Abstract
Haemophilus influenzae Rd is a gram-negative natural transformer. A mutant strain, RJ248, that has normal DNA uptake and translocation but whose transformation frequency is 300 times lower than that of wild-type H. influenzae and whose phage recombination is 8 times lower was isolated. The affected gene, comM, is induced during competence development in wild-type H. influenzae but not in RJ248.
Haemophilus influenzae Rd is a gram-negative bacterium capable of natural DNA transformation. It achieves low-level competence in late log phase and can be induced to high levels of competence by a nutritional downshift into MIV medium (12) or by transient anaerobic growth (10).
Transformation involves several steps. The first is the uptake of naked DNA into a membrane-protected compartment (13). Uptake of DNA is facilitated by an uptake signal sequence (20). Following uptake, the DNA translocates slowly into the cytoplasm, where the 5′ strand is completely degraded and the 3′ strand is partially degraded (2). If homology exists between the incoming DNA and the chromosome of the cell, the entering 3′ strand will invade and replace the chromosomal DNA by homologous recombination (2).
The comABCDEF operon is essential for transformation and is induced during competence development but not during exponential growth (21, 22). A 26-bp palindromic competence-regulatory element (CRE) (also known as the dyad symmetry element) is required for induction of the operon (9, 14, 22). This element has been found upstream of three other known competence genes (14, 23) (pilABCD [8, 23], rec2 [7], and dprA [14]), all of which have been shown to be induced during competence development (14, 23). In all cases, the element is positioned about one turn of the helix upstream of either a known or a putative promoter (14, 22). It is likely that this sequence is the binding site for a competence-specific, positively acting transcriptional regulator. A known regulator of competence which could fill this role is the product of the gene sxy (also known as tfoX). This gene is required for competence development and for the induction of both dprA and comF (14, 24, 26). In addition, certain point mutations of this gene which allow cells to be competent all of the time exist (18). It is not known whether the effect of sxy on expression of competence genes is direct or indirect (14).
Strains with mutations in rec2, dprA, and comF have normal DNA uptake but are transformation deficient (7, 14, 23, 26). In rec2 mutants, the DNA remains trapped in the membrane compartment and never translocates into the cytoplasm (4). The H. influenzae homolog of the Escherichia coli recA gene, rec1, is required for homologous recombination of donor DNA into the chromosome (16).
Here we report on RJ248, a new transformation mutant of H. influenzae Rd that has a transformation frequency 300-fold lower than that of wild-type H. influenzae and a deficiency in the competence-induced increase in phage recombination. However, it has normal DNA uptake and translocation. The affected gene has been identified and named comM. It has a CRE site upstream of its putative promoter. It is induced during competence development in wild-type cells but not in RJ248.
Growth, transformation, and β-galactosidase activity.
H. influenzae was grown as described by Barcak et al. (3). E. coli was grown as described by Sambrook et al. (19). The MIV nutritional-downshift procedure was used to measure transformation frequencies (3, 12). DNA uptake was measured as described by Gwinn (11). β-Galactosidase activity was measured with a permeabilized-cell assay (17).
Mutagenesis and isolation of RJ248.
MIV-competent wild-type cells were transformed with a plasmid carrying Tn916 (6, 15). Transposition events were selected for by tetracycline resistance, which is encoded by Tn916. Tetracycline-resistant colonies were screened by the cyclic AMP transformation plate assay (25). One strain, RJ24, was consistently transformation deficient. Genomic DNA from RJ24 was transformed into wild-type cells to isolate a pure strain with only one Tn916 insertion, resulting in RJ248. Southern and MIV transformation analysis confirmed that RJ248 carries one Tn916 insertion responsible for the transformation phenotype and that the insertion was not in a known transformation gene.
UV sensitivity.
RJ248, KW20 (wild type), and DB117 (rec1 mutant; negative control) were grown and exposed to a GE 615T8 germicidal lamp for 0, 3, or 6 s at a distance of 37 cm. Plates were incubated at 37°C for 24 h, and survivors were counted.
Phage recombination.
Exponential-phase cells and competent cells were separately coinfected with ts1 and ts3, temperature-sensitive point mutants of the H. influenzae phage Hp1c1. Recombinant progeny were identified by their ability to grow at the nonpermissive temperature (40°C) (5).
Translocation.
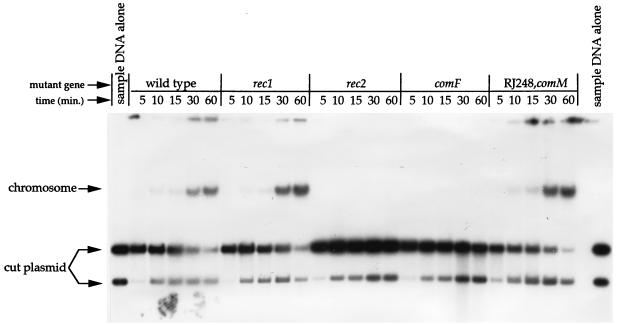
A plasmid containing an H. influenzae genomic PstI fragment was cut with PstI and end labeled with [32P]dCTP by using T4 polymerase. Cells which had been made competent by the MIV method were incubated with labeled plasmid. At 5, 10, 15, 30, and 60 min, samples of the cells were removed and washed with MIV medium by centrifugation. Total DNA was isolated. DNA samples were electrophoresed on a 0.8% agarose gel and autoradiographed (4).
RNA isolation and Northern analysis.
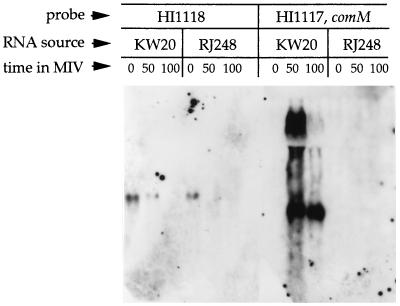
All buffers used in RNA isolation were made with diethyl pyrocarbonate-treated water. KW20 (wild type) and RJ248 were made competent by the MIV method. At 0, 50, and 100 min in MIV medium, 35 ml of cells was removed. Cells were lysed, and RNA was isolated by hot-phenol extraction. Northern analysis was performed with the Gene Images CDP-Start labeling and detection kits (Amersham, Indianapolis, Ind.). Probes were gene-internal fragments of HI1118 and HI1117 DNA.
Phenotype of RJ248.
RJ248 has a transformation frequency that is 300-fold lower than that of the wild type. It has a wild-type DNA uptake level and grows at the wild-type rate. Its resistance to UV light is the same as that seen in the wild type (data not shown).
To test whether the mutation in RJ248 affected regulation, we transformed RJ248 with genomic DNA from MGH100 (a strain containing lacZ kan in an operon fusion with comA), which resulted in a strain with the RJ248 mutation and a β-galactosidase transcription indicator in comA. We found that induction of comA, as measured by β-galactosidase activity, is unaffected in the RJ248 background (data not shown).
Competent cells exhibit a higher rate of phage recombination than cells in exponential phase (5). We found that with wild-type cells there is about a 40-fold increase in the recombination rate in competent cells over that in cells in exponential phase, while with RJ248 the increase is only 5-fold.
To test whether DNA translocation is normal in RJ248, we compared the strain to several other mutant strains. Samples of competent cells were given radioactively labeled, digested plasmid DNA (two fragments). In wild-type cells, labeled donor DNA was taken up and became associated with the chromosome over time (Fig. 1). As expected, rec2 mutants did not translocate DNA into the cytoplasm, as indicated by the absence of accumulation of label in the chromosome. Cells with a mutation in rec1 take DNA into the cytoplasm; however, since the DNA cannot undergo homologous recombination, it is degraded in the cytoplasm and the labeled nucleotides are incorporated into the chromosome through subsequent reuse (4). We found that the accumulations of label in the chromosome in the wild type and RJ248 were indistinguishable, indicating that translocation in RJ248 is normal (Fig. 1). It is theoretically possible that in RJ248 the DNA becomes trapped in the membrane compartment, where it undergoes degradation and subsequent entry via another mechanism. However, we feel this to be unlikely, since there is substantial evidence indicating that large-scale degradation of donor DNA does not occur until it reaches the cytoplasm. In addition, although slow DNA degradation has been observed in the membrane compartment, this has not been shown to allow the DNA to then enter the cytoplasm (2, 4).
FIG. 1.
Results of a translocation experiment. Translocation is indicated by the appearance in the chromosome of labeled plasmid DNA.
Identification of the affected gene.
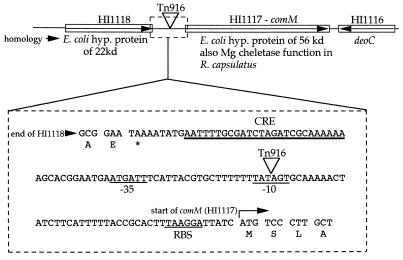
Sequence analysis of clones carrying the region of insertion revealed that Tn916 was inserted between the HI1117 and HI1118 genes (9), which are in the same orientation (Fig. 2). The predicted products of both open reading frames have significant homology to hypothetical proteins in E. coli (see the H. influenzae database at www.tigr.org). In addition, HI1117 has homology to a magnesium chelatase gene of Rhodobacter capsulatus, bchI, involved in bacteriochlorophyll biosynthesis (1) and to related genes from other photosynthetic organisms. The putative promoter region of HI1117 contains a CRE sequence about one turn of the helix upstream of a putative promoter (Fig. 2). The insertion point of Tn916 is in the −10 hexamer region of the possible promoter. This insertion is beyond the stop codon of HI1118. We conclude that HI1117 is affected by the disruption of its promoter and have named it comM.
FIG. 2.
Sequence around the point of insertion of Tn916 with the putative comM promoter indicated. hyp., hypothetical. The asterisk denotes a stop codon. RBS, ribosomal binding site.
To test whether comM is induced during competence development, as would be expected based on the presence of the CRE, and whether induction is prevented by the presence of Tn916 in the mutant, total RNA from both KW20 (wild type) and RJ248 before and after competence induction was probed with a DNA fragment from within comM and with a DNA fragment from within HI1118. We found that in the wild type, comM is strongly induced during competence development; however, in RJ248, there is no detectable induction of comM (Fig. 3). Expression of HI1118 diminishes in both backgrounds and is unaffected by the presence of Tn916 (Fig. 3).
FIG. 3.
Northern analysis of RJ248. Times are given in minutes.
The comM gene is clearly not induced in RJ248 during competence development. This is due to the presence of Tn916, which disrupts the putative promoter. In addition to its transformation deficiency, the only competence-associated defect we have found in RJ248 is a reduction in competence-dependent phage recombination. This phenotype and the ability of RJ248 to translocate DNA normally imply that comM acts very late in transformation, presumably at a step in the recombination of the donor DNA into the chromosome.
Acknowledgments
This work was funded by grant NP838C from the American Cancer Society and by grant GM48251 from the National Institutes of Health. H. O. Smith is an American Cancer Society Research Professor.
REFERENCES
- 1.Armstrong G A, Cook D N, Ma D, Alberti M, Burke D H, Hearst J E. Regulation of carotenoid and bacteriochlorophyll biosynthesis genes and identification of an evolutionarily conserved gene required for bacteriochlorophyll accumulation. J Gen Microbiol. 1993;139:897–906. doi: 10.1099/00221287-139-5-897. [DOI] [PubMed] [Google Scholar]
- 2.Barany F, Kahn M E, Smith H O. Directional transport and integration of donor DNA in Haemophilus influenzae transformation. Proc Natl Acad Sci USA. 1983;80:7274–7278. doi: 10.1073/pnas.80.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcak G J, Chandler M S, Redfield R J, Tomb J-F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 4.Barouki R, Smith H O. Reexamination of phenotypic defects in rec-1 and rec-2 mutants of Haemophilus influenzae Rd. J Bacteriol. 1985;163:629–634. doi: 10.1128/jb.163.2.629-634.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boling M E, Setlow J K. Dependence of vegetative recombination among Haemophilus influenzae bacteriophage on the host cell. J Virol. 1969;4:240–243. doi: 10.1128/jvi.4.3.240-243.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell D B, Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- 7.Clifton S W, McCarthy D, Roe B A. Sequence of the rec-2 locus of Haemophilus influenzae: homologies to comE-ORF3 of Bacillus subtilis and msbA of Escherichia coli. Gene. 1994;146:95–100. doi: 10.1016/0378-1119(94)90840-0. [DOI] [PubMed] [Google Scholar]
- 8.Dougherty, B. A. Unpublished data.
- 9.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenny K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelly J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Goodgal S H, Herriott R M. Studies on transformations of Haemophilus influenzae. I. Competence. J Gen Physiol. 1961;44:1201–1227. doi: 10.1085/jgp.44.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gwinn M L. Ph.D. thesis. Baltimore, Md: Johns Hopkins University School of Medicine; 1997. [Google Scholar]
- 12.Herriott R M, Meyer E M, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahn M E, Smith H O. Transformation in Haemophilus: a problem in membrane biology. J Membr Biol. 1984;81:89–103. doi: 10.1007/BF01868974. [DOI] [PubMed] [Google Scholar]
- 14.Karudapuram S, Barcak G J. The Haemophilus influenzae dpr ABC genes constitute a competence-inducible operon that requires the product of the tfoX (sxy) gene for transcriptional activation. J Bacteriol. 1997;179:4815–4820. doi: 10.1128/jb.179.15.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauc L, Goodgal S H. Introduction of transposon Tn916 DNA into Haemophilus influenzae and Haemophilus parainfluenzae. J Bacteriol. 1989;171:6625–6628. doi: 10.1128/jb.171.12.6625-6628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kooistra J, Setlow J K. Similarity in properties and mapping of three Rec mutants of Haemophilus influenzae. J Bacteriol. 1976;127:327–333. doi: 10.1128/jb.127.1.327-333.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Redfield R J. sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence. J Bacteriol. 1991;173:5612–5618. doi: 10.1128/jb.173.18.5612-5618.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Smith H O, Tomb J-F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 21.Tomb J-F, El-Hajj H, Smith H O. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene. 1991;104:1–10. doi: 10.1016/0378-1119(91)90457-m. [DOI] [PubMed] [Google Scholar]
- 22.Tomb, J.-F., M. L. Gwinn, H. Amitai, S. Abuchakra, and H. O. Smith. Unpublished results.
- 23.Tomb, J.-F. Unpublished data.
- 24.Williams P M, Bannister L A, Redfield R J. The Haemophilus influenzae sxy-1 mutation is in a newly identified gene essential for competence. J Bacteriol. 1994;176:6789–6794. doi: 10.1128/jb.176.22.6789-6794.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise E M, Alexander S P, Powers M. Adenosine 3′:5′-cyclic monophosphate as a regulator of bacterial transformation. Proc Natl Acad Sci USA. 1973;70:471–474. doi: 10.1073/pnas.70.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zulty J J, Barcak G J. Identification of a DNA transformation gene required for com101A+ expression and supertransformer phenotype in Haemophilus influenzae. Proc Natl Acad Sci USA. 1995;92:3616–3620. doi: 10.1073/pnas.92.8.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]