Abstract
Background:
The prenatal environment influences lifetime health; epigenetic mechanisms likely predominate. In 2016, the first international consortium paper on cigarette smoking during pregnancy and offspring DNA methylation identified extensive, reproducible exposure signals. This finding raised expectations for epigenome-wide association studies (EWAS) of other exposures.
Objective:
We review the current state-of-the-science for DNA methylation associations across prenatal exposures in humans and provide future recommendations.
Methods:
We reviewed 134 prenatal environmental EWAS of DNA methylation in newborns, focusing on 51 epidemiological studies with meta-analysis or replication testing. Exposures spanned cigarette smoking, alcohol consumption, air pollution, dietary factors, psychosocial stress, metals, other chemicals, and other exogenous factors. Of the reproducible DNA methylation signatures, we examined implementation as exposure biomarkers.
Results:
Only 19 (14%) of these prenatal EWAS were conducted in cohorts of 1,000 or more individuals, reflecting the still early stage of the field. To date, the largest perinatal EWAS sample size was 6,685 participants. For comparison, the most recent genome-wide association study for birth weight included more than 300,000 individuals. Replication, at some level, was successful with exposures to cigarette smoking, folate, dietary glycemic index, particulate matter with aerodynamic diameter and , nitrogen dioxide, mercury, cadmium, arsenic, electronic waste, PFAS, and DDT. Reproducible effects of a more limited set of prenatal exposures (smoking, folate) enabled robust methylation biomarker creation.
Discussion:
Current evidence demonstrates the scientific premise for reproducible DNA methylation exposure signatures. Better powered EWAS could identify signatures across many exposures and enable comprehensive biomarker development. Whether methylation biomarkers of exposures themselves cause health effects remains unclear. We expect that larger EWAS with enhanced coverage of epigenome and exposome, along with improved single-cell technologies and evolving methods for integrative multi-omics analyses and causal inference, will expand mechanistic understanding of causal links between environmental exposures, the epigenome, and health outcomes throughout the life course. https://doi.org/10.1289/EHP12956
Introduction
Prenatal Exposures That Impact Health and Epigenetics
Environmental exposures, including chemicals and other exogenous factors, are prevalent and heterogenous. The United States produces and uses more than 85,000 different chemicals,1 including more than 100 regularly detected in pregnant people’s biospecimens.2 Levels of numerous chemicals are higher among pregnant Black and Hispanic people and those of lower educational attainment,3 representing an equity issue. Many chemicals cross the placenta, influencing fetal cellular function (Figure 1A). The prenatal period of rapid and exquisitely timed cellular differentiation and expansion confers heightened susceptibility to exposures. For example, in utero exposure to lead impairs neurodevelopment,7 arsenic is associated with impaired lung function and increased cardiovascular mortality,8 cigarette smoke causes reduced birth weight and reduced infant lung function,9 and folic acid deficiency causes neural tube defects.10 Exposures during pregnancy may impact the developing embryo/fetus with consequences for postnatal life,11 known as the Developmental Origins of Health and Disease (DOHaD) hypothesis.12 Investigating the totality of diverse prenatal exposures, termed the exposome,13 with later health and disease is an evolving area of etiological, prevention, and policy research. Specific and accurate retrospective markers of exposures experienced years prior to health outcomes are unfortunately rare, limiting available data for most prenatal exposures to prospective collection efforts.
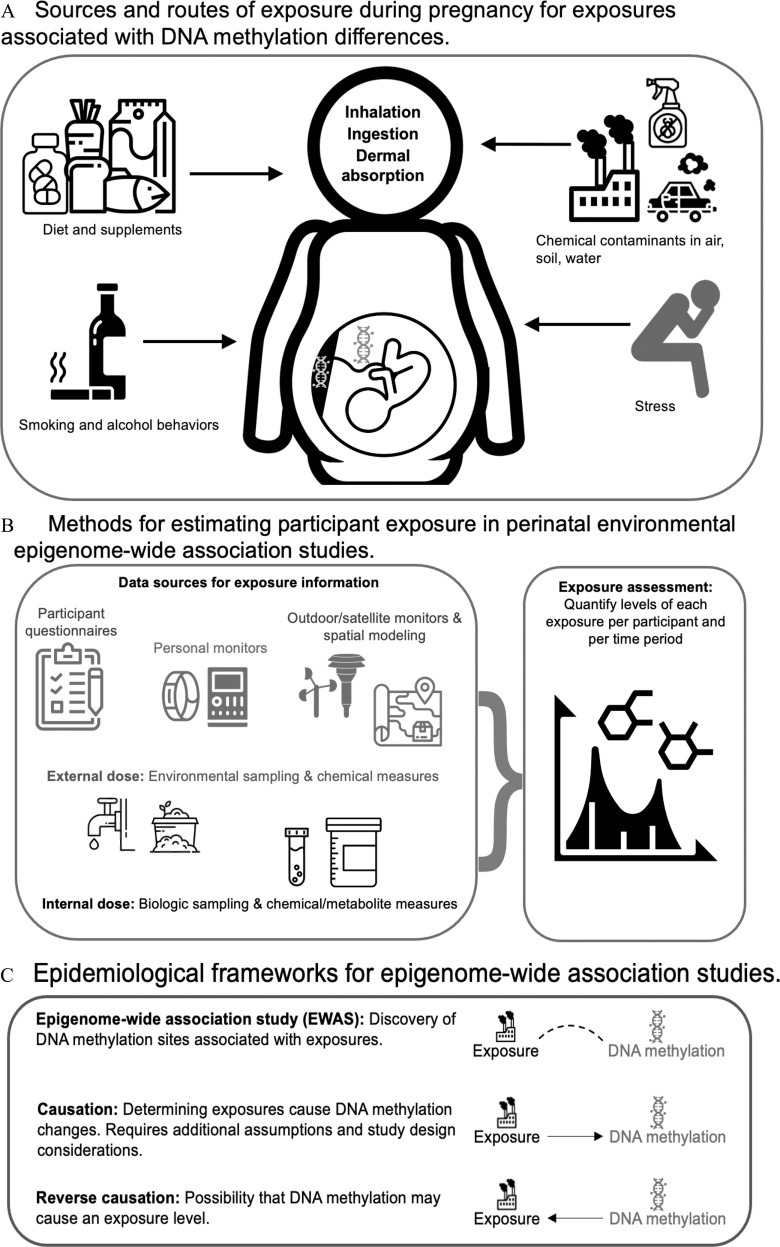
Figure 1.
(A) Sources of prenatal exposures can include (clockwise from top left) dietary intake from food and supplements, pollution, psychosocial stress, and smoking and alcohol drinking behaviors. Exposures enter the pregnant person’s body typically through inhalation, ingestion, or dermal absorption. Once in the body, exposures are distributed and metabolized. They can interact with DNA in various tissues, including placental and fetal tissues. These are the major categories of exposure during pregnancy that were assessed for association with DNA methylation in this review article. (B) Common sources of information for exposure assessment include questionnaires, spatial sensors linked to participant residential history generally followed by exposure modeling, as well as blood- and urine-based exposure biomarkers. These methods are used by environmental health scientists to quantify individual levels of exposure to chemicals and to estimate the relevant time frame the exposure measure is applicable. (C) When we test for a relationship between an environmental exposure and DNA methylation, typically using multivariable regression methods, this is a test of association. Association tests do not indicate causation,4 because causal inference requires additional assumptions.5 In the case of environmental exposures and DNA methylation, a reverse causation scenario would mean the DNA methylation influences the exposure level. Although it is possible that for a given exposure level, differential methylation at genes essential for metabolism could influence measured concentrations of the contaminant (internal dose), methylation is unlikely to be causally related to being exposed. For this reason, reverse causation is less of a concern in studies of exogenous environmental exposures than in EWAS of disease or physiologic traits.6
Epigenetic marks may represent one such useful persistent biomarker of exposure, and potentially a mechanistic link between exposures and lagged health effects. Epigenetic mechanisms are essential for normal development: Errors in epigenetic processes can result in serious developmental disorders.14 DNA methylation, a type of epigenetic mark, is typically observed at cytosine residues upstream of guanine residues (CpG sites). Although other molecules, such as histones, can be methylated, henceforth we refer to DNA methylation as “methylation.” During normal reproduction and development, two major waves of methylation reprogramming occur shortly after fertilization and during gametogenesis.15 Cells maintain their differentiated lineage in part because of their methylation patterns.16 Prenatal environmental exposures can leave epigenetic marks.
Environmental epigenetics was pioneered through clever model system research.17 For example, in the Agouti mouse model,18 prenatal exposures including bisphenol A,19 dietary methyl donors,20 and lead21 caused persistent methylation changes in offspring, demonstrating that diverse prenatal environmental exposures can impact offspring epigenetics. In humans, the Dutch Hunger Winter is a classic example linking prenatal exposures to postnatal health and methylation. Nazi blockades of food supplies produced famine in the Netherlands during the years 1944–1945. People pregnant then bore children with increased rates of cardiovascular, metabolic, and psychiatric disorders.22 Timing and degree of prenatal exposure to this famine were also associated with adult methylation differences.23 Early-life epigenetic changes that persist into later life may provide biomarkers of prenatal exposures and/or a potential mechanism of later health effects.
Scope of This State-of-the-Science Review
This review of the current state of the science is motivated by exciting recent advances in both exposure and methylation assessment investigations in the field of prenatal environmental epigenetic epidemiology. Environmental exposures during pregnancy are estimated through multiple methods with varying accuracy, reliability, and cost (Figure 1B). The availability of high-throughput, low-sample-input arrays with reasonable epigenomic coverage24 enables measurement of methylation in pregnancy cohorts.25,26 Epigenome-wide association studies (EWAS) using these arrays test for associations between hundreds of thousands of individual CpG sites and environmental exposures (Figure 1C). Promising findings from groundbreaking studies in modest sample sizes have led to large, international, and collaborative consortia, which facilitate rigor and reproducibility of EWAS findings.26,27 Recent mapping of the epigenome across tissues and cell types,28 more frequent availability of genomic or other omic data in the same samples, and evolving bioinformatic tools are enabling stronger biological inferences on EWAS findings. Chemical exposure assessment and statistical methods advances are starting to allow evaluation of many environmental exposures simultaneously to comprehensively assess the exposome.29,30 Together, these efforts are increasing the identification of reproducible epigenetic effects of the prenatal environment.30
Here, we review the state of the science on associations between the prenatal environment and methylation. We take an epidemiological perspective to evaluate population-level studies. First, we summarize literature on perinatal environmental EWAS meta-analyses and studies with attempted replication. Consortium efforts have identified reproducible methylation signatures of some prenatal exposures, most notably smoking. Second, we evaluate the implementation of exposure biomarkers for those perinatal exposures with robust and reproducible methylation signatures, a major advance for identifying health impacts of exposures going forward. Third, we provide epidemiological considerations that can impact the power to detect associations in the EWAS framework. Fourth, we make recommendations for new cohorts considering prenatal exposure EWAS. Fifth, we discuss putative mechanisms linking exposures to methylation and challenges in determining their role in exposure-related health outcomes. Finally, we describe areas of needed future development and recommend approaches for implementing emerging tools to provide insights to advance the field. With these analyses and recommendations, this article illustrates the current strengths and weaknesses of perinatal environmental epigenetics research and highlights its future potential.
Methods
We retrieved EWAS of prenatal exogenous exposures with designated search terms identified via PubMed, Web of Science, and Embase searches through 10 May 2022 (search terms in Supplemental Material, “Supplementary Methods”). Because a global term such as environmental exposures may fail to capture specific environmental exposures, we included a broad collection of search terms. Abstracts for all retrieved studies were screened using specified inclusion and exclusion criteria based on the research question of interest. To be included, studies had to be conducted in human populations evaluating the relationship between prenatal exogenous exposures and epigenome-wide methylation. To avoid including duplicated studies, we included only original research articles, excluding reviews and commentaries. Because we focused on epigenome-wide studies, we excluded studies that reported only associations with summary DNA methylation measures (e.g., global methylation, epigenetic clocks) or candidate CpGs, genes, or regions. Given our focus on exogenous exposures, we excluded EWAS evaluating nonexternal exposures, for example, maternal health constructs such as maternal depression, anxiety, or body mass index. All studies meeting inclusion criteria after abstract screening were briefly reviewed to determine the following key characteristics: the genetic ancestry of the population; the exposure; the tissue and method of DNA methylation measurement; the sample size; the number of significant CpGs, genes, or regions reported; the method of correcting for multiple testing; and the use of meta-analysis or replication. Abstract screening and brief review were conducted by F.B., with oversight by K.M.B. and S.J.L. In high-dimensional analyses such as EWAS, the myriad statistical tests require large sample sizes to limit false positive findings, which can also be minimized by replication testing in independent studies or meta-analyses across studies, which also provides evidence of consistency. From the earliest days of genome-wide association studies (GWAS), attempted replication has been regarded as an essential design element.31,32 Although replication does not guarantee causation, it provides stronger statistical evidence for association, makes artifactual association less likely, and improves effect estimates. Associations might not replicate from one population to another for reasons related to true sources of heterogeneity, including genetic ancestry and exposure distribution differences, and these cannot be evaluated without attempted replication.31,32 Therefore, as a final inclusion criterion, we restricted in-depth review to only studies that were either meta-analyses or single studies that attempted replication in an independent study (regardless of replication success).
Results
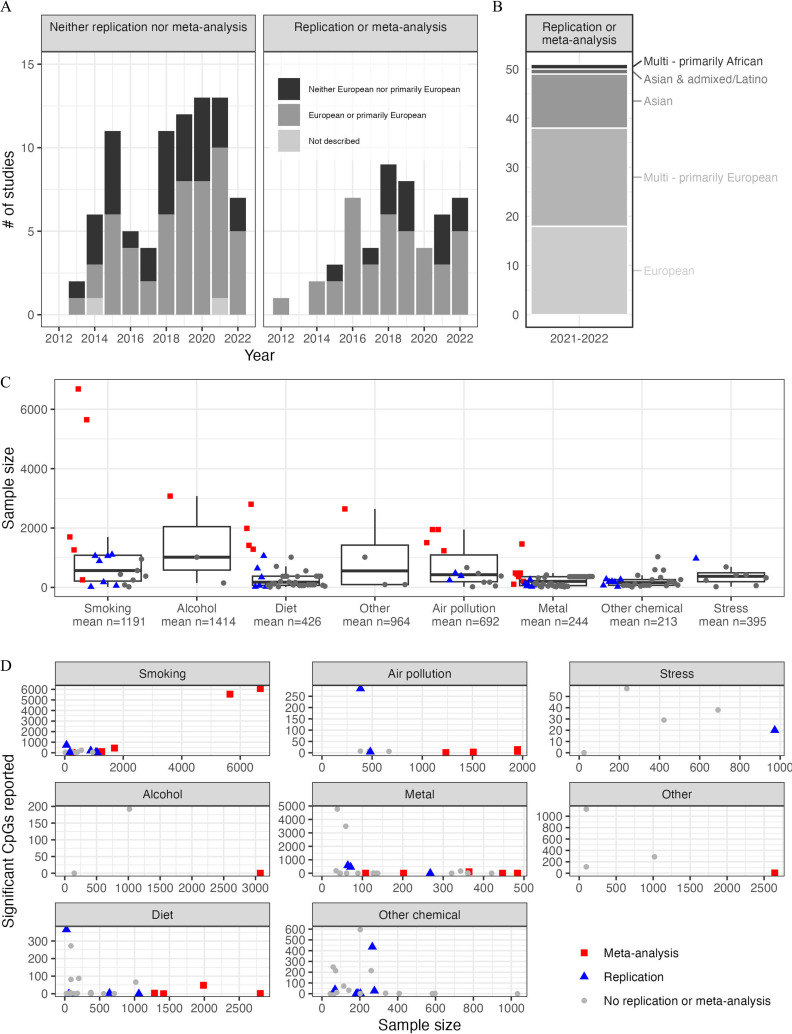
Our literature search identified 360 peer-reviewed publications. After excluding studies with only global or candidate methylation positions or examining nonexogenous exposures (Figure S1), we performed further review for 134 exogenous exposure EWAS conducted from 2012 to 2022. The vast majority () of these EWAS used the Illumina450K or EPIC arrays, and most measured DNA methylation in cord blood, peripheral blood, or placental tissue. Most of these EWAS were conducted in European or primarily European ancestry populations (Figure 2A–B). Meta-analyses or replication attempts were slightly more common among EWAS conducted in European or primarily European ancestry populations (43%) than in other ancestries (30%), likely reflecting greater availability of European ancestry cohorts in which to perform replication analyses, a situation that was also observed in GWAS.31 Sample size of EWAS varied across exposure categories, with the largest EWAS for prenatal maternal cigarette smoking (), but with many EWAS reporting sample sizes in the hundreds (Figure 2C; Table S1). Of 134 reviewed studies, only 19 (14%) have been conducted in cohorts of 1,000 or more individuals. Some studies with smaller sample sizes that did not attempt replication reported high numbers of significant differentially methylated positions (Figure 2D). Of the 134 reviewed studies, 51 reported a replication attempt or were a meta-analysis (Table 1). These studies were reviewed in depth and summarized below.
Figure 2.
(A) The number of epigenome-wide association studies of external exposures identified in this review ( studies; one study appears twice in bar graphs because performed replication for one exposure tested but not the other) by year of publication, reported genetic ancestry, and replication or meta-analysis status. (B) The number of epigenome-wide association studies of external exposures with meta-analysis or replication ( studies reviewed in depth) by reported genetic ancestry. (C) Sample sizes of prenatal environmental epigenome-wide association studies (; studies involving multiple exposures represented as multiple points) by exposure category (x-axis) among meta-analyses (red squares), studies incorporating replication (blue triangles), and other studies (gray dots). Midline: median sample size, lower whisker: smallest sample size that is greater than or equal to the 25% – IQR, upper whisker: largest sample size that is less than or equal to the IQR, box limits: 25% and 75%; these statistics are reported in Table S1. (D) The number of significant differentially methylated positions reported in each study graphed against the sample size of each study and faceted by the category of exogenous exposure considered. Only studies using EPIC or 450K arrays that reported significance at either a or Bonferroni correction represented ( of original 134 studies, some studies represented time, because they tested multiple exposures). The y-axis range varies across panel of exposure category. Data used to generate Figure 2 is provided in Table S2. Note: FDR, false discovery rate; IQR, interquartile range.
Table 1.
Prenatal environmental exposure epigenome-wide association study literature review results. Studies () were included if they performed replication or meta-analysis.
| Reference | Cohort/consortia | a | Ancestry | Exposure | Exposure tissue | Epigenetic measure | Epigenetic tissue | Significant DMPs | Criteria for significance | Validation type | Replication successful yes/no |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cigarette smoking | |||||||||||
| Joubert et al.33 | PACE | 6,685 | Multi – primarily European | Prenatal maternal smoking | NA | 450K | Cord blood | FDR – 6,073; Bonferroni - 568 | Bonferroni | Meta-analysis & replication | Yes, Kolmogorov for replication look-up in older children’s blood |
| Sikdar et al.34 | PACE | 5,648 | Multi – primarily European | Prenatal maternal smoking | NA | 450K | Cord blood | 5,547 | Meta-analysis | NA | |
| Everson et al.35 | PACE | 1,700 | Multi – primarily European | Prenatal maternal smoking | NA | 450K | Placenta | 443 | Bonferroni | Meta-analysis | NA |
| Hannon et al.36 | MINERVA | 1,263 | European | Prenatal maternal smoking | NA | 450K | Blood spots | 110 | Bonferroni | Meta-analysis & replication | Yes, 102 of 110 DMPs, sign test |
| Vives-Usano37 | HELIX | 1,105 | Multi – primarily European | Prenatal maternal smoking | NA | 450K | Child peripheral blood | 41 | Replication | Yes, 17 of 18 loci | |
| Joubert et al.38 | MoBa | 1,062 | Multi – primarily European | Prenatal maternal smoking | NA | 450K | Cord blood | 26 | Bonferroni | Replication | Yes |
| Rotroff et al.39 | MoBA | 1,062 | European | Prenatal maternal smoking | Maternal plasma (cotinine) | 450K | Cord blood | 27 | Bonferroni | Replication | Mixed, 9 of 15 genes |
| Markunas et al.40 | Norway facial clefts study | 889 | European | Prenatal maternal smoking | NA | 450K | Heel prick blood spots | 185 | Replication | Mixed, 22 of 26 DMPs | |
| Küpers et al.41 | GECKO Drenthe | 255 | European | Prenatal maternal smoking | NA | 450K | Cord blood | 35 | Meta-analysis & replication | Mixed, 3 of 8 DMPs | |
| Morales et al.42 | INMA | 179 | European | Prenatal maternal smoking | Survey and maternal urine cotinine | 450K | Placenta | 50 | Replication | Yes, 4 of 4 DMPs | |
| Shorey-Kendrick et al.43 | NA | 59 | Multi – primarily European | Prenatal maternal smoking | NA | EPIC | Placenta | 726 | Replication | Mixed, 105 of 726 DMPs | |
| Howe et al.44 | MACHS | 20 | European | Prenatal maternal Smoking | NA | Whole-Genome bisulfite sequencing | Cord blood | 10,381 | Replication | Mixed, 4 of 9 DMRs Replicated in adult | |
| Maternal alcohol and diet | |||||||||||
| Sharp et al.45 | PACE | 3,075 | Multi – primarily European | Maternal alcohol | NA | 450K | Cord blood | 0 | Meta-analysis | NA | |
| Küpers et al.46 | PACE | 2,802 | Multi – primarily European | Mediterranean diet | NA | 450K | Cord blood | 1 | Meta-analysis | No, 0 of 1 DMPs | |
| Joubert et al.47 | PACE | 1,988 | European | Folate | NA | 450K | Cord blood | 48 | Bonferroni | Meta-analysis | NA |
| Gonseth et al.48 | CCLS | 343 | Multi – primarily European | Folate | NA | 450K | Blood spots | 4 | Permutation-based values test | Replication | Mixed, 3 of 4 DMPs |
| Amarasekera et al.49 | NA | 23 | European | Folate | Maternal serum in last trimester of pregnancy | 450K | Cord blood ( T-cells & antigen-presenting cells) | No DMPs/DMPs reported, only DMR | NA | Replication | Mixed, DMR only |
| Suderman et al.50 | MoBa; ALSPAC | 1,416 | European | 25-hydroxyvitamin D | Maternal plasma | 450K | Cord blood | 0 | Meta-analysis | NA | |
| Tauebert et al.51 | INMA and Generation R | 1,286 | European | Iron | Maternal serum; cord blood | 450K | Cord blood | 3 | Meta-analysis | NA | |
| Størdal et al.52 | MoBA | 1,062 | European | Iron supplements | NA | 450K | Cord blood | 0 | Bonferroni | Replication | NA |
| Caramaschi et al.53 | ALSPAC | 641 | European | B12 | Genotype (proxy) | 450K | Cord blood | 3 | Replication | Mixed, 1 of 3 DMPs | |
| Geraghty et al.54 | ROLO | 60 | European | Glycemic index diet intervention | NA | EPIC | Cord blood serum | 0 | Replication | NA | |
| Yan et al.55 | NA | 24 | Asian | Glycemic index dietary intervention | NA | 450K | Placenta | 365 | Replication | Mixed, 4 of 10 DMPs | |
| Air pollution | |||||||||||
| Gruzieva et al.56 | PACE | 1,949 | Multi – primarily European | NA | 450K | Cord blood | 6 | Meta-analysis & replication | Mixed, 0 of 6 loci in newborns, 4 of 6 DMPs in older children | ||
| Isaevska et al.57 | Piccolipin | 384 | European | NA | EPIC | Cord blood | 284 | Replication | Mixed, 10 of 151 DMPs tested | ||
| Gruzieva et al.56 | PACE | 1,551 | Multi – primarily European | NA | 450K | Cord blood | 14 | Meta-analysis & replication | No, 0 of 14 DMPs | ||
| Plusquin et al.58 | ALSPAC and EXPOsOMICS (ENVIRONAGE, INMA, Piccolipiù and Rhea) | 850 | Multi – primarily European | PM | NA | 450K | Cord blood; peripheral blood | 0 – cross-sectional; 1 – longitudinal | Meta-analysis | NA | |
| Breton et al.59 | NA | 240 | Multi – primarily European | Air pollution PM | NA | 450K | Blood spots | 31 | Replication | Mixed, 1 of 2 DMPs | |
| Gruzieva et al.60 | PACE | 1,508 | Multi – primarily European | NA | 450K | Cord blood | 3 | Meta-analysis & replication | Mixed, 1 of 3 DMPs | ||
| Peng et al.61 | Project Viva | 482 | Multi – primarily European | Proximity to roadways | NA | 450K | Cord blood | 4 | Replication | No, 0 of 4 DMPs | |
| Metals and other elements | |||||||||||
| Lozano et al.62 | PACE | 1,462 | Multi – primarily European | Methylmercury | Maternal blood; cord blood; maternal hair | 450K and EPIC | Cord blood; childhood peripheral blood | 2 | Meta-analysis | No, 0 of 2 DMPs | |
| Bakulski et al.63 | Baltimore THREE | 141 | Multi – primarily African | Mercury | Cord blood | CHARM 2.0 | Cord blood | 0 | Replication | Yes, 1 of 1 DMR region replicated | |
| Tian et al.64 | NHBCS and RICHS | 484 | Multi – primarily European | Selenium | Placenta | 450K | Placenta | 5 | Meta-analysis | NA | |
| Everson et al.65 | NHBCS and RICHS | 484 | Multi – primarily European | Cadmium | Placenta | 450K | Placenta | 3 | Meta-analysis | NA | |
| Park et al.66 | MOCEH | 384 | Asian | Cadmium | Maternal venous blood; cord blood | EPIC | Cord blood | 2 | Meta-analysis | NA | |
| Gliga et al.67 | NA | 71 | Asian | Cadmium | Maternal blood and child urine (age 9 y) | 450K | PBMCs – cord blood and peripheral blood | Cord blood – 458; child blood age 9–6 y | Replication | Yes, 1 of 1 DMR tested | |
| Kennedy et al.68 | NA | 447 | European | Copper | Placenta | 450K | Placenta | 0 | Bonferroni | Meta-analysis | NA |
| Park et al.69 | NA | 364 | Asian | Lead | Maternal blood and cord blood | EPIC | Cord blood | 0 (111 in male-only analysis) | Meta-analysis | NA | |
| Wu et al.70 | Project Viva | 268 | Multi – primarily European | Lead | Maternal blood | 450K | Cord blood | 4 | Replication | No | |
| Bozack et al.71 | NA | 120 | Asian & admixed/Latino | Arsenic | Natural experiment and water samples | EPIC & 450K | Blood; buccal cells | PBMCs – 1; PBMCs & buccal cells – 3 | Meta-analysis | NA | |
| Kaushal et al.72 | NA | 64 | Asian | Arsenic | Maternal urine | 450K | Cord blood | 579 | Replication | No, 0 of 553 DMPs tested | |
| Bozack et al.73 | NA | 44 | Asian | Arsenic | Drinking water; maternal toenails | 450K | Cord blood | 380 | Replication | Mixed, 2 of 3 genes tested using pyrosequencing | |
| Zeng et al.74 | NA | 24 | Asian | E-waste exposure (heavy metals) | Prenatal maternal blood (validate heavy metal exposure) | 450K | Cord blood | 125 | Replication | Yes, 2 of 2 DMPs | |
| Other chemicals | |||||||||||
| Liu et al.75 | HOME | 266 | Multi – primarily European | PFAS | Maternal serum | EPIC | Cord blood; peripheral blood at 12 y of age | 2 DMPs for PFOS, 12 for PFOA, 8 for PFHxS, and 413 for PFNA | Replication | Mixed, 6 of 315 DMPs | |
| Miura et al.76 | Hokkaido study | 190 | Asian | PFAS | Maternal blood | 450K | Cord blood | 4 | Replication | No, 0 of 4 DMPs | |
| Miura et al.77 | Hokkaido study | 203 | Asian | di-2-ethylhexyl phthalate | Maternal blood | 450K | Cord blood | 2 | Replication | No, 0 of 2 DMPs | |
| Vilahur et al.78 | NA | 181 | European | Xenoestrogens | Placenta | 450K | Placenta | 0 | Replication | No, 0 of 2 DMPs | |
| Miura et al.79 | Hokkaido study | Male – 123; female – 154 | Asian | BPA | Cord blood | 450K | Cord blood | Male – 27; female – 16 | Replication | No, 0 of 14 DMPs in females, 0 of 26 in males | |
| McCabe et al.80 | MMIP | 69 | Multi – primarily European | BPA | Maternal urine | EPIC | Umbilical cord blood leukocytes | 38 | Replication | No, non-significant and slight correlation between effect estimates of DMPSs in independent study | |
| Yu et al.81 | NA | 24 | Asian | DDT pesticide | Cord blood | 450K | Cord blood | 1,131 | Replication | Yes, 2 of 2 DMPs | |
| Social stressors | |||||||||||
| Alfano et al.82 | ALSPAC | 973 | European | Socio-economic position | NA | 450K | Cord blood; whole blood | Birth:4/childhood:0/adolescence:20 | Replication | No, 0 of 4 DMPs | |
| Other exposures | |||||||||||
| Caramaschi et al.83 | ALSPAC and MoBa | 2,644 | European | Medically assisted reproduction | NA | 450K | Cord blood | 5 | Meta-analysis | No, 0 of 5 DMPs | |
Note: ALSPAC, Avon Longitudinal Study of Parents And Children; BPA, bisphenol A; CCLS, California Childhood Leukemia Study; DDT, dichlorodiphenyltrichloroethane; DMP, differentially methylated position; DMR, differentially methylated region; ENVIRONAGE, ENVIRonmental influence ON early AGEing; FDR, false discovery rate; HOME, Health Outcomes and Measures of the Environment; GECKO, Groningen Expert Center for Kids with Obesity; HELIX, Human Early Life Exposome; HOME, Health Outcomes and Measures of the Environment; INMA, INfancia y Medio Ambiente; MACHS, Moderate Alcohol and Cardiovascular Health; MINERVA, MINimizE Right Ventricular pacing to prevent Atrial fibrillation and heart failure; MMIP, Michigan Mother and Infant Pairs Cohort; MoBa, The Norwegian Mother & Child Cohort Study; MOCEH, Mothers and Children’s Environmental Health; NHBCS, New Hampshire Birth Cohort Study; NA, Not Applicable; PACE, Pregnancy And Childhood Epigenetics consortium; PBMCs, peripheral blood mononuclear cell; PFAS, perfluoroalkyl substances; PFHxS, perfluorohexane sulfonate; PFNA, perfluorononanoate; PFOA, perfluorooctanoate; PFOS, perfluorooctane sulfonate; PM, particulate matter; RICHS, Rhode Island Child Health Study; ROLO, Randomised cOntrol trial of LOw glycaemic index diet to prevent macrosomia; THREE, Tracking Health Related to Environmental Exposures.
Largest reported when multiple exposure tissues, sum of all study for meta-analyses.
Maternal Smoking
Given smoking’s known impacts on offspring health,9 most birth cohort studies collect some smoking data, mostly via questionnaires. Thus, smoking EWAS were among the most frequent and largest of the EWAS we identified. Smoking EWAS have successfully identified robust and well-replicated methylation differences in newborn blood (Table 1), with larger studies revealing more differentially methylated CpGs (DM-CpGs). Smoking EWAS have considered both timing of exposure and of DNA methylation measurement. Changes to methylation appear to reflect sustained smoking across the pregnancy, not smoking limited to early pregnancy.84 Many methylation differences observed at birth persisted into later childhood.33,85 Substantial overlap existed in DM-CpGs from EWAS meta-analyses of sustained prenatal exposure to maternal cigarette smoking in newborns33 and personal current cigarette smoking in adults.86 However, many genes were uniquely differentially methylated in newborns,34 underscoring the importance of timing on exposure influences and epigenetic plasticity.
The placenta provides oxygen, nutrients, and hormones to the fetus. Placental abnormalities have been linked to later health outcomes in offspring.87 Maternal blood perfuses the placenta, exposing it to circulating environmental chemicals. An EWAS meta-analysis (, seven studies) identified widespread impacts of prenatal smoking on placental methylation.35 Overlap with the most prominent DM-CpGs in newborn blood was minimal; reasons for this tissue specificity are unclear. Similar to results in newborn blood, placental findings were much more extensive for sustained smoking than for any smoking in pregnancy, supporting the importance of exposure timing.
Maternal Diet
Maternal nutrition can impact fetal development and child health.88 Assessing food and nutrient intake requires detailed dietary assessments that are less frequently collected than smoking information. Some nutrients are more reliably assessed by blood measurements than by questionnaires, and these more expensive measures requiring appropriate samples are available in even fewer birth cohorts. Thus, fewer EWAS of maternal prenatal diet with smaller sample sizes have been conducted to date than smoking EWAS (Table 1).
Folic acid supplementation in pregnancy is associated with fewer neural tube defects.10 Widespread differential methylation by maternal blood folate levels was reported in an EWAS meta-analysis.47 Prenatal alcohol consumption,89 vitamin D levels,90 iron levels, and Mediterranean diet91 are also associated with fetal and child health outcomes. However relatively large EWAS meta-analyses (n range 1,062–3,075) of these exposures have found few (range: 0–3) DM-CpGs.45,46 For most nutrients and dietary intakes measured by questionnaires, substantial measurement error in dietary assessment will require large sample sizes to adequately test for reproducible associations.
Environmental Contaminants
Prenatal air pollution exposure has been associated with shorter gestation and impaired fetal growth.92 EWAS meta-analyses of and particulate matter (n range 850–1,949) reported between 1 and 14 DM-CpGs (Figure 2). As with diet, air pollution is heterogeneous and generally measured differently and with error across locations, decreasing power.
Prenatal exposures to metals have been associated with adverse offspring health outcomes.93 The largest metals EWAS meta-analysis () of prenatal methylmercury exposure and cord blood methylation reported 2 CpGs []; neither replicated. EWAS meta-analyses of other metals and elements have been small ( range 120–484) and reported few significantly differentially methylated sites (range 0–5). Various other prenatal chemical contaminants (see Table 1) have been examined in EWAS of newborn blood or placenta in single studies, without successful replication.
Social Stressors
Psychosocial stressors before and during pregnancy can impact offspring health.94 One EWAS of maternal socioeconomic position in a United Kingdom cohort () identified differential methylation in blood at 4 CpGs at birth, none in childhood, and 20 in adolescence, but no sites replicated.82 Exposure to psychosocial stressors can be challenging to harmonize across studies. Very large sample sizes will likely be required to find true associations. An ongoing Pregnancy And Childhood Epigenetics consortium (PACE) EWAS project is examining carefully harmonized prenatal stress measures across multiple cohorts.
Other Exposures
A large meta-analysis () comparing children conceived using medical assistance to those conceived without assistance identified 5 DM-CpGs; none successfully replicated.83
Summary of Prenatal EWAS Meta-Analyses and Potential Application as Biomarkers
When extensive, replicable, differentially methylated sites are available, a supremely useful application is the development of methylation biomarkers for prenatal exposures.95 (See Figure 4) For example, robust methylation signals of exposure to prenatal cigarette smoke enabled creation of methylation biomarkers of prenatal smoke exposure in newborns,98 older children,99 and young adults.99 For methylation biomarker generation, it is important to have a gold standard of exposure. In studies of prenatal smoke exposure, measurements of cotinine levels, a biomarker of recent smoking, enable identification of pregnant individuals who falsely report as nonsmokers.98 Objective methylation biomarkers are a major advance for identifying exposure health effects.95 For example, on questionnaires, some smokers falsely deny smoking, with more pronounced underreporting in pregnant persons,100 leading to information bias. Smoking data may also be missing, and missingness is likely nonrandom—another source of information bias. Methylation exposure biomarkers avoid these sources of bias. In addition, methylation biomarkers in offspring offer information on both dose and duration of smoking across pregnancy that are difficult for respondents to accurately report.101 Epidemiological questionnaires often only considered any smoking during pregnancy. However, half of women smoking at conception quit soon thereafter.33 Exposure timing is critical for health impacts, because sustained smoking across pregnancy reduces birth weight, whereas smoking ending early does not.102 This finding matches EWAS findings, where methylation signals reflect sustained smoking across pregnancy, not transient early smoking.33,84 In contrast, cotinine, the previous primary biomarker of smoking, reflects only very recent exposure. By capturing additional parameters of exposure that are difficult to collect on questionnaires and identifying smokers who would be classified as nonsmokers, methylation biomarkers should improve researchers’ ability to accurately estimate health impacts of exposure. Because some methylation biomarkers of prenatal smoke exposure persist,99 they are useful in childhood studies. Smoking strongly correlates with many harmful exposures. Methylation biomarkers allow thorough adjustment for confounding introduced by smoking exposure when studying other exposures. Clearly methylation biomarkers greatly improve on previous assessments of prenatal smoke exposure.
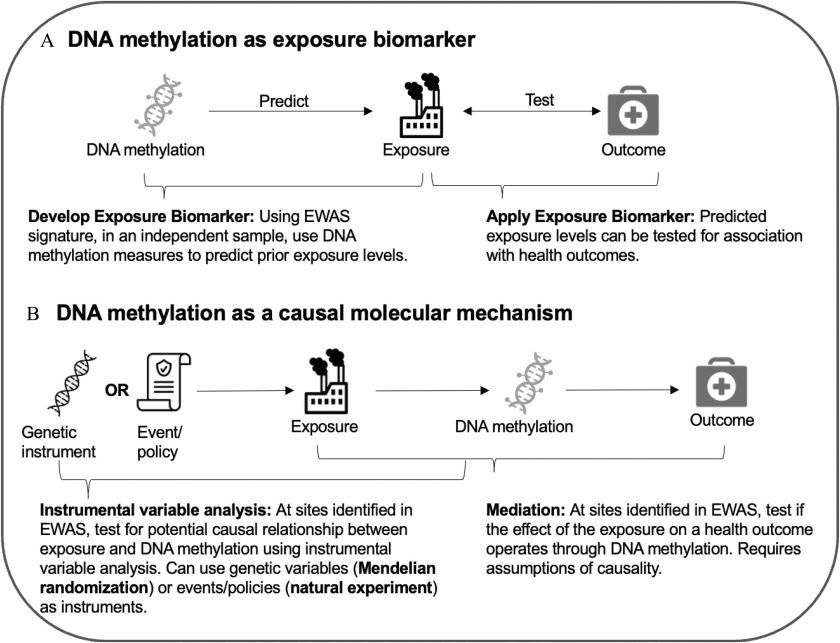
Figure 4.
Prenatal exposure EWAS findings can lead to multiple downstream research opportunities. First, prenatal exposure EWAS findings can be used to develop and test a DNA methylation-based exposure biomarker. Active methods development work is optimizing the DNA methylation site selection procedures and improving biomarker signals.95 Second, once a DNA methylation exposure biomarker has been developed, this tool can be used to test associations between exposures and health outcomes. This approach is particularly useful in situations where other exposure measures are not feasible. Another potential application of prenatal exposure EWAS findings is to establish causal relationships. Instrumental variable analysis can be used to test potential causality between the exposure and DNA methylation. Mendelian randomization uses genetic factors as an instrumental variable, but this approach may be challenging for environmental exposures where genetic factors that contribute to exposure absorption, distribution, metabolism, and excretion are poorly characterized. Randomized controlled trials are feasible for nonhazardous dietary factors. Natural experiments featuring an event or policy that alter exposure levels may be a more realistic instrumental variable for prenatal exposures.96,97 However, these would need to be drastic events, such as a policy banning a commonly used chemical. Last, prenatal exposure EWAS findings can be used in mediation analyses of exposures and health outcomes. In this case, DNA methylation is tested as a mediator linking exposure to a health outcome. This scenario is challenging when DNA methylation is a strong biomarker of the exposure, leading to overestimation of mediation effects. Note: EWAS, epigenome-wide association study.
Success of EWAS in generating biomarkers of maternal smoking provides a proof of principle for other pregnancy exposures. Routinely stored newborn blood spots become a goldmine of information about the prenatal environment for studies of later childhood health outcomes.103 Even for exposures that can be queried in later childhood, methylation biomarkers avoid biased recall influenced by disease diagnosis.
Standard and novel statistical and computational approaches to biomarker generation from EWAS findings have recently been reviewed.95 Reliable biomarker generation is possible only when substantial and reproducible differential methylation is identified. To date, only smoking and folate meet this criterion (Figure 2). In the “Discussion” section, we highlight important factors influencing EWAS power and suggest advances that will lead to better-powered EWAS of many exposures and thus novel biomarker generation.
Discussion
Our review suggests that although the number of prenatal EWAS conducted has increased over recent years, reproducible DNA methylation signatures for prenatal exposures besides cigarette smoking and folate are currently lacking. We hold that this reflects a lack of power, rather than a failure of the approach. To expand on current research and potentially identify additional effects of prenatal exposure on methylation, better-powered EWAS are necessary.
The genome-wide nature of EWAS impacts power. In any high-dimensional genomic analysis, the large number of statistical tests results in greater testing burden and requires larger sample sizes. The burden of multiple tests creates a trade-off between higher coverage of the methylome and reduced power. As technology advances, more sites are tested for differential methylation. We saw this happen with the shift from the earlier Illumina450K array ( CpGs) to the higher coverage EPIC ( CpG) and the EPIC version 2 ( CpGs) arrays and it may happen again with the implementation of reduced representation or whole genome sequence bisulfite sequencing. Each time, the multiple testing burden correspondingly increases. When holding all other factors constant, larger sample sizes increase power. Yet the largest EWAS conducted to date are small in comparison with genome-wide analyses. For example, the most recent birth weight GWAS has a sample size of 321,223,104 whereas the largest EWAS of birth weight has a sample size of 8,825.105 In GWAS, larger sample sizes have generally led to identification of many additional genetic variants with smaller effect sizes. The larger the expected magnitude of the association, the higher the power to detect it. For maternal smoking and newborn methylation, only a few genes, such as AHRR, GFI1, and MYO1G, have CpGs with large effect sizes—10%–15% differences by exposure group.33 However, the average smoking effect sizes across all significantly differentially methylated sites have been small, in the range of 0.5% differences34 but nonetheless are highly reproducible across studies. In EWAS meta-analyses of other exposures to date, effect sizes have generally been small but are potentially important for child health.25 For studies to identify these small differences, or reliably conclude that they do not exist, samples sizes need to be large.
The power of any exposure effect study is also a function of the exposure prevalence (for a binary exposure) or variability (for a continuous exposure) and of the precision and validity of the measures of both outcome and exposure.106 Optimizing these factors is often more difficult to achieve than simply increasing the overall sample size. We reflect on some of these factors and give recommendations for addressing them below.
New pregnancy cohort studies seeking to perform prenatal environmental EWAS studies in the future can take advantage of lessons learned in the field and prepare themselves to take advantage of emerging approaches. Each study will have specific research questions and resource constraints. Below, we walk through general recommendations to improve study power and validity at each stage of the sample and data collection process. Rapid developments are being made in environmental epigenetics. The greatest improvements in the ability of EWAS to detect prenatal exposure effects will come from advances in inclusion, methods, and biological interpretation.
Diversity, Equity, and Inclusion
Study participants are the foundation of every epidemiological research study. Most prenatal (Figure 2) and adult EWAS identified in this review were from participants of European genetic ancestry.107 GWAS faced a similar limitation and took steps to increase coverage of diverse populations.108 Multiethnic studies enable confirmation of signals in diverse groups, while identifying those unique to one group who would otherwise escape discovery. Methylation is partially heritable and levels at specific loci may differ across populations.109 As in GWAS, inclusion of diverse populations will extend the informativeness of EWAS.107,110 Partnering with communities to ensure research participants are represented from diverse backgrounds is another essential area. We believe that expanding the participation in epigenetic research will allow for the identification of signals that are either generalizable across groups or specific within groups, which is important for biomarker validation and mechanistic investigation.107 Similarly, it will be important to expand the representation of participants included in reference databases that are used for functional annotation and interpretation of EWAS findings, such as the Genotype Tissue Expression (GTEx) database.111
For environmental epidemiology studies, it is important to recruit participants who may have a wide range of exposure levels. Environmental health disparities across populations112 can influence exposure patterns relevant to prenatal EWAS.113,114 Including diverse populations who may have greater exposure variability will increase power to detect exposure related differential methylation. Understanding complex factors underlying exposure variation across geographic and racial/ethnic groups is critical to reducing disparities115 and should inform future EWAS study designs and interpretation of findings.
More diverse, interdisciplinary research teams lead to more creative, innovative, different, and broad research questions116 and increase equity in the field.117 Reducing structural barriers in training and funding, as supported by the recent National Institutes of Health UNITE initiative,118 will enhance prenatal epigenetics research.119
Journals should require authors to deposit entire EWAS meta-analysis results in public databases such as the EWAS Catalog120 and should be annotated for ancestry. Deposition will support replication testing in new studies, enable creation of methylation risk scores, and stimulate mechanistic studies.
Tissue Selection and Biospecimen Storage for Epigenetic Measures Including Cell Type–Specific Samples
Most prenatal EWAS use cord blood because it is easily collected at birth and may be a surrogate tissue. To promote harmonization of epigenetic measures with existing cohorts, we recommend that new perinatal epidemiology studies collect newborn cord blood, regardless of the additional samples they are collecting. Collection of other available tissues should be based on health outcomes of interest and feasibility. For example, placental tissue has recently been an effective model of neurological and cardiovascular disorders and is an emerging tissue type in environmental EWAS.35 If the newborns in a birth or pregnancy cohort will be followed into childhood, additional accessible tissues should be considered. In children, blood collection is challenging, and thus buccal cells or saliva are often used.121 Sampling disease-relevant target tissues can improve EWAS inferences.122 Nasal epithelium, a proxy for the lower respiratory epithelium,123 is easily collected in children and has been used for EWAS of asthma and rhinitis124 but could also help identify local effects of inhaled exposures. Skin cells can be obtained noninvasively in children using tape and could be useful for EWAS of eczema or sun exposure.125 Follicles from plucked hairs can provide a source of stem cells.126 Fibroblasts can be reprogrammed into induced pluripotent stem cells for further toxicological investigation.127 Blood can be challenging to collect in population-based studies of children. However, for some exposures or health outcomes, blood may indeed be the relevant target tissue or an excellent surrogate.122 The postnatal tissues of interest may influence the selection of tissues at birth to enable longitudinal epigenetic measures within a common tissue type.
DNA methylation analyses can be performed on whole blood or whole tissue specimens, which can be stored frozen using standard practices. If the study’s goal is assessment of epigenetic marks beyond DNA methylation, assessment of specific types of cells (for example, mesenchymal stem cells128) or measurement of single cells, careful investigation of the type of collection tubes and storage and processing is essential. Standard freezing lyses cell membranes; thus cell culture approaches or traditional cell counting methods, such as complete blood counts, or cell sorting with fluorescence-assistance, are possible only with fresh or specially cryopreserved samples. These analyses may mandate the use of cryopreservation tubes for blood collection. Use of cryopreservation tubes require centrifugation to isolate peripheral blood mononuclear cells (PBMCs) within a few hours of collection and aliquoting into cryovials containing freezing media. Sampling processing steps may be less feasible during field sample collection, such as during home visits. Smaller studies may be able to invest in the processing to focus on specific cell types, whereas larger studies may opt to use bulk tissue specimens such as whole blood. If biospecimens will also be used to measure other epigenetic factors, additional consideration is required. If RNA will be analyzed, RNA stabilizing solutions will be required. Ensuring higher quality RNA samples at collection and storage can save money during sequencing. Better-quality RNA samples can use more affordable poly(A) tail selection and library preparation methods than the more expensive ribosome depletion methods required for poorer quality samples. In our experience, routinely discarded term human placental tissue often has lower RNA quality, as expected, based on the physiology of pregnancy and labor,129 relative to other tissues such as blood, with implications for RNA sequencing approaches. Many standard RNA extraction protocols omit small RNAs, and additional care must be taken if these are of interest. Epigenetic measures requiring chromatin (histone modifications, ATAC-seq) must also be considered at the sample-collection phase. Sample collection and storage methods can influence stability of epigenetic measures.130 Samples can be processed fresh for these measures, and archiving recommendations for either snap freezing or slow freezing vary by tissue and lab.
Most tissues are complex mixtures of cell types with distinctive methylation patterns. Including cell type proportions in EWAS regression models can improve precision.131 Many exposures influence cell composition,132 however, which might mediate associations of exposures with methylation, and conditioning on a mediator can introduce spurious effects. Cell proportions in bulk tissue can be estimated from methylation measures using reference panels for many tissues including cord blood,133 saliva,134 and placenta.135 Cell proportions estimated from methylation provide another relevant outcome measure.131 Newer methods leverage single-cell RNA sequencing (scRNA-seq) data to deconvolute bulk tissue methylation data.136 ScRNA-seq identifies untargeted cells in the tissue, including cells in various states of activation or proliferation that might be influenced by exposure.137 Improved cell estimation will also enable investigators to discern whether exposure–methylation associations are driven by specific cell types.138 For example, smoking effects might be partially mediated by activating subpopulations of immune cells not captured in published reference panels.139
Improved and Standardized DNA Methylation Measurements
As noted above, current methylation arrays cover only a fraction of the DNA methylome (935,000 CpGs on the IlluminaEPIC version 2.0 array vs. 28.3 million in the genome). Beyond the coverage-burden trade-off discussed above, the validity of DNA methylation measurements can also impact the ability to reliably detect associations with prenatal exposures. As expected, associations with exposures or outcomes have been disproportionately observed at CpGs that are measured more reproducibly.140 Further, both arrays include many sites largely invariant in blood in general populations.141 At these invariant CpGs, even the most precise measurements will not be sufficient to reliably capture the tiny differences between individuals. Reproducibility of measurement tends to be lower at CpGs with either very low () or very high () levels of methylation.142 Removal of invariant probes prior to analysis has been suggested to decrease statistical testing burden and therefore increase power.143 However, even at some loci with high interindividual variability, reproducibility has been shown to be low.140 In the meta-analysis setting, it would be important to show that these probes are uniformly invariant or poorly reproducible across most populations, but such data are lacking. Increased precision of DNA methylation measurements will increase power. Illumina is developing a methylation screening array intended to be sold at a lower price with far fewer CpGs than the current EPIC version 2, targeting CpGs differentially methylated in prior EWAS plus other informative content. We anticipate that as the price of methylation arrays come down, using methylation exposure biomarkers in population studies will become more feasible and widespread. Like newer genetic arrays tailored for population-based studies (e.g., Illumina Global Diversity Array), methylation arrays tailored for using blood from diverse populations would increase power of EWAS.
Whole genome bisulfite sequencing is increasingly being recommended to increase genomic coverage. Although the increased genomic coverage enhances completeness of environmental methylation signatures, it also increases the statistical testing burden, decreasing power and requiring larger sample sizes. To improve the feasibility of implementing sequencing in epidemiological cohorts, decreases in sample input requirements and cost are needed. Alternative and more affordable sequencing approaches, including reduced representation bisulfite sequencing or sequencing following TruSeq Methyl Capture EPIC library preparation, can limit the genomic areas sequenced, thereby maintaining power. The TruSeq approach is designed to be compatible with microarray approaches and users have observed high technical correlations.144 High sequencing depth is recommended for methylation quantification.145 Low sequencing depths can result in measurement error, also decreasing power.
Like most laboratory assays, methylation arrays are subject to batch effects and lab drift. In early EWAS, expectation of similarity to genotyping, which is relatively robust to batch effects, contributed to underappreciating the importance of randomizing cases and controls, or exposed and unexposed, across plates and batches. Batch effects in sequencing data can be an even larger issue than for array-based measures.146 A recent comparison of split sample reproducibility in measurements of methylation in newborn blood samples between the EPIC array and bisulfite sequencing found the array to be more reproducible.147 Improved preprocessing and analytic methods for sequencing data will be needed to incorporate these into consortia. With either array- or sequencing-based methods to assess methylation, appropriate control for technical sources of variation is needed and improves power.148
Even though sequencing approaches are becoming cheaper and offer better coverage, to promote replication and meta-analyses, investigators may strongly consider including an Illumina DNA methylation array for greatest harmonization opportunity with other cohorts. The explosion of software for all aspects of processing and analysis of array data makes them more accessible to those with less-specialized bioinformatic assistance or training. For the widest genome-wide discovery, investigators may select whole genome bisulfite sequencing approaches. Because there are few epidemiological studies with sequencing measures available for replication testing, investigators may need to adopt a testing and replication sample set within their study population. To balance costs, investigators may opt for an enriched sequencing approach, such as reduced representation bisulfite sequencing, but this approach may have limited overlap of coverage with previous studies. When performing laboratory epigenetic measures, we strongly recommend that investigators group multiple longitudinal specimens for measurement at the same time to avoid batch effects. This methodology may require waiting to measure specimens from an earlier collection period until specimens from a later collection period are available. This approach will allow for the analysis of longitudinal epigenetic associations, which enhances the ability to make causal inference. Stacking multiple omics measures on the same participants improves the biological interpretability of findings. For example, we recommend measuring DNA methylation, genetics, and RNA expression in the same participants.
Improved and Standardized Considerations for Exposure Assessment
Issues in exposure assessment for environmental epidemiology, including special considerations in pregnancy, have been extensively discussed elsewhere.149–152 Here we briefly touch on points of special relevance to prenatal epigenetic studies. The power to detect exposure effects on an outcome, including differential methylation, depends on both its prevalence (binary exposures) or variability (continuous exposures) and on how well the exposure is measured. If the exposure is very rare or hardly varies, even the largest study will be poorly powered. For most exposures, populations differ in the prevalence and/or variability of any exposure, emphasizing the importance of diversity in populations as discussed above. Regardless of the exposure’s prevalence/variability, if an exposure is poorly measured, exposure misclassification can impact validity and power. Generally, if misclassification is nondifferential with respect to the outcome (as is often the case for prenatal exposures and DNA methylation levels), the resulting bias is toward the null.153 In the setting of meta-analysis (or pooled analysis), exposure misclassification will be reduced when the exposure metric can be well harmonized across studies. For prenatal exposure EWAS, considerations of persistence of the chemical are important. For nonpersistent chemicals with shorter half-lives (such as phthalates with a urinary half-life h), repeated measures during pregnancy better characterize exposure.151,154 For some prenatal exposures, timing during gestation of exposure may matter. For example, for maternal smoking during pregnancy, methylation signatures in newborns predominantly reflect exposure that lasts throughout the pregnancy rather than smoking that ends early in the pregnancy.33,84 About half of women smoking at the beginning of pregnancy quit early during the pregnancy, but it is very uncommon to take up smoking de novo later in pregnancy.33,84 Thus, we cannot be certain that sustained smoking across pregnancy is required or whether new smoking late in pregnancy would lead to similar methylation patterns in newborns. Exquisitely detailed measurement of exposure will not increase the power to identify differential methylation if the timing captured during the pregnancy is not the relevant one. We recommend careful consideration of timing, frequency, and method of exposure measurement because these can impact the power and validity of an EWAS.
The decision of what method to use to measure an exposure often relates to pragmatic issues of availability, expense, and participant burden as well as the research question at hand. Some exposures, such as prenatal smoking, are feasible to assess using questionnaires, albeit with caveats expressed earlier, which decreases costs of data collection, facilitating larger sample sizes. When collecting exposure data via questionnaire, prospective data collection is important because after development or diagnosis of the condition, recall bias can influence participants’ exposure reports. Many prenatal exposures of interest cannot be reliably measured via questionnaire.
Sometimes multiple sources of exposure data can be collected and combined to improve measurement reliability. For example, outdoor air pollution exposure can be estimated by combining residential history with measured exposure levels at routine monitoring stations and/or satellite data, using modeling with land use features, point sources of exposure, and spatiotemporal factors, and that can incorporate corrections for measurement error.155,156 Measures relevant to climate change, including flooding and heat islands, can also be obtained.157 Residential history-based measures are least invasive for participants (no participant contact or biospecimens required) and can be selected for various periods of time to test windows of susceptibility. Additional information on how and where participants spend their time can be incorporated with additional participant burden. These strategies may not reflect what enters the body or a given target tissue.
External doses to chemical exposures can be measured using an emerging exposure assessment approach to monitor human contact. Silicon bracelets are worn by participants for a period (generally a week), and chemicals deposited on the bracelets can be quantified.158 Bracelet approaches are appealing to many new studies for their ease of use, though exposure levels do not reflect the internal dose of an exposure, because the body’s barriers prevent absorption of a portion of external doses.
Internal exposure doses are generally measured in biospecimens from participants, such as blood, urine, hair, nails, or teeth. The feasibility of collecting each type of biospecimen may vary by participant group, and the half-life of exposure measures may vary by sample type, such as urine vs. blood. Exposure to many chemicals varies with calendar time due to secular trends in industrial, policy, or personal practices. This variation should be considered in deciding when to initiate or complete data collection. Accounting for time of day of collection, time since last consuming food, and delay from collection to freezing as potential confounders is often needed in environmental epidemiology even in cross-sectional studies. Exposure assessment in urinary specimens requires a correction for hydration status or dilution, which is most often done through measurement of urinary specific gravity or creatinine levels. Urinary exposure measure interpretation may be further impacted by a number of physiological changes that occur during pregnancy, including changes to glomerular filtration rate, urinary output, creatinine excretion.159–161 Biomarker concentrations may also correlate with a participant’s physiology; for example, 1,1-dichloro-2,2-bis(p-chlorophenyl) ethylene (DDE), a pesticide metabolite, is correlated with adiposity,162 and numerous xenobiotics that are excreted in the urine are correlated with kidney function.163 Genetic factors may influence absorption, metabolism, or excretion of xenobiotics, which influence measured biomarker concentrations.164 Based on the target window of susceptibility of the chemical under investigation, different tissues may be selected. Laser ablation inductively coupled–mass spectrometry (ICP-MS) of shed baby teeth in children can simultaneously quantify exposure to multiple metals during the in utero period with precise timing information.165 This method has been adapted for additional tissues, including placenta and brain, allowing for cell type specific exposure resolution.166 Consideration must also be given to avoiding contamination in collection. For example, if trace metals will be measured, samples must be collected in metal-free tubes. Some analytes are impacted by freeze–thaw cycles, which influence the size of aliquots for storage. Whether the analytes of interest are stable in freezer storage needs to be ascertained in advance. If no prior data are available, pilot studies will be required.
When combining exposure data across cohorts for an EWAS meta-analysis, divergent exposure estimation methods can increase misclassification and reduce power. The Environmental Influences on Child Health Outcomes (ECHO) consortium combines U.S. birth cohorts to examine health effects of prenatal environmental exposures.27 ECHO is applying common exposure estimation techniques for air pollution across birth cohort studies167 and is measuring prenatal environmental exposures in central laboratories using common technologies. There is a similar European cohort harmonization project, Advancing Tools for Human Early Lifecourse Exposome Research and Translation (ATHLETE).168 Refined exposure harmonization should reduce measurement error and increase power.
Improved exposure assessment (Figure 1B) should decrease exposure misclassification, thus increasing power to identify prenatal exposure-related differential methylation. Even for persistent chemicals where single measurements might be acceptable, most chemical exposures are currently measured individually at high cost. Technologies to measure hundreds of compounds simultaneously, including nuclear magnetic resonance and mass spectrometry,169 enable assessment of the exogenous exposome. We believe that improvements in techniques to capture time-integrated multiple early-life exposures with high precision and low cost will revolutionize the field.
Replication, Meta-Analyses, and the Need for Improved Statistical Methods
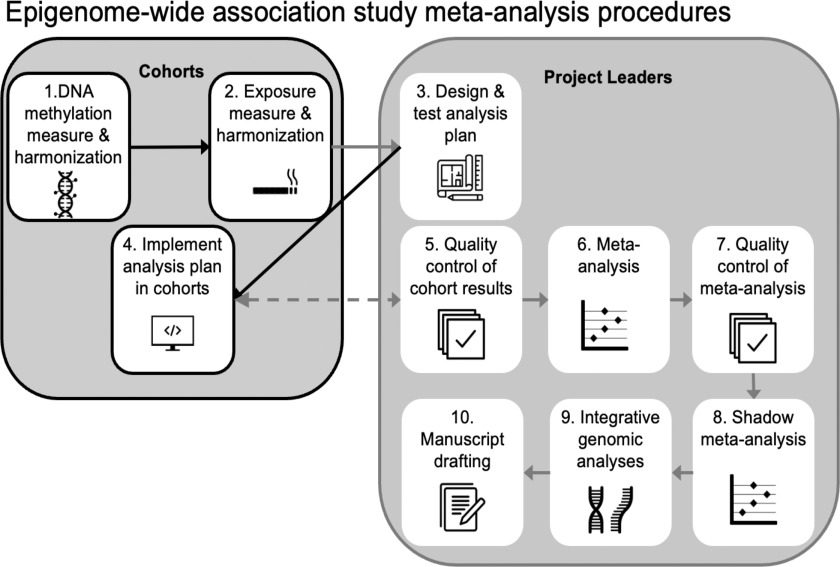
Of note, the prenatal EWASs with the largest sample sizes in our review were meta-analyses or pooled analyses conducted through collaboration via consortia. An international consortium examining the prenatal environment’s impact on early-life methylation, and subsequent impacts on later child health outcomes is the PACE consortium.26 PACE was modeled on highly successful GWAS consortia that have produced a wealth of reproducible and reliable results linking genetic variants to numerous phenotypes and diseases.170,171 PACE and other EWAS consortia apply a multistep meta-analysis approach (Figure 3). This multistep approach can improve replicability (by verifying an association exists across multiple populations), reproducibility (by increasing sample size and implementing quality controls on analyses), and rigor (for example, by developing harmonized definition of exposure included in clear analysis plans that are tested before distributing to participating cohorts, by performing an independent shadow repeat of the meta-analysis from scratch). We note that some prenatal EWAS of relatively small sample size, which were neither performed as meta-analyses nor attempted replication, nevertheless reported large numbers of significant findings (Figure 2D), which may reflect false positives or otherwise nonreplicable findings. We believe this underscores the importance of meta-analysis and replication attempts. To increase the rigor and reproducibility of the research, we recommend that new studies partner with existing consortia where possible to take advantage of existing expertise and processes as illustrated in Figure 3 to enable better-powered, higher-quality studies. Continued increases in power will enable detection of weaker differential methylation signatures, as we might expect for exposures measured less precisely than smoking or those that vary less. Lower costs of both exposure assessment and methylation measures will also facilitate larger EWAS.
Figure 3.
Overview of the suggested process for conducting a prenatal exposure EWAS meta-analysis in the consortium setting. Note: EWAS, epigenome-wide association study.
The standard method to combine study-specific results in genome-wide analyses is fixed-effects meta-analysis,172 which weights by the inverse of the variances to produce an overall effect estimate and standard error. Although it is often stated that random effects models should be used when there might be between-study heterogeneity, Rice et al.172 show that inverse variance–weighted average estimates a reasonable and interpretable parameter, even under the assumption that effect sizes differ. They further point out that a fixed-effects meta-analysis does not require the assumption of homogeneity. Rather than relying on tests of homogeneity, it is important to evaluate meta-analysis effect estimates along with visualization both of study-specific estimates and leave-one-out analyses whereby the meta-analysis is repeated after leaving out each study. These more convincingly demonstrate whether meta-analysis results are driven by a single study than statistical tests of heterogeneity.172 In the setting of environmental EWAS, the exposure may have been measured in greater detail and with higher precision in some studies than others. Studies with more accurate characterization of the harmonized exposure metric will tend to have larger effect sizes and smaller standard errors, which would make them more influential in inverse variance–weighted meta-analysis.
Pooled analyses167 are an alternative to meta-analysis that enable more flexibility in the analysis, including ability to perform additional analyses that might be suggested by reviewers. However, pooled analyses require sharing of underlying data, which requires more administrative person-time to coordinate human subjects issues and data transfer agreements as well as extensive data harmonization.
New studies should identify others with the most similar designs and sample-collection protocols to facilitate future collaboration and replication. Studies can partner for coordinated epigenetic and exposure measures, which makes pooling of data across cohorts more feasible.173 If appropriate partnerships cannot be found, new studies performing prenatal environmental EWAS should look up their results in comparison with previously published results, a routine step even in consortium meta-analyses. Prior studies may differ in their exposure assessment approaches or tissue of DNA methylation measure, but comparing results across studies will help readers understand which findings are generalizable and which may either be population specific or potentially false positives.
The exposome is a complex mixture. Statistical mixture methods are currently computationally intensive, even for a few analytes, and thus not applicable for high-dimensional epigenomic analyses.174 We expect that the development of computationally efficient methods will improve the ability to capture associations between the exposome, methylation, and health outcomes.
Statistical advances have improved EWAS, including consideration of bias and inflation across genome-wide models175 and dealing with extreme methylation values prior to analysis, such as by winsorizing.176 Improved methods are needed to address potential heterogeneity from incorporating multiple genetic ancestries in EWAS meta-analyses. New methods are also needed to optimize biomarker development and validation in the EWAS meta-analysis setting.
Many methods papers propose novel methods to address an EWAS limitation. However, when these methods are tested only in one or few publicly available data sets, which may vary in quality control, it becomes difficult for EWAS researchers to evaluate how widely applicable they may be. EWAS consortia facilitate testing new methods in multiple data sets. We believe that finding a superior method to standard approaches across many different studies is most informative.
Epidemiologic Approaches to Investigate Persistence and Causality of DNA Methylation Signals
Although it is possible that differential methylation at genes involved in metabolism of a xenobiotic could influence measured concentration, in general methylation is unlikely to influence the likelihood of exposure. Therefore EWAS of environmental exposure are less likely to result from reverse causation than EWAS of health conditions, where the direction of association can be difficult to determine. For nonharmful dietary exposures, sufficiently powered randomized controlled trials could be used to confirm causality of the relationships between the exposure and DNA methylation. Air pollution epidemiology studies have also used short-term controlled exposure chambers,177 which have been deployed in a randomized design for DNA methylation research.178 Natural experiments, whereby a drastic change in exposure occurs within a population because of policy changes or other unusual events, can provide an opportunity for exploring causal exposure effects.96,97
When methylation data are available from both newborns and older children, persistence of signals present at birth can be studied. Assessment of the same exposure in childhood is required to assess whether persistent signatures reflect postnatal, rather than prenatal, exposure. Currently much more EWAS data are available regarding prenatal exposures than for later in childhood. The Avon Longitudinal Study of Parents And Children (ALSPAC) study measured methylation at birth, childhood, and adolescence in the same analytic runs. Accounting for postnatal smoke exposure, ALSPAC found persistence of prenatal exposure differential methylation varied substantially across loci,85 a result mirrored in later cross-sectional meta-analysis.33 Future studies with careful repeated measures are necessary to identify persistent signatures of other prenatal exposures. We expect that in addition to being useful biomarkers in later life of prenatal exposure, loci retaining these signatures might be more likely to be involved in the etiology of childhood and adult health outcomes. Based on the likely relationship of exposure influencing methylation, the next step is to determine if exposure-related methylation contributes to causation of exposure-related health outcomes.
Although prenatal exposure biomarkers are a major advance, even without elucidating mechanisms,95 determining whether methylation signals are on the causal pathway to exposure-related health outcomes is of primary interest. Mediation analysis, a statistical technique developed to assess causal relationships between exposures and outcomes in social psychology research,179 is commonly used. Methylation is tested as a mediator of the exposure–disease relationship (Figure 4). For maternal smoking, large-effect size CpGs41 in three genes were tested as mediators of the well-established association between smoking and reduced birth weight. These CpGs were estimated to mediate nearly half of the effect of self-reported any smoking during pregnancy on birth weight. However, any smoking during pregnancy has a much weaker association with birth weight than sustained smoking,98,102 and these smoking-related CpGs capture sustained smoking much better than self-report. One such CpGs is so powerful a biomarker of lifetime personal smoking history that it was patented for use in the insurance industry.180 Because smoking CpGs are such strong biomarkers, false positive evidence of mediation between self-reported exposure and the outcome can result.181 Further, it seems biologically implausible that one, or few, CpGs in blood, among the thousands differentially methylated by maternal smoking,33 could mediate so much of smoking’s impact on birth weight.41,182
Another statistical technique used to assess whether exposure-related methylation causes exposure-related health outcomes is Mendelian randomization (MR). To evaluate whether exposure-related methylation is causal for an exposure-related health outcome, MR uses genetic variants related to the exposure-associated methylation and to the outcome only through the pathway of methylation.183 Even when many CpGs are differentially methylated by exposure, there may be few genetic instruments meeting these criteria, and most genetic instruments for exposure CpGs are weak predictors. Fully half of CpGs on the Illumina450K platform are related to nearby genetic variants, but the variation in methylation explained is exceedingly low.184 Further, local genetic correlation can lead to noncausal associations between genetic variants and methylation in cis,185 obscuring inferences. Pleiotropy assumptions of MR can be difficult to verify.6 Another limitation of MR for interpreting blood EWAS findings is that genetic instruments predict methylation of the CpG site in blood, which is likely a proxy for epigenetic processes in the relevant target tissue.186 Along with mediation, MR requires strong casual inference assumptions that are generally difficult to meet or verify.187 Understanding whether exposure-related differential methylation causes exposure-related disease will require identification of the underlying epigenetic mechanisms.185
Laboratory Approaches for Mechanistic Studies of DNA Methylation Signals
We currently lack insight into basic biological mechanisms linking exposures to sequence-specific methylation. Nutrients, such as folic acid, vitamin B12, and choline, provide the methyl substrate used in methylation.188 Chemicals that cause oxidative DNA damage may inhibit DNA methyltransferase enzyme binding with DNA, resulting in hypomethylation.189,190 Although one might predict that these chemicals would produce global, random shifts in methylation, they are often associated with altered sequence-specific methylation. Mechanisms for the sequence specificity remain largely unknown, limiting our ability to understand whether they cause health outcomes.191 Some hypotheses regarding mechanisms of exposure impacts on DNA methylation, as observed in EWAS investigations, have been proffered. The sequence-specificity suggests involvement of transcription factors.191 Exposures may activate or repress transcription factors that either hinder or facilitate gene-specific differential methylation.192 Alternatively, or additionally, exposure-related methylation changes may be proxies for histone modifications that alter gene function and contribute to exposure-related disease pathogenesis.186 Indeed, during normal development, histone modifications often precede methylation changes.193 Thus, differential methylation might be downstream impacts of exposure-induced histone changes rather than direct exposure effects.
Controlled, laboratory-based validation of population-based findings will provide important mechanistic insights. One approach involves examining downstream effects after experimentally modulating methylation levels. Traditional in vitro methods to modulate methylation use compounds such as 5-azacytidine,194 or alter levels of methyl donors195; both influence methylation genome-wide and often have off-target effects. Like CRISPR methods to modify genotypes at specific positions,196 new epigenetic editing methods allow for sequence-specific alteration of methylation levels in animal and cell culture models.197 Although current epigenetic editing approaches are less accurate than genetic methods, we observe that epigenetic editing is already revolutionizing investigation of the effects of methylation differences by helping identify the downstream biological processes resulting from exposure-induced differential methylation.
Precisely how methylation alters gene expression is poorly understood.185 Methylation upstream of a gene in cis is generally assumed to be associated with reduced gene expression. In several EWAS of exposures or health outcomes, this canonical inverse association (higher DNA methylation with lower gene expression) holds for about two-thirds of DM-CpGs.33,47,198 The correlation with gene expression depends on genomic context,199 and many exceptions to the canonical association exist. By combining modern causal inference methods with genome-wide analysis,185 specific and directional effects of gene expression on trans methylation ( from gene) were identified at 818 genes. Most of these genes were neither transcription factors nor previously known to regulate methylation, pointing toward future research avenues. Understanding the fundamental epigenetic mechanisms influencing gene expression will enable meaningful interpretation of EWAS findings.
Multi-omic analyses for functional annotation, biological interpretation, and discovery.
Methylation is only one aspect of the epigenome. Additional levels include regulatory RNA and histone modifications. Storing samples for measurement requires steps currently too labor-intensive to be practical for large studies. Modified sample storage in future studies could enable better epigenome assessment. Each type of histone modification requires separate measurement.200 Development of high-throughput, low sample input methods to interrogate multiple epigenomic features on routinely stored samples will be a major advance.
Incorporating multiple types of omics on the same samples (e.g., gene expression, genetics, metabolomics, proteomics) can improve biological interpretation and mechanistic insights from EWAS findings.175,201–204 A common first level of integrated omics analyses with EWAS (Figure 3) is to look at correlation between gene expression and DNA methylation at differentially methylated CpGs. There are relatively few publicly available data sets with both Illumina methylation array data and gene expression in cord blood.205,206 For this reason, prenatal epigenetic studies often also look up key findings in data sets with paired blood methylation and gene expression from up to a few hundred children207 or much larger studies of adults.208,209 Given the tissue specificity of both methylation and gene expression, of considerable interest is the recent addition of Illumina EPIC methylation data to the GTEx gene expression resource across nine different tissues from 987 adults.210 Colocalization methods211–214 and integrative epigenomic215 tools are increasingly aiding in EWAS biological interpretation. For example, eFORGE integrates multiple layers of epigenetic data to identify blood EWAS findings with potential functional impact in disease-relevant target tissues.215 Such tools will become more valuable as the underlying epigenomic sample sets amalgamated by the International Human Epigenome Consortium216 become larger and incorporate more diverse populations.110
Development of methods to use multi-omics data to discover novel loci or intermediate omic phenotypes, as opposed to interpreting EWAS204 loci, is an active area of research173 and well-validated methods have not been established or widely employed in epidemiology. Multi-omics methods were reviewed, and their limitations were discussed in 2020.217 A more recent approach is multi-set correlation and factor analysis, an unsupervised integration method to enable novel inference from multi-omic data.218 Widespread future implementation of computationally efficient methods for multi-omics discovery, including epigenomic data, will help understand mechanisms of health impacts of prenatal exposures.
Summary
The early-life environment impacts health throughout the life course, and epigenetic mechanisms play a key role in programming responses to these exposures. Methylation is the epigenetic mechanism best epidemiologically studied to date, thanks to stable platforms with reasonable epigenome-wide coverage. Prenatal environmental exposure to smoking is associated with highly reproducible and specific methylation signatures that have great utility as biomarkers of early-life exposure. Improvements to study design could result in robust biomarkers for other exposures. Best practices for EWAS include careful study design, attention to tissue- and cell-type heterogeneity, rigorous exposure measurement, large and diverse study populations, and replication. Environmental epigenetics is an emerging area with promising research and clinical implications. Mechanisms underlying the specificity of exposure methylation signatures and consequences of the methylation differences are currently unclear but are a crucial future research area essential for developing clinical applications.
Supplementary Material
Acknowledgments
The authors appreciate S. Mantooth, from National Institute of Environmental Health Sciences (NIEHS) library for conducting the literature review for this manuscript. The authors acknowledge K. A. Campbell of the University of Michigan and P. Wade and D. Bell of NIEHS for comments on the manuscript. The authors also acknowledge the use of the Noun Project Pro software for sourcing figure icons.
K.B. was supported by grants from the National Institutes of Health (NIH) (R01 AG067592, R35 ES031686, R01 MD013299). S.L. is supported by the Intramural Research Program of the NIH, NIEHS (ZO1 ES49019). F.B. was supported by grants from the NIH (F31DE029992).
References
- 1.Institute of Medicine. 2014. Identifying and Reducing Environmental Health Risks of Chemicals in Our Society. Workshop Summary. Washington, DC: The National Academies Press, PMID: , 10.17226/18710. [DOI] [PubMed] [Google Scholar]
- 2.Woodruff TJ, Zota AR, Schwartz JM. 2011. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ Health Perspect 119(6):878–885, PMID: , 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley JP, Kuiper JR, Bennett DH, Barrett ES, Bastain T, Breton CV, et al. . 2022. Exposure to contemporary and emerging chemicals in commerce among pregnant women in the United States: Environmental influences on Child Health Outcome (ECHO) program. Environ Sci Technol 56(10):6560–6573, PMID: , 10.1021/acs.est.1c08942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breslow NE. 1996. Statistics in epidemiology: the case-control study. J Am Stat Assoc 91(433):14–28, PMID: , 10.1080/01621459.1996.10476660. [DOI] [PubMed] [Google Scholar]
- 5.Fedak KM, Bernal A, Capshaw ZA, Gross S. 2015. Applying the Bradford Hill criteria in the 21st century: how data integration has changed causal inference in molecular epidemiology. Emerg Themes Epidemiol 12:14, PMID: , 10.1186/s12982-015-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battram T, Gaunt TR, Relton CL, Timpson NJ, Hemani G. 2022. A comparison of the genes and genesets identified by GWAS and EWAS of fifteen complex traits. Nat Commun 13(1):7816, PMID: , 10.1038/s41467-022-35037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandjean P, Landrigan PJ. 2014. Neurobehavioural effects of developmental toxicity. Lancet Neurol 13(3):330–338, PMID: , 10.1016/S1474-4422(13)70278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boekelheide K, Blumberg B, Chapin RE, Cote I, Graziano JH, Janesick A, et al. . 2012. Predicting later-life outcomes of early-life exposures. Environ Health Perspect 120(10):1353–1361, PMID: , 10.1289/ehp.1204934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. 2014. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. Atlanta (GA): Centers for Disease Control and Prevention (US). PMID: . [PubMed] [Google Scholar]
- 10.CDC (U.S. Centers for Disease Control and Prevention). 1991. Effectiveness in disease and injury prevention use of folic acid for prevention of spina bifida and other neural tube defects – 1983–1991. MMWR Weekly 40:513–516. [PubMed] [Google Scholar]
- 11.Haugen AC, Schug TT, Collman G, Heindel JJ. 2015. Evolution of DOHaD: the impact of environmental health sciences. J Dev Orig Health Dis 6(2):55–64, PMID: , 10.1017/S2040174414000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker DJ. 2007. The origins of the developmental origins theory. J Intern Med 261(5):412–417, PMID: , 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 13.DeBord DG, Carreon T, Lentz TJ, Middendorf PJ, Hoover MD, Schulte PA. 2016. Use of the “exposome” in the practice of epidemiology: a primer on -omic technologies. Am J Epidemiol 184(4):302–314, PMID: , 10.1093/aje/kwv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoghbi HY, Beaudet AL. 2016. Epigenetics and human disease. Cold Spring Harb Perspect Biol 8(2):a019497, PMID: , 10.1101/cshperspect.a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faulk C, Dolinoy DC. 2011. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics 6(7):791–797, PMID: , 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khavari DA, Sen GL, Rinn JL. 2010. DNA methylation and epigenetic control of cellular differentiation. Cell Cycle 9(19):3880–3883, PMID: , 10.4161/cc.9.19.13385. [DOI] [PubMed] [Google Scholar]
- 17.Cavalli G, Heard E. 2019. Advances in epigenetics link genetics to the environment and disease. Nature 571(7766):489–499, PMID: , 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 18.Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8(12):1463–1472, PMID: , 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 19.Dolinoy DC, Huang D, Jirtle RL. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA 104(32):13056–13061, PMID: , 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson OS, Sant KE, Dolinoy DC. 2012. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem 23(8):853–859, PMID: , 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC. 2013. Early-life lead exposure results in dose-and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5(5):487–500, PMID: , 10.2217/epi.13.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz LC. 2010. The Dutch Hunger Winter and the developmental origins of health and disease. Proc Natl Acad Sci USA 107(39):16757–16758, PMID: , 10.1073/pnas.1012911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. . 2014. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 5:559–2, PMID: , 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. . 2016. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17(1):208, PMID: , 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breton CV, Marsit CJ, Faustman E, Nadeau K, Goodrich JM, Dolinoy DC, et al. . 2017. Small-Magnitude effect sizes in epigenetic end points are important in children’s environmental health studies: the Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environ Health Perspect 125(4):511–526, PMID: , 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felix JF, Joubert BR, Baccarelli AA, Sharp GC, Almqvist C, Annesi-Maesano I, et al. . 2018. Cohort profile: Pregnancy And Childhood Epigenetics (PACE) consortium. Int J Epidemiol 47(1):22–23u, PMID: , 10.1093/ije/dyx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaisdell CJ, Park C, Hanspal M, Roary M, Arteaga SS, Laessig S, et al. . 2022. The NIH ECHO program: investigating how early environmental influences affect child health. Pediatr Res 92(5):1215–1216, 10.1038/s41390-021-01574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ENCODE Project Consortium. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583:699–710, PMID: , 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen R, Schymanski EL, Barabási AL, Miller GW. 2020. The exposome and health: where chemistry meets biology. Science 367(6476):392–396, PMID: , 10.1126/science.aay3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M, Guan Y, Huang R, Duan J, Zhou J, Chen T, et al. . 2022. Associations between the maternal exposome and metabolome during pregnancy. Environ Health Perspect 130(3):37003, PMID: , 10.1289/EHP9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson TA, Manolio TA. 2008. How to interpret a genome-wide association study. JAMA 299(11):1335–1344, PMID: , 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 32.Kraft P, Zeggini E, Ioannidis JP. 2009. Replication in genome-wide association studies. Stat Sci 24(4):561–573, PMID: , 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. . 2016. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 98(4):680–696, PMID: , 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikdar S, Joehanes R, Joubert BR, Xu C-J, Vives-Usano M, Rezwan FI, et al. . 2019. Comparison of smoking-related DNA methylation between newborns from prenatal exposure and adults from personal smoking. Epigenomics 11(13):1487–1500, PMID: , 10.2217/epi-2019-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everson TM, Vives-Usano M, Seyve E, Cardenas A, Lacasaña M, Craig JM, et al. . 2021. Placental DNA methylation signatures of maternal smoking during pregnancy and potential impacts on fetal growth. Nat Commun 12(1):5095, PMID: , 10.1038/s41467-021-24558-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannon E, Schendel D, Ladd-Acosta C, Grove J, Hansen CS, Hougaard DM, et al. . 2019. Variable DNA methylation in neonates mediates the association between prenatal smoking and birth weight. Philos Trans R Soc Lond B Biol Sci 374(1770):20180120, PMID: , 10.1098/rstb.2018.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vives-Usano M, Hernandez-Ferrer C, Maitre L, Ruiz-Arenas C, Andrusaityte S, Borràs E, et al. . 2020. In utero and childhood exposure to tobacco smoke and multi-layer molecular signatures in children. BMC Med 18(1):243, PMID: , 10.1186/s12916-020-01686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. . 2012. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120(10):1425–1431, PMID: , 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rotroff DM, Joubert BR, Marvel SW, Håberg SE, Wu MC, Nilsen RM, et al. . 2016. Maternal smoking impacts key biological pathways in newborns through epigenetic modification in utero. BMC Genomics 17(1):976, PMID: , 10.1186/s12864-016-3310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markunas CA, Xu Z, Harlid S, Wade PA, Lie RT, Taylor JA, et al. . 2014. Identification of DNA methylation changes in newborns related to maternal smoking during pregnancy. Environ Health Perspect 122(10):1147–1153, PMID: , 10.1289/ehp.1307892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Küpers LK, Xu X, Jankipersadsing SA, Vaez A, la Bastide-van Gemert S, Scholtens S, et al. . 2015. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int J Epidemiol 44(4):1224–1237, PMID: , 10.1093/ije/dyv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morales E, Vilahur N, Salas LA, Motta V, Fernandez MF, Murcia M, et al. . 2016. Genome-wide DNA methylation study in human placenta identifies novel loci associated with maternal smoking during pregnancy. Int J Epidemiol 45(5):1644–1655, PMID: , 10.1093/ije/dyw196. [DOI] [PubMed] [Google Scholar]
- 43.Shorey-Kendrick LE, McEvoy CT, O’Sullivan SM, Milner K, Vuylsteke B, Tepper RS, et al. . 2021. Impact of vitamin C supplementation on placental DNA methylation changes related to maternal smoking: association with gene expression and respiratory outcomes. Clin Epigenetics 13(1):177, PMID: , 10.1186/s13148-021-01161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howe CG, Zhou M, Wang X, Pittman GS, Thompson IJ, Campbell MR, et al. . 2019. Associations between maternal tobacco smoke exposure and the cord blood CD4(+) DNA methylome. Environ Health Perspect 127(4):47009, PMID: , 10.1289/EHP3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp GC, Arathimos R, Reese SE, Page CM, Felix J, Küpers LK, et al. . 2018. Maternal alcohol consumption and offspring DNA methylation: findings from six general population-based birth cohorts. Epigenomics 10(1):27–42, PMID: , 10.2217/epi-2017-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Küpers LK, Fernández-Barrés S, Nounu A, Friedman C, Fore R, Mancano G, et al. . 2022. Maternal mediterranean diet in pregnancy and newborn DNA methylation: a meta-analysis in the PACE consortium. Epigenetics 17(11):1419–1431, 10.1080/15592294.2022.2038412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, et al. . 2016. Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 7:10577, PMID: , 10.1038/ncomms10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonseth S, Roy R, Houseman EA, de Smith AJ, Zhou M, Lee S-T, et al. . 2015. Periconceptional folate consumption is associated with neonatal DNA methylation modifications in neural crest regulatory and cancer development genes. Epigenetics 10(12):1166–1176, PMID: , 10.1080/15592294.2015.1117889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amarasekera M, Martino D, Ashley S, Harb H, Kesper D, Strickland D, et al. . 2014. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. FASEB J 28(9):4068–4076, PMID: , 10.1096/fj.13-249029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suderman M, Stene LC, Bohlin J, Page CM, Holvik K, Parr CL, et al. . 2016. 25-Hydroxyvitamin D in pregnancy and genome wide cord blood DNA methylation in two pregnancy cohorts (MoBa and ALSPAC). J Steroid Biochem Mol Biol 159:102–109, PMID: , 10.1016/j.jsbmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taeubert MJ, de Prado-Bert P, Geurtsen ML, Mancano G, Vermeulen MJ, Reiss IKM, et al. . 2022. Maternal iron status in early pregnancy and DNA methylation in offspring: an epigenome-wide meta-analysis. Clin Epigenetics 14(1):59, PMID: , 10.1186/s13148-022-01276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Størdal K, McArdle HJ, Hayes H, Tapia G, Viken MK, Lund-Blix NA, et al. . 2018. Prenatal iron exposure and childhood type 1 diabetes. Sci Rep 8(1):9067, PMID: , 10.1038/s41598-018-27391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caramaschi D, Sharp GC, Nohr EA, Berryman K, Lewis SJ, Davey Smith G, et al. . 2017. Exploring a causal role of DNA methylation in the relationship between maternal vitamin B12 during pregnancy and child’s IQ at age 8, cognitive performance and educational attainment: a two-step mendelian randomization study. Hum Mol Genet 26(15):3001–3013, PMID: , 10.1093/hmg/ddx164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geraghty AA, Sexton-Oates A, O’Brien EC, Alberdi G, Fransquet P, Saffery R, et al. . 2018. A low glycaemic index diet in pregnancy induces DNA methylation variation in blood of newborns: results from the ROLO randomised controlled trial. Nutrients 10(4):, PMID: , 10.3390/nu10040455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan W, Zhang Y, Wang L, Yang W, Li C, Wang L, et al. . 2019. Maternal dietary glycaemic change during gestation influences insulin-related gene methylation in the placental tissue: a genome-wide methylation analysis. Genes Nutr 14:17, PMID: , 10.1186/s12263-019-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruzieva O, Xu C-J, Yousefi P, Relton C, Merid SK, Breton CV, et al. . 2019. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ Health Perspect 127(5):57012, PMID: , 10.1289/EHP4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Isaevska E, Fiano V, Asta F, Stafoggia M, Moirano G, Popovic M, et al. . 2022. Prenatal exposure to PM10 and changes in DNA methylation and telomere length in cord blood. Environ Res 209:112717, PMID: , 10.1016/j.envres.2022.112717. [DOI] [PubMed] [Google Scholar]
- 58.Plusquin M, Chadeau-Hyam M, Ghantous A, Alfano R, Bustamante M, Chatzi L, et al. . 2018. DNA methylome marks of exposure to particulate matter at three time points in early life. Environ Sci Technol 52(9):5427–5437, PMID: , 10.1021/acs.est.7b06447. [DOI] [PubMed] [Google Scholar]
- 59.Breton CV, Gao L, Yao J, Siegmund KD, Lurmann F, Gilliland F. 2016. Particulate matter, the newborn methylome, and cardio-respiratory health outcomes in childhood. Environ Epigenet 2(2):dvw005, PMID: , 10.1093/eep/dvw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruzieva O, Xu C-J, Breton CV, Annesi-Maesano I, Antó JM, Auffray C, et al. . 2017. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect 125(1):104–110, PMID: , 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng C, den Dekker M, Cardenas A, Rifas-Shiman SL, Gibson H, Agha G, et al. . 2018. Residential proximity to major roadways at birth, DNA methylation at birth and midchildhood, and childhood cognitive test scores: Project Viva (Massachusetts, USA). Environ Health Perspect 126(9):97006, PMID: , 10.1289/EHP2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lozano M, Yousefi P, Broberg K, Soler-Blasco R, Miyashita C, Pesce G, et al. . 2022. DNA methylation changes associated with prenatal mercury exposure: a meta-analysis of prospective cohort studies from PACE consortium. Environ Res 204(pt B):112093, PMID: , 10.1016/j.envres.2021.112093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bakulski KM, Lee H, Feinberg JI, Wells EM, Brown S, Herbstman JB, et al. . 2015. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. Int J Epidemiol 44(4):1249–1262, PMID: , 10.1093/ije/dyv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian F-Y, Everson TM, Lester B, Punshon T, Jackson BP, Hao K, et al. . 2020. Selenium-associated DNA methylation modifications in placenta and neurobehavioral development of newborns: an epigenome-wide study of two U.S. birth cohorts. Environ Int 137:105508, PMID: , 10.1016/j.envint.2020.105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, Chen J, et al. . 2018. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two U.S. birth cohorts. Environ Health Perspect 126(1):017010, PMID: , 10.1289/EHP2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park J, Kim J, Kim E, Won S, Kim WJ. 2022. Association between prenatal cadmium exposure and cord blood DNA methylation. Environ Res 212(pt B):113268, PMID: , 10.1016/j.envres.2022.113268. [DOI] [PubMed] [Google Scholar]
- 67.Gliga AR, Malin Igra A, Hellberg A, Engström K, Raqib R, Rahman A, et al. . 2022. Maternal exposure to cadmium during pregnancy is associated with changes in DNA methylation that are persistent at 9 years of age. Environ Int 163:107188, PMID: , 10.1016/j.envint.2022.107188. [DOI] [PubMed] [Google Scholar]
- 68.Kennedy E, Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, et al. . 2020. Copper associates with differential methylation in placentae from two US birth cohorts. Epigenetics 15(3):215–230, PMID: , 10.1080/15592294.2019.1661211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park J, Kim J, Kim E, Kim WJ, Won S. 2021. Prenatal lead exposure and cord blood DNA methylation in the Korean exposome study. Environ Res 195:110767, PMID: , 10.1016/j.envres.2021.110767. [DOI] [PubMed] [Google Scholar]
- 70.Wu S, Hivert M-F, Cardenas A, Zhong J, Rifas-Shiman SL, Agha G, et al. . 2017. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: an epigenome-wide association study. Environ Health Perspect 125(8):087019, PMID: , 10.1289/EHP1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bozack AK, Boileau P, Wei L, Hubbard AE, Sillé FCM, Ferreccio C, et al. . 2021. Exposure to arsenic at different life-stages and DNA methylation meta-analysis in buccal cells and leukocytes. Environ Health 20(1):79, PMID: , 10.1186/s12940-021-00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaushal A, Zhang H, Karmaus WJJ, Everson TM, Marsit CJ, Karagas MR, et al. . 2017. Genome-wide DNA methylation at birth in relation to in utero arsenic exposure and the associated health in later life. Environ Health 16(1):50, PMID: , 10.1186/s12940-017-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bozack AK, Cardenas A, Quamruzzaman Q, Rahman M, Mostofa G, Christiani DC, et al. . 2018. DNA methylation in cord blood as mediator of the association between prenatal arsenic exposure and gestational age. Epigenetics 13(9):923–940, PMID: , 10.1080/15592294.2018.1516453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng Z, Huo X, Zhang Y, Hylkema MN, Wu Y, Xu X. 2019. Differential DNA methylation in newborns with maternal exposure to heavy metals from an e-waste recycling area. Environ Res 171:536–545, PMID: , 10.1016/j.envres.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Liu Y, Eliot MN, Papandonatos GD, Kelsey KT, Fore R, Langevin S, et al. . 2022. Gestational perfluoroalkyl substance exposure and DNA methylation at birth and 12 years of age: a longitudinal epigenome-wide association study. Environ Health Perspect 130(3):37005, PMID: , 10.1289/EHP10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miura R, Araki A, Miyashita C, Kobayashi S, Kobayashi S, Wang S-L, et al. . 2018. An epigenome-wide study of cord blood DNA methylations in relation to prenatal perfluoroalkyl substance exposure: the Hokkaido study. Environ Int 115:21–28, PMID: , 10.1016/j.envint.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 77.Miura R, Ikeda-Araki A, Ishihara T, Miyake K, Miyashita C, Nakajima T, et al. . 2021. Effect of prenatal exposure to phthalates on epigenome-wide DNA methylations in cord blood and implications for fetal growth: the Hokkaido Study on Environment and Children’s Health. Sci Total Environ 783:147035, PMID: , 10.1016/j.scitotenv.2021.147035. [DOI] [PubMed] [Google Scholar]
- 78.Vilahur N, Bustamante M, Morales E, Motta V, Fernandez MF, Salas LA, et al. . 2016. Prenatal exposure to mixtures of xenoestrogens and genome-wide DNA methylation in human placenta. Epigenomics 8(1):43–54, PMID: , 10.2217/epi.15.91. [DOI] [PubMed] [Google Scholar]
- 79.Miura R, Araki A, Minatoya M, Miyake K, Chen M-L, Kobayashi S, et al. . 2019. An epigenome-wide analysis of cord blood DNA methylation reveals sex-specific effect of exposure to bisphenol A. Sci Rep 9(1):12369, PMID: , 10.1038/s41598-019-48916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCabe CF, Padmanabhan V, Dolinoy DC, Domino SE, Jones TR, Bakulski KM, et al. . 2020. Maternal environmental exposure to bisphenols and epigenome-wide DNA methylation in infant cord blood. Environ Epigenet 6(1):dvaa021, PMID: , 10.1093/eep/dvaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu X, Zhao B, Su Y, Zhang Y, Chen J, Wu W, et al. . 2018. Association of prenatal organochlorine pesticide-dichlorodiphenyltrichloroethane exposure with fetal genome-wide DNA methylation. Life Sci 200:81–86, PMID: , 10.1016/j.lfs.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 82.Alfano R, Guida F, Galobardes B, Chadeau-Hyam M, Delpierre C, Ghantous A, et al. . 2019. Socioeconomic position during pregnancy and DNA methylation signatures at three stages across early life: epigenome-wide association studies in the ALSPAC birth cohort. Int J Epidemiol 48(1):30–44, PMID: , 10.1093/ije/dyy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caramaschi D, Jungius J, Page CM, Novakovic B, Saffery R, Halliday J, et al. . 2021. Association of medically assisted reproduction with offspring cord blood DNA methylation across cohorts. Hum Reprod 36(8):2403–2413, PMID: , 10.1093/humrep/deab137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joubert BR, Håberg SE, Bell DA, Nilsen RM, Vollset SE, Midttun O, et al. . 2014. Maternal smoking and DNA methylation in newborns: in utero effect or epigenetic inheritance? Cancer Epidemiol Biomarkers Prev 23(6):1007–1017, PMID: , 10.1158/1055-9965.EPI-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, et al. . 2015. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents And Children (ALSPAC). Hum Mol Genet 24(8):2201–2217, PMID: , 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. . 2016. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 9(5):436–447, PMID: , 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barker DJ, Thornburg KL. 2013. Placental programming of chronic diseases, cancer and lifespan: a review. Placenta 34(10):841–845, PMID: , 10.1016/j.placenta.2013.07.063. [DOI] [PubMed] [Google Scholar]
- 88.Koletzko B, Godfrey KM, Poston L, Szajewska H, van Goudoever JB, de Waard M, et al. . 2019. Nutrition during pregnancy, lactation and early childhood and its implications for maternal and long-term child health: the early nutrition project recommendations. Ann Nutr Metab 74(2):93–106, PMID: , 10.1159/000496471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Riley EP, Infante MA, Warren KR. 2011. Fetal alcohol spectrum disorders: an overview. Neuropsychol Rev 21(2):73–80, PMID: , 10.1007/s11065-011-9166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perez-Lopez FR, Pilz S, Chedraui P. 2020. Vitamin D supplementation during pregnancy: an overview. Curr Opin Obstet Gynecol 32(5):316–321, PMID: , 10.1097/GCO.0000000000000641. [DOI] [PubMed] [Google Scholar]
- 91.Amati F, Hassounah S, Swaka A. 2019. The impact of Mediterranean dietary patterns during pregnancy on maternal and offspring health. Nutrients 11(5):, PMID: , 10.3390/nu11051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bekkar B, Pacheco S, Basu R, DeNicola N. 2020. Association of air pollution and heat exposure with preterm birth, low birth weight, and stillbirth in the US: a systematic review. JAMA Netw Open 3(6):e208243, PMID: , 10.1001/jamanetworkopen.2020.8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vrijheid M, Casas M, Gascon M, Valvi D, Nieuwenhuijsen M. 2016. Environmental pollutants and child health - a review of recent concerns. Int J Hyg Environ Health 219(4–5):331–342, PMID: , 10.1016/j.ijheh.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 94.Hobel CJ, Goldstein A, Barrett ES. 2008. Psychosocial stress and pregnancy outcome. Clin Obstet Gynecol 51(2):333–348, PMID: , 10.1097/GRF.0b013e31816f2709. [DOI] [PubMed] [Google Scholar]
- 95.Yousefi PD, Suderman M, Langdon R, Whitehurst O, Davey Smith G, Relton CL. 2022. DNA methylation-based predictors of health: applications and statistical considerations. Nat Rev Genet 23(6):369–383, PMID: , 10.1038/s41576-022-00465-w. [DOI] [PubMed] [Google Scholar]
- 96.Craig P, Katikireddi SV, Leyland A, Popham F. 2017. Natural experiments: an overview of methods, approaches, and contributions to public health intervention research. Annu Rev Public Health 38:39–56, PMID: , 10.1146/annurev-publhealth-031816-044327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rich DQ. 2017. Accountability studies of air pollution and health effects: lessons learned and recommendations for future natural experiment opportunities. Environ Int 100:62–78, PMID: , 10.1016/j.envint.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reese SE, Zhao S, Wu MC, Joubert BR, Parr CL, Håberg SE, et al. . 2017. DNA methylation score as a biomarker in newborns for sustained maternal smoking during pregnancy. Environ Health Perspect 125(4):760–766, PMID: , 10.1289/EHP333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rauschert S, Melton PE, Heiskala A, Karhunen V, Burdge G, Craig JM, et al. . 2020. Machine learning-based DNA methylation score for fetal exposure to maternal smoking: development and validation in samples collected from adolescents and adults. Environ Health Perspect 128(9):97003, PMID: , 10.1289/EHP6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dietz PM, Homa D, England LJ, Burley K, Tong VT, Dube SR, et al. . 2011. Estimates of nondisclosure of cigarette smoking among pregnant and nonpregnant women of reproductive age in the United States. Am J Epidemiol 173(3):355–359, PMID: , 10.1093/aje/kwq381. [DOI] [PubMed] [Google Scholar]
- 101.Maas SCE, Vidaki A, Wilson R, Teumer A, Liu F, van Meurs JBJ, et al. . 2019. Validated inference of smoking habits from blood with a finite DNA methylation marker set. Eur J Epidemiol 34(11):1055–1074, PMID: , 10.1007/s10654-019-00555-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larsen S, Haavaldsen C, Bjelland EK, Dypvik J, Jukic AM, Eskild A. 2018. Placental weight and birthweight: the relations with number of daily cigarettes and smoking cessation in pregnancy. A population study. Int J Epidemiol 47(4):1141–1150, PMID: , 10.1093/ije/dyy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lupo PJ, Petrick LM, Hoang TT, Janitz AE, Marcotte EL, Schraw JM, et al. . 2021. Using primary teeth and archived dried spots for exposomic studies in children: exploring new paths in the environmental epidemiology of pediatric cancer. Bioessays 43(9):e2100030, PMID: , 10.1002/bies.202100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, et al. . EGG Consortium. 2019. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet 51(5):804–814, PMID: , 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Küpers LK, Monnereau C, Sharp GC, Yousefi P, Salas LA, Ghantous A, et al. . 2019. Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat Commun 10(1):1893, PMID: , 10.1038/s41467-019-09671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Faul F, Erdfelder E, Lang AG, Buchner A. 2007. G*power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 39(2):175–191, PMID: , 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 107.Breeze CE, Wong JYY, Beck S, Berndt SI, Franceschini N. 2022. Diversity in EWAS: current state, challenges, and solutions. Genome Med 14(1):71, PMID: , 10.1186/s13073-022-01065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quick C, Anugu P, Musani S, Weiss ST, Burchard EG, White MJ, et al. . 2020. Sequencing and imputation in GWAS: cost-effective strategies to increase power and genomic coverage across diverse populations. Genet Epidemiol 44(6):537–549, PMID: , 10.1002/gepi.22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rizzardi LF, Hickey PF, Idrizi A, Tryggvadóttir R, Callahan CM, Stephens KE, et al. . 2021. Human brain region-specific variably methylated regions are enriched for heritability of distinct neuropsychiatric traits. Genome Biol 22(1):116, PMID: , 10.1186/s13059-021-02335-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Breeze CE, Beck S, Berndt SI, Franceschini N. 2022. The missing diversity in human epigenomic studies. Nat Genet 54(6):737–739, PMID: , 10.1038/s41588-022-01081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.GTEx Consortium. 2015. Human genomics. The Genotype-Tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660, PMID: , 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gee GC, Payne-Sturges DC. 2004. Environmental health disparities: a framework integrating psychosocial and environmental concepts. Environ Health Perspect 112(17):1645–1653, PMID: , 10.1289/ehp.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Salas LA, Peres LC, Thayer ZM, Smith RW, Guo Y, Chung W, et al. . 2021. A transdisciplinary approach to understand the epigenetic basis of race/ethnicity health disparities. Epigenomics 13(21):1761–1770, PMID: , 10.2217/epi-2020-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Olden K, Lin YS, Gruber D, Sonawane B. 2014. Epigenome: biosensor of cumulative exposure to chemical and nonchemical stressors related to environmental justice. Am J Public Health 104(10):1816–1821, PMID: , 10.2105/AJPH.2014.302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jones MR, Diez-Roux AV, Hajat A, Kershaw KN, O’Neill MS, Guallar E, et al. . 2014. Race/ethnicity, residential segregation, and exposure to ambient air pollution: the Multi-Ethnic Study of Atherosclerosis (MESA). Am J Public Health 104(11):2130–2137, PMID: , 10.2105/AJPH.2014.302135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jackson CL. 2020. Food for thought: opportunities to improve diversity, inclusion, representation, and participation in epidemiology. Am J Epidemiol 189(10):1016–1022, PMID: , 10.1093/aje/kwaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ðoàn LN, Bacong AM, Ma KPK, Morey BN. 2020. Epidemiologists count: the role of diversity and inclusion in the field of epidemiology. Am J Epidemiol 189(10):1033–1036, PMID: , 10.1093/aje/kwaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bernard MA, Johnson AC, Hopkins-Laboy T, Tabak LA. 2021. The US National Institutes of Health approach to inclusive excellence. Nat Med 27(11):1861–1864, PMID: , 10.1038/s41591-021-01532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Collins FS, Adams AB, Aklin C, Archer TK, Bernard MA, Boone E, et al. . 2021. Affirming NIH’s commitment to addressing structural racism in the biomedical research enterprise. Cell 184(12):3075–3079, PMID: , 10.1016/j.cell.2021.05.014. [DOI] [PubMed] [Google Scholar]
- 120.Battram T, Yousefi P, Crawford G, Prince C, Sheikhali Babaei M, Sharp G, et al. . 2022. The EWAS Catalog: a database of epigenome-wide association studies. Wellcome Open Res 7:41, PMID: , 10.12688/wellcomeopenres.17598.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Odintsova VV, Rebattu V, Hagenbeek FA, Pool R, Beck JJ, Ehli EA, et al. . 2021. Predicting complex traits and exposures from polygenic scores and blood and buccal DNA methylation profiles. Front Psychiatry 12:688464, PMID: , 10.3389/fpsyt.2021.688464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD. 2016. Epigenetic research in neuropsychiatric disorders: the “Tissue Issue”. Curr Behav Neurosci Rep 3(3):264–274, PMID: , 10.1007/s40473-016-0083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brugha R, Lowe R, Henderson AJ, Holloway JW, Rakyan V, Wozniak E, et al. . 2017. DNA methylation profiles between airway epithelium and proxy tissues in children. Acta Paediatr 106(12):2011–2016, PMID: , 10.1111/apa.14027. [DOI] [PubMed] [Google Scholar]
- 124.Qi C, Jiang Y, Yang IV, Forno E, Wang T, Vonk JM, et al. . 2020. Nasal DNA methylation profiling of asthma and rhinitis. J Allergy Clin Immunol 145(6):1655–1663, PMID: , 10.1016/j.jaci.2019.12.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Castelo-Soccio L. 2019. Stripping away barriers to find relevant skin biomarkers for pediatric atopic dermatitis. JAMA Dermatol 155(12):1342–1343, PMID: , 10.1001/jamadermatol.2019.2792. [DOI] [PubMed] [Google Scholar]
- 126.Li H, Masieri FF, Schneider M, Kottek T, Hahnel S, Yamauchi K, et al. . 2020. Autologous, non-invasively available mesenchymal stem cells from the outer root sheath of hair follicle are obtainable by migration from plucked hair follicles and expandable in scalable amounts. Cells 9(9):, PMID: , 10.3390/cells9092069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weltner J, Balboa D, Katayama S, Bespalov M, Krjutškov K, Jouhilahti E-M, et al. . 2018. Human pluripotent reprogramming with CRISPR activators. Nat Commun 9(1):2643, PMID: , 10.1038/s41467-018-05067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Todtenhaupt P, van Pel M, Roest AAW, Heijmans BT. 2022. Mesenchymal stromal cells as a tool to unravel the developmental origins of disease. Trends Endocrinol Metab 33(9):614–627, PMID: , 10.1016/j.tem.2022.06.002. [DOI] [PubMed] [Google Scholar]
- 129.Nelson DM, Kay H, Wang Y. 2011. The Placenta: from Development to Disease. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 130.Jarmasz JS, Stirton H, Davie JR, Del Bigio MR. 2019. DNA methylation and histone post-translational modification stability in post-mortem brain tissue. Clin Epigenetics 11(1):5, PMID: , 10.1186/s13148-018-0596-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Campbell KA, Colacino JA, Park SK, Bakulski KM. 2020. Cell types in environmental epigenetic studies: biological and epidemiological frameworks. Curr Envir Health Rpt 7(3):185–197, 10.1007/s40572-020-00287-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Middleton LYM, Nguyen VK, Dou J, Park SK, Colacino JA, Bakulski KM. 2022. Environmental chemical-wide associations with immune biomarkers in the US: a cross-sectional analysis. medRxiv 2022.2003.2022.22272789, 10.1101/2022.03.22.22272789. [DOI] [PubMed] [Google Scholar]
- 133.Gervin K, Salas LA, Bakulski KM, van Zelm MC, Koestler DC, Wiencke JK, et al. . 2019. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin Epigenet 11(1):1–15, 10.1186/s13148-019-0717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Middleton LYM, Dou J, Fisher J, Heiss JA, Nguyen VK, Just AC, et al. . 2022. Saliva cell type DNA methylation reference panel for epidemiological studies in children. Epigenetics 17(2):161–177, 10.1080/15592294.2021.1890874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yuan V, Hui D, Yin Y, Peñaherrera MS, Beristain AG, Robinson WP. 2021. Cell-specific characterization of the placental methylome. BMC Genomics 22(1):1–20, 10.1186/s12864-020-07186-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu T, Liu J, Beck S, Pan S, Capper D, Lechner M, et al. . 2022. A pan-tissue DNA methylation atlas enables in silico decomposition of human tissue methylomes at cell-type resolution. Nat Methods 19(3):296–306, PMID: , 10.1038/s41592-022-01412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Teschendorff AE, Feinberg AP. 2021. Statistical mechanics meets single-cell biology. Nat Rev Genet 22(7):459–476, PMID: , 10.1038/s41576-021-00341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rahmani E, Schweiger R, Rhead B, Criswell LA, Barcellos LF, Eskin E, et al. . 2019. Cell-type-specific resolution epigenetics without the need for cell sorting or single-cell biology. Nat Commun 10(1):3417, PMID: , 10.1038/s41467-019-11052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Martos SN, Campbell MR, Lozoya OA, Wang X, Bennett BD, Thompson IJB, et al. . 2020. Single-cell analyses identify dysfunctional CD16+ CD8 T cells in smokers. Cell Rep Med 1(4):, PMID: , 10.1016/j.xcrm.2020.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sugden K, Hannon EJ, Arseneault L, Belsky DW, Corcoran DL, Fisher HL, et al. . 2020. Patterns of reliability: assessing the reproducibility and integrity of DNA methylation measurement. Patterns (NY) 1(2):, PMID: , 10.1016/j.patter.2020.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Edgar RD, Jones MJ, Robinson WP, Kobor MS. 2017. An empirically driven data reduction method on the human 450K methylation array to remove tissue specific non-variable CpGs. Clin Epigenetics 9:11, PMID: , 10.1186/s13148-017-0320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu Z, Taylor JA. 2021. Reliability of DNA methylation measures using Illumina methylation BeadChip. Epigenetics 16(5):495–502, PMID: , 10.1080/15592294.2020.1805692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen J, Just AC, Schwartz J, Hou L, Jafari N, Sun Z, et al. . 2016. CpGFilter: model-based CpG probe filtering with replicates for epigenome-wide association studies. Bioinformatics 32(3):469–471, PMID: , 10.1093/bioinformatics/btv577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shu C, Zhang X, Aouizerat BE, Xu K. 2020. Comparison of methylation capture sequencing and Infinium MethylationEPIC array in peripheral blood mononuclear cells. Epigenetics Chromatin 13(1):51, PMID: , 10.1186/s13072-020-00372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ziller MJ, Hansen KD, Meissner A, Aryee MJ. 2015. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods 12(3):230–232, PMID: , 10.1038/nmeth.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. . 2010. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11(10):733–739, PMID: , 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heiss JA, Brennan KJ, Baccarelli AA, Téllez-Rojo MM, Estrada-Gutiérrez G, Wright RO, et al. . 2020. Battle of epigenetic proportions: comparing Illumina’s EPIC methylation microarrays and TruSeq targeted bisulfite sequencing. Epigenetics 15(1–2):174–182, PMID: , 10.1080/15592294.2019.1656159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang J, Bai L, Cui B, Wu L, Wang L, An Z, et al. . 2020. Leveraging biological and statistical covariates improves the detection power in epigenome-wide association testing. Genome Biol 21(1):88, PMID: , 10.1186/s13059-020-02001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Patel CJ, Kerr J, Thomas DC, Mukherjee B, Ritz B, Chatterjee N, et al. . 2017. Opportunities and challenges for environmental exposure assessment in population-based studies. Cancer Epidemiol Biomarkers Prev 26(9):1370–1380, PMID: , 10.1158/1055-9965.EPI-17-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee G, Kim S, Park H, Lee J, Lee JP, Kho Y, et al. . 2021. Variability of urinary creatinine, specific gravity, and osmolality over the course of pregnancy: implications in exposure assessment among pregnant women. Environ Res 198:110473, PMID: , 10.1016/j.envres.2020.110473. [DOI] [PubMed] [Google Scholar]
- 151.Braun JM, Smith KW, Williams PL, Calafat AM, Berry K, Ehrlich S, et al. . 2012. Variability of urinary phthalate metabolite and bisphenol a concentrations before and during pregnancy. Environ Health Perspect 120(5):739–745, PMID: , 10.1289/ehp.1104139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nieuwenhuijsen MJ, ed. 2015. Exposure Assessment in Environmental Epidemiology. Oxford, UK: Oxford University Press. [Google Scholar]
- 153.Delpizzo V, Borghesi JL. 1995. Exposure measurement errors, risk estimate and statistical power in case-control studies using dichotomous analysis of a continuous exposure variable. Int J Epidemiol 24(4):851–862, PMID: , 10.1093/ije/24.4.851. [DOI] [PubMed] [Google Scholar]
- 154.Kamai EM, McElrath TF, Ferguson KK. 2019. Fetal growth in environmental epidemiology: mechanisms, limitations, and a review of associations with biomarkers of non-persistent chemical exposures during pregnancy. Environ Health 18(1):43, PMID: , 10.1186/s12940-019-0480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu W, Li S, Ye T, Xu R, Song J, Guo Y. 2022. Deep ensemble machine learning framework for the estimation of PM2.5 concentrations. Environ Health Perspect 130(3):37004, PMID: , 10.1289/EHP9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wei Y, Qiu X, Yazdi MD, Shtein A, Shi L, Yang J, et al. . 2022. The impact of exposure measurement error on the estimated concentration–response relationship between long-term exposure to PM2.5 and mortality. Environ Health Perspect 130(7):77006, PMID: , 10.1289/EHP10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.White-Newsome JL, Brines SJ, Brown DG, Dvonch JT, Gronlund CJ, Zhang K, et al. . 2013. Validating satellite-derived land surface temperature with in situ measurements: a public health perspective. Environ Health Perspect 121(8):925–931, PMID: , 10.1289/ehp.1206176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Samon SM, Hammel SC, Stapleton HM, Anderson KA. 2022. Silicone wristbands as personal passive sampling devices: current knowledge, recommendations for use, and future directions. Environ Int 169:107339, PMID: , 10.1016/j.envint.2022.107339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cheung KL, Lafayette RA. 2013. Renal physiology of pregnancy. Adv Chronic Kidney Dis 20(3):209–214, PMID: , 10.1053/j.ackd.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Davison JM, Dunlop W, Ezimokhai M. 1980. 24-hour creatinine clearance during the third trimester of normal pregnancy. Br J Obstet Gynaecol 87(2):106–109, PMID: , 10.1111/j.1471-0528.1980.tb04501.x. [DOI] [PubMed] [Google Scholar]
- 161.Davison JM, Noble MC. 1981. Serial changes in 24 hour creatinine clearance during normal menstrual cycles and the first trimester of pregnancy. Br J Obstet Gynaecol 88(1):10–17, PMID: , 10.1111/j.1471-0528.1981.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 162.Moriceau M-A, Cano-Sancho G, Kim MJi, Coumoul X, Emond C, Arrebola J-P, et al. . 2022. Partitioning of persistent organic pollutants between adipose tissue and serum in human studies. Toxics 11(1):, PMID: , 10.3390/toxics11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weaver VM, Kotchmar DJ, Fadrowski JJ, Silbergeld EK. 2016. Challenges for environmental epidemiology research: are biomarker concentrations altered by kidney function or urine concentration adjustment? J Expo Sci Environ Epidemiol 26(1):1–8, PMID: , 10.1038/jes.2015.8. [DOI] [PubMed] [Google Scholar]
- 164.Aronica L, Ordovas JM, Volkov A, Lamb JJ, Stone PM, Minich D, et al. . 2022. Genetic biomarkers of metabolic detoxification for personalized lifestyle medicine. Nutrients 14(4):, PMID: , 10.3390/nu14040768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hare D, Austin C, Doble P, Arora M. 2011. Elemental bio-imaging of trace elements in teeth using laser ablation-inductively coupled plasma-mass spectrometry. J Dent 39(5):397–403, PMID: , 10.1016/j.jdent.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 166.Pozebon D, Scheffler G, Dressler V. 2017. Recent applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) for biological sample analysis: a follow-up review. J Anal at Spectrom 32(5):890–919, 10.1039/C7JA00026J. [DOI] [Google Scholar]
- 167.Volk HE, Perera F, Braun JM, Kingsley SL, Gray K, Buckley J, et al. . 2021. Prenatal air pollution exposure and neurodevelopment: a review and blueprint for a harmonized approach within ECHO. Environ Res 196:110320, PMID: , 10.1016/j.envres.2020.110320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vrijheid M, Basagaña X, Gonzalez JR, Jaddoe VWV, Jensen G, Keun HC, et al. . 2021. Advancing tools for human early lifecourse exposome research and translation (ATHLETE): project overview. Environ Epidemiol 5(5):e166, PMID: , 10.1097/EE9.0000000000000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Labine LM, Simpson MJ. 2020. The use of nuclear magnetic resonance (NMR) and mass spectrometry (MS)–based metabolomics in environmental exposure assessment. Curr Opin Environ Sci Health 15:7–15, 10.1016/j.coesh.2020.01.008. [DOI] [Google Scholar]
- 170.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. . 2009. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2(1):73–80, PMID: , 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, et al. . 2018. Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum Mol Genet 27(4):742–756, PMID: , 10.1093/hmg/ddx429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rice K, Higgins J, Lumley T. 2018. A re-evaluation of fixed effect(s) meta-analysis. J R Stat Soc Ser A Stat Soc 181(1):205–227, 10.1111/rssa.12275. [DOI] [Google Scholar]
- 173.Maitre L, Bustamante M, Hernández-Ferrer C, Thiel D, Lau C-HE, Siskos AP, et al. . 2022. Multi-omics signatures of the human early life exposome. Nat Commun 13(1):7024, PMID: , 10.1038/s41467-022-34422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hoang TT, Qi C, Paul KC, Lee M, White JD, Richards M, et al. . BIOS Consortium. 2021. Epigenome-wide DNA methylation and pesticide use in the Agricultural Lung Health Study. Environ Health Perspect 129(9):97008, PMID: , 10.1289/EHP8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van Iterson M, van Zwet EW, BIOS Consortium, Heijmans BT. 2017. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol 18(1):19, PMID: , 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Davidov O, Jelsema CM, Peddada S. 2018. Testing for inequality constraints in singular models by trimming or winsorizing the variance matrix. J Am Stat Assoc 113(522):906–918, PMID: , 10.1080/01621459.2017.1301258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Alexis N, Urch B, Tarlo S, Corey P, Pengelly D, O’Byrne P, et al. . 2000. Cyclooxygenase metabolites play a different role in ozone-induced pulmonary function decline in asthmatics compared to normals. Inhal Toxicol 12(12):1205–1224, PMID: , 10.1080/08958370050198548. [DOI] [PubMed] [Google Scholar]
- 178.Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C. 2014. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol 11:71, PMID: , 10.1186/s12989-014-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Baron RM, Kenny DA. 1986. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol 51(6):1173–1182, PMID: , 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 180.Andersen AM, Ryan PT, Gibbons FX, Simons RL, Long JD, Philibert RA. 2018. A droplet digital PCR assay for smoking predicts all-cause mortality. J Insur Med 47(4):220–229, PMID: , 10.17849/insm-47-4-1-10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Valeri L, Reese SL, Zhao S, Page CM, Nystad W, Coull BA, et al. . 2017. Misclassified exposure in epigenetic mediation analyses. Does DNA methylation mediate effects of smoking on birthweight? Epigenomics 9(3):253–265, PMID: , 10.2217/epi-2016-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cardenas A, Lutz SM, Everson TM, Perron P, Bouchard L, Hivert MF. 2019. Mediation by placental DNA methylation of the association of prenatal maternal smoking and birth weight Am J Epidemiol 188(11):1878–1886, PMID: , 10.1093/aje/kwz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jamieson E, Korologou-Linden R, Wootton RE, Guyatt AL, Battram T, Burrows K, et al. . 2020. Smoking, DNA methylation, and lung function: a Mendelian randomization analysis to investigate causal pathways. Am J Hum Genet 106(3):315–326, PMID: , 10.1016/j.ajhg.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Min JL, Hemani G, Hannon E, Dekkers KF, Castillo-Fernandez J, Luijk R, et al. . 2021. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet 53(9):1311–1321, PMID: , 10.1038/s41588-021-00923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hop PJ, Luijk R, Daxinger L, van Iterson M, Dekkers KF, Jansen R, et al. . 2020. Genome-wide identification of genes regulating DNA methylation using genetic anchors for causal inference. Genome Biol 21(1):220, PMID: , 10.1186/s13059-020-02114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tobi EW, van Zwet EW, Lumey LH, Heijmans BT. 2018. Why mediation analysis trumps mendelian randomization in population epigenomics studies of the Dutch famine. bioRxiv:362392, 10.1101/362392. [DOI] [Google Scholar]
- 187.Pingault JB, O’Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. 2018. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet 19(9):566–580, PMID: , 10.1038/s41576-018-0020-3. [DOI] [PubMed] [Google Scholar]
- 188.Mahmoud AM, Ali MM. 2019. Methyl donor micronutrients that modify DNA methylation and cancer outcome. Nutrients 11(3):, PMID: , 10.3390/nu11030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pogribny IP, Tryndyak VP, Woods CG, Witt SE, Rusyn I. 2007. Epigenetic effects of the continuous exposure to peroxisome proliferator WY-14,643 in mouse liver are dependent upon peroxisome proliferator activated receptor alpha. Mutat Res 625(1–2):62–71, PMID: , 10.1016/j.mrfmmm.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. 2004. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 32(14):4100–4108, PMID: , 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lappalainen T, Greally JM. 2017. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet 18(7):441–451, PMID: , 10.1038/nrg.2017.32. [DOI] [PubMed] [Google Scholar]
- 192.Martin EM, Fry RC. 2018. Environmental influences on the epigenome: exposure – associated DNA methylation in human populations. Annu Rev Public Health 39:309–333, PMID: , 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- 193.Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S-I, et al. . 2015. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev 29(23):2449–2462, PMID: , 10.1101/gad.271353.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Christman JK. 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21(35):5483–5495, PMID: , 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 195.Zeisel S. 2017. Choline, other methyl-donors and epigenetics. Nutrients 9(5):445, PMID: , 10.3390/nu9050445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Barrangou R, Doudna JA. 2016. Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34(9):933–941, PMID: , 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 197.Waryah CB, Moses C, Arooj M, Blancafort P. 2018. Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing. Methods Mol Biol 1767:19–63, PMID: . [DOI] [PubMed] [Google Scholar]
- 198.Hoang TT, Sikdar S, Xu C-J, Lee MK, Cardwell J, Forno E, et al. . 2020. Epigenome-wide association study of DNA methylation and adult asthma in the Agricultural Lung Health Study. Eur Respir J 56(3):, PMID: , 10.1183/13993003.00217-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492, PMID: , 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 200.Millan-Zambrano G, Burton A, Bannister AJ, Schneider R. 2022. Histone post-translational modifications – cause and consequence of genome function. Nat Rev Genet 23(9):563–580, PMID: , 10.1038/s41576-022-00468-7. [DOI] [PubMed] [Google Scholar]
- 201.Ebrahim A, Brunk E, Tan J, O’Brien EJ, Kim D, Szubin R, et al. . 2016. Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun 7:13091, PMID: , 10.1038/ncomms13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hasin Y, Seldin M, Lusis A. 2017. Multi-omics approaches to disease. Genome Biol 18(1):83, PMID: , 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC, et al. . 2018. Multi-omics factor analysis – a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol 14(6):e8124, PMID: , 10.15252/msb.20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tekola-Ayele F, Zeng X, Chatterjee S, Ouidir M, Lesseur C, Hao K, et al. . 2022. Placental multi-omics integration identifies candidate functional genes for birthweight. Nat Commun 13(1):2384, PMID: , 10.1038/s41467-022-30007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mukherjee N, Arathimos R, Chen S, Kheirkhah Rahimabad P, Han L, Zhang H, et al. . 2021. DNA methylation at birth is associated with lung function development until age 26 years. Eur Respir J 57(4):, PMID: , 10.1183/13993003.03505-2020. [DOI] [PubMed] [Google Scholar]
- 206.Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, et al. . 2015. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci 143(1):97–106, PMID: , 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Merid SK, Bustamante M, Standl M, Sunyer J, Heinrich J, Lemonnier N, et al. . 2021. Integration of gene expression and DNA methylation identifies epigenetically controlled modules related to PM2.5 exposure. Environ Int 146:106248, PMID: , 10.1016/j.envint.2020.106248. [DOI] [PubMed] [Google Scholar]
- 208.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. . 2017. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet 49(1):131–138, PMID: , 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 209.Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, et al. . 2019. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun 10(1):4267, PMID: , 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Oliva M, Demanelis K, Lu Y, Chernoff M, Jasmine F, Ahsan H, et al. . 2023. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat Genet 55(1):112–122, PMID: , 10.1038/s41588-022-01248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Giambartolomei C, Zhenli Liu J, Zhang W, Hauberg M, Shi H, Boocock J, et al. . 2018. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics 34(15):2538–2545, PMID: , 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. . 2014. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10(5):e1004383, PMID: , 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gatev E, Gladish N, Mostafavi S, Kobor MS. 2020. CoMeBack: DNA methylation array data analysis for co-methylated regions. Bioinformatics 36(9):2675–2683, PMID: , 10.1093/bioinformatics/btaa049. [DOI] [PubMed] [Google Scholar]
- 214.Lee M, Huan T, McCartney DL, Chittoor G, de Vries M, Lahousse L, et al. . 2022. Pulmonary function and blood DNA methylation: a multiancestry epigenome-wide association meta-analysis. Am J Respir Crit Care Med 206(3):321–336, PMID: , 10.1164/rccm.202108-1907OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Breeze CE, Reynolds AP, van Dongen J, Dunham I, Lazar J, Neph S, et al. . 2019. eFORGE v2.0: updated analysis of cell type-specific signal in epigenomic data. Bioinformatics 35(22):4767–4769, PMID: , 10.1093/bioinformatics/btz456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bujold D, Morais DADL, Gauthier C, Côté C, Caron M, Kwan T, et al. . 2016. The international human epigenome consortium data portal. Cell Syst 3(5):496–499.e2, PMID: , 10.1016/j.cels.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 217.Subramanian I, Verma S, Kumar S, Jere A, Anamika K. 2020. Multi-omics data integration, interpretation, and its application. Bioinform Biol Insights 14:1177932219899051, PMID: , 10.1177/1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Brown BC, Wang C, Kasela S, Aguet F, Nachun DC, Taylor KD, et al. . 2022. Multiset correlation and factor analysis enables exploration of multi-omic data. bioRxiv:2022.2007.2018.500246, 10.1101/2022.07.18.500246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.