Abstract
The papillomavirus transcriptional activator, E2, is involved in key functions of the viral life cycle. These include transcriptional regulation, viral DNA replication, and viral genome segregation. The transactivation domain of E2 is required for each of these functions. To identify the regions of the domain that mediate binding to mitotic chromosomes, a panel of mutations has been generated and their effect on various E2 functions has been analyzed. A structural model of the bovine papillomavirus type 1 (BPV1) E2 transactivation domain was generated based on its homology with the solved structure of the human papillomavirus type 16 (HPV16) domain. This model was used to identify distinct surfaces of the domain to be targeted by point mutation to further delineate the functional region of the transactivation domain responsible for mitotic chromosome association. The mutated E2 proteins were assessed for mitotic chromosome binding and, in addition, transcriptional activation and transcriptional repression activities. Mutation of amino acids R37 and I73, which are located on a surface of the domain that in HPV16 E2 is reported to mediate self-interaction, completely eliminated mitotic chromosome binding. Mitotic chromosome binding activity was found to correlate well with the ability to interact with the cellular chromosomal associated factor Brd4, which has recently been proposed to mediate the association between BPV1 E2 and mitotic chromosomes.
Papillomaviruses are small DNA viruses that infect the squamous epithelium of the skin and mucosa, causing benign tumors or papillomas (22). A key step in the viral life cycle occurs upon infection of basal epithelial cells, when the viral genome must be maintained as an extrachromosomal element to ensure persistence of the infection in dividing cells. Transient viral DNA replication requires the E2 transcriptional transactivator, the E1 helicase protein, and the origin of replication (54, 55, 57, 61). Long-term, persistent replication requires, in addition, numerous high-affinity E2 binding sites in cis with the replication origin (47). The E2 transcriptional transactivator protein maintains the viral genomes as multicopy extrachromosomal elements by tethering them to mitotic chromosomes to ensure their nuclear retention and segregation to daughter cells (24, 32, 51).
The E2 transactivator protein can be divided into three domains. The amino-terminal ∼200 amino acids comprise the transactivation domain of E2, and the carboxy-terminal ∼100 amino acids mediate DNA binding and dimerization. These domains are linked by a flexible region called the hinge. The transactivation domain and DNA binding domain are conserved among E2 proteins of different papillomaviruses, whereas the length and amino acid sequence of the hinge region vary greatly (reviewed in reference 42). The transactivation domain is critical for the segregation, DNA replication, and transcriptional regulatory functions of the E2 protein (5, 41, 51, 58).
Several groups have mapped residues important for the transactivation and replication functions of the E2 N-terminal domain by site-directed mutational analysis (3, 8-10, 15, 49). Most of these studies targeted amino acid residues that are conserved among all papillomavirus E2 proteins and were performed prior to the solution of the human papillomavirus type 16 (HPV16) X-ray crystal structure (4). In most cases, individual residues were either replaced conservatively or mutated to alanine. Several residues important for the transactivation and replication functions were identified, but the studies yielded little delineation of functional regions within the transactivation domain. Two more recent studies have used these series of mutated E2 proteins to examine the role of specific residues in the association of E2 with mitotic chromosomes (2, 63). A recent study from our laboratory examined the chromosome binding function of E2 proteins mutated in the highly conserved residues of the bovine papillomavirus type 1 (BPV1) E2 transactivation domain (63). However, it was found that mutations that eliminated chromosome binding did so by disrupting conformation of the domain, sometimes in a temperature-sensitive manner. Abroi et al. also extended an earlier analysis of point mutations within the transactivation domain to examine effects on chromatin attachment of E2 and tethering of E2-binding site containing plasmids to mitotic chromosomes (2). These authors were able to identify several single-amino-acid substitutions that eliminated the ability of the E2 protein to bind mitotic chromosomes.
Here, a different approach was used for a mutational analysis of the transactivation domain. The primary aim was to determine the regions of the domain that are involved in the process of mitotic chromosome binding and to provide a better delineation of the functional regions within the domain. To do this, the structure of the bovine papillomavirus type 1 (BPV1) E2 transactivation domain was modeled based on its homology with the HPV16 domain and used to identify regions on the surface of the domain that have potential for interaction with other proteins. These regions were targeted by mutation, and the resulting mutated proteins were tested for mitotic chromosome binding activity and transcriptional regulatory functions. In addition, the series of mutated E2 proteins was used to determine whether their interaction with the chromosomal associated factor Brd4 (recently identified as one of the proteins responsible for tethering E2 to chromosomes) (62) correlated with the chromosome binding ability of the E2 proteins.
MATERIALS AND METHODS
Plasmids.
The parent plasmid for generation of site-specific E2 transactivation domain mutations, pTZE2kz, has been described previously (40). Site-directed mutagenesis was carried out with this plasmid, and the mutated E2 genes were excised by HindIII/BamHI digestion and cloned into the episomal expression vector pMEP4 (Invitrogen) for inducible expression in mammalian cells. The E2-responsive luciferase reporter plasmid pBS1073 contains the firefly luciferase gene downstream of four E2 binding sites and the herpes simplex virus type 1 thymidine kinase promoter (a gift from B. Sugden) (21).
Site-directed mutagenesis.
All mutated E2 genes, with the exception of the mutations in the acidic amphipathic helix, were generated by using Transformer site-directed mutagenesis (Clontech). Mutagenic primers were designed to mutate residues D24, K25, and D28; Q12, Q15, E39, and R68; R47, K48, K49, R58, and H61; R37, K70, I73, E74, Q76, and Q80; E86, D89, and E90; R172, D175, E176, and R179; and R189, D190, D192, and R193. Partially mutated targets were generated during the cloning process and were also further analyzed. The selection primer used for the Transformer mutagenesis converted a unique XmnI site within the beta-lactamase gene of pTZE2kz to an EcoRV site. pTZE2kz plasmids encoding the E2 gene with a mutated acidic amphipathic helix were generated by double-stranded oligonucleotide reconstruction of the 5′ end of the E2 gene from the HindIII site upstream of the start codon to the unique internal AvrII site, introducing mutations into the codons for amino acids E2, E6, E13, and E20. The minimal number of base changes was made for each mutated codon.
Cell culture.
CV-1 and HeLa cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and were grown in a 10% CO2 incubator. Sf9 insect cells were maintained as a suspension culture in Sf900 II serum-free medium at 27°C.
Establishment of stable pMEP cell lines.
CV-1-derived lines were generated by transfection with the appropriate pMEP4 or pMEP-E2 plasmid. Cells were plated at a density of 3 ×105 cells per 10-cm-diameter dish. The next day, each dish was transfected with 6 μg of pMEP4 or with 6 μg of wild-type or mutated pMEP-E2 plasmid, using 30 μl of Superfect transfection reagent (QIAGEN) per dish. One day posttransfection, cells were selected with 200 μg of hygromycin B (Roche)/ml. After selection was complete, colonies were pooled and cultures were expanded.
E2 protein expression.
CV-1-derived pMEP-E2 cell lines were plated onto 10-cm-diameter dishes. Expression from the metallothionein promoter was induced by the addition of 0.5 to 1.25 μM CdSO4 for 3 h. Cellular proteins were extracted in modified radioimmunoprecipitation assay buffer (20 mM HEPES [pH 7.0], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing Complete (Roche) protease inhibitor cocktail and Staphylococcus aureus protein A (Pansorbin; Calbiochem) to preclear. Total protein concentrations in the extracts were determined using a bicinchoninic acid protein assay kit (Pierce).
Western blotting.
For each sample, 5 μg of protein was heated in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer at 100°C for 5 min. Samples were separated by SDS-PAGE on a 10% polyacrylamide gel and electrotransferred onto Immobilon-P polyvinylidene fluoride membranes (Millipore). Western blots were performed by following standard protocols with the anti-E2 antibody B201 (provided by Elliot Androphy) followed by horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Pierce). E2 protein bands were detected by using the chemiluminescence reagent SuperSignal West Dura (Pierce).
Immunofluorescence.
CV-1-derived pMEP-E2 cell lines were plated onto glass slides and blocked in S phase by the addition of 2 mM thymidine for 14 to 16 h. The thymidine block was released, and cells were cultured for 9 h. Expression from the metallothionein promoter was induced by the addition of 1 μM CdSO4 for the last 3 h. Cells were quickly fixed for 20 min in 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.1% Triton X-100 in PBS. E2 was detected with monoclonal antibody B201 (1:10 dilution; provided by Elliot Androphy) and goat anti-mouse immunoglobulin G conjugated to fluorescein isothiocyanate (FITC) (1:100 dilution; Jackson Immunochemicals). Slides were mounted in Vectashield mounting medium containing 20 μg of propidium iodide/ml. Immunofluorescent staining was detected, and digital images were captured with a Leica TCS-SP2 laser scanning confocal imaging system. At least 50 mitotic cells were assessed for E2 chromosomal binding.
Transactivation assay.
Transactivation assays were performed in one of two ways. (i) pMEP-E2 cell lines were plated onto 6-cm-diameter dishes at a density of 2.5 × 105/dish. One day later, the cells were transfected with 2 μg of luciferase reporter plasmid pBS1073 by using 6 μl of Fugene (Roche) transfection reagent/dish. Cells were harvested about 48 h posttransfection. After washing with PBS, cells were lysed with 1 ml of 1× cell culture lysis reagent (Promega)/dish. Cell extracts were assayed for firefly luciferase activity with Promega's luciferase assay system. Luciferase activity was measured in a Zylux Femtomaster FB12 luminometer, and measurements were carried out in triplicate for each sample. The luciferase activity, measured as relative light units per second, was normalized to the protein concentration of the lysate and is expressed relative to the activity of wild-type E2, which was set at 100% activity. (ii) CV-1 cells (2.5 × 105/dish) were plated onto 6-cm-diameter dishes. One day later, the cells were cotransfected with 100 ng of wild-type or mutated pMEP-E2 expression plasmid and 2 μg of pBS1073 by using 6 μl of Fugene transfection reagent per dish. Cells were harvested at 48 h posttransfection and were processed as described above.
HeLa cell growth suppression assay.
HeLa cells were plated at a density of 3 × 105 cells per 10-cm-diameter dish. The next day, each dish was transfected with 6 μg of pMEP4 or with 6 μg of wild-type or mutated pMEP-E2 plasmid by using 30 μl of Superfect transfection reagent/dish. One day posttransfection, cells were split 1:9. One day later, selection was begun with 200 μg of hygromycin B/ml and E2 expression was induced with 0.1 μM CdSO4. Cultures were grown until the pMEP4 control plate approached confluence. Cells were fixed with formalin and stained with methylene blue.
E2-Brd4 coimmunoprecipitation.
Wild-type and mutated E2 proteins were expressed in vitro from pTZE2 plasmids by using Promega's TNT T7 quick-coupled reticulocyte lysate system. Parallel [35S]methionine-labeled and unlabeled reactions were performed, and the amount of E2 in each reaction mixture was normalized to wild-type E2 by using the labeled reaction mixtures. [35S]methionine-labeled murine Brd4 was expressed in vitro from pBSK-Brd4 (38) by using Promega's TNT T3 coupled reticulocyte lysate system. Equivalent amounts of unlabeled E2 reaction products were mixed with Brd4 reaction product and incubated at room temperature for 1 h with mixing. Protein A-Sepharose beads containing prebound anti-E2 antibody B202 (provided by Elliot Androphy), suspended in PBS-10% glycerol-0.1% Triton X-100, were added to the mixture and incubated for 30 min with mixing. Immune complexes were washed three times with PBS-10% glycerol-0.1% Triton X-100. Proteins were eluted in SDS-PAGE sample buffer, boiled for 5 min, and separated by SDS-PAGE on 7.5% polyacrylamide gels. Gels were fixed and dried, and Brd4 protein bands were detected and quantitated by using a Typhoon phosphorimager with ImageQuant software (Molecular Dynamics). Brd4 bands were also detected by autoradiography.
Brd4-E2 coimmunoprecipitation.
FLAG-tagged Brd4 was expressed in Sf9 insect cells from a previously described recombinant baculovirus (38). Three days postinfection, cells were harvested. Soluble proteins were extracted by three cycles of freeze-thawing in extraction buffer (20 mM Tris [pH 8.0], 10% glycerol, 0.5 M KCl, 0.2 mM EDTA, 0.1% Tween 20, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 1× Complete EDTA-free protease inhibitor cocktail [Roche]), followed by ultracentrifugation (35,000 rpm in an SW41 rotor for 30 min at 4°C) and recovery of the supernatant. Five microliters of protein extract was mixed with equivalent amounts of [35S]methionine-labeled in vitro-translated E2 protein (wild type or mutated, expressed as described above) in 100 μl of PBS-10% glycerol-0.1% Triton X-100 containing 1× Complete EDTA-free protease inhibitor cocktail and incubated at room temperature for 1 h. Anti-FLAG M2-agarose affinity gel (Sigma), suspended in PBS-10% glycerol-0.1% Triton X-100 with 1× Complete EDTA-free protease inhibitor cocktail, was added to the mixture and incubated for 1 h at room temperature with mixing. Immune complexes were washed three times with PBS-10% glycerol-0.1% Triton X-100. Proteins were eluted in SDS-PAGE sample buffer and separated by SDS-PAGE. E2 protein bands were detected and quantitated by using a Typhoon phosphorimager with ImageQuant software (Molecular Dynamics) and by autoradiography.
RESULTS
Modeling of BPV1 E2 transactivation domain and identification of targets for mutation.
The sequence of the transactivation domain of the BPV1 E2 protein has 34% identity with that of the HPV16 E2 protein. This similarity was used to model the structure of the BPV1 transactivation domain against the X-ray crystal structure of the HPV16 domain (4) with the programs Look and RasMol. The BPV E2 model was scanned for patches of surface residues that could be targeted by mutation. The rationale was to primarily target clusters of charged residues for mutation with the intention of disrupting protein-protein interactions. It has been shown that interactive surfaces are enriched for arginine and aspartate residues (7). Additionally, mutation of surface residues is less likely to cause global impacts on the protein structure. Eight potential regions of protein-protein interaction were identified on the E2 domain. Residues within these regions were replaced either conservatively with similar residues or with alanines, to minimize potential structural effects of each type of mutation. Each mutated protein sequence was analyzed with the program Protean from the DNASTAR sequence analysis software suite to ensure that it resulted in minimal disruption of secondary structure. Five of the charged clusters were designated groups B, D, G, H, and I. Three additional regions that were targeted have been implicated in various functions of E2. In group A, an acidic amphipathic alpha-helix at the amino terminus of the domain was mutated. These acidic residues are highly conserved, and it has previously been proposed that the amphipathic helical structure would be important for transcriptional activation (18, 31). For target group F, the face of the transactivation domain containing arginine 37 and isoleucine 73 was mutated. These residues have been implicated in the transactivation function of E2 (15, 49, 60) and lie along the putative self-interactive surface of the HPV16 transactivation domain observed in the crystal structure (4). Finally, group C surrounds glutamate 39, which has been found by some to be required for the initiation of DNA replication (3, 9, 49) and, in the HPV16 protein, is required for interaction with the viral E1 helicase (44). In several groups, various subsets of these mutations were also generated. The eight groups are shown on a model of the BPV1 transactivation domain in Fig. 1, and the list of specific mutated amino acids is indicated in Table 1.
FIG. 1.
Structural model of BPV1 E2 transactivation domain. Shown are opposing views of a structural representation of the BPV1 E2 transactivation domain. The BPV transactivation domain (amino acids 1 to 203) was modeled on the structure of the HPV16 transactivation domain by using the programs Look and RasMol. The model is displayed in space-filling mode, with the eight mutation target groups (A to I) indicated in different colors. The locations of the amino (N) and carboxyl (C) termini of the transactivation domain are indicated in both views of the structural model.
TABLE 1.
Mutated amino acids
| Group (description) | E2 Protein | Mutation(s)a |
|---|---|---|
| A (acidic amphipathic alpha-helix) | A1 | E2A, E6A, E13A, E20A |
| A2 | E2Q, E6Q, E13Q, E20Q | |
| B (charge cluster) | B1 | D28N |
| B2 | D24N, K25Q, D28N | |
| B3 | D24A, K25A, D28A | |
| C (replication/E1 interaction) | C1 | R68A |
| C2 | E39A | |
| C3 | E39A, R68A | |
| C4 | Q12A, Q15A, E39A | |
| C5 | Q12A, Q15A, E39A, R68A | |
| C6 | R68Q | |
| C7 | E39Q | |
| C8 | E39Q, R68Q | |
| C9 | Q12N, R68Q | |
| C10 | Q12N, Q15N, E39Q | |
| C11 | Q12N, Q15N, E39Q, R68Q | |
| D (basic cluster) | D1 | R47A, K48A, K49A, R58A, H61A |
| D2 | R47Q, K48Q, K49Q, R58Q, H61N | |
| F (transactivation/self-interaction) | F1 | R37A |
| F2 | I73A | |
| F3 | R37A, I73A | |
| F4 | R37A, K70A, E74A, Q76A, Q80A | |
| F5 | R37A, K70A, I73A, E74A, Q76A, Q80A | |
| F6 | R37Q, K70Q, E74Q, Q76A, Q80A | |
| G (acidic cluster) | G1 | D89A, E90A |
| G2 | E86A, D89A, E90A | |
| G3 | E86Q, D89N, E90Q | |
| H (charge cluster) | H1 | R172A, D175A, E176A |
| H2 | R172A, D175A, E176A, R179A | |
| H3 | R172Q, D175N, E176Q, R179Q | |
| I (charge cluster) | I1 | R189A, D190A, D192A, R193A |
| I2 | R189Q, D190N, D192N, R193Q |
Mutations are listed as wild-type E2 amino acid, residue number, mutated amino acid.
Expression of mutated E2 proteins.
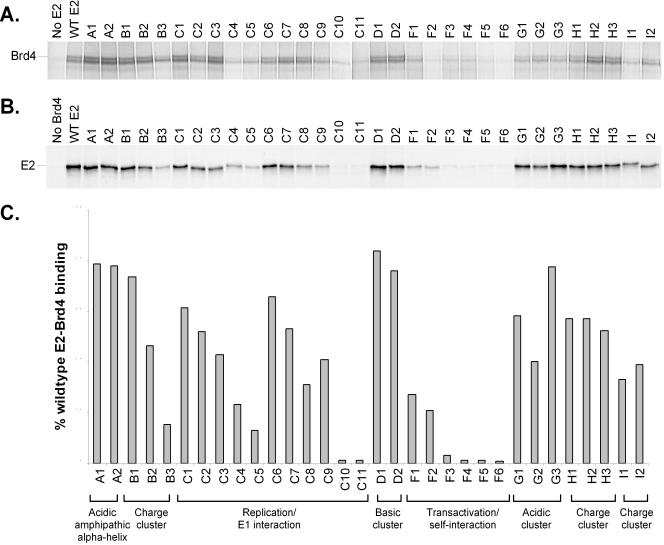
Stably transfected CV-1 cell lines containing the series of pMEP-E2 plasmids were established and tested for expression and stability of the mutated E2 proteins. E2 expression was induced from the metallothionein promoter with 0.5 μM CdSO4 for 3 h. The levels of the various E2 proteins were analyzed in cell lysates by Western blotting with the anti-E2 monoclonal antibody B201. As shown in Fig. 2, all of the cell lines expressed full-length E2 proteins. The only cell line that consistently produced lower steady-state expression levels of E2 than the others was G2 (E86A, D89A, and E90A). Notably, this protein also had growth-inhibitory properties. Cells expressing this protein grew much slower than the other cell lines.
FIG. 2.
Expression of mutated E2 proteins. Shown is a Western blot of protein extracts from CV-1 cell lines stably transfected with pMEP4 (No E2), wild-type pMEP-E2 (WT E2), or mutated pMEP-E2 plasmids (A1 to I2) (specific mutations are listed in Table 1). E2 proteins were detected with the anti-E2 monoclonal antibody B201.
Identification of a region of E2 required for mitotic chromosome binding.
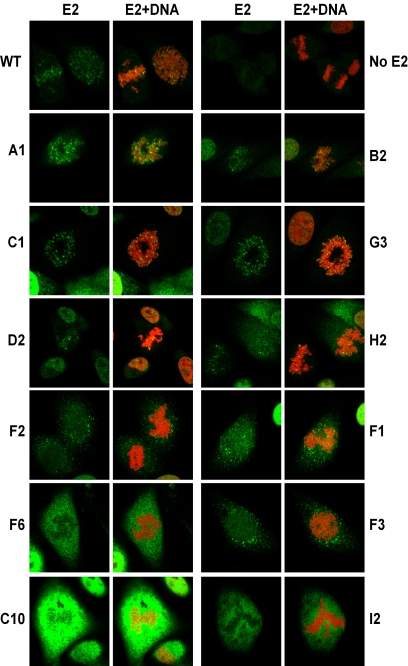
After ensuring that all mutated E2 proteins were expressed, their intracellular localization in mitotic cells was examined by indirect immunofluorescence with the B201 antibody. Wild-type E2 shows a characteristic speckled pattern of staining that colocalizes with mitotic chromosomes (Fig. 3). For the mutated proteins, E2 expression patterns observed ranged from E2 protein found exclusively bound to mitotic chromosomes, E2 protein found both colocalized with chromosomes and in the cytosol, E2 protein completely excluded from chromosomes and localized in the cytosol, and finally, no E2 expression. To quantitate chromosome binding for each of the mutated proteins, approximately 50 mitotic cells from each cell line were assessed. Selected images of individual mitotic cells expressing the mutated E2 proteins are shown in Fig. 3, and all of the data are summarized in Table 2. The percentage of mitotic cells containing E2 protein on mitotic chromosomes was calculated as 85% for wild-type E2. Mutations in most of the target groups did not completely disrupt E2 chromosome binding. However, no chromosomal association could be detected for proteins in group I, most of the proteins in group F, several in group C, and the protein B3.
FIG.3.
Mitotic chromosome binding of mutated E2 proteins. E2 localization was analyzed in stably transfected CV-1 cell lines by confocal fluorescence microscopy. Shown are representative mitotic cells for the indicated samples (mutations are listed in Table 1). The sample without E2 (No E2) was transfected with the pMEP4 vector. E2 was detected by indirect immunofluorescence with an FITC-conjugated secondary antibody and is shown in green (E2), while the merged image shows E2 staining plus cellular DNA counterstained in red with propidium iodide (E2+DNA). The images are ordered from those that bind chromosomes at the top to those that don't at the bottom. WT, wild type
TABLE 2.
Summary of mitotic chromosome binding datab
| E2a | No. of cells with:
|
Total no. of cells | % | |||
|---|---|---|---|---|---|---|
| ON | Both | OFF | No E2 | |||
| WT | 38 | 9 | 8 | 3 | 58 | 85 |
| A1 | 36 | 5 | 1 | 0 | 42 | 98 |
| A2 | 40 | 0 | 0 | 2 | 42 | 100 |
| B1 | 19 | 1 | 6 | 6 | 32 | 77 |
| B2 | 38 | 1 | 7 | 9 | 55 | 85 |
| B3 | 0 | 0 | 47 | 0 | 47 | 0 |
| C1 | 35 | 8 | 9 | 2 | 54 | 83 |
| C2 | 26 | 14 | 9 | 1 | 50 | 82 |
| C3 | 29 | 5 | 16 | 7 | 57 | 68 |
| C4 | 2 | 2 | 35 | 5 | 44 | 10 |
| C5 | 2 | 0 | 26 | 8 | 36 | 7 |
| C6 | 20 | 18 | 6 | 2 | 46 | 86 |
| C7 | 27 | 8 | 5 | 0 | 40 | 88 |
| C8 | 25 | 9 | 6 | 0 | 40 | 85 |
| C9 | 0 | 0 | 30 | 16 | 46 | 0 |
| C10 | 0 | 0 | 35 | 0 | 35 | 0 |
| C11 | 0 | 0 | 30 | 0 | 30 | 0 |
| D1 | 15 | 13 | 14 | 0 | 42 | 67 |
| D2 | 16 | 10 | 21 | 0 | 47 | 55 |
| F1 | 1 | 15 | 17 | 2 | 35 | 48 |
| F2 | 0 | 3 | 31 | 5 | 39 | 9 |
| F3 | 0 | 0 | 33 | 7 | 40 | 0 |
| F4 | 0 | 0 | 38 | 8 | 46 | 0 |
| F5 | 0 | 0 | 30 | 15 | 45 | 0 |
| F6 | 0 | 0 | 38 | 6 | 44 | 0 |
| G1 | 35 | 10 | 8 | 4 | 57 | 85 |
| G2 | 6 | 0 | 0 | 7 | 13 | 100 |
| G3 | 35 | 9 | 2 | 1 | 47 | 96 |
| H1 | 16 | 9 | 18 | 4 | 47 | 58 |
| H2 | 25 | 13 | 11 | 0 | 49 | 78 |
| H3 | 18 | 23 | 5 | 2 | 48 | 89 |
| I1 | 0 | 0 | 31 | 12 | 43 | 0 |
| I2 | 0 | 0 | 36 | 13 | 49 | 0 |
Refer to Table 1 for mutations. WT, wild type.
Mitotic chromosome binding activity is expressed as ability to bind to mitotic chromosomes. ON, exclusive colocalization with mitotic chromosomes; Both, E2 protein localized both on and off mitotic chromosomes; OFF, exclusion from mitotic chromosomes; No E2, no E2 expression; %, percentage of mitotic cells with E2 bound to mitotic chromosome (ON and Both).
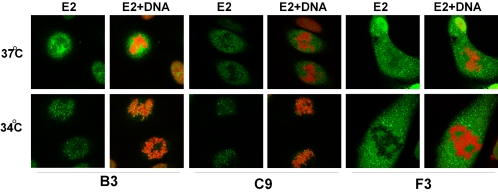
We have previously found that many point mutations in the E2 transactivation domain give rise to a conditional phenotype for mitotic chromosome binding (63). E2 proteins that are slightly misfolded at 37°C are unable to bind to mitotic chromosomes. However, incubation at 34°C, hypotonic swelling, or treatment of E2-expressing cells with agents that promote protein folding results in association of these proteins with mitotic chromosomes (63). Therefore, the E2 proteins described above, which were defective for chromosome binding, were analyzed for their ability to bind mitotic chromosomes in cells cultured at 34°C. As shown in Table 3, E2 proteins B3, C4, C5, C9, I1, and I2 could be rescued for their ability to associate with mitotic chromosomes (Fig. 4, B3 and C9). Even the percentage of mitotic cells with wild-type E2 protein bound to mitotic chromosomes increased from 85 to 100% at 34°C. Notably, temperature-sensitive E2 proteins that had been cultured at 37°C could also be induced to associate with chromosomes by washing cells in PBS before fixation or even by allowing cells to sit in the culture dish for several minutes at room temperature before fixation. Therefore, an accurate assessment of chromosome binding activity could only be made by direct and rapid fixation of cultured E2 cells in paraformaldehyde.
TABLE 3.
Summary of data from binding, transactivation, repression, and replication experiments
| E2 protein | % Mitotic cells with chromosome binding at:
|
BRD4 binding in vitro (% of wild type) | Transactivation (% of wild type) ata:
|
HeLa repression atb:
|
DNA replicationc | E1 binding in vitroc | |||
|---|---|---|---|---|---|---|---|---|---|
| 37°C | 34°C | 37°C | 34°C | 37°C | 34°C | ||||
| Wild type | 85 | 100 | 100 | 100 | 100 | +++ | +++ | +++ | +++ |
| A1 | 98 | 79 | 79 | 72 | +++ | +++ | ± | + | |
| A2 | 100 | 78 | 88 | 81 | +++ | +++ | ± | + | |
| B1 | 77 | 74 | 34 | 64 | ± | ++ | + | +++ | |
| B2 | 85 | 46 | 36 | 59 | ± | ++ | ++ | +++ | |
| B3 | 0 | 88 | 15 | 8 | 10 | ± | ± | − | +++ |
| C1 | 83 | 61 | 86 | 106 | ++ | +++ | + | +++ | |
| C2 | 82 | 52 | 81 | 93 | ++ | ND | ++ | +++ | |
| C3 | 68 | 43 | 36 | 68 | ± | ++ | ++ | +++ | |
| C4 | 10 | 78 | 23 | 62 | 99 | + | +++ | − | ++ |
| C5 | 7 | 19 | 13 | 18 | 49 | ± | ++ | − | + |
| C6 | 86 | 66 | 84 | 102 | +++ | +++ | ± | +++ | |
| C7 | 88 | 53 | 125 | 111 | +++ | +++ | ++ | +++ | |
| C8 | 85 | 31 | 50 | 87 | ± | ++ | + | +++ | |
| C9 | 0 | 36 | 41 | 8 | 15 | ± | ± | − | ++ |
| C10 | 0 | 0 | 1 | 7 | 7 | + | + | − | ± |
| C11 | 0 | 0 | 1 | 8 | 6 | ± | ± | − | ± |
| D1 | 67 | 84 | 120 | 121 | +++ | +++ | ± | ++ | |
| D2 | 55 | 76 | 99 | 104 | +++ | +++ | + | ++ | |
| E1 | 48 | 46 | 27 | 64 | 61 | + | + | + | +++ |
| F2 | 9 | 8 | 21 | 41 | 40 | + | + | ++ | +++ |
| F3 | 0 | 0 | 3 | 30 | 34 | ± | ± | +++ | +++ |
| F4 | 0 | 0 | 1 | 16 | 17 | ± | ± | − | +++ |
| F5 | 0 | 0 | 1 | 19 | 24 | ± | ± | ± | ++ |
| F6 | 0 | 0 | 1 | 10 | 6 | ± | ± | − | +++ |
| G1 | 85 | 58 | 95 | 104 | ++ | +++ | + | +++ | |
| G2 | 100 | 40 | 12 | 26 | +++ | +++ | ± | +++ | |
| G3 | 96 | 78 | 103 | 103 | +++ | +++ | ++ | +++ | |
| H1 | 58 | 57 | 108 | 127 | + | +++ | ++ | +++ | |
| H2 | 78 | 57 | 93 | 119 | + | +++ | ± | ++ | |
| H3 | 89 | 52 | 80 | 92 | + | +++ | ± | ++ | |
| I1 | 0 | 25 | 33 | 53 | 79 | ± | ++ | ± | ++ |
| I2 | 0 | 67 | 39 | 47 | 76 | + | ++ | ± | +++ |
The E2-responsive luciferase reporter plasmid pBS1073 was cotransfected with the indicated pMEP-E2 constructs. Luciferase expression is expressed relative to wild-type E2. Each mutation was tested in at least three independent transfections.
HeLa growth suppression activity, as described in Fig. 6. ND, not done.
DNA replication activity and cooperative origin binding activity with the E1 protein are expressed as a percentage of wild-type E2 activity. +++, >60% activity; ++, >30 to 60%; +, >10 to 30% activity; ±, ≤10% activity. Data are from reference 6.
FIG. 4.
The chromosome binding phenotype of several E2 proteins is temperature sensitive. E2 localization was analyzed in stably transfected CV-1 cell lines that were cultured at 34 or 37°C by confocal fluorescence microscopy. Shown are representative mitotic cells for the B3, C9, and F3 proteins. E2 was detected by indirect immunofluorescence with an FITC-conjugated secondary antibody and is shown in green (E2), while the merged image shows E2 staining plus cellular DNA counterstained in red with propidium iodide (E2+DNA).
Only six E2 proteins were unable to bind mitotic chromosomes at either 34 or 37°C. These proteins are all contained within the C and F groups. C10 (Q12N, Q15N, and E39Q) and C11 (Q12N, Q15N, E39Q, and R68Q) were unable to bind mitotic chromosomes. F3 (R37A and I73A), F4 (R37A, K70A, E74A, Q76A, and Q80A), F5 (R37A, K70A, I73A, E74A, Q76A, and Q80A), and F6 (R37Q, K70Q, E74Q, Q76A, and Q80A) were completely defective for chromosome binding, and even E2 proteins with single amino acid substitutions in this region, F1 (R37A) and F2 (I73A), showed greatly reduced chromosome binding of 48 and 8%, respectively.
Ability of E2 to bind to mitotic chromosomes correlates with interaction with the cellular mitotic chromosome associated factor BRD4.
Several cellular proteins have been proposed to interact with the analogous herpesvirus proteins, EBNA1 and LANA, to tether their genomes to mitotic chromosomes (26, 27, 29, 48). One of the proteins involved in the interaction of the LANA protein with mitotic chromosomes is the bromodomain-containing protein Ring-3 (Brd2) (48), a member of the BET family of transcriptional regulators (16). To determine whether the E2 protein could also bind this family of proteins, both Ring-3 (Brd2) and a related mitotic chromosome binding family member, MCAP (Brd4), were tested for E2 interaction. Brd4, but not Brd2, showed specific interaction with the E2 protein (Fig. 5 and data not shown). Brd4 has been shown to associate with mitotic chromosomes at all stages of mitosis (11, 12) and in this respect is similar to E2 (5). The cellular bromodomain protein Brd4 was also recently proposed to be the cellular mitotic chromosomal protein to which E2 is tethered to segregate viral genomes (62). Additionally, we have shown that expression of Brd4 can reconstitute E2-mediated plasmid segregation in yeast (7a).
FIG. 5.
Brd4 interaction of mutated E2 proteins. (A) In vitro-translated E2 proteins were mixed with 35S-labeled in vitro-translated Brd4 and immunoprecipitated with the E2-specific antibody B202. Shown is a representative autoradiograph of the SDS-PAGE analysis of these proteins. The location of the full-length Brd4 band is indicated on the left. (B) 35S-labeled in vitro-translated E2 proteins were mixed with a protein extract containing baculovirus-expressed FLAG-Brd4 and immunoprecipitated with anti-FLAG M2 agarose affinity gel. Shown is an autoradiograph of SDS-PAGE analysis of the precipitated proteins. The location of the E2 band is indicated on the left. (C) Quantitation of Brd4-E2 binding experiments. The average of binding data is derived from experiments as described for panel B. Brd4 binding activity is expressed relative to that of wild-type (WT) E2, which has been set at 100%.
Therefore, the ability of the panel of mutated E2 proteins to interact with Brd4 was examined by two different coimmunoprecipitation assays. The first assay used both Brd4 and E2 proteins that were generated by expression in vitro in reticulocyte lysates. Lysates containing unlabeled E2 were mixed with lysates containing 35S-labeled Brd4 and immunoprecipitated with the anti-E2 monoclonal antibody B202. A specific interaction was detected as evidenced by the coprecipitation of Brd4 only in the presence of E2 (Fig. 5A). The ability of the entire series of mutated E2 proteins to bind to Brd4 was then assayed. The autoradiograph of immunoprecipitation results is shown in Fig. 5A. Because the in vitro expression plasmid for Brd4 produced low levels of full-length protein, the amount of Brd4 signal that was coprecipitated with E2 was relatively low. To overcome this limitation, and to corroborate the results obtained by anti-E2 immunoprecipitation, a reciprocal coimmunoprecipitation experiment was performed. For this assay, a FLAG epitope-tagged Brd4 protein was expressed from a recombinant baculovirus in Sf9 cells. A protein extract from Brd4-expressing infected cells was mixed with 35S-labeled, in vitro-translated E2 protein (wild type or mutated) and immunoprecipitated with the anti-FLAG M2 monoclonal antibody. The immunoprecipitation results for the panel of mutated E2 proteins is shown in Fig. 5B and C. Quantitation and comparison of the autoradiographs in panels A and B show good agreement between the results obtained by immunoprecipitation with antibodies directed against either E2 or Brd4.
All proteins that were unable to bind mitotic chromosomes (C10, C11, F3, F4, F5, and F6) were also unable to bind to Brd4. E2 proteins that were temperature sensitive for chromosome binding (B3, C4, C5, C9, I1, and I2) showed significant but reduced Brd4 binding (15 to 41% of wild-type E2). The in vitro binding experiments were carried out at room temperature (23°C), which could enhance Brd4-E2 binding, but the lower temperature may also not be ideal for this interaction. Nevertheless, there is a strong correlation between the ability to interact with Brd4 and the ability to bind to mitotic chromosomes.
Mitotic chromosome binding and DNA replication functions of the E2 protein are separable.
Six mutated proteins have been identified (C10, C11, F3, F4, F5, and F6) that are unable to bind to mitotic chromosomes or the Brd4 protein. It is important to determine whether these six proteins are specifically defective in these functions or are completely defective because of a global disruption of conformation. In a separate study, this series of E2 proteins was analyzed for their ability to interact with the viral E1 protein and to support papillomavirus DNA replication (6). These data are summarized in Table 3. In these experiments, of the six proteins defective for chromosomal binding, only the F3 (R37A and I73A) E2 protein had significant DNA replication activity, and it functioned at a level that approached that of wild-type E2 (83%). Thus, the defect in the binding of the F3 (R37A and I73A) protein to mitotic chromosomes is not due to a global disruption of transactivation domain structure but is specific to the chromosome binding and Brd4 binding functions of E2. Although the other proteins with mutations in this region (F4, F5, and F6) were defective in DNA replication, they could cooperatively bind to the origin with the E1 protein at levels that were at least 64% of that of the wild-type E2 protein. Therefore, they are also unlikely to have a complete disruption of conformation. However, the C10 and C11 proteins were unable to cooperatively bind with the viral E1 protein and may have a more global disruption of protein structure. Therefore, the F3 protein (R37A and I73A) is specifically defective for interaction with mitotic chromosomes and the Brd4 protein.
Conversely, proteins in the A group were shown to be specifically defective for DNA replication, at least in part due to their inability to associate with the E1 protein (6). However, the A1 and A2 proteins were able to associate with mitotic chromosomes and the Brd4 protein with wild-type efficiency, so the replication function of E2 is not required for these functions.
Transcriptional activation function of the mutated E2 proteins shows a general correlation with mitotic chromosome association.
The R37 and I73 residues have previously been implicated in the transactivation function of E2. To assess the transcriptional activity of the series of E2 proteins and to determine whether this function correlates with mitotic chromosomal and Brd4 binding, the E2 proteins were tested for their ability to transcriptionally activate an E2-responsive promoter expressing the luciferase reporter gene. E2 activates transcription by binding directly to cis elements consisting of multiple E2 binding sites upstream from promoters (52). For this assay, the reporter plasmid, pBS1073, was either cotransfected into CV-1 cells with the E2 expression plasmids or transfected into pMEP-E2 cell lines that already expressed E2. Luciferase activities were measured 2 days posttransfection, and the averaged results from several such experiments are shown in Table 3. The E2 proteins showed a range of transcriptional activities, but those that were either defective or temperature sensitive for chromosome binding had the lowest activities. To test whether any of the mutated proteins were also temperature sensitive for the transactivation function of E2, the experiments were also carried out at 34°C.
Mutations in the A, D, and G groups had minimal effect on the transactivation ability of the E2 protein and did not show an increase in activity at 34°C. All of these proteins bind to mitotic chromosomes and the Brd4 protein. However, all three groups of proteins are relatively defective for DNA replication (6). (The G2 protein had reduced activity in transactivation and replication, but this protein has growth-inhibitory effects that could indirectly result in reduced transactivation.) The A region is composed of an N-terminal acidic amphipathic helix that had previously been postulated to be important for transcriptional activation (31, 41). However, our results showed that mutation of the four acidic surface amino acids, E2, E6, E13, and E20, to either alanines or to glutamines had no effect on the ability of the protein to transactivate E2-responsive promoters or to bind to mitotic chromosomes. In summary, the A, D, and H regions appear to be more important for replication functions of E2 rather than transcriptional regulation, chromosome binding, or Brd4 interaction.
Many of the proteins are temperature sensitive for transcriptional repression and activation. Group H proteins can also bind mitotic chromosomes and have almost wild-type levels of transactivation; however, transactivation is increased even further at 34°C. Group I proteins, which are completely temperature sensitive for chromosome binding, can activate transcription at about 50% of wild-type levels, and this activity is increased to 75 to 80% at 34°C. In the B group, B1 and B2 proteins bind mitotic chromosomes efficiently but have only about 35% of wild-type transactivation activity, which is almost doubled at 34°C. The B3 protein is completely temperature sensitive for chromosome binding and has low transactivation activity (<10%). However, transactivation is not significantly increased at 34°C. Therefore, while there is a general correlation between chromosomal association and transactivation activities, the temperature sensitivity of these functions is not always paralleled.
As was observed for mitotic chromosomal association, mutations within the C and F groups had the most effect on transcriptional activation. Almost all C group proteins, with the exception of those found to be globally defective (C10 and C11), showed an increase in transactivation at 34°C. C4, C5, and C9 were also temperature sensitive for chromosome binding. Therefore, this region of the protein seems to be very sensitive to amino acid substitution, which results in subtle conformational defects and temperature-sensitive phenotypes.
Within group F, the putative self-interactive interface, a trend was observed between transactivation and chromosome binding activities. As seen with the mitotic chromosome binding results, mutation of residue R37 or I73 alone did not disrupt the transactivation function as severely as did the double mutation, with the single mutations resulting in proteins that retained 64 and 41% of wild-type activity, respectively (Table 3) (F1 and F2), and the R37A and I73A protein having only 30% of wild-type activity (Table 3) (F3). Mutation of additional residues decreased transactivation activity to 10% of that of wild-type E2. Notably, however, none of the proteins were temperature sensitive. Despite an inability to bind mitotic chromosomes and almost undetectable Brd4 binding, the F3 protein could still activate transcription at low levels. This may reflect the sensitivity of the transactivation assay compared with the former two assays. However, while there is a parallel in the ability to bind mitotic chromosomes and transactivation, it is not absolute. For example, the B3 and C9 proteins can efficiently associate with mitotic chromosomes at 34°C, yet they still only activate transcription at 10 to 15% of wild-type E2 levels at this temperature.
Mutational effects on E2-mediated transcriptional activation and repression functions correlate strongly.
The E2 transactivator can repress transcription by binding to E2 binding sites that overlap essential elements in the HPV18 P105 promoter (53). The P105 promoter expresses the HPV18 E6 and E7 oncoproteins, and HeLa cells depend on the functions of these proteins for continuous growth (14, 23). Expression of E2 in HeLa cells down-regulates the expression of the integrated HPV18 E6 and E7 genes, resulting in reactivation of the p53 and pRB pathways that control cellular proliferation and progression through the cell cycle and leading to growth arrest and senescence (14, 19, 20, 23, 53).
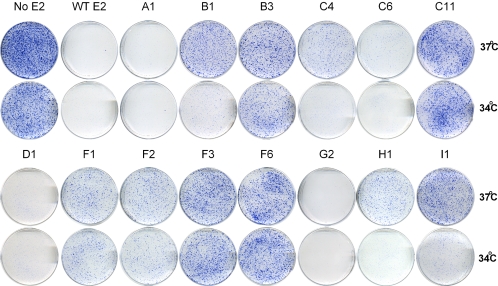
To examine the effects of the panel of mutations on E2-mediated transcriptional repression, the ability of the mutated E2 proteins to suppress the growth of HeLa cells was assayed by transfection with pMEP-E2 expression plasmids followed by selection with hygromycin for maintenance of the episomal plasmid. After 2 weeks of selection at either 34 or 37°C, the plate of cells transfected with the control pMEP4 vector contained hundreds of healthy colonies and approached confluence. Almost no drug-resistant colonies survived on plates transfected with wild-type E2 (Fig. 6). The cells were fixed and stained with methylene blue. An example of the repression observed for a selection of the mutated E2 proteins is shown in Fig. 6. Growth suppression was expressed as the reduction in stained area on the E2 plate relative to the control, and the data for the entire panel of E2 mutants are summarized in Table 3. Similar results were obtained with or without CdSO4 induction because of the leakiness of the metallothionein promoter, indicating that very low levels of E2 are required for growth suppression in this assay (data not shown).
FIG. 6.
HeLa cell growth suppression activity of mutated E2 proteins. HeLa cells were transfected with pMEP4 (No E2), wild-type pMEP-E2 (WT E2), or mutated pMEP-E2 constructs as indicated. Transfected cells were selected with 200 μg of hygromycin B/ml and cultured at either 34 or 37°C, as indicated. When the No E2 plate approached confluence, the cells were fixed and stained with methylene blue. Shown are plates from several groups.
The correlation between transcriptional activation and transcriptional repression functions is quite remarkable. There is a very good parallel between transcriptional activation and repression levels. For example, the transactivation-defective F3 (R37A and I73A) protein showed a marked reduction in the ability to suppress HeLa cell growth (Fig. 6 and Table 3), whereas the individual F1 (R37A) and F2 (I73A) mutations, which retained significantly higher transactivation activity, were more effective in transcriptional repression. Likewise, mutations in the A, D, and G groups were all able to repress HeLa cell growth as well as the wild-type E2 protein. These proteins were also able to bind mitotic chromosomes and activate transcription. Several of the groups (groups B, H, and I) as well as some proteins in the C group (C3, C5, and C8) were temperature sensitive for transcriptional repression. Each of these proteins also activated transcription at correspondingly higher levels at 34°C. The only protein that appeared to separate transcriptional activation and repression is the G2 (E86A, D89A, and E90A) protein. G2 activates transcription at 12% of wild-type E2 but was extremely effective in the HeLa repression assay. However, this is most likely due to unique growth-inhibitory effects of this mutated protein that augment the repression assay but are detrimental to the transactivation assay.
In summary, there is a very strong correlation between transcriptional activation and repression functions of the E2 protein, despite the fact that these activities are measured by very different assays and in different cell lines. There is a trend in the ability of the E2 protein to regulate transcription and bind mitotic chromosomes, but the relative levels and temperature sensitivity of these functions do not always correspond exactly. However, all of these functions are completely separable from DNA replication.
DISCUSSION
The BPV1 E2 protein mediates several key functions in the life cycle of the virus. One of these functions is the tethering of viral genomes to mitotic chromosomes to enable stable genome segregation. The segregation function requires the interaction of the amino-terminal transactivation domain of E2 with mitotic chromosomes (5). In this study, a set of E2 proteins with mutations in the transactivation domain were generated to further characterize the functional architecture of the domain (Table 1). The mutated E2 proteins were analyzed in several different functional assays, including transcriptional activation, transcriptional repression, and mitotic chromosome binding. Target group F defined a region of the transactivation domain that is crucial for the mitotic chromosome binding function of E2. The amino acids within group F lie along the putative self-interactive surface identified by Antson and colleagues (4). Mutation of just two amino acids on this surface, R37 and I73, to alanines eliminates the chromosome binding activity of E2. The R37A and I73A protein is also reduced in transactivation and transcriptional repression functions but retains the ability to very efficiently support DNA replication in cooperation with the viral E1 helicase, indicating that the mutation is not globally disruptive to E2 function (6).
The LANA protein of the Kaposi's sarcoma herpesvirus, human herpesvirus 8, interacts with Ring3 (Brd2) in its association with mitotic chromosomes (39, 48). In this study, we examined whether Ring3 and family member MCAP (Brd4) also interacted with E2. Only E2 and Brd4 were able to form a complex. Brd4 has recently been shown to be a likely candidate for the mitotic chromosome-associated factor that mediates the chromosome binding of E2 (62). This is further supported by the Brd4-E2 binding studies presented here; Brd4-E2 binding correlated well with the mitotic chromosome binding function in our series of E2 proteins. All proteins that associate with mitotic chromosomes could also bind Brd4. Conversely, mutations that disrupted chromosome binding (C10, C11, F3, F4, F5, and F6) eliminated the interaction with Brd4. We have further substantiated the Brd4-E2 interaction results obtained in vitro by in vivo colocalization studies with several of the key mutated E2 (M. G. McPhillips and A. A. McBride, submitted for publication).
In the recent study by You et al., the BRD4-E2 complex was purified from asynchronous cells (62). Since mitotic cells would only constitute a small fraction of these cells, the E2 and Brd4 complex must exist at other stages of the cell cycle. Therefore, Brd4 is likely to be involved in additional E2 functions. Our study indicates that the E2-Brd4 interaction may also be important for the transcriptional regulatory properties of E2 but it is not required for the DNA replication function.
The E2 protein forms a stable dimer through its carboxy-terminal DNA binding domain (13, 41). Antson and colleagues have reported that the HPV16 transactivation domain also self-associates (4) and have suggested that interactions between dimers of E2 bound to distant binding sites on DNA could allow formation of DNA loops. Such E2 bound DNA loops, mediated by the BPV1 E2 protein, have been observed by electron microscopy (28). Such interdimer interactions and DNA loops would allow transcription factors that are bound near E2 at distal sites to be brought into the proximity of a transcription initiation site. However, recent reports of the structure of the HPV11 E2 transactivation domain (56) or HPV18 E2 transactivation domain (1) found no evidence of self-interaction or dimerization through the transactivation domain, so it is unclear whether the BPV1 E2 protein can also participate in interdimer interactions. Notably, the Epstein-Barr virus EBNA1 protein, which has analogous genome maintenance and segregation functions to those of E2 and associates with mitotic chromosomes, also mediates the looping of DNA (25, 34-36, 59). In fact, two of the three regions of EBNA1 that are responsible for the DNA looping activity can also bind to mitotic chromosomes (25, 30, 34-37, 59). Whether this similarity in protein function is significant for mitotic chromosome tethering remains to be determined.
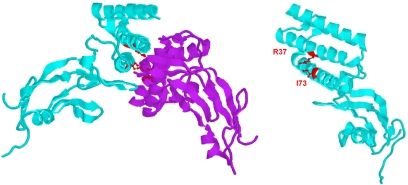
If it is assumed that the transactivation domain of BPV1 E2 is able to self-interact as has been reported for HPV16 E2, then our chromosome binding results leave open the question of how the essential residues R37 and I73 are involved. One possible answer is that these residues are directly involved in the interaction with chromosomal factor(s), such as Brd4, to mediate attachment to mitotic chromosomes. An alternative explanation is that R37 and I73 are required for the self-interaction of the transactivation domain and that self-interaction creates a new protein interaction interface composed of both monomers of the domain. Figure 7 shows a diagram of the positions of these residues on a monomer and dimer of the transactivation domain.
FIG. 7.
Location of key mutations on the transactivation domain. The positions of the R37 and I73 residues, which are key for mitotic chromosome binding, are shown on models of both a monomer and homodimer of the BPV1 E2 transactivation domain.
In a recent report, Abroi and colleagues also examined the effects of individual point mutations within the BPV1 E2 transactivation domain on chromosome binding (2). In this study, the authors assessed chromosome binding by using a hypotonic treatment prior to immunofluorescence, washing away unbound E2. The effect of mutations on chromosome binding was assessed by comparing the percentage of cells that had E2 remaining on the chromosomes. They concluded that the surface of the transactivation domain opposite the putative self-interactive surface, which includes residues in target group C of our study (Q12, E39, and R68), is important for chromosome binding. In our study, we also observed that various combinations of mutations in these residues did eliminate binding to chromosomes. However, single-amino-acid substitutions in these residues did not eliminate binding in our experiments. Abroi et al. did find that, although the percentage of mitotic cells containing chromosome-associated E2 was greatly reduced with these mutations, at least some mitotic cells had E2 localized on the chromosomes for all of the single point mutations that they examined. It is possible that these proteins have reduced affinity for mitotic chromosomes which is altered by the hypotonic treatment. In our study, mutations that eliminated chromosome binding, such as R37A and I73A, yielded proteins that were present in mitotic cells but were excluded from the chromosomes.
The transactivation domain of E2 has been shown to be necessary and sufficient for the association of the protein with mitotic chromosomes (5). E2 differs from other viral proteins that mediate genome maintenance and segregation, such as EBNA1 of Epstein-Barr virus and LANA of human herpesvirus 8, in that a structurally intact domain is required for chromosomal interaction. For EBNA1 and LANA, the chromosome binding function has been mapped to short fragments of the protein (50). For E2, however, no fragment shorter than the full transactivation domain has been shown to retain the ability to associate with mitotic chromosomes. The inability to more finely map the binding motif suggests that the structural integrity of the transactivation domain is crucial for the chromosome binding function. The HPV16 transactivation domain has been shown to have two distinct subdomains that are highly structured (4). A nonlinear binding surface, comprising residues from different segments of the primary sequence of the domain, would be highly sensitive to even small deletions. Additionally, if self-interaction of the domain plays a role in formation of the binding site for a chromosomal factor such as Brd4, deletions that disrupt self-interaction will also indirectly eliminate chromosome binding.
Several studies, including this one, have shown that many point mutations in the E2 transactivation domain result in temperature-sensitive phenotypes (15, 63). The efficiency of chromosome binding activity of the wild-type E2 protein is even increased at 34°C. The BPV1 E2 protein is encoded by a virus that infects the cutaneous epithelium of bovine skin, which is unlikely to be maintained at 37°C, and this may explain why the protein is so conformationally sensitive and fragile at this temperature. This feature of the E2 protein necessitates that all mutated proteins be tested for temperature sensitivity before concluding that a particular region is directly involved in a specific function or interaction. In addition, we have found that the mitotic chromosomal association of the BPV1 E2 protein can be changed by a reduction in temperature in the few minutes before fixation; remarkably, E2 proteins that are not associated with chromosomes at 37°C quickly associate if the temperature is reduced just a few degrees before they are fixed.
The E2 protein can strongly repress the HPV18 P105 promoter, and it was initially assumed that this was due primarily to competitive binding of E2 to sites that overlapped with essential promoter elements. However, several studies have shown that DNA binding is not sufficient for repression and that the transactivation domain is also required (14, 20, 45). The repression function of E2 could not be replaced by a different transactivation domain such as that of VP16 (20). Similar findings are presented in this paper with the new series of mutations that we have developed. All of the mutated E2 proteins have very similar activities in transcriptional activation and repression. There are many mechanisms of transcriptional repression (reviewed in reference 17), and it is not yet clear how E2 down-regulates transcription. However, it has been shown that the transactivation domain of E2 is required for nucleosomal exclusion of chromatin-based replication templates (33) and that E2 interacts with histone acetyltransferases, such as p300 (43, 46). Therefore, interaction with chromatin modifiers could be important to enable the E2 protein to gain access to its binding sites as well as to modify the adjacent transcriptional environment to repress transcription.
In summary, this study has produced a novel panel of mutations in the E2 transactivation domain that are able to discriminate among and provide insight into the various functions of this key papillomavirus regulatory protein.
Acknowledgments
We thank Carl Baker and Jaquelline Oliveira for critical comments on the manuscript and Jon Spindler for expert technical assistance.
REFERENCES
- 1.Abbate, E. A., J. M. Berger, and M. R. Botchan. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18:1981-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abroi, A., I. Ilves, S. Kivi, and M. Ustav. 2004. Analysis of chromatin attachment and partitioning functions of bovine papillomavirus type 1 E2 protein. J. Virol. 78:2100-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abroi, A., R. Kurg, and M. Ustav. 1996. Transcriptional and replicational activation functions in the BPV1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antson, A. A., J. E. Burns, O. V. Moroz, D. J. Scott, C. M. Sanders, I. B. Bronstein, G. G. Dodson, K. S. Wilson, and N. J. Maitland. 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403:805-809. [DOI] [PubMed] [Google Scholar]
- 5.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 6.Baxter, M. K., and A. A. McBride. 2005. An acidic amphipathic helix in the bovine papillomavirus E2 protein is critical for DNA replication and interaction with the E1 protein. Virology 332:78-88. [DOI] [PubMed] [Google Scholar]
- 7.Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280:1-9. [DOI] [PubMed] [Google Scholar]
- 7a.Brannon, A. R., J. A. Maresca, J. D. Boeke, M. A. Basrai, and A. A. McBride. 2005. Reconstitution of papillomavirus E2-mediated plasmid maintenance in Saccharomyces cerevisiae by the Brd4 bromodomain protein. Proc. Natl. Acad. Sci. USA 102:2998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breiding, D. E., M. J. Grossel, and E. J. Androphy. 1996. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology 221:34-43. [DOI] [PubMed] [Google Scholar]
- 9.Brokaw, J. L., M. Blanco, and A. A. McBride. 1996. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J. Virol. 70:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, C. S., S. N. Upmeyer, and P. L. Winokur. 1998. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions. Virology 241:312-322. [DOI] [PubMed] [Google Scholar]
- 11.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostatni, N., F. Thierry, and M. Yaniv. 1988. A dimer of BPV-1 E2 containing a protease resistant core interacts with its DNA target. EMBO J. 7:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowhanick, J. J., A. A. McBride, and P. M. Howley. 1995. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 69:7791-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson, M. F., and M. R. Botchan. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florence, B., and D. V. Faller. 2001. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 6:D1008-D1018. [DOI] [PubMed] [Google Scholar]
- 17.Gaston, K., and P. S. Jayaraman. 2003. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell. Mol. Life Sci. 60:721-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giniger, E., and M. Ptashne. 1987. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature 330:670-672. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin, E. C., and D. DiMaio. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 97:12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin, E. C., L. K. Naeger, D. E. Breiding, E. J. Androphy, and D. DiMaio. 1998. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 72:3925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heino, P., J. Zhou, and P. F. Lambert. 2000. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology 276:304-314. [DOI] [PubMed] [Google Scholar]
- 22.Howley, P. M. 1995. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Lippincott-Raven, Philadelphia, Pa.
- 23.Hwang, E. S., D. J. Riese II, J. Settleman, L. A. Nilson, J. Honig, S. Flynn, and D. DiMaio. 1993. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J. Virol. 67:3720-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapoor, P., and L. Frappier. 2003. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 77:6946-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight, J. D., R. Li, and M. Botchan. 1991. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc. Natl. Acad. Sci. USA 88:3204-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laine, A., and L. Frappier. 1995. Identification of Epstein-Barr virus nuclear antigen 1 protein domains that direct interactions at a distance between DNA-bound proteins. J. Biol. Chem. 270:30914-30918. [DOI] [PubMed] [Google Scholar]
- 31.Lambert, P. F., N. Dostatni, A. A. McBride, M. Yaniv, P. M. Howley, and B. Arcangioli. 1989. Functional analysis of the papilloma virus E2 trans-activator in Saccharomyces cerevisiae. Genes Dev. 3:38-48. [DOI] [PubMed] [Google Scholar]
- 32.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, R., and M. R. Botchan. 1994. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl. Acad. Sci. USA 91:7051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackey, D., T. Middleton, and B. Sugden. 1995. Multiple regions within EBNA1 can link DNAs. J. Virol. 69:6199-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackey, D., and B. Sugden. 1997. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J. Biol. Chem. 272:29873-29879. [DOI] [PubMed] [Google Scholar]
- 36.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maruyama, T., A. Farina, A. Dey, J. Cheong, V. P. Bermudez, T. Tamura, S. Sciortino, J. Shuman, J. Hurwitz, and K. Ozato. 2002. A mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 22:6509-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattsson, K., C. Kiss, G. M. Platt, G. R. Simpson, E. Kashuba, G. Klein, T. F. Schulz, and L. Szekely. 2002. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 83:179-188. [DOI] [PubMed] [Google Scholar]
- 40.McBride, A. A., J. B. Bolen, and P. M. Howley. 1989. Phosphorylation sites of the E2 transcriptional regulatory proteins of bovine papillomavirus type 1. J. Virol. 63:5076-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride, A. A., and G. Myers. 1997. The E2 proteins: an update, p. III54-III98. In G. Myers, C. Baker, K. Munger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomaviruses 1997. Los Alamos National Laboratory, Los Alamos, N.M.
- 43.Muller, A., A. Ritzkowsky, and G. Steger. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76:11042-11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakahara, T., A. Nishimura, M. Tanaka, T. Ueno, A. Ishimoto, and H. Sakai. 2002. Modulation of the cell division cycle by human papillomavirus type 18 E4. J. Virol. 76:10914-10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura, A., T. Ono, A. Ishimoto, J. J. Dowhanick, M. A. Frizzell, P. M. Howley, and H. Sakai. 2000. Mechanisms of human papillomavirus E2-mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J. Virol. 74:3752-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng, Y. C., D. E. Breiding, F. Sverdrup, J. Richard, and E. J. Androphy. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74:5872-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 48.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakai, H., T. Yasugi, J. D. Benson, J. J. Dowhanick, and P. M. Howley. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 70:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spalholz, B. A., Y. C. Yang, and P. M. Howley. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183-191. [DOI] [PubMed] [Google Scholar]
- 53.Thierry, F., and M. Yaniv. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 6:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Y., R. Coulombe, D. R. Cameron, L. Thauvette, M. J. Massariol, L. M. Amon, D. Fink, S. Titolo, E. Welchner, C. Yoakim, J. Archambault, and P. W. White. 2004. Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J. Biol. Chem. 279:6976-6985. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, V. G., and J. Ludes-Meyers. 1991. A bovine papillomavirus E1-related protein binds specifically to bovine papillomavirus DNA. J. Virol. 65:5314-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winokur, P. L., and A. A. McBride. 1996. The transactivation and DNA binding domains of the BPV-1 E2 protein have different roles in cooperative origin binding with the E1 protein. Virology 221:44-53. [DOI] [PubMed] [Google Scholar]
- 59.Wu, H., P. Kapoor, and L. Frappier. 2002. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J. Virol. 76:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yabe, Y., A. Sakai, Y. Tanimura, M. Kuramitsu, T. Hitsumoto, K. Ishii, and H. Ueki. 1988. Two human papillomavirus DNAs molecularly cloned from a patient with epidermodysplasia verruciformis: restriction maps. Acta Med. Okayama 42:243-245. [DOI] [PubMed] [Google Scholar]
- 61.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papillomavirus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus e2 protein with brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, P.-S., J. L. Brokaw, and A. A. McBride. 2005. Conditional mutations in the mitotic chromosome binding function of the BPV1 E2 protein. J. Virol. 79:1500-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]