnosis in individual 1 led to the clinical investigation and genetic examination of her older sister, individual 2, who also was diagnosed with LPI. At that time, she was 48 years old. Delayed age at diagnosis of both individuals might not only be due to phenotypic variability but also due to their atypical presentation. Short stature and multiple fractures in childhood might remind of osteogenesis imperfecta. In addition, skeletal changes in LPI resemble those in homocystinuria; juvenile osteoporosis, biconcave vertebrae, scoliosis, abnormal metaphysis, and genu valgus are features of both diseases [11]. Therefore, especially for this kind of metabolic disease, exome sequencing might be the best approach to establish an accurate diagnosis. Clinical exome sequencing (CES) is a valuable tool to reveal an even rarer underlying disease that might be overlooked. In the case presented here, the diagnosis of the patient was achieved by CES, although it was a virtual hemochromatosis panel. It is crucial to accurately interpret any incidental pathogenic variants that may appear outside of targeted genes. Furthermore, we recognize the benefits of conducting CNV analysis on the CES in the diagnosis of rare diseases. Although the MLPA technique is widely recognized as the gold standard for targeted gene deletion and duplication analysis, due to the absence of suitable probes, the microarray method at 850 K resolution was performed to identify exons 3–10 by NGS. Although there were some technical problems in the analysis program, we detected heterozygous deletion in the parents as suspicious. Nonetheless, there were highly intense loss of heterozygosity (LOH) regions in the microarray data of both patients, which might be an indication of unknown distance consanguinity. In addition, we checked the transposable elements near the breakpoints for a possible effect on the deletion (Fig. 4). Currently, it is unclear if these transposable elements were involved in the formation of this structural alteration. By designing suitable qPCR probes, we confirmed the homozygous deletion in both affected individuals and showed the heterozygous carrier state for the SLC7A7 deletion in the parents. The homozygous deletion comprising exons 3 to 10 has not been reported yet in any database [14]. This report presents the first case of lysinuric protein intolerance in a Turkish family with a gross unidentified deletion in the SLC7A7 gene.
Fig. 4.
SLC7A7 genomic position and gene structure. SLC7A7 (NM_003982.4) is located on chromosome 14 and has 10 exons on the minus strand. The deletion region (grey bar) and transposable elements (blue bars) close to the deletion are marked. SINEs near breaking point from left to right are as the following: AluSx, AluY, AluJb, AluJb, AluSz6, AluJb, MIR, AluSg4, MIR, AluSq, AluSx, AluJb, AluSg, AluJb, AluSx1, MIR3, MIRb (https://genome.ucsc.edu/).
In summary, we report two individuals with an atypical presentation of LPI including multiple bone fractures and hyperferritinemia leading to delayed diagnosis of their underlying condition. Due to its broad phenotypic variability, LPI has to be included in the differential diagnosis of various metabolic disorders such as urea cycle disorders, lysosomal storage diseases, hemochromatosis, and osteoporosis. Up to date, this is the first report of lysinuric protein intolerance in a Turkish family with a novel gene deletion causing severe hyperferritinemia and osteoporosis.
Declaration of Competing Interest
None.
Acknowledgement
We thank all of the family members for their collaboration. We would like to convey special thanks to Yun Li for the support of the qPCR analysis. We are highly grateful to Ipek Ilgin Gönenc for the figures and Karin Boss for critically reading the manuscript.
Data availability
Data will be made available on request.
References
- 1.Carpentieri D., Barnhart M.F., Aleck K., Miloh T., deMello D. Lysinuric protein intolerance in a family of Mexican ancestry with a novel SLC7A7 gene deletion. Case report and review of the literature. Mol. Genet. Metab. Rep. 2015;2:47–50. doi: 10.1016/j.ymgmr.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko J.M., Shin C.H., Yang S.W., Seong M.W., Park S.S., Song J. The first Korean case of lysinuric protein intolerance: presented with short stature and increased somnolence. J. Korean Med. Sci. 2012;27(8):961–964. doi: 10.3346/jkms.2012.27.8.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperandeo M.P., Andria G., Sebastio G. Lysinuric protein intolerance: update and extended mutation analysis of the SLC7A7 gene. Hum. Mutat. 2008;29(1):14–21. doi: 10.1002/humu.20589. [DOI] [PubMed] [Google Scholar]
- 4.Puomila K., Simell O., Huoponen K., Mykkänen J. Two alternative promoters regulate the expression of lysinuric protein intolerance gene SLC7A7. Mol. Genet. Metab. 2007;90(3):298–306. doi: 10.1016/j.ymgme.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Mykkänen J., Toivonen M., Kleemola M., et al. Promoter analysis of the human SLC7A7 gene encoding y+L amino acid transporter-1 (y+LAT-1) Biochem. Biophys. Res. Commun. 2003;301(4):855–861. doi: 10.1016/s0006-291x(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 6.Nunes V., Niinikoski H. GeneReviews® [Internet] University of Washington; Seattle: 2018. Lysinuric protein intolerance.https://www.ncbi.nlm.nih.gov/books/NBK1361/ Accessed July 27, 2023. [Google Scholar]
- 7.Oyanagi K., Miura R., Yamanouchi T. Congenital lysinuria: a new inherited transport disorder of dibasic amino acids. J. Pediatr. 1970;77(2):259–266. doi: 10.1016/s0022-3476(70)80333-x. [DOI] [PubMed] [Google Scholar]
- 8.Contreras J.L., Ladino M.A., Aránguiz K., et al. Immune dysregulation mimicking systemic lupus erythematosus in a patient with Lysinuric protein intolerance: case report and review of the literature. Front. Pediatr. 2021;9 doi: 10.3389/fped.2021.673957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parto K., Penttinen R., Paronen I., Pelliniemi L., Simell O. Osteoporosis in lysinuric protein intolerance. J. Inherit. Metab. Dis. 1993;16(2):441–450. doi: 10.1007/BF00710296. [DOI] [PubMed] [Google Scholar]
- 10.Lukkarinen M., Näntö-Salonen K., Ruuskanen O., et al. Varicella and varicella immunity in patients with lysinuric protein intolerance. J. Inherit. Metab. Dis. 1998;21(2):103–111. doi: 10.1023/a:1005335423939. [DOI] [PubMed] [Google Scholar]
- 11.Rajantie J., Rapola J., Siimes M.A. Ferritinemia with subnormal iron stores in lysinuric protein intolerance. Metabolism. 1981;30(1):3–5. doi: 10.1016/0026-0495(81)90211-0. [DOI] [PubMed] [Google Scholar]
- 12.Intermittent hemophagocytic lymphohistiocytosis is a regular feature of lysinuric protein intolerance - PubMed. November 2, 2023. https://pubmed.ncbi.nlm.nih.gov/9931537/ [DOI] [PubMed]
- 13.Rajantie J., Simell O., Perheentupa J., Siimes M.A. Changes in peripheral blood cells and serum ferritin in lysinuric protein intolerance. Acta Paediatr. Scand. 1980;69(6):741–745. doi: 10.1111/j.1651-2227.1980.tb07143.x. [DOI] [PubMed] [Google Scholar]
- 14.Genomic Variants in Human Genome (Build GRCh37: Feb. 2009, hg19) 9.343 kbp from chr14:23,241,787..23,251,130. July 27, 2023. http://dgv.tcag.ca/gb2/gbrowse/dgv2_hg19/?name=chr14%3A23241787-23251130;search=Search
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.