Abstract
Noroviruses are single-stranded RNA viruses with high genomic variability. They have emerged in the last decade as a major cause of acute gastroenteritis. It remains so far unclear whether norovirus evolution is driven by sequence mutation and/or recombination. In this study, we have assessed the occurrence of recombination in the norovirus capsid gene. For this purpose, 69 complete capsid sequences of norovirus strains accessible in GenBank as well as 25 complete capsid sequences generated from norovirus-positive clinical samples were examined. Unreported recombination was detected in about 8% of norovirus strains belonging to genetic clusters I/1 (n = 1), II/1 (n = 1), II/3 (n = 1), II/4 (n = 3), and II/5 (n = 1). Recombination breakpoints were mainly located at the interface of the putative P1-1 and P2 domains of the capsid protein and/or within the P2 domain. The recombination region displayed features such as length, sequence composition (upstream and downstream GC- and AU-rich sequences, respectively), and predicted RNA secondary structure that are characteristic of homologous recombination activators. Our results suggest that recombination in the norovirus capsid gene may naturally occur, involving capsid domains presumably exposed to immunological pressure.
Noroviruses (NV) are small nonenveloped viruses with a 7.8-kb positive-strand RNA genome. They belong to the family Caliciviridae (13). Based upon their genomic heterogeneity, human NV are subdivided into two genogroups termed I and II. Viral strains belonging to NV genogroup I have been further subclassified in genetic clusters I/1 to I/7, and NV genogroup II strains have been subclassified into genetic clusters II/1 to II/7 (11). NV are an important cause of acute gastroenteritis, being second only to rotavirus as causative agents of nonbacterial gastroenteritis in infants (18). They are also important agents of nonbacterial food-borne diseases (33).
NV evolution has been so far poorly understood. It remains to date unclear whether NV evolution depends upon accumulation of mutational changes and/or homologous recombination, involving structural and nonstructural proteins. The NV capsid protein (ranging from 535 to 549 amino acids) is subdivided into two major domains, termed shell (S) and protruding (P) domains (39, 40). The protruding domain is in turn subdivided into P1-1, P1-2, and P2 domains, the last being located at the exterior surface of the capsid. The P2 domain is therefore predicted to bear antigenic determinants of the immunological response of the host (30). Recently, a putative receptor binding pocket was identified within the P2 domain (48).
Recombination is a common event in RNA viruses (54). It has been reported in retroviruses (17, 42), flaviviruses (12, 16, 50, 54-56), enteroviruses (44), plant viruses (1), and, to a much lesser extent, in negative-sense RNA viruses (5). In human caliciviruses, the first evidence of homologous recombination was shown in 2000, as Jiang et al. characterized an NV recombinant bearing open reading frames (ORFs) belonging to NV genetic clusters II/3 and II/4 (15). This strain, Arg320/1995/AR (GenBank accession number AF190817), has since been reported to circulate in the community (14). Recombination is also a major determinant of viral virulence, being implicated in the emergence of new viral strains (54). Its implications for the development of vaccines and virus control programs are essential. In this study, we have attempted to assess the extent of homologous recombination in the NV capsid gene. For this purpose, the entire capsid gene of NV strains belonging to genogroups I and II was examined for evidence of recombination motifs within and between NV genetic clusters.
MATERIALS AND METHODS
Viral strains.
Complete capsid sequences available from GenBank were used. Criteria for selection were (i) the presence of amino acid sequences MMMASK(D/G)A and MKMAS(S/N)DA at the beginning of ORF-2 for NV genogroup I and II strains, respectively, and (ii) the presence of amino acid sequences (R/G)RLG(I/V/L)RR(I/S/V) and GR(R/V)R(V/A/I/F)(Q/L) at the end of ORF-2 for NV genogroup I and II strains, respectively.
Generation of NV capsid sequences from clinical samples.
Stool samples were collected from children under 4 years of age presenting with acute gastroenteritis in 20 pediatric practices from five German cities over a 14-month period (34). In the first step, NV RNA extraction and amplification were performed as described previously (43). Those samples testing positive for NV were selected for complete capsid sequence characterization. Therefore, NV RNA was extracted from a pea-sized stool sample (approximately 100 mg or 200 μl of watery stool) with the viral RNA kit (Qiagen) according to the instructions of the manufacturer. After elution in a final volume of 250 μl of RNase-DNase free water, RNasin (1 U/μl; Promega) was added and the eluate was directly used in the amplification step.
The viral nucleic acid extraction, preparation of reaction mix, and amplification reaction were performed in separate rooms. The reverse transcription-PCR (RT-PCR) assay for detection of NV was performed in a single step with the Superscript One-Step RT-PCR with Platinum Taq kit (Invitrogen). The RT-PCR mix (50 μl) consisted of 13 μl of RNA template (ranging from 10 to 100 pg); 23 μl of 2× reaction mix containing 0.4 mM (each) dATP, dCTP, dGTP, and dTTP and 2.4 mm MgSO4; 1 μM (each) primers Calman-1 (5′-GCACACTGTGTTACACTTCC-3′, forward) and Calman-6 (5′-GCTGAAGAACCTAGTCTCG-3′, reverse); and 1 μl of RT Platinum Taq mix. Reverse transcription was carried out for 45 min at 50°C. This was followed by denaturation for 2 min at 94°C and 40 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and extension for 3 min at 72°C. A final extension step was carried out for 7 min.
For nested PCR amplification of cDNA, the reaction mix (50 μl) consisted of 2 μl of cDNA template; 5 μl of 10× Herculase polymerase reaction buffer (Stratagene); 0.2 mM (each) dATP, dCTP, dGTP, and dTTP; 5% dimethyl sulfoxide; 1 μM (each) primers Calman-3 (5′-GCAGAAGACCTTCTATCTCC-3′, forward) and Calman-8 (5′-TCTTTATCATGTTGGAAAGAAGC-3′, reverse) or Calman-10 (5′-AAGAAGTGGCAGTAGGAGC-3′, reverse) or Calman-12 (5′-TTGATGGCTCCAGCTCCAGCATTG-3′, reverse); and 5 U of Herculase Hotstart polymerase (Stratagene). Initial denaturation was carried out for 5 min at 94°C. This was followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and extension for 3 min at 72°C. A final extension step was carried out for 7 min. PCR products were separated by 1% agarose gel electrophoresis in the presence of ethidium bromide, and the gel was visualized under UV light.
Amplicons were cloned into pCR4Blunt-Topo vector (Invitrogen) and sequenced. Sequencing was done using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA Polymerase FS according to the manufacturer's protocol (Applied Biosystems). Samples were run on an ABI PRISM 310 sequencer. Data were collected and analyzed with ABI 310 software. Sequences were generated in both directions for each amplified viral nucleic acid. Sequences were aligned in CLUSTAL X with the default settings (49). Maximum likelihood (ML) phylogenetic trees were drawn in PAUP (47) with the models of substitution, a gamma distribution of among-site variation (with eight rate categories) as well as correction for invariant sites, and base composition estimated from the data. The appropriate model of evolution was selected according to the likelihood ratio test and/or Akaike's information criterion generated in MODELTEST (37) (alignments are available upon request at www.calicilab.de).
Detecting recombination by statistical, phylogenetic, and structural analysis.
For the detection of recombination, a stepwise strategy was used. In the first step, we focused on the detection of recombination between and within NV genogroup I and II strains, independently from the identification of the location of recombination breakpoints. This was achieved by screening of the alignments with Sawyer's test, exploratory tree analysis, and similarity plots. Sawyer's test is a polymorphic site-based method that has proven to be reliable in its power to detect recombination in simulated studies (38, 53). Exploratory tree analysis and similarity plots have been previously used to detect recombination in RNA viruses (5, 12, 50). In the second step, localization of recombinational breakpoints was identified by likelihood ratio analysis. Evidence of recombination was further assessed by phylogenetic analysis of the capsid regions located at either side of the breakpoint, as defined by others (5, 50). Finally, the predictive RNA secondary structure of the recombination region was determined. Strong evidence of recombination was given by congruence between the above-mentioned methods.
Detecting recombination under Sawyer's test.
Sawyer's test, a rather conservative test, looks in a given alignment for the appearance of long tracts of nucleotide identities between a pair of sequences that may be suggestive of gene conversion (46). It has proven to be a powerful method to detect recombination, and its performance has been validated in computational studies (53). Multiple sequence alignments of NV genogroup I and genogroup II were generated in CLUSTAL X with the default settings (49). Alignments were run in GENECONV (45). Here and in all runs, the default settings with 10,000 permutations were used and the statistical significance of gene conversion events was defined as a global permutation P value of <0.05.
Exploratory tree analysis and similarity plots.
For exploratory tree analysis, phylogenetic trees were generated by sliding a window of 300 nucleotides along the capsid gene alignment in 150-nucleotide increments. For each region, ML trees were reconstructed using the appropriate model of nucleotide substitution, a gamma distribution of among-site variation (with eight rate categories) as well as correction for invariant sites, and base composition estimated from the data. The appropriate model of evolution was selected according to the likelihood ratio test and/or Akaike's information criterion generated in MODELTEST (37). A recombination event was suspected when a strain changed phylogenetic position over different capsid regions. This was further examined by similarity plot analysis in Simplot, as previously described by others (17). The pairwise percent difference between the query sequence (putative recombinant) and other sequences in the alignment was determined by sliding a window of 200 nucleotides along the capsid alignment in 20-nucleotide increments. Strains displaying alternate sequence identities (ranging from 90 to 95%) to the putative recombinant strain were identified as the closest possible parental strains, as proposed by others (50).
Breakpoint estimation.
Once the closest parental sequences were identified, we estimated the recombination breakpoints using the ML method followed by a likelihood ratio test as described by others (LARD program, version 1.0) (5, 12, 50). The statistical significance of the likelihood ratio was determined with a χ2 test and a Monte Carlo simulation in Seq-Gen (41) as described by others (8, 29, 32).
Phylogenetic trees and bootstrapping.
After breakpoints were identified, separate ML trees were constructed for each putative recombinant sequence with the appropriate model of evolution as described above. Phylogenetic conflicts were assessed using a bootstrapping approach (1,000 replicates). In order to minimize the chance of false-positive results due to rate variation heterogeneities, rate variation among sites was estimated from the data and taken into account for phylogenetic incongruence analysis. Under these conditions, a strong support for phylogenetic conflict was evidenced by a bootstrap value of ≥80% of 1,000 replications at phylogenetically relevant nodes as previously defined (5).
Structural characterization of recombination region.
Characterization of recombination hotspots at either side of the breakpoints was based upon sequence composition (length and percentage of nucleotide similarity in both parental strains) and predictive RNA secondary structure of the recombination region (≥50 nucleotides) as defined elsewhere (20, 21, 26, 28). The predictive RNA secondary structure of the recombination region was computed using the Mfold web server for nucleic acid folding and hybridization prediction (57).
Nucleotide sequence accession numbers.
The complete capsid sequences of the strains collected in Germany from March 1997 to April 1998 are accessible in GenBank under the accession numbers AY532111 to AY532135.
RESULTS
Phylogenetic analysis of NV strains based upon complete capsid sequences available from GenBank.
Sixty-nine complete capsid sequences from NV genogroup I and II strains accessible in GenBank were used. In addition, 25 new NV capsid sequences were generated from clinical samples collected in a defined geographic area (five German cities) over a defined period of time (14 months, from March 1997 to April 1998). After sequence alignment, ML trees were drawn and a bootstrap analysis was performed as previously described. The sequences clustered as follow: NV genetic clusters I/1 (n = 3), I/2 (n = 2), I/3 (n = 5), I/4 (n = 4), I/5 (n = 1), I/6 (n = 1), and I/7 (n = 1) and NV genetic clusters II/1 (n = 8), II/2 (n = 2), II/3 (n = 17), II/4 (n = 37), II/5 (n = 3), II/6 (n = 3), and II/7 (n = 2). All 25 new NV strains clustered within genetic cluster II/4. In addition, five NV strains grouped in a thus-far-unclassified cluster. All in all, 15 distinct NV genetic clusters were identified as phylogenetically distinct groups supported by high bootstrap values (>98% of 1,000 replicates) at relevant nodes (Table 1 and Fig. 1 and 2).
TABLE 1.
Capsid sequences of NV strains used in this study
| Strain | Origin | Genetic cluster | GenBank accession no. |
|---|---|---|---|
| Norwalk/1968/US | United States | I/1 | M87661 |
| KY-89/1989/JP | Japan | I/1 | L23828 |
| Aichi124/1989/JP | Japan | I/1 | AB031013 |
| Southampton/1991/UK | United Kingdom | I/2 | L07418 |
| Whiterose/1996/UK | United Kingdom | I/2 | AJ277610 |
| Desert Shield/1990/SA | Saudi Arabia | I/3 | U04469 |
| Birmingham/1993/UK | United Kingdom | I/3 | AJ277612 |
| Stav/1995/NO | Norway | I/3 | AF145709 |
| Potsdam196/2000/GE | Germany | I/3 | AF439267 |
| Winchester/1994/UK | United Kingdom | I/3 | AJ277609 |
| Chiba407/1987/JP | Japan | I/4 | AB022679 |
| Valetta/1995/Malta | Malta | I/4 | AJ277616 |
| Thistlehall/1990/UK | United Kingdom | I/4 | AJ277621 |
| Koblenz433/2000/GE | Germany | I/4 | AF394960 |
| Musgrove/1989/UK | United Kingdom | I/5 | AJ277614 |
| Hesse3/1997/GE | Germany | I/6 | AF093797 |
| Sindlesham/1995/UK | United Kingdom | I/7 | AJ277615 |
| Hawaii/1971/US | United States | II/1 | U07611 |
| Girlington/1993/UK | United Kingdom | II/1 | AJ277606 |
| Schwerin003/2000/GE | Germany | II/1 | AF397905 |
| Wortley/1990/UK | United Kingdom | II/1 | AJ277618 |
| Aichi76-96Chitta/1996/JP | Japan | II/1 | AB032758 |
| Port Canaveral301/1994/US | United States | II/1 | AF414421 |
| Richmond283/1994/US | United States | II/1 | AF414419 |
| Honolulu314/1994/US | United States | II/1 | AF414420 |
| Melksham/1994/UK | United Kingdom | II/2 | X81879 |
| Snow Mountain/1976/US | United States | II/2 | U75682 |
| Toronto24/1991/CA | Canada | II/3 | U02030 |
| Mexico/1989/MX | Mexico | II/3 | U22498 |
| OTH-25/1989/JP | Japan | II/3 | L23830 |
| Arg320/1995/AR | Argentina | II/3 | AF190817 |
| Rbh/1993/UK | United Kingdom | II/3 | AJ277617 |
| Bham132/1995/UK | United Kingdom | II/3 | AJ277611 |
| Auckland/1994/NZ | New Zealand | II/3 | U46039 |
| Bitburg289/2001/GE | Germany | II/3 | AF427112 |
| Berlin226/2001/GE | Germany | II/3 | AF427111 |
| Herzberg385/2001/GE | Germany | II/3 | AF539439 |
| Oberhausen455/2001/GE | Germany | II/3 | AF539440 |
| MD134-10/1987/US | United States | II/3 | AY030313 |
| MD101-2/1987/US | United States | II/3 | AY030312 |
| Lionville247/1993/FR | France | II/3 | AF414411 |
| Brattleboro321/1995/US | United States | II/3 | AF414415 |
| Towson313/1994/US | United States | II/3 | AF414414 |
| Montgomery312/1994/US | United States | II/3 | AF414413 |
| Parkroyal/1995/UK | United Kingdom | II/4 | AJ277613 |
| Grimsby/1995/UK | United Kingdom | II/4 | AJ004864 |
| Bristol/1993/UK | United Kingdom | II/4 | X6716 |
| Lorsdale/1993/UK | United Kingdom | II/4 | X86557 |
| Camberwell/1994/AU | Australia | II/4 | AF145896 |
| 345/96002726/1996/US | United States | II/4 | AF080549 |
| Symgreen/1995/UK | United Kingdom | II/4 | AJ277619 |
| MD134-7/1987/US | United States | II/4 | AY030098 |
| UK3-17/12700/1992/UK | United Kingdom | II/4 | AF414417 |
| Miami Beach326/1995/US | United States | II/4 | AF414424 |
| Mora/1997/SE | Sweden | II/4 | AY081134 |
| Burwash Landing331/1995/US | United States | II/4 | AF414425 |
| Freiburg057/1997/GE | Germany | II/4 | AY532111 |
| Freiburg204/1997/GE | Germany | II/4 | AY532112 |
| Freiburg253/1998/GE | Germany | II/4 | AY532113 |
| Erlangen195/1997/GE | Germany | II/4 | AY532114 |
| Dresden153/1997/GE | Germany | II/4 | AY532115 |
| Dresden245/1997/GE | Germany | II/4 | AY532116 |
| Dresden267/1997/GE | Germany | II/4 | AY532117 |
| Dresden319/1997/GE | Germany | II/4 | AY532118 |
| Bochum024/1998/GE | Germany | II/4 | AY532119 |
| Bochum026/1997/GE | Germany | II/4 | AY532120 |
| Bochum031/1997/GE | Germany | II/4 | AY532121 |
| Bochum108/1997/GE | Germany | II/4 | AY532122 |
| Bochum136/1998/GE | Germany | II/4 | AY532123 |
| Bochum220/1997/GE | Germany | II/4 | AY532124 |
| Bochum224/1998/GE | Germany | II/4 | AY532125 |
| Bochum272/1998/GE | Germany | II/4 | AY532126 |
| Bochum339/1997/GE | Germany | II/4 | AY532127 |
| Hamburg048/1997/GE | Germany | II/4 | AY532128 |
| Hamburg135/1998/GE | Germany | II/4 | AY532129 |
| Hamburg137/1997/GE | Germany | II/4 | AY532130 |
| Hamburg139/1997/GE | Germany | II/4 | AY532131 |
| Hamburg180/1997/GE | Germany | II/4 | AY532132 |
| Hamburg189/1997/GE | Germany | II/4 | AY532133 |
| Hamburg236/1997/GE | Germany | II/4 | AY532134 |
| Hamburg316/1998/GE | Germany | II/4 | AY532135 |
| Hillingdon/1990/UK | United Kingdom | II/5 | AJ277607 |
| White River290/1994/US | United States | II/5 | AF414423 |
| MOH/1999/HU | Hungary | II/5 | AF397156 |
| Florida269/1993/US | United States | II/6 | AF414407 |
| Seacroft/1990/US | United States | II/6 | AJ277620 |
| Miami292/1994/US | United States | II/6 | AF414410 |
| Leeds/1990/UK | United Kingdom | II/7 | AJ277608 |
| Gwynedd273/1994/US | United States | II/7 | AF414409 |
| Amsterdam/1998/NL | The Netherlands | NAa | AF195848 |
| M7/1999/US | United States | NA | AY130761 |
| Alphatron/1998/NL | The Netherlands | NA | AF195847 |
| Saint Cloud624/1998/FR | France | NA | AF414427 |
| Fort Lauderdale560/1998/US | United States | NA | AF414426 |
NA, not assigned.
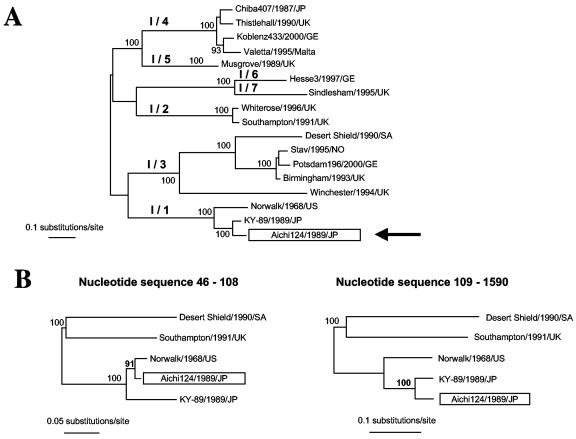
FIG. 1.
Evolutionary relationships among NV genogroup I strains based upon the entire capsid sequence. (A) An ML tree was drawn using a general time-reversible model of nucleotide substitution and a gamma distribution of among-site variation (with eight rate categories) as well as correction for invariant sites and base composition estimated from the data. Bootstrap values (expressed as percentages of 1,000 replications) are shown. Recombinant strain Aichi124/1989/JP is highlighted and indicated by an arrow. The tree is midpoint rooted for purposes of clarity, and all branch lengths are drawn to scale. (B) NV genetic cluster I/1, recombinant strain Aichi124/1989/JP, with NV strains Southampton/1991/UK and Desert Shield/1990/SA as outgroup.
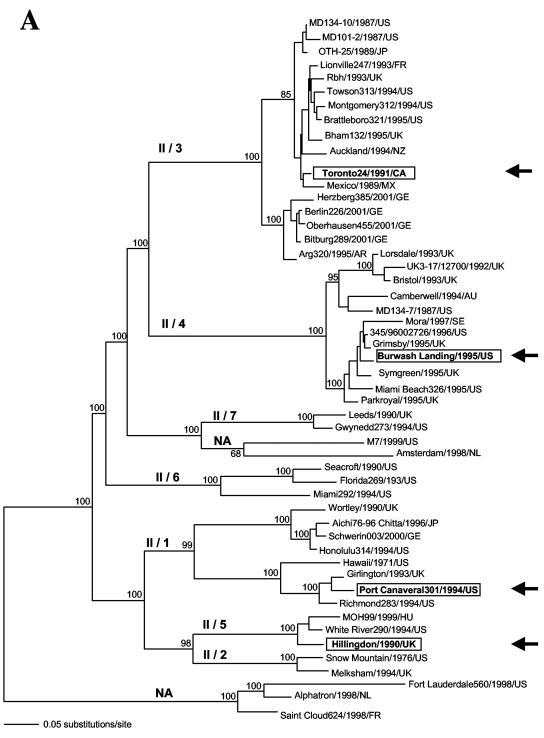
FIG. 2.
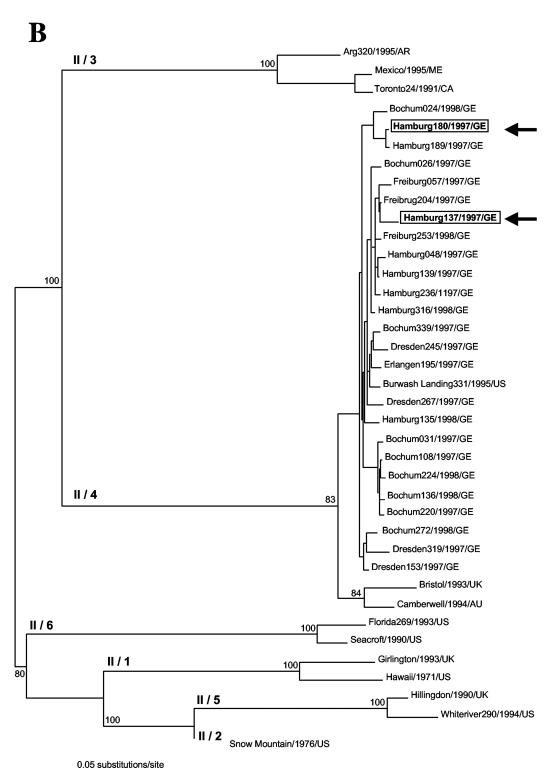
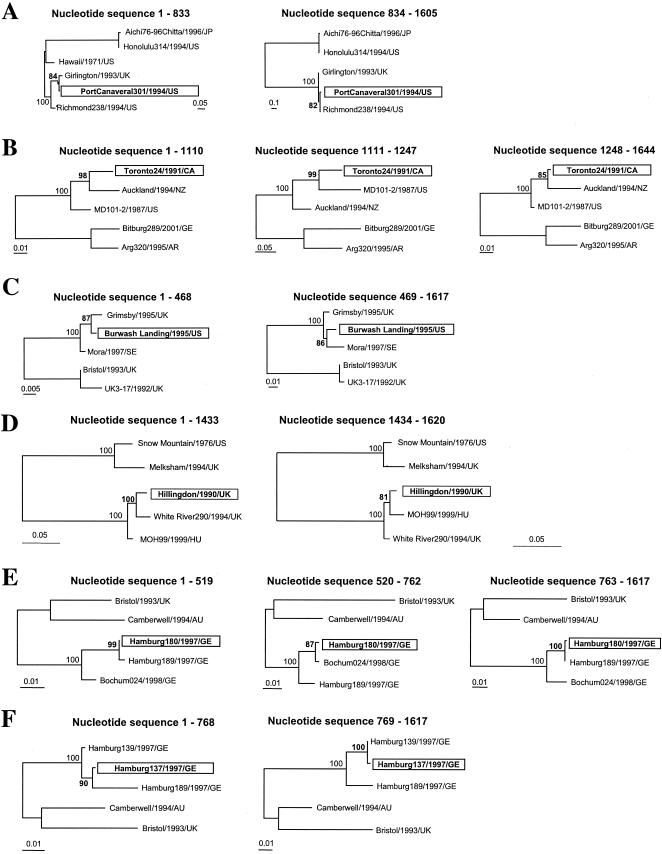
Evolutionary relationships among NV genogroup II strains based upon the entire capsid sequence. An ML tree was drawn using a general time-reversible model of nucleotide substitution and a gamma distribution of among-site variation (with eight rate categories) as well as correction for invariant sites and base composition estimated from the data. Bootstrap values (expressed as percentages of 1,000 replications) are shown. The tree is midpoint rooted for purposes of clarity, and all branch lengths are drawn to scale. (A) Recombinant strains Port Canaveral/301/1994/US, Toronto24/1991/CA, Burwash Landing331/1995/US, and Hillingdon/1990/UK are highlighted and indicated by arrows. NA, not assigned. (B) Recombinant strains Hamburg180/1997/GE and Hamburg137/1997/GE are highlighted and indicated by arrows.
Evidence of recombination in the capsid gene of NV genogroup I strains.
After alignment of the 17 NV genogroup I capsid sequences, Sawyer's test and exploratory tree analysis detected recombination in NV strain Aichi124/1989/JP belonging to genetic cluster I/1 (Fig. 1B) (Table 1). In this strain, two breakpoints were identified (nucleotide positions 46 and 108; Table 2). The strain Aichi124/1989/JP displays conflicting phylogenies at either side of the breakpoints to strain Norwalk/1968/USA in a short capsid region at the beginning of the shell domain and to strain KY-89/1989/JP in the capsid region encoding P1-1, P2, and P1-2 domains. These conflicting phylogenies across the breakpoints are supported by high bootstrap values at phylogenetically relevant nodes (Fig. 1B). Likelihood ratio analysis identified also a unique breakpoint at position 615 (Table 3). Phylogenetic analysis of the capsid regions at either side of this breakpoint was not supported by strong bootstrap values (60%). We therefore located the recombinational breakpoints for strain Aichi124/1989/JP at positions 46 and 108, in accordance with phylogenetic analysis results.
TABLE 2.
Evidence of recombination in the norovirus capsid gene under Sawyer's testa
| Genetic cluster | Recombinant strains | Capsid region (nt) | Length (nt) | P valueb |
|---|---|---|---|---|
| I/1 | Norwalk/1968/US, Aichi124/1989/JP | 46-108 | 63 | 0.0091 |
| II/1 | Girlington/1993/UK, Port Canaveral301/1994/US | 1439-1471 | 33 | 0.0010 |
| II/3 | Toronto24/1991/CA, MD101-2/1987/US | 1037-1184 | 148 | 0.0090 |
| II/4 | Mora/1997/SE, Burwash Landing331/1995/US | 790-906 | 117 | 0.0310 |
| II/4 | Hamburg180/1997/GE, Bochum024/1998/GE | 580-761 | 182 | 0.0007 |
| II/4 | Hamburg137/1997/GE, Hamburg139/1997/GE | 1166-1696 | 531 | 0.0100 |
| II/5 | Hillingdon/1990/UK, MOH/1999/HU | 1433-1606 | 174 | 0.0048 |
nt, nucleotides.
P value is the global P value for the alignment of interest.
TABLE 3.
Breakpoint analysis of the NV recombinants and their putative parental strainsa
| Genetic cluster | Recombinant strain | Putative parental strain | Capsid region(s) (nt) | Breakpoint (nt) | LRb | P valuec |
|---|---|---|---|---|---|---|
| I/1 | Aichi124/1989/JP | Norwalk/1968/US | 1-615 | 615 | 22.7 | <0.001 |
| KY-89/1989/JP | 616-1590 | |||||
| II/1 | Port Canaveral301/1994/US | Girlington/1993/UK | 1-833 | 833 | 5.276 | <0.025 |
| Richmond283/1994/US | 834-1605 | |||||
| II/3 | Toronto24/1991/CA | Auckland/1994/NZ | 1-1110, 1248-1644 | 1110 | 6.820 | <0.010 |
| MD101-2/1987/US | 1111-1247 | 1247 | 5.720 | <0.010 | ||
| II/4 | Burwash Landing331/1995/US | Grimsby/1995/UK | 1-468 | 468 | 5.140 | <0.025 |
| Mora/1997/SE | 469-1617 | |||||
| II/4 | Hamburg180/1997/GE | Hamburg189/1997/GE | 1-519, 763-1617 | 519 | 8.355 | <0.001 |
| Bochum024/1998/GE | 520-762 | 762 | 5.858 | <0.010 | ||
| II/4 | Hamburg137/1997/GE | Hamburg189/1997/GE | 1-768 | 768 | 5.607 | <0.010 |
| Hamburg139/1997/GE | 769-1617 | |||||
| II/5 | Hillingdon/1990/UK | White River290/1994/US | 1-1201 | 1201 | 6.551 | <0.010 |
| MOH/1999/HU | 1201-1620 |
nt, nucleotides.
LR, likelihood ratio.
P value was determined by a χ2 test with 3 df.
Evidence of recombination in the capsid gene of NV genogroup II strains.
In NV genetic clusters II/1, II/3, II/4, and II/5, gene conversion events were detected by Sawyer's test (Table 2) and exploratory tree analysis that showed swapping of putative recombinants over different capsid regions. The similarity plots and breakpoint analysis identified the putative recombinants, their parental strains, and the breakpoint of recombination in each case (Table 3). Representative examples of similarity plots for three putative recombinant strains are shown in Fig. 3. Further evidence of recombination in the capsid sequence was given by ML trees of sequences located at either side of the breakpoint (Fig. 4). High bootstrap values (≥80% of 1,000 replications) confirmed the localization of the breakpoints and supported the resulting conflicting phylogenies. In all, 16 ML trees of seven putative recombinants and their parental strains were constructed, involving regions of variable length (174 to 1,110 nucleotides).
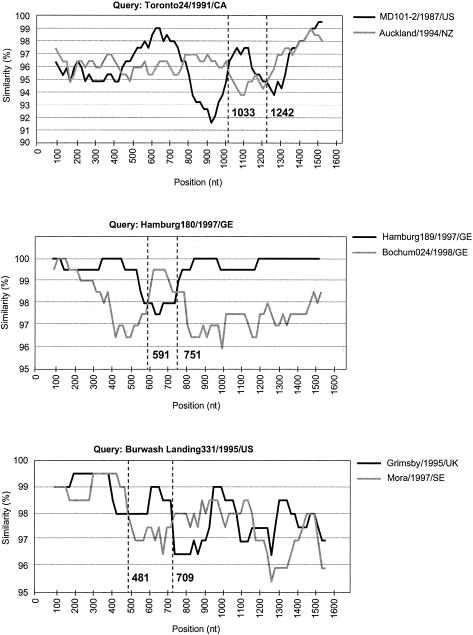
FIG. 3.
Similarity plots of recombinant NV genogroup II strains (Toronto24/1991/CA, Hamburg180/1997/GE, and Burwash Landing331/1995/US) and their putative parental strains. The analysis shows the percentage of nucleotide identity (vertical axis) along the capsid gene sequence (horizontal axis) with a sliding window of 200 nucleotides (nt) with 20-nucleotide increments. The estimated position of breakpoints is indicated.
FIG.4.
ML trees of the entire capsid sequence of NV genogroup II strains showing phylogenetic incongruence between recombinant and parental strains within four different genetic clusters of NV. Swapping of the branches on either side of the breakpoint brings definitive evidence of recombination. ML trees were drawn using a general time-reversible model of nucleotide substitution and a gamma distribution of among-site variation (with eight rate categories) as well as correction for invariant sites and base composition estimated from the data. The trees are unrooted, and all branch lengths are drawn to scale. Bootstrap values (expressed as percentages of 1,000 replications) are shown at relevant nodes. (A) NV genetic cluster II/1, recombinant strain Port Canaveral301/1994/US, with NV strains Aichi76-96Chitta/1996/JP, Honolulu314/1994/US, and Hawaii/1971/US as outgroup. (B) NV genetic cluster II/3, recombinant strain Toronto24/1991/CA, with NV strains Bitburg289/2001/GE and Arg320/1995/AR as outgroup. (C) NV genetic cluster II/4, recombinant strain Burwash Landing331/1995/US, with NV strains Bristol/1993/UK and UK3-17/1992/GB as outgroup. (D) NV genetic cluster II/5, recombinant strain Hillingdon/1990/UK, with NV strains Snow Mountain/1976/US and Melksham/1994/UK as outgroup. (E and F) NV genetic cluster II/4, recombinant strains Hamburg180/1997/GE (E) and Hamburg137/1997/GE (F), with NV strains Bristol/1993/UK and Camberwell/1994/AU as outgroup.
Within NV genetic cluster II/1, we identified the recombinant strain Port Canaveral301/1994/US. This strain was detected in the United States in 1994 (2). In its capsid sequence, the breakpoint is located at the beginning of the P2 domain (Fig. 5). Conflicting phylogenies at either side of the breakpoint were evidenced and were strongly supported by bootstrapping (Fig. 4A). The capsid sequence of the recombinant strain Port Canaveral301/1994/US shows alternate identities to strain Richmond234/1994/US (nucleotides 834 to 1605, encoding the putative P2 and P1-2 domains, respectively) and strain Girlington/1993/UK (nucleotides 1 to 833, encoding the putative S and P1-1 domains). NV strain Richmond234/1994/US was detected in 1994 in the United States (2), whereas strain Girlington/1993/UK was detected in 1993 in the United Kingdom (9).
FIG. 5.
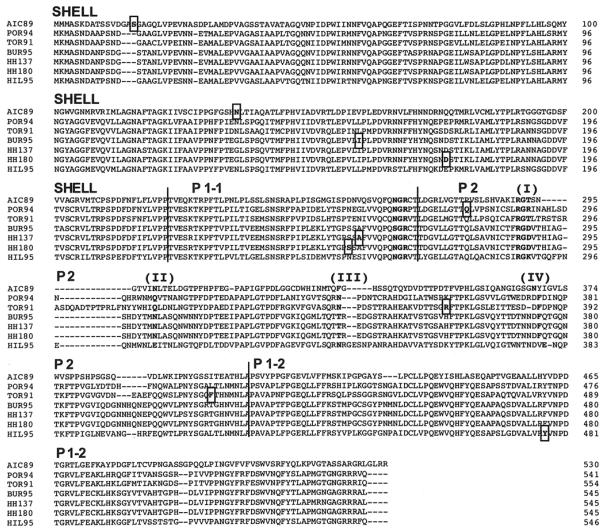
Amino acid alignment of the complete capsid sequence of NV genogroup I and II putative recombinants. Localization of the breakpoints at predictive codons is highlighted. Domains I to IV surrounding the predicted binding pocket for histo-blood group antigens are shown in boldface, as well as RGD and NGR motifs (48). AIC89, Aichi124/1989/JP; POR94, Port Canaveral301/1994/US; TOR91, Toronto24/1991/CA; BUR95, Burwash Landing331/1995/US; HH137, Hamburg137/1997/GE; HH180, Hamburg180/1997/GE; HIL95, Hillingdon/1990/UK.
Within NV genetic cluster II/3, we identified the putative recombinant strain Toronto24/1991/CA. In this strain's capsid sequence, two breakpoints are located at nucleotide positions 1110 and 1247 and strongly supported by ML tree analysis and bootstrapping (Fig. 4B). Both of them are located at the end of the P2 domain, encompassing a 45-amino-acid region, downstream of the N383 motif (Fig. 5). This motif has been predicted to bind specifically to human histo-blood group antigens (48). The similarity plots showed that different capsid regions of the recombinant strain Toronto24/1991/CA display alternate identities to strains Auckland/1994/NZ and MD101-2/1987/US (Fig. 3). Strain Auckland/1994/NZ was detected in 1994 in New Zealand, whereas the strain MD101-2/1987/US was detected from acute gastroenteritis outbreaks in 1987 in Maryland (United States) (10).
Within NV genetic cluster II/4, we identified the putative recombinant strain Burwash Landing331/1995/US. In this strain, alternate identities were found between the capsid regions encoding the shell domain on one side and the P1-1 and P2 domains on the other side. The breakpoint is located near the end of the shell domain (Fig. 5). Interestingly, similarity plot analysis indicated an additional putative breakpoint in the capsid sequence at position 709 (Fig. 3). This breakpoint was, however, not significantly supported by likelihood ratio and phylogenetic analysis. In contrast, conflicting phylogenies across the other identified breakpoint were supported by high bootstrap values (Fig. 4C). Putative parental strains were collected from geographically and chronologically unrelated gastroenteritis outbreaks (Grimsby/1995/UK and Mora/1997/SE).
Within NV genetic cluster II/5, the putative recombinant strain Hillingdon/1990/UK was identified. This strain was detected in 1990 in the United Kingdom. In this strain, one breakpoint was identified by Sawyer's test (nucleotide position 1433, Table 2). It is located near the end of the P1-2 domain (Fig. 5). This strain displays high nucleotide identities to strain White River290/1994/US in the capsid region encoding the shell, P1-1, and P2 domains and high identities to strain MOH/1999/HU in the capsid region encoding the C-terminal half of the P1-2 domain. These conflicting phylogenies across the breakpoint are supported by high bootstrap values at phylogenetically relevant nodes (Fig. 4D). Likelihood ratio analysis identified also a unique breakpoint at position 1201. Phylogenetic analysis of the capsid regions at either side of this breakpoint was not supported by strong bootstrap values (63%). We therefore located the recombinational breakpoint for strain Hillingdon/1990/UK at position 1433, in accordance with phylogenetic analysis results.
We have also assessed the extent of recombination in the NV capsid sequences amplified from clinical samples in Germany. In this set of sequences, Sawyer's test, exploratory tree analysis, and similarity plots identified two putative recombinants and their parental strains collected from different patients in two different years (1997 or 1998). The recombinant strain Hamburg180/1997/GE showed highest nucleotide identity to strain Hamburg189/1997/GE in the region predicted to encode the shell, P2, and P1-2 domains (nucleotides 1 to 519 and 763 to 1617, respectively, Fig. 5). Two breakpoints located at either side of the P1-1 domain (nucleotides 520 to 762) were significantly supported by likelihood ratio analysis (Table 3). NV strain Hamburg180/1997/GE closely resembles strain Bochum024/1998/GE in the region predicted to encode the P1-1 domain. The conflicting phylogenies at either side of the P1-1 domain were supported by high bootstrap values (>85% of 1,000 replications) (Fig. 4E). The same observation was made for strain Hamburg137/1997/GE, where conflicting phylogeny with strains Hamburg139/1997/GE and Hamburg189/1997/GE was observed. The breakpoint at nucleotide position 768 is located at the interface between domains P1-1 and P2 (Fig. 5). Likelihood ratio analysis significantly supported the breakpoint's position (Table 3), and phylogenetic analysis brought strong evidence of recombination (Fig. 4F).
DISCUSSION
In this study, we have examined the extent of recombination in the capsid gene of NV, which has to our knowledge not been reported so far. Our results suggest that homologous recombination in the NV capsid gene is likely to occur.
Recombination of viral RNA is known to depend upon various immunological and intracellular constraints that may allow the emergence of viable recombinants (54). These constraints imply (i) successful coinfection of the host and further on of a single cell by two parental strains, (ii) efficient and simultaneous replication of both parental viral genomes, and (iii) purifying selection that allows only viable recombinants to be transmitted. Recent observations on circulating recombinant NV strains in the community suggest that NV theoretically fulfill all prerequisites for recombination (14, 19, 31).
In this study and in a first step, classification of available NV genogroup I and II strains was performed. This was in accordance with a previous classification based upon the complete sequence of the capsid gene (11). For the detection of recombination and the correct identification of putative recombinants and their parental strains as well as the location of recombinational breakpoints, a stepwise strategy was used. It relies on statistical, phylogenetic, and structural analysis of the sequences. The advantage of this approach is that it combines the power of incompatibility methods, i.e., Sawyer's test (53), with the robustness of phylogenetic analysis (5), assuming the appropriate evolutionary model. Indeed, as shown in simulation and analytical studies of empirical data, no single method is sufficiently reliable by itself to detect recombination (35, 36, 38). In addition, rate heterogeneity among sites may be confounded with recombination, leading sometimes to false positives (35, 36, 38). The combination of various methods under the appropriate model of evolution brings, in case of congruent results, strong evidence of recombination. This strategy has proven to be powerful for the detection of recombination in RNA viruses (5, 12, 50).
With this approach, the putative recombinant strain Aichi124/1989/JP belonging to NV genetic cluster I/1 was identified. In NV genogroup II, seven strains belonging to genetic clusters II/1, II/3, II/4, and II/5 displayed evidence of recombination in the capsid gene. Among NV strains belonging to genetic cluster II/4, two recombinant capsid sequences were amplified from clinical samples collected in Germany during the years 1997 and 1998. In all but two of the examined strains, congruence between the methods strongly supported recombination. In recombinant strains Aichi124/1989/JP and Burwash Landing/1995/US, disagreements among Sawyer's test, similarity plots, and likelihood ratio analysis values were found. Disagreement among the methods is possibly due to rate heterogeneities among sites located at either side of the recombination breakpoint. Because recombination was detected between closely related strains, similarity plots and likelihood ratio analysis may have possibly been misled by rate variations at or around the recombination breakpoints (35, 36, 38). For this reason, identification of recombination breakpoints was also performed by phylogenetic analysis based upon ML trees that were drawn using explicit assumptions on the rate and pattern of nucleotide substitution. ML trees have been widely used in RNA virus recombination studies (5, 12, 50), probably because they are usually consistent and because of their good performance (accuracy and precision) under the appropriate evolution model (8, 29, 32). Indeed, the ML method uses the character state information at all sites (8, 29, 32). It requires, however, explicit assumptions on the rate and pattern of nucleotide substitution as well as the model being used, in order to maximize the likelihood of the best tree given the data. In this study, phylogenetic trees were drawn using the most appropriate models and parameters of evolution given by likelihood ratio test and/or Akaike's information criterion. Accordingly, violation of assumptions did not presumably occur. In this context, phylogenetic incongruence is considered to be a major indicator of recombination. It was used to rule in and out the location of recombination breakpoints in case of disagreement between other methods. The validity of this approach was recently discussed by others (5, 12, 50).
In recombinant capsid sequences, one or two recombination breakpoints were present on average. The breakpoints were distributed between the S domain and the interface between the P1-1 and P2 domains or within the P2 domain. This observation supports the hypothesis of an immune-driven evolution of NV depending upon recombination in the capsid gene. A similar hypothesis was recently proposed, based upon an observation on accumulating mutations in the P2 domain of an NV strain presumably submitted to immune pressure (30). The implications of recombination involving immunogenic viral domains have already been discussed for other RNA viruses (4, 5, 42, 54, 56). The genomic variability in the NV capsid gene, i.e., after recombination, may play a key role in virulence, allowing new recombinants to evade immune response. The capsid gene is predicted to be well suited for detection of recombination in circulating NV strains. This also suggests that phylogenetic classification of NV based upon partial nucleotide and amino acid sequences from the capsid gene is rather questionable. Therefore, in the absence of a cell culture system for virus isolation and serotyping (6) and because of the natural occurrence of recombination in the NV capsid gene, we suggest that classification of NV strains should rely on the complete capsid sequence.
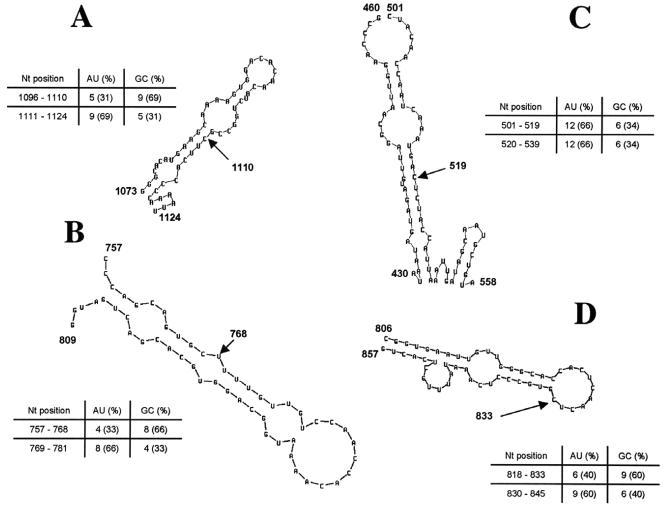
Homologous recombination based upon copy choice and template switching mechanisms has been reported to strongly depend upon nucleotide composition and RNA secondary structure of the recombination region. As proposed by Nagy et al. and based upon homologous recombination models in brome mosaic bromovirus (20, 22, 24), tomato bushy stunt tombusvirus (51, 52), and turnip crinkle carmovirus (27, 28), viral sequences having near-equal G/C and A/U contents or being GC rich (≥60% G/C content) may increase homologous recombination when located upstream of an AU-rich region (≥60% A/U). In addition, comparable lengths of AU- and GC-rich sequences at either side of the breakpoint were reported to efficiently support recombination. Sequences displaying all of these features were termed homologous recombination activators (25). Interestingly, the authors postulated that sequence similarity may not by itself support homologous recombination in brome mosaic bromovirus (23) but may require additional structural features in the recombination region. The presence of a local conformation fluctuation and/or a stable hairpin structure in the AU-rich segment was reported to favor RNA-dependent RNA polymerase docking by facilitating local base pairing between the donor and acceptor RNA strands (28). In this study, the composition and structure of the recombination region (≥50 nucleotides) at either side of identified breakpoints were determined. Sequences located upstream of the recombination breakpoint displayed high GC contents, whereas downstream 3′-end sequences displayed high AU contents (Fig. 6). In one strain (Hamburg180/1997/GE), AU-rich sequences were found at the 5′ and the 3′ ends of the recombination region (Fig. 6C). In all cases, both GC- and AU-rich sequences were similar in length. RNA secondary structure analysis predicted stable hairpin structures at the 3′ end of the recombination region in most strains. Thus, most of the recombinant strains displayed characteristics of homologous recombination activators. Representative examples are shown in Fig. 6.
FIG. 6.
Characterization of sequence composition and RNA secondary structure of the putative recombination region. Breakpoint position is highlighted and indicated by an arrow. Nucleotide composition at either side of the putative breakpoint is shown. (A) Strain Toronto24/1991/CA, nucleotides 1073 to 1124. (B) Strain Hamburg137/1997/GE, nucleotides 757 to 809. (C) Strain Hamburg180/1997/GE, nucleotides 430 to 460 and 501 to 558. (D) Strain Port Canaveral301/1994/US, nucleotides 806 to 857.
All matters considered, our study suggests that recombination in NV may naturally occur. Indeed, 7.5% (7 of 94) of the sequences examined displayed recombination patterns. In most cases, the accordance of various methods used to assess recombination (Sawyer's test, similarity plots, split-tree analysis [data not shown], likelihood ratio analysis, and structural analysis) strongly indicates that recombination has occurred in the NV capsid gene. However, the extent and rate of recombination in the NV genome remain unclear. There is increasing evidence that recombination in NV may not be limited to the capsid gene, as observed in recombinant strain Arg320/1995/AR. This strain displays conflicting identities and phylogenies in the polymerase gene (ORF1) and capsid gene (ORF2) to parental strains Lorsdale/1993/UK and Mexico/1989/MX, respectively (14). In this strain, recombination was postulated to be located at the ORF1-ORF2 interface. It was supported by similarity plots and phylogenetic incongruence. However, no recombination breakpoint has been so far precisely identified. Likelihood ratio analysis located the recombination breakpoint at nucleotide position 26 from the 5′ end of the capsid gene (likelihood ratio = 53.59, P < 0.0001). Again, RNA secondary structure of the recombination region displayed nucleotide composition as well as hairpin motifs characteristic of homologous recombination activators (data not shown). This strongly suggests that recombination in the NV genome is a probable event.
In this study, no recombination was identified between NV genogroups I and II, although coinfections with strains belonging to both genogroups may theoretically occur, as observed with other gastroenteritis viruses (3, 31). The reason for this may presumably be an increase in immunological and intracellular constraints with the divergence of parental strains, favoring recombination in similar strains in comparison to more divergent ones (54). Furthermore, recombination was detected in four different genetic clusters of NV genogroup II but in only one cluster of NV genogroup I. This discrepancy is possibly due to sampling bias, as a larger number of capsid sequences from NV genogroup II strains was analyzed. It may also be a consequence of the higher frequency of NV genogroup II strains causing gastroenteritis outbreaks, in particular strains belonging to NV genetic cluster II/4. This is supported by a recent observation suggesting that density of geographical and temporal sampling increases the probability of identifying recombinant sequences (50). NV genogroup II strains, and in particular those belonging to genetic cluster II/4, are recognized to be responsible for about 80% of human calicivirus outbreaks (7, 18). It remains unclear, however, whether the frequency of recombination seen in this genogroup is driven by an increasing virulence of NV strains, or vice versa. It is possible that the higher frequency of NV genogroup II strains detected in different countries depends upon biological factors related to NV tropism, replication, and pathogenicity that still have to be elucidated.
Acknowledgments
We are grateful to Wolfram Rudolph and Donna Rowe for critical reading of the manuscript. We thank the RoMoD study group (J. Henker, J. Forster, and G. Petersen) for providing us with clinical samples.
This work was supported by a start-up grant of the Medical Faculty Carl Gustav Carus of Dresden, Germany (MeDDrive 2001) and the Jürgen-Manchot-Stiftung.
REFERENCES
- 1.Aaziz, R., and M. Tepfer. 1999. Recombination in RNA viruses and in virus-resistant transgenic plants. J. Gen. Virol. 80:1339-1346. [DOI] [PubMed] [Google Scholar]
- 2.Ando, T., S. S. Monroe, J. S. Noel, and R. I. Glass. 1997. A one-tube method of reverse transcription-PCR to efficiently amplify a 3-kilobase region from the RNA polymerase gene to the poly(A) tail of small round-structured viruses (Norwalk-like viruses). J. Clin. Microbiol. 35:570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, D. S. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg. Infect. Dis. 3:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chare, E. R., E. A. Gould, and E. C. Holmes. 2003. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 84:2691-2703. [DOI] [PubMed] [Google Scholar]
- 6.Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79-87. [DOI] [PubMed] [Google Scholar]
- 7.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 8.Graur, D., and W.-H. Li. 2000. Fundamentals of molecular evolution. Sinauer Associates, Sunderland, Mass.
- 9.Green, J., J. Vinje, C. I. Gallimore, M. Koopmans, A. Hale, D. W. Brown, J. C. Clegg, and J. Chamberlain. 2000. Capsid protein diversity among Norwalk-like viruses. Virus Genes 20:227-236. [DOI] [PubMed] [Google Scholar]
- 10.Green, K. Y., G. Belliot, J. L. Taylor, J. Valdesuso, J. F. Lew, A. Z. Kapikian, and F. Y. Lin. 2002. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 185:133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. .In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Holmes, E. C., M. Worobey, and A. Rambaut. 1999. Phylogenetic evidence for recombination in dengue virus. Mol. Biol. Evol. 16:405-409. [DOI] [PubMed] [Google Scholar]
- 13.International Committee on the Taxonomy of Viruses. 2002, posting date. ICTVdB; the universal virus database of the International Committee on the Taxonomy of Viruses. [Online.] http://www.ncbi.nlm.nih.gov/ICTVdb.
- 14.Iritani, N., Y. Seto, H. Kubo, T. Murakami, K. Haruki, M. Ayata, and H. Ogura. 2003. Prevalence of Norwalk-like virus infections in cases of viral gastroenteritis among children in Osaka City, Japan. J. Clin. Microbiol. 41:1756-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall, J. A., S. Salamone, L. Yuen, M. G. Catton, and J. P. Wright. 2001. High level excretion of Norwalk-like virus following resolution of clinical illness. Pathology 33:50-52. [PubMed] [Google Scholar]
- 20.Nagy, P. D., and J. J. Bujarski. 1995. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 69:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy, P. D., and J. J. Bujarski. 1997. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 71:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy, P. D., and J. J. Bujarski. 1992. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 66:6824-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy, P. D., and J. J. Bujarski. 1996. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 70:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy, P. D., and J. J. Bujarski. 1993. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc. Natl. Acad. Sci. USA 90:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy, P. D., C. Ogiela, and J. J. Bujarski. 1999. Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots. Virology 254:92-104. [DOI] [PubMed] [Google Scholar]
- 26.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, P. D., and A. E. Simon. 1998. In vitro characterization of late steps of RNA recombination in turnip crinkle virus. I. Role of motif1-hairpin structure. Virology 249:379-392. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, P. D., C. Zhang, and A. E. Simon. 1998. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 17:2392-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom.
- 30.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh, D. Y., G. Gaedicke, and E. Schreier. 2003. Viral agents of acute gastroenteritis in German children: prevalence and molecular diversity. J. Med. Virol. 71:82-93. [DOI] [PubMed] [Google Scholar]
- 32.Page, R. D. M., and E. C. Holmes. 1998. Molecular evolution: a phylogenetic approach. Blackwell Science, Oxford, United Kingdom.
- 33.Parashar, U. D., and S. S. Monroe. 2001. ‘Norwalk-like viruses’ as a cause of foodborne disease outbreaks. Rev. Med. Virol. 11:243-252. [DOI] [PubMed] [Google Scholar]
- 34.Poppe, M., B. Ehlken, A. Rohwedder, S. Lugauer, H. D. Frank, K. Stehr, C. H. Rieger, G. Petersen, G. Lorkowski, W. Karmaus, H. Werchau, J. Henker, J. Forster, and the RoMoD-Study Group. 2002. Epidemiologie und Klinik von Rotavirus-Gastroenteritiden bei hospitalisierten Säuglingen und Kleinkindern in Deutschland. Monatsschr. Kinderheilkd. 150:491-496. [Google Scholar]
- 35.Posada, D. 2002. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol. Biol. Evol. 19:708-717. [DOI] [PubMed] [Google Scholar]
- 36.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 38.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 39.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 40.Prasad, B. V., R. Rothnagel, X. Jiang, and M. K. Estes. 1994. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 68:5117-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut, A., and N. C. Grassly. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13:235-238. [DOI] [PubMed] [Google Scholar]
- 42.Robertson, D. L., P. M. Sharp, F. E. McCutchan, and B. H. Hahn. 1995. Recombination in HIV-1. Nature 374:124-126. [DOI] [PubMed] [Google Scholar]
- 43.Rohayem, J., S. Berger, T. Juretzek, O. Herchenroder, M. Mogel, M. Poppe, J. Henker, and A. Rethwilm. 2004. A simple and rapid single-step multiplex RT-PCR to detect norovirus, astrovirus and adenovirus in clinical stool samples. J. Virol. Methods 118:49-59. [DOI] [PubMed] [Google Scholar]
- 44.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawyer, S. 1999, posting date. GENECONV: a computer package for the statistical detection of gene conversion. [Online.] http://www.math.wustl.edu/sawyer.
- 46.Sawyer, S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 47.Swofford, D. L. 2002. PAUP: phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 48.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twiddy, S. S., and E. C. Holmes. 2003. The extent of homologous recombination in members of the genus Flavivirus. J. Gen. Virol. 84:429-440. [DOI] [PubMed] [Google Scholar]
- 51.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, K. A., and T. J. Morris. 1994. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl. Acad. Sci. USA 91:3642-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiuf, C., T. Christensen, and J. Hein. 2001. A simulation study of the reliability of recombination detection methods. Mol. Biol. Evol. 18:1929-1939. [DOI] [PubMed] [Google Scholar]
- 54.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 55.Worobey, M., and E. C. Holmes. 2001. Homologous recombination in GB virus C/hepatitis G virus. Mol. Biol. Evol. 18:254-261. [DOI] [PubMed] [Google Scholar]
- 56.Worobey, M., A. Rambaut, and E. C. Holmes. 1999. Widespread intra-serotype recombination in natural populations of dengue virus. Proc. Natl. Acad. Sci. USA 96:7352-7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]