Background
Although many studies reported a close relationship between depression and Alzheimer’s disease (AD), the underlying pathophysiological mechanism remains unclear. The present study aimed to investigate the mechanism of AD and major depressive disorder (MDD). Method: The datasets were downloaded from the Gene Expression Omnibus. After screening differentially expressed genes (DEGs), gene ontology and pathway analysis were performed and protein–protein interaction, TF-target gene, and miRNA-target gene networks were established. Results: 171 DEGs of AD-related datasets and 79 DEGs shared by AD and MDD were detected. Functional analysis revealed that AD and MDD common genes were significantly enriched in circadian entrainment and long-term depression signaling pathways. Five hub genes were identified after construction of networks and validation of hub gene signatures. In conclusion, DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL may be potential targets for the diagnosis and treatment of AD and MDD.
Keywords: Alzheimer’s disease, major depressive disorder, differentially expressed genes, pathophysiology association, hub gene
Significance Statement
● The differentially expressed genes of AD-related datasets were significantly enriched in the long-term depression pathway.
● DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL might play essential roles in the pathophysiological mechanism of Alzheimer’s disease and major depressive disorder.
Introduction
Alzheimer’s disease (AD) is a central nervous system disease characterized by anterograde amnesia, accounting for 60 to 80 percent of all types of dementia. 1 Alzheimer’s Disease International (ADI) estimates that the number of people with dementia worldwide will increase to 152 million by 2050. The annual cost of dementia is estimated to be $2 trillion in 2030. 2 Alzheimer’s disease severely damages the ability of daily life and quality of life of patients and brings a substantial care burden and economic losses to caregivers and society. The US Food and Drug Administration (FDA) approved six drugs to alleviate the symptom development of AD patients,3,4 but the drug treatment effects were not satisfactory.
Major depressive disorder (MDD) is a complex disease characterized by mood depression and various physiological disorders, with high morbidity and suicide mortality. More than 264 million people worldwide suffer from depressive disorders, 5 and nearly 1 million people commit suicide each year due to mental disorder. 6 Globally, 163 million have MDD 5 ; the lifetime prevalence of MDD is approximately 15–18% and the 12-month MDD prevalence is 6%. 7 Genetic factors account for approximately 40% of the causes of MDD, 8 and chronic or nervous system diseases (e.g., AD, 9 Parkinson’s disease, 9 Huntington’s disease, 9 migraine, 10 glioblastoma multiforme, 11 and so forth) are also important factors affecting MDD. A recent network meta-analysis study has reported that MDD does exist in approximately 40% of AD cases. 12 Nevertheless, when AD coexists with MDD, the number of hospitalizations is considerably higher when compared to either MDD or AD alone.13,14 Most importantly, however, the mortality rate is increased which is experienced by individuals with comorbid MDD and AD. 15 Therefore, there is a high risk of comorbidities between MDD and AD.
A considerable amount of literature has revealed that depression may be a risk factor for dementia or part of the prodromal symptoms of dementia.16,17 Older patients with MDD exhibit significant cognitive deficits, which have been demonstrated in various cognitive domains in more than half of patients with late-life depression. 18 It has been reported that the prevalence of depression in patients with dementia is 29%.16,19 In addition, venlafaxine, a commonly used drug for the treatment of depression, has been reported to effectively improve the behavioral performance of rats in the Morris water maze test. 20 Although depression plays a significant role in the occurrence of AD, the pathological mechanism of the two comorbidities remains unclear.
In this study, we downloaded the gene expression profiles of AD, MDD, and MDD patients treated with venlafaxine for eight weeks. After determining the differentially expressed genes (DEGs) between diseased samples and healthy subjects, cluster analysis and functional enrichment analysis were carried out to explore the biological pathways in the processes of AD and MDD. Finally, the protein–protein interaction (PPI) network was constructed to find the potentially related genes between AD and MDD, and the hub genes were identified in MDD patients treated with venlafaxine. The identified hub genes may play an essential role in the genetics underpinning AD and MDD. These findings provide a reference for explaining the relationship between AD and MDD, but the specific pathological mechanism remains to be further studied.
Materials and Methods
Data Collection
Gene expression profiles of GSE48350, GSE5281, GSE18309, GSE98793, and GSE32280 were obtained from the GPL570 platform (HG‐U133_Plus_2; Affymetrix Human Genome U133 Plus 2.0 Array) of the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). Because of the different functions of each brain region and the complexity of AD pathology, the AD specimens in this study include hippocampal tissue, frontal lobe tissue, and whole blood. The GSE48350 and GSE5281 datasets contain 155 brain tissue samples, including 73 (29 hippocampal tissues and 44 frontal lobe tissues) AD brain tissue samples and 82 (38 hippocampal tissues and 44 frontal lobe tissues) standard brain tissue samples. Another dataset, GSE18309, includes three AD patients and three healthy subjects. The MDD dataset GSE98793 includes 128 peripheral blood samples, 64 from MDD patients and 64 from healthy people, while the GSE32280 dataset includes 8 MDD samples before treatment and 8 MDD samples after venlafaxine treatment. The detailed information for these datasets is listed in Table 1. Probes corresponding to none or multiple genes were deleted. When multiple probes corresponded to the same gene, the average expression value was considered its final expression. 21
Table 1.
Description of data used in this study.
| GEO Accession | Platforms | Control | AD/treat | Type |
|---|---|---|---|---|
| GSE48350 | GPL570 | 58 ① Normal | 40 ② AD | Brain tissue |
| GSE5281 | GPL570 | 24 ③ Normal | 33 ④ AD | Brain tissue |
| GSE18309 | GPL570 | 3 Normal | 3 AD | Whole blood |
| GSE98793 | GPL570 | 64 Normal | 64 MDD | Whole blood |
| GSE32280 | GPL570 | 8 MDD untreated | 8 ⑤ treated | Whole blood |
①58 healthy human brain tissues included 25 hippocampal tissues and 33 frontal lobe tissues;
②40 brain tissues of AD patients included 19 hippocampal tissues and 21 frontal lobe tissues; ③24 healthy human brain tissues included 13 hippocampal tissues and 11 frontal lobe tissues; ④33 brain tissues of AD patients included 10 hippocampal tissues and 23 frontal lobe tissues; ⑤Patients with MDD were only treated with venlafaxine for 8 weeks.
Identification of Differentially Expressed Genes
We carried out a comprehensive bioinformatics analysis of the GEO dataset. The identification of DEGs was performed to compare the differences in gene expression between two conditions. The limma package 22 was used in GEO2R 23 to screen DEGs in the GEO dataset. To screen out appropriate number of DEGs and high-quality molecular markers of AD and MDD, we used different |log base 2 Fold Change| to screen DEGs and compared their results. When P < .05 and |log base 2 Fold Change | > 1, which identified 6 DEGs of AD-related datasets, 0 DEGs shared by AD and MDD were identified. When P < .05 and |log base 2 Fold Change | > .5, which identified 66 DEGs of AD-related datasets, 9 DEGs shared by AD and MDD were identified. When P < .05 and |log base 2 Fold Change | > .3, which identified 171 DEGs of AD-related datasets, 79 DEGs shared by AD and MDD were identified. Finally, we selected P < .05 and |log base 2 Fold Change | > .3 as the criteria for DEGs. 24 For visualization, the Venn diagram and the intersection plot of DEGs were generated using two web tools: (http://bioinformatics.psb.ugent.be/webtools/Venn/) and (http://www.bioinformatics.com.cn/plot_basic_upsetR_plot_009).
Functional Enrichment Analysis
To explore more comprehensive biological information for the above DEGs, GO enrichment analysis, including biological processes (BP), cellular component (CC), and molecular function (MF) terms, was performed on the R package cluster profiler (V3.16.0). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment was performed in the R packages cluster profiler (V3.16.0) and DOSE (V3.14.0). 25 The bubble map was drawn using the ggplot2 R package for the top 10 significant KEGG pathways based on p. adjust <.05. The GO enrichment results were divided into three functional categories, BP, CC, and MF, and the top 10 results of BP, CC, and MF (p. adjust <.05.) were visualized using the GO plot R package (V1.0.2).
Construction of the PPI Network and Identification of Hub Genes
The STRING 26 (http://string-db.org) online database was used to determine the interaction genes, and the interaction parameters were set to a maximum confidence of >.4. 27 To fully understand the functional interactions between proteins and select essential hub genes, the PPI network was visualized and analyzed with Cytoscape (version 3.7.2) software (http://www.cytoscape.org/), and hub genes were screened with cytoHubba, 28 a plug-in of Cytoscape software.
CytoHubba was used to rank genes in the PPI network from highest to lowest according to the maximum neighborhood component (MNC), density of maximum neighborhood component (DMNC), maximal clique centrality (MCC), and degree method scores (degree). The top 10 genes were selected in each algorithm, and the common genes were identified as hub genes. Finally, nine hub genes were obtained. 29
Validation of Hub Gene Signatures
To evaluate the clinical value of these 9 hub genes, we used one independent GEO cohort as a validation dataset. The GSE32280 dataset, including peripheral blood samples from 8 MDD patients before treatment and 8 MDD patients treated with venlafaxine for eight weeks, was used to demonstrate the expression of 9 hub genes. To show the expression patterns of 9 hub genes between MDD patients before treatment and MDD patients treated with venlafaxine, differences between each group were analyzed by T-test, and violin plot using the ggplot2 (V3.3.1) package in R (V4.0.0) was used for visualization.
Construction of Target Genes—Transcription Factor Regulatory Network
The miRNet database (https://www.mirnet.ca/) was used to predict TFs of significant target genes. Finally, Cytoscape was used to visualize the regulatory network of TF-target genes, which described the interaction between TFs and their potential targets in AD and MDD. 30
Construction of Target Genes-miRNA Regulatory Network
The miRNet database (https://www.mirnet.ca/) was used to predict miRNAs of significant target genes. Finally, Cytoscape was used to visualize the regulatory network of miRNA-target genes, which described the interaction between miRNAs and their potential targets in AD and MDD. 30
Receiver Operating Characteristic Curve Analysis
ROC curves were used to evaluate classifiers in bioinformatics applications. In order to further evaluate the predictive accuracy of hub genes, ROC analysis was performed to distinguish AD from normal control and MDD from normal control. Based on the expression profile by high-throughput sequencing data of hub genes, the pROC in R was used to generate the ROC curves of hub genes. The diagnostic value of hub gene was compared by measuring the area under ROC curve and AUC. 30
Results
Identification of Differentially Expressed Genes
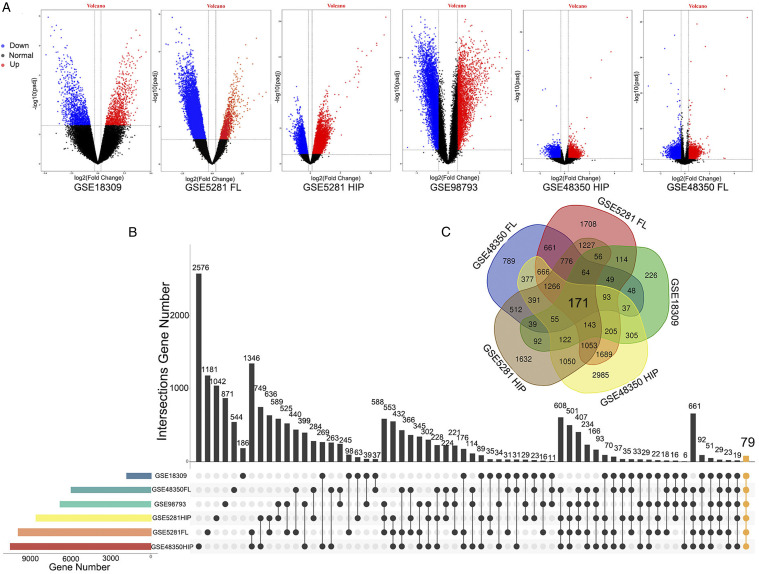
In the hippocampal tissue of AD, there were 11188 DEGs (GSE48350 HIP, including 8128 upregulated genes and 3060 downregulated genes) in GSE48350 and 9539 DEGs (GSE5281 HIP, including 5605 upregulated genes and 3934 downregulated genes) in GSE5281. In the frontal lobe tissue of GSE48350, there were 6099 DEGs (GSE48350 FL, including 2279 upregulated genes and 3820 downregulated genes) and 10127 DEGs (GSE5281 FL, including 748 upregulated genes and 9379 downregulated genes) in GSE5281. There were 1866 DEGs (765 upregulated genes and 1101 downregulated genes) in GSE18309. After screening the DEGs of the GSE98793 dataset, a dataset related to MDD patients, including 64 MDD patients and 64 standard samples, we obtained 6846 DEGs, including 1904 upregulated genes and 4942 downregulated genes. Figure 1A shows the volcano plots of DEGs in each dataset drawn using the ggplot2 (V3.3.1) package in R (V4.0.0).
Figure 1.
Identification of DEGs among each GEO dataset. (A) The volcano plots present the expression pattern of DEGs in each dataset. Red: upregulated; Blue: downregulated; Black: normal. (B) The intersection plot of DEGs among these datasets. The vertical yellow line represents the intersection of DEGs (79 DEGs shared in AD and MDD datasets) from six datasets. (C) The Venn plot of DEGs (171 DEGs of AD-related datasets) among these datasets. HIP: hippocampal tissue; FL: frontal lobe tissue.
As shown in Figure 1B, the 79 DEGs (orange bar chart), the intersection of all the datasets (GSE48350 HIP, GSE5281 HIP, GSE48350 FL, GSE5281 FL, GSE18309, and GSE98793) in this study, were common to AD and MDD. Herein, 26 upregulated genes and 53 downregulated genes were found in AD, while 40 upregulated genes and 39 downregulated genes were found in MDD. In Figure 1C, the 171 DEGs are in common among all the AD datasets.
Functional Enrichment Analysis of AD-Related Differentially Expressed Genes
According to KEGG pathway enrichment analysis, AD-related DEGs were significantly enriched in the Rap1 signaling pathway, adrenergic signaling in cardiomyocytes, retrograde endocannabinoid signaling, glutamatergic synapse, cholinergic synapse, circadian entrainment, dilated cardiomyopathy (DCM), salivary secretion, and nicotine addiction. The long-term depression pathway was also enriched (Figures S1A and Table S1).
The DEG GO enrichment analysis results were divided into three functional categories (BP, CC, and MF). For BP, the DEGs were mainly enriched in adult behavior, synaptic vesicle cycle, regulation of membrane potential, regulation of postsynaptic membrane potential, synapse organization, hindbrain development, adult locomotory behavior, cognition, positive regulation of morphogenesis of an epithelium, and locomotory behavior (Figures S1B and Table S2). In terms of CC, the genes were mainly enriched in the glutamatergic synapse, presynapse, synaptic membrane, GABAergic synapse, ion channel complex, axon terminus, postsynaptic density, transmembrane transporter complex, asymmetric synapse, and transporter complex (Figures S1C and Table S2). In the MF group, the DEGs were significantly enriched in calmodulin-binding, ligand-gated anion channel activity, ion channel activity, substrate-specific channel activity, channel activity, passive transmembrane transporter activity, gated channel activity, cell adhesion molecule binding, extracellular ligand-gated ion channel activity, and alpha-actinin binding (Figures S1D and Table S2).
Functional Enrichment Analysis for the Differentially Expressed Genes Related to AD and Major Depressive Disorder
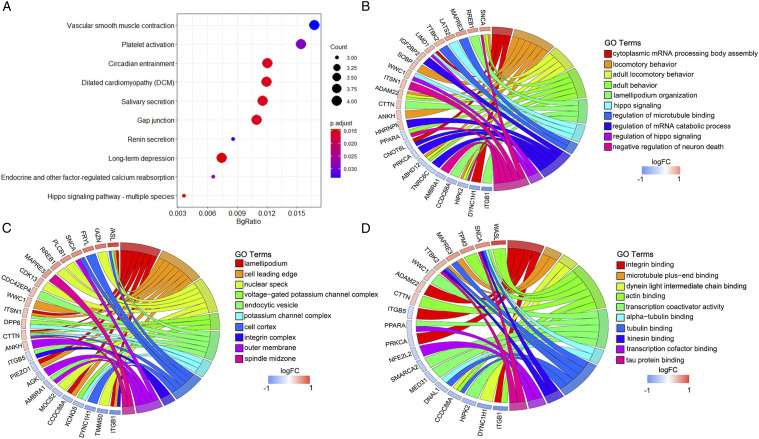
The top 10 KEGG pathways of the DEGs with overlapping AD and MDD were mainly enriched in vascular smooth muscle contraction, platelet activation, circadian entrainment, dilated cardiomyopathy (DCM), salivary secretion, gap junction, renin secretion, long-term depression, endocrine and other factor-regulated calcium reabsorption, and Hippo signaling pathway-multiple species (Figures 2A and Table S3).
Figure 2.
Functional characteristics analysis for the DEGs related to AD and MDD. (A) Top 10 KEGG pathway enrichment results. (B) The BP category of GO enrichment analyses. (C) The CC category of GO enrichment analyses. (D) The MF category of GO enrichment analyses.
The BP category of the GO analysis results showed that DEGs were significantly enriched in cytoplasmic mRNA processing body assembly, locomotory behavior, adult locomotory behavior, adult behavior, lamellipodial organization, Hippo signaling, regulation of microtubule-binding, regulation of mRNA catabolic process, regulation of Hippo signaling, and negative regulation of neuron death (Figures 2B and Table S4). For CC, these genes were enriched in the lamellipodial, cell leading edge, nuclear speck, voltage-gated potassium channel complex, endocytic vesicle, potassium channel complex, cell cortex, integrin complex, outer membrane, and spindle midzone (Figures 2C and Table S4). Moreover, they were significantly enriched in integrin binding, microtubule plus-end binding, dynein light intermediate chain binding, actin binding, transcription coactivator activity, alpha-tubulin binding, tubulin binding, kinesin binding, transcription cofactor binding, and tau protein binding in the MF categories (Figures 2D and Table S4).
PPI Network Analysis
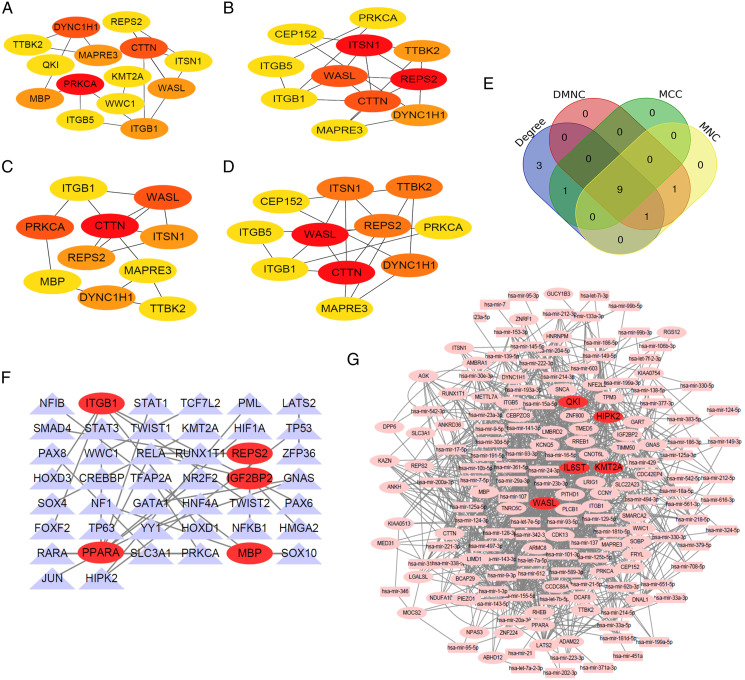
To further explore the biological interaction between AD and MDD, the PPI network of DEGs was constructed based on the STRING database. The separated and partially connected nodes were removed. The PPI network was composed of 76 nodes and 44 edges, with an average local clustering coefficient of .38, and the PPI enrichment P-value was <.011 (Figure S2). The intersection of the four algorithms cytoHubba (MNC, DMNC, MCC, and Degree; Table S5) was used to determine hub genes (Figure 3E). Finally, nine hub genes were identified, namely, DYNC1H1, MAPRE3, TTBK2, ITGB1, WASL, PRKCA, ITSN1, CTTN, and REPS2 (Figures 3A–3E).
Figure 3.
Establishment of the protein-protein interaction network, TF-target gene network and miRNA-target gene network. (A-D) The hub genes were identified using four models (Degree, DMNC, MCC, and MNC) with the Cytoscape plug-in cytoHubba. The darker the red color of a point is, the higher the score of the gene in the algorithm is, indicating the gene is more important in the network. (E) A Venn diagram was used to identify the nine hub genes in AD and MDD (DYNC1H1, MAPRE3, TTBK2, ITGB1, WASL, PRKCA, ITSN1, CTTN, and REPS2). (F). Target gene-TF regulatory network between target genes and TFs. target genes are marked in red. (G). Target gene-miRNA regulatory network between target genes and miRNAs. target genes are marked in red.
Construction of TF—Target Regulatory Network
After combining the consequences of TF-target genes with the interactive network of TFs, the TFs that may control the target genes are shown in Figure 3F. The top five target genes were PPARA, ITGB1, IGF2BP2, MBP, and REPS2 in the TF-target regulatory network. PPARA interacts with 9 TFs such as RELA and others; ITGB1 interacts with 6 TFs such as FOXF2 and others; IGF2BP2 interacts with 3 TFs such as HMGA2 and others; MBP interacts with 3 TFs such as SOX10 and others; and REPS2 interacts with 2 TFs such as NFKB1 and others (Table S6).
Construction of miRNA—Target Regulatory Network
After combining the consequences of miRNA-target genes with the interaction network of miRNAs, the genes and miRNAs are shown in Figure 3G. The top five target genes were QKI, HIPK2, KMT2A, WASL, and IL6ST in the miRNA-target regulatory network. QKI interacts with 45 miRNAs such as hsa-mir-16-5p and others; HIPK2 interacts with 39 miRNAs such as hsa-mir-124-3p and others; KMT2A interacts with 33 miRNAs such as hsa-mir-155-5p and others; WASL interacts with 32 miRNAs such as hsa-mir-1-3p and others; and IL6ST interacts with 31 miRNAs such as hsa-mir-23b-3p and others (Table S7).
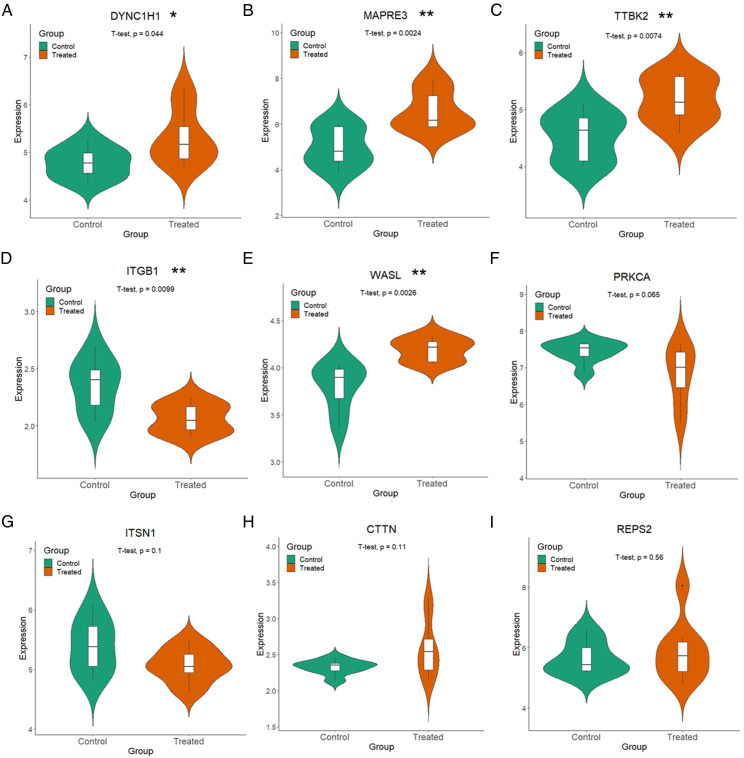
Changes in the Gene Expression Values of the Nine Hub Genes After Treatment With Venlafaxine in Major Depressive Disorder
To further validate the 9 potential hub genes in the relationship between AD and MDD, the expression of these genes was demonstrated in GSE32280 in a group of MDD patients treated with venlafaxine for eight weeks. The results showed that five of the nine hub genes, including DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL (Figures 4A–E), changed significantly in MDD patients after venlafaxine treatment (P<.05, T-test), while the gene expression values of PRKCA, ITSN1, CTTN, and REPS2 (Figures 4F–I) were not significantly different in MDD patients after treatment with venlafaxine (P>.05, T-test). As illustrated in Figure 4, compared with untreated control group, in the five hub genes whose expression changed significantly after venlafaxine treatment, the expression values of four hub genes were increased significantly and tended to return to normal levels, namely, DYNC1H1, MAPRE3, TTBK2, and WASL; and the expression value of ITGB1 was decreased significantly and tended to return to normal levels. More details of the five hub genes are listed in Table 2. This further supported the possibility that these five genes play an essential role in the process of AD and MDD.
Figure 4.
Changes in gene expression values of nine hub genes after treatment of MDD patients with venlafaxine. (A-I) represents the expression of DYNC1H1, MAPRE3, TTBK2, ITGB1, WASL, PRKCA, ITSN1, CTTN, and REPS2 genes in eight venlafaxine treated MDD patients (red) and eight control MDD patients (green).
Table 2.
The hub genes in AD and MDD shared network.
| Gene | Genecards identifier a | Full name of the gene | Gene-related diseases a |
|---|---|---|---|
| DYNC1H1 MAPRE3 TTBK2 ITGB1 WASL |
GC14P104635 GC02P026935 GC15M042738 GC10M032900 GC07M123681 |
Dynein cytoplasmic 1 heavy chain 1 Microtubule associated protein RP/EB family member 3 Tau tubulin kinase 2 Integrin subunit beta 1 WASP like actin Nucleation promoting factor |
Charcot-marie-tooth disease, axonal, type 2O (CMT2O), spinal muscular atrophy, lower extremity-predominant 1, autosomal dominant (SMA-LED1), mental retardation, autosomal dominant 13 (MRD13), Neuronal migration disorders, neuropathy, hereditary, with liability to pressure palsies (HNPP) Neuronopathy, distal hereditary motor, type viib (HMN7B), distal hereditary motor neuronopathy type 7 (DHMN7), perry syndrome (PERRYS) Spinocerebellar ataxia 11 (SCA11), autosomal dominant cerebellar ataxia (SCA), autosomal dominant cerebellar ataxia type iii (ADCA3), spinocerebellar ataxia 25 (SCA25), spinocerebellar ataxia 30 (SCA30) Neural tube defects (NTDs), Neuroblastoma (NB) Wiskott-aldrich syndrome (WAS), ataxia, early-onset, with oculomotor apraxia and hypoalbuminemia (EAOH), shigellosis |
afrom the GeneCards database (www.genecards.org).
Receiver Operating Characteristic Curve Analysis
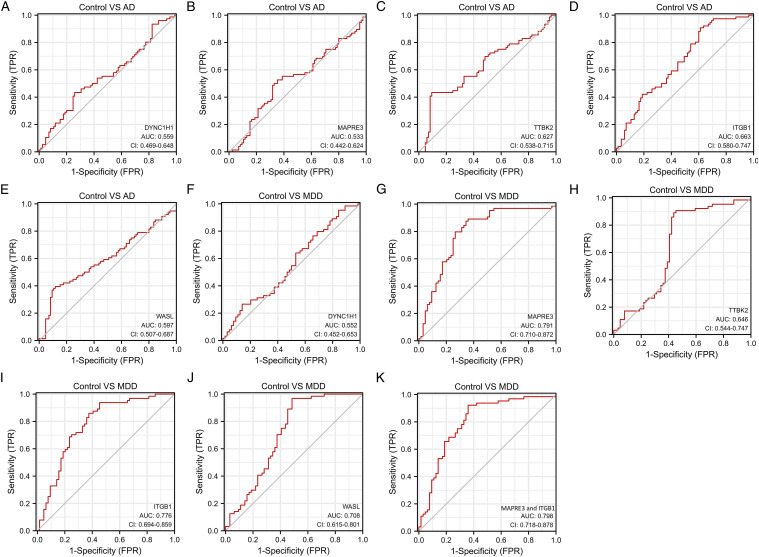
In addition, ROC curve analysis was implemented to calculate the capacity of the expression levels of hub genes to distinguish AD from normal control and MDD from normal control. The AUC value of DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL in AD indicated a moderate diagnostic efficiency (.7 > AUC >.5) (Figures 5A–E). The AUC value of DYNC1H1, and TTBK2 in MDD indicated a moderate diagnostic efficiency (.7 > AUC >.5) (Figures 5F and H). The results of ROC curve analysis suggested five hub genes exhibited a moderate diagnostic efficiency in AD and MDD. The AUC value of MAPRE3, ITGB1, and WASL in MDD suggested good diagnostic efficiency (.9 > AUC >.7) (Figures 5G, I, and J). The AUC value of combining MAPRE3 and ITGB1 was the highest in MDD and suggested good diagnostic efficiency (.9 > AUC >.7) (Figure 5K).
Figure 5.
ROC curve validated the sensitivity, specificity of five hub genes as a predictive biomarker for AD and MDD prognosis. (A). DYNC1H1 in AD; (B). MAPRE3 in AD; (C). TTBK2 in AD; (D). ITGB1 in AD; (E). WASL in AD; (F). DYNC1H1 in MDD; (G). MAPRE3 in MDD; (H). TTBK2 in MDD; (I). ITGB1 in MDD; (J). WASL in MDD.
Discussion
AD and MDD are common and frequently occurring diseases.2,31 At present, there is mounting evidence of a close relationship between AD and depression. It has been reported that the severity of depressive symptoms is positively correlated with whole brain cognitive impairment in patients with mild cognitive impairment (MCI), 32 while the elderly with severe depression and increasing depression symptoms have a higher risk of dementia. 33 The co-disease relationship between MDD and AD is exceptionally complex. At present, most of the relationship between AD and MDD is about clinical symptoms and co-disease, and more research on the common pathological mechanism of AD and MDD is needed. Therefore, the purpose of this study was to reveal the potential pathophysiological relationship between AD and MDD.
In this study, in order to mitigate the impact of sequencing batch effects and screen out molecular markers that can be shared by brain and blood tissues, 34 we restricted our choice of datasets to those produced from the same platform (GPL570 platform) and four gene expression profile datasets (GSE48350, GSE5281, GSE18309, and GSE98793) were used. Since there really is not suitable dataset related to brain tissue in MDD patients, we can only use the dataset related to blood tissue (https://www.ncbi.nlm.nih.gov/geo/). The samples of GSE18309 are blood samples with AD patients. GSE18309 acted as a bridge between AD and MDD’s samples. Although GSE18309 provides gene expression data from fewer samples compared to the other datasets, the data also provide a strong evidence for screening hub genes. For researchers, blood samples are the easiest to obtain, and we will examine the expression of hub genes in the blood tissues of patients with AD and MDD in the next experiment.
We used bioinformatics methods to integrate four gene expression profile datasets, which identified 171 DEGs (Table S8) of AD-related datasets; 79 DEGs (Table S9) shared by AD and MDD were identified. The main enrichment result of KEGG in AD-related DEGs was enriched in the long-term depression pathway provides a powerful reference in the cellular process for the relationship between AD and depression, which is indirectly consistent with previous studies showing that depression is closely related to AD.35-37 Seventy-nine DEGs shared by AD and MDD were mainly enriched in signaling pathways such as circadian entrainment, dilated cardiomyopathy (DCM), gap junction, and long-term depression, and GO enrichment results were enriched in adult behavior, endocytic vesicle, and protein binding. The role of circadian entrainment36,37 and gap junction38,39 in the KEGG pathway in AD and MDD has been reported, while the roles of dilated cardiomyopathy (DCM) 40 and the Hippo signaling pathway 41 in AD have been reported, but the roles of these pathways in MDD remain to be elucidated.
Subsequently, through the establishment of the PPI network by the STRING database and module analysis, nine hub genes were identified. Furthermore, after validating the expression of nine hub genes in MDD patients treated with venlafaxine, we found five genes (DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL) with significant changes in expression values (P < .05, T-test), which makes the expression level of these five hub genes in MDD patients closer to that in healthy people. Finally, we conducted the ROC curve analysis for five hub genes, and we found the ROC curves of MAPRE3 and ITGB1 looked good and the ROC curve of the combination of MAPRE3 and ITGB1 only improved slightly compared to either gene alone. We surmise that MAPRE3 and ITGB1 are in the same pathway and thus highly correlated in expression. The results indicate that these five hub genes had critical biological functions in the development of AD and MDD, and also suggest that antidepressant therapy may play a decisive role in the treatment of AD.
Dynein heavy chain 1 (DYNC1H1) dimer is the core of the cytoplasmic dynein 1 complex. The dynamic protein complex with housekeeping function is involved in the localization of the mitotic spindle, nuclear localization, golgi apparatus maintenance, and endosomal dynamics,42-44 while in the nervous system, the complex affects gene expression, development and regeneration, and the return of misfolded proteins and organelles from synapses to the cell body. 45 It has been reported that compared with the control group, the expression of DYNC1H1 in the CA1 region of the hippocampus in AD patients was significantly decreased, which may be due to the facts that the hyperphosphorylation of tau protein in the hippocampus of AD patients destroyed the stability of microtubules and affected the retrograde transport of DYNC1H1 along microtubules. 46 In addition, it has been reported that DYNC1H1 is associated with lower limb dominant spinal muscular atrophy (SMA-LED), 47 cortical dysplasia (MCD), 48 epileptic encephalopathy, 49 Parkinson’s disease, and hereditary spastic paraplegia. 50 Consequently, DYNC1H1 is likely to play an important role in AD and MDD.
MAPRE3 (microtubule-associated protein RP/EB family member 3) binds microtubule plus-ends to promote mature mushroom spines over immature thin spines in cultured neurons. 51 MAPRE3 has been reported to be associated with neuroblastoma. 52 The ADNP-SIRT1 complex consists of ADNP and SIRT1 and one of two major interaction points between ADNP and SIRT1 is at the level of EB1 (MAPRE1)/EB3 (MAPRE3)/Tau (MAPT) binding, shown to be significant for microtubule dynamics, axonal transport, synapse formation, and protection against tauopathy. The ADNP-SIRT1 complex expression is significantly reduced in brain of AD patients. So, MAPRE3 may participate in the occurrence of AD through the ADNP-SIRT1 complex. 53 Although MAPRE3 has not been reported in MDD, the expression value of MAPRE3 changed most obviously after the treatment of MDD patients with venlafaxine, which shows that MAPRE3 may be a crucial factor in the development of AD and MDD.
TTBK2, a kinase known to phosphorylate tau and tubulin, acts as a critical regulator of ciliogenesis. 54 TTBK2 is involved in tau/tubulin phosphorylation 55 and TDP-43 accumulation. 56 A study revealed that TTBK2 was directly or potentially associated with AD and neural development. 57 Although TTBK2 can phosphorylate two AD-related sites of tau, the primary phenotype of the mutant TTBK2 has nothing to do with AD but is associated with severe neurodegenerative disease, autosomal dominant spinocerebellar ataxia type 11 (SCA11). 58 TTBK2 is also associated with cancer progression.59,60 Combined with these previous reports, the results of our research further confirmed the association between the dysregulation of TTBK2 and the development of AD at the gene level. Nevertheless, to explain how this gene functions in MDD or the treatment of MDD, more experimental research is needed.
ITGB1 is a member of the integrin family. ITGB1 is also known as CD29. In mammals, most integrin subunits are expressed in different regions of the brain and are highly expressed in developing neurons. 61 Previous studies have shown that integrins, especially ITGB1 and ITGB3, play a role in nervous system development by regulating processes related to neuronal connections, such as axon growth and guidance, dendritic spine and synaptic formation and maintenance, and synaptic plasticity. 62 ITGB1 mRNA expression was changed in hippocampus and peripheral blood mononuclear cells (PBMCs) of AD patients, and researchers suggested using ITGB1 as an AD biomarker.34,63,64 In hippocampal neurons, ITGB1 and its downstream signaling pathway signals through the nonreceptor tyrosine kinase Arg (also known as Abelson tyrosine-protein kinase 2; ABL2) to regulate dendritic branches, synaptic plasticity, and behavior in the postnatal mouse hippocampus. 65 Furthermore, changes in the genetic code of integrin and other cell adhesion molecules (CAM) or proteins that mediate CAM signal transduction affect neural connections and are closely related to neurodevelopmental disorders, such as schizophrenia 66 and autism.67,68 Stress-associated epigenetic change of ITGB1 was correlated with depression. 69 In addition, ITGB1 70 is also associated with cancer progression.
WASL is also known as neural Wiskott–Aldrich syndrome protein (N-WASP). The expression of N-WASP is highest in nerve tissue. In developing hippocampal neurons, N-WASP is phosphorylated by FAK (focal adhesion kinase) to promote axonal growth 71 and is located in the growth cone by Nck1 and Cdc42 to promote expansion. 72 In addition to regulating axonal growth, N-WASP seems to be involved in the differentiation of neural stem cells by promoting an increase in filamentous foot processes. 73 In addition, N-WASP has been found to affect the myelin growth of Schwann cells, which wrap axons by using a giant lamellipodial sheet-like structure. 74 N-WASP, which regulates the reorganization and rearrangement of actin assembly, is highly expressed in brain tissue from AD patients, and may participate in the neurodegenerative aberrant sprouting in AD neurons. 75 Further research is required to determine the importance of WASL in AD and MDD.
Furthermore, the five hub genes in AD have been previously reported, while their roles in psychiatric disorders have been less reported. The five genes were novel DEGs identified in brain and blood samples from AD and MDD patients. Taken together, we speculate that AD and MDD are related not only in common genes and pathways but also in the function of proteins such as DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL.
Conclusions
In this study, bioinformatics analysis methods were used to screen and verify the shared and potential pathophysiologically related network hub genes in AD and MDD. DYNC1H1, MAPRE3, TTBK2, ITGB1, and WASL were identified as hub genes and they all associated with the progression of AD and MDD. Our findings may provide a reference for further exploration of the treatment of AD and MDD.
Supplemental Material
Supplemental Material, sj-pdf-1-aja-10.1177_15333175211046123 for Identification of Hub Genes Related to Alzheimer’s Disease and Major Depressive Disorder by Yajing Cheng, Meiyue Sun, Feng Wang, Xin Geng and Fei Wang in American Journal of Alzheimer's Disease & Other Dementias
Appendix
List of abbreviations
- AD
Alzheimer’s disease
- ADI
Alzheimer’s Disease International
- CTTN
Cortactin
- DEGs
Differentially expressed genes
- DYNC1H1
Dynein heavy chain 1
- MDD
Major depressive disorder
- MAPRE3
Microtubule Associated Protein RP/EB Family Member 3
- ITGB1
Integrin Subunit Beta 1
- ITSN1
Intersectin 1
- PRKCA
Protein Kinase C Alpha
- REPS2
RALBP1 Associated Eps Domain Containing 2
- TTBK2
Tau Tubulin Kinase 2
- WASL
WASP Like Actin Nucleation Promoting Factor
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by a Chinese National Natural Science Foundation Grant (No. 81671054, No. 81771135, No. 31771520, No. 91649102), the key project of Tianjin Natural Science Grant (19JCZDJC35600), Tianjin Natural Science Grant (19JCJQJC63500), and the Training program for young and middle-aged key Innovative Personnel (303078100407).
Supplemental Material: Supplemental material for this article is available online.
ORCID iD
Yajing Cheng https://orcid.org/0000-0002-9851-1559
References
- 1.Association As.. Alzheimer's disease facts and figures. Alzheimers Dement. 2016;12(4):459-509. 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 2.ADI. World . Alzheimer Report 2019: Attitudes to dementia. London: Alzheimer’s Disease International; 2019. [Google Scholar]
- 3.Patnode CD, Perdue LA, Rossom RC, et al. Rockville (MD): Agency for Healthcare Research and Quality (US). Screening for Cognitive Impairment in Older Adults: An Evidence Update for the U.S. Preventive Services Task Force [Internet]. 2020 Feb. Report No.: 19-05257-EF-1. PMID: 32129963 [PubMed] [Google Scholar]
- 4.Mullard A. FDA approval for Biogen's aducanumab sparks Alzheimer disease firestorm. Nat Rev Drug Discov. 2021. .1038/d41573-021-00099-3. [DOI] [PubMed] [Google Scholar]
- 5.Disease GBD. Injury i, prevalence c. global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease Study 2017. Lancet. Nov 10 2018;392(10159):1789-1858. 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisholm D. Investing in mental health. E Mediterr Health J. 2015;21(7):531-534. [PubMed] [Google Scholar]
- 7.Malhi GS, Mann JJ. Depression. Lancet. Nov. 2018;24(10161):2299-2312. 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- 8.Kendler KS, Gatz M, Gardner CO, Pedersen NL. A swedish national twin study of lifetime major depression. Am J Psychiatry. 2006;163(1):109-114. 10.1176/appi.ajp.163.1.109. [DOI] [PubMed] [Google Scholar]
- 9.Galts CPC, Bettio LEB, Jewett DC, et al. Depression in neurodegenerative diseases: Common mechanisms and current treatment options. Neurosci Biobehav Rev. Jul. 2019;102:56-84. doi: 10.1016/j.neubiorev.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Jahangir S, Adjepong D, Al-Shami HA, Malik BH. Is there an association between migraine and major depressive disorder? A narrative review. Cureus. 2020;12(6):e8551. 10.7759/cureus.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mugge L, Mansour TR, Crippen M, Alam Y, Schroeder J. Depression and glioblastoma, complicated concomitant diseases: a systemic review of published literature. Neurosurg Rev. Apr. 2020;43(2):497-511. 10.1007/s10143-018-1017-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhao QF, Tan L, Wang HF, et al. The prevalence of neuropsychiatric symptoms in Alzheimer's disease: systematic review and meta-analysis. J Affect Disord. 2016;190:264-271. 10.1016/j.jad.2015.09.069. [DOI] [PubMed] [Google Scholar]
- 13.Petersen JD, Waldorff FB, Siersma VD, Phung TKT, Bebe A, Waldemar G. Major depressive symptoms Increase 3-Year mortality rate in patients with mild dementia. Int J Alzheimer's Dis. 2017;2017:7482094. 10.1155/2017/7482094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davydow DS, Zivin K, Katon WJ, et al. Neuropsychiatric disorders and potentially preventable hospitalizations in a prospective cohort study of older Americans. J Gen Intern Med. Oct. 2014;29(10):1362-1371. 10.1007/s11606-014-2916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lara E, Haro JM, Tang MX, Manly J, Stern Y. Exploring the excess mortality due to depressive symptoms in a community-based sample: the role of Alzheimer's disease. J Affect Disord. 2016;202:163-170. 10.1016/j.jad.2016.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reus GZ, Titus SE, Abelaira HM, et al. Neurochemical correlation between major depressive disorder and neurodegenerative diseases. Life Sci. 2016;158:121-129. 10.1016/j.lfs.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Green RC, Cupples LA, Kurz A, et al. Depression as a risk factor for Alzheimer disease - The MIRAGE study. Arch Neurol-Chicago. 2003;60(5):753-759. 10.1001/archneur.60.5.753. [DOI] [PubMed] [Google Scholar]
- 18.Vermeulen T, Lauwers T, Van Diermen L, Sabbe BG, van der Mast RC, Giltay EJ. Cognitive deficits in older adults With psychotic depression: a meta-analysis. Am J Geriatr Psychiatry. 2019;27(12):1334-1344. 10.1016/j.jagp.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Steinberg M, Shao H, Zandi P, et al. Point and 5-year period prevalence of neuropsychiatric symptoms in dementia: the Cache County Study. Int J Geriatr Psychiatry. 2008;23(2):170-177. 10.1002/gps.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowakowska E, Kus K, Chodera A. Comparison of behavioural effects of venlafaxine and imipramine in rats. Arzneimittelforschung. 2003;53(4):237-242. 10.1055/s-0031-1297102. [DOI] [PubMed] [Google Scholar]
- 21.Peng WF, Bai F, Shao K, Shen LS, Li HH, Huang S. The key genes underlying pathophysiology association between the type 2-diabetic and colorectal cancer. J Cell Physiol. 2018;233(11):8551-8557. 10.1002/jcp.26440. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MX, Jin LT, Wang TJ, et al. Identification of potential core genes in triple negative breast cancer using bioinformatics analysis. Onco Targets Ther. 2018;11:4105-4112. 10.2147/OTT.S166567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SM, Wang XJ, Huang SH, et al. Construction of artificial neural network model for predicting the efficacy of first-line FOLFOX chemotherapy for metastatic colorectal cancer. Zhonghua Zhong Liu Za Zhi. 2021;43(2):202-206. 10.3760/cma.j.cn112152-20200419-00355. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y, Ding X, She Z, et al. Exploring shared pathogenesis of Alzheimer's disease and type 2 diabetes mellitus via co-expression networks analysis. Curr Alzheimer Res. 2020;17(6):566-575. 10.2174/1567205017666200810164932. [DOI] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Wang Y, Kong F, et al. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J Cell Physiol. 2020;235(3):3056-3068. 10.1002/jcp.29210. [DOI] [PubMed] [Google Scholar]
- 28.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan H, Zhang C, Zhang T, He Y, Su Y, Zhou L. Identification of key prognostic biomarker and Its correlation with immune infiltrates in Pancreatic Ductal Adenocarcinoma. Dis Markers. 2020;2020:8825997. 10.1155/2020/8825997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devarbhavi P, Telang L, Vastrad B, Tengli A, Vastrad C, Kotturshetti I. Identification of key pathways and genes in polycystic ovary syndrome via integrated bioinformatics analysis and prediction of small therapeutic molecules. Reprod Biol Endocrinol. 2021;19(1):31. 10.1186/s12958-021-00706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otte C, Gold SM, Penninx BW, et al. Major depressive disorder. Nat Rev Dis Primers. 2016;2:16065. 10.1038/nrdp.2016.65. [DOI] [PubMed] [Google Scholar]
- 32.Yatawara C, Lim L, Chander R, Zhou J, Kandiah N. Depressive symptoms influence global cognitive impairment indirectly by reducing memory and executive function in patients with mild cognitive impairment. J Neurol Neurosurg Psychiatry. 2016;87(12):1375-1383. 10.1136/jnnp-2016-314191. [DOI] [PubMed] [Google Scholar]
- 33.Kaup AR, Byers AL, Falvey C, et al. Trajectories of depressive symptoms in older adults and risk of dementia. JAMA Psychiatry. 2016;73(5):525-531. 10.1001/jamapsychiatry.2016.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma G, Liu M, Du K, et al. Differential expression of mRNAs in the brain tissues of patients with alzheimer's disease based on GEO expression profile and its clinical significance. BioMed Res Int. 2019;2019:8179145. 10.1155/2019/8179145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daviglus ML, Bell CC, Berrettini W, et al. National institutes of health state-of-the-science conference statement: preventing alzheimer disease and cognitive decline. Ann Intern Med. 2010;153(3):176-181. 10.7326/0003-4819-153-3-201008030-00260. [DOI] [PubMed] [Google Scholar]
- 36.Deibel SH, Young B, Mohajerani MH, McDonald RJ. Activity rhythms are largely intact in APPNL-G-F alzheimer's disease mice. J Alzheimers Dis. 2019;71(1):213-225. 10.3233/JAD-190102. [DOI] [PubMed] [Google Scholar]
- 37.Spulber S, Conti M, Elberling F, et al. Desipramine restores the alterations in circadian entrainment induced by prenatal exposure to glucocorticoids. Transl Psychiatry. 2019;9(1):263. 10.1038/s41398-019-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarrouilhe D, Mesnil M, Dejean C. Targeting gap junctions: new insights into the treatment of major depressive disorder. Curr Med Chem. 2019;26(20):3775-3791. 10.2174/0929867325666180327103530. [DOI] [PubMed] [Google Scholar]
- 39.He JT, Li XY, Yang L, Zhao X. Astroglial connexins and cognition: memory formation or deterioration?. Biosci Rep. 2020;40(1):1-10. 10.1042/BSR20193510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tublin JM, Adelstein JM, Del Monte F, Combs CK, Wold LE. Getting to the heart of alzheimer disease. Circ Res. 2019;124(1):142-149. 10.1161/CIRCRESAHA.118.313563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irwin M, Tare M, Singh A, et al. A positive feedback loop of hippo- and c-jun-amino-terminal kinase signaling pathways regulates amyloid-beta-mediated neurodegeneration. Front Cell Dev Biol. 2020;8:117. 10.3389/fcell.2020.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bader JR, Vaughan KT. Dynein at the kinetochore: timing, interactions and functions. Semin Cell Dev Biol. 2010;21(3):269-275. 10.1016/j.semcdb.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19(7):347-355. 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riera J, Lazo PS. The mammalian NudC-like genes: a family with functions other than regulating nuclear distribution. Cell Mol Life Sci. 2009;66(14):2383-2390. 10.1007/s00018-009-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiavo G, Greensmith L, Hafezparast M, Fisher EMC. Cytoplasmic dynein heavy chain: the servant of many masters. Trends Neurosci. 2013;36(11):641-651. 10.1016/j.tins.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrotter A, Oberhaus A, Kolbe K, et al. LMD proteomics provides evidence for hippocampus field-specific motor protein abundance changes with relevance to alzheimer's disease. Biochim Biophys Acta Proteins Proteom. 2017;1865(6):703-714. 10.1016/j.bbapap.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Scoto M, Rossor AM, Harms MB, et al. Novel mutations expand the clinical spectrum of DYNC1H1-associated spinal muscular atrophy. Neurology. 2015;84(7):668-679. 10.1212/WNL.0000000000001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Donato N, Timms AE, Aldinger KA, et al. Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet Med. 2018;20(11):1354-1364. 10.1038/gim.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Z, Liu Z, Li X, et al. Whole-exome sequencing identifies a novel de novo mutation in DYNC1H1 in epileptic encephalopathies. Sci Rep. 2017;7(1):258. 10.1038/s41598-017-00208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tey S, Ahmad-Annuar A, Drew AP, Shahrizaila N, Nicholson GA, Kennerson ML. Mutation analysis of genes within the dynactin complex in a cohort of hereditary peripheral neuropathies. Clin Genet. 2016;90(2):127-133. 10.1111/cge.12712. [DOI] [PubMed] [Google Scholar]
- 51.Jaworski J, Kapitein LC, Gouveia SM, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61(1):85-100. 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Tong CW, Wang JL, Jiang MS, Hsu CH, Chang WT, Huang AM. Novel genes that mediate nuclear respiratory factor 1-regualted neurite outgrowth in neuroblastoma IMR-32 cells. Gene. 2013;515(1):62-70. 10.1016/j.gene.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 53.Hadar A, Kapitansky O, Ganaiem M, et al. Introducing ADNP and SIRT1 as new partners regulating microtubules and histone methylation. Mol Psychiatry. 2021. 10.1038/s41380-021-01143-9. [DOI] [PubMed] [Google Scholar]
- 54.Goetz SC, Liem KF, Jr., Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151(4):847-858. 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ikezu S, Ikezu T. Tau-tubulin kinase. Front Mol Neurosci. 2014;7:33. 10.3389/fnmol.2014.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liachko NF, McMillan PJ, Strovas TJ, et al. The tau tubulin kinases TTBK1/2 promote accumulation of pathological TDP-43. PLoS Genet. 2014;10(12):e1004803. 10.1371/journal.pgen.1004803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Godini R, Pocock R, Fallahi H. Caenorhabditis elegans hub genes that respond to amyloid beta are homologs of genes involved in human alzheimer's disease. PloS One. 2019;14(7):e0219486. doi: 10.1371/journal.pone.0219486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Houlden H, Johnson J, Gardner-Thorpe C, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39(12):1434-1436. 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]
- 59.Ha ES, Choi S, In KH, et al. Identification of proteins expressed differently among surgically resected stage I lung adenocarcinomas. Clin Biochem. 2013;46(4-5):369-377. 0.1016/j.clinbiochem.2012.11.014 [DOI] [PubMed] [Google Scholar]
- 60.Bender C, Ullrich A. PRKX, TTBK2 and RSK4 expression causes Sunitinib resistance in kidney carcinoma- and melanoma-cell lines. Int J Cancer. 2012;131(2):E45-E55. 10.1002/ijc.26486. [DOI] [PubMed] [Google Scholar]
- 61.Pinkstaff JK, Detterich J, Lynch G, Gall C. Integrin subunit gene expression is regionally differentiated in adult brain. J Neurosci. 1999;19(5):1541-1556. 10.1523/JNEUROSCI.19-05-01541.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park YK, Goda Y. Integrins in synapse regulation. Nat Rev Neurosci. 2016;17(12):745-756. 10.1038/nrn.2016.138. [DOI] [PubMed] [Google Scholar]
- 63.Sullivan SE, Liao M, Smith RV, et al. Candidate-based screening via gene modulation in human neurons and astrocytes implicates FERMT2 in Abeta and TAU proteostasis. Hum Mol Genet. 2019;28(5):718-735. 10.1093/hmg/ddy376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z, Xiong Z, Manor LC, Cao H, Li T. Integrative computational evaluation of genetic markers for Alzheimer's disease. Saudi J Biol Sci. 2018;25(5):996-1002. 10.1016/j.sjbs.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warren MS, Bradley WD, Gourley SL, et al. Integrin beta1 signals through Arg to regulate postnatal dendritic arborization, synapse density, and behavior. J Neurosci. 2012;32(8):2824-2834. 10.1523/JNEUROSCI.3942-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Dushlaine C, Kenny E, Heron E, et al. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16(3):286-292. 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- 67.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898-907. 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368-372. 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park C, Rosenblat JD, Brietzke E, et al. Stress, epigenetics and depression: a systematic review. Neurosci Biobehav Rev. 2019;102:139-152. 10.1016/j.neubiorev.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 70.Sun Q, Zhou C, Ma R, et al. Prognostic value of increased integrin-beta 1 expression in solid cancers: a meta-analysis. Onco Targets Ther. 2018;11:1787-1799. 10.2147/OTT.S155279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chacon MR, Navarro AI, Cuesto G, et al. Focal adhesion kinase regulates actin nucleation and neuronal filopodia formation during axonal growth. Development. 2012;139(17):3200-3210. 10.1242/dev.080564. [DOI] [PubMed] [Google Scholar]
- 72.Shekarabi M, Moore SW, Tritsch NX, Morris SJ, Bouchard JF, Kennedy TE. Deleted in colorectal cancer binding netrin-1 mediates cell substrate adhesion and recruits Cdc42, Rac1, Pak1, and N-WASP into an intracellular signaling complex that promotes growth cone expansion. J Neurosci. 2005;25(12):3132-3141. 10.1523/JNEUROSCI.1920-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liebau S, Steinestel J, Linta L, et al. An SK3 channel/nWASP/Abi-1 complex is involved in early neurogenesis. PloS One. 2011;6(3):e18148. 10.1371/journal.pone.0018148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Novak N, Bar V, Sabanay H, et al. N-WASP is required for membrane wrapping and myelination by Schwann cells. J Cell Biol. 2011;192(2):243-250. 10.1083/jcb.201010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kitamura Y, Tsuchiya D, Takata K, et al. Possible involvement of wiskott-aldrich syndrome protein family in aberrant neuronal sprouting in alzheimer's disease. Neurosci Lett. Aug 7 2003;346(3):149-152. 10.1016/s0304-3940(03)00506-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-pdf-1-aja-10.1177_15333175211046123 for Identification of Hub Genes Related to Alzheimer’s Disease and Major Depressive Disorder by Yajing Cheng, Meiyue Sun, Feng Wang, Xin Geng and Fei Wang in American Journal of Alzheimer's Disease & Other Dementias