Abstract
Although the DNA cleavage mechanism of Type I restriction–modification enzymes has been extensively studied, the mode of cleavage remains elusive. In this work, DNA ends produced by EcoKI, EcoAI and EcoR124I, members of the Type IA, IB and IC families, respectively, have been characterized by cloning and sequencing restriction products from the reactions with a plasmid DNA substrate containing a single recognition site for each enzyme. Here, we show that all three enzymes cut this substrate randomly with no preference for a particular base composition surrounding the cleavage site, producing both 5′- and 3′-overhangs of varying lengths. EcoAI preferentially generated 3′-overhangs of 2–3 nt, whereas EcoKI and EcoR124I displayed some preference for the formation of 5′-overhangs of a length of ∼6–7 and 3–5 nt, respectively. A mutant EcoAI endonuclease assembled from wild-type and nuclease-deficient restriction subunits generated a high proportion of nicked circular DNA, whereas the wild-type enzyme catalyzed efficient cleavage of both DNA strands. We conclude that Type I restriction enzymes require two restriction subunits to introduce DNA double-strand breaks, each providing one catalytic center for phosphodiester bond hydrolysis. Possible models for DNA cleavage are discussed.
INTRODUCTION
Type I restriction–modification (R–M) systems are multifunctional complexes composed of three different subunits, HsdS (S), HsdM (M) and HsdR (R), which are present in a stoichiometry of 1:2:2 (1–3). This heteromeric complex can act as a DNA methyltransferase, a DNA-dependent ATPase, a DNA translocase and a restriction endonuclease. HsdS is responsible for the recognition of a specific DNA sequence. The complex M2S has a methylation function and the addition of HsdR to the methylase complex is essential for the endonuclease activity. Type I R–M systems form several related families of enzymes called IA, IB, IC and more recently ID (4). Each family comprises members with considerable amino acid sequence homology and family members can genetically complement each other. Members of different families show very little amino acid sequence homology and, hence, genetic complementation does not occur (2).
The Type I R–M enzymes require Mg2+, S-adenosylmethionine and ATP hydrolysis for the endonuclease activity (2,3). After binding to a non-modified DNA recognition site, the enzyme complex translocates DNA towards itself from both directions in a reaction coupled to extensive ATP hydrolysis, consistent with the presence of two HsdR subunits. DNA cleavage occurs when this translocation reaction is blocked, for example, upon collision with a second enzyme complex translocating from another recognition site or by an unusual DNA structure (5,6). DNA translocation allows the cleavage to occur several hundreds to thousands of base pairs away from the recognition sequence. Type I restriction enzymes do not display turn-over in the DNA cleavage reaction, but massive ATP hydrolysis and, presumably, DNA translocation activity continues for a long time after the cleavage (7).
On circular DNA containing a single recognition site, cleavage can occur when the enzyme stalls due to bi-directional translocation of the entire DNA circle (6). In contrast, linear DNA molecules containing a single recognition site are refractory to cleavage as they lack a block to DNA translocation. Linear DNA molecules can be cleaved by Type I enzymes only if they contain at least two unmodified recognition sites, with cleavage occurring in the region between the recognition sites (5,8,9).
Despite significant progress in understanding the mechanism of how Type I restriction enzymes select DNA cleavage sites, the mode of cleavage of double-stranded DNA remains elusive. Also, little is known about the nature of the DNA ends produced by these enzymes. Here, we analyze the DNA ends resulting from the cleavage of a circular DNA substrate by EcoKI, EcoAI and EcoR124I, representatives of the Type IA, IB and IC families, respectively. We demonstrate that these enzymes cleave DNA to produce both 5′- and 3′-single-stranded protrusions of varying lengths. To gain deeper insight into the restriction mechanism, an EcoAI HsdR subunit mutated in the nuclease catalytic center was prepared and used to assemble a mutant endonuclease containing both the wild-type and the nuclease-deficient HsdR subunits. We have found that the presence of wild-type and mutant HsdR subunits in the enzyme complex resulted in the formation of a nicked circular DNA, suggesting that two cleavage-proficient HsdR subunits are required to catalyze the breakage of a DNA duplex.
MATERIALS AND METHODS
DNA manipulations
The plasmid pJP25 containing a single recognition site for EcoAI has been described previously (10). The AflIII–HindIII fragment of pJP25 including the EcoAI recognition site was inserted into the corresponding sites in pDRM.2R (11). The resulting plasmid, named pARK, contains a single recognition site each for EcoKI, EcoAI and EcoR124I.
A DNA fragment encoding a chloramphenicol resistance gene (Cm) was amplified by PCR from the pACYC184 plasmid (NEB) using Pfu polymerase (Stratagene), and CmF (5′-aaaaaagccggcttggcgaaaatgagacgttga-3′) and CmR (5′-aaaaaagccggcggcaccaataactgccttaaa-3′) primers (Microsynth).
The linear products from the cleavage reactions of EcoKI, EcoAI and EcoR124I with pARK were purified from agarose gel using a gel extraction kit (Qiagen) and blunt-ended by incubation with T4 DNA polymerase (NEB) in the presence of all four dNTPs. The fragments generated using EcoKI and EcoAI were dephosphorylated by Shrimp alkaline phosphatase (Promega), while EcoR124I products were phosphorylated by T4 polynucleotide kinase (NEB). The resulting DNA fragments were ligated with the chloramphenicol resistance marker (see above) and cloned in ElecroTen-Blue cells (Stratagene). Plasmid DNA was isolated from the individual clones, and the regions around the pARK/Cm junctions were sequenced using an ABI PRISM™ 310 sequence analyzer (Perkin-Elmer) and primers originating in the Cm gene (Seq1, 5′-attgggatatatcaacggtgg-3′; and Seq2, 5′-aaatattataggcaaggcgac-3′). The resulting sequences were aligned with the sequence of pARK using AutoAssembler 2.0.1 software (Perkin-Elmer).
Protein purifications
EcoKI endonuclease was overexpressed from the plasmid pVMC3 and purified as described previously (12). The EcoAI methylase (Mtase), the HsdR subunit and its HsdRD61A mutant were produced from the plasmids pJP21, pJP22 and pJP41D61A, respectively, and purified as described previously (10,13). The Mtase and HsdR subunit of EcoR124I were produced from the plasmids pJS4M and pTrcR124, respectively, and purified as described elsewhere (11,14).
DNA cleavage assays
Type I DNA cleavage reactions of EcoKI, EcoAI and EcoR124I were performed under the conditions described previously (10–12). Reaction products were analyzed on the agarose gels run in 0.5× TBE buffer. DNA was visualized by ethidium bromide staining.
For experiments with the mutant EcoAI endonuclease, wild-type HsdR (HsdRwt) and its D61A mutant (HsdRD61A) were mixed in the following molar ratios: 1:0, 0.9:0.1, 0.8:0.2, 0.7:0.3, 0.6:0.4, 0.5:0.5, 0.4:0.6, 0.3:0.7, 0.2:0.8, 0.1:0.9 and 0:1. EcoAI nuclease was reconstituted by mixing the Mtase complex (M2S1) with the appropriate HsdR mixture at a molar ratio of 1:6. Restriction reactions were carried out under the conditions described previously for the wild-type EcoAI enzyme (10).
RESULTS
Cloning and sequencing of the EcoKI, EcoAI and EcoR124I restriction products
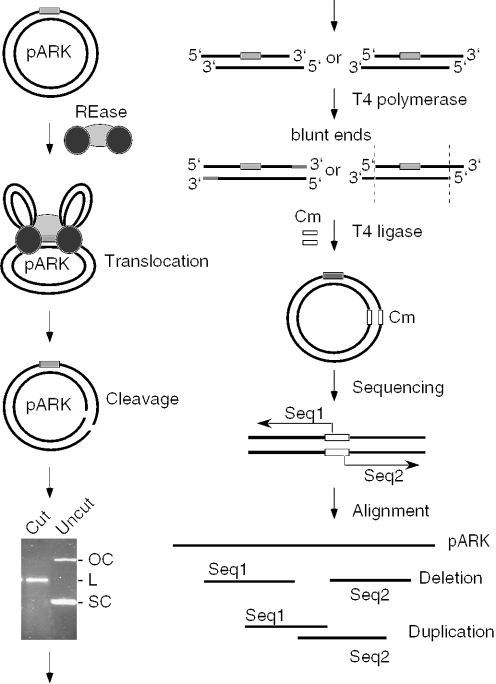
In order to characterize the structure of DNA ends created by Type I R–M enzymes, we sought to clone and sequence the cleavage products from the reactions with plasmid DNA containing a single recognition site, which is cleaved by Type I enzymes only once at random positions throughout the DNA circle. For each of the three major Type I families we have chosen one representative member to be studied: EcoKI for Type IA, EcoAI for Type IB and EcoR124I for Type IC. The supercoiled form of the pARK plasmid (3 kb), which contains a single recognition site for each of these enzymes served as a DNA substrate in these experiments (Figure 1). The products of these reactions were treated with T4 DNA polymerase to produce blunt ends and then cloned as described in Materials and Methods. The sequences surrounding the pARK/Cm junctions in individual clones were determined using primers originating in the Cm gene to map the positions of the putative strand breaks (Figure 1). A terminal deletion in the cloned fragment (a result of 3′–5′ exonuclease activity of T4 polymerase) indicates the generation of 3′-overhangs while duplicated sequences at the pARK/Cm junctions (a result of the T4 DNA polymerase activity) indicate the formation of 5′-overhangs. Thus, the lengths of deletions and duplications would define the lengths of 3′- and 5′-overhangs, respectively.
Figure 1.
Experimental strategy for the analysis of the DNA ends produced by Type I restriction enzymes. Circular plasmid DNA (black lines) containing a single recognition site (gray box) is cleaved by a Type I restriction enzyme (REase) that is represented as one light gray oval (methylase) and two dark gray ovals (HsdR subunits). The linearized DNA is purified from an agarose gel and treated with T4 DNA polymerase to remove possible 3′-overhangs or to fill in 5′-overhangs. The resulting blunt-ended DNA fragments are ligated with a fragment carrying a chloramphenicol (Cm) resistance marker (open box) and transformed into bacteria. Plasmid DNA from individual clones is isolated and the sequence of the ends of cloned Type I restriction products is determined using primers originating in the Cm gene. The obtained sequences are aligned with the original pARK sequence. Deletions at the ends of cloned DNA fragments suggest the presence of 3′-overhangs, while sequence duplications indicate 5′-overhangs. No change in the sequence with respect to pARK indicates the presence of blunt ends.
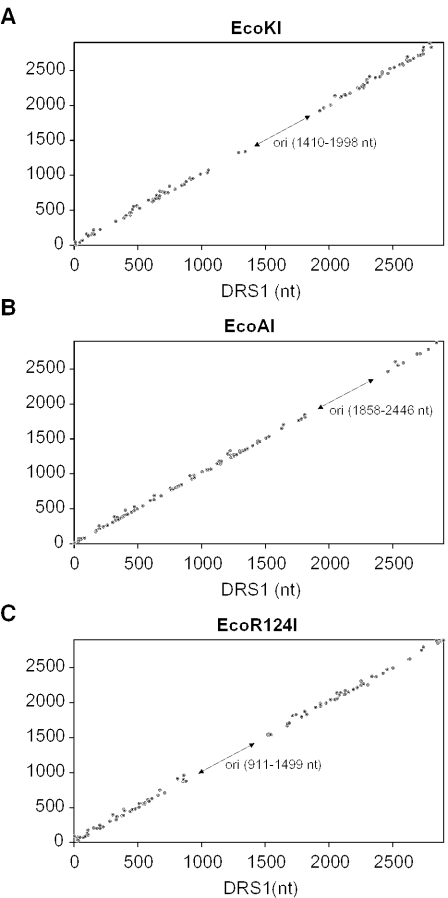
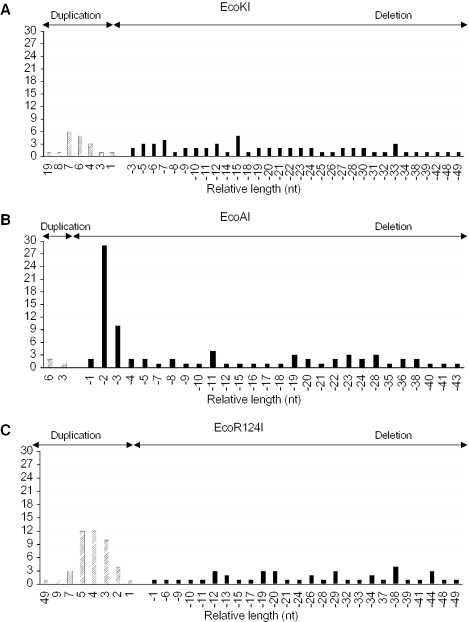
In total, we analyzed 146 clones for EcoKI, 136 clones for EcoAI and 155 clones for EcoR124I (Table 1). Clones without a deletion or duplication were not found for any of the enzymes, excluding the formation of blunt ends. For each enzyme, ∼30% of clones contained deletions >100 bp. Since these deletions could result from restriction or from illegitimate recombination during cloning, they were not further analyzed. The 50–100 bp deletions were found in 15% of EcoKI clones, 6% of EcoAI clones and 10% of EcoR124I clones, while deletions <50 bp were found in 43% of EcoKI clones, 57% EcoAI clones and 27% of EcoR124I clones. Finally, duplications of up to 50 bp were found in 13% of EcoKI clones, 2% of EcoAI clones and 28% of EcoR124 clones. Duplications longer than 50 bp were not detected. As evident from Figure 2, DNA cleavage produced by all three enzymes occurred randomly throughout the whole DNA molecule, with no striking preference for any particular DNA site. Analysis of the nucleotide sequence around the identified cleavage sites revealed neither a consensus sequence nor a significant preference for a particular A/T content (data not shown). Furthermore, the analysis of the clones with deletions or duplications up to 50 bp indicated that EcoKI showed some preference for the formation of 5′-overhangs of 6–7 nt (14.5% of clones) although 3′-overhangs of similar lengths were also quite frequent (Figure 3A). In contrast, EcoAI displayed strong preference for the generation of 3′-overhangs of 2–3 nt (45% of clones) (Figure 3B). In the EcoR124I reaction, 5′-overhangs of 3–5 nt were preferentially formed (40% of clones) (Figure 3C).
Table 1.
Summary of the clones analyzed
| Total | Number of clones | ||||||
|---|---|---|---|---|---|---|---|
| >1000 bp deletion | 1000–1500 bp deletion | 500–100 bp deletion | 100–150 bp deletion | <50 bp deletion | <50 bp duplication | ||
| EcoKI | 136 | 17 (12%) | 9 (7%) | 14 (10%) | 20 (15%) | 58 (43%) | 18 (13%) |
| EcoAI | 146 | 23 (16%) | 12 (8%) | 16 (11%) | 9 (6%) | 83 (57%) | 3 (2%) |
| EcoR124I | 155 | 12 (8%) | 14 (9%) | 28 (18%) | 16 (10%) | 41 (27%) | 44 (28%) |
Figure 2.
Plot of the distance between the recognition site and the break in the top strand (DRS2) against the distance between the recognition site and the break in the bottom strand (DRS1). The distances were measured from the 5′ end of the asymmetric recognition sequence. Data obtained from clones with deletions up to 100 bp and all clones with duplications are plotted: 79 clones for EcoKI (A), 89 clones for EcoAI (B) and 101 clones for EcoR124I (C). DRS1 and DRS2 are given in nucleotides (nt). The arrows indicate the position of the origin of replication (ori) located in the substrate DNA. DNA molecules cleaved in this region could not be cloned, giving an area free of points. For all three enzymes cleavage was random and there were no preferred sites.
Figure 3.
Distribution of deletions and duplications in selected clones. The lengths of deletions (black bars) and duplications (hatched bars) are plotted against the number of clones for EcoKI (A), EcoAI (B) and EcoR124 (C). Deletions indicate the presence of 3′-overhangs, while the duplications indicate the presence of 5′-overhangs. Clones with deletions up to 50 nt were considered for this analysis.
Stoichiometry of the DNA cleavage complex of EcoAI
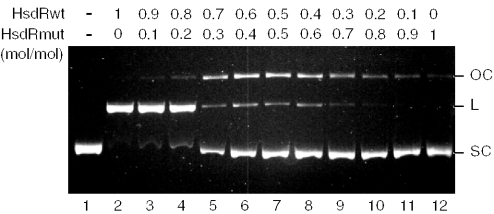
In the current model proposed for Type I restriction enzymes, which is based on several lines of experimental evidence, the two HsdR subunits of the enzyme complex simultaneously translocate along DNA from the recognition site in opposite directions (2). This mode of action would seem to preclude their cooperation in DNA cleavage. Considering such a mechanism, the question arises of how Type I restriction enzymes introduce double-strand DNA breaks if their HsdR subunits contain only one catalytic center for phosphodiester bond hydrolysis. One possibility is that the HsdR subunit can rearrange its catalytic center for the sequential cleavage of both DNA strands. Alternatively, a complex of higher order is assembled at the site of cleavage, providing two nuclease catalytic centers associated with two HsdR subunits. To address these possibilities, we have exploited a mutant of the EcoAI HsdR subunit containing a single alanine substitution at the first acidic residue (Asp-61 → Ala) in the conserved N-terminal P-D… (D/E)-X-K motif, which forms the active site for DNA cleavage (13). This mutation completely abolishes the endonuclease activity of the enzyme without affecting its DNA translocation activity (13,15). We have mixed this mutant subunit with the wild-type HsdR subunit in various molar ratios, and used these mixtures to assemble EcoAI restriction enzyme. The resulting enzymes were then used to cleave a supercoiled DNA substrate containing a single recognition site for EcoAI. If EcoAI requires two HsdR subunits to introduce a double-stranded break, the presence of both wild-type and mutant subunits in the cleavage complex should result in the formation of a nicked circular DNA. If only one HsdR subunit was sufficient for the DNA cleavage, the replacement of the wild-type HsdR subunit with the D61A mutant in the cleavage complex would simply result in the inhibition of DNA cleavage activity, with no nicked DNA being formed. The result of this experiment is demonstrated in Figure 4. At low-HsdRD61A concentrations (<20% of total), only a slight effect on double-strand cleavage efficiency was observed (Figure 4, lanes 2 and 3). However, in the presence of 30% mutated subunit, accumulation of nicked DNA was apparent, while the formation of linear DNA was dramatically inhibited (Figure 4, lane 3). The concentration of nicked DNA peaked when HsdRwt and HsdRD61A were present in equimolar amounts. Further increase in the proportion of mutated subunit in the mixture resulted in the gradual inhibition of DNA cleavage (Figure 4, lanes 8–12). Thus, these experiments suggest a requirement for two HsdR subunits for breakage of the DNA duplex.
Figure 4.
Cleavage of pJP25 plasmid DNA by EcoAI assembled from wild-type and mutant HsdR subunits. HsdRD61A carries a mutation in the nuclease active center that inactivate DNA cleavage activity of the EcoAI enzyme while it has no effect on its ATP-dependent DNA translocation activity. Reactions contained 10 nM DNA, 40 nM Mtase and 250 nM HsdRwt/HsdRD61A. Relative molar ratios between HsdRwt and HsdRmut in individual reactions were: 1:0, 0.9:0.1, 0.8:0.2, 0.7:0.3, 0.6:0.4, 0.5:0.5, 0.4:0.6, 0.3:0.7, 0.2:0.8, 0.1:0.9 and 0:1. Reactions were incubated for 15 min under conditions described in Materials and Methods. Reaction products were resolved in 1% agarose gel and visualized by ethidium bromide staining.
DISCUSSION
In this work, we have characterized the structure of the DNA ends generated by the Type I restriction endonucleases EcoKI, EcoAI and EcoR124I in order to gain further insight into their reaction mechanism. These enzymes belonging to three different families all proved to have a similar cleavage mechanism. Our data indicate that these enzymes can generate both 5′- and 3′-overhangs of various lengths, whereas blunt ends were not detected. EcoKI showed some preference for the formation of 5′-overhangs of 6–7 nt. EcoAI displayed a strong preference for 3′-overhangs of 2–3 nt and EcoR124I preferentially formed 5′-overhangs of 3–5 nt. Formation of short overhangs is consistent with a mechanism similar to that of Type II restriction enzymes that involves the introduction of two adjacent nicks, one in each strand of the DNA duplex (16). However, a number of restriction products of all three enzymes were also found to have much longer terminal deletions, suggesting the formation of long 3′-overhangs, which cannot be explained by a Type II-like mechanism. How do Type I restriction enzymes produce long single-stranded termini? Formation of 3′-overhangs up to 100 nt long has already been described for the Type IA restriction enzyme EcoBI, which appears to generate DNA gaps during the cleavage reaction rather than nicks, with concomitant appearance of acid-soluble oligonucleotides of an average size of 7 nt (17,18). A model for the restriction mechanism of EcoBI, in which the enzyme first generates a gap in one strand and then excises nucleotides from the opposite strand starting from the position corresponding to the 3′-side of the gap, removing ∼70–100 nt, has been proposed (18). This would result in the formation of long 3′ single-stranded tails. However, such a mechanism would require an exonuclease activity that has not been observed with any of the enzymes used in this study. An alternative explanation for the presence of the long deletions (up to 100 bp) in the cloned restriction products could be that the enzyme introduces two DNA double-strand breaks into the contracting DNA loop at the positions where the two convergently translocating HsdR subunits stall due to structural constraints in the enzyme–DNA complex. Some support for this proposal is provided by our previous observation that on a circular DNA molecule containing a Holliday junction, EcoR124II endonuclease could indeed introduce two DNA double-strand breaks, one on each side of the junction (6). Finally, the possibility that the long deletions result from illegitimate recombination in the host cells during the cloning of the restriction products cannot readily be excluded.
An important question concerning the DNA cleavage mechanism of Type I enzymes that remained unanswered was the number of HsdR subunits involved in the generation of a DNA double-strand break. The HsdR subunit contains only a single catalytic center for phosphodiester bond cleavage of the same type as that present in the classical Type II restriction enzymes, such as EcoRI and EcoRV (13,19). These Type II enzymes act as homodimers where each subunit introduces a single-strand break (16). However, there are examples of Type II enzymes, such as BfiI, that can introduce double-strand breaks via a rearrangement of a single catalytic center (20). Our finding that mutant EcoAI endonuclease assembled from wild-type and nuclease-deficient HsdR subunits converted a supercoiled single-site substrate into its nicked circular form, whereas the wild-type enzyme efficiently catalyzed double-strand breakage clearly suggests that at least two HsdR subunits of EcoAI are required for the introduction of one double-strand break.
According to the current model proposed for DNA cleavage by Type I restriction enzymes, the enzyme's methylase core binds to the recognition site while its two HsdR subunits translocate non-specific DNA in opposite directions with the recognition site remaining associated with the methylase–DNA complex during this process (6,21). DNA cleavage can result at any site where the translocation process is halted (6). The two translocating HsdR subunits from a single enzyme–DNA complex are not likely to cooperate in the DNA cleavage reaction since they are bound to distant DNA sites. Instead, we propose that double-strand breaks resulting from the cooperation between translocating HsdR subunit and excess HsdR subunit, either free in solution or as a part of an assembled enzyme, in a manner similar to the mechanism of Type II restriction enzymes, such as FokI. FokI appears to cleave DNA after dimerization of the nuclease domains of the enzyme monomer bound to the recognition sequence and another enzyme monomer from the solution (22). The possibility of a similar restriction mechanism being employed by Type I endonucleases is also supported by our previous studies, indicating that the stoichiometry of the cleavage reaction between EcoR124I and a circular DNA substrate containing a single recognition site is one molecule of methylase and six molecules of HsdR per one cleavage event (23).
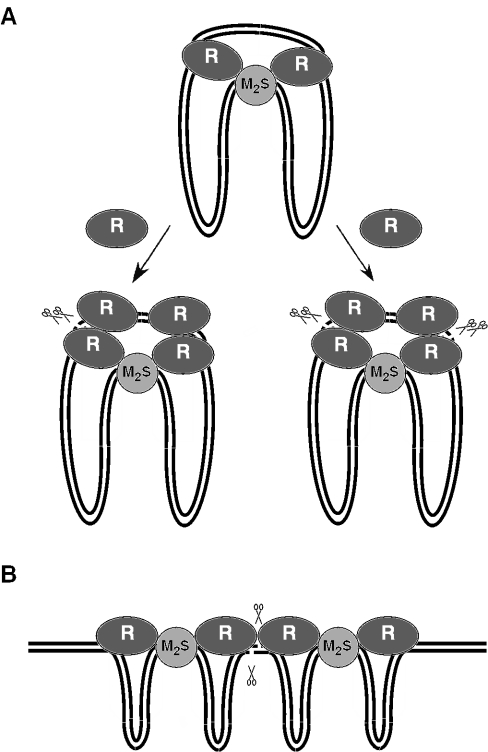
Our revised model for the cleavage of circular DNA molecules by Type I restriction enzymes is depicted in Figure 5A. We propose that the enzyme binds the circular DNA substrate in the form of a R2M2S1 complex and then translocates the DNA towards itself from both directions until the translocation process stalls due to structural constraints in the complex. Our previous work suggested that the enzyme can relieve DNA supercoiling in the contracting and expanding DNA loops associated with the translocation process, which would allow translocation of the entire DNA circle (6,15). The stalled complex interacts with two additional HsdR subunits (free or as a part of an assembled enzyme) to yield two cleavage units. In each unit, two nuclease catalytic centers are brought to close proximity on the DNA—one provided by the translocating HsdR subunit and the other by HsdR subunit from the solution. Since Type I enzymes cleave at non-specific DNA sites, the local DNA sequence can produce different cleavage rates so that one HsdR dimer would introduce double-strand break faster than the other (Figure 5A, left outcome). After this event, the cleavage complex would dissociate as the translocating HsdR subunits are free to reinitiate tracking on DNA. It is also possible that occasionally both HsdR dimers in the cleavage complex introduce double-strand break (Figure 5A, right outcome), which would explain the small number of long deletions observed in this study. However, the EcoKI endonuclease has been shown to introduce nicks into covalently closed circular DNA when the translocation process was impaired by DNA condensation or non-specific DNA-binding ligands (24). Therefore, an alternative model in which double-strand breaks result from cooperation between the two translocating HsdR subunits cannot be excluded at least for this enzyme. A similar scenario would occur on linear DNA substrates containing more than one recognition site where the nuclease catalytic centers of the translocating HsdR subunits of two converging R2M2S1 complexes can readily get to close proximity to mediate double-strand cleavage (Figure 5B). Nevertheless, we cannot exclude an assembly of a higher order complex similar to that proposed for circular substrates. Such a complex can be formed when the enzyme collides with a non-specific block such as the secondary DNA structure that can also trigger DNA cleavage (6).
Figure 5.
Proposed models for DNA duplex breakage by Type I restriction enzymes on circular (A) and linear (B) DNA substrates. For more explanation see Discussion. DNA is represented as two black lines. The methylase (M2S1) and HsdR (R) subunits are represented as light gray and dark gray ovals. The pairs of scissors indicate the position of DNA double-strand break.
Acknowledgments
We thank Mark Folcher for help with protein preparations, and Patrick Garcia and Aude Bourniquel for their comments on the manuscript. This work was supported by the Swiss National Science Foundation. Funding to pay the Open Access publication charges for this article was provided by the Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Redaschi N., Bickle T.A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd edn. Neidhardt F.C., Curtiss R. III, Ingraham J.L., Brooks Low K.B., Magasanik B., Reznikoff W.S., Riley R., Schaechter M., Umbarger H.E., editors. Washington, DC: ASM Press; 1996. pp. 773–781. [Google Scholar]
- 2.Murray N.E. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourniquel A.A., Bickle T.A. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 4.Titheradge A.J., King J., Ryu J., Murray N.E. Families of restriction enzymes: an analysis prompted by molecular and genetic data for type ID restriction and modification systems. Nucleic Acids Res. 2001;29:4195–4205. doi: 10.1093/nar/29.20.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Studier F.W., Bandyopadhyay P.K. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janscak P., MacWilliams M.P., Sandmeier U., Nagaraja V., Bickle T.A. DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J. 1999;18:2638–2647. doi: 10.1093/emboj/18.9.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eskin B., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. III. Studies of the restriction adenosine triphosphatase. J. Biol. Chem. 1972;247:6192–6196. [PubMed] [Google Scholar]
- 8.Dreier J., MacWilliams M.P., Bickle T.A. DNA cleavage by the type IC restriction–modification enzyme EcoR124II. J. Mol. Biol. 1996;264:722–733. doi: 10.1006/jmbi.1996.0672. [DOI] [PubMed] [Google Scholar]
- 9.Szczelkun M.D., Janscak P., Firman K., Halford S.E. Selection of non-specific DNA cleavage sites by the type IC restriction endonuclease EcoR124I. J. Mol. Biol. 1997;271:112–123. doi: 10.1006/jmbi.1997.1172. [DOI] [PubMed] [Google Scholar]
- 10.Janscak P., Bickle T.A. The DNA recognition subunit of the type IB restriction–modification enzyme EcoAI tolerates circular permutations of its polypeptide chain. J. Mol. Biol. 1998;284:937–948. doi: 10.1006/jmbi.1998.2250. [DOI] [PubMed] [Google Scholar]
- 11.Janscak P., Abadjieva A., Firman K. The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J. Mol. Biol. 1996;257:977–991. doi: 10.1006/jmbi.1996.0217. [DOI] [PubMed] [Google Scholar]
- 12.Weiserova M., Janscak P., Benada O., Hubacek J., Zinkevich V.E., Glover S.W., Firman K. Cloning, production and characterization of wild type and mutant forms of the R.EcoK endonucleases. Nucleic Acids Res. 1993;21:373–379. doi: 10.1093/nar/21.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janscak P., Sandmeier U., Bickle T.A. Single amino acid substitutions in the HsdR subunit of the type IB restriction enzyme EcoAI uncouple the DNA translocation and DNA cleavage activities of the enzyme. Nucleic Acids Res. 1999;27:2638–2643. doi: 10.1093/nar/27.13.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor I., Patel J., Firman K., Kneale G. Purification and biochemical characterisation of the EcoR124 type I modification methylase. Nucleic Acids Res. 1992;20:179–186. doi: 10.1093/nar/20.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janscak P., Bickle T.A. DNA supercoiling during ATP-dependent DNA translocation by the type I restriction enzyme EcoAI. J. Mol. Biol. 2000;295:1089–1099. doi: 10.1006/jmbi.1999.3414. [DOI] [PubMed] [Google Scholar]
- 16.Pingoud A., Jeltsch A. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem. 1997;246:1–22. doi: 10.1111/j.1432-1033.1997.t01-6-00001.x. [DOI] [PubMed] [Google Scholar]
- 17.Kimball M., Linn S. The release of oligonucleotides by the Escherichia coli B restriction endonuclease. Biochem. Biophys. Res. Commun. 1976;68:585–591. doi: 10.1016/0006-291x(76)91185-2. [DOI] [PubMed] [Google Scholar]
- 18.Endlich B., Linn S. The DNA restriction endonuclease of Escherichia coli B. II. Further studies of the structure of DNA intermediates and products. J. Biol. Chem. 1985;260:5729–5738. [PubMed] [Google Scholar]
- 19.Davies G.P., Martin I., Sturrock S.S., Cronshaw A., Murray N.E., Dryden D.T.F. On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol. 1999;290:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 20.Sasnauskas G., Halford S.E., Siksnys V. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl Acad. Sci. USA. 2003;100:6410–6415. doi: 10.1073/pnas.1131003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seidel R., van Noort J., van der Scheer C., Bloom J.G., Dekker N.H., Dutta C.F., Blundell A., Robinson T., Firman K., Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nature Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 22.Bitinaite J., Wah D.A., Aggarwal A.K., Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janscak P., Dryden D.T.F., Firman K. Analysis of the subunit assembly of the type IC restriction–modification enzyme EcoR124I. Nucleic Acids Res. 1998;26:4439–4445. doi: 10.1093/nar/26.19.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keatch S.A., Su T.J., Dryden D.T. Alleviation of restriction by DNA condensation and non-specific DNA binding ligands. Nucleic Acids Res. 2004;32:5841–5850. doi: 10.1093/nar/gkh918. [DOI] [PMC free article] [PubMed] [Google Scholar]