Abstract
Polyamines, such as putrescine, spermidine and spermine, have indirectly been linked with the regulation of gene expression, and their concentrations are typically increased in cancer cells. Although effects on transcription factor binding to cognate DNA targets have been demonstrated, the mechanisms of the biological action of polyamines is poorly understood. Employing uranyl photo-probing we now demonstrate that polyamines at submillimolar concentrations bind preferentially to bent adenine tracts in double-stranded DNA. These results provide the first clear evidence for the sequence-specific binding of polyamines to DNA, and thereby suggest a mechanism by which the cellular effects of polyamines in terms of differential gene transcriptional activity could, at least partly, be a direct consequence of sequence-specific interactions of polyamines with promoters at the DNA sequence level.
INTRODUCTION
The naturally occurring polyamines, such as putrescine, spermidine and spermine, are ubiquitous cellular cations that play multifunctional roles in cell growth and differentiation (1,2). The polyamine concentration is higher in cancer cells when compared with normal cells and, hence, linked with the initiation and progression of cancer. Furthermore, the concentration of polyamines has been demonstrated to be critical for the cell deciding between cell progression and apoptosis (3,4).
The level of polyamines in the cell directly affects the range of genes expressed in response to both growth stimulatory and growth inhibitory agents (5–8). It has been demonstrated that polyamines affect gene expression at the transcriptional level and this effect is most probably due to the direct interaction of polyamine with DNA and/or transacting protein factors (5,9). For instance, it has been shown that polyamines modulate sequence-specific protein–DNA interactions involved in transcription and also enhance the binding of some gene regulatory proteins to their DNA targets, while binding of other proteins is inhibited (10–13). Specifically, it was shown that polyamines facilitate the binding of estrogen receptor α(ERα) and nuclear factor κB (NF-κB) proteins, the two transcription factors implicated in breast cancer cell proliferation and cell survival, to the estrogen response element and to the NF-κB response element, respectively. The specific effect exerted by the polyamines was demonstrated by the use of polyamine analogues, which inhibited the transcription of NF-κB and ERα responsive genes and initiated the onset of apoptosis (12,14).
Several studies have concluded that polyamines interact with DNA in a non-sequence-specific manner (15–17). Furthermore, polyamines stabilize both duplex and triplex B-DNA structures (18), as well as A-form DNA and also promote B- to Z-DNA transitions of certain sequences (19,20). The close proximity of interstrand phosphates in Z-DNA has been suggested as the reason for strong binding to Z-DNA (21–24). Finally, polyamines may have an important role in the formation of nucleosome structures by interacting with and condensing DNA (15,25).
Although polyamines cause DNA condensation in general, the molecular details of polyamine interactions with DNA are poorly understood. In particular, NMR and X-ray crystallographic studies have so far not demonstrated sequence-specific interactions between polyamines and DNA (26). Theoretical studies have suggested spermine binding in the major groove of GC-rich regions and in the minor groove of AT-rich regions, thereby replacing water in the grooves (27–30). Indeed, several lines of evidence suggest that the sequence and the structure of the DNA play a significant role for the polyamine–DNA interactions (15,31). Especially, it has been demonstrated that the binding of polyamines causes macroscopic curvature and DNA bending (32–35). Thus, the action of the polyamine could bring sequence-specific transcription factors and basal transcription factors in close proximity through DNA structure modifications.
Uranyl photo-probing can be employed for analysing protein–DNA interactions and drug–DNA interactions (36,37), for the detection of metal-ion-binding sites in nucleic acids (38,39), as well as for probing the structure and conformation of double-stranded DNA, including A-tract-induced DNA bending (40–43). This makes uranyl photo-probing more versatile in comparison with the other probing techniques. Thus, we decided to study polyamine–DNA interactions by employing uranyl photo-probing and from these studies we conclude that polyamines bind preferentially to bent A-tracts in the double-stranded DNA and may exert sequence-specific modulation of gene expression.
MATERIALS AND METHODS
Plasmids
The plasmid pA1-8 was constructed by cloning a 75 bp fragment containing adenine tracts with 1–8 adenines separated by CGGC in the BamHI site of pUC19. Similarly, pAT1-8 was constructed by cloning a 75 bp fragment containing mixed A–T tracts of 1–8 A/T base pairs in the BamHI site of pUC19. Plasmid pA5N15A5 was constructed by the insertion of a 24 bp fragment in the BamHI site of pUC19. The kinetoplast pCAT was a gift from Paul Englund. Plasmid pKMp27 contains the tyrT promoter (44). The human cytomegalovirus (CMV) promoter is included in the pCMV-Script Vector from Stratagene.
Purification of DNA fragments
An aliquot of 10 μg of each plasmid was cleaved by restriction enzymes (pUC derivatives with EcoRI and PvuII, tyrT with AvaI and EcoRI, kinetoplast with SacI and HindIII, and the CMV promoter with Eco1051 and NheI) using standard techniques and 32P-labelled either at the 3′ end using [α-32P]dATP (Amersham) and the Klenow fragment from DNA polymerase I (Invitrogen) or at the 5′ end using [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Fermentas).
DNA fragments containing the relevant sequences were isolated on the 5% native polyacrylamide gels and eluted from the gel slice by diffusion in 90 mM Tris-borate and 1 mM EDTA, pH 7.5.
Uranyl photocleavage
The purified DNA fragments (∼0.2 pmol DNA) were subject to uranyl photocleavage in a volume of 100 μl containing 50 mM NaAc, pH 6.2 and 1 mM uranyl nitrate and the concentration of polyamines as indicated in the text. The samples were incubated for 20 min at 23°C and then irradiated in open tubes placed just below a 40W/03 Phillips fluorescent light tube with maximum emission at 420 nm. After 20 min of irradiation, NaAc at pH 4.5 was added to a final concentration of 0.2 M together with 2.5 vol of 96% ethanol. The tubes were placed on ice for 15 min and thereafter centrifuged for 15 min. The dried pellet was dissolved in 6 μl formamide, 90 mM Tris-borate and 1 mM EDTA, pH 8.3 containing xylene cyanol and bromophenol blue, and the samples were heated at 90°C before loading onto a 6, 8 or 10% denaturing polyacrylamide gel (19:1 acrylamide/methylenebisacrylamide). The autoradiograms were obtained by overnight exposure using intensifying screens.
DNase I cleavage
The purified DNA fragments (∼0.2 pmol DNA) were subject to DNase I cleavage in a volume of 100 μl containing 50 mM Tris–HCl, pH 7.5 and the concentration of polyamines as indicated in the text. The samples were incubated for 20 min at 23°C before digestion with 2 μl DNase I (0.01 U/ml, dissolved in 1 mM MgCl2, 1 mM MnCl2 and 20 mM NaCl). The digestion was stopped by placing the samples in 96% ethanol with dry ice for 5 min. The samples were then precipitated by adding 96% ethanol and centrifuged at 15 000 g for 20 min. The dried pellet was dissolved in 6 μl formamide, 90 mM Tris-borate and 1mM EDTA, pH 8.3, containing xylene cyanol and bromophenol blue, and the samples were heated at 90°C before loading onto a 6, 8 or 10% denaturing polyacrylamide gel (19:1 acrylamide/methylenebisacrylamide). The autoradiographs were obtained by overnight exposure using intensifying screens.
Ligation ladders
DNA oligonucleotides were purchased from DNA Technology A/S (Denmark). Single-stranded oligonucleotides were phosphorylated with [γ-32P]ATP and T4 polynucleotide kinase (Fermentas) according to the manufacturer's instructions. Then, equal amounts (1 μg) of complementary 32P-labelled oligonucleotides were mixed and heated at 90°C for 2 min and cooled gradually to room temperature (23°C). The annealed oligonucleotides were then assembled into ligation reactions containing 800 U T4 DNA ligase (Fermentas) and incubated at 4°C for 12–14 h. After incubation, the samples were analysed by using PAGE on native gels. The autoradiographs were obtained by exposure for 15–60 min depending on ligation efficiency. Autoradiographs were used to determine the relative mobility of multimers of the decamer A5N5 (5′-CCGAAAAAG-3′) and the migration was compared to a defined standard sequence (5′-CCGTGTCTGG-3′). A linear interpolation from the defined standard was used to determine the ‘apparent size’ of the fragments. The R-value was calculated as the ratio of the ‘apparent size’ and the ‘real size’ of each fragment. A set of restriction enzyme fragments from pUC19 was used in the migration experiments and these consistently yielded R-values very close to 1.0.
Data analysis
Autoradiographs of uranyl cleavage and DNase I digestion patterns were scanned using a Molecular Dynamics computing densitometer. In order to quantify the gels, baseline-corrected scans were analysed by integrating all the densities between two selected boundaries using ImageQuant software version 5.1. The area under the peak was integrated by simple addition of the pixels under the curve. The data were then imported into GraphPad Prism, version 2.01, spreadsheet program.
Cleavage data are presented as autoradiographs or densitometric scans of the autoradiographs or as differential cleavage plots. Differential cleavage plots in which the data are presented in the form ln(fa) − ln(fc) [where(fa) is the fractional cleavage at any bond in the presence of polyamine and (fc) is the fractional cleavage of the same bond in the control] represent the differential cleavage at each bond relative to that in the control. The results are displayed on a logarithmic scale and positive values indicate enhanced cleavage whereas negative values indicate ligand-protected sites. Each band in the gel was assigned to a particular bond within the respective DNA fragment by comparison of its position relative to standard Maxam and Gilbert sequencing standards (A/G reaction).
RESULTS AND DISCUSSION
Strong binding of polyamines in curved A-tracts of a tyrT promoter fragment
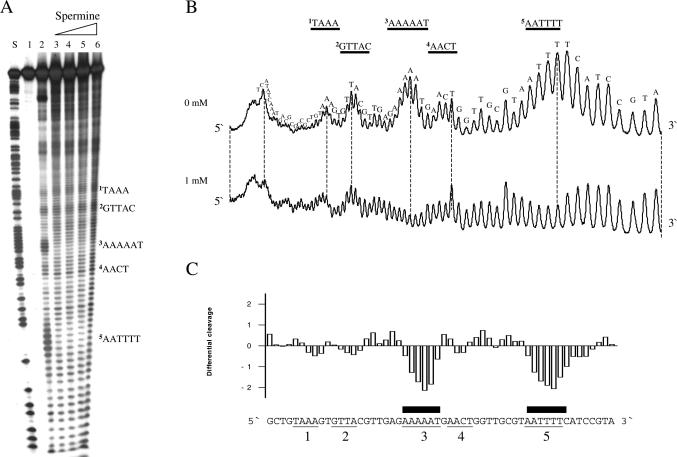
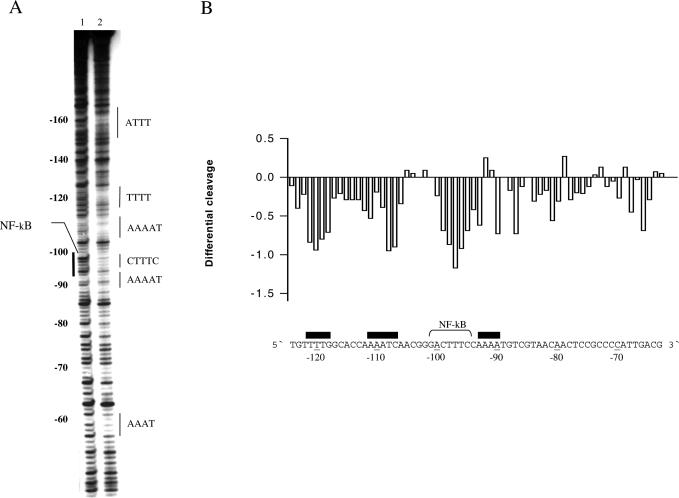
The effects of polyamines were first analysed on a 160 bp DNA fragment containing the Escherichia coli tyrT promoter. This fragment has been thoroughly studied for drug-binding and protein-binding abilities and has furthermore been the subject of analysis by uranyl photocleavage (45). Uranyl cleavage was performed in the presence of putrescine, spermidine or spermine, but since the results were qualitatively similar for the three polyamines only the results for spermine are shown. As established previously, uranyl cleavage when performed at slightly acidic pH is strongest in the specific regions of this DNA fragment (Figure 1, lane 2 and upper scan). It is well known that uranyl cleavage is most effective in DNA sequence regions containing four or more A/T base pairs and this preference has been ascribed to a particular narrow minor groove in such DNA regions [possibly in combination with increased DNA flexibility (42)]. We chose to focus on five uranyl hypersensitive regions: 1, TAAA; 2, GTTAC; 3, AAAAAT; 4, AACT; and 5, AATTTT. Regions 1, 3 and 5 contain four or more consecutive A/T base pairs suggesting that a narrow minor groove is present in these regions. Although a narrow minor groove may exist in regions 2 and 4, the observed uranyl hyperreactivity most probably reflects a different recognition parameter (such as increased flexibility).
Figure 1.
Effect of spermine on uranyl cleavage of the tyrT promoter from E.coli. (A) Autoradiograph showing uranyl cleavage of the E.coli tyrT promoter in the absence and presence of 1, 2, 4 and 8 mM spermine (lanes 2–6). Lane 1 is the uncleaved free DNA and lane S is an A/G sequence reaction. Boldface letters on the right-hand side of the autoradiograph indicate the position of A-tracts and A/T-rich sequences. (B) Densitometric scans of the autoradiograph in (A) showing uranyl cleavage in the absence and presence of 1 mM spermine. The DNA sequence is annotated on the scan. Regions with A-tracts and AT-rich sequences are indicated and underlined. The orientation of the DNA is indicated with 5′ and 3′. (C) Differential cleavage plot comparing the susceptibility of the tyrT promoter DNA fragment to uranyl cleavage in the absence and in the presence of 1 mM spermine. Negative values correspond to a ligand-protected site, and positive values represent enhanced cleavage. The vertical scale is in units of ln(fa) − ln(fc), where fa is the fractional cleavage at any bond in the presence of polyamine and fc is the fractional cleavage of the same band in the control. Black boxes indicate the position of inhibition of uranyl cleavage in the presence of spermine. Regions with A-tracts and AT-rich sequences are indicated and underlined as in (A and B).
Interestingly, spermine only affects uranyl cleavage in specific DNA regions (lanes 3–6 in Figure 1A and the corresponding scan in Figure 1B and the differential cleavage plot in Figure 1C). The strongly cleaved regions 3 and 5 are highly affected, whereas regions 1, 2 and 4 are either weakly or not affected by polyamines. Interestingly, both regions 3 and 5 contain at least four consecutive adenines (or thymines), indicative of a possible bent DNA conformation. The corresponding differential cleavage plot in Figure 1C show the exact location of the reduced cleavage (‘footprints’, negative values) as well as regions with enhanced cleavage (positive values). Therefore, this result could imply that polyamines have preference for binding to curved A-tract DNA. We, therefore, decided to study such DNA in more detail.
Polyamines bind specifically in curved A-tract DNA
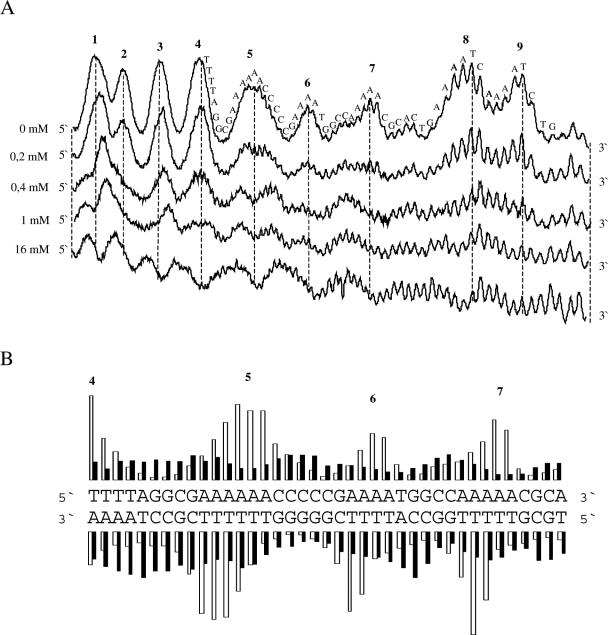
The phenomenon of intrinsic DNA curvature has mainly been developed on the basis of analysis of the kinetoplast DNA of the trypanosomatids (46–49). Therefore, we chose to use the kinetoplast DNA to further analyse whether polyamines preferentially bind to or distort the structure of curved A-tracts as was suggested by the result of the tyrT fragment. As expected in the absence of polyamine, the uranyl cleavage intensity in each A-tract of this fragment is increasing from the 5′ end towards the 3′ end (40,42) (Figure 2A). However, in the presence of spermidine the strongly cleaved positions in the 3′ end of each A-tract are dramatically reduced. An analysis of both strands shows that uranyl cleavage hyperreactivity is still staggered ∼2–3 bp towards the 3′ end between the two strands, indicating that uranyl cleavage occurs via the minor groove (40,42,43) (Figure 2B). These results suggest that spermidine binds to or distorts the structure in the A-tract regions (where uranyl cleavage is strongest in the absence of spermidine). A differential cleavage plot confirms this conclusion since the reduced uranyl cleavage is located exactly at the pure A-tracts (Figure 2C). Although most of the adenine tracts are in phase, tracts 8 and 9 are not in phase with the helical pitch. Therefore, the observed effect is hardly a result of DNA condensation.
Figure 2.
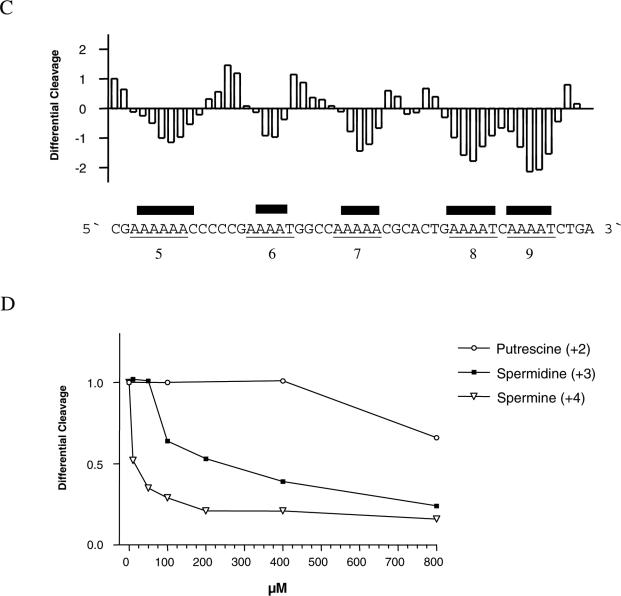
Effect of polyamine on uranyl cleavage of kinetoplast DNA. (A) Selected densitometric scans of a titration experiment showing the uranyl cleavage in the absence and presence of 0.2, 0.4, 1.0 and 16 mM spermidine. The DNA sequence is annotated on the scan. Numbers on the top of the scan represent cleavage maxima also shown in (B and C). (B) Uranyl cleavage of both strands of kinetoplast DNA in the absence (white bars) and presence (black bars) of 1 mM spermine. The cleavage intensities were measured by densitometric scanning and are plotted as a function of sequence. (C) Differential cleavage plot comparing the susceptibility of the kinetoplast DNA fragment to uranyl cleavage in the absence and presence of 1 mM spermidine. Black boxes indicate the position of inhibition of uranyl cleavage in the presence of spermidine. Regions with A-tracts are indicated and underlined. (D) Comparison of the effect of putrescine, spermidine and spermine on uranyl cleavage. The effect of polyamine on uranyl cleavage of A-tract number 5 in (A) has been analysed. Uranyl cleavage efficiency without polyamine has been set to 1.0.
The effect of two other polyamines (putrescine and spermine with different charges) on uranyl cleavage of the kinetoplast fragment was also studied (Figure 2D). Qualitatively, the effect is similar for all three polyamines, but the effect is dependent on the charge of the polyamine. Spermine with four charges has the strongest effect on uranyl cleavage, whereas putrescine has the weakest effect. We found that the concentration of the different polyamines needed to reduce the uranyl cleavage to 50% was as follows: spermine = ∼10 μM; spermidine = ∼250 μM and putrescine = >1000 μM).
The results of tyrT and the kinetoplast DNA indicate that polyamines have differential effects on pure A-tracts (as found in kinetoplast DNA) as compared with the mixed TA sequences. However, we have previously shown that uranyl photocleavage at mixed TA sequences (TA-tracts) shows significant resemblance to the cleavage of pure A-tracts (41,42). Therefore, we chose to analyse systematically the effect of polyamines on A-tracts, and TA-tracts containing different numbers of A/T base pairs. This was performed using DNA fragments from two plasmids, one containing five pure A-tracts (pA1-8) and the other containing five TA-tracts (pAT1-8). The intervening sequences between the A and the TA-tracts are identical in the two fragments, which make a direct comparison between the two types of sequences possible.
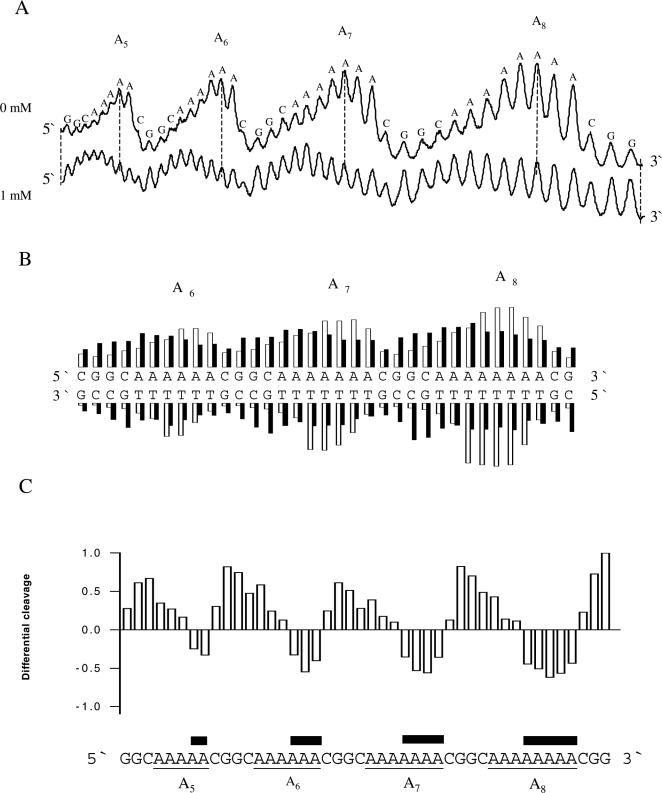
Not surprisingly, the cleavage pattern of the A-tracts in pA1-8 is changed in the presence of 1 mM spermidine (Figure 3A). As in the A-tracts of the kinetoplast fragment, the cleavage maxima are shifted approximately half a helical turn towards the 5′ end. Uranyl cleavage of both strands of the A-tracts of pA1-8 containing the A6, A7 and A8 sequences was analysed (Figure 3B). The cleavage intensities in the absence of polyamine are shown in the histogram as white bars for three pure A-tracts (A6, A7 and A8). The analysis shows that the most strongly cleaved positions are staggered between the two strands ∼2 nt towards the 3′ end. However, the cleavage pattern completely changes in the presence of spermidine (black bars). The strong uranyl cleavage in the 3′ end of the A-tracts has disappeared and instead cleavage is stronger at the 5′ end of the A-tracts, in complete accordance with the results obtained with the kinetoplast DNA. The corresponding differential cleavage plot in Figure 3C elegantly emphasizes this conclusion.
Figure 3.
Effect of spermidine on uranyl cleavage of plasmid pA1-8. (A) Densitometric scans of an autoradiograph presented in Figure 1A in the absence and presence of 1 mM spermidine. Numbers of adenines in the different A-tracts and the DNA sequence is annotated on the scan. (B) Uranyl cleavage of both strands of plasmid pA1-8 in the absence (white bars) and presence (black bars) of 1 mM spermidine. The cleavage intensities were measured with a densitometric scanning. (C) Differential cleavage plot comparing the susceptibility of the plasmid pA1-8 DNA fragment to uranyl cleavage in the absence and presence of 1 mM spermidine. Black boxes indicate the position of inhibition of uranyl cleavage in the presence of spermidine. Regions with A-tracts are indicated and underlined.
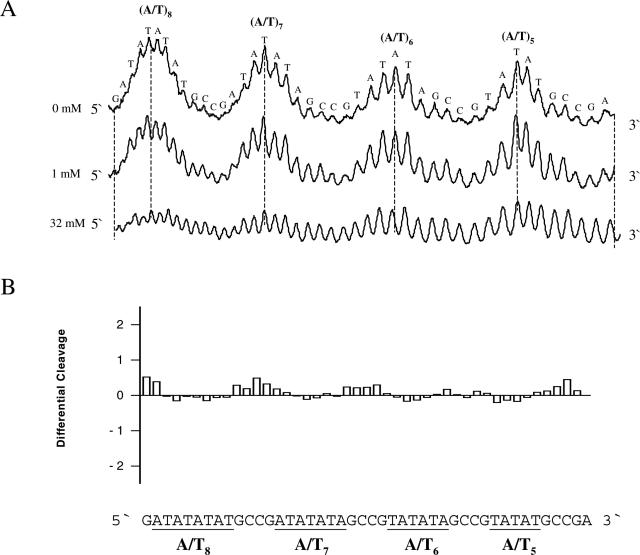
Uranyl cleavage of TA-tracts from pTA1-8 also exhibits the expected, characteristic cleavage in the A/T-rich regions (Figure 4A). However, in contrast to the effect on pure A-tracts, spermine has only modest quantitative (and not qualitative) effect on the uranyl cleavage pattern. A differential cleavage plot for pTA1-8 clearly shows this (Figure 4B).
Figure 4.
Effect of spermine on uranyl cleavage of pT/A1-8. (A) Densitometric scans of an autoradiograph in the absence and presence of 1 and 32 mM spermine. The DNA sequence is annotated on the scan. (B) Differential cleavage plot comparing the susceptibility of plasmid pT/A1-8 DNA fragments to uranyl cleavage in the absence and presence of 1 mM spermine. Regions with AT-rich sequences are indicated and underlined.
This clearly demonstrates a very specific effect of polyamines on curved A-tracts as compared with A/T-rich sequences with TA steps. In support of this conclusion, we find that A-tracts in non-bent DNA (T4A4) binds polyamines very poorly in comparison with A-tracts in bent DNA (A4T4) (see Supplementary Material). Indeed, these results would be compatible with uranyl footprinting of polyamine bound preferentially to bent A-tracts.
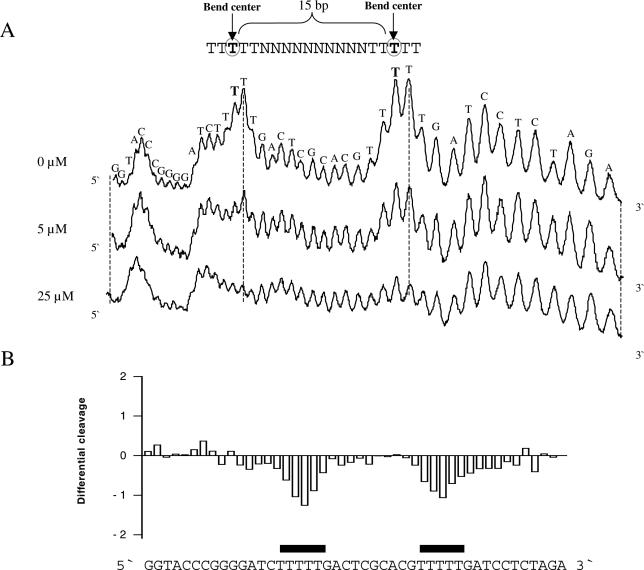
Most of the DNA fragments analysed appear to contain A-tracts spaced ∼10–11 bp from each other, especially in the kinetoplast fragment. Although the effect of polyamine is observed at low concentration (10 μM for the kinetoplast DNA shown in Figure 2D), one might speculate that the observed effect of the polyamines could be caused by DNA condensation (50–52). To exclude this possible interpretation of the results, we constructed a DNA fragment where two A-tracts are separated by 15 bp (plasmid pA5N15A5). Uranyl photo-probing of this DNA shows a clear decrease of cleavage in both A-tracts in the presence of polyamine (Figure 5A and B) and, therefore, the observed effect cannot simply be caused by DNA condensation.
Figure 5.
Effect of polyamine on uranyl cleavage of plasmid pA5N15A5. (A) Selected densitometric scans of a titration experiment showing the uranyl cleavage in the absence and presence of 5, 25 and 500 μM spermine. The DNA sequence is annotated on the scan and the bend centre for each A-tract is indicated. (B) Differential cleavage plot comparing the susceptibility of the DNA fragment to uranyl cleavage in the absence and in the presence of 25 μM spermine. Black boxes indicate the position of the inhibition of uranyl cleavage in the presence of spermine.
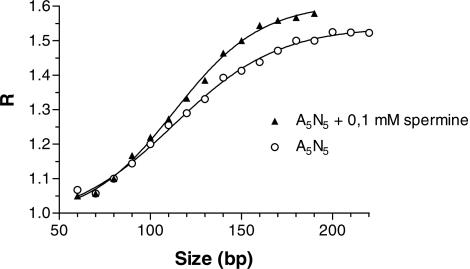
Gel migration studies have shown that polyamines slightly increase the anomalous migration of A-tract containing DNA fragments indicating increased bending (53–55). As the published effect appeared very minor compared with the dramatic effect indicated by the uranyl probing experiments, we decided to confirm this effect. In essence our results are compatible with those of Ussery et al. (55). However, in contrast to these earlier studies, we see a weaker effect of polyamines and it is only observed at 37°C (Figure 6). With 150 bp fragments, we observe a difference between R-values in the absence and presence of 0.1 mM spermine of ∼0.1. Ussery et al. (55) has a difference of ∼0.2 but at 40°C. Thus, the specific interaction of polyamines with A-tracts does not drastically modulate the DNA curvature although the uranyl cleavage is dramatically changed. This would argue that the polyamine is in fact binding to the A-tracts without significantly affecting the DNA structure, and that the uranyl cleavage results, therefore, predominantly represent a footprint of polyamine on the DNA (vide infra).
Figure 6.
Effect of spermine on curvature. Graph showing the R-values for multimers of A5N5 in the absence or presence of 0.1 mM spermine in the gel and running buffer at 37°C plotted as a function of actual length. The sequence of the decamer used in the migration experiment is 5′-CCGCAAAAAG-3′.
Polyamine effect in a eukaryotic promoter
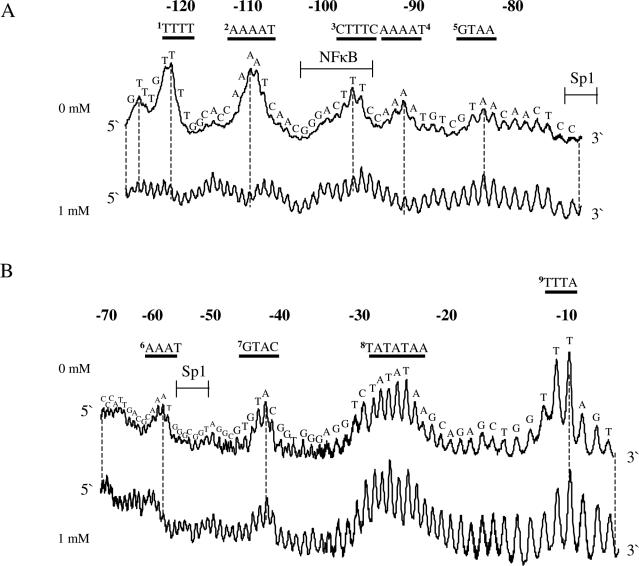
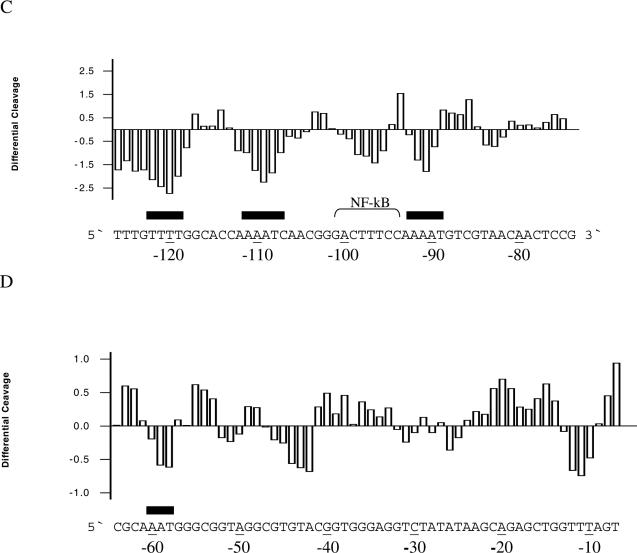
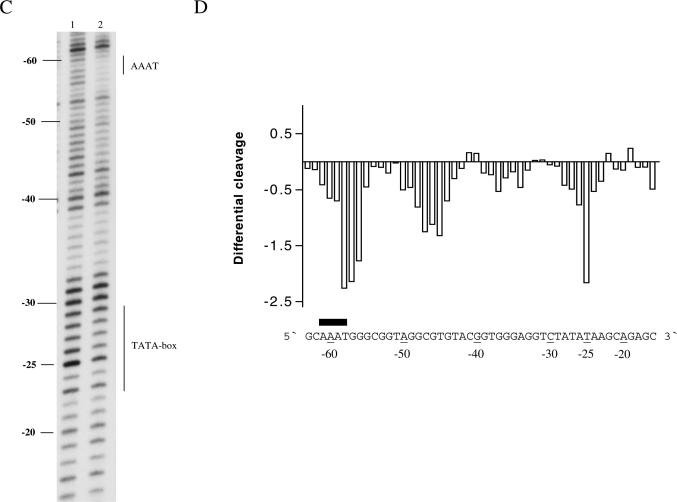
A multitude of reports show that spermine has profound direct or indirect effects on transcription either through interference and/or interaction with transcription factors or through structural changes in the DNA. Therefore, we chose to analyse a DNA fragment from pCMV-Script Vector containing the human CMV promoter in terms of polyamine effects on uranyl cleavage. The results of uranyl photocleavage show that hypersensitivity is mainly observed in TTTA (−10), TATATAA (−25), GTAC (−42), AAAT (−60), GTAA (−85), AAAAT (−92), CTTTC (−97), AAAAT (−110) and TTTT (−122) (the positions indicated in parentheses are relative to the transcription start). As expected, the addition of spermidine significantly modifies the uranyl cleavage pattern in the A-tracts at positions −122, −110, −92 and −60 (Figure 7A–D). However, uranyl cleavage is also modified in regions other than A-tracts. Especially, the cleavage in the CTTTC sequence, which is part of the binding site for the NF-κB protein, is clearly reduced in the presence of spermidine (Figure 7C). Furthermore, it is noted that minor changes take place in the TATA-box and some A/T-rich sequences are protected (−45 and −10 in Figure 7D).
Figure 7.
Uranyl cleavage of human cytomegalovirus: effect of spermidine. (A) Densitometric scans of an autoradiograph showing uranyl cleavage of the human CMV promoter in the absence and presence of 1 mM spermidine. The DNA sequence is annotated on the scans and covers from −80 to −120 bp and (B) same content as in (A), but the densitometric scan covers from −10 to −70 bp upstream from the transcription start site. Binding sites for transcription factor Sp1 and NF-κB are indicated above the sequence in (A and B). (C) Differential cleavage plot comparing the susceptibility of the human CMV promoter to uranyl cleavage in the absence and presence of 1 mM spermidine. The analysis covers from −80 to −120 bp upstream from the transcription start site. Black boxes indicate the position of A-tracts. (D) Same as in (C) but the differential cleavage plot analysis covers from −10 to −60 bp upstream from the transcription start site of the human CMV promoter. Black boxes indicate the position of A-tracts.
In order to further address whether the spermine-induced changes of uranyl photocleavage is due to direct binding or indirect induction of changes in DNA conformation, a DNase I footprinting experiment was performed (Figure 8A–D). This experiment shows that spermine has a clear effect on the DNase I cleavage pattern. The differential cleavage plots show that spermine decrease DNase I reactivity strongly in the curved A-tract regions, in the CTTTC region (NF-κB binding site) and in the region around −45 (Figure 8B and D). In addition the reactivity pattern is changed in the TATA-box (shown at a concentration of 8 mM of spermine, Figure 8D). These results are consistent with specific, strong binding of spermine in bent A-tract regions, and the lack of strong hypersensitive DNase I sites in the presence of spermine indicates that major structural changes in the promoter DNA are not likely. Furthermore, a good correlation between the uranyl and the DNase I probing results is noted (cf. Figure 7C with 8B and 7D with 8D), although the effect of the polyamine in the TATA-box is much stronger in the DNase I cleavage. Nevertheless, the DNase I cleavage results of the CMV promoter support that polyamines bind in A-tracts and, furthermore, demonstrates that the polyamine-induced changes in sequences other than pure A-tracts do take place. Interestingly, both uranyl and DNase I probing show that spermine binds in the NF-κB binding site. Previous experiments have shown that polyamines increase the binding of the NF-κB protein to a recognition site containing an A-tract (12). Therefore, it is tempting to suggest that a mechanism may involve specific polyamine binding in the recognition sequence. Likewise, it is noted that DNase I cleavage is changed in the TATA-box in the presence of spermine. Indirect readout has been shown to be most important for the recognition of the TATA-box by the TATA-binding protein and polyamines may, thus, influence this recognition by modulating the DNA structure in a yet unclear way. However, this observation needs to be further analysed. Moreover, despite the absence of a clear distortion of the DNA structure in A-tracts as analysed by gel migration, other subtle structural changes may still take place. For instance, it has been demonstrated by the use of UV spectroscopy, electric and circular dichroism that A/T- and G/C-rich sequences are differently affected by polyamines and, in particular, that binding in A/T-rich sequences modulates the structure of the DNA helix (53,56).
Figure 8.
Effect of spermine on DNase I cleavage of the human CMV promoter DNA. (A) A typical autoradiograph is shown in the absence and in the presence of 8 mM spermine (lanes 1 and 2, respectively). The autoradiograph covers from −60 to −160 bp upstream from the transcription start site. Boldface numerals on the left-hand side indicate base positions with respect to transcription start site. The position of AT-rich sequences and NF-κB binding site are also shown. Black boxes indicate the position of A-tracts. (B) Differential cleavage plot comparing the susceptibility of the human CMV promoter to DNase I in the absence and presence of 8 mM spermine. The analysis covers from −70 to −120 bp upstream from the transcription start site. Black boxes indicate the position of A-tracts. (C) Same content as in (A) but the autoradiograph covers from −20 to −60 bp upstream from the transcription start site. The TATA-box and one A-tract is indicated. (D) Differential cleavage plot comparing the susceptibility of the human CMV promoter to DNase I in the absence and presence of 8 mM spermine. The analysis covers from −20 to −60 bp upstream from the transcription start site and includes the TATA-box and one A-tract. Black boxes indicate the position of A-tracts.
In conclusion, the present results show that polyamines bind preferentially to bent A-tracts. The polyamines are present in millimolar concentrations in vivo and thus the binding is observed at biologically relevant concentrations of the polyamines (57,58). How (and if) the polyamine binding manifests itself in biological effects at the transcriptional level is still unclear. However, in the CMV promoter two bent A-tracts (both AAAAT sequences, Figure 7C) are prominently present proximal to the binding site of the transcription factor NF-κB, the DNA binding of which is affected by spermine in vitro (12). These two A-tracts are highly affected by the presence of the polyamine. Although the gel migration results do not indicate any major structural changes of the A-tracts upon polyamine binding, one may speculate whether sliding of the protein from non-specific DNA to the specific binding site of a protein, such as NF-κB, is affected by polyamine binding in the nearby A-tracts. This theory could warrant further scrutiny. In addition one should consider effects on a higher level of gene/chromatin organization, such as changes in nuclear matrix attachment points and, consequently, transcriptional domains. Additional work is needed to clarify this, but the present data allow formulation of the new specific hypotheses to be tested.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the Novo Nordisk Foundation and the Lundbeck Foundation. Funding to pay the Open Access publication charges for this article was provided by the Augustinus Foundation and the Danish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pegg A.E., McCann P.P. Polyamine metabolism and function. Am. J. Physiol. 1982;243:212–221. doi: 10.1152/ajpcell.1982.243.5.C212. [DOI] [PubMed] [Google Scholar]
- 2.Thomas T., Thomas T.J. Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol. Life Sci. 2001;58:244–258. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hibshoosh H., Johnson M., Weinstein I.B. Effects of overexpression of ornithine decarboxylase (ODC) on growth control and oncogene-induced cell transformation. Oncogene. 1991;6:739–743. [PubMed] [Google Scholar]
- 4.Bachrach U. Polyamines and neoplastic transformation. In: Nishioka K., editor. Polyamines in Cancer: Basic Mechanisms and Clinial Approaches. Austin, TX: Landes Bioscience Publishers; 1996. pp. 45–74. [Google Scholar]
- 5.Wang Y., Devereux W., Stewart T.M., Casero R.A., Jr Cloning and characterization of human polyamine-modulated factor-1, a transcriptional cofactor that regulates the transcription of the spermidine/spermine N(1)-acetyltransferase gene. J. Biol. Chem. 1999;274:22095–22101. doi: 10.1074/jbc.274.31.22095. [DOI] [PubMed] [Google Scholar]
- 6.Wang J.Y., McCormack S.A., Viar M.J., Wang H., Tzen C.Y., Scott R.E., Johnson L.R. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am. J. Physiol. 1993;265:331–338. doi: 10.1152/ajpgi.1993.265.2.G331. [DOI] [PubMed] [Google Scholar]
- 7.Celano P., Baylin S.B., Giardiello F.M., Nelkin B.D., Casero R.A., Jr Effect of polyamine depletion on c-myc expression in human colon carcinoma cells. J. Biol. Chem. 1988;263:5491–5494. [PubMed] [Google Scholar]
- 8.Celano P., Baylin S.B., Casero R.A., Jr Differential response to treatment with the bis(ethyl)polyamine analogues between human small cell lung carcinoma and undifferentiated large cell lung carcinoma in culture. Cancer Res. 1989;49:639–643. [PubMed] [Google Scholar]
- 9.Wang Y., Devereux W., Stewart T.M., Casero R.A., Jr Polyamine-modulated factor 1 binds to the human homologue of the 7a subunit of the Arabidopsis COP9 signalosome: implications in gene expression. Biochem. J. 2002;366:79–86. doi: 10.1042/BJ20020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panagiotidis C.A., Artandi S., Calame K., Silverstein S.J. Polyamines alter sequence-specific DNA–protein interactions. Nucleic Acids Res. 1995;23:1800–1809. doi: 10.1093/nar/23.10.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas T., Gallo M.A., Klinge C.M., Thomas T.J. Polyamine-mediated conformational perturbations in DNA alter the binding of estrogen receptor to poly(dG-m5dC).poly(dG-m5dC) and a plasmid containing the estrogen response element. J. Steroid Biochem. Mol. Biol. 1995;54:89–99. doi: 10.1016/0960-0760(95)00126-k. [DOI] [PubMed] [Google Scholar]
- 12.Shah N., Thomas T., Shirahata A., Sigal L.H., Thomas T.J. Activation of nuclear factor kappaB by polyamines in breast cancer cells. Biochemistry. 1999;38:14763–14774. doi: 10.1021/bi991291v. [DOI] [PubMed] [Google Scholar]
- 13.Kuramoto N., Inoue K., Gion K., Takano K., Sakata K., Ogita K., Yoneda Y. Modulation of DNA binding of nuclear transcription factors with leucine-zipper motifs by particular endogenous polyamines in murine central and peripheral excitable tissues. Brain Res. 2003;28:170–180. doi: 10.1016/s0006-8993(02)04268-3. [DOI] [PubMed] [Google Scholar]
- 14.Shah N., Thomas T.J., Lewis J.S., Klinge C.M., Shirahata A., Gelinas C., Thomas T. Regulation of estrogenic and nuclear factor kappa B functions by polyamines and their role in polyamine analog-induced apoptosis of breast cancer cells. Oncogene. 2001;29:1715–1729. doi: 10.1038/sj.onc.1204247. [DOI] [PubMed] [Google Scholar]
- 15.Braunlin W.H., Strick T.J., Record M.T., Jr Equilibrium dialysis studies of polyamine binding to DNA. Biopolymers. 1982;21:1301–1314. doi: 10.1002/bip.360210704. [DOI] [PubMed] [Google Scholar]
- 16.Plum G.E., Bloomfield V.A. Effects of spermidine and hexaamminecobalt(III) on thymine imino proton exchange. Biochemistry. 1990;29:5934–5940. doi: 10.1021/bi00477a008. [DOI] [PubMed] [Google Scholar]
- 17.Burton D.R., Forsen S., Reimarsson P. The interaction of polyamines with DNA: a 23Na NMR study. Nucleic Acids Res. 1981;9:1219–1228. doi: 10.1093/nar/9.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musso M., Thomas T., Shirahata A., Sigal L.H., Van Dyke M.W., Thomas T.J. Effects of chain length modification and bis(ethyl) substitution of spermine analogs on purine–purine–pyrimidine triplex DNA stabilization, aggregation, and conformational transitions. Biochemistry. 1997;36:1441–1449. doi: 10.1021/bi9624494. [DOI] [PubMed] [Google Scholar]
- 19.Garriga P., Garcia-Quintana D., Sagi J., Manyosa J. An A-form of poly(amino2dA-dT)·poly(amino2dA-dT) induced by polyamines. Biochemistry. 1993;32:1067–1071. doi: 10.1021/bi00055a011. [DOI] [PubMed] [Google Scholar]
- 20.Thomas T.J., Gunnia U.B., Thomas T. Polyamine-induced B-DNA to Z-DNA conformational transition of a plasmid DNA with (dG-dC)n insert. J. Biol. Chem. 1991;266:6137–6141. [PubMed] [Google Scholar]
- 21.van Dam L., Korolev N., Nordenskiold L. Polyamine–nucleic acid interactions and the effects on structure in oriented DNA fibers. Nucleic Acids Res. 2002;30:419–428. doi: 10.1093/nar/30.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tippin D.B., Sundaralingam M. Nine polymorphic crystal structures of d(CCGGGCCCGG), d(CCGGGCCm5CGG), d(Cm5CGGGCCm5CGG) and d(CCGGGCC(Br)5CGG) in three different conformations: effects of spermine binding and methylation on the bending and condensation of A-DNA. J. Mol. Biol. 1997;267:1171–1185. doi: 10.1006/jmbi.1997.0945. [DOI] [PubMed] [Google Scholar]
- 23.Egli M., Tereshko V., Teplova M., Minasov G., Joachimiak A., Sanishvili R., Weeks C.M., Miller R., Maier M.A., An H., et al. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers. 1998;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.Bancroft D., Williams L.D., Rich A., Egli M. The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry. 1994;33:1073–1086. doi: 10.1021/bi00171a005. [DOI] [PubMed] [Google Scholar]
- 25.Basu H.S., Smirnov I.V., Peng H.F., Tiffany K., Jackson V. Effects of spermine and its cytotoxic analogs on nucleosome formation on topologically stressed DNA in vitro. Eur. J. Biochem. 1997;243:247–258. doi: 10.1111/j.1432-1033.1997.0247a.x. [DOI] [PubMed] [Google Scholar]
- 26.Korolev N., Lyubartsev A.P., Nordenskiold L., Laaksonen A. Spermine: an ‘invisible’ component in the crystals of B-DNA. A grand canonical Monte Carlo and molecular dynamics simulation study. J. Mol. Biol. 2001;308:907–917. doi: 10.1006/jmbi.2001.4642. [DOI] [PubMed] [Google Scholar]
- 27.Haworth I.S., Rodger A., Richards W.G. A molecular mechanics study of spermine complexation to DNA: a new model for spermine-poly(dG-dC) binding. Proc. R. Soc. Lond. B Biol. Sci. 1991;244:107–116. doi: 10.1098/rspb.1991.0058. [DOI] [PubMed] [Google Scholar]
- 28.Feuerstein B.G., Williams L.D., Basu H.S., Marton L.J. Implications and concepts of polyamine–nucleic acid interactions. J. Cell. Biochem. 1991;46:37–47. doi: 10.1002/jcb.240460107. [DOI] [PubMed] [Google Scholar]
- 29.Zakrzewska K., Pullman B. Spermine–nucleic acid interactions: a theoretical study. Biopolymers. 1986;25:375–392. doi: 10.1002/bip.360250302. [DOI] [PubMed] [Google Scholar]
- 30.Korolev N., Lyubartsev A.P., Laaksonen A., Nordenskiold L. On the competition between water, sodium ions, and spermine in binding to DNA: a molecular dynamics computer simulation study. Biophys. J. 2002;82:2860–2875. doi: 10.1016/S0006-3495(02)75628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raspaud E., Olvera de la Cruz M., Sikorav J.L., Livolant F. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys. J. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feuerstein B.G., Pattabiraman N., Marton L.J. Spermine–DNA interactions: a theoretical study. Proc. Natl Acad Sci. USA. 1986;83:5948–5952. doi: 10.1073/pnas.83.16.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feuerstein B.G., Pattabiraman N., Marton L.J. Molecular dynamics of spermine–DNA interactions: sequence specificity and DNA bending for a simple ligand. Nucleic Acids Res. 1989;17:6883–6892. doi: 10.1093/nar/17.17.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu B., Liang X., Scott G.K., Chang C.H., Baldwin M.A., Thomas T., Benz C.C. Polyamine inhibition of estrogen receptor (ER) DNA-binding and ligand-binding functions. Breast Cancer Res. Treat. 1998;48:243–257. doi: 10.1023/a:1005949319064. [DOI] [PubMed] [Google Scholar]
- 35.Thomas T., Balabhadrapathruni S., Gallo M.A., Thomas T.J. Development of polyamine analogs as cancer therapeutic agents. Oncol. Res. 2002;13:123–135. [PubMed] [Google Scholar]
- 36.Møllegaard N.E., Rasmussen P.B., Valentin-Hansen P., Nielsen P.E. Characterization of promoter recognition complexes formed by CRP and CytR for repression and by CRP and RNA polymerase for activation of transcription on the Escherichia coli deoP2 promoter. J. Biol. Chem. 1993;268:17471–17477. [PubMed] [Google Scholar]
- 37.Møllegaard N.E., Nielsen P.E. Applications of uranyl cleavage mapping of RNA structure. Methods Mol. Biol. 1997;90:43–49. doi: 10.1016/s0076-6879(00)18043-7. [DOI] [PubMed] [Google Scholar]
- 38.Møllegaard N.E., Murchie A.I., Lilley D.M., Nielsen P.E. Uranyl photoprobing of a four-way DNA junction: evidence for specific metal ion binding. EMBO J. 1994;13:1508–1513. doi: 10.1002/j.1460-2075.1994.tb06412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassi G.S., Møllegaard N.E., Murchie A.I., von Kitzing E., Lilley D.M. Ionic interactions and the global conformations of the hammerhead ribozyme. Nature Struct. Biol. 1995;2:45–55. doi: 10.1038/nsb0195-45. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen P.E., Møllegaard N.E., Jeppesen C. DNA conformational analysis in solution by uranyl mediated photocleavage. Nucleic Acids Res. 1991;18:3847–3851. doi: 10.1093/nar/18.13.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnichsen S.H., Nielsen P.E. Enhanced uranyl photocleavage across the minor groove of all (A/T)4 sequences indicates a similar narrow minor groove conformation. J. Mol. Recognit. 1996;9:219–227. doi: 10.1002/(sici)1099-1352(199605)9:3<219::aid-jmr242>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 42.Møllegaard N.E., Nielsen P.E. Increased temperature and 2-methyl-2,4-pentanediol change the DNA structure of both curved and uncurved adenine/thymine-rich sequences. Biochemistry. 2003;42:8587–8593. doi: 10.1021/bi0343267. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen P.E., Hiort C., Sonnichsen S.H., Buchardt O., Dahl O., Norden B. DNA binding and photocleavage by uranyl(VI)(UO22) salts. J. Am. Chem. Soc. 1992;114:4967–4975. [Google Scholar]
- 44.Drew H., Travers A.A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984;37:491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- 45.Bailly C., Møllegaard N.E., Nielsen P.E., Waring M.J. The influence of the 2-amino group of guanine on DNA conformation. Uranyl and DNase I probing of inosine/diaminopurine substituted DNA. EMBO J. 1995;14:2121–2131. doi: 10.1002/j.1460-2075.1995.tb07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkhoff A.M., Tullius T.D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 47.Marini J.C., Levene S.D., Crothers D.M., Englund P.T. A bent helix in kinetoplast DNA. Cold Spring Harb. Symp. Quant. Biol. 1983;47:279–283. doi: 10.1101/sqb.1983.047.01.033. [DOI] [PubMed] [Google Scholar]
- 48.Wu H.M., Crothers D.M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 49.Levene S.D., Wu H.M., Crothers D.M. Bending and flexibility of kinetoplast DNA. Biochemistry. 1986;25:3988–3995. doi: 10.1021/bi00362a003. [DOI] [PubMed] [Google Scholar]
- 50.Wilson R.W., Bloomfield V.A. Counterion-induced condensation of deoxyribonucleic acid. A light-scattering study. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 51.Gosule L.C., Schellman J.A. DNA condensation with polyamines I. Spectroscopic studies. J. Mol. Biol. 1978;121:311–326. doi: 10.1016/0022-2836(78)90366-2. [DOI] [PubMed] [Google Scholar]
- 52.Chattoraj D.K., Gosule L.C., Schellman J.A. DNA condensation with polyamines I. Electron microscopic studies. J. Mol. Biol. 1978;121:327–337. doi: 10.1016/0022-2836(78)90367-4. [DOI] [PubMed] [Google Scholar]
- 53.Bordin F., Cacchione S., Savino M., Tufillaro A. Different interactions of spermine with a ‘curved’ and a ‘normal’ DNA duplex: (CA4T4Gn) and (CT4A4G)n. Gel electrophoresis and circular dichroism studies. Biochem. Int. 1992;27:891–901. [PubMed] [Google Scholar]
- 54.Bordin F., Cacchione S., Savino M., Tufillaro A. Different superstructural features of the complexes between spermine and the light responsive elements of the two pea genes rbcS-3A and rbcS-3.6. Gel electrophoresis and circular dichroism studies. Biophys. Chem. 1992;44:99–112. doi: 10.1016/0301-4622(92)85042-3. [DOI] [PubMed] [Google Scholar]
- 55.Ussery D.W., Higgins C.F., Bolshoy A. Environmental influences on DNA curvature. J. Biomol. Struct. Dyn. 1999;16:811–823. doi: 10.1080/07391102.1999.10508294. [DOI] [PubMed] [Google Scholar]
- 56.Marquet R., Houssier C. Different binding modes of spermine to A-T and G-C base pairs modulate the bending and stiffening of the DNA double helix. J. Biomol. Struct. Dyn. 1988;6:235–242. doi: 10.1080/07391102.1988.10507710. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe S., Kusama-Eguchi K., Kobayashi H., Igarashi K. Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver. J. Biol. Chem. 1991;266:20803–20809. [PubMed] [Google Scholar]
- 58.Igarashi K., Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.